Abstract
Background
The global over-reliance on non-renewable fossil fuels has led to the emission of greenhouse gases, creating a critical global environmental challenge. There is an urgent need for alternative solutions like biofuels. Advanced biofuel is a renewable sustainable energy generated from lignocellulosic plant materials, which can significantly contribute to mitigating CO2 emissions. Microbial Carbohydrate Active Enzymes (CAZymes) are the most crucial enzymes for the generation of sustainable biofuel energy. The present study designed shotgun metagenomics approaches to assemble, predict, and annotate, aiming to gain an insight into the taxonomic diversity, annotate CAZymes, and identify carbohydrate hydrolyzing CAZymes from microbiomes in Menagesha suba forest soil for the first time.
Results
The microbial diversity based on small subunit (SSU) rRNA analysis revealed the dominance of the bacterial domain representing 81.82% and 92.31% in the studied samples. Furthermore, the phylum composition result indicated the dominance of the phyla Proteobacteria (23.08%, 27.27%), Actinobacteria (11.36%, 20.51%), and Acidobacteria (10.26%, 15.91%). The study also identified unassigned bacteria which might have a unique potential for biopolymer hydrolysis. The metagenomic study revealed that 100,244 and 65,356 genes were predicted from the two distinct samples. A total number of 1806 CAZyme genes were identified, among annotated CAZymes, 758 had a known enzyme assigned to CAZymes. Glycoside hydrolases (GHs) CAZyme family contained most of the CAZyme genes with known enzymes such as β-glucosidase, endo-β-1,4-mannanase, exo-β-1,4-glucanase, α-L-arabinofuranosidase and oligoxyloglucan reducing end-specific cellobiohydrolase. On the other hand, 1048 of the identified CAZyme genes were putative CAZyme genes with unknown enzymatical activity and the majority of which belong to the GHs family.
Conclusions
In general, the identified putative CAZymes genes open up an opportunity for the discovery of new enzymes responsible for hydrolyzing biopolymers utilized for biofuel energy generation. This finding is used as a first-hand piece of evidence to serve as a benchmark for further and comprehensive studies to unveil novel classes of bio-economically valuable genes and their encoded products.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03436-9.
Keywords: Carbohydrate-active enzyme, CAZyme genes, Forest soil, Shotgun metagenomic sequencing
Introduction
Microorganisms are highly diversified and enormously represent living things in the biosphere. Especially, the soil environment is highly diversified and significantly a wide variety of microbial niches [1, 2]. As a matter of fact, microbial diversity in soils exceeds that of other environments like water, air, and unique environmental niches. Notably, the forest soil habitat is one of the largest and most suitable environments in our ecosystem and is home to diversified soil-dwelling microbes. Microbial community inhabiting forest soil is highly rich and diversified due to the accumulation of organic matter of plant waste recalcitrant biopolymers in their soil habitat, such as the complex biomass of wood, roots, litter, and other organic matter, which leads to the influx of carbon compounds into the forest soil [3]. This environment creates an opportunity for diversified microorganisms in the forest soil.
The culture-dependent approaches have led to wasteful efforts in the identification of just cultured microbes from the environment and unexplored hidden microbes available in the environment. Recently, in the era of next-generation sequencing technology, the development of bioinformatics as an independent field of specialization with well-structured biological databases is the cornerstone for the metagenomic approaches. Shotgun metagenomic approaches are the most powerful method for the discovery of most microorganisms with their genetic potential from the environment via directly extracting the metagenome [4]. It has a key role in driving biotechnological application via facilitating the discovery of new enzymes, which is essential to addressing emerging challenges in various fields such as bioenergy generation, for drug development, and agriculture.
Nowadays, the overreliance on fossil fuel energy is an emerging problem, the utilization of fossil fuel energy contributing to greenhouse gas emissions is the main environmental challenge in the world [5, 6]. An alternate fuel production approach will be needed from renewable biomass and sustainable energy production. Biofuel is one of the promising renewable and sustainable energy resources to help reduce the environmental challenges of CO2 emission and the depletion of hydrocarbon [7].
Potential enzymes produced by microorganisms play a great role in degrading the lignocellulosic biopolymer plant materials, especially the microbial community that survives in the forest soil have a potential ability. The process of biopolymer conversion via microbial potential enzymes is called Bioconversion. Bioconversion of recalcitrant plant material is a complex process, it requires the action of a diverse group of microbial enzymes consortia to convert into fermentable sugar for bioenergy generation [8]. Microbial enzymes have the primary potential role in the production of biofuel from forest material.
Microbial diversity is highly abundant, particularly in the forest soil, and plays the central role of hydrolyzing the complex lignocellulosic biopolymer for producing renewable biofuel energy. The core microbial enzymes responsible for this process are microbial carbohydrate-active enzymes. CAZymes are a group of enzymes that are associated with the complex carbohydrate build-up or synthesis, breakdown, and modification of the glycosidic bonds. CAZymes can be classified into six classes in the CAZy database based on sequence similarity and substrate specificity, such as Glycoside Hydrolase, GlycosylTransferase, Polysaccharide Lyase, Carbohydrate Esterase, Auxiliary Activities, and Carbohydrate-Binding Module. From these classes of enzymes, Glycoside Hydrolase is more diversified and responsible for the breakdown of glycosidic bonds that link the complex carbohydrate as well as non-carbohydrate molecules that play a vital role in the generation of biofuel energy [9]. Those groups of enzymes are organized and structured into families and subfamilies in the CAZy database.
This research aimed to gain a comprehensive overview of the microbial diversity, abundance, and functional potential of the microbiome in Menagesha Suba forest soil through the application of shotgun metagenomics approaches. In addition, the study specifically focused on identifying CAZymes genes responsible for hydrolyzing carbohydrate substrates. As the study utilizes a shotgun metagenomic approach it can fill the gap of a culture-dependent method which introduces the problem of discovering the same enzyme over and over again. In addition, the study also facilitates the utilization of lignocellulosic biopolymer potential using microbial enzymes for the production of biofuels. In the long run, this research can address the emerging issue of greenhouse gas emissions by functionally testing the identified CAZymes genes which are important in the production of biofuels.
Materials and methods
Description of the study area
The sampling site for this study was Menagesha Suba Natural Forest, which is recognized as one of the oldest and largest natural rainforests in Ethiopia. This forest is located southwest of the Oromia region and approximately 55 km from the capital city, Addis Ababa. The forest has a latitude of 8°56’12” N and a longitude of 38°50’54” E. It spans an elevation range of 2,300 to 3,000 m above sea level/m. a.s. l/ and experiences an annual rainfall ranging between 900 mm and 1500 mm, whereas the mean average temperature of the forest is estimated to be 16.5 °C. the forest area is known to have a Loam and Clay loam soil textural class. The menagesha suba forest is covered with various array of tree species and has preserved indigenous natural trees, among these trees are Juniper procera locally known as (Ted in Amharic), Hygenia abyssinica (kosso in Amharic), Podocarpus gracilior (zigba in Amharic), African olive (weyira in Amharic), Prunus Africana (red stinkwood), and Cordia Africana (wanza in Amharic). Among the different tree species, Juniperus procera, Olea europea, subsp cuspidate are the top three dominant species in the forest [10]. The thriving ecosystem within Menagesha Suba Forest sustains a wide array of plant and animal life. For this study, two soil samples were collected explicitly from different locations within Menagesha Suba Forest; The first sample, coded as ‘AMF1’, was obtained from the northern part of the forest, while the second sample, coded as ‘AMF2’, was collected from the southern part. Twenty (20) sites were selected from each sampling site and the soil was collected aseptically.
Soil sample collection
From the AMF1 and AMF2 Menagesha Suba Natural Forest sites, a total of 20 soil samples were collected from randomly selected points for both sites. Before collecting the soil samples from each site, the soil surface litter or layer was removed using a sterilized spatula until clean soil. The soil samples were obtained using a cone cutter with a diameter of 10 cm and a depth of 15 cm from the randomly selected sampling places at 20 points in each site. To create composite soil samples, the collected soil samples from each site were pooled, mixed in separate sterile plastic containers, and thoroughly homogenized. Adequate composite soil samples were collected separately from the two forest sites and placed in sterile polyethylene plastic bags and appropriately labeled, which were then transported to the laboratory in an ice box to maintain low temperatures. The soil samples were filtered through a 0.2 mm sieve to remove larger particles and stored at -20 ºC until DNA extraction.
Physicochemical analysis of soil
The soil samples’ physicochemical characteristics were analyzed, and parameters such as pH, electrical conductivity (EC), and total dissolved solids (TDS) were measured using a HANNA HI98194 Multiparameter Portable Meter., and the temperature of soil samples was measured using a thermometer. The organic carbon (OC) was measured using Walkley–Black titration method by TITREX 2000 instrument [11]. The total nitrogen of soil samples was analyzed using the Kjeldahal digestion method by Gerhardt, KT 40s instrument [12]. The carbon-nitrogen ratio was calculated. The proportion of Sand, Silt, and Clay was measured using Hydro Meter (H152 model) [13], and the soil texture classification was analyzed using USDA Soil Triangle. Furthermore, the particle density of soil samples was determined using a Pycnometer [14].
Metagenomic DNA extraction, purification, and quantification
Metagenomic DNA from two forest soil samples was extracted by using an improved metagenomic DNA extraction method, developed by [15]. The extracted metagenomic DNA was purified using a mini spin column [EXgene TM] and the quality of the purified metagenomic DNA was evaluated on a 0.8% agarose gel and visualized using a gel documentation system [UVITEC], and the purity and quantity of the extracted DNA were determined using Nanodrop [Thermo Scientific 2000 instrument]. Subsequently, the triplicate extracted and purified metagenomic DNA samples were pooled into one for each soil sample.
Metagenome library preparation and sequencing
The purified environmental DNA was sent to NovogeneAIT Genomics Singapore for sequencing. The metagenomic DNA was sheared into short fragments, which involved the process of library preparation. The obtained fragments were end-repaired, A-tailed, and further ligated with an Illumina adapter. The fragments with adapters were PCR amplified, size selected, and purified. The library was assessed using Qubit and real-time PCR for quantification and a bioanalyzer for size distribution detection. Quantified libraries were pooled and whole metagenome shotgun sequencing was carried out in an Illumina NovaSeq 6000 with the running mode of PE 150 (paired-end 150 bp) conducted.
Metagenomic data preprocessing
Metagenomics sequences data, including forward read (F1) and reverses read (F2) sequences were uploaded and analyzed for both preprocessing and downstream sequences analysis using the Galaxy Europe server [16]. The quality assessment of the raw reads metagenomic sequences was performed using the FASTQC tool (Galaxy version 0.73, default parameter). To eliminate the influence of host genomes, both plant and human genome sequences were removed from the metagenomic dataset by applying the following steps. Initially, the human reference genome were mapped to the metagenomics sequences data sets using BWA MEM alignment tool. Then read pairs not mapped to the human genome sequences were filtered and extracted their identifiers using Samtools fastx (Galaxy version 1.9 + galaxy1). Then use the non-human read identifiers to extract the reads of interest from the original inputs using seqtk_subseq (Galaxy version 1.3.1) tool. Similarly to remove the plant host genome, Arabidopsis thaliana references genome sequences were mapped with the filtered metagenome sequences as an input data set and the subsequent process is similar to removing the human host genome sequences.
De novo assembly and quality control
The preprocessed raw paired end metagenomic sequences data was assembled using two different assemblers, metaSPAdes (Galaxy version 3.15.4, default parameters) and MEGAHIT (Galaxy version 1.2.9, default parameters) tools. Then, the quality assessment of assembled contigs was performed using MetaQUAST (Galaxy version 5.2.0) to evaluate various metrics. Based on the evaluation metrics, contigs were selected from the output of the metaSPAdes assembler as input for the subsequent steps.
Microbial taxonomic diversity analysis
A pipeline was implemented to evaluate the microbial diversity in forest soil samples. RNA prediction was performed on the assembled contigs using cmsearch tool (Galaxy version 1.1.4) with the help of the Rfam (RNA family) Database. Then, the coordinates and sequences of small subunit (SSU) were extracted using Easel software (version 0.48). Taxonomic assignment was determined using the SILVA database via the MAPseq tool [17]. Finally, the resulting taxonomic distribution of domain and phylum composition was visualized using a pie chart, generated through the online plotting tool, Plotly Chart Studio [18].
Metagenome gene prediction
The de novo assembled metagenomes were used for gene prediction. The FragGeneScan tool (Galaxy version 1.30.0) was utilized with default parameters to predict putative genes. The tool generated both nucleotide sequences and protein sequences in file formats for further analysis.
Functional annotation and pathway analysis
The functional diversity within these metagenomes was explored by analyzing the predicted coding sequences using the InterPro database via the InterProScan tool (Galaxy version 5.59-91.0). This analysis allowed for the extraction of functional annotations from InterPro summary results associated with protein family (Pfam) and identifying gene products assigned to the Gene Ontology (GO) terms, providing insights into the molecular functions, biological processes, and cellular components represented in the metagenomic data.
Additionally, to annotate KEGG Orthology (KO) and perform KEGG Module pathway analysis, the study accessed the KEGG database. The KAAS (KEGG Automatic Annotation Server, Ver. 2.1) available at [19] was utilized. This powerful tool automatically assigns KEGG Orthology numbers to genes and offers valuable insights into metabolic pathways. The result is represented using a pie chart and bar graph design in Plotly Chart Studio.
CAZymes annotation and identification of carbohydrate-degrading enzymes
In this study, dbCAN3 was used to predict and annotate genes encoding CAZymes from metagenome contig sequences. The dbCAN3 server [20] is a web-based tool specifically designed for automated annotation of CAZymes and substrates. By using three integrated tools/databases, the CAZymes were annotated into families, subfamilies, and enzymes, along with respective substrates. After annotating CAZymes, specific CAZyme families were manually selected to establish a link between the genes and the decomposition of plant biomass carbon.
Results
Physicochemical properties of soil
The physicochemical properties of the soil sample of both sites were measured in triplicate and the mean and standard deviation for both samples are indicated in supplementary Table 1. In this study, the analysis of the properties of the two soil sample sites didn’t show significant differences. The organic carbon of the soil was 4.49% and 3.76% in the AMF1 and AMF2 samples, respectively and the soil pH was indicated neutral for both study areas. The parameters of the chemical analysis result included the measurement of organic carbon and total nitrogen expressed with (%), Electrical conductivity (EC, µS/cm), pH, T (°C), and total dissolved solids (TDS, mg/L) and the results of the physical analysis of both AMF 1 and AMF 2 soil samples including the assessment of soil texture and particle density (g/cm3) are also indicated in supplementary Table 1.
Metagenome raw sequence statistics
In total, 6.8 Gb and 7.1 Gb raw data were generated by the Illumina sequencing platform, for AMF1 and AMF2 soil metagenome, respectively. The total amount of raw reads generated for AMF1 was 45,400,974 and 47,408,332 for the AMF2 sample. The raw read metagenome sequence had a length of 150 bp. The raw sequence statistics showed the GC content 63.42% and 62.52%, effectiveness 99.81% and 99.78%, and error rate 0.03% and 0.03%, for AMF1 and AMF2, respectively.
Metagenome assembled contigs
From metagenome de novo assembly, there were significant differences in contigs numbers. In total, 68,265 contigs from AMF1 and 44,782 contigs from AMF2 sequence were generated with the minimum, average, and maximum contigs length. The minimum contig length in both samples were 500 bp, the average contig length were 674.35 bp and 681.06 bp, and the maximum contig length were 9919 bp and 8824 bp for the AMF1 and AMF2 sample, respectively. The GC content indicated 62.78% and 62.29%, and the AT content were 37.22% and 37.71% for the AMF1 and AMF2 sample, respectively.
Microbial diversity
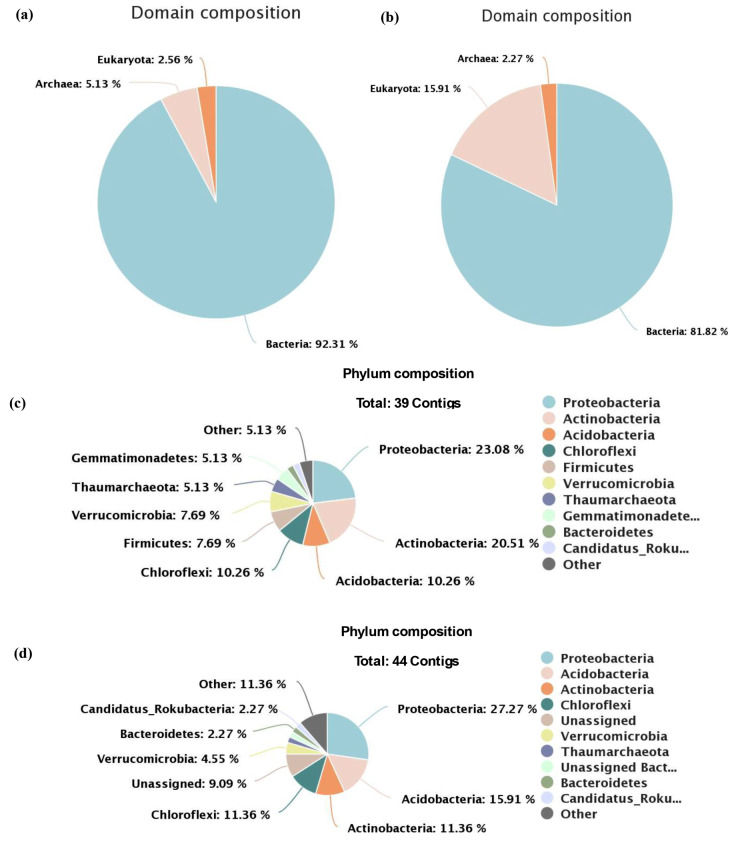
Taxonomic annotation of predicted contigs, a total of 533 and 432 RNAs were generated in the AMF1 and AMF2 samples, respectively. From the total predicted RNA 39 contigs were small subunit (SSU) rRNA obtained from AMF1 samples and 44 contigs were small subunit (SSU) rRNA generated for AMF2 samples. The taxonomic compositions of the AMF1 sample domain were dominated by 92.31% bacteria, 5.13% Archaea, and 2.56% Eukaryota obtained based on SSU rRNA analysis. The Domain compositions of AMF2 were dominated by 81.82% Bacteria, 15.91% Eukaryota, and 2.27% Archaea obtained based on SSU rRNA analysis. The domain composition based on SSU rRNA analysis is illustrated in Fig. 1, (a) and (b) representing the AMF1 and AMF2 samples, respectively.
Fig. 1.
Illustrates the domain composition and the top ten taxonomic analyses of microbial diversity at the Phylum level composition based on SSU rRNA analysis, (a) and (b) represent the domain composition for AMF1 and AMF2 samples, respectively. The bacteria domain is the more dominant domain in both samples than the Archaea and Eukaryota domains. The pie chart (c) and (d) represents the phylum composition of AMF1 and AMF2 samples, respectively. From the microbial taxonomic analyses, proteobacteria were more abundant phyla in both samples
The microbial diversity at the phylum level composition of both samples retrieved did not reveal significant differences. The relative abundances of microbes at the phylum level of both samples generated based on SSU rRNA analysis were 23.08% Proteobacteria, 20.51% Actinobacteria, 10.26% Acidobacteria, 10.26% Chloroflexi for sample AMF1 and 27.27% Proteobacteria, 15.91% Acidobacteria, 11.36% Actinobacteria, and 11.36% Chloroflexi for sample AMF2 were the most abundant bacteria phyla. The pie chart represents the top ten taxonomic analyses of microbial diversity at the phylum level based on SSU rRNA analysis for the AMF1 and AMF2 samples in Fig. 1, (c) and (d) respectively. Based on SSU rRNA analysis, the phylum composition both samples showed that Proteobacteria were more abundant phyla followed by Actinobacteria, and Acidobacteria in both studied areas.
Predicted CDS and functional annotation
From the trimmed contigs, 68,070 and 44,447 contigs with predicted CDS were identified in the AMF1 and AMF2 samples, respectively. In the functional annotation of both samples, there were slightly significant differences. In total 100,244 and 65,356 predicted CDS were generated from the AMF1 and AMF2 samples, respectively. From those Predicted CDS with InterProSan match, 64,406 for AMF1 and 41,119 for AMF2 samples were identified.
InterPro annotation
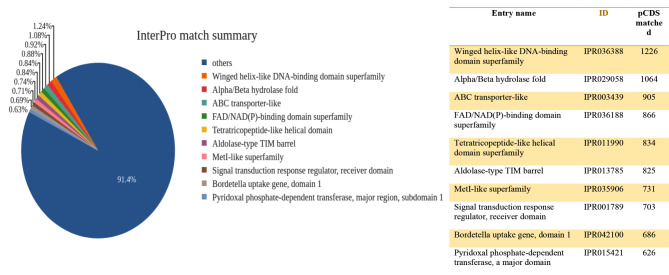
From the total number of predicted CDS, the predicted CDS matched with InterPro entries were 98,802, and 63,353 IntrerPro matches generated from the AMF1 and AMF2 samples, respectively. The AMF1 InterPro matches contained diverse annotated genes compared to the second sample AMF2. The AMF1 sample contained the more abundant matched in winged helix-like DNA-binding domain superfamily, with 1226 counts InterPro matched followed by 1064 counts for the Alpha/Beta hydrolase fold and 905 counts for the ABC transporter-like were also identified matched predicted sequences as depicted in Fig. 2.
Fig. 2.
The pie chart illustrates the InterPro match summary with pCDS matches and description for the AMF1 sample
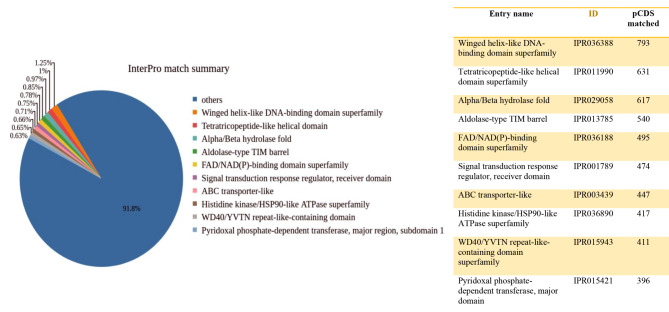
In the AMF2 sample, the winged helix-like DNA-binding domain superfamily was more abundant InterPro matched with 793 counts followed by 631 counts of Tetratricopeptide-like helical domain superfamily, 617 counts of Alpha/Beta hydrolase fold, and 540 counts of Aldolase-type TIM barrel predicted coding sequences matched were identified in the sample (Fig. 3).
Fig. 3.
The pie chart illustrates the InterPro match summary with pCDS matches and description for the AMF2 sample
Protein family (Pfam) annotation
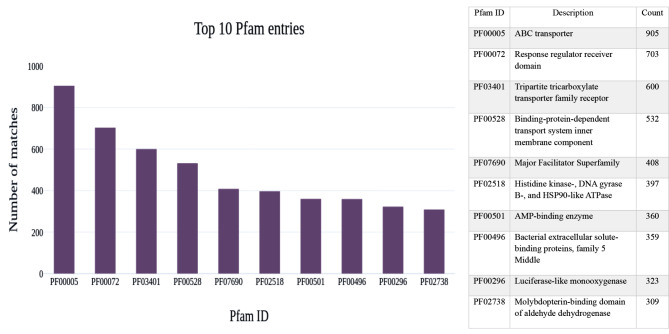
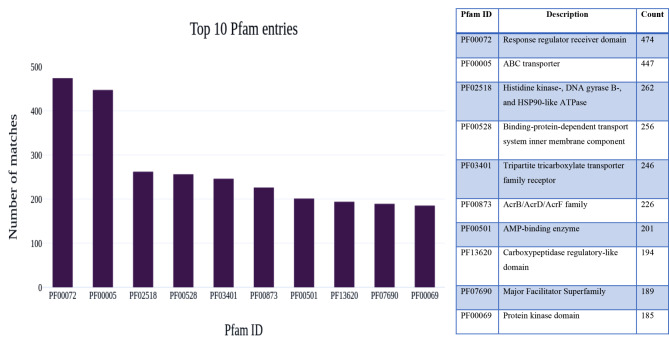
The extracted protein family (Pfam) annotations result from the InterPro annotation were reported in the top ten Pfam entries with the descriptions for the AMF1 and AMF2 samples. The count of annotated protein families present in both samples was significantly different. Among the protein family annotation, the ABC transporter with 905 counts was the most abundant Pfam matched followed by the Response regulator receiver domain with 703, and the Tripartite tricarboxylate transporter family receptor with 600 pfam matched was identified in the AMF1 sample. In Fig. 4, the bar graph shows the Pfam entries with the number of matches present in the AMF1 sample. In the AMF2 sample, among the protein family annotation, the response regulator receiver domain with 474 counts was the most abundant Pfam entry matched, followed by the ABC transporter with 447 counts. In addition, Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase with 262 counts Pfam matches were abundant and identified in the sample (Fig. 5).
Fig. 4.
The bar graph shows the Pfam entries with the number of matches with descriptions present in the AMF1 sample
Fig. 5.
The bar graph shows the Pfam entries with the number of matches with descriptions present in the AMF2 sample
Gene Ontology (GO) annotation
A total of 31,673, 44,142, and 7446 contigs were assigned to biological processes, molecular functions, and cellular component annotations respectively, from the AMF1 sample. On the other hand, 20,391, 28,455, and 4852 contigs originating from sample AMF2 were assigned to biological processes, molecular functions, and cellular components respectively.
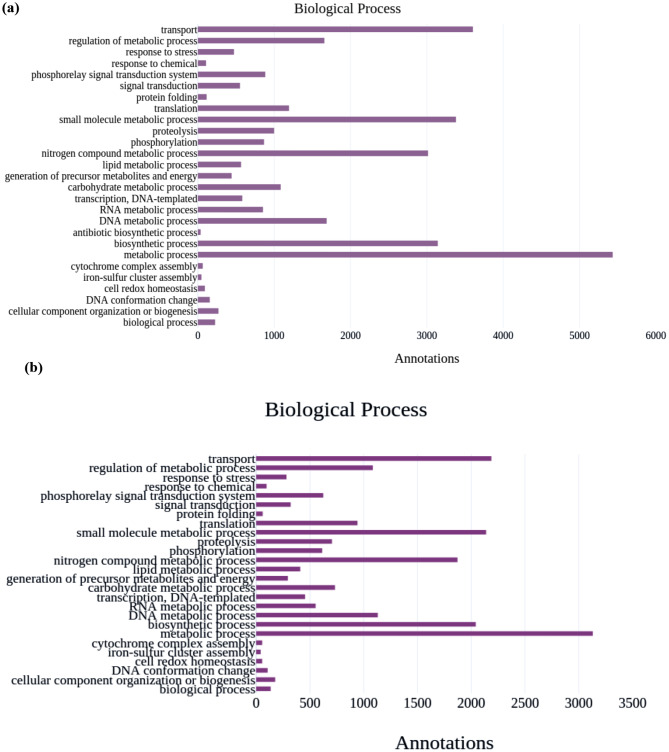
From the biological process, the most abundance were 5434 metabolic processes, which include 1085 carbohydrate metabolic processes, 3603 transport, and 3381 small molecules metabolic processes in the AMF1 sample, and in the AMF2 sample, 3130 metabolic processes which include 733 carbohydrate metabolic processes, 2187 transport, and 2138 small molecules metabolic process, were the abundance of biological processes. The biological process of GO terms illustrated in the bar graph, (a) represents the GO annotation for the AMF1 and (b) represents the GO annotation for the AMF2 sample (Fig. 6).
Fig. 6.
Biological process of GO terms. The bar graph, (a) represents the GO annotation for the AMF1 and (b) represents the GO annotation for the AMF2 sample
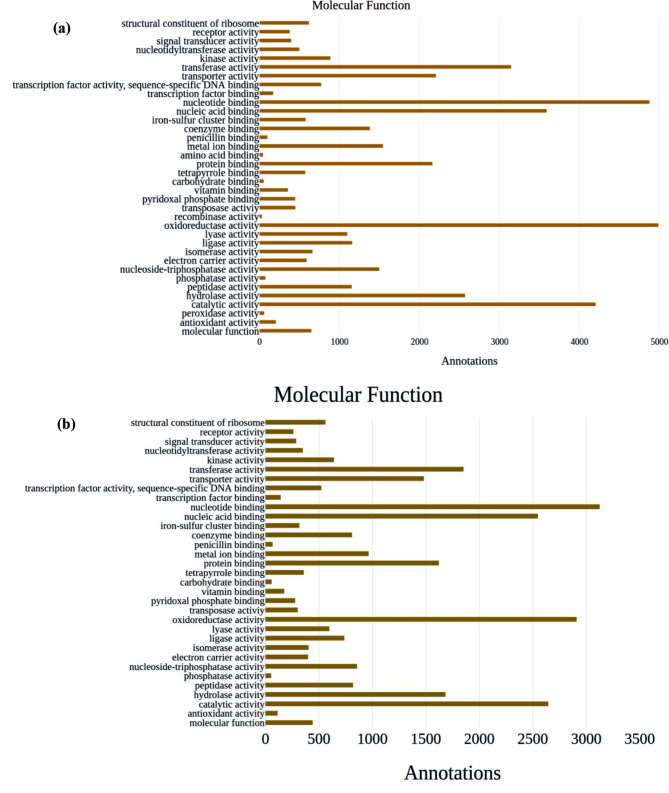
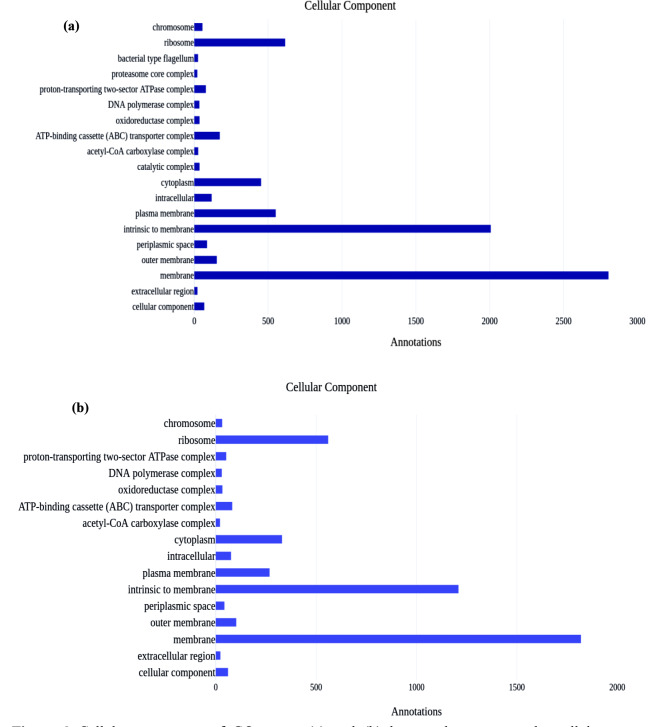
In the molecular function, 4987 oxidoreductases activity, 4875 nucleotide binding, and 4202 catalytic activities were the most abundant molecular function, and specifically, 2569 hydrolase activity, and 51 carbohydrate-binding were available in the AMF1 sample, and 3125 nucleotide binding, 2910 oxidoreductases activity, and 2644 catalytic activities were the most diversified molecular function, and 1683 genes assigned in hydrolase activity and 57 assigned in carbohydrate-binding were present in the AMF2 sample. The molecular function of GO terms shown in Fig. 7, (a) and (b) bar graph represents the molecular function of GO annotation for the AMF1 and AMF2 samples, respectively. In a cellular component, 2804 membranes, 2007 intrinsic to membrane, and 615 ribosomes were the most abundant processes in the AMF1 sample, and 1819 membranes, 1209 intrinsic to membrane, 560 ribosomes were the most abundances annotation in the AMF2 samples (Fig. 8). The Cellular component of GO terms is illustrated in. (a) and (b) bar graph represents the cellular component of GO annotation for the AMF1 and AMF2 samples, respectively.
Fig. 7.
Molecular function of GO terms. (a) and (b) bar graph represents the molecular function of GO annotation for the AMF1 and AMF2 samples, respectively
Fig. 8.
Cellular component of GO terms. (a) and (b) bar graph represents the cellular component of GO annotation for the AMF1 and AMF2 samples, respectively
KEGG orthology annotation and KEGG module analysis
The KASS (KEGG Automatic Annotation) Server provided the KEGG orthology (KO) annotation of the AMF1 and AMF2 samples and KO entry numbers were identified annotated protein assigned in the orthology groups present for each sample. Among annotated proteins assigned to the orthology groups, RNA polymerase sigma-70 factor, ECF subfamily with 174 counts was the most abundant KEGG orthology group followed by 166 putative transposase and 160 Transposase were identified in the AMF1 sample. From the KO annotation in the AMF2 sample, the annotated proteins assigned to the orthology groups in the RNA polymerase sigma-70 factor, ECF subfamily with 142 counts was the most abundant KEGG orthology group followed by 113 Transposase and 112 putative transposases were identified in the sample.
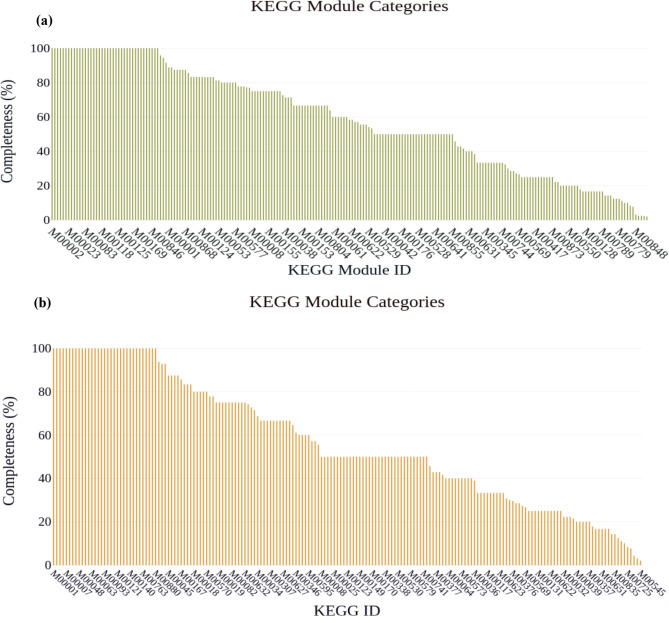
In this study, the KEGG module was analyzed for both samples to gain insight into their biological pathway and functional annotations. In total, 215 and 185 KEGG pathways module identifiers were detected from AMF1 and AMF2 samples, respectively. From those 39 modules in the AMF1 and 34 modules in the AMF2 samples were completed KEGG module pathway classes and analyzed with 100% completeness or annotated all KO identifiers associated with the KEGG pathway module system of both samples. The KEGG pathway module identifiers with completeness illustrated in Fig. 9: (a) and (b) represents bar graph of the AMF1 and AMF2 samples, respectively.
Fig. 9.
Illustrate KEGG pathway module identifiers with completeness: (a) and (b) represents bar graph of the AMF1 and AMF2 samples, respectively
Carbohydrate active enzymes (CAZymes) annotation
The dbCAN3 server was used for accurate prediction and annotation of the CAZymes’ genes from the contigs and identified the putative CAZyme genes and aligned with the manually curated CAZy database set in dbCAN3. The six classes of CAZymes were predicted and annotated to identify the putative CAZyme genes and genes that had been assigned with EC numbers for both samples. The most abundant enzyme classes were GHs followed by GTs in each sample. The total number of CAZymes genes identified in the AMF1 and AMF2 samples were 1006 and 800, respectively.
From those total predicted CAZymes genes, 412 GHs belong to 52 families, 390 GTs belong to 28 families, 96 CEs belong to 12 families, 27 PLs belong to 9 families, 40 AAs belong to 8 families and 41 CBMs belong to 9 families identified in the AMF1 sample and from the AMF2 sample, 268 GHs belong to 61 families, 253 GTs belong to 20 families, 108 CEs belong to 12 families, PLs belong to 9 families, 44 AAs belong to 7 families, and 42 CBMs belong from 9 families were generated. Among identified CAZymes classes, the Glycoside Hydrolases were the most abundant CAZymes families, and subfamily enzymes with EC numbers were summarized in Table 1 for the AMF1 sample and Table 2 for the AMF2 sample. From those identified matched enzymes with the CAZy database, the GH13 enzymes were the abundant enzymes in both samples like 1,4-alpha-glucan branching enzyme, and β-glucosidase enzymes belonging to different GH families responsible for cellulose hydrolyzing were identified in both studies areas.
Table 1.
dbCAN predicted CAZymes genes match with enzyme EC numbers for the AMF1 sample
| CAZyme Classes | Family | Sub-Family | Enzyme | EC |
|---|---|---|---|---|
| GH | GH23 | - | peptidoglycan lyase | 4,2.2. n1 |
| GH13 | GH13_26 | malto-oligosyltrehalose synthase, -alpha-D-glucan 1-alpha-D-glucosylmutase | 5.4.99.15 | |
| GH13_29 | alpha, alpha-phosphotrehalase | 3.2.1.93 | ||
| GH13_9 | 1,4-alpha-glucan branching enzyme | 2.4.1.18 | ||
| GH13_11 | Isoamylase | 3.2.1.68 | ||
| GH13_16 | maltose glucosylmutase | 5.4.99.16 | ||
| GH13_23 | alpha-glucosidase, oligo-1,6-glucosidase | 3.2.1.20, 3,2,1.10 | ||
| GH13_10 | maltooligosyltrehalose trehalohydrolase | 3.2.1.141 | ||
| GH3 | - | β-glucosidase | 3.2.1.21 | |
| GH5 | GH5_7 | endo-β-1,4-mannanase | 3.2.1.78 | |
| GH74 | - | oligoxyloglucan reducing end-specific cellobiohydrolase | 3.2.1.150 | |
| GH1 | - | exo-β-1,4-glucanase | 3.2.1.74 | |
| GH51 | GH51_2 | xylan β-1,4-xylosidase | 3.2.1.37 | |
| GH43 | GH43_10 | α-L-arabinofuranosidase | 3.2.1.55 | |
| GH77 | - | Amylomaltase | 2.4.1.25 | |
| GH15 | - | alpha, alpha-trehalase | 3.2.1.28 | |
| - | glucan 1,4-alpha-glucosidase | 3.2.1.3 | ||
| GH95 | - | 1,2-alpha-L-fucosidase | 3.2.1.63 | |
| GH27 | - | alpha- galactosidase | 3.2.1.22 | |
| GH29 | - | Alpha-Lfucosidase, alpha-1,2-L-fucosidase | 3.2.1.51, 3.2.1.63 |
Table 2.
dbCAN predicted CAZymes genes match with enzyme EC numbers for the AMF2 sample
| CAZyme Classes | Family | Sub-Family | Enzyme | EC |
|---|---|---|---|---|
| GH | GH2 | - | beta-galactosidase | 3.2.1.23 |
| GH20 | - | beta-hexosaminidase | 3.2.1.52 | |
| GH37 | - | alpha, alpha-trehalase | 3.2.1.28 | |
| GH171 | - | peptidoglaycan beta-N-acetylmuramidase | 3.2.1.92 | |
| GH44 | - | Endoglucanase | 3.2.1.4 | |
| GH29 | - | 1,3-alpha-L-fucosidase, alpha-L-fucosidase | 3.2.1.111, 3.2.1.51 | |
| GH51 | GH51_1 | alpha-L-arabinofuranosidase | 3.2.1.55 | |
| GH3 | - | beta-glucosidase, xylan 1,4-beta-xylosidase | 3.2.1.21, 3.2.1.37 | |
| GH67 | - | alpha-glucuronidase, xylan alpha-1,2-glucuronidase | 3.2.1.139, 3.2.1.131 | |
| GH10 | - | endo-1,4-β-xylanase | 3.2.1.8 | |
| GH116 | - | β-glucosidase | 3.2.1.21 | |
| GH1 | - | β-glucosidase | 3.2.1.21 | |
| GH13 | GH13_9 | 1,4-alpha-glucan branching enzyme | 2.4.1.18 |
Putative carbohydrate-active enzyme genes that do not have EC numbers were derived from the CAZy database. From the total CAZymes annotation, 503 and 545 putative CAZymes genes were obtained in the AMF1 and AMF2 samples, respectively. Forty-six (46) GHs families, 26 GTs families, 11 CEs families, 8 AAs families, 7 PLs families, and 8 CBM CAZymes families were obtained in the AMF1 sample and 53 GHs families, 18 GTs families, 12 CEs families, 8 PLs families, 7 AAs families, and 9 CBMs families were acquired from AMF2 sample. A high relative abundance (39.2% and 31.2%) of CAZymes belonging to Glycoside Hydrolases (GHs) involved in hydrolyzing the glycosidic bond of carbohydrate compounds and coding genes for putative Glycosyltransferases (GTs) ( 30.4% and 28.6%), Carbohydrate Esterases (CEs) (14.9% and 16.3%), Carbohydrate-binding Module (CBM) ( 8.15% and 7.7%), Auxiliary Activities (AAs) (5.36% and 5.5%) and Polysaccharide Lyases (PLs) (2% and 10.6%) have been identified for AMF1 and AMF2 samples, respectively.
Detection of carbohydrate-degradation CAZymes
The main families of CAZymes responsible for the decomposition of plant-drive biomass have been identified. GH5, GH3, GH8, GH9, GH12 and GH1 were utilized for cellulose, GH109, GH30, GH39, GH74, GH10, GH51, GH43, CE4, CE1, CE7, CE15, and CE3 were used for hemicellulose breakdown and AA3, AA6, AA1, AA5, AA4, and AA2 were utilized for lignin degradation were detected in the AMF1 sample. However, GH3, GH5, GH9, GH1, GH116, and GH12 were utilized for cellulose breakdown, GH43, GH51, GH36, GH39, GH95, GH67, GH10, GH2, CE4, CE1, CE3, CE12, CE7, and CE15 were used for hemicellulose degradation, and AA3, AA5, AA1, AA2, and AA4 were used for lignin decomposition obtained in the AMF2 sample. The genes encoding for carbohydrate binding modules were obtained in both studied areas. In the AMF1 sample, CBM32, CBM9, CBM50, CBM66, CBM48, CBM5, CBM57, and CBM6 were identified, however in the AMF2 sample, CBM16, CBM6, CBM67, CBM2, CBM91, CBM50, CBM48, CBM9, and CBM 32 were present in the sample.
Discussion
Soil physicochemical property and microbial diversity analysis
From the results of the physicochemical properties of the Menagesha suba forest, soil pH was indicated as neutral for both samples, 7.70 in the AMF1 and 7.39 in the AMF2. Neutral pH environmental condition provides a suitable environment for high microbial growth and diversity. Organic carbon in the soil was available in both samples, but the AMF1 sample was higher compared with the AMF2 sample. The availability of organic carbon in the study area indicated the long-term accumulation of plant biomass. This may be associated with a strong correlation between carbon content and microbial diversity and richness. According to previous studies, high organic carbon content can be related to high microbial growth and diversity [21]. In contrast to C, the total nitrogen content was 0.49% in the AMF1 and 0.43% in the AMF2 sample indicates extremely low in both samples. This leads to a suitable C: N ratio (9 in both samples) environment for microbial composition inhabiting it [22]. Additionally, the forest soil texture was loam and clay loam in AMF1 and AMF2 samples, respectively. This gives the soil property to absorb water and organic matter to provide suitable inhabiting habitats for microbes [23]. The electrical conductivity (EC) indicating salinity or salt concentration in the soil were measured to be 151 µS/cm in the AMF1 and 173 µS/cm in the AMF2 samples, this indicates low salt concentration in both samples. Lower concentration of salt in the soil can be related with favorable environment for microbial growth.
The microbial diversity analysis of the Menagesha suba forest was revealed to have a highly remarkable richness of bacterial domain compositions inhabiting the soil based on the results of small subunit (SSU) rRNA analysis than archaea and Eukaryota domain in both sample sites. Similarly, previous studies reported that bacteria domain composition was higher than archaea and Eukaryota in the forest soil [24, 25]. The higher bacteria domain in the study areas indicated the functional diversity of the bacteria community. The forest soil in this area plays a vital role in microbial diversity due to the accumulation of plant litter and organic matter. The identified Bacterial phyla in the studied area play diversified ecological roles including organic matter decomposition, Nutrient cycling, Soil health maintenance, plant-pathogen interaction, and pathogen suppression [3]. The most relative abundance of bacteria at the phylum level was observed to be Proteobacteria in both samples and this was followed by Actinobacteria, Acidobacteria, and Chloroflexi. The relative abundance of Proteobacteria at the phylum level was aligned with previous studies reported in forest soil [25], forest, and grassland [26] and a comparative study of two forest sites [27]. As a result of their high adaptability nature and their association with plant roots, Proteobacteria are highly diversified phyla in forest soil and play key ecological functional role in organic matter decomposing and nutrient cycling. Among their versatile metabolic activities, carbon metabolism, provides them with the ability to synthesize CAZymes that are highly efficient for the degradation of cellulose, hemicellulose, and lignin plant biomass [28, 29]. Actinobacteria, Acidobacteria, and Chloroflexi phyla were also abundant in the current study. This finding aligned with similar studies where, Actinobacteria, Acidobacteria, and Chloroflexi phyla were observed abundantly in the three tropical forest soil and forest ecosystems [25, 30]. The influx of carbon in the soil from plant material provides an opportunity for these phyla to metabolize the carbon compounds with the ability to synthesize genes encoding for carbohydrate-active enzyme activity [31]. Bacteroidetes phylum has a unique ability to secrete extracellular enzymes and contains Polysaccharide Utilization genes responsible for the degradation of polysaccharide carbohydrate compounds and have the potential to synthesize diverse groups of CAZymes [32].
Based on the analysis of small subunit rRNA, unassigned bacteria were identified in both study areas, referring to the identification of microbial domain composition in the present study. However, the taxonomic classification was unassigned to the known bacteria taxonomic group present in the reference databases. These unassigned bacteria obtained in the Menagesha Suba forest soil may be indicated as an undiscovered or novel species distinct from the exciting bacteria. This leads to the representation of some microbes in low abundances and is difficult to analyze at the species level. In general, the physicochemical properties of the soil samples including neutral pH, high carbon content, low total nitrogen, clay loam and loam soil textural classes provide a suitable environmental condition for microbial growth and their metabolic activity. The results of microbial diversity analysis revealed the bacteria as a dominant domain and Proteobacteria as the most abundant phylum in the studied samples. Bacteroidetes is a unique phylum that secretes enzyme activity. Owing to the accumulation of recalcitrant plant biomass in the forest and the adaptability of microbes in this environment it provides the potential to synthesize diverse groups of CAZymes.
Functional annotation
The result of predicted coding sequences (pCDS) match with InterPro annotation highlighted the diverse group of protein domains and protein superfamilies that were identified in the studied samples. The winged helix-like DNA-binding domain superfamily was more diversified in both samples. The occurrences of this domain indicate potential involvement in nucleic acid binding and are responsible for DNA metabolism [33]. Alpha/Beta hydrolase fold was present in the current study areas, this protein superfamily suggests the involvement of diverse enzyme activity. Especially, hydrolytic reactions responsible for different substrates [34]. The presence of Alpha/Beta hydrolase fold in our dataset indicates that the microbial community may have an enzyme responsible for the hydrolysis of complex organic matter present in the environment and participate in a different metabolic pathway. The signal transduction response regulator, receiver domain was present in the studied areas, this Signal transduction response regulator, receiver domain is present in the response regulator protein, which is involved in signaling pathways that allow microbes to sense and respond the environmental changes [35].
The protein families that have been identified in the investigated samples from the annotation of InterPro match, ABC transporter, and Response regulator receiver domain were more diversified groups of the protein family in both study areas. The findings suggest that the forest soil microbial community allows sufficient nutrient acquisition and the ability to receive and respond to the environmental signal and this domain is responsible for the survival of the microbial community in this environment [36]. AMP-binding enzyme is a domain of diverse enzymes that have a specific binding site for AMP (Adenosine monophosphate). The findings of those families revealed that the microbial community is used for the ecological role and adaptation of the soil environment. In summary, ABC transporter, Response regulator receiver domain, and AMP-binding enzyme domain play interconnected ecological roles in nutrient uptake, environmental sensing, metabolic adaptation, and regulation. These roles are extremely important in the survival, adaptation, and functioning of the soil microbes.
The annotated gene products were identified as assigned to the biological process, molecular function, and cellular component present in the current study. From the assigned gene products in the biological process, the most abundant were gene products for metabolic processes, which include carbohydrate metabolic processes. The finding of these matched gene products for metabolic processes in our dataset revealed that the microorganisms involved in the breakdown and synthesis of a component utilizing organic matter in the environment. Specifically, the carbohydrate metabolic processes are the breakdown and utilization of complex carbohydrates which is important for the decomposition of plant biomass material and the recycling of carbon sources. The finding of genes responsible for carbohydrate metabolic processes indicates that microorganisms play a vital role by utilizing carbon sources and energy production in the soil ecosystem [37]. These findings provide insight into microorganisms that are associated with the synthesis of CAZymes for the breakdown of recalcitrant plant material and the synthesis of carbohydrates present in the forest soil. From the assigned gene products at the molecular function, the genes or gene products involved in hydrolase activity and carbohydrate binding were present in both samples. The annotated genes that were assigned in hydrolase activity revealed, the microorganisms present in this environment that have the potential to catalyze the hydrolysis of chemical bonds of protein, lipids, carbohydrates, etc. [38], and the carbohydrate-binding revealed that the gene products specifically interact and are recognized with carbohydrates which gives insights into the presence of CAZymes. Some CAZymes have a specific carbohydrate-binding domain to interact and recognize carbohydrate structures which are called carbohydrate-binding modules (CBMs) responsible for activating the process of breaking the carbohydrate by providing a specific position [39]. In general, the hydrolase activity and carbohydrate-binding domain play interconnected ecological roles such as; organic matter decomposition, where hydrolase activity catalyzes the breakdown of complex organic matter and the carbohydrate-binding domain facilitates the attachment of microbial enzyme to specific carbohydrate substrate. Nutrient cycling is another ecological role, where hydrolase activity liberates nutrients from organic compounds and carbohydrate-binding domains targets complex carbohydrates facilitating the degradation process.
From the result of KEGG Orthology (KO) annotation, the orthology proteins were identified in the current study area. From those, RNA polymerase sigma-70 factor, ECF subfamily were the significantly more abundant orthologues group in both samples. The Bacteria Sigma factor is a core element of RNA polymerase to direct promoter specificity and determine the transcription process [40]. The findings of these genes related to RNA polymerase sigma-70 factor belonging to the ECF subfamily in both samples with the high count revealed the microbial community can survive under environmental stress conditions. Transposase and Putative transposase were the most abundant identified KO groups in both samples. These enzymes are responsible for the genetic diversity and evolution of microorganisms [41]. The finding of genes encoding for transposase enzyme suggests that there has been genetic rearrangement within microbial communities. Genetic rearrangement is responsible for driving microbial genetic diversity and adaptation by introducing genetic variation which enables microbes to adapt to various and changing environmental conditions facilitating the acquisition of advantageous traits.
The KEGG Module annotation determined the specific biological pathway present in the studied areas. From those modules’ results, 39 modules in the AMF1 and 34 modules in the AMF2 samples were complete KEGG module pathway classes. The completeness 100% revealed that all gene sets and enzymes involved in the processes of specific biological processes were identified in the shotgun metagenomics data. From all annotated 100% complete module pathway results, specifically, the Pathway modules; Carbohydrate metabolism; Central carbohydrate metabolism were present in the samples. Which includes Glycolysis, core module involving three-carbon compounds identified in both study areas. Gluconeogenesis, oxaloacetate to fructose-6P were identified in both samples, the occurrence of this module pathway in the samples revealed the microbial community has the potential to synthesize glucose from non-carbohydrate compounds. This may be used for different nutrient conditions available in the environment to survive in the environment. The pentose phosphate pathway (Pentose phosphate cycle), Pentose phosphate pathway, oxidative phase, glucose 6P to ribulose 5P and Pentose phosphate pathway, non-oxidative phase, fructose 6P to ribose 5P under the classes of central carbohydrate metabolism were identified in both samples. The existence of the glycolysis/ glycogenesis pathway and the pentose phosphate pathways indirectly indicate the occurrences of CAZymes in the soil metagenome [42]. In summary, KEGG annotation of shotgun metagenomic sequence data gives an insight into the microbial metabolic capability in the environment by identifying KO identifiers and mapping diverse KO identifiers responsible for module pathway systems to understand microorganism potential. The identified KEGG modules such as carbohydrate metabolism, central carbohydrate metabolism, and glycolysis/ glycogenesis pathway play a vital ecological role in nutrient cycling, energy production, and microbial interaction influencing microbial diversity, production, and adaptation.
Carbohydrate active enzymes (CAZymes) annotation
From the annotation of CAZymes results, the most abundant CAZymes classes were Glycoside Hydrolases (GHs) followed by Glycosyltransferases (GTs) in the current study. From the identified Glycoside Hydrolases (GHs), GH23, GH13, and GH109 were the most abundant Glycoside Hydrolases in both samples. Similarly, previous studies showed that Glycoside Hydrolases were the most abundant in forest soil [43]. The GH23, GH13, and GH109 have diverse enzymes that are involved in hydrolyzing the glycosidic bond of peptidoglycan, starch, and hemicelluloses, respectively. In the current study CAZymes annotation result, CAZymes belonging to the GH13 family were the more abundant with different subfamilies.
From the identified Glycoside Hydrolases with EC numbers, β-glucosidase belongs to GH3, GH119, and GH1 families, endo-β-1,4-mannanfamily, αs to GH5_7 subfamily within the GH5 family, exo-β-1,4-glucanase belong to GH1 family, α-L-arabinofuranosidase belong to GH43_10 and oligoxyloglucan reducing end-specific cellobiohydrolase belong to GH74 were most diversified enzymes in both samples responsible for cellulose and hemicellulose degradation. β-glucosidase is an essential enzyme that can hydrolyze the terminal, nonreducing β-D-glucosyl residues by hydrolyzing the β-1,4 glycosidic bond of various glycoconjugates [44]. According to the CAZy database, the GH5 family is one of the diversified from all glycoside hydrolases and have different specificity including endoglucanase (cellulase), and endo-mannanase, in addition to exo-mannanases, exo-glucanases, and β-glucosidase and β-mannosidase. endo-β-1,4-mannanase plays a vital role in specifically targeting and cleavage of the β-D-1,4-mannopyranosyl linkage in mannan, this enzyme has a crucial role for their potential biotechnological applications in biofuel production [45]. The exo-β-1,4-glucanase is one of the cellulase enzymes used to hydrolyze cellulose that acts on the end of the chain at the β-1,4-glucan glycosidic bond linkage. The α-L-arabinofuranosidase is a hydrolytic enzyme that is used to cleavage of α-L-arabinose linked arabinoxylans and arabinogalactans essential components of hemicellulose. This enzyme is used as a debranching enzyme to remove arabinose from the hemicelluloses component and enhance the bioconversion lignocellulosic biomass [46].
The other enzyme was oligoxyloglucan reducing end-specific cellobiohydrolase this enzyme is an exoglucanse responsible for the hydrolysis of cellobiose from the end of the xyloglucan subset of hemicellulose plant biomass. endo-1,4-β-xylanase enzyme belonging to the GH10 family was identified, this enzyme gets attention used to degrade the major component of hemicellulose called xylan [47].
Plant biomass degradation is a sophisticated process that transforms complex carbohydrates into simple sugars. However, the collaborative role of the CAZymes family in specific substrates is a pivotal and essential process in the microbial ecosystem. Different CAzymes families contribute synergistically to hydrolise specific substrates. Some GH families like endo-glucanase responsible for the breakdown of glycosidic bonds within the cellulose chain and the other GH family is used to hydrolyze cellulose that acts on the end of the chain. Additionally, the GH families collaborate in the degradation of hemicellulose compounds. Enzymes like endo-mannanase, exo-mannanases, α-L-arabinofuranosidase, and xylanase enzymes belong to different CAZymes families that hydrolyzed various components of hemicellulose synergistically. The collaborative role of CAZymes breaking various substrates provided insight into the microbial dynamics and ecological balances through nutrient cycling and organic matter decomposition in the ecosystems. Additionally, the synergistic role of CAZymes for plant biomass conversion into simple sugar plays a central role in the application of biofuel production. Carbohydrate Esterases (CEs), Auxiliary Activities (AAs), Polysaccharide Lyases (PLs), and Carbohydrate-binding Module (CBM) CAZymes classes were identified with diversified families in both samples. Similarly, previous studies indicate the presence of those CAZymes classes in the top and deep forest soil [24]. From the result of Carbohydrate Esterases (CEs), the CE4 family was the most abundant followed by CE1 and CE14 in both samples. These CEs are a diverse group of enzymes that are used for the hydrolysis of ester linkage bonds including xylan and chitin [48]. The acetyl xylan esterase belonging to the CE1 family was the abundant enzyme in both samples. This enzyme which catalyzes the hydrolysis of xylan and xylan-oligosaccharides present in the plant material has an important role in the degradation of hemicellulose to produce biofuel. In forest soil, CEs enzymes contribute to the subsequences process to other CAZyme for the breakdown of xylan and chitin. Enzymes belonging to CEs family have a crucial role in nutrient cycling and organic matter decomposition in the ecosystem. From the result of Auxiliary Activities, AA7 was abundant followed by AA3 and AA6 in the AMF1, however, in the AMF2 sample AA3 was abundant followed by AA5 and AA7. These Auxiliary Activities families are essential for degrading lignin (AA3, AA5, and AA6) and chitooligosaccharides (AA7). The Laccase belongs to the subfamily of AA1_1 and was the identified enzyme. Bioconversion of lignocellulose is difficult due to the primary component of lignin complex polymer, this enzyme plays a crucial role in oxidizing the complex polymer [49]. Auxiliary Activities play a pivotal role in unlocking cellulose and hemicellulose plant material through degradation of lignin compound. This CAZyme class is responsible for organic matter turnover and nutrient dynamics in the forest ecosystem. The other identified enzyme classes were Polysaccharide Lyases present in both samples. From those families, PL38 followed by PL9 was the abundant family in both samples. Most of the Polysaccharide lyase enzymes cleave the glycosidic bond within pectin. The degradation of pectin through polysaccharide lyases is essential by increasing nutrient availability and nutrient cycling in the soil environment. Overall, the CAZymes classes are shaping the forest environmental ecosystem and activating microbial growth. Finally, genes for the Carbohydrate-binding Module (CBM) family were identified in our samples, these associated modules are essential for correctly binding carbohydrate substrate to activate the enzymatical process. From the identified CBM family, CBM9, CBM16, CBM6, and CBM2 for cellulose binding, CBM91 for xylan binding, and CBM32 for pectin binding were identified in both studied areas.
Additionally, the more abundant putative CAZymes genes were identified in the current study areas, interestingly, the Glycoside Hydrolases (GHs) families were the more abundant predicted putative genes compared to the other carbohydrate-degrading enzyme genes. The classification of CAZymes is based on sequence similarity and functional domain. The putative CAZymes that do not have EC numbers were derived from the CAZy database and predicted based on the sequences, however, the specific catalytical activity for the substrate is not determined. Overall, CAZymes play a crucial role in the ecological environment by contributing hydrolysis of carbon-based complex plant material and nutrient recycling. The identified CAZymes responsible for degradation have distinct roles in the hydrolyzing process like catalysts activity, transforming complex material into simpler ones by breaking the bonds within complex material. The putative CAZymes genes may have unique catalytical activity for substrate and they may be used for discovering novel enzymes responsible for conversion of lignocellulosic plant material utilized for biofuel production. Further experimental validation will be needed to determine the catalytical activity of putative carbohydrate-degrading enzyme genes that were identified from the Menagesha Suba forest soil using shotgun metagenomics approaches. The prediction of putative CAZymes genes has opened up the discovery of novel enzymes that offer innovative solutions for lignocellulosic biomass conversion to address the challenges of biofuel production. These putative CAZymes genes are needed to be experimentally validated using approaches such as, primer design and functional metagenomics to confirm substrate specificity and enzymatic activity to ensure the potential application of novel CAZymes and to advance our understanding of microbial diversity and enzymatic capability for efficient biomass utilization for biofuel generation.
Conclusion
In this study, a shotgun metagenome approach was utilized to explore microbial and functional diversity and identify CAZymes genes present in the menagesha suba forest soil samples. The results of microbial diversity analysis revealed, Proteobacteria as the dominant bacterial phylum in both samples. The presence of Proteobacteria might contribute to the bioconversion of the forest residues. Furthermore, the identified unassigned bacteria might lead to the finding of novel species. The functional annotation analysis results indicated the potential of the microbial community for, hydrolysis of complex organic matter and different enzymatic activity. These results revealed that the microbial community present in the forest soil participates in different groups of metabolic activity. The main achievement of this study was the successful identification of six classes of CAZymes genes. Among them, GHs were the most abundant and diversified families. Enzymes responsible for cellulose and hemicellulose degradation including, β-glucosidase, endo-β-1,4-mannanase, exo-β-1,4-glucanase, α-L-arabinofuranosidase and oligoxyloglucan reducing end-specific cellobiohydrolase were the most abundant GHs enzymes in both samples. These enzymes are crucial in the sustainable biofuel energy generation and in turn address the problem of greenhouse gas emission. Additionally, the identification of putative CAZymes genes opens up an opportunity for the discovery of new enzymes responsible for hydrolyzing biopolymers.The findings from this study can be used as a first-hand piece of evidence to serve as a benchmark for further and comprehensive studies to unveil novel classes of bio-economically valuable genes and their encoded products.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like thank Biotechnology Department of AASTU and its research and academic support staff members for availing the necessary lab consumables, and reagent for this work.
Abbreviations
- AAs
Auxiliary Activities
- ABC
transporter-ATP-binding cassette (ABC) transporters
- AMF1 and AMF2
Northern and Southern part sampling sites
- CAZymes
Carbohydrate Active Enzymes
- CBMs
Carbohydrate-Binding Modules
- CEs
Carbohydrate Esterase
- dbCAN
Data Base for automated Carbohydrate-active enzyme Annotation
- EC
Electrical Conductivity/ Enzyme commission
- ECF
Extra Cytoplasmic Function
- ENA
European Nucleotide Archive
- GHs
Glycoside Hydrolases
- GO
Gene Ontology
- GTs
GlycosylTransferase
- KAAS
KEGG Automatic Annotation Server
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KO
KEGG Orthology
- pCDS
Predicted Coding Sequences
- PLs
Polysaccharide Lyases
- SSU
Small Subunit
- TDS
Total Dissolved Solids
Author contributions
AM: Wrote the research project proposal, collected samples, conducted laboratory experiments, documented and analyzed data, and drafted the manuscript. BA: Assisted in laboratory technical procedures. AA: Co- research advising, financial support, and idea formulation. MT: Idea formulation, research design and advising, laboratory and financial support. All authors reviewed the manuscript.
Funding
This work was supported by Addis Ababa Science and Technology University’s internal small grant optained for the sequencing of a shotgun metagenome. The funds were mainly used for the purchase of laboratory reagents and gene sequencing purposes. The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and also in writing the manuscript.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author and first author upon reasonable request. The raw shotgun sequence data of both sites generated in this study were deposited to the European Nucleotide Archive (ENA). The studies have been made public with all associated data and it is recently reflected in the ENA browser as study ID PRJEB57985 and Secondary Study Accession number of ERP143008 which could be accessed online by using the link: https://www.ebi.ac.uk/ena/browser/view/PRJEB57985.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rosselló-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25(1):39–67. 10.1016/S0168-6445(00)00040-1 [DOI] [PubMed] [Google Scholar]
- 2.Aislabie J, Deslippe JR, Dymond J. Soil microbes and their contribution to soil services. Ecosyst Serv New Zealand–conditions Trends Manaaki Whenua Press Linc New Zeal. 2013;1(12):143–61. [Google Scholar]
- 3.Lladó S, López-Mondéjar R, Baldrian P. Forest soil bacteria: diversity, involvement in ecosystem processes, and response to global change. Microbiol Mol Biol Rev. 2017;81(2):e00063–16. 10.1128/MMBR.00063-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma J, Prince A, Aagaard KM. Use of whole genome shotgun metagenomics: a practical guide for the microbiome-minded physician scientist. Seminars in reproductive medicine. Thieme Medical; 2014. pp. 5–13. [DOI] [PubMed]
- 5.Höök M, Tang X. Depletion of fossil fuels and anthropogenic climate change—A review. Energy Policy. 2013;52:797–809. 10.1016/j.enpol.2012.10.046 [DOI] [Google Scholar]
- 6.Yi S, Abbasi KR, Hussain K, Albaker A, Alvarado R. Environmental concerns in the United States: can renewable energy, fossil fuel energy, and natural resources depletion help? Gondwana Res. 2023;117:41–55. 10.1016/j.gr.2022.12.021 [DOI] [Google Scholar]
- 7.Yadav AK, Pandey S, Tripathi AD, Paul V. Role of enzymes in biofuel production. Bioenergy Res Eval Strateg Commer Sustain. 2021;1–18.
- 8.Kumar R, Singh S, Singh OV. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J Ind Microbiol Biotechnol. 2008;35(5):377–91. 10.1007/s10295-008-0327-8 [DOI] [PubMed] [Google Scholar]
- 9.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(D1):D490–5. 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemi T, Eshete A, Seid G, Mulugeta S, Egeta D, Teshome M. (2023). Aboveground Biomass Models for Indigenous Tree Species in the Dry Afromontane Forest, Central Ethiopia. International Journal of Forestry Research, 2023.
- 11.Poudel S. Organic Matter determination (Walkley-Black method). 2020.
- 12.Bremner JM. Nitrogen-total. Methods soil Anal Part 3 Chem Methods. 1996;5:1085–121. [Google Scholar]
- 13.Ashworth J, Keyes D, Kirk R, Lessard R. Standard procedure in the hydrometer method for particle size analysis. Commun Soil Sci Plant Anal. 2001;32(5–6):633–42. 10.1081/CSS-100103897 [DOI] [Google Scholar]
- 14.Flint AL, Flint LE. 2.2 particle density. Methods soil Anal Part 4 Phys Methods. 2002;5:229–40. [Google Scholar]
- 15.Verma SK, Singh H, Sharma PC. An improved method suitable for isolation of high-quality metagenomic DNA from diverse soils. 3 Biotech. 2017;7:1–7. 10.1007/s13205-017-0847-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galaxy Europe server. (http://usegalaxy.eu). Accessed on 15 May 2022.
- 17.Matias Rodrigues JF, Schmidt TSB, Tackmann J, von Mering C. MAPseq: highly efficient k-mer search with confidence estimates, for rRNA sequence analysis. Bioinformatics. 2017;33(23):3808–10. 10.1093/bioinformatics/btx517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plotly Chart Studio. (https://chart-studio.plotly.come). Accessed on 15 September 2022.
- 19.KEGG Automatic Annotation Server. (www.genome.jp/tools/kaas/). Accessed on 3 November 2022.
- 20.Zheng J, Ge Q, Yan Y, Zhang X, Huang L, Yin Y. dbCAN3: automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023;gkad328. [DOI] [PMC free article] [PubMed]
- 21.Bastida F, Eldridge DJ, García C, Kenny Png G, Bardgett RD, Delgado-Baquerizo M. Soil microbial diversity–biomass relationships are driven by soil carbon content across global biomes. ISME J. 2021;15(7):2081–91. 10.1038/s41396-021-00906-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan X, Huang Z, He Z, Yu Z, Wang M, Davis MR, Yang Y. Soil C: N ratio is the major determinant of soil microbial community structure in subtropical coniferous and broadleaf forest plantations. Plant Soil. 2015;387:103–16. 10.1007/s11104-014-2277-4 [DOI] [Google Scholar]
- 23.Hamarashid NH, Othman MA, Hussain MAH. Effects of soil texture on chemical compositions, microbial populations and carbon mineralization in soil. Egypt J Exp Biol (Bot). 2010;6(1):59–64. [Google Scholar]
- 24.Frey B, Varliero G, Qi W, Stierli B, Walthert L, Brunner I. Shotgun metagenomics of deep forest soil layers show evidence of altered microbial genetic potential for biogeochemical cycling. Front Microbiol. 2022;606. [DOI] [PMC free article] [PubMed]
- 25.Onyango LA, Ngonga FA, Karanja EN, Kuja JO, Boga HI, Cowan DA, et al. The soil microbiomes of forest ecosystems in Kenya: their diversity and environmental drivers. Sci Rep. 2023;13(1):7156. 10.1038/s41598-023-33993-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao D, Zhang N, Liu S, Ning C, Wang X, Feng S. Urbanization imprint on Soil Bacterial communities in forests and grasslands. Forests. 2023;14(1):38. 10.3390/f14010038 [DOI] [Google Scholar]
- 27.Wei H, Peng C, Yang B, Song H, Li Q, Jiang L, et al. Contrasting soil bacterial community, diversity, and function in two forests in China. Front Microbiol. 2018;9:1693. 10.3389/fmicb.2018.01693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Talia P, Sede SM, Campos E, Rorig M, Principi D, Tosto D, et al. Biodiversity characterization of cellulolytic bacteria present on native Chaco soil by comparison of ribosomal RNA genes. Res Microbiol. 2012;163(3):221–32. 10.1016/j.resmic.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Liu Q, Yan L, Gao Y, Wang Y, Wang W. A novel lignin degradation bacterial consortium for efficient pulping. Bioresour Technol. 2013;139:113–9. 10.1016/j.biortech.2013.04.033 [DOI] [PubMed] [Google Scholar]
- 30.Kenya E, Kinyanjui G, Kipnyargis A, Kinyua F, Mwangi M, Khamis F, et al. Amplicon-based assessment of bacterial diversity and community structure in three tropical forest soils in Kenya. Heliyon. 2022;8(11):e11577. 10.1016/j.heliyon.2022.e11577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tláskal V, Baldrian P. Deadwood-inhabiting bacteria show adaptations to changing carbon and nitrogen availability during decomposition. Front Microbiol. 2021;12:685303. 10.3389/fmicb.2021.685303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larsbrink J, McKee LS. Bacteroidetes bacteria in the soil: glycan acquisition, enzyme secretion, and gliding motility. Adv Appl Microbiol. 2020;110:63–98. 10.1016/bs.aambs.2019.11.001 [DOI] [PubMed] [Google Scholar]
- 33.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10(1):110–6. 10.1016/S0959-440X(99)00057-3 [DOI] [PubMed] [Google Scholar]
- 34.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, et al. The α/β hydrolase fold. Protein Eng Des Sel. 1992;5(3):197–211. 10.1093/protein/5.3.197 [DOI] [Google Scholar]
- 35.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3(10):e334. 10.1371/journal.pbio.0030334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao R, Mack TR, Stock AM. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci. 2007;32(5):225–34. 10.1016/j.tibs.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durica-Mitic S, Göpel Y, Görke B. Carbohydrate utilization in bacteria: making the most out of sugars with the help of small regulatory RNAs. Regulating with RNA in Bacteria and Archaea; 2018. pp. 229–48. [DOI] [PMC free article] [PubMed]
- 38.Sharma A, Sharma T, Sharma T, Sharma S, Kanwar SS. (2019). Role of microbial hydrolases in bioremediation. Microbes Enzymes Soil Health Bioremediat, 149–64.
- 39.Duan C-J, Feng Y-L, Cao Q-L, Huang M-Y, Feng J-X. Identification of a novel family of carbohydrate-binding modules with broad ligand specificity. Sci Rep. 2016;6(1):19392. 10.1038/srep19392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pátek M, Manganelli R, Toyoda K. Role of sigma factors of RNA polymerase in bacterial physiology, II. Front Microbiol. 2023;14:1220519. 10.3389/fmicb.2023.1220519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reznikoff WS. Tn5 as a model for understanding DNA transposition. Mol Microbiol. 2003;47(5):1199–206. 10.1046/j.1365-2958.2003.03382.x [DOI] [PubMed] [Google Scholar]
- 42.Sharmin F. (2012). Metabolic and ecological study of environmental pentose utilizing bacteria (E-PUB) (Doctoral dissertation, Queensland University of Technology).
- 43.Chen L, Wang J, He L, Xu X, Wang J, Ren C et al. Metagenomic highlight contrasting elevational pattern of bacteria-and fungi-derived compound decompositions in forest soils. Plant Soil. 2023;1–13.
- 44.Sengupta S, Datta M, Datta S. β-Glucosidase: structure, function and industrial applications. Glycoside Hydrolases. Elsevier; 2023. pp. 97–120.
- 45.Chauhan PS, Puri N, Sharma P, Gupta N. Mannanases: microbial sources, production, properties and potential biotechnological applications. Appl Microbiol Biotechnol. 2012;93:1817–30. 10.1007/s00253-012-3887-5 [DOI] [PubMed] [Google Scholar]
- 46.Poria V, Saini JK, Singh S, Nain L, Kuhad RC. Arabinofuranosidases: characteristics, microbial production, and potential in waste valorization and industrial applications. Bioresour Technol. 2020;304:123019. 10.1016/j.biortech.2020.123019 [DOI] [PubMed] [Google Scholar]
- 47.Mendonça M, Barroca M, Collins T. Endo-1, 4-β-xylanase-containing glycoside hydrolase families: characteristics, singularities and similarities. Biotechnol Adv. 2023;108148. [DOI] [PubMed]
- 48.Nakamura AM, Nascimento AS, Polikarpov I. Structural diversity of carbohydrate esterases. Biotechnol Res Innov. 2017;1(1):35–51. 10.1016/j.biori.2017.02.001 [DOI] [Google Scholar]
- 49.Zhang, Q., Miao, R., Liu, T., Huang, Z., Peng, W., Gan, B., ... & Tan, H. (2019). Biochemical characterization of a key laccase-like multicopper oxidase of artificially cultivable Morchella importuna provides insights into plant-litter decomposition. 3 Biotech, 9, 1–10. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author and first author upon reasonable request. The raw shotgun sequence data of both sites generated in this study were deposited to the European Nucleotide Archive (ENA). The studies have been made public with all associated data and it is recently reflected in the ENA browser as study ID PRJEB57985 and Secondary Study Accession number of ERP143008 which could be accessed online by using the link: https://www.ebi.ac.uk/ena/browser/view/PRJEB57985.