SUMMARY PARAGRAPH
The scarcity of malignant Hodgkin and Reed–Sternberg cells hampers tissue-based comprehensive genomic profiling of classic Hodgkin lymphoma (cHL). By contrast, liquid biopsies show promise for molecular profiling of cHL due to relatively high circulating tumour DNA (ctDNA) levels.1–4 Here we show that the plasma representation of mutations exceeds the bulk tumour representation in most cases, making cHL particularly amenable to noninvasive profiling. Leveraging single-cell transcriptional profiles of cHL tumours, we demonstrate Hodgkin and Reed–Sternberg ctDNA shedding to be shaped by DNASE1L3, whose increased tumour microenvironment- derived expression drives high ctDNA concentrations. Using this insight, we comprehensively profile 366 patients, revealing two distinct cHL genomic subtypes with characteristic clinical and prognostic correlates, as well as distinct transcriptional and immunological profiles. Furthermore, we identify a novel class of truncating IL4R mutations that are dependent on IL-13 signalling and therapeutically targetable with IL-4Rα-blocking antibodies. Finally, using PhasED-seq5, we demonstrate the clinical value of pretreatment and on-treatment ctDNA levels for longitudinally refining cHL risk prediction and for detection of radiographically occult minimal residual disease. Collectively, these results support the utility of noninvasive strategies for genotyping and dynamic monitoring of cHL, as well as capturing molecularly distinct subtypes with diagnostic, prognostic and therapeutic potential.
The rapid evolution of high-throughput technologies over the last two decades has transformed our understanding of human malignancies. The genomic landscapes of many tumors have now been comprehensively characterized, leading to the identification of molecularly defined disease subtypes and targeted therapies.6–9 While this certainly applies to non-Hodgkin lymphomas (NHL),10–13 the genomic landscape of classic Hodgkin lymphoma (cHL) is less well defined. Published cohorts in cHL are relatively small,1,2,4,14–20 and the profiling methods employed have been limited in both genomic breadth and depth. A major reason for these limitations is the relative paucity of malignant cells within cHL tumors, which typically account for ~1% of the bulk tumor tissue cellularity. Indeed, cHL tumors primarily consist of infiltrating immune cells, thus making it technically challenging to apply state-of-the-art high-throughput techniques to interrogate Hodgkin and Reed-Sternberg (HRS) cells. While methods to enrich individual tumor cells by microdissection or sorting techniques can be useful,14–16,20–22 these are time-consuming, labor intensive, and generally require intact cells, thus limiting large-scale analyses.
Circulating tumor DNA (ctDNA) has emerged as a useful biomarker in diverse cancers, including B-cell lymphomas. Applications of liquid biopsies include early detection, noninvasive genotyping, tissue-of-origin and cell-of-origin classification,23,24 quantification of pretreatment ctDNA levels as a measure of tumor burden as well as detection of minimal residual disease (MRD) and relapse.2,25–29 Prior cHL studies have shown that somatic copy number aberrations (SCNAs) in HRS cells can be retrieved from ctDNA,4 and that plasma ctDNA mutational allelic fractions (AFs) are relatively high, despite the paucity of HRS cells in bulk tumor tissues.1–3
In a large international cohort comprising 366 patients including pediatric and adult patients of all ages (Table 1, Extended Data Fig. 1A, Supplementary Table 1 and Supplementary Note [Tables 1-3]), we here demonstrate that cHL can be comprehensively profiled from plasma, and that noninvasive profiling effectively overcomes the limitations of formalin-fixed, low HRS-burden tissue specimens. We further describe distinct cHL subgroups as defined by molecular subtypes, specific genotypes, ctDNA levels, and MRD with diagnostic, prognostic, and therapeutic potential.
Table 1:
Noninvasive study cohort.
| Number of patients | 366 | |
|
| ||
| Frontline | 361/366 (99%) | |
|
| ||
| Age: median (range) | 32 (4–88) | |
|
| ||
| Histological Subtype | ||
| cHL | 366 (100%) | |
| - NS | 242/295 (82%) | |
| - MC | 43/295 (15%) | |
| - LR | 8/295 (3%) | |
| - LD | 2/295 (1%) | |
|
| ||
| Stage | ||
| I/II | 183/351 (52%) | |
| - Favorable Risk | 15/166 (9%) | |
| - Unfavorable Risk | 151/166 (91%) | |
| III/IV | 168/351 (48%) | |
|
| ||
| Treatment center | ||
| Stanford, CA | 33 (9%) | |
| BREACH trial, FR/BE | 102 (28%) | |
| AHL2011 trial, FR/BE | 40 (11%) | |
| PVAB trial, FR/BE | 58 (16%) | |
| Leuven, BE | 29 (8%) | |
| Bellinzona, CH | 30 (8%) | |
| PAVD trial, WA | 30 (8%) | |
| Pediatric studies, USA | 44 (12%) | |
cHL: Classic Hodgkin lymphoma; NS: nodular-sclerosis; MC: mixed cellularity; LR: lymphocyte rich; LD: lymphocyte depleted.
Liquid biopsies in cHL
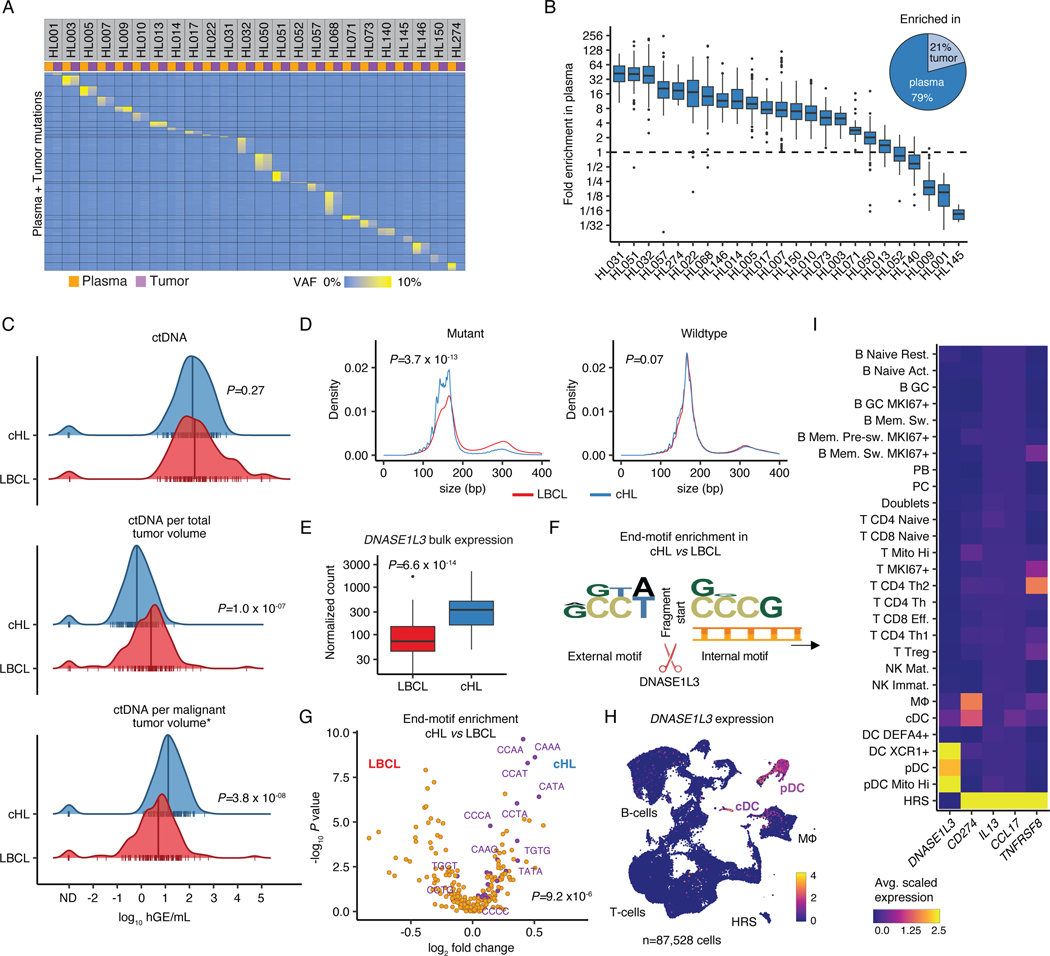
Liquid biopsies have demonstrated potential to explore cHL genetics,1–3 yet systematic characterization of mutational representation in the blood and tissue have been limited. In 24 cases evaluable for paired tumor and blood specimens, we compared the allelic abundance of mutations called in either specimen using enhanced CAPP-Seq,30,31 and found that mutations called from plasma samples were highly patient- and tumor-specific (Fig. 1A, Supplementary Table 2). Analysis of shared mutations detected in both analytes revealed plasma variant AFs (VAFs) to exceed tumor VAFs in 79% of cases (Fig. 1B). Strikingly, the median enrichment exceeded 7-fold (median 7.2, range 0.054–42.2) and was associated with improved genotyping when compared with bulk tumor tissue (Supplementary Note [Fig. 1-2]). Overall, these findings suggest that plasma genotyping in cHL is not only feasible, but superior to bulk tissue-based methods for most patients.
Figure 1: Liquid biopsies facilitate molecular profiling of cHL.
(A) Heatmap visualizing plasma and tumor representation (VAF) of cHL SNVs (rows) called in plasma or tumor samples (columns, n=24 pairs). (B) Boxplot depicts the fold enrichment of plasma over tumor VAF for SNVs called in either specimen (n=3,171; median per patient 124 [range 10–335]). The pie chart shows the fraction of sample pairs enriched in either tissue. (C) Comparison of ctDNA levels between previously untreated patients with cHL (n=142) and LBCL28 (n=114) visualized by density ridge plots with Wilcoxon p-values (two-sided). The top graph shows absolute ctDNA levels (log10 hGE/mL), the middle graph log10 ctDNA levels per TMTV (mL) and the bottom graph log10 ctDNA levels per inferred malignant tumor volume (calculated as TMTV [mL] multiplied by estimated tumor fraction [cHL: 0.05; LBCL: 0.5]). (D) Density plots with Wilcoxon p-values (two-sided) comparing cfDNA fragment length for mutant and wildtype molecules between cHL (n=300) and LBCL32 (n=53). (E) Boxplot with Wilcoxon p-value (two-sided) comparing DNASE1L3 bulk expression visualized as normalized counts (n=86 cHL, n=66 LBCL32). (F) Logo plot summarizing overrepresentation of external and internal 4-mer motifs at fragment ends in mutant cHL over mutant LBCL molecules (n=294 cHL, n=48 LBCL32). (G) Volcano plot depicting enrichment of end-motifs in mutant cHL vs LBCL molecules (x-axis: log2 median fold change, y-axis: -log10 p-value [Wilcoxon, two-sided]). Wilcoxon p-value (two-sided) comparing the abundance of previously described 25 end-motifs associated with DNASE1L3 digestion34 (purple) among mutant molecules in cHL vs LBCL patients is provided. (H) scRNA-Sequencing UMAP with DNASE1L3 expression (heat). (I) Average scaled expression of selected HRS genes and DNASE1L3 by cell type. Panels B,E: each box represents the interquartile range (the range between the 25th and 75th percentile) with the median of the data, whiskers indicate the upper and lower value within 1.5 times the IQR.
We next compared ctDNA levels found in cHL to those in aggressive B-cell NHL. Compared to untreated large B-cell lymphomas (LBCL),27,28 unadjusted ctDNA levels were not different (P=0.27, Fig. 1C top). To account for differences in disease burden, we calculated ctDNA concentrations per mL total metabolic tumor volume (TMTV) and found that LBCL tumors shed higher levels of ctDNA per mL tumor volume than cHL tumors (P=1.0e-7, Fig. 1C middle). However, this effect was reversed when taking the malignant cell content within each tumor type into account. Conservatively assuming 50% and 5% cancer cell fractions for LBCL and cHL respectively (Supplementary Note [Fig. 3]), we estimated that HRS cells shed at least 2.5x more ctDNA per mL malignant tumor volume (median 13.1 vs 5.0 haploid genome equivalents [hGE], P=3.8e-8, Fig. 1C bottom) than LBCL cells. These findings collectively suggest that the increased shedding of ctDNA by HRS cells compensates for the low cancer cell fraction in tumor biopsies, thus making cHL particularly amenable to noninvasive profiling using ctDNA.
DNASE1L3 shapes cHL cfDNA
Given the surprisingly high level of ctDNA shedding in cHL, we next explored differences in ctDNA fragmentation features between cHL and NHL. We first compared fragment size distributions between cHL and LBCL.32 Fragments harboring lymphoma-specific mutations from cHL patients were significantly shorter than those of LBCL patients (P=3.7e-13, Fig. 1D). This suggests a differential role of tissue endonucleases, especially because no such difference was evident in corresponding wildtype (wt) cfDNA fragments expected to originate predominantly from non-tumor tissue (P=0.07; Fig. 1D). Indeed, among DNases known to play crucial roles in cfDNA fragmentation,33 we found significantly higher DNASE1L3 expression in bulk cHL tumors (n=86) compared to LBCL tumors (n=66, P=6.6e-14, Fig. 1E, Supplementary Note [Figs. 4-6]).
To further characterize the unique role of DNASE1L3 in shaping the distinctive cHL ctDNA fragmentation profiles, we analyzed internal sequence motifs at cfDNA fragment ends. We observed an enrichment of CCNN-motifs in mutant cHL molecules as compared to their LBCL counterparts (Fig. 1F), resembling DNASE1L3 preferred end-motifs. Indeed, motifs described to be associated with DNASE1L3 digestion34 were significantly enriched among end-motifs overrepresented in mutant cHL molecules (P=9.2e-6, Fig. 1G), but not wildtype molecules (P=0.83). Given DNASE1L3’s role in cleaving multi-nucleosomal DNA molecules and its established role in cfDNA shedding,34,35 these data suggest that there is higher DNASE1L3 activity in cHL than LBCL and that this may contribute to increased ctDNA shedding by HRS cells.
To elucidate the cellular source of DNASE1L3, we performed single-cell RNA-Sequencing (scRNA-Seq) after careful presorting to enrich for intact, live HRS cells from 7 cHL tumors, with 2 normal reactive lymph nodes as controls (Methods; n=87,528 cells total from of n=9 biopsies from n=8 donors; Supplementary Table 3, Supplementary Note [Fig. 7]). Unlike previous scRNA-Seq studies in cHL, we reliably identified a robust HRS cell cluster after dimension reduction. Notably, there was no appreciable DNASE1L3 expression detectable in HRS cells. Strikingly, the expression of DNASE1L3 was instead confined to plasmacytoid and conventional dendritic cells (pDC/cDC, Fig. 1H-I), recently shown to spatially co-localize with HRS cells in cHL tumors.36
Noninvasive genomic profiles of cHL
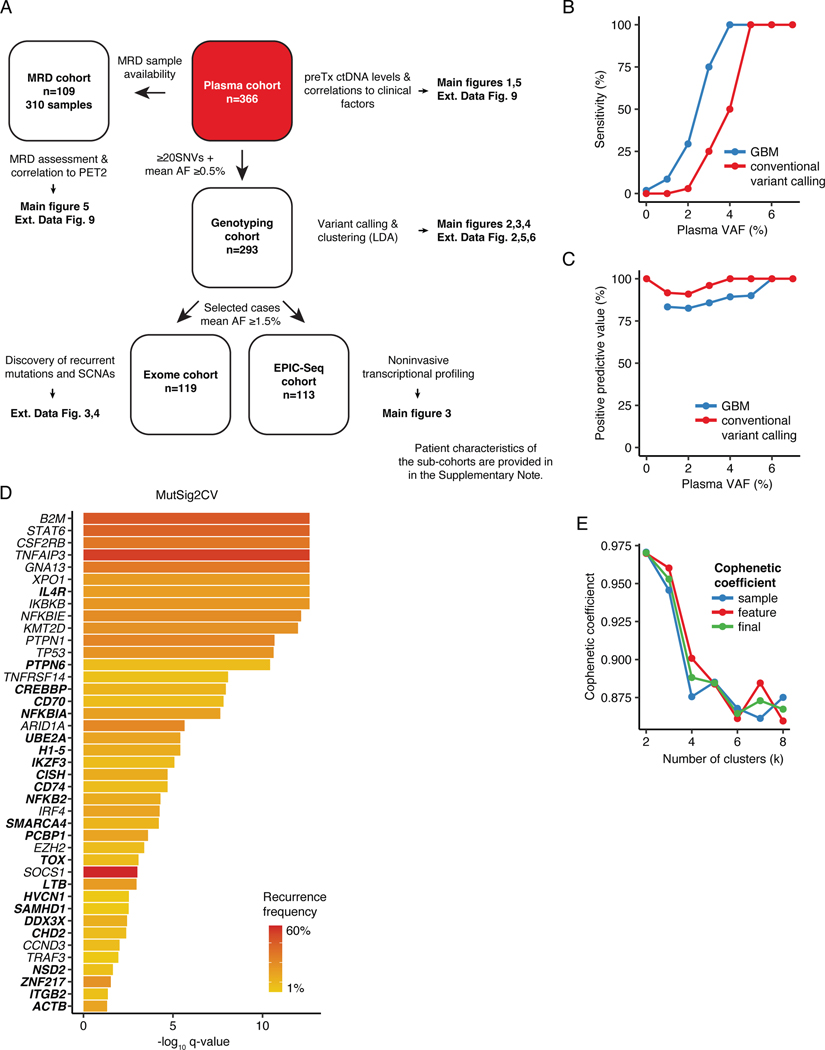
Having confirmed that efficient plasma shedding of HRS-derived mutations can obviate the need for technically challenging and time-consuming enrichment steps from bulk tumors, we set out to comprehensively and noninvasively genetically profile cHL. We prospectively collected pretreatment plasma samples from a total of 366 patients (Table 1, Supplementary Table 4). We used a hierarchical genotyping strategy to serially characterize the molecular landscape of cHL across the genome at multiple levels of resolution. We first profiled all pretreatment samples targeting 576-kb of the genome selected to genotype diverse B-cell lymphomas, and broadly considering existing cHL literature. In addition to targeting hypermutated regions across several B-cell lymphoma subtypes,5 we also included full coding region coverage for 151 genes in this panel (Supplementary Table 5). Because the fidelity of noninvasive genotyping is a function of ctDNA concentration,37 we used empiric heuristics to restrict all genotyping analyses to 293 patients (80%) with sufficient ctDNA burden (i.e., ≥0.5% mean AF and ≥20 single nucleotide variants [SNVs], Extended Data Fig. 1A).
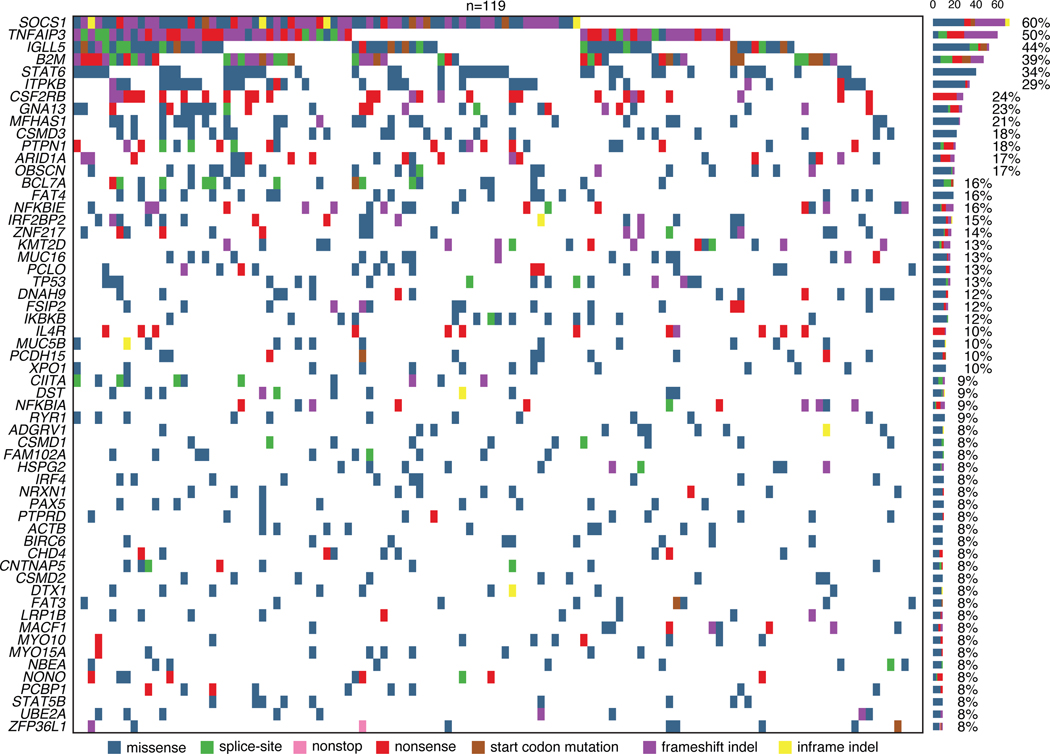
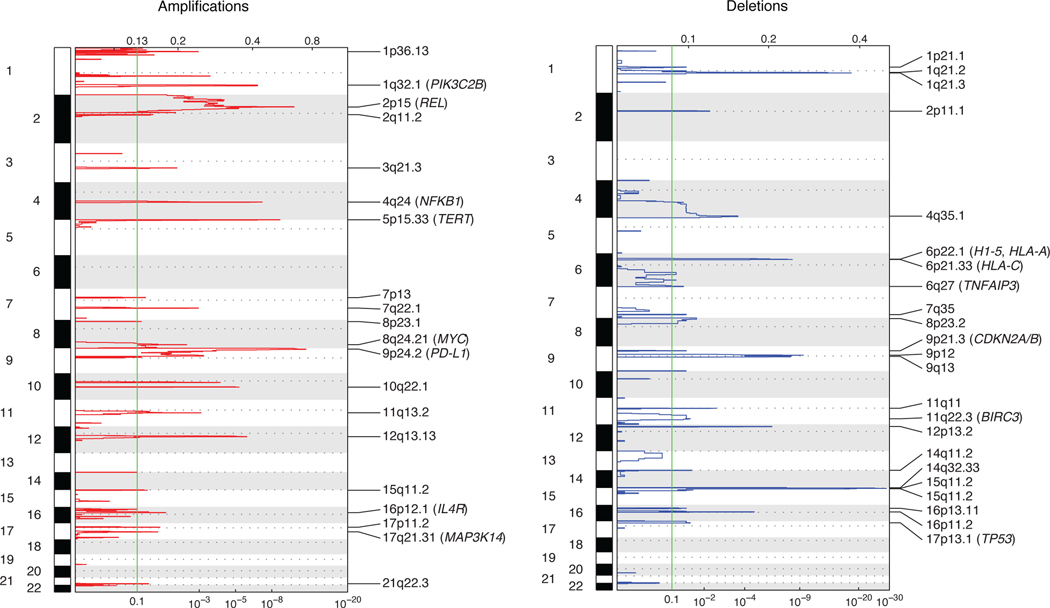
To characterize the broader landscape of somatic alterations in cHL, we separately used whole exome sequencing (WES) to additionally profile a subset of patients (n=119; 41%) enriched for samples with higher plasma AF (median ~5%, Extended Data Fig. 1A). We used an adaptive sequencing strategy to optimize WES genotyping with the use of machine learning (ML). Specifically, to enable robust and comparable WES genotyping across the range of ctDNA burdens, cases with mean AFs between 1.5–3% were sequenced at ~510x mean unique coverage depth, while cases with ≥3% were profiled at ~360x (Supplementary Table 6, Supplementary Note [Fig. 8], Methods, Extended Data Figs. 1B-C). Using MutSig2CV, we identified 41 total genes as being significantly mutated in cHL including SOCS1 (60%), TNFAIP3 (50%), B2M (39%), STAT6 (34%), CSF2RB (24%), GNA13 (23%), PTPN1 (18%) and ARID1A (17%; Extended Data Fig. 1D, 2–3; Supplementary Table 7, Supplementary Note [Fig. 9]). When considering genes not widely established as recurrently mutated in prior cHL next-generation-sequencing studies, we identified somatic mutations in ZNF217 (14%), IL4R (10%), NFKBIA (9%), ACTB (9%), PCBP1 (8%), CISH (6%), NFKB2 (6%), linker histone H1–5 (6%) and CD74 (3%). We also identified several recurrent amplifications and deletions using WES, including amplifications involving 2p15 (REL), 9p24.1–9p24.2 (CD274/PD-L1), 5p15.33 (TERT), and 17q21.31 (MAP3K14/NIK), as well as deletions involving 6q27 (TNFAIP3), 17p13.1 (TP53), 9p21.3 (CDKN2A/B), 11q22.3 (BIRC3), and 6p21–22 (H1–5, HLA-A, HLA-C, Methods, Extended Data Fig. 4, Supplementary Table 8).
Distinct genetic cHL subtypes
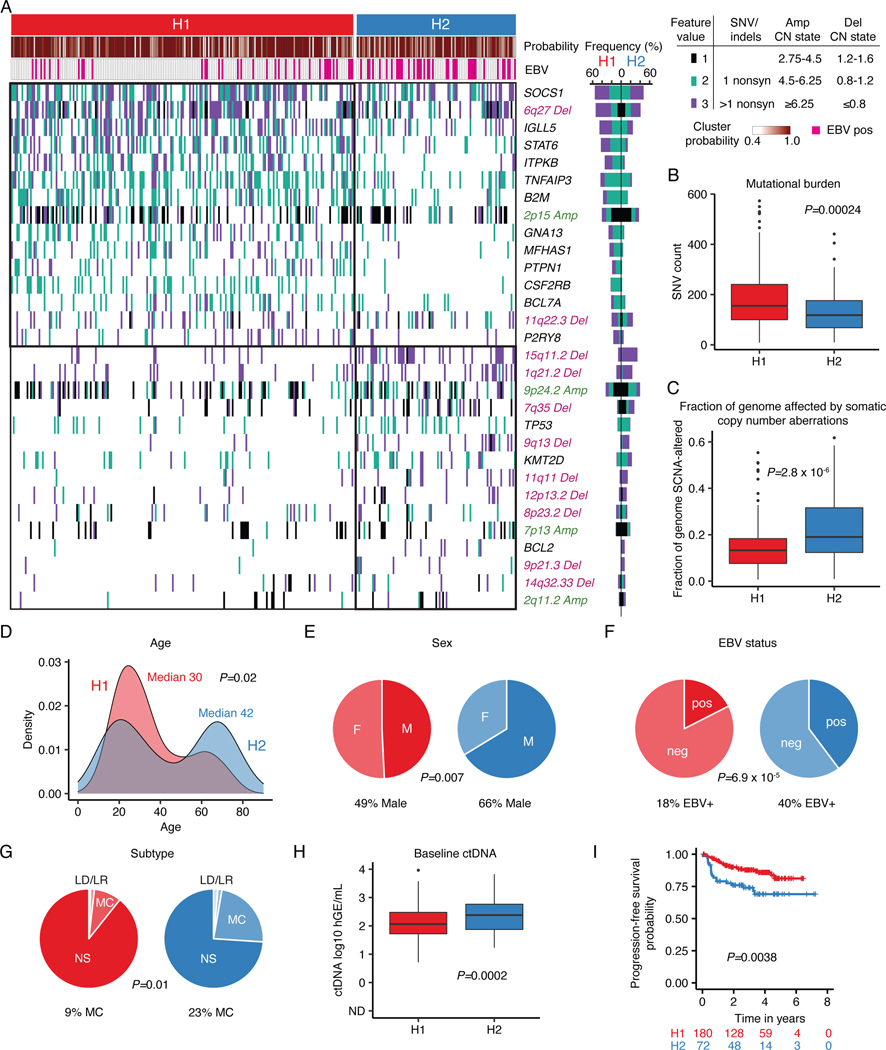
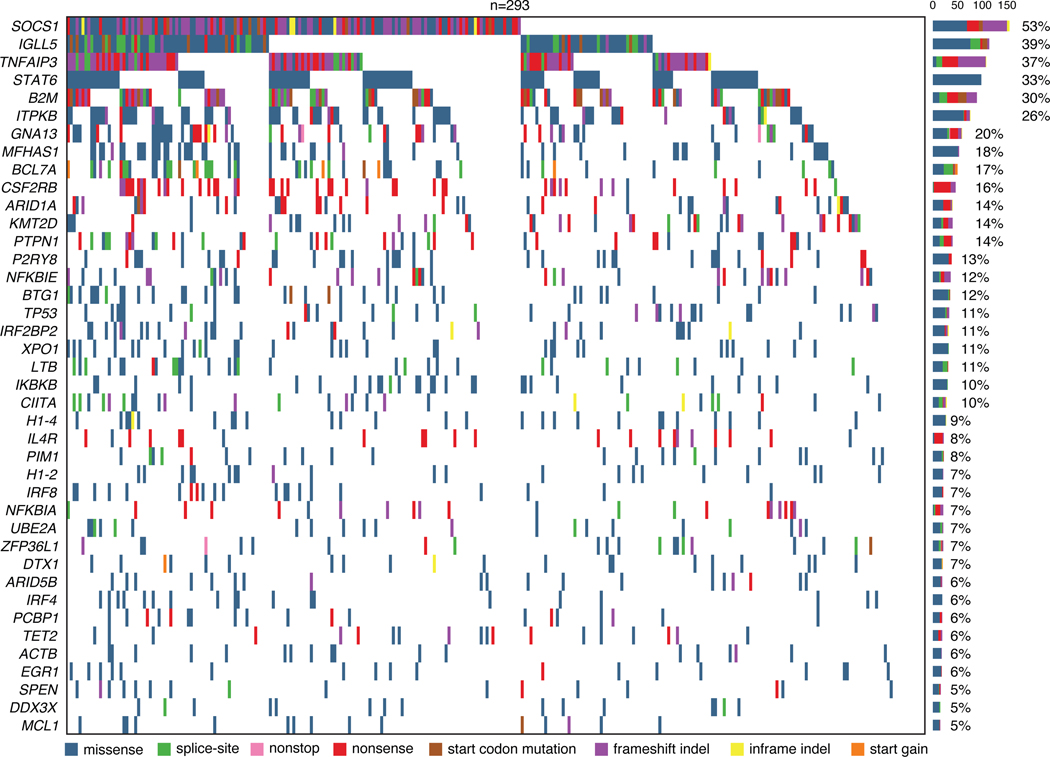
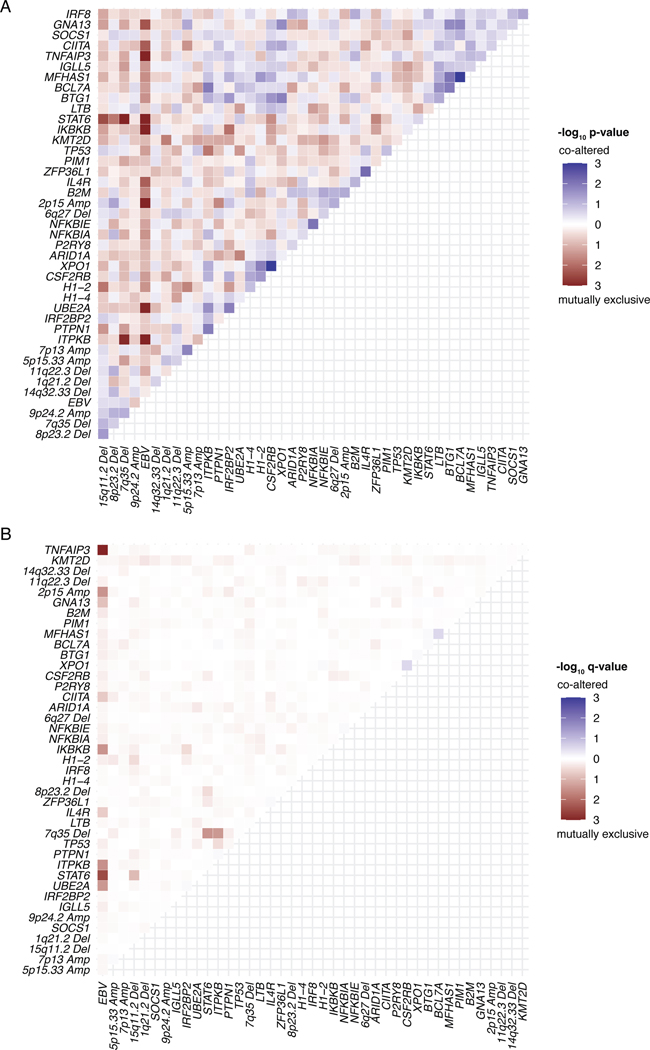
We next used an unsupervised approach to explore genotypically defined clusters in cHL, by adapting Latent Dirichlet Allocation (LDA), a probabilistic generative model in natural language processing.38 Specifically, we integrated SCNAs with non-silent somatic mutation calls as weighted features to define dominant genetic subtypes using LDA, where two dominant, highly-stable cHL clusters were identified (Methods, Fig. 2A, Extended Data Figs. 1E, 5, 6A, Supplementary Table 9). Cluster H1 tumors comprised ~68% of cases and were dominated by somatic mutations in genes canonically involved in NFkB, JAK/STAT, and PI3K signaling pathways. Conversely, cluster H2 tumors, which comprised ~32% of cases, were primarily characterized by a variety of SCNA events as well as mutations in TP53 and KMT2D (Fig. 2A, Supplementary Note [Table 4]). Importantly, all genes found to be significantly mutated by WES and differentially mutated between clusters were also covered by our targeted capture panel.
Figure 2: Genetic subtypes of cHL identified by LDA clustering.
(A) Heatmap summarizing non-silent mutations and SCNAs (rows) identified through noninvasive profiling of 293 pretreatment plasma samples (columns). Unsupervised clustering identified two distinct genetic subtypes denoted as H1 (n = 200) and H2 (n = 93). The top annotations visualize cluster assignment probability and EBV status. A legend summarizing the definition of feature values as used for clustering is provided in the top right. Alteration recurrence frequencies within each subtype by feature value are provided as a stacked bar plot. The top 15 features associated with each cluster are visualized. Indel, insertion and deletion; CN, copy number; SNV, single-nucleotide variant. (B-C) Boxplots with Wilcoxon p-values (two-sided) summarizing the targeted SNV burden (B), and fraction of the genome affected by SCNAs (C) by genetic subtype (n=293). (D) Density plot and Wilcoxon p-value (two-sided) summarizing age by genetic subtype (n=292 [n=200 H1, n=92 H2]). Median values are provided in the graph. (E-G) Pie charts and two-sided Fisher’s exact test p-values summarizing distributions of sex (E, n=288 [n=199 H1, n=89 H2]), EBV status (F, n=293) and histological subtype (G, n=241 [n=168 H1, n=73 H2], NS: nodular sclerosis, MC: mixed cellularity, LR: lymphocyte rich, LD: lymphocyte depleted subtypes) distribution by genetic subtype. (H) Boxplots and Wilcoxon p-value (two-sided) summarizing pretreatment ctDNA levels by genetic subtype (n=293). (I) Kaplan-Meier curves showing progression-free survival (PFS) stratified by genetic subtype. Logrank p-value and numbers at risk are provided in the graph. Only previously untreated, adult cHL patients were included in this analysis (n=252). Panels B,C,H: each box represents the interquartile range (the range between the 25th and 75th percentile) with the median of the data, whiskers indicate the upper and lower value within 1.5 times the IQR.
Comparing the two clusters, we observed H1 tumors to have a significantly higher somatic SNV mutational burden (P=0.00024), and H2 tumors to have a significantly larger fraction of their genome affected by SCNAs (P=2.8e-6, Fig. 2B-C). Of note, described associations with mutational burden and SCNAs were independent of the EBV status (Extended Data Figs. 6B-C). When considering clinical associations, cHL patients with H2 tumors demonstrated the expected bimodal age distribution with an early peak in the 20s and a second peak at >60 years. In contrast, H1 tumors predominantly occurred in younger patients (P=0.02, Fig. 2D), with less pronounced bimodality. Patients with an H2 genotype had a modest male predominance (P=0.007, Fig. 2E), were enriched for EBV positive tumors (P=6.9e-5, Fig. 2F) and mixed cellularity subtype (P=0.01, Fig. 2G). Patients with the H2 subtype also had higher ctDNA levels (P=0.0002, Fig. 2H), and inferior clinical outcomes (P=0.0038, Fig. 2I). Importantly, the negative prognostic implication of H2 tumors persisted when adjusting for high ctDNA levels (Hazard ratio 2.0 [95% Confidence interval 1.1–3.6], P=0.027).
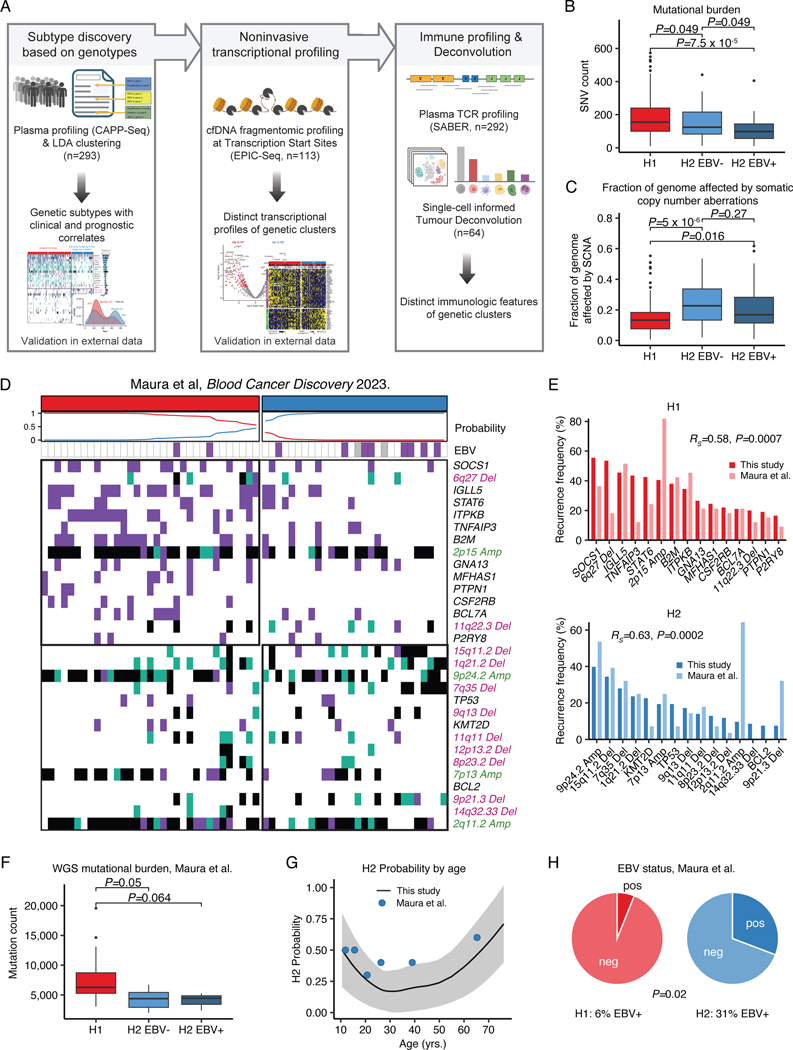
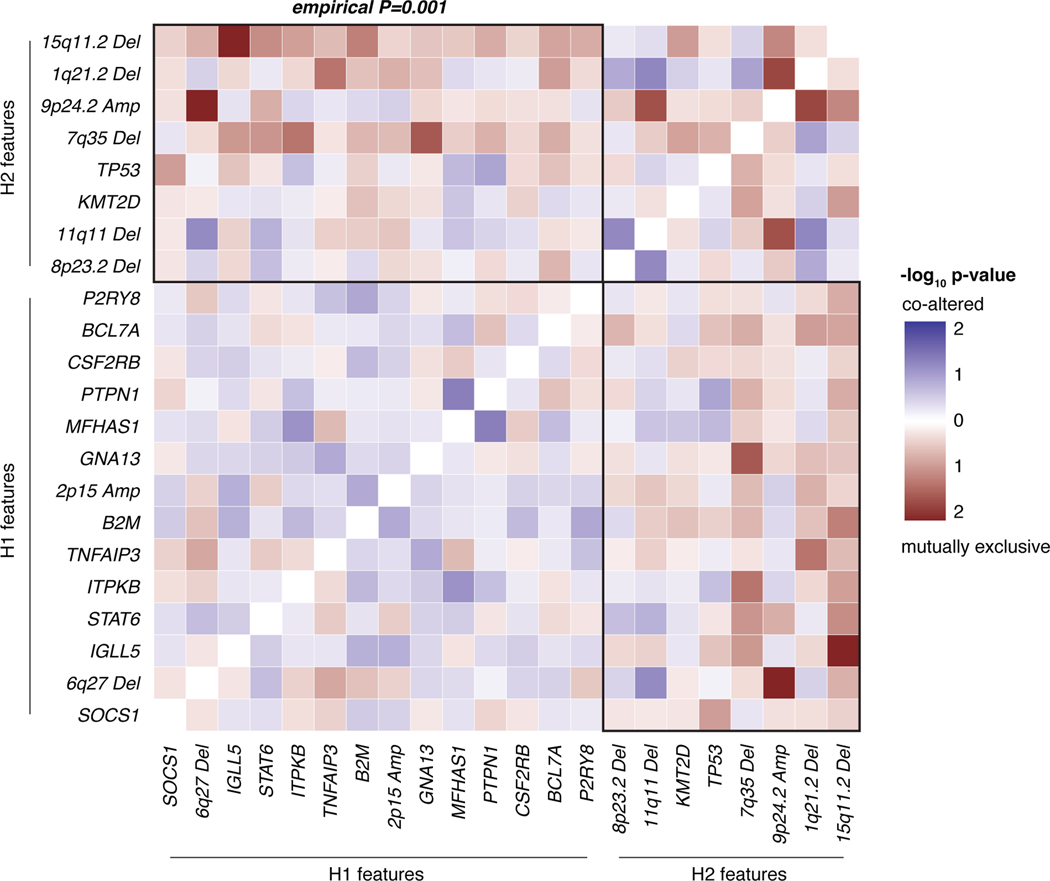
To validate our clusters in an external dataset, we then leveraged public whole genome sequencing (WGS) and WES genotypes of flow-sorted HRS cells, from 61 cHL patients enriched for pediatric cases.20 We assigned the H1/H2 subtype for each case using our previously defined probabilistic classifier generated through LDA from plasma sequencing, as described above (https://hodgkin.stanford.edu). Again, H1 was found to be more prevalent in comprising 54% of tumors, while 46% were classified as H2 tumors (Extended Data Fig. 6D). Recurrence frequencies of genetic features were comparable to and significantly correlated with those from our plasma discovery cohort (Extended Data Fig. 6E). Notably, we observed significant mutual exclusivity between H1- and H2-defining features as defined in the plasma cohort (P=0.001, Extended Data Fig. 7). When considering the whole genome space, we validated the higher mutational burden of H1 tumors, and confirmed this association to be independent of tumor EBV status (Extended Data Fig. 6F). Similarly, the bimodal age distribution and increased EBV positivity in the H2 subtype were also recapitulated (Extended Data Fig. 6G-H). Collectively, these results serve to validate the robustness of H1 and H2 subtypes of cHL and the characteristic genotypes defining them, as well as their distinctive associations with key clinical and pathological variables, including age, EBV status, and mutation burden.
Immuno-transcriptomic profiling
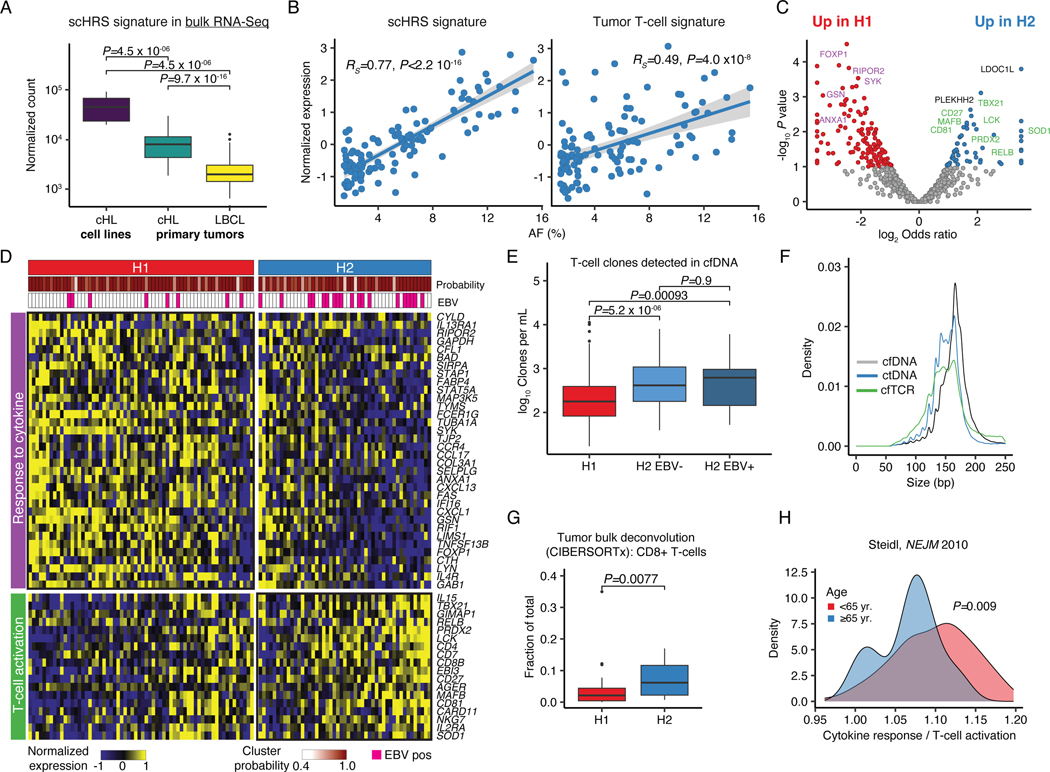
To explore transcriptional differences between genetic subtypes and to take advantage of the plasma enrichment of cHL, we leveraged EPIC-Seq, which allows for noninvasive gene expression profiling from cfDNA fragmentation patterns at transcription start sites (TSS, Supplementary Table 10).24 Using scRNA-Seq data, we first identified expression signatures of malignant HRS cells and their tumor infiltrating T-cell counterparts. When correlating plasma AF to these two signatures, we observed a high correlation with the HRS signature (Fig. 3A-B, RS=0.77, P<2.2e-16) and the Tumor T-cell signature (RS=0.49, P=4.0e-8, Fig. 3B), suggesting successful noninvasive profiling of both malignant HRS cells and the cHL tumor microenvironment (TME) by EPIC-Seq. In differential gene expression analysis, we found a substantial enrichment of a cytokine response signature in H1 tumors, while T-cell activation was among the top upregulated signatures in H2 tumors (Fig. 3C-D, Supplementary Table 11).
Figure 3: Genetic cHL subtypes are transcriptionally distinct.
(A) Boxplot and Wilcoxon p-values (two-sided) visualizing expression of a single-cell derived HRS (scHRS) gene signature in bulk RNA-Sequencing of cHL cell lines (n=8), and primary tumors (n=86 cHL, n=66 LBCL). (B) Scatter plot with Spearman rho and p-value (algorithm AS 89) delineating the correlation between mutational AF and inferred expression of the scHRS and a tumor T-cell signature using EPIC-Seq (n=113). (C) Volcano plot showing differentially expressed genes between clusters H1 (n=64) and H2 (n=49) as assessed by EPIC-Seq. (colored dots: unadjusted P<0.1 two-sided Fisher’s exact test). Genes from pathways visualized in (D) are highlighted in purple or green, respectively. (D) Heatmap visualization of 2 top differentially expressed pathways between H1 and H2 (GO:0034097, GO:0042110) at case and gene level. Top annotations denote cluster assignment probability and EBV status. (E) Boxplots and unadjusted Wilcoxon p-values (two-sided) visualizing T-cell counts per mL plasma detected from cfDNA using SABER (n=199 H1, n=56 H2 EBV-, n=37 H2 EBV+). (F) Length density plots of fragments supporting rearranged TCRs (cfTCR) as compared to wildtype (cfDNA) and mutant (ctDNA) supporting molecules (n=292, same as E). (G) CIBERSORTx deconvolution of 64 bulk cHL profiled by RNA-Sequencing using a scRNA-Sequencing derived signature matrix. Boxplot and Wilcoxon p-value (two-sided) comparing the CD8 T-cell content by genetic subtype (n=47 H1, n=17 H2). (H) Density plot and Wilcoxon p-value (two-sided) visualizing the ratio of the average normalized expression of cytokine response and T-cell activation genes visualized in (D) in gene expression data generated in prior studies.40 Patients with ≥65 years (yr., n=19) and those <65 yr. (n=111) are visualized separately. Panels A,E,G: each box represents the interquartile range (the range between the 25th and 75th percentile) with the median of the data, whiskers indicate the upper and lower value within 1.5 times the IQR.
We then used SABER to enumerate T-cell receptor (TCR) rearrangements in cfDNA (cfTCR) from plasma samples.32,39 Strikingly, we found significantly more T-cell clones in the plasma from H2 patients, independent of the tumor EBV status (P=5.2e-6 and P=0.00093, Fig. 3E). Notably, cfTCR fragment length profiles resembled the mutant ctDNA profiles, strongly suggesting a tumor origin of the TCR rearrangements detected in plasma (Fig. 3F). To confirm this finding, we applied immune cell deconvolution using CIBERSORTx to bulk RNA-Seq specimens with inferable genetic subtype. Indeed, we observed a higher abundance of CD8+ T-cells in H2 tumors (P=0.0077, Fig. 3G), which generally lack B2M mutations and thus is expected to be enriched for tumors with preserved MHC-I expression. As additional line of evidence supporting the generalizability of our findings, we observed lower ratios of cytokine response over T-cell activation signature in older patients (≥65 years) expected to be enriched for the H2 subtype (P=0.009, Fig. 3H) in an external gene expression dataset.40
IL4R mutations enhance IL13 signaling
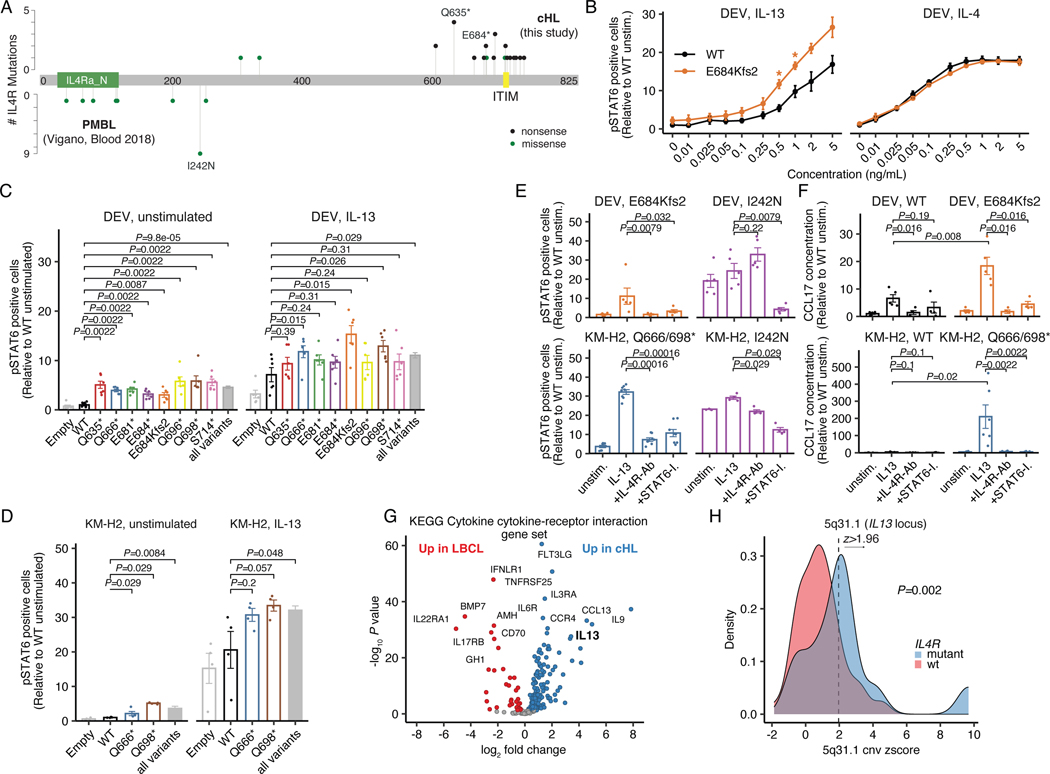
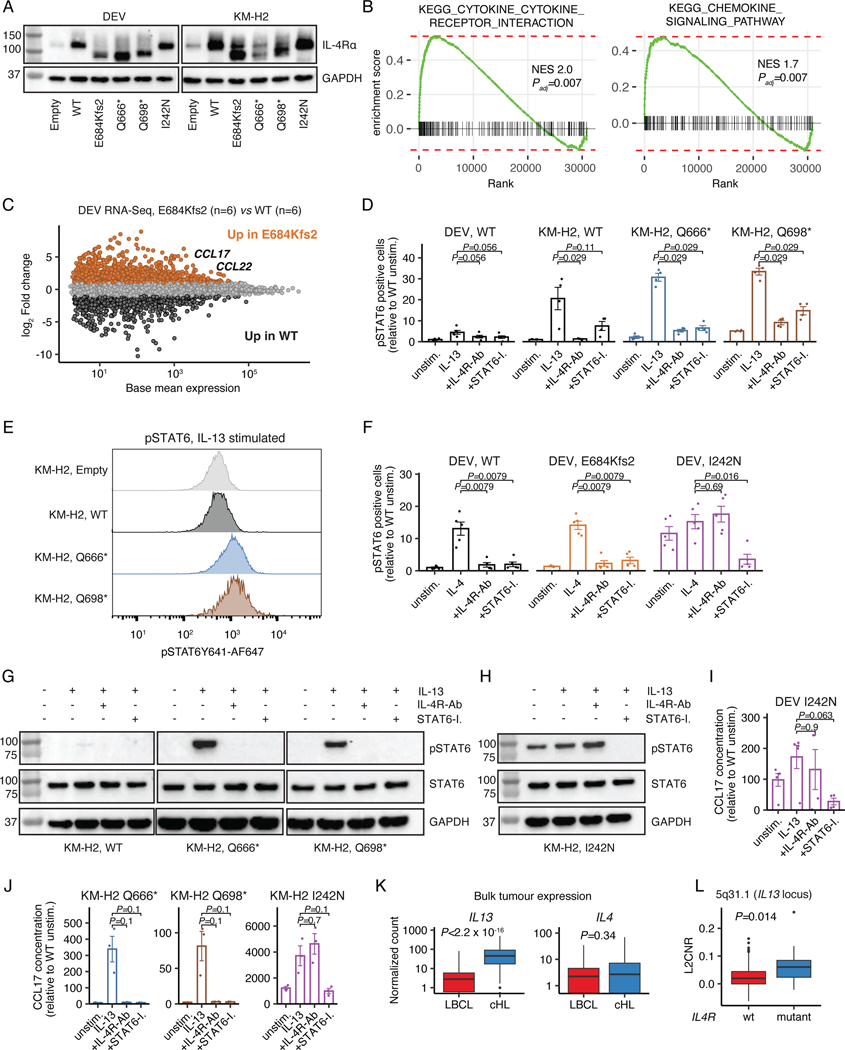
Among the recurrently and significantly mutated genes in cHL, we identified Interleukin-4 receptor (IL4R) lesions in 10% of cases profiled by plasma WES (Supplementary Table 12). IL4R mediates both interleukin 4 (IL4) and interleukin 13 (IL13) signaling,41 with gain-of-function hotspot IL4R mutations previously described in the extracellular and transmembrane domains in primary mediastinal large B-cell lymphoma (PMBL)42 and Diffuse Large B-cell Lymphoma (DLBCL).43 Rare IL4R mutations have recently also been described in cHL patients in small case series.19 Strikingly, the 26 IL4R mutations observed in the larger sequencing cohort were distinct from those observed in PMBL and DLBCL, with ~85% of cHL mutations being truncating and clustering at the cytosolic C-terminus of the IL4R protein. Moreover, most of these truncating lesions were nonsense mutations or frameshifting indels generating stop-codons that immediately preceded and disrupted IL4R’s immunoreceptor tyrosine-based inhibitory motif (ITIM) domain (Fig. 4A), with one additional missense mutation directly affecting the conserved Tyr residue within the ITIM domain itself. Of note, IL4R mutations were not restricted to either of the genetic subtypes.
Figure 4: Truncating IL4R mutations enhance IL13 signaling and are targetable through IL4R antibodies.
(A) Lollipop plot summarizing IL4R mutations identified in cHL patients (n=26, top) as compared to PMBL patients (n=16, bottom42). Nonsense (black) and missense mutations (green). (B) Dose-response curves for IL13/IL4 in transduced DEV cells using phospho-STAT6 (flow cytometry) as readout. WT and E684Kfs2: n=3 each. * P<0.05 (t-test, two-sided; P=0.013 at 0.5ng/mL and P=0.028 at 1ng/mL). (C) Phospho-STAT6 levels in unstimulated and IL13-stimulated transduced DEV cells (n=6 each; grey bar: all mutants [all variants, n=48]). Unadjusted Wilcoxon p-values (two-sided) compared to WT are provided. (D) Phospho-STAT6 levels in unstimulated and IL13-stimulated transduced KM-H2 cells (n=4 each; grey bar: all mutants [all variants, n=8]). Unadjusted Wilcoxon p-values (two-sided) compared to WT are provided. (E) Phospho-STAT6 levels in transduced DEV (n=5 each) and KM-H2 (n=4 each) cells under unstimulated, IL13-stimulated, IL13-stimulated + IL4R-antibody (IL4R-Ab) treated as well as IL13-stimulated + STAT6-Inhibitor (STAT6-I.) treated conditions. Unadjusted Wilcoxon p-values (two-sided) compared to IL13 stimulation alone are provided. (F) CCL17 (TARC) concentrations with unadjusted Wilcoxon p-values (two-sided) in supernatant of transduced DEV (n=5 for unstimulated/IL13; n=4 for IL4R-Ab/STAT6-I.) and KM-H2 cells (n=3 each) under unstimulated, IL13-stimulated, IL13-stimulated + IL4R-Ab treated as well as IL13-stimulated + STAT6-I. treated conditions. (G) Volcano plot summarizing differentially expressed genes from the KEGG Cytokine cytokine-receptor interaction gene set between cHL (n=86) and LBCL (n=66) in bulk RNA-Sequencing. (H) Copy-number (CN) z-score (density plot) of the 5q31.1 cytoband harboring IL13 stratified by IL4R mutation status in patients with plasma exome sequencing [IL4R mutant: n=12; IL4R WT: n=107]. 7/12 (58%, IL4R mutant) and 16/107 (15%, IL4R WT) cases were found to have a 5q31.1 amplification when considering a z-score of 1.96 as threshold. The corresponding two-sided Fisher’s exact test p-value is provided. Panels B-F: mean +/− standard error (se).
We next hypothesized that this novel class of IL4R mutations might confer a gain-of-function phenotype and sought to functionally validate this hypothesis. We stably transduced the Hodgkin lymphoma-derived cell lines DEV and KM-H2 with mutant IL4R constructs (Extended Data Fig. 8A) and measured phosphorylation levels of Signal transducer and activator of transcription 6 (STAT6), a downstream mediator of IL4-R signaling. In vitro soluble cytokine titration experiments revealed the E684Kfs2 truncating mutation enhanced IL13-, but not IL4-signaling in DEV cells (n=3, P=0.013 at 0.5ng/mL and P=0.028 at 1 ng/mL IL13, respectively, Fig. 4B and Supplementary Note [Fig. 10]). We then verified this gain-of-function phenotype across a broad variety of IL4R mutants identified from cHL patient samples in DEV cells (n=48, P=9.8e-5 [unstimulated] and P=0.029 [IL13], respectively, Fig. 4C) as well as in KM-H2 cells (n=8, P=0.0084 [unstimulated] and P=0.048 [IL13], respectively, Fig. 4D and Supplementary Note [Fig. 10]). In addition, we found an enrichment of cytokine and chemokine gene expression signatures in DEV cells expressing mutant IL4R (E684Kfs2 [n=6] vs WT [n=6]) as well as increased expression of downstream genes that are transcriptionally regulated by STAT6, including CCL17 (TARC) and CCL22 (Extended Data Figs. 8B-C).
Therapeutic targeting of IL4R mutations
We next assessed whether cells harboring these novel IL13 cytokine-dependent IL4R mutations could be therapeutically targeted. While gain-of-function phenotypes of both hotspot IL4R mutations in PMBL (I242N) and cHL-type mutations could be reversed by downstream STAT6 inhibition using a small molecule (DEV: n=5, P=0.032 [E684Kfs2] and P=0.0079 [I242N]; KM-H2: P=0.00016 [Q666*/Q698*, n=8] and P=0.029 [I242N, n=4]), only truncating cHL mutations responded to blockade with antibodies targeting surface IL4R (DEV: n=5, P=0.0079 [E684Kfs2]; KM-H2: n=8, P=0.00016 [Q666*/Q698*], Fig. 4E, Extended Data Fig. 8D-H and Supplementary Note [Fig. 10]). Importantly, both the gain-of-function phenotype (DEV: P=0.008 [E684Kfs2, n=5]; KM-H2: P=0.02 [Q666*/Q698*, n=3 each]) as well as its reversal through IL4R blockade (DEV: P=0.016 [E684Kfs2, n=4]; KM-H2: P=0.0022 [Q666*/Q698*, n=3 each]) were confirmed by quantifying supernatant levels of CCL17 (TARC), a STAT6 transcriptional target and key chemokine produced by HRS cells that shapes the cHL TME HRS cells depend on44 (Fig. 1I, 4F and Extended Data Fig. 8I-J). These findings suggest that C-terminal truncating IL4R mutations found in cHL, but not transmembrane domain hotspot mutations found in PMBL, mediate IL13 cytokine-dependent gain-of-function phenotypes.
We next wondered why this novel class of IL4R mutations has not been observed in related entities including LBCL subtypes or Follicular Lymphoma (FL), in which IL4 secreted by T follicular helper cells is known to contribute to tumor pathogenesis.45,46 Interestingly, bulk RNA-Seq revealed that IL13, but not IL4, was among the top upregulated genes in cHL (n=86) as compared to LBCLs (n=66; Log2 fold change [IL13] 3.4, adjusted p-value=2.6e-28, Fig. 4G, Extended Data Fig. 8K and Supplementary Note [Fig. 11-12]). Further, we found amplifications of the IL13 locus (5q31.1) to be enriched in IL4R mutant cases (P=0.002 [Fisher, copy-number (CN) z-score] and P=0.014 [L2CNR], Fig. 4H, Extended Data Fig. 8L). These findings suggest that HRS cells are genetically poised to produce IL13 (Fig. 1I), and that IL13 plays a key role in cHL growth and pathogenesis within an autocrine loop. Importantly, our findings are in line with previous reports describing IL13 as a hallmark of cHL.47–50 We here show that mutations leading to loss of the ITIM domain of IL4R further enhances this mechanism. The special TME in cHL may therefore promote the emergence of truncating IL4R mutations and apply the unique selective pressure which confines this class of mutations to cHL.
Pretreatment ctDNA levels in cHL
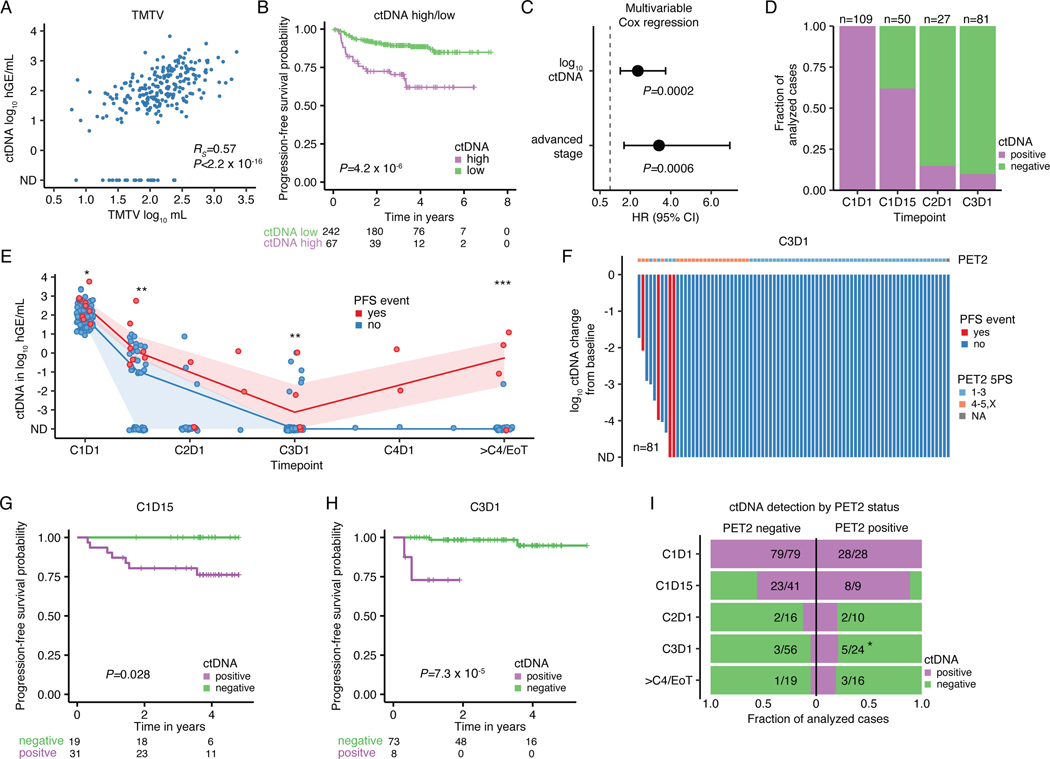
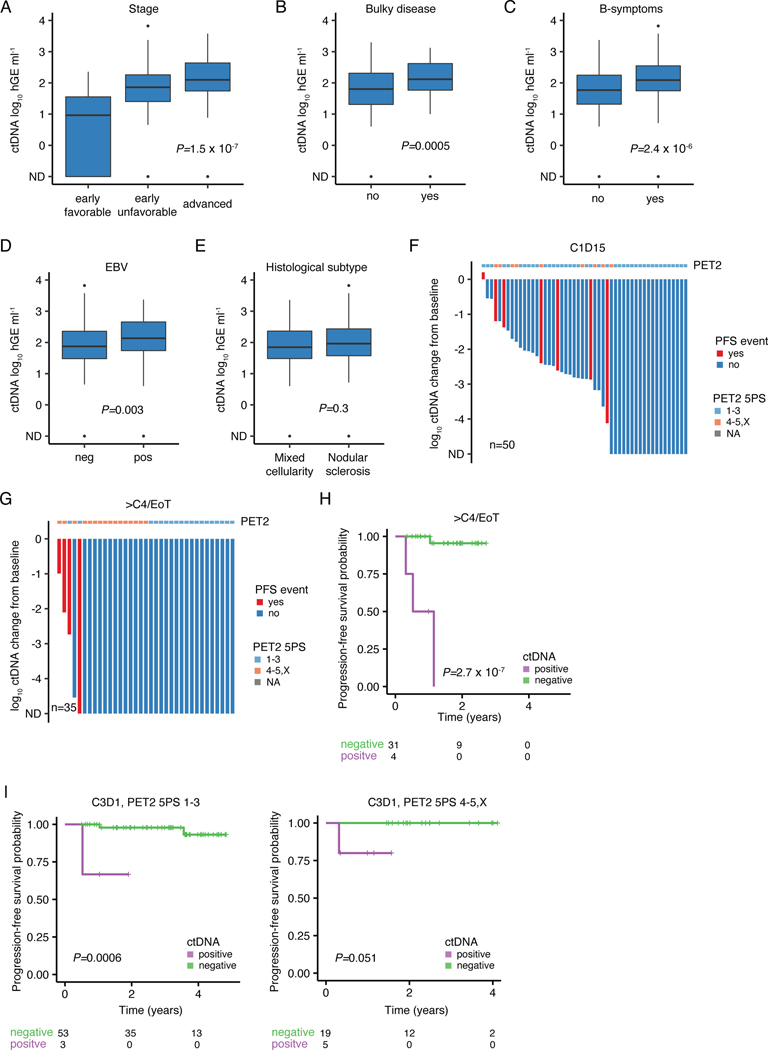
Pretreatment ctDNA levels are known to largely reflect tumor volume, and are associated with several adverse risk factors capturing disease burden in non-Hodgkin lymphomas.27,28 We also observed TMTV to be highly correlated with ctDNA levels in cHL (Spearman rho=0.57; Fig. 5A), and with other established adverse risk factors (Extended Data Fig. 9A-E). When stratified into groups of high and low ctDNA levels using a threshold previously established in DLBCL,27,28 patients with high pretreatment ctDNA levels had significantly shorter progression-free survival (PFS, P=4.2e-6, Fig. 5B). Similarly, in univariable Cox regression analysis, continuous ctDNA levels were strongly predictive of PFS (HR 2.9 [1.8–4.6], P=4.3e-6). Remarkably, the prognostic impact of ctDNA was independent of disease stage in a multivariable Cox regression (HR 2.4 [1.5–3.7], P=0.0002, Fig. 5C).
Figure 5: Pretreatment ctDNA levels and ctDNA minimal residual disease to prognosticate cHL.
Previously untreated, adult cHL patients were considered for associations between pretreatment ctDNA levels and clinical variables in panels A-C (n=309). (A) Scatter plot and Spearman rho and p-value (algorithm AS 89) correlating TMTV and pretreatment ctDNA levels (n=241). (B) Kaplan-Meier curves showing PFS stratified by pretreatment ctDNA levels binarized using a previously defined threshold in DLBCL27 (2.5 log10 hGe/mL). Numbers at risk and logrank p-value are provided. (C) Forest plot summarizing hazard ratios (HR) and 95%-confidence interval (CI) derived from multivariable Cox regression including ctDNA and disease stage as a categorical variable (early favorable, early unfavorable, and advanced). Early favorable stage (n=14, no event) were omitted from visualization. Patients with at least one profiled on-treatment sample were evaluated for MRD assessment in panels D-I (n=109). (D) Stacked bar plot visualizing the fraction of evaluable cases with detectable ctDNA by PhasED-seq at various milestones. CxDy, cycle x day y. (E) Line plot summarizing median ctDNA levels and the interquartile range along the course of treatment. * P<0.05; ** P<0.01; *** P<0.001 (Wilcoxon test, two-sided; C1D1: P=0.03, C1D15: P=0.005, C3D1: P=0.007, ≥4 cycles: P=2.0e-5). (F) Waterfall plot showing log10 ctDNA changes from baseline at C3D1. Bars are colored by PFS event status with PET2 readings according to 5-point scale (5PS Deauville) as top annotation. (G-H) Kaplan-Meier curves showing PFS stratified by ctDNA detection at (G) C1D15 and at (H) C3D1. Logrank p-values are provided. (I) Stacked bar plot summarizing the fraction of evaluable cases with detectable/undetectable ctDNA by PET2 status. PET2 positive cases (5PS 4–5,X) are plotted on the right, PET2 negative cases (5PS 1–3) on the left. * P<0.05 (two-sided Fisher’s exact test; P=0.048 at C3D1). 2/109 patients with missing PET2 status were excluded.
Minimal residual disease, outcomes & PET
While MRD has demonstrated prognostic utility in diverse cancers including B-cell lymphomas,1–3,27,29,51 the role of ctDNA MRD in cHL is less clear. We therefore used ultrasensitive PhasED-Seq ctDNA assessment5 to measure MRD in adult cHL patients with available serial on-treatment plasma samples during curative-intent therapy (310 samples from 109 patients, Methods, Extended Data Fig. 1A, Supplementary Note [Tables 1-3, Fig. 13]), representing the largest cHL cohort profiled to date.
In line with cHL’s distinctive early chemosensitivity, we observed rapid molecular responses to frontline induction regimens. Indeed, molecular remission by ctDNA rose with each successive cycle of therapy, yielding corresponding MRD negativity rates of 38%, 85%, and 90% at (C)ycle 1 (D)ay 15, C2D1 and C3D1 (Fig. 5D). Patients with durable remissions experienced more precipitous and sustained ctDNA decreases compared with patients ultimately experiencing disease progression. Specifically, patients with durable remissions achieved significantly lower ctDNA levels at various milestones throughout treatment including at C1D1 (P=0.03), C1D15 (P=0.005), C3D1 (P=0.007), and after ≥4 cycles (P=2.0e-5, Fig. 5E). Accordingly, ctDNA detection was significantly associated with relapse risk, even at very low levels (Figs. 5F, Extended Data Fig. 9F-G). Indeed, patients with detectable ctDNA had inferior progression-free survival (PFS) at C1D15 (P=0.028, Fig. 5G), C3D1 (P=7.3e-5, Fig. 5H) and after ≥4 cycles (P=2.7e-7, Extended Data Fig. 9H).
While most interim PET/CT (iPET) “positive” patients were negative for ctDNA MRD at the corresponding landmark and remained disease-free, we observed an enrichment for ctDNA MRD positivity in patients with positive PET/CT scans after two cycles (PET2, P=0.048 at C3D1, Fig. 5I). Of note, ctDNA MRD demonstrated independence of PET2 when correlated with treatment outcome (Extended Data Fig. 9I).
Discussion
In the most comprehensive analysis thus far, we demonstrate that cHL mutations are enriched in blood plasma compared to corresponding bulk tumor specimens in most patients. While validating prior studies that have suggested this phenomenon,2,52 our work also extends on these prior data and elucidates the etiology of this surprising and paradoxical observation. Specifically, increased ctDNA shedding in cHL could be driven by several distinct mechanisms. Such mechanisms might include (1) the active secretion of mutant ctDNA by HRS cells, e.g. as extrachromosomal circular DNA (eccDNA),53,54 (2) the more rapid division of HRS cells than other adjacent cell types in the cHL TME resulting in their higher ctDNA shedding, (3) the presence of HRS progenitors outside of biopsied tumors (e.g. in radiographically occult anatomic compartments) that nevertheless contribute to the ctDNA pool, (4) the active protection or cryptic sequestration of wildtype cfDNA molecules by cHL tumors, or (5) some combination of these factors with contributions from HRS cells and/or other cells in the cHL TME.
Notably, cHL appears distinct from most other human tumor types in expressing higher levels of DNASE1L3 compared to matched normal tissue.55 Accordingly, we demonstrate that the exceptionally high DNASE1L3 expression levels in cHL appear to drive both the distinct fragmentation patterns and end-motif sequences of mutant ctDNA molecules and TME-derived cfDNA, capturing this endonuclease’s substrate preferences. In line with our findings, DNASE1L3 has been shown to mediate fragmentation of circulating multi-nucleosomal DNA molecules and enhanced cfDNA shedding in a murine liver necrosis model.34,35 Among distinct cell types in the cHL tumor microenvironment, DNASE1L3 expression was primarily restricted to pDCs and cDCs, while HRS cells were not found to express this enzyme. Collectively, these findings suggest that DNASE1L3 cleaves tumor-derived molecules extracellularly, and may play a role in increased ctDNA shedding and maintaining the supportive TME that is characteristic of cHL.56
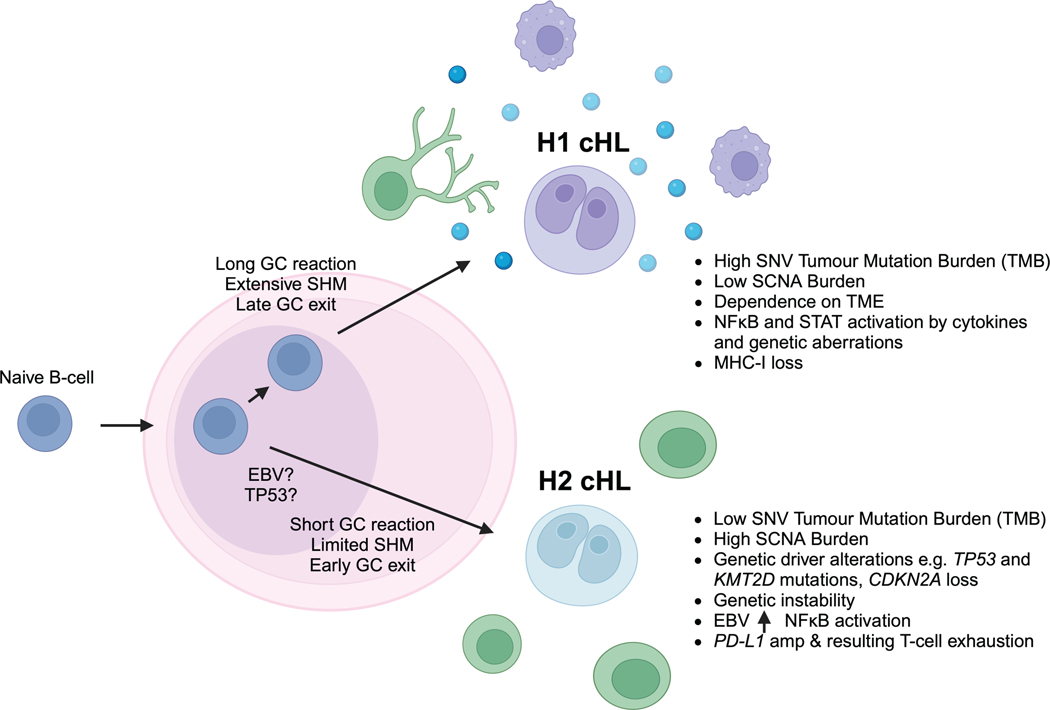
Given the especially robust representation of circulating cHL tumor genomes in blood plasma, noninvasive cfDNA profiling allowed us to study the genomic landscape of the largest cHL cohort to date, comprising 366 pretreatment plasma samples, which we profiled by targeted sequencing and/or whole-exome sequencing. This rich genomic landscape of somatic variants allowed us to apply clustering methods to identify two distinct cHL genetic subtypes, H1 and H2. Importantly, we validated the genetic classes in an external dataset, and also characterized their distinct biological features through several orthogonal methods, including noninvasive transcriptional profiling, as well as by immunological profiling of plasma and tumor tissue. Integrating these orthogonal observations, H1 tumors appear more driven by NF-κB signaling, crosstalk signaling within the cHL TME, cytokine receptors, and downstream STAT signaling. This cytokine-driven phenotype is paired with frequent MHC-I loss in H1 tumors, as an immune escape mechanism. In contrast, H2 tumors appear driven by more “conventional” lymphoma drivers such as KMT2D and TP53, as well as genetic instability similar to the A53 subtype in DLBCL, with these somatic changes paired with additional mechanisms including PD-L1 amplifications driving T-cell exhaustion (Extended Data Fig. 10).
While checkpoint inhibitors appear broadly effective in inducing high remission rates cHL, our observations suggest that mechanisms of response to such blockade could vary between subtypes. For example, we speculate that checkpoint inhibitors might mainly act in H1 tumors by disrupting CD4 T-cell mediated nursing through the TME, while in H2 tumors, reversing CD8 T-cell exhaustion may be their dominant mechanism of action. Accordingly, future studies are required to assess the predictive value of genetic subtypes and implications for personalized therapy selection in the context of modern regimens that include checkpoint inhibitors. Importantly, LDA, as a generative probabilistic model, allows the assignment of any given new sample to one of the 2 genetic subtypes, thus facilitating the incorporation of these cHL subtypes into future studies.
Among the newly discovered recurrent cHL variants defined by ctDNA profiling, we identified a novel class of truncating, IL13 cytokine-dependent IL4R gain-of-function mutations. Through in vitro and in silico analyses, we demonstrate an autocrine IL13 loop to be a hallmark of cHL that drives the selection of such mutations, thus largely confining them to these tumors. Importantly, unlike transmembrane mutations identified in Non-Hodgkin lymphomas,42,43 this novel class of mutations appears uniquely susceptible to drugs targeting IL4R. Antibodies targeting IL4R or related proteins, which are currently used to treat atopic disease,57 could therefore have a future role in the treatment of cHL, perhaps as part of abbreviated chemotherapeutic strategies relying on immunological blockade. It is noteworthy, that truncating IL4R mutations are the only genetic alterations described to date that seem to exclusively occur in cHL, and therefore discriminate between Hodgkin and non-Hodgkin lymphomas.
Finally, we demonstrate the significant prognostic potential of pre- and on-treatment ctDNA levels in cHL. We envision that ctDNA levels will refine staging procedures, complement existing risk stratification tools such as iPET and guide therapy selection. For example, early MRD timepoints may help guide treatment de-escalation in favorable subgroups, while later timepoints could be used to guide treatment intensification due to the higher positive predictive value. Analysis of ctDNA MRD may be particularly beneficial in the context of checkpoint inhibitor treatment, given the higher uncertainty of interim PET in this setting.58,59
Collectively, this study contributes to the broader understanding of cHL biology, offers opportunities to improve risk stratification and response assessment, and describes novel targets for precision therapy.
Methods
cHL patients & samples
We included pediatric and adult patients diagnosed with cHL with available plasma and/or tumor specimens (Supplementary Table 1 and Supplementary Note [Tables 1-3]). All patients were treated at cancer centers across Europe and North America between 2011 and 2020. Patients treated in or outside of clinical trials at 3 cancer centers, namely Stanford (CA, USA), Bellinzona (CH) and Leuven (BE) were eligible. Additionally, clinical trial collectives were included in this study. Pediatric patients enrolled in the Phase III trial AHOD1331 trial (NCT02166463)60 and the Phase II cHOD17 study (NCT03755804)61 were included. Pediatric patients were not considered for associations with outcome, yet included in genotyping analyses to characterize the full spectrum of the disease. Further, patients enrolled in the Phase III AHL2011 (NCT01358747),62 the Phase II BREACH trial (NCT02292979),52,63 a pilot study evaluating concurrent Pembrolizumab-AVD (pembrolizumab and doxorubicin, vinblastine, and dacarbazine, NCT03331341)64 as well as a Phase II study evaluating PVAB (Prednisone, Vinblastine, Doxorubicin and Bendamustine, NCT02414568)65 in elderly patients were included. Patients were consented either for observational studies approved by the local institutional review board (IRB) at Stanford University (Stanford, CA), the University of Bellinzona (CH), UC Leuven (BE) and St. Jude Children’s Research Hospital (Memphis, TN), or IRB-approved protocols (i.e. NCT02166463, NCT03755804, NCT01358747, NCT02292979, NCT03331341, NCT02414568).
Large B-cell lymphoma (LBCL) plasma samples
We compared both unadjusted and TMTV-adjusted ctDNA levels in cHL to those of untreated LBCL patients (n=114) previously published by our group.28 Patients were selected based on the availability of ctDNA levels using matched normal samples and TMTV measurements. We performed fragment length and end-motif analyses using pretreatment samples from a recently reported cohort of patients with relapsed/refractory LBCL (n=53) treated with CD19 CAR-T cells.32
Nucleic acid isolation & processing
Plasma samples were collected in EDTA, BCT or Streck tubes, and processed according to local standards. Cell-free DNA (cfDNA) was isolated using the QIAamp Circulating Nucleic Acid Kit (Qiagen). Cellular DNA from either plasma-depleted whole blood or peripheral blood mononuclear cells was isolated using the DNeasy Blood and Tissue Kit (Qiagen). Tumor-derived DNA and RNA was isolated from 2–4, 10 μm-thick, formalin-fixed, paraffin-embedded (FFPE) scrolls of tumor tissue using the RNAstorm/DNAstorm Combination Kit (Cell Data Sciences, Fremont, CA). Following isolation, cellular DNA was fragmented to a target size of 170-bp using a Covaris S2 sonicator. DNA was quantified using the Qubit dsDNA High Sensitivity Kit (Thermo Fisher Scientific, Waltham, MA) and fragment length was assessed using a Fragment Analyzer System (Agilent, Santa Clara, CA). RNA was quantified by Nanodrop.
DNA library preparation & sequencing
DNA sequencing libraries were prepared as previously described.31 Barcoded libraries underwent hybrid capture using biotinylated oligonucleotides targeting selected regions of the hg19 reference genome. All pretreatment samples were captured using custom 608 kb (SeqCap EZ Choice, Roche) or 772 kb (Discovery Pool, IDT) oligonucleotide panels targeting genomic regions known to be recurrently mutated or otherwise biologically significant in B cell-derived neoplasms. For genotyping purposes, mutation calls were restricted to the 576 kb overlap between the two panels to avoid batch effects. On-treatment samples were sequenced with smaller panels targeting regions with the highest mutational density in lymphoma and known to be enriched for phased variants. The smaller panels were designed as subsets of their respective parent panels, and covered 144 kb (SeqCap EZ Choice, Roche) and 268 kb (xGen, IDT), respectively. Pretreatment samples were additionally captured using oligonucleotides targeting oncogenic DNA viruses (Discovery Pool, IDT) including Epstein–Barr virus (EBV). A subset of the pretreatment libraries was subjected to whole-exome sequencing (WES) using the NimbleGen SeqCap EZ MedExome panel (Roche) or the EPIC-Seq panel targeting TSS (Twist Bioscience). All captures were performed according to the manufacturers’ instructions with minor optimizations.
Libraries were sequenced on Illumina HiSeq4000/X-ten or NovaSeq6000 machines with 2×150bp paired-end reads. FASTQ files were demultiplexed using 8-bp dual sample barcodes and reads were mapped to the human reference genome (hg19) using BWA ALN and BWA MEM (the latter was only used for small insertion/deletion and fusion calling). Molecules were deduplicated with error-suppression using a custom pipeline as previously described.30,31
CAPP-Seq variant calling
Single nucleotide variants
‘Cancer Personalized Profiling by Deep Sequencing’ (CAPP-Seq) was performed to identify somatic mutations from plasma or tumor specimens. Prior to variant calling, stereotypical background noise was removed from plasma sequencing data using cfDNA from ≥16 healthy donors, as previously described.30,66 Variant calling was performed with or without matched leukocyte sequencing (‘germline’) using an adaptive variant calling strategy that takes into account the specific base change observed, the local depth, and both global and local error rates, as previously described.30 Variant calls were further filtered using the following heuristics to increase specificity: deduplicated depth ≥100; ≥4 variant supporting reads; VAF >1% or duplex VAF ≥0.1%; variant read support ≥0.1% VAF in either (1) <10% of panel of normal (PON) samples or (2) <30%, provided sample VAF >20-fold maximum read support in the PON samples. In cases with matched germline sequencing, we required a normal deduplicated depth of ≥20 and a normal VAF of either (1) <0.25% or (2) <1%, provided sample VAF >20-fold normal VAF. In cases with variant support ≥0.1% in any PON sample, we applied a stricter normal VAF threshold of 0.1% unless sample VAF was >20 fold higher than the maximum VAF in the matched normal and PON samples (read support in <30% of PON samples required instead). COSMIC-annotated variants with sample VAF >20-fold normal VAF were rescued from PON filtering steps. In cases without matched germline, we restricted variant calls to those with VAF <35% to exclude single nucleotide polymorphisms, and filtered gnomAD-annotated variants with a population frequency of ≥0.015%. Funcotator (GATK) was used to annotate single nucleotide variants.
Small insertions/deletions (indels)
We employed an adaptive indel calling strategy analogous to our SNV workflow.30 Variant read support thresholds for indel calling were determined based on the error profiles of different indel categories. For this purpose, insertions and deletions were considered separately and grouped based on indel length and proximity to a homopolymer. To model the error distribution for each group, a log-linear regression was fit to the number of times we observed 1 through 5 variant-supporting reads. Candidate indels with >5 supporting reads were automatically included since they were expected to be enriched for true-positive variants. Each regression function was then solved to identify the minimum number of reads required to yield 0 false-positive indels. This number was rounded up to the nearest integer and used as the threshold for calling a particular indel category. If there was insufficient data for a regression fit, the threshold was chosen as the first point (1–5) where no detected reads were observed. Thresholds were then refined based on the number of error positions in a given gene, as previously described.30 Only candidate indels with read support greater than or equal to the final threshold were considered and subjected to additional filtering steps.
Only indels supported by both forward- and reverse-orientation reads were included. Indels ≤1% were required to have duplex read support. Indel calls were further filtered by restricting calls to positions with unique sequencing depth ≥25% of the median sample depth. We further removed indels with >2 variant reads in ≥10% PON samples. If a matched normal was available, we removed variants with either (1) read support >0.25% in the matched normal or (2) >2 variant reads in the matched normal and any PON sample. If a germline sample was unavailable, we restricted calls to VAF <35% and removed gnomAD-annotated variants with a population frequency of ≥ 0.015%.
Genotyping & non-silent mutations
For genotyping, calls were restricted to mutations with a minimum VAF of 0.5%. No VAF threshold was applied when calculating mean AF of a sample. Missense, splice-site, nonsense, nonstop, start codon and start-gain SNVs as well as splice-site, exonic in-frame and frame-shift indels were considered “non-silent”. Co-association/mutual exclusivity analysis were performed using the DISCOVER R-package.67
Tumor-plasma pair variant calling and heatmap visualization
SNVs called in either plasma or tumor samples were considered. Variant read support for each mutation was evaluated in both tissues and expressed as VAF. For visualization as a heatmap, SNVs called in >1 patient (i.e., hotspot mutations), variants with a VAF ≥0.25% in any germline sample of the 24 patients included in this analysis, or with VAFs ≥35% in cases without matched normal (i.e. somatic mutations that reflect germline polymorphisms in an unrelated patient) were removed prior to visualization.
ctDNA quantification and monitoring
ctDNA levels were measured in haploid genome equivalents per milliliter of plasma (hGE/mL), calculated as the product of total cfDNA concentration and the mean AF of somatic mutations. Pretreatment samples without matched germline and <20 SNV calls were considered negative for pretreatment ctDNA.
Plasma exome variant calling
Feature annotation
Candidate SNV calls in the exome space were generated from samtools mpileup considering Q30 bases using minimal filtering steps: variant support ≥3; plasma VAF ≥0.5%; deduplicated plasma depth ≥20; read support ≥0.5% VAF of specific base change in <30% of panel of normal (PON) samples; in cases with matched normal: normal deduplicated depth ≥10 and normal VAF<0.5%; in cases without matched normal: plasma VAF <35%. Candidate variants were annotated using GATK Funcotator (gatk-4.1.9.0) and ANNOVAR. Various features from each candidate variant were fed into a machine learning classifier. For each candidate variant, we annotated the copy number z-score of the respective cytoband, as well as the distance to the closest transcriptional start site (ensembl75). The variant trinucleotide sequence context was determined using the R package ‘MutationalPatterns’. Genomic loci were annotated with the following UCSC tracks: EncodeDukeMapabilityUniqueness35bp, EncodeCrgMapabilityAlign36mer, and RepeatMasker. We summarized read-level data of candidate supporting reads such as average mapping quality and Phred score, as well as the mean number of non-reference bases. Internal fragment end-motifs (4bp motif) from both ends of a given DNA fragment were extracted from deduplicated BAM files, similar to a previous publication68. Only fragments with reads mapped to chromosomes chr1–22,X,Y in proper pair orientation and within the selector space were considered for motif analysis. Additionally, we required a perfect cigar string (all M), mapping quality of ≥25 of at least one of the reads, and the absence of N/n bases in the reference genome at each candidate genomic locus. Motifs were derived from the reference genome rather than the actual read sequence. Fragment and end-motif counts were then summarized by whether they a) supported a given variant in read1 and/or read2, b) whether 1 or both mates were spanning the variant, and c) template length (1: <160; 2: ≥160–230, 3: ≥230–310, 4: ≥310bp) as previously described.31 We selected 15 end-motifs that significantly differentiated wild-type from mutant fragments in a separate set of 306 pretreatment plasma samples from non-Hodgkin lymphoma27 and lung cancer31 patients subjected to targeted plasma sequencing (ACTG, AGGG, CAGA, CCAC, CCTA, CCTG, CTCC, CTGC, CTGG, GCAT, TCGC, TTCA, TTCC, TTGG, TTGT; data not shown).
Tumor variant classification
Variants with mean mapping quality of supporting reads <25, variants supported by reads containing an average of ≥4 non-reference bases, and variants called in genomic regions with zero uniqueness (EncodeDukeMapabilityUniqueness35bp) or containing repeats (RepeatMasker) were censored prior to model training. Features with missing values were replaced by zero. The model was trained on samples from 120 patients using ground truth labels from ultra-deep targeted (n=117, cHL) or exome-wide plasma (n=2, cHL) or tumor sequencing (n=1, DLBCL, Supplementary Table 6), respectively. Since most labeled variants were part of the targeted panel, and hence potentially biased towards certain genomic regions, a deep neural network-based autoencoder was implemented. In this model, the feature matrix of all variants (labeled and unlabeled) was input to the model. The autoencoder consists of three encoding & decoding layers with dimensions from feature size 68 to 64, (Rectified Linear Unit) ReLU, 64 to 32, ReLU, 32 to 16, ReLU, and from 16 to 10 ‘encoded reduced features’, and on the decoder side, from 10 to 16, ReLU, 16 to 32, ReLU, 32 to 64, ReLU, and from 64 to the feature size, followed by a Sigmoid. An L1-loss was used with Adam optimizer, with 50 epochs, and batch size of 10,000. The 10 reduced representations were then augmented to the initial feature set. A Gradient Boosting Machine (GBM) with 100 estimators was then trained to label each putative variant as tumor- vs non-tumor-derived. A series of learning rates were tested from the GBM (0.05, 0.1, 0.15, 0.2, 0.25, and 0.3). The model performance was evaluated using 5-fold cross-validation with 5 repetitions. Within each fold/repeat, we performed a nested cross-validation (stratified 10-fold CV) to find the optimal learning rate as evaluated by the average precision. All unlabeled variants were then scored by all 25 (5×5) models and the average scores were calculated. To make binary calls, the cross-validation false-positive rate was set to 1% using the labeled variants.
GBM classifier performance assessment
The performance of the GBM classifier was validated in 2 withheld plasma samples, collected at two different time points from a LBCL patient. For both samples, matched deep plasma and tumor sequencing were generated in addition to the “regular” depth (Supplementary Table 6). Tumor mutation calls were generated using an ensemble of 3 variant callers (mutect2, strelka2, varscan2), only considering SNVs called by at least 2 callers with a VAF ≥10%. Deep plasma variant calls were generated using the standard CAPP-Seq workflow as described above. Tumor mutations were considered ground truth for sensitivity estimation, while the union of tumor and deep plasma calls were considered ground truth for positive predictive value (PPV) estimation. The performance of the GBM classifier was compared to the conventional CAPP-Seq workflow.
Indel calling from exome profiles.
No machine learning model was trained for indels due to the limited data available for training (SNVs>>indels). Indel variant calling in the exome space was therefore done as described in the targeted variant calling section with the exception that duplex read support was not considered. Exome indel calls were combined with those of CAPP-Seq and restricted to genes with non-silent SNVs in ≥2.5% of cases.
Significantly mutated genes
We used MutSig2CV (v3.11) to identify significantly mutated genes. Both SNV and indel calls were included in the analysis. C2CD3 and CLEC2D mutations were removed after manual review. Lollipop plots were generated using the Mutation Mapper Tool in cBioPortal69,70 and subsequently modified. MutSig2CV was run on both targeted and exome-wide mutation calls. For genes covered in the targeted sequencing panel, MutSig2CV results from CAPP-Seq were reported.
Somatic copy number aberrations (SCNA)
Genome-wide somatic copy number alterations (SCNAs) were ascertained using both on-target (~60–80% of reads) and off-target (~20–40%) sequencing reads with an in-house method, as previously described.31 To select recurrently copy number-altered regions to use as features for the identification of genetic subtypes, we used CNVkit v0.9.8 and GISTIC2 to define amplification and deletion peaks. To maximize sensitivity, we restricted this analysis to samples which were plasma exome profiled and had a mean AF ≥5% (n=61). GISTIC2 was run using the following flags: -js 85 -qvt 0.1 -conf 0.99 -maxseg 2000 -broad 0 -armpeel 1 -savegene 1 -gcm extreme (amplifications) or -js 150 -qvt 0.1 -conf 0.99 -maxseg 2000 -broad 0 -armpeel 1 -savegene 1 -gcm extreme (deletions).
A total of 20 amplification and 23 deletion autosomal peaks were identified with this approach (Supplementary Table 8). Log2 copy number ratios and z-scores were summarized for each peak from cytoband level segmentation data. If a given peak spanned more than one cytoband, data was summarized by the mean. As a final step, z-scores from each cytoband/peak were readjusted using the distribution of z-scores of control samples analyzed in the same manner. Noisy cytobands/peaks (i.e. absolute value of 1st or 3rd quartile of z-scores >1.96 or absolute delta of median of z-scores between sequencing panels >1.96) were removed and not considered as features for genetic clustering.
Unsupervised clustering of cHL using genetic variants
Clustering feature matrix
SNVs and copy number alterations were first converted into an integer-valued matrix, with the following definitions: non-synonymous mutation (<=1): 2, non-synonymous mutation (>=2): 3; allele fraction (AF)-corrected CNV state: (2.75<=CN state>=4.5): 1, (4.5<=CN state<6.25): 2, (CN state>=6.25): 3; (1.2<CN state<=1.6): 1, (0.8<CN state<=1.2): 2, (CN state<0.8): 3. Features were excluded if observed in less than 2.5% of plasma cases (Supplementary Table 9).
Latent Dirichlet Allocation model
The matrix data is integer valued, and linear models such as non-negative matrix factorization are not suitable. We therefore used a Latent Dirichlet Allocation (LDA). LDA is a popular generative probabilistic model used in natural language processing. Several formulations have been described, but here we use the notation from Blei and colleagues38. In this model, each variant (e.g., SOCS1 mutation) is encoded as a “word” ( words: ), the entire patient genotype is encoded as a “document”, and the cohort of patients is encoded as a “corpus”. The total number of words (i.e., sum of all features in the genotype) is Poisson-distributed, and the LDA is used to extract the “topics”, i.e., clusters. To fully characterize the LDA model, three matrices , , and are inferred, with the following definitions:
A given genotype, g is then assigned as . The fitting was done by Gibbs sampler, with 250 ‘burn-in’ iterations, 2,000 iterations and optimized (prior of clusters over genomes). Here the implementation from the R package textmineR was used.
Optimal number of clusters
To find the optimal number of clusters, a custom metric based on cophenetic correlation coefficients was implemented, where both sample and feature robustness were assessed. We used n=25 initializations and 250 burn-in samples for the LDA fitting. Varying the number of clusters from k=2 to k=8, the cophenetic coefficients achieved their maximum for both sample and feature curves at k=2 (Extended Data Fig. 1E).
LDA clusters heatmap visualization
To visualize the feature matrix after the LDA unsupervised clustering as shown in Fig. 2A, first genetic variants are assigned to the clusters via the inferred matrix. Then, samples are sorted based on their fidelity to the assigned cluster via the inferred matrix. For visualization, the maximum number of cluster-defining features is capped at 15. The final heatmap is visualized by the R package ComplexHeatmap. The same features were visualized in Extended Data Fig. 6D, however samples were sorted by cluster assignment probability.
Cell-free DNA fragment length determination
Only samples with ≥20 SNVs were included in this analysis. ctDNA fragment length distributions in each plasma sample were determined as follows: (A) Variant calls were restricted to SNVs ≥0.5% VAF to ensure high confidence in the calls. (B) Fragments where either read mate spanned mutations in (A) were extracted from the BAM files. (C) Fragments supporting mutations in (A) were identified, and fragments with both reads spanning the position of a given mutation yet disagreeing on mutation status were excluded. (D) Fragment lengths were extracted from field 9 of each BAM file, corresponding to the mapping distance between read 1 and read 2 for each read pair. Wildtype cfDNA fragment length distributions were calculated from fragments spanning mutations in A) supporting the reference allele. Only properly-paired reads aligned to chromosomes chr1–22,X,Y with perfect CIGAR strings were considered for analysis. We further only included fragments with mapping quality of ≥25 for at least one of the read mates.
For cfTCR fragment length estimation, mapping based template length was considered, if possible. For cfTCR molecules that did not properly map given their CDR3 rearrangement, fragment length was obtained by merging overlapping paired-end reads.
Internal fragment end-motif analysis
We extracted 4bp reference (hg19) internal end-motifs of cfDNA fragments from both ends of a given DNA molecules, similarly to a previous publication.68 Only fragments with reads mapped to chromosomes chr1–22,X,Y in proper pair orientation within the selector space were considered for this analysis. Additionally, we required a perfect CIGAR string, mapping quality of ≥25 for at least one of the read mates, and the absence of N/n bases in the reference genome at each genomic locus. Per a previous publication34, the following DNASE1L3-associated, 4bp internal motifs were considered: CCCA, CCTG, CCAG, CCAA, CCAT, CCTC, CAAA, TGTG, TGTT, CCTA, TATT, CCAC, TCTT, CCCC, TGAG, CAAG, CATG, TATA, GGAA, TGTA, CATA, TACA, TCTG, CAAT, TGCT.
When comparing fragment length of mutant and wiltype molecules between cHL and LBCL (Fig. 1D), only pretreatment samples with at least 20 SNVs were included in the analysis (n=300 cHL, n=53 LBCL32).
Sequence logo plot
Samples with at least 20 SNVs and at least 100 evaluable mutant fragments were included in the analysis (cHL: n=294; LBCL: n=48). To avoid overrepresentation of any particular patient’s fragments, 11,892 ctDNA end-motifs (median in cHL patients) were randomly sampled, with replacement, from each sample. After pooling and filtering, 3,492,532 and 570,816 ctDNA end-motifs were evaluable for downstream analysis for cHL and LBCL, respectively. Pooled cHL motifs were downsampled to the same count as LBCL (n=570,816). FASTA files were generated from both motif datasets, and fed as input to pLogo71 with cHL ctDNA motifs as the ‘foreground’ and LBCL ctDNA motifs as the ‘background’. The pLogo plot was subsequently modified for visualization in Fig. 1F.
Noninvasive determination of tumor Epstein–Barr virus (EBV) status
Pretreatment samples were captured using oligonucleotides targeting Epstein–Barr virus (EBV). Human sequencing reads were subtracted before alignment to the EBV reference genome NC_007605.1. Read coverage and plasma cfDNA concentration were used to calculate absolute viral abundance, and expressed as genome copies per mL plasma. Using 10-fold cross validation in 100 cases, we trained a threshold for plasma EBV reads separating EBV positive and negative tumors using clinical in situ hybridization (EBER)/immunohistochemistry (LMP1) as ground truth. The chosen threshold of 1.5 log10 EBV reads per mL plasma, achieved a 90% accuracy in predicting the tumor EBV status in 101 withheld samples. Specimens positive either by clinical in situ hybridization (EBER)/immunohistochemistry (LMP1) or the noninvasive method were considered EBV positive for analyses.
Genetic subtype validation in external sequencing data
We assigned genetic subtypes to patients from Maura et al20 (n=61) utilizing the probabilistic model generated by LDA in the plasma discovery cohort. Non-silent mutation calls were used as published in the data supplement of the original publication. Case level SCNAs were kindly shared by the authors. The tabulated segmented SCNA data provided by the authors was intersected to GISTIC2 peaks identified in our cohort to transfer SCNAs into the same space.
Modelling of probability of assignment to H2 subtype by age
To model the association between age and the probability of assignment to the H2 subtype, we calculated the fraction of patients assigned to the H2 subtype in groups of 10 patients after sorting by age (n=292 patients). We recorded the mean age and the fraction of H2 cases for each group as x and y value, respectively, and fit a loess regression to the data using the msir R-package [loess.sd(out, nsigma = 1.96)]. Data is visualized in Extended Data Fig. 6G. The H2 assignment probability in Maura et al20 was also estimated in groups of 10 after sorting by age analogous to the training cohort.
Single-cell RNA-Sequencing (scRNA-Seq)
The single-cell study was reviewed and approved by the St. Jude Institutional Review Board (IRB) (IRB# 21–0841). LN excisional biopsy samples obtained at St. Jude Children’s Research Hospital with cryopreserved cell suspension(s) available in an institutional biorepository were identified. Additional data, including biopsy site and diagnosis, were available in institutional biorepository records and were recorded (Supplementary Table 3). LN samples were obtained between 2000 and 2023.
Excisional biopsy specimen processing
Freshly resected excisional biopsies were processed immediately for cryopreservation in the institutional biorepository. Tissue was placed in a petri dish with 2mL of Hank’s Balanced Salt Solution with HEPES and heparin (HHH solution). The tissue was cut into half or into quarters, depending on size, with a number 10 blade scalpel. The cut edge of the lymph node tissue was then scraped with the blunt edge of the scalpel, releasing cells into the HHH solution in the petri dish. The suspension was then pipetted with a transfer pipette from the petri dish to a 15mL conical tube. Cells were counted and assessed for viability using a hemocytometer before cryopreservation in liquid nitrogen in freezing medium (Roswell Park Memorial Institute 1640 [RPMI 1640] media supplemented with glutamine, 20% heat inactivated fetal calf serum (FCS), and 10% dimethyl sulfoxide [DMSO]).
Cell suspension processing and labeling
Cell suspensions cryopreserved as described above were then thawed and sorted using fluorescence-activated cell sorting (FACS) as described previously (Reichel et al Blood 2015), with minor modifications. Cells were thawed quickly from liquid nitrogen in a 37°C water bath then added dropwise to 45mLs of pre-warmed thawing medium (RPMI 1640 [Life Technologies] with 20% FCS [Atlas Biologicals] and 100ug/ml DNASE I (Worthington Biochemical Corp]). The cells were incubated at room temperature (RT) for 15 minutes (mins) and then centrifuged at 500g for 10mins. The cell pellet was then resuspended in 1mL of thawing medium and cells were counted using a Countess 3FL automated cell counter (Invitrogen) with trypan blue staining for dead cell identification. After counting, cells were centrifuged at 500g for 5 mins and then resuspended in 100ul RPMI 1640 with 20% FCS per 100,000–500,000 cells. Ten microliters each of unconjugated CD2 clone RPA 2.10 (300202, BioLegend), CD54 clone HA58 (353102, BioLegend), CD58 clone TS2/9 (330902, BioLegend), CD11a/CD18 clone m24 (363402, BioLegend) antibodies per 100ul RPMI 1640 with 20% FCS (1:10 dilution for each antibody per 100ul) were then added and the cells were incubated on ice for one hour. After incubation, cells were washed in 5ml of RPMI with 20% FCS and centrifuged at 500g for 5 mins and the supernatant was removed. An antibody cocktail (dilutions are per 145ul stock cocktail) was then prepared, which consisted of 10ul Tonbo Ghost Dye Violet 510 Viability Dye (1:14.5 dilution; 13–0870-T100, Cytek Biosciences), 10ul CD31 APC/Cy7 clone WM59 (1:14.5 dilution; 303119, BioLegend), 10ul CD326 APC/Cy7 clone 9C4 (1:14.5 dilution; 324245, BioLegend), 10ul CD45 BV785 clone HI30 (1:14.5 dilution; 304048, BioLegend), 5ul CD90 PerCP/Cy 5.5 clone 5E10 (1:29 dilution; 328117, BioLegend), 10ul CD49e PE clone NKI-SAM-1 (1:14.5 dilution; 328009, BioLegend), 10ul CD20 AF700 clone 2H7 (1:14.5 dilution; 3020322, BioLegend), 20uL CD15 FITC clone HI98 (1:7.25 dilution; 301904, BioLegend), 20ul CD30 PE/Cy7 clone BY88 (1:7.25 dilution; 333918, BioLegend), 5ul CD40 PE-Dazzle 594 clone 5C3 (1:29 dilution; 334342, BioLegend), 5ul CD95 APC clone DX2 (1:29 dilution; 305612, BioLegend), 20ul CD64 BV421 Clone 10.1 (1:7.25 dilution; 305020, BioLegend and 562872, BD Biosciences), and 10ul CD5 BV605 Clone L17F12 (1:14.5 dilution; 364019, BioLegend). After preparation, 100uL of the stock antibody cocktail was added for every 100,000–500,00 cells and incubated at RT for 15 mins. After incubation, 3mLs of sorting medium (phosphate buffered saline [PBS] with 2mM EDTA and 0.5% bovine serum albumin [BSA, Miltenyi Biotech]) was added and the sample was centrifuged at 500g for 10 mins. The cell pellet was then resuspended in 300ul sorting medium and filtered through a 70uM 5ml Falcon cell strainer (352235, Corning).
Purification of cell populations by fluorescence-activated cell sorting (FACS)
Using BD FACSDiva (v9.0.1), labeled cells were then sorted using a FACSAria Fusion special order research product (SORP, BD Biosciences), using the 130μm nozzle at 10p.s.i. Compensation was performed using UltraComp eBeads and ArC Amine Reactive compensation beads (01–2222-41/A10346, Invitrogen). Cell populations were collected in 300μL of 100% FCS (Atlas Biologicals). The gating strategy used was based on the work by Reichel and colleagues14 with minor modifications, with the overall goal of enriching for HRS cells, which are a rare population in the tumor microenvironment relative to phenotypically normal T and B cells, in a single cell transcriptomics pipeline. Debris and dead cells were excluded by FSC-A and viability dye, respectively, and doublets were excluded by plotting FSC-H against FSC-A on viable cells. This gate was expanded to include high FSC-H and FSC-A events as per Reichel et al.14 A second doublet exclusion gate, FSC-A against SSC-W, was added to remove RBCs and debris. After doublet, viability, and debris exclusion, we collected phenotypically normal T and B lymphocytes (CD45+, CD20+, CD5+) as an individual population from all samples. With early samples, we collected HRS cells (CD5-, CD20-, CD30+, CD40+, CD95+) as an individual population (Supplementary Note [Fig. 14A]). However, we found that the absolute number of HRS cells sorted was limited by the absolute total number of cells in the cryopreserved aliquot and the cell viability, which varied between samples. Thus, we made modifications over time to the sorting strategy to collect the phenotypically normal T and B lymphocytes (CD45+, CD20+, CD5+) along with macrophages (CD64+) and the rest of cells not phenotypically consistent with T and B lymphocytes or macrophages cells as three separate populations (Supplementary Note [Fig. 14B], Supplementary Table 3).
Single cell transcriptomics from FACS purified cell populations
FACS purified cell populations were processed immediately for single cell transcriptomic assays using the 10X Genomics solution. From samples of cHL patients, two populations of cells per lymph node cell suspension specimen were used for individual gel bead in emulsion (GEM) preparations: FACS purified T and B cells and a pooled combination of other purified populations, to which sorted T and B cells were added to increase absolute cell number for use in the 10X Genomics pipeline (Supplementary Table 3). For samples from patients not diagnosed with cHL, one GEM was prepared from a pooled population of T and B cells along with other sorted populations (Supplementary Table 3). Once cell populations were pooled after FACS, they were centrifuged at 400g for 5mins and resuspended in 0.04% ultrapure BSA (Invitrogen) in PBS. Each of the two cellular fractions were then counted using a hemocytometer and the cellular concentration of each fraction was adjusted (median 680 cells/μL; range 300–1000 cells/μL) for GEM preparation. Once at appropriate concentrations, GEMs (median 8000; range 3000–10000 goal cell recovery) were generated using the 10X Genomics Chromium or Chromium X Controllers (10X Genomics; PNs 1000204 or 1000331) and the Chromium Single-Cell V(D)J 5’ v2 reagent kits (10X Genomics; PN 1000265). Gene expression libraries were generated using library construction kits (10X Genomics; PN 1000190). cDNA and gene expression library quality assessment and quantification were performed using Agilent TapeStation High Sensitivity (HS) D5000 reagents and HS D5000 ScreenTape (Agilent; PNs 5067–5593 and 5067–5592 respectively) on an Agilent TapeStation (Agilent; PN G2992AA). Gene expression libraries were sequenced on the Illumina NovaSeq platform using a 26–10–10–90 (R1-I7-I5-R2) configuration with a 150bp paired end read length and type.
Single cell gene expression analyses
Raw sequencing data were processed using CellRanger (v6.0.1; 10X Genomics) and aligned to the GRCh38 transcriptome. Independent libraries were aggregated using CellRanger, without normalization, into a single matrix. Key quality control metrics after CellRanger aggregation included an estimated 91,297 recovered cells from which 9,460,308,019 reads were obtained for a mean of 103,621 reads per cell. An estimated 88.4% of reads were within cells and there were a median of 1,911 genes per cell and a median of 5,113 unique molecular identifier (UMI) counts per cell. After aggregation without normalization, downstream analyses were then performed using the Seurat (v4.0.3)72 framework in the R statistical computing environment (v4.0.2).
We created a Seurat object from the aggregated data matrix, setting min.cells = 3. We then filtered cells with >20% mitochondrial gene expression and performed an “integrated” analysis on the remaining 87,528 cells to remove potential batch effects by grouping cells by 10X Genomics NextGEM Chip (in total there were n=9 independent chips used to generate the n=16 libraries included in the aggregated dataset) using Seurat’s reciprocal principal component analysis (RPCA). After integration, we proceeded with unsupervised clustering analysis using Seurat’s FindNeighbors and FindClusters with reduction set to ‘pca’, dims set to 1:30, and resolution set to 0.5. Guided by canonical markers, differential gene expression (DGE) analysis (using Seurat’s FindMarkers function in default settings), and prior publications in this area,73–76 we then performed cell cluster annotation manually. The DefaultAssay for DGE analysis and labeling of cells using DotPlots was set to “RNA”. Cell-type specific gene signatures for correlation in bulk/plasma samples and deconvolution were generated from a subset of cells, and evaluated for consistency in the remainder of cells.
T-cell receptor profiling from cfDNA
We used SABER (Sequence Affinity capture & analysis By Enumeration of cell-free Receptors) as a technique for TCR enrichment and analysis of fragmented rearrangements shed in cfDNA as previously described.32,39 In brief, sequencing libraries were enriched for the TCR locus using custom oligos. After deduping and error-correction, unique CDR3 sequences are resolved to quantify T-cells. TCR counts per mL plasma were calculated as the product of total TCR count divided by unique depth and cfDNA concentration, and expressed as clones per mL.
Noninvasive gene expression profiling from fragment length profiles
We leveraged ‘Epigenetic expression inference from cell-free DNA-Sequencing’ (EPIC-Seq) to noninvasively infer gene expression profiles of 28 plasma samples from healthy controls, and a total of 113 cHL patients selected from the larger genotyping cohort. Plasma libraries were therefore recaptured using a 2.6 MB panel targeting transcriptional start sites of 1,676 genes (Twist Biosciences). Gene expression was inferred as previously described.24 For genes with multiple transcription start sites (TSSs) in the panel, we used the TSS with maximal inferred expression. We then normalized the expression of each gene by the average expression of housekeeping genes in the panel (n=25 genes). We denote this normalized inferred expression by for gene . We then used the healthy control data to estimate the mean and standard deviation of the “baseline” expression for each gene. For differential gene expression analysis, inferred expression was converted to discrete data.
Three levels were defined based on the inferred expression as follows:
Enrichment for high and low expression in each gene were separately assessed using 2-sided Fisher’s exact tests. Odds ratios (OR) for low expression were converted as 1/OR to integrate them with analyses of high gene expression and visualized as volcano plot. Genes with P<0.1 and log2 OR>0 or log2 OR<0 were separately used as inputs for the ToppGene Suite (https://toppgene.cchmc.org). For heatmap visualization, inferred gene expression was used as a continuous variable. Inferred expression for each gene was first transformed to a z-score relative to controls, and then z-normalized across samples.
The following gene signatures derived from scRNA-Seq were used to explore transcriptional representation of HRS and T-cells in plasma selected by upregulation in the respective cell type and low expression levels in other cell types: HRS: BATF3, CCL17, CCL22, CD86, ETS2, FSCN1, IL6, LTA, PERP, TFPI2; Tumor T-cell : APOBEC3G, CCL3, CCL4, CCL4L2, CCL5, CD8A, CD8B, CMC1, CTSW, CXCR3, CXCR6, EOMES, FXYD2, GNLY, GZMA, GZMH, GZMK, HAVCR2, IFNG, KLRG1, KLRK1, LAG3, LYST, NKG7, PRF1, PTMS.
RNA Sequencing
RNA from pretreatment tumor biopsies was available for RNA sequencing for n=86 cHL patients and the following cHL cell lines: KM-H2, L591, L1236, SUPHD1, HDLM2, L428, UH01 and L540. cHL was compared to LBCL bulk gene expression (n=66) using data from a recently published cohort.32 RNA was isolated from 2–4 10μm FFPE scrolls using the CELLDATA RNAstorm Kit and quantified using Nanodrop. RNA-Sequencing (RNA-Seq) libraries were prepared from 50ng RNA using the SMARTer Stranded Total RNA-Seq v2 Kit (Takara). Fragmentation steps were omitted for FFPE source RNA as recommended by the vendor. Libraries were evaluated using Qubit and Agilent TapeStation, and sequenced on HiSeq4000/X-ten or NovaSeq6000 machines targeting ≥35 million paired-end reads per sample. After demultiplexing, reads were quality filtered and trimmed using ‘afterqc’ (version 0.9.6) using the following flags (-q 30 -u 50 -p 30 -a 3 --trim_front=9 --trim_tail=50). Transcript abundance was quantified using salmon (version 0.8.2). We used the DESeq2 R-package to normalize transcript abundance and perform differential gene expression. The deconvolution method CIBERSORTx (https://cibersortx.stanford.edu) was used to estimate fractions of the following cell types as previously described77 using a gene signature matrix derived from cHL scRNA-Sequencing: Early B cells, CD4 T cells, HRS cells, Memory B cells, CD8 T cells, NK Cells, Plasma cells, Dendritic Cells, Regulatory T Cells, Germinal Center B cells, Macrophages.
Laser microdissection
Consecutive slides from FFPE blocks were cut at 7 μm thickness using a microtome, mounted on polyethylene naphthalate membranes (Thermo Fisher Scientific LCM0522), and stored overnight in nitrogen atmosphere. Subsequently, slides were stained with hematoxylin as per published methods78, and dissected after staining using the ArcturusXT LCM System. In general, 2–3 slides representing consecutive sections were used to microdissect at least 200 Reed Sternberg cells using ultraviolet laser to cut and infrared laser to adhere to CapSure HS LCM Cap (Thermo Fisher Scientific LCM0215). Dissected cells were incubated for 16 hours with isolation of DNA using PicoPure DNA Extraction Kit (Thermo Fisher Scientific KIT0103). After incubation, a 3X bead wash was performed followed by enzymatic fragmentation. Isolated DNA was quantified by qRT-PCR with subsequent library preparation performed according to our general CAPP-Seq protocol with at minimum 1ng DNA input.
Total Metabolic Tumor Volume (TMTV) measurement
TMTV was measured from functional imaging by positron emission tomography with 2-deoxy-2-[fluorine-18]fluoro-D-glucose integrated with computed tomography (PET-CT). TMTV was quantified using semiautomated software tools (e.g. Beth Israel Fiji (http://petctviewer.org)79, PETRA accurate tool80 and Metavol81), as previously described.28 Regional volumes were automatically identified by the software and assessed by an expert to confirm accurate inclusion of pathological lesions only.
Phased variant enrichment and detection sequencing (PhasED-Seq)
We used ‘Phased Variant Enrichment and Detection Sequencing’ (PhasED-seq) to monitor ctDNA in on-treatment plasma samples. This technology maximizes the analytical sensitivity and reduces background error rates by tracking two or more variants (‘phased variants’) on a single cfDNA molecule.5 Phased variants with a VAF ≥0.5% were identified from pretreatment genotyping using plasma samples (n=109), and restricted to variants targeted by the smaller capture panels used for sequencing of on-treatment samples (i.e. 144 kb [SeqCap EZ Choice, Roche] and 268 kb [xGen, IDT] panels). Phased reporters were filtered against matched germline sequencing, if applicable. A panel of normals was employed, consisting of leukocyte sequencing from two cohorts of 135 and 107 patients, respectively, as well as separate sets of 16 and 19 plasma specimens from healthy individuals, as previously described5. Using a previously reported Monte Carlo framework, the phased variant lists identified from pretreatment samples were used to monitor ctDNA levels in matched on-treatment specimens.5,30
Recursive Phylon Nomination, Enumeration, and Recovery (RePhyNER) to allow for germline-free minimal residual disease monitoring using PhasED-Seq
MRD assays like PhasED-Seq track tumor-specific reporters over time. False positive reporters, e.g., through germline artifacts, may lead to false positive MRD results. MRD assays therefore usually rely on matched germline samples, which were not available for all samples presented here. To allow for MRD measurements in samples without matched germline (n=65), we developed and employed a novel technique to remove potential germline artifacts from phased reporter lists. We named the method Recursive Phylon Nomination, Enumeration, and Recovery (RePhyNER). The basic idea behind RePhyNER is to use 2 non-germline samples with non-zero AF gradient to remove germline artifacts instead of using a matched sample-germline pair. RePhyNER identifies outlier reporters (i.e., phased variants) not following the sample AF trend using a Poisson test (Supplementary Note [Fig. 13A]). We first assessed the performance of this filtering algorithm in simulations. When applying a static filter, i.e. removing all putative false variants at once, simulations revealed a high probability of correctly removing false variants, even at relatively modest AF trends between the samples. However, with increasing noise rate, i.e. an increasing fraction of false positive reporters, the probability of wrongfully removing true variants increased dramatically. This could be overcome by implementing the filter in an iterative manner, where outlier reporters are recursively removed one by one (Supplementary Note [Fig. 13B]). After each iteration (i.e., one variant removal), RePhyNER updates the mean AF trend between the 2 samples based on the partially filtered variant list, so that the estimated mean AF change becomes more reliable with each iteration. Once no outlier variant is present, results are considered final. The algorithm underlying RePhyNER is summarized below:
Input: two nx3 arrays ( and )corresponding to two samples, columns being PV ID, numRef, numAlt
Initialize: iter=0; mFDR = 0; removedPVs = {}; ;
Set parameters: maxIter = 50; FDR = 0.1
While and ; do
; ; ;
-
For in , ; do
Define two vectors corresponding to :
Remove the variant from lists
Calculate the mean AF in the new lists with each AF being
Estimate the change in mean AFs
Calculate the individual expected AF in sample 2:
-
Perform Poisson significance test and return p-value
Test parameters rate as , and observed events:
Update :
Convert the P-value vector to FDR values
Define
If ; then
Update removedPVs:
We benchmarked RePhyNER in 108 sample pairs of 47 cHL patients with available matched germline. We compared the performance of the germline-informed assay with a germline-free methods with and without RePhyNER (Supplementary Note [Fig. 13C]). Of note, the matched germline samples were not used for any filtering step within the workflow. Using RePhyNER, we achieved a sensitivity of 91%, specificity of 99% and precision of 96% compared to MRD monitoring using PhasED-Seq with matched germline. Performance metrics for the germline-free approach without RePhyNER were significantly worse: Sensitivity 100%, Specificity 68%, Precision 46% (Supplementary Note [Fig. 13D]). We conclude that RePhyNER allows for specific MRD assessment in samples without matched germline while preserving tumor signal.
In-vitro cell line studies to functionally characterize IL4R mutations
Cell lines
293FT (R70007) and KM-H2 (ACC 8) were purchased from Thermo Fisher and DSMZ, respectively. DEV was a gift from Dr. Diepstra (University Medical Center Groningen, Groningen, The Netherlands). All cell lines were authenticated by short tandem repeats (STR) profiling (The Centre for Applied Genomics, Toronto, Canada) and confirmed to be negative for mycoplasma contamination using a PCR-based method (VenorGeM Mycoplasma Detection kit, Sigma-Aldrich).
IL4R site-directed mutagenesis and viral transduction
Q635*, Q666*, E681*, E684Kfs*2, E684*, Q696*, Q698* and S714* mutations were introduced into the pRETRO-TO-CMV-IL4R-PuroGFP retroviral plasmid vector (pRETRO-IL4R) using the GENEART site-directed mutagenesis system (Thermo Fisher Scientific) according to the manufacturer’s instructions. The pRETRO-IL4R I242N mutant construct was obtained from a previous publication.42 Viral particles were generated in 293FT cells (Thermo Fisher) and concentrated using the Lenti-X concentrator (TakaraBio). DEV cells were sequentially engineered with: 1) feline endogenous virus expressing the ecotropic retroviral receptor, 2) retrovirus expressing the tetracycline repressor, 3) retrovirus containing the pRETRO-IL4R mutants. KM-H2 cells were engineered using retrovirus containing the pRETRO-IL4R mutants. Retroviral transduction of DEV and KM-H2, and GFP-positive fraction cell sorting followed by puromycin selection was performed as previously described.43
Dose response curves for IL13 and IL4 stimulation
DEV cells were pre-treated with doxycycline (100 ng/ml) for 24hr to induce expression of IL4R, then seeded at 0.5 × 106 c/ml with serial dilutions of recombinant human IL4 for 15min (R&D Systems), or IL13 (Peprotech) for 24hr. Cells were washed twice with cold PBS, then prepared for flow cytometry antibody staining.
Phospho-STAT6/IL4R inhibition experiments
DEV cells were pre-treated with doxycycline and prepared for flow cytometry as described above. IL4Rα blocking antibody (5μg/ml at 1:100 dilution, R&D Systems) or phospho-STAT6 inhibitor (0.5μM AS1517499, Selleckchem) were added to DEV and KM-H2 (24hr). For flow cytometry experiments, cell lines were stimulated with IL4 (0.1ng/ml for 15min) or IL13 (0.5ng/ml for 24h) where indicated. For immunoblotting and ELISA experiments, cells were stimulated with IL13 (2.5ng/ml for 24hr).
Flow cytometry
Flow cytometry for surface IL4R (CD124, dilution 1:25) and intracellular STAT6 phospho-Y641 (dilution 1:25) was performed as previously described (Supplementary Note [Fig. 15]).43 Fixed cells were pre-incubated in Human Fc Block (Invitrogen, 1:100 dilution) in 2% fetal bovine serum in PBS (v/v) for 15mins at room temperature to prevent unspecific staining. Phospho-STAT6 expression in IL4R mutants was normalized as fold-change to wild-type treated with vehicle control.
Immunoblotting
Whole cell lysates were prepared using RIPA buffer (Invitrogen) supplemented with Protease Inhibitor Cocktail (Sigma) and Phosphatase Inhibitor Cocktail (New England Biolabs). Protein levels were determined using the BCA protein assay kit (Invitrogen). 10μg protein for each sample was loaded onto NU-PAGE 4–12% Bis-Tris gels (Invitrogen) and separated using electrophoresis settings of 100V for 120min. Semi-dry blotting transfer was performed using the TransBlot Turbo system (Bio-Rad). Immunoblotting was performed using the following primary antibodies: P-Stat6 (Y641) mAb clone C11A12 (dilution 1:1000), Stat6 mAb clone D3H4 (dilution 1:1000), IL-4Rα mAb H-4 (dilution 1:200), GAPDH clone 14C10 (dilution 1:20000), followed by secondary staining using anti-Mouse or Rabbit IgG (H+L) HRP conjugate (Promega, dilution 1:10000), and detected using ECL Western Blotting Detection Reagent (Amersham). Cropped images were included in manuscript figures, full scans are provided as Supplementary Note [Fig. 16].
Human CCL17 ELISA
Sandwich enzyme linked immunosorbent assay (ELISA) was performed to quantitate CCL17 secretion in the DEV and KM-H2 cell lines. Briefly, fresh cell line supernatant was prepared by centrifuging cell cultures and assayed using the Human CCL17/TARC DuoSet ELISA kit (R&D Systems).
RNA-Sequencing
RNA from cell lines was isolated using the RNeasy Mini Kit (Qiagen) and quantified using Nanodrop. After DNAse digestion, RNA-Sequencing libraries were prepared, and analyzed using DESeq2 as described above. For the differential gene expression analyses comparing mutant (E684Kfs2) and WT expressing DEV cells, the following culture conditions were included in the analysis: WT unstim., WT + IL13, E684Kfs2 unstim. and E684Kfs2 +IL13 (n=3 each). Differential gene expression was assessed using DESeq2 for mutant vs WT using IL13 stimulation as covariate. Gene set enrichment analyses were performed using the fgsea R package.
Statistical analysis
Statistical analyses were performed using R (version 4.2.1) and Python (2.7.5). The following R packages were used for the analyses: ‘reshape2’ (1.4.4), ‘data.table’ (1.14.0), ‘MASS’ (7.3.51.6), ‘zoo’ (1.8.8), ‘DescTools’ (0.99.36), ‘gtools’ (3.8.2), ‘matrixStats’ (0.58.0), ‘glmnet’ (4.0.2), ‘survival’ (3.2.3), ‘ggplot2’ (3.3.2), ‘ComplexHeatmap’ (2.4.2), and ‘optparse’ (1.6.6), ‘discover’ (0.9.4), ‘DESeq2’ (1.36.0), ‘fgsea’ 1.22.0, ‘textmineR’ (3.0.5) and ‘MCMCpack’ (1.6.3), ‘patchwork’ (1.1.1), Seurat (4.0.3) and dplyr (1.0.7). The python library ‘torch’ (2.0.1) was used. In boxplot graphs, each box represents the interquartile range (IQR, the range between the 25th and 75th percentile) with the median of the data, whiskers indicate the upper and lower value within 1.5 times the IQR. Outliers are plotted individually. Continuous variables were compared using Student’s t-test, Wilcoxon rank-sum tests or one-way ANOVA on ranks as indicated. 2-sided tests were used throughout the manuscript unless indicated otherwise. Pearson and nonparametric Spearman correlations were used to correlate continuous variables as noted. Time-to-event variables were visualized using the Kaplan-Meier method; log-rank tests were applied to compare survival between cohorts. Cox proportional hazards regression was used to assess the impact of risk factors on outcome variables. For regression analyses, units were defined as follows: ctDNA: log10 hGE/mL and stage as categorical variable. PFS was calculated from time of treatment initiation. PFS events were defined as progression, relapse, or death resulting from any cause. Outcome analyses throughout the manuscript were restricted to previously untreated patients ≥18 years not treated in pediatric studies.
Extended Data
Extended Data Figure 1: Noninvasive profiling of cHL.
(A) Study overview. MRD: Minimal residual disease; PET2: Positron emission tomography after 2 cycles of chemotherapy; preTx: pretreatment; AF: allelic fraction; SNV: single nucleotide variants; LDA: Latent Dirichlet Allocation; SCNA: somatic copy number aberration; EPIC-Seq: epigenetic expression inference from cell-free DNA-sequencing. (B) Line plot summarizing sensitivity of the Gradient Boosting Machine (GBM) model to call exome-wide SNVs as a function of plasma VAF in 2 non-Hodgkin lymphoma validation samples as compared with a conventional workflow. Sensitivity was calculated considering mutations with VAFs ranging from x-1 to x+1%. Tumor mutation calls were considered ground truth for sensitivity estimation. (C) Line plot summarizing positive predictive values (PPV) of the GBM model as a function of plasma VAF in 2 non-Hodgkin lymphoma validation samples as compared with a conventional workflow. PPV was calculated considering mutations with VAFs ranging from x-1 to x+1%. The union of tumor and deep plasma exome mutation calls were considered ground truth for PPV estimation. (D) Bar plot summarizing MutSig2CV -log10 q-values of 41 genes found to be significantly mutated in targeted or whole exome sequencing (q-value <0.05). The heat of the bars reflects mutation recurrence frequency. Genes that have not been recurrently described in the cHL literature (i.e. ≤1 study comprising at least 50 patients) are highlighted in bold. (E) Cophenetic coefficients (sample, feature and final) by number of clusters (k) when applied to the Latent Dirichlet Allocation (LDA) framework to identify genetic subtypes in 293 cHL cases.
Extended Data Figure 2: Oncoprint visualizing targeted SNV and indel variant calls.
Genes with mutation frequencies ≥5% are visualized. If more than 1 variant per gene was identified in a patient, color coding was derived using the following hierarchy: nonsense > start codon mutation > frameshift indel > nonstop > splice-site > missense > inframe indel. N=293 pretreatment plasma samples were included in the analysis.
Extended Data Figure 3: Oncoprint visualizing whole exome SNV and indel variant calls.
Genes with mutation frequencies ≥7.5% are visualized. If more than 1 variant per gene was identified in a patient, color coding was derived using the following hierarchy: nonsense > start codon mutation > frameshift indel > nonstop > start gain > splice-site > missense > inframe indel. N=119 cases profiled by plasma exome sequencing were included in the analysis.
Extended Data Figure 4: GISTIC2 peaks identified in 61 plasma samples profiled by whole exome sequencing with AFs ≥5%.
Selected gene symbols of genes of interest falling within a peak are annotated with the cytobands. Top x-axis: G-score, bottom x-axis: q-value.
Extended Data Figure 5: Mutual exclusivity/ co-association analysis using the DISCOVER R-package.
Heatmaps summarizing (A) unadjusted -log10 p-values or (B) -log10 q-values for mutual exclusivity/ co-association, respectively. Alterations that tend to co-occur are visualized in blue, while those with a tendency for mutual exclusivity are depicted in red colors. Non-silent mutations and SCNA as summarized in the LDA clustering matrix observed in >20 patients among 293 cases were included in the analysis. Statistics were performed using the DISCOVER R-package.
Extended Data Figure 6: Genetic subtypes are independent of EBV status and validate in external data.
(A) Overview figure summarizing genetic cHL subtype discovery and validation. LDA: Latent Dirichlet Allocation; cfDNA: cell-free DNA; TCR: T-cell receptor. (B-C) Boxplots and Wilcoxon p-values (two-sided) summarizing the targeted SNV burden (B), and fraction of the genome affected by SCNAs (C) in cluster H1 (n=200), H2 EBV- (n=56) and H2 EBV+ (n=37) in the plasma sequencing cohort. (D) Heatmap summarizing non-silent mutations and SCNAs (rows) of 61 patients with cHL (columns) as published in Maura et al.20 Clusters were assigned using the probabilistic model generated by LDA from the plasma discovery cohort as shown in Figure 2. (E) Bar plot visualizing recurrence frequencies of features associated with subtype H1 (top) and H2 (bottom) as presented in panel D. Dark colors denote frequencies from plasma genotyping (H1: n=200; H2: n=93, as visualized in Figure 2) while light colors reflect frequencies as described in Maura et al (H1: n=33; H2: n=28). Spearman rhos and p-values (algorithm AS 89) provided in the graphs describe the correlation of recurrence frequencies from all 30 features visualized in D between this study and Maura et al. within H1 and H2, respectively. (F) Boxplots summarizing the whole genome mutational burden in cluster H1 (n=16), H2 EBV- (n=5) and H2 EBV+ (n=3) in patients with available whole genome sequencing and known EBV status from Maura et al. Wilcoxon p-values (two-sided) are provided. (G) Loess regression describing the association of age and the probability of assignment to the H2 subtype in n=292 patients from the plasma genotyping cohort (black line: mean; ribbon: standard deviation*1.96). Each dot represents a group of 10 patients from Maura et al (n=60 total) with x and y illustrating average age, and the fraction of H2 cases within the group, respectively. Patients were sorted by age prior to grouping. (H) Pie chart summarizing EBV status of patients from Maura et al. assigned to the H1 (n=33) and H2 (n=26) clusters. Two-sided Fisher’s exact test p-value is provided.
Panels B,C,F: each box represents the interquartile range (the range between the 25th and 75th percentile) with the median of the data, whiskers indicate the upper and lower value within 1.5 times the IQR.
Extended Data Figure 7: Mutual exclusivity analysis in the validation cohort.20.
Mutual exclusivity/ co-association analysis using the DISCOVER R-package. The heatmap summarizes -log10 p-values for mutual exclusivity/ co-association, respectively. The mutual exclusivity of H1- and H2- defining features (black boxes) was compared to a null distribution constructed by random shuffling of the matrix while maintaining the number of cluster-defining features. To provide a single measure of mutual exclusivity, p-values (one-sided, mutual exclusivity) were combined using Fisher’s method. The empirical p-value provided on the top of the heatmap was defined by comparing the observed combined p-value with the null distribution. Features from Extended Data Fig. 6D with greater than 4 occurrences were visualized. In addition, only one cytoband per chromosome was visualized selected by their rank in the feature list of H1/H2.
Extended Data Figure 8: Truncating IL4R mutations enhance STAT6 signaling.
(A) Immunoblot showing protein levels of IL4R and GAPDH in transduced DEV and KM-H2 cells. (B) Gene set enrichment plots from RNA-Sequencing showing enrichment of canonical KEGG pathways in mutant (E684Kfs2, n=6) vs WT (n=6) expressing DEV cells. Normalized enrichment scores (NES) and adjusted p-values (fgsea R-package) are provided. (C) Scatter plot visualizing base mean expression and log2 fold change comparing gene expression in mutant (E684Kfs2, n=6) vs WT (n=6) expressing DEV cells. Absolute log2 fold changes >1 are highlighted in orange or black, respectively. (D) Phospho-STAT6 levels (flow) in transduced DEV (n=5 each) and KM-H2 (n=4 each) cells under unstimulated, IL13-stimulated, IL13-stimulated + IL4R-Ab treated as well as IL13-stimulated + STAT6-I. treated conditions. Unadjusted Wilcoxon p-values (two-sided) compared to IL13 stimulation alone are provided. (E) Representative phospho-STAT6 flow raw data for KM-H2 Empty, WT, Q666* and Q698* constructs under IL13-stimulated conditions. (F) Phospho-STAT6 levels in WT, E684Kfs2 and I242N (PMBL hotspot) DEV cells under unstimulated, IL4-stimulated, IL4-stimulated + IL4R-Ab treated as well as IL4-stimulated + STAT6-I. treated conditions (n=5 each). Unadjusted Wilcoxon p-values (two-sided) compared to IL4 stimulation alone are provided. (G) Representative immunoblot showing protein levels of pSTAT6, STAT6 and GAPDH in KM-H2 cells as a function on IL4R construct expression under unstimulated, IL13-stimulated, IL13-stimulated + IL4R-Ab treated as well as IL13-stimulated + STAT6-I. treated conditions. All conditions were run on the same gel. Ladders run between IL4R constructs were cropped and are not shown. (H) Immunoblot showing protein levels of pSTAT6, STAT6 and GAPDH in IL4R I242N expressing KM-H2 cells under unstimulated, IL13-stimulated, IL13-stimulated + IL4R-Ab treated as well as IL13-stimulated + STAT6-I. treated conditions. (I-J) CCL17 (TARC) concentrations in supernatant of transduced (I) DEV (n=5 for unstimulated and IL13; n=4 for IL4R-Ab and STAT6-I.) and (J) KM-H2 (n=3 each) cells under unstimulated, IL13-stimulated, IL13-stimulated + IL4R-Ab treated as well as IL13-stimulated + STAT6-I. treated conditions. Unadjusted Wilcoxon p-values (two-sided) are provided. (K) Boxplots and Wilcoxon p-values (two-sided) comparing IL13 and IL4 expression in RNA-Sequencing of primary bulk tumor specimens visualized as normalized counts (n=86 cHL, n=66 LBCL32). (L) Log2 copy number ratio (L2CNR, boxplot and Wilcoxon p-value, two-sided) of the 5q31.1 cytoband harboring IL13 stratified by IL4R mutation status in n=119 patients with plasma exome sequencing [IL4R mutant: n=12; IL4R WT: n=107].
Panels A,G-H: At least 2 independent experiments were performed for each condition. Panels D,F,I,J: mean +/− standard error (se). Panels K-L: each box represents the interquartile range (the range between the 25th and 75th percentile) with the median of the data, whiskers indicate the upper and lower value within 1.5 times the IQR.
Extended Data Figure 9: Pretreatment ctDNA correlates with clinical risk factors and MRD is an independent prognostic factor in cHL.
(A-E) Previously untreated, adult cHL patients were considered for associations between pretreatment ctDNA levels and clinical variables (n=309). Boxplots summarizing pretreatment ctDNA levels by (A) stage (n=309, Kruskal-Wallis p-value), (B) bulky disease (n=184, Wilcoxon p-value), (C) B-symptoms (n=308, Wilcoxon p-value), (D) EBV status (n=309, Wilcoxon p-value) and (E) histological subtype (n=247, Wilcoxon p-value). Patients with lymphocyte rich and lymphocyte depleted subtypes are not visualized due to small numbers (n=9). All p-values were derived from two-sided tests. (F-G) Waterfall plot showing log10 ctDNA changes from baseline at (F) C1D15 and (G) the >C(ycle)4/EoT (End of Treatment) timepoint. Bars are colored by PFS event status. Top annotation visualizes PET2 readings according to 5-point scale (5PS Deauville). (H) Kaplan-Meier curves and logrank p-value showing PFS stratified by ctDNA detection at >C4/EoT. (I) Kaplan-Meier curves and logrank p-values showing progression-free survival (PFS) stratified by ctDNA detection at C3D1 in PET2 negative and PET2 positive patients.
Panels A-E: each box represents the interquartile range (the range between the 25th and 75th percentile) with the median of the data, whiskers indicate the upper and lower value within 1.5 times the IQR.
Extended Data Figure 10: Model depicting the potential pathogenesis of H1 and H2 cHL subtypes.
GC: Germinal center; SHM: Somatic hypermutation; SNV: Single nucleotide variant; SCNA: Somatic copy number aberrations. Created with BioRender.com.
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Institute (R01CA257655 and R01CA233975 to A.A.A. and M.D., the Department of Defense (CA210872, Award Number W81XWH-22-1-0336) the Virginia and D.K. Ludwig Fund for Cancer Research (A.A.A. and M.D.), Hanna and Michael Murphy family gift, the Stanford Cancer Institute (A.A.A.), the Damon Runyon Cancer Research Foundation (DR-CI#71-14 to A.A.A.), the American Society of Hematology Scholar Award (A.A.A.), V Foundation for Cancer Research Abeloff Scholar Award (A.A.A.), the Emerson Collective Cancer Research Fund (A.A.A.), the Stinehart/Reed Award (A.A.A.), the CRK Faculty Scholar Fund (M.D.), Lung Cancer Research Foundation (M.S.E.), K08CA241076 (D.M.K.) and the SDW/DT, Shanahan Family Foundations (A.A.A. and D.M.K.), the Terry Fox Research Institute (Grants #1061 and #1108 to C.S.) and the Canadian Institutes of Health Research (#180613 to C.S.). S.K.A. is a scholar of the German Cancer Aid (Deutsche Krebshilfe, Dr. Mildred Scheel scholarship 57406718), A.A.A. of the Leukemia & Lymphoma Society. M.Y.L. is a PhD candidate at the University of British Columbia, Canada and is supported by an Elizabeth C. Watters Research Fellowship. T.F. and J.E.F. were supported by the American Syrian Lebanese Associated Charities and the National Institutes of Health Cancer Support Core grant (R03CA21765). The Children Oncology Group (COG) was supported by NCTN Operations Center Grant U10CA180886, NCTN Statistics & Data Center Grant U10CA180899 and NCTN HM-ITSC Grant (UG1CA233249).
S.K.A reports speaker honoraria from Takeda. M.S.E. reports consultancy for Foresight Diagnostics. J.E.F. reports research funding from Seattle Genetics. B.J.S. reports consultancy for Foresight Diagnostics. A.S. is currently employed by Foresight Diagnostics. Y.N. reports consultancy for Roche & Leica Biosystems as well as research funding from Kite Pharma. D.R. reports consultancy, honoraria, research funding and travel support from AstraZeneca, Janssen, AbbVie, Gilead, MSD, BMS and BeiGene. R.L. reports research funding from TG Therapeutics, Incyte, Bayer, Cyteir, Genentech, SeaGen and Rapt as well as consultancy for Cancer Study Group, SeaGen, Foresight Diagnostics and Abbvie. D.M.K. reports consultancy for Roche, Adaptive Biotechnologies, and Genentech, equity ownership interest in Foresight Diagnostics, and patent filings, including patent issued, licensed, and with royalties paid from Foresight. M.A. reports consultancy for Takeda, Bristol-Myers-Squibb, Karyopharm, Gilead and Incyte, research funding from Roche, Johnson & Johnson and Takeda as well as travel support from Roche, Bristol-Myers-Squib, Celgene, Gilead, Abbvie, Astra-Zeneca and Takeda. C.S. has performed consultancy for Bayer, and has received research funding from Epizyme and Trillium Therapeutics. M.D. reports research funding from AstraZeneca, Genentech, Varian Medical Systems and Illumina, ownership interest in CiberMed and Foresight Diagnostics, consultancy from AstraZeneca, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Gritstone Oncology, Illumina, Regeneron, and Roche, and multiple issued and pending patents including patents licensed to Foresight Diagnostics and Roche. A.A.A. reports consultancy for Celgene, Chugai, Genentech, Gilead, Janssen, Pharmacyclics and Roche, Scientific Advisory Board Membership in the Lymphoma Research Foundation, and Professional Affiliations with the American Society of Hematology, American Society of Clinical Oncology, American Society of Clinical Investigation, Leukemia & Lymphoma Society, Research Funding from the National Cancer Institute, National Heart, Lung, and Blood Institute, National Institutes of Health, Celgene, Bristol Myers Squibb, and Pfizer, patent filings including patent issued, licensed, and with royalties paid from FortySeven, a patent pending and Licensed to Foresight, a patent pending relating to MARIA, a patent issued and licensed to CiberMed, a patent issued, a patent pending to CiberMed, a patents issued to Idiotype Vaccines, and a patent issued, licensed, and with royalties paid From Roche, and equity ownership interests in CiberMed Inc., ForeSight Diagnostics, FortySeven Inc., and CARGO Therapeutics. S.K.A., M.S.E., M.Y.L., B.J.S., D.M.K., C.S., M.D. and A.A.A. also report patent filings related to cancer biomarkers.
Footnotes
Disclaimer: The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interests
The remaining authors declare no competing interests.
Compound numbering
1) IL4
2) IL13
3) IL4Rα blocking antibody (R&D Systems)
4) phospho-STAT6 inhibitor (AS1517499, Selleckchem)
5) Doxycycline
Supplementary information
Supplementary information is available for this manuscript.
Data availability
The sequencing data is available in the database of Genotypes and Phenotypes dbGaP (study accession phs003435.v1.p1) in compliance with European General Data Protection Regulation (GDPR) and US Health Insurance Portability and Accountability Act (HIPAA) as applicable. In addition, an online tool to assign new samples to the genetic subtypes H1 and H2 is available at https://hodgkin.stanford.edu. Mutation calls can be downloaded from the same website.
Code availability
The code for LDA clustering and RePhyNER is also available at https://hodgkin.stanford.edu.
REFERENCES
- 1.Sobesky S et al. In-depth cell-free DNA sequencing reveals genomic landscape of Hodgkin’s lymphoma and facilitates ultrasensitive residual disease detection. Med (N Y) 2, 1171–1193 e1111 (2021). 10.1016/j.medj.2021.09.002 [DOI] [PubMed] [Google Scholar]
- 2.Spina V et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood 131, 2413–2425 (2018). 10.1182/blood-2017-11-812073 [DOI] [PubMed] [Google Scholar]
- 3.Desch AK et al. Genotyping circulating tumor DNA of pediatric Hodgkin lymphoma. Leukemia 34, 151–166 (2020). 10.1038/s41375-019-0541-6 [DOI] [PubMed] [Google Scholar]
- 4.Vandenberghe P et al. Non-invasive detection of genomic imbalances in Hodgkin/Reed-Sternberg cells in early and advanced stage Hodgkin’s lymphoma by sequencing of circulating cell-free DNA: a technical proof-of-principle study. The Lancet Haematology 2, e55–e65 (2015). 10.1016/s2352-3026(14)00039-8 [DOI] [PubMed] [Google Scholar]
- 5.Kurtz DM et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat Biotechnol 39, 1537–1547 (2021). 10.1038/s41587-021-00981-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flaherty KT et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363, 809–819 (2010). 10.1056/NEJMoa1002011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lievre A et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol 26, 374–379 (2008). 10.1200/JCO.2007.12.5906 [DOI] [PubMed] [Google Scholar]
- 8.Paez JG et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004). 10.1126/science.1099314 [DOI] [PubMed] [Google Scholar]
- 9.Soda M et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007). 10.1038/nature05945 [DOI] [PubMed] [Google Scholar]
- 10.Alizadeh AA et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000). 10.1038/35000501 [DOI] [PubMed] [Google Scholar]
- 11.Chapuy B et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 24, 679–690 (2018). 10.1038/s41591-018-0016-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitz R et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med 378, 1396–1407 (2018). 10.1056/NEJMoa1801445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Treon SP et al. Ibrutinib in previously treated Waldenstrom’s macroglobulinemia. N Engl J Med 372, 1430–1440 (2015). 10.1056/NEJMoa1501548 [DOI] [PubMed] [Google Scholar]
- 14.Reichel J et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 125, 1061–1072 (2015). 10.1182/blood-2014-11-610436 [DOI] [PubMed] [Google Scholar]
- 15.Tiacci E et al. Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood 131, 2454–2465 (2018). 10.1182/blood-2017-11-814913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wienand K et al. Genomic analyses of flow-sorted Hodgkin Reed-Sternberg cells reveal complementary mechanisms of immune evasion. Blood Adv 3, 4065–4080 (2019). 10.1182/bloodadvances.2019001012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang WS et al. Comprehensive Genomic Profiling of Hodgkin Lymphoma Reveals Recurrently Mutated Genes and Increased Mutation Burden. Oncologist 24, 219–228 (2019). 10.1634/theoncologist.2018-0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mata E et al. Analysis of the mutational landscape of classic Hodgkin lymphoma identifies disease heterogeneity and potential therapeutic targets. Oncotarget 8, 111386–111395 (2017). 10.18632/oncotarget.22799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomez F et al. Ultra-Deep Sequencing Reveals the Mutational Landscape of Classical Hodgkin Lymphoma. medRxiv, 2021.2006.2025.21258374 (2021). 10.1101/2021.06.25.21258374 [DOI] [PMC free article] [PubMed]
- 20.Maura F et al. Molecular Evolution of Classic Hodgkin Lymphoma Revealed Through Whole-Genome Sequencing of Hodgkin and Reed Sternberg Cells. Blood Cancer Discovery 4, 208–227 (2023). 10.1158/2643-3230.Bcd-22-0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiacci E et al. Analyzing primary Hodgkin and Reed-Sternberg cells to capture the molecular and cellular pathogenesis of classical Hodgkin lymphoma. Blood 120, 4609–4620 (2012). 10.1182/blood-2012-05-428896 [DOI] [PubMed] [Google Scholar]
- 22.Weniger MA & Küppers R NF-κB deregulation in Hodgkin lymphoma. Seminars in Cancer Biology 39, 32–39 (2016). 10.1016/j.semcancer.2016.05.001 [DOI] [PubMed] [Google Scholar]
- 23.Jamshidi A et al. Evaluation of cell-free DNA approaches for multi-cancer early detection. Cancer Cell 40, 1537–1549 e1512 (2022). 10.1016/j.ccell.2022.10.022 [DOI] [PubMed] [Google Scholar]
- 24.Esfahani MS et al. Inferring gene expression from cell-free DNA fragmentation profiles. Nat Biotechnol 40, 585–597 (2022). 10.1038/s41587-022-01222-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roschewski M et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol 16, 541–549 (2015). 10.1016/S1470-2045(15)70106-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scherer F et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med 8, 364ra155 (2016). 10.1126/scitranslmed.aai8545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurtz DM et al. Circulating Tumor DNA Measurements As Early Outcome Predictors in Diffuse Large B-Cell Lymphoma. J Clin Oncol 36, 2845–2853 (2018). 10.1200/JCO.2018.78.5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alig S et al. Short Diagnosis-to-Treatment Interval Is Associated With Higher Circulating Tumor DNA Levels in Diffuse Large B-Cell Lymphoma. J Clin Oncol 39, 2605–2616 (2021). 10.1200/JCO.20.02573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meriranta L et al. Molecular features encoded in the ctDNA reveal heterogeneity and predict outcome in high-risk aggressive B-cell lymphoma. Blood 139, 1863–1877 (2022). 10.1182/blood.2021012852 [DOI] [PubMed] [Google Scholar]
- 30.Newman AM et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 34, 547–555 (2016). 10.1038/nbt.3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chabon JJ et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 580, 245–251 (2020). 10.1038/s41586-020-2140-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sworder BJ et al. Determinants of resistance to engineered T-cell therapies targeting CD19 in large B-cell lymphomas. Cancer Cell In Press (2023). [DOI] [PMC free article] [PubMed]
- 33.Han DSC et al. The Biology of Cell-free DNA Fragmentation and the Roles of DNASE1, DNASE1L3, and DFFB. Am J Hum Genet 106, 202–214 (2020). 10.1016/j.ajhg.2020.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serpas L et al. Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc Natl Acad Sci U S A 116, 641–649 (2019). 10.1073/pnas.1815031116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe T, Takada S & Mizuta R Cell-free DNA in blood circulation is generated by DNase1L3 and caspase-activated DNase. Biochemical and biophysical research communications 516, 790–795 (2019). 10.1016/j.bbrc.2019.06.069 [DOI] [PubMed] [Google Scholar]
- 36.Stewart BJ et al. Spatial and molecular profiling of the mononuclear phagocyte network in classic Hodgkin lymphoma. Blood 141, 2343–2358 (2023). 10.1182/blood.2022015575 [DOI] [PubMed] [Google Scholar]
- 37.Husain H et al. Tumor Fraction Correlates With Detection of Actionable Variants Across > 23,000 Circulating Tumor DNA Samples. JCO Precis Oncol 6, e2200261 (2022). 10.1200/PO.22.00261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blei DM, Ng AY & Jordan MI Latent dirichlet allocation. Journal of machine Learning research 3, 993–1022 (2003). [Google Scholar]
- 39.Shukla ND et al. Profiling T-Cell Receptor Diversity and Dynamics during Lymphoma Immunotherapy Using Cell-Free DNA (cfDNA). Blood 136, 49–50 (2020). 10.1182/blood-2020-141655 [DOI] [Google Scholar]
- 40.Steidl C et al. Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N Engl J Med 362, 875–885 (2010). 10.1056/NEJMoa0905680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wills-Karp M & Finkelman FD Untangling the complex web of IL-4- and IL-13-mediated signaling pathways. Sci Signal 1, pe55 (2008). 10.1126/scisignal.1.51.pe55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vigano E et al. Somatic IL4R mutations in primary mediastinal large B-cell lymphoma lead to constitutive JAK-STAT signaling activation. Blood 131, 2036–2046 (2018). 10.1182/blood-2017-09-808907 [DOI] [PubMed] [Google Scholar]
- 43.Duns G et al. Characterization of DLBCL with a PMBL gene expression signature. Blood 138, 136–148 (2021). 10.1182/blood.2020007683 [DOI] [PubMed] [Google Scholar]
- 44.Skinnider BF & Mak TW The role of cytokines in classical Hodgkin lymphoma. Blood 99, 4283–4297 (2002). 10.1182/blood-2002-01-0099 [DOI] [PubMed] [Google Scholar]
- 45.Rawal S et al. Cross talk between follicular Th cells and tumor cells in human follicular lymphoma promotes immune evasion in the tumor microenvironment. J Immunol 190, 6681–6693 (2013). 10.4049/jimmunol.1201363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pangault C et al. Follicular lymphoma cell niche: identification of a preeminent IL-4-dependent T(FH)-B cell axis. Leukemia 24, 2080–2089 (2010). 10.1038/leu.2010.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kapp U et al. Interleukin 13 is secreted by and stimulates the growth of Hodgkin and Reed-Sternberg cells. J Exp Med 189, 1939–1946 (1999). 10.1084/jem.189.12.1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Natoli A et al. Targeting the IL-4/IL-13 signaling pathway sensitizes Hodgkin lymphoma cells to chemotherapeutic drugs. Int J Cancer 133, 1945–1954 (2013). 10.1002/ijc.28189 [DOI] [PubMed] [Google Scholar]
- 49.Skinnider BF, Kapp U & Mak TW Interleukin 13: a growth factor in hodgkin lymphoma. Int Arch Allergy Immunol 126, 267–276 (2001). 10.1159/000049523 [DOI] [PubMed] [Google Scholar]
- 50.Skinnider BF et al. Interleukin 13 and interleukin 13 receptor are frequently expressed by Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood 97, 250–255 (2001). 10.1182/blood.v97.1.250 [DOI] [PubMed] [Google Scholar]
- 51.Kurtz DM et al. Leveraging phased variants for personalized minimal residual disease detection in localized non-small cell lung cancer. Journal of Clinical Oncology 39, 8518–8518 (2021). 10.1200/JCO.2021.39.15_suppl.8518 [DOI] [Google Scholar]
- 52.Buedts L et al. The landscape of copy number variations in classical Hodgkin lymphoma: a joint KU Leuven and LYSA study on cell-free DNA. Blood Adv 5, 1991–2002 (2021). 10.1182/bloodadvances.2020003039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu Z, Chen H, Long Y, Li P & Gu Y The main sources of circulating cell-free DNA: Apoptosis, necrosis and active secretion. Crit Rev Oncol Hematol 157, 103166 (2021). 10.1016/j.critrevonc.2020.103166 [DOI] [PubMed] [Google Scholar]
- 54.Sin STK et al. Identification and characterization of extrachromosomal circular DNA in maternal plasma. Proc Natl Acad Sci U S A 117, 1658–1665 (2020). 10.1073/pnas.1914949117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng Z et al. DNASE1L3 as a Prognostic Biomarker Associated with Immune Cell Infiltration in Cancer. Onco Targets Ther 14, 2003–2017 (2021). 10.2147/OTT.S294332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi G, Abbott KN, Wu W, Salter RD & Keyel PA Dnase1L3 Regulates Inflammasome-Dependent Cytokine Secretion. Front Immunol 8, 522 (2017). 10.3389/fimmu.2017.00522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang HY & Nadeau KC IL-4Ralpha Inhibitor for Atopic Disease. Cell 170, 222 (2017). 10.1016/j.cell.2017.06.046 [DOI] [PubMed] [Google Scholar]
- 58.Bauckneht M, Piva R, Sambuceti G, Grossi F & Morbelli S Evaluation of response to immune checkpoint inhibitors: Is there a role for positron emission tomography? World J Radiol 9, 27–33 (2017). 10.4329/wjr.v9.i2.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferrari C et al. Early Evaluation of Immunotherapy Response in Lymphoma Patients by 18F-FDG PET/CT: A Literature Overview. J Pers Med 11 (2021). 10.3390/jpm11030217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castellino SM et al. Brentuximab Vedotin with Chemotherapy in Pediatric High-Risk Hodgkin’s Lymphoma. N Engl J Med 387, 1649–1660 (2022). 10.1056/NEJMoa2206660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.ClinicalTrials.gov Identifier: NCT03755804. https://clinicaltrials.gov/ct2/show/NCT03755804.
- 62.Casasnovas RO et al. PET-adapted treatment for newly diagnosed advanced Hodgkin lymphoma (AHL2011): a randomised, multicentre, non-inferiority, phase 3 study. Lancet Oncol 20, 202–215 (2019). 10.1016/S1470-2045(18)30784-8 [DOI] [PubMed] [Google Scholar]
- 63.Fornecker LM et al. Brentuximab Vedotin Plus AVD for First-Line Treatment of Early-Stage Unfavorable Hodgkin Lymphoma (BREACH): A Multicenter, Open-Label, Randomized, Phase II Trial. J Clin Oncol 41, 327–335 (2023). 10.1200/JCO.21.01281 [DOI] [PubMed] [Google Scholar]
- 64.Lynch RC et al. Concurrent pembrolizumab with AVD for untreated classic Hodgkin lymphoma. Blood 141, 2576–2586 (2023). 10.1182/blood.2022019254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghesquieres H et al. Prednisone, Vinblastine, Doxorubicin and Bendamustine (PVAB) Regimen in First Line Therapy for Older Patients with Advanced-Stage Classical Hodgkin Lymphoma: Results of a Prospective Multicenter Phase II Trial of the Lymphoma Study Association (LYSA). Blood 134, 2832–2832 (2019). 10.1182/blood-2019-129016 [DOI] [Google Scholar]
- 66.Newman AM et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 20, 548–554 (2014). 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Canisius S, Martens JW & Wessels LF A novel independence test for somatic alterations in cancer shows that biology drives mutual exclusivity but chance explains most co-occurrence. Genome Biol 17, 261 (2016). 10.1186/s13059-016-1114-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang P et al. Plasma DNA End-Motif Profiling as a Fragmentomic Marker in Cancer, Pregnancy, and Transplantation. Cancer Discov 10, 664–673 (2020). 10.1158/2159-8290.CD-19-0622 [DOI] [PubMed] [Google Scholar]
- 69.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012). 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6, pl1 (2013). 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Shea JP et al. pLogo: a probabilistic approach to visualizing sequence motifs. Nat Methods 10, 1211–1212 (2013). 10.1038/nmeth.2646 [DOI] [PubMed] [Google Scholar]
- 72.Hao Y et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021). 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aoki T et al. Single-Cell Transcriptome Analysis Reveals Disease-Defining T-cell Subsets in the Tumor Microenvironment of Classic Hodgkin Lymphoma. Cancer Discov 10, 406–421 (2020). 10.1158/2159-8290.CD-19-0680 [DOI] [PubMed] [Google Scholar]
- 74.Sanz I et al. Challenges and Opportunities for Consistent Classification of Human B Cell and Plasma Cell Populations. Front Immunol 10, 2458 (2019). 10.3389/fimmu.2019.02458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Holmes AB et al. Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome. J Exp Med 217 (2020). 10.1084/jem.20200483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang L et al. Integrated single-cell RNA sequencing analysis reveals distinct cellular and transcriptional modules associated with survival in lung cancer. Signal Transduction and Targeted Therapy 7, 9 (2022). 10.1038/s41392-021-00824-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Newman AM et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nature Biotechnology 37, 773–782 (2019). 10.1038/s41587-019-0114-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Foley JW et al. Gene expression profiling of single cells from archival tissue with laser-capture microdissection and Smart-3SEQ. Genome Res 29, 1816–1825 (2019). 10.1101/gr.234807.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grossiord E et al. in 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI). 1118–1121 (IEEE; ). [Google Scholar]
- 80.Boellaard R Quantitative oncology molecular analysis suite: ACCURATE. Journal of Nuclear Medicine 59, 1753–1753 (2018). [Google Scholar]
- 81.Hirata K et al. A semi-automated technique determining the liver standardized uptake value reference for tumor delineation in FDG PET-CT. PLoS One 9, e105682 (2014). 10.1371/journal.pone.0105682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data is available in the database of Genotypes and Phenotypes dbGaP (study accession phs003435.v1.p1) in compliance with European General Data Protection Regulation (GDPR) and US Health Insurance Portability and Accountability Act (HIPAA) as applicable. In addition, an online tool to assign new samples to the genetic subtypes H1 and H2 is available at https://hodgkin.stanford.edu. Mutation calls can be downloaded from the same website.
The code for LDA clustering and RePhyNER is also available at https://hodgkin.stanford.edu.