Abstract
Complex three-dimensional in vitro organ-like models, or organoids, offer a unique biological tool with distinct advantages over two-dimensional cell culture systems, which can be too simplistic, and animal models, which can be too complex and may fail to recapitulate human physiology and pathology. Significant progress has been made in driving stem cells to differentiate into different organoid types, though several challenges remain. For example, many organoid models suffer from high heterogeneity, and it can be difficult to fully incorporate the complexity of in vivo tissue and organ development to faithfully reproduce human biology. Successfully addressing such limitations would increase the viability of organoids as models for drug development and preclinical testing. On April 3–6, 2022, experts in organoid development and biology convened at the Keystone Symposium “Organoids as tools for fundamental discovery and translation” to discuss recent advances and insights from this relatively new model system into human development and disease.
Keywords: development, differentiation, inflammatory bowel disease, microfluidics, kidney disease, organoids, single-cell sequencing
Graphical abstract

Introduction
Organoids are three-dimensional in vitro organ-like models generated from pluripotent stem cells (PSC) or primary donor tissues. Protocols have been developed to drive the differentiation of PSCs toward multiple different organoid systems that recapitulate the structural, molecular, and functional characteristics of in vivo tissues and organs, including those associated with the gastrointestinal (GI) tract, central nervous system (CNS), liver, kidney, and skeletal muscle. These organs-in-a-dish hold immense potential as a model for human development and disease. They offer more complexity than traditional two-dimensional cell culture systems while being easier to control, manipulate, and quantify than in vivo models. Regarding development, organoids are more accessible than developing embryos. Human organoids may also recapitulate human biology more faithfully than animal models. That said, it can be difficult to reproducibly generate organoids and to incorporate the complexity so important for proper function of native tissues and organs. Organoids often lack characteristic cellular organization or important organ-supportive tissues, for example, immune, vasculature, lymphatic system, stroma, and innervation tissues.
On April 3–6, 2022, experts in organoid development met for the Keystone Symposium “Organoids as tools for fundamental discovery and translation” to discuss recent advances and insights using organoid systems to study human development and disease. This was a joint meeting held concurrently with “Engineering Multi-Cellular Living Systems”.
Speakers discussed efforts to benchmark organoid systems against their in vivo counterparts, to control organoid development via bioengineering approaches, and to increase organoid complexity and enhance their organization and functions by incorporating multiple cell and tissue types. They also highlighted insights organoids have provided on human development and disease, including identifying possible drug targets for various diseases.
Engineering epithelial organoids
Matthias Lutolf from Roche Institute for Translational Bioengineering and Ecole Polytechnique Fédérale de Lausanne presented work on using advanced bioengineering to generate organoids that more closely resemble in vivo structures, with a focus on intestinal epithelial organoids. Pioneering work in Hans Clevers’s lab at the Hubrecht Institute showed that the crypt-villus structure of the mouse intestinal epithelium could be recapitulated in vitro from even a single stem cell. However, the resulting organoids were heterogeneous in size, shape, and cellular composition1, which is a common problem with most conventional organoids. Using bioengineering approaches, Lutolf’s group showed that confining these organoids in hydrogel microcavities can reproducibly generate organoids of a predefined shape and size; the organoids also show the characteristic cell type patterning seen in the intestinal epithelium. Lutolf explained that the geometry of the crypt creates differential cell crowding and cell shape that lead to differences in YAP and NOTCH signaling and the spatially controlled emergence of Paneth cells, the signaling source for the crypt domain.2 Lutolf’s group is now using this principle to create macroscopic tissues that faithfully recapitulate features of the native tissue. The group has developed crypt-villus substrates that mimic the intestine, generating tissues that incorporate both cell patterning and the macroscopic crypt–villus structures of the intestine. They are using these organoids to study tissue-dynamic processes like cell shedding that occurs at the tip of the villus.
Lutolf also described work to generate tubular perfusable mouse organoids on biomicrofluidic chips. The system allows dead cells to be continually removed, creating a homeostatic culture condition in which organoids can be maintained for months. Fluid can be delivered to the apical side of the lumen to model processes like drug delivery, bacteria colonization, or viral infection. These “mini-gut” tubes possess several qualities of natural tissue that are not often seen in classical organoids, including rare cell types and the capacity to regenerate after injury.3 Lutolf’s group is now working on building reliable human epithelial organoids of the intestine, bladder, respiratory tract, and other tissues to model disease as well as increasing the complexity of the organoids by incorporating other tissue compartments, like vasculature and immune cells. Lutolf also presented unpublished data on using mouse colon organoids to better understand tumorigenesis and the potential for this tumor-on-a-chip to delineate the spatiotemporal dynamics involved in tumorigenesis as well as the impact of genetic and environmental factors.
Embryoids and Gastruloids for Early Development
Understanding post-implantation with synthetic human embryo-like structures
While pre-implantation embryonic development has been relatively well characterized, it is more difficult to study post-implantation development because implanted embryos are difficult to observe or manipulate in vivo.
Jianping Fu from the University of Michigan discussed his work using human PSCs (hPSC) to model peri-implantation human development. In collaboration with Deb Gumucio at the University of Michigan, Fu’s group showed that when grown in 3D culture, a subset of hPSC colonies organize and develop into 3D structures that mimic early post-implantation human developmental events, including tissue morphogenesis, lineage diversification, and tissue-tissue interaction. Some colonies even undergo symmetry breaking and gastrulation-related events. The gastrulating cells upregulated canonical markers for gastrulation. Fu’s group used this system to identify the molecular pathways involved in these early developmental events and cell fate decisions, identifying BMP activity as a key pathway driving amniogenesis from pluripotent hPSCs.4,5
Fu’s group has also developed a microfluidic model to investigate amnion development in early human development. They showed that hPSCs loaded into this microfluidic system display synchronous development and undergo lumenogenesis, forming an epiblast-like sac. Exogenous signals can be introduced into the microfluidic device to establish asymmetric chemical stimulation. Fu showed, for example, that asymmetric BMP stimulation induced embryonic-like sac formation—amnion-like cells developed at the pole directly exposed to BMP4 stimulation while the opposite pole remained pluripotent. Continuous development of the embryonic-like sac led to gastrulation-related events at the pluripotent pole. They also looked at the emergence of primordial germ cells in the model, as there has been some controversy in the literature over where they develop. In addition, expression of BRACHYURY, a key gene marking the onset of gastrulation, at the junction of the amniotic ectoderm and epiblast compartments suggested an important role for tissue–tissue interactions. Co-culture of amnion and epiblast cells revealed that the amnion induces gastrulation via non-canonical WNT secretion. Fu’s group is currently investigating other molecular players and pathways involved.6 His group is also characterizing their embryoids using single-cell sequencing methods and comparing the embryoids to both human and non-human primate embryos.
OCTOPUS: Engineering organoid cultures to enhance organogenesis
Sunghee Estelle Park from Dongeun Huh’s lab at the University of Pennsylvania presented work on a new platform called organoid culture-based three-dimensional organogenesis platform with unrestricted supply of soluble signals (OCTOPUS). Huh’s lab focuses on developing micro-engineered cell culture devices to grow human organs on a chip.7 They have previously established models of human muscle8, eye9, and placenta.10 More recently, the OCTOPUS platform was designed to increase the lifespan and maturity of organoids beyond what can be achieved with traditional Matrigel drop culture methods. In the OCTOPUS platform, stem cells are equally distributed between eight culture chambers, though different chamber designs and sizes are available. Nutrients diffuse freely throughout the culture chambers, obviating the necrotic core that drop-based cultures develop. Park showed that intestinal enteroids grown in the OCTOPUS system were highly viable after 14 days of culture and could grow up to 2 to 3 mm in size. By comparison, Matrigel-drop enteroids showed significant decrease in viability at 14 days. Single-cell RNAseq showed that the cellular diversity of OCTOPUS-grown enteroids more closely resembled in vivo tissue than Matrigel-grown enteroids. Park also described how they are using OCTOPUS-developed enteroids to model disease, specifically inflammatory bowel disease (IBD). Enteroids grown from intestinal stem cells from IBD patients are smaller than those grown from normal cells, and they have markers of decreased cell proliferation, increased apoptosis, and impaired tight junction formation. Single-cell RNAseq of the IBD enteroids confirmed upregulation of several IBD genes. Co-culture of human enteroids with blood vessels produced vascularized organoids that enabled Park to assess vascular abnormalities in IBD and their ability to recruit immune cells. These data provide proof of concept that OCTOPUS can serve as in vitro platform to engineer vascularized organoids and expand the capabilities of conventional organoid culture systems.
High Content Screening with Organoids
Developing an organ-on-a-chip for drug discovery
Athanasia Apostolou from Emulate Inc. described the company’s work to generate ex vivo experimental models that better predict human physiology and could improve the effectiveness of the drug discovery pipeline. The Human Emulation System® developed by Emulate is a complete organ-on-a-chip system that enables an automated fluidic culture and incorporates in vivo relevant mechanical forces. Apostolou described the use of the Colon Intestine-Chip to model mechanisms that elicit the collapse of the intestinal epithelial barrier integrity in pathophysiological conditions, such as IBD. Emulate has developed Intestine-Chips from several cell sources, including Caco2 cells, human tissue biopsies, and iPSC-derived organoids.11–13 Apostolou showed that the transcriptome profiles of both Duodenum- and Colon-Chips grown in their system are more similar to human tissue than to organoids grown in suspension culture.12,14 During her talk, Apostolou focused on the phenotypic and functional validation of the Colon Intestine-Chip, developed from human biopsy-derived colonoids co-cultured with endothelium cells. She showed that co-culture of endothelial and epithelial cells recreated human colon physiology more closely than do classical organoids, with enhanced formation of a tight junction network and epithelial polarity (Fig. 1). Applying mechanical force to the Colon-Chips altered transcription, increasing expression of genes involved in ion, lipoprotein, and water transportation.15 Apostolou also showed how the Colon Intestine-Chip responds to factors known to play a role in IBD, while providing additional insights into their effects on the colon. Addition of IFN-γ, a cytokine that disrupts the epithelial barrier16, resulted in time-dependent collapse of the epithelial barrier, compromised epithelial morphology, disruption of tight junctions, and restructuring of the actin cytoskeleton. The Colon Intestine-Chip also recapitulated the polarized secretion of cytokines as well as interindividual variability observed in the clinic. Addition of IL-22, which has been implicated in the pathogenesis of IBD and is generally thought to support the epithelial barrier17–19, showed, as anticipated, that IL-22 induced STAT3 activation in a concentration- and time-dependent manner. However, unexpectedly, IL-22 compromised epithelial morphology and induced apoptosis.15 These data show the importance of using more in vivo-like and species-specific experimental models to understand human diseases.
Figure 1.
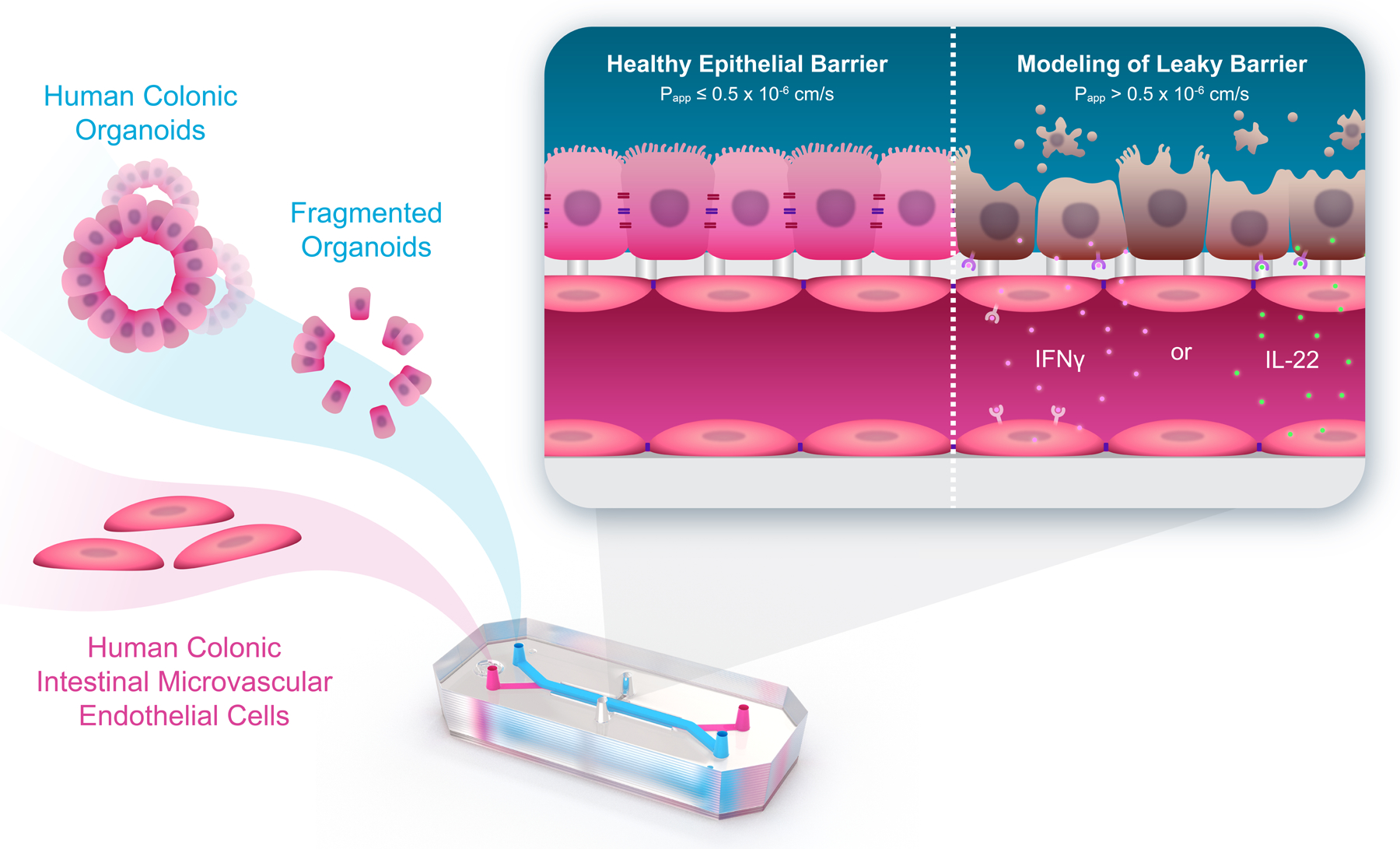
Colon Intestine-Chip, seeded with human colon crypt–derived epithelial and primary microvascular endothelial cells, used to investigate leaky gut. This ex vivo platform recapitulates the effects of proinflammatory cytokines in the intestinal epithelial barrier and identified novel mechanisms of action. From Apostolou et al. (2021) DOI: https://doi.org/10.1016/j.jcmgh.2021.07.004. By CC BY-NC-ND 4.0. license.
A multi-organoid platform to study SARS-CoV-2 infection and drug screening
While the lung is a major target for SARS-CoV-2, the virus has effects on multiple organs. Shuibing Chen from Weill Cornell Medical College discussed work using organoids to better understand SARS-CoV-2 infection and to screen for drugs that inhibit infection. Chen and colleagues are leveraging multiple cell and organoid models derived from hPSCs, including macrophages, cardiomyocytes, endothelial cells, alveolar organoids, airway organoids, small intestine organoids, colon organoids, pancreatic endocrine organoids, liver organoids, and neurons. Chen showed that a SARS-CoV-2 pseudovirus was able to infect various cell types known to be susceptible to infection, such as colon, lung, and liver, as well as additional cell types, including cardiomyocytes, pancreatic endocrine cells, and dopaminergic neurons.20 Single-cell RNAseq of infected cells showed that infection affects different cell types differently. For example, infection induces pacemaker cells to undergo ferroptosis; cardiomyocytes and alveolar and airway organoids show signs of apoptosis; while pancreatic endocrine cells undergo transdifferentiation.21–23
In addition, Chen’s group, in collaboration with David Ho at Columbia University and Ben tenOever at New York University, has used these organoid models to screen for compounds that inhibit SARS-CoV-2 infection. In a high-throughput chemical screen in which alveolar organoids were infected with a SARS-CoV-2 pseudovirus, three small molecules were identified to block viral entry. Chen showed that these molecules also inhibited live SARS-CoV-2 infection in a humanized mouse model in which human alveolar organoids are xenografted into mice. This system provides an in vivo model with which to evaluate drug activity on human cells.21 In a separate drug screen using airway organoids and authentic SARS-CoV-2 virus, the compound GW6471 was shown to block viral replication. Mechanistic studies indicate that GW6471 blocks the HIF1ɑ pathway, which inhibits downstream fatty acid synthesis. Small molecules that block fatty acid synthesis inhibited viral replication in the organoid model. These data demonstrate the importance of the HIF1ɑ-glycolysis axis in mediating SARS-CoV-2 infection in the human airway.22
Chen’s group has also used these organoid models to understand immune-mediated host damage during SARS-CoV-2 infection. In a co-culture of cardiomyocytes and macrophages, SARS-CoV-2 infection resulted in an increase in reactive oxygen species (ROS) and apoptotic cells, which was not seen without macrophages. Additional data and mechanistic studies support a model in which SARS-CoV-2 infection of cardiomyocytes induces the secretion of the cytokine CCL-2, which recruits monocytes to the site of infection. The monocytes differentiate into macrophages that secrete pro-inflammatory cytokines IL-6 and TNFɑ, which can damage cardiomyocytes. A high-throughput drug screen showed that a JAK inhibitor that blocks downstream events in TNFɑ signaling can block macrophage-mediated cardiac damage.24,25
A mini-kidney organoid to model genetic disease
Cheng Jack Song from Andrew McMahon’s group at the University of Southern California presented work on developing mini-kidney organoids to model the genetic disease autosomal dominant polycystic kidney disease (ADPKD). While various protocols have been developed to generate kidney organoids, creating a unified, quantifiable system has been challenging.26–31 Song has developed a protocol to create more unified mini kidney organoids from hPSCs.a Single-cell RNAseq shows that these mini organoids differentiate like other kidney organoids and human fetal kidney. Knocking out the genes responsible for ADPKD in the mini organoids recapitulates the cyst formation indicative of the disease. Song has used this model to screen for small molecules that prevent cyst formation. During the talk, he focused on two compounds: celastrol, which had previously been shown to inhibit cyst formation in in vitro and in vivo models, and QNZ, a novel compound that potently blocks cyst formation in a dose-dependent manner. While high amounts of QNZ negatively impact nephrogenesis, it appears that there may be a therapeutic window in which cyst formation is blocked without kidney toxicity. Song is using the model to understand the mechanism of ADPKD and other kidney diseases.
Increasing Complexity in Organoids by Leveraging Development
Increasing organoid complexity is a key challenge that, as it is overcome, will improve the utility of these systems to model disease. Several speakers spoke about their work to leverage insights into human development to increase organoid complexity, primarily by understanding and increasing cell diversity.
Using organoids to identify and characterize new cell types
Jason Spence from the University of Michigan described unpublished work on characterizing a newly identified secretory cell in the fetal human lung. Spence’s group is broadly interested in translational embryology, i.e., leveraging insights from developmental biology to differentiate PSCs toward certain lineages. Spence’s group has observed that iPSCs sometimes give rise to previously unknown cell types, underscoring the need for a comprehensive single-cell reference atlas of developing human tissues. To address this gap, Spence collaborated with J. Gray Camp at the University of Basel and Barbara Treutlein at ETH Zurich to characterize the lung epithelial cell types present in the developing human lung over time and space32 and to generate an endoderm atlas of multiple organ systems for benchmarking organoid models.33 The scRNAseq data of lung epithelial cells revealed a previously unidentified secretory progenitor cell type, fetal airway secretory (FAS) cells. Fairly abundant within the fetal lung, FAS cells are enriched in the middle airway at 15 to 18 weeks post conception and display a unique molecular profile.32 Spence showed unpublished data using organoid models to understand the function of FAS cells. This work helps to demonstrate how organoids can be used to identify new cell types that are relevant in vivo.
Understanding symmetry breaking in intestinal organoids
Prisca Liberali from the Friedrich Miescher Institute discussed efforts to understand how symmetry breaking is achieved during intestinal organoid development. Liberali’s group is broadly interested in how genetically identical cells coordinate across short- and long-range scales to generate multicellular systems and structures. Liberali’s group is using mouse intestinal organoids as a model system. Regarding symmetry breaking, Liberali put forth a model in which genetically identical pluripotent cells within a population must inherently acquire meta-stable, transient cellular states that “interpret” and respond to their environment differently. Some cells, therefore, have a higher probability to irreversibly differentiate, leading to symmetry breaking.34,35 Multiplexed time-course imaging of mouse intestinal organoids showed that organoid formation is very plastic—both Lgr5+ cells (representing the stem cell population) and Lgr5– cells form organoids via similar development pathways. In addition, some organoids develop without breaking symmetry—these consist entirely of enterocytes and do not contain the Paneth cells found in the intestinal crypt. Liberali found that organoid formation depends on transient activation of the transcription factor Yap, and that cell-to-cell variability in Yap1 expression is required for symmetry breaking.36
Newer work on the function of Yap1 showed that tissue geometry can affect Yap1 signaling. Specifically, symmetry breaking occurs at areas of higher curvature and higher polarity. Single-cell RNAseq conducted at different timepoints during the early stages of symmetry breaking revealed decreased Stretch genes, increased ERK and WNT target genes, and glycolytic activity. These data indicate that symmetry breaking requires the emergence of cell states that differ in their mechanical and polarization state, signaling state, and metabolic profile. Liberali put forth a model in which symmetry breaking occurs during a select time window, between 16 and 32 cells, when the cell population contains the proper distribution of these cell states.2
The impact of tissue architecture on brain organoid development
Madeline Lancaster from the Medical Research Council Laboratory of Molecular Biology showed how her lab is using organoids to understand human brain development. Using brain organoids from human and apes, Lancaster’s group elucidated the mechanisms that drive brain size. Compared with ape brain organoids, human brain organoids display delayed neuroepithelial transitions that result in slower elongation of neuroepithelial cells, which correlates with a short cell cycle length. In other words, in human brain organoids, neuroepithelial cells divide more, giving rise to more neurons and thus bigger brains. Because these events happen early in development, they impact all the cortical neuron layers evenly, which is consistent with morphological differences between human and ape brains.37
Other studies Lancaster discussed focus on the impact of sex steroids on neurogenesis. Lancaster showed that androgens increase proliferation of radial glial progenitors and increase the neurogenetic potential of excitatory neurons, but not inhibitory progenitors. These findings may have implications for neurodevelopmental conditions like autism spectrum disorder, which is more prevalent in males and shows differences in the balance and maturity of excitatory and inhibitory neurons.38
Lancaster also presented unpublished work on the impact of tissue architecture on brain organoid development. Previous work suggests that fate determination in neurogenesis is not dependent on tissue architecture and that developmental programs are at least partially hard wired.39,40 More recent studies using scRNAseq showed that brain organoids more closely recapitulate the in vivo transcriptome profile than do cells grown in 2D culture, suggesting a role for tissue architecture and geometry.41 Other studies are mixed on whether organoids are more similar to in vivo models than 2D culture.42,43 Lancaster’s group is working to reconcile these contradictory findings to better understand how tissue architecture affects fate determination and temporal development during brain development.
Generating epicardioids to understand heart development and disease
Anna Meier from the Technical University of Munich presented work on creating human heart organoid models that incorporate both the epicardium and myocardium. The heart has proven relatively recalcitrant to the development of self-organizing organoids. It is still not possible to generate organoids that spontaneously form all three layers of the heart—the epicardium, myocardium, and endocardium. Researchers have developed protocols to create cardiogenic gastruloids that imitate gastrulating embryos44, multilineage organoids that form more mature tissues45,46, and cardioids that contain the myocardium and endocardium.47 Despite this progress, incorporating the epicardium into these models has been difficult. Meier noted that the epicardium plays important roles in the developing embryo—where epicardial cells give rise to multiple cardiac lineages—and promote myocardial development and repair. In species that can regenerate the adult heart, like zebrafish, the epicardium plays an important role in regenerating the myocardium.48 Understanding the cross-talk between the epicardium and myocardium could therefore have significant implications in mitigating the effects of heart disease. Meier presented unpublished work on developing self-organizing organoids of the myocardium and epicardium, with the aim of using these epicardioids to understand how epicardial cells give rise to different cardiac cell lineages during development, the crosstalk between the myocardium and epicardium, and how to model heart disease.
An organoid model to study alveologenesis
Nicole Pek from Mingxia Gu’s lab at the University of Cincinnati presented work on using blood vessel organoids to understand how the pulmonary vasculature influences alveologenesis. Disrupted alveologenesis underlies a group of congenital lung diseases, including alveolar capillary dysplasia (ACD). ACD is associated with malposition of the pulmonary vein; most patients die during the first few weeks of life due to respiratory failure. Mutations in the gene FOXF1 and the surrounding chromosome have been associated with ACD.49,50 Pek described unpublished work being done in collaboration with Darrell Kotton at Boston University and Robbert Rottier at Erasmus University Medical Center to generate blood vessel organoids generated from ACD patient iPSCs that contain three unique mutations in FOXF1. They plan to use these models to understand the impact of FOXF1 mutation on vascular development and its role in alveolar/capillary defects in patients with ACD.
Improvements in Organoid Maturation
Adding complexity via co-development and separate development
Jim Wells from Cincinnati Children’s Hospital Medical Center discussed two approaches his lab is taking to introduce more complexity into organoids—by co-developing different cell types within an organoid and by combining separately generated progenitor populations. Wells is ultimately interested in using organoids to understand more complex organ-related functions and crosstalk. In the first example, which is unpublished, Wells focused on work to characterize the cell types of a PSC-derived human colonic organoid model developed in his lab after observing a population of co-developing immune cells.51 In the second example, Wells showed how they have generated functional gastric organoids by combining the three progenitor cell populations52—the endoderm which forms the epithelium, glands, and endocrine cells; the mesoderm, which forms vascular, smooth muscle, and immune cells; and the ectoderm, which forms the enteric nervous system. In this process, hPSCs are separately differentiated into ectoderm, endoderm, and mesoderm. Using previously established protocols, endoderm cells are differentiated into foregut epithelium, and ectoderm cells are differentiated into neural crest stem cells.53,54 Wells’s collaborators at Cincinnati Children’s devised a new protocol to differentiate mesoderm cells into splanchnic mesenchyme via signaling pathways identified by Han et al. who developed a single-cell atlas of the developing mouse foregut.55 Combining these three cell types led to self-assembly of gastric organoids with the expected cell diversity, morphology, and function.52
A suspension-based culture system for human intestinal organoids
Meghan Capeling from Jason Spence’s lab at the University of Michigan presented work on developing a suspension-based culture system for intestinal organoids. Typically, hPSC-derived human intestinal organoids are embedded in a Matrigel matrix; these organoids often suffer from limited reproducibility, are expensive, and are generally immature—they do not recapitulate the full cell diversity seen in vivo and must be transplanted into mice to give rise to more mature intestinal tissue.56,57 Alternative, Matrigel-free culture systems could offer greater experimental control and better recapitulate the diversity seen in vivo.58,59 Capeling showed that human intestinal organoids can be grown in suspension culture on a similar timescale to Matrigel-based organoids as a cheaper, simpler, and more scalable alternative without the poorly defined composition of Matrigel. Since the organoids contain both an epithelium and mesenchyme, they form their own niche without any external cues from Matrigel. Suspension-grown organoids contained an epithelium comparable to Matrigel-grown organoids with the expected epithelial cell types, proper polarization, and mature intestinal cell types like enterocytes and goblet cells. One key difference between the two organoid types was the mesenchymal layer. In Matrigel-grown organoids, the mesenchyme is disorganized and lacks the serosal mesothelium, the outermost layer, which contributes to mesenchyme and vascular smooth muscle development and is implicated in adhesions that form after abdominal surgery. In contrast, the mesenchyme in suspension-grown organoids was better organized, more closely resembled the circular architecture of human tissue, and formed a defined outer serosal mesothelium. An scRNAseq analysis revealed molecular similarities between suspension-grown intestinal organoid serosa and human fetal serosa. Capeling is using these organoids as a model to understand how the serosa forms, which has been previously difficult to address. Using small molecule inhibitors that target different signaling pathways involved in intestinal development, she has identified Hedgehog and WNT signaling as important regulators of serosa development.60
Understanding cortical cell diversity
Ana Uzquiano from Paola Arlotta’s lab at Harvard University presented work using human cortical organoids to understand how cell diversity emerged during cortical development. Uzquiano has used the cortical organoids to build a single-cell resolution map of human cortical development; the map consists of both transcriptional and chromatin accessibility information of more than 600,000 cells from 83 organoids representing 8 time points from 23 days to 6 months of development. Uzquiano showed that organoids reproducibly recapitulated the processes of cell diversification of the developing human cortex;61 data defining the lineage relationships and longitudinal molecular trajectories of cortical cell types during development in organoids; and that cell diversification in the organoids correlated with human fetal datasets.61–63 Uzquiano showed that the identity of the majority of cortical cell types present in this cortical organoid model is not affected by diverging metabolic states, which affects only two cell types, one of which does not have a fetal counterpart (‘unspecified projection neurons’). These two cell types localize to the inner region of the organoid and display an altered metabolic profile characterized by upregulation of glycolysis and hypoxia-related pathways. With the exception of these cell types, cell identity was not affected by metabolic state. Finally, by inferring the molecular trajectories associated with human cortical lineages and comparing this information to data from the developing mouse cortex64, Uzquiano identified novel regulators of neurogenesis and cell identity acquisition in humans.61
Organoids for Disease Modeling
Integrating stem cell and organ chip technologies to model and understand human kidney disease
Samira Musah from Duke University presented work on developing stem cell-derived organ chips as in vitro platforms to model human diseases and for therapeutic drug development. Mush described models that incorporate molecular and mechanical forces, depending on the organ of interest, to mimic the in vivo environment as faithfully as possible.65,66 She stressed that while they are very early in terms of being able to model organs, they believe that in vitro platforms will be able to model disease in a patient-specific manner.
Musah’s group has developed several methodologies to promote pluripotency in vitro using synthetic matrices,67,68 as well as to drive cell fate commitment to different lineages including neuronal cells69, endothelial cells70, and kidney epithelial cells.71–73 During her talk, Musah focused on their efforts in developing a glomerulus-on-a-chip. The filtering unit of the kidney, the glomerulus is often the site of damage in patients with kidney disease and drug-related kidney toxicity. The absence of physiologically relevant models has made it difficult to identify biomarkers and drug targets for kidney disease; consequently, there are still no targeted therapies in this area despite its prevalence.
Musah’s group developed a novel method to direct differentiation of kidney podocytes from human iPSCs. They incorporated these cells into a microfluidic device that recapitulates the structure of the glomerulus. Musah showed that this glomerulus-on-a-chip mimics the tissue-tissue structure of the glomerulus, with iPS-derived podocytes and a glomerular endothelium. The system can filter out small molecules while retaining larger proteins––similar to the kidney––and is susceptible to genetic and exogenous signals that mimic disease. The system can also mimic different stages of development by introducing mechanical strain, i.e., adding strain resulted in tissue that more closely resembles the tissue-tissue interface of the intact glomerulus while lack of strain produced a tissue structure that mimics earlier stages of development.71 Musah showed how her group has used this platform to demonstrate that SARS-CoV-2 can directly infect podocytes,74 as well as to identify possible therapeutic targets for kidney disease.75 In addition to these efforts, her group is working to use the platform for drug screening.
Developing organoids to investigate mechanisms of liver regeneration and disease
Meritxell Huch from the Max Planck Institute of Molecular Cell Biology and Genetics presented work on liver organoids derived from adult tissue. For the past 10 years, the Huch lab has been developing organoids from healthy and diseased, human and mouse, and adult and embryonic tissues for a range of organs including stomach76, liver77, and pancreas.78 The work of the Huch lab has shown how adult liver organoids can be used to investigate mechanisms of liver regeneration and cancer at different biological scales, from molecules to cells and tissues. In the first part of her talk, Huch focused on molecular mechanisms of cellular plasticity during regeneration and cancer. The group had shown that liver organoids recapitulate many aspects of liver regeneration in a dish.79 In her talk, Huch showed that in vivo and in mouse models, both liver organoid formation and liver regeneration require transient, genome-wide transcriptional and epigenetic reprogramming to switch on regenerative programs. These transcriptional and epigenetic changes enable the cellular plasticity required to license liver differentiated cells for organoid formation and in vivo regeneration. Huch also showed preliminary data on how the lab is transferring this knowledge to study cellular plasticity in cancer using patient-derived liver cancer organoids the lab had previously established in 2017.80
In the second part of her talk, Huch elaborated on how a novel organoid co-culture system her lab has developed has enabled the study of cellular mechanisms of liver regeneration. She focused on a mesenchymal population that resides near the ductal epithelium. She first showed that the numbers between both populations dynamically change during the damage-repair response in vivo. To investigate whether dynamic changes in these cellular interactions could regulate the repair response, the group developed a co-culture system of ductal epithelial and portal mesenchymal cells by microencapsulating both cell types to facilitate cellular interactions. Huch showed that the tissue architecture and direct cell-cell interactions between both populations can be recapitulated in vitro in this novel co-culture system (Fig. 2).81 Interestingly, this organoid co-culture system was crucial to underscore a very interesting paradox (that would otherwise had remained unnoticed) that mesenchymal cell contact inhibits epithelial cell proliferation while paracrine signaling promotes it. Notably, non-physiological numbers of mesenchymal cells resulted in loss of ductal epithelial integrity and collapse of the epithelial organoid structure. This effect could not be rescued by supplementing the cultures with growth factors, indicating that cell contact inhibition is dominant over growth factor presence. These organoid-based observations could potentially reconcile the dichotomy between a homeostatic/pro-quiescent niche and a proliferating/pro-regenerative niche, suggesting the number of cellular interactions, not the total number of cells, is the critical parameter during tissue regeneration and, consequently, during organoid formation. Overall, Huch provided an overview of how adult tissue-derived organoid cultures represent excellent in vitro models to gain mechanistic understanding of basic biological principles of tissue regeneration and cancer across different biological scales.
Figure 2.
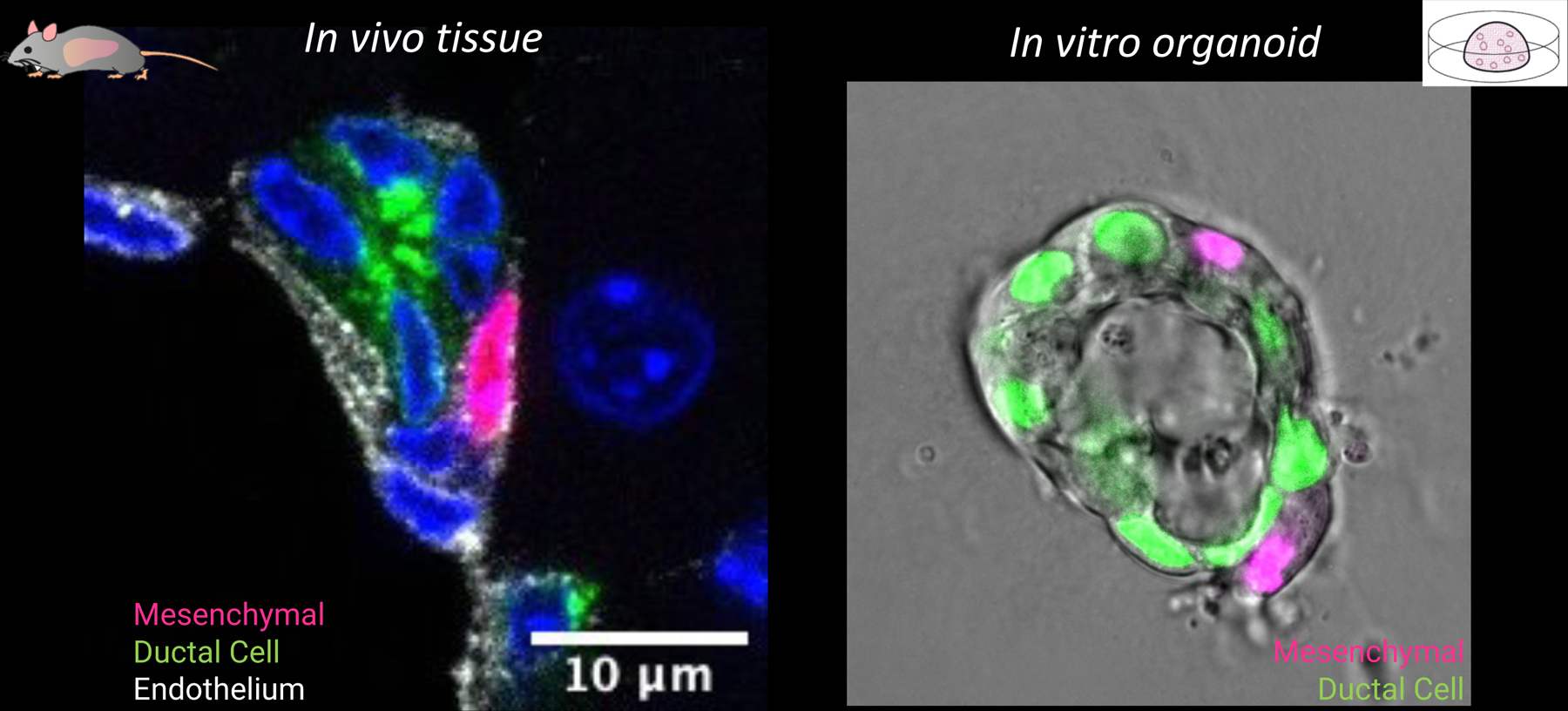
Tissue architecture and direct cell-cell interactions between ductal epithelial and portal mesenchymal cells can be recapitulated in vitro in a novel co-culture system.
Developing a model of the neuromuscular system
Mina Gouti from the Max Delbruck Center for Molecular Medicine presented work on developing human neuromuscular organoids from PSCs. Insights from developmental biology show that the spinal cord motor neurons that enervate skeletal muscle express different types of Hox genes based on where they are found in the body. Gouti’s work has been instrumental in defining the developmental origins of the posterior and anterior neuromuscular system. In brief, the anterior and posterior regions have distinct development origins. Anterior neuroprogenitors generate the brain and hindbrain, while neuromesodermal progenitors (NMP) generate spinal neurons and the posterior somites that generate the musculoskeletal system.82–84 Therefore, to study development of the neuromuscular system, it is necessary to understand the development and differentiation of NMPs.
Gouti has previously developed a protocol to generate mouse and human NMPs from PSCs in vitro.82 Goutís group has pioneered the generation of human neuromuscular organoids (NMOs) from NMPs, which are the building blocks of the posterior neuromuscular system. When grown in a nonadherent cell culture system, NMPs generate both spinal cord and muscle tissues that self-organize into two distinct regions within the organoid (Fig. 3).85,86 Single-cell sequencing of the neuromuscular organoids revealed two primary differentiation trajectories—a neural lineage and skeletal muscle lineage. After approximately two months of culture, the organoids consist of skeletal muscle cells surrounded by spinal cord neurons and contain other mature cell types and features (such as glia, interneurons, myelinated axons, and terminal Schwann cells) and functional neuromuscular junctions.86 Gouti’s group has successfully cultured these neuromuscular organoids for over two years and are now using them to model different neuromuscular diseases. For example, addition of autoantibodies derived from patients with myasthenia gravis, an autoimmune disease in which autoantibodies destroy acetylcholine receptors, reduced the number of neuromuscular junctions and affected contractions in the organoid model.86 Her group is also using these neuromuscular organoids to model ALS and SMA. Gouti showed preliminary data on the effect of mutations responsible for SMA on neuromuscular organoid function and structure. Ultimately, Gouti plans to use these organoids as a platform for drug screening for these diseases.
Figure 3.
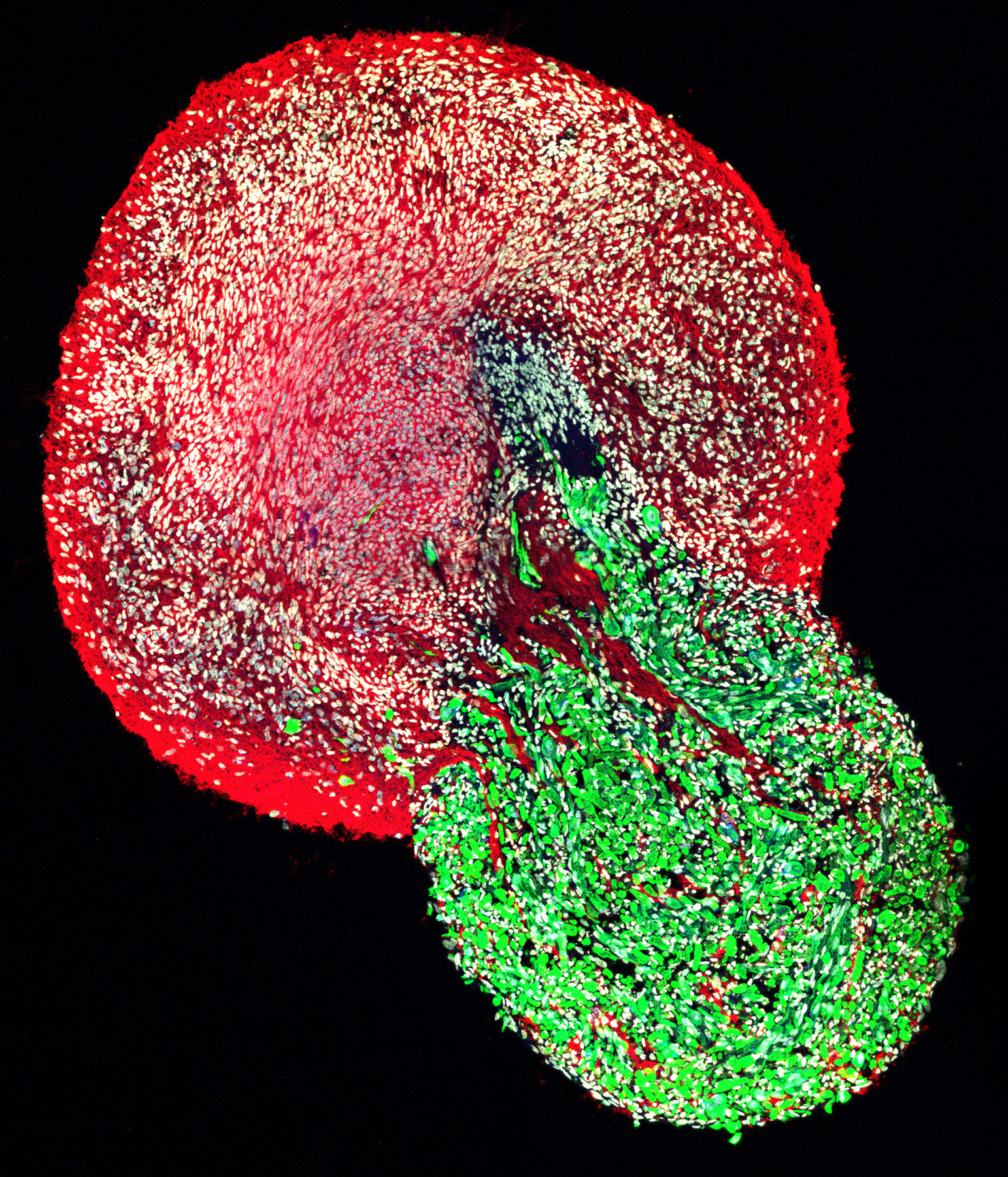
Neuromesodermal progenitors generate both spinal cord and muscle tissues that self-organize into two distinct regions within the organoid.
Tumor-immune cell organoids
Pleun Hombrink from Hubrecht Organoid Technology (HUB) presented the company’s approach to developing personalized organoid co-cultures for use in drug development for immunotherapies. Immunotherapy has revolutionized the field of oncology by leveraging the body’s T cells to kill cancer cells. Several modalities are either currently in use or under investigation, including monoclonal antibodies that regulate T cell activity, cell-based therapies, cancer vaccines, oncolytic viruses, and bispecific antibodies. HUB has created several organoid biobanks to serve as preclinical models for drug development.
During his talk, Hombrink focused on patient-derived colorectal cancer (CRC) organoids. The platform consists of colon organoids from patient tumors and healthy colon tissue and patient-derived tumor infiltrating lymphocytes (TILs). Hombrink showed that they could isolate and expand TILs in a scalable manner and enrich tumor-reactive T cells. They plan to use this organoid–TILs co-culture model to investigate personalized tumor-specific immune responses and as a screening platform for immunotherapies.
Increasing connectivity in brain organoids
Several speakers throughout the meeting showed how brain organoids can recapitulate much of the cell diversity of brain tissue, but several macroscopic features are still lacking. During development, the regions of the brain develop simultaneously forming macrocircuit connectivities that link different brain regions. In organoid models, however, different brain regions develop consecutively. Organoids thus lack macrocircuit connectivity and activity-dependent maturation.
Giorgia Quadrato from the University of Southern California presented work on modeling SYNGAP1 related disorders with human cortical organoids. SYNGAP1 is a top autism spectrum disorder (ASD) risk-gene and one of the most abundant proteins found at the postsynaptic density of excitatory synapses. Cortical organoids haploinsufficient for Syngap1 revealed novel information about the expression pattern and functionality of this gene initially thought to be expressed only in neurons underscoring the importance of dissecting the role of genes associated with ASD in distinct cell types and across developmental stages. Quadrato also described the establishment of a method for generating functional 3D human cerebellar organoids that can reproducibly generate the cellular diversity of the human cerebellum within and across multiple cell lines.
Acknowledgements
The work in Jianping Fu’s lab is supported by the Michigan-Cambridge Collaboration Initiative, the University of Michigan Mcubed Fund, the 21st Century Jobs Trust Fund received through the Michigan Strategic Fund from the State of Michigan (Grant CASE-315037), the National Science Foundation (I-Corps 2112458, CBET 1901718, and CMMI 1917304), and the National Institutes of Health (R21 NS113518 and R21 HD100931).
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Sato T, Vries RG, Snippert HJ, et al. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265. [DOI] [PubMed] [Google Scholar]
- 2.Gjorevski N, Nikolaev M, Brown TE, et al. 2022. Tissue geometry drives deterministic organoid patterning. Science 375: eaaw9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikolaev M, Mitrofanova O, Broguiere N, et al. 2020. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 585: 574–578. [DOI] [PubMed] [Google Scholar]
- 4.Shao Y, Taniguchi K, Gurdziel K, et al. 2017. Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat. Mater 16: 419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao Y, Taniguchi K, Townshend RF, et al. 2017. A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat. Commun 8: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Y, Xue X, Shao Y, et al. 2019. Controlled modelling of human epiblast and amnion development using stem cells. Nature 573: 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park SE, Georgescu A & Huh D 2019. Organoids-on-a-chip. Science 364: 960–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mondrinos MJ, Alisafaei F, Yi AY, et al. 2021. Surface-directed engineering of tissue anisotropy in microphysiological models of musculoskeletal tissue. Sci. Adv 7: eabe9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo J, Byun WY, Alisafaei F, et al. 2019. Multiscale reverse engineering of the human ocular surface. Nat. Med 25: 1310–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park JY, Mani S, Clair G, et al. 2022. A microphysiological model of human trophoblast invasion during implantation. Nat. Commun 13: 1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HJ, Huh D, Hamilton G, et al. 2012. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab. Chip 12: 2165–2174. [DOI] [PubMed] [Google Scholar]
- 12.Kasendra M, Luc R, Yin J, et al. 2020. Duodenum Intestine-Chip for preclinical drug assessment in a human relevant model. eLife 9: e50135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Workman MJ, Gleeson JP, Troisi EJ, et al. 2018. Enhanced Utilization of Induced Pluripotent Stem Cell-Derived Human Intestinal Organoids Using Microengineered Chips. Cell. Mol. Gastroenterol. Hepatol 5: 669–677.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manatakis DV, VanDevender A & Manolakos ES 2021. An information-theoretic approach for measuring the distance of organ tissue samples using their transcriptomic signatures. Bioinforma. Oxf. Engl 36: 5194–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apostolou A, Panchakshari RA, Banerjee A, et al. 2021. A Novel Microphysiological Colon Platform to Decipher Mechanisms Driving Human Intestinal Permeability. Cell. Mol. Gastroenterol. Hepatol 12: 1719–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarry A, Malard F, Bou-Hanna C, et al. 2017. Interferon-Alpha Promotes Th1 Response and Epithelial Apoptosis via Inflammasome Activation in Human Intestinal Mucosa. Cell. Mol. Gastroenterol. Hepatol 3: 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parks OB, Pociask DA, Hodzic Z, et al. 2015. Interleukin-22 Signaling in the Regulation of Intestinal Health and Disease. Front. Cell Dev. Biol 3: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muñoz M, Eidenschenk C, Ota N, et al. 2015. Interleukin-22 induces interleukin-18 expression from epithelial cells during intestinal infection. Immunity 42: 321–331. [DOI] [PubMed] [Google Scholar]
- 19.Sabat R, Ouyang W & Wolk K 2014. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat. Rev. Drug Discov 13: 21–38. [DOI] [PubMed] [Google Scholar]
- 20.Yang L, Han Y, Nilsson-Payant BE, et al. 2020. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 27: 125–136.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han Y, Duan X, Yang L, et al. 2021. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589: 270–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan X, Tang X, Nair MS, et al. 2021. An airway organoid-based screen identifies a role for the HIF1α-glycolysis axis in SARS-CoV-2 infection. Cell Rep 37: 109920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Y, Zhu J, Yang L, et al. 2022. SARS-CoV-2 Infection Induces Ferroptosis of Sinoatrial Node Pacemaker Cells. Circ. Res 130: 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, Nilsson-Payant BE, Han Y, et al. 2021. Cardiomyocytes recruit monocytes upon SARS-CoV-2 infection by secreting CCL2. Stem Cell Rep 16: 2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang L, Han Y, Jaffré F, et al. 2021. An Immuno-Cardiac Model for Macrophage-Mediated Inflammation in COVID-19 Hearts. Circ. Res 129: 33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morizane R, Lam AQ, Freedman BS, et al. 2015. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol 33: 1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freedman BS, Brooks CR, Lam AQ, et al. 2015. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun 6: 8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Czerniecki SM, Cruz NM, Harder JL, et al. 2018. High-Throughput Screening Enhances Kidney Organoid Differentiation from Human Pluripotent Stem Cells and Enables Automated Multidimensional Phenotyping. Cell Stem Cell 22: 929–940.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Przepiorski A, Sander V, Tran T, et al. 2018. A Simple Bioreactor-Based Method to Generate Kidney Organoids from Pluripotent Stem Cells. Stem Cell Rep 11: 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar SV, Er PX, Lawlor KT, et al. 2019. Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells. Dev. Camb. Engl 146: dev172361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takasato M, Er PX, Chiu HS, et al. 2015. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526: 564–568. [DOI] [PubMed] [Google Scholar]
- 32.Miller AJ, Yu Q, Czerwinski M, et al. 2020. In Vitro and In Vivo Development of the Human Airway at Single-Cell Resolution. Dev. Cell 53: 117–128.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Q, Kilik U, Holloway EM, et al. 2021. Charting human development using a multi-endodermal organ atlas and organoid models. Cell 184: 3281–3298.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayr U, Serra D & Liberali P 2019. Exploring single cells in space and time during tissue development, homeostasis and regeneration. Dev. Camb. Engl 146: dev176727. [DOI] [PubMed] [Google Scholar]
- 35.Xavier da Silveira Dos Santos A & Liberali P 2019. From single cells to tissue self-organization. FEBS J 286: 1495–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serra D, Mayr U, Boni A, et al. 2019. Self-organization and symmetry breaking in intestinal organoid development. Nature 569: 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benito-Kwiecinski S, Giandomenico SL, Sutcliffe M, et al. 2021. An early cell shape transition drives evolutionary expansion of the human forebrain. Cell 184: 2084–2102.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelava I, Chiaradia I, Pellegrini L, et al. 2022. Androgens increase excitatory neurogenic potential in human brain organoids. Nature 602: 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaspard N, Bouschet T, Hourez R, et al. 2008. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature 455: 351–357. [DOI] [PubMed] [Google Scholar]
- 40.van den Ameele J, Tiberi L, Vanderhaeghen P, et al. 2014. Thinking out of the dish: what to learn about cortical development using pluripotent stem cells. Trends Neurosci 37: 334–342. [DOI] [PubMed] [Google Scholar]
- 41.Luo C, Lancaster MA, Castanon R, et al. 2016. Cerebral Organoids Recapitulate Epigenomic Signatures of the Human Fetal Brain. Cell Rep 17: 3369–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhaduri A, Andrews MG, Mancia Leon W, et al. 2020. Cell stress in cortical organoids impairs molecular subtype specification. Nature 578: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka Y, Cakir B, Xiang Y, et al. 2020. Synthetic Analyses of Single-Cell Transcriptomes from Multiple Brain Organoids and Fetal Brain. Cell Rep 30: 1682–1689.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossi G, Broguiere N, Miyamoto M, et al. 2021. Capturing Cardiogenesis in Gastruloids. Cell Stem Cell 28: 230–240.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drakhlis L, Biswanath S, Farr C-M, et al. 2021. Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol 39: 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silva AC, Matthys OB, Joy DA, et al. 2021. Co-emergence of cardiac and gut tissues promotes cardiomyocyte maturation within human iPSC-derived organoids. Cell Stem Cell 28: 2137–2152.e6. [DOI] [PubMed] [Google Scholar]
- 47.Hofbauer P, Jahnel SM, Papai N, et al. 2021. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 184: 3299–3317.e22. [DOI] [PubMed] [Google Scholar]
- 48.Quijada P, Trembley MA & Small EM 2020. The Role of the Epicardium During Heart Development and Repair. Circ. Res 126: 377–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geddes GC, Dimmock DP, Hehir DA, et al. 2015. A novel FOXF1 mutation associated with alveolar capillary dysplasia and coexisting colobomas and hemihyperplasia. J. Perinatol. Off. J. Calif. Perinat. Assoc 35: 155–157. [DOI] [PubMed] [Google Scholar]
- 50.Sen P, Yang Y, Navarro C, et al. 2013. Novel FOXF1 mutations in sporadic and familial cases of alveolar capillary dysplasia with misaligned pulmonary veins imply a role for its DNA binding domain. Hum. Mutat 34: 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Múnera JO, Sundaram N, Rankin SA, et al. 2017. Differentiation of Human Pluripotent Stem Cells into Colonic Organoids via Transient Activation of BMP Signaling. Cell Stem Cell 21: 51–64.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eicher AK, Kechele DO, Sundaram N, et al. 2022. Functional human gastrointestinal organoids can be engineered from three primary germ layers derived separately from pluripotent stem cells. Cell Stem Cell 29: 36–51.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCracken KW, Catá EM, Crawford CM, et al. 2014. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516: 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Workman MJ, Mahe MM, Trisno S, et al. 2017. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med 23: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han L, Chaturvedi P, Kishimoto K, et al. 2020. Single cell transcriptomics identifies a signaling network coordinating endoderm and mesoderm diversification during foregut organogenesis. Nat. Commun 11: 4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spence JR, Mayhew CN, Rankin SA, et al. 2011. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470: 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watson CL, Mahe MM, Múnera J, et al. 2014. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med 20: 1310–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Capeling MM, Czerwinski M, Huang S, et al. 2019. Nonadhesive Alginate Hydrogels Support Growth of Pluripotent Stem Cell-Derived Intestinal Organoids. Stem Cell Rep 12: 381–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cruz-Acuña R, Quirós M, Farkas AE, et al. 2017. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol 19: 1326–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Capeling MM, Huang S, Childs CJ, et al. 2022. Suspension culture promotes serosal mesothelial development in human intestinal organoids. Cell Rep 38: 110379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uzquiano A, Kedaigle AJ, Pigoni M, et al. 2022. Single-cell multiomics atlas of organoid development uncovers longitudinal molecular programs of cellular diversification of the human cerebral cortex. 2022.03.17.484798 [Google Scholar]
- 62.Polioudakis D, de la Torre-Ubieta L, Langerman J, et al. 2019. A Single-Cell Transcriptomic Atlas of Human Neocortical Development during Mid-gestation. Neuron 103: 785–801.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trevino AE, Müller F, Andersen J, et al. 2021. Chromatin and gene-regulatory dynamics of the developing human cerebral cortex at single-cell resolution. Cell 184: 5053–5069.e23. [DOI] [PubMed] [Google Scholar]
- 64.Di Bella DJ, Habibi E, Stickels RR, et al. 2021. Molecular logic of cellular diversification in the mouse cerebral cortex. Nature 595: 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bonner MG, Gudapati H, Mou X, et al. 2022. Microfluidic systems for modeling human development. Dev. Camb. Engl 149: dev199463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mou X, Shah J, Bhattacharya R, et al. 2022. A Biomimetic Electrospun Membrane Supports the Differentiation and Maturation of Kidney Epithelium from Human Stem Cells. Bioeng. Basel Switz 9: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Musah S, Morin SA, Wrighton PJ, et al. 2012. Glycosaminoglycan-binding hydrogels enable mechanical control of human pluripotent stem cell self-renewal. ACS Nano 6: 10168–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derda R, Musah S, Orner BP, et al. 2010. High-throughput discovery of synthetic surfaces that support proliferation of pluripotent cells. J. Am. Chem. Soc 132: 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Musah S, Wrighton PJ, Zaltsman Y, et al. 2014. Substratum-induced differentiation of human pluripotent stem cells reveals the coactivator YAP is a potent regulator of neuronal specification. Proc. Natl. Acad. Sci. U. S. A 111: 13805–13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roye Y, Bhattacharya R, Mou X, et al. 2021. A Personalized Glomerulus Chip Engineered from Stem Cell-Derived Epithelium and Vascular Endothelium. Micromachines 12: 967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Musah S, Mammoto A, Ferrante TC, et al. 2017. Mature induced-pluripotent-stem-cell-derived human podocytes reconstitute kidney glomerular-capillary-wall function on a chip. Nat. Biomed. Eng 1: 0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Musah S, Dimitrakakis N, Camacho DM, et al. 2018. Directed differentiation of human induced pluripotent stem cells into mature kidney podocytes and establishment of a Glomerulus Chip. Nat. Protoc 13: 1662–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burt M, Bhattachaya R, Okafor AE, et al. 2020. Guided Differentiation of Mature Kidney Podocytes from Human Induced Pluripotent Stem Cells Under Chemically Defined Conditions. J. Vis. Exp. JoVE [DOI] [PubMed] [Google Scholar]
- 74.Kalejaiye TD, Bhattacharya R, Burt MA, et al. 2021. BSG/CD147 and ACE2 receptors facilitate SARS-CoV-2 infection of human iPS cell-derived kidney podocytes. BioRxiv Prepr. Serv. Biol. 2021.11.16.468893 [Google Scholar]
- 75.Burt MA, Kalejaiye TD, Bhattacharya R, et al. 2021. Adriamycin-Induced Podocyte Injury Disrupts the YAP-TEAD1 Axis and Downregulates Cyr61 and CTGF Expression. ACS Chem. Biol [DOI] [PubMed] [Google Scholar]
- 76.Barker N, Tan S & Clevers H 2013. Lgr proteins in epithelial stem cell biology. Dev. Camb. Engl 140: 2484–2494. [DOI] [PubMed] [Google Scholar]
- 77.Huch M, Dorrell C, Boj SF, et al. 2013. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature 494: 247–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huch M, Bonfanti P, Boj SF, et al. 2013. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J 32: 2708–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aloia L, McKie MA, Vernaz G, et al. 2019. Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat. Cell Biol 21: 1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Broutier L, Mastrogiovanni G, Verstegen MM, et al. 2017. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med 23: 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cordero-Espinoza L, Dowbaj AM, Kohler TN, et al. 2021. Dynamic cell contacts between periportal mesenchyme and ductal epithelium act as a rheostat for liver cell proliferation. Cell Stem Cell 28: 1907–1921.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gouti M, Tsakiridis A, Wymeersch FJ, et al. 2014. In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol 12: e1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gouti M, Delile J, Stamataki D, et al. 2017. A Gene Regulatory Network Balances Neural and Mesoderm Specification during Vertebrate Trunk Development. Dev. Cell 41: 243–261.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Metzis V, Steinhauser S, Pakanavicius E, et al. 2018. Nervous System Regionalization Entails Axial Allocation before Neural Differentiation. Cell 175: 1105–1118.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martins JMF, Nguyen LVN & Gouti M 2021. Humane neuromuskuläre Organoide — Anwendung und Perspektive. BIOspektrum 27: 135–138. [Google Scholar]
- 86.Faustino Martins J-M, Fischer C, Urzi A, et al. 2020. Self-Organizing 3D Human Trunk Neuromuscular Organoids. Cell Stem Cell 26: 172–186.e6. [DOI] [PubMed] [Google Scholar]


