Abstract
Mouse hepatitis virus (MHV) RNA transcription is regulated mainly by the leader and intergenic (IG) sequences. However, a previous study has shown that the 3′ untranslated region (3′-UTR) of the viral genome is also required for subgenomic mRNA transcription; deletion of nucleotides (nt) 270 to 305 from the 3′-UTR completely abolished subgenomic mRNA transcription without affecting minus-strand RNA synthesis (Y.-J. Lin, X. Zhang, R.-C. Wu, and M. M. C. Lai, J. Virol. 70:7236–7240, 1996), suggesting that the 3′-UTR affects positive-strand RNA synthesis. In this study, by UV-cross-linking experiments, we found that several cellular proteins bind specifically to the minus-strand 350 nucleotides complementary to the 3′-UTR of the viral genome. The major protein species, p55, was identified as the polypyrimidine tract-binding protein (PTB, also known as heterogeneous nuclear RNP I) by immunoprecipitation of the UV-cross-linked protein and binding of the recombinant PTB. A strong PTB-binding site was mapped to nt 53 to 149, and another weak binding site was mapped to nt 270 to 307 on the complementary strand of the 3′-UTR (c3′-UTR). Partial substitutions of the PTB-binding nucleotides reduced PTB binding in vitro. Furthermore, defective interfering (DI) RNAs harboring these mutations showed a substantially reduced ability to synthesize subgenomic mRNA. By enzymatic and chemical probing, we found that PTB binding to nt 53 to 149 caused a conformational change in the neighboring RNA region. Partial deletions within the PTB-binding sequence completely abolished the PTB-induced conformational change in the mutant RNA even when the RNA retained partial PTB-binding activity. Correspondingly, the MHV DI RNAs containing these deletions completely lost their ability to transcribe mRNAs. Thus, the conformational change in the c3′-UTR caused by PTB binding may play a role in mRNA transcription.
Mouse hepatitis virus (MHV) belongs to the Coronaviridae family. Its RNA is a single-stranded, positive-sense RNA of 31 kb (12, 13, 22) which encodes seven to eight genes, depending on virus strains. Viral proteins are translated from six to seven subgenomic mRNAs as well as from the genome (10). These RNAs have a 3′-coterminal nested-set structure (9) and contain a leader sequence of approximately 70 nucleotides (nt) at the 5′ end (11, 26). All the mRNAs, except the mRNA coding for the N protein, are structurally polycistronic, and all these mRNAs can translate only the 5′-most open reading frame, except mRNA 5, which translates the E protein from a downstream open reading frame (10).
The transcription of these subgenomic mRNAs is an important step during the life cycle of MHV. Previous studies have shown that there are several cis-acting elements on the RNA genome of MHV that are involved in the regulation of this important step. These cis-acting elements include the intergenic sequence (IG) (20), leader sequence (16, 28), and a sequence at the 3′ untranslated region (3′-UTR) of the MHV genome (18). The IG could direct subgenomic mRNA synthesis when it was inserted in a defective interfering (DI) RNA that contains a leader sequence (20). The leader sequences can act both in cis and in trans to influence subgenomic mRNA synthesis (6, 16, 28). Deletion of the leader sequence significantly impairs subgenomic mRNA synthesis (16). Also, the leader sequence on the subgenomic mRNAs can be derived from either the same or a different RNA (in cis or in trans) (6, 16, 28). However, the discovery of the existence of a third cis-acting signal for subgenomic mRNA synthesis at the 3′-UTR was unexpected. This cis-acting signal was first identified as being located in the 3′-UTR (350 nt) of the viral genome by an MHV DI-RNA deletion study (18). This signal does not regulate minus-strand RNA synthesis since the cis-acting signal for the synthesis of minus-strand RNA had been determined to be the last 55 nucleotides plus the poly(A) tail at the 3′ end of the MHV genome (19). Therefore, the 3′-UTR sequence that regulates subgenomic mRNA synthesis most likely acts to affect positive-strand RNA synthesis and thus likely resides on the strand complementary to the 3′-UTR (c3′-UTR).
In order for this cis-acting signal on the c3′-UTR to regulate synthesis of the subgenomic mRNA, it must probably interact with the other two cis-acting signals {i.e., the negative-strand IG [(−)IG] and the negative-strand leader sequence on the template RNA} directly or indirectly during transcription. Since there is no significant sequence complementarity between the c3′-UTR and (−)IG or the negative-strand leader, this interaction very probably is mediated through protein-RNA and protein-protein interactions. The heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) has previously been reported to interact specifically with the (−)IG and the negative-strand leader of the minus strand of the MHV genome (15), potentially bringing these two cis-acting sequences together. In this study, we aim to identify the protein that can specifically interact with the cis-acting signal at the c3′-UTR. We found that polypyrimidine tract-binding protein (PTB) binds to two stretches of sequence within this region; the stronger binding site of the two contains two polypyrimidine tracts. Mutations (substitutions or deletions) of the pyrimidine nucleotides in these two regions reduced PTB binding by various degrees in vitro. When these mutations were introduced into a DI RNA, transcription of subgenomic mRNA from these DI RNAs was substantially reduced. Interestingly, PTB induces a conformational change in the RNA and deletion of either of the polypyrimidine tracts abolished this conformational change. These deletions also abolished subgenomic mRNA transcription from a DI RNA. This study thus suggests that the PTB-induced conformational change at c3′-UTR may be one of the mechanisms of regulating mRNA transcription.
MATERIALS AND METHODS
Viruses and cells.
The plaque-cloned A59 strain (21) of MHV was used throughout this study. Viruses were propagated in DBT cells (a mouse astrocytoma cell line) (3).
PCR primers.
The primer sequences used for PCR amplification are listed in Table 1. The nucleotide numbers on the c3′-UTR are assigned from the 5′ end (e.g., nt 1 on the c3′-UTR is complementary to the last nucleotide on the genomic strand). MHV DI cDNA constructs 25CAT (16) and 25HE (17) were used as the templates for PCR amplification.
TABLE 1.
Primers used for PCR amplification of the cDNA constructs
| Primera | Sequenceb |
|---|---|
| Sense | |
| SP6-350 | 5′-ATTTAGGTGACACTATAGATAGATGATGGCGTAGTGCCAGATGGGTTAG-3′ |
| 242(−) | 5′-ACACTCTCTATC-3′ |
| 149(−) | 5′-AGAGAATGTGTG-3′ |
| 52(−) | 5′-GCAATGCCCTAG-3′ |
| Antisense | |
| T7-0 | 5′-TAATACGACTCACTATAGGTGATTCTTCCAATTGGCCATGATCAACTTC-3′ |
| T7-53 | 5′-TAATACGACTCACTATAGGGAATAGTACCCTGATGTGAG |
| T7-149 | 5′-TAATACGACTCACTATAGGGATTTTGCAATCTCTTTCAAC |
| T7-240 | 5′-TAATACGACTCACTATAGGTCCTATCCCGACTTTCTCG |
| T7-308 | 5′-TAATACGACTCACTATAATTAGAGTCATCTTCTAACC |
The numbers in the primer names correspond to the nucleotide positions of the first MHV nucleotide sequence of each primer on the c3′-UTR (from the 5′ end).
T7 or SP6 promoter sequences are in boldface type.
In vitro transcription of PCR products.
PCR products made by primers containing either T7 or SP6 promoter sequences were directly used in in vitro transcription reaction mixtures without further purification. Briefly, PCR-amplified double-stranded DNA fragments (0.5 μg) were used as templates to transcribe the 32P-labeled RNA probes in a standard in vitro transcription reaction with T7 or SP6 RNA polymerase (Promega). All positive-sense RNAs were transcribed by SP6 polymerase, and all negative-sense RNAs were transcribed by T7 polymerase.
UV-cross-linking assay.
A UV-cross-linking assay was performed as described previously (2). Briefly, an in vitro-transcribed RNA probe was incubated with DBT cell extracts (30 μg) for 10 min at 30°C. The RNA-protein complex was then UV irradiated in a UV Stratalinker 2400 for 10 min. RNase A (20 μg) was added to the reaction mixture, and the mixture was incubated for 15 min at 37°C.
Immunoprecipitation.
DBT cell cytoplasmic extract which had been UV cross-linked to 32P-labeled RNA was diluted to 500 μl with NETS buffer (50 mM Tris-HCl [pH 7.4], 5 mM EDTA, 1 mM dithiothreitol, 100 mM NaCl, 0.05% NP-40) and mixed with various antibodies. The immunocomplexes were immobilized on protein A-Sepharose 4B beads (Pharmacia) and washed five to six times with NETS buffer. Protein sample loading buffer was then added to the beads, and the mixture was boiled for 3 min. The supernatant was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE). The monoclonal anti-PTB antibody was kindly provided by E. Wimmer (State University of New York, Stony Brook). The monoclonal anti-TATA-binding protein antibody and anti-Sam68 antibody were purchased from Calbiochem.
Plasmid constructions.
The 25CAT plasmid used in this study is as previously described (16). The PCR product containing the stretch a deletion (ΔA) mutation was amplified by using the primers 5′-CCTACGTCTAACCATAAGAACGGCGATAGGCGCCCCCTGG*CTCACATCAGG-3′ (* indicates the location of deleted nucleotides) (nt 120 to 63 of c3′-UTR) and T7-0. The PCR product containing the ΔC mutation was amplified by using the primers 5′-TT CTTATGGTTAGACGTAGGACCTTGCTAA*CTCACACATTCTCTATTTT GC-3′ (* indicates the location of deleted nucleotides) (nt 100 to 156) and SP6-350. The two PCR products were gel purified and combined as the template in a third PCR with the primers T7-0 and SP6-350. The resulting ΔAΔC PCR product (370 nt) was gel purified and doubly digested with the restriction enzymes BstEII and BclI (New England Biolabs). The PCR product containing the A substitution (subsA) mutation was amplified by using the primers 5′-CTAAC CATAAGAACGGCGATAGGCGCCCCCTGGGATTTTCTCACATCAG G-3′ (boldface letters are the substituted nucleotides) (nt 113 to 63) and T7-0. The PCR product containing the substitution (subsC) mutation was amplified by using the primers 5′-GCCGTTCTTATGGTTAGACGTAGGACCTTGCTAAAATCTCTCACACATTC-3′ (boldface letters are the substituted nucleotides) (nt 96 to 146) and SP6-350. The two PCR products were gel purified and combined as the template in a third PCR with the primers T7-0 and SP6-350. The resulting subsAsubsC PCR product (370 nt) was gel purified and doubly digested with the restriction enzymes BstEII and BclI (New England Biolabs). The 25CAT plasmid was also doubly digested with the restriction enzymes BstEII and BclI, and the large fragment was recovered from agarose gel. The medium-sized fragment (BstEII site at both ends) was also purified and saved for future use. The 25CAT large BstEII/BclI restriction fragment and ΔAΔC or subsAsubsC PCR product BstEII/BclI restriction fragments were ligated. The resulting plasmids were digested with BstEII and ligated with the 25CAT/BstEII medium-sized fragment. The resulting plasmids were 25CAT/ΔAΔC and 25CAT/subsAsubsC, respectively. 25CAT/ΔAΔC, 25CAT/subsAsubsC, and 25CAT were all digested with the restriction enzymes PpuMI and XbaI. Ligation of the 25CAT PpuMI/XbaI small fragment and the 25CAT/ΔAΔC PpuMI/XbaI large fragment resulted in the 25CAT/ΔA plasmid. Ligation of the 25CAT PpuMI/XbaI large fragment and the 25CAT/ΔAΔC PpuMI/XbaI small fragment resulted in the 25CAT/ΔC plasmid. Ligation of the 25CAT PpuMI/XbaI small fragment and the 25CAT/ subsAsubsC PpuMI/XbaI large fragment resulted in the 25CAT/subsA plasmid. Ligation of the 25CAT PpuMI/XbaI large fragment and the 25CAT/subsAsubsC PpuMI/XbaI small fragment resulted in the 25CAT/subsC plasmid. All the mutations were confirmed by DNA sequencing with Sequenase version 2.0 (Amersham).
DI RNA transcription and transfection.
The 25CAT, 25CAT/ΔA, 25CAT/ΔC, 25CAT/ΔAΔC, 25CAT/subsA, 25CAT/subsC, and 25CAT/subsAsubsC plasmids were first linearized with XbaI and then used as templates in in vitro RNA transcription reaction mixtures with T7 RNA polymerase. RNA transfection was performed according to the N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP) method (Boehringer Mannheim). Briefly, approximately 80%-confluent DBT cells were infected with A59 virus in 6-cm-diameter petri dishes at a multiplicity of infection of 10. At 1 h postinfection, cells were washed with serum-free Eagle’s minimal essential medium (MEM). In vitro-transcribed RNA (5 μg) was dissolved in a final volume of 200 μl of 15% (vol/vol) DOTAP mixture (Boehringer Mannheim). After the RNA-DOTAP mixture was incubated for 15 min at room temperature, it was added to 5 ml of prewarmed serum-free MEM and applied to the A59-infected cells. Transfection was carried out at 37°C for 1 h with gentle shaking every 15 min. Then the medium containing the RNA-DOTAP mixture was replaced by MEM containing 2% newborn calf serum and incubated at 37°C for the desired time.
CAT assay.
Cells were harvested at 8 h postinfection and lysed three times by freezing in ethanol-dry ice and thawing at 37°C. After centrifugation at 12,000 rpm for 10 min in a microcentrifuge (Beckman), the supernatant was used in the chloramphenicol acetyltransferase (CAT) assay as described previously (18). The CAT reaction products were resolved by chromatography on thin-layer chromatography plates (J. T. Baker). The plates were air dried and analyzed with the AMBIS Systems Radioanalytic Imaging System.
RNP complex preparation.
In vitro-transcribed RNA was labeled at its 5′ end with polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Dupont NEN) and purified by PAGE. Approximately 104 cpm of gel-purified, end-labeled RNA was incubated with purified glutathione S-transferase (GST)-PTB fusion protein (1 μg) in the binding buffer (25 mM KCl, 5 mM HEPES [pH 7.4], 2 mM MgCl2, 0.1 mM EDTA, 0.2% glycerol, 2 mM dithiothreitol) for 30 min at room temperature to form RNP complex. Glutathione-Sepharose 4B beads (5 to 10 μl; Pharmacia) were added and incubated with the RNP complex at room temperature for another 15 min. The beads were centrifuged down and washed with the binding buffer at least five times until the supernatant contained less than 500 cpm.
RNA secondary-structure probing.
RNA or RNP complex prepared as described above was diluted to 20 μl with the binding buffer. Equal counts of different mutant RNAs were used in each experiment. Nuclease probing was performed by treating the sample with RNase A (specific for unpaired pyrimidines, 0.5 ng per reaction mixture) on ice for 30 min or RNase V1 (specific for double-stranded region, 0.05 U per reaction mixture) at room temperature for 30 min. Pb2+ probing was performed in a solution containing 50 mM HEPES (pH 7.4), 5 mM magnesium acetate, and 50 mM potassium acetate in the presence of 10 mM lead acetate at room temperature for 15 min (24). The reaction was stopped by adding EDTA to a final concentration of 50 mM. After the reaction, all the samples were extracted with acid-phenol (Ambion) and precipitated in the presence of 1 μg of carrier tRNA for 5 min at −20°C. RNA was recovered by centrifugation at 12,000 rpm for 15 min in a microcentrifuge (Beckman) and analyzed on an 8% polyacrylamide gel containing 7 M urea.
RESULTS
PTB binds to the MHV c3′-UTR.
To study protein-RNA interaction at the previously identified cis-acting signal for transcription in the 3′-UTR of MHV RNA, a UV-cross-linking assay was first performed to detect the protein species that can interact with this RNA fragment. We reasoned that the cis-acting sequence at the 3′-UTR most likely resides on the negative strand, since this sequence regulates positive-strand RNA synthesis (18). The 350-nt RNA, corresponding to the negative strand of the entire MHV 3′-UTR of 305 nt and the last 45 nt of the coding region of the N protein (termed the c3′-UTR) was used. This region has previously been shown to be required for MHV mRNA transcription (18). The RNA was used in the UV-cross-linking assay with nuclear or cytoplasmic extracts from uninfected DBT cells. Results in Fig. 1 show that two major protein species of 70 and 55 to 57 kDa (referred to as p55) from both nuclear and cytoplasmic extracts were UV-cross-linked to the c3′-UTR of MHV RNA. As a comparison, the 70-kDa protein was also cross-linked to the positive-strand 3′-UTR, but p55 did not, indicating that p55 specifically binds the c3′-UTR. Several other proteins (e.g., the 35-kDa protein) bound the positive-strand 3′-UTR. The nature of these proteins was not further investigated in this study.
FIG. 1.
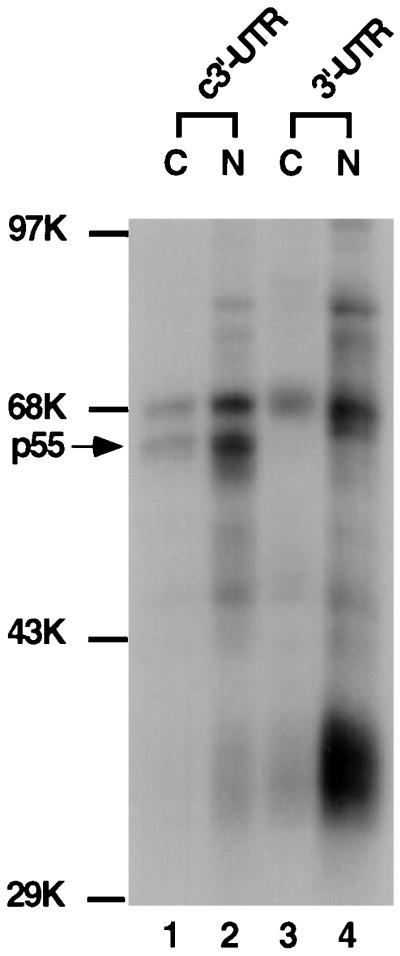
UV-cross-linking assay of DBT cell extract with the MHV 3′-UTR and its complementary strand. The 32P-labeled 3′ 350 nt of the positive strand of the 3′-UTR and c3′-UTR (104 cpm) were UV cross-linked to either cytoplasmic (C) or nuclear (N) extracts from DBT cells. The arrow points to the p55 protein. Molecular mass markers (in kilodaltons) are indicated. K, thousand.
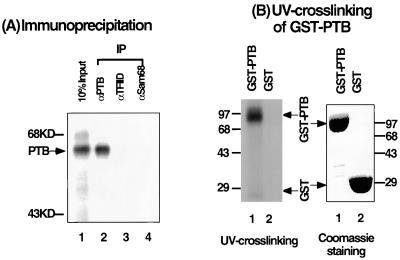
Previous studies from our laboratory have shown that a 55- or 57-kDa protein, PTB, binds the leader sequence of the MHV RNA genome (14). To investigate whether the p55 shown in Fig. 1 is also PTB, an immunoprecipitation experiment was performed as shown in Fig. 2A. Results showed that p55 was precipitated only by the monoclonal antibody against PTB and not the antibodies against TFIID or Sam68. These results indicate that this p55 is probably PTB. To confirm the immunoprecipitation result, purified recombinant GST-PTB was used in the UV-cross-linking assay with the c3′-UTR. Figure 2B shows that GST-PTB, but not GST, was UV cross-linked to the c3′-UTR. Combined, these results clearly show that it is PTB that binds to the c3′-UTR.
FIG. 2.
Identification of the p55 protein UV cross-linked to the c3′-UTR. (A) Immunoprecipitation of DBT cell extract, which was UV cross-linked to the 32P-labeled c3′-UTR by the indicated antibodies. Lane 1 contains 10% of the input UV-cross-linked cell extract. IP, immunoprecipitate. (B) UV-cross-linking assay of the recombinant GST-PTB. The right panel shows Coomassie brilliant blue staining of the purified GST and GST-PTB proteins used in the left panel. The left panel shows the results of a UV-cross-linking assay of these recombinant proteins to the c3′-UTR. Molecular mass standards (in kilodaltons [KD]) are shown.
The interaction between PTB and the MHV c3′-UTR is specific.
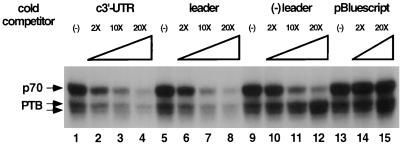
A competition assay was performed to determine the specificity of the interaction between PTB and the c3′-UTR. UV-cross-linking experiments between the 32P-labeled c3′-UTR and DBT cell lysates were performed in the presence of increasing amounts of the cold homologous and heterologous competitor RNAs. Results in Fig. 3 show that the PTB–c3′-UTR interaction was competed away by increasing amounts of the unlabeled homologous c3′-UTR (lanes 1 to 4) and also by the leader sequence of MHV RNA, which has previously been shown to contain a PTB-interacting domain (lanes 5 to 8) (14). In contrast, negative-strand leader RNA (lanes 9 to 12), which does not interact with PTB, or an unrelated RNA transcribed from the pBluescript plasmid (lanes 13 to 15) did not compete with PTB binding at all. This competition assay result clearly showed that the interaction between PTB and the c3′-UTR is specific. Significantly, p70 binding to the c3′-UTR was competed away by all three MHV RNA segments, since all of them have been shown to bind p70 (14, 15). Thus, the binding of p70 to MHV RNA is less specific. The nature of p70 was not further determined in this study.
FIG. 3.
Competition assay by UV cross-linking. DBT cell extract was incubated with different amounts (fold excess) of the various unlabeled RNAs before UV cross-linking to the 32P-labeled c3′-UTR. Lanes 1 to 4, c3′-UTR; lanes 5 to 8, MHV positive-strand leader sequence; lanes 9 to 12, MHV negative-strand leader sequence; lanes 13 to 15, unrelated RNA transcribed from plasmid pBluescript. (−) leader, negative-strand leader.
Mapping of PTB-binding sites within the c3′-UTR.
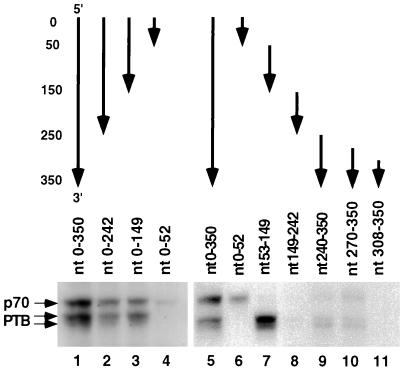
To define the sequence of the c3′-UTR that is important for PTB binding, several RNA segments corresponding to different regions of the c3′-UTR were used for the UV-cross-linking assay to test their abilities to interact with PTB. The experiments using 3′-end-truncation mutants first identified a PTB-binding site within nt 53 to 149 (lanes 1 to 4 of Fig. 4). This conclusion was confirmed by using four RNA fragments corresponding to four consecutive regions of the c3′-UTR (lanes 6 to 9), which revealed a strong PTB-binding site within nt 53 to 149. In addition, a weak binding site was found within nt 240 to 350 and was further mapped within nt 270 to 307 (lanes 10 and 11). Thus, we conclude that all of the PTB-binding sequences are within the noncoding region of the c3′-UTR (305 nt).
FIG. 4.
Mapping of PTB-binding sites on the c3′-UTR. The structures of the various fragments of the c3′-UTR RNAs used are indicated on the top. The arrows point from the 5′ end to the 3′ end of each RNA. Nucleotide positions from the 5′ end of the c3′-UTR are indicated at the left of the figure.
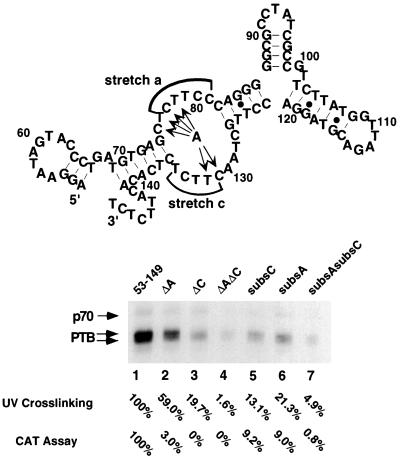
To further identify the nucleotides important for PTB binding within the strong PTB-binding site (nt 53 to 149), the secondary structure of this RNA was first predicted with the Mulfold2 computer program (4, 5, 29). As shown in Fig. 5, there are two obvious polypyrimidine tracts on the predicted single-stranded regions of the folded RNA from nt 53 to 149, one at nt 77 to 82 (stretch a) and the other at nt 132 to 136 (stretch c). This predicted structure was largely confirmed by enzymatic and chemical probing (see Fig. 6 and 7). Furthermore, this structure remained the same when the full-length c3′-UTR (300 nt) was used for computer analysis (data not shown), suggesting that this structure likely exists as an independent unit. We predicted that these two unpaired polypyrimidine tracts are responsible for PTB binding to this RNA fragment. To test this possibility, we constructed several deletion and substitution mutation RNAs (with deletion of either stretch a or c or both or replacement of either stretch c or a or both with adenosine residues) (Fig. 5) for the UV-cross-linking assay. The mutations of these sequences did not result in substantial alteration of the overall RNA structure, as was predicted by computer modeling and confirmed by enzymatic and chemical probing (see below). Results in Fig. 5 show that the deletion or substitution of stretch a or stretch c reduced the UV-cross-linking of PTB to the mutant RNAs to 59.0, 19.7, 21.3, or 13.1% relative to that of the wild-type RNA (nt 53 to 149). The deletion or substitution of both stretch a and stretch c almost completely abolished PTB binding to the RNA. Taken together, these results clearly showed that both of the two polypyrimidine tracts at nt 77 to 82 and 132 to 136 are responsible for the efficient binding of PTB to this RNA fragment.
FIG. 5.
Identification of nucleotides required for PTB binding on nt 53 to 149. The top of the figure shows a computer-predicted secondary structure of nt 53 to 149. The two polypyrimidine tracts are marked by stretch a and stretch c. ΔA, ΔC, and ΔAΔC deletion mutants and subsC, subsA, and subsAsubsC substitution mutants were used in the UV-cross-linking assay with the DBT cell extract. The amounts of PTB binding relative to that of the wild-type RNA are indicated (in percentages). MHV DI RNAs (25CAT) (16) containing the ΔA, ΔC, ΔAΔC, subsA, subsC, and subsAsubsC mutations were used for transfection into A59-infected cells, and CAT activity was determined at 7 h posttransfection. The CAT activities of various RNAs relative to that of the wild type are indicated (in percentages).
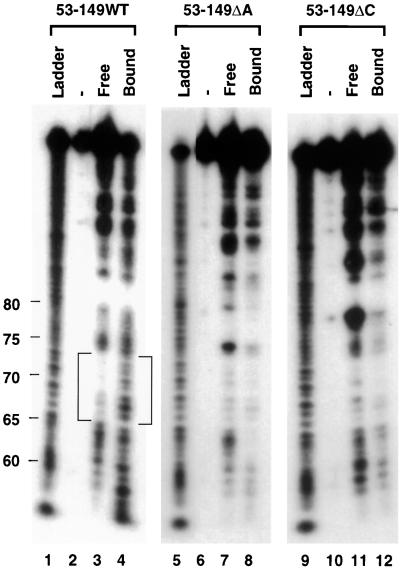
FIG. 6.
Secondary-structure analysis of free and PTB-bound nt 53 to 149 of the wild-type RNA (wt) and ΔA and ΔC mutants by lead probing. In lanes 1 to 4, free and PTB-bound wild-type RNA from nt 53 to 149 32P labeled at the 5′ end (WT) was subjected to Pb2+-induced hydrolysis, and the products were analyzed by 8% PAGE, with the polyacrylamide gel containing 7 M urea. Lanes −, an undigested control; lanes Ladder, partially alkaline-hydrolyzed wild-type RNA (nt 53 to 149) 32P labeled at its 5′ end (the positions of nucleotides [from the 5′ end of the negative-strand RNA] in the wild-type RNA from nt 53 to 149 are indicated on the left of the ladder in lane 1); lanes 5 to 8, ΔA RNA (nt 53 to 149); lanes 9 to 12, ΔC RNA (nt 53 to 149). The brackets at the left of the figure represent the region that changed from double-stranded to single-stranded after PTB binding in the wild-type RNA (nt 53 to 149).
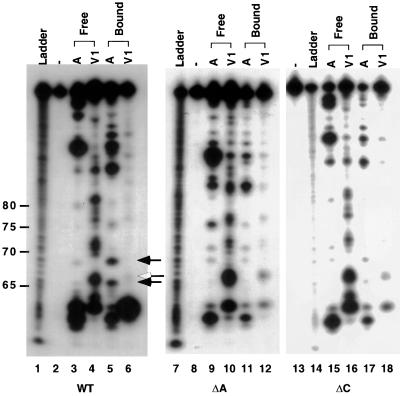
FIG. 7.
Secondary-structure analysis of free and PTB-bound wild-type RNA (nt 53 to 149) and ΔA and ΔC mutants by nuclease digestion. Lanes 3 to 6, free and PTB-bound wild-type RNA (WT) (nt 53 to 149) 32P labeled at its 5′ end was subjected to limited digestion with RNase A (lanes A) and RNase V1 (lanes V1), and the products were analyzed by 8% PAGE with the polyacrylamide gel containing 7 M urea. The minus sign in lane 2 represents an undigested control; the ladder in lane 1 represents partially alkaline-hydrolyzed wild-type RNA labeled at its 5′ end with 32P. The positions of the nucleotides are shown on the left of the ladder. Lanes 7 to 12, ΔA RNA (nt 53 to 149); lanes 13 to 18, ΔC RNA (nt 53 to 149).
Mutations of the polypyrimidine tracts in the c3′-UTR of a DI RNA reduced subgenomic mRNA transcription.
To address the biological significance of PTB binding to the c3′-UTR, we first used MHV DI RNA (25CAT) mutants that contain substitutions of stretch a, c, or both to assess the effects of these mutations on subgenomic mRNA transcription from this DI RNA. This DI vector has been shown to express CAT activity, which reflects the transcription activity of the DI RNA (16). Since these mutants have intact 55 nt at the 3′ end, they should retain the ability to synthesize negative-strand RNA (19). Thus, the CAT activity should reflect positive-strand subgenomic mRNA synthesis. The wild-type and the mutant DI RNAs were transfected into A59-infected DBT cells at 1 h postinfection, and cell lysates were harvested 7 h posttransfection and assayed for CAT activity. Our results showed that when pyrimidine nucleotides in either stretch a or c were replaced with adenosine (indicated by arrows in Fig. 5), the CAT activity was reduced to about 9%, which corresponded roughly with the results of the in vitro UV-cross-linking assay (21.3 and 13.1%, respectively, as shown in Fig. 5, lanes 5 and 6). When both stretch a and stretch c were replaced, the CAT activity was almost undetectable, corresponding to the lack of PTB binding in the UV-cross-linking assay (Fig. 5, lane 7). These results suggest that there is a correlation between PTB binding and subgenomic mRNA transcription.
When the ΔA, ΔC, and ΔAΔC deletion mutants were introduced into the MHV DI RNA, all three of these mutants lost almost entirely their ability to express CAT activity even though two of these mutants (ΔA and ΔC mutants) still bound PTB to different extents (59.0 and 19.7%, respectively) (Fig. 5, lanes 2 to 4). This result suggests that deletions in these PTB-binding sites may cause some other changes that result in the complete loss of the transcriptional activities of these RNAs. Therefore, PTB binding is not sufficient for mRNA transcription. Nevertheless, this result indicates that these PTB-binding sites are important for mRNA transcription.
PTB binding to nt 77 to 82 and 132 to 136 converts nt 66 to 74 from a double-stranded region to a single-stranded region.
To determine the possible molecular mechanism by which the deletion of the PTB-binding site abolished transcription, despite the fact that these mutants still retained some PTB-binding activity, we examined the possibility that PTB binding may induce some structural changes in these RNAs and that the deletion of the PTB-binding site abolished these PTB-induced structural changes. For this purpose, we used both chemical and enzymatic methods to probe the secondary structure of the RNA from nt 53 to 149, which contains the strong PTB-binding site, before and after PTB binding. As shown in Fig. 6, the study using Pb2+-induced RNA cleavage, which cleaves at single-stranded RNA regions, showed a double-stranded region at nt 66 to 74 (Fig. 6) in the unbound wild-type RNA (nt 53 to 149) (lane 3). In contrast, in the PTB-bound RNA, this double-stranded region was converted to the single-stranded region, which was sensitive to Pb2+-induced cleavage (lane 4). The change in secondary structure induced by PTB binding was further confirmed by RNase digestion assay (Fig. 7). The result shows that the wild-type RNA from nt 53 to 149 has several RNase V1 cleavage signals at nt 66 and nt 70 to 74 (lane 4) but no RNase A cleavage sites in the same region (lane 3). However, after PTB binding, the RNase V1 signals disappeared; instead, RNase A cleavage signals appeared at nt 66 and 69, confirming that this region was converted from a double-stranded region to a single-stranded structure. It is notable that the efficiency of V1 digestion of the PTB-bound RNA was overall reduced compared to that of the wild-type RNA; nevertheless, only the region from nt 68 to 75 showed a corresponding increase in RNase A digestion.
Similar experiments were performed on the ΔA and ΔC mutants. The overall secondary structures of ΔA and ΔC mutants were very similar to that of the wild-type RNA, with the exception that there was a shorter single-stranded region as a result of the deleted sequences. However, PTB binding to ΔA and ΔC mutants did not induce the same kind of conversion of the double-stranded region to a single-stranded region as was observed for the wild-type RNA (Fig. 6 and 7), even though the ΔA mutant still bound a considerable amount of PTB. Similar experiments could not be performed on the ΔAΔC double mutant because it does not bind PTB at all. Based on this result, we propose that PTB needs to contact both of the polypyrimidine tracts (stretch a and stretch c) to induce the conversion of the double-stranded region at nt 68 to 75 into a single-stranded structure to facilitate subgenomic mRNA synthesis. When one of the polypyrimidine tracts is deleted, the double-stranded region can not be converted to the single-stranded region, although PTB still binds to the remaining polypyrimidine tract. As a result, all these deletion mutants are unable to synthesize subgenomic mRNA.
DISCUSSION
In this study, we found that PTB interacts specifically with the complementary strand of the 3′-UTR of MHV RNA. The PTB-binding sites appear to be important for the transcription of subgenomic mRNA from a DI vector. Several cellular proteins have now been shown to bind to either the genomic RNA or its complementary strand in different viruses (8), including the mosquito homolog of La autoantigen in binding to Sindbis virus (23), La autoantigen in binding to human immunodeficiency virus (1, 27), calreticulin in binding to rubella virus (25), and hnRNP A1 and PTB in binding to MHV (14, 15). Some of these cellular proteins have been shown to be important for virus replication. In MHV, hnRNP A1 specifically binds to the minus-strand leader and IG (15), both of which are important regulatory elements in the synthesis of subgenomic mRNAs. Furthermore, PTB binds specifically to the leader sequence of viral RNA (14). Both PTB and hnRNP A1 are splicing factors and exist as part of a splicing complex in uninfected cells (7), suggesting that these two proteins interact with each other directly or indirectly. Indeed, interaction between hnRNP A1 molecules and between hnRNP A1 and PTB have been demonstrated in vitro (unpublished data). The present finding that PTB binds specifically to the c3′-UTR, which is the 5′ end of the minus strand of MHV RNA, conceivably allows this end of the template RNA to interact with the leader sequence and IG of the template RNA through an hnRNP A1-PTB interaction. This may provide a mechanism by which the 3′ end of the viral genome (corresponding to the 5′ end of the template RNA) can regulate subgenomic mRNA transcription.
Our results have shown that there are two PTB-binding sites on the c3′-UTR, one at nt 53 to 149 and the other at nt 270 to 307. Both of these sites are located exclusively in the 3′-UTR. Previous results have shown that nt 270 to 305 of the 3′ end of the viral genome is required for subgenomic mRNA synthesis, and the sequence requirement is stringent because the replacement of this region with the similar region from bovine coronavirus completely abolishes subgenomic mRNA synthesis (18). Thus, there is a good correlation between PTB binding and transcriptional activity. In this study, we have further shown that nt 77 to 82 and 132 to 136 are also important for mRNA transcription. Partial substitution of the PTB-binding sites reduced both PTB binding and subgenomic mRNA transcription. Although, at this time, we could not establish unequivocally that PTB binding to these regions is required for mRNA transcription, the strong correlation between PTB binding and transcriptional activity suggests a functional role for PTB binding. Nevertheless, an in vitro transcriptional assay system will probably be required to establish such a role for PTB.
Surprisingly, partial deletion of the major PTB-binding site in the c3′-UTR resulted in the complete abolition of transcriptional activity, even when substantial amounts of PTB still bound to the c3′-UTR. Thus, the association of PTB to a site on the c3′-UTR was not sufficient to confer the transcriptional activity. Instead, we found that PTB binding induced a change in the secondary structure of c3′-UTR RNA and that all the deletion mutations studied inhibited this induced structural change. This induced opening up of a double-stranded region might expose some important regulatory sequence which is otherwise buried in the double-stranded region and enable this sequence to participate in the synthesis of subgenomic mRNA. Although evidence obtained from a correlation between the transcription activity and the PTB-induced structural changes in RNA is not sufficient to establish the importance of such a structural change, it does suggest a potential mechanism for the 3′-UTR to regulate mRNA transcription. Thus, PTB may potentially participate in the regulation of MHV RNA synthesis either by providing protein-protein interactions to allow various cis-acting RNA sequences to form a transcription complex or by facilitating the formation of a critical RNA structure. Increasing evidence has suggested the importance of the 3′-UTR sequence in the regulation of viral RNA synthesis for many different viruses (reviewed in reference 8). The MHV 3′-UTR falls into such a category. Further identification of this regulatory sequence will be very helpful in our understanding of the mechanism of RNA synthesis of MHV.
ACKNOWLEDGMENTS
We thank Daphne Shimoda for editorial assistance.
This work was supported by a research grant, AI19244, from the National Institutes of Health. M.M.C. Lai is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Chang Y N, Kenan D J, Keene J D, Gatignol A, Jeang K T. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J Virol. 1994;68:7008–7020. doi: 10.1128/jvi.68.11.7008-7020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furuya T, Lai M M C. Three different cellular proteins bind to complementary sites on the 5′-end-positive and 3′-end-negative strands of mouse hepatitis virus RNA. J Virol. 1993;67:7215–7222. doi: 10.1128/jvi.67.12.7215-7222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirano N, Fujiwara K, Hino S, Matsumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44:298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- 4.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaeger J A, Turner D H, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1989;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 6.Jeong Y S, Makino S. Evidence for coronavirus discontinuous transcription. J Virol. 1994;68:2615–2623. doi: 10.1128/jvi.68.4.2615-2623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 8.Lai M M C. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 9.Lai M M C, Brayton P R, Armen R C, Patton C D, Pugh C, Stohlman S A. Mouse hepatitis virus A59: messenger RNA structure and genetic localization of the sequence divergence from the hepatotropic strain MHV 3. J Virol. 1981;39:823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai M M C, Patton C D, Stohlman S A. Further characterization of mRNAs of mouse hepatitis virus: presence of common 5′-end nucleotides. J Virol. 1982;41:557–656. doi: 10.1128/jvi.41.2.557-565.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai M M C, Stohlman S A. The RNA of mouse hepatitis virus. J Virol. 1978;26:236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee H-J, Shieh C-K, Gorbalenya A E, Koonin E V, La Monica N, Tuler J, Bagdzyahdzhyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H-P, Huang P, Park S, Lai M M C. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J Virol. 1999;73:772–777. doi: 10.1128/jvi.73.1.772-777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H-P, Zhang X, Duncan R, Comai L, Lai M M C. Heterogeneous nuclear ribonucleoprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao C-L, Lai M M C. Requirement of the 5′-end genomic sequence as an upstream cis-acting element for coronavirus subgenomic mRNA transcription. J Virol. 1994;68:4727–4737. doi: 10.1128/jvi.68.8.4727-4737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao C-L, Zhang X M, Lai M M C. Coronavirus defective-interfering RNA as an expression vector: the generation of a pseudorecombinant mouse hepatitis virus expressing hemagglutinin-esterase. Virology. 1995;208:319–327. doi: 10.1006/viro.1995.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin Y-J, Zhang X, Wu R-C, Lai M M C. The 3′ untranslated region of coronavirus RNA is required for subgenomic mRNA transcription from a defective interfering RNA. J Virol. 1996;70:7236–7240. doi: 10.1128/jvi.70.10.7236-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Y-J, Liao C-L, Lai M M C. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makino S, Joo M, Makino J K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991;65:6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manaker R A, Piczak C V, Miller A A, Stanton M F. A hepatitis virus complicating studies with mouse leukemia. J Natl Cancer Inst. 1961;27:29–51. [PubMed] [Google Scholar]
- 22.Pachuk C J, Bredenbeek P J, Zoltick P W, Spaan W J M, Weiss S R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus strain A59. Virology. 1989;171:141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardigon N, Strauss J H. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J Virol. 1996;70:1173–1181. doi: 10.1128/jvi.70.2.1173-1181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlegl J, Gegout V, Schlager B, Hentze M W, Westhof E, Ehresmann C, Ehresmann B, Romby P. Probing the structure of the regulatory region of human transferring receptor messenger RNA and its interaction with iron regulatory protein-1. RNA. 1997;3:1159–1172. [PMC free article] [PubMed] [Google Scholar]
- 25.Singh N K, Atreya C D, Hakhasi H L. Identification of calreticulin as a rubella virus RNA binding protein. Proc Natl Acad Sci USA. 1994;91:12770–12774. doi: 10.1073/pnas.91.26.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spaan W J M, Delius H, Skinner M, Armstrong J, Rottier P, Smeekens S, van der Zeijst B A M, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svitikin Y V, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J Virol. 1994;69:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Liao C-L, Lai M M C. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription, both in trans and in cis. J Virol. 1994;68:4738–4746. doi: 10.1128/jvi.68.8.4738-4746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]