Abstract
Salivary gland cancers (SGCs) are rare, aggressive cancers without effective treatments when metastasized. We conducted a phase 2 trial evaluating nivolumab (nivo, anti-PD-1) and ipilimumab (ipi, anti-CTLA-4) in 64 patients with metastatic SGC enrolled in two histology-based cohorts (32 patients each): adenoid cystic carcinoma (ACC; cohort 1) and other SGCs (cohort 2). The primary efficacy endpoint (≥4 objective responses) was met in cohort 2 (5/32, 16%) but not in cohort 1 (2/32, 6%). Treatment safety/tolerability and progression-free survival (PFS) were secondary endpoints. Treatment-related adverse events grade ≥3 occurred in 24 of 64 (38%) patients across both cohorts, and median PFS was 4.4 months (95% confidence interval (CI): 2.4, 8.3) and 2.2 months (95% CI: 1.8, 5.3) for cohorts 1 and 2, respectively. We present whole-exome, RNA and T cell receptor (TCR) sequencing data from pre-treatment and on-treatment tumors and immune cell flow cytometry and TCR sequencing from peripheral blood at serial timepoints. Responding tumors universally demonstrated clonal expansion of pre-existing T cells and mutational contraction. Responding ACCs harbored neoantigens, including fusion-derived neoepitopes, that induced T cell responses ex vivo. This study shows that nivo+ipi has limited efficacy in ACC, albeit with infrequent, exceptional responses, and that it could be promising for non-ACC SGCs, particularly salivary duct carcinomas. ClinicalTrials.gov identifier: NCT03172624.
Salivary gland cancers (SGC) are rare, lethal malignancies comprising ~5% of all head and neck cancers and less than 0.5% of cancers overall1. They arise in the major (parotid, submandibular or sublingual) salivary glands, in the minor salivary glands throughout the upper aerodigestive tract or as salivary gland-type cancers of the breast, trachea, lung, skin or cervix2. The World Health Organization recognizes 24 histologic SGC subtypes3, including adenoid cystic carcinoma (ACC) and salivary duct carcinoma (SDC). Upon recurrence or distant metastasis (R/M), SGC is incurable, and patients are treated with palliative intent4,5. There is no consensus standard or FDA-approved treatment for R/M SGC.
The effort to develop biologically rational therapies has been stymied by a limited understanding of SGC biology and paucity of preclinical models. In this context, taking on the inherent challenges of conducting clinical trials for rare diseases is critical not only for evaluating clinical endpoints but also as an opportunity for serial biospecimen collection, to study how SGC tumors change upon interventions and identify potential therapeutic vulnerabilities.
Immune checkpoint blockade (ICB) with antibodies targeting the T cell checkpoint programmed cell death protein 1 (PD-1) or its ligand PD-L1 has activity in multiple R/M tumor types6. However, objective response rates (ORRs) in patients with SGC have been low (4–12% across four trials7–10), perhaps due to the low rate of PD-L1 positivity (23%) in SGCs11. In some solid tumor types, concurrent blockade of the PD-1 and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) checkpoints yields durable responses12–14. The 55% ORR observed upon dual ICB in patients with PD-L1-negative melanoma14 is notable, considering the low PD-L1 expression in most SGCs. Hence, we hypothesized that anti-PD-1/anti-CTLA-4 dual ICB would be efficacious in patients with R/M SGC.
We performed a phase 2 trial evaluating nivolumab (nivo, anti-PD-1) and ipilimumab (ipi, anti-CTLA-4) in patients with R/M SGC. Patients with ACC (cohort 1, n = 32) and non-ACC (cohort 2, n = 32) SGCs were enrolled and analyzed in two cohorts, given the distinct clinical behavior and immunological and genomic profiles of different SGC histologies15–20. The primary endpoint was objective response per Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1 (ref. 21); secondary endpoints were tolerability and progression-free survival (PFS). A key aim for the trial was the serial collection of pre-treatment and on-treatment tumor and peripheral blood biospecimens to investigate the mechanistic basis for ICB efficacy in SGC. This approach allowed us to conduct a comprehensive, integrated analysis linking the clinical efficacy of nivo+ipi with immunogenomic features, patient-specific neoantigens and T cell dynamics. Together, these results shed light on the clinical and molecular determinants of response to nivo+ipi for patients with SGC and may help guide the rational development of more effective therapies for this orphan disease.
Results
Study treatment efficacy and toxicity
From 16 June 2017 to 21 July 2020, 64 patients with R/M SGC were enrolled: 32 in cohort 1 (ACC) and 32 in cohort 2 (non-ACC SGC). Patient characteristics and tumor histologies are shown in Table 1 and Supplementary Table 1. All patients had distant metastases.
Table 1 |. Baseline characteristics of cohort 1 (ACC) and cohort 2 (non-ACC SGC) patients.
| Cohort 1 (n = 32) | Cohort 2 (n = 32) | Overall (n = 64) | |
|---|---|---|---|
| Age, years (min–max) | 58.0 (31–80) | 64.5 (30–87) | 61.5 (30–87) |
| Sex, n (%) | |||
| Female | 19 (59) | 12 (38) | 31 (48) |
| Male | 13 (41) | 20 (63) | 33 (52) |
| Histology, n (%) | |||
| Adenoid cystic carcinoma | 32 (100) | 0 | 32 (50) |
| Salivary duct carcinoma | 0 | 12 (38) | 12 (19) |
| Acinic cell carcinoma | 0 | 7 (22) | 7 (11) |
| Myoepithelial carcinoma | 0 | 3 (9) | 3 (5) |
| Mucoepidermoid carcinoma | 0 | 2 (6) | 2 (3) |
| Unclassified carcinoma ex pleomorphic adenoma | 0 | 2 (6) | 2 (3) |
| Epithelial-myoepithelial carcinoma | 0 | 1 (3) | 1 (2) |
| Cribriform adenocarcinoma of minor salivary gland | 0 | 1 (3) | 1 (2) |
| Secretory carcinoma | 0 | 1 (3) | 1 (2) |
| SWI/SNF-deficient carcinoma with myoepithelial features | 0 | 1 (3) | 1 (2) |
| AR+ high-grade carcinoma NOS | 0 | 1 (3) | 1 (2) |
| Adenocarcinoma NOS | 0 | 1 (3) | 1 (2) |
| ECOG, n (%) | |||
| 0 | 15 (47) | 21 (66) | 36 (56) |
| 1 | 17 (53) | 11 (34) | 28 (44) |
| Primary origin, n (%) | |||
| Major salivary gland | 10 (31) | 28 (88) | 38 (59) |
| Minor salivary gland | 15 (47) | 4 (13) | 19 (30) |
| Non-salivary gland | 7 (22) | 0 | 7 (11) |
| Brain metastases, n (%) | |||
| No | 31 (97) | 27 (84) | 58 (91) |
| Yes, treated | 1 (3) | 1 (3) | 2 (3) |
| Yes, untreated | 0 | 4 (13) | 4 (6) |
| Previous palliative chemotherapy, n (%) | |||
| No | 22 (69) | 22 (69) | 44 (69) |
| Yes | 10 (31) | 10 (31) | 20 (31) |
| Previous targeted therapya, n (%) | |||
| No | 20 (63) | 24 (75) | 44 (69) |
| Yes | 12 (38) | 8 (25) | 20 (31) |
| Previous anti-AR therapy, n (%) | |||
| No | 32 (100) | 24 (75) | 56 (88) |
| Yes | 0 | 8 (25) | 8 (13) |
| Previous anti-HER2 therapy, n (%) | |||
| No | 32 (100) | 31 (97) | 63 (98) |
| Yes | 0 | 1 (3) | 1 (2) |
| Prior lines of systemic, palliative therapy, n (%) | |||
| 0 | 14 (44) | 15 (47) | 29 (45) |
| 1–2 | 16 (50) | 12 (38) | 28 (44) |
| ≥3 | 2 (6) | 5 (16) | 7 (11) |
Percentages may not add up to 100 due to rounding.
Excluding therapies targeting AR or HER2. Primary subsites are listed in Supplementary Table 1.
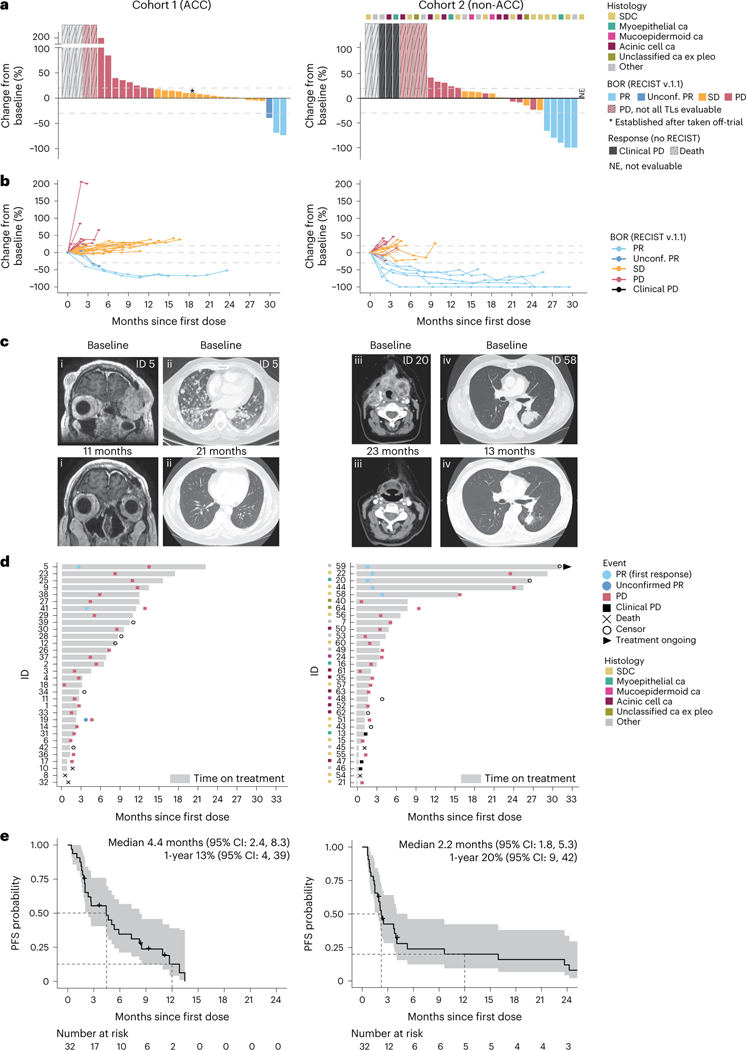
One of the 18 patients with ACC enrolled first (cohort 1, stage 1) had a confirmed partial response (cPR), leading to the accrual of 14 more patients in stage 2 in which one more cPR was seen—yielding a total ORR of two of 32 (6%), which failed to meet the pre-specified primary endpoint (Fig. 1a,b and Extended Data Table 1). However, the two patients with cPRs experienced deep and durable tumor regressions, indicated by marked (maximal 73% and 69%) reductions in target lesions (TLs; Fig. 1c) and PFS intervals of 13.5 months and 12.8 months (Fig. 1d). The 17 patients in cohort 1 with stable disease (SD) as best objective response (BOR) included one patient with an unconfirmed PR (39% TL regression) whose therapy was held for grade 2 treatment-related nephritis and who subsequently developed new brain metastases after the scan showing PR. Another patient was designated SD after being taken off-trial for toxicity and before subsequent treatment was started. Thirteen (41%) patients in cohort 1 had progressive disease (PD) as best response, including three patients who died of disease before evaluable imaging was obtained. Fourteen patients were treated with study medication beyond PD, which yielded no additional responses. Ten patients (31%) experienced clinical benefit (defined as PR or SD >6 months), a post hoc analysis. The median PFS for cohort 1 was 4.4 months (95% confidence interval (CI): 2.4, 8.3; Fig. 1e).
Fig. 1 |. Clinical outcomes for patients with advanced ACC (cohort 1, left) and patients with non-ACC SGC (cohort 2, right) treated with concurrent nivo+ipi.
a, Waterfall plots demonstrating the maximum change from baseline TL sum diameter (%ΔTL) for cohort 1 patients (n = 32) and cohort 2 patients (n = 31; one unevaluable patient was marked ‘NE’ but counted as a non-responder for primary endpoint). Thresholds for PD (+20%) and PR (–30%) are indicated. Bar colors represent BOR (RECIST version 1.1) (ref. 21). Five patients (light gray bars; three in cohort 1, two in cohort 2) who died before RECIST evaluation; three patients (all cohort 2, dark gray bars) with clinically evident PD but no evaluable imaging; and six patients (two in cohort 1, four in cohort 2) with RECIST-PD but undeterminable %ΔTL are visualized with a fictitious high value and diagonal stripes. One cohort 1 patient’s imaging was obtained off-trial but before the start of subsequent treatment (asterisk). Note the interruption of the y axis. Top row of squares marks tumor histology in cohort 2. b, Spaghetti plot visualizing the longitudinal %ΔTL for patients with one or more evaluable follow-up scan (27 in cohort 1, 22 in cohort 2). The thresholds for RECIST-PD (+20%), RECIST-PR (–30%) and the 0% horizontal are indicated. Line color represents BOR. c, Scans of one responding patient with ACC in cohort 1 (panels i and ii) and two unique responders in cohort 2 (iii (myoepithelial carcinoma) and iv (SDC)) obtained at baseline (top) and on-treatment (bottom). Depicted tumors show marked regressions upon treatment. Patient trial IDs are printed. The on-treatment image in panel ii was obtained while the patient was on study treatment beyond disease progression. d, Swimmer plot overview showing time on study treatment, first PR and PD events, censors (only in the absence of a PD event) and whether treatment is ongoing at data cutoff. Colors mark the histological subtype in cohort 2. e, PFS estimates for cohort 1 and cohort 2, calculated from start of study treatment. Dotted lines intersect curves at median and 1-year PFS (also printed). Shaded areas show 95% CI; crosses mark a censor. ca, carcinoma.
In cohort 2 (non-ACC SGCs), three of 18 patients in stage 1 and two of 14 patients in stage 2 developed a cPR, leading to an ORR of 16%, meeting the primary endpoint (Fig. 1a–c and Extended Data Table 1). The cPR rate was 25% in tumors of salivary duct histology (3/12). Two cohort 2 patients had 100% TL regression but did not meet complete response criteria due to the persistence of non-TLs. Of the five patients with cPR in cohort 2, three had a PFS of 15.9–24.2 months before developing PD; one was censored without PD after a PFS of 26.7 months (continued nivo+ipi locally, off-trial); and one remained on-study at data cutoff with a PFS of 28.0 months (Fig. 1d). The BOR was SD in eight (25%) patients and PD in 18 (56%) patients, including two with deaths due to disease and three with clinically evident, symptomatic PD before imaging was obtained. One patient in cohort 2 went off-study for toxicity 1.4 months after treatment start and received subsequent off-study treatment before response imaging was acquired; this patient was not evaluable for BOR and, per the statistical plan, was counted as a non-responder for the primary endpoint. Median PFS for cohort 2 was 2.2 months (95% CI: 1.8, 5.3) (Fig. 1e). The clinical benefit rate was 19% (6/32). Ten patients were treated beyond progression, with no responses.
Sixty patients (94%) across both cohorts developed an adverse event (AE) of any grade per the Common Terminology Criteria for Adverse Events (CTCAE) version 4.0 (ref. 22) that was deemed at least possibly treatment related (TR). Twenty-four patients (38%) had at least one TRAE categorized as ≥grade 3 (Table 2 and Supplementary Table 2). Although the main reason for study treatment discontinuation was disease progression (47/64, 73%), 11 patients (eight in cohort 1, three in cohort 2) came off-trial due to TRAEs and —one each in cohorts 1 and 2 came off-trial with simultaneous PD and a TRAE (Supplementary Table 3a,b).
Table 2 |. Most common TRAEs.
| AE | Any grade, n (%) | Grades 3–5, n (%) |
|---|---|---|
| Fatigue | 28 (44) | 3 (5) |
| Diarrhea | 17 (27) | 4 (6) |
| Aspartate aminotransferase increased | 15 (23) | 4 (6) |
| Pruritus | 15 (23) | 0 |
| Alanine aminotransferase increased | 14 (22) | 0 |
| Rash maculo-papular | 12 (19) | 0 |
| Anemia | 11 (17) | 1 (2) |
| Weight loss | 10 (16) | 0 |
| Hypothyroidism | 9 (14) | 0 |
| Dry skin | 8 (13) | 0 |
| Nausea | 8 (13) | 0 |
| Arthralgia | 7 (11) | 2 (3) |
| Pain | 7 (11) | 2 (3) |
| Anorexia | 6 (9) | 0 |
| Alkaline phosphatase increased | 5 (8) | 1 (2) |
| Dyspnea | 4 (6) | 0 |
| Lymphocyte count decreased | 4 (6) | 1 (2) |
| White blood cell count decreased | 4 (6) | 0 |
| Blurred vision | 3 (5) | 0 |
| Cough | 3 (5) | 0 |
| Fever | 3 (5) | 0 |
| Headache | 3 (5) | 0 |
| Hyperglycemia | 3 (5) | 2 (3) |
| Platelet count decreased | 3 (5) | 1 (2) |
| Pneumonitis | 3 (5) | 0 |
| Urticaria | 3 (5) | 0 |
| Vomiting | 3 (5) | 0 |
| Neutrophil count decreased | 3 (5) | 1 (2) |
| Colitis | 2 (3) | 2 (3) |
| Hypophysitis | 2 (3) | 1 (2) |
| Mucositis oral | 2 (3) | 1 (2) |
| Adrenal insufficiency | 1 (2) | 1 (2) |
| Arthritis | 1 (2) | 1 (2) |
| Bone marrow hypocellular | 1 (2) | 1 (2) |
| Creatine phosphokinase increased | 1 (2) | 1 (2) |
| Death NOSa | 1 (2) | 1 (2) |
| Heart failure | 1 (2) | 1 (2) |
| Hyponatremia | 1 (2) | 1 (2) |
| Sepsis | 1 (2) | 1 (2) |
| Syncope | 1 (2) | 1 (2) |
Only TRAEs occurring in more than two total patients and/or grade >2 are included.
Unwitnessed death of a patient who had increasing liver function tests and liver metastases in cycle 1 of therapy. This death was designated as PD, but, because it was unwitnessed, it was also attributed as possibly related to study drugs.
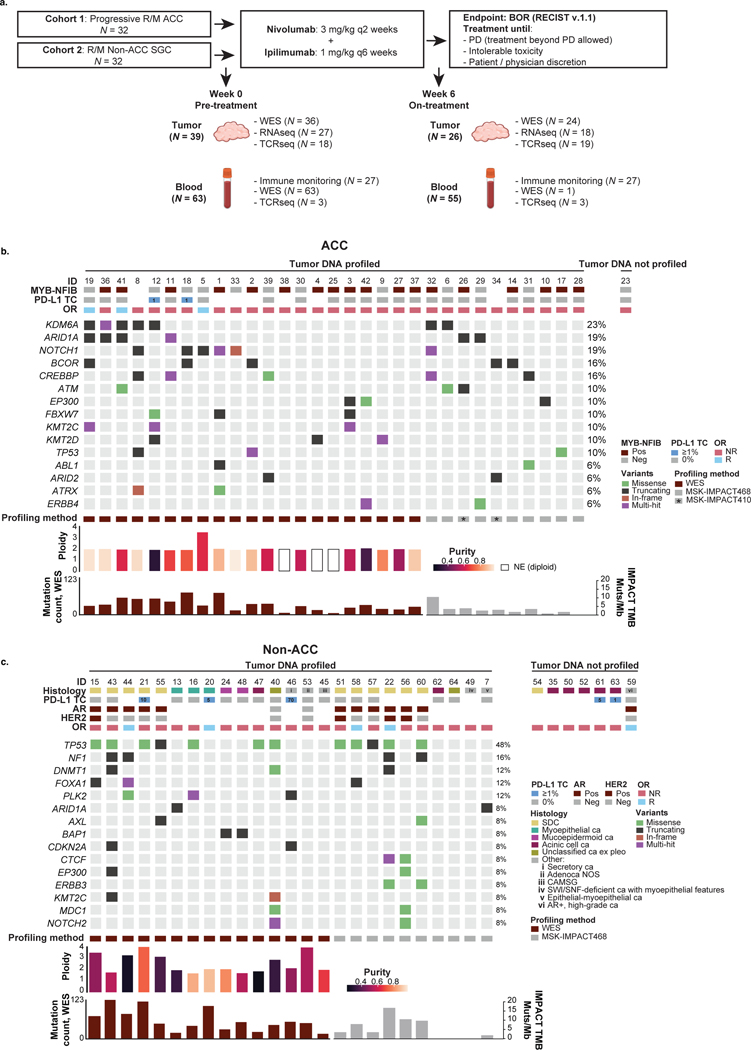
Patients consented to the collection of tumor biopsies at pre-treatment (week 0) and on-treatment (week 6) timepoints as well as serial blood draws (Extended Data Fig. 1a).
Distinct immunogenomic landscape of ACCs and non-ACC SGCs
Pre-treatment tumors were profiled using whole-exome sequencing (WES; median target coverage 197×) of snap-frozen samples from 36 patients. An additional 20 tumors were analyzed with targeted next-generation sequencing (tNGS) with MSK-IMPACT23 (median target coverage 560×; Supplementary Table 4).
Data from WES, tNGS and immunohistochemistry (IHC) for cohort 1 (ACCs) and cohort 2 (non-ACC SGCs) were consistent with our previous findings (Extended Data Fig. 1b,c)19,20. One responding ACC tumor demonstrated whole-genome duplication (Supplementary Fig. 1). Six of 31 (19%) genetically profiled ACC tumors harbored NOTCH1 mutations, of which four (13%) had NOTCH1 variants predicted to be activating and associated with poor prognosis in ACC (Supplementary Fig. 2)19,24–26. MYB–NFIB gene fusions, putative oncogenic drivers in ACCs16,19,27, were found in a majority (60%, 18/30).
Among the non-ACC tumors, 12 samples were androgen receptor (AR) positive by IHC: 11 of the 12 SDCs (the remaining SDC had insufficient material for investigation) and one unclassified, high-grade SGC not otherwise specified (NOS). Four samples (all SDC) had HER2 overexpression by IHC (3+) or amplification by fluorescence in situ hybridization (FISH) or tNGS (Extended Data Fig. 1c).
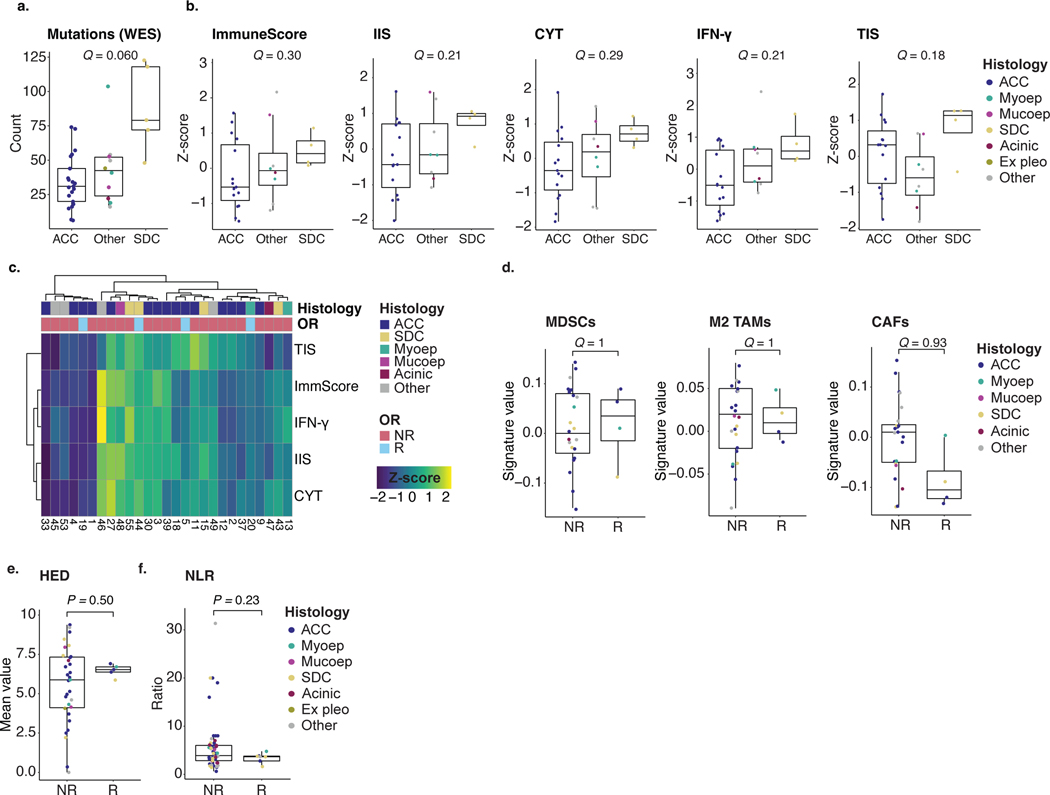
PD-L1 expression on tumor cells (≥1%) was seen in two of 25 ACCs (8%) and five of 22 (23%) non-ACC SGCs (Extended Data Fig. 1b,c). The WES-based mutation count was lowest in ACCs and highest in SDC tumors (median 31 and 79, respectively; q = 0.06; Extended Data Fig. 2a). SDC samples had the lowest median tumor purity (49%), followed by non-ACC/SDC tumors (55%) and, finally, ACCs (68%; q = 0.13; Supplementary Fig. 3a). In addition, a hyperploid state, defined as a mean ploidy greater than 2.5 (ref. 28), was often seen in SDCs (4/5 (80%)), whereas it was rare in ACC (1/21 (5%)) and non-ACC/SDC (2/10 (20%)) tumors (Supplementary Fig. 3b).
Bulk RNA sequencing (RNA-seq) was performed in 27 pre-treatment tumors and analyzed using several orthogonal tools (Methods) to assess immune infiltration in the tumor microenvironment (TME). Deconvolved immune signatures (ImmuneScore29, IIS30, CYT31, TIS30 and REACTOME IFN-γ geneset32) were consistently highest in SDC and low in ACC, although not statistically significant (q = 0.18–0.30; Extended Data Fig. 2b,c), consistent with previous studies20. Similar directionality was seen for an antigen presentation RNA signature30 and checkpoint gene expression (Supplementary Fig. 3c,d). Analyzing RNA signatures associated with the presence of individual immune cell populations33, we found that non-ACC tumors clustered in the more intensely T-cell-infiltrated subgroup (Supplementary Fig. 3e). Signature values of three immune populations previously correlated with poor ICB response34—cancer-associated fibroblasts (CAFs), myeloid-derived suppressor cells (MDSCs) and M2-polarized tumor-associated macrophages (M2 TAMs)—were not significantly different among ACC, non-ACC/SDC and SDC tumors (Supplementary Fig. 3f).
Taken together, these immunogenomic characteristics suggest that ACCs would be less poised to respond to nivo+ipi (low tumor mutational burden (TMB) and immune infiltration) compared to SDC tumors (higher TMB and more immune infiltration)—in line with the results of this trial.
Immunogenomic predictors of treatment response
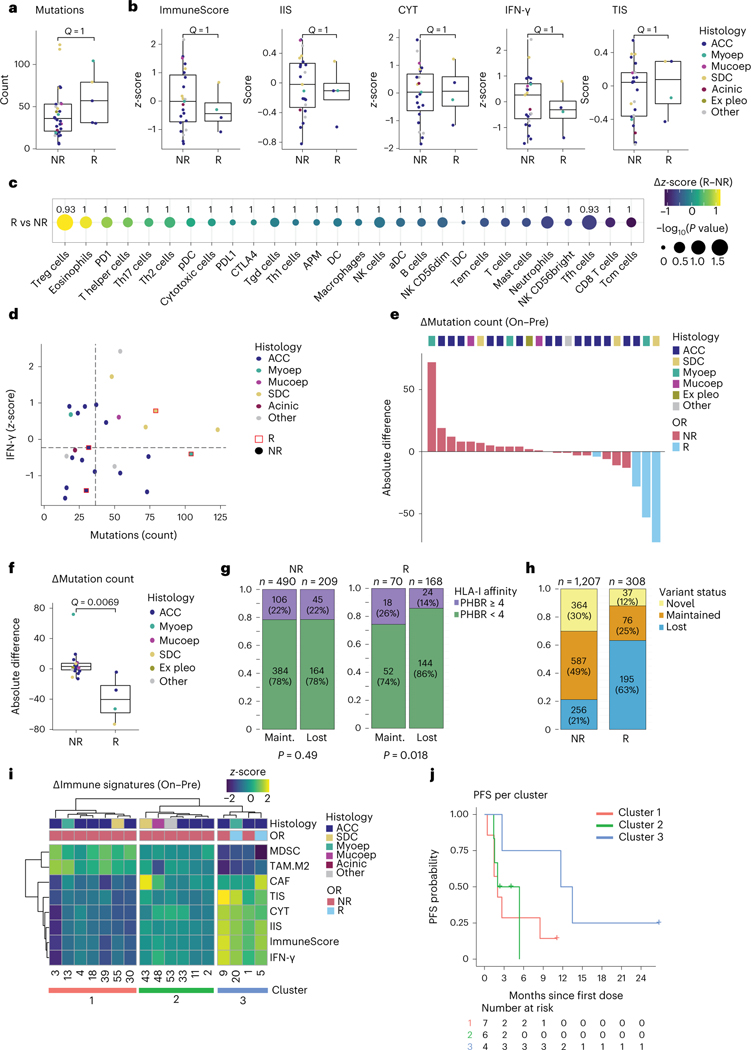
We next evaluated if previously described biomarkers predicting ICB response in other tumor types are relevant for SGCs, by comparing responding (CR or PR) to non-responding (SD or PD) tumors. No significant differences were observed in the pre-treatment mutation count (Fig. 2a) or any RNA-seq immune signature (Fig. 2b). Similarly, no statistically significant differences were observed in RNA signatures for individual immune cells (including inhibitory MDSCs, M2 TAMs and CAFs)33,34, the antigen presentation signature30, immune checkpoint gene expression, germline HLA class I diversity (HED)35, peripheral blood neutrophil-to-lymphocyte ratio (NLR) or chemokine genes CXCL9, CXCL10 and CXCL11 (Fig. 2c, Extended Data Fig. 2d–f and Supplementary Fig. 3g). When plotting each tumor according to its mutation count and IFN-γ score, two critical biomarkers of ICB response36, responding samples were found in all quadrants (Fig. 2d).
Fig. 2 |. Pre-treatment and on-treatment immunogenomic profiles of SGCs in the context of treatment response.
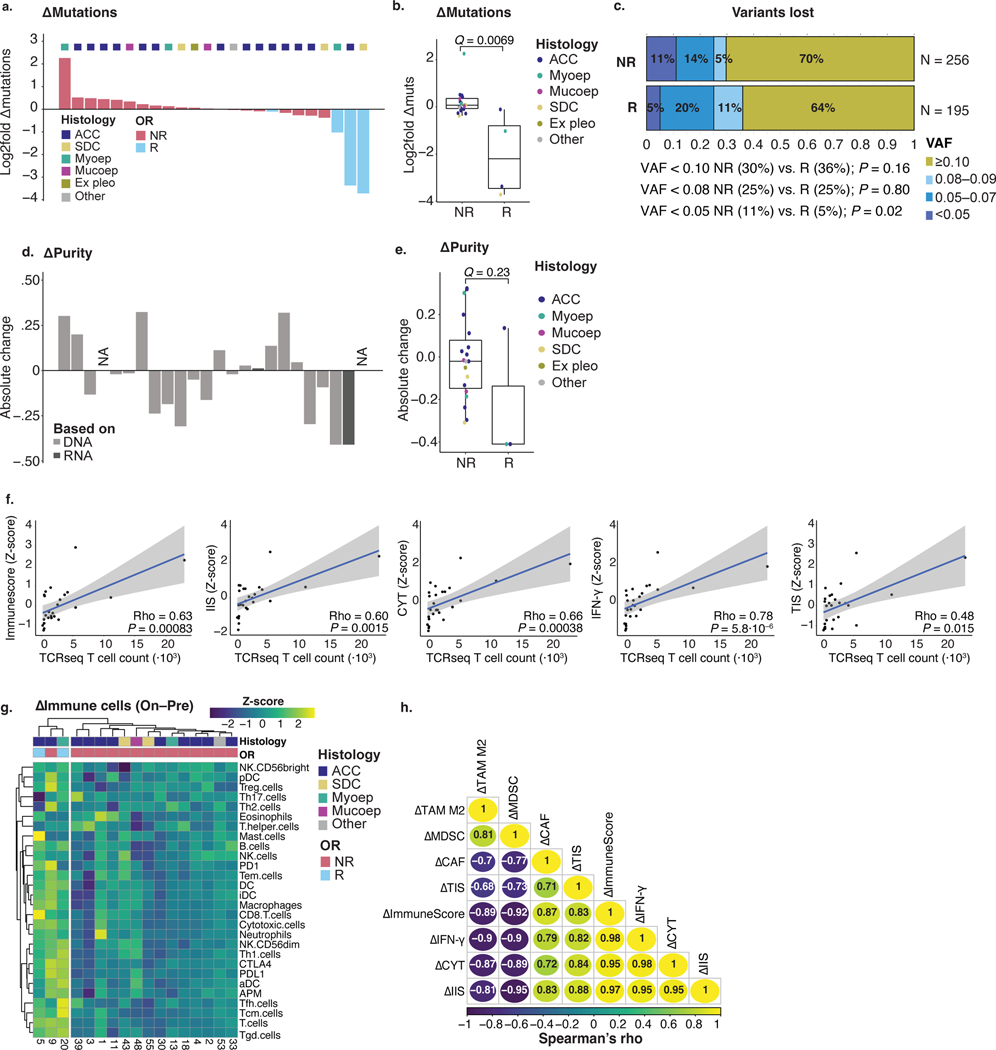
a,b, Box plots are defined in Methods. Dot colors in a,b,d,f indicate tumor histology. Exact, nominal P values in a,b,f were calculated using two-sided Wilcoxon rank-sum tests. P values in a,b,c,f were adjusted for multiplicity (Methods), yielding q values. a, Pre-treatment mutations per exome in non-responding (NR, n = 31) and responding (R, n = 5) patients. b, Pre-treatment values of the ImmuneScore29,30, IIS30, CYT31, IFN-γ pathway32 and TIS30 RNA signatures in NR (n = 23) and R (n = 4) patients. c, Absolute difference in z-score (R minus NR, visualized by color gradient) between R (n = 4) and NR (n = 23) patients for 24 immune cell subsets, APM signature and individual PD-1, PD-L1 and CTLA-4 checkpoint gene expression30,33. Dot size represents the −log10 of the nominal P value, obtained through a two-sided Wilcoxon rank-sum test comparing R with NR. q values are printed. d, Expression of IFN-γ pathway genes plotted against mutation count (n = 26). Responding samples (squares; red outline) and non-responders (dots) are indicated. Median values (dotted lines) distinguish quarters. e, Waterfall plot representing samples’ absolute change in WES-based mutation count from pre-treatment to on-treatment. Bar color represents objective response. Top track shows tumor histology. f, Absolute change in mutation count for NR (n = 20) and R (n = 4) tumors. g, Fraction of pre-treatment mutations with a PHBR41 considered lower (<4, green) or higher (≥4, purple) among mutations lost or maintained upon treatment, in NR and R tumors. Non-productive variants were excluded. Proportions were compared using a χ2 test. P values are one-sided. h, Proportion of variants lost, maintained or novel (present on-treatment only) upon treatment, for NR and R samples. Denominator is the sum of unique variants in a sample pair, per patient, in each response group; maintained variants were counted once. i, Heat map visualizing change in MDSC, M2 TAM, CAF, TIS, CYT, IIS, ImmuneScore and IFN-γ immune RNA signatures (on-treatment minus pre-treatment). Top tracks represent tumor histology and objective response. j, PFS estimates for the three clusters obtained from the heat map in h. Acinic, acinic cell carcinoma; Ex pleo, carcinoma ex pleomorphic adenoma; OR, objective response; Maint., maintained; Mucoep, mucoepidermoid carcinoma; Myoep, myoepithelial carcinoma.
Based on recent data in mice suggesting that viruses, including enteric viruses, may replicate and persist in salivary glands37, we examined the presence of viral reads among non-aligning reads from RNA-seq (Supplementary Table 5). Interestingly, we discovered viral reads in three pre-treatment tumors: two with a cPR and one with rapid progression (%ΔTL + 201%; Supplementary Table 6). No viral reads were detected in matched on-treatment samples. However, we caution that these numbers are small, and further investigation is required.
Although the low number of responding patients limits analytical power, biomarkers developed in other cancer types lacked predictive value for nivo+ipi-efficacy in this SGC cohort, highlighting the need for biomarkers specific to SGC biology.
Responding tumors show mutational contraction upon treatment
Mutational contraction—a decrease in somatic mutation count observed during ICB treatment—was previously described in ICB-treated skin cancers profiled with longitudinal biopsies38,39 and attributed to immunologic ‘editing’ of immunogenic tumor cells. In the 24 SGC samples with WES available at both timepoints, a decrease in tumor mutation count among responding tumors was observed (median Δmutations −40.5 (interquartile range (IQR) −58.0 to −22.0)) but not in most non-responding samples (Δmutations +3.0 (IQR −1.5 to +7.3), q = 0.0069; Fig. 2e,f and Extended Data Fig. 3a,b). Next, we predicted the binding affinity for every 8–11-mer neopeptide derived from non-synonymous, pre-treatment, protein-coding mutations (single-nucleotide variants (SNVs) and indels) to each patient’s HLA-I molecules (Methods)40. We calculated the previously developed patient harmonic-mean best rank (PHBR) metric, which measures neoepitope presentation across all of a patient’s HLA-I alleles41. In responding tumors, mutations eliminated during treatment were more likely to have a low PHBR (by a previously defined threshold of high presentation by HLA41; 86% versus 74%, P = 0.018) but not in non-responding patients (both 78%, P = 0.49; Fig. 2g). The preferential loss of variants with stronger HLA-I binding affinity during ICB treatment response is supportive of a process of mutational contraction through immunologic selection pressure.
Because sequencing of these samples was performed at high depth (197× at baseline, 205× on-treatment), it is unlikely that the observed mutational contraction was attributable only to low variant allelic fraction (VAF) mutations escaping detection due to changes in tumor purity. Indeed, most mutations lost upon treatment had a VAF >0.10—well above the detection limit for our sample coverage38,42—and this proportion was not significantly higher in responders than in non-responders (64% versus 70%, P = 0.16), nor did proportions of variants at lower VAF (below lowest quartile (0.08) or <0.05) differ between responders and non-responders (Extended Data Fig. 3c). The change in purity during treatment did not differ significantly between responding and non-responding samples29,43 (q = 0.23; Extended Data Fig. 3d,e).
Focusing on the unique mutations in the paired pre-treatment and on-treatment samples per patient, we found a higher proportion of novel, treatment-emerging variants in non-responding tumors (30% versus 12%), suggestive of neutral drift or the emergence of new clones in these ICB-resistant tumors. Conversely, responding tumors had a higher proportion of lost variants (63% versus 21%), suggesting immunoediting in responding tumors (Fig. 2h). We note that these data are observational and, although consistent with previous findings in serially sampled skin cancers, do not definitively prove that all mutational contraction results from immunologic selection pressure.
Responding tumors show an increase in immune infiltration
To profile changes in the immune TME during nivo+ipi, we performed RNA-seq in paired pre-treatment and on-treatment samples. Although immune cell deconvolution from bulk RNA-seq data is indirect, the direct T cell count from T cell receptor β-chain sequencing (TCR-seq; details below) correlated strongly with the RNA-seq-derived T cell signatures (ρ = 0.48−0.78, P values 5.8 × 10−6 to 0.015; Extended Data Fig. 3f) and closely with IHC in our previous SGC study20. Overall, the aggregate immune infiltration signatures, and signatures of most immune cell subsets, T cell checkpoints and antigen-presenting machinery (APM), increased from pre-treatment to on-treatment timepoints in responding samples (Fig. 2i and Extended Data Fig. 3g). Clustering samples by changes in immune signatures, we observed one cluster characterized by on-treatment increase of all immune signatures and simultaneous, correlated decrease of the signatures for MDSCs and M2 TAMs (ρ = –0.68 to –0.95, P values 7.5 × 10–9 to 0.0026; Fig. 2i and Extended Data Fig. 3h), whereas such a pattern was absent from the other two clusters. Although the low number of observations limits analytical power, PFS (Fig. 2j) was longer for the four patients (including two cPRs) in the immune-upregulated cluster 3 (median 12.6 months, 95% CI: 2.7, not available (NA)) than for patients in clusters 1 or 2 (respective median 2.0, 95% CI: 1.5, NA and 3.7, 95% CI: 1.5, NA).
The TCR repertoire is clonally skewed in responding tumors
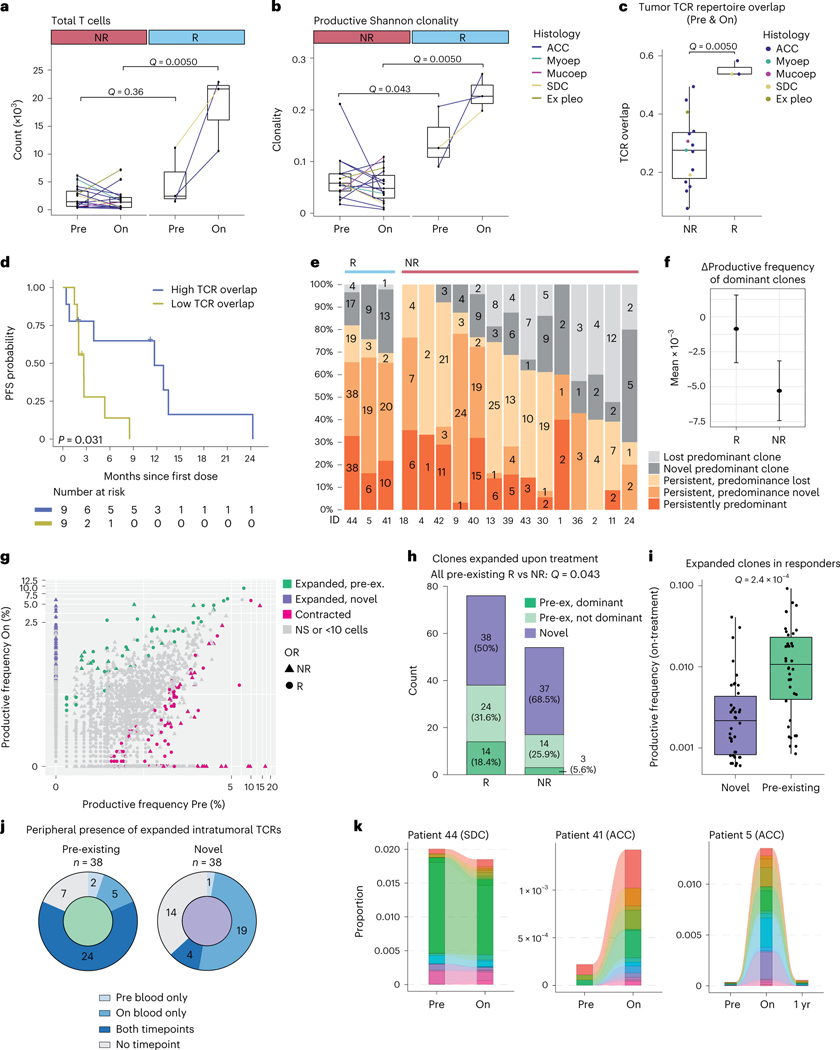
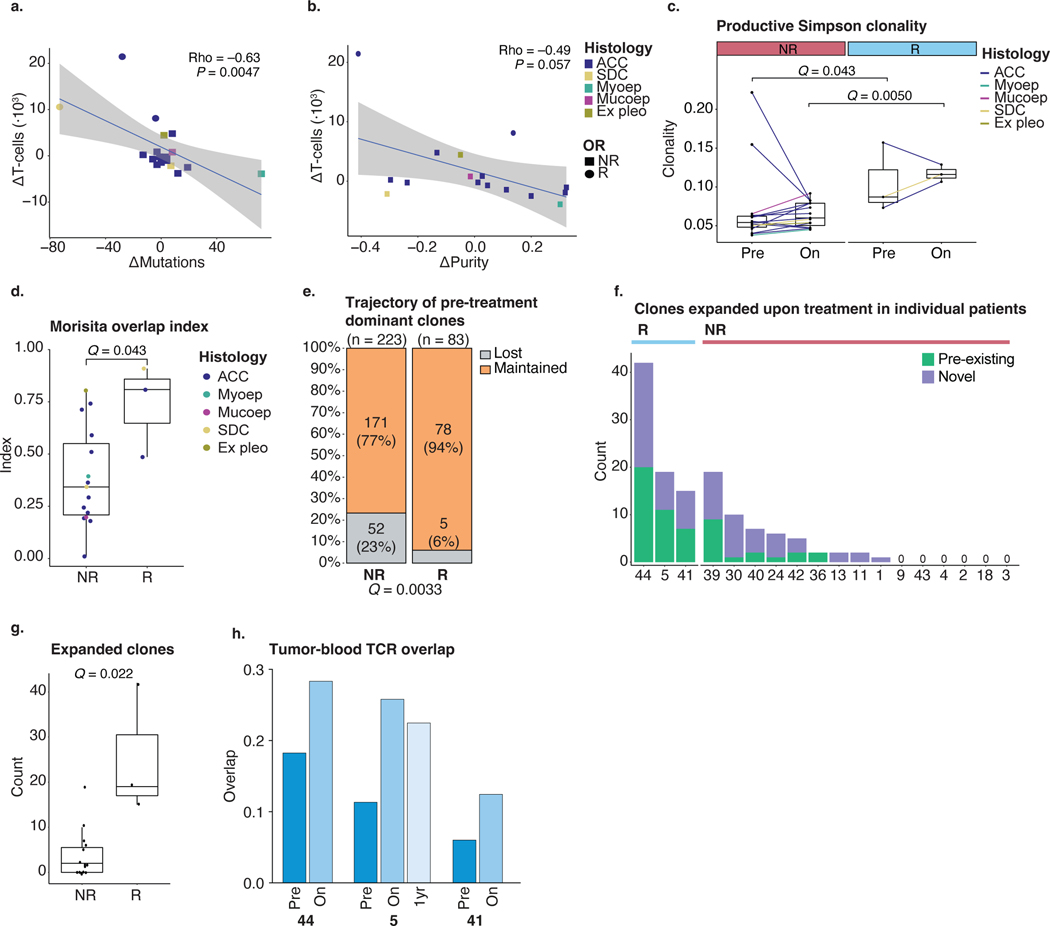
A pre-existing, clonally expanded TCR repertoire has been associated with response to anti-PD-1 (±anti-CTLA-4) in melanoma44–46. To examine the TCR repertoire in SGC tumors treated with nivo+ipi, TCR-seq of pre-treatment and on-treatment tumors was performed. The number of tumor-infiltrating T cells enumerated by TCR-seq strongly increased in responders (median 4.3×) and was significantly higher than non-responding samples at the on-treatment timepoint (q = 0.0050; Fig. 3a). Furthermore, the change in T cells correlated significantly with mutational contraction (ρ = –0.63, P = 0.0047; Extended Data Fig. 4a) and showed a trend in correlating with the change in purity (ρ = –0.49, P = 0.057; Extended Data Fig. 4b), consistent with T cell influx and T-cell-mediated cancer cell clearance.
Fig. 3 |. Pre-treatment and on-treatment tumor and peripheral blood TCR repertoire analyses.
a–c, Box plots are defined in Methods. Lines connect paired pre-treatment and on-treatment samples in a,b. Line (a,b) and dot colors (c) indicate histology. Exact P values in a–c,i were calculated using a two-sided Wilcoxon rank-sum test. Nominal P values in a–c,g–i were multiplicity adjusted (Methods), yielding q values. TCR-seq data were available for 18 patients (15 non-responders and three responders). a, Change in the TCR-seq-based T cell count upon treatment, per objective response. b, Change in the TCR repertoire Shannon clonality upon treatment, per objective response. c, Longitudinal TCR overlap47, per objective response. d, PFS of patients with high/low TCR repertoire overlap (c), using the cohort median (0.30) as threshold. The P value was calculated using a log-rank test. e, Proportion of predominant T cell clones (Methods) considered persistent or novel upon treatment, per patient. Denominator is the sum of unique clones in a patient’s longitudinal samples—persistent clones were counted once. Colors represent persistence/replacement. Numbers refer to clonotype counts per bar section. f, Mean absolute change in productive frequency upon treatment for pre-treatment dominant clones, per objective response. Error bars represent 95% CI. g, Overlay plot of patients showing TCR clones’ productive frequency at pre-treatment versus on-treatment. Statistical testing was performed per patients’ sample pair. Colors highlight significantly contracting or expanding clones (Methods). Expanding clones are further colored based on their pre-treatment detection. Shapes indicate (non-)response. h, Proportion of clones considered significantly expanded (g), per objective response. Novel clonotypes are indicated in purple, pre-existing clones in green. Pre-existing clonotypes are separated based on dominance in pre-treatment tumors. Proportions of expanding clones that were pre-existing in responding and non-responding tumors were compared using a χ2 test. i, Productive frequencies of novel (n = 38) and pre-existing (n = 38) clonotypes from responsive tumors. j, Fraction of expanding clonotypes considered novel or pre-existing in responding tumors’ TME, identified in the blood at pre-treatment and/or on-treatment. k, Longitudinal productive frequencies of intratumorally expanding clones (g) in the blood of responders. Sample ‘1 yr’ was obtained 336 d after treatment start. Color legends are provided in Supplementary Fig. 4. Ex pleo, carcinoma ex pleomorphic adenoma; OR, objective response; Maint., maintained; Mucoep, mucoepidermoid carcinoma; Myoep, myoepithelial carcinoma; NS, not significant; pre-ex., pre-existing.
At baseline, productive Shannon and Simpson TCR repertoire clonality metrics were significantly higher in responding tumors (median 0.13/0.09) than in non-responding tumors (median 0.06/0.05; both q = 0.043; Fig. 3b and Extended Data Fig. 4c). TCR clonality increased further, and was markedly higher, in responding tumors than in non-responding tumors upon treatment (Shannon clonality 0.23 versus 0.05, Simpson clonality 0.12 versus 0.06, both q = 0.0050). This suggests that tumor response is associated with a pre-existing, clonally skewed intratumoral TCR repertoire and further immunotherapy-mediated clonal expansion of TCRs.
Responding SGCs maintain and expand pre-existing TCR clones
In some cancer types, tumor regression is associated with the recruitment of novel T cell clones from the periphery (‘clonal replacement’39). However, it is unknown if this is a pan-cancer phenomenon or if nivo+ipi reinvigorates pre-existing T cells in the SGC TME. In responding SGC tumors, the TCR overlap index47 (between paired baseline and on-treatment TCR repertoires) was significantly greater than in non-responders (q = 0.0050, Fig. 3c; similar results with the Morisita–Horn index48, Extended Data Fig. 3d). Patients with a high TCR overlap index (>median, 0.30) had significantly longer PFS (median 11.7 months versus 2.7 months, P = 0.031; Fig. 3d).
Of the most predominant 1% of TCR clonotypes detected before treatment49,50, few (6%) were lost on-treatment in responding tumors. In contrast, a significantly larger fraction (23%) was lost in non-responders (q = 0.0033; Fig. 3e and Extended Data Fig. 4e). Similarly, the mean productive frequency of the top 1% predominant clones decreased more profoundly upon treatment in non-responders (−5.3 × 10−3; 95% CI: −7.4 × 10−3, −3.2 × 10−3) than in responding patients (−8.7 × 10−4; 95% CI: −3.3 × 10−3, 1.5 × 10−3) (Fig. 3f). These results indicate T cell clonal maintenance in responding tumors.
Next, we focused on the intratumoral TCR clonotypes that expanded most markedly upon nivo+ipi (Methods): 130 unique clonotypes showed a significant increase in productive frequency upon treatment (Fig. 3g and Extended Data Fig. 4f). The median number of expanding clones was significantly higher in responding than non-responding tumors (19 versus two, q = 0.022; Extended Data Fig. 4g). In fact, in responding tumors, 50% (38/76) of expanding clones were pre-existing (14 were among the top 1% pre-treatment), which is a significantly higher proportion than in non-responders (17/54; 31%), three of which were among the top 1% pre-treatment, q = 0.043) (Fig. 3h). Among expanding clones in responding tumors, pre-existing clones had a significantly higher on-treatment productive frequency than newly emergent clonotypes (q = 2.4 × 10−4; Fig. 3i), further highlighting the importance of pre-existing clonotypes in the anti-tumor immune response.
We performed additional TCR-seq of pre-treatment and on-treatment peripheral blood samples from three responding patients (two with ACCs and one with SDC) with matching tumor-TCR-seq (Methods). As expected, we observed an overlap in the TCR repertoires in tumor and blood samples at the pre-treatment timepoint, which considerably increased (1.5–2.3×) early during treatment and remained persistently high as far out as 336 d (Extended Data Fig. 4h). Focusing on the pre-existing intratumoral TCR clonotypes that significantly expanded upon treatment in these responders, most were detected in both pre-treatment and on-treatment blood (63%; Fig. 3j). Only 13% of newly emergent intratumoral clonotypes were detected in pre-treatment blood, suggesting that novel T cell recruitment from the periphery into the TME does not appear to be common in these responding SGC tumors39.
When tracking each responder’s expanded intratumoral TCR clonotypes in longitudinal blood samples (Fig. 3k and Supplementary Fig. 4), we observed a marked peripheral expansion of these clones in the two patients with ACC (patient 5 and patient 41). In the responding patient with SDC (patient 44), expanded intratumoral TCR clones comprised a strikingly high proportion of the pre-treatment peripheral, productive TCR repertoire (up to 2%; Fig. 3k) and remained high, perhaps consistent with the more immunogenic state of SDCs at baseline compared to ACCs. Of note, 58% of expanded intratumoral clones could still be identified in the peripheral blood of patient 5 obtained 336 d on-treatment (response still ongoing), although they did contract over time (Fig. 3k).
These TCR-seq data show that responding and non-responding salivary tumors have different T cell clonotype profiles and on-treatment trajectories. Responding SGCs are characterized by greater maintenance and expansion of pre-existing TCR clonotypes—an observation reflected in all three responders’ peripheral blood. In contrast, the expansion of novel T cell clonotypes was not strongly associated with ICB response or with origin in the peripheral blood. However, we caution that the low sample number precludes any strong conclusions. Still, these data are consistent with the hypothesis that, at an early on-treatment timepoint (week six), an immunotherapy response—particularly in SGCs lacking a high neoantigen burden—depends on pre-existing tumor-infiltrating TCR clonotypes undergoing clonal expansion rather than replacement by a new repertoire of TCRs. The ‘clonal replacement’ hypothesis of immune-mediated tumor regression, described in other cancers, does not seem universally applicable across all cancer types, although we cannot exclude the occurrence of this phenomenon at later timepoints.
Peripheral T cell reinvigoration and checkpoint upregulation
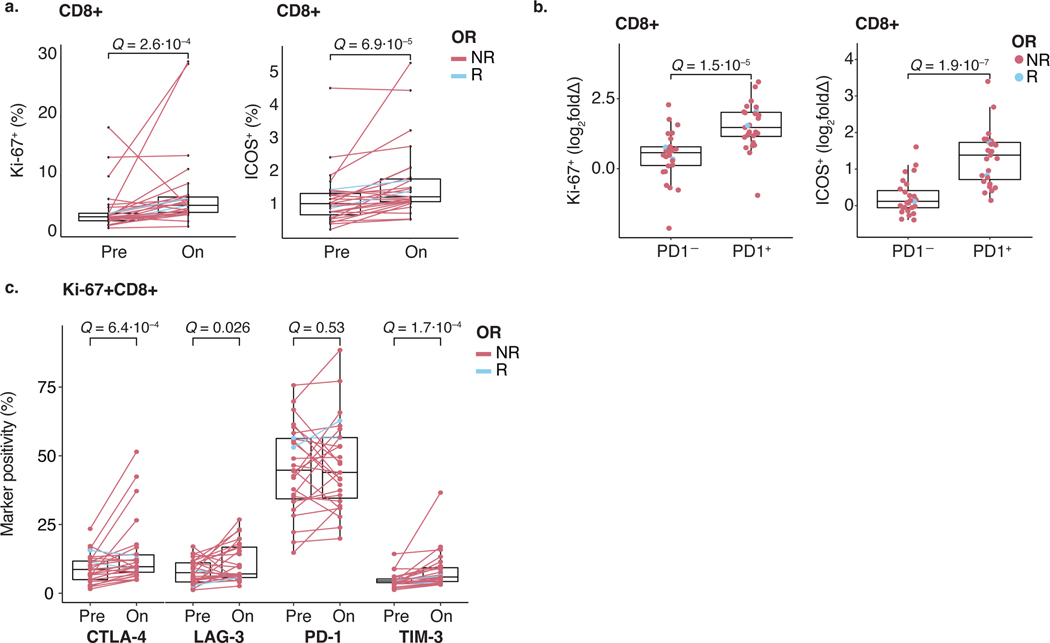
Using a T cell activation and exhaustion flow cytometry panel (Supplementary Table 7 and Supplementary Fig. 5), we assessed if response to nivo+ipi was mirrored by T cell frequency and surface marker changes in the blood of 27 patients with ACC during treatment. In line with previous reports51,52, CD8+ T cells significantly increased proliferation markers Ki-67 and ICOS (Extended Data Fig. 5a) in all but one (non-responding) patient more markedly on PD-1+CD8+ versus PD-1−CD8+ T cells (q = 1.5 × 10−5 and 1.9 × 10−7, respectively; Extended Data Fig. 5b). This suggests that nivo+ipi re-invigorates a phenotypically exhausted T cell population, which is likely necessary, but not sufficient, for tumor response in SGC. Proliferating Ki-67+CD8+ T cells demonstrated simultaneous upregulation of CTLA-4, LAG-3 and TIM-3 upon treatment (Extended Data Fig. 5c), suggesting that these adaptive mechanisms may be relevant to nivo+ipi efficacy.
Immunogenicity of MYB–NFIB fusion and other variants in ACCs
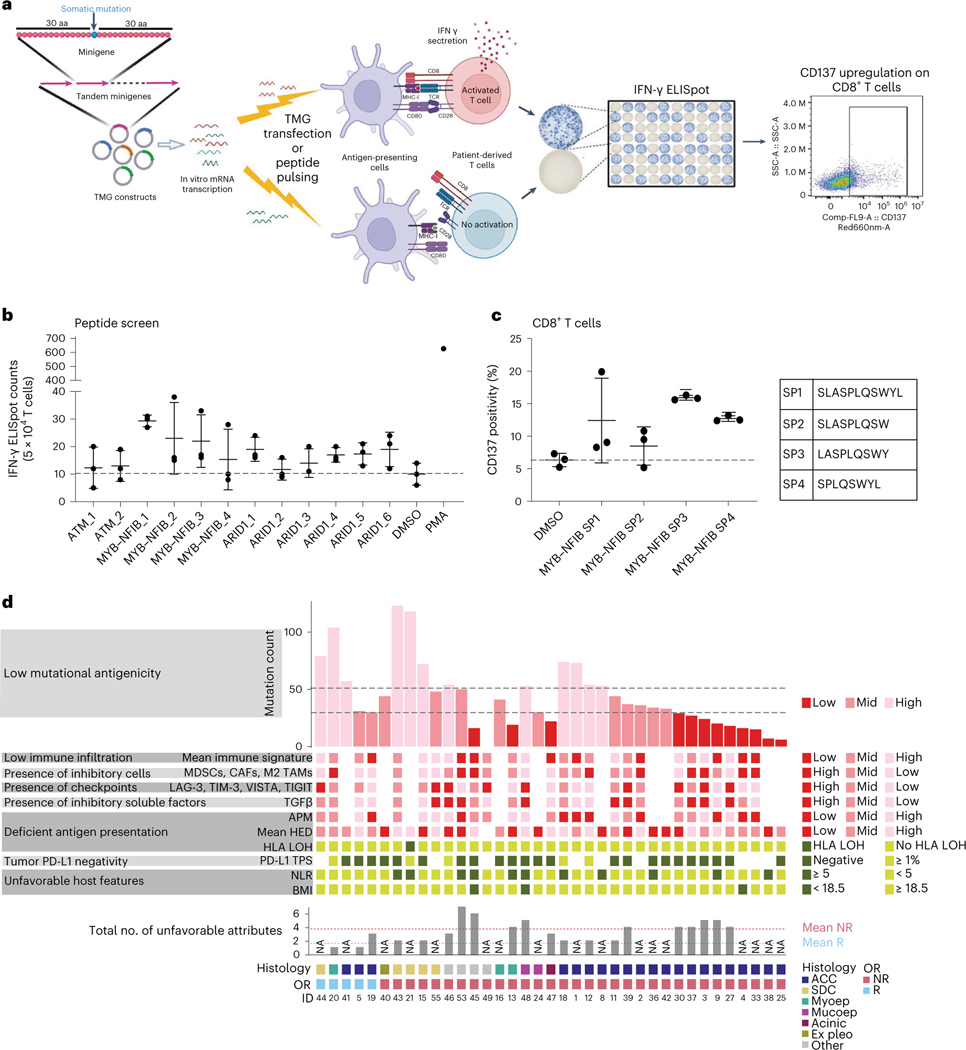
To gain insight into the mechanistic underpinnings of nivo+ipi-mediated responses achieved in genomically quiet and poorly infiltrated ACCs, we screened potentially immunogenic neoantigens in the two patients with ACC with deep and durable responses (Fig. 1).
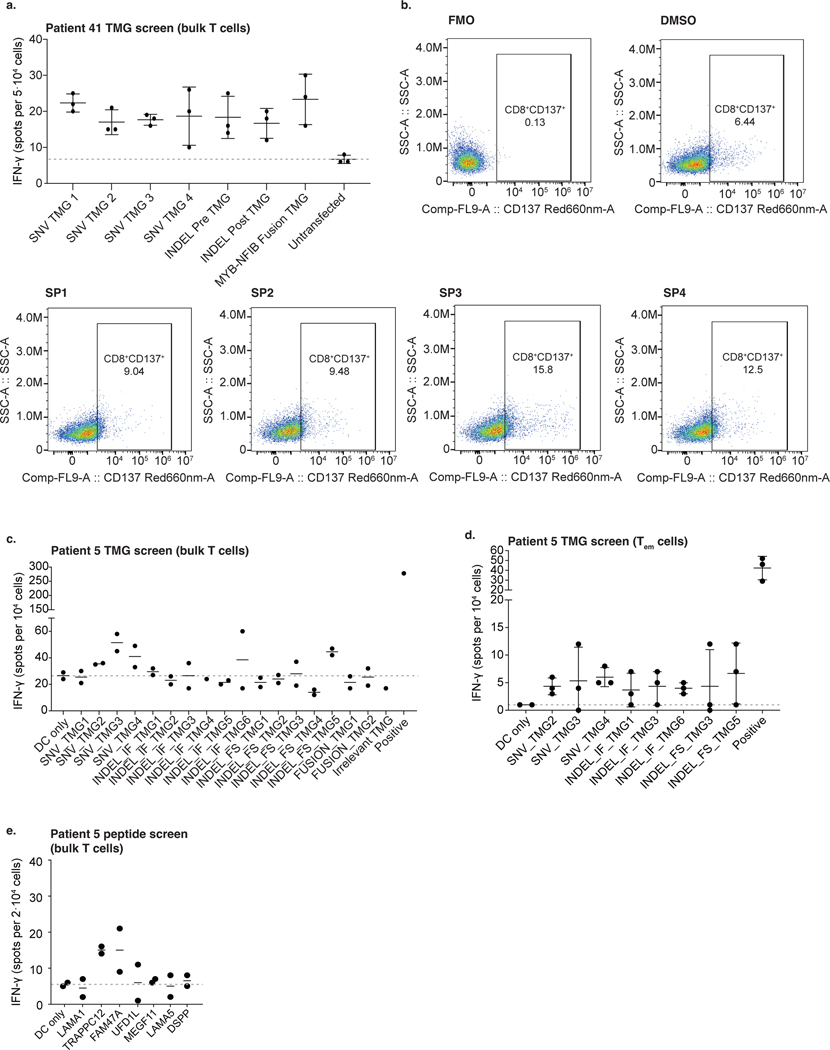
For patient 41, WGS was performed. Variant sequences—SNVs with 30 flanking amino acids and the entire 3′ sequence downstream of indels and fusion breakpoints until the first stop codon—were cloned into a series of tandem minigenes (TMGs) (Fig. 4a)53: four TMGs corresponding to 39 SNVs, two TMGs containing 12 indels and one TMG for the MYB–NFIB fusion breakpoint (Supplementary Table 8a). Because this patient ultimately died, peripheral blood mononuclear cell (PBMC) samples and, consequently, professional antigen-presenting cells (APCs) were limited. Therefore, we expressed TMG constructs with patient-specific HLA alleles in COS-7 (monkey kidney fibroblast-like) cells, co-cultured with patient-autologous T cells54. Multiple TMGs and the fusion minigene elicited T cell activation (ELISpot assay; Extended Data Fig. 6a). We subsequently focused our neoantigen identification on testing the immunogenicity of recurrent (‘public’) genetic alterations in this ACC, shared across patients: the MYB–NFIB fusion and mutations in ATM and ARID1A. Several 8–11-mer peptides derived from the MYB–NFIB fusion gene and ARID1A mutation consistently stimulated T cell responses in an ELISpot assay (Fig. 4b and Supplementary Table 8b). Notably, MYB–NFIB fusion peptides induced activation marker CD137 (4–1BB) expression on patient-autologous CD8+ T cells upon restimulation (Fig. 4c and Extended Data Fig. 6b).
Fig. 4 |. Neoantigen identification in responding patients with ACC and potential immune-evasion mechanisms in SGC.
a, Neoantigen identification workflow. Based on WES-called mutations, TMGs were constructed and electroporated into autologous DCs (patient 5) or co-transfected with the patient’s HLA (patient 41) into COS-7 cells as APCs. Autologous T cells from PBMCs were co-cultured with the TMG-electroporated APCs for primary screening and subsequently with APCs pulsed with peptides for exact neoantigen identification. T cell activation was measured by IFN-γ production in an ELISpot assay and CD137 upregulation by flow cytometry. This cartoon was created using BioRender. b,c, Data in b and c are representative of two independent experiments with n = 3 technical replicates. Horizontal bars and whiskers represent the mean and s.d., respectively, of triplicate experiments, and the gray dotted lines indicate the mean value in the negative control (DMSO). b, Autologous T cells expanded from PBMCs (patient 41) were co-cultured with autologous HLA-electroporated T2 cells, pulsed with 8-mer or 9-mer short peptides corresponding to the mutations in the listed genes. IFN-γ production (ELISpot) is shown. T2 pulsed with DMSO (no peptide) was the negative control. PMA-stimulated T cell response was the positive control. c, CD137 expression assessed by flow-cytometry at the end of MYB–NFIB fusion peptide-specific T cell expansion, in patient 41. After two rounds of stimulation, T cells were restimulated with fusion breakpoint-derived short peptides (MYB–NFIB SP1–4). DMSO stimulation was the negative control. The percentage of CD137+CD8+ T cells is plotted per peptide. Peptide sequences are shown on the right. d, Exploratory overview of eight immune-evasion mechanisms in 37 patients (Methods). Bottom tracks show response and tumor histology. Dotted lines in the mutation count chart (top) represent the 1st and 2nd tertile thresholds. Darker color shades for the boxes or bars indicate a phenotype expected to be unfavorable for nivo+ipi response. Bottom bar chart shows the number of unfavorable attributes—if not all parameters were available, a sample is labeled ‘NA’. Dotted horizontal lines show the mean number of unfavorable attributes in R (blue) and NR (red) patients. aa, amino acids; Acinic, acinic cell carcinoma; Ex pleo, carcinoma ex pleomorphic adenoma; OR, objective response; Mucoep, mucoepidermoid carcinoma; Myoep, myoepithelial carcinoma.
In the second responding patient with ACC (patient 5), 17 TMGs corresponding to alterations (20 SNVs, four indels and 11 fusions) were constructed (Supplementary Table 9a). Autologous dendritic cells (DCs) were electroporated with individual TMGs before co-culture with autologous T cells. Eight TMGs elicited T cell activation on ELISpot (Extended Data Fig. 6c), of which three showed consistently higher activation of effector memory T cells (CD3+CD45RA−CCR7−) in replicate experiments (Extended Data Fig. 6d). To deconvolve positive TMG hits, autologous DCs were pulsed with 25-mers (Supplementary Table 9b) and co-cultured with autologous T cells. An ELISpot assay confirmed that two SNVs (in TRAPPC12 and FAM47A) consistently elicited T cell activation (Extended Data Fig. 6e), underlining the presence of immunogenic neoantigens in this responding patient with ACC.
Certainly, data from two tumors limit definitive conclusions about the broader landscape of shared neoantigens. Nevertheless, these analyses provide critical mechanistic insights into exceptional nivo+ipi-generated responses of ACC tumors, which are nearly always resistant to ICB. Neoantigen immunogenicity screens provide proof of principle that several genetic alterations in ACC—the MYB–NFIB gene fusion in particular—can generate neoantigens associated with ICB responses. If MYB–NFIB fusion-derived neoepitopes are mediators of tumor regression, this fusion’s high prevalence (~60%) in ACCs16,27 makes it an attractive therapeutic target.
Potential immune evasion mechanisms leveraged by SGCs
Cancers may leverage any of multiple tumor-intrinsic or host factors to evade immune rejection. Based on a previously described framework55, we mapped eight immune evasion mechanisms (Fig. 4d and Methods). The distribution of immune-evasion mechanisms was diverse in non-responding tumors, which tended to have more involved mechanisms (3.8 (95% CI: 3.0, 4.5)) than responding tumors (1.7 (95% CI: −1.2, 4.5)).
Finally, we profiled a tumor biopsy obtained upon progression after PFS of 12.8 months from ACC patient 41. In this tumor, a new, truncating B2M mutation was acquired at progression, anecdotally showing that ACCs may evolve to escape rejection by compromising antigen presentation, even in the context of an initial response.
These analyses are limited by their exploratory nature but together suggest that SGCs leverage a diversity of immune-evasion mechanisms. Although this reveals a clinical challenge, common mechanisms (immunosuppressive TME and upregulation of other T cell checkpoints) are areas of ongoing clinical investigation and may be promising avenues to pursue.
Discussion
An incomplete understanding of the unique lineage-specific biology of rare cancers hinders therapeutic advances. For this reason, correlative analyses of patient samples from clinical trials can be critical to developing new effective treatments for rare cancers. Here we report the results of a phase 2 trial of nivo+ipi in patients with R/M SGC. These data and our analyses of serially collected tumor and blood samples delineate the efficacy of this therapeutic strategy and correlates of response in this rare, aggressive cancer type. This trial met its primary endpoint in cohort 2 (non-ACC SGC histologies) with a 16% ORR but not in cohort 1 (ACC; 6% ORR). Efficacy appeared enriched in SDCs (25% ORR). These responses highlight the therapeutic potential of ICB in SDC and the need for novel strategies to bring this benefit to more patients with other SGC subtypes. However, 48% of patients experienced progression of disease as BOR, and 13 (20%; two with simultaneous PD) discontinued therapy due to toxicity—underlining the need for biomarkers to limit ICB therapy to those who may experience efficacy and spare others from the toxicities of ineffective treatment.
In other cancer types, anti-PD-1 ICB is more effective in patients with highly T-cell-infiltrated, TMB-high tumors36. The data presented here show that, among SGCs, ACC and SDC tumors represent opposite ends of this spectrum. ACC tumors have a low TMB and an immunologically depleted TME; thus, nivo+ipi is expected to have a low success rate—as shown in this trial. However, this does not guarantee that ICB will universally be ineffective. Notably, two patients with ACC tumors experienced deep and long-lasting tumor regressions not previously observed with other classes of therapeutics. The neoantigen screen performed in two ACC responders demonstrates that native immune recognition of neoepitopes can be leveraged by nivo+ipi to induce therapeutic responses. Notably, several neoantigens identified were derived from recurrent mutations and fusions. These ‘public neoantigens’ are promising therapeutic targets, particularly MYB–NFIB fusions in ACC. As prevalent and central oncogenic drivers, they are less likely to be lost under selective pressure. Our findings argue that personalized, neoantigen-directed strategies may be effective for ACC. The low TMB of ACC tumors minimizes the number of neoantigens to target, potentially supporting this approach’s feasibility. In contrast to ACC, the TMB and degree of pre-existing immune cell infiltration were higher in SDCs. The variability in clinical efficacy observed with T-cell-directed therapies may be driven by the distinct immunogenomic profiles that characterize different SGC histologies56.
TCR-seq data revealed that nivo+ipi-responsive tumors were characterized by a significantly more clonally expanded pre-treatment T cell infiltrate, suggesting a pre-existing, tumor-educated expansion of infiltrating T cells. Although the low TCR-seq sample count precludes definitive conclusions, this finding recapitulates reports in other cancers and could, upon validation, offer a pre-treatment biomarker44,46,49. Approaches to estimate the TCR repertoire diversity and clonality from bulk RNA-seq may make this approach clinically feasible57,58.
In concert with the reinvigoration of exhausted T cells in the blood, intratumoral T cell clonotypes were observed to expand markedly in responding tumors—possibly provoked by successful antigenic stimulation—akin to the phenomenon of TCR ‘clonotype expansion’ described in renal cell and lung tumors49,59. ‘Clonal replacement’ (recruitment of novel T cell clones from the periphery into the tumor) was not clearly observed in SGC tumors at week six and is unlikely to be a universal phenomenon in cancer. However, this phenomenon could still occur later on-treatment.
The reinvigoration of pre-existing T cell clonotypes could be a critical component of ICB response44,46,49. Combined with the observed mutational contraction and increase in immune-related RNA signature values, it suggests a model of immunoediting in which nivo+ipi shifts the TME toward a tumor rejection phenotype in treatment-sensitive SGCs, producing an immunologic selection pressure that leads to the loss of mutated tumor cell clones and tumor regression. This model is further supported by the preferential contraction of mutations predicted to be stronger HLA-I binders in responding tumors and aligns with what has been seen in melanomas treated with adoptive cell transfer60 or ICB38. Resistant tumors showed upregulation of inhibitory MDSCs and M2 TAMs or other T cell checkpoints, possibly posing targetable adaptive resistance mechanisms12,61.
Limitations of this study in a rare cancer type are the low number of responding tumors available for analyses and, consequently, the modest analytical power. With this sample size, only more robust molecular determinants of response were identifiable.
A clinical trial evaluating whether the multi-targeted tyrosine kinase inhibitor (TKI) lenvatinib can modify the SGC TME to facilitate ICB responses (NCT04209660) is currently underway. Recently, another trial testing this hypothesis using axitinib plus avelumab (anti-PD-L1) met its primary endpoint with an ORR of 18% in patients with R/M ACC62. Our data suggest that other strategies—currently undergoing clinical testing—aiming to enhance intratumoral immune infiltration and antigen presentation may be promising, such as combining ICB with chemotherapy (pemetrexed (NCT04895735), docetaxel (NCT03360890) or radiation8 (NCT03749460)).
In conclusion, our clinical results show that meaningful responses to nivo+ipi, although uncommon in SGC, can be profound and durable, even in poorly immune-infiltrated and TMB-low ACCs. Other SGC histologies—particularly SDC—seem more susceptible, justifying further development of ICB approaches in these histologies. Empirically testing all potential ICB combinations is not a feasible drug development strategy for this rare disease. The ability to use data presented herein to prioritize the most biologically rational approaches will enhance the likelihood of improving immunotherapy strategies for this understudied cancer.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41591-023-02518-x.
Methods
Patients
Patients were enrolled in two cohorts: cohort 1 included patients with R/M ACC, and cohort 2 consisted of patients with R/M SGC of any histology but ACC. All eligible patients were at least 18 years of age, had an Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1, minimal bone marrow and end-organ function and a histological or cytological diagnosis of ACC (cohort 1) or non-ACC SGC (cohort 2). Willingness to undergo two research tumor biopsies and the presence of RECIST version 1.1-measurable disease not amenable to potentially curative surgery or radiotherapy were required. Pre-treatment with any number of local or (non-immunotherapeutic) systemic treatments was allowed. For cohort 1, patients with a non-salivary gland primary ACC site were allowed. All cohort 1 patients and cohort 2 patients with acinic cell carcinoma histology were required to have radiographic evidence of disease progression on an imaging study performed within 6 months of study enrollment or new/worsening disease-related symptoms during that same period (or both). Critical exclusion criteria for both cohorts were: the presence of symptomatic central nervous system (CNS) metastases (asymptomatic or treated CNS tumors were allowed); prior treatment with immunotherapy targeting PD-(L)1, PD-L2 or CTLA-4 or any other drug targeting T cell co-stimulation or immune checkpoint pathways; active autoimmune disease within the past 2 years; known history of HIV or known AIDS; and the concurrent use of systemic immunosuppressive medications within 2 weeks of study drug administration (including corticosteroids equivalent to >10 mg of prednisone per day). This phase 2 trial was not designed to identify any influence of sex/gender on the efficacy of the study treatment, owing to a lack of evidence for such an interaction in previous studies; in this trial, patient sex was recorded, but no additional gender data were recorded. Further details on eligibility criteria are detailed in the trial protocol (Supplementary Information).
Trial oversight and approval
The trial protocol and all its amendments were approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board (MSKCC IRB) under registration number 17–219. This trial was registered on ClinicalTrials.gov under identifier NCT03172624 and was performed in accordance with the Declaration of Helsinki and International Conference on Harmonization Good Clinical Practice guidelines. All patients provided written informed consent before study enrollment. Enrolled patients were not financially compensated for participation. All correlative tissue analyses were conducted with MSKCC IRB approval under a Morris Lab biospecimen protocol with registration number 11–195.
Trial design and treatment
This is an open-label, non-randomized phase 2 study of nivo+ipi in patients with R/M ACC (cohort 1) or non-ACC SGC (cohort 2), conducted at MSKCC. This manuscript reports on the results of both cohorts. Enrolled patients in either cohort were treated with intravenous nivolumab 3 mg kg−1 every 2 weeks and ipilimumab 1 mg kg−1 every 6 weeks; one complete treatment cycle consisted of 6 weeks. Patients were continued on therapy until unacceptable toxicity, withdrawal of consent or disease progression. Treatment beyond radiographic progression was allowed at the treating investigator’s discretion if the patient was tolerating study treatment; treatment was discontinued if follow-up imaging (performed 4–8 weeks later and then at 8-week or 12-week intervals subsequently) showed an additional ≥10% tumor burden increase from the time of initial progression. Concomitant, palliative radiotherapy of non-target lesions was permitted; dose modifications of study treatment were not. Treatment was delayed based on TRAEs per the trial protocol (Supplementary Information). At the principal investigator’s (PI) discretion, continued treatment with just nivolumab or ipilimumab alone due to AEs was allowed. Treatment was permanently discontinued after a delay of more than 6 weeks per the PI’s discretion or when a patient met the discontinuation criteria specified in the trial protocol (Supplementary Information).
Trial objectives, endpoints and assessments
The primary objective of this study was to determine the efficacy of nivo+ipi in patients with R/M ACC and non-ACC SGC. The primary endpoint was ORR, defined as a confirmed complete response or a confirmed partial response (cCR or cPR) documented by RECIST version 1.1 (ref. 21). The primary endpoint would be met if four or more responses were observed in the 32 patients (13%) in each cohort. Secondary endpoints were PFS and the safety/tolerability of nivo+ipi in patients with R/M ACC or non-ACC SGC. An exploratory objective was to investigate tumor and peripheral blood tissues for potential biologic correlates of nivo+ipi efficacy. The clinical benefit rate—a post hoc analysis because it was not a pre-specified endpoint—was defined as the proportion of patients with cCR, cPR or SD lasting at least 6 months.
Tumor assessments per RECIST version 1.1 (computed tomography (CT) or magnetic resonance imaging (MRI)) were performed at baseline and every 12 weeks (±1 week) after treatment initiation (or every two cycles), using consistent imaging modality—objective PRs or CRs required confirmation with a second assessment performed at least 4 weeks later. AEs were monitored from the start of nivo+ipi until 30 d after the last dose and registered according to CTCAE version 4.0. Any AE deemed at least possibly related to nivolumab or ipilimumab was considered a TRAE. Peripheral blood was drawn at baseline (within 3 d before treatment initiation), within the first weeks of cycles 2 and 3 and then off-study. Tumor specimens were to be obtained at baseline and before administering the second dose of ipilimumab, as per protocol. Exceptions regarding the timing of the second biopsy could be made at the PI’s discretion, and patients were exempt from biopsy if no tumor was safely accessible or if the only tumor accessible for biopsy was also the sole RECIST version 1.1-evaluable lesion.
In total, tissues for genomic analysis were available for 31 of 32 enrolled patients with ACC (cohort 1) and 25 of 32 non-ACC patients (cohort 2). Tissues comprised pre-treatment biopsies in 39 patients (22 cohort 1, 17 cohort 2) and on-treatment biopsies, which were successfully collected in 26 patients (15 cohort 1, 11 cohort 2) at a median of 35 d (IQR 29–40) after treatment start. All samples were snap-frozen upon collection. In an additional 20 patients (10 in each cohort) without dedicated research biopsies, clinical formalin-fixed paraffin-embedded (FFPE) specimens (pre-treatment or on-treatment) were used for genomic profiling. Pre-treatment PBMCs were collected from 63 patients (31 cohort 1, 32 cohort 2); on-treatment PBMCs were obtained a median of 42 d (IQR 42–43) after the start of nivo+ipi in 55 patients (29 cohort 1, 26 cohort 2) (Extended Data Fig. 1a).
Sex-based and gender-based analyses were not pre-specified in the trial protocol and were not performed. Further details on all study assessments are provided in the trial protocol (Supplementary Information).
DNA extraction and WES
DNA was extracted from tumor biopsies, which had been snap-frozen and stored at −80 °C, using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s protocol, modified by replacing AW2 buffer with 80% ethanol. DNA was eluted in 50 μl of 0.5× Buffer AE heated to 55 °C. Matched germline DNA was extracted from whole blood. Exome capture and sequencing were performed at MSKCC and the Broad Institute.
At MSKCC, PicoGreen quantification and quality control by Agilent BioAnalyzer were performed. Then, 199–250 ng of DNA was used to prepare libraries using the KAPA HyperPrep Kit (Kapa Biosystems) with eight cycles of polymerase chain reaction (PCR). After sample barcoding, 100–500 ng of the library was captured by hybridization using SureSelectXT Human All Exon V4 (Agilent) or xGen Exome Research Panel version 1.0 (IDT) according to the manufacturer’s protocol. PCR amplification of the post-capture libraries was carried out for eight cycles. Samples were run on a HiSeq 4000 in a PE100 run, using the HiSeq 3000/4000 SBS Kit (Illumina).
At the Broad Institute, library construction was performed as previously described63, with some modifications. Initial DNA input into shearing was reduced from 3 μg to 10–100 ng in 50 μl of solution. For adapter ligation, Illumina paired-end adapters were replaced with palindromic forked adapters (IDT). KAPA HyperPrep reagents in 96-reaction kit format were used for end repair/A-tailing, adapter ligation and library enrichment PCR. During the post-enrichment SPRI cleanup, the elution volume was reduced to 30 μl to maximize library concentration, and a vortexing step was added to maximize the amount of template eluted. Hybridization and capture were performed using Illumina’s TruSeq Rapid Exome Kit and following the manufacturer’s protocol, with the following modifications: (1) all libraries within a library construction plate were pooled before hybridization; and (2) the Midi plate was replaced with a skirted PCR plate to facilitate automation. All hybridization and capture steps were automated on the Agilent Bravo liquid handling system. Library pools were quantified using quantitative PCR (qPCR) (on the Agilent Bravo) using a Kapa Biosystems kit. Based on qPCR quantification, libraries were normalized and denatured. Next, libraries were diluted to 20 pM using hybridization buffer (Illumina). Cluster amplification was performed according to the manufacturer’s protocol (Illumina) using exclusion amplification cluster chemistry and HiSeq X flow cells. Flow cells were sequenced on version 2.5 sequencing-by-synthesis chemistry for HiSeq X flow cells. The flow cells were then analyzed using RTA version 2.7.0 or later. Each pool of whole-exome libraries was run on paired 76-bp runs, reading the dual-indexed sequences to identify molecular indices and sequenced across the number of lanes needed to meet coverage for all libraries in the pool.
Mutation analysis
The Burrows–Wheeler Aligner (BWA, 0.7.17) (ref. 64) was used to align reads in FASTQ format to the hg37 reference genome. Indel realignment, base quality score recalibration and removal of duplicate reads were done with the Genome Analysis Toolkit (GATK, 3.8–1-0) (ref. 65), following raw reads alignment guidelines66. Five mutation callers were used for SNV detection: MuTect2 (part of GATK 3.8–1-0) (ref. 67,68), SomaticSniper (1.0.5.0) (ref. 69), Strelka (2.9.2) (ref. 70), Varscan (2.4.3) (ref. 71) and Platypus (0.8.1) (ref. 72). Indel calls were generated with VarScan, Strelka, Platypus and MuTect2. Mutations were annotated using SnpEffect & SnpSift (4.3) (ref. 73). All tools were used with the parameters recommended by the authors. Variants were reported if they were called by two or more callers.
A validated custom pipeline17,18 was used to filter variants and minimize the number of false-positive calls. Filters included: a coverage depth of ≥10×; >4% variant nucleotide allelic fraction in tumor DNA; and ≥99% normal allelic fraction in normal DNA. Variants with normal allelic fraction <1%, coverage depth <20× or variant nucleotide allelic fraction in tumor DNA <4% were considered low confidence and manually reviewed using the Integrative Genomics Viewer (IGV, 2.8) (ref. 74).
Indel calls (called by ≥2 callers) were subjected to additional optimization and filtering. Indels in blocklisted (https://www.encodeproject.org/annotations/ENCSR636HFF/) and low-mappability regions (such as repeat maskers) were excluded. Those included in the Catalogue Of Somatic Mutations In Cancer (COSMIC)75, OncoKB76 or the MSK-IMPACT tNGS panel (Supplementary Table 4) were put back in. Common single-nucleotide polymorphisms (SNPs) were eliminated by comparison to snp142.vcf.
Copy number, purity and ploidy analysis
FACETS software (0.6) (ref. 77) was used to acquire allele-specific copy number data from WES. HLA zygosity was assessed by manually reviewing the copy number at the HLA locus on chromosome 6. Estimations for sample tumor purity and mean ploidy were also determined using FACETS. In two patients for whom the change in tumor purity from pre-treatment to on-treatment could not be assessed using FACETS, an RNA-seq-based Δpurity obtained using the ESTIMATE algorithm29 was reported instead. We acknowledge that this algorithm was developed using Affymetrix microarray data.
HLA-I genotyping and neopeptide binding affinity analysis
Germline WES data were analyzed using Polysolver (version 4) (ref. 78) with default settings to determine each patient’s HLA-I alleles in silico. All protein-coding, non-synonymous point mutations and indels (identified as described above) in pre-treatment tumors were translated into strings of amino acids with lengths of 8–11 using a previously described, in-house-written R package38. For SNVs, this was done using a sliding window method. For INDELs, the insertion or deletion of the reported DNA sequence was performed and, using the new sequence, peptides were extracted from the reported amino acid position to the first stop codon or transcript end. The binding affinity of the 38 resulting 8–11-mer peptides for a patient’s HLA-I alleles was tested using NetMHCPan4.0 (ref. 40). Following methodology previously developed by Marty et al.41, the k-mer with the best rank score was selected for each of the patient’s HLA-I alleles. Next, the harmonic mean of these best rank scores was calculated, yielding the PHBR, an aggregate score suggested to reflect a patient’s ability to present a particular mutation. Following Marty et al.41, a PHBR <4 was considered a variant with a relatively high presentation, whereas a PHBR ≥4 was considered relatively poorly presented.
HLA-I evolutionary diversity analysis
HED was calculated as previously described35,79. In short, the protein sequence of the HLA-I peptide-binding groove (exons 2 and 3) was obtained from the ImMunoGeneTics/HLA database80, and the divergence between the allele sequences for HLA-A, -B and -C was calculated using Grantham’s distance metric81. Grantham’s distance accounts for functional differences between amino acids and is defined as the sum of all amino acid differences along the domain of interest (the peptide-binding groove); divergence has a value of 0 in case of homozygosity at that locus, whereas a higher score means a greater evolutionary divergence. Mean HED was calculated per patient as the mean of the pairwise divergences at the HLA-A, -B and -C loci.
Targeted NGS on the MSK-IMPACT platform
For 20 patients whose tumor was not genomically profiled using WES, tNGS data generated using the MSK-IMPACT platform were available, from more variable timepoints: 16 patients before study treatment initiation (median 363 d (IQR 463)) and four patients after study treatment initiation (median 88 d (IQR 159)). MSK-IMPACT is a validated and FDA-approved, capture-based NGS platform targeting 468 genes (410 for two samples in which an older panel was used; Supplementary Table 4). The tissue processing, sequencing and downstream analysis methodology was extensively described in previous publications23,82. The median target coverage of tNGS was 560×. Due to the likely incomparability of NGS-based values obtained at diverse timepoints as opposed to those calculated from WES-biopsies obtained per the study protocol, tNGS-based estimates for parameters such as TMB, purity and ploidy were not statistically compared in concert with WES-based values.
WGS
Additional WGS was performed at the Broad Institute for one sample. An aliquot of genomic DNA (350 ng in 50 μl) was used as the input into DNA fragmentation. Shearing was performed acoustically using a Covaris focused ultrasonicator, targeting 385-bp fragments. After fragmentation, additional size selection was performed using an SPRI cleanup. Library preparation was performed using a commercially available kit provided by Kapa Biosystems (KAPA HyperPrep without amplification module, product KK8505) and with palindromic forked adapters with unique 8-base index sequences embedded within the adapter (purchased from Roche). After sample preparation, libraries were quantified using a qPCR (kit purchased from Kapa Biosystems), with probes specific to the ends of the adapters. This assay was automated using Agilent’s Bravo liquid handling platform. Based on qPCR quantification, libraries were normalized to 2.2 nM and pooled into 24-plexes. Sample pools were combined with NovaSeq Cluster Amp Reagents DPX1, DPX2 and DPX3 and loaded into single lanes of a NovaSeq 6000 S4 flow cell using the Hamilton STARlet liquid handling system. Cluster amplification and sequencing were performed on NovaSeq 6000 instruments using sequencing-by-synthesis kits to produce 151-bp paired-end reads. Output from Illumina software was processed by the Picard data processing pipeline to yield CRAM or BAM files containing demultiplexed, aggregated aligned reads. All sample information tracking was performed by automated LIMS messaging.
RNA extraction and transcriptome sequencing
Phase separation in cells lysed or tissue homogenized in TRIzol reagent (Thermo Fisher Scientific) was induced with chloroform. RNA was precipitated with isopropanol and linear acrylamide and washed with 75% ethanol. The samples were resuspended in 13–35 μl of nuclease-free water. Total RNA was quantified using the Quant-iT RiboGreen RNA Assay Kit (Thermo Fisher Scientific). Transcriptome sequencing was performed at MSKCC under two protocols (ribodepletion and poly(A)) and at the Broad Institute.
For samples run at MSKCC under the ribodepletion protocol, 0.2–1 μg of total RNA with DV200 percentages varying from 54–95% underwent ribosomal depletion and library preparation using the TruSeq Stranded Total RNA LT Kit (Illumina) per the manufacturer’s instructions with eight cycles of PCR. For samples following the poly(A) protocol, 110–150 ng of total RNA with RNA integrity number (RIN) values of 2.5–9.8 underwent poly(A) selection and TruSeq library preparation according to Illumina’s instructions (TruSeq Stranded mRNA LT Kit) with eight cycles of PCR. Both ribodepletion and poly(A) samples were barcoded and run on a HiSeq 2500 or HiSeq 4000 in a PE100 run using the HiSeq Rapid SBS Kit version 2 or HiSeq 3000/4000 SBS Kit (Illumina). For ribodepletion samples, an average of 91 million paired reads were generated per sample, and 23% of the data mapped to the transcriptome. For poly(A) samples, an average of 126 million paired reads were generated per sample, and mRNA bases averaged 56%.
For samples sequenced at the Broad Institute, 2 ul of External RNA Controls Consortium (ERCC) controls was spiked into each plated sample. Then, 200 ng of the sample was transferred into library preparation, which uses an automated variant of the Illumina TruSeq Stranded mRNA Sample Preparation Kit, using oligo dT beads to select mRNA from the total RNA sample. It is followed by heat fragmentation and cDNA synthesis from the RNA template. The resultant 400-bp cDNA then goes through dual-indexed library preparation: ‘A’ base addition, adapter ligation using P7 adapters and PCR enrichment using P5 adapters. Enriched libraries were quantified using Quant-iT PicoGreen. The set was pooled and quantified using the KAPA Library Quantification Kit. Pooled libraries were normalized (to 2 nM) and denatured (using 0.1 N NaOH). Flow cell cluster amplification and sequencing were performed using the NovaSeq 6000. Each run was a 101-bp paired-end with an 8-base index barcode read.
Gene expression analysis
STAR (STAR_2.5.3a) two-pass alignment83 was used to align RNA-seq reads to the hg19 genome. Quality control metrics, such as general sequencing statistics, gene feature and body coverage, were calculated based on the alignment result through RseQC (2.6.4) (ref. 84. Next, RNA-seq gene-level count values were computed using the R package GenomicAlignments (1.14.2) (ref. 85) over aligned reads with UCSC KnownGene86 in hg19 as the base gene model. The Union counting mode was used, and only mapped paired reads after alignment quality filtering were considered. For both the ACC and non-ACC samples, regularized logarithm transformation was performed with the rlog function of the R package DeSeq2 (1.18.1) (ref. 87). Finally, gene-level FPKM (fragments per kilobase of transcript per million mapped reads) and raw read count values were computed by DESeq2.
Immune infiltration assessed using bulk RNA-seq data
Levels of immune infiltration and activity were assessed using multiple previously published scores and signatures. We used ESTIMATE29, a single-sample gene set enrichment analysis (ssGSEA)-based algorithm88, to determine the ImmuneScore. ssGSEA takes the sample FPKM expression values as the input and computes an enrichment score for a given gene list compared to all the other genes in the sample transcriptome. ESTIMATE was also used to infer tumor purity in the samples for which purity could not be assessed with FACETS using WES data. Analyses of individual immune cells were exploratory. The individual immune cell populations were deconvoluted using signatures previously described by Bindea et al.33. The immune infiltration score (IIS, an aggregate score from Bindea et al.’s adaptive and innate immune population scores), T cell infiltration score (TIS, based on nine T cell scores from Bindea et al.) and APM score (based on expression of HLA-A/B/C, B2M and TAP1/2/BP expression) were calculated using ssGSEA methodology according to Şenbabaoğlu et al.30. Immune cytolytic activity was assessed using the CYT score31, calculated from the geometric mean of TPM of GZMA and PRF1 transcript levels. IFN-γ pathway activation was assessed using ssGSEA enrichment of the Reactome IFN-γ pathway (http://reactome.org/)32. The self-mean normalized matrices for ACC and non-ACC were merged and used as input into the Tumor Immune Dysfunction and Exclusion (TIDE) algorithm to determine the MDSC, CAF and M2 TAM signatures34. Finally, Mariathasan et al.’s pan-fibroblast TGF-β response signature was calculated89.
Detection of viral reads
To assess the potential presence of viruses in SGCs, we used the publicly available workflow VirDetect90 (version 1), which is described in detail at https://github.com/dmarron/virdetect. In brief, RNA-seq reads were aligned against the human genome hg38 assembly, including unmapped reads in the result BAM file. Unmapped reads were then aligned to a collection of 1,894 viral genomes (full list in Supplementary Table 5), including human endogenous retrovirus K113 as a positive control. The viral read counts per sample were then captured using the countStarViralAlignments functionality in VirDetect, creating a final viral read count matrix.
MYB–NFIB gene fusion detection
The presence of MYB–NFIB fusions was investigated in cohort 1 patients with sufficient material available using FISH. FISH was performed on paraffin sections using custom probes developed from bacterial artificial chromosomes covering and flanking the MYB and NFIB genes. In total, 200 successive nuclei were examined. Detection of a sufficient break-apart signal was considered a positive score.
Gene fusions were investigated in five tumors with an RNA-based custom fusion panel (MSK-Fusion)91 based on Archer FusionPlex (Invitae) per the manufacturer’s instructions92. If available, bulk RNA-seq data were analyzed for MYB–NFIB gene fusions using the NeoFuse pipeline (version 1.1.2) (ref. 93). This manuscript only reports the Arriba results obtained from NeoFuse implementation. A tumor was declared positive for the MYB–NFIB fusion if any one of the assays was positive.
TCR-seq and analysis
Genomic DNA extracted from tumor biopsies and, for three responding patients, peripheral blood, were submitted to Adaptive Biotechnologies for bulk sequencing of the TCR β-chain on the immunoSEQ platform. Amplicon libraries were prepared by PCR with primers annealing to TCR β-chain V and J gene segments94, which were then sequenced on the HiSeq platform (Illumina). Only those receptor rearrangements that encode for a functional protein (productive rearrangements) were included in downstream analyses. One patient’s (ACC, NR) TCR-seq data were excluded from analysis owing to insufficient pre-treatment sample input (58 sequenced T cells; the trial mean was 3,503 cells).
For tumor samples, the productive Shannon95 and Simpson96 clonality measures were obtained from the ImmunoSEQ Analyzer version 3.0 online software, in which Shannon’s clonality is defined as the inverse of the normalized version of Shannon’s entropy or
and Simpson’s clonality as
where is the total number of productive rearrangements, and is the productive frequency of rearrangement . Both clonality measures have a value between 0 and 1; higher values represent a more clonally skewed sample. The similarity between a patient’s baseline and on-treatment sample was quantified using the TCR overlap index47 and the Morisita–Horn index of similarity97 obtained from the ImmunoSEQ Analyzer, where higher values indicate greater similarity between the two samples. The total number of T cells in a sample was also obtained from the Analyzer platform. We selected the top 1% of the empirical productive frequency distribution for each sample to focus on the most predominant T cell clones in a sample, following methodology in previous reports on TCR-seq analyses49,50.
Intratumoral TCR clonotypes that significantly expanded or contracted upon treatment were identified by comparing their productive frequencies in each pre-treatment and on-treatment sample pair using a binomial model in the ImmunoSEQ Analyzer. Clones were matched in pre-treatment and on-treatment samples based on their unique nucleotide rearrangement. Clones with a combined abundance of fewer than 10 cells in the pre-treatment and on-treatment sample pair were considered very low-frequency TCRs and excluded from statistical testing. The resulting P values were corrected for multiple testing using the Bonferroni–Holm method within each longitudinal tumor sample pair separately. Clones with a corrected P < 0.01 were considered to have undergone significant expansion or contraction.
The peripheral presence of TCR clonotypes considered undergoing expansion based on TCR-seq of tumor samples was investigated using their unique nucleotide rearrangement in the pre-treatment and on-treatment (week 6, same time as on-treatment tumor-TCR-seq) blood samples of three responding patients. For one of these patients, an additional blood sample obtained 336 d on-treatment was available for TCR-seq. The clonotypes of interest were tracked in the blood, and figures were created using Immunarch version 0.9.0 (ref. 98).
Peripheral blood flow cytometry and NLR
Patients’ PBMCs were isolated from a peripheral blood draw at baseline and 6 weeks after the first dose of nivo+ipi by centrifugation in CPT tubes at 1,500g for 20 min without brakes and stored in liquid nitrogen. Upon analysis, PBMCs were thawed, washed and counted. Staining was performed using a fixable Aqua viability dye (Invitrogen, L34957, 1:250) and a cocktail of antibodies directed at six surface markers: CD8, Qdot 605 (Invitrogen, clone 3B5, Q10009, 1:250); CD4, Qdot 655 (Invitrogen, clone S3.5, Q10007, 1:500); PD-1, PE (BD Biosciences, clone MIH4, 557946, 1:2.5); LAG-3, FITC (Enzo Life Sciences, clone 17B4, ALX-804–806F-C100, 1:100); ICOS, PE-Cy7 (eBioscience, clone ISA-3, 25–9948-42, 1:200); and TIM-3, APC (R&D Systems, clone 344823, FAB2365A, 1:20). Next, cells were fixed and permeabilized with FoxP3/Ki-67 Fixation/Permeabilization Concentrate and Diluent (eBioscience, 00–5123 and 00–5223) before intracellular staining with CD3, BV570 (BioLegend, clone UCHT1, 300436, 1:200); Ki-67, Alexa Fluor 700 (BD Biosciences, clone B56, 561277, 1:83); FoxP3, eFluor450 (eBioscience, clone PCH101, 48–4776-42, 1:125); and CTLA-4, PerCP-eFluor710 (eBioscience, clone 14D3, 46–1529-42, 1:50). Stained cells were acquired on a BD LSRFortessa and analyzed with FlowJo software (version 10.5). Isotype control stains were performed to establish positivity gates for PD-1, LAG-3, ICOS, TIM-3, FoxP3 and CTLA-4. The gating strategy is visualized in Supplementary Fig. 5, and the panel is summarized in Supplementary Table 7.
The NLR was defined as the neutrophil count (per nanoliter) divided by the lymphocyte count (per nanoliter) using routine clinical laboratory blood testing obtained at trial enrollment.
IHC
Staining for PD-L1 was performed using a rabbit anti-human PD-L1 antibody (Cell Signaling Technology, 13684, clone E1L3N, 1:400). The percentage of positive tumor cells (TCs) was calculated as the surface area of TCs showing partial or complete membranous staining of any intensity divided by the surface area of the tumor.
Staining for AR (Abcam, 105225, clone SP107, 1:125) and HER2 (Roche, 790–100, clone 4B5, ready-to-use) was performed as part of routine pathology evaluation of SDCs. HER2 immunostaining was performed as follows: HER2 scoring of 0+ was defined as either no staining observed or incomplete membrane staining that is faint or barely perceptible within ≤10% of invasive tumor cells; HER2 IHC scoring of 1+ was defined as incomplete faint or barely perceptible membrane staining within >10% of invasive tumor cells; HER2 scoring of 2+ was defined as weak to moderate complete membrane staining observed in >10% of invasive tumor cells; and HER2 scoring of 3+ was defined as complete, intense circumferential membranous staining in >10% of invasive tumor cells. Samples with a score of 3+ were considered HER2 positive by IHC.
Neoantigen identification experiments
Cells and media.
PBMCs were cultured in complete IMDM (supplemented with GlutaMAX and 10% human serum, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin). Immature DCs were isolated from PBMCs using the plastic-adherence method, as described previously53, in AIM-V media supplemented with GM-CSF (1,000 U ml−1) and IL-4 (200 U ml−1) over 6 d. Cell media were replenished on the third day. T cells were isolated from PBMCs by pan-expansion using anti-CD3/anti-CD28 Dynabeads, IL-2 (30 U ml−1) and IL-15 (50 ng ml−1) and cultured in complete IMDM media. Cytokines were replenished every 3 d. T cells were expanded for 10–12 d before cryopreservation. COS-7 (a monkey fibroblast cell line) and T2 cells were cultured in RPMI media supplemented with 10% FBS, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin and 2 mM L-glutamine. T2 is a lymphoma-derived, HLA-A2-expressing cell line deficient for the transporter associated with antigen processing (TAP) protein and, hence, lacks endogenous antigen presentation. T2 cells can bind peptides exogenously and are routinely used to study major histocompatibility complex class I (MHC-I)-restricted peptide presentation and T cell activation in co-culture assays.
TMG construction.
For the TMG construction, each non-synonymous SNV was constructed as a minigene encoding the mutant amino acid flanked by 30 amino acids of the wild-type (WT) sequence and fused in-frame in tandem. For other variants (indels and fusions), the entire out-of-frame gene fragments were fused in-frame until the truncation by the first stop codon. Five to 10 TMGs were cloned into pcRNA2SL using EcoRI and BamHI. The constructs were linearized by Not I (New England Biolabs) and purified using a PCR purification kit (Qiagen). One microgram of linearized DNA was used as a template for in vitro transcription of TMG mRNA with a 5′-cap analog and 3′-polyadenylation using the HiScribe T7 ARCA mRNA with tailing kit (New England Biolabs) as per the manufacturer’s instructions. RNA was precipitated using LiCl2 and resuspended at 1 μg μl−1 in nuclease-free water.
Co-culture assay in the ELISpot plates.
TMG mRNA-electroporated or peptide-pulsed DCs were used as target APCs for neoantigen screening in patient 5. One million DCs were resuspended in 200 μl of Opti-MEM with 2 μg of in vitro synthesized TMG mRNA and electroporated with a single pulse of 150 V for 10 ms. Electroporated DCs were allowed to recover in complete IMDM media supplemented with 200 U ml−1 IL-4 at 37 °C overnight and then washed with serum-free RPMI before co-culturing with expanded T cells. For patient 41 neoantigen screening, TMG-transfected COS-7 or peptide-pulsed T2 cells were used as APCs—autologous DCs could not be generated due to the limited availability of PBMCs for this patient. Every TMG vector was co-transfected in COS-7 cells with each of the patient’s autologous HLA-encoding plasmids (A0201, B4402, B0801, C0501 and C0701) individually in a 1:1 ratio using Lipofectamine LTX reagent in a six-well tissue culture treated plate. Co-transfected cells were pooled for each TMG clone after 24 h of incubation and used as APCs in the TMG screen. T2 cells were electroporated using the Neon Transfection system with a single pulse of 1,375 V for 20 ms with the autologous HLA mRNAs of the patient other than HLA A2 (naturally expressed in T2) and incubated for 12 h before peptide pulsing. For peptide pulsing, autologous DCs or HLA-transfected T2 cells were incubated with 20 μM peptides and β2-microglobulin (5 μg ml−1) for 18 h at 37 °C and then irradiated with 50 Gy before co-culturing with the T cells.
Freshly expanded or cryopreserved T cells were rested for 1–3 d before using them as effector cells in the co-culture assay. Additionally, T effector memory cells were FACS sorted from PBMCs (patient 5) to use as effector cells for a replicate experiment (Extended Data Fig. 6d) to confirm neoantigen immunogenicity of selected TMGs in this patient. Freshly thawed PBMCs were stained with fluorescently labeled antibodies for CD3, CD45RA and CCR7 surface markers, followed by sorting of T effector memory (CD3+CD45RA−CCR7−) cells, which were used as effector cells in the ELISpot assay. In total, 10,000–50,000 T cells (as indicated) were co-cultured with the APCs in a 1:1 ratio in complete IMDM overnight at 37 °C on the IFN-γ ELISpot plates (R&D Systems) without cytokines. Duplicate or triplicate wells were set up for each condition. IFN-γ ELISpot plates were developed according to the manufacturer’s instructions.
Expansion and activation of fusion peptide-specific T cells.
The antigen-specific T cell expansion was performed as reported previously99, with some modifications. Cryopreserved PBMCs derived from patient 41 were revived in AIM-V media without serum. One million PBMCs were seeded per well in a 24-well plate and supplemented with 1,000 IU ml−1 GM-CSF and 500 IU ml−1 IL-4 for 24 h. On day 2, the cells were stimulated with MYB–NFIB fusion long peptide MYB–NFIB LP (10 μM) in the presence of IL-β (10 ng ml−1) and LPS (100 ng ml−1) for effective activation of APCs and, subsequently, T cells. One well was stimulated with DMSO (no peptide) as negative control. Starting on day 2, IL-2 (10 IU ml−1) and IL-15 (10 ng ml−1) cytokines were supplemented every 3 d in complete IMDM media to feed the T cells. On day 6, specific short fusion peptides (10 μM) were added to the respective wells for stimulation to expand the peptide-specific T cell population. On day 11, the last round of short peptide stimulation was given in the absence of cytokines, and, 24 h after stimulation, T cells were analyzed with flow cytometry for the upregulation of activation marker CD137. DMSO stimulation was given in the respective well to estimate background activation. The cells were stained with FITC-CD3 (BioLegend, 300406, clone UCHT1, 1:200), BUV395-CD4 (BD Biosciences, 563552, clone SK3, 1:200), PE/Dazzle 594-CD8 (BioLegend, 344743, clone SK1, 1:200) and APC-CD137 (BioLegend, 309809, clone 4b4–1, 1:200) surface marker antibodies after staining with LIVE/DEAD Aqua dye (Invitrogen, L34957 1:500) and Human TruStain FcX Fc receptor block (BioLegend, 422302). CD3+CD8+ gated cells were analyzed for CD137 surface expression. The percentage of CD137+ cells in the CD8+ T cell population was plotted. CD137 FMO was used to set the gate for the CD137+ population. Triplicate wells were set up for each condition.
Exploratory analysis of immune-evasion mechanisms in SGCs
The presence of eight well-described immune-evasion mechanisms was explored in our dataset: (1) low mutational antigenicity (based on the tumor mutation count); (2) low immune cell infiltration (based on the arithmetic mean of the z-scores for the TIS, IIS, ImmuneScore, CYT and IFN-γ RNA signatures); (3) the presence of T cell checkpoints (based on the arithmetic mean of the gene expression z-scores of the non-targeted checkpoint genes coding for LAG-3, TIM-3, VISTA and TIGIT); (4) the presence of immunosuppressive cellular populations (based on the arithmetic mean of the gene expression signature scores for MDSCs, CAFs and M2 TAMs); (5) soluble inhibitory factors (based on a previously developed TGF-β RNA signature89); (6) deficient antigen presentation (based on the APM RNA signature, the presence of HLA loss of heterozygosity (LOH) and the mean LOH); (7) the absence of the PD-L1 checkpoint on the tumor surface (based on the PD-L1 tumor proportion score (TPS)); and (8) unfavorable host features (based on a patient’s body mass index (BMI) and NLR).
The results of this exploratory analysis are summarized in Fig. 4d. For the mutation count, mean immune signature, inhibitory cells, immune checkpoints, TGF-β, APM and mean germline HED, the tertiles of the data distribution were used as thresholds to classify values as low, intermediate (mid) or high. HLA LOH was dichotomized between present and absent. PD-L1 TPS was dichotomized between negative (TPS 0) and non-negative (TPS ≥1). NLR was dichotomized using a widely used value of 5 as threshold100. For BMI, patients were dichotomized between underweight (<18.5 kg m−2) and not underweight (≥18.5 kg m−2). In Fig. 4d, a darker color shade for the boxes or bars represents a phenotype for that mechanism expected to be unfavorable for response to nivo+ipi. The total number of unfavorable attributes was calculated by summing all the scores considered unfavorable and is shown in a bar chart; samples for which not all parameters were available were labeled ‘NA’. Dotted horizontal lines show the mean number of unfavorable attributes in responding (blue) and non-responding (red) patients.
Statistical analysis
Clinical data for cohorts 1 and 2 were analyzed separately. Adopting an ORR (CR or PR) of 5% as the null hypothesis and a 20% ORR as desirable, a minimax two-stage design was used for each cohort. To detect a difference between an unacceptable 5% and a desirable 20% ORR with an alpha of 10% and 90% power, one or more response(s) detected within six cycles of study treatment was required in the first 18 patients of the first stage. After achieving this, an additional 14 patients were accrued in the second stage, for a total of 32 per cohort. At the end of the trial, four or more responses among the 32 patients (13%) in a cohort would be needed to meet the primary endpoint.
All patients who received at least one dose of either nivo or ipi were included in the evaluation of the primary efficacy evaluation. Patients removed from the study before the first response assessment for disease progression or toxicity were classified as non-responders. All patients who received at least one dose of either nivo or ipi were assessed for the secondary safety endpoint using CTCAE version 4.0. PFS was estimated using Kaplan–Meier methodology and defined as the time from treatment start until death or disease progression, whichever occurred first. In the absence of disease progression or death, patients were censored for PFS on the date they came off-study or, for the one patient still on study treatment, on the date of their most recent RECIST evaluation. Clinical disease progression was defined as any clinical event that, in the investigator’s judgment, constitutes PD apart from RECIST PD.
Translational correlates were compared among histologic subtypes (ACC versus SDC versus non-ACC/SDC) or between responders (CR or PR) and non-responders (SD or PD) from both cohorts grouped together, unless expressly stated otherwise. The cohort 2 patient unevaluable for the primary ORR endpoint was included as a non-responder, whereas the cohort 1 patient with an unconfirmed PR was grouped with the responders. Median values were compared between groups using Wilcoxon rank-sum or Kruskal–Wallis tests. Paired values (for example, baseline and on-treatment within the same patient) were compared using Wilcoxon signed-rank tests. Frequency data cross-classified according to two categorical variables were statistically compared using a χ2 test. In consideration of multiplicity when performing multiple tests under the same hypothesis, the Benjamini–Hochberg method was used to control for the false discovery rate (FDR) at an alpha of 10% when comparing (1) immunogenomic variables in pre-treatment tumors between responding and non-responding tumors (Fig. 2a–c, Extended Data Fig. 2d and Supplementary Fig. 3g); (2) immunogenomic variables among ACCs, SDCs and other SGC histologies (Extended Data Fig. 2a,b and Supplementary Fig. 3a,c,d,f); (3) the degree of on-treatment change in mutation count and purity, comparing baseline to on-treatment (Fig. 2f and Extended Data Fig. 3b,e); (4) TCR repertoire metrics derived from TCR-seq between responding and non-responding tumors (Fig. 3a–c,h,i and Extended Data Fig. 4c–e,g); and (5) peripheral blood immune frequency and marker changes (Extended Data Fig. 5a–c). In these figures, FDR-controlled q values are reported. In all other analyses, a nominal P value less than 0.05 was considered statistically significant. We report two-sided P values in all figures except Fig. 2g (PHBR analysis), where we validated a directional hypothesis based on prior literature using a one-sided hypothesis test. Linear correlations between continuous variables were calculated using Spearman’s method. Exact CIs (95%) were calculated using the Clopper–Pearson method. Where appropriate, samples were hierarchically clustered using the complete linkage method. All box plots in this manuscript visualize the median (center bar) and IQR, with the whiskers extending from the first and third quartile to the minimal and maximal value, respectively, but no further than 1.5× IQR. The trial protocol (Supplementary Information) details the full statistical analysis plan.
Data visualization was performed in RStudio version 1.4.1717, using the following packages: tidyverse (1.3.1), survival (3.2–11), survminer (0.4.9), corrplot (0.85), pheatmap (1.0.12), table1 (1.4.2), swimplot (1.2.0), ggstatsplot (0.9.1), Immunarch (0.9.0) and maftools (2.8.05) (ref. 101).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. Schematic trial overview and the genomic landscapes of ACC and non-ACC SGCs.
In b and c, all 64 patients are shown. Patients are grouped per cohort and according to the molecular profile method used (WES or targeted next-generation sequencing (tNGS) with the MSK-IMPACT panel); non-ACC patients are further grouped by histologic subtype. Tracks for genes were limited to show only the genes included in the MSK-IMPACT468 panel (Supplementary Table 4). a. Trial overview flowchart. The numbers for WES, RNAseq, and TCRseq refer to the samples that were subject to these investigations and passed quality control. Cartoons representing tumor and blood samples were created using BioRender.com. b. From top to bottom: patient trial ID, MYB-NFIB fusion gene presence, percentage of tumor cells (TC) staining positive for PD-L1, objective response (OR), mutation status for the top 15 most frequently mutated genes, the molecular profiling method used for each sample, the WES-based (FACETS) ploidy and purity estimate, and number of mutations per exome (WES-based) or TMB score (tNGS-based). c. From top to bottom: patient trial ID, the histologic subtype per the WHO classification, PD-L1 %TC staining, androgen and HER2-receptor status (performed as part of routine clinical care; only on suspected salivary duct carcinomas), OR, status for the top 15 most frequently mutated genes, the molecular profiling method used for that sample, the FACETS-based ploidy and purity estimate, and number of mutations per exome (WES-based) or TMB score (tNGS-based). Pos, positive; Neg, negative; OR, objective response; R, response; NR, no response; NE, not evaluable; SDC, salivary duct carcinoma; ca, carcinoma; NOS, not otherwise specified; CAMSG, cribriform adenocarcinoma of the minor salivary gland; SWI/SNF, SWItch/Sucrose Non-Fermentable; ex pleo, ex pleomorphic adenoma; AR, androgen receptor; WES, whole-exome sequencing; tNGS, targeted next-generation sequencing.
Extended Data Fig. 2 |. Pre-treatment immunogenomic features of ACCs and non-ACC SGCs in the context of treatment (non-)response.
Box plots defined in Methods. Individual dot colors in a, b, d–f indicate SGC histology. A Kruskal-Wallis (a, b) or two-sided Wilcoxon rank-sum test (d–f) was used to calculate exact P-values. P-values in a, b, d were adjusted for multiplicity (Methods), yielding q-values. a. Non-synonymous mutation count per exome in ACC (n = 21), SDC (n = 5), and SGCs of other histologies (n = 10). b. Z-scores for the ESTIMATE T cell and immune infiltration score (TIS and IIS), ImmuneScore, Reactome interferon gamma (IFN-γ), and cytolytic activity (CYT) RNA signatures in ACC (n = 15), SDC (n = 4), and SGCs of other histologies (n = 8). c. Heatmap of the signatures shown in b. Top tracks represent sample histology and objective response of n = 27 samples. d. Values for the MDSC, M2 TAM, and CAF RNA signatures in NR (n = 23) and R tumor samples (n = 4). e. Mean evolutionary divergence of germline HLA (HED), obtained from healthy control WES data, in NR (n = 31) and R patients (n = 5). f. Peripheral blood neutrophil-to-lymphocyte ratio (NLR) in NR (n = 56) and R patients (n = 8).
Extended Data Fig. 3 |. On-treatment trajectories of the mutational and microenvironmental profiles of SGCs, in the context of treatment (non-) response.
Box plots defined in Methods. Dot colors in b, e indicate histology and two-sided Wilcoxon rank-sum tests were used to calculate exact P-values. Panel f shows linear models with regression lines flanked by 95% CIs. Spearman’s rho and two-tailed P-values are printed in f. P-values in b, e were multiplicity-adjusted (Methods), yielding q-values. a. Waterfall plot of the log2-fold change in mutation count from pre-treatment to on-treatment. Bar colors represent response. Top track shows histology. b. Log2-fold change in mutation count from pre-treatment to on-treatment in sample pairs for in R (n = 4) and NR patients (n = 20). c. Proportion of lost mutations with a variant allele frequency of ≥0.10, 0.08–0.09, 0.05–0.07, and <0.05. The denominator is the sum of variants that were lost upon treatment, in NR and R patients. Comparisons of proportions between NR and R patients are printed to the right of the plot; P-values were calculated using a χ2 test. d. Waterfall plot showing the absolute change in tumor purity from pre-treatment to on-treatment. Color indicates if the WES-based FACETS or RNAseq-based ESTIMATE tool was used. The order of samples in the plot is identical to the waterfall plot in a. Two samples are marked NA (no RNAseq and a diploid copy number precluding purity estimation from FACETS). e. Absolute change in tumor purity for sample pairs from pre- to on-treatment (see d) in R (n = 3) and NR patients (n = 19). f. Linear regression of the TCR-enumerated T cell count versus the ImmuneScore, IIS, CYT, IFN-γ, and TIS RNA signatures (n = 13 pre-treatment and n = 13 on-treatment combined). g. Heatmap of the change (on-treatment minus pre-treatment) in immune cell populations, checkpoints, and antigen presentation machinery (APM) RNA signatures. h. Correlation matrix of the change in immune-related RNA signatures. Circle color represents Spearman’s rho (also printed). All correlations were statistically significant (P < 0.05).
Extended Data Fig. 4 |. Additional T cell receptor sequencing data from pre-treatment and on-treatment tumors and peripheral blood.
Boxes are defined in Methods. Panels a and b show linear models with regression lines flanked by 95% CIs; Spearman’s rho and exact, two-tailed P-values are printed. In a and b, squares and circles represent NR and R samples, respectively. Dot colors in a, b, d, and line colors in c indicate SGC histology. P-values in c, d, and g were calculated using two-sided Wilcoxon rank-sum tests and adjusted for multiplicity (Methods), yielding q-values. a. Absolute change in mutation count per exome versus the absolute change in TCR-enumerated T cell count (n = 18). b. Absolute change in WES-based sample purity estimates versus the absolute change in TCR-enumerated T cell count (n = 16). c. Productive Simpson TCR repertoire clonality of pre-treatment and on-treatment R (n = 3) and NR (n = 15) samples. Lines connect a sample pair. d. Morisita-Horn similarity index between the pre-treatment and on-treatment TCR repertoires in R (n = 3) and NR (n = 15) patients. e. On-treatment trajectories of T-cell clonotypes considered predominant (top 1% of the productive frequency distribution in that sample) in pre-treatment tumors, for R (n = 3) and NR (n = 15) samples. Clones maintained upon treatment are shown in orange, lost clones in gray. P-value was calculated using a χ2-test and adjusted for multiplicity (Methods). f. Total number of TCR clones considered significantly expanded (see Fig. 3g,h) in individual patients. The fraction that is pre-existing or novel is indicated. Patients’ trial IDs are printed. g. Total number of expanded TCR clonotypes in responders (n = 3) and non-responders (n = 15). h. TCR repertoire overlap between the tumor and peripheral blood for three responding patients (44, 5, and 41) at pre-treatment and on-treatment. For patient 5 with an additional, 336 d blood sample available (‘1 yr’), the TCR overlap between the 336 d sample and the early on-treatment (week 6) tumor was calculated.
Extended Data Fig. 5 |. Peripheral blood immune monitoring using flow cytometry.
Gating strategy available in the Supplementary Information. Boxes are defined in the Methods. Line and dot colors indicate response. Two-tailed, exact P-values were calculated using a Wilcoxon signed rank (a, c) or rank sum test (b). Nominal P-values were adjusted for multiplicity (Methods). a. The percentage of peripheral CD8+ T cells expressing Ki-67 (left plot) or ICOS (right plot) at the pre-treatment and week 6 on-treatment time point for 27 ACC patients. b. Log2-fold change in Ki-67 (left plot) and ICOS (right plot) surface expression in peripheral CD8+PD1– and CD8+PD1+ T cells for 27 ACC patients. c. Box plots showing the percentage of peripheral CD8+Ki67+ T cells expressing immune checkpoints CTLA-4, LAG-3, PD-1, or TIM-3 at the pre-treatment and on-treatment time points for 27 ACC patients.
Extended Data Fig. 6 |. Extended data from the neoantigen identification experiments for patients 41 and 5.
Data in a and c–e are representative of two independent experiments with either n = 3 (a, d) or n = 2 (c, e) technical replicates. In a and c–e, the black horizontal bars indicate the mean of replicate experiments, and the dotted gray lines represent the mean of the negative control experiment. In a and d, the whiskers represent the standard deviation. a. Autologous T cells from patient 41 were co-cultured with a pool of HLA and individual TMG co-transfected COS-7 cells. T cell responses were measured by IFN-γ ELISpot assay. Untransfected COS-7 cells (no TMG) were co-cultured with T cells for background response determination. b. Representative flow cytometry plots showing CD137 upregulation on CD8+ T cells as an activation marker after restimulation with MYB-NFIB fusion breakpoint-derived short peptides (SP1–4). The DMSO-stimulated T cell response was used to estimate the background activity. Fluorescence minus one (FMO) control was used to set the gate for CD137 expression. c. Interferon-γ production from T cells in ELISpot assay after co-culture with autologous DCs electroporated with TMGs in patient 5. The negative control consisted of co-culture with DCs only. d. Repeat IFN-γ ELISpot from peripheral blood effector memory T cells (CD3+CD45RA–CCR7–) in patient 5, after co-culture with autologous DCs electroporated with TMG constructs. The negative control consisted of co-culture with DCs only. e. IFN-γ ELISpot from patient 5’s peripheral blood T cells upon co-culture with autologous DCs pulsed with peptides translated from the variants from two SNV TMGs and one in-frame INDEL TMG. The negative control consisted of co-culture with DCs only.
Extended Data Table 1 |. BORs for cohort 1 (ACC) and cohort 2 (non-ACC) patients.
| BOR | Cohort 1 | Cohort 2 | Overall |
|---|---|---|---|
| (n = 32) | (n = 32) | (n = 64) | |
| Confirmed CR, n (%) | 0 | 0 | 0 |
|
| |||
| Confirmed PR, n (%) | 2(6) | 5(16) | 7(11) |
|
| |||
| SD, n (%) | 17(53) | 8(25) | 25(39) |
| RECIST SD | 16 | 8 | 24 |
| Unconfirmed PR | 1 | 0 | 1 |
|
| |||
| PD. n (%) | 13(41) | 18 (56) | 31 (48) |
| RECIST PD | 10 | 13 | 23 |
| Clinical PD | 0 | 3 | 3 |
| Death | 3 | 2 | 5 |
|
| |||
| Unevaluable, n (%) | 0 | 1(3)* | 1(2) |
|
| |||
| ORR, n (%) | 2(6) | 5(16) | 7(11) |
Percentages may not add up to 100 due to rounding.
Counted as non-responder for the primary endpoint.
Supplementary Material
Acknowledgements
We are grateful to our patients and their families for their bravery and support of cancer research. This study was supported, in part, by National Institutes of Health (NIH) grant R01 DE027738 (to L.G.T.M., A.L.H. and T.A.C.); the Geoffrey Beene Cancer Research Center (to L.G.T.M. and A.L.H.); the Jayme and Peter Flowers Fund; the Sebastian Nativo Fund; Congressionally Directed Medical Research Programs Award CA210784 and the MSK Population Science Research Program (to L.G.T.M.); the Overman Fund (to A.L.H.); NIH grants R37 CA259177, R01 CA269733 and P50 CA217694 (to C.A.K.); and NIH/NCI Cancer Center Support Grant P30 CA008748 (institutional, to MSKCC). Bristol Myers Squibb provided the study drugs and funding (institutional, to MSKCC) to support conduct of the clinical trial, including research biopsies, but was not involved in data analysis,manuscript writing or the decision to submit this manuscript. In addition, we acknowledge using the MSKCC Integrated Genomics Operation Core, funded by NCI Cancer Center Support Grant P30 CA08748, Cycle for Survival and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Competing interests
A.L.H. reports research funding for clinical trials from Allos Therapeutics, Astellas Pharma, AstraZeneca, Bayer, Ayala Pharmaceuticals, Bristol Myers Squibb, Genentech, Celldex Therapeutics, Daiichi Sankyo, Eisai., Elevar Therapeutics, Eli Lilly & Company, Genentech/Roche, Hoikpia, Kolltan Pharmaceuticals, Kura Oncology, Merck, Novartis, Pfizer, Poseida and Verastem; service in consulting/advisory roles for AffyImmune Therapeutics, AstraZeneca, Ayala Pharmaceuticals, Bristol Myers Squibb, Cellestia Biotech, Coherus, CureVac, Eisai, Elevar Therapeutics, Exelixis, Expert Conncet, Genzyme, InxMed, Kura Oncology, McGivney Global Advisors, Merck, Novartis, CureVac, Prelude Therapeutics, Regeneron, Rgenta, Remix Therapeutics, Sanofi, Sun Pharma, the Chemotherapy Foundation and TRM Oncology; service on speakers’ bureaus for Medscape, Omniprex America, Novartis and Physician Education Resource; and receipt of travel/accommodations expenses from Janssen Oncology, Merck, Kura Oncology, Ignyta, Ayala Pharmaceuticals and KLUS Pharma, outside the submitted work. A.L.H. is also inventor on a patent for the use of lesional dosimetry methods for tailoring targeted radiotherapy in cancer. L.G.T.M. is listed as an inventor on intellectual property held by MSK on using tumor mutation burden to predict immunotherapy response, with pending patent, which has been licensed to Personal Genome Diagnostics. T.A.C. reports, all outside the submitted work, being a co-founder of Gritstone Oncology and holding equity; holding equity in An2H and acknowledging grant funding from Bristol Myers Squibb, AstraZeneca, Illumina, Pfizer, An2H and Eisai; having served as an advisor for Bristol Myers, MedImmune, Squibb, Illumina, Eisai, AstraZeneca and An2H; andd being an inventor on intellectual property held by MSK on using tumor mutation burden to predict immunotherapy response, with pending patent, which has been licensed to Personal Genome Diagnostics. C.A.K. is a scientific co-founder and equity holder of Affini-T Therapeutics; is a compensated member of the scientific and/or clinical advisory boards for Achilles Therapeutics, Affini-T Therapeutics, Aleta BioTherapeutics, Bellicum Pharmaceuticals, Catamaran Bio, Obsidian Therapeutics and T-knife; has consulted for Bristol Myers Squibb, Decheng Capital, PACT Pharma and Roche/Genentech; and has patents broadly related to cell and gene therapy outside the scope of this work. N.R. reports research funding from ArcherDx and Repare Therapeutics and personal fees from Illumina, PaigeAI and Pfizer Canada, outside the submitted work. E.J.S. reports institutional research funding from Merck, outside the submitted work, and personal fees from Eli Lilly & Company, Blueprint Medicines Corporation, Regeneron Pharmaceuticals, Loxo Oncology and Eisai, outside the submitted work. L.A.D. reports research funding and personal fees from CUE-101, Eisai, CUE-101, Replimune Group and Regeneron Pharmaceuticals and service on an advisory board at Merck, outside the submitted work. V.T. reports holding stock in Infinity Pharmaceuticals, Bluebird Bio and Mersana Therapeutics. C.L.Z. is linked to investigator-initiated clinical trials in collaboration with Bristol Myers Squibb, outside the submitted work. D.G.P. reports grants from the National Institutes of Health (NIH) and the Philanthropy-Serra Fund; research support from Hookipa Pharma; and personal fees from Nykode and Hookipa Pharma, outside the submitted work. B.B. is an employee of AstraZeneca. Z.N. is an employee of PPD, part of Thermo Fisher Scientific. W.Y. is an employee of Eli Lilly & Company. V.M. is listed as an inventor on a patent assigned to MSK broadly related to determinants of cancer response to immunotherapy. J.L.V., S.J., C.W.R.F., F.K., C.Y.H., M.P., N.K., R.M.S., I.O., A.K., L.S.M., J.V.F., K.K.N., J.E. and S.H. declare no competing interests.
Footnotes
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41591-023-02518-x.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41591-023-02518-x.
Data availability
A de-identified dataset—containing the clinical features and processed data that underlie results reported in this article derived from tumor whole-exome, RNA and TCR sequencing—has been made available on Zenodo (https://doi.org/10.5281/zenodo.8180441). The relevant sections of the trial protocol, including the statistical analysis plan, have been made available in the Supplementary Information. Raw whole-exome and RNA sequencing data generated in this study have been made publicly available at the Sequence Read Archive under Bio-Project number PRJNA940989. The hg37 and hg19 reference genomes are available from https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000001405.13/, and the hg38 assembly is available from https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000001405.26/.
References
- 1.Speight PM & Barrett AW Salivary gland tumours. Oral. Dis 8, 229–240 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Bennett AK, Mills SE & Wick MR Salivary-type neoplasms of the breast and lung. Semin. Diagn. Pathol 20, 279–304 (2003). [DOI] [PubMed] [Google Scholar]
- 3.WHO Classification of Head and Neck Tumours 4th edn (eds EI-Naggar AK, Chan JK, Grandis JR, Takata T. & Slootweg P) (IARC, 2017). [Google Scholar]
- 4.Laurie SA, Ho AL, Fury MG, Sherman E. & Pfister DG Systemic therapy in the management of metastatic or locally recurrent adenoid cystic carcinoma of the salivary glands: a systematic review. Lancet Oncol. 12, 815–824 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Laurie SA & Licitra L. Systemic therapy in the palliative management of advanced salivary gland cancers. J. Clin. Oncol 24, 2673–2678 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Ribas A. & Wolchok JD Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen RB et al. Pembrolizumab for the treatment of advanced salivary gland carcinoma: findings of the phase 1b KEYNOTE-028 study. Am. J. Clin. Oncol 41, 1083–1088 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahmood U. et al. A randomized phase 2 study of pembrolizumab with or without radiation in patients with recurrent or metastatic adenoid cystic carcinoma. Int. J. Radiat. Oncol. Biol. Phys 109, 134–144 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Even C. et al. Evaluation of pembrolizumab monotherapy in patients with previously treated advanced salivary gland carcinoma in the phase 2 KEYNOTE-158 study. Eur. J. Cancer 171, 259–268 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Fayette J. et al. NISCAHN: a phase II, multicenter nonrandomized trial aiming at evaluating nivolumab (N) in two cohorts of patients (pts) with recurrent/metastatic (R/M) salivary gland carcinoma of the head and neck (SGCHN), on behalf of the Unicancer Head & Neck Group. J. Clin. Oncol 37, 6083–6083 (2019). [Google Scholar]
- 11.Mukaigawa T. et al. Programmed death ligand-1 expression is associated with poor disease free survival in salivary gland carcinomas. J. Surg. Oncol 114, 36–43 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Motzer RJ et al. Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med 378, 1277–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ready N. et al. First-line nivolumab plus ipilimumab in advanced non-small-cell lung cancer (CheckMate 568): outcomes by programmed death ligand 1 and tumor mutational burden as biomarkers. J. Clin. Oncol 37, 992–1000 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larkin J. et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med 373, 23–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mimica X. et al. Distant metastasis of salivary gland cancer: incidence, management, and outcomes. Cancer 126, 2153–2162 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Persson M. et al. Recurrent fusion of MYB and NFIB transcription factor genes in carcinomas of the breast and head and neck. Proc. Natl Acad. Sci. USA 106, 18740–18744 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalin MG et al. Comprehensive molecular characterization of salivary duct carcinoma reveals actionable targets and similarity to apocrine breast cancer. Clin. Cancer Res. 22, 4623–4633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalin MG et al. Multi-dimensional genomic analysis of myoepithelial carcinoma identifies prevalent oncogenic gene fusions. Nat. Commun 8, 1197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho AS et al. Genetic hallmarks of recurrent/metastatic adenoid cystic carcinoma. J. Clin. Invest 129, 4276–4289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linxweiler M. et al. The immune microenvironment and neoantigen landscape of aggressive salivary gland carcinomas differ by subtype. Clin. Cancer Res. 26, 2859–2870 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenhauer EA et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer 45, 228–247 (2009). [DOI] [PubMed] [Google Scholar]
- 22.National Cancer Institute. Common Terminology Criteria for Adverse Events (CTCAE) v4.0 (2010).
- 23.Zehir A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med 23, 703–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weng AP et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Breit S. et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood 108, 1151–1157 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Ferrarotto R. et al. Activating NOTCH1 mutations define a distinct subgroup of patients with adenoid cystic carcinoma who have poor prognosis, propensity to bone and liver metastasis, and potential responsiveness to Notch1 inhibitors. J. Clin. Oncol 35, 352–360 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho AS et al. The mutational landscape of adenoid cystic carcinoma. Nat. Genet 45, 791–798 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciani Y. et al. Allele-specific genomic data elucidate the role of somatic gain and copy-number neutral loss of heterozygosity in cancer. Cell Syst. 13, 183–193 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshihara K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun 4, 2612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Şenbabaoğlu Y. et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 17, 231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rooney MS, Shukla SA, Wu CJ, Getz G. & Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160, 48–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jassal B. et al. The reactome pathway knowledgebase. Nucleic Acids Res. 48, D498–D503 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bindea G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Jiang P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med 24, 1550–1558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chowell D. et al. Evolutionary divergence of HLA class I genotype impacts efficacy of cancer immunotherapy. Nat. Med 25, 1715–1720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cristescu R. et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade–based immunotherapy. Science 362, eaar3593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh S. et al. Enteric viruses replicate in salivary glands and infect through saliva. Nature 607, 345–350 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riaz N. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yost KE et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med 25, 1251–1259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurtz V. et al. NetMHCpan-4.0: improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol 199, 3360–3368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marty R. et al. MHC-I genotype restricts the oncogenic mutational landscape. Cell 171, 1272–1283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shin HT et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat. Commun 8, 1377 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carter SL et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol 30, 413–421 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tumeh PC et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valpione S. et al. The T cell receptor repertoire of tumor infiltrating T cells is predictive and prognostic for cancer survival. Nat. Commun 12, 4098 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roh W. et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med 9, eaah3560 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emerson RO et al. High-throughput sequencing of T-cell receptors reveals a homogeneous repertoire of tumour-infiltrating lymphocytes in ovarian cancer. J. Pathol 231, 433–440 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horn HS Measurement of ‘overlap’ in comparative ecological studies. Am. Nat 100, 419–424 (1966). [Google Scholar]
- 49.Au L. et al. Determinants of anti-PD-1 response and resistance in clear cell renal cell carcinoma. Cancer Cell 39, 1497–1518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barennes P. et al. Benchmarking of T cell receptor repertoire profiling methods reveals large systematic biases. Nat. Biotechnol 39, 236–245 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Huang AC et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 545, 60–65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamphorst AO et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1–targeted therapy in lung cancer patients. Proc. Natl Acad. Sci. USA 114, 4993–4998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tran E. et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344, 641–645 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chandran SS et al. Immunogenicity and therapeutic targeting of a public neoantigen derived from mutated PIK3CA. Nat. Med 28, 946–957 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blank CU, Haanen JB, Ribas A. & Schumacher TN The ‘cancer immunogram’. Science 352, 658–660 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Sharma P. & Allison JP The future of immune checkpoint therapy. Science 348, 56–61 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Song L. et al. TRUST4: immune repertoire reconstruction from bulk and single-cell RNA-seq data. Nat. Methods 18, 627–630 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolotin DA et al. Antigen receptor repertoire profiling from RNA-seq data. Nat. Biotechnol 35, 908–911 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu B. et al. Temporal single-cell tracing reveals clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nat. Cancer 3, 108–121 (2022). [DOI] [PubMed] [Google Scholar]
- 60.Verdegaal EME et al. Neoantigen landscape dynamics during human melanoma–T cell interactions. Nature 536, 91–95 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Li K. et al. Myeloid-derived suppressor cells as immunosuppressive regulators and therapeutic targets in cancer. Signal Transduct. Target. Ther 6, 362 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferrarotto R. et al. Phase II clinical trial of axitinib and avelumab in patients with recurrent/metastatic adenoid cystic carcinoma. J. Clin. Oncol 41, 2843–2851 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fisher S. et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 12, R1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H. & Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McKenna A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cibulskis K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benjamin D. et al. Calling somatic SNVs and indels with Mutect2. Preprint at bioRxiv 10.1101/861054 (2019). [DOI] [Google Scholar]
- 69.Larson DE et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 28, 311–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15, 591–594 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Koboldt DC et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rimmer A. et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet 46, 912–918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cingolani P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robinson JT et al. Integrative genomics viewer. Nat. Biotechnol 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tate JG et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chakravarty D. et al. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol 2017, PO.17.00011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen R. & Seshan VE FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shukla SA et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol 33, 1152–1158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pierini F. & Lenz TL Divergent allele advantage at human MHC genes: signatures of past and ongoing selection. Mol. Biol. Evol 35, 2145–2158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robinson J. et al. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res. 43, D423–D431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grantham R. Amino acid difference formula to help explain protein evolution. Science 185, 862–864 (1974). [DOI] [PubMed] [Google Scholar]
- 82.Cheng DT et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn 17, 251–264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang L, Wang S. & Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 85.Lawrence M. et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rosenbloom KR et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43, D670–D681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Love MI, Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barbie DA et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mariathasan S. et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554, 544–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Selitsky SR et al. Virus expression detection reveals RNA-sequencing contamination in TCGA. BMC Genomics 21, 79 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Benayed R. et al. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin. Cancer Res. 25, 4712–4722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zheng Z. et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med 20, 1479–1484 (2014). [DOI] [PubMed] [Google Scholar]
- 93.Fotakis G, Rieder D, Haider M, Trajanoski Z. & Finotello F. NeoFuse: predicting fusion neoantigens from RNA sequencing data. Bioinformatics 36, 2260–2261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carlson CS et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat. Commun 4, 2680 (2013). [DOI] [PubMed] [Google Scholar]
- 95.Shannon CE A mathematical theory of communication. Bell Syst. Tech. J 27, 379–423 (1948). [Google Scholar]
- 96.Simpson EH Measurement of diversity. Nature 163, 688–688 (1949). [Google Scholar]
- 97.Morisita M. Measuring of the dispersion of individuals and analysis of the distributional patterns. Mem. Fac. Sci. Kyushu Univ. Ser. E Biol. 2, 215–235 (1959). [Google Scholar]
- 98.Nazarov V. et al. Immunarch: bioinformatics analysis of T-cell and B-cell immune repertoires. GitHub; https://immunarch.com/, https://github.com/immunomind/immunarch (2023). [Google Scholar]
- 99.Cimen Bozkus C, Blazquez AB, Enokida T. & Bhardwaj N. A T-cell-based immunogenicity protocol for evaluating human antigen-specific responses. STAR Protoc. 2, 100758 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Valero C. et al. Pretreatment neutrophil-to-lymphocyte ratio and mutational burden as biomarkers of tumor response to immune checkpoint inhibitors. Nat. Commun 12, 729 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mayakonda A, Lin DC, Assenov Y, Plass C. & Koeffler HP Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 28, 1747–1756 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A de-identified dataset—containing the clinical features and processed data that underlie results reported in this article derived from tumor whole-exome, RNA and TCR sequencing—has been made available on Zenodo (https://doi.org/10.5281/zenodo.8180441). The relevant sections of the trial protocol, including the statistical analysis plan, have been made available in the Supplementary Information. Raw whole-exome and RNA sequencing data generated in this study have been made publicly available at the Sequence Read Archive under Bio-Project number PRJNA940989. The hg37 and hg19 reference genomes are available from https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000001405.13/, and the hg38 assembly is available from https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_000001405.26/.