Abstract
Rapidly increasing antibiotic-resistant bacterial strains in Bangladesh’s food and farm animals stem from the excessive and inappropriate use of antibiotics. To assess the prevalence of multi-drug resistant (MDR) Gram-negative bacteria in poultry chicks, we sought to isolate and identify strains carrying antimicrobial resistance genes. Isolation and identification involved biochemical tests, 16S rRNA sequencing, and PCR screening of species-specific genes. MDR patterns were evaluated using CLSI guidelines with seventeen antibiotics across twelve classes. Targeted gene sequences were amplified for the detection of Extended-spectrum β-Lactamase (ESBL), carbapenem, tetracycline, sulfonamide, and colistin resistance genes. Common isolates, such as Escherichia coli, Klebsiella pneumoniae, Proteus penneri, and Enterobacter hormaechei, exhibited average Multiple Antimicrobial Resistance (MAR) indices of 0.66, 0.76, 0.8, 0.84, and 0.81, 0.76, 0.84, 0.41 for broiler and layer chicken, respectively. Providencia stuartii and Salmonella enterica, exclusive to broiler samples, had MAR indices of 0.82 and 0.84, respectively. Additional isolates Morganella morganii, Aeromonas spp., and Wohlfahrtiimonas chitiniclastica were found in layers (Average MAR indices: 0.73, 0.71, and 0.91). Notably, M. morganii, E. hormaechei and W. chitiniclastica were identified for the first time in Bangladeshi poultry chicken, although their evolution is yet to be understood. In this study, Pan-drug resistance was observed in one P. stuartii (broiler) and one Aeromonas spp. (layer) with a MAR index 1, while all isolates exhibited MAR indices >0.2, indicating MDR. Antimicrobial resistance (AMR) gene screening identified blaTEM, blaSHV, tetA, and sul1 in a majority of the MDR strains. Interestingly, E. coli (lactose positive and negative) and E. hormaechei were exclusively found to possess the tetB gene. In addition, E. coli (lactose negative), Klebsiella pneumoniae, Enterobacter hormaechei, M. morganii, and P. stuartii were observed to carry the colistin-resistant mcr-1 gene, whereas sul2 was detected in E. coli (lactose positive and negative), E. hormaechei, P. stuartii, and P. penneri. These findings emphasize the health risk of our consumers of both broiler and layer chickens as they have turned into a potent reservoir of various AMR gene carrying MDR and Pan-drug resistant bacteria.
Introduction
More recently, antibiotic resistant (AR) bacteria are considered one of the biggest threats to global health, food security, and development [1]. Bacteria are becoming stronger day by day against diverse group of antibiotics [2, 3]. The impact of antibiotic resistance is indiscriminate, posing a threat to individuals of all ages, regardless of geographical location. As a consequence, a growing list of infections–such as pneumonia, tuberculosis, blood poisoning, gonorrhea, and foodborne diseases–are becoming harder, and sometimes impossible, to treat as antibiotics become less effective [4]. Antibiotics are used for therapeutic purposes and disease prevention in veterinary medicine and are commonly added to animal feed at sub-therapeutic levels for growth promotion. According to reports, Asia consumed the largest quantity of antibiotics, totaling approximately 57,167 tonnes for non-therapeutic purposes in 2017. Projections indicate that this figure is expected to rise to 63,062 tonnes by the year 2030 [5]. With global consumption of chicken as a protein source and the projected rapid growth of agro-based poultry industries, this trend is set to intensify [6]. According to the Department of Livestock Services 2020–21, there are more than 350 million poultry in Bangladesh which contributes to the growth of the national economy and the creation of job opportunities. Furthermore, many people benefit from the poultry industry since it provides a cheaper and more conveniently accessible source of nutrition and protein in the form of eggs and meat [7, 8]. Due to the concern about the care and maintenance of chicken health, poultry breeders use antibiotics at therapeutic and non-therapeutic levels to avoid disease and improve feed utilization and growth performance to meet consumer demand [6].
The Enterobacteriaceae family comprises a diverse group of Gram-negative bacteria found in water, decaying waste, soil, and the gastrointestinal tracts of both humans and animals. These bacteria are often responsible for diarrhea, primarily transmitted through contaminated food or water [9]. Notably, pathogenic bacteria like Salmonella spp., Escherichia coli, Yersinia spp., Klebsiella species, and Shigella spp. are commonly linked to gastrointestinal complications in humans and are frequently identified in chickens and their associated food products [10–13]. Enterobacteriaceae frequently serve as carriers of extended-spectrum β-lactamase (ESBL)-encoding genes and are frequently found in poultry and their surrounding environment, contributing to the prevalence of ESBL-producing strains [14]. The predominant cause of antibiotic resistance in Enterobacteriaceae is the production of ESBLs, AmpC-lactamases, and carbapenemases, rendering them resistant to a diverse array of antibiotics. Apart from penicillins and first, second, and third generation cephalosporins, these bacteria exhibit resistance to multiple other antimicrobials. Notably, tetracycline, fluoroquinolone, and trimethoprim resistance have rapidly spread worldwide, driven by the proliferation of ESBL genes [15, 16]. The presence of blaCTX-M, blaSHV, and blaTEM genes encoding CTX-M, TEM, and SHV β-lactamases, respectively, empowers these bacteria with resistance against penicillins, first, second, and third-generation cephalosporins, as well as aztreonam [17, 18]. Notably, studies by Kluytmans et al. [19] and Leverstein-van Hall et al. [20] have highlighted strong genetic similarities between ESBL-producing E. coli isolated from chicken meat and humans. The identification of CTX-M-1 and TEM-52 genetic traits on similar plasmids in E. coli from these distinct sources further supports the potential transmission of ESBL genes through food pathways [19, 20]. The presence of diverse ESBL and carbapenemase genetic elements, along with numerous other antibiotic resistance determinants, on mobile genetic elements presents an ongoing challenge. This scenario has the potential to lead to the emergence of bacteria resistant to all available antibacterial agents [21–23]. Colistin (polymyxin E) usage for therapeutic, preventative, and growth promotion purposes in food animals has been prevalent in several Asian countries, including India, Japan, Korea, and Vietnam. In Bangladesh, colistin is already allowed as a veterinary therapeutic agent and a dietary supplement, which is used to treat infections caused by multi-drug resistant Gram-negative microbes as a penultimate resource [24–27]. Moreover, the mobilization of the colistin resistance gene variant-1 (mcr-1) represents a plasmid-mediated mechanism, and additional variants of the mcr gene (mcr-1 to mcr-9) have been detected in Enterobacteriaceae species across various countries, including Bangladesh. These findings indicate that these genes are present in natural environments, livestock, and humans, posing a significant threat to public health [28–32]. Following this, a subsequent epidemic of colistin-resistant bacteria emerged among humans in China, resulting in a notable increase in mortality. Colistin-resistant Enterobacteriaceae were identified as human pathogens, in line with earlier reports [27, 33–35].
Bangladesh, despite being a developing economy, features a spectrum of poultry and food-animal farming systems that span from traditional family farms to medium- and large-scale commercial enterprises. The absence of an effective government-led animal healthcare system has led farm proprietors to depend on unskilled and informal healthcare practitioners for animal treatment. Consequently, the prevalent misuse, overuse, and suboptimal administration of antibiotics on farms have been exacerbated by unrestricted access to these drugs [36, 37]. As a result, MDR bacteria are persistently emerging within the poultry industry, and the characteristics of these MDR strains are continually changing each year. Consequently, it has become imperative to assess the present situation of MDR bacteria in the poultry sector to implement appropriate precautionary measures. For the reasons mentioned above, this study aims to isolate and characterize multi-drug resistant (MDR) bacteria from the gut and rectal swabs of broiler and layer chickens raised in poultry farms within the Noakhali region. Additionally, the study seeks to detect resistance genes (ESBL, carbapenem, tetracycline, trimethoprim, colistin) within these isolated bacteria. A comparative analysis was conducted between MDR bacteria from broiler and layer chickens, with a focus on identifying potential reasons for any observed differences. Ultimately, this research endeavor aims to enhance our understanding of multi-drug resistant bacterial species and their resistance patterns against a diverse range of antibiotics.
Material and methods
Ethical approval and area of study
This study received approval from the Ethics and Research Review Committee of Noakhali Science and Technology University Faculty of Sciences (Approval No. NSTU/SCI/EC/2023/186). All methods adhered to guidelines and regulations. We obtained farm and chicken information with informed consent from farm owners, ensuring their privacy and commercial data protection. We utilized sample codes during data collection to maintain accuracy and protect privacy.
We chose three distinct small-scale commercial broiler and layer farms for this study situated in Noakhali Sadar. Information collected included farm size, total chicken population, age and weight of chickens, feeding habits, disease prevalence, medications administered for treatment, and the prevalent antibiotics used within these farms (S1 and S2 Tables).
Selection and criteria of the study site
Samples were collected from local small-scale commercial poultry farms in Noakhali Sadar upazila. Details regarding the farms and the collected samples can be found in the S1 and S2 Tables. The farms, run by 4–6 workers, handle tasks such as feeding, medication, and overall chicken care. These chickens are sold in nearby markets like Sonapur Bazar, Maijdi Pouro Bazar, and Maijdi bazar. Nearby residents often purchase eggs and poultry directly from these farms. For our study, we gathered fresh stool and rectal swab samples from Mousumi Poultry and Nur-hossain Agro (two broiler and two layer chickens each), and from Shohag Poultry, Poultry Farms, and Dipto Poultry (one broiler and one layer chicken each).
Sample collection
We conducted this study by procuring rectal swab and stool samples from three distinct farms situated in Noakhali Sadar. To achieve this, we selected two chickens from each farm for the collection of rectal swab and stool samples. The rectal swab specimens were meticulously collected using sterile cotton buds and were then placed in sterile falcon tubes. Stool samples, on the other hand, were gathered using sterile forceps and deposited into sterile petri plates. These Falcon tubes and petri plates were airtight sealed using Parafilm and securely stored in separate zipper bags at 4 ˚C, ensuring protection against contamination from collection to laboratory transportation. Stringent safety protocols were upheld throughout the sample collection process, and visits were limited to one farm per day to prevent any potential cross-contamination.
Isolation and presumptive identification of bacteria
Gram-negative bacterial isolation was accomplished using the standard serial dilution plate technique, slightly modified from Bushen et. al. (2021) [38]. Initially, 1 g of stool sample or a rectal swab cotton bud was introduced into 9 ml of peptone water broth media and incubated at 37°C for 18 hours. Subsequently, 1 ml of peptone water broth with the sample was transferred into 9 ml of sterile 0.9% saline water and thoroughly mixed by vortex agitation. Serial dilutions were performed, spanning up to a 10−5-fold reduction. A 0.1 ml inoculum was extracted from each dilution and spread across the surfaces of MacConkey agar, Eosin methylene blue (EMB) agar, and Xylose Lysine Deoxychocolate (XLD) agar plates. Incubation of the plates occurred at 37°C for 24 hours. Following incubation, individual colonies were selected and streaked onto a MacConkey agar plate to achieve pure cultures. Subsequently, each bacterial isolate from the pure culture was subjected to presumptive identification using a series of tests, including the Lactose test, Triple sugar iron test, Motility test, Urease test, Indole test, and Oxidase test, in accordance with Bergey’s Manual of Determinative Bacteriology [39].
Molecular identification of bacterial isolates
Bacterial DNA extraction was conducted using the boiling method, as per [40] with some modifications. The complete extraction procedure consisted of the following steps: i) Transfer 1 ml of pre-enrichment culture (peptone water) into a sterile microcentrifuge tube. ii) Centrifuge at 16,000 g for 15 minutes and carefully remove the supernatant. iii) Repeat steps (i) and (ii). iv) Resuspend the pellet in 400 μl of DNase and RNase free water through vortexing. v) Centrifuge at 16,000 g for 10 minutes and discard the supernatant. vi) Resuspend the pellet in 200 μl of DNase-RNase free water through vortexing. vii) Incubate at 100°C for 15 minutes and promptly chill on ice for 10 minutes. viii) Centrifuge for 5 minutes at 16,000 g at 4°C. ix) Carefully transfer the supernatant to a new microcentrifuge tube. x) Use an aliquot of 2–5 μl of the supernatant as the template DNA for PCR.
Following the bacterial DNA extraction, presumptively identified isolates of Klebsiella spp. and Aeromonas spp. underwent PCR screening for molecular level identification. Specifically, the PCR screening for K. pneumoniae identification targeted the rcsA gene [41], while for Aeromonas spp. identification, the gyrB gene was the focus of PCR screening [42]. S3–S5 Tables outlines Primers details, PCR mixture preparation and conditions for K. pneumoniae and Aeromonas spp. detection. Further, 16S rRNA gene amplification was performed on presumptively identified bacteria using the 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) primer as forward and the 1492R (5’-GGTTACCTTGTTACGACTT-3’) primer as reverse. Primer details, PCR mixture preparation and PCR conditions are provided in S3, S6 and S7 Tables. Amplified 16S rRNA products were purified and sequenced commercially at the National Institute of Biotechnology using Sanger di-deoxy sequencing. Sequenced DNA data were analyzed with SnapGene Viewer 6.0.5 (https://www.snapgene.com/snapgene-viewer) and subjected to individual Basic Local Alignment Search Tool (BLAST) analysis against the National Center for Biotechnology Information (NCBI) blastn server for database comparison (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_SPEC=GeoBlast&PAGE_TYPE=BlastSearch). In 16S rRNA sequencing, two bacterial isolates per genus were selected, validating biochemical test outcomes.
Phylogenetic analysis of 16S rRNA gene sequenced DNA data
Phylogenetic analysis of the bacterial isolates identified through 16S rRNA gene sequencing was conducted using the online web tool MAFFT version 7 (https://mafft.cbrc.jp/alignment/server/phylogeny.html) [43, 44]. For DNA analysis, a scoring matrix of 1PAM/K = 2 was employed, alongside default settings for other parameters.
Detection of phenotypic antibiotic resistance
Phenotypic antibiotic resistance profiling of identified bacterial isolates was done by Kirby-Bauer disk-diffusion method according to Hudzicki, (2009) [45]. Bacterial isolates identified through 16S rRNA gene sequencing, as well as those exhibiting similar biochemical profiles, were selected for antibiotic resistance testing. A panel of seventeen antibiotics from eight classes, recommended by the Center for Disease Prevention and Control (CDC) for Enterobacteriaceae family bacterial infections was utilized. These antibiotics encompassed Ampicillin (AMP25), Amoxicillin-clavulanic acid (AMC 30), Cefotaxime (CTX 30), Cefoxitin (CX 30), Aztreonam and Imipenem from β-lactams; Ciprofloxacin (CIP 5) and Norfloxacin (NX 10) from fluoroquinolones; Gentamicin (GEN 10) and Kanamycin (K 30) from aminoglycosides; Azithromycin (AZM 30) and Erythromycin (E 10) from macrolides; Chloramphenicol (C 30) from phenicol; Trimethoprim-Sulfamethoxazole (COT 25) from sulphonamides; Tetracycline (TE 30) from tetracyclines; Colistin (CL 10) and Polymyxin B (PB 300) from polymyxins (Hi-media, India). Zone of inhibition was measured in millimeters (mm) and interpreted as sensitive (S), intermediate (I), or resistant (R) according to Clinical and Laboratory Standards Institute (CLSI) standards [46]. For colistin resistance analysis, we followed the studies by Uwizeyimana et al (2020) and Fadare et. al. (2021) [47, 48]. Additionally, a zone of inhibition <12 mm considered as resistance.
Evaluation of Multiple antimicrobial resistance phenotype (MARP) and multiple antimicrobial resistance index (MARI)
Bacterial species demonstrating resistance to one or more antibiotics from distinct antibiotic classes are categorized as exhibiting multi-drug resistance (MDR) [49]. In accordance with this principle, bacterial isolates in this study manifesting resistance across three antibiotic classes are classified as MDR. Furthermore, the Multiple Antibiotic Resistance Index (MARI) of each isolate, an assessment of antimicrobial resistance, was calculated using an equation elucidated by [44], as follows:
Here, “AR” signifies the cumulative count of antibiotics to which bacterial isolates displayed resistance, while “AU” denotes the total count of antibiotics employed. If the MARI value of any isolate surpasses 0.2, it is categorized as demonstrating multi-drug resistance (MDR) [48, 50, 51].
Genotypic identification of antimicrobial resistant genes (ARG)
Subsequent to the identification of bacterial isolates and their patterns of multi-drug resistance, we conducted a comprehensive screening for nine distinct antibiotic resistance genes (ARGs). These encompassed Extended Spectrum β-lactamases (ESBLs) genes including blaCTX-M, blaTEM, and blaSHV, alongside the New Delhi Metallo β-lactamase (blaNDM), a metallo-carbapenemase gene. Additionally, non-β-lactamase genes such as tetA and tetB were targeted for tetracycline resistance assessment, while sul1 and sul2 were examined for Sulfonamides resistance. Moreover, among the globally described ten mcr variants encoding colistin resistance, only mcr-1 gene was investigated by PCR since it is more prevalent worldwide. The selection criteria for ARG screening encompassed bacterial isolates demonstrating resistance or intermediate phenotypes to the aforementioned antibiotics. Primers for the ARGs mentioned above were chosen from prior studies listed in S3 Table and were synthesized commercially. The annealing temperature was determined using the web-based Tm calculator tool (https://tmcalculator.neb.com/#!/main). Detailed information regarding PCR mixture preparation and PCR conditions for ARG screening is provided in S8–S12 Tables.
Result
Isolation and identification of bacterial isolates
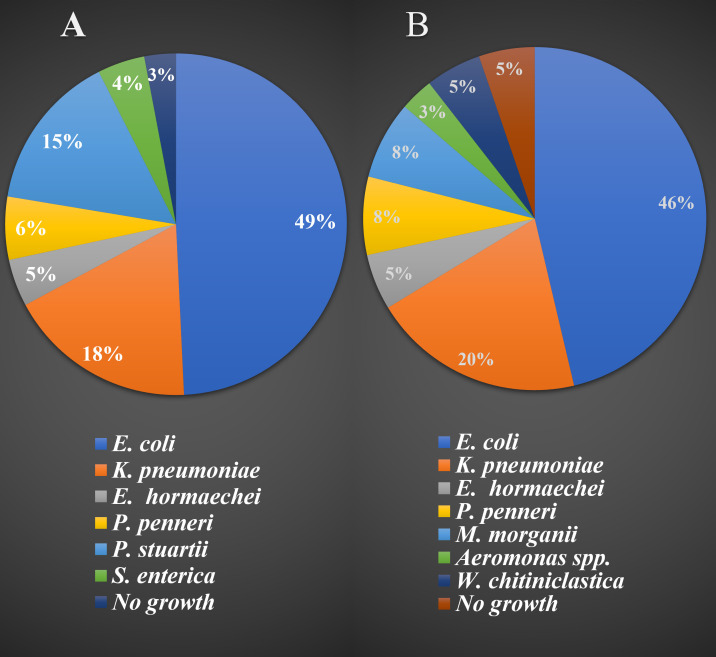
A total of 12 samples were examined, including 6 stool samples each from broiler chickens and layer chickens, as well as 6 rectal swab samples (3 from broiler chickens and 3 from layer chickens). These samples were cultured on MacConkey agar, EMB agar, and XLD agar plates. A total of 160 bacterial isolates were identified from the twelve samples. The occurrence and distribution of these isolated bacterial strains in both broiler and layer chickens are presented in Fig 1.
Fig 1. Identified bacterial isolates from broiler and layer chicken stool and rectal swab.
(A represents the bacterial isolates from broiler chicken and B represents the bacterial isolates from layer chickens).
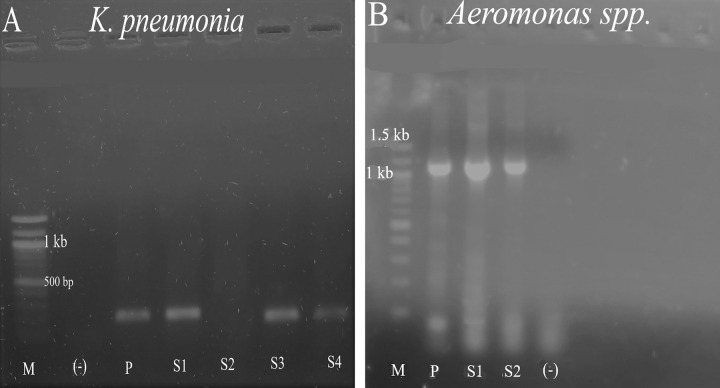
The isolated bacteria underwent presumptive identification through conventional microbiological and biochemical techniques. As a result, nine distinct bacterial genera emerged from the pool of 160 isolates. Among these, E. coli, Klebsiella spp., Salmonella spp., Providencia spp., Enterobacter spp., Proteus spp., and Morganella spp. were identified within the family Enterobacteriaceae. Additionally, Aeromonas spp. from the Aeromonadaceae family and Pseudomonas spp. from the Pseudomonadaceae family constituted the remaining two. Following this preliminary identification, K. pneumoniae and Aeromonas spp. were pinpointed to the species level via housekeeping gene amplification (refer to Fig 2), while 16S rRNA sequencing was employed to validate the species-level identification of other bacterial isolates.
Fig 2.
Detection of K. pneumoniae (A) and Aeromonas spp. (B) by PCR amplification of rcsA and gyr-B gene targeted segment respectively. (M = 100 bp ladder, (-) = negative control, P = positive control, S = Sample).
Consequently, 16S rRNA sequencing confirmed the species identities of E. coli, K. pneumoniae, Salmonella enterica, Providencia stuartii, Enterobacter hormaechei, Proteus penneri, and Morganella morganii (S1 File). Surprisingly, our 16S rRNA gene sequencing revealed a strain of Wohlfahrtiimonas chitiniclastica, which had initially been presumptively identified as Pseudomonas spp. through conventional microbiological and biochemical techniques. A detailed breakdown of the identity match percentages and query coverage, as compared to the NCBI database, can be found in Table 1.
Table 1. 16S rRNA sequencing result of bacterial isolates.
| Isolates Name | Query coverage % | Identity in percentage | Accession length in base pair | Bacterial family |
|---|---|---|---|---|
| E. coli (Lactose positive) | 94 | 98 | 1439 | Enterobacteriaceae |
| E. coli (Lactose negative) | 100 | 100 | 1443 | Enterobacteriaceae |
| K. pneumoniae | 100 | 99.40 | 1372 | Enterobacteriaceae |
| P. stuartii | 100 | 99.64 | 1545 | Enterobacteriaceae |
| S. enterica | 100 | 99.88 | 1069 | Enterobacteriaceae |
| E. hormaechei | 100 | 99.38 | 1196 | Enterobacteriaceae |
| P. penneri | 99 | 98.80 | 1365 | Enterobacteriaceae |
| M. morganii | 99 | 99.06 | 1464 | Enterobacteriaceae |
| W. chitiniclastica | 100 | 98.80 | 1423 | Incertae sedis |
During our investigation, we identified lactose-negative E. coli isolates through 16S rRNA sequencing, leading us to hypothesize that these non-lactose fermenters belonged to bacterial groups outside the Enterobacteriaceae family. Notably, the culture characteristics of E. coli on MacConkey agar exhibited variation in response to lactose utilization, as illustrated in S1 Fig. Among the 160 bacterial isolates, E. coli (33 isolates from broiler and 44 isolates from layer), K. pneumoniae (12 isolates from broiler and 19 isolates from layer), E. hormaechei (3 isolates from broiler and 5 isolates from layer), and P. penneri (4 isolates from broiler and 7 isolates from layer) were commonly encountered in both broiler and layer chickens respectively. Conversely, P. stuartii (10 isolates) and S. enterica (4 isolates) were present solely in broiler chicken samples, while M. morganii (7 isolates), Aeromonas spp. (3 isolates), and W. chitiniclastica (4 isolates) were exclusively isolated from layer chicken samples. A comprehensive depiction of the occurrence and frequency of these bacterial isolates in chicken samples is provided in Fig 1.
Phylogenetic analysis of identified bacterial isolates
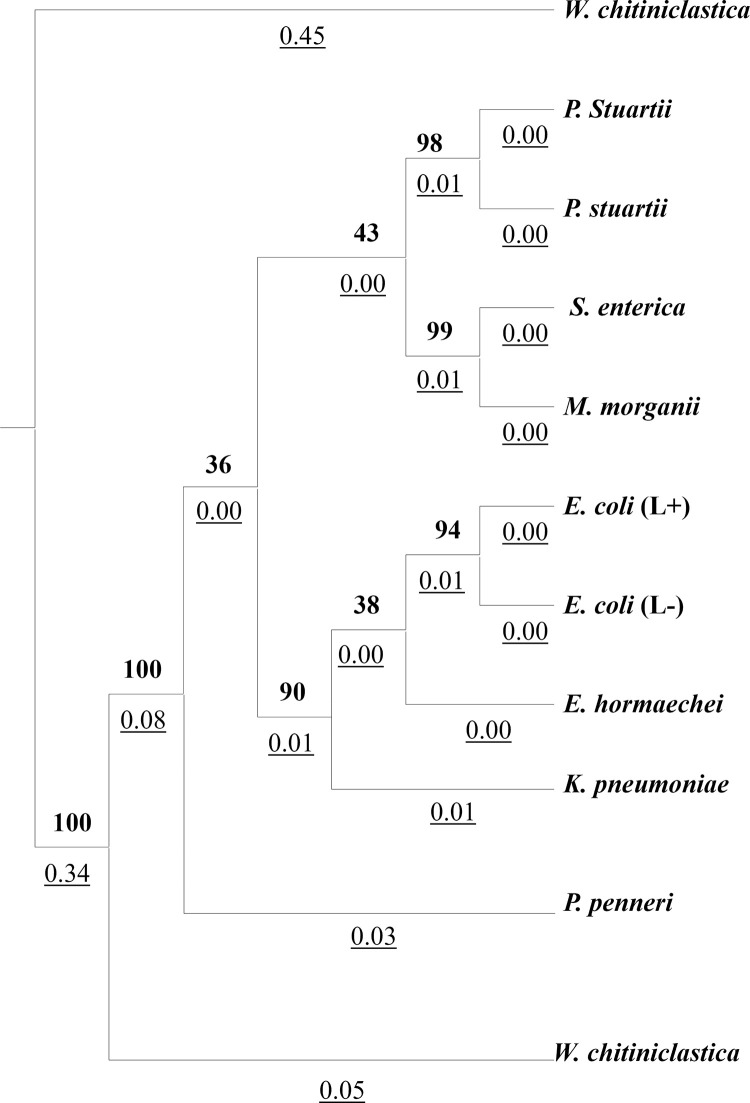
Phylogenetic analysis utilizing the 16S rRNA sequencing data of the bacterial isolates reveals distinct clustering patterns (Fig 3). Notably, P. stuartii, S. enterica, and M. morganii isolates exhibit a close relationship, forming a tightly grouped cluster. Similarly, E. coli (lactose positive), E. coli (lactose negative), K. pneumoniae, and E. hormaechei isolates also form a cohesive cluster, indicating their close relatedness. In contrast, P. penneri isolates from the Enterobacteriaceae family display a greater genetic distance from the aforementioned bacteria. Additionally, the species W. chitiniclastica, belonging to the Incertae sedis family, demonstrates a notable divergence from other Enterobacteriaceae family members, displaying a distant relationship in the phylogenetic analysis.
Fig 3. Phylogenetic tree of identified bacterial isolates (Numeric values with bottom border represent the branch length and numeric values in bold represent the boot strap value).
Phenotypic characterization of antibiotic resistant pattern
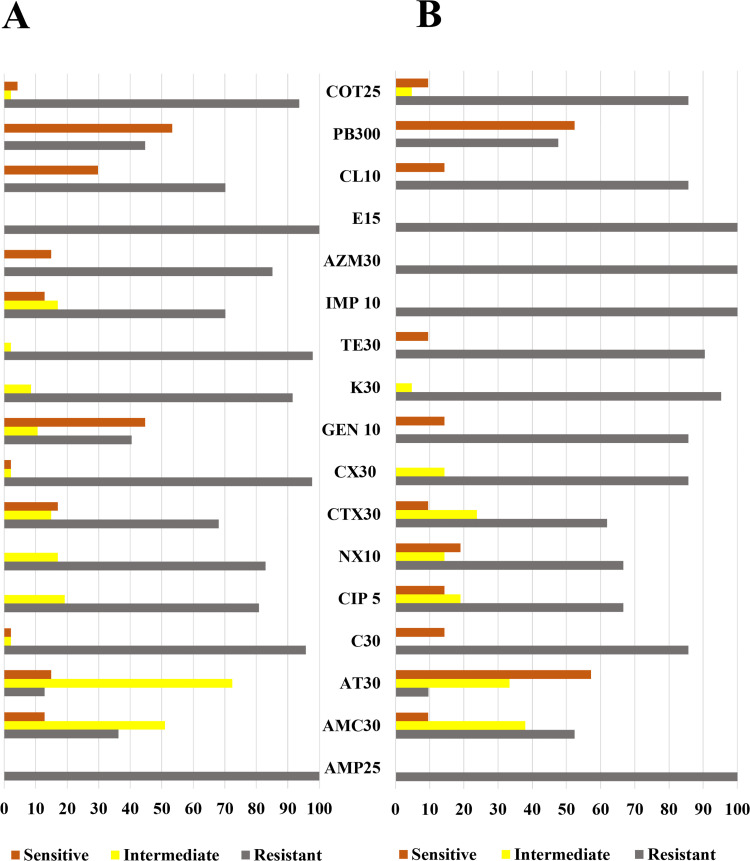
Upon scrutinizing the antibiogram results, a noteworthy trend emerged. All tested 47 isolates from broiler chickens exhibited resistance to both ampicillin and erythromycin. Among the layer chickens, all tested bacterial isolates demonstrated complete resistance to ampicillin, imipenem, azithromycin, and erythromycin. In the case of broiler chickens, a unique pattern emerged whereby all bacterial isolates displayed sensitivity to ciprofloxacin, norfloxacin, kanamycin, and tetracycline. However, an intermediate state was observed with respect to these drugs, ranging from 19.15% to 97.87% for ciprofloxacin (9 isolates), norfloxacin (8 isolates), kanamycin (4 isolates), and tetracycline (1 isolates), respectively (Fig 4).
Fig 4.
Phenotypic identification of antibiotic resistant of total bacterial isolates found in broiler (A) and layer (B) chickens. (Ampicillin (AMP25), Amoxicillin-clavulanic acid (AMC 30), Cefotaxime (CTX 30) Cefoxitin (CX 30) Ciprofloxacin (CIP 5) and Norfloxacin (NX 10), Aztreonam (AT 30), Gentamicin (GEN 10), Kanamycin (K 30), Azithromycin (AZM 30) Erythromycin (E 10), Imipenem (IMP 10), Chloramphenicol (C 30), Trimethoprim-Sulfamethoxazole (COT 25), Tetracycline (TE 30), Colistin (CL 10) and Polymyxin B (PB 300)).
Meanwhile, bacterial isolates derived from layer chickens showcased a distinctive pattern of sensitivity to cefoxitin and kanamycin but exhibited intermediate effects in 3 isolates (14.29%) and in 1 isolate (4.76%) towards these drugs, respectively. Remarkably, among the bacterial isolates from broiler chickens, gentamicin (21 isolates, 44.68%) and polymyxin B (26 isolates, 52.38%) demonstrated notably higher efficacy compared to other drugs employed in this study. Notably, this study did not uncover any bacterial group exhibiting sensitivity to only one type of drug utilized in this study. The phenotypic antimicrobial traits of bacterial isolates found in both broiler and layer chickens, along with a comparison of antimicrobial characteristics between the two groups, are elucidated in Fig 4. This study reveals a concerning trend: E. coli, K. pneumoniae, P. stuartii, and P. penneri isolates from both broiler and layer chicken samples displayed heightened resistance levels compared to those found exclusively in broiler chicken samples. Notably, isolates of W. chitiniclastica, newly identified in Bangladeshi poultry chicken samples through this study, exhibited a similarly elevated level of resistance to commonly used antibiotics. We also observed potential polymyxin (colistin and polymyxin-B) resistance among isolates based on their zone of inhibition, measuring less than 12 mm. The implications of resistance to these drugs are grave, as colistin serves as the last resort when other antibiotics fail—a concerning development for the Noakhali region.
Among E. coli isolates, AT 30 (12 isolates, 63.18%), GEN 10 (10 isolates, 52. 63%), CL 10 (13 isolates, 68.42%), PB 300 (18 isolates, 92.74%) demonstrated effectiveness in broiler isolates, while AMC 30 (2 isolates, 40%), AT 30 (3 isolates, 60%), PB 300 (5 isolates, 100%) observed high efficacy in layer chicken isolates S2 Fig. Conversely, all tested K. pneumoniae isolates from broiler chickens exhibited medium level of sensitivity to AT 30 (6 isolates, 81.82%), GEN 10 (4 isolates, 57.14%), PB 300 (4 isolates, 57.14%). Intriguingly, K. pneumoniae isolates from layer chickens displayed sensitivity at low to medium level to AMC 30 (1 isolates, 25%), AT 30 (3 isolates, 75%), NX 10 (1 isolates, 25%) drugs only as depicted in S3 Fig. Similarly, all E. hormaechei isolates from broiler chickens were sensitive to AT 30 (2 isolates, 50%), TE 30 (1 isolates, 25%), PB 300 (3 isolates, 75%) whereas layer chicken isolates showed sensitivity to C 30 (2 isolates, 100%), CIP 5 (2 isolates, 100%), NX 10 (2 isolates, 100%), GEN 10 (2 isolates, 100%), TE 30 (2 isolates, 100%), PB 300 (2 isolates, 100%), COT 25 (2 isolates, 100%) demonstrated in S4 Fig. Furthermore, P. penneri isolates from broiler chicken demonstrated sensitivity to AMC 30 (2 isolates, 66.67%), AT 30 (2 isolates, 66.67%), CTX 30 (2 isolates, 66.67%), GEN 10 (1 isolates, 33.33%), and from layer chicken isolates demonstrated sensitivity to AT 30 (3 isolates, 100%), CTX 30 (1 isolates, 33.33%) (S5 Fig). One P. stuartii and one Aeromonas spp. isolate from this study found pan-drug resistant and other isolates show high level of resistance to the used antibiotics. Antibiogram result data of P. stuartii, S. enterica, M. morganii, W. chitiniclastica, and Aeromonas spp. illustrated in S6–S8 Figs.
Analyzing MAR phenotype and MAR index in bacterial isolates from broiler and layer chickens
The bacterial species identified in this study exhibited diverse MAR phenotypes, each characterized by distinct MAR indices. Among E. coli isolates from broiler chicks, 14 distinct MAR phenotypes were observed, whereas E. coli isolates from layer chicks displayed 2 different MAR phenotypes. K. pneumoniae isolates from broiler and layer chicks demonstrated 7 and 3 distinct MAR phenotypes, respectively. Similarly, E. hormaechei and P. penneri isolates from both broiler and layer chick samples exhibited 3 and 2 MAR phenotypes, respectively. Notably, P. stuartii and S. enterica isolates from broiler chicks displayed 9 and 4 different MAR phenotypes, respectively. Intriguingly, a single P. stuartii and Aeromonas spp. isolate exhibited resistance to all 17 drugs tested in this study, classifying it as pan-drug resistant. In contrast, M. morganii, W. chitiniclastica, and Aeromonas spp. isolates from layer chicks displayed 2, 3, and 3 different MAR phenotypes, respectively. Particularly concerning is the high level of resistance exhibited by W. chitiniclastica isolates, underscoring a worrisome state of antibiotic resistance. Detailed information regarding MAR phenotypes and MAR indices of the isolated bacterial strains can be found in Table 2.
Table 2. MAR phenotypes and MAR index profile of isolated bacterial isolates from broiler and layer chicken samples.
| Antibiotic resistant profile | No. of Isolates | MAR index |
|---|---|---|
| E. coli isolate from broiler chickens | ||
| AMP 25, AMC 30, AT 30, C 30, CIP 5, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, E 15, CL 10, PB300, COT 25 | 1 | 0.82 |
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, IMP10, AZM 30, E 15, CL 10, COT 25 | 1 | 0.82 |
| AMP 25, AMC 30, C 30, CIP 5, NX 10, CTX 30, CX30, K 30, TE 30, IMP10, AZM 30, E 15, COT 25 | 1 | 0.76 |
| AMP 25, AT 30, C 30, CIP 5, NX 10, CTX 30, CX30, K 30, TE 30, AZM 30, E 15, COT 25 | 1 | 0.71 |
| AMP 25, C 30, CIP 5, NX 10, CX30, GEN 10, K 30, TE 30, AZM 30, E 15, CL 10, COT 25 | 2 | 0.71 |
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX30, K 30, TE 30, IMP 10, E 15, CL 10, COT 25 | 1 | 0.71 |
| AMP 25, C 30, CIP 5, NX 10, CX30, GEN 10, K 30, TE 30, E 15, CL 10, COT 25 | 2 | 0.65 |
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX30, K 30, TE 30, AZM 30, E 15, COT 25 | 1 | 0.65 |
| AMP 25, C 30, CIP 5, NX 10, CTX 30, K 30, TE 30, IMP10, AZM 30, E 15, COT 25 | 2 | 0.65 |
| AMP 25, C30, CIP 5, NX 10, CX 30, CTX30, K 30, TE 30, E 15, COT 25 | 1 | 0.59 |
| AMP 25, CIP 5, NX 10, CX 30, TE 30, IMP10, AZM 30, E 15, COT 25 | 1 | 0.53 |
| AMP 25, C30, CIP 5, NX 10, CX 30, K 30, TE 30, AZM 30, E 15. | 1 | 0.53 |
| AMP 25, AT 30, C30, NX 10, CTX 30, CX 30, K 30, E 15 | 1 | 0.47 |
| AMP 25, AT 30, C30, CIP5, NX 10, K 30, TE 30, AZM 30, E 15, | 1 | 0.47 |
| E. coli isolates from layer chickens | ||
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, COT 25 | 3 | 0.82 |
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, COT 25 | 1 | 0.76 |
| K. pneumoniae isolates from broiler chickens | ||
| AMP 25, AMC 30, C 30, CIP 5, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.88 |
| AMP 25, AMC 30, C 30, CIP 5, NX 10, CTX 30, CX30, TE 30, IMP10, AZM 30, E 15, CL 10, PB 300, COT 25 | 1 | 0.82 |
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, COT 25 | 1 | 0.82 |
| AMP 25, AMC 30, C 30, CIP 5, CTX 30, CX30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB 300, COT 25 | 1 | 0.76 |
| AMP 25, C 30, CIP 5, NX 10, CX30, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, COT 25 | 1 | 0.71 |
| AMP 25, C 30, CIP 5, NX 10, CX30, K 30, TE 30, AZM 30, E 15, CL 10, COT 25 | 1 | 0.65 |
| AMP 25, C 30, CIP 5, NX 10, CX30, GEN 10, K 30, TE 30, AZM 30, E 15, COT 25 | 1 | 0.65 |
| K. pneumoniae isolates from layer chickens | ||
| AMP 25, AMC 30, C 30, CIP 5, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.94 |
| AMP 25, AMC 30, C 30, CTX 30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL10, PB 300, COT 25 | 2 | 0.76 |
| AMP 25, AMC 30, CX 30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL10, PB 300 | 1 | 0.64 |
| E. hormaechei isolates from broiler chickens | ||
| AMP 25, AMC 30, C 30, CIP 5, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.94 |
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX 30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL10, COT 25 | 2 | 0.82 |
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX 30, GEN 10, TE 30, IMP 10, AZM 30, E 15, CL10, COT 25 | 1 | 0.76 |
| E. hormaechei isolates from layer chickens | ||
| AMP 25, AMC 30, CTX 30, CX30, IMP 10, AZM 30, E 15, CL 10 | 1 | 0.47 |
| AMP 25, AMC 30, CTX 30, CX30, E 15, CL 10 | 1 | 0.35 |
| P. penneri isolates from broiler chickens | ||
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX30, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.82 |
| AMP 25, AMC 30, C 30, CIP 5, NX 10, CX30, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.82 |
| AMP 25, CIP 5, NX 10, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.76 |
| P. penneri isolates from layer chickens | ||
| AMP 25, AMC 30, C 30, CIP 5, NX 10, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.88 |
| AMP 25, C 30, CIP 5, NX 10, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 2 | 0.82 |
| P. stuartii isolates from broiler chickens | ||
| AMP 25, AMC30, AT 30, C 30, CIP 5, NX 10, CTX 30, CX30, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 1.0 |
| AMP 25, AMC 30, C 30, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.88 |
| AMP 25, AMC 30, C 30, CIP 5, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.88 |
| AMP 25, AMC 30, C 30, CIP 5, NX 10, CTX 30, CX30, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.88 |
| AMP 25, AMC 30, AT 30, C 30, CTX, CX30, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0,82 |
| AMP 25, AMC 30, C 30, CTX, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.82 |
| AMP 25, AMC 30, C 30, CTX, CX30, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 2 | 0.76 |
| AMP 25, C 30, NX 10, CX 30, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.71 |
| AMP 25, AMC 30, C 30, CX 30, K 30, TE 30, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.64 |
| S. enterica isolates from broiler chickens | ||
| AMP 25, AMC 30, C 30, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.88 |
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.88 |
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 0.82 |
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX30, K 30, TE 30, IMP10, AZM 30, E 15, CL 10, COT 25 | 1 | 0.76 |
| M. morganii isolates from layer chickens | ||
| AMP 25, C 30, CIP 5, NX 10, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, COT 25 | 1 | 0.82 |
| AMP 25, C 30, CX30, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB 300, COT 25 | 1 | 0.65 |
| W. chitiniclastica isolates from layer chickens | ||
| AMP 25, AMC 30, AT 30, C 30, CIP 5, NX 10, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, COT 25 | 1 | 0.88 |
| AMP 25, AMC 30, C 30, CIP 5, NX 10, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, COT 25 | 1 | 0.82 |
| AMP 25, AMC 30, C 30, CIP 5, NX 10, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, COT 25 | 1 | 0.76 |
| Aeromonas spp. isolates from layer chickens | ||
| AMP 25, AMC30, AT 30, C 30, CIP 5, NX 10, CTX 30, CX30, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB300, COT 25 | 1 | 1.0 |
| AMP 25, C 30, CTX 30, CX30, GEN 10, K 30, TE 30, IMP 10, AZM 30, E 15, CL 10, PB 300, COT 25 | 1 | 0.76 |
| AMP 25, AMC 30, K 30, IMP 10, AZM 30, E 15 | 1 | 0.35 |
Genotypic characterization of bacterial isolates from broiler and layer chicken samples
Upon evaluating ESBL gene presence and other genes encoding resistance markers in collected isolates, we observed that lactose-positive and lactose-negative E. coli carried blaTEM, tet A, tet B, sul 1, and sul 2 genes, with the latter also harboring blaSHV (Fig 5A–5C). These E. coli strains displayed resistance to β-lactam combination agents, penicillin, cephalosporins, tetracycline, and folate pathway antagonists. Similarly, K. pneumoniae isolates from broiler and layer chickens showed the presence of blaTEM, blaSHV, sul 1, and tet A genes (Fig 5D). A significant portion of K. pneumoniae isolates from both groups exhibited resistance to β-lactam combination agents, penicillin, cephalosporins, tetracycline, and folate pathway antagonists.
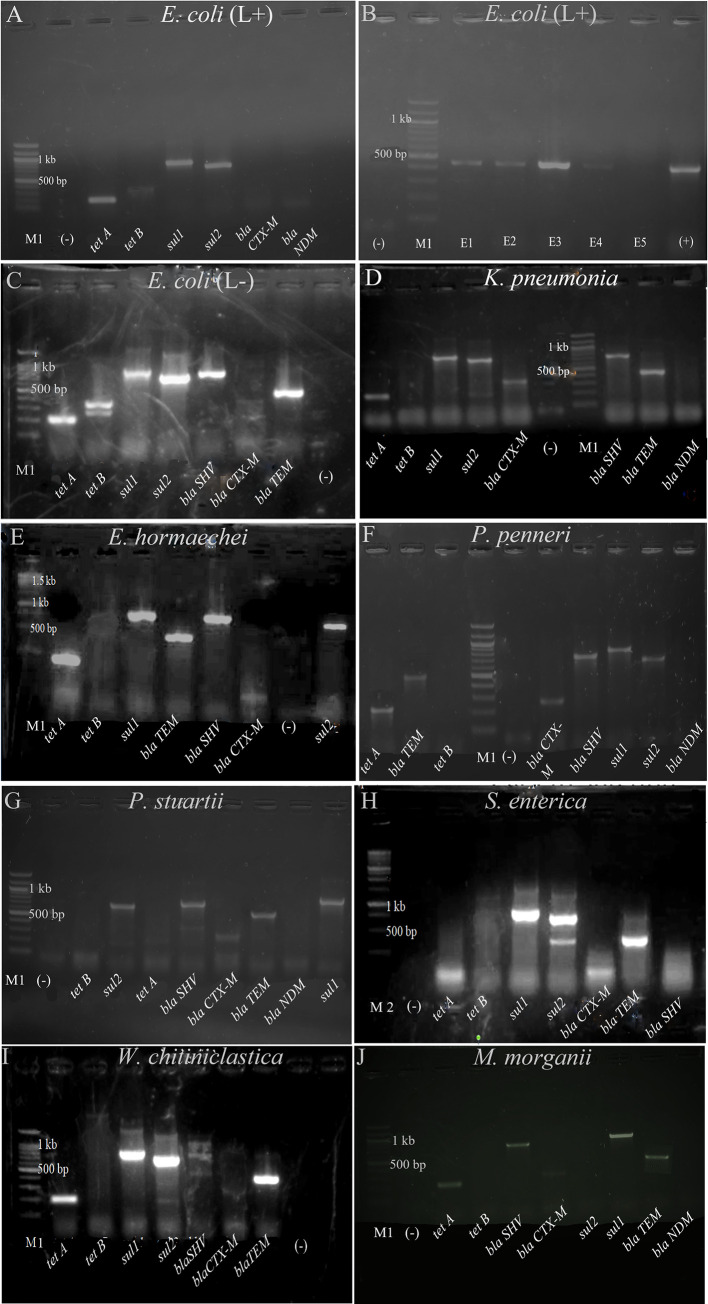
Fig 5.
Detection of antibiotic resistant genes from A) E. coli (lactose positive) B) blaTEM gene for lactose positive E. coli C) E. coli (lactose negative) D) K. pneumoniae, E) E. hormaechei, F) P. penneri, G) P. stuartii, H) S. enterica, I) W. chitiniclastica, and J) M. morganii, (M1 = 100 bp DNA ladder, M2 = 250 bp ladder for E and H, E = E. coli, (-) = negative control, (L+) = Lactose positive, (L-) = Lactose negative, tet A = 201 bp, tet B = 359 bp, sul1 = 822 bp, sul2 = 625 bp, blaTEM = 445 bp, blaCTX-M = 593 bp, blaSHV = 747 bp, blaNDM = 621 bp. We have found blaCTX-M in K. pneumoniae, P. stuartii, M. morganii, and P. penneri.at 350 bp approximately).
Among the isolates, E. hormaechei, P. penneri, P. stuartii, W. chitiniclastica, and M. morganii carried tet A, sul 1, blaSHV, and blaTEM genes (Fig 5E–5G, 5I, 5J) aligning with their phenotypic resistance.
Lastly, S. enterica isolates carried sul 1, sul 2, blaTEM genes (Fig 5H) and most of the S. enterica isolates shown resistance to β-lactam combination agents, penicillin, cephalosporins, tetracycline, and folate pathway antagonists. As those isolates showed resistance to tetracycline without the presence of both tet A and tet B genes indicated that the presence of other genes encoding tetracycline resistance, which were not investigated in this study, in S. enterica isolates.
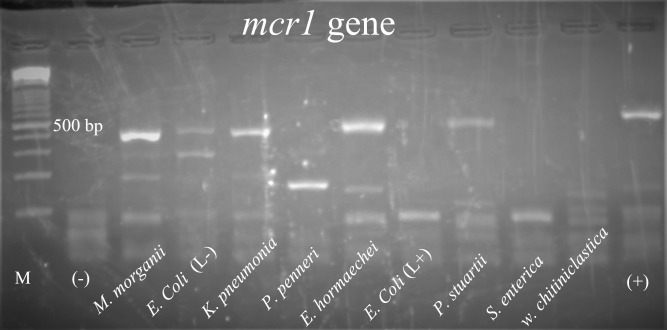
Along with those eight different types of antibiotic-resistant genes, isolates of lactose-negative E. coli, K. pneumoniae, E. hormaechei, P. stuartii, and M. morganii were also found to carry the mcr-1 variant of the colistin-resistant gene (Fig 6).
Fig 6. Detection of mcr-1 gene from E. coli (lactose positive), E. coli (lactose negative), K. pneumoniae, E. hormaechei, P. penneri, P. stuartii, S. enterica, W. chitiniclastica, and M. morganii isolates.
(M = 100 bp DNA ladder, (-) = negative control, (L+) = Lactose positive, (L-) = Lactose negative, (+) = Positive control, band size for mcr-1 gene was 309 bp).
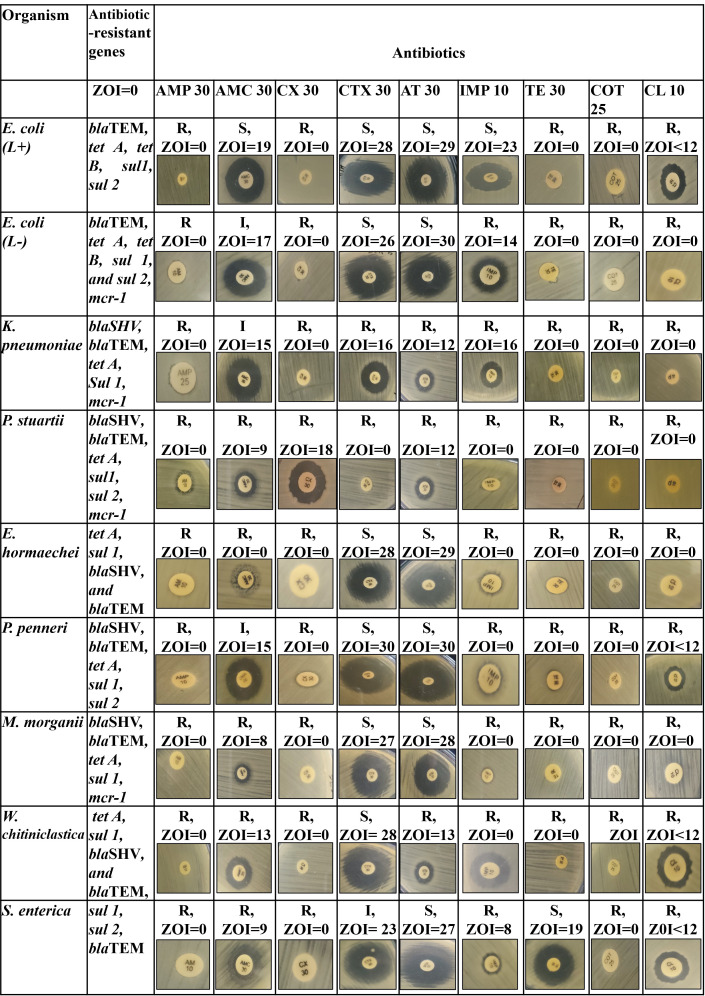
Co-relation between phenotypic and genotypic antibiotic resistant showed that identified resistant genes were strongly associated with the phenotypic effect to show resistant against different drugs illustrated in Fig 7. Standard zone of inhibition (ZOI) value to interpret the antibiogram result given in S13 Table. Moreover, E. coli (lactose positive) S. enterica and W. chitiniclastica showed resistance to colistin but didn’t carry mcr-1 gene (Fig 7). This may be due to presence of other variant of mcr gene.
Fig 7. Phenotypic and genotypic co-relation of bacterial isolates.
(Ampicillin (AMP25), Amoxicillin-clavulanic acid (AMC 30), Cefotaxime (CTX 30) Cefoxitin (CX 30), Aztreonam (AT 30), Imipenem (IMP 10), Tetracycline (TE 30), Trimethoprim-Sulfamethoxazole (COT 25), Colistin (CL 10), ZOI<10 = Zone of inhibition is less than 10 mm).
Discussion
Poultry production is a highly significant food industry, accounting for approximately 90 billion tons of chicken meat produced annually worldwide [52]. To promote poultry growth, many countries rely on a diverse range of antimicrobials [53–55], some of which are considered essential in human medicine [56, 57]. However, the unrestricted use of these critical antimicrobials in animal production contributes to the rise of antimicrobial resistance (AR) in both commensal and pathogenic microbes. Besides, AR characteristics of pathogenic bacteria are continuously changing every year. This alarming situation can lead to treatment failures, financial losses, and the potential transmission of resistant genes to humans. Furthermore, concerns about human health arise due to the presence of antibiotic residues in meat, eggs, and other animal products [58, 59]. To address this issue, we conducted this study, and our focus was on evaluating multi-drug resistant (MDR) Enterobacteriaceae and Gram-negative non-Enterobacteriaceae family bacteria from poultry chickens from Noakhali region of Bangladesh, known to be causative agents of different diseases in both humans and animals.
The prevalent bacteria included E. coli (Lactose positive and lactose negative), K. pneumoniae, E. hormaechei, and P. penneri. Additionally, Providencia spp. and S. enterica were found exclusively in broiler chickens, while M. morganii, W. chitiniclastica, and Aeromonas spp. were detected in stool and rectal swab samples from layer chickens. These isolates have been previously recognized as pathogens that can cause diseases in both humans and animals and align with previous research conducted in Bangladesh and globally [60–68]. To the best of our knowledge, Lactose negative E. coli, M. morganii, E. hormaechei and W. chitiniclastica were identified for the first time in Bangladeshi poultry chicken Gut sample. MDR Lactose negative E. coli isolates found from human gut can cause different types of intestinal and non-intestinal infections as reported by one study conducted in Bangladesh [69].
After identifying the isolates, we conducted a phenotypic analysis of antibiotic resistance in the bacterial samples obtained from both broiler and layer chickens. Our findings revealed, in broiler chickens all E. coli isolates demonstrated resistance to ampicillin, norfloxacin, and erythromycin. Additionally, E. coli isolates exhibited intermediate effects towards ciprofloxacin (1 isolate, 94.74%), kanamycin (1 isolate, 94.74%), and tetracycline (1 isolate, 94.74%) with 0% sensitivity which means those isolates can become resistant in near future. However, we also identified colistin-resistant E. coli (mostly in lactose negative) isolates, which were primarily assessed using the agar disk diffusion method, showing either no zone of inhibition or a zone of inhibition below 12 mm according to Uwizeyimana JD. et al (2020) and Fadare FT et. al. (2021) [47, 48]. All E. coli isolates from the layer chicken samples exhibited resistance to multiple antibiotics, including ampicillin, chloramphenicol, ciprofloxacin, norfloxacin, cefotaxime, cefoxitin, gentamicin, kanamycin, tetracycline, imipenem, azithromycin, erythromycin, and Co-Trimoxazole. In comparison to broiler, (4 isolates, 80%) of E. coli isolates from layer chickens were resistant to colistin, and some of these isolates (mostly lactose negative E. coli) showed no zone of inhibition. The results from our study indicating a higher percentage of E. coli isolates showing resistance to colistin in both broiler (6 isolates, 38%) and layer (4 isolates, 80%) chickens compared to the previous study conducted in Bangladesh are indeed concerning [70–72].
All the K. pneumoniae isolates of this study from broiler chickens exhibited resistance to multiple antibiotics, including ampicillin, chloramphenicol, ciprofloxacin, cefoxitin, tetracycline, azithromycin, erythromycin, and Co-Trimoxazole, classifying them as multi-drug resistant (MDR). Among the tested antibiotics, Aztreonam (6 isolates, 86%), gentamicin (4 isolates, 57%), and Polymyxin B (4 isolates, 57%) were found to be the most effective against K. pneumoniae isolates in broiler chickens. In contrast, K. pneumoniae isolates from layer chicken samples demonstrated resistance to ampicillin, gentamicin, kanamycin, tetracycline, imipenem, azithromycin, erythromycin, colistin, and polymyxin B. Additionally, 1 isolate (25%) of K. pneumoniae showed intermediate effects, and 3 isolates (75%) were resistant to Co-Trimoxazole, suggesting that Co-Trimoxazole resistance might become more prevalent in the near future. These findings indicate a substantial increase in antibiotic-resistant K. pneumoniae compared to previously reported studies on poultry chickens and antibiotic-resistant bacteria [73–76].
In our study, E. hormaechei isolates from both broiler and layer samples exhibited resistance to seven and four classes of antibiotics, respectively, which was previously unreported in Bangladesh. Interestingly, E. hormaechei from layer samples displayed higher sensitivity to chloramphenicol, ciprofloxacin, norfloxacin, gentamicin, tetracycline, and polymyxin B compared to broiler samples. On the other hand, all P. penneri isolates from both broiler and layer samples demonstrated resistance to multiple antibiotics, including ampicillin, ciprofloxacin, norfloxacin, cefoxitin, kanamycin, tetracycline, imipenem, azithromycin, erythromycin, colistin, polymyxin B, and Co-Trimoxazole. In broiler samples, 2 isolates (67%) of P. penneri were sensitive to amoxicillin-clavulanic acid and aztreonam, whereas in layer samples, no isolates were sensitive to amoxicillin-clavulanic acid, and 2 isolates (67%) showed intermediate effects, while all isolates were resistant to aztreonam. Additionally, 2 (66.67%) of P. penneri isolates from broiler samples showed sensitivity to cefotaxime, while in layer samples, 1 (33%) of isolates exhibited sensitivity to this antibiotic.
Several studies on MDR S. enterica and P. stuartii have reported resistance patterns in broiler samples from Dhaka, Gazipur, Sherpur, Mymensingh, and Chattogram. In these regions, S. enterica isolates exhibited resistance to various antibiotics, such as Penicillin-g (90–100%), ampicillin (82.85–100%), amoxicillin (90–98%), cephalexin (70%), streptomycin (77.14%), tetracycline (93–97.14%), chloramphenicol (94.28%), cotrimoxazole (80%), nitrofurantoin (50–78%), sulfamethoxazole (60%), gentamicin (40–46%), erythromycin (80%), nalidixic acid (40–66.6%), kanamycin (40–80%), doxycycline (66.66%), ciprofloxacin (20–40%), and imipenem (83.33%) [63, 77–82]. In comparison, the S. enterica isolates obtained in our study exhibited higher levels of resistance to the antibiotics tested compared to those in previous studies. All P. stuartii isolates displayed resistance to ten antibiotics from nine different classes out of the seventeen antibiotics tested across twelve classes. In our study, 7 (70%), 1 (10%), and 5 isolates (50%) of P. stuartii isolates exhibited resistance to aztreonam, cefotaxime, and gentamicin, respectively. Regarding S. enterica isolates, all of them demonstrated resistance to nine antibiotics from twelve classes tested, and 2 (50%) of these isolates were also resistant to azithromycin. However, all S. enterica isolates in this study were sensitive to aztreonam.
M. morganii, Aeromonas spp., and W. chitiniclastica isolates were exclusively detected in layer chicken samples, with M. morganii and W. chitiniclastica being isolated for the first time in Bangladesh. Previous studies reported concerning rates of multi-drug resistance (MDR) in M. morganii, with 54% in poultry chicken meat from Tennessee [62] and 52% in poultry chickens in Nigeria [83]. In our study, we found 2 isolates (100%) of M. morganii to be resistant to ampicillin, cefoxitin, kanamycin, tetracycline, imipenem, azithromycin, erythromycin, colistin, and Co-Trimoxazole, posing an alarming situation for the Noakhali region of Bangladesh.
In our study, W. chitiniclastica strains isolated from layer chicken samples displayed resistance to all antibiotics used, except aztreonam and polymyxin B. Moreover, 2 isolates (66.7%) of these showed intermediate effects towards aztreonam. W. chitiniclastica has been recognized as an emerging zoonotic pathogen by the United States Centers for Disease Control and Prevention (CDC) [84]. Kopf et al. suggested the use of trimethoprim/sulfamethoxazole, levofloxacin, and cephalosporins (e.g., ceftazidime) antibiotics for W. chitiniclastica infections [65]. However, in our study, we found that all isolates of W. chitiniclastica had already developed resistance to these drugs, indicating that these antibiotics may not be effective for treating W. chitiniclastica infections.
Alam et al. (2010) reported on Aeromonas spp. isolated from poultry chicken samples, showing resistance to erythromycin, (16%), norfloxacin (16%), nalidixic acid (15%), tetracycline (15%), ampicillin (10%), and gentamicin (20%), while being sensitive to streptomycin (30%), chloramphenicol (65%), ciprofloxacin (18%), norfloxacin (83%), nalidixic acid (85%), tetracycline (20%), rifampicin (25%), and gentamicin (80%) [85]. Moreover, one other study from Igbinosa et. al. (2014) found that all Aeromonas spp. from poultry chicken fecal samples were sensitive to ciprofloxacin, gentamicin, and tetracycline [86]. Conversely, in a study on poultry feed used in Bangladesh’s poultry farms, Aeromonas spp. isolates displayed resistance to rifampin (30%), gentamicin (40%), erythromycin (100%), ceftriaxone (10%), kanamycin (10%), novobiocin (40%), nalidixic acid (10%), amoxicillin (30%), and ciprofloxacin (45%) [87]. In comparison, the isolates from our study exhibited a higher rate of resistance to antibiotics. We observed that all Aeromonas spp. isolates showed resistance to ampicillin, kanamycin, imipenem, azithromycin, and erythromycin. Additionally, Aeromonas spp. showed resistance to amoxicillin-clavulanic acid (2 isolates, 66.67%), aztreonam (1 isolate, 33.33%), chloramphenicol (2 isolates, 66.67%), ciprofloxacin (2 isolates, 66.67%), norfloxacin (1 isolate, 33.33%), cefotaxime (2 isolates, 66.67%), gentamicin (2 isolates, 66.67%), tetracycline (2 isolates, 66.67%), colistin (2 isolates, 66.67%), polymyxin B (2 isolates, 66.67%), and Co-Trimoxazole (2 isolates, 66.67%).
Chicken farms serve as reservoirs of multi-drug resistant genes worldwide, including Bangladesh [14, 17, 18, 28, 29, 88–91]. MDR genes, such as ESBL genes, AmpC producing genes, carbapenem-resistant genes, colistin-resistant genes, tetracycline, and sulfonamide-resistant genes, have been reported in poultry and poultry products in different regions of Bangladesh [8, 60, 61, 77, 79, 81, 92]. In our study, we screened for ESBL and carbapenem-resistant genes (blaCTX-M, blaSHV, blaTEM, blaNDM), tetracycline-resistant genes (tetA, tetB), and sulfonamide-resistant genes (sul1, sul2) in the isolates from poultry chicken samples to understand their genotypic characteristics. The identified genes included blaSHV, blaTEM, tetA, and sul2 in all isolates, while E. coli, K. pneumoniae, and P. penneri isolates contained tetB genes. However, the blaCTX-M gene was not found in any isolates, but an unwanted band approximately 350 bp was observed, with the actual band size for blaCTX-M being 593 bp [93]. The presence of ESBL, tetracycline, and sulfonamide resistant genes in bacterial isolates led to resistance against ampicillin, amoxicillin-clavulanic acid, aztreonam, cefotaxime, cefoxitin, tetracycline, and Co-Trimoxazole. Notably, ESBL genes (blaCTX-M, blaSHV, blaTEM) found on sizable conjugative plasmids are responsible for resistance to other classes of antibiotics like fluoroquinolones, aminoglycosides, and trimethoprim-sulfamethoxazole [94–96]. This highlights the importance of monitoring and controlling the spread of antibiotic resistance in poultry settings to mitigate its impact on public health.
Conclusion
Poultry and poultry products, particularly in the Noakhali region of Bangladesh, have become reservoirs of multi-drug resistant (MDR) isolates, joining clinical sources in this role. Studies in Bangladesh have focused on MDR isolates such as E. coli, K. pneumoniae, Salmonella spp., Campylobacter spp., and Citrobacter spp. from poultry chickens and farms. Our study identified Enterobacteriaceae isolates, including M. morganii, showing higher resistance to common drugs and acting as pathogenic agents for humans and animals, contributing to the spread of antibiotic resistance genes in the region. Additionally, this study assessed antibiotic resistance in Aeromonas spp., and W. chitiniclastica from the non-Enterobacteriaceae family.
However, the study has limitations. Firstly, the sample collection was limited to four small-scale farms in Sadar Upazila of Noakhali, potentially limiting the comprehensive understanding of MDR bacteria in poultry chickens. Fourth-generation cephalosporins were also not part of the study. Only one variant of the mcr gene was evaluated, and other genes responsible for carbapenem, aminoglycoside, and macrolide resistance were not included. Nonetheless, the study highlights the rapid development of antibiotic resistance in both Enterobacteriaceae and non-Enterobacteriaceae bacterial strains. The high antibiotic use in the poultry industry remains a major concern, posing economic and health risks to both humans and animals.
Supporting information
((+) = Lactose Positive, (-) = Lactose negative Mac = MacConkey agar).
(TIFF)
(Ampicillin (AMP25), Amoxicillin-clavulanic acid (AMC 30), Cefotaxime (CTX 30) Cefoxitin (CX 30) Ciprofloxacin (CIP 5) and Norfloxacin (NX 10), Aztreonam (AT 30), Gentamicin (GEN 10), Kanamycin (K 30), Azithromycin (AZM 30) Erythromycin (E 10), Imipenem (IMP 10), Chloramphenicol (C 30), Trimethoprim-Sulfamethoxazole (COT 25), Tetracycline (TE 30), Colistin (CL 10) and Polymyxin B (PB 300)).
(TIF)
(Ampicillin (AMP25), Amoxicillin-clavulanic acid (AMC 30), Cefotaxime (CTX 30) Cefoxitin (CX 30) Ciprofloxacin (CIP 5) and Norfloxacin (NX 10), Aztreonam (AT 30), Gentamicin (GEN 10), Kanamycin (K 30), Azithromycin (AZM 30) Erythromycin (E 10), Imipenem (IMP 10), Chloramphenicol (C 30), Trimethoprim-Sulfamethoxazole (COT 25), Tetracycline (TE 30), Colistin (CL 10) and Polymyxin B (PB 300)).
(TIF)
(Ampicillin (AMP25), Amoxicillin-clavulanic acid (AMC 30), Cefotaxime (CTX 30) Cefoxitin (CX 30) Ciprofloxacin (CIP 5) and Norfloxacin (NX 10), Aztreonam (AT 30), Gentamicin (GEN 10), Kanamycin (K 30), Azithromycin (AZM 30) Erythromycin (E 10), Imipenem (IMP 10), Chloramphenicol (C 30), Trimethoprim-Sulfamethoxazole (COT 25), Tetracycline (TE 30), Colistin (CL 10) and Polymyxin B (PB 300)).
(TIF)
(Ampicillin (AMP25), Amoxicillin-clavulanic acid (AMC 30), Cefotaxime (CTX 30) Cefoxitin (CX 30) Ciprofloxacin (CIP 5) and Norfloxacin (NX 10), Aztreonam (AT 30), Gentamicin (GEN 10), Kanamycin (K 30), Azithromycin (AZM 30) Erythromycin (E 10), Imipenem (IMP 10), Chloramphenicol (C 30), Trimethoprim-Sulfamethoxazole (COT 25), Tetracycline (TE 30), Colistin (CL 10) and Polymyxin B (PB 300)).
(TIF)
(Ampicillin (AMP25), Amoxicillin-clavulanic acid (AMC 30), Cefotaxime (CTX 30) Cefoxitin (CX 30) Ciprofloxacin (CIP 5) and Norfloxacin (NX 10), Aztreonam (AT 30), Gentamicin (GEN 10), Kanamycin (K 30), Azithromycin (AZM 30) Erythromycin (E 10), Imipenem (IMP 10), Chloramphenicol (C 30), Trimethoprim-Sulfamethoxazole (COT 25), Tetracycline (TE 30), Colistin (CL 10) and Polymyxin B (PB 300)).
(TIF)
(Ampicillin (AMP25), Amoxicillin-clavulanic acid (AMC 30), Cefotaxime (CTX 30) Cefoxitin (CX 30) Ciprofloxacin (CIP 5) and Norfloxacin (NX 10), Aztreonam (AT 30), Gentamicin (GEN 10), Kanamycin (K 30), Azithromycin (AZM 30) Erythromycin (E 10), Imipenem (IMP 10), Chloramphenicol (C 30), Trimethoprim-Sulfamethoxazole (COT 25), Tetracycline (TE 30), Colistin (CL 10) and Polymyxin B (PB 300)).
(TIF)
(Ampicillin (AMP25), Amoxicillin-clavulanic acid (AMC 30), Cefotaxime (CTX 30) Cefoxitin (CX 30) Ciprofloxacin (CIP 5) and Norfloxacin (NX 10), Aztreonam (AT 30), Gentamicin (GEN 10), Kanamycin (K 30), Azithromycin (AZM 30) Erythromycin (E 10), Imipenem (IMP 10), Chloramphenicol (C 30), Trimethoprim-Sulfamethoxazole (COT 25), Tetracycline (TE 30), Colistin (CL 10) and Polymyxin B (PB 300)).
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the ‘Noakhali Science and Technology University- Research Cell’ Teachers’ grant of the budget year 2021-2022 (Project ID: NSTU/RC-BG-06/T-23/32). The funders contributed solely by providing financial support for this study. They were not involved in the study's design, data collection, analysis, the decision to publish, or the preparation of the manuscript.
References
- 1.Schwartz T, Kohnen W, Jansen B, Obst U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS microbiology ecology. 2003;43(3):325–35. doi: 10.1111/j.1574-6941.2003.tb01073.x [DOI] [PubMed] [Google Scholar]
- 2.Siddiqui MK, Khatoon N, Roy PC. Untreated liquid hospital waste: potential source of multidrug resistant bacteria. Bangladesh Journal of Microbiology. 2015:21–4. [Google Scholar]
- 3.Le T-H, Ng C, Chen H, Yi XZ, Koh TH, Barkham TMS, et al. Occurrences and characterization of antibiotic-resistant bacteria and genetic determinants of hospital wastewater in a tropical country. Antimicrobial agents and chemotherapy. 2016;60(12):7449–56. doi: 10.1128/AAC.01556-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chitanand M, Kadam T, Gyananath G, Totewad N, Balhal D. Multiple antibiotic resistance indexing of coliforms to identify high risk contamination sites in aquatic environment. Indian Journal of Microbiology. 2010;50(2):216–20. doi: 10.1007/s12088-010-0042-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiseo K, Huber L, Gilbert M, Robinson TP, Van Boeckel TP. Global trends in antimicrobial use in food animals from 2017 to 2030. Antibiotics. 2020;9(12):918. doi: 10.3390/antibiotics9120918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olatoye O. Antibiotics use and resistance patterns of Salmonella species in poultry from Ibadan, Nigeria. Tropical Veterinarian. 2011;29(1):28–35. [Google Scholar]
- 7.Economy L. Livestock Economy at a Glance 2019–2020. In: Services DoL, editor. 2021. [Google Scholar]
- 8.Islam MS, Sabuj AAM, Haque ZF, Pondit A, Hossain MG, Saha S. Seroprevalence and risk factors of avian reovirus in backyard chickens in different areas of Mymensingh district in Bangladesh. Journal of Advanced Veterinary and Animal Research. 2020;7(3):546. doi: 10.5455/javar.2020.g452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talaro K, Talaro A. Drugs, microbes, host-The elements of chemotherapy. Foundations in Microbiology 4th ed Mc Grow Hill, New York. 2002:348–79. [Google Scholar]
- 10.Apata D. Antibiotic resistance in poultry. International Journal of Poultry Science. 2009;8(4):404–8. [Google Scholar]
- 11.Helmy YA, El-Adawy H, Abdelwhab EM. A comprehensive review of common bacterial, parasitic and viral zoonoses at the human-animal interface in Egypt. Pathogens. 2017;6(3):33. doi: 10.3390/pathogens6030033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafez HM, Hauck R. Zoonoses with public health relevance in poultry. Zoonoses-Infections Affecting Humans and Animals: Springer; 2015. p. 103–23. [Google Scholar]
- 13.Kang J, Sickbert-Bennett EE, Brown VM, Weber DJ, Rutala WA. Relative frequency of health care-associated pathogens by infection site at a university hospital from 1980 to 2008. American journal of infection control. 2012;40(5):416–20. doi: 10.1016/j.ajic.2011.06.013 [DOI] [PubMed] [Google Scholar]
- 14.Saliu E-M, Vahjen W, Zentek J. Types and prevalence of extended–spectrum beta–lactamase producing Enterobacteriaceae in poultry. Animal health research reviews. 2017;18(1):46–57. doi: 10.1017/S1466252317000020 [DOI] [PubMed] [Google Scholar]
- 15.Ojer-Usoz E, González D, Vitas AI, Leiva J, García-Jalón I, Febles-Casquero A, et al. Prevalence of extended-spectrum β-lactamase-producing Enterobacteriaceae in meat products sold in Navarra, Spain. Meat Science. 2013;93(2):316–21. [DOI] [PubMed] [Google Scholar]
- 16.Carattoli A. Animal reservoirs for extended spectrum β‐lactamase producers. Clinical Microbiology and Infection. 2008;14:117–23. [DOI] [PubMed] [Google Scholar]
- 17.Boonyasiri A, Tangkoskul T, Seenama C, Saiyarin J, Tiengrim S, Thamlikitkul V. Prevalence of antibiotic resistant bacteria in healthy adults, foods, food animals, and the environment in selected areas in Thailand. Pathogens and global health. 2014;108(5):235–45. doi: 10.1179/2047773214Y.0000000148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tekiner İH, Özpınar H. Occurrence and characteristics of extended spectrum beta-lactamases-producing Enterobacteriaceae from foods of animal origin. brazilian journal of microbiology. 2016;47:444–51. doi: 10.1016/j.bjm.2015.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kluytmans JA, Overdevest IT, Willemsen I, Kluytmans-Van Den Bergh MF, Van Der Zwaluw K, Heck M, et al. Extended-spectrum β-lactamase–producing Escherichia coli from retail chicken meat and humans: comparison of strains, plasmids, resistance genes, and virulence factors. Clinical Infectious Diseases. 2013;56(4):478–87. [DOI] [PubMed] [Google Scholar]
- 20.Leverstein-van Hall M, Dierikx C, Stuart JC, Voets G, Van Den Munckhof M, van Essen-Zandbergen A, et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clinical Microbiology and Infection. 2011;17(6):873–80. doi: 10.1111/j.1469-0691.2011.03497.x [DOI] [PubMed] [Google Scholar]
- 21.Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrobial agents and chemotherapy. 2010;54(3):969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitout JD. Extraintestinal pathogenic Escherichia coli: an update on antimicrobial resistance, laboratory diagnosis and treatment. Expert review of anti-infective therapy. 2012;10(10):1165–76. doi: 10.1586/eri.12.110 [DOI] [PubMed] [Google Scholar]
- 23.Hussain A, Ranjan A, Nandanwar N, Babbar A, Jadhav S, Ahmed N. Genotypic and phenotypic profiles of Escherichia coli isolates belonging to clinical sequence type 131 (ST131), clinical non-ST131, and fecal non-ST131 lineages from India. Antimicrobial agents and chemotherapy. 2014;58(12):7240–9. doi: 10.1128/AAC.03320-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kempf I, Jouy E, Chauvin C. Colistin use and colistin resistance in bacteria from animals. International journal of antimicrobial agents. 2016;48(6):598–606. doi: 10.1016/j.ijantimicag.2016.09.016 [DOI] [PubMed] [Google Scholar]
- 25.Lim S-K, Kang HY, Lee K, Moon D-C, Lee H-S, Jung S-C. First detection of the mcr-1 gene in Escherichia coli isolated from livestock between 2013 and 2015 in South Korea. Antimicrobial agents and chemotherapy. 2016;60(11):6991–3. doi: 10.1128/AAC.01472-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islam S, Urmi UL, Rana M, Sultana F, Jahan N, Hossain B, et al. High abundance of the colistin resistance gene mcr-1 in chicken gut-bacteria in Bangladesh. Scientific reports. 2020;10(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Sun J, Ding Y, Li X-P, Liu Y-H, Feng Y. Genomic insights into mcr-1-positive plasmids carried by colistin-resistant Escherichia coli isolates from inpatients. Antimicrobial agents and chemotherapy. 2017;61(7):e00361–17. doi: 10.1128/AAC.00361-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hembach N, Schmid F, Alexander J, Hiller C, Rogall ET, Schwartz T. Occurrence of the mcr-1 colistin resistance gene and other clinically relevant antibiotic resistance genes in microbial populations at different municipal wastewater treatment plants in Germany. Frontiers in microbiology. 2017;8:1282. doi: 10.3389/fmicb.2017.01282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang X, Yu L, Chen X, Zhi C, Yao X, Liu Y, et al. High prevalence of colistin resistance and mcr-1 gene in Escherichia coli isolated from food animals in China. Frontiers in Microbiology. 2017;8:562. doi: 10.3389/fmicb.2017.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H-W, Zhang T, Ma J-H, Fang Y, Wang H-Y, Huang Z-X, et al. Occurrence of plasmid-and chromosome-carried mcr-1 in waterborne Enterobacteriaceae in China. Antimicrobial agents and chemotherapy. 2017;61(8):e00017–17. doi: 10.1128/AAC.00017-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance. 2018;23(6):17–00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gharaibeh MH, Shatnawi SQ. An overview of colistin resistance, mobilized colistin resistance genes dissemination, global responses, and the alternatives to colistin: a review. Veterinary World. 2019;12(11):1735. doi: 10.14202/vetworld.2019.1735-1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian G-B, Doi Y, Shen J, Walsh TR, Wang Y, Zhang R, et al. MCR-1-producing Klebsiella pneumoniae outbreak in China. The Lancet Infectious Diseases. 2017;17(6):577. doi: 10.1016/S1473-3099(17)30266-9 [DOI] [PubMed] [Google Scholar]
- 34.Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, et al. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Eurosurveillance. 2015;20(49):30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085 [DOI] [PubMed] [Google Scholar]
- 35.Beyrouthy R, Robin F, Lessene A, Lacombat I, Dortet L, Naas T, et al. MCR-1 and OXA-48 in vivo acquisition in KPC-producing Escherichia coli after colistin treatment. Antimicrobial agents and chemotherapy. 2017;61(8):e02540–16. doi: 10.1128/AAC.02540-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roess AA, Winch PJ, Akhter A, Afroz D, Ali NA, Shah R, et al. Household animal and human medicine use and animal husbandry practices in rural Bangladesh: risk factors for emerging zoonotic disease and antibiotic resistance. Zoonoses and public health. 2015;62(7):569–78. doi: 10.1111/zph.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.BBS. Yearbook of Agricultural Statistics-2021: Bangladesh Bureau of Statistics; 2021. Available from: http://www.bbs.gov.bd/site/page/3e838eb6-30a2-4709-be85-40484b0c16c6/(2021). [Google Scholar]
- 38.Bushen A, Tekalign E, Abayneh M. Drug-and multidrug-resistance pattern of Enterobacteriaceae isolated from droppings of healthy chickens on a poultry farm in Southwest Ethiopia. Infection and drug resistance. 2021:2051–8. doi: 10.2147/IDR.S312185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.American Society for M, Bergey DH, Breed RS. Bergey’s manual of determinative bacteriology. 7th ed. ed. Baltimore: Williams & Wilkins Co; 1957. [Google Scholar]
- 40.De Medici D, Croci L, Delibato E, Di Pasquale S, Filetici E, Toti L. Evaluation of DNA extraction methods for use in combination with SYBR green I real-time PCR to detect Salmonella enterica serotype enteritidis in poultry. Applied and environmental microbiology. 2003;69(6):3456–61. doi: 10.1128/AEM.69.6.3456-3461.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong D, Liu W, Li H, Wang Y, Li X, Zou D, et al. Survey and rapid detection of Klebsiella pneumoniae in clinical samples targeting the rcsA gene in Beijing, China. Frontiers in microbiology. 2015;6:519. doi: 10.3389/fmicb.2015.00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanez M, Catalán V, Apráiz D, Figueras M, Martínez-Murcia A. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. International journal of systematic and evolutionary microbiology. 2003;53(3):875–83. doi: 10.1099/ijs.0.02443-0 [DOI] [PubMed] [Google Scholar]
- 43.Kuraku S, Zmasek CM, Nishimura O, Katoh K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic acids research. 2013;41(W1):W22–W8. doi: 10.1093/nar/gkt389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in bioinformatics. 2019;20(4):1160–6. doi: 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hudzicki J. Kirby-Bauer disk diffusion susceptibility test protocol. American society for microbiology. 2009;15:55–63. [Google Scholar]
- 46.CLSI. Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing: 20th informational supplement. CLSI document M100-S20. 2018. [Google Scholar]
- 47.Uwizeyimana JD, Kim D, Lee H, Byun JH, Yong D. Determination of Colistin Resistance by Simple Disk Diffusion Test Using Modified Mueller-Hinton Agar. Ann Lab Med. 2020;40(4):306–11. Epub 2020/02/19. doi: 10.3343/alm.2020.40.4.306 ; PubMed Central PMCID: PMC7054692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fadare FT, Okoh AI. Distribution and molecular characterization of ESBL, pAmpC β-lactamases, and non-β-lactam encoding genes in Enterobacteriaceae isolated from hospital wastewater in Eastern Cape Province, South Africa. Plos one. 2021;16(7):e0254753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ateba CN, Bezuidenhout CC. Characterisation of Escherichia coli O157 strains from humans, cattle and pigs in the North-West Province, South Africa. International Journal of Food Microbiology. 2008;128(2):181–8. doi: 10.1016/j.ijfoodmicro.2008.08.011 [DOI] [PubMed] [Google Scholar]
- 50.Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Applied and environmental microbiology. 1983;46(1):165–70. doi: 10.1128/aem.46.1.165-170.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osundiya O, Oladele R, Oduyebo O. Multiple antibiotic resistance (MAR) indices of Pseudomonas and Klebsiella species isolates in Lagos University Teaching Hospital. African Journal of Clinical and Experimental Microbiology. 2013;14(3):164–8. [Google Scholar]
- 52.Agyare C, Boamah VE, Zumbi CN, Osei FB. Antibiotic use in poultry production and its effects on bacterial resistance. Antimicrobial resistance—A global threat. 2018:33–51. [Google Scholar]
- 53.Landers TF, Cohen B, Wittum TE, Larson EL. A review of antibiotic use in food animals: perspective, policy, and potential. Public health reports. 2012;127(1):4–22. doi: 10.1177/003335491212700103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sahoo KC, Tamhankar AJ, Johansson E, Lundborg CS. Antibiotic use, resistance development and environmental factors: a qualitative study among healthcare professionals in Orissa, India. BMC Public health. 2010;10(1):1–10. doi: 10.1186/1471-2458-10-629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boamah V, Agyare C, Odoi H, Dalsgaard A. Practices and factors influencing the use of antibiotics in selected poultry farms in Ghana. Journal of antimicrobial Agents. 2016;2(2):2–8. [Google Scholar]
- 56.Wajid M, Mushtaq A, Ahmad M, Jabeen Q, Rasheed A, Bashir K, et al. World Journal of Pharmaceutical Sciences. [Google Scholar]
- 57.Mathew AG, Liamthong S, Lin J, Hong Y. Evidence of class 1 integron transfer between Escherichia coli and Salmonella spp. on livestock farms. Foodborne Pathogens and Disease. 2009;6(8):959–64. doi: 10.1089/fpd.2009.0263 [DOI] [PubMed] [Google Scholar]
- 58.Aalipour F, Mirlohi M, Jalali M. Prevalence of antibiotic residues in commercial milk and its variation by season and thermal processing methods. International Journal of Environmental Health Engineering. 2013;2(1):41. [Google Scholar]
- 59.Darwish WS, Eldaly EA, El-Abbasy MT, Ikenaka Y, Nakayama S, Ishizuka M. Antibiotic residues in food: the African scenario. Japanese Journal of Veterinary Research. 2013;61(Supplement):S13–S22. [PubMed] [Google Scholar]
- 60.Saifuddin A, Isalm SA, Anwar MN. Molecular characterization and antimicrobial resistance patterns of spp. and Escherichia coli of laying chicken. Microbes and Health. 2016;5(1):4–6. [Google Scholar]
- 61.Nandi SP, Sultana M, Hossain MA. Prevalence and characterization of multidrug-resistant zoonotic Enterobacter spp. in poultry of Bangladesh. Foodborne pathogens and disease. 2013;10(5):420–7. doi: 10.1089/fpd.2012.1388 [DOI] [PubMed] [Google Scholar]
- 62.Kilonzo-Nthenge A, Rotich E, Nahashon SN. Evaluation of drug-resistant Enterobacteriaceae in retail poultry and beef. Poultry science. 2013;92(4):1098–107. doi: 10.3382/ps.2012-02581 [DOI] [PubMed] [Google Scholar]
- 63.Shahjada Z, Hussain K, Islam MM, Majumder S, Hasan I, Rahman M, et al. Bacteria causing omphalitis in newly hatched chicks from broiler and layer flocks and their antibiotic profiles. Int J Natl Soc Sci. 2017;4(2):73–81. [Google Scholar]
- 64.Matos J, Queiroga AP, Ribeiro RL, Teixeira LM, Albano RM, de Freitas-Almeida AC, et al. First report of the emerging zoonotic agent Wohlfahrtiimonas chitiniclastica isolated from a retail frozen chicken in Rio de Janeiro, Brazil. Antonie Van Leeuwenhoek. 2016;109(5):729–34. doi: 10.1007/s10482-016-0673-x [DOI] [PubMed] [Google Scholar]
- 65.Kopf A, Bunk B, Coldewey SM, Gunzer F, Riedel T, Schröttner P. Identification and Antibiotic Profiling of Wohlfahrtiimonas chitiniclastica, an Underestimated Human Pathogen. Frontiers in microbiology. 2021;12. doi: 10.3389/fmicb.2021.712775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Almuzara MN, Palombarani S, Tuduri A, Figueroa S, Gianecini A, Sabater L, et al. First case of fulminant sepsis due to Wohlfahrtiimonas chitiniclastica. Journal of clinical microbiology. 2011;49(6):2333–5. doi: 10.1128/JCM.00001-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shukla S, Mishra P. Pseudomonas aeruginosa infection in broiler chicks in Jabalpur. International J Ext Res. 2015;6:37–9. [Google Scholar]
- 68.BADR H, ROSHDY H, ABD EL-HAFEZ AE-H S, M FARGHALY E. Prevalence, pathogenicity and antibiogram sensitivity of Pseudomonas aeruginosa isolated from diseased chickens. Assiut Veterinary Medical Journal. 2016;62(151):119–26. [Google Scholar]
- 69.Mazumder R, Hussain A, Phelan JE, Campino S, Haider S, Mahmud A, et al. Non-lactose fermenting Escherichia coli: Following in the footsteps of lactose fermenting E. coli high-risk clones. Frontiers in Microbiology. 2022;13:4242. doi: 10.3389/fmicb.2022.1027494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sobur MA, Ievy S, Haque ZF, Nahar A, Zaman SB, Rahman MT. Emergence of colistin-resistant Escherichia coli in poultry, house flies, and pond water in Mymensingh, Bangladesh. Journal of Advanced Veterinary and Animal Research. 2019;6(1):50. doi: 10.5455/javar.2019.f311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amin MB, Sraboni AS, Hossain MI, Roy S, Mozmader TAU, Unicomb L, et al. Occurrence and genetic characteristics of mcr-1-positive colistin-resistant E. coli from poultry environments in Bangladesh. Journal of Global Antimicrobial Resistance. 2020;22:546–52. doi: 10.1016/j.jgar.2020.03.028 [DOI] [PubMed] [Google Scholar]
- 72.Ahmed S, Das T, Islam MZ, Herrero-Fresno A, Biswas PK, Olsen JE. High prevalence of mcr-1-encoded colistin resistance in commensal Escherichia coli from broiler chicken in Bangladesh. Scientific reports. 2020;10(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hasan B, Swedberg G. Molecular Characterization of Clinically Relevant Extended-Spectrum β-Lactamases bla CTX-M-15-Producing Enterobacteriaceae Isolated from Free-Range Chicken from Households in Bangladesh. Microbial Drug Resistance. 2022;28(7):780–6. [DOI] [PubMed] [Google Scholar]
- 74.Savin M, Alexander J, Bierbaum G, Hammerl JA, Hembach N, Schwartz T, et al. Antibiotic-resistant bacteria, antibiotic resistance genes, and antibiotic residues in wastewater from a poultry slaughterhouse after conventional and advanced treatments. Scientific Reports. 2021;11(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saud B, Paudel G, Khichaju S, Bajracharya D, Dhungana G, Awasthi MS, et al. Multidrug-resistant bacteria from raw meat of buffalo and chicken, Nepal. Veterinary medicine international. 2019;2019. doi: 10.1155/2019/7960268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hayati M, Indrawati A, Mayasari NLPI, Istiyaningsih I, Atikah N. Molecular detection of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates of chicken origin from East Java, Indonesia. Veterinary World. 2019;12(4):578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alam SB, Mahmud M, Akter R, Hasan M, Sobur A, Nazir K, et al. Molecular detection of multidrug resistant Salmonella species isolated from broiler farm in Bangladesh. Pathogens. 2020;9(3):201. doi: 10.3390/pathogens9030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Momtaz S, Saha O, Usha MK, Sultana M, Hossain MA. Occurrence of pathogenic and multidrug resistant Salmonella spp. in poultry slaughter-house in Bangladesh. Bioresearch Communications-(BRC). 2018;4(2):506–15. [Google Scholar]
- 79.Sultana S, Islam MA, Khatun MM, Nasrin S. Multidrug Resistant Bacteria in the Respiratory Tract of Apparently Healthy Quails. Microbes and Health. 2012;1(2):46–9. [Google Scholar]
- 80.Al-Salauddin AS, Hossain MF, Dutta A, Mahmud S, Islam MS, Saha S, et al. Isolation, identification, and antibiogram studies of Salmonella species and Escherichia coli from boiler meat in some selected areas of Bangladesh. Int J Basic Clin Pharmacol. 2015;4(5):999–1003. [Google Scholar]
- 81.Mahmud MS, Bari ML, Hossain MA. Prevalence of Salmonella serovars and antimicrobial resistance profiles in poultry of Savar area, Bangladesh. Foodborne pathogens and disease. 2011;8(10):1111–8. doi: 10.1089/fpd.2011.0917 [DOI] [PubMed] [Google Scholar]
- 82.Parvej MS, Nazir KNH, Rahman MB, Jahan M, Khan MFR, Rahman M. Prevalence and characterization of multi-drug resistant Salmonella Enterica serovar Gallinarum biovar Pullorum and Gallinarum from chicken. Veterinary World. 2016;9(1):65. doi: 10.14202/vetworld.2016.65-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akinbami OR, Olofinsae S, Ayeni FA. Prevalence of extended spectrum beta lactamase and plasmid mediated quinolone resistant genes in strains of Klebsiella pneumonia, Morganella morganii, Leclercia adecarboxylata and Citrobacter freundii isolated from poultry in South Western Nigeria. PeerJ. 2018;6:e5053. doi: 10.7717/peerj.5053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oakley BB, Melgarejo T, Bloom PH, Abedi N, Blumhagen E, Saggese MD. Emerging Pathogenic Gammaproteobacteria Wohlfahrtiimonas chitiniclastica and Ignatzschineria Species in a Turkey Vulture (Cathartes aura). Journal of Avian Medicine and Surgery. 2021;35(3):280–9. doi: 10.1647/19-00033 [DOI] [PubMed] [Google Scholar]
- 85.Alam S, Hassan S, Shirin M, Akond MA. Sensitivity of Aeromonas obtained from poultry sources of Dhaka, Bangladesh. Bangladesh Journal of Botany. 2010;39(1):123–5. [Google Scholar]
- 86.Igbinosa IH. Antibiogram profiling and pathogenic status of Aeromonas species recovered from Chicken. Saudi Journal of Biological Sciences. 2014;21(5):481–5. doi: 10.1016/j.sjbs.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Roy CR, Ahmed T, Uddin MA. Microbiological analysis of poultry feeds along with the demonstration of the antibiotic susceptibility of the isolates and the antibacterial activity of the feeds. Bangladesh Journal of Microbiology. 2017;34(2):103–7. [Google Scholar]
- 88.Dandachi I, Fayad E, Sleiman A, Daoud Z, Rolain J-M. Dissemination of multidrug-resistant and mcr-1 gram-negative bacilli in broilers, farm workers, and the surrounding environment in Lebanon. Microbial Drug Resistance. 2020;26(4):368–77. doi: 10.1089/mdr.2019.0137 [DOI] [PubMed] [Google Scholar]
- 89.Rahman M, Husna A, Elshabrawy HA, Alam J, Runa NY, Badruzzaman A, et al. Isolation and molecular characterization of multidrug-resistant Escherichia coli from chicken meat. Scientific Reports. 2020;10(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McNeece G, Naughton V, Woodward MJ, Dooley JS, Naughton PJ. Array based detection of antibiotic resistance genes in Gram negative bacteria isolated from retail poultry meat in the UK and Ireland. International journal of food microbiology. 2014;179:24–32. doi: 10.1016/j.ijfoodmicro.2014.03.019 [DOI] [PubMed] [Google Scholar]
- 91.Das A, Dhar PK, Dutta A, Jalal MS, Ghosh P, Das T, et al. Circulation of oxytetracycline-and ciprofloxacin-resistant commensal Escherichia coli strains in broiler chickens and farm environments, Bangladesh. Veterinary world. 2020;13(11):2395. doi: 10.14202/vetworld.2020.2395-2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hasan B, Faruque R, Drobni M, Waldenström J, Sadique A, Ahmed KU, et al. High prevalence of antibiotic resistance in pathogenic Escherichia coli from large-and small-scale poultry farms in Bangladesh. Avian diseases. 2011;55(4):689–92. doi: 10.1637/9686-021411-Reg.1 [DOI] [PubMed] [Google Scholar]
- 93.Monstein HJ, Östholm‐Balkhed Å, Nilsson M, Nilsson M, Dornbusch K, Nilsson L. Multiplex PCR amplification assay for the detection of blaSHV, blaTEM and blaCTX‐M genes in Enterobacteriaceae. Apmis. 2007;115(12):1400–8. doi: 10.1111/j.1600-0463.2007.00722.x [DOI] [PubMed] [Google Scholar]
- 94.Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clinical microbiology reviews. 2001;14(4):933–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Eurosurveillance. 2008;13(47):19044. [PubMed] [Google Scholar]
- 96.Gniadkowski M. Evolution and epidemiology of extended-spectrum β-lactamases (ESBLs) and ESBL-producing microorganisms. Clinical Microbiology and Infection. 2001;7(11):597–608. [DOI] [PubMed] [Google Scholar]