Abstract
The region of the human cytomegalovirus (CMV) genome between the UL127 open reading frame and the major immediate-early (MIE) enhancer is referred to as the unique region. DNase I protection analysis with human cell nuclear extracts demonstrated multiple protein binding sites in this region of the viral genome (P. Ghazal, H. Lubon, C. Reynolds-Kohler, L. Hennighausen, and J. A. Nelson, Virology 174:18–25, 1990). However, the function of this region in the context of the viral genome is not known. In wild-type human CMV-infected human fibroblasts, cells permissive for viral replication, there is little to no transcription from UL127. We determined that the unique region prevented transcription from the UL127 promoter but had no effect on the divergent MIE promoter. In transient-transfection assays, the basal level of expression from the UL127 promoter increased significantly when the wild-type unique sequences were mutated. In recombinant viruses with similar mutations in the unique region, expression from the UL127 promoter occurred only after de novo viral protein synthesis, typical of an early viral promoter. A 111-bp deletion-substitution of the unique sequence caused approximately a 20-fold increase in the steady-state level of RNA from the UL127 promoter and a 245-fold increase in the expression of a downstream indicator gene. This viral negative regulatory region was also mutated at approximately 50-bp regions proximal and distal to the UL127 promoter. Although some repressive effects were detected in the distal region, mutations of the region proximal to the UL127 promoter had the most significant effects on transcription. Within the proximal and distal regions, there are potential cis sites for known eucaryotic transcriptional repressor proteins. This region may also bind unknown viral proteins. We propose that the unique region upstream of the UL127 promoter and the MIE enhancer negatively regulates the expression from the UL127 promoter in permissive human fibroblast cells. This region may be a regulatory boundary preventing the effects of the very strong MIE enhancer on this promoter.
Infections by human cytomegalovirus (CMV) are associated with congenital neurological complications in newborns and pneumonitis, retinitis, hepatitis, and gastrointestinal diseases in immunocompromised patients (1, 32). The virus replicates in fibroblasts, endothelial and epithelial cells, smooth muscle cells, neurons, glial cells, and macrophages (64, 69, 70, 89, 90). Productive infection is associated with terminal differentiation of the cell. During viremia, primitive hematopoietic cell populations of the bone marrow are also infected (28). Infected myeloid cell progenitors harbor both the latent viral genome and viral latency-associated transcripts (28, 38, 39). These transcripts originate within the major immediate-early (MIE) enhancer at −356 and −292 relative to the transcription start site for productive infection (+1) and are terminated at the immediate-early 1 (IE1) or IE2 polyadenylation signals (38, 39). Only a small portion of bone marrow mononuclear cells from naturally infected individuals express viral latency-associated transcripts, but the role of the viral proteins encoded by these transcripts is presently not understood (38, 39). Peripheral blood monocytes isolated from CMV-seropositive individuals also carry latent viral genomes (53, 66, 67, 85), but latency-associated transcripts have not been demonstrated in these cells.
Viral transcripts from the MIE promoter at +1 are associated with productive infection (77). Inflammatory cytokines such as gamma interferon can convert nonpermissive monocytes to permissive macrophages (71). They may also stimulate tissue dendritic cells to a permissive state for viral replication (28). In addition, monocyte-derived macrophages from seropositive individuals can be activated and differentiated by inflammatory cytokines plus allogeneic T cells to produce infectious virus (72). The switch from latent transcription to productive infection transcription is not understood, but it may be related to the state of cellular differentiation. As the monocyte differentiates, repressive chromatin structures may become less restrictive for productive viral transcripts and facilitate the switch from latent transcription to productive viral transcripts. A better understanding of the viral chromatin structure in this region of the viral genome may help elucidate viral reactivation mechanisms.
The first class of viral genes to be expressed after reactivation from latency or primary infection is the IE genes. There are two IE genes downstream of the MIE promoter, IE1 and IE2 (75, 77, 78). These genes code for proteins that regulate viral and cellular gene expression (31, 48, 74–76). The IE1 gene encodes a 491-amino-acid nuclear phosphoprotein of approximately 72 kDa (IE72). The viral gene is necessary for efficient viral replication at low multiplicities of infection, but it is not essential at high multiplicities (27, 54). The IE2 gene encodes several viral gene products (75). The largest viral protein is a 579-amino-acid nuclear phosphoprotein of approximately 86 kDa (IE86) which is presumably essential for viral replication because the viral gene cannot be deleted. These and the ancillary IE genes, UL36 through UL38 and IRS1/TRS1, affect the level of early gene expression (13). The early viral genes encode viral proteins involved in ori-Lyt-mediated viral DNA synthesis (2). The late viral genes are expressed after viral DNA replication and encode many of the viral structural proteins (23).
A genetic target for reactivation from latency or for efficient replication after primary infection is the MIE enhancer (8, 52, 80). The human (6, 86), simian (9), rat (63), and murine (17) CMV MIE enhancers are regarded as critical regulatory elements for reactivation from latency to productive infection. With murine CMV, the efficiency of viral replication in tissue culture or in the host is greatly impaired when the MIE enhancer is deleted, but infectious viruses can be produced, and consequently, the enhancer is not essential (3).
The human CMV MIE enhancer and the flanking regions were assigned arbitrary boundaries to better discuss this complicated regulatory region (52, 80). Downstream of the MIE +1 start site is a positive regulatory control region from +8 to +112 referred to as leader exon 1 (22). The MIE promoter between −50 and +8 contains a TATA box, a cis-repression sequence (crs) between −13 and −1, and an initiator-like sequence between +1 and +7 (46, 47, 52, 80). After expression of the downstream IE1-IE2 genes, the MIE promoter is negatively autoregulated by the IE86 protein (11, 44, 60). The IE86 protein binds to the crs and may interfere with the binding of a 150-kDa cellular transcription initiation factor (46). The IE86 protein and possibly the UL84 gene product (62), which interacts with IE86 at early and late times after infection, repress RNA polymerase II initiation at the MIE promoter (47, 91). The MIE enhancer between −550 and −50 has cis-acting elements in repetitive motifs which bind factors such as CREB/ATF, NF-κB/rel, and SP-1 and in nonrepetitive motifs which bind AP-1, RA, SRE, and ETS. These cis-acting elements respond to a variety of signal transduction events which occur early after infection (4, 8, 52, 80). Upstream of the MIE enhancer between approximately −750 and −550 is a region referred to as the unique region (21, 52, 80). The region contains a cluster of CTF/NF-1 binding sites (29), but no function, to date, has been assigned to these sites or to this region. Deletion of the modulator region between −1108 and −750 had no effect on transcription from the MIE promoter in either infected undifferentiated or infected differentiated cells (51). This region is conserved in all human CMVs sequenced to date and is present in chimpanzee CMV (8). The region contains a 393-bp open reading frame (ORF) designated UL127, which is divergent from the MIE promoter and genes (10). A TATA box sequence is positioned 38 bp upstream of UL127. Although the viral gene product may be important to either viral latency or pathogenesis, it is nonessential for replication in cell culture (51).
The genomes of viruses such as simian virus 40, adenovirus, and adeno-associated virus do not exist in vivo as naked DNA but are in the form of nucleosomal structures separated by 40- to 50-bp stretches of naked DNA (14, 40, 49, 88). Little is known about the chromatin structure of herpesviruses. Although the genomes of herpes simplex virus and Epstein-Barr virus are in a nucleosome structure during latency (15, 18), this structure is altered during productive infection.
All herpesviruses encode virus-specific transcriptional regulatory proteins. These viral proteins function either by directly binding to the viral DNA or by interacting with other viral or cellular proteins. The net effect of these interactions could be a remodeling of viral chromatin to facilitate the expression of early viral genes. It has been proposed that the human CMV IE86 protein might activate viral as well as cellular promoters by inducing chromatin remodeling (37).
In this report, we describe a repressive sequence between the UL127 promoter and the MIE enhancer of human CMV that blocks transcription from the UL127 promoter at early and late times after infection of permissive human fibroblasts. When this regulatory viral DNA sequence in the unique region was replaced with nonregulatory stuffer DNAs and the UL127 ORF was replaced with the chloramphenical acetyltransferase (CAT) gene, there was very strong early transcription and downstream reporter gene expression from the wild-type UL127 promoter. Within the unique region, there are several cis sites with consensus or near-consensus sites for the binding of transcriptional repressor proteins. Mutation of the unique region between the UL127 promoter and the MIE enhancer may affect the binding of cellular proteins. Reasons why the expression of the UL127 gene is repressed in wild-type virus in permissive human fibroblasts during productive infection are discussed.
MATERIALS AND METHODS
Cells and viruses.
Primary human foreskin fibroblasts (HFFs) were grown in Eagle’s minimal essential medium (Life Technologies, Gaithersburg, Md., or Mediatech, Herndon, Va.) supplemented with 10% newborn bovine serum (Sigma, St. Louis, Mo.), 100 U of penicillin per ml, and 100 μg of streptomycin per ml. The same conditions were used for the growth of fibroblasts with hypoxanthine guanine phosphoribosyltransferase deficiency (GM02291), which were obtained from the Coriell Institute for Medical Research (Coriell Cell Repositories, Camden, N.J.). Human CMV strain Towne and all recombinant viruses made from the Towne strain were propagated as described previously (81). All recombinant viruses grew as well as did the parent recombinant virus RdlMSVgpt(r1) (51). Titers of infectious virus in the extracellular fluid were determined on HFFs by plaque assay as described previously (51).
Enzymes.
All DNA-modifying enzymes were used according to the manufacturers’ specifications and were obtained from either New England Biolabs, Inc. (Beverly, Mass.), Bethesda Research Laboratories, Inc. (Gaithersburg, Md.), Boehringer Mannheim Biochemicals (Indianapolis, Ind.), or Promega (Madison, Wis.).
Plasmids.
Since the transcription start site from the UL127 promoter had not been mapped precisely, the region of the viral genome between the putative UL127 promoter and the MIE promoter was designated relative to the transcription start site (+1) of the MIE promoter (52, 80). By using plasmid pLUX/CAT (36), HindIII restriction endonuclease sites were located downstream of the UL127 promoter at −706 and downstream of the MIE promoter at +171 to generate plasmid pLC3+. The resultant plasmid contains wild-type viral DNA sequence between the divergent UL127 and MIE promoters with the luciferase (LUX) and CAT indicator genes downstream, respectively. The following deletions upstream of the UL127 promoter were introduced into plasmid pLC3+. (i) An SpeI site was inserted at −694, and the region from −694 to −583 was deleted by restriction endonuclease digestion with SpeI. The ends were ligated with T4 DNA ligase to generate plasmid pLCdl−694/−583. (ii) To maintain the relative spacing between the UL127 promoter and the enhancer, heterologous DNA fragments of 116, 121, and 134 bp were isolated from plasmid pCAT-basic (Promega) after restriction endonuclease digestion with either ScaI/StyI, SalI/BanI, or BamHI/HincII, respectively. After each DNA fragment was made blunt with Klenow polymerase, the DNA fragments were ligated to the SpeI site at −583 of pLCdl−694/−583, which was made blunt with Klenow polymerase, to generate plasmids pLCdl−694/−583+116bp, pLC−694/−583+121bp, and pLCdl−694/−583+134bp, respectively. All plasmid constructions were sequenced by the dideoxynucleotide method.
Recombinant viruses.
All recombinant viruses were made from the parental recombinant virus RdlMSVgpt(r1), which has a simian virus 40 early kinetic-class transcription unit containing the bacterial guanyl phosphoribosyltransferase (gpt) ORF in place of the deleted modulator region at −640 to −1108 relative to the MIE promoter start site (51). The shuttle vector pdlMCAT was constructed as described for pdlMSVgpt (46), except that the gpt gene was replaced by the CAT gene and the wild-type sequence (−706 to +171) containing the UL127 putative promoter was located upstream of the CAT gene. A deletion upstream of the UL127 promoter between −694 and −583 was introduced into plasmid pdlMCAT as follows. Two oligonucleotides, 5′-CTAGTGCTATATAACCAAGCTTGCGATCAAGCTTATCGACT-3′ and 5′-CTAGAGTCGATAAGCTTGATCGCAAGCTTGGTTATATAGCA-3′, were annealed and ligated into the XbaI site (−726) downstream of the UL127 promoter and the SpeI site (−583) upstream of the UL127 promoter to generate shuttle vector pdlMCATdl−694/−583. DNA fragments of 116, 121, or 134 bp were isolated from pCAT-basic as described above. The DNA fragments were made blunt with Klenow polymerase and ligated into the SpeI site (−583) of pdlMCATdl−694/−583, which was made blunt by Klenow polymerase, to generate shuttle vectors pdlMCAT+116, pdlMCAT+121, and pdlMCAT+134, respectively. Shuttle vectors pdlMCATTheta, pdlMCATdl694-640, and pdlMCATdl640-583 were constructed as follows. Plasmid pdlMCATTheta had a deletion between the XbaI site at −726 and the BsrGI site at −640 which caused a deletion of the UL127 promoter. Deletions between −694 and −640 and −640 and −583 were made by restriction endonuclease digestion with SpeI at −694 and BsrGI at −640 and digestion with BsrGI (−640) and SpeI at −583, respectively.
Shuttle vectors (5 or 10 μg) were cotransfected into HFFs with infectious RdlMSVgpt(r1) viral DNA by calcium phosphate precipitation (25). Three to four days after 100% cytopathic effect, the extracellular virus was harvested; passed through a 0.45-μm-pore-size filter; and diluted 1:20, 1:50, or 1:100. Lesch-Nyhan human fibroblasts (Coriell Institute for Medical Research) were infected. Selection against parental viruses containing the gpt gene was carried out with hypoxanthine guanine phosphoribosyltransferase-deficient fibroblasts with medium containing 50 μg of 6-thioguanine (Sigma) per ml as described previously (26). After approximately 14 days, the extracellular virus was harvested, filtered, and diluted for plaque purification. Recombinant virus plaques were isolated on either normal HFFs or Lesch-Nyhan HFFs, and after approximately 14 days, the plaques were picked and transferred to 12-well HFF culture units. Three to four days after the appearance of 100% cytopathic effect, cell-free virus from each well was mixed with an equal volume of 100% newborn calf serum and stored at −70°C. Infected cells were also harvested, and the cell lysates were assayed for CAT activity. After identification of CAT gene-containing recombinant viruses, the viruses were further plaque purified on HFF cultures as described above. Two or three recombinant viruses were isolated and characterized from at least two independent transfections. Recombinant viruses designated A and B were from one transfection, and those designated C and D were from another transfection.
Southern blots.
Virus was isolated from the culture medium as described previously (81). The viral envelope was solubilized with 1% Sarkosyl, and viral proteins were digested with 200 μg of proteinase K per ml in 0.1% sodium dodecyl sulfate. Viral DNAs were digested with restriction endonucleases EcoRI and NdeI and fractionated by electrophoresis in 1.5% agarose gels. DNA probes were radioactively labeled by the Rediprime 32P-label method of Amersham Pharmacia Biotech (Piscataway, N.J.). Consecutive hybridizations for Southern blot analysis were done as described previously (43, 51). Specific bands were visualized by autoradiography on Hyperfilm MP (Amersham).
Northern blots.
Whole-cell RNA was isolated according to the method of Chomczynski and Sacchi (12). RNAs isolated from infected cells treated with cycloheximide (CH) (200 μg/ml) for 24 h or phosphonoacetic acid (PAA) (200 μg/ml) for 48 h or untreated for 48 h were designated IE, early, and late RNAs, respectively. RNAs were fractionated by electrophoresis in 1.5% agarose gels containing 6% formaldehyde and blotted onto Nytran as described previously (51). A gel-purified SpeI-EcoRV (174,310 to 174,645 map units) fragment was multi-prime 32P-labeled for use as a UL127-specific probe with bidirectional hybridization potential. This same fragment was used to PCR generate, by the method of Bednarczuk et al. (5), the UL127-specific unidirectional 32P-labeled probe from an oligonucleotide, 5′-CGCTATAAACGCTGTGTGTC-3′, starting at 174,580 map units. Hybridization of 32P-labeled probes was done at 68°C overnight as described previously (51). Probes were stripped from blots by boiling in 0.1× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) containing 0.5% sodium dodecyl sulfate. Sense UL127 RNA was synthesized by cloning the UL127-containing template into pBluescript and using phage polymerase for in vitro RNA synthesis according to the instructions of Promega.
RNase protection assay.
Riboprobe synthesis was performed by the method of Krieg and Melton (42) and as described previously (30). Cytoplasmic RNA was harvested from mock-infected or infected HFFs as described previously (87). The multiplicity of infection was approximately 5 PFU per cell. Twenty micrograms of RNA was hybridized to the 32P-labeled riboprobes at room temperature overnight. The specific activities of the riboprobes were similar. After digestion with 100 U of RNase T1 (Boehringer Mannheim) at 37°C for 1 h and then 65 μg of proteinase K for 15 min at 37°C, the RNAs protected from RNase digestion were fractionated in denaturing 6% polyacrylamide-urea gels. Signals were visualized by autoradiography on Hyperfilm MP (Amersham Pharmacia Biotech) and quantitated by image acquisition analysis.
Indicator gene product assays.
All transfections were done three times in quadruplicate on 100-mm-diameter plates of confluent HFFs by the calcium phosphate precipitation method of Graham and van der Eb (25). All infections with recombinant viruses were done in triplicate on 100-mm-diameter plates of HFFs. CAT activities were determined in substrate excess as described by Gorman et al. (24). The acetylated and nonacetylated [14C]chloramphenicol was separated by thin-layer chromatography in a chloroform-methanol (95:5) solvent. The amount of [14C]chloramphenicol acetylation was determined by image acquisition analysis, and the protein concentration was determined by the Bradford method (Bio-Rad Laboratories, Richmond, Calif.). The mean percent acetylation of [14C]chloramphenicol was determined per microgram of protein lysate.
LUX assays were performed by the method of De Wet et al. (16). Luminescence was detected in an Anthos Lucy 1 luminometer (Salzburg, Austria). The mean LUX units per microgram of protein lysate was determined.
RESULTS
Lack of UL127 gene transcription.
The approximately 200-bp region between the 5′ end of the MIE enhancer (−550 relative to the IE1 transcription start site) and the UL127 ORF (−741) is referred to as the unique region (21, 52). Within this region is a consensus TATA box sequence that is identical to the TATA box of the divergent MIE promoter (Fig. 1A). The TATA box sequence is 38 bp upstream of the UL127 ORF in a position typical for a viral promoter. When approximately 100 bp of the unique region proximal to the UL127 ORF was deleted in the construction of recombinant virus RdlMSVgpt(r1) (Fig. 1A), there was no detectable effect on transcription from the MIE promoter (51). Transcription from the minimal simian virus 40 promoter at −640 allowed for expression of the downstream gpt gene substituted for the viral UL127 ORF (51). In addition, the proposed effect of the modulator, which was based on transient-transfection experiments, was not demonstrable in the context of the viral genome. Deletion of the modulator did not affect transcription from the MIE promoter in undifferentiated or differentiated cells (51). We were interested in determining if and when transcription from the putative UL127 promoter occurred in terminally differentiated HFF cells infected with wild-type human CMV Towne. HFFs were infected with either wild-type Towne virus or recombinant virus RdlMSVgpt(r1) at a multiplicity of infection of approximately 5 PFU/cell. We constructed bidirectional or unidirectional probes that span the junction between the unique region and the UL127 ORF as diagramed in Fig. 1A. Whole-cell RNA was isolated at various times after infection, fractionated by denaturing gel electrophoresis, and analyzed by Northern blot hybridization. The sensitivity of the Northern blot assay was evaluated by analyzing 2, 4, and 10 pg of in vitro-synthesized sense RNA from the UL127 ORF as described in Materials and Methods.
FIG. 1.
Northern blotting of viral transcripts from the unique region and the UL127 gene at various times after infection. (A) Diagram of the unique long (UL) and unique short (US) components of the human CMV (HCMV) genome flanked by terminal repeat (TR) or inverted repeat (IR) sequences containing an a sequence. An EcoRI restriction endonuclease map of the viral genome and the EcoRI region of the wild-type (WT) virus and recombinant virus RdlMSVgpt(r1) is diagramed. Mock, IE, early, and late total cell RNAs were isolated and subjected to Northern blot hybridization with either a 32P-labeled bidirectional probe (B) or a 32P-labeled unidirectional probe (C). To determine the sensitivity of this method, RNA was synthesized in vitro from the UL127 gene template and analyzed. Standard molecular size markers are indicated in kilobases. SV40, simian virus 40.
The radioactively labeled bidirectional probe did not detect IE viral RNA, but viral RNA was detected at early and late times after infection (Fig. 1B). Since the viral RNAs were detected with both wild-type virus and RdlMSVgpt(r1), the RNAs must have been initiated from adjacent regions because the putative UL127 promoter and ORF were deleted in RdlMSVgpt(r1). The bidirectional probe detected viral RNAs running through a common 57-bp sequence in the unique region. The unidirectional probe, which would detect sense RNAs from the UL127 ORF, did not detect any viral RNA to the 2-pg level originating from the unique region, the putative UL127 promoter, or the downstream UL127 ORF at any time after infection (Fig. 1C). These data suggested that the putative TATA box sequence upstream of the UL127 ORF was either nonfunctional, very inefficient, or repressed in HFFs.
Activation of the UL127 promoter.
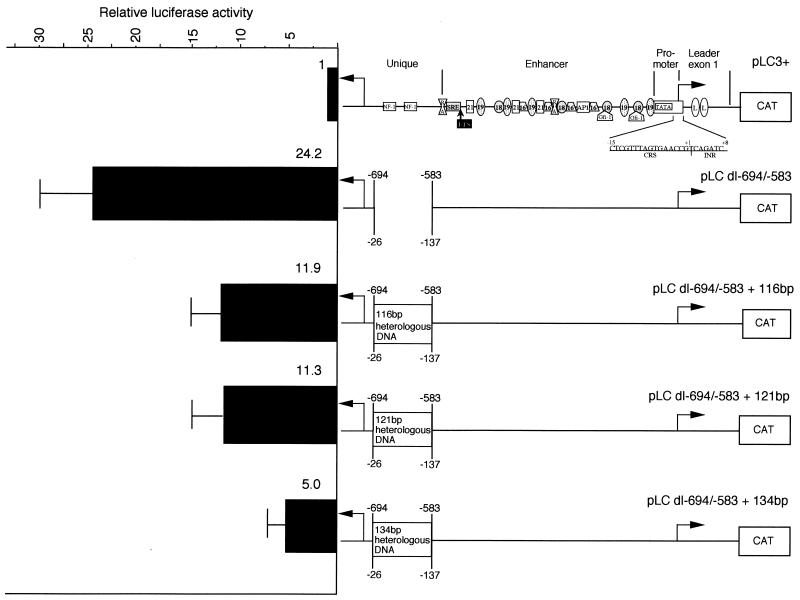
We constructed expression plasmids containing the MIE enhancer and unique region flanked by the divergent MIE promoter and putative UL127 promoter, respectively, for the expression of the CAT gene or the LUX gene, respectively (Fig. 2). The cis-acting transcription elements in repeat motifs of 18, 19, and 21 bp, which contain NF-κB/rel, CREB/ATF, and SP-1 binding sites, respectively, and the upstream RA, SRE, and ETS sites have been described previously (52, 80). The CTF/NF-1 binding sites in the unique region have also been described elsewhere (29). Since the start site of the MIE promoter was mapped precisely, all designations were relative to this start site, which was designated +1 (77). The TATA box sequence upstream of the UL127 ORF was between −703 and −697. We deleted 111 bp from −694 to −583 of the unique region and replaced the 111-bp region with nonregulatory stuffer DNAs of 116, 121, or 134 bp as diagramed in Fig. 2. The transcription start site from the putative UL127 promoter had not been mapped, and consequently, the region from −694 to −583 was estimated to be between approximately −26 and −137 upstream of the UL127 start site based on the data obtained for Fig. 4A. HFFs were transfected with 10 μg of the various expression plasmids, and the level of CAT from the MIE promoter or of LUX from the putative UL127 promoter was determined per microgram of protein lysate as described in Materials and Methods.
FIG. 2.
Relative LUX activity from the UL127 promoter in transiently transfected HFFs. CAT or LUX activity per microgram of protein was measured from the MIE promoter or the divergent UL127 promoter, respectively, as described in Materials and Methods. The cis-acting transcription factor binding sites in repeat and nonrepeat sequences have been described elsewhere (52, 79, 80, 82, 83). Plasmids containing wild-type viral DNA sequence, deleted DNA sequence, or substituted nonregulatory stuffer DNA sequence are indicated relative to the MIE promoter transcription start site. The approximate positions of the mutations relative to the putative UL127 transcription start are also indicated. CAT activity was similar for all expression plasmids as described in the text.
FIG. 4.
Transcription from the MIE promoter or the UL127 promoter at various times after infection with recombinant viruses. Cytoplasmic RNA was isolated from either mock-infected or infected cells treated with CH or PAA or untreated for IE, early, or late times after infection, respectively. (A) RNase protection assay of IE1 or UL127-CAT RNA harvested at 6 h p.i. from nontreated or CH-treated cells. Lanes: 1, standard molecular weight markers (std) (in thousands); 2, mock-infected cell RNA; 3 to 7 and 8 to 12, infected cell RNA from RdlMCATWT, -116, -121, -134, and -Theta, respectively. A map of the riboprobe and the protected fragment is shown. nt, nucleotide; AS, antisense riboprobe. (B) RNase protection assay of IE1 or UL127-CAT RNA harvested at 24 or 48 h p.i. from nontreated or PAA-treated cells. Lanes: 1, IE1 probe without RNase; 2, standard molecular weight markers (std) (in thousands); 3, CAT-UL127 probe without RNase; 4, mock-infected cell RNA; 5 to 7, 8 to 10, and 11 to 13, infected cell RNA from RdlMCATWT, -121, and -Theta, respectively. CAT-UL127 and IE1 RNase-protected RNA are indicated.
All deletions or substitutions between −694 and −583 had no detectable effect on CAT expression from the MIE promoter (data not shown). In contrast, LUX activity increased significantly after the 111-bp deletion or heterologous DNA replacement of 116, 121, or 134 bp (Fig. 2). In each case, the level of LUX expression was significantly higher relative to expression from plasmid pLC3+, which contained wild-type DNA sequence upstream of the putative UL127 promoter. We conclude that the UL127 promoter-like element is functional in transient-transfection experiments in HFFs when sequences within the unique region are deleted or replaced with heterologous DNA.
Recombinant viruses lacking 111 bp of the unique region.
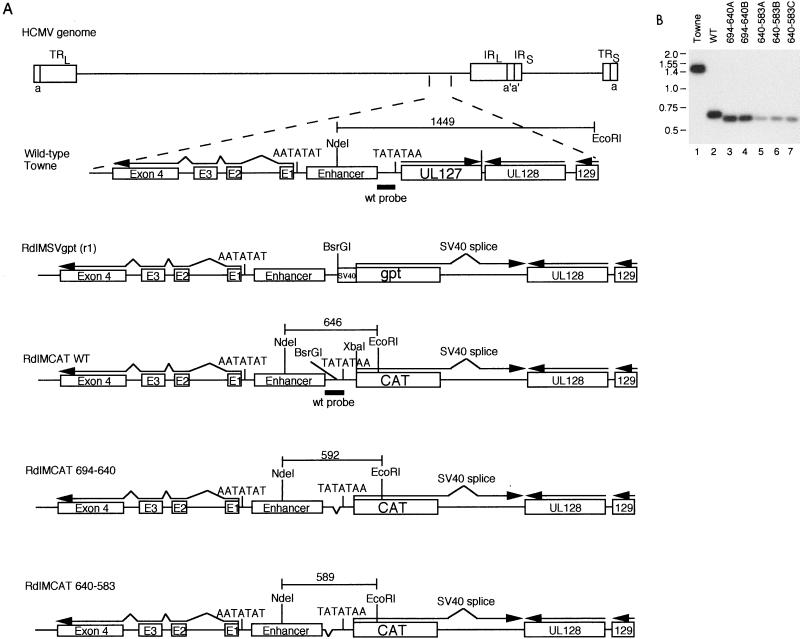
To test whether the 111 bp of the wild-type unique region had an effect on transcription from the UL127 promoter in the context of the viral genome, recombinant viruses with deletions and replacements of this 111-bp region were isolated and characterized. To facilitate recombinant virus characterization and in the event that overexpression of the UL127 ORF was deleterious to viral replication, the UL127 gene was replaced with the CAT gene. All recombinant viruses were derived from RdlMSVgpt(r1) (Fig. 1) by homologous recombination with the shuttle vectors described in Materials and Methods. We selected against the gpt gene product by using Lesch-Nyhan human fibroblasts and 6-thioguanine-containing medium as described previously (26). To characterize the recombinant viruses, viral DNAs were digested with restriction endonucleases EcoRI and NdeI and analyzed by Southern blot hybridization. The sizes of the expected DNA fragments are indicated in Fig. 3A. The radioactively labeled probes were either wild-type DNA from the unique region or heterogeneous DNA from plasmid pCAT-basic. The black bars in Fig. 3A indicate the region to which the 32P-DNA probes hybridize. DNA probes of 116 and 134 bp, which were isolated from the CAT gene of pCAT-basic, were expected to hybridize to the region of substitution between −694 and −583 as well as to the CAT gene inserted into the virus by homologous recombination. The heterogeneous DNA isolated from the region upstream of the CAT gene in plasmid pCAT-basic for the 121-bp DNA was expected to hybridize only to the region of heterogeneous DNA substitution between −694 and −583. The wild-type DNA probe was expected to hybridize to DNA fragments of approximately 1,400, 650, and 550 bp after restriction endonuclease digestion of wild-type Towne strain, RdlMCATWT, and RdlMCATTheta DNAs. RdlMCATTheta has the UL127 promoter deleted. All DNA fragments obtained by EcoRI and NdeI restriction endonuclease digestion were of the appropriate size (Fig. 3B) as predicted in the diagram of Fig. 3A. A longer exposure of the autoradiogram was required to detect hybridization of the 116-bp probe to the CAT gene DNA fragment from RdlMCAT121. From two separate transfections with the shuttle vectors and the infectious RdlMSVgpt(r1) DNA, two to three recombinant viruses of each type were plaque purified and characterized as described above. The heterologous DNA substitutions caused no discernible phenotypic differences in viral growth or plaque size (data not shown).
FIG. 3.
Recombinant viruses with wild-type (wt or WT) or mutant DNA sequences upstream of the UL127 promoter. All recombinant viruses containing the CAT gene were derived from RdlMSVgpt(r1) by selection against the gpt gene product as described in Materials and Methods. (A) Diagram of the recombinant viruses with the MIE promoter and the downstream IE1 gene and the divergent UL127 promoter and the downstream CAT gene. Both promoters have identical TATA boxes. Sequences upstream of the UL127 promoter with either wild type or deleted and replaced with nonregulatory DNA sequence of either 116, 121, or 134 bp. The DNA fragment containing the UL127 promoter is deleted in recombinant virus RdlMCATTheta. The origins of probes for detection of wild-type and 116-, 121-, or 134-bp heterologous DNAs are designated. The predicted sizes of DNA fragments after EcoRI and NdeI restriction endonuclease digestion of recombinant viral DNAs are indicated in base pairs. HCMV, human CMV. SV40, simian virus 40. (B) Southern blot hybridization of wild-type (WT) Towne strain DNA and recombinant virus DNA digested with restriction endonucleases EcoRI and NdeI. 32P-labeled probes of wild-type and 116-, 121-, and 134-bp heterologous DNA sequences are indicated. Lanes: 1 to 6, viral DNAs from human CMV Towne strain and RdlMCATWT, -Theta, -116, -121, and -134, respectively; 1 to 4 (at right), viral DNA from human CMV Towne strain and RdlMCATWT, -Theta, and -134, respectively. Standard molecular size markers are indicated in base pairs.
Effect of a 111-bp deletion and replacement of the unique region in recombinant viruses.
To test the effect of the unique region between −694 and −583 on transcription from the MIE promoter or the putative UL127 promoter in the context of the viral genome, HFFs were infected at approximately 5 PFU per cell with recombinant viruses RdlMCATWT, -116, -121, -134, and -Theta. Cytoplasmic RNA was harvested at various times after infection from cells treated or not treated with CH or PAA. The viral RNAs were detected by RNase protection assay with antisense IE1 or CAT-UL127 riboprobes as described in Materials and Methods.
The mutation between −694 and −583 had little to no effect on IE1 transcription (Fig. 4A, lanes 3 to 7) or on IEUS3 transcription (data not shown). In the presence of CH, there was no transcription from the UL127 promoter with or without substitutions in the unique region (Fig. 4A, lanes 3 to 7). In contrast, there was significant transcription in the absence of CH from the UL127 promoter at 6 h postinfection (p.i.) with RdlMCAT116, -121, and -134 when the wild-type sequence between −694 and −583 was not present (Fig. 4A, lanes 9 to 11). There was little to no detectable transcription from RdlMCATWT containing wild-type sequence, and as expected, there was no transcription from RdlMCATTheta, which has the UL127 promoter deleted (Fig. 4A, lanes 8 and 12). Assuming that transcription initiation was approximately 25 bp downstream of the putative UL127 promoter, a protected RNA band of approximately 175 nucleotides was expected (Fig. 4A). Only the recombinant viruses with the 111-bp deletion of the unique region had a protected RNA of approximately 175 nucleotides. Although the transcription start site downstream of the UL127 promoter was not precisely mapped, the data suggest that a deletion in the unique region between −694 and −583 allows for transcription from the UL127 promoter at early times after infection. The increase in the steady-state level of RNA from the UL127 promoter was approximately 20-fold or greater at 6 h after infection relative to the wild-type recombinant virus RdlMCATWT. Why IE transcription from the UL127 promoter in the presence of CH was blocked requires further investigation.
Cytoplasmic RNA was also harvested at early and late times after infection. Transcription from the UL127 promoter was detected only when the unique region of 111 bp was deleted and replaced with heterologous DNA of 121 bp (Fig. 4B, lanes 6, 9, and 12) or 116 or 134 bp (data not shown). There was little to no detectable transcription from the UL127 promoter with RdlMCATWT (Fig. 4B, lanes 5, 8, and 11). As expected, there was also little to no detectable transcription when the promoter was deleted (Fig. 4B, lanes 7, 10, and 13). We conclude that the 111-bp unique region upstream of the UL127 promoter has a repressive effect on transcription from the UL127 promoter in HFFs at all times after infection.
Expression of the CAT gene downstream of the UL127 promoter.
We had demonstrated previously that the UL127 gene is nonessential for replication in HFFs (51). The above results demonstrate that UL127 is not expressed in HFFs. After infection of HFFs with RdlMCATWT, -116, -121, -134, and -Theta at approximately 5 PFU per cell, infected cells were harvested and analyzed for CAT expression at various times after infection. CAT activity was determined as percent acetylation per microgram of protein as described in Materials and Methods.
There were very low levels of CAT activity with either RdlMCATWT or RdlMCATTheta that did not increase with time after infection (Fig. 5). In contrast, the level of CAT activity with the recombinant viruses lacking the 111 bp of wild-type DNA sequence in the unique region was approximately 90- to 245-fold higher at 48 h p.i. Since the CAT mRNA is relatively unstable in mammalian cells and the CAT protein is very stable (24), these fold increases presumably represent the cumulative activity of the UL127 promoter in the infected HFFs. The different levels of CAT activity with the recombinant viruses containing different replacement DNAs reflect slight differences in the multiplicities of infection that are amplified days after infection. We conclude that the viral UL127 promoter can express a downstream gene in infected HFFs when the wild-type DNA in the unique region is not present.
FIG. 5.
Expression of the CAT gene downstream of the UL127 promoter at various times after infection with recombinant viruses containing wild-type (WT), promoterless, or substituted heterologous DNA sequence upstream of the UL127 promoter. The recombinant viruses are as described for Fig. 3. CAT activity was determined as percent acetylation per microgram of protein-infected cell lysate as described in Materials and Methods.
Effect of approximately 50-bp deletions in the unique region.
To further characterize the repressive region, deletions from −694 to −640 or −640 to −583 were made. These viruses were derived by homologous recombination between shuttle vectors and infectious viral DNA from recombinant virus RdlMSVgpt(r1) and by selection against gpt-expressing virus in infected Lesch-Nyhan cells as described in Materials and Methods. After plaque purification of the recombinant viruses, the viral DNAs were isolated and digested with restriction endonucleases EcoRI and NdeI and analyzed by Southern blot hybridization. Figure 6A is a diagram of the recombinant viruses, and the expected viral DNA fragment sizes produced after digestion with EcoRI and NdeI are indicated. The approximately 50-bp deletions should generate DNA fragments of 592 bp for the −694 to −640 deletion and 589 bp for the −640 to −583 deletion. Wild-type DNA probe from −686 to −583 should hybridize to either DNA fragment after EcoRI and NdeI digestion of the recombinant viral DNAs.
FIG. 6.
Recombinant viruses with approximately 50-bp deletions upstream of the UL127 promoter. All recombinant viruses containing the CAT gene were derived from RdlMSVgpt(r1) by selection against the gpt gene product as described in Materials and Methods. (A) Diagram of the recombinant viruses with the MIE promoter and the downstream IE1 gene and the divergent UL127 promoter and the downstream CAT gene. Approximately 50-bp deletions between −694 and −640 or −640 and −583 are indicated. The origin of a probe for detection of wild-type (WT or wt) DNA is designated. The predicted sizes of DNA fragments after EcoRI and NdeI restriction endonuclease digestion of recombinant viral DNAs are indicated in base pairs. HCMV, human CMV. SV40, simian virus 40. (B) Southern blot hybridization of wild-type Towne strain DNA and recombinant viral DNAs digested with restriction endonucleases EcoRI and NdeI and fractionated by agarose gel electrophoresis as described in Materials and Methods. Lanes: 1, human CMV Towne strain; 2, RdlMCATWT; 3, RdlMCAT694-640A; 4, RdlMCAT694-640B; 5, RdlMCAT640-583A; 6, RdlMCAT640-583B; 7, RdlMCAT640-583C. Numbers at left indicate molecular sizes in kilobases.
Figure 6B is the result of the Southern blot hybridization. The data suggest that the recombinations occurred as predicted. Because the region between −694 and −640 is AT rich, the probe did not hybridize as efficiently to the DNA fragments generated from recombinant viruses RdlMCAT640-583A, -B, and -C. Southern blot hybridizations of RdlMCAT694-640C, RdlMCAT694-640D, and RdlMCAT640-583D are not shown.
After infection of HFFs with recombinant viruses RdlMCATWT, RdlMCAT694-640, and RdlMCAT640-583 at approximately 5 PFU per cell, cytoplasmic RNA was harvested at 6 h p.i. and analyzed by RNase protection assay as described in Materials and Methods. Again, deletions in this region of the viral genome did not substantially alter transcription from the MIE promoter (Fig. 7, lanes 3 to 9). Recombinant virus RdlMCATWT, which has wild-type DNA sequence in the unique region, had little to no detectable RNA from the UL127 promoter (Fig. 7, lane 3). Deletion of the proximal region (−694 to −640) resulted in a significant increase (17- to 25-fold) in steady-state RNA from the UL127 promoter at 6 h after infection (Fig. 7, lanes 4 and 7). Deletions in the distal region (−640 to −583) also increased UL127 promoter activity, but the increase was significantly less (two- to threefold) at 6 h after infection (Fig. 7, lanes 8 and 9). These data suggest that both the proximal and the distal regions have a repressive effect on the UL127 promoter activity but that the major effect is due to the proximal region.
FIG. 7.
Effect of either the proximal or distal deletions of approximately 50 bp on transcription from the UL127 promoter at early times after infection. Infected cell RNA was isolated at 6 h p.i. and analyzed as described in the legend to Fig. 4. Lanes: 1, standard molecular weight markers (std) (in thousands); 2, mock-infected cell RNA; 3, infected cell RNA from RdlMCATWT; 4 to 7, infected cell RNA from RdlMCAT694-640A, -B, -C, and -D, respectively; 8 and 9, infected cell RNA from RdlMCAT640-583A and -C, respectively; 10, IE1 probe not treated with RNase; 11, CAT-UL127 probe not treated with RNase; CAT-UL127 and IE1 RNase-protected RNAs are indicated.
The cumulative effect of the approximately 50-bp deletions in the unique region on downstream expression at various times after infection was tested by determining the amount of CAT gene product. After infection of HFFs with RdlMCATWT, RdlMCAT694-640A, RdlMCAT694-640B, RdlMCAT640-583A, and RdlMCAT640-583B at approximately 5 PFU per cell, infected cells were harvested and analyzed for CAT expression at various times after infection as described in Materials and Methods.
The proximal deletion of the negative regulatory region between −694 and −640 increased the cumulative level of CAT expression from approximately 75- to 118-fold at various times after infection (Fig. 8). The distal deletion of the negative regulatory region between −640 and −583 increased the cumulative level of CAT expression from approximately 9- to 12-fold at various times after infection (Fig. 8). The cumulative level of CAT expression increased with time after infection with either RdlMCAT694-640 or RdlMCAT640-583. The proximal deletion had approximately a 10-fold-more significant effect on downstream CAT expression. The data suggest that both the proximal and the distal regions repress transcription but that the proximal region has the strongest repressive effect on expression from the UL127 promoter.
FIG. 8.
Expression of the CAT gene downstream of the UL127 promoter at various times after infection with recombinant viruses containing proximal or distal deletions of approximately 50 bp upstream of the UL127 promoter. The recombinant viruses are as described for Fig. 6. CAT activity was determined as percent acetylation per microgram of protein of infected cell lysate as described in Materials and Methods.
DISCUSSION
In the human CMV genome, like the murine CMV genome, the MIE enhancer is flanked by divergent promoters (Fig. 1). With human CMV, efficient transcription occurs from only the MIE promoter at +1 in permissive HFFs and not from the divergent promoter upstream of the UL127 gene. In contrast, the murine CMV MIE enhancer influences transcription from both divergent promoters in permissive cells. We hypothesized that human CMV is different from murine CMV because human CMV has regions that prevent divergent transcription in permissive cells. After extensive deletion analysis and transient-transfection assays in HFFs, we located a regulatory region between the UL127 TATA box and the MIE enhancer. When the wild-type sequences between −694 and −583 were deleted or replaced with nonregulatory stuffer DNAs, we found that the UL127 promoter was now driving transcription. Deletion of this region caused a significant increase in the basal activity of the divergent UL127 promoter in transient-transfection assays but had no effect on the MIE promoter.
In cells infected with recombinant viruses, the UL127 promoter was repressed by the wild-type sequences at all times after infection and was activated only when the wild-type sequence was replaced with different nonregulatory stuffer DNAs. Although the UL127 promoter was not converted to an IE promoter, it was a highly inducible early promoter that required viral protein synthesis for activation. Subsequent deletion analysis indicated that the region immediately upstream of the UL127 TATA box, between −694 and −640, had the major negative regulatory effect on transcription from the UL127 promoter at early times after infection.
DNase I footprinting assays of the unique region that lies upstream of the MIE enhancer have detected the binding of cellular factors present in transcriptionally active nuclear extracts (21). Since chromatin structure is known to have a role in transcriptional regulation, one or more of these cellular DNA binding proteins may repress transcription from the UL127 promoter at early times after infection. As a result, the region could constitute a boundary between the UL127 promoter and the MIE enhancer that functions to repress transcription from the UL127 promoter at early times after infection of permissive HFFs. Deletion of 54 bp between −694 and −640 had the most significant effect on transcription from the UL127 promoter at early times. Deletion of 57 bp between −640 and −583 had less of an effect on transcription from the UL127 promoter. These two regions were referred to as the proximal and distal negative regulatory regions, respectively.
Whether expression of the UL127 ORF is repressed when wild-type virus infects cells that favor latency is presently not known. It is tempting to speculate that expression of the UL127 gene is latency specific like that of the UL126 gene (39). In this regard, our analysis also detects little to no initiation of transcription from the −356 and −292 latency-associated transcription start sites for expression of the UL126 gene during productive infection of HFFs (50). An examination of the proximal and distal regions upstream of the UL127 promoter identified several cis sites for the binding of known transcriptional activator or repressor proteins. Within both the proximal and distal human CMV unique regions are NF-1 binding sites (Fig. 2). The NF-1 DNA binding protein family has been implicated in both positive and negative regulation of cellular and viral promoters (19, 35, 58, 59, 61). Our additional deletion analysis indicates that the NF-1 sites do not regulate repression of transcription from the UL127 promoter (data not shown). Immediately upstream of the UL127 TATA box between −690 and −680 is a consensus binding site for members of the winged-helix transcriptional regulatory protein family (Fig. 9). Winged-helix proteins are a family of transcriptional activator and repressor proteins found in mammalian cells. Winged-helix proteins are preferentially expressed in undifferentiated stem cells and not in differentiated cells (84). Therefore, it is possible that the winged-helix transcriptional activator may induce expression of the UL127 gene in the myeloid cell-committed progenitors. There are also winged-helix repressor proteins. The winged-helix repressor protein termed genesis (HFH-2) is a critical mammalian cell-regulatory factor in embryonic differentiation (84). HFH-2 protein binding sites are found upstream of a number of developmentally regulated promoters, and HFH-2 expression is restricted to primitive embryonic stem cells (84). Since winged-helix activators and repressors are found preferentially in undifferentiated cells, whether this site has a role in repression of the UL127 promoter in differentiated HFF cells remains to be determined. There is also a near-consensus suppressor of hairy wing (suHW) binding site in the proximal region (Fig. 9). In Drosophila melanogaster, suHW can block the effect of an enhancer on a promoter (73). Although there are mammalian cell proteins related to suHW, their role as repressors of transcription is not understood. In addition, within both the proximal and the distal unique regions, there are human papillomavirus type 16 (HPV-16) silencing motifs (Fig. 9). This element was identified as a 9-bp sequence with a critical core sequence that binds an unknown human transcriptional repressor protein (57). The element is located within the HPV-16 origin of DNA replication between the HPV-16 enhancer and the E6/E7 promoter. In transient-transfection assays, two elements are necessary for efficient repression of the E6/E7 promoter (57).
FIG. 9.
Diagram of the region between the UL127 promoter and the MIE enhancer demonstrating putative binding sites for winged-helix transcriptional repressor, NF-1, suHW, and the HPV silencing motif (PSM).
It is possible that a combination of different host cell repressor proteins binds cooperatively to the unique region of human CMV to prevent transcription from the UL127 promoter in permissive HFFs. This region of the human CMV genome may serve as a boundary to suppress the transcription-activating effects of the extremely strong MIE enhancer region from the UL127 promoter and to inhibit the effects of the viral IE transactivators on this promoter.
When the human CMV unique region was deleted, the UL127 promoter behaved like an early viral promoter. The viral IE1 and IE2 gene products are regulatory proteins that enhance transcription from both cellular and viral promoters (7, 20, 31, 33, 48, 55, 76). In recombinant virus-infected cells, we assume that the viral gene products of IE1 and IE2 and possibly UL36- to -38 and TRS1/IRS1 were necessary for the activation of this promoter since the UL127 promoter was not activated without de novo viral protein synthesis.
A viral factor that controls the activity of the MIE promoter is the IE2 gene product, IE86 (46, 47). The viral autoregulatory protein binds to the crs (Fig. 2) and inhibits transcription from the MIE promoter (46, 47). Maximum expression of the IE1 and IE2 genes occurs within 5 to 6 h after infection. In the recombinant viruses with wild-type sequence upstream of the UL127 promoter, the autoregulation at the crs of the MIE promoter was not sufficient to shift the effects of the MIE enhancer to the divergent UL127 promoter. In contrast, recombinant viruses lacking the wild-type regulatory sequence had very strong transcription from the UL127 promoter at 5 to 6 h after infection. Compared to IE1 transcription, which served as an internal control, the UL127 promoter was strongly activated relative to other early viral promoters such as UL4 (data not shown). Our data suggest that elements at the 5′ end of the MIE enhancer and the unique region bind repressor proteins. The putative viral or cellular repressor proteins that bind to the unique region may dominate both the effects of the MIE enhancer and the viral transactivators that activate early viral promoters.
There are several proposed repressor elements within the MIE enhancer that were detected by transient-transfection assays. These include the YY1 sites (41, 45, 68), Gfi-1 sites (92), and the RA sites (80) (Fig. 2). One or more of these elements could affect the IE transcription from the MIE promoter as well as the UL127 promoter. However, the functionality of these elements in the context of the viral genome has not been tested. In addition, the region between −1140 and −750 was termed the modulator because the region differently affected transcription from the MIE promoter in various cell types in transient-transfection experiments (34, 56, 65). However, when this region was deleted in the context of the viral genome, there was no detectable effect on transcription from the MIE promoter in different cell types infected with the recombinant viruses (51). With the recombinant viruses used in this study, the modulator, which also contains the UL127 ORF, was replaced with an indicator gene to facilitate characterization of the regulatory elements upstream of the UL127 promoter. Therefore, it was not determined whether the modulator also affects transcription from the UL127 promoter at IE or early times after infection.
We propose that the unique region of the viral genome between the UL127 promoter and MIE enhancer contains sites for the binding of either viral or cellular repressor proteins. While we have discussed possible cellular protein binding sites, we have not eliminated viral protein binding sites. Viral proteins could be bound to the unique region as the viral genome enters the nucleus, or cellular proteins could bind to the unique region as the viral DNA enters the nucleus. Alternatively, the unique region could serve to anchor the human CMV genome to matrix proteins associated with transcription domains within the nucleus. The type of cellular proteins that bind to this region may differ depending on the cell type. The nature of the viral or cellular proteins that bind to this regulatory region remains to be determined.
ACKNOWLEDGMENTS
We thank members of the laboratories for helpful discussion and Richard Roller for critical reading of the manuscript. We are grateful to Philip Lashmit and Jonathan Pruessner for assistance.
This work was supported by grants AI-13562 (M.F.S.) and AI-40130 (J.L.M.) from the National Institutes of Health and by a Burroughs Wellcome Young Investigator Award (J.L.M.).
REFERENCES
- 1.Alford C A, Britt W J. Cytomegalovirus. In: Fields B N, Knipe D M, et al., editors. Fields virology. New York, N.Y: Raven Press, Ltd.; 1990. pp. 1981–2010. [Google Scholar]
- 2.Anders D G, McCue L A. The human cytomegalovirus genes and proteins required for DNA synthesis. Intervirology. 1996;39:378–388. doi: 10.1159/000150508. [DOI] [PubMed] [Google Scholar]
- 3.Angulo A, Messerle M, Koszinowski U H, Ghazal P. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J Virol. 1998;72:8502–8509. doi: 10.1128/jvi.72.11.8502-8509.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angulo A, Suto C, Boehm M F, Heyman R A, Ghazal P. Retinoid activation of retinoic acid receptors but not of retinoid X receptors promotes cellular differentiation and replication of human cytomegalovirus in embryonal cells. J Virol. 1995;69:3831–3837. doi: 10.1128/jvi.69.6.3831-3837.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bednarczuk T A, Wiggins R C, Konat G W. Generation of high efficiency DNA hybridization probes by PCR. BioTechniques. 1991;10:478. [PubMed] [Google Scholar]
- 6.Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 7.Burns L J, Waring J F, Reuter J J, Stinski M F, Ginder G D. Only the HLA class I gene minimal promoter elements are required for transactivation by human cytomegalovirus immediate early genes. Blood. 1993;81:1558–1566. [PubMed] [Google Scholar]
- 8.Chan Y-J, Chiou C-J, Huang Q, Hayward G S. Synergistic interactions between overlapping binding sites for the serum response factor and ELK-1 proteins mediate both basal enhancement and phorbol ester responsiveness of primate cytomegalovirus major immediate-early promoters in monocyte and T-lymphocyte cell types. J Virol. 1996;70:8590–8605. doi: 10.1128/jvi.70.12.8590-8605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang Y N, Crawford S, Stall J, Rawlins D R, Jeang K T, Hayward G S. The palindromic series I repeats in the simian cytomegalovirus major immediate-early promoter behave as both strong basal enhancers and cyclic-AMP response elements. J Virol. 1990;64:264–277. doi: 10.1128/jvi.64.1.264-277.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chee M S, Bankier S, Beck S, Bohni R, Brown C R, Horsnell T, Hutchison III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 11.Cherrington J M, Khoury E L, Mocarski E S. Human cytomegalovirus ie2 negatively regulates α gene expression via a short target sequence near the transcription start site. J Virol. 1991;65:887–896. doi: 10.1128/jvi.65.2.887-896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 13.Colberg-Poley A M. Functional roles of immediate early proteins encoded by the human cytomegalovirus UL36-38, UL115-119, TRS1/IRS1 and US3 loci. Intervirology. 1996;39:350–360. doi: 10.1159/000150506. [DOI] [PubMed] [Google Scholar]
- 14.Dery C V, Toth M, Brown M, Horvath J, Allaire S, Weber J M. The structure of adenovirus chromatin in infected cells. J Gen Virol. 1985;66:2671–2684. doi: 10.1099/0022-1317-66-12-2671. [DOI] [PubMed] [Google Scholar]
- 15.Deshmane S L, Fraser N W. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J Virol. 1989;63:943–947. doi: 10.1128/jvi.63.2.943-947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Wet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorsch-Hasler K, Keil G M, Weber F, Schaffner J M, Koszinowski U H. A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate early mRNA in murine cytomegalovirus. Proc Natl Acad Sci USA. 1985;82:8325–8329. doi: 10.1073/pnas.82.24.8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyson P J, Farrell P J. Chromatin structure of Epstein Barr virus. J Gen Virol. 1985;66:1931–1940. doi: 10.1099/0022-1317-66-9-1931. [DOI] [PubMed] [Google Scholar]
- 19.Eskild W, Simard J, Hansson V, Guerin S. Binding of a member of the NF1 family of transcription factors to two distinct cis-acting elements in the promoter and 5′-flanking region of the human cellular retinol binding protein 1 gene. Mol Endocrinol. 1994;8:732–745. doi: 10.1210/mend.8.6.7935489. [DOI] [PubMed] [Google Scholar]
- 20.Geist L J, Monick M M, Stinski M F, Hunninghake G W. The immediate early genes of human cytomegalovirus upregulate expression of the interleukin-2 and interleukin-2 receptor genes. Am J Respir Cell Mol Biol. 1991;5:292–296. doi: 10.1165/ajrcmb/5.3.292. [DOI] [PubMed] [Google Scholar]
- 21.Ghazal P, Lubon H, Reynolds-Kohler C, Hennighausen L, Nelson J A. Interactions between cellular regulatory proteins and a unique sequence region in the human cytomegalovirus major immediate-early promoter. Virology. 1990;174:18–25. doi: 10.1016/0042-6822(90)90049-w. [DOI] [PubMed] [Google Scholar]
- 22.Ghazal P, Nelson J A. Enhancement of RNA polymerase II initiation complexes by a novel DNA control domain downstream from the cap site of the cytomegalovirus major immediate-early promoter. J Virol. 1991;65:2299–2307. doi: 10.1128/jvi.65.5.2299-2307.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson W. Structure and assembly of the virion. Intervirology. 1996;39:389–400. doi: 10.1159/000150509. [DOI] [PubMed] [Google Scholar]
- 24.Gorman C M, Moffatt L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham F L, van der Eb A J. A new technique for the assay of infectivity of adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 26.Greaves R F, Brown J M, Vieira J, Mocarski E S. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using the E. coli guanosine phosphoribosyl transferase (gpt) gene. J Gen Virol. 1995;76:2151–2160. doi: 10.1099/0022-1317-76-9-2151. [DOI] [PubMed] [Google Scholar]
- 27.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn G, Jores R, Mocarski E S. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci USA. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hennighausen L, Fleckenstein B. Nuclear factor 1 interacts with five DNA elements in the promoter region of the human cytomegalovirus major immediate early gene. EMBO J. 1986;5:1367–1371. doi: 10.1002/j.1460-2075.1986.tb04368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermiston T W, Malone C L, Stinski M F. Human cytomegalovirus immediate-early two protein region involved in negative regulation of the major immediate-early promoter. J Virol. 1990;64:3532–3536. doi: 10.1128/jvi.64.7.3532-3536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho M. Cytomegalovirus: biology and infection. New York, N.Y: Plenum Publishing Corp.; 1991. [Google Scholar]
- 33.Huang L, Malone C L, Stinski M F. A human cytomegalovirus early promoter with upstream negative and positive cis-acting elements: IE2 negates the effect of the negative element, and NF-Y binds to the positive element. J Virol. 1994;68:2108–2117. doi: 10.1128/jvi.68.4.2108-2117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang T H, Oka T, Asai T, Okada T, Merrills B W, Gerston P N, Whitson R N, Itakura K. Repression by a differentiation-specific factor of the human cytomegalovirus enhancer. Nucleic Acids Res. 1996;24:1695–1701. doi: 10.1093/nar/24.9.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jahroudi N, Ardekani A M, Greenberger J S. An NF1-like protein functions as a repressor of the von Willebrand factor promoter. J Biol Chem. 1996;271:21413–21421. doi: 10.1074/jbc.271.35.21413. [DOI] [PubMed] [Google Scholar]
- 36.Jain V K, Morgan I T, Shimada T. Dual reporter vectors for determination of activity of bidirectional promoters. BioTechniques. 1992;12:681–683. [PubMed] [Google Scholar]
- 37.Klucher K M, Sommer M, Kadonaga J T, Spector D H. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol Cell Biol. 1993;13:1238–1250. doi: 10.1128/mcb.13.2.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondo K, Kaneshima H, Mocarski E S. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondo K, Xu J, Mocarski E S. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc Natl Acad Sci USA. 1996;93:11137–11142. doi: 10.1073/pnas.93.20.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kondoleon S K, Kurkinen N A, Hallick L M. The SV40 nucleosome-free region is detected throughout the virus life cycle. Virology. 1989;173:129–135. doi: 10.1016/0042-6822(89)90228-6. [DOI] [PubMed] [Google Scholar]
- 41.Kothari S K, Baillie J, Sissons J G P, Sinclair J H. The 21 bp repeat element of the human cytomegalovirus major immediate early enhancer is a negative regulator of gene expression in undifferentiated cells. Nucleic Acids Res. 1991;19:1767–1771. doi: 10.1093/nar/19.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krieg P A, Melton D A. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–414. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- 43.Lashmit P E, Stinski M F, Murphy E A, Bullock G C. A cis repression sequence adjacent to the transcription start site of the human cytomegalovirus US3 gene is required to down regulate gene expression at early and late times after infection. J Virol. 1998;72:9575–9584. doi: 10.1128/jvi.72.12.9575-9584.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu B, Hermiston T W, Stinski M F. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu R, Baillie J, Sissons J G P, Sinclair J H. The transcription factor YY1 binds to negative regulatory elements in the human cytomegalovirus major immediate early enhancer/promoter and mediates repression in nonpermissive cells. Nucleic Acids Res. 1994;22:2453–2459. doi: 10.1093/nar/22.13.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macias M P, Huang L, Lashmit P E, Stinski M F. Cellular and viral protein binding to a cytomegalovirus promoter transcription initiation site: effects on transcription. J Virol. 1996;70:3628–3635. doi: 10.1128/jvi.70.6.3628-3635.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macias M P, Stinski M F. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc Natl Acad Sci USA. 1993;90:707–711. doi: 10.1073/pnas.90.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marcus-Sekura C J, Carter B J. Chromatin-like structure of adeno-associated virus DNA in infected cells. J Virol. 1983;48:79–87. doi: 10.1128/jvi.48.1.79-87.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meier, J. L., and J. A. Pruessner. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 51.Meier J L, Stinski M F. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J Virol. 1997;71:1246–1255. doi: 10.1128/jvi.71.2.1246-1255.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meier J L, Stinski M F. Regulation of human cytomegalovirus immediate-early gene expression. Intervirology. 1996;39:331–342. doi: 10.1159/000150504. [DOI] [PubMed] [Google Scholar]
- 53.Minton E J, Tysoe C, Sinclair J H, Sissons J G. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mocarski E S, Kemble G, Lyle J, Greaves R F. A deletion mutant in the human cytomegalovirus gene encoding IE1 491aa is replication defective due to a failure in autoregulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monick M M, Geist L J, Stinski M F, Hunninghake G W. The immediate early genes of human cytomegalovirus upregulate expression of the cellular genes myc and fos. Am J Respir Cell Mol Biol. 1992;7:251–256. doi: 10.1165/ajrcmb/7.3.251. [DOI] [PubMed] [Google Scholar]
- 56.Nelson J A, Reynolds-Kohler C, Smith B. Negative and positive regulation by a short segment in the 5′-flanking region of the human cytomegalovirus major immediate-early gene. Mol Cell Biol. 1987;7:4125–4129. doi: 10.1128/mcb.7.11.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Connor M J, Stunkel W, Zimmermann H, Koh C-H, Bernard H-U. A novel YY1-independent silencer represses the activity of the human papillomavirus type 16 enhancer. J Virol. 1998;72:10083–10092. doi: 10.1128/jvi.72.12.10083-10092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osada S, Daimon S, Ikeda T, Nishihara T, Yano K, Yamasaki M, Imagawa M. Nuclear factor 1 family proteins bind to the silencer element in the rat glutathione transferase P gene. J Biochem. 1997;121:355–363. doi: 10.1093/oxfordjournals.jbchem.a021595. [DOI] [PubMed] [Google Scholar]
- 59.Osada S, Ikeda T, Xu M, Nishihara T, Imagawa M. Identification of the transcriptional repression domain of nuclear factor 1-A. Biochem Biophys Res Commun. 1997;238:744–747. doi: 10.1006/bbrc.1997.7382. [DOI] [PubMed] [Google Scholar]
- 60.Pizzorno M C, Mullen M A, Chang Y N, Hayward G S. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J Virol. 1991;65:3839–3852. doi: 10.1128/jvi.65.7.3839-3852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy R J, Gosselin P, Anzivino M J, Moore D D, Guerin S L. Binding of a nuclear protein to the rat growth hormone silencer element. Nucleic Acids Res. 1992;20:401–408. doi: 10.1093/nar/20.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samaniego L A, Tevethia M J, Spector D J. The human cytomegalovirus 86-kilodalton immediate-early 2 protein: synthesis as a precursor polypeptide and interaction with a 75-kilodalton protein of probable viral origin. J Virol. 1994;68:720–729. doi: 10.1128/jvi.68.2.720-729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandford G R, Burns W H. Rat cytomegalovirus has a unique immediate early gene enhancer. Virology. 1996;222:310–317. doi: 10.1006/viro.1996.0428. [DOI] [PubMed] [Google Scholar]
- 64.Schmidbauer M, Budka H, Ulrich W, Ambros P. Cytomegalovirus (CMV) disease of the brain in AIDS and connatal infection: a comparative study by histology, immunocytochemistry, and in situ DNA hybridization. Acta Neuropathol (Berlin) 1989;79:286–293. doi: 10.1007/BF00294663. [DOI] [PubMed] [Google Scholar]
- 65.Shelbourn S L, Kothari S K, Sissons J G P, Sinclair J H. Repression of human cytomegalovirus gene expression associated with a novel immediate early regulatory region binding factor. Nucleic Acids Res. 1989;17:9165–9171. doi: 10.1093/nar/17.22.9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinclair J, Sissons P J G. Latent and persistent infections of monocytes and macrophages. Intervirology. 1996;39:293–301. doi: 10.1159/000150501. [DOI] [PubMed] [Google Scholar]
- 67.Sinclair J, Sissons P J G. Human cytomegalovirus: pathogenesis and models of latency. Semin Virol. 1994;53:249–258. [Google Scholar]
- 68.Sinclair J H, Baillie J, Bryant L A, Taylor-Wiedeman J A, Sissons P J G. Repression of human cytomegalovirus major immediate early gene expression in a monocytic cell line. J Gen Virol. 1992;73:433–435. doi: 10.1099/0022-1317-73-2-433. [DOI] [PubMed] [Google Scholar]
- 69.Sinzger C, Grefte A, Plachter B, Gouw A S H, The T H, Jahn G. Fibroblasts, epithelial cells, endothelial cells, and smooth muscle cells are the major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J Gen Virol. 1995;76:741–750. doi: 10.1099/0022-1317-76-4-741. [DOI] [PubMed] [Google Scholar]
- 70.Sinzger C, Plachter B, Grefte A, Gouw A S H, The T H, Jahn G. Tissue macrophages are infected by human cytomegalovirus. J Infect Dis. 1996;173:240–245. doi: 10.1093/infdis/173.1.240. [DOI] [PubMed] [Google Scholar]
- 71.Soderberg-Naucler C, Fish K N, Nelson J A. Interferon-gamma and tumor necrosis factor-alpha specifically induce formation of cytomegalovirus-permissive monocyte-derived macrophages that are refractory to the antiviral activity of these cytokines. J Clin Investig. 1997;100:3154–3163. doi: 10.1172/JCI119871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soderberg-Naucler C, Fish K N, Nelson J A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 73.Spana C, Harrison D A, Corces V G. The Drosophila melanogaster suppressor of hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 1988;2:1414–1423. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- 74.Spector D H. Activation and regulation of human cytomegalovirus early genes. Intervirology. 1996;39:361–377. doi: 10.1159/000150507. [DOI] [PubMed] [Google Scholar]
- 75.Stenberg R M. The human cytomegalovirus major immediate-early gene. Intervirology. 1996;39:343–349. doi: 10.1159/000150505. [DOI] [PubMed] [Google Scholar]
- 76.Stenberg R M, Depto A S, Fortney J, Nelson J. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J Virol. 1989;63:2699–2708. doi: 10.1128/jvi.63.6.2699-2708.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stenberg R M, Thomsen D R, Stinski M F. Structural analysis of the major immediate early gene of human cytomegalovirus. J Virol. 1984;49:190–199. doi: 10.1128/jvi.49.1.190-199.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stenberg R M, Witte P R, Stinski M F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stinski M F. Cytomegalovirus and its replication. In: Knipe D, editor. Virology. 2nd ed. New York, N.Y: Raven Press, Ltd.; 1990. pp. 1959–1980. [Google Scholar]
- 80.Stinski M F. Cytomegalovirus promoter for expression in mammalian cells. In: Fernandez J M, Hoeffler J P, editors. Gene expression systems: using nature for the art of expression. San Diego, Calif: Academic Press, Inc.; 1999. pp. 211–233. [Google Scholar]
- 81.Stinski M F. Human cytomegalovirus: glycoproteins associated with virions and dense bodies. J Virol. 1976;19:594–609. doi: 10.1128/jvi.19.2.594-609.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stinski M F, Macias M P, Malone C L, Thrower A R, Huang L. Regulation of transcription from the cytomegalovirus major immediate early promoter by cellular and viral proteins. In: Michelson S, Plotkin S A, editors. Multidisciplinary approach to understanding cytomegalovirus. Amsterdam, The Netherlands: Elsevier Science Publishers; 1993. pp. 3–12. [Google Scholar]
- 83.Stinski M F, Malone C L, Hermiston T W, Liu B. Regulation of human cytomegalovirus transcription. In: Wagner E K, editor. Herpesvirus transcription and its control. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 240–260. [Google Scholar]
- 84.Sutton J, Costa R, Klug M, Field L, Xu D, Largaespada D A, Fletcher C F, Jenkins N A, Copeland N G, Klemsz M, Hormas R. Genesis, a winged helix transcriptional repressor with expression restricted to embryonic stem cells. J Biol Chem. 1996;271:23126–23133. doi: 10.1074/jbc.271.38.23126. [DOI] [PubMed] [Google Scholar]
- 85.Taylor-Wiedeman J, Sissons J G, Borysiewicz L K, Sinclair J H. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 86.Thomsen D R, Stenberg R M, Goins W F, Stinski M F. Promoter-regulatory region of the major immediate early gene of human cytomegalovirus. Proc Natl Acad Sci USA. 1984;81:659–663. doi: 10.1073/pnas.81.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thrower A R, Bullock G C, Bissell J E, Stinski M F. Regulation of a human cytomegalovirus immediate-early gene (US3) by a silencer-enhancer combination. J Virol. 1996;70:91–100. doi: 10.1128/jvi.70.1.91-100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vayda M E, Rogers A E, Flint S J. The structure of the nucleoprotein cores released from adenovirions. Nucleic Acids Res. 1983;11:441–460. doi: 10.1093/nar/11.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wiley C A, Nelson J A. Role of human immunodeficiency virus and cytomegalovirus in AIDS encephalitis. Am J Pathol. 1988;133:73–81. [PMC free article] [PubMed] [Google Scholar]
- 90.Wiley C A, Schrier R D, Denaro F J, Nelson J A, Lampert P W, Oldstone M B. Localization of cytomegalovirus proteins and genome during fulminant central nervous system infection in an AIDS patient. J Neuropathol Exp Neurol. 1986;45:127–139. doi: 10.1097/00005072-198603000-00003. [DOI] [PubMed] [Google Scholar]
- 91.Wu J, Jupp R, Stenberg R M, Nelson J A, Ghazal P. Site-specific inhibition of RNA polymerase II preinitiation complex assembly by human cytomegalovirus IE86 protein. J Virol. 1993;67:7547–7555. doi: 10.1128/jvi.67.12.7547-7555.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zweidler-McKay P A, Grimes H L, Flubacher M M, Tsichlis P N. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol Cell Biol. 1996;16:4024–4034. doi: 10.1128/mcb.16.8.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]