Abstract
Expression of the Epstein-Barr virus (EBV) latency-associated genes activates cell cycle progression and drives immortalization of the infected cell. In contrast, progression of the EBV replication program occurs most efficiently in growth-arrested cells. Previous studies showed that the EBV-encoded immediate-early transcription factor, Zta, can induce expression of the cyclin-dependent kinase inhibitors, p21 and p27, the tumor suppressor, p53, and cell growth arrest. Moreover, Zta-mediated induction of growth arrest occurs independently of its transcriptional transactivation function. Here we show that substitution of Zta’s basic DNA binding domain with the analogous region of the Zta homologue, c-Fos, abrogates Zta’s ability to induce growth arrest and to induce p21, p27, or p53 expression, suggesting that protein-protein interactions between this region of Zta and key cell cycle control proteins are involved in signaling cell cycle arrest. We also show that despite the crucial role for Zta’s basic domain in eliciting cell growth arrest, its amino terminus is required for efficient induction of p27 and it modulates the level of p53 induction. Last, we provide evidence that Zta-mediated inductions of p21, p27, and p53 occur, at least in part, through distinct pathways. Therefore, Zta interacts with multiple growth arrest pathways, a property which may have evolved partly as a means to ensure that lytic replication occurs in a growth-arrested setting in multiple different tissues in various states of differentiation.
Epstein-Barr virus (EBV) is a human gammaherpesvirus which is associated with several B-cell and epithelial cell malignancies, including the endemic form of Burkitt’s lymphoma, posttransplantation lymphoproliferative diseases, AIDS-associated lymphomas, Hodgkin’s disease, and undifferentiated nasopharyngeal carcinoma (31, 42, 65).
EBV employs a variety of distinct genetic programs which are utilized at various stages of its life cycle (31, 32). Each program is categorized by the unique set of viral genes that are expressed, and these programs can be grouped into either the latency-associated programs or the replicative program. Latency-associated gene expression is limited to only small subsets of the viral genes whose corresponding gene products perform various functions, including replication of the viral plasmid genome and activation of cell cycle pathways (2, 11, 24–26, 28, 29, 32, 40, 43, 54, 59, 63).
In B cells, EBV exists primarily in a latent state; however, a switch from latency to the lytic replication program can be triggered by agents that mimic terminal differentiation signals and/or which cause cell cycle arrest (31, 42). In contrast to B lymphocytes, epithelial cells are highly permissive for EBV replication (55, 56, 62). In the oral epithelium, full EBV replication appears to be highly dependent on the state of differentiation, as it is observed primarily in the upper spinous layer, which contains cells that have stopped dividing, but not in the basal, mitotically active layer (4, 62, 64). Consistent with these observations, Shadan et al. (52) showed that in contrast to small DNA tumor viruses such as simian virus 40 and papillomavirus, replication of EBV, as well as other herpesviruses, occurs readily in cells that are treated with agents that arrest cell cycle progression. Furthermore, Takase et al. (58) have shown that forced progression of cells into the S phase of the cell cycle inhibits EBV replication.
It is clear that unlike many small viruses which require host cell proliferation for viral replication, EBV and other herpesviruses favor a nonproliferating host cell status for viral replication. Because herpesviruses have a much greater genomic complexity than many tumor viruses, and therefore encode proteins required for DNA synthesis (DNA polymerase, DNA metabolic enzymes, and other replication factors), they can replicate independently of host cell replication. Moreover, we (6, 7, 57) and others (38, 49) have suggested that herpesviruses have evolved active mechanisms to block cell cycle progression so that viral replication takes place in noncycling cells where cellular resources can be more readily diverted for replication of the viral genome and ultimately virus production.
We have previously shown that the EBV-encoded immediate-early transcription/replication factor, Zta (also referred to as ZEBRA and EB1), elicits a G0/G1 cell cycle arrest in several different EBV-negative and EBV-positive epithelial tumor cell lines (6, 7). Furthermore, infection of primary epithelial cells with a Zta-expressing adenovirus results in a G0/G1 cell cycle arrest (41). Other groups have reported that infection of fibroblast cells with another herpesvirus, cytomegalovirus, causes a cell cycle arrest (5, 13, 37, 38, 49), and it has recently been shown that the cytomegalovirus-encoded UL69 protein specifically induces an arrest in the G1 phase of the cell cycle (38).
The immediate-early transcription factor, Zta, plays a crucial role in initiating the lytic cycle since ectopic expression of Zta in latently infected B-lymphocyte cell lines triggers progression through the entire lytic cycle (12, 22). Zta is a member of the basic leucine zipper (bZIP) family of DNA binding transcription factors and exhibits extended homology to the cellular oncogene product c-Fos (14). Moreover, Zta interacts with AP-1 promoter elements (as well as other AP-1-related elements) with high affinity (14, 17, 30, 36, 60). Sequences carboxyl terminal to the basic DNA binding region encode a dimerization domain which forms a coiled-coil structure despite a lack of a heptad repeat of leucine residues (9, 18, 34). Amino terminal to the bZIP sequence of Zta is a region which is likely to interact with various transcriptional coactivators to elicit effective transcriptional activation (10, 16, 20, 35). In addition to its transactivator function, Zta is an essential replication factor which functions through binding to the EBV lytic origin of replication (15, 50, 51).
Although Zta can activate transcription through cellular AP-1 elements (8, 14), our previous studies showed that it can induce growth arrest independently of its ability to bind DNA (7). In the present study, we have extended our genetic analysis of Zta-mediated growth arrest and provide evidence that Zta stimulates multiple distinct growth arrest pathways. Furthermore, although DNA binding is not required for Zta’s growth arrest functions, sequences within its DNA binding domain are involved in eliciting signaling to cell cycle control pathways. We propose that protein-protein interactions between Zta’s basic/DNA binding region and multiple cellular proteins play a role in modulating Zta’s various functions.
MATERIALS AND METHODS
Cell culture.
All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Life Technologies), 2 mM glutamine, 100 μg/ml streptomycin, and 100 U of penicillin per ml in a humidified atmosphere at 37°C with 5% CO2–95% air. Stable inducible cell lines were propagated in DMEM supplemented with 2 μg of tetracycline per ml. Induction experiments were carried out as indicated in the relevant figure legends.
Transient transfection experiments were performed by using a modified version of the calcium phosphate precipitation procedure. Approximately 90% confluent cell cultures were split between 1/12 and 1/20 onto 100-mm-diameter tissue culture dishes. The following day, the medium was replaced with 8 ml of fresh supplemented DMEM; 4 h later, DNA precipitates were added dropwise to the cells. The cells were left in contact with the precipitate for 16 h, until the medium was replaced. Precipitates were generated by mixing 0.5 ml of 1× HEPES-buffered saline (0.5% HEPES, 0.8% NaCl, 0.1% dextrose, 0.01% anhydrous Na2HPO4, 0.37% KCl [pH adjusted to 7.05]) with the appropriate DNAs (a total of 30 μg) followed by the addition of 30 μl of 2.5 M CaCl2. Precipitates were allowed to form at room temperature for 20 min before addition to the cells.
Stable cell lines were generated by cotransfecting cells (ca. 0.5 × 106 in a 100-mm-diameter plate) with 1.5 μg of the indicated tetracycline-regulated pUHD10-Zta expression vector plus 29 μg of a plasmid containing a hygromycin-selectable marker (pREP4; Invitrogen). Two days after transfection, cells were split into 10 100-mm-diameter plates containing 250 U of hygromycin B per ml. Approximately 25 colonies were isolated for each plasmid transfection, and clones were assessed for low basal expression, comparable induced expression levels, and the percentage of cells which express the protein of interest following induction. Surviving clones were expanded only enough to perform the indicated experiments.
CAT assay.
For chloramphenicol acetyltransferase (CAT) analysis, 100-mm-diameter plates containing approximately 5 × 105 cells were transfected with a total of 30 μg of plasmid DNA (typically, 2 μg of reporter plasmid, between 1 and 5 μg of effector plasmid, and 23 to 27 μg of the carrier plasmid, pGL3 Basic [Promega], were used). Twenty four hours after removal of the precipitates (and replacement with fresh medium), cells were harvested, suspended in 100 μl of 0.25 M Tris (pH 7.5), and subjected to three rounds of freeze-thawing. Samples were clarified by centrifugation, and between 2 and 75 μl of extract was used for CAT reactions. CAT reactions were performed with using 2 μl of 60 mM acetyl coenzyme A, 2 μl of [14C]chloramphenicol (New England Nuclear), and a total of 96 μl of cell extract plus 0.25 M Tris (pH 7.5). Reactions were carried out for 1 h at 37°C, after which the [14C]chloramphenicol was extracted with 300 μl of ethyl acetate; the samples were then dried in a Speed Vac, resuspended in 20 μl of ethyl acetate, and spotted onto thin-layer chromatography (TLC) plates. Chromatography was carried out for approximately 45 min in a TLC chamber containing a solution of CHCl3-methanol (95:5). The TLC plates were then analyzed by autoradiography.
Cell cycle analysis.
For cell cycle analysis, cells were collected, washed once with 1× phosphate-buffered saline (PBS), suspended in cold (4°C) 0.5 ml of 1× PBS–0.1% glucose, fixed with 5 ml of 70% cold ethanol for at least 45 min at 4°C, washed with 1× PBS, and treated for 30 min at 37°C with RNase A (0.1 mg/ml) in a propidium iodide (69 mM; Sigma) sodium Na citrate (38 mM) solution. Cell cycle analysis was carried out with a fluorescence-activated cell sorting (FACS) (FACScan; Becton Dickinson). For transient transfection studies, cells were cotransfected with the green fluorescent protein (GFP) expression vector pGFP-SP (27), and the cell cycle profiles were determined for the GFP-gated cells.
Western blot analysis.
After a single 1× PBS wash, a fraction of cells harvested for cell cycle analysis were separated for Western blot analysis. Cells were immediately suspended in 15 pellet volumes of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading (Laemmli) buffer (39) and boiled for 20 min to shear the DNA. Cell lysates were subjected to SDS-PAGE separation and transferred to nitrocellulose membranes. The blots were blocked for 30 min in Tris-buffered saline containing 5% low-fat powdered milk and 1% fetal bovine serum and then incubated with the indicated primary antibody (in blocking buffer) for either 1 h at room temperature or overnight at 4°C. The blots were washed once with 1× PBS, once with 1× PBS buffer containing 0.5% (vol/vol) Tween 20, and twice with 1× PBS buffer alone (each wash was carried out for approximately 15 min). The blots were then incubated with peroxidase-conjugated secondary antibody in blocking buffer for 1 to 2 h at room temperature. Blots were washed as described above and analyzed with an enhanced chemiluminescence detection system (Amersham) according to manufacturer’s recommendations, and filters were exposed to Kodak XR film. Antibodies used for each experiment are indicated in the relevant figure legends.
Plasmid construction.
pMARK-Zta wt contains the genomic BZLF1 sequences under the control of a simian virus 40 promoter and was described previously (6). The amino-terminal Zta deletion mutants were generated by introducing a restriction site immediately upstream from the indicated starting amino acid in plasmid pSV40-BZLF1 (17), using site-directed mutagenesis (Muta-Gene; Bio-Rad). The respective portions of Zta were then excised from plasmid pSV40-BZLF1 and ligated in frame with a translation initiation sequence in plasmid pMARK. 2×(ZIIIB)BG-CAT has been described elsewhere (16). Plasmid pUHD10 (21) was used to generate tetracycline-inducible expression plasmids. pUHD10-Zta was described previously (8). pUHD10-Z(S186-A) was generated by first introducing the S186A mutation into plasmid pSV40-BZLF1 via site-directed mutagenesis. Zta-encoding sequences were then transferred to pUHD10. The pUHD10-Zta(129-245) and pUHD10-Zta(159-245) were generated by transferring the Zta sequences from the corresponding pMARK plasmid to the pUHD10 vector. The Z/Fos(basic) chimeric gene has been described elsewhere (33); this sequence was cloned into pUHD10 to generate the inducible vector, pUHD10-Z/Fos(basic). Detailed maps of the above-mentioned plasmids are available upon request.
RESULTS
Involvement of Zta amino-terminal sequences in Zta-mediated growth arrest and induction of p53, p21, and p27.
Using transient transfection experiments, we have previously shown that a Zta point mutant which is defective for binding to DNA retains the ability to induce growth arrest and to induce expression of the cyclin-dependent kinase (CDK) inhibitor p21 (7). Furthermore, truncation of a large portion of Zta’s activation domain (amino acids 1 to 128) did not abrogate its ability to induce growth arrest or p21 expression (7). These experiments indicated that Zta’s transactivation function is not essential for its growth arrest function and that Zta’s carboxyl-terminal region is crucial for eliciting growth arrest (Fig. 1A). In this study, however, we also noted that the Zta amino-terminal truncation mutant, Zta(128–245), induces G0/G1 growth arrest less effectively than wild-type Zta. In an attempt to address the molecular basis for this difference, we sought to further explore the role of Zta’s amino-terminal sequences in the induction of p21, p27, and p53.
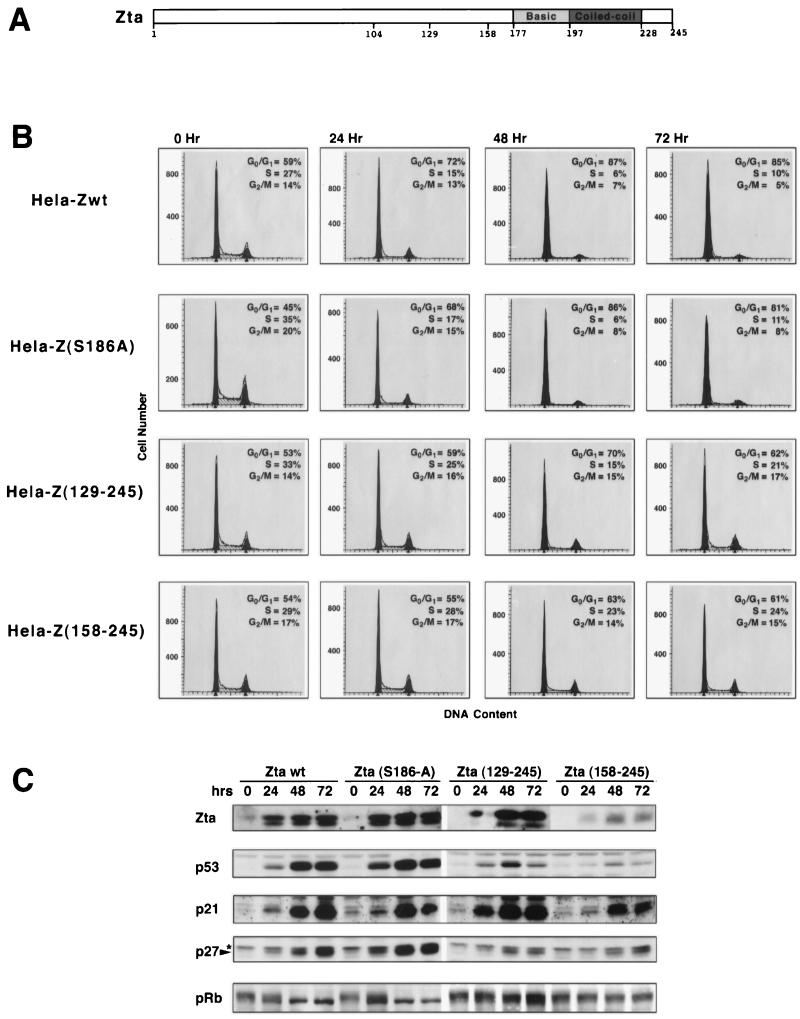
FIG. 1.
Involvement of amino-terminal Zta sequences in growth arrest and induction of p53, p21, and p27. (A) Schematic representation of Zta structure. (B and C) Cell cycle analysis (B) and Western blot analysis of the cell cycle regulatory proteins p53, p21, p27, and pRb (C) following induction of the indicated Zta proteins in stably transfected HeLa cells. The indicated cell lines were generated simultaneously, and induction experiments were performed in parallel. Cells were expanded in the presence of tetracycline (2 μg/ml). Cells were then trypsinized, washed twice with 1× PBS, and either collected (for 0-h time point) or plated in medium with no tetracycline for the indicated times. A portion of collected cells were stained with propidium iodide and subjected to FACS analysis to determine the DNA content (B). (C) The remaining cells were suspended in Laemmli buffer and boiled for 20 min to reduce the viscosity of the samples. A portion of these samples were subjected to SDS-PAGE, transferred to nitrocellulose, and subjected to sequential Western blot analysis (with a stripping step between probings) using anti-Zta (M47), anti-p53 (DO-1; Santa Cruz), anti-p21 (C19G; Santa Cruz), anti-Kip1/p27 (Transduction Laboratories), and anti-pRb (G3-245; PharMingen) antibodies. wt, wild type.
While transient transfection studies are a common means of addressing the cell cycle-related functions of gene products, we have recently reported that transient transfection of DNA into cells elicits signaling of cell cycle regulatory pathways (48). We therefore carried out experiments to first demonstrate that our previous results, which were obtained from transient transfections, were not due to cooperation between Zta (or Zta mutants) and DNA transfection-based signaling. To this end, stable tetracycline-regulated HeLa cell lines which allowed for inducible expression of wild-type Zta, Zta(129–245) or Zta(158–245) were generated. Positive cell clones were grown to equal cell densities and induced in parallel for 0, 24, 48, or 72 h (Fig. 1). In addition to wild-type Zta, Zta(129–245) and Zta(158–245), a Zta mutant containing a single amino acid substitution in the basic region which was shown previously to be defective for the capacity to induce of the EBV lytic cycle (19), were also analyzed in this experiment.
As expected, induction of wild-type Zta expression effectively induced growth arrest and expression of p53, p21, and p27 and elicited hypophosphorylation of the tumor suppressor protein pRb (Fig. 1B and C). The Zta point mutant, Zta(S186A), was equally effective for inducing growth arrest, p53, p21, and p27 expression and hypophosphorylation of pRb.
Analysis of Zta(129–245) yielded results similar to those obtained previously in transient transfection studies: Zta(129–245) induced a lesser degree of G0/G1 growth arrest and nearly the same levels of p21 expression as wild-type Zta (Fig. 1 and reference 7). In contrast to the efficient induction of p21 by Zta(129–245), induction of p53 and that of p27 are significantly compromised. Moreover, we observed significantly less hypophosphorylation of pRb, which may be due to the observed lower level of p27 induction (Fig. 1C). Therefore, despite efficient induction of p21 expression, Zta(129–245) induces p53 and p27 less well, and this decrease in p53 and/or p27 induction may explain the reduced efficiency with which Zta(129–245) elicits G0/G1 growth arrest.
Despite the significantly lower expression of the mutant Zta(158–245), it is clear that deletion of amino-terminal residues 1 to 157 does not abrogate Zta’s ability to induce p21, p27, or p53 (Fig. 1). Moreover, some apparent low-level growth arrest activity is observed in induced Zta(158–245) cells at days 2 and 3. This indicates that although the amino terminal region of Zta contributes to the induction of p21, p27, and p53, carboxyl-terminal sequences are likely to play a role in the induction of these factors.
The DNA binding domain of Zta is required for its growth arrest function.
The data shown in Fig. 1 indicate that the amino terminus of Zta contributes to the efficacy of Zta- mediated growth arrest. However, they also indicate that sequences carboxyl terminal to amino acid 158 affect alterations in the cell cycle control machinery. Deletion of sequences carboxyl terminal to Zta’s dimerization domain does not effect its ability to induce p21, p27, or growth arrest (although a lower level of p53 induction is observed) [see Fig. 4, Z(1–227)]. We were therefore interested in assessing the contribution of Zta’s DNA binding and/or dimerization domain in its growth arrest function. We generated stable inducible cell lines that express a Zta chimera in which Zta’s basic/DNA binding domain is replaced by the analogous basic/DNA binding region of the Zta homologue, c-Fos (33) (Fig. 2A) (attempts to generate stable inducible cell lines that express significant levels of a Zta chimera in which the dimerization domain is replaced by the corresponding region of the yeast transactivator, GCN4, were unsuccessful [data not shown]). This Zta/c-Fos chimera, Z/Fos(basic), is expressed as well as wild-type Zta, is localized to the nucleus, and transactivates reporter plasmids containing AP-1 binding elements (Fig. 2C, reference 33, and data not shown). While we have previously used Zta genomic sequences to generate stable inducible cell lines (e.g., Fig. 1), the Z/Fos(basic) chimeric gene is cDNA based, and we therefore simultaneously generated new wild-type Zta-inducible cell lines which were similarly made by using a cDNA-based wild-type Zta gene.
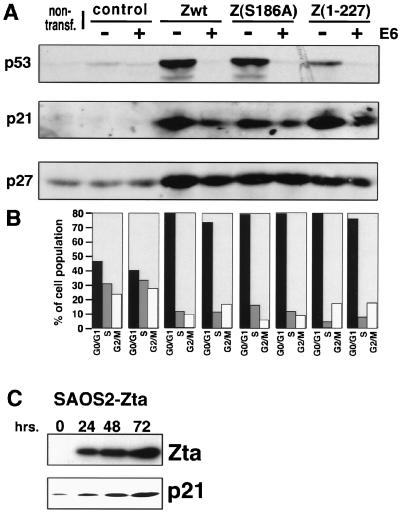
FIG. 4.
Uncoupling of Zta-mediated p21 and p27 induction from p53 induction. (A and B) Zta-mediated induction of p27 and p21 in p53-suppressed HeLa cells. Cells were nontransfected (non-transf.) or transfected with 5 μg of the indicated pMARK expression vector and 25 μg of either a control vector (RSV) or a HPV E6 expression vector (RSV-E6). Cells were analyzed as indicated in the legend to Fig. 3. wt, wild type. (C) A stable inducible SAOS2-Zta cell line was generated, and the induction experiment was carried out as described for Fig. 1. Cells were harvested at the indicated times following tetracycline withdrawal, and extracts were subjected to Western blot analysis using anti-Zta (M47) and anti-p21 (C19G; Santa Cruz) antibodies.
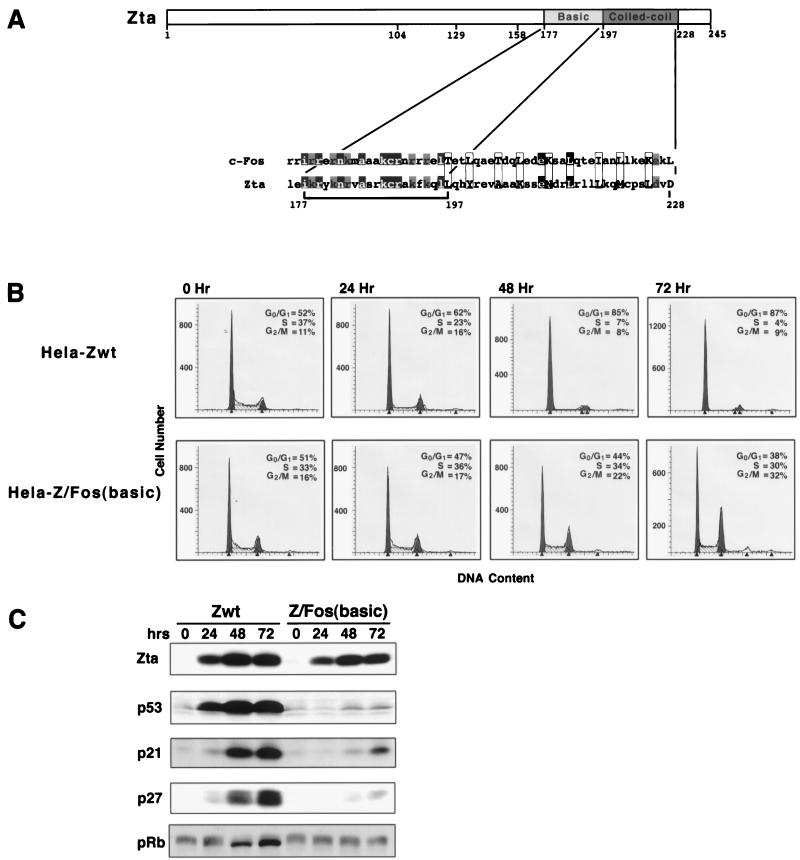
FIG. 2.
The DNA binding domain of Zta is required for induction of p21, p27, and p53 and growth arrest. (A) Schematic representation of Zta structure and alignment of Zta and c-Fos basic/dimerization domains. The bracket indicates the region of Zta which is replaced in the Z/Fos(basic) domain swap mutant. Black boxes indicate homologous amino acids, and gray boxes indicate related amino acids. The aligned 3-4 repeats of hydrophobic residues for c-Fos and Zta are enclosed by rectangles. (B and C) Cell cycle analysis (B) and Western blot analysis of the cell cycle regulatory proteins p53, p21, p27, and pRb (C) following induction of the indicated Zta proteins in stably transfected HeLa cells. Cell lines were generated simultaneously, and induction experiments were performed in parallel. Cells were expanded in the presence of tetracycline (2 μg/ml). Cells were then trypsinized, washed twice with 1× PBS, and either collected (for 0-h time point) or plated in medium with no tetracycline for the indicated times. A portion of collected cells were stained with propidium iodide and subjected to FACS analysis to determine the DNA content (B). (C) The remaining cells were suspended in Laemmli buffer and boiled for 20 min to reduce the viscosity of the samples. A portion of these samples were subjected to SDS-PAGE, transferred to nitrocellulose, and subjected to sequential Western blot analysis (with a stripping step between probings) using anti-hemmagglutinin (HA) (HA11.1; Boehringer Mannheim) (to detect HA-tagged Zta proteins), anti-p53 (DO-1; Santa Cruz), anti-p21 (C19G; Santa Cruz), anti-Kip1/p27 (Transduction Laboratories), and anti-pRb (G3-245; PharMingen) antibodies. wt, wild type.
Zta- and Z/Fos(basic)-inducible cell cultures were scaled up and induced in parallel for 0, 24, 48, or 72 h (Fig. 2). As expected, induction of wild-type Zta resulted in efficient growth arrest, induction of p53, p21, and p27, and a decrease in the phosphorylation status of pRb (Fig. 2). In contrast, the Z/Fos(basic) chimera failed to elicit any of these effects (although a slight delayed increase in p53, p21, or p27 is observed). These results indicate that although the DNA binding function of Zta is not required for inducing growth arrest, sequences within the basic region of Zta are essential. Importantly, the inability of Z/Fos(basic) to induce growth arrest or to induce p53, p21, or p27 is not due to a gain of function elicited by the c-Fos DNA binding domain since cotransfection of increasing amounts of a Z/Fos(basic) expression vectors with a wild-type Zta expression vector does not affect the ability of wild-type Zta to induce growth arrest (data not shown).
Overexpression of Zta amino-terminal deletion mutants increases the induction of p53 and p21 but not p27.
The efficient induction of p21 expression by Zta(129–245) despite Zta(129–245)’s compromised p53 and p27 signaling suggests that induction of p21 can be, at least in part, mechanistically uncoupled from the induction of p53 and p27 (see below). On the other hand, the coordinate reduction of p53 and p27 induction that is observed upon deletion of amino acids 1 to 128 suggests the possibility that a common pathway which is modulated by Zta’s amino terminus is involved in augmenting both p27 and p53 induction (although dual signaling of distinct pathways by this region is also a reasonable possibility).
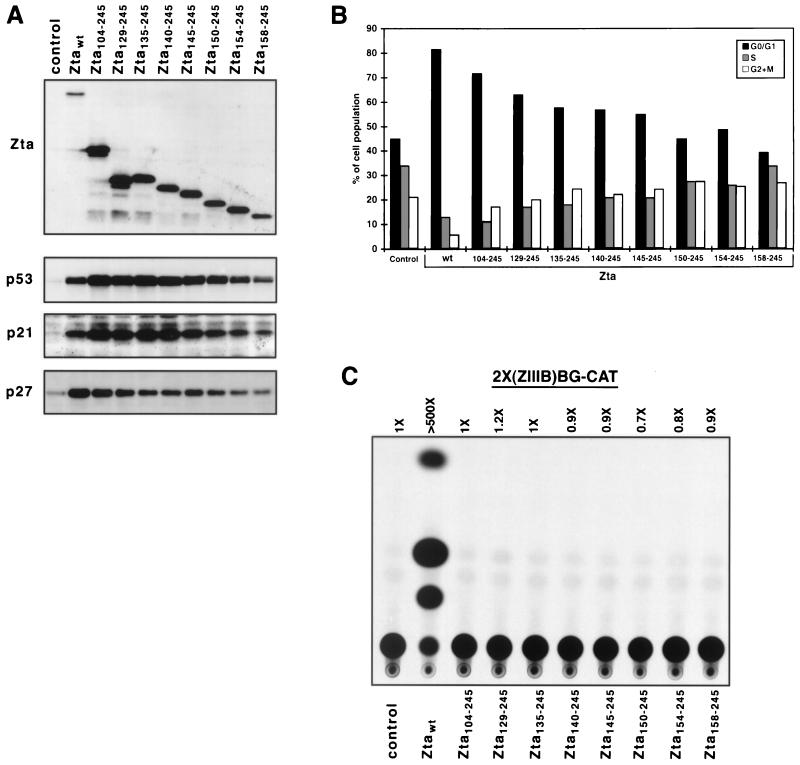
In an effort to gain insight into the relationship between Zta-mediated induction of p27 and p53, we reasoned that distinct pathways may be expected to be uniquely sensitive to effector concentration. Therefore, we investigated whether high-level Zta expression differentially affects the levels of p27 and p53 induction. Transient transfection experiments typically yield significantly higher levels of effector gene expression per cell than stably transfected cell lines. Therefore, we transiently transfected cells with a panel of Zta amino-terminal truncation mutants and performed Western blot and cell cycle analyses (Fig. 3). In such transient transfection experiments, we consistently observe significantly higher expression of amino-terminal deletion mutants relative to wild-type Zta (Fig. 3A). Interestingly, this higher expression level of some of the amino-terminal truncation mutants [Zta(104–245), Zta(129–245), Zta(135–245), and Zta(140–245)] appears to translate into a higher level of not only p21 expression but also p53 expression. This finding indicates that overproduction of carboxyl-terminal Zta sequences can compensate for the deficiency in p53 induction caused by deleting Zta’s amino terminus. In contrast, the higher expression level observed with these deletion mutants does not compensate for, or overcome, the defect in the p27 induction pathway that is incurred by the deletion of amino-terminal Zta sequences. This finding indicates that Zta-mediated induction of p27 is, at least in part, mechanistically distinct from that of p21 and p53.
FIG. 3.
Analysis of amino-terminal Zta mutants by transient transfectin analysis. HeLa cells were transfected with the indicated pMARK-Zta expression vector (5 μg), pGFP-SP (1 μg), and the carrier plasmid, pGL3 Basic (24 μg). Twenty-four hours after transfection, cells were harvested, expression of Zta derivatives p53, p21, and p27 was assessed by Western blot analysis (A), and the cell cycle distribution of GFP-positive cells was assessed by FACS analysis (following propidium iodide staining) (B). Western blot analyses were performed with the anti-Zta (M47), anti-p21 (C19G; Santa Cruz), anti-Kip1/p27 (Transduction Laboratories), and anti-p53 (DO-1; Santa Cruz) antisera. (C) Five micrograms of each of the indicated pMARK-Zta expression plasmids was cotransfected into HeLa cells with 2 μg of the Zta-responsive reporter, 2×(ZIIIB)BG-CAT (23 μg of pGL3 Basic was also used as a carrier). Cells were harvested 24 h later, and CAT analysis was performed. The level of acetylated chloramphenicol was quantitated with a Molecular Dynamics PhosphorImager. Control, pMARK (empty vector)-transfected cells. wt, wild type.
As shown in Fig. 3A and B, despite the increased level of p53 and p21 which is observed with several Zta amino-terminal truncation mutants that are expressed at a high level [e.g., Zta(104–245), Zta(129–245), Zta(135–245), and Zta(140–245)], this does not reverse the decrease in the percentage of G0/G1 cells that is observed by these mutants. It is possible, therefore, that even though Zta induces p53 and p21 in this cell system, the level of p27, and/or some other unknown factor, is a dominant factor in Zta-mediated growth arrest.
Analysis of the ability of Zta amino-terminal truncation mutants to activate a reporter plasmid containing Zta response elements in HeLa cells helps further affirm the dissociation of p21, p27, and p53 induction from Zta’s transactivation property (Fig. 3C).
Uncoupling of Zta-mediated p21 and p27 induction from induction of p53.
Previous studies have demonstrated that activation of p21 expression can occur through either a p53-dependent pathway or a p53-independent pathway (53). Results in Fig. 1 showing efficient induction of p21 but not p53 by Zta(129–245) suggest that Zta-mediated induction of p21 expression may occur, at least in part, through a p53-independent mechanism.
To further address the possible dissociation of Zta-mediated p21 induction from the observed induction of p53, we suppressed p53 protein levels by cotransfecting Zta expression vectors with a Rous sarcoma virus (RSV)-based human papillomavirus (HPV) E6 expression vector, the product of which targets p53 for degradation. As shown in Fig. 4A, overexpression of HPV E6 effectively prevented the induction of p53 by Zta or by two Zta mutants that also induce growth arrest efficiently [Z(S186A) and Z(1–227)]. In the absence of Zta, HPV E6 suppresses basal p53 expression below background levels (Fig. 4A, control). (Note that HeLa cells constitutively express low levels of p53 due to the expression of an integrated E6 gene.) As expected, Zta-mediated induction of p27 is unaffected by cotransfections with the HPV E6 expression vector (Fig. 4A). In addition, as we had previously shown (6), overexpression of E6 reduces the level of Zta-mediated p21 induction (Fig. 4A). However, it is clear that in the absence of any p53 induction, significant p21 expression is observed, suggesting that Zta-mediated p21 induction may occur, at least in part, through a p53-independent pathway. Moreover, in line with the induction of p27 and p21 in the presence of HPV E6, full Zta-mediated growth arrest is observed, indicating that efficient Zta-mediated growth arrest can be elicited in the absence of induced p53 levels (Fig. 4B).
We also considered the possibility that despite a lack of an increase in p53 levels in the context of HPV E6 overexpression, Zta might still be able to induce p53’s transactivation function. However, in transient reporter studies, we have been unable to demonstrate that Zta-mediated p53 induction results in the activation of the p21 promoter or artifical promoters containing consensus p53 binding sites (48a). These data are consistent with previous studies showing cross-inhibition of transactivation by Zta and p53 in transient transfection assays (67).
Finally, we tested whether Zta can induce p21 in a p53−/− cell line. As shown in Fig. 4C, expression of Zta in a stable inducible SAOS2 cell line, SAOS2-Zta, results in induction of p21 (Fig. 4C). Together, these data support a model whereby the induction of p21 occurs, at least to some extent, through a p53-independent pathway.
DISCUSSION
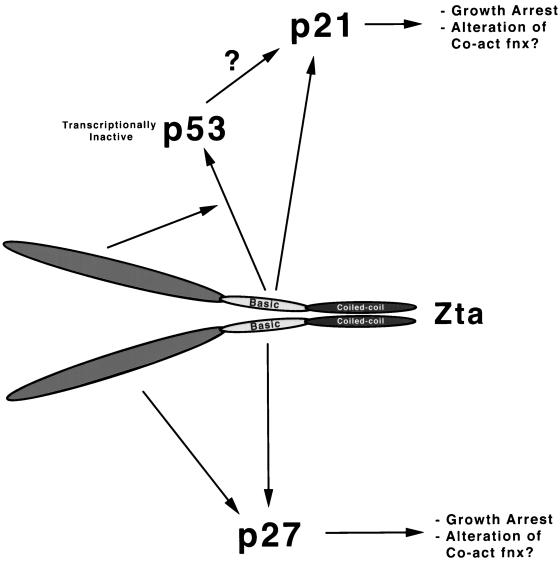
We have previously demonstrated that Zta mediates the induction of three key cell cycle regulatory factors, p21, p27, and p53, each of which is known to play a role in eliciting cell growth arrest in various cell systems (Fig. 5). Here, we provide evidence that Zta mediates induction of each of these target proteins, in part, through distinct pathways. Replication of EBV can be triggered in multiple tissues in various states of differentiation, and accumulating evidence indicates that the induction of growth arrest in different tissues is elicited through distinct pathways. Therefore, it is possible that Zta has evolved a way to stimulate growth arrest through multiple points in the cell cycle control machinery, thereby increasing the chances that viral replication will take place in a nondividing cellular environment regardless of the infected cell type.
FIG. 5.
Model for Zta-mediated induction of the cell cycle regulatory proteins p21, p27, and p53 and their possible roles in altering cellular signal transduction pathways. Co-act fnx, coactivator function.
The results presented in this report show a strong correlation between Zta-mediated growth arrest and the induction of p27. For example, amino-terminal Zta genetic studies show the best correlation between the level of p27 induction and the degree of G0/G1 arrest observed. Second, the higher level of p21 expression that is induced by some amino-terminal truncation mutants in the transient transfection-based analysis cannot compensate for the decrease in growth arrest which accompanies the lower level of p27 induction. Last, while the expression of HPV E6 decreased the level of p21 induction, it had no apparent effect on either the level of p27 induction or the degree of G0/G1 arrest observed. Therefore, it is possible that in the system used here, p27 plays a key role in Zta-induced growth arrest but p21 induction does not (or perhaps plays another unknown role). Recent studies, however, suggest an alternative, more complex possibility. We have recently found that Zta down-regulates the expression of c-Myc (48a), and other studies have shown that c-Myc destabilizes p27 (44–46). Since c-Myc also induces p21 and p27 sequestration (61), rendering them nonfunctional, c-Myc expression should have a dominant impact on the activity of p21 and p27. In such a scenario, growth arrest in the presence of elevated p21 and/or p27 would be dictated by c-Myc expression levels, and the link to p27 would be due to the cause-effect relationship between c-Myc and p27 expression. Dissecting the exact contribution of each of these factors in Zta-mediated growth arrest will require further studies.
In addition to possible roles in triggering cell growth arrest, the induction of p21 and p27 may perform other functions in the viral life cycle. Recent studies have shown that the activities of the transcriptional coactivator proteins p300 and CREB binding protein are modulated by CDKs and the CDK inhibitor p21 (1, 47). This raises the possibility that through the induction of p21 and/or p27, Zta might regulate the activity or specificity of these coactivator proteins in a way that favors the expression of viral genes over the bulk of cellular genes. Moreover, recent studies have shown that Zta itself activates transcription in part through binding to CREB binding protein (66), suggesting that Zta may modulate its own coactivator through the induction of p21 and/or p27.
Previous studies have shown that Zta inhibits p53’s ability to activate transcription (67), and we have observed that despite an increase in p53 levels, Zta cannot activate a reporter plasmids containing the p21 promoter or minimal reporter plasmids containing p53 binding elements (data not shown). Despite increasing p53 levels, it is likely that Zta does not induce p53-mediated transcriptional activation but instead probably interferes with this activity. Importantly, while stimulation of p53 function is known to lead to either growth arrest or induction of apoptosis, the induction of p21 or p27 function leads primarily to a growth arrest response. Therefore, the capacity to induce p21 and p27 without activating p53 transactivation function (and perhaps inhibiting it) offers an excellent mechanism through which a growth-arrested cellular environment can be established without the risk of triggering of a much more energy-consuming p53-dependent apoptotic response.
We have provided evidence that Zta’s basic region elicits cell growth arrest signaling, and we propose that this occurs through protein-protein interactions with key cell cycle regulatory factors. It seems reasonable that substitution of the analogous region of c-Fos for Zta’s basic/DNA binding region should maintain the structural integrity (or secondary structure) of this region of Zta, and previous studies demonstrating that this fusion protein binds strongly to AP-1 promoter elements (33) support this idea. Moreover, Z/Fos(basic) is functional for activating some reporter constructs in vivo (reference 33 and data not shown). Despite the likely structural similarity and the high degree of sequence homology between the basic regions of Zta and c-Fos (Fig. 2A), replacement of Zta’s basic region with that of c-Fos severely compromises Zta’s ability to induce p21, p27, and p53 and growth arrest. Therefore, specific residues within the basic region of Zta which are not conserved with respect to c-Fos may specify a unique interaction surface which communicates with cell cycle regulatory factors. The small number of amino acid differences between c-Fos and Zta in this region (Fig. 2A), therefore, genetically defines key residues in Zta’s growth control function.
Interestingly, a recent study has shown that protein kinase C phosphorylates serine 186 of Zta and activates Zta’s transactivation function (3). Further, these investigators provided evidence that phosphorylation of serine 186 of Zta facilitates the binding of a cellular factor to this region, which modulates its transactivation function. Although protein-protein interactions between the basic region of Zta and unknown cellular factors are likely to play a role in Zta-mediated growth arrest function, the cell factor(s) involved in this signaling is likely to be distinct from the factor which binds to serine 186-phosphorylated Zta since the phospho-site mutant, Z(S186A), effectively induces p21, p27, and p53 and growth arrest (Fig. 2). The genetically defined proximity of sequences involved in these interactions, however, suggests that an interesting interplay between these distinct factors may be controlled by phosphorylation of serine 186.
A previous study has reported the utility of using Zta in a gene therapy approach to reverse the immortalizing functions of EBV latency-associated gene expression (23). A drawback to the application of this approach to treating human disorders is the concomitant production of high titers of infectious EB virions. We show here, however, that a mutant which does not induce lytic EBV replication, Z(S186A), is fully functional for induction of growth arrest as well as the induction of p21, p27, and p53. Therefore, this mutant may provide a safer alternative to wild-type Zta in such gene therapy schemes.
ACKNOWLEDGMENTS
We thank George Miller for providing the Zta/Fos cDNA plasmid, and we thank Bert Vogelstein for the p21 reporter plasmids, WWP-Luc and DM-Luc. Special thanks go to Jesus Martin for special assistance during the course of this work. We also thank Miguel Campanero, Maria Escudero, and Eun Joo Jung for helpful advice and for reading the manuscript.
This work was supported by American Cancer Society grant RPG-97-065-VM (E.F.), National Institutes of Health grant (R01 GM48045) (E.F.), and a Lady Tata Postdoctoral fellowship (A.R.).
REFERENCES
- 1.Ait-Si-Ali S, Ramirez S, Barre F-X, Dkhissi F, Magnaghi-Jaulin L, Girault F A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- 2.Alday M J, Sinclair A, Parker G, Crawford D H, Farrell P J. Epstein-Barr virus efficiently immortalizes human B cells without neutralizing the function of p53. EMBO J. 1995;14:1382–1391. doi: 10.1002/j.1460-2075.1995.tb07124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumann M, Mischak H, Dammeier S, Kolch W, Gires O, Pich D, Zeidler R, Delecluse H-J, Hammerschmidt W. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-tetradecanolyphorbol-13-acetate-induced phosphorylation. J Virol. 1998;72:8105–8114. doi: 10.1128/jvi.72.10.8105-8114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker J, Leser U, Marschall M, Langford A, Jilg W, Gelderblom H, Reichart P, Wolf H. Expression of proteins encoded by Epstein-Barr virus transactivator genes depends on differentiation of epithelial cells in oral hairy leukoplakia. Proc Natl Acad Sci USA. 1991;88:8332–8336. doi: 10.1073/pnas.88.19.8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bresnahan W, Boldogh P, Thompson E, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1997;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 6.Cayrol C, Flemington E. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 7.Cayrol C, Flemington E K. G0/G1 growth arrest mediated by a region encompassing the bZIP domain of the Epstein-Barr virus transactivator Zta. J Biol Chem. 1996;271:31799–31802. doi: 10.1074/jbc.271.50.31799. [DOI] [PubMed] [Google Scholar]
- 8.Cayrol C, Flemington E K. Identification of cellular target genes of the Epstein-Barr virus transactivator Zta: activation of transforming growth factor βigh3 (TGF-βigh3) and TGF-β1. J Virol. 1995;69:4206–4212. doi: 10.1128/jvi.69.7.4206-4212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang Y-N, Dong D L-Y, Hayward G S, Hayward S D. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA binding specificity and dimerization domain that lacks the characteristic heptad leucine zipper motif. J Virol. 1990;64:3358–3369. doi: 10.1128/jvi.64.7.3358-3369.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi T, Carey M. The ZEBRA activation domain: modular organization and mechanism of action. Mol Cell Biol. 1993;13:7045–7055. doi: 10.1128/mcb.13.11.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Countryman J, Jensen H, Seibel R, Wolf H, Miller G. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J Virol. 1987;61:3672–3679. doi: 10.1128/jvi.61.12.3672-3679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dittmer D, Mocarski E. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrell P J, Rowe D T, Rooney C M, Kouzarides T. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 1989;8:127–132. doi: 10.1002/j.1460-2075.1989.tb03356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fixman E D, Hayward G S, Hayward S D. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and a dependence on Zta in cotransfection assays. J Virol. 1995;69:2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flemington E, Borras A M, Lytle J P, Speck S. Characterization of the Epstein-Barr virus BZLF1 protein trans-activation domain. J Virol. 1992;66:922–929. doi: 10.1128/jvi.66.2.922-929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flemington E, Speck S. Autoregulation of the Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1227–1232. doi: 10.1128/jvi.64.3.1227-1232.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flemington E, Speck S H. Evidence for coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc Natl Acad Sci USA. 1990;87:9459–9463. doi: 10.1073/pnas.87.23.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francis A, Bradoville L, Miller G. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J Virol. 1997;71:3051–3061. doi: 10.1128/jvi.71.4.3054-3061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giot J-F, Mikaelian I, Buisson M, Manet E, Joab I, Nicolas J-C, Sergeant A. Transcriptional interference between the EBV transcription factors EB1 and R: both DNA binding and activations of EB1 are required. Nucleic Acids Res. 1991;19:1251–1258. doi: 10.1093/nar/19.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grogan E J, Jenson J, Countryman J, Heston L, Gradoville L, Miller G. Transfection of a rearranged viral DNA fragment, WZhet, stably converts latent Epstein-Barr virus infection to productive infection in lymphoid cells. Proc Natl Acad Sci USA. 1987;84:1332–1336. doi: 10.1073/pnas.84.5.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutierrez M I, Judde J-G, MaGrath I T, Bhatia K G. Switching viral latency to viral lysis: a novel therapeutic approach for Epstein-Barr virus-associated neoplasia. Cancer Res. 1996;56:969–972. [PubMed] [Google Scholar]
- 24.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature (London) 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 25.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jk. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh J J-D, Hayward S D. Masking of the CBF/RBPJκ transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 27.Kalejta R, Shenk T, Beavis A. Use of a membrane-localized green fluorescent protein allows simultaneous identification of transfected cells and cell cycle analysis by flow cytometry. Cytometry. 1997;29:286–291. doi: 10.1002/(sici)1097-0320(19971201)29:4<286::aid-cyto4>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 28.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kempkes B, Spitkovsky D, Jansen-Durr P, Ellwart J W, Kremmer E, Delecluse H-J, Rottenberger C, Bornkamm G W, Hammerschmidt W. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 1995;14:88–96. doi: 10.1002/j.1460-2075.1995.tb06978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenney S, Holley-Guthrie E, Mar E C, Smith M. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J Virol. 1989;63:3878–3883. doi: 10.1128/jvi.63.9.3878-3883.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kieff E, editor. Epstein-Barr virus and its replication. New York, N.Y: Lippincott-Raven Press; 1996. [Google Scholar]
- 32.Klein G. Epstein-Barr virus strategy in normal and Neoplastic B cells. Cell. 1994;77:791–793. doi: 10.1016/0092-8674(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 33.Kolman J L, Taylor N, Gradoville L, Countryman J, Miller G. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J Virol. 1996;70:1493–1504. doi: 10.1128/jvi.70.3.1493-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouzarides T, Packham G, Cook A, Farrell P. The BLZF1 protein of EBV has a coiled coil dimerisation domain without a heptad leucine repeat but with homology to the C/EBP leucine zipper. Oncogene. 1991;6:195–204. [PubMed] [Google Scholar]
- 35.Lieberman P, Berk A. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J Virol. 1990;64:2560–2568. doi: 10.1128/jvi.64.6.2560-2568.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lieberman P M, Hardwick J M, Hayward S D. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J Virol. 1989;63:3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu M, Shenk T. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J Virol. 1999;73:676–683. doi: 10.1128/jvi.73.1.676-683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 40.Mannick J, Cohen J I, Birkenbach M, Marchini A, Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mauser A, Kenny S. The Epstein-Barr virus BZLF1 IE protein inhibits cellular differentiation and cell cycle progression. Epstein-Barr Virus Symposium; 1998. [Google Scholar]
- 42.Miller G. Epstein-Barr virus. In: Fields B N, Knipe D M, editors. Virology. New York, N.Y: Raven Press; 1990. pp. 1921–1958. [Google Scholar]
- 43.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 44.Muller D, Bouchard C, Rudolph B, Steiner P, Stuckmann I, Saffrich R, Ansorge W, Huttner W, Eilers M. Cdk2-dependent phosphorylation of p27 facilitates its Myc-induced release from cyclin E/cdk2 complexes. Oncogene. 1997;15:2561–2576. doi: 10.1038/sj.onc.1201440. [DOI] [PubMed] [Google Scholar]
- 45.Nass S, Dickson R. Epidermal growth factor-dependent cell cycle progression is altered in mammary epithelial cells that overexpress c-myc. Clin Cancer Res. 1998;4:1813–1822. [PubMed] [Google Scholar]
- 46.Perez-Roger I, Solomon D, Sewing A, Land H. Myc activation of cyclinE/Cdk2 kinase involves induction of cyclin E gene transcription and inhibition of p27(Kip1) binding to newly formed complexes. Oncogene. 1997;14:2373–2381. doi: 10.1038/sj.onc.1201197. [DOI] [PubMed] [Google Scholar]
- 47.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-kB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez A, Flemington E. Transfection-mediated cell-cycle signaling: considerations for transient transfection-based cell-cycle studies. Anal Biochem. 1999;272:171–181. doi: 10.1006/abio.1999.4156. [DOI] [PubMed] [Google Scholar]
- 48a.Rodriguez, A., and E. Flemington. Unpublished data.
- 49.Salvant B S, Fortunato E A, Spector D H. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J Virol. 1998;72:3729–3741. doi: 10.1128/jvi.72.5.3729-3741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarisky R T, Gao Z, Lieberman P M, Fixman E D, Hayward G S, Hayward S D. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J Virol. 1996;70:8340–8347. doi: 10.1128/jvi.70.12.8340-8347.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schepers A, Pich D, Mankertz J, Hammerschmidt W. cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J Virol. 1993;67:4237–4245. doi: 10.1128/jvi.67.7.4237-4245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shadan F F, Cowsert L M, Billarreal L P. n-Butyrate, a cell cycle blocker, inhibits the replication of polyomaviruses and papillomaviruses but not that of adenoviruses and herpesviruses. J Virol. 1994;68:4785–4796. doi: 10.1128/jvi.68.8.4785-4796.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheikh M, Li X-S, Chen J-C, Shao Z-M, Ordonez J, Fontana J. Mechanisms of regulation of WAF1/Cip1 gene expression in human breast carcinoma: role of p53-dependent and independent signal transduction pathways. Oncogene. 1994;9:3407–3415. [PubMed] [Google Scholar]
- 54.Sinclair A J, Palmero I, Peters G, Farrell P J. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 1994;13:3321–3328. doi: 10.1002/j.1460-2075.1994.tb06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sixbey J W, Lemon S M, Pagano J S. A second site for Epstein-Barr virus shedding: the uterine cervix. Lancet. 1986;ii:1122–1124. doi: 10.1016/s0140-6736(86)90531-3. [DOI] [PubMed] [Google Scholar]
- 56.Sixbey J W, Nedrud J G, Raab-Traub N, Hanes R A, Pagano J S. Epstein-Barr virus replication in oropharyngeal epithelial cells. N Engl J Med. 1984;310:1225–1230. doi: 10.1056/NEJM198405103101905. [DOI] [PubMed] [Google Scholar]
- 57.Speck S H, Chatila T, Flemington E K. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 1997;5:399–405. doi: 10.1016/S0966-842X(97)01129-3. [DOI] [PubMed] [Google Scholar]
- 58.Takase K, Kelleher C A, Terada N, Jones J F, Lucas J J, Gelfand E W. Dissociation of EBV genome replication and host cell proliferation in anti-IgG-stimulated Akata cells. Clin Immunol Immunopathol. 1996;8:168–174. doi: 10.1006/clin.1996.0173. [DOI] [PubMed] [Google Scholar]
- 59.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urier G, Buisson M, Chambard P, Sergeant A. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 1989;8:1447–1453. doi: 10.1002/j.1460-2075.1989.tb03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vlach J, Hennecke S, Alevizopoulos K, Conti D, Amati B. Growth arrest by the cyclin dependent kinase inhibitor p27kip1 is abrogated by c-Myc. EMBO J. 1996;15:6595–6604. [PMC free article] [PubMed] [Google Scholar]
- 62.Wolf H, Haus M, Wilmes E. Persistence of Epstein-Barr virus in the parotid gland. J Virol. 1984;51:795–798. doi: 10.1128/jvi.51.3.795-798.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in a variety of mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 64.Young L S, Lau R, Rowe M, Niedobotek G, Packham G, Shanahan F, Rowe D T, Greenspan D, Greenspan J S, Rickinson A B, Farrell P J. Differentiation associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J Virol. 1991;65:2868–2874. doi: 10.1128/jvi.65.6.2868-2874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Young L S, Rowe M. Epstein-Barr virus, lymphomas and Hodgkin’s disease. Semin Cancer Biol. 1992;3:273–284. [PubMed] [Google Scholar]
- 66.Zerby D, Chen C-J, Poon E, Lee D, Shiekhattar R, Lieberman P. The amino-terminal C/H1 domain of CREB binding protein mediates Zta transcriptional activation of latent Epstein-Barr virus. Mol Cell Biol. 1999;19:1617–1626. doi: 10.1128/mcb.19.3.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Z, Gutsch D, Kenney S. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. J Virol. 1994;14:1929–1938. doi: 10.1128/mcb.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]