Abstract
Objective - Addressing the challenges that come with identifying and delineating brain tumours in intraoperative ultrasound. Our goal is to both qualitatively and quantitatively assess the interobserver variation, amongst experienced neuro-oncological intraoperative ultrasound users (neurosurgeons and neuroradiologists), in detecting and segmenting brain tumours on ultrasound. We then propose that, due to the inherent challenges of this task, annotation by localisation of the entire tumour mass with a bounding box could serve as an ancillary solution to segmentation for clinical training, encompassing margin uncertainty and the curation of large datasets. Methods - 30 ultrasound images of brain lesions in 30 patients were annotated by 4 annotators - 1 neuroradiologist and 3 neurosurgeons. The annotation variation of the 3 neurosurgeons was first measured, and then the annotations of each neurosurgeon were individually compared to the neuroradiologist’s, which served as a reference standard as their segmentations were further refined by cross-reference to the preoperative magnetic resonance imaging (MRI). The following statistical metrics were used: Intersection Over Union (IoU), Sørensen-Dice Similarity Coefficient (DSC) and Hausdorff Distance (HD). These annotations were then converted into bounding boxes for the same evaluation. Results - There was a moderate level of interobserver variance between the neurosurgeons and a larger level of variance when compared against the MRI-informed reference standard annotations by the neuroradiologist, mean across annotators . After converting the segments to bounding boxes, all metrics improve, most significantly, the interquartile range drops by . Conclusion - This study highlights the current challenges with detecting and defining tumour boundaries in neuro-oncological intraoperative brain ultrasound. We then show that bounding box annotation could serve as a useful complementary approach for both clinical and technical reasons.
Keywords: Intraoperative ultrasound, Tumour margin uncertainty, Segmentation
Introduction
Maximal-safe resection of brain tumours is a key pillar of modern neuro-oncology management, improving symptoms, quality of life and overall survival [11]. Accurate delineation of a lesion from surrounding normal functional brain tissue remains challenging but is critical to ensure optimal resection. Ultrasound (US) has been employed intraoperatively (intraoperative US iUS) in neurosurgery for 70 years [9]. Applied for surgical guidance, as it allows real-time detection, characterisation and outlining of tumours. Unlike Magnetic resonance imaging (MRI) - specifically intraoperative MRI (iMRI) - it integrates easily into the surgical workflow and is relatively affordable. With the availability of high-quality US machines and navigated US co-registered with preoperative MRI and computed tomography (CT) [5, 10, 13, 18, 19], its adoption has increased over the last two decades.
However, factors associated with the quality of the modality as well as standardisation of training has effected universal adoption [4]. There remains a perceived steep learning curve secondary to limited fields of view with unfamiliar topographical representation, artefacts, the unique visuo-tactile task and the difficulty with gaining experience outside of the intraoperative setting. There are also concerns regarding the imaging accuracy and granularity. These factors can make learning iUS difficult, with the potential to impair tumour and tumour boundary detection with the risk of leaving unintended residuum or causing inadvertent damage. This challenge is further compounded by the inherent great variation in types of brain lesions, their appearances, the degree of infiltration and the intraoperative changes (such as oedema and haemorrhage) which can further confound.
Presently, there have been only a few meta-analyses looking at the impact of iUS in glioma surgery. One pooled series reported an average 77% gross total resection rate in 739 patients undergoing iUS-guided resection (71.9% in HGG compared to 78.1% in LGG) which was comparable to other forms of navigation. A recent meta-analysis of 409 diffuse gliomas compared the accuracy of iUS to the reference standard post-operative MRI. They found that iUS was an effective technique in assessing diffuse glioma resection, with a 72.2% pooled sensitivity and a 93.5% pooled specificity [30]. Whilst these results are encouraging the current evidence supports a need to improve iUS accuracy if it is to become part of the standard of care in brain tumour surgery. Trials assessing the role of US in neurosurgery, such as the randomized controlled trial Functional and Ultrasound-Guided Resection of Glioblastoma (FUTURE-GB), and refining of US techniques are therefore needed [23].
There are three aims of this study. Firstly, to assess whether tumour boundary detection on iUS is challenging, we measure interobserver variation between regular iUS operators in segmenting US images of brain lesions. Secondly, we model the pixel intensities of the segmented tumour boundaries to mathematically model the clarity and blurriness - factors relating to issues of low resolution and low signal-to-noise ratio - of tumour boundaries. Finally, we evaluate the interobserver variation of bounding boxes to assess whether this annotation method has a role as an alternative, complimentary simplified method for outlining lesion margins. Additional evaluation is performed to determine if these bounding boxes can be used as a guide to improve the accuracy of the segmentation.
To improve the utility of iUS in neurosurgery, the understanding of the limitations of tumour-margins delineation capabilities needs to be understood and measured. With standardised training, new supporting techniques as well as the development of new tools required to reduce uncertainty and error. By formally addressing the issue of tumour segmentation error and uncertainty, we hope to highlight a fundamental challenge with iUS that has hindered universal adoption. Until now, this issue has largely been an implicit challenge recognised by experienced iUS operators.
Materials and methods
A preliminary study was conducted using 4 annotators (AN) experienced with iUS - a neuroradiologist and 3 neurosurgeons - to determine the foundation of our hypothesis. The neuroradiologist and the three neurosurgeons are all post-training doctors. All clinicians involved have extensive research backgrounds and familiarity with segmentation tools and protocols. Specifically: Neuroradiologist - 10 years of US and neuroimaging experience; An1 - 10 years of neuro-oncology experience with 10 years of 2-3 cases per week using iUS; An2 - 8 years of neuro-oncology experience with 6 years of 1 case per week using iUS; An3 - 9 years of neuro-oncology experience with 9 years of 2 cases per week using iUS. The order of the annotators has been randomised.
Data
The dataset consists of 30 images, from 30 patients, taken during brain surgery at Imperial College NHS Trust London. The images were retrospectively selected by the neuroradiologist from cine clips of US sweeps that captured the entire tumour. Images with a field of view which covered the boundaries of the tumour and at least 2cm of surrounding normal brain were selected with reference to the preoperative MRI (Magnetom Vera 3T, Siemens) to ensure accuracy. All images were captured before tumour resection. For patient information please see Table 1. Images were acquired using a Canon i900 US machine (Canon Medical Systems, Japan) with the 8MHz i8MCX1 microconvex probe, and were stored using the Digital Imaging and Communications in Medicine (DICOM) format. For all images, the dynamic range was fixed at 70dB. The aperture was set to either 1 or 3. The power level was set to either 5 or 7. The gain varied between 79 to 98. The maximum depth was varied between 5cm to 11cm, with the depth focus between 1.8cm to 8.1cm. All images are of size .
Table 1.
Patient information. KPS = Karnofsky Performance Status, ASA = American Society of Anesthesiologists Grade, SCC = Squamous Cell Carcinoma, SS = Supratentorial Subependymoma, DNET = Dysembryoplastic Neuroepithelial Tumor, TB = Tuberculosis, Lt = Left, Rt = Right
| ID | G | Age | Lesion | Location | Histology | KPS | ASA |
|---|---|---|---|---|---|---|---|
| 1 | F | 60-69 | Metastasis | Rt Parietal | SCC - lung | 80 | 3 |
| 2 | M | 30-39 | Glioma | Rt Frontal | Astrocytoma | 90 | 1 |
| 3 | M | 50-59 | Glioma | Rt Parietal | Glioblastoma | 80 | 2 |
| 4 | F | 40-49 | Glioma | Lt Frontal, Parietal | Astrocytoma | 90 | 2 |
| 5 | M | 20-29 | Glioma | Lt Parietal | Astrocytoma | 80 | 2 |
| 6 | M | 60-69 | Glioma | Lt Parietal | Glioblastoma | 70 | 2 |
| 7 | M | 50-59 | Glioma | Lt Frontal | Oligodendroglioma | 90 | 3 |
| 8 | M | 50-59 | Glioma | Rt Frontal | Glioblastoma | 90 | 2 |
| 9 | M | 60-69 | Glioma | Lt Parietal | Glioblastoma | 90 | 3 |
| 10 | M | 50-59 | Glioma | Rt Frontal | Glioblastoma | 90 | 2 |
| 11 | M | 40-49 | Glioma | Lt Parietal | Glioblastoma | 80 | 2 |
| 12 | M | 60-69 | Glioma | Rt Parietal | Glioblastoma | 70 | 3E |
| 13 | M | 70-79 | Glioma | Rt Frontal | Glioblastoma | 70 | 2 |
| 14 | M | 10-19 | Granuloma | Lt Temporal | Granuloma TB | 100 | 1 |
| 15 | F | 60-69 | Glioma | Rt Parietal | Glioblastoma | 70 | 2 |
| 16 | M | 60-69 | Glioma | Rt Parietal | Glioblastoma | 80 | 2 |
| 17 | M | 40-49 | Glioma | Lt Temporal | Glioblastoma | 100 | 2 |
| 18 | F | 60-69 | Glioma | Lt Temporal | Glioblastoma | 70 | 2 |
| 19 | M | 40-49 | Glioma | Lt Frontal | Glioblastoma | 80 | 2 |
| 20 | F | 50-59 | Ependymal | Lt Temporal | SS | 90 | 2 |
| 21 | M | 30-39 | Glioma | Post Fossa | Pilocytic astrocytoma | 90 | 1 |
| 22 | M | 60-69 | Glioma | Rt Parietal, Occipital | Glioblastoma | 70 | 2 |
| 23 | F | 60-69 | Glioma | Lt Frontal | Oligodendroglioma | 90 | 2 |
| 24 | M | 30-39 | Glioma | Lt Parietal | Glioblastoma | 90 | 1 |
| 25 | M | 70-79 | Glioma | Rt Parietal | Oligodendroglioma | 90 | 2 |
| 26 | F | 20-29 | Glioma | Rt Frontal | Astrocytoma | 90 | 2 |
| 27 | M | 40-49 | Glioma | Rt Frontal | Astrocytoma | 90 | 2 |
| 28 | F | 10-19 | Glioneuronal | Lt Frontal | DNET | 90 | 1 |
| 29 | F | 40-49 | Glioma | Lt Temporal, Occipital | Oligodendroglioma | 90 | 2 |
| 30 | F | 30-39 | Glioma | Lt Parietal | Astrocytoma | 90 | 2 |
The study had full local ethical approval by the HRA and Health and Care Research Wales (HCRW) authorities. Study title - US-CNS: Multiparametric Advanced Ultrasound Imaging of the Central Nervous System Intraoperatively and Through Gaps in the Bone, IRAS project ID: 275556, Protocol number: 22CX7609, REC reference: 22/WA/0259, Sponsor: Research Governance and Integrity Team (RGIT).
Annotation protocol
The boundaries of tumours were annotated, for both segmentations and bounding boxes, using 3D Slicer (5.4.0) [8]. The annotations provided by the neuroradiologist were made using the benefit of the full US dataset, cross-registration with the preoperative MRI and patient metadata; and we define these as the reference standard/ground truth for this study. The other annotators were given only the individual 2D US images - these annotations were used to evaluate the consistency and accuracy of the annotation capability, comparing inter-neurosurgeon variation and dissimilarity against the annotations of the neuroradiologist. Whilst this is unlike normal clinical practice (where the MRI and full real-time US would be available and employed by the operator) the aim of this study is to assess the ability of B-mode US to delineate brain lesion margins in isolation. The utility of bounding boxes as a guide to refine tumour boundary segmentation was then assessed by An1 repeating their segmentations 3 months later (to mitigate bias) with the reference standard bounding boxes produced by the neuroradiologist overlaid. The bounding box annotations are created by taking the maximum and minimum, x and y coordinates from the annotators’ segments, fitting a box around the outer limits of the segment - as opposed to re-annotating.
Statistical analysis
Three statistical metrics were used to compare the similarity between annotations. Within the mathematical framework, let A and B represent two annotations from the same image i.e. A could be the neuroradiologist’s annotation and B, one of the neurosurgeon’s annotations. The first two metrics that we define are methods to quantify the degree of overlap - indirectly the degree of similarity in the shape and volume - between A and B. The intersection over union (IoU) [26] , which is defined as the set of image coordinates occupied in the intersection of the two segmentations, divided by the union of the two segmentations, the total set of image coordinates occupied by both segmentations. The Sørensen-Dice Similarity Coefficient (DSC) [12] , which is defined as twice the intersection of the two segmentations divided by the cardinalities of the two segmentations. For both IoU and DSC, the closer the value to 1 the better the score.
For evaluation of the uncertainty of the boundary delineation, the contours of the segmentations were compared using the Hausdorff Distance (HD) [14] , , which determines the maximum Euclidean distance (pixels) between all closest point pairs between the two contours sets - the subscript c denotes the contours of the corresponding segmentations, where the contours are the lines intersecting the endpoints of the segmentations. For this metric, the closer the value is to 0 the better.
This analysis was performed using Python, pynrrd [7] for reading the segmentation files and scikit-image [33] and SciPy [34] for the quantitative analysis.
Tumour border pixel dispersion substudy
To assess the visible distinguishability of the tumour from the surrounding normal tissue, the dispersion of pixel intensities around the neuroradiologist’s tumour margins are evaluated. By measuring the pixels along the segmentation’s contour plus a 10 pixels border, perpendicular to, and on both sides of the contour. To calculate the properties of the distributions, a local maxima peak-finding method is implemented. Where one peak = unimodal, two = bimodal and three or more = other (could be either multimodal or uniform).
Results
Inter-Neurosurgeon annotation variance
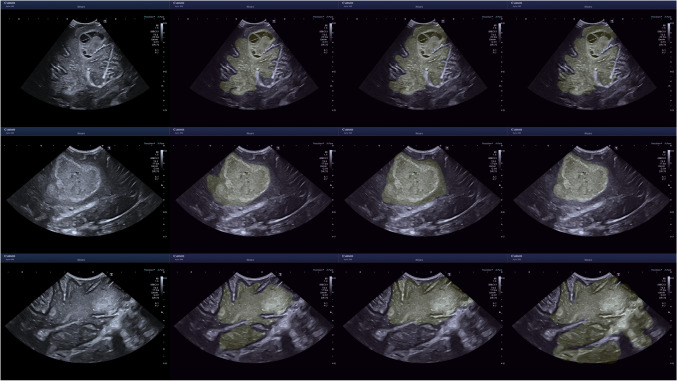
All results are presented in the following format - Metric[Mean:interquartile range (IQR)]. The similarity measure between the neurosurgeons’ annotations is as follows - IoU[0.789:0.115], DSC[0.876:0.072], HD[103.227:73.071]. Although the overlap scores indicate general similarity between the annotations, there are still inconsistencies, which when considering the precision required for tumour resection, this marginal difference can be considered impactful. The average HD, on the other hand, is a significant result. Showing that there is frequent disagreement on at least one point along the tumour boundary. Figure 4 highlights 3 examples of the annotation variance. All figures show cases with minor and severe variance, to visually highlight the features and general causes of uncertainty.
Fig. 4.
Inter neurosurgeon annotation variance. From top to bottom - ID 14, ID 16, ID 18. ID is the image index
Tumour border pixel dispersion substudy
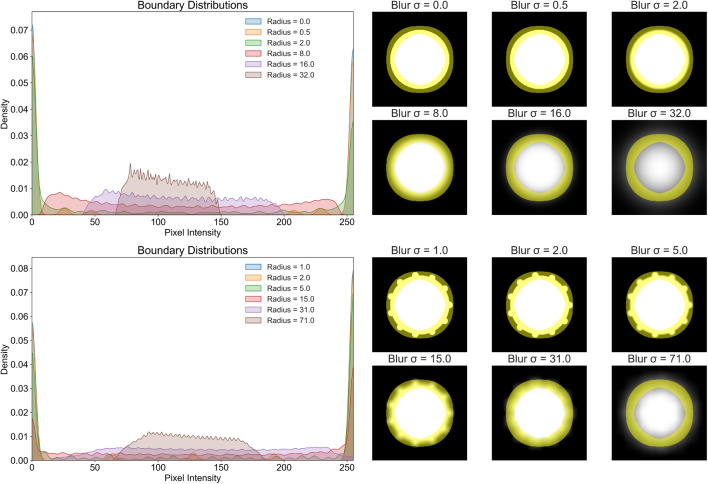
First, we provide a simplified example to illustrate how smoothing affects pixel distributions along a binary boundary (perfect separability). In Fig. 1 a binary circle is created and both Gaussian and box filters are applied with increasing amplification. For the case of the binary circle without smoothing, the boundary is perfectly defined and as such the pixel distribution is bimodal. However, as the smoothing factor increases, the distribution tends towards uniformity or multimodal and under severe blurring, becomes unimodal.
Fig. 1.
The top row/example shows the effect on the boundary of a binary circle when Gaussian blurring is applied - using a kernel density estimate plot. The bottom row/example shows the same but using a box filter, where the circle also now has protrusions, unaffecting the defined boundary represented by the yellow mask
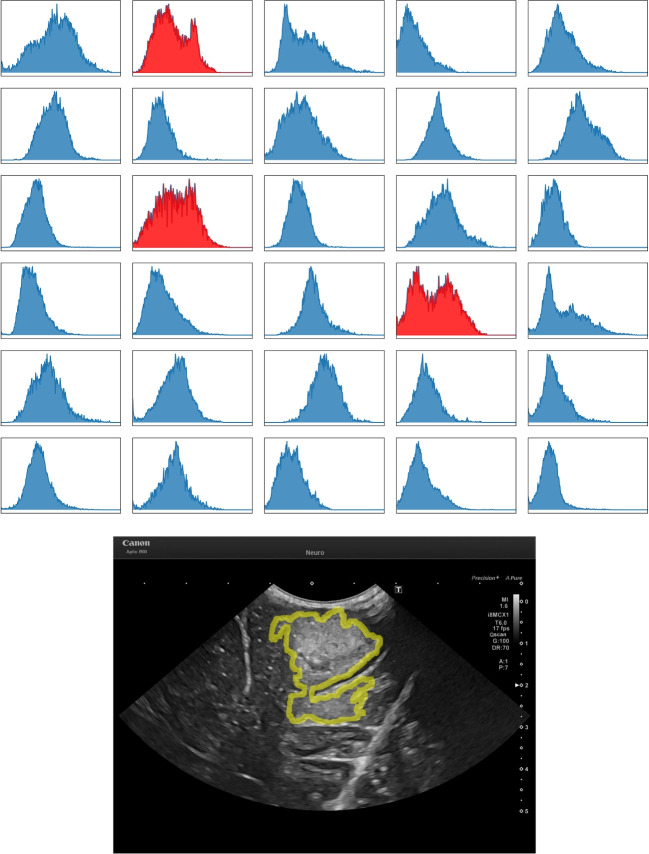
From evaluating the tumour boundaries, from the 30 images, 27 were classified as unimodal and 3 as other Fig. 2. This result is strong evidence of the severity of the pixel intensity variation along the tumour boundary, which provides mathematical evidence for the ambiguity in defining the discrete, definite boundary points.
Fig. 2.
Top is a plot of all tumour boundary pixel intensities distributions. Blue highlights unimodal distributions and red highlights other. Bottom is an example of tumour boundary is shown using ID 009. ID is the image index
Neuroradiologist-Neurosurgeon annotation variance
The results of the similarity comparison between the reference standard annotations produced by the neuroradiologist (which benefited from correlation with the preoperative MRI and the full US dataset) and the neurosurgeons is tabulated in Tables 2 and 3, and visualised using a box and whiskers plot in Fig. 3. This showed a moderate interobserver variance between the reference standard segmentations’ and the annotations performed by the neurosurgeon on the single slice B-mode images alone, highlighting the potential limitations and uncertainties of isolated B-mode in defining tumour boundaries.
Table 2.
Shown are the IoU and DSC similarity results. The closer the value to 1 the more similar the annotations are. IoU is Intersection Over Union, DSC is Sørensen-Dice Similarity Coefficient
| Image | An1 | An2 | An3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Seg | BBox | Seg | BBox | Seg | BBox | |||||||
| IoU | DSC | IoU | DSC | IoU | DSC | IoU | DSC | IoU | DSC | IoU | DSC | |
| 1 | 0.952 | 0.976 | 0.954 | 0.976 | 0.875 | 0.933 | 0.862 | 0.926 | 0.941 | 0.970 | 0.971 | 0.985 |
| 2 | 0.908 | 0.952 | 0.962 | 0.981 | 0.911 | 0.954 | 0.957 | 0.978 | 0.880 | 0.936 | 0.847 | 0.917 |
| 3 | 0.888 | 0.941 | 0.907 | 0.951 | 0.906 | 0.951 | 0.914 | 0.955 | 0.891 | 0.943 | 0.955 | 0.977 |
| 4 | 0.902 | 0.948 | 0.926 | 0.962 | 0.848 | 0.918 | 0.886 | 0.940 | 0.389 | 0.560 | 0.230 | 0.374 |
| 5 | 0.905 | 0.950 | 0.940 | 0.969 | 0.856 | 0.923 | 0.912 | 0.954 | 0.893 | 0.943 | 0.856 | 0.922 |
| 6 | 0.570 | 0.726 | 0.536 | 0.698 | 0.811 | 0.895 | 0.854 | 0.921 | 0.876 | 0.934 | 0.893 | 0.943 |
| 7 | 0.578 | 0.733 | 0.734 | 0.846 | 0.781 | 0.877 | 0.916 | 0.956 | 0.819 | 0.901 | 0.940 | 0.969 |
| 8 | 0.889 | 0.941 | 0.863 | 0.927 | 0.957 | 0.978 | 0.954 | 0.977 | 0.965 | 0.982 | 0.971 | 0.985 |
| 9 | 0.591 | 0.743 | 0.830 | 0.907 | 0.693 | 0.819 | 0.746 | 0.854 | 0.679 | 0.809 | 0.724 | 0.840 |
| 10 | 0.902 | 0.948 | 0.960 | 0.980 | 0.788 | 0.881 | 0.733 | 0.846 | 0.913 | 0.955 | 0.943 | 0.971 |
| 11 | 0.644 | 0.783 | 0.581 | 0.735 | 0.800 | 0.889 | 0.919 | 0.958 | 0.670 | 0.803 | 0.777 | 0.875 |
| 12 | 0.833 | 0.909 | 0.776 | 0.874 | 0.872 | 0.932 | 0.887 | 0.940 | 0.923 | 0.960 | 0.976 | 0.988 |
| 13 | 0.823 | 0.903 | 0.816 | 0.899 | 0.809 | 0.895 | 0.843 | 0.915 | 0.865 | 0.928 | 0.870 | 0.931 |
| 14 | 0.265 | 0.419 | 0.217 | 0.357 | 0.219 | 0.359 | 0.199 | 0.332 | 0.206 | 0.342 | 0.199 | 0.333 |
| 15 | 0.130 | 0.230 | 0.148 | 0.257 | 0.125 | 0.221 | 0.159 | 0.275 | 0.129 | 0.228 | 0.143 | 0.251 |
| 16 | 0.885 | 0.939 | 0.903 | 0.949 | 0.809 | 0.894 | 0.855 | 0.922 | 0.802 | 0.890 | 0.763 | 0.865 |
| 17 | 0.706 | 0.828 | 0.702 | 0.825 | 0.548 | 0.708 | 0.649 | 0.787 | 0.616 | 0.762 | 0.780 | 0.876 |
| 18 | 0.168 | 0.287 | 0.139 | 0.245 | 0.167 | 0.286 | 0.172 | 0.293 | 0.083 | 0.154 | 0.078 | 0.145 |
| 19 | 0.901 | 0.948 | 0.875 | 0.933 | 0.790 | 0.883 | 0.805 | 0.892 | 0.835 | 0.910 | 0.616 | 0.762 |
| 20 | 0.956 | 0.977 | 0.943 | 0.971 | 0.870 | 0.930 | 0.819 | 0.901 | 0.839 | 0.912 | 0.747 | 0.855 |
| 21 | 0.873 | 0.932 | 0.902 | 0.948 | 0.452 | 0.623 | 0.475 | 0.644 | 0.853 | 0.920 | 0.881 | 0.937 |
| 22 | 0.849 | 0.918 | 0.850 | 0.919 | 0.854 | 0.921 | 0.888 | 0.940 | 0.876 | 0.934 | 0.877 | 0.934 |
| 23 | 0.402 | 0.573 | 0.208 | 0.345 | 0.624 | 0.769 | 0.746 | 0.854 | 0.534 | 0.696 | 0.233 | 0.378 |
| 24 | 0.743 | 0.853 | 0.785 | 0.880 | 0.632 | 0.775 | 0.723 | 0.839 | 0.694 | 0.820 | 0.740 | 0.850 |
| 25 | 0.838 | 0.912 | 0.883 | 0.938 | 0.904 | 0.950 | 0.919 | 0.958 | 0.751 | 0.858 | 0.744 | 0.853 |
| 26 | 0.619 | 0.765 | 0.773 | 0.872 | 0.561 | 0.718 | 0.698 | 0.822 | 0.572 | 0.728 | 0.707 | 0.829 |
| 27 | 0.873 | 0.932 | 0.857 | 0.923 | 0.813 | 0.897 | 0.764 | 0.866 | 0.804 | 0.891 | 0.836 | 0.911 |
| 28 | 0.803 | 0.891 | 0.874 | 0.933 | 0.773 | 0.872 | 0.832 | 0.908 | 0.756 | 0.861 | 0.908 | 0.952 |
| 29 | 0.791 | 0.883 | 0.867 | 0.929 | 0.620 | 0.766 | 0.754 | 0.859 | 0.622 | 0.767 | 0.807 | 0.893 |
| 30 | 0.821 | 0.902 | 0.810 | 0.895 | 0.878 | 0.935 | 0.816 | 0.899 | 0.832 | 0.908 | 0.879 | 0.936 |
| Mean | 0.734 | 0.821 | 0.751 | 0.827 | 0.719 | 0.812 | 0.755 | 0.837 | 0.717 | 0.807 | 0.730 | 0.808 |
Table 3.
Shown are the HD similarity results. The closer the value to 0 pixels the better aligned the annotation margins are. HD is Hausdorff Distance
| Image | An1 | An2 | An3 | |||
|---|---|---|---|---|---|---|
| Seg | BBox | Seg | BBox | Seg | BBox | |
| 1 | 14.765 | 8.544 | 43.000 | 36.620 | 14.142 | 5.000 |
| 2 | 22.204 | 4.123 | 25.000 | 5.000 | 52.802 | 46.271 |
| 3 | 33.734 | 16.492 | 32.558 | 14.142 | 23.324 | 9.220 |
| 4 | 29.069 | 8.246 | 42.438 | 33.061 | 371.389 | 379.120 |
| 5 | 51.614 | 13.153 | 57.245 | 19.416 | 40.162 | 23.345 |
| 6 | 162.926 | 163.515 | 56.223 | 35.511 | 52.924 | 17.804 |
| 7 | 74.632 | 40.311 | 32.650 | 7.280 | 42.579 | 6.000 |
| 8 | 52.000 | 48.010 | 33.121 | 7.616 | 28.636 | 6.403 |
| 9 | 137.773 | 49.477 | 74.686 | 47.265 | 79.202 | 48.836 |
| 10 | 25.807 | 4.243 | 55.803 | 44.598 | 18.682 | 6.403 |
| 11 | 117.652 | 119.620 | 362.627 | 15.000 | 138.105 | 69.231 |
| 12 | 66.000 | 70.000 | 68.447 | 47.170 | 43.600 | 7.071 |
| 13 | 46.174 | 28.844 | 62.000 | 30.000 | 40.522 | 23.324 |
| 14 | 331.724 | 341.264 | 329.524 | 336.265 | 331.965 | 345.307 |
| 15 | 263.610 | 294.703 | 218.563 | 247.746 | 254.342 | 288.043 |
| 16 | 36.770 | 36.000 | 81.006 | 44.000 | 90.554 | 68.710 |
| 17 | 309.015 | 110.000 | 330.510 | 102.000 | 377.922 | 62.169 |
| 18 | 429.439 | 419.640 | 323.303 | 328.056 | 404.901 | 524.675 |
| 19 | 27.295 | 23.000 | 58.052 | 36.401 | 419.715 | 112.058 |
| 20 | 30.887 | 23.000 | 64.938 | 58.694 | 87.727 | 84.000 |
| 21 | 42.012 | 15.000 | 119.549 | 111.973 | 33.377 | 14.866 |
| 22 | 68.447 | 28.425 | 70.264 | 25.000 | 43.600 | 24.000 |
| 23 | 415.473 | 290.493 | 57.079 | 44.721 | 415.120 | 302.154 |
| 24 | 71.007 | 48.703 | 89.022 | 63.789 | 80.000 | 53.488 |
| 25 | 56.511 | 20.809 | 22.627 | 14.213 | 63.600 | 35.468 |
| 26 | 156.259 | 45.188 | 171.047 | 70.767 | 175.026 | 57.271 |
| 27 | 15.000 | 10.440 | 24.000 | 24.413 | 29.547 | 21.024 |
| 28 | 34.670 | 30.067 | 36.056 | 30.017 | 35.228 | 13.601 |
| 29 | 53.254 | 38.949 | 180.049 | 79.649 | 171.234 | 54.406 |
| 30 | 67.231 | 40.792 | 26.926 | 27.731 | 59.641 | 24.739 |
| Mean | 108.098 | 79.702 | 104.94 | 66.270 | 133.986 | 91.134 |
Fig. 3.
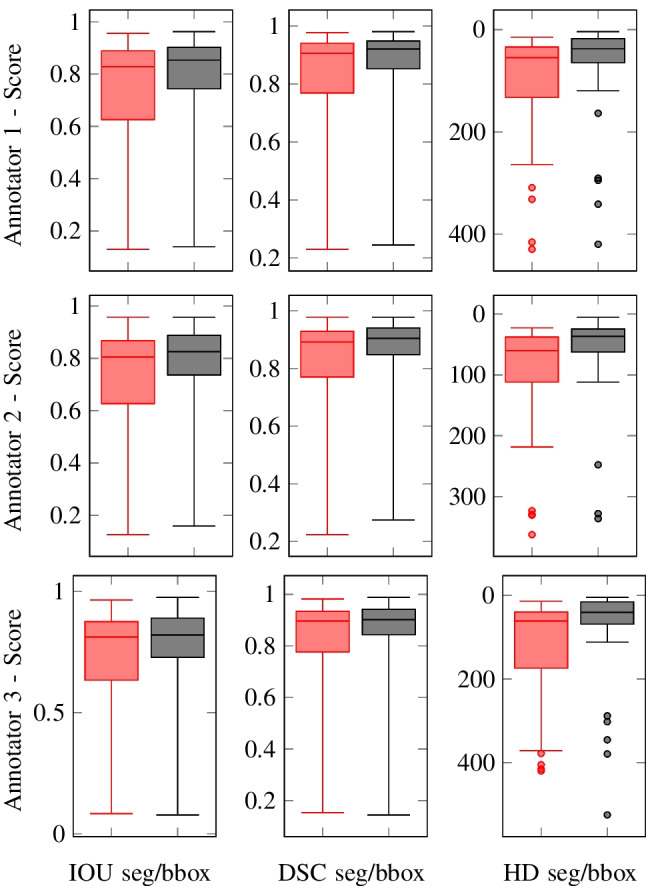
Box and Whisker plots of the annotators IoU, DSC and HD scores on the 30 images - red = segmentation, black = bounding box. What is highlighted is the greatly reduced median score and IQR when using the bounding box method. IoU is Intersection Over Union, DSC is Sørensen-Dice Similarity Coefficient, HD is Hausdorff Distance and IQR is Interquartile Range
With conversion of the segments to bounding boxes there was a noticeable improvement in annotation similarity with a sizable decrease in the IQR. For all annotators, the conversion to a bounding box, overall, increases the annotation similarity. The decrease in the IQR, as a percentage, for [IoU, DSC, HD] are as follows - An1=[40%, 44%, 52%], An2=[37%, 42%, 49%], An3=[33%, 37%, 60%].
Further evaluation of the bounding box proposal is conducted by measuring individually for each image, the percentage of the neuroradiologist’s segmentation contained within the neurosurgeon’s corresponding bounding box. The average results per annotator are - An1:98.387%, An2:98.833%, An3:99.052%. From the results, it can be concluded that the bounding box approach is usable for localising the entire tumour mass, and (Fig. 4) is a suitable method for reducing inter-observer annotation variance.
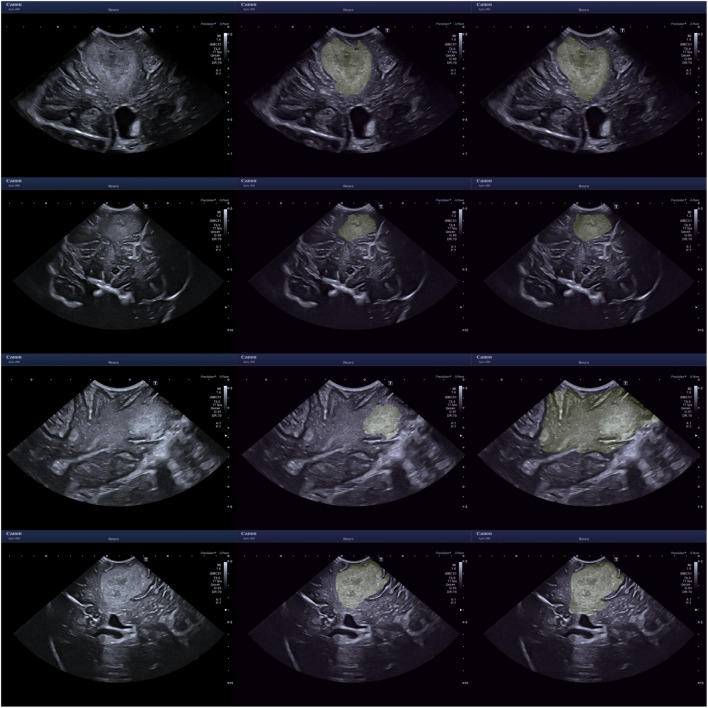
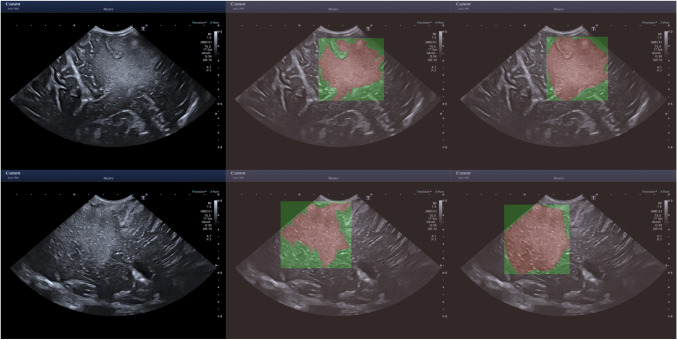
Sample images are displayed in Figs. 5 and 6. An assortment of different sources of boundary aleatoric uncertainty are shown in Fig. 5. Including images containing fuzzy borders and continued hyperechogenicity extending beyond the tumour. Shown in Fig. 6 are example cases where using bounding boxes has substantially improved the annotation similarity.
Fig. 5.
Uncertainty caused by unclear boundary. The left images are the original images, the middle from the neuroradiologist and the right from a neurosurgeon. From top to bottom - ID 13 An3, ID 07 An3, ID 18 An2, ID 19 An2. ID is the image index
Fig. 6.
Examples for where the segments have large dissimilarity whilst the bounding boxes don’t. The left images are the original images, the middle from the neuroradiologist and the right from a neurosurgeon. From top to bottom - ID 11 AN 2, the bottom from ID 26 AN 1. ID is the image index
Improving segmentation accuracy using overlaid bounding boxes as a guide
In the substudy looking at the impact of overlaid reference bounding boxes on An1 segmentation accuracy, we found a substantial improvement in segmentation similarity between the neurosurgeon and reference standard. The previous results compared to the new results are as follows - without bounding box [IoU:0.734, DSC:0.821, HD:108.098], with bounding box [IoU:0.858, DSC:0.922, HD:52.072]. This result shows that the bounding box can be used as a visual anchor to minimise the uncertainty when segmenting.
Discussion
Historical background and clinical landscape
Several intraoperative imaging devices have been developed to help neurosurgeons localizing the tumour during surgery. Aside from CT scan and MRI scans, introduced in clinical practice during the ’70s and in the ’80s, neuro-navigation frameless devices were introduced during the ’90s, and they were the first tools allowing intraoperative localization of the tumour [6] , although they rely on pre-operative images rather than real time acquisition. The challenging problem of having a real time tool for tumour location and visualization has been partially sorted by the gradual introduction of iUS in the armoury of neurosurgical equipment [5] . Interestingly, in several cases the signal obtained from iUS was different when compared to that obtained using traditional imaging technologies (CT or MRI scan), thus sparking attention on the fact that the iUS could be used not only as an intraoperative aid, but also as a complementary tool for those lesions of unclear nature or margins [5] . Moreover, coupling of the iUS with MRI based neuro-navigation has greatly improved the possibility of midline shift adjustment during surgery [16, 17, 19] and has been proven useful to improve tumour demarcation when close to eloquent areas [25] . In more recent years, further technological advancement has led to the development of 3D iUS [2, 28, 31, 32] . The 3D reconstruction is automatically generated by the neuro-navigation software after an intraoperative acquisition through a single spatial plane. The images can be integrated with doppler angiography when required, so that vessel encasement by an intra-cranial mass or aneurysm can also be detected [24, 29] . Recent research has also been focusing on the possibility of integrating iUS with contrast [1, 36].
Findings, challenges, and future perspectives
In this study, we highlight the specific challenge of tumour detection and tumour boundary delineation in cranial iUS. Here we demonstrate that there remains moderate to high interobserver variation in the identification and segmentation of tumours on B-mode images acquired on a modern, current-generation US scanner, between four individuals with experience in iUS-guided brain surgery. Three broad elements likely contribute to this variance, 1) the specific qualities of brain tumours, 2) technical ultrasound factors and 3) operator influences.
Firstly, there are inherent features of brain tumours themselves which can make them difficult to delineate. Gliomas, in particular, are well-known to be infiltrative tumours, meaning that they spread cancer cells beyond their obvious radiological margins [20, 27]. Moreover, both high-grade gliomas and low-grade gliomas often show a degree of surrounding oedema which is, to this day, challenging to interpret in terms of differential diagnosis between reactive inflammatory tissue or actually infiltrated brain, even using more established neuroimaging tools such as MRI. Moreover, the intraoperative imaging quality varies based on the stage of surgical resection [18, 32] . Superficial, small sized lesions are typically very well visualized by the acquisition and during resection. However, even modest to moderate amount of bleeding causes a visible artifact that can hamper surgical view beyond the limits of resection. To detect residual tumour, it is crucial to perform accurate haemostasis and remove all haemostatic material to the surgical cavity.
One of the main points of discrepancy in the present series was the definition of tumour margins. For example, in some cases, the lack of clarity over whether the highlighted hyperechogenicity of the tissue was caused by a continuation of the tumour mass or just reactive oedema, introduces significant heteroscedastic aleatoric uncertainty when defining the exact margin. This challenge is further emphasised by how there remains no reference standard imaging technique that absolutely defines tumour extent, nor is it usually possible to remove tumours en bloc in intra-axial neurosurgery, precluding accurate histological correlation of tumour margins.
Secondly, there are unique challenges that US presents. There are numerous ways that an ultrasound image can be altered, including changes in settings (such as gain and frequency), ultrasound machine, probe type, probe contact and probe angle. In most cases, several of these parameters need to be intentionally tailored to the particular tumour being imaged. For instance, using a low-frequency probe, which has a trade-off in reduced spatial resolution, to visualise a deep tumour versus a high-frequency probe, with high spatial resolution, to image a superficial cortical tumour. This wide range of US options can greatly alter the final image creating another source of variance and in turn, aleatoric uncertainty which is arguably greater than typically seen with other established imaging modalities such as CT and MRI.
US is also vulnerable to several unique artefacts, such as acoustic shadowing [35] and acoustic enhancement, which can alter and obscure the image creating a further source of uncertainty. These issues could be mitigated somewhat by the establishment of a standardised protocol for US settings and image acquisition. However, even then, it would be impossible to fully account for all scenarios due to the wide spectrum of tumours and anatomical locations. The potential for confounding artefacts and uncertainty regarding tumour boundaries further increases as surgery progresses due to increased deformation, oedema and potential obscuring blood products. This uncertainty could also be reduced with operator experience, which links to operator factors which are the final source of variance. Currently, neurosurgical training in US is predominantly experiential based on exposure to live cases in theatres. Whilst this is an essential aspect of learning a new surgical skill this can greatly prolong the learning phase due to a relatively low rate of exposure to the imaging technique as it necessitates an intraoperative setting with a craniotomy window. This is in contrast to CT and MRI, which neurosurgeons are much more comfortable with interpreting, owing, in part, to these being readily available and performed regularly on most patients.
There are several complementary ways that iUS accuracy could be improved and the steep learning curve could be flattened and shortened. This includes the use of advanced multimodal US (including contrast-enhanced US) and navigated US. In addition, dedicated courses employing US phantoms with brain tumour models can greatly help, by providing both applied formal training in US theory in addition to hands-on time with US scanning. In all cases, however, these approaches still require significant time investment and training. In this context, automated, computer-assisted, detection and segmentation of brain tumours on ultrasound would be a desirable and useful additional tool. Automated segmentation of brain tumours on MRI has been well explored with several robust and open source tools now available [21]. In contrast, development in automated segmentation of iUS images is in its infancy with no applications yet available. There are several reasons for this. As illustrated in this study it is challenging to establish a ground truth dataset using manual segmentation due to high interobserver variance - especially as most published data usually contains only individual frames or volumes. Furthermore, unlike MRI there is a paucity of neuro-oncology iUS imaging datasets. This is likely due to the relatively low number of iUS scans acquired, the greater logistical challenges with saving and downloading scans, the potentially large file sizes of video acquisitions, the greater variation of iUS across sites and the need for highly experienced annotators to perform the time-consuming segmentations. Considering these many boundaries, we are far off the realisation of a reliable system to rapidly automatically and accurately segment iUS brain tumour images. To bridge this gap, here we assess the utility of bounding boxes as an additional complementary tool for simplified tumour detection and delineation.
Unsurprisingly, we found much lower interobserver variation when using bounding boxes to define tumour location and margins compared to segmentations. The advantage of our proposed use of bounding boxes is a step in the direction of overcoming the above-mentioned challenges. From a clinical perspective, the bounding box would be useful for training purposes and immediate identification of the tumour mass. While this system is expected to be lower in specificity, the high sensitivity should assist inexperienced surgeons in detecting tumours and providing an area to focus on. Further, the whole signal change (fuzzy margin) would be included in the bounding box, thus making sure that there is no missing tumour from the targeted area. The process of annotation is also much quicker for bounding boxes although this will be annotator-dependent, from our experience, bounding box annotation may take as little as 1/3 of the time of segmentation, reducing the manual labour cost.
In computer vision and AI, segmentation methods will typically define, assume that the segmentation task can be framed as separating an image into sub-regions with defined and complete boundaries. For example, explicitly through the definition of a mathematical optimisation framework, or implicitly by training a neural network on common datasets. Because of this, the task of semantically segmenting brain tumours in US becomes uniquely difficult. From an engineering perspective, there are a large number of benefits to using bounding boxes. First and foremost, the reduced annotation complexity should facilitate the collection of large datasets. Accelerating technical and clinical research into this topic. For technical development, the primary benefit is the reduced complexity of the estimation task, which should lead to highly accurate systems. The bounding boxes can also be used for different tasks such as: representing the segment as a probabilistic heat map [22] to account for the tumour infiltration [20, 27], prompting large/powerful models such as Segment Anything Model (SAM) [15], tracking algorithms [3].
Conclusion
What has been highlighted in this paper is our thoughts on the challenges of segmenting brain tumours in 2D US images, with a preliminary study conducted to corroborate our hypothesis. For future work, larger curated and consensus annotated datasets of iUS brain tumour images and volumes are needed to develop more accurate computer assisted boundary detection tools. This is likely to only be achieved through multi-site collaboration and pooling of data.
There are a few limitations of this study which we hope to be addressed in future work. Firstly, the number of annotated cases was small which is why we have withheld from identifying correlates - such as whether certain tumour types are more accurately segmented. Secondly, only single slices were used for segmentation as opposed to volumes. This is unlike the real-world use of iUS where assessment of boundaries is based on live 3D sweeps of tumours and adjacent anatomy, plus often cross-correlation with preoperative MRI, both of which would help refine the accuracy of segmentations. However, we do comment that even though 3D information may be available, decisions are still biased by what is visible, which invariably would still be a 2D slice. Extending from this, there remains the recurring issue in neuro-oncology of there not being an accepted gold-standard ground truth for tumour boundaries and our use of integrated multimodal MRI and US to create the reference standard segmentations has to be an accepted compromise. Finally, whilst bounding boxes may serve as an efficient method to improve the detection of tumours, this is at the expense of specificity, which is important for the prevention of inadvertent removal of normal, functional brain tissue.
Acknowledgements
This research was supported by the UK Research and Innovation (UKRI) Centre for Doctoral Training in AI for Healthcare (EP/S023283/1), the Royal Society (URFR2 01014]), the NIHR Imperial Biomedical Research Centre, Canon Medical Systems and Brain Tumour Research (BTR) charity.
Author Contributions
- Alistair Weld: Co-first author - Luke Dixon: Co-first author - Giulio Anichini: Co-first author - Neekhil Patel: Data annotation for the study - Amr Nimer: Data annotation for the study - Michael Dyck: Editing and reviewing - Kevin O’Neill: Supervision - Adrian Lim: Supervision - Stamatia Giannarou: Supervision - Sophie Camp: Supervision
Funding
All funding has been declared.
Code Availibility
Code may be made available via the first author on reasonable request.
Availability of data and material
Data may be made available via the first author on reasonable request.
Declarations
Conflicts of interest
There are no conflicts of interest.
Ethics approval
All required ethics approvals were attained and have been declared.
Consent to participate
All patients gave consent and the information presented is anonymised.
Consent for publication
All patients gave consent and the information presented is anonymised.
Approval
This paper has been approved by all authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Comments
Dr. Weld and his colleagues presented an interesting study on a relevant topic and important diagnostic tool what should not be missing in the neurosurgical armamentarium. However, although the advantages of the use of intraoperative ultrasound are obvious, the examinations themselves are often challenging and their quality depends on many different factors such as the anatomical and technical conditions, the sonographic accessibility of the structures and the morphological characteristics of the pathologies, and the skills and experiences of the respective examiners. Furthermore, structured training and standardized workflows are unusual and frequently considerable learning curves must be considered. In the present study, the challenges of intraoperative ultrasound examinations of brain tumors were addressed and potential interobserver variations amongst ultrasound users assessed. Moreover, bounding box annotation as a promising approach was evaluated and proposed as a complementary both clinical and technical useful method. I congratulate the authors on this exciting study which results can contribute to facilitate the application of intraoperative ultrasound and to improve the quality of the examination. Markus Florian Oertel. Zurich, Switzerland.
Alistair Weld, Luke Dixon and Giulio Anichini contributed equally to this work.
References
- 1.Arlt F, Chalopin C, Müns A, Meixensberger J, Lindner D (2016) Intraoperative 3d contrast-enhanced ultrasound (ceus): a prospective study of 50 patients with brain tumours. Acta Neurochirurgica 158:685–694 [DOI] [PubMed] [Google Scholar]
- 2.Bastos DCDA, Juvekar P, Tie Y, Jowkar N, Pieper S, Wells WM, Bi WL, Golby A, Frisken S, Kapur T (2021) Challenges and opportunities of intraoperative 3d ultrasound with neuronavigation in relation to intraoperative mri. Front Oncol 11:656519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cartucho J, Weld A, Tukra S, Xu H, Matsuzaki H, Ishikawa T, Kwon MS, Jang YE, Kim K-J, Lee G, Bai B, Kahrs LA, Boecking L, Allmendinger S, Muller L, Zhang Y, Jin Y, Sophia B, Vasconcelos F, Reiter W, Hajek J, Silva BLB, Buschle LR, Lima E, Vilaça JL, Queirós S, Giannarou S (2023) Surgt challenge: Benchmark of soft-tissue trackers for robotic surgery. Medical Image Anal 91:102985. https://api.semanticscholar.org/CorpusID:257232585 [DOI] [PubMed]
- 4.Dixon L, Lim A, Grech-Sollars M, Nandi D, Camp SJ (2022) Intraoperative ultrasound in brain tumor surgery: A review and implementation guide. Neurosurgical Rev 45:2503–2515. https://api.semanticscholar.org/CorpusID:247781613 [DOI] [PMC free article] [PubMed]
- 5.Dohrmann GJ, Rubin JM (2001) History of intraoperative ultrasound in neurosurgery. Neurosurgery Clinics North America 12(1):155–66 [PubMed] [Google Scholar]
- 6.Enchev Y (2009) Neuronavigation: geneology, reality, and prospects. Neurosurgical Focus 27(3):E11 [DOI] [PubMed] [Google Scholar]
- 7.Everts M, Elliott A, Braun-Jones T, Court R, Johnson H, Norton I, Warner J, Fischer P, Meine H, Ghayoor A, Billah T, Ekström S, Leinweber K, JcNils, van de Gronde J, Brown D (2019) mhe/pynrrd: v0.4.1 released. 10.5281/zenodo.3544486
- 8.Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R (2012) 3d slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30:1323–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.French LA, Wild JJ, Neal D (1950) Detection of cerebral tumors by ultrasonic pulses. pilot studies on postmortem material. Cancer 3(4):705–708 [DOI] [PubMed]
- 10.Hata N, Dohi T, Iseki H, Takakura K (1997) Development of a frameless and armless stereotactic neuronavigation system with ultrasonographic registration. Neurosurgery 41(3):608–13 [DOI] [PubMed] [Google Scholar]
- 11.Hervey-Jumper SL, Berger MS (2016) Maximizing safe resection of low- and high-grade glioma. J Neuro-Oncol 130:269–282. https://api.semanticscholar.org/CorpusID:9403758 [DOI] [PubMed]
- 12.Hofmanninger J, Prayer F, Pan J, Röhrich S, Prosch H, Langs G (2020) Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. European Radiol Exper 4 [DOI] [PMC free article] [PubMed]
- 13.Ji S, Roberts DW, Hartov A, Paulsen KD (2012) Intraoperative patient registration using volumetric true 3d ultrasound without fiducials. Med Phys 39(12):7540–7552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karimi D, Salcudean SE (2019) Reducing the hausdorff distance in medical image segmentation with convolutional neural networks. IEEE Trans Med Imaging 39:499–513 [DOI] [PubMed] [Google Scholar]
- 15.Kirillov A, Mintun E, Ravi N, Mao H, Rolland C, Gustafson L, Xiao T, Whitehead S, Berg AC, Lo W-Y, Dollár P, Girshick RB (2023) Segment anything. arXiv:2304.02643
- 16.Lindner D, Trantakis C, Renner C, Arnold S, Schmitgen A, Schneider J, Meixensberger J (2006) Application of intraoperative 3d ultrasound during navigated tumor resection. min-Minimally Invasive Neurosurgery 49(04):197–202 [DOI] [PubMed]
- 17.Lindseth F, Kaspersen JH, Ommedal S, Langø T, Bang J, Hokland J, Unsgaard G, Nagelhus Hemes TA (2003) Multimodal image fusion in ultrasound-based neuronavigation: improving overview and interpretation by integrating preoperative mri with intraoperative 3d ultrasound. Comput Aided Surgery 8(2):49–69 [DOI] [PubMed]
- 18.Mahboob SO, McPhillips R, Qiu Z, Jiang Y, Meggs C, Schiavone G, Button TW, Desmulliez MPY, Demore C, Cochran S, Eljamel S (2016) Intraoperative ultrasound-guided resection of gliomas: A meta-analysis and review of the literature. World Neurosurgery 92:255–263 [DOI] [PubMed] [Google Scholar]
- 19.Ohue S, Kumon Y, Nagato S, Kohno S, Harada H, Nakagawa K, Kikuchi K, Miki H, Ohnishi T (2010) Evaluation of intraoperative brain shift using an ultrasound-linked navigation system for brain tumor surgery. Neurologia Medico-chirurgica 50(4):291–300 [DOI] [PubMed] [Google Scholar]
- 20.Pallud J, Audureau E, Blonski M, Sanai N, Bauchet L, Fontaine D, Mandonnet E, Dezamis E, Psimaras D, Guyotat J et al (2014) Epileptic seizures in diffuse low-grade gliomas in adults. Brain 137(2):449–462 [DOI] [PubMed] [Google Scholar]
- 21.Pati S, Baid U, Edwards B et.al (2022) Federated learning enables big data for rare cancer boundary detection. Nature Commun 13. https://api.semanticscholar.org/CorpusID:248366603 [DOI] [PMC free article] [PubMed]
- 22.Petsiuk V, Jain R, Manjunatha V, Morariu VI, Mehra A, Ordonez V, Saenko K (2020) Black-box explanation of object detectors via saliency maps. 2021 IEEE/CVF Conference on computer vision and pattern recognition (CVPR), pages 11438–11447
- 23.Plaha P, Camp S, Cook J, McCulloch P, Voets N, Ma R, Taphoorn MJB, Dirven L, Grech-Sollars M, Watts C, Bulbeck H, Jenkinson MD, Williams M, Lim A, Dixon L, Price SJ, Ashkan K, Apostolopoulos V, Barber VS, Taylor A, FUTURE-GB collaborators, Nandi D (2022) Future-gb: functional and ultrasound-guided resection of glioblastoma – a two-stage randomised control trial. BMJ Open 12(11). ISSN 2044-6055. 10.1136/bmjopen-2022-064823. https://bmjopen.bmj.com/content/12/11/e064823 [DOI] [PMC free article] [PubMed]
- 24.Podlesek D, Meyer T, Morgenstern U, Schackert G, Kirsch M (2015) Improved visualization of intracranial vessels with intraoperative coregistration of rotational digital subtraction angiography and intraoperative 3d ultrasound. PLoS One 10(3):e0121345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rasmussen I-A, Lindseth F, Rygh OM, Berntsen EM, Selbekk T, Xu J, Nagelhus Hernes TA, Harg E, Håberg A, Unsgaard G (2007) Functional neuronavigation combined with intraoperative 3d ultrasound: initial experiences during surgical resections close to eloquent brain areas and future directions in automatic brain shift compensation of preoperative data. Acta Neurochirurgica 149:365–378 [DOI] [PubMed] [Google Scholar]
- 26.Rezatofighi SH, Tsoi N, Gwak JY, Sadeghian A, Reid ID, Savarese S (2019) Generalized intersection over union: A metric and a loss for bounding box regression. 2019 IEEE/CVF Conference on computer vision and pattern recognition (CVPR), pages 658–666
- 27.Sahm F, Capper D, Jeibmann A, Habel A, Paulus W, Troost D, Von Deimling A (2012) Addressing diffuse glioma as a systemic brain disease with single-cell analysis. Archives Neurology 69(4):523–526 [DOI] [PubMed] [Google Scholar]
- 28.Saß B, Zivkovic D, Pojskic M, Nimsky C, Bopp MH (2022) Navigated intraoperative 3d ultrasound in glioblastoma surgery: analysis of imaging features and impact on extent of resection. Front Neurosci 16:883584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Šteňo A, Buvala J, Toma D, Jezberová M, Šteňo J (2022) Navigated 3d-ultrasound power doppler and visualization of lenticulostriate arteries during resections of insular gliomas. Brain & Spine 2 [DOI] [PMC free article] [PubMed]
- 30.Trevisi G, Barbone P, Treglia G, Mattoli MV, Mangiola A (2019) Reliability of intraoperative ultrasound in detecting tumor residual after brain diffuse glioma surgery: a systematic review and meta-analysis. Neurosurgical Rev 43:1221–1233. https://api.semanticscholar.org/CorpusID:199551095 [DOI] [PubMed]
- 31.Unsgaard G, Ommedal S, Muller T, Gronningsaeter A, Nagelhus Hernes TA (2002) Neuronavigation by intraoperative three-dimensional ultrasound: initial experience during brain tumor resection. Neurosurgery 50(4):804–812 [DOI] [PubMed]
- 32.Unsgård G, Lindseth F (2019) 3d ultrasound-guided resection of low-grade gliomas: principles and clinical examples. Neurosurgical Focus 47(6):E9 [DOI] [PubMed]
- 33.Van Der Walt S, Schönberger JL, Nunez-Iglesias J, Boulogne F, Warner JD, Yager N, Gouillart E, Yu T, the scikit-image contributors (2014) scikit-image: image processing in Python. PeerJ, 2:e453, 6 ISSN 2167-8359. 10.7717/peerj.453 [DOI] [PMC free article] [PubMed]
- 34.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat İ, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, SciPy 1.0 Contributors, (2020) SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nature Methods 17:261–272. 10.1038/s41592-019-0686-2 [DOI] [PMC free article] [PubMed]
- 35.Weld A, Dixon L, Anichini G, Dyck M, Ranne A, Camp SJ, Giannarou S (2023) Identifying visible tissue in intraoperative ultrasound images during brain surgery: A method and application. arXiv:2306.01190. https://api.semanticscholar.org/CorpusID:259064174
- 36.Zhang X, Shi Z, Xie Y, Wang Y, Shen C, Qi Z, Zhang L, Yang B, Yu J, Ding H (2023) Quantitative analysis using intraoperative contrast-enhanced ultrasound in adult-type diffuse gliomas with isocitrate dehydrogenase mutations: association between hemodynamics and molecular features. Ultrasonography 42(4):561 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Code may be made available via the first author on reasonable request.
Data may be made available via the first author on reasonable request.