Abstract
Translation in plants is highly cap dependent, and the only plant mRNAs known to naturally lack a cap structure (m7GpppN) are viral in origin. The genomic RNA of tobacco etch virus (TEV), a potyvirus that belongs to the picornavirus superfamily, is a polyadenylated mRNA that is naturally uncapped and yet is a highly competitive mRNA during translation. The 143-nucleotide 5′ leader is responsible for conferring cap-independent translation even on reporter mRNAs. We have carried out a deletion analysis of the TEV 5′ leader to identify the elements responsible for its regulatory function and have identified two centrally located cap-independent regulatory elements (CIREs) that promote cap-independent translation. The introduction of a stable stem-loop structure upstream of each element demonstrated that CIRE-1 is less 5′ end dependent in function than CIRE-2. In a dicistronic mRNA, the presence of the TEV 5′ leader sequence in the intercistronic region increased expression of the second cistron, suggesting that the viral sequence can function in a 5′-distal position. Interestingly, the introduction of a stable stem-loop upstream of the TEV leader sequence or upstream of either CIRE in dicistronic constructs markedly increased their regulatory function. These data suggest that the TEV 5′ leader contains two elements that together promote internal initiation but that the function of one element, in particular, is facilitated by proximity to the 5′ end.
Virtually all eukaryotic cellular mRNAs contain a 5′ cap structure [m7G(5′)ppp(5′)N] that is required for maintaining mRNA integrity and promoting efficient translation. In an early step during translation initiation, the cap is bound by the eukaryotic initiation factor 4F (eIF4F), which is composed of eIF4E, the cap-binding subunit, and eIF4G, a considerably larger subunit. eIF4G serves as a multiadapter protein that, in addition to eIF4E, binds eIF4A, eIF3, the poly(A)-binding protein, and RNA (9, 15, 22, 23, 34, 38). The binding of a 40S ribosomal subunit to an mRNA is mediated by eIF3. As a result, the binding of eIF4E to the cap structure and the interaction between eIF4E, eIF4G, and eIF3 direct the binding of 40S subunits preferentially to the 5′ ends of capped mRNAs.
The only known mRNAs that naturally lack a cap are viral in origin. The picornaviral superfamily includes viruses that infect animals, e.g., poliovirus and encephalomyocarditis virus, and those that use plants as their host, e.g., tobacco etch virus (TEV). Regardless of the difference in host species, the genomic architectures of these viruses are highly similar. In each case, the single-stranded, positive-sense RNA genome functions as a monocistronic mRNA for a single polyprotein which, once produced, is processed by virus-encoded proteases into capsid and noncapsid proteins that are required to carry out the viral life cycle. Moreover, the viral RNA, which is polyadenylated but lacks a 5′ cap structure, contains a virus protein genome (VPg) linked to the 5′ terminus (5). However, the VPg is removed prior to recruitment of the viral RNA into polysomes, at least for poliovirus (13, 25, 26, 33). Consequently, it is the 5′ leader sequences of these viral mRNAs that confer on them the ability to be translated in a cap-independent manner (3, 11, 17, 30). For poliovirus and encephalomyocarditis virus, the 5′ leader varies in length from 650 to 1,300 nucleotides (nt), is highly structured, and contains multiple AUGs upstream of the initiation codon of the polyprotein-coding region (reviewed in reference 27). A region within these leaders serves as an internal ribosome entry site (IRES) that allows 40S ribosomal subunits to bind at or upstream of the true initiation codon (reviewed in reference 6). 40S subunit binding to the IRES can be mediated by a proteolytic fragment of eIF4G that contains the RNA-binding and eIF3 interaction domains (31, 32). Consequently, the 5′ leaders of picornaviral mRNAs promote cap-independent translation by recruiting 40S subunits via an internal initiation mechanism.
In plants, it follows that those viral mRNAs that naturally lack a 5′ cap structure must be translated through a cap-independent mechanism. The 5′ leaders of several plant viral mRNAs, including members of the tobamoviral, potyviral, comoviral, and luteoviral families, are responsible for conferring cap-independent translation (3, 11, 16, 37, 39). However, whether the translation of any plant viral mRNA occurs through internal initiation has been controversial (2, 39). A 351-nt region upstream of an AUG distal to the first initiation codon within the cowpea mosaic virus middle component RNA was reported to allow internal initiation at the distal AUG (39), although this has been disputed (2). Because both of these studies used animal cells or lysate for their analyses, neither could conclude whether internal initiation occurs in plants. The 5′ leader of turnip mosaic potyvirus RNA or the sequence upstream of the 3′-proximal coat protein subgenomic mRNA of a crucifer-infecting tobamovirus was reported to allow internal initiation (1, 16); however, the use of capped mRNAs in plant cells in the former example or uncapped mRNAs in in vitro translation lysates failed to demonstrate internal initiation of uncapped mRNAs in vivo.
Previous studies have demonstrated that the TEV 5′ leader mediates cap-independent translation (3) and is functionally analogous to a cap in that it interacts with the poly(A) tail to promote efficient translation (11) similar to that observed between a cap and a poly(A) tail (10). Precisely how the TEV 5′ leader substitutes for the cap to confer cap-independent translation has not been investigated. In this study, we have identified the elements within the TEV 5′ leader that are required to direct cap-independent translation. Two distinct cap-independent regulatory elements (CIREs) are present within the central region of the 143-base leader. The 5′-proximal element (CIRE-1) is more 5′ end independent than is the 5′-distal element (CIRE-2), and their combinatorial effects are approximately multiplicative, suggesting that the two elements are not functionally redundant in promoting cap-independent translation. Moreover, the TEV leader sequence or each CIRE promoted the translation of a second cistron when the viral leader sequence was present in the intercistronic region of a dicistronic mRNA. These observations suggest that specific CIREs are present within the TEV 5′ leader and that these CIREs function optimally in a 5′-proximal position but can also promote translation when positioned internally within a dicistronic mRNA.
MATERIALS AND METHODS
Plasmid and mRNA constructs.
The full-length TEV 5′ leader and subsequences were synthesized by PCR, and each DNA fragment was flanked by a HindIII and SalI restriction site. Each PCR product was introduced into the HindIII and SalI restriction sites upstream of the luciferase (luc) gene in the pT7-luc-A50 construct, a construct that has been described previously (10) to result in either TEV1–143-luc-A50 (i.e., the full-length TEV leader upstream of the luc coding region) or the following TEV 5′ leader deletion constructs: TEV1–118-luc-A50, TEV1–65-luc-A50, TEV28–143-luc-A50, TEV28–118-luc-A50, TEV28–65-luc-A50, TEV66–143-luc-A50, and TEV66–118-luc-A50.
Control leader constructs were designed to contain one or two copies (both in a forward orientation) of a 60% AT-rich, 72-nt sequence (AATATCTTATTG CCGGGAAAAGTGTACGTATCACCGTTTGTGTGAACAACGAACTGAA CTGGCAGACTATAA) introduced into the HindIII and SalI sites of pT7-luc-A50, resulting in Con72-luc-A50 or Con144-luc-A50 mRNAs. The free energy (ΔG) calculated by the fold algorithm for these mRNA leaders is −11.5 kcal/mol, which is approximately equal to the free energy of the 5′ leader of the TEV1–143-luc-A50 mRNA construct (ΔG = −10.7 kcal/mol) (41). A third control mRNA, Con17-luc-A50, was constructed with the 17-nt 5′ leader sequence GCCTAAGCTTGTCGACC, representing a free energy of −0.9 kcal/mol.
The above control 72-nt sequence was used to replace the 5′-terminal 65 nt of the TEV 5′ leader to result in Con72-TEV66–143-luc-A50 mRNA or was used to replace the 3′-terminal 78 nt of the TEV 5′ leader to result in TEV1–65-Con72-luc-A50 mRNA.
Introduction of a stable secondary structure was carried out by inserting a palindromic sequence (AAGCTTGGGCCCAGATCTACGCGTACGTACGCGTAGATCTGGGCCCAAGCTT) into the HindIII site 4 nt downstream of the T7 promoter transcriptional start site, producing a stem-loop (SL) composed of a 24-bp stem close to the 5′ terminus of the TEV-leader mRNA constructs SL-TEV1–143-luc-A50, SL-TEV1–65-luc-A50, SL-TEV66–143-luc-A50, and SL-TEV66–118-luc-A50 and the control mRNA constructs SL-Con7-luc-A50, SL-Con62-luc-A50, and SL-Con134-luc-A50. The calculated free energy of this SL structure is −42.9 kcal/mol.
Dicistronic constructs were generated by inserting the uidA gene (composed of the coding region for β-glucuronidase [GUS] and 73 nt of sequence 3′ to the uidA termination codon) upstream of the TEV-luc and control-luc constructs, resulting in the following mRNA constructs: GUS-TEV1–143-luc-A50, GUS-SL-TEV1–143-luc-A50, GUS-TEV1–65-luc-A50, GUS-SL-TEV1–65-luc-A50, GUS-TEV66–143-luc-A50, GUS-SL-TEV66–143-luc-A50, GUS-Con17-luc-A50, GUS-SL-Con7-luc-A50, GUS-Con72-luc-A50, GUS-SL-Con62-luc-A50, GUS-Con144-luc-A50, and GUS-SL-Con134-luc-A50.
In vitro transcription.
RNAs were synthesized with template plasmids linearized immediately downstream of the poly(A)50 sequence by NdeI to produce polyadenylated mRNAs or template plasmids linearized upstream of the poly(A)50 sequence by BamHI to produce poly(A)− mRNAs. Uncapped mRNAs were synthesized in vitro as described previously (40) with 10 μg of template DNA in a solution consisting of 40 mM Tris-HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 100 μg of bovine serum albumin/ml, 500 μM (each) ATP, CTP, UTP, and GTP, 10 mM dithiothreitol, 0.5 U of RNase inhibitor RNasin (Promega), and 0.5 U of T7 RNA polymerase (New England Biolabs) per ml. Capped RNAs were synthesized with 8 μg of template in the same reaction mixture as that described above except that GTP was used at 160 μM and 1 mM m7GpppG was included in the reaction mixture. Under these conditions more than 95% of the mRNA is capped.
mRNA delivery to plant protoplasts.
Protoplasts were isolated from a carrot (RCWC) cell suspension (used previously in the analysis of the translational regulatory function of the TEV 5′ leader as well as for other viral translation studies [11, 12]) by digestion with 0.25% CELF cellulase, 1% cytolase, 0.05% pectolyase Y23, 0.5% bovine serum albumin, and 7 mM β-mercaptoethanol in protoplast isolation buffer (12 mM sodium acetate [pH 5.8], 50 mM CaCl2, 0.25 M mannitol) for 90 to 120 min. Protoplasts were washed with protoplast isolation buffer followed by electroporation buffer (10 mM HEPES [pH 7.2], 130 mM KCl, 10 mM NaCl, 4 mM CaCl2, 0.2 M mannitol) and resuspended in electroporation buffer to approximately 106 cells/ml. Equal amounts of mRNAs (approximately 2.5 μg) were added to 400 μl of cell suspension immediately before electroporation (250 μF, 300 V, 0.2-mm electrode) with an IBI GeneZapper. The electroporated cells were incubated in protoplast growth medium (MS salts [pH 5.8] and 30 g of sucrose, 100 mg of myo-inositol, 0.1 mg of 2,4-dichlorophenoxyacetic acid, 1.3 mg of niacin, 0.25 mg of thiamine, 0.25 mg of pyridoxine, and 0.25 mg of calcium pentothenate per liter) supplemented with 20% cultured medium (protoplast growth medium conditioned with carrot cells for 3 days) overnight prior to the assay for reporter gene activity. For each experiment, an mRNA was delivered to triplicate samples of protoplasts and each sample was assayed in duplicate. Each experiment was repeated a minimum of three times. The average value and standard deviation for the constructs of a typical experiment are reported. For the monocistronic luc mRNA constructs, capped GUS-A50 mRNA was codelivered to serve as an internal control. For dicistronic mRNAs, GUS served as the 5′-proximal cistron. Luciferase in vivo activity normalized to GUS specific activity is reported.
In vitro translation.
Equal amounts of mRNA were translated with wheat germ extract as described by the manufacturer (Promega) except that all amino acids were unlabeled. The reaction mixtures were incubated for 2 h at 22°C, and aliquots of 3 μl were assayed. Each mRNA construct was translated in triplicate, and each in vitro translation was assayed in duplicate for luciferase activity. The average value and standard deviation for each construct are reported.
Luciferase and GUS assays.
Carrot protoplast extracts in luciferase assay buffer (25 mM Tricine [pH 8], 5 mM MgCl2, 0.1 mM EDTA supplemented with 33.3 mM dithiothreitol, 270 μM coenzyme A, and 500 μM ATP) were assayed for luciferase activity following injection of 0.5 mM luciferin with a Monolight 2010 luminometer (Analytical Luminescence Laboratory).
GUS activity in a 100-μl reaction mixture was assayed as described previously (7) with 1 mM 4-methylumbelliferyl-β-d-glucuronide as the substrate. The assay was performed for 30 min at 37°C, whereupon the reaction was terminated by addition of 900 μl of 0.2 M NaCO2. The amount of the fluorescent product produced in each assay was measured in a TKO 100 fluorometer (Hoefer Scientific, Inc.), with excitation at 365 nm and emission at 455 nm.
RESULTS
Identification of the CIREs within the TEV 5′ leader.
Previous work demonstrated that the TEV 5′ leader enhances the in vivo translation of reporter mRNAs in a cap-independent manner (3, 11). Enhancement was observed in tobacco (3), a host for TEV, and carrot (11), which, as a nonhost, permits the analysis of the translational regulatory function of the TEV 5′ leader independent of any potential host factor influence. To identify the region within the leader that is responsible for conferring cap-independent translation, a series of deletions was introduced throughout the TEV leader sequence and the resulting mutant leaders were tested for their ability to enhance the translation of luciferase (luc) mRNA in vivo following mRNA delivery to carrot protoplasts or in vitro by translating each mRNA in wheat germ lysate. The context of the luc initiation codon (GUCGACCAUGG, where the underlined AUG indicates the luc start codon) for all constructs used in this study was identical. The degree of cap-independent translation conferred by the full-length construct or each truncated TEV leader construct was measured relative to a control luc mRNA that contained a 5′ leader of similar length and degree of secondary structure (as described in Materials and Methods) but was unrelated in sequence. As previous work demonstrated that in vivo translation increases moderately as the length of the 5′ leader increases (12), it was necessary to employ control mRNAs that approximate the length of the TEV leader in order to establish that the enhancement conferred by the TEV 5′ leader was the result of specific regulation associated with this viral leader and not a consequence of its length.
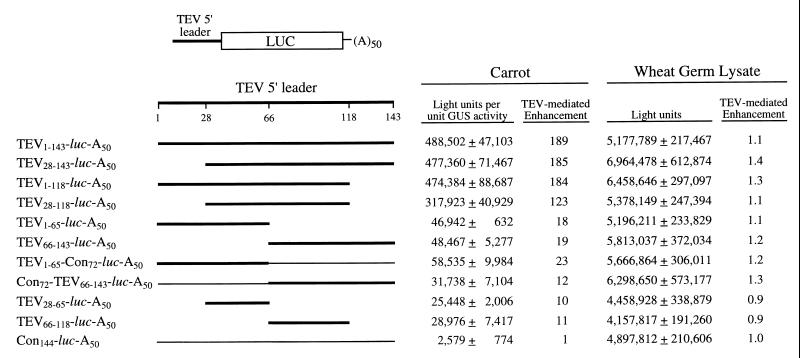
The presence of the TEV 5′ leader upstream of the luc coding region (i.e., TEV1–143-luc-A50) resulted in a 189-fold increase in translation of the uncapped mRNA relative to the uncapped control luc mRNA containing a 5′ leader of similar length (i.e., Con144-luc-A50) (Fig. 1), demonstrating that the increased translation conferred by the TEV leader was not merely a consequence of its length. Deletion of the 5′-terminal 27 nt (i.e., TEV28–143-luc-A50) or 3′-terminal 25 nt (i.e., TEV1–118-luc-A50) had little effect on the ability of the TEV 5′ leader to confer cap-independent translation (Fig. 1). The TEV leader in which both terminal regions were deleted (i.e., TEV28–118-luc-A50) exhibited only a small decrease in its regulatory function, suggesting that neither terminal region was essential for the TEV leader-mediated cap-independent translation. However, deletion of the 3′-terminal 78 nt (i.e., TEV1–65-luc-A50) did result in a reduction in cap-independent translation, i.e., TEV1–65-luc-A50 mRNA was translated to an 18-fold-greater degree than the control mRNA, compared to the 189-fold enhancement associated with the full-length TEV leader construct, i.e., TEV1–143-luc-A50 (Fig. 1). Similarly, the deletion of the 5′-terminal 65 nt (i.e., TEV66–143-luc-A50) resulted in a reduction in the degree of cap-independent translation to 19-fold (relative to the control mRNA). It should be noted that although the separation of the 5′ and 3′ halves of the TEV 5′ leader resulted in a 10-fold reduction in cap-independent translation, the TEV1–65 and TEV66–143 subsequences still retain substantial regulatory function (18- and 19-fold increases, respectively, in cap-independent translation relative to the control mRNA) (Fig. 1).
FIG. 1.
Identification of the two elements required for cap-independent translation. The full-length TEV 5′ leader or various subsequences were introduced upstream of the luc coding region in a T7 promoter-based construct that permitted the in vitro synthesis of mRNA terminating in a poly(A)50 tail. The region of the TEV 5′ leader present in each construct is indicated by a thick line, and the portion of the TEV leader sequence included in each construct is indicated as a subscript. The control sequence is indicated by a thin line, and the length of control sequence (Con) included in each construct is indicated as a subscript. Expression from an mRNA following delivery to carrot protoplasts by electroporation or following translation in wheat germ lysate is shown to the right of each construct. Each uncapped mRNA construct was delivered to triplicate samples of protoplasts or translated in triplicate in vitro, and each sample was assayed in duplicate. The experiment was repeated a minimum of three times, and the average value and standard deviation for the constructs of a typical experiment are reported.
One possible explanation for the reduction of cap-independent translation of the TEV1–65-luc-A50 and TEV66–143-luc-A50 mRNAs could be the change in position with respect to either the initiation codon or the 5′ terminus, respectively. To test this possibility, a 72-nt control sequence (one of two copies present in the leader of the Con144-luc-A50 mRNA) was substituted for the 3′-terminal 78 nt of the TEV leader sequence, resulting in the construct TEV1–65-Con72-luc-A50. Introduction of this control sequence positioned the TEV1–65 subsequence relative to the luc initiation codon with approximately the same spacing as that in the TEV1–143-luc-A50 mRNA construct. The degree of cap-independent regulatory function from the TEV1–65-Con72-luc-A50 mRNA was not significantly different (23-fold enhancement relative to the control construct, Con144-luc-A50) (Fig. 1) from that observed for the TEV1–65-luc-A50 mRNA (18-fold enhancement relative to the control construct); these data suggest that the change in spacing had little effect on the regulatory function of the TEV1–65 subsequence. Replacing the 5′-terminal 65 nt with the 72-nt control sequence (i.e., Con72-TEV66–143-luc-A50) positioned the TEV66–143 subsequence relative to the 5′ terminus with approximately the same spacing as that in the TEV1–143-luc-A50 mRNA construct. The regulatory function from Con72-TEV66–143-luc-A50 did not increase relative to that observed for TEV66–143-luc-A50 (12-fold versus 19-fold, respectively, relative to the Con144-luc-A50 control mRNA) (Fig. 1), suggesting that maintaining the spacing of the 5′ terminus relative to the TEV66–143 subsequence is not required for the regulatory function of this subsequence.
The regulatory elements present within the TEV1–65 and TEV66–143 subsequences were further delineated following the deletion of the 5′-terminal 27 nt from the TEV1–65-luc-A50 construct (resulting in TEV28–65-luc-A50) or 3′-terminal 25 nt from the TEV66–143-luc-A50 construct (resulting in TEV66–118-luc-A50). Translation from TEV28–65-luc-A50 mRNA was 10-fold higher than that from the control mRNA, and translation from TEV66–118-luc-A50 mRNA was 11-fold higher than that from the control. Consequently, deletion of the terminal sequences reduced cap-independent translation by only a small extent relative to that observed from the TEV1–65-luc-A50 and TEV66–143-luc-A50 constructs. These data suggest that a CIRE resides within the TEV28–65 subsequence (referred to as CIRE-1) and that a second element is present within the TEV66–118 subsequence (referred to as CIRE-2). The multiplicative effect of the individual contributions of CIRE-1 and CIRE-2 (i.e., 10-fold × 11-fold = 110-fold relative to the control mRNA) produced a degree of cap-independent translation that was similar to the actual degree of translation conferred when both elements were present together in the TEV28–118-luc-A50 construct (123-fold relative to the control mRNA), suggesting independence of function. The failure of the full-length TEV 5′ leader or any TEV deletion construct to enhance translation in vitro (Fig. 1) suggested that CIRE-1 and CIRE-2 do not function in vitro. The in vitro results are in good agreement with previous observations with the full-length TEV 5′ leader (11).
CIRE-1 and CIRE-2 promote optimal cap-independent translation largely through a 5′-end-dependent mechanism.
eIF4F, when bound to a 5′ cap structure, directs 40S subunit binding to the 5′ end of a capped mRNA. The absence of a cap or a reduction in functional eIF4F reduces 5′-end-dependent binding of 40S subunits and increases internal binding in Saccharomyces cerevisiae (35). The presence of secondary structure or RNA-protein complexes positioned close to the 5′ cap structure can function as an effective barrier to 5′-end-dependent translation by blocking 40S subunit binding to the mRNA (19, 20, 24, 28, 29). Secondary structure is less effective in impeding the translation of uncapped mRNA in yeast because 40S subunit binding is not directed to the 5′ terminus (35, 36). However, in rabbit reticulocyte lysate, translation of an uncapped mRNA remains 5′ end dependent (4), suggesting that a difference between yeast and mammalian translation may exist.
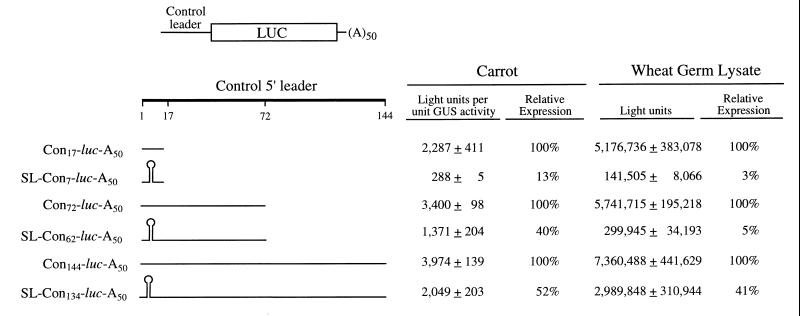
To examine whether the cap-independent translation conferred by the TEV 5′ leader requires an unstructured 5′ terminus, a stable SL structure (with a calculated free energy of −42.9 kcal/mol) was introduced 4 nt downstream of the 5′ terminus of the control and TEV-containing mRNA constructs used in the previous experiment. Translation of uncapped mRNAs with or without the SL structure was assayed following their delivery to carrot protoplasts or in wheat germ lysate. In the absence of an SL, expression from the constructs containing the control sequence as the 5′ leader increased moderately with the length of the leader in protoplasts (Fig. 2). Translation of the SL-Con7-luc-A50 construct (in which the SL was introduced into the control construct Con17-luc-A50) was reduced to just 13% of that observed for Con17-luc-A50 (Fig. 2). In contrast, the introduction of the SL 62 nt upstream of the luc cistron, i.e., SL-Con62-luc-A50, resulted in a level of translation that was 40% of that for the corresponding control mRNA (i.e., Con72-luc-A50) in protoplasts (Fig. 2). Introduction of the SL 134 nt upstream of the luc cistron had an even smaller effect on in vivo expression: translation from SL-Con134-luc-A50 was 52% that of Con144-luc-A50. Translation of the same mRNAs in vitro yielded a similar trend: the presence of the SL 7 nt upstream of the luc initiation codon reduced translation to 3% of the control, whereas translation was 41% of the corresponding control when the SL was positioned 134 nt upstream of the initiation codon (Fig. 2). One exception to the trend observed in vitro was with the SL-Con62-luc-A50 mRNA construct, which was translated to just 5% of the corresponding control in vitro but was translated to 40% of the control in vivo. These results suggest that the spacing between the secondary structure and the initiation codon determines the extent of translational repression at the appropriate start codon of uncapped mRNAs. Moreover, these data demonstrate that although the repressive effect of an SL on the translation of an uncapped mRNA can be reduced by increasing the distance between it and the initiation codon, a difference in the distance required is observed for translation in vivo versus in vitro.
FIG. 2.
Analysis of the translational efficiency of uncapped mRNAs with a free or inaccessible 5′ terminus. The effect of increasing the length of the 5′ leader on the translational efficiency of an uncapped mRNA with or without an SL present at the 5′ terminus was examined in carrot protoplasts and in wheat germ lysate. Control 5′ leaders 17, 72, or 144 nt in length were introduced upstream of the luc coding region, and each mRNA was synthesized in vitro to terminate in a poly(A)50 tail. The 144-nt leader contains two copies of the 72-nt sequence present in the 72-nt leader construct. The length of the control sequence (Con) included in each construct is indicated as a subscript. The 24-bp SL introduced into each construct is indicated diagrammatically. Expression from each mRNA following delivery to carrot protoplasts by electroporation or following translation in wheat germ lysate is shown to the right of each construct. Each uncapped mRNA construct was delivered to triplicate samples of protoplasts or translated in triplicate in vitro, and each sample was assayed in duplicate. The experiment was repeated a minimum of three times, and the average value and standard deviation for the constructs of a typical experiment are reported.
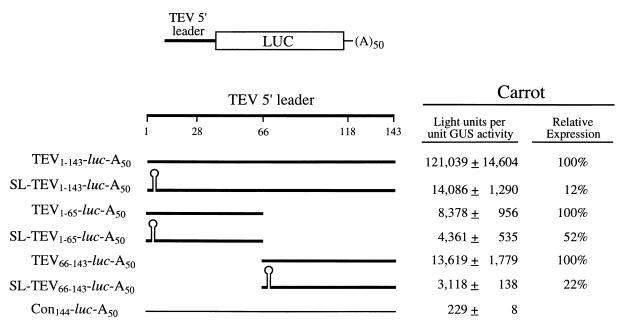
The same SL was then introduced upstream of the TEV 5′ leader in order to examine whether the cap-independent translation mediated by this leader is 5′ end dependent or 5′ end independent. Translation of SL-TEV1–143-luc-A50 in protoplasts (in which the SL was introduced into the control construct TEV1–143-luc-A50) was reduced to just 12% of that observed for TEV1–143-luc-A50 (Fig. 3). As seen above, translation from SL-Con134-luc-A50 was 52% of that observed for the Con144-luc-A50 control construct. Consequently, the repressive effect of the SL when present upstream of the TEV 5′ leader was disproportionately greater than what would have been expected from its position with respect to the luc cistron, suggesting that the TEV 5′ leader requires an accessible 5′ terminus for optimal regulatory function. However, even with the SL positioned upstream of the TEV 5′ leader, translation was still more than 60-fold greater than that from the Con144-luc-A50 construct (Fig. 3), suggesting that the TEV 5′ leader retained substantial function even in the absence of a free 5′ terminus.
FIG. 3.
Analysis of the 5′-end dependence of the TEV 5′ leader and CIRE-1 and CIRE-2. The 5′-end dependence of the full-length TEV leader and of each CIRE individually was examined by introducing each sequence upstream of the luc coding region. Each uncapped mRNA, with or without an SL present at the 5′ terminus, was synthesized in vitro to terminate in a poly(A)50 tail. The region of the TEV 5′ leader present in each construct is indicated by a thick line, and the portion of the TEV leader sequence included in each construct is indicated as a subscript. The 24-bp SL introduced into each construct is indicated diagrammatically. The control sequence is indicated by a thin line, and the length of the control sequence (Con) included in the control construct is indicated as a subscript. Expression from each mRNA following delivery to carrot protoplasts by electroporation is shown to the right of each construct.
To examine the effect of the SL on cap-independent translation conferred by each of the two CIREs within the TEV 5′ leader, the SL was introduced upstream of each CIRE individually. Translation from SL-TEV1–65-luc-A50 (in which the SL was introduced into the construct TEV1–65-luc-A50, which contains CIRE-1) in protoplasts was 52% of that observed for TEV1–65-luc-A50 (Fig. 3). The effect of the SL was similar to its effect on the control leader of similar length (compare the degree of repression from SL-Con62-luc-A50 relative to that from Con72-luc-A50 [Fig. 2]). In contrast, the introduction of the SL upstream of CIRE-2, i.e., SL-TEV66–143-luc-A50, had a disproportionately repressive effect in that it reduced translation to 22% of that observed for TEV66–143-luc-A50 (Fig. 3). Consequently, the cap-independent translation conferred by the region of the TEV 5′ leader containing CIRE-2 is more 5′ end dependent than that conferred by the region containing CIRE-1. It should be noted that the effect of the SL was reproducibly greater for the full-length TEV 5′ leader than it was for either CIRE individually, indicating that the two elements exhibit greater 5′ end dependence when together than when separate. Additionally, it should be noted that the introduction of the SL upstream of either CIRE failed to abolish entirely the cap-independent translation conferred by either element, suggesting that although CIRE-2 may be more 5′ dependent than CIRE-1, both retain substantial regulatory function in the absence of a free 5′ end.
The poly(A) tail promotes cooperativity between CIRE-1 and CIRE-2.
Previous work demonstrated that the TEV 5′ leader functionally interacts with the poly(A) tail to promote cap-independent translation (11). This interaction is analogous to that observed between a cap and a poly(A) tail (10), suggesting that one or more elements within the TEV 5′ leader are responsible for the functional interaction with the poly(A) tail. To examine the poly(A) tail dependence of each CIRE and the degree of 5′-end dependence of each CIRE in the absence of a poly(A) tail, those constructs used in Fig. 3 were introduced as poly(A)− mRNAs into carrot protoplasts and their relative levels of translation were determined (Fig. 4). The presence of the SL upstream of the full-length TEV 5′ leader construct (i.e., SL-TEV1–143-luc) reduced translation to 33% of that observed for TEV1–143-luc (Fig. 4). However, as noted above, the introduction of the SL upstream of the full-length TEV 5′ leader had a greater repressive effect when the mRNA was polyadenylated (i.e., SL-TEV1–143-luc translated at 12% of the level observed for TEV1–143-luc [Fig. 3]), suggesting that one functional consequence of the interaction between the TEV 5′ leader and the poly(A) tail is to increase the 5′-end dependence of the full-length TEV 5′ leader.
FIG. 4.
The poly(A) tail is required for CIRE-1 and CIRE-2 regulatory function. The regulatory function and 5′-end dependence of the full-length TEV 5′ leader and each CIRE individually were examined by introducing each sequence upstream of the luc coding region. Each uncapped mRNA, with or without an SL present at the 5′ terminus, was synthesized in vitro without a poly(A) tail. The region of the TEV 5′ leader present in each construct is indicated by a thick line, and the portion of the TEV leader sequence included in each construct is indicated as a subscript. The 24-bp SL introduced into each construct is indicated diagrammatically. Control sequence is indicated by a thin line, and the length of the control sequence (Con) included in the control construct is indicated as a subscript. Expression from each mRNA following delivery to carrot protoplasts by electroporation is shown to the right of each construct.
Each CIRE, when tested individually in poly(A)− mRNAs, i.e., as either TEV1–65-luc (containing CIRE-1) or TEV66–143-luc (containing CIRE-2), retained the ability to confer cap-independent translation (Fig. 4), demonstrating that neither element is wholly dependent on the poly(A) tail for its function. As observed for the full-length TEV 5′ leader, introduction of the SL affected the function of each CIRE to a lesser extent in poly(A)− mRNAs (Fig. 4) than in polyadenylated mRNAs (Fig. 3).
The absence of a poly(A) tail also affected the combinatorial effect of CIRE-1 and CIRE-2 on cap-independent translation. When mRNA lacked a poly(A) tail, translation from TEV1–65-luc (containing CIRE-1) and TEV66–143-luc (containing CIRE-2) was 3.8- and 3.2-fold, respectively, less than that from TEV1–143-luc (containing both CIRE-1 and CIRE-2), as calculated from the data in Fig. 4, but was 14.4- and 8.9-fold, respectively, less than that from TEV1–143-luc-A50 when the mRNAs were polyadenylated (calculated from the data in Fig. 3). Because separation of the CIREs resulted in a greater loss of translational efficiency when the mRNA was polyadenylated, these data suggest that another consequence of the interaction between the TEV 5′ leader and the poly(A) tail is to increase the combinatorial effect of CIRE-1 and CIRE-2 on cap-independent translation.
The TEV 5′ leader sequence can increase translation when positioned internally in a dicistronic construct.
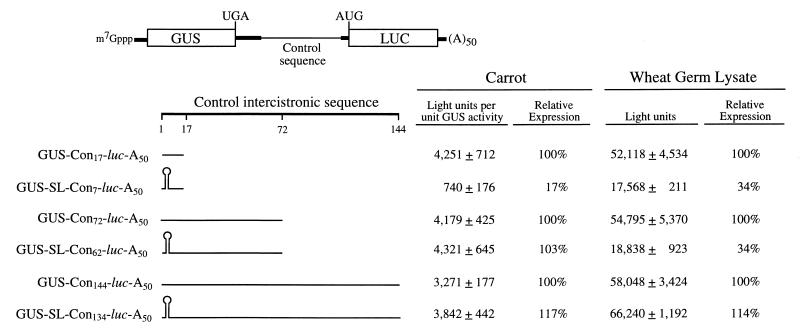
To examine whether the TEV 5′ leader sequence can promote the translation of the second cistron of a dicistronic mRNA when present in an intercistronic position, a series of dicistronic constructs in which TEV leader or control sequences were introduced between the region encoding GUS as the 5′-proximal cistron and the luc coding region as the distal cistron were made (see Fig. 5 and 6 for construct design). In the first series of constructs to be examined, the control 5′-leader sequences used in Fig. 2 were tested as intercistronic sequences. These included the control sequence of 17 (i.e., GUS-Con17-luc-A50), 72 (i.e., GUS-Con72-luc-A50), and 144 nt (i.e., GUS-Con144-luc-A50). An additional 73 nt from the GUS 3′-untranslated region contributed to the intercistronic region of each construct. Each construct was introduced as a capped mRNA into carrot protoplasts, and the degree of translation from the 5′-distal luc cistron was normalized to the amount of GUS produced from the 5′-proximal cistron. Levels of translation from these three control mRNAs in carrot protoplasts and in wheat germ lysate were not significantly different (Fig. 5), suggesting that the difference in the length of the intercistronic region in the range represented by these constructs did not influence initiation at the distal cistron. This is in contrast to the moderate increase in translation observed when the length of the 5′ leader was increased for a monocistronic mRNA (Fig. 2).
FIG. 5.
The effect of intercistronic length on the translation of the 5′-distal cistron in a dicistronic mRNA. Dicistronic mRNAs with GUS as the 5′-proximal cistron and luc as the 5′-distal cistron were constructed with intercistronic regions containing control sequence 17, 72, or 144 nt in length. Each capped mRNA construct was synthesized in vitro to terminate in a poly(A)50 tail. The 144-nt intercistronic region contains two copies of the 72-nt sequence present in the 72-nt intercistronic construct. The length of the control sequence (Con) included in each construct is indicated as a subscript. The 24-bp SL introduced into each construct is indicated diagrammatically. Expression from each mRNA following delivery to carrot protoplasts by electroporation or following translation in wheat germ lysate is shown to the right of each construct. Each mRNA construct was delivered to triplicate samples of protoplasts or translated in triplicate in vitro, and each sample was assayed in duplicate. The experiment was repeated a minimum of three times. The average value and standard deviation for the constructs of a typical experiment are reported.
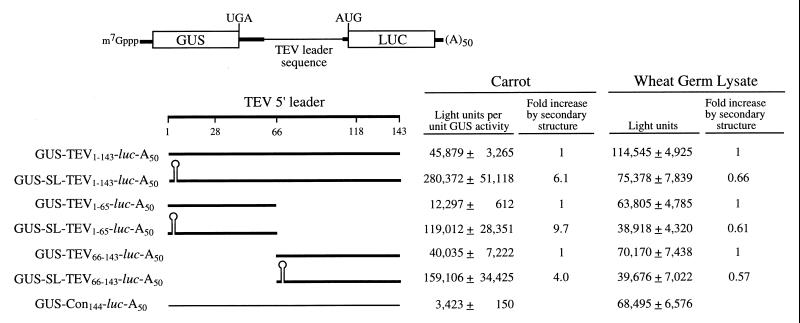
FIG. 6.
The TEV 5′ leader can promote translation from an intercistronic position. Dicistronic mRNAs with GUS as the 5′-proximal cistron and luc as the 5′-distal cistron were constructed with intercistronic regions containing the full-length TEV leader sequence or each CIRE individually. The region of the TEV leader present in each construct is indicated by a thick line, and the region of the TEV leader sequence included in each construct is indicated as a subscript. The 24-bp SL introduced into each construct is indicated diagrammatically. The control sequence is indicated by a thin line, and the length of the control sequence (Con) included in the control construct is indicated as a subscript. Each capped mRNA construct was synthesized in vitro to terminate in a poly(A)50 tail. Expression from each mRNA following delivery to carrot protoplasts by electroporation or following translation in wheat germ lysate is shown to the right of each construct. Each mRNA construct was delivered to triplicate samples of protoplasts or translated in triplicate in vitro, and each sample was assayed in duplicate. The experiment was repeated a minimum of three times, and the average value and standard deviation for the constructs of a typical experiment are reported.
The translational regulation of GCN4 in yeast involves several short upstream open reading frames (uORFs) present in the 5′ leader of the mRNA (reviewed in reference 14). Translation initiation at the main cistron occurs by those 40S ribosomal subunits that have resumed scanning following translational termination of the short uORFs (14). Whether 40S subunits can resume scanning following translational termination of a coding region of typical length has not been established in plants. However, in order to prevent the possibility of any 40S subunits reaching the intercistronic region through the resumption of scanning following translational termination of the upstream GUS coding region, the SL used in Fig. 2 to 4 was introduced into the 17-, 72-, and 144-nt control intercistronic sequences. The 73-nt GUS 3′-untranslated region upstream of the SL provided sufficient distance between the termination codon of the GUS coding region and the SL so that translational termination from the 5′-proximal cistron would not be negatively affected (24a). Introduction of the SL 7 nt upstream of the distal cistron (i.e., GUS-SL-Con7-luc-A50) resulted in a level of translation that was just 17% of that observed for the corresponding control (i.e., GUS-Con17-luc-A50) in protoplasts (Fig. 5). Introduction of the SL into the 72- or 144-nt intercistronic region (i.e., GUS-SL-Con62-luc-A50 or GUS-SL-Con134-luc-A50, respectively) had no effect on translation from the distal cistron in vivo, and introduction of the SL into the 144-nt intercistronic region had no effect in vitro (Fig. 5). The presence of the SL 62 nt upstream of the luc cistron in the dicistronic construct (i.e., GUS-SL-Con62-luc-A50) reduced luc expression in vitro (relative to that for the control GUS-Con62-luc-A50 construct [Fig. 5]) just as it had when the SL was positioned the same distance upstream of luc in the analogous monocistronic construct (i.e., SL-Con62-luc-A50) during in vitro translation (Fig. 2), confirming that 62 nt between an SL and an initiation codon is not a sufficiently long distance to permit optimal expression of the dicistronic construct in vitro. These data indicate that any ribosomal subunits that may resume scanning from the upstream GUS coding region do not contribute significantly to the translation of the 5′-distal luc cistron and suggest that the in vivo translation of the distal cistron in the 72- or 144-nt intercistronic region constructs results from internal initiation.
The presence of the TEV sequence in the intercistronic region increased translation from the distal luc cistron 13.4-fold relative to that from the construct containing the 144-nt control sequence in the intercistronic region (i.e., GUS-Con144-luc-A50) in protoplasts (Fig. 6), demonstrating that the TEV leader sequence can function to promote translation when in an intercistronic position. As above, an SL was introduced immediately upstream of the TEV sequence to exclude the possibility of any ribosomal subunits reaching the TEV leader sequence. Surprisingly, instead of a negative or neutral effect on luc expression, as observed when the control sequence constituted the intercistronic region, the presence of the SL upstream of the TEV leader sequence in a dicistronic mRNA resulted in an additional 6.1-fold increase in the translation of the distal luc cistron (Fig. 6). This increase was specific to expression from the downstream luc cistron because expression from the upstream GUS cistron was not significantly affected by the introduction of the SL (data not shown). Consequently, the TEV leader sequence positioned downstream of the SL resulted in an 82-fold increase in expression from the luc cistron relative to that from the GUS-Con144-luc-A50 control construct (Fig. 6).
Similar results were observed when the region of the TEV leader sequence containing either CIRE-1 or CIRE-2 was introduced as the intercistronic region (i.e., GUS-TEV1–65-luc-A50 or GUS-TEV66–143-luc-A50, respectively). The presence of either CIRE-1 or CIRE-2 in the intercistronic region increased translation from the 5′-distal luc cistron by 3.6- and 11.7-fold, respectively, relative to that from the GUS-Con144-luc-A50 control construct (Fig. 6). Similar to the results obtained with the full-length TEV leader, the introduction of the SL upstream of either CIRE-1 or CIRE-2 (i.e., GUS-SL-TEV1–65-luc-A50 or GUS-SL-TEV66–143-luc-A50, respectively) resulted in a further 9.7- or 4.0-fold increase, respectively, in the translation of the distal luc cistron (Fig. 6). Consequently, CIRE-1 or CIRE-2 positioned downstream of the SL resulted in a 35- or 46-fold increase, respectively, in the expression from the luc cistron relative to that from the GUS-Con144-luc-A50 control construct. No SL-associated increase was observed when these same constructs were translated in vitro, data consistent with the lack of a TEV leader-mediated enhancement in translation lysate as observed in Fig. 1. These results suggest that the TEV leader sequence or the individual CIRE elements can promote translation when positioned in the intercistronic region and that the presence of an SL upstream of these elements in a dicistronic construct assists in maintaining their function as translational regulators.
DISCUSSION
In the present study, we identified the regulatory elements within the TEV 5′ leader responsible for promoting cap-independent translation and determined the extent to which their function was dependent on proximity to the 5′ end. We observed that the TEV 5′ leader contained two elements (i.e., CIRE-1 and CIRE-2) that were required for cap-independent translation. The 5′-proximal CIRE-1 (present within a 39-nt region) was less 5′ end dependent than the distal CIRE-2 (present within a 53-nt region), suggesting that the two elements are functionally distinct. Moreover, as the combinatorial effect of CIRE-1 and CIRE-2 was approximately multiplicative, both elements appear to be required to confer full cap-independent translation and may be considered as components of a single regulatory locus.
The TEV 5′ leader confers cap-independent translation, in part, through a functional interaction with the poly(A) tail (11). Analysis of the function of CIRE-1 and CIRE-2 in poly(A)+ and poly(A)− mRNAs suggests that the combinatorial effect of CIRE-1 and CIRE-2 is regulated by the presence of the poly(A) tail. Separation of CIRE-1 and CIRE-2 in polyadenylated mRNAs resulted in a 9- to 14-fold decrease in cap-independent translation. In contrast, the separation of CIRE-1 and CIRE-2 resulted in only a 3- to 4-fold decrease in the cap-independent translation of nonpolyadenylated mRNAs. These data suggest that one function of the poly(A) tail on the expression from TEV mRNA is to increase the combined effect of CIRE-1 and CIRE-2 in order to promote cap-independent translation.
The TEV leader sequence (or either CIRE) also promoted translation of the second cistron of a dicistronic mRNA when present in the intercistronic region. Interestingly, the introduction of an SL downstream of the first cistron but immediately upstream of the TEV leader or either CIRE substantially increased the regulatory ability of the TEV leader sequence (Fig. 6). As no similar increase was observed when the SL was introduced upstream of a control sequence of equivalent length, these observations suggest that the SL promoted the TEV regulatory function. Because the presence of the SL decreased the regulatory function of the TEV sequence when the latter was present as the 5′ leader of monocistronic mRNAs, the stimulatory effect of the SL on TEV regulatory function is specific to the dicistronic constructs. We postulate that the SL prevented interference with the TEV regulatory function by ribosomal subunits that may have remained associated with the mRNA following translational termination of the first cistron. Translation of GCN4 mRNA in yeast requires that 40S subunits resume scanning following translational termination of the short uORFs in the 5′ leader. Such 40S subunits are initially incompetent following translation termination and cannot participate in a subsequent round of initiation until a new ternary complex, composed of eIF2, Met-tRNAi, and GTP, has bound (14, 21). As a minimum distance between a short uORF and the main ORF is necessary, this has been interpreted to mean that the restoration of translational competence of a 40S subunit by means of binding a new ternary complex is time dependent. Evidence that ribosomal subunits remain associated following the translation of a cistron of normal length has been obtained with electron micrographs of polysomes (8, 18), but the extent to which these subunits regain competency for translational initiation while associated with an mRNA is not known. If, following translational termination of a full-sized 5′-proximal cistron, a 40S subunit is at least as incompetent for reinitiation as it is following termination from a minicistron, its scanning through the TEV leader sequence (as the intercistronic region) may inhibit its ability to recruit translationally competent 40S subunits and thereby reduce the extent to which the TEV leader can enhance translation of the second cistron. Consequently, the introduction of an SL immediately upstream of the TEV leader sequence might stall incompetent 40S subunits from scanning into the TEV leader sequence and thereby allow the TEV leader sequence to function unimpeded to recruit translationally competent 40S subunits. Thus, the introduction of an SL upstream of the TEV leader sequence when present in the intercistronic region of a dicistronic mRNA would act to increase its function. The inhibitory effect of the SL on the TEV sequence when present as the 5′ leader is consistent with this conclusion: the SL would be expected to reduce but not abolish the regulatory function of the TEV leader sequence because (i) there is no upstream cistron from which incompetent 40S subunits may scan and (ii) the TEV leader exhibits a functional preference for proximity to a free 5′ terminus.
As mentioned above, CIRE-1 is functionally less 5′ end dependent than CIRE-2, suggesting that the two elements are functionally distinct. The multiplicative increase in cap-independent translation when both elements are present suggests a concerted mechanism to recruit 40S subunits or to stabilize the association of 40S subunits recruited by one CIRE through interaction with the other CIRE. 40S subunit recruitment may involve direct interaction with a CIRE or, alternatively, each CIRE may serve as a binding site for trans-acting factors that mediate 40S subunit recruitment. The latter possibility is supported by the observation that the TEV 5′ leader forms specific complexes with proteins in a gel retardation assay (37a). Such a trans-acting factor(s) may preferentially associate with a CIRE when positioned proximal to an accessible 5′ end and may be prevented from binding efficiently to an intercistronic CIRE if translationally incompetent ribosomal subunits are transiting between cistrons in a dicistronic mRNA. The increased function of an intercistronic CIRE positioned downstream of an SL could be explained if the SL excluded incompetent ribosomal subunits and thus permitted more-efficient trans-acting factor binding. These data suggest that significant differences between the translational regulatory mechanism of those members of the picornaviral superfamily that infect plants and that of those that infect animals exist. Those that infect animals contain a highly structured 5′ leader, which can be several hundred nucleotides in length, whereas the TEV 5′ leader is not long or highly structured. And yet both types of viral leaders direct 40S subunit binding in the absence of a 5′ cap structure. How these different members of the same viral superfamily achieve the efficient recruitment of the translational machinery in their respective host species will provide insight into the similarities and differences in translation in plants and animals.
ACKNOWLEDGMENT
This work was supported by U.S. Department of Agriculture grant NRICGP 96-35301-3144 (to D.R.G.).
REFERENCES
- 1.Basso J, Dallaire P, Charest P J, Devantier Y, Laliberte J-F. Evidence for an internal ribosome entry site within the 5′ non-translated region of turnip mosaic potyvirus RNA. J Gen Virol. 1994;75:3157–3165. doi: 10.1099/0022-1317-75-11-3157. [DOI] [PubMed] [Google Scholar]
- 2.Belsham G J, Lomonossoff G P. The mechanism of translation of cowpea mosaic virus middle component RNA: no evidence for internal initiation from experiments in an animal cell transient expression system. J Gen Virol. 1991;72:3109–3113. doi: 10.1099/0022-1317-72-12-3109. [DOI] [PubMed] [Google Scholar]
- 3.Carrington J C, Freed D D. Cap-independent enhancement of translation by a plant potyvirus 5′ nontranslated region. J Virol. 1990;64:1590–1597. doi: 10.1128/jvi.64.4.1590-1597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Gregorio E, Preiss T, Hentze M W. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA. 1998;4:828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dougherty W G, Carrington J C. Expression and function of potyviral gene products. Annu Rev Phytopathol. 1988;26:123–143. [Google Scholar]
- 6.Ehrenfeld E. Initiation of translation by picornavirus RNAs. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 549–574. [Google Scholar]
- 7.Everett J G, Gallie D R. RNA delivery in Saccharomyces cerevisiae using electroporation. Yeast. 1992;8:1007–1014. doi: 10.1002/yea.320081203. [DOI] [PubMed] [Google Scholar]
- 8.Francke C, Edstrom J-E, McDowall A W, Miller O L., Jr Electron microscopic visualization of a discrete class of giant translation units in salivary gland cells of Chironomus tentans. EMBO J. 1982;1:59–62. doi: 10.1002/j.1460-2075.1982.tb01124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser C S, Pain V W, Morley S J. The association of initiation factor 4F with poly(A)-binding protein is enhanced in serum-stimulated Xenopus kidney cells. J Biol Chem. 1999;274:196–204. doi: 10.1074/jbc.274.1.196. [DOI] [PubMed] [Google Scholar]
- 10.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 11.Gallie D R, Tanguay R, Leathers V. The tobacco etch viral 5′ leader and poly(A) tail are functionally synergistic regulators of translation. Gene. 1995;165:233–238. doi: 10.1016/0378-1119(95)00521-7. [DOI] [PubMed] [Google Scholar]
- 12.Gallie D R, Walbot V. Identification of the motifs within the tobacco mosaic virus 5′-leader responsible for enhancing translation. Nucleic Acids Res. 1992;20:4631–4638. doi: 10.1093/nar/20.17.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hewlett M J, Rose J K, Baltimore D. 5′-terminal structure of poliovirus polyribosomal RNA is pUp. Proc Natl Acad Sci USA. 1976;73:327–330. doi: 10.1073/pnas.73.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinnebusch A. Translational control of GCN4: gene-specific regulation of phosphorylation of eIF-2. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 199–244. [Google Scholar]
- 15.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivanov P A, Karpova O V, Skulachev M V, Tomashevskaya O L, Rodionova N P, Dorokhov Y L, Atabekov J G. A tobamovirus genome that contains an internal ribosome entry site function in vitro. Virology. 1997;232:32–43. doi: 10.1006/viro.1997.8525. [DOI] [PubMed] [Google Scholar]
- 17.Jang S K, Krausslich H-G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiseleva E V. Secretory protein synthesis in Chironomus salivary gland cells is not coupled with protein translocation across endoplasmic reticulum membranes. FEBS Lett. 1989;257:251–253. doi: 10.1016/0014-5793(89)81545-5. [DOI] [PubMed] [Google Scholar]
- 19.Koloteva N, Muller P P, McCarthy J E G. The position dependence of translational regulation via RNA-RNA and RNA-protein interactions in the 5′-untranslated region of eukaryotic mRNA is a function of the thermodynamic competence of 40 S ribosomes in translational initiation. J Biol Chem. 1997;272:16531–16539. doi: 10.1074/jbc.272.26.16531. [DOI] [PubMed] [Google Scholar]
- 20.Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci USA. 1986;83:2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak M. Effect of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases—implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 23.Le H, Tanguay R L, Balasta M L, Wei C-C, Browning K S, Metz A M, Goss D J, Gallie D R. The translation initiation factors eIFiso4G and eIF-4B interact with the poly(A)-binding protein to increase its RNA binding affinity. J Biol Chem. 1997;272:16247–16255. doi: 10.1074/jbc.272.26.16247. [DOI] [PubMed] [Google Scholar]
- 24.Muckenthaler M, Gray N K, Hentze M W. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol Cell. 1998;2:383–388. doi: 10.1016/s1097-2765(00)80282-8. [DOI] [PubMed] [Google Scholar]
- 24a.Niepel, M., and D. Gallie. Unpublished data.
- 25.Nomoto A, Lee Y F, Wimmer E. The 5′ end of poliovirus mRNA is not capped with m7G(5′)ppp(5′)Np. Proc Natl Acad Sci USA. 1976;73:375–380. doi: 10.1073/pnas.73.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomoto A, Kitamura N, Golini F, Wimmer E. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc Natl Acad Sci USA. 1977;74:5345–5349. doi: 10.1073/pnas.74.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmenberg A C. Comparative organization and genome structure in picornaviruses. UCLA Symp Mol Cell Biol. 1987;54:25–34. [Google Scholar]
- 28.Paraskeva E, Gray N K, Schläger B, Wehr K, Hentze M W. Ribosome pausing and scanning arrest as mechanisms of translational regulation from cap-distal iron-responsive elements. Mol Cell Biol. 1999;19:807–816. doi: 10.1128/mcb.19.1.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985;40:515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- 30.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 31.Pestova T V, Hellen C U T, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pestova T V, Hellen C U T, Shatsky I N. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersson R F, Flanegan J B, Rose J K, Baltimore D. 5′-terminal nucleotide sequences of polio virus polyribosomal RNA and virion RNA are identical. Nature. 1977;268:270–272. doi: 10.1038/268270a0. [DOI] [PubMed] [Google Scholar]
- 34.Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Preiss T, Hentze M. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 36.Preiss T, Muckenthaler M, Hentze M W. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tacke E, Prufer D, Salamini F, Rohde W. Characterization of a potato leafroll luteovirus subgenomic RNA: differential expression by internal translation initiation and UAG suppression. J Gen Virol. 1990;71:2265–2272. doi: 10.1099/0022-1317-71-10-2265. [DOI] [PubMed] [Google Scholar]
- 37a.Tanguay, R., and D. Gallie. Unpublished data.
- 38.Tarun S Z, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas A A M, Ter Haar E, Wellink J, Voorma H O. Cowpea mosaic virus middle component RNA contains a sequence that allows internal binding of ribosomes and that requires eukaryotic initiation factor 4F for optimal translation. J Virol. 1991;65:2953–2959. doi: 10.1128/jvi.65.6.2953-2959.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ysraeli J K, Melton D A. Synthesis of long, capped transcripts in vitro by SP6 and T7 RNA polymerases. Methods Enzymol. 1989;180:42–50. doi: 10.1016/0076-6879(89)80090-4. [DOI] [PubMed] [Google Scholar]
- 41.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]