Abstract
The final stage of poliovirus assembly is characterized by a cleavage of the capsid precursor protein VP0 into VP2 and VP4. This cleavage is thought to be autocatalytic and dependent on RNA encapsidation. Analysis of the poliovirus empty capsid structure has led to a mechanistic model for VP0 cleavage involving a conserved histidine residue that is present in the surrounding environment of the VP0 cleavage site. Histidine 195 of VP2 (2195H) is hypothesized to activate local water molecules, thus initiating a nucleophilic attack at the scissile bond. To test this hypothesis, 2195H mutants were constructed and their phenotypes were characterized. Consistent with the requirement of VP0 cleavage for poliovirus infectivity, all 2195H mutants were nonviable upon introduction of the mutant genomes into HeLa cells. Replacement of 2195H with threonine or arginine resulted in the assembly of a highly unstable 150S virus particle. Further analyses showed that these particles contain genomic RNA and uncleaved VP0, criteria associated with the provirion assembly intermediate. These data support the involvement of 2195H in mediating VP0 cleavage during the final stages of virus assembly.
Poliovirus is a plus-stranded RNA virus belonging to the picornavirus family. The viral genome is protected by an icosahedrally symmetric capsid that is made of 60 copies of capsid proteins VP1 and VP3 and an average of 58 to 59 copies of capsid proteins VP2 and VP4, plus 1 to 2 copies of their capsid precursor protein, VP0. Morphogenesis of the poliovirus virion is initiated by proteolytic processing of the myristoylated capsid precursor protein, P1, to form the cleaved protomer. This cleaved protomer is a heterotrimer composed of VP0-VP3-VP1. The self-assembly of five protomers leads to the formation of 14S pentamers, an obligate intermediate in the virion assembly pathway (17). Encapsidation of the genomic RNA by 12 pentamers forms the provirion with the final cleavage of VP0, to yield VP2 and VP4, occurring during the formation and maturation of the provirion. VP0 cleavage, also called maturation cleavage, is hypothesized to be an autocatalytic cleavage that is dependent on viral RNA encapsidation (3, 17, 32) and is crucial for virion stability and infectivity (2, 4, 10).
The residues forming the VP0 cleavage site (VP4 Asn 68 and VP2 Ser 1) are in a significantly different location in the poliovirus empty capsid than they are after VP0 cleavage in the mature native virus structure (4, 17). Because VP0 is uncleaved in the empty capsid, the local environment surrounding the cleavage site in the empty capsid structure is thought to be most similar to that found in the provirion just prior to cleavage and suggests a potential cleavage mechanism (4). Two conserved amino acids (histidine 195 and proline 194 of VP2 [2195H and 2194P, respectively]) as well as two bound water molecules are found in the immediate vicinity of the VP0 scissile bond. A tight bend in the main chain, formed by the presence of 2194P, places the imidazole ring of 2195H in the appropriate spatial configuration to activate the local water molecules. Activation of these local water molecules by 2195H could lead to a nucleophilic attack of the scissile bond by the water molecules. Polarization of the scissile bond due to the coordination of the carbonyl oxygens with RNA bases or with an RNA-metal ion complex would increase the efficiency of the hydrolysis reaction, thus accounting for the cleavage dependence on RNA encapsidation.
Several mutants were constructed at 2195H by site-directed mutagenesis to test this mechanism. Consistent with the predictions of this model, we report here that these 2195H substitutions are lethal. Furthermore, several of the 2195H mutants assemble empty capsids and encapsidate viral RNA to form 150S provirion intermediates. These 150S particles fail to cleave VP0 and are highly unstable.
MATERIALS AND METHODS
Cells and media.
HeLa-S3 cells were maintained in suspension (3 × 105 to 4 × 105 cells/ml) in Joklik’s modified minimal essential medium supplemented with 7.5% horse serum and 1 mM sodium pyruvate. Prior to cDNA transfection, HeLa cells in suspension were grown at 37°C in the presence of Joklik’s modified minimal essential medium supplemented with 5% fetal calf serum and 1 mM sodium pyruvate for 24 h. HeLa and CV-1 cell monolayers were maintained in Dulbecco’s modified Eagle’s medium supplemented with 5% fetal calf serum, 50 U of penicillin per ml, 50 μg of streptomycin per ml, and 1 mM sodium pyruvate (DMEM–5% FCS) in a 5% CO2 incubator.
Bacterial strains, plasmids, and virus mutant nomenclature.
Escherichia coli CJ236 (dut negative ung negative) was used to generate phage stocks containing uridylated genomes. E. coli XL1-Blue was used to amplify all plasmids and phage stocks. Plasmid cDNAs were purified on CsCl gradients (5). Plasmid PVM contains the wild-type poliovirus infectious cDNA (serotype 1, Mahoney strain) that was originally obtained from P. Sarnow (33).
Mutant construction.
The wild-type poliovirus sequence encoding nucleotides 70 to 2474 was subcloned into M13mp19. Synthetic oligonucleotides corresponding to the region of the poliovirus genome between nucleotides 1519 and 1543 were generated at the Harvard Medical School biopolymer facility with a random mixture of nucleotides at positions 1532 to 1534. These oligonucleotides were used to generate mutations at the desired site (19, 28). Amino acid substitutions were identified by sequencing the recovered phage genomes with an fMol kit (Promega) or a Perkin-Elmer automated sequencer (ABI model 377). Full-length poliovirus genomic cDNAs for each mutant were reconstructed from the mutant subclone.
cDNA and RNA transfections.
Mutant RNA transcripts were synthesized in vitro from the linearized cDNA plasmid by using T7 RNA polymerase. HeLa or CV-1 cell monolayers (70 to 80% confluent) in 60-mm-diameter plates were transfected with the poliovirus mutant cDNA (0.5 μg) or RNA transcripts by using DEAE-dextran (35) or DOTAP (Boehringer Mannheim), respectively. After incubation at 32, 37, or 39°C for 8 h, the cells were overlaid with DMEM-5% FCS containing 0.8% agarose. The monolayers were then incubated at the above-mentioned temperatures for 5 to 6 days. Plaques were visualized by staining with 1.0% crystal violet.
Coupled vaccinia infection and poliovirus transfections.
HeLa cells (5 × 106/ml) were infected with recombinant vaccinia virus (VVT7-3; multiplicity of infection, 6), which expresses the T7 polymerase gene (12). At 1.5 h postinfection, poliovirus mutant cDNAs (20 μg/ml) with carrier DNA (60 μg/ml) were electroporated into the VVT7-3-infected HeLa cells by using 2-mm gap cuvettes and the BTX Electro cell manipulator 600 with settings at 150 V, 13 Ω, and 850 μF. Dactinomycin (5 μg/ml) was added after 2 h postelectroporation. [35S]methionine (final concentration, 20 μCi/ml; NEN Dupont; specific activity, 1,175 Ci/mMol) was added at 3 h postelectroporation, and the transfected cells were labeled continuously for 4 h (34). Cells were lysed with 1 ml of buffer containing 10 mM Tris-HCl [pH 7.2]–10 mM NaCl–1.5 mM MgCl2–0.1% Nonidet P-40 (TNM–0.1% NP-40), passed twice through a 25-gauge needle, and frozen and thawed twice. The nuclei and cell debris were removed by centrifugation for 3 min at 13,000 × g in an Eppendorf centrifuge at 4°C. When transfections were grown in the presence of Win51711 or 5-(3,4-dichlorophenyl) methylhydantoin (referred to as hydantoin), the drug was added immediately postelectroporation at final concentrations of 1 and 50 μg/ml, respectively.
Sucrose gradient analyses.
Radiolabeled cell lysates (500 μl) were analyzed immediately after lysis on 15 to 30% or 6 to 25% (wt/vol) linear sucrose gradients in TNM–0.1% NP-40. The gradients were centrifuged at 4°C in a Beckman SW40.1 rotor for 2.5 h at 39,000 rpm (15 to 30% gradients) or 16.5 h at 40,000 rpm (6 to 25% gradients). When viral lysates contained Win51711, Win51711 (10 μg/ml) was present throughout the gradients. Gradients were fractionated from the top (300 μl/fraction), and the radioactivity in each fraction (50 μl) was determined by liquid scintillation counting. Fractions at the desired sedimentation values were analyzed by electrophoresis on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels (20).
In vitro assembly assays: dissociation of empty capsids in vitro into pentamers.
Sucrose-purified radiolabeled empty capsids were added to TNM buffer at either pH 7.4 or 8.3 and incubated for 20 min at 4°C. Samples were loaded onto 6 to 25% sucrose gradients (in TNM buffer at the appropriate pH), sedimented at 40,000 rpm for 3 h in a Beckman SW40.1 rotor at 4°C, and fractionated.
In vitro assembly of pentamers to empty capsids.
Sucrose fractions containing radiolabeled pentamers were incubated at either 37 or 4°C for 1 h. Samples were layered onto 15 to 30% sucrose gradients (in TNM, pH 7.2), and the gradients were centrifuged in a Beckman SW40.1 rotor at 40,000 rpm for 4.5 h at 4°C and fractionated.
Immunoprecipitation of provirions with monoclonal antibodies.
Monoclonal antibodies specific for poliovirus antigenic sites (3A, 3B, and 2) were grown from hybridoma cell lines, and supernatants (250 μl) were added to sucrose-purified radiolabeled provirions (5,000 cpm) in RSB buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 1.5 MgCl2)–1% NP-40 and incubated for 2 h at 0°C. StaphA beads (100 μl; Sigma) were added and incubated at 4°C for 2 h. The precipitates were washed with RIPA buffer (1 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% deoxycholate, 1% Triton X-100, 1% SDS) three times and analyzed by SDS–10% polyacrylamide gel electrophoresis (PAGE). Viral proteins were visualized by autoradiography.
RT-PCR analyses.
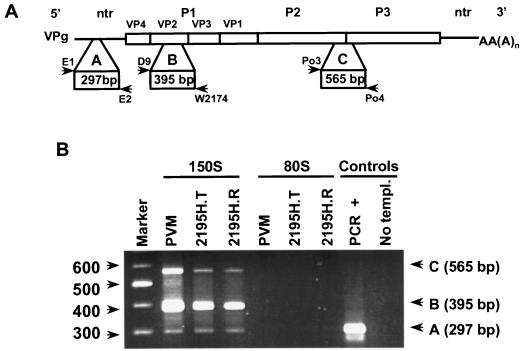
RNA was extracted from sucrose gradient fractions by using TRIzol (Gibco-BRL) according to the manufacturer’s recommendations. Briefly, the sucrose fractions (250 μl) were sequentially incubated with TRIzol (750 μl) and chloroform (200 μl) at room temperature, and the aqueous phases were recovered. The RNA was precipitated at 4°C with isopropanol (700 μl) in the presence of glycogen (5 μg/ml). The RNA pellets were resuspended in diethylpyrocarbonate-treated water (5 μl), incubated at 55°C for 10 min, and then stored at −20°C. Reverse transcription-PCR (RT-PCR) was performed with previously described primers (9). Primers E1 (5′-AAGCACTTCCTGTTTCC-3′) and E2 (5′-CATTCAGGGGCCGGAGGA-3′) were used to amplify a 297-bp fragment from the 5′ nontranslated region, and primers Po3 (5′-GAAATGTGTAAGAACTGTCA-3′) and Po4 (5′-GTAACAATGTTTCTTTTAGCC-3′) were used to amplify a 565-bp product from the P2-P3 region of the viral genome. The primers used to amplify a 395-bp fragment from the VP2 region of the genome were D9 (5′-GGCACACGCGACGGA-3′) and 2074 (5′-TCACCTGCCCGCAGGTTCTGCCCG-3′).
RESULTS
2195H is conserved in all picornaviruses sequenced to date. To assess the role of this amino acid residue in VP0 cleavage, it was replaced by several amino acids with different side chain chemistries: threonine (2195H.T), arginine (2195H.R), glycine (2195H.G), and aspartic acid (2195H.D). The viabilities of these mutants were assessed in different cell lines and at different temperatures by transfection of the cDNAs or in vitro-synthesized RNA transcripts. No plaques were observed for any mutant under any condition. Thus, all 2195H substitutions were lethal. The nonviability of these mutants is consistent with the conserved nature of the histidine residue and with previous studies which indicated that VP0 cleavage was necessary for picornaviruses’ viability (6, 8, 18, 21).
Characterization of 2195H mutants.
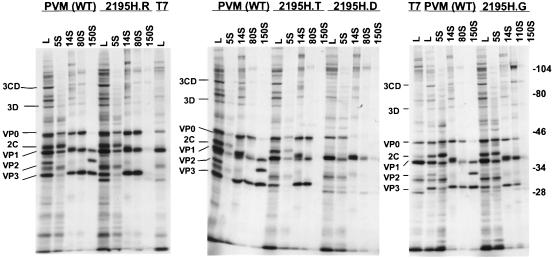
Capsid assembly and replication of nonviable poliovirus mutants can be studied by using a coupled vaccinia virus-T7 infection and poliovirus cDNA transfection system (12, 34). With this system, levels of viral protein expression in wild-type and mutant-transfected cell lysates were compared (Fig. 1, lanes L). All viral proteins, except for capsid protein VP2, were observed with normal stoichiometries in 2195H.G-, 2195H.T-, and 2195H.R-transfected cells. Thus, overall viral protein synthesis and processing appeared unaffected in these mutants. However, capsid protein VP2 was absent in these 2195H mutants and suggested that VP0 cleavage was defective in these mutants.
FIG. 1.
Protein composition of the assembly intermediates for the 2195H mutants. Lysates (lanes L) from vaccinia virus-infected cDNA-transfected cells and sucrose gradient fractions corresponding to the different assembly intermediates (5S protomers, 14S pentamers, 80S empty capsid, 110S encapsidation intermediate, and 150S virion) were analyzed by SDS–10% PAGE for the wild-type poliovirus and 2195H mutants. T7, vaccinia virus-T7-infected cell lysates; PVM (WT), vaccinia virus-infected cells transfected with the wild-type poliovirus cDNA plasmid. The migration of the poliovirus proteins and the prestained molecular mass markers (in kilodaltons; Bio-Rad) are indicated.
The assembly intermediates were analyzed for their sedimentation behavior and protein composition to determine if the 2195H mutants accumulated provirions with uncleaved VP0. Several different assembly phenotypes were observed (Fig. 2). No assembly intermediates (pentamer intermediates, empty capsids, or mature virion particles) were detected in lysates from 2195H.D-transfected cells (Fig. 2A and B). SDS-PAGE analyses indicated that expression of the viral nonstructural proteins, as well as the structural proteins, was significantly reduced in lysates from 2195H.D-transfected cells (Fig. 1). Other studies suggested that protein folding and processing were severely altered in this mutant (data not shown). This is consistent with previous studies which have shown that inappropriate folding of the P1 capsid precursor interferes with proteolytic processing (37). Thus, the 2195H.D mutant was not characterized further.
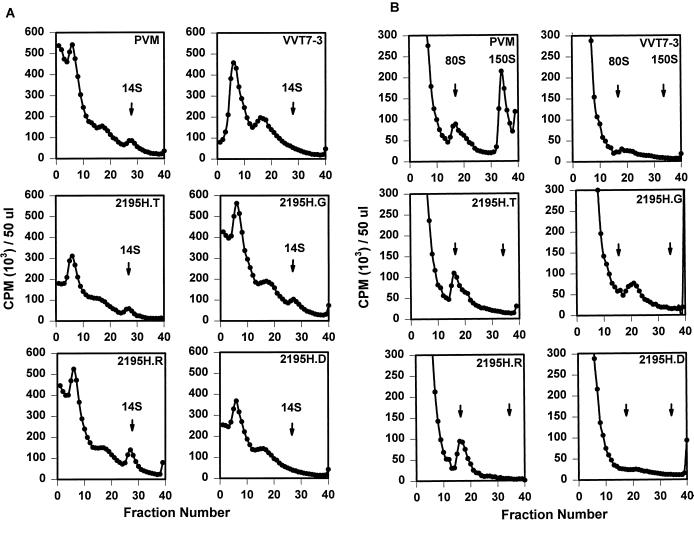
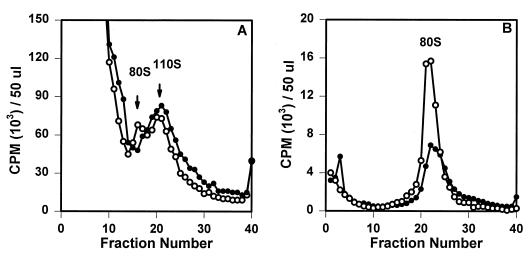
FIG. 2.
Sucrose gradient analyses of wild-type poliovirus (PVM) and 2195H mutant assembly intermediates. Equal volumes (500 μl) of [35S]methionine-labeled viral lysates were sedimented on either 6 to 25% (A) or 15 to 30% (B) linear sucrose gradients. Gradients were fractionated, and the radioactivity was counted.
Sucrose gradient analyses indicated that pentamer intermediates (14S) were present in the transfected lysates of the remaining 2195H mutants at levels similar to that in the wild-type poliovirus (Fig. 2A). However, the assembly of higher-order intermediates from the pentamers appeared different for the 2195H.G mutant. No empty capsid intermediates (80S) were observed in 2195H.G-transfected cell lysates; rather, an intermediate that broadly sedimented in the 110-to-120S region of the gradient was consistently observed (Fig. 2B). SDS-PAGE analysis of this complex showed that VP0, VP3, and VP1 were present in normal proportions, indicating that this 110S species was an assembly product (Fig. 1). The position of the 2195H.G 110-to-120S intermediate was identical with the recently identified 110-to-120S intermediate which reversibly accumulates in wild-type infected cells in the presence of a novel antiviral drug, 5-(3,4-dichlorophenyl) methylhydantoin (Fig. 3A). The properties of the wild-type 110S intermediate are consistent with that predicted for the poliovirus encapsidation intermediate; accumulation of the 110S intermediate in the presence of the drug indicated that the drug targeted the encapsidation process and that the mechanism of action for this drug was to inhibit the completion of the encapsidation process (36). Thus, accumulation of a 110S intermediate in 2195H.G lysates (in the absence of drug) suggested that viral encapsidation may be similarly inhibited in this mutant. Further analyses indicated that the 2195H.G 110S intermediate was highly unstable and, within hours after isolation, would decay to an 80S empty capsid intermediate, thus preventing further characterization (Fig. 3B).
FIG. 3.
(A) Equal volumes of [35S]methionine-labeled viral lysates from wild-type poliovirus grown in the presence of the drug hydantoin (open circle) or 2195H.G viral cell lysates grown in the absence of the drug (solid circle) were loaded onto 15 to 30% sucrose gradients and analyzed. Samples were sedimented at 39,000 rpm in a Beckman SW40.1 rotor for 3 h at 4°C, fractionated, and counted. (B) 2195H.G 110S assembly intermediates from gradient A (solid circle) were pooled immediately and rerun along with a wild-type 80S empty capsid marker (open circle) on 15 to 30% sucrose gradients as described above.
The remaining two mutants, 2195H.T and 2195H.R, assembled empty capsids (80S) in amounts similar to that in the wild-type poliovirus (Fig. 2B). The protein compositions of pentamer and empty capsid assembly intermediates for these mutants were indistinguishable from those of the wild-type virus (Fig. 1). However, although low levels of capsid proteins were observed in the 150S region in the gradient for each mutant by SDS-PAGE, a peak of 150S particles could not be detected (Fig. 2B). Because no obvious assembly defects could be identified for 2195H.T and 2195H.R, these mutants were characterized further.
2195H.T and 2195H.R assembly intermediates are assembly active in vitro.
The levels of wild-type pentamers and empty capsids appear to be in dynamic equilibrium within an infected cell; wild-type assembly-competent empty capsid intermediates will reversibly disassemble in vivo into pentamers. Previous studies have identified two species of purified empty capsids which differ in antigenic properties and their dissociability in vitro at pH 8.3 (29–31). Antigenically native empty capsids are dissociable to pentamers at pH 8.3, and the resultant dissociated pentamers reassemble into empty capsids at 37°C upon pH neutralization. This form of empty capsids and pentamers is considered to be analogous to the assembly-competent intermediates observed in vivo. Nonnative antigenic empty capsids are not dissociable into pentamers and appear to be antigenically analogous to the heat-inactivated virus particle. Similarly, the pentamers fail to assemble in vitro into empty capsids and are assembly incompetent. To determine whether the lack of 150S particles was due to the inability of 2195H.T and 2195H.R mutants to form assembly-competent pentamers and empty capsid intermediates, the in vitro assembly activities of these intermediates were examined (Fig. 4).
FIG. 4.
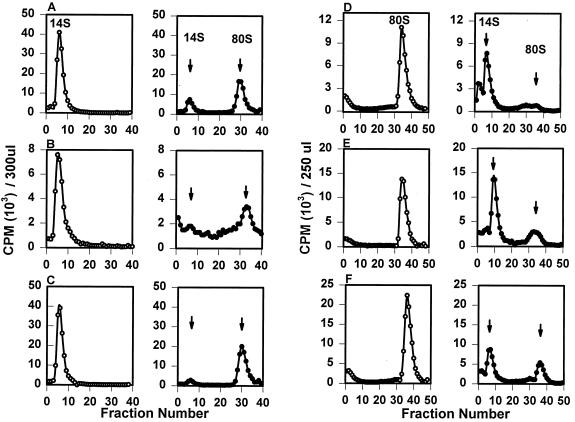
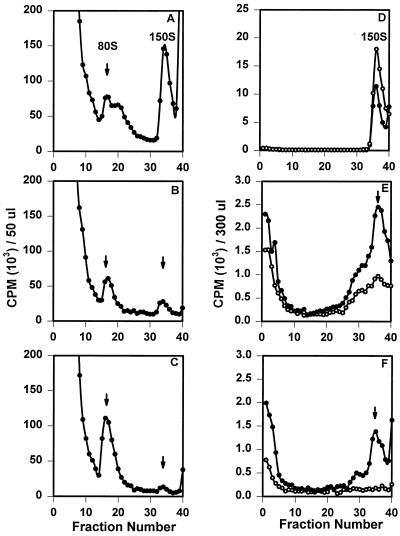
Sucrose-purified [35S]methionine-labeled wild-type poliovirus (A), 2195H.T (B), or 2195H.R (C) pentamers were pooled and incubated for 1 h at either 4°C (open circle) or 37°C (solid circle). Samples were then analyzed on 15 to 30% linear sucrose gradients. Radiolabeled sucrose-purified wild-type poliovirus (D), 2195H.T (E), or 2195H.R (F) empty capsids were incubated in TNM (pH 7.2) (open circle) or TNM (pH 8.7) (solid circle) for 20 min and then analyzed on 6 to 25% linear sucrose gradients.
Mutant 2195H.T and 2195H.R pentamers assembled into empty capsids upon incubation at 37°C (Fig. 4B and C). When the total amount of radioactivity present in the pentamer and empty capsid fractions was measured, approximately 75 and 87% of the labeled 2195H.T and 2195H.R pentamers, respectively, assembled into empty capsids and were comparable with that measured for wild-type pentamer intermediates (71%). Thus, both 2195H.T and 2195H.R pentamers were as assembly active in vitro as the wild-type poliovirus pentamers. Similarly, mutant 2195H.T and 2195H.R empty capsids dissociated into pentamers at pH 8.3 (Fig. 4E and F). Although the fractions of dissociable mutant 2195H.T (71%) and 2195H.R (61%) empty capsids were not as high as those observed with wild-type empty capsids (82%), this reduction in assembly-competent empty capsids was not sufficient to account for the absence of 150S particles in mutant transfected cells.
Win51711 stabilizes 2195H.T and 2195H.R 150S particles.
The instability of the 2195H.G 110S complex as well as the transient nature of the wild-type VP0-containing provirion structure suggested that if 2195H mutants encapsidated RNA to form 150S provirion particles, the mutant provirions might be highly unstable and thus escape detection. The capsid binding drug, Win51711, has been shown previously to stabilize in vivo and in vitro the structures of both pentamer and empty capsid intermediates as well as the mature infectious virion (11, 26, 38). Thus, mutants 2195H.T and 2195H.R were grown in the presence of Win51711 to see whether any 150S mutant particles could be detected in the presence of a capsid-stabilizing drug (Fig. 5B and C). A 150S sedimenting species was detected in both 2195H.T and 2195H.R mutant-transfected cells, albeit at much lower levels than that observed for the wild-type virus. Win51711-stabilized 2195H.G particles were also detected in the 150S region when fractions from the gradients were scanned on SDS–10% polyacrylamide gels, although in amounts approximately 5- to 10-fold less than those of the other mutants (data not shown). In all cases, the resultant 150S particles are highly unstable. Similar to the instability of the 110S 2195H.G complex, the assembled mutant particles were unstable upon storage at 4°C; in the absence of Win51711, these particles decayed within a week to particles with sedimentation values equivalent to those of empty capsids. Furthermore, if lysates from 2195H.T-transfected cells were made by using a standard lysis buffer containing higher detergent concentrations (TNM–1.0% NP-40), then mutant 150S particles were not detected, whereas wild-type 150S particles are stable in this lysis buffer (data not shown).
FIG. 5.
Cell lysates from wild-type poliovirus (PVM) (A), 2195H.T (B), and 2195H.R (C) cDNA-transfected cells grown in the presence of Win51711 (1 μg/ml) were loaded and analyzed on 15 to 30% sucrose gradients. Lysates were made from PVM (D)-, 2195H.T (E)-, and 2195H.R (F)-transfected cells grown in the absence of Win51711. The lysates were then incubated in the presence (solid circle) or absence (open circle) of Win51711 (10 μg/ml) overnight at 4°C. The lysates were loaded onto 15 to 30% sucrose gradients, and the 150S regions (fractions 33 to 35) were subsequently pooled and immediately rerun on another 15 to 30% sucrose gradient.
The ability to detect 150S particles when the mutant virus is grown in the presence of Win51711 may be because assembly of mutant 150S particles is drug dependent or because the drug increases the half-life of the particles by stabilizing the 150S particle after assembly. Thus, the mutant histidine viruses were grown in the absence of Win51711, and cell lysates were subsequently incubated overnight in the presence or absence of Win51711 to allow drug binding (Fig. 5D to F). Assembly of 2195H.T and 2195H.R 150S particles was clearly observed after incubation with the drug, and for mutant 2195H.T, the presence of 150S particles could be detected even in absence of drug if samples were rerun to remove background levels of unincorporated label. Thus, to varying extents for each mutant, complete assembly of a virion particle occurs in the presence of Win51711, and for 2195H.T, virion morphogenesis could be detected even in the absence of drug.
Characterization of 2195H mutant 150S particles.
The mutant 2195H.T and 2195H.R 150S provirions have external structural antigenic properties similar to those of the mature wild-type virus particle. The antigenic determinants (sites 3A, 3B, and 2) of 2195H.T- and 2195H.R-assembled 150S provirions were recognized by monoclonal antibodies directed against these sites (data not shown).
Polarization of the VP0 scissile bond by the encapsidated RNA is proposed to be important for mediating VP0 cleavage (4). To confirm that the 150S particles had indeed encapsidated viral RNA sequences, RNA was extracted from mutants and wild-type 150S particles, and the presence of regions across the poliovirus genome was analyzed by RT-PCR with multiplexed virus-specific primers (Fig. 6A) (9). Three fragments of the expected sizes were amplified from 2195H.T or 2195H.R 150S particles as well as wild-type particles (Fig. 6B). Sequence analyses of the VP2 region from the encapsidated RNA genomes showed that the histidine substitution was maintained.
FIG. 6.
(A) Schematic representation of the poliovirus genome and the primers used to amplify fragments across poliovirus mutant genomes. Three fragments were amplified. Fragment A was amplified from the 5′ terminus with primers E1 and E2, fragment B was amplified from the P1 region with primers D9 and W2074, and fragment C was amplified from the P2-P3 region with primers Po3 and Po4. (B) RT-PCR products were analyzed on a 2.0% agarose gel. RNA was isolated from 150S particles and the empty capsids (80S) of the wild-type poliovirus (PVM) and Win51711-stabilized 2195H.T- and 2195H.R-transfected cell lysates. RT-PCR positive (supplied by the manufacturer) and negative (No templ.) controls are shown. A 100-bp ladder, used as a molecular weight marker, is shown on the left.
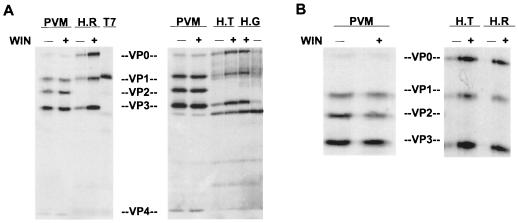
Effects of 2195H substitutions on VP0 cleavage could be examined in the assembled 2195H.R, 2195H.T, and 2195H.G 150S particles (Fig. 7). Irrespective of whether the assembled mutant particles were stabilized by incubation with Win51711 or growth in the presence of Win51711, none of the mutants cleaved VP0 into VP2 and VP4. In addition, the trace amounts of assembled mutant virions in cell lysates that were grown in the absence of drug (as suggested by the presence on SDS-polyacrylamide gels of a peak of viral proteins in the 150S region of the gradient) also did not show any evidence of VP0 cleavage. Thus, substitutions at 2195H prevent VP0 cleavage.
FIG. 7.
Absence of VP0 cleavage in 2195H mutant 150S particles. (A) Autoradiogram of SDS–10% polyacrylamide gel of the wild-type poliovirus (PVM) 150S mature virions and 2195H.T (H.T), 2195H.G (H.G), and 2195H.R (H.R) 150S particles grown in the absence (−) or presence (+) of Win51711. The samples were matched for counts per minute. The 150S region from sucrose gradients of vaccinia virus–T7-infected cell lysates is also shown. (B) Autoradiogram of SDS–10% polyacrylamide gel of PVM, 2195H.T, and 2195H.R 150S particles stabilized by the addition of Win51711 to cell lysates grown in the absence of Win51711.
DISCUSSION
Based on the poliovirus and rhinovirus mature virion structures, it was previously proposed that serine 10 of VP2 (which is found in the immediate vicinity of the carboxyl terminus of VP4) might be responsible for mediating VP0 cleavage (3, 17, 32). This autocatalytic cleavage was proposed to occur via a serine-protease type mechanism, where an RNA base could abstract a proton from the serine, thus activating the serine residue to catalyze VP0 cleavage. However, replacement of serine 10 of VP2 with either alanine or cysteine did not inhibit VP0 cleavage in poliovirus and provided concrete evidence that VP2 serine 10 could not be involved in the VP0 cleavage mechanism (14). In addition, comparison of the poliovirus empty capsid (containing uncleaved VP0) and mature native virion (containing cleaved VP0) structures revealed that the residues forming the uncleaved VP0 scissile bond in the empty capsid structure are approximately 20 Å away from their final location within the mature infectious virus (4).
Analyses of the environment surrounding the cleavage site in the empty capsid showed that two highly conserved amino acids, 2195H and 2194P, were in the immediate vicinity of the VP0 cleavage site. In this study, we tested a model proposed by Basavappa et al. which involved the participation of these highly conserved amino acids in VP0 cleavage (4). The 2194P was proposed to lock the main chain in a tight bend, thus placing 2195H in position to activate bound local water molecules. The activated water molecules initiate a nucleophilic attack at the VP0 cleavage site, thus mediating VP0 cleavage into VP2 and VP4.
All substitutions introduced at 2195H were lethal. This is consistent with earlier reports which showed that VP0 cleavage was required for viral infectivity (6, 10, 18, 21). The phenotypes of the block resulting in the lethality were all in the capsid assembly pathway but varied depending on the nature of the amino acid substitution. An aspartic acid substitution led to misfolding of the P1 capsid precursor. The 2195H.G mutant accumulates an assembly intermediate with sedimentation properties identical to that of a recently reported 110S packaging intermediate for poliovirus, suggesting that assembly is blocked at the packaging stage (36). This packaging block could be overcome inefficiently by growing the mutant in the presence of the capsid stabilizing drug Win51711. This indicates that the 2195H.G 110S intermediate is not a dead-end product but, upon capsid stabilization, can assemble into 150S particles. Interestingly, in the earlier study wherein a hydantoin drug was used to trap and identify a 110S packaging intermediate in the wild-type virus-infected cells, the wild-type poliovirus 110S intermediate contained cleaved VP0, suggesting that VP0 cleavage occurs concurrently as the RNA is being packaged (36). However, 2195H.G 110S intermediates and the drug-stabilized 2195H.G 150S particles did not contain any VP2 and VP4, consistent with the VP0 cleavage-minus phenotypes of the other histidine substitutions.
Two mutants, 2195H.T and 2195H.R, assembled 150S particles that failed to cleave VP0. The absence of VP0 cleavage in these two mutants did not appear to be due to defects at earlier stages of the assembly pathway. Indeed, in vivo, these mutants assembled similar amounts of pentamers and empty capsids as the wild-type poliovirus, and in vitro, both pentamers and empty capsid were assembly active. Moreover, antigenic determinants (sites 3A, 3B, and 2) present on the mutant 150S particles were recognized by monoclonal antibodies. Several of these monoclonal antibodies, in particular those recognizing site 3B, are highly specific for the native structure of the virus, indicating that the 2195H substitutions did not grossly affect the external structure of the 150S particles. In addition, modeling studies with the poliovirus empty capsid structure showed that residue 2195H is located in a region that can accommodate all substitutions made with no obvious steric hindrance problems (16). Based on this analysis, 2195H substitutions are not predicted to cause localized conformational changes in this region of the peptide backbone, which would potentially alter the location of residues surrounding the VP0 cleavage site. Although it remains possible that these substitutions lead to subtle, generalized conformational changes within the mutant particles, given the existing data, we favor the model that the 2195H is directly involved in VP0 cleavage.
Further evidence supporting the proposed cleavage mechanism was obtained upon characterizing 2194P mutants, 2194P.S and 2194P.G (15). Both 2194P substitutions cleaved VP0 and were viable. However, the content of VP0 in these mutant virions is greater than that in the wild-type particle, indicating that VP0 cleavage is inefficient in these mutants. Inefficient cleavage of VP0 in 2194P mutants is also consistent with the proposed mechanism. Substitution of this proline residue would lead to increased degrees of rotational freedom for the main chain in the vicinity of 2195H and consequently reduce the ability of the imidazole ring of 2195H to activate the water molecules mediating VP0 cleavage.
The location of the histidine residue is conserved in all picornavirus structures known to date (1, 13, 17, 22, 23, 25, 32). In addition, structure-based sequence alignments demonstrate that the histidine is conserved at the analogous position in all picornaviruses (27). This evidence suggests that VP0 cleavage might occur through a common mechanism mediated by the conserved histidine residue.
Mutations in 2145H of foot-and-mouth disease virus (FMDV; the analogous conserved histidine residue) have also been constructed to test its role in VP0 cleavage. Replacement of 2145H with lysine generated a nonviable FMDV mutant that assembled empty capsids but failed to assemble 150S particles (24). This phenotype is similar to that of the poliovirus 2195H.R mutant, where only empty capsids were detected in the absence of Win51711. Interestingly, when FMDV 2145H was replaced with serine, provirions accumulated. In contrast, when poliovirus 2195H was replaced with threonine, empty capsids accumulated and provirions were detected only once a capsid-stabilizing drug was used. It is possible that the additional methyl group in the threonine side chain generally results in a more unstable provirion structure. Alternatively, the wild-type FMDV provirion structure may be inherently more stable than that of the wild-type poliovirus.
Mutations at the VP0 scissile bond also have been characterized to study the mechanism of VP0 cleavage. Ansardi and Morrow reported that upon mutating the poliovirus VP0 cleavage site from asparagine-serine (amino acid 68 of VP4 and amino acid 1 of VP2, respectively) to glutamine-glycine, assembly appeared to terminate with the assembly of the empty capsids. Thus, it was proposed that because of the nature of the mutations generated at the cleavage site, capsid assembly could not progress to form provirions (2). Given the phenotypes of the 2195H mutants reported here and the fact that VP0 cleavage significantly increases the stability of the particle, an alternative explanation could be that this scissile bond mutant is actually capable of assembling provirions, which in the absence of capsid binding drugs are highly unstable and dissociate into empty capsids. Consistent with this alternative interpretation is the fact that provirion particles are observed in poliovirus and rhinovirus mutants which have replaced only the asparagine residue with a threonine. Interestingly, these provirions slowly cleaved VP0 upon incubation at 37°C (2, 21). This indicates that the presence of asparagine at the cleavage site is not required for VP0 cleavage. Indeed, other substitutions replacing this residue of the VP0 scissile bond in rhinovirus with serine or alanine did not inhibit VP0 cleavage. The phenotypes observed with the VP0 scissile bond mutants are also consistent with the proposed 2195H-mediated cleavage mechanism. At the cleavage site, coordination of an RNA base with the carbonyl oxygens is required to polarize the scissile bond, rendering it susceptible to nucleophilic attack by the activated water molecules. Thus, the identity of the specific amino acids forming this bond may not be critical so long as the side chains do not interfere with the orientation of the carbonyl oxygens.
The accumulation of a 110S packaging intermediate in the 2195H.G mutant and phenotypes of the 2194P mutants also suggest that residues 2194G and 2195H, indeed this region of the particle, may be involved in an additional stage of the poliovirus assembly process, namely, in RNA synthesis and packaging. Analysis of total RNA synthesis in 2194P.S-infected cells showed that mutant viral RNA synthesis was reduced at late but not early stages of infection (15). The VP0 cleavage site is located at the rim of a large trefoil-shaped hydrophobic depression present in the inner surface of the poliovirus empty capsid. This depression is similar to a site in bean pod mottle virus that has been reported to bind icosahedrally ordered segments of viral RNA in the virion (7). Thus, it has been proposed that this depression, centered around the threefold axes, may also be the site of RNA binding in the poliovirus particle (4). If so, then, due to the proximity of these residues to this putative RNA binding site, substitutions at 2195H and 2194G could also affect RNA packaging and/or concurrent RNA genome synthesis. Therefore, these residues appear to affect directly two processes in the virus life cycle: RNA packaging and VP0 cleavage after RNA encapsidation. The participation of these two amino acids in such dissimilar aspects of the poliovirus life cycle may account for the conservation of these residues in picornaviruses.
ACKNOWLEDGMENTS
We thank P. Mason, H. Wang, W. Stroop, T. Kelly, and X. Zhang for helpful discussions and Y. Huang for technical assistance. We also thank R. A. Grant for assistance in the modeling studies.
This work was supported by Public Health Service grant AI122627 from the National Institutes of Health.
REFERENCES
- 1.Acharya R, Fry E, Stuart D, Fox G, Rowlands D, Brown F. The three-dimensional structure of foot-and-mouth disease virus at 2.9Å resolution. Nature. 1989;337:709–716. doi: 10.1038/337709a0. [DOI] [PubMed] [Google Scholar]
- 2.Ansardi D, Morrow C. Amino acid substitutions in the poliovirus maturation cleavage site affect assembly and result in accumulation of provirions. J Virol. 1995;69:1540–1547. doi: 10.1128/jvi.69.3.1540-1547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold E, Luo M, Vriend G, Rossmann M G, Palmenberg A C, Parks G D, Nicklin M J H, Wimmer E. Implications of picornavirus capsid structure for polyprotein processing. Proc Natl Acad Sci USA. 1987;84:21–25. doi: 10.1073/pnas.84.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basavappa R, Syed R, Flore O, Icenogle J P, Filman D J, Hogle J M. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9Å resolution. Protein Sci. 1994;3:1651–1669. doi: 10.1002/pro.5560031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim H, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1512–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop N, Anderson D A. RNA-dependent cleavage of VP0 capsid protein in provirions of hepatitis A virus. Virology. 1993;197:616–623. doi: 10.1006/viro.1993.1636. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Stauffacher C, Ynge L, Schmidt T, Bomu W, Kamer G, Shanks M, Lomonossoff G, Johnson J E. Protein-RNA interactions in an icosahedral virus at 3.0 Å resolution. Science. 1989;245:154–168. doi: 10.1126/science.2749253. [DOI] [PubMed] [Google Scholar]
- 8.Compton S R, Nelson B, Kirkegaard K. Temperature-sensitive poliovirus mutant fails to cleave VP0 and accumulates provirions. J Virol. 1990;64:4067–4075. doi: 10.1128/jvi.64.9.4067-4075.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egger D, Pasamontes L, Ostermayer M, Bienz K. Reverse transcription multiplex PCR for differentiation between polio- and enteroviruses from clinical and environmental samples. J Clin Microbiol. 1995;33:1442–1447. doi: 10.1128/jcm.33.6.1442-1447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Tomas C, Baltimore D. Morphogenesis of poliovirus. II. Demonstration of a new intermediate, the provirion. J Virol. 1973;12:1122–1130. doi: 10.1128/jvi.12.5.1122-1130.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fox M P, Otto J J, McKinlay M A. Prevention of rhinovirus uncoating by Win51711, a new antiviral drug. Antimicrob Agents Chemother. 1986;30:110–116. doi: 10.1128/aac.30.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant R A, Filman D J, Fujinami R S, Icenogle J P, Hogle J M. Three-dimensional structure of Theiler virus. Proc Natl Acad Sci USA. 1992;89:2061–2065. doi: 10.1073/pnas.89.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harber J J, Bradley J, Anderson C W, Wimmer E. Catalysis of poliovirus VP0 maturation cleavage is not mediated by serine 10 of VP2. J Virol. 1991;65:326–334. doi: 10.1128/jvi.65.1.326-334.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hindiyeh M. Ph.D. dissertation. Little Rock: University of Arkansas for Medical Sciences; 1998. [Google Scholar]
- 16.Hindiyeh, M., and M. Chow. 1999. Unpublished results.
- 17.Hogle J M, Chow M, Filman D J. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 18.Knipe T, Rieder E, Baxt B, Ward G, Mason P. Characterization of synthetic foot-and-mouth disease virus provirions separates acid-mediated disassembly from infectivity. J Virol. 1997;71:2851–2856. doi: 10.1128/jvi.71.4.2851-2856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lee W-M, Monroe S S, Rueckert R R. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J Virol. 1993;67:2110–2122. doi: 10.1128/jvi.67.4.2110-2122.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo M, He C, Toth K S, Zhang C X, Lipton H L. Three-dimensional structure of Theiler murine encephalomyelitis virus (BeAn strain) Proc Natl Acad Sci USA. 1992;89:2409–2413. doi: 10.1073/pnas.89.6.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo M, Vriend G, Kamer G, Minor I, Arnold E, Rossmann M G, Boege U, Scraba D G, Duke G M, Palmenberg A C. The atomic structure of Mengo virus at 3.0 Å resolution. Science. 1987;235:182–191. doi: 10.1126/science.3026048. [DOI] [PubMed] [Google Scholar]
- 24.Mason, P. 1999. Personal communication.
- 25.Muckelbauer J, Kremer M, Minor I, Diana G, Dutko F, Groarke J, Pevear D, Rossmann M. The structure of coxsackievirus B3 at 3.5 Å resolution. Structure. 1995;3:653–667. doi: 10.1016/s0969-2126(01)00201-5. [DOI] [PubMed] [Google Scholar]
- 26.Otto M, Fox M P, Fancher M J, Kuhrt M F, Diana G D, McKinlay M A. In vitro activity of WIN 51711, a new broad-spectrum antipicornavirus drug. Antimicrob Agents Chemother. 1985;27:883–886. doi: 10.1128/aac.27.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmenberg A. Sequence alignments of picornaviral capsid proteins. In: Semler B L, Ehrenfeld E, editors. Molecular aspects of picornaviral infection and detection. Washington, D.C: American Society for Microbiology; 1989. pp. 211–241. [Google Scholar]
- 28.Reynolds C, Birnby D, Chow M. Folding and processing of the capsid protein precursor P1 is kinetically retarded in neutralization site 3B mutants of poliovirus. J Virol. 1992;66:1641–1648. doi: 10.1128/jvi.66.3.1641-1648.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rombaut B, Boeye A. In vitro assembly of poliovirus 14S subunits: disoxaril stabilization as a model for the antigenicity conferring activity of infected cell extracts. Virology. 1991;180:788–792. doi: 10.1016/0042-6822(91)90092-p. [DOI] [PubMed] [Google Scholar]
- 30.Rombaut B, Foriers A, Boeye A. In vitro assembly of poliovirus 14S subunits: identification of the assembly promoting activity of infected cell extracts. Virology. 1991;180:781–787. doi: 10.1016/0042-6822(91)90091-o. [DOI] [PubMed] [Google Scholar]
- 31.Rombaut B, Vrijsen R, Brioen P, Boeye A. A pH-dependent antigenic conversion of empty capsids of poliovirus studied with the aid of monoclonal antibodies to N and H antigen. Virology. 1982;122:215–218. doi: 10.1016/0042-6822(82)90393-2. [DOI] [PubMed] [Google Scholar]
- 32.Rossmann M G, Arnold E, Erickson J W, Frankenberger E A, Griffith J P, Hecht H-J, Johnson J E, Kamer G, Luo M, Mosser A G, Rueckert R R, Sherry B, Vriend G. The structure of a human common cold virus (rhinovirus 14) and its functional relationships to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 33.Sarnow P. Role of the 3′ end sequences in infectivity of poliovirus transcripts made in vitro. J Virol. 1989;63:467–470. doi: 10.1128/jvi.63.1.467-470.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simons J, Rogove A, Moscufo N, Reynolds C, Chow M. Efficient analysis of nonviable poliovirus capsid mutants. J Virol. 1993;67:1734–1738. doi: 10.1128/jvi.67.3.1734-1738.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sompayrac L M, Danna K J. Efficient infection of Monkey cells with DNA of simian virus 40. Proc Natl Acad Sci USA. 1981;78:7575–7578. doi: 10.1073/pnas.78.12.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vance L, Moscufo N, Chow M, Heinz B. Poliovirus 2C region functions during encapsidation of viral RNA. J Virol. 1997;71:8759–8765. doi: 10.1128/jvi.71.11.8759-8765.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ypma-Wong M F, Filman D J, Hogle J M, Semler B L. Structural domains of the poliovirus polyprotein are major determinants for proteolytic cleavage at gln-gly pairs. J Biol Chem. 1988;263:17846–17856. [PubMed] [Google Scholar]
- 38.Zeichhardt H, Otto M, McKlinlay M, Willingmann P, Habermehl K-O. Inhibition of poliovirus uncoating by disoxaril (WIN 51711) Virology. 1987;160:281–285. doi: 10.1016/0042-6822(87)90075-4. [DOI] [PubMed] [Google Scholar]