Abstract
Gut microbial imbalance is noted in Crohn’s disease (CD), but the specific bacteria associated with CD vary between studies. Chen et al.1 pair CD patients with their healthy first-degree relatives to mitigate some of the environmental and genetic effects.
Gut microbial imbalance is noted in Crohn’s disease (CD), but the specific bacteria associated with CD vary between studies. Chen et al. pair CD patients with their healthy first-degree relatives to mitigate some of the environmental and genetic effects.
Main text
Environmental factors such as household, country, hygiene level, exposure, diet, and, to a lesser degree, host genetics influence the human gut microbiome. For example, there are significant similarities in the compositions of the microbiomes of genetically unrelated individuals who share a household, and over 20% of inter-person microbiome variability is associated with factors related to diet, drugs, and anthropometric measurements.2 Studies in twins provide models to explore the influences of heritable and environmental factors and, as such, further show that similarity between twins is defined mainly by household sharing and aging rather than host genetics.3
Crohn’s disease (CD) burden has increased with westernization, and its rapid rise in prevalence in the last decade is likely linked to environmental changes. Epidemiological and, more recently, dietary and multi-omics studies (i.e., Braun et al.4) aim to capture the contribution of exposure to CD pathogenesis and the related gut microbial alterations. Many previous studies have shown alterations in the gut microbiome, but the specific bacteria involved have varied between studies. These gaps may be partly due to differences in sample processing protocols and the inclusion of patients with different geographic locations, diets, and exposures. Additionally, they can be attributed to the CD patients’ characterization—for example, the use of CD medications (if they are not newly diagnosed, treatment-naive patients) or the presence of gut inflammation that may change the physical and chemical environment—as well as to the selection of controls without inflammatory bowel disease (i.e., relatives vs. nonrelatives of CD patients). In addition, a small overlap between features (i.e., bacteria) reaching statistical significance between two groups of patients is an inherent property of this type of statistical analysis due to the large differences between individual microbiomes and the relatively small number of samples, as has been shown for human RNA microarrays.5 One approach to overcome these limitations of a single study is to systematically compare independent studies from different geographies in a metanalysis, using cohort studies rather than samples as basic units, and identifying bacterial signal patterns across studies. Such an approach, for example, identified predominant shared microbial responses within CD and across various diseases and found more specific signals of primarily increased taxa, enriched for oral bacteria that dislocated to the gut in CD and ulcerative colitis.6
Using siblings or cohabiting relatives provides another promising approach, controlling for environmental and genetic factors. Recently, the Genetic, Environmental, Microbial project research team discovered that relatives of patients with CD who later go on to develop CD themselves have different gut microbiome risk scores, based on a nonlinear nonparametric model, from relatives who remain healthy.7 However, no significant differences in the abundance of any genus were observed between the groups. In their recent study, to reduce the effect of environmental and genetic factors, Chen et al.1 compared 27 CD patients with active inflammation and their paired siblings and 25 CD patients in remission and their paired siblings. An independent control group of 44 healthy subjects unrelated to the CD patients was also included. Here, the selection of relative controls and the comparison to CD patients in remission resulted in the findings that (1) three taxa were reduced in quiescent CD vs. control; (2) these identified three taxa were not associated with CRP, a systemic inflammatory marker; and (3) the models based on these microbial markers demonstrated improved diagnostic capability that was validated across multiple cohorts.
Despite this, the overall microbial compositions and functions in Chen et al.1 were similar between the relatives and the nonrelative control group. These observations were consistent with the notion that the number of shared taxa between relatives and CD patients did not differ significantly from the number between nonrelatives and patients with CD. Therefore, it is still questionable how the authors’ approach led to the improved prediction model. By contrast, another study that included both unaffected relatives of patients with CD and nonrelated controls indicated differences in bacteria between these two control groups.8 Furthermore, all three taxa used in Chen et al.’s diagnostic model are reduced in CD. Several studies have shown that the reduction of taxa is not specific to one disease but is shared between different pathogenic conditions6,9 and that an increase, rather than a decrease, in specific taxa is more specifically linked with CD6 and colon cancer,9 for example. Testing for the specificity of the three taxa used in Chen et al.’s diagnostic model demonstrates that its performance in predicting disease state was significantly better for CD than for other tested diseases. Furthermore, using the Integrative Human Microbiome Project dataset,10 the authors demonstrated that demographics and lifestyle, particularly age and dietary habits, likely impact the diagnostic performance of their model.
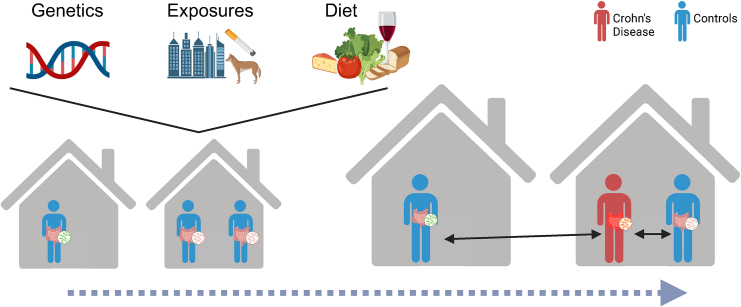
In summary, family history is a known risk factor for CD. However, even in monozygotic twins, disease penetrance is far from 100%. In addition, CD involves complex interactions among attenuated epithelial barrier function, microbial alterations, immune activation, and environmental factors. This indicates a potential theory of a succession of hits required for CD manifestation. Focusing on microbial differences between patients and their first-degree relatives, as Chen et al. have done,1 may enable the elucidation of the involvement of bacteria in the later, pathogenic hits required for CD development (Figure 1).
Figure 1.
Using healthy relatives to define Crohn’s gut microbiome alterations
While family members share similar genetics, environmental exposures, and diet, not all relatives will eventually develop CD. Focusing on the microbial differences between CD cases and healthy controls from the same family may enable the detection of disease-development-associated bacteria without confounding household-related factors.
Acknowledgments
The Haberman lab is funded by ERC starting grant 758313; the Helmsley Charitable Trust; the Israel Science Foundation (785/22); Tel Aviv University’s Colton Center for Autoimmunity; Israel’s Science, Culture, and Sport Ministry (4361); and a Crohn’s & Colitis Foundation Litwin IBD Pioneers award (1165359).
Declaration of interests
The authors declare no competing interests.
References
- 1.Chen W., Li Y., Wang W., Gao S., Hu J., Xiang B., Wu D., Jiao N., Xu T., Zhi M., et al. Enhanced microbiota profiling in patients with quiescent Crohn’s disease through comparison with paired healthy first-degree relatives. Cell Rep. Med. 2024;5:101624. doi: 10.1016/j.xcrm.2024.101624. [DOI] [PubMed] [Google Scholar]
- 2.Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D., Costea P.I., Godneva A., Kalka I.N., Bar N., et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 3.Vilchez-Vargas R., Skieceviciene J., Lehr K., Varkalaite G., Thon C., Urba M., Morkūnas E., Kucinskas L., Bauraite K., Schanze D., et al. Gut microbial similarity in twins is driven by shared environment and aging. EBioMedicine. 2022;79 doi: 10.1016/j.ebiom.2022.104011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun T., Feng R., Amir A., Levhar N., Shacham H., Mao R., Hadar R., Toren I., Algavi Y., Abu-Saad K., et al. Diet-omics in the Study of Urban and Rural Crohn disease Evolution (SOURCE) cohort. Nat. Commun. 2024;15 doi: 10.1038/s41467-024-48106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ein-Dor L., Zuk O., Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc. Natl. Acad. Sci. USA. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbas-Egbariya H., Haberman Y., Braun T., Hadar R., Denson L., Gal-Mor O., Amir A. Meta-analysis defines predominant shared microbial responses in various diseases and a specific inflammatory bowel disease signal. Genome Biol. 2022;23 doi: 10.1186/s13059-022-02637-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raygoza Garay J.A., Turpin W., Lee S.-H., Smith M.I., Goethel A., Griffiths A.M., Moayyedi P., Espin-Garcia O., Abreu M., Aumais G.L., et al. Gut Microbiome Composition Is Associated With Future Onset of Crohn’s Disease in Healthy First-Degree Relatives. Gastroenterology. 2023;165:670–681. doi: 10.1053/j.gastro.2023.05.032. [DOI] [PubMed] [Google Scholar]
- 8.Joossens M., Huys G., Cnockaert M., De Preter V., Verbeke K., Rutgeerts P., Vandamme P., Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 9.Duvallet C., Gibbons S.M., Gurry T., Irizarry R.A., Alm E.J. Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-01973-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd-Price J., Arze C., Ananthakrishnan A.N., Schirmer M., Avila-Pacheco J., Poon T.W., Andrews E., Ajami N.J., Bonham K.S., Brislawn C.J., et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]