Abstract
During lytic herpes simplex virus (HSV) infections, the HSV virion host shutoff protein (UL41) accelerates the turnover of host and viral mRNAs. Although the UL41 polypeptides from HSV type 1 (HSV-1) strain KOS and HSV-2 strain 333 are 87% identical, HSV-2 strains generally shut off the host more rapidly and completely than HSV-1 strains. In a previous study, we identified three regions of the HSV-2 UL41 polypeptide (amino acids 1 to 135, 208 to 243, and 365 to 492) that enhance the activity of KOS when substituted for the corresponding portions of the KOS protein (D. N. Everly, Jr., and G. S. Read, J. Virol. 71:7157–7166, 1997). These results have been extended through the analysis of more than 50 site-directed mutants of UL41 in which selected HSV-2 amino acids were introduced into an HSV-1 background and HSV-1 amino acids were introduced into the HSV-2 allele. The HSV-2 amino acids R22 and E25 were found to contribute dramatically to the greater activity of the HSV-2 allele, as did the HSV-2 amino acids A396 and S423. The substitution of six HSV-2 amino acids between residues 210 and 242 enhanced the HSV-1 activity to a lesser extent. In most cases, individual substitutions or the substitution of combinations of fewer than all six amino acids reduced the UL41 activity to less than that of KOS. The results pinpoint several type-specific amino acids that are largely responsible for the greater activity of the UL41 polypeptide of HSV-2. In addition, several spontaneous mutations that abolish detectable UL41 activity were identified.
Controls of the rate of mRNA turnover play an important role in eukaryotic gene expression (4, 37, 48). During lytic herpes simplex virus (HSV) infections, the HSV virion host shutoff (vhs) protein (UL41) negatively regulates the half-lives of viral and cellular mRNAs (33). Immediately after infection, copies of the vhs (UL41) polypeptide, which enter the cell as components of infecting virions, destabilize host mRNAs in the cytoplasm (14, 38, 46). This, together with the inhibition of pre-mRNA splicing by the immediate-early polypeptide ICP27 (16, 17), plays an important role in redirecting the cell from synthesis of cellular to viral proteins. Following the onset of viral transcription, the vhs (UL41) protein accelerates the turnover of viral mRNAs belonging to all kinetic classes (23, 28, 29, 46). In this role, it helps determine viral mRNA levels and facilitates the sequential transition between expression of different classes of viral genes (33).
While not lethal, mutations that inactivate the vhs polypeptide result in a 5- to 10-fold reduction in the production of progeny virus in cell culture (34, 35), and wild-type virus rapidly outgrows vhs mutants in mixed infections (24). In addition, recent studies indicate that the vhs function may play a significant role in HSV pathogenesis (43–45). HSV type 2 (HSV-2)-infected fibroblasts are poorly lysed by autologous HSV-specific cytotoxic T lymphocytes (18, 21, 32, 49). This appears to be due, at least in part, to a block in antigen presentation by class I major histocompatibility complex molecules resulting from inhibition of the TAP antigen transporter by the immediate-early protein ICP47, combined with the vhs-mediated inhibition of major histocompatibility complex synthesis (49, 50, 52). Furthermore, in several animal models, vhs mutants have been reported to replicate more poorly than wild-type virus and to be less pathogenic (2, 26, 43–45).
UL41 homologues have been identified in a number of alphaherpesviruses, including HSV-1 and HSV-2 (8, 27), varicella-zoster virus (6), equine herpesvirus 1 (11, 47), pseudorabies virus (3), and bovine (40), gallid (5), canine (36), and feline (51) herpesviruses. Of these, the most extensively studied have been the UL41 homologues of HSV-1 and HSV-2. Although the UL41 polypeptides of HSV-1 and HSV-2 are 87% identical (8), HSV-2 strains generally shut off the host more rapidly and completely than HSV-1 strains (12, 15), and the transfer of UL41 alleles between strains transfers the host shutoff phenotype (13). Further evidence that strain-specific differences in host shutoff reflect differences in the UL41 polypeptides comes from studies using a transient-expression assay in which vhs activity was measured by the ability of a transfected UL41 allele to inhibit expression of a cotransfected reporter gene (9). Besides demonstrating that UL41 is the only viral polypeptide required to induce mRNA degradation, this assay has the advantage that UL41 alleles can be compared in vivo in the absence of other viral gene products (19, 30). This circumvents potential complications due to differences between the proteins with regard to interactions with other viral polypeptides, packaging, or release from virions. In this assay, both HSV-1 and HSV-2 alleles inhibited reporter gene expression in a dose-dependent fashion. However, 40-fold less of the HSV-2 allele was required to inhibit reporter expression to the same extent as the HSV-1 allele, indicating that the HSV-2 polypeptide has considerably greater mRNA degradative activity (9).
The existence of naturally occurring UL41 variants that have similar sequences but different activities offers an attractive system for identifying residues that modulate vhs activity. In an earlier study, we used the cotransfection assay to compare the activities of a series of chimeric UL41 alleles containing various mixtures of HSV-1 and HSV-2 sequences (9). The results identified three regions (amino acids 1 to 135, 208 to 243, and 365 to 492) of the UL41 polypeptide from HSV-2 strain 333 which significantly enhance the activity of HSV-1 KOS when substituted for the corresponding amino acids of the KOS protein. In this study, we extend these results through the analysis of more than 50 site-directed mutants of UL41 in which selected HSV-2 amino acids were introduced into an HSV-1 background and HSV-1 amino acids were introduced into the HSV-2 allele. The results pinpoint several type-specific amino acids that are largely responsible for the greater activity of the UL41 polypeptide of HSV-2.
MATERIALS AND METHODS
Cells.
Vero cells were purchased from the American Type Culture Collection and maintained in Eagle’s minimum essential medium (GIBCO) supplemented with 10% (vol/vol) calf serum and antibiotics as described previously (9, 29, 30).
Plasmids.
The plasmids pKOS and p333, containing the UL41 open reading frames from HSV-1 strain KOS and HSV-2 strain 333 cloned into the vector pcDNAI (Invitrogen), have been described previously (9). The plasmids p3/K(135), pK/3/K(208,243), and pK/3(365) encode chimeric UL41 alleles containing various combinations of HSV-1 and HSV-2 sequences and have also been described previously (9). For this study, the UL41-containing inserts were excised from pKOS, p333, p3/K(135), pK/3/K(208,243), and pK/3(365) by digestion with HindIII and XbaI and recloned between the corresponding sites of the vector pcDNA1.1amp (Invitrogen) to yield pKOSamp, p333amp, p3/K(135)amp, pK/3/K(208,243)amp, and pK/3(365)amp, respectively. Each of these plasmids contains a UL41 open reading frame cloned downstream from the cytomegalovirus (CMV) immediate-early promoter as well as a promoter for T7 RNA polymerase. In each case, sequence analysis confirmed that the authentic UL41 start codon is the first AUG from the 5′ end of mRNAs produced in vivo from the CMV immediate-early promoter or by in vitro transcription with T7 RNA polymerase. These pcDNA1.1amp-derived plasmids were the parental plasmids for the construction of all site-directed mutants.
Site-directed mutagenesis.
UL41 alleles were mutagenized by using the Chameleon Double-Stranded, Site-Directed Mutagenesis Kit (Stratagene) according to a modification of the manufacturer’s protocol. Synthetic oligonucleotide primers, modified by 5′ phosphorylation, were purchased from Integrated DNA Technologies (Coralville, Iowa). Briefly, the UL41-containing plasmids were denatured and annealed with two synthetic primers, both of which were complementary to the same DNA strand. The first, termed the mutagenic primer, contained the desired nucleotide changes within UL41. Typically, these changes caused an alteration of one or more amino acids of UL41 and created or destroyed a restriction site within the gene. The second primer, termed the selection primer, contained nucleotide changes which inactivated a unique restriction site (the selection site) outside UL41. The primers were extended with T7 DNA polymerase, and the resulting plasmids were treated with T4 DNA ligase. These were digested with the restriction enzyme that cleaves the starting plasmid at the selection site. Due to a mismatch at the selection site, plasmids containing the annealed selection primer were not cleaved and remained circular. The DNA mixture was then used to transform the XLmutS strain of Escherichia coli (Stratagene) that is deficient in mismatch repair. Because transformation is more efficient for circular than linear DNA molecules, this enriched for plasmids containing alterations at the selection site. A batch preparation of plasmid DNA was made from the transformants, digested with the selection site endonuclease, and used to transform E. coli TOP10F′ (Invitrogen), resulting in a second step of enrichment. Plasmids were prepared from individual transformants and screened for resistance to the selection site endonuclease, as well as for the creation or destruction of the restriction site within UL41. Candidate mutants were sequenced to confirm that they contained the desired mutations within UL41 and evaluated by in vitro transcription and translation to confirm that they encoded a UL41 polypeptide of the expected molecular mass.
Multiple mutations were created in UL41 by a combination of two strategies. In the first strategy, selection primers were designed in pairs such that the first primer destroyed one unique restriction site while creating a unique site for another enzyme. The second selection primer destroyed the second site and recreated the first. For example, mutagenesis near the 5′ end of UL41 was accomplished with a pair of selection primers, one of which destroyed an EcoRI site in the polylinker and created a unique SalI site. The other selection primer changed the SalI site back to one cleaved by EcoRI. In the first round of mutagenesis, a mutagenic primer was paired with the first selection primer, and the transformants were screened for plasmids that were resistant to EcoRI and sensitive to SalI. Subsequently, additional mutations were added to UL41, using another mutagenic primer paired with the second selection primer, and screening the transformants for plasmids that were resistant to SalI and sensitive to EcoRI. Using this approach, it was possible to construct multiple mutations in UL41 through the successive use of mutagenic primers paired alternately with selection primers that changed the selection site back and forth between an EcoRI site and a SalI site. A similar strategy was used to mutagenize the 3′ end of UL41, using a pair of selection primers which alternated between creating and destroying sites for XbaI and XhoI.
The second way that multiple mutations were introduced into UL41 was by using multiple mutagenic primers along with one selection primer in a single round of mutagenesis. Provided the mutagenic primers did not overlap, several mutations could be introduced at the same time. By using a combination of the two strategies, UL41 alleles were constructed containing as many as eight amino acid differences from the parent allele.
The structures of the parent alleles that were used for site-directed mutagenesis are shown in Fig. 1, and the structures of the various mutants are shown in Fig. 2 to 7. Each mutant was given a name that reflects the amino acids at which it differs from its parent. Thus, the mutant RRE19 was derived from the KOS allele and differs from it by the presence of an arginine at position 19, an arginine at position 22, and a glutamic acid at position 25. Construction of many of the mutants involved multiple mutagenic steps, which are summarized in Tables 1 and 2. For example, RRE19 (mutant 1 in Table 1) was constructed in a two-step process in which the KOS allele was first mutated to RSE19 (mutant 4 in Table 1) and then RSE19 was mutated to RRE19. Several of the mutants (mutant 5 in Table 1 and mutants 54, 55, and 56 in Table 2) contain both deliberately constructed mutations and spontaneous mutations that occurred during mutant construction, which were identified by sequencing of the mutant alleles.
FIG. 1.
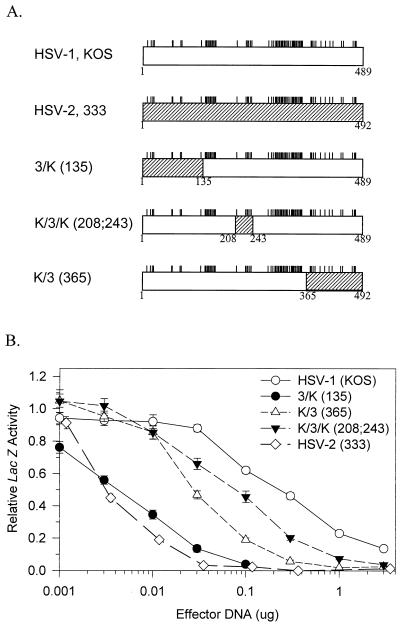
Structures and activities of chimeric UL41 alleles. (A) The UL41 polypeptide encoded by HSV-1 (strain KOS) is represented by the open rectangle, and that encoded by HSV-2 (strain 333) is represented by the hatched rectangle. The short vertical lines above the rectangles indicate the sites where the KOS and 333 polypeptides differ (9). The polypeptides encoded by several chimeric UL41 alleles are shown, with the portion contributed by KOS represented by an open rectangle and that contributed by strain 333 represented by a hatched rectangle. The junctions between KOS and 333 sequences are indicated by the coordinates of the KOS amino acids. (B) Replicate Vero cell cultures were transfected with 3 μg of the reporter plasmid pSV-β-Galactosidase and the indicated amounts of UL41-expressing effector plasmids. lacZ expression was determined 40 to 48 h after transfection and expressed as a fraction of that observed for transfections involving the pcDNA1.1amp vector and no UL41 effector plasmid. Error bars represent standard errors of the means. For datum points where no error bars are visible, the error bars were smaller than the datum points.
FIG. 2.
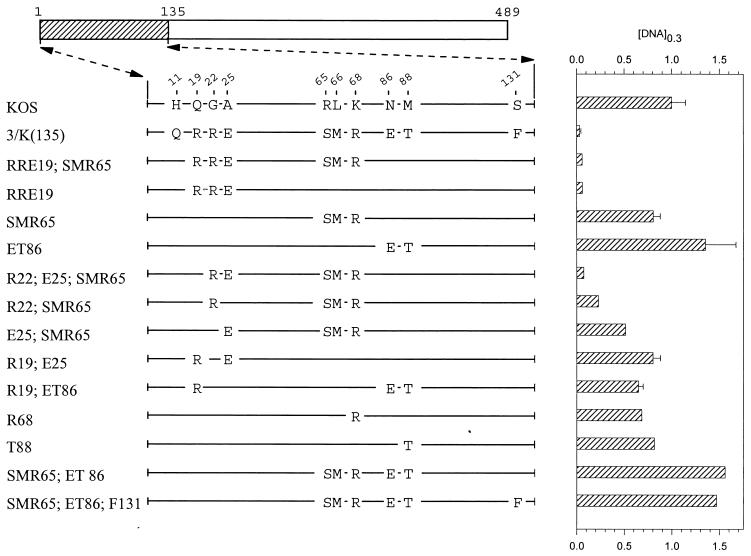
UL41 alleles with mutations in amino acids 1 to 135. The UL41 polypeptide of HSV-1 (strain KOS) is depicted by the open rectangle, with the portion (amino acids 1 to 135) containing site-directed mutations indicated by hatching. The structures of the UL41 polypeptides encoded by HSV-1(KOS) and the 3/K(135) chimera are shown, with only those amino acids that differ indicated. The UL41 polypeptides encoded by a number of mutant alleles are indicated, with only those amino acids at which the mutants differ from KOS indicated. Amino acid coordinates are those for the KOS allele. The [DNA]0.3 for each allele is shown at the right of the figure, expressed as a fraction of the [DNA]0.3 for KOS. The error bars represent standard errors of the means.
FIG. 7.
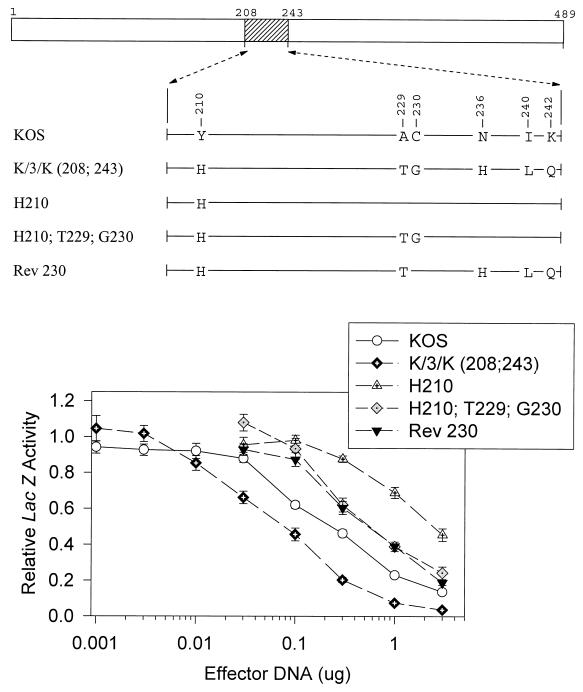
UL41 alleles with mutations in amino acids 208 to 243. The UL41 polypeptide of HSV-1 (strain KOS) is depicted by the open rectangle, with the portion (amino acids 208 to 243) containing site-directed mutations indicated by hatching. The structures of the UL41 polypeptides encoded by HSV-1(KOS) and the K/3/K(208;243) chimera are shown, with only those amino acids that differ indicated. The UL41 polypeptides encoded by a number of mutant alleles are shown, with only those amino acids at which the mutants differ from KOS indicated. Amino acid coordinates are those for the KOS allele. Dose-response curves for the inhibition of reporter gene expression by the KOS, K/3/K(208;243), and mutant alleles are shown at the bottom. Error bars represent standard errors of the means. For datum points where no error bars are visible, the error bars were smaller than the datum points.
TABLE 1.
Construction of site-directed mutations in region I (amino acids 1 to 135)
| Mutant no. | Mutant allele | Steps in constructiona |
|---|---|---|
| 1 | RRE19 | KOS→4→1 |
| 2 | RRE19;SMR65 | KOS→29→26→13→2 |
| 3 | Q11;RSE19 | KOS→4→3 |
| 4 | RSE19 | KOS→4 |
| 5 | Q11;RSE19;H435 | KOS→4→directed Q11 and spontaneous mutagenesis |
| 6 | R22;E25;SMR65 | KOS→29→26→13→9→6 |
| 7 | R19;S22 | KOS→4→7 |
| 8 | R19;E25 | KOS→4→8 |
| 9 | S22;E25;SMR65 | KOS→29→26→13→9 |
| 10 | S22;SMR65 | KOS→29→26→13→9→10 |
| 11 | R22;SMR65 | KOS→29→26→13→9→11 |
| 12 | E25;SMR65 | KOS→29→26→13→9→12 |
| 13 | RSE19;SMR65 | KOS→29→26→13 |
| 14 | RSE19;R68 | KOS→29→14 |
| 15 | RSE19;S65 | KOS→4→15 |
| 16 | RSE19;M66 | KOS→4→15 |
| 17 | RSE19;S65;M66 | KOS→29→26→13→17 |
| 18 | RSE19;SMR65;ET86 | KOS→29→26→21→18 |
| 19 | RSE19;SMR65;E86 | KOS→29→26→13→19 |
| 20 | RSE19;SMR65;T88 | KOS→29→26→13→20 |
| 21 | SMR65;ET86 | KOS→29→26→21 |
| 22 | Q11;SMR65;ET86 | KOS→29→26→21→22 |
| 23 | SMR65;ET86;F131 | KOS→29→26→21→23 |
| 24 | RSE19;T88 | KOS→30→24 |
| 25 | RSE19;R68;T88 | KOS→30→25 |
| 26 | SMR65 | KOS→29→26 |
| 27 | ET86 | KOS→30→27 |
| 28 | R19;ET86 | KOS→30→27→28 |
| 29 | R68 | KOS→29 |
| 30 | T88 | KOS→30 |
Steps by which the parent allele was altered by site-directed mutagenesis to yield the mutant allele. Mutant numbers are used.
TABLE 2.
Construction of site-directed mutations in region II (amino acids 208 to 243) and region III (amino acids 365 to 489)
| Mutant no. | Mutant allele | Parent allele | Steps in constructiona |
|---|---|---|---|
| 31 | H210 | KOS | KOS→31 |
| 32 | H210;T229;G230 | KOS | KOS→31→32 |
| 33 | H210;H236 | KOS | KOS→31→33 |
| 34 | H210;Q242 | KOS | KOS→31→34 |
| 35 | H210;T229;G230;H236 | KOS | KOS→39→35 |
| 36 | T229;G230 | KOS | KOS→36 |
| 37 | T229 | KOS | KOS→36→37 |
| 38 | G230 | KOS | KOS→36→38 |
| 39 | H236 | KOS | KOS→39 |
| 40 | L240 | KOS | KOS→40 |
| 41 | Q242 | KOS | KOS→41 |
| 42 | A396 | KOS | KOS→42 |
| 43 | S423 | KOS | KOS→43 |
| 44 | A396;S423 | KOS | KOS→43→44 |
| 45 | A396;WAH470 | KOS | KOS→48→45 |
| 46 | S423;WAH470 | KOS | KOS→48→46 |
| 47 | A396;S423;WAH470 | KOS | KOS→48→46→47 |
| 48 | WAH470 | KOS | KOS→48 |
| 49 | Rev 210 | K/3/K(208;243) | K/3/K(208;243)→49 |
| 50 | Rev 230 | K/3/K(208;243) | K/3/K(208;243)→50 |
| 51 | Rev 396 | K/3(365) | K/3(365)→51 |
| 52 | Rev 423 | K/3(365) | K/3(365)→52 |
| 53 | Rev 396;423 | K/3(365) | K/3(365)→52→53 |
| 54 | Rev 423;FrSh425 | K/3(365) | K/3(365)→52 + spontaneous mutagenesis |
| 55 | Rev 396;423;FrSh425 | K/3(365) | K/3(365)→52→53 + spontaneous mutagenesis |
| 56 | RSE19;F131;RLQ200 | K/3(365) | KOS→4→directed F131 + spontaneous mutagenesis |
Steps by which the parent allele was altered by site-directed mutagenesis to yield the mutant allele. Mutant numbers are used.
DNA isolation and sequencing.
Plasmids for transfections and sequencing were prepared from bacterial lysates by using the MidiPrep and MaxiPrep systems as recommended by the manufacturer (Qiagen Corp.). Sequencing of the UL41 alleles was accomplished with an Applied Biosystems model 377 DNA Sequencer in the Molecular Biology Core Facility of the University of Missouri—Kansas City.
Transient-expression assay for vhs activity.
vhs activity was measured, as described previously (9), by determining the ability of a transfected UL41 allele to inhibit expression of a cotransfected reporter plasmid containing the E. coli lacZ gene under the control of the simian virus 40 early promoter and enhancer. Transfections were performed by using the Profection Mammalian Transfection System (Promega) according to the manufacturer’s instructions. Briefly, Vero cells were plated the day before transfection in 60-mm-diameter petri dishes at a density of 2.5 × 104 cells/cm2. Three hours before transfection, the medium was replaced with fresh Eagle’s minimum essential medium containing 10% (vol/vol) calf serum. The cultures were transfected with 0.6-ml aliquots containing calcium phosphate coprecipitates of 3 μg of the reporter plasmid pSV-β-Galactosidase (Promega) and various amounts of a UL41-containing effector plasmid. Each transfection mixture also contained enough vector to maintain the amount of CMV promoter sequences (effector plasmid plus vector) equal to 0.73 pmol (3.36 μg of pKOSamp is 0.73 pmol), and enough salmon sperm carrier DNA to bring the total amount of DNA to 12 μg.
Cell extracts were prepared 40 to 48 h after transfection and assayed for reporter gene expression by using a β-galactosidase enzyme assay system purchased from Promega (Madison, Wis.). Briefly, cell extracts were aliquoted into 96-well trays and mixed with substrate, and the absorbance at 405 nm was determined at various times over a 60-min interval by using a Thermo Max Microplate Reader (Molecular Devices, Sunnyvale, Calif.). The β-galactosidase activity in each well was determined from the initial velocity of the enzyme reaction. In each experiment, triplicate cultures were transfected with each concentration of effector plasmid. For each culture, the amount of β-galactosidase activity was expressed as a fraction of that observed in transfections involving 0.73 pmol of vector without any UL41-containing effector plasmid. For each effector allele, the data were plotted to yield a dose-response curve showing inhibition of reporter gene expression as a function of the concentration of effector DNA. A curve was fitted to the data by third-order regression analysis with SigmaPlot version 2.02 (Jandel Scientific, San Rafael, Calif.) and used to determine the concentration of effector DNA required to reduce the reporter gene expression to 30% of the control value ([DNA]0.3). Replicate experiments were performed on different days, each yielding a separate dose-response curve and value of [DNA]0.3. These values were then averaged to yield the values of [DNA]0.3 and to calculate the standard deviations from the means.
Homology searches and alignments.
Searches for UL41 homologues were performed by comparing the sequence of the UL41 protein from HSV-1 KOS to other known protein sequences with the BLAST search program (1). UL41 homologues from the various alphaherpesviruses were aligned by using the ClustalW alignment algorithm of MacVector version 6.0 (Oxford Molecular, Campbell, Calif.).
RESULTS
Activities of wild-type and chimeric UL41 alleles.
In an earlier study (9), we compared the activities of UL41 alleles from HSV-1 (strain KOS) and HSV-2 (strain 333) by using an assay in which cells were transfected with a constant amount of a lacZ reporter gene and increasing amounts of a UL41 allele. vhs activity was determined by the ability of the transfected UL41 allele to inhibit lacZ expression (9). This assay allows the activities of UL41 polypeptides to be compared in vivo in the absence of other viral gene products. This is important because during virus infections, interactions with other viral proteins are likely to influence the activity of the UL41 polypeptide. vhs has been shown to interact with VP16 in vitro and in the yeast two-hybrid system (39, 41), and interactions with VP16 have been implicated as important in controlling the activity of newly synthesized copies of the vhs (UL41) polypeptide at late times during virus infections (25). Interactions with VP16 and/or other viral proteins may be important for packaging of the vhs (UL41) protein into virus, and disruption of these interactions may be required for its release from incoming virions. Mutations that prevent vhs packaging or its release from infecting virions would result in virus lacking virion host shutoff activity, even if the UL41 protein still retained mRNA degradative activity. For these reasons, it was desirable to assay the activity of UL41 alleles in the absence of other viral proteins.
In the cotransfection assay, both HSV-1 and HSV-2 alleles inhibited reporter gene expression over a range of transfected vhs DNA concentrations. However, 40-fold less of the HSV-2 allele was required to yield the same level of inhibition as HSV-1, indicating that the HSV-2 allele encodes a significantly more active UL41 polypeptide than its HSV-1 homologue (Fig. 1) (9). The assay also was used to examine a series of chimeric UL41 alleles containing various combinations of HSV-1 and HSV-2 sequences (9). The structures of key chimeric alleles and their dose-response curves are shown in Fig. 1. The chimera 3/K(135) encodes a UL41 polypeptide with the first 135 amino acids from HSV-2 (strain 333) fused to amino acids 136 through 489 of KOS. This allele inhibited reporter gene expression at vhs DNA concentrations that were 40-fold less than those required for the KOS allele (Fig. 1) and was only slightly less active, in this assay, than the wild-type strain 333 allele. The chimera K/3(365) encodes a polypeptide with the first 365 amino acids from KOS fused to amino acids 366 through 492 from strain 333. This allele did not inhibit reporter expression as well as 3/K(135) but still was significantly more active than KOS, requiring 10-fold-lower vhs DNA concentrations to yield the same level of inhibition. The chimera K/3/K(208;243) encodes a polypeptide with amino acids 208 to 243 from strain 333 sandwiched between amino acids 1 to 135 and 244 to 489 from KOS. It was the least active of the chimeras; however, it was still significantly more active than KOS, requiring approximately 2.5-fold-lower DNA concentrations to yield the same level of inhibition. Examination of these chimeric UL41 alleles identified three regions of the strain 333 polypeptide (amino acids 1 to 135, 208 to 243, and 365 to 492) that increase the activity of KOS when substituted for the corresponding amino acids of the KOS protein. To extend these results, we turned to site-directed mutagenesis of the strain KOS and 333 alleles.
Mutations in region 1 (amino acids 1 to 135).
Within the first 135 amino acids of UL41, the strain KOS and 333 polypeptides differ at 10 locations (see Fig. 2). To determine which of these differences contribute to the greater activity of the strain 333 polypeptide, the wild-type KOS allele was mutated to introduce selected strain 333 amino acids into an otherwise KOS background. Dose-response curves were determined for each of the alleles and used to determine the concentration of the mutant allele ([DNA]0.3) that reduced lacZ expression to 30% of the control value. These values are shown in Fig. 2 and 3 along with the structures of the mutant alleles.
FIG. 3.
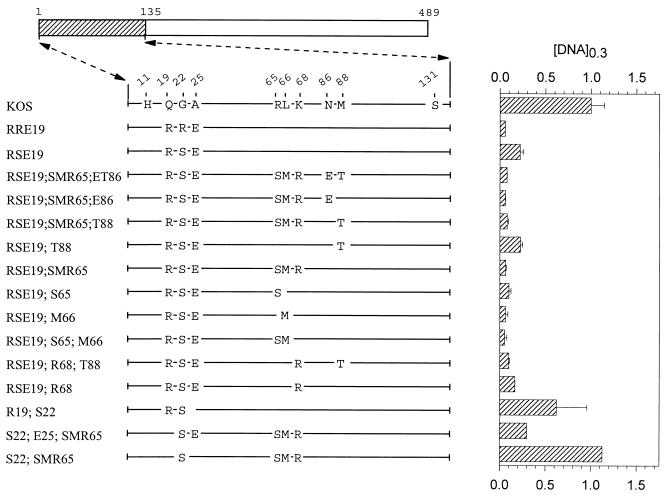
UL41 alleles with mutations in amino acids 1 to 135. The UL41 polypeptide encoded by HSV-1 (strain KOS) is depicted by the open rectangle, with the portion (amino acids 1 to 135) containing site-directed mutations indicated by hatching. The structure of the KOS polypeptide is shown, with only those amino acids at which it differs from the 3/K(135) chimera shown. The UL41 polypeptides encoded by a number of mutant alleles are shown, with only those amino acids that differ from KOS indicated. Amino acid coordinates are those for the KOS allele. The [DNA]0.3 for each allele is shown at the right of the figure, expressed as a fraction of the [DNA]0.3 for KOS. The error bars represent standard errors of the means.
The alleles SMR65;ET86 and SMR65;ET86;F131 (Fig. 2) both contained clusters of strain 333-specific amino acids located between residues 65 and 131. Neither allele was more active than KOS, indicating that the introduction of these HSV-2 residues, by themselves, did not enhance the KOS activity. Interestingly, the allele RRE19;SMR65, which encodes strain 333-specific amino acids at positions 19, 22, 25, 65, 66, and 68, inhibited reporter gene expression almost as much as the 3/K(135) chimera (Fig. 2). SMR65 (Fig. 2), which contains the cluster of changes at positions 65, 66, and 68, was not appreciably more active than KOS. In contrast, RRE19, which contains the strain 333-specific cluster R19, R22, and E25, was just as active as RRE19;SMR65 and the 3/K(135) chimera (Fig. 2). Thus, the presence of one or more of the strain 333-specific amino acids at positions 19, 22, and 25 was able to greatly enhance UL41 activity when introduced into an otherwise KOS polypeptide. This effect was due primarily to R22 and E25, since the mutant R22;E25;SMR65 was just as active as RRE19;SMR65 or RRE19 (Fig. 2). In addition, although the mutants R22;SMR65 and E25;SMR65 were both more active than KOS, neither was as active as R22;E25;SMR65 (Fig. 2) indicating that the combination of R22;E25 was necessary for the full effect. The results indicate that the combination of strain 333-specific amino acids R22 and E25 is able to greatly enhance UL41 activity when introduced into an otherwise KOS polypeptide.
During mutagenesis of region 1, an error in the design of a mutagenic primer led to the construction of the mutant RSE19, which contains the amino acids R19, S22, and E25 in an otherwise KOS background (Fig. 3). This mutant was significantly more active than the parental KOS allele, although it did not inhibit reporter gene expression quite as well as the RRE19 allele (Fig. 3). Interestingly, the UL41 homologues encoded by varicella-zoster and pseudorabies viruses contain a serine at position 22, while the UL41 polypeptides of equine herpesvirus 1 and bovine and gallid herpesviruses contain a threonine at residue 22 (see Fig. 8). The effect of the RSE19 cluster in enhancing the activity of the KOS allele was augmented somewhat by the presence of the cluster of strain 333-specific amino acids S65, M66, and R68. Thus, the mutant RSE19;SMR65 (Fig. 3) inhibited reporter gene expression just as much as RRE19. This augmentation was also caused by just S65, just M66, or a combination of the two (Fig. 3). Taken together, the data indicate that, in an otherwise KOS background, the activity of UL41 is greatly affected by the identities of amino acids 22 and 25. The combination of strain 333-specific residues R22 and E25 is primarily responsible for the enhanced activity of the chimeric 3/K(135) allele. The cluster R19, S22, and E25 also enhances the KOS activity, an effect which is augmented by strain 333-specific residues at positions 65 and 66.
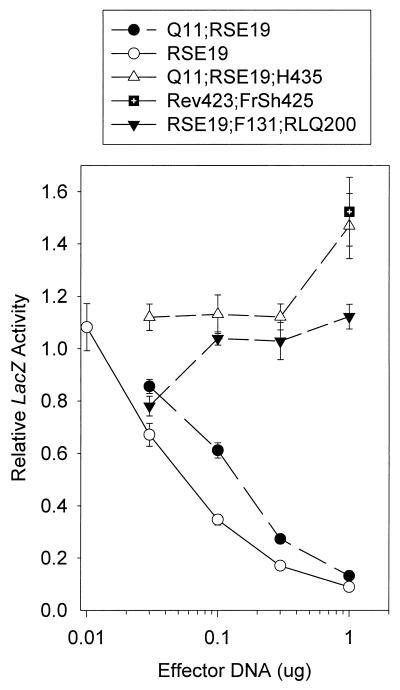
FIG. 8.
Dose-response curves for the inhibition of reporter gene expression by spontaneous and site-directed mutants of UL41. Replicate Vero cell cultures were transfected with 3 μg of the reporter plasmid pSV-β-Galactosidase and the indicated amounts of UL41-expressing effector plasmids. lacZ expression was determined 40 to 48 h after transfection and expressed as a fraction of that observed for transfections involving the pcDNA1.1amp vector and no UL41 effector plasmid. Error bars represent standard errors of the means. For datum points where no error bars are visible, the error bars were smaller than the datum points.
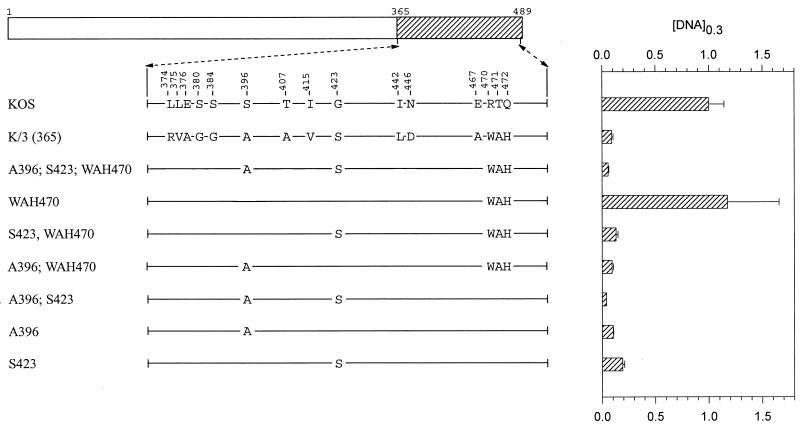
Mutations in region 3 (amino acids 365 to 489).
The chimera K/3(365) differs from KOS at 15 codons scattered between positions 374 and 472 (Fig. 4). K/3(365) was significantly more active than KOS, requiring only 10% as much vhs DNA to cause equal inhibition of lacZ expression (Fig. 1 and Fig. 4). Initially, site-directed mutagenesis was used to introduce strain 333-specific amino acids into an otherwise KOS background. The mutant A396;S423;WAH470 differs from KOS at only five locations (Fig. 4). Nevertheless, it inhibited lacZ expression, if anything, slightly better than the K/3(365) chimera. The mutant WAH470 contains the triplet of strain 333-specific amino acids W470, A471, and H472, yet it inhibited reporter expression no better than KOS (Fig. 4). In contrast, the mutants A396 and S423, each of which is identical to KOS except for one amino acid, were as active as the K/3(365) chimera (Fig. 4), and the double mutant A396;S423 was even more active (Fig. 5 and Fig. 4).
FIG. 4.
UL41 alleles with mutations in amino acids 365 to 489. The UL41 polypeptide of HSV-1 (strain KOS) is depicted by the open rectangle, with the portion (amino acids 365 to 489) containing site-directed mutations indicated by hatching. The structures of the UL41 polypeptides encoded by HSV-1(KOS) and the K/3(365) chimera are shown, with only those amino acids that differ indicated. The UL41 polypeptides encoded by a number of mutant alleles are shown, with only those amino acids that differ from KOS indicated. Amino acid coordinates are those for the KOS allele. The [DNA]0.3 for each allele is shown at the right of the figure, expressed as a fraction of the [DNA]0.3 for KOS. The error bars represent standard errors of the means.
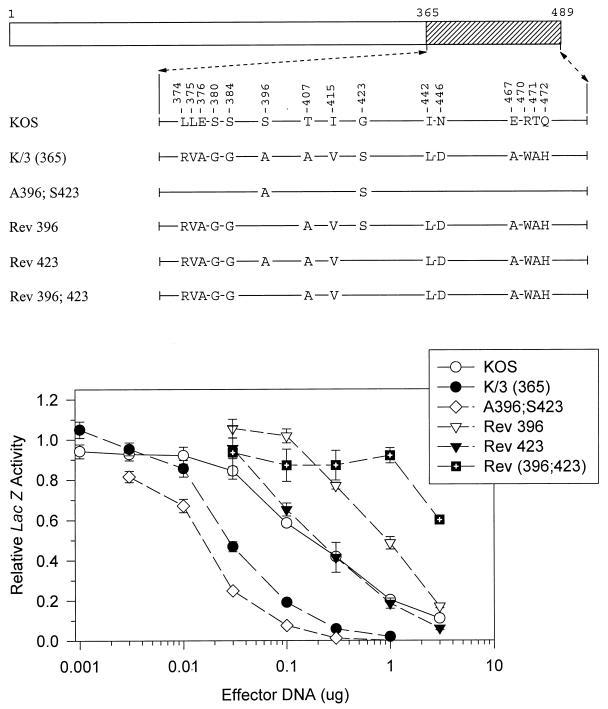
FIG. 5.
UL41 alleles with mutations in amino acids 365 to 489. The UL41 polypeptide of HSV-1 (strain KOS) is depicted by the open rectangle, with the portion (amino acids 365 to 489) containing site-directed mutations indicated by hatching. The structures of the UL41 polypeptides encoded by HSV-1(KOS) and the K/3(365) chimera are shown, with only those amino acids that differ indicated. The UL41 polypeptides encoded by a number of mutant alleles are shown, with only those amino acids that differ from KOS indicated. Amino acid coordinates are those for the KOS allele. Dose-response curves for the inhibition of reporter gene expression by the KOS, K/3(365), and mutant alleles are shown at the bottom. Error bars represent standard errors of the means. For datum points where no error bars are visible, the error bars were smaller than the datum points.
To further investigate the implication that A396 and S423 are important to the greater activity of the strain 333 allele, the K/3(365) chimera was mutated to revert these residues to their KOS counterparts. Rev 423 is identical to K/3(365) except for the presence of a KOS-specific glycine at residue 423 (Fig. 5). This allele was indistinguishable from KOS in the cotransfection assay, supporting the conclusion that S423 is key to the enhanced activity of strain 333. The mutant Rev 396 is identical to K/3(365) except for a serine at position 396 (Fig. 5). Interestingly, this allele was less active than KOS in the cotransfection assay. Even more striking was the observation that Rev 396;423, which has KOS-specific amino acids at both positions, was even less active than Rev 396 (Fig. 5). Taken together, the data indicate that alanine 396 and serine 423 are key to the greater UL41 activity of HSV-2 (strain 333). Furthermore, in the absence of these two strain 333-specific amino acids, a UL41 polypeptide containing the 13 other strain 333-specific residues within region 3 is actually less active than the KOS protein.
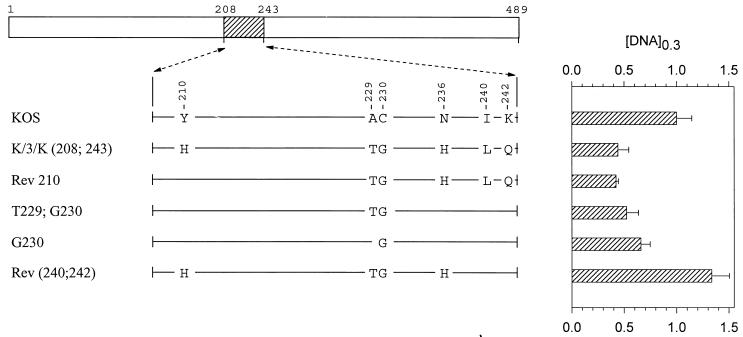
Mutations in region 2 (amino acids 208 to 243).
The last chimera that was found to be significantly more active than KOS in the transient-expression assay is K/3/K(208;243). This allele encodes a polypeptide that differs from KOS at six locations scattered between amino acids 210 and 242 (Fig. 6). While more active than KOS, it was less active than either 3/K(135) or K/3(365), requiring 40% as much vhs DNA to yield the same level of inhibition as KOS (Fig. 1, 6, and 7). Rev 210 contains five of the six strain 333-specific amino acids, lacking only histidine 210, and inhibited lacZ expression as readily as the K/3/K(208;243) chimera (Fig. 6). This suggests that histidine 210 is not required for the greater activity of strain 333. The mutant G230 is identical to KOS except for a glycine at position 230. Nevertheless, it was significantly more active, requiring only two-thirds as much vhs DNA to yield equivalent inhibition (Fig. 6). vhs activity was accentuated even more for the double mutant T229;G230 (Fig. 6), indicating that this pair of strain 333-specific amino acids enhances vhs activity in an otherwise KOS background.
FIG. 6.
UL41 alleles with mutations in amino acids 208 to 243. The UL41 polypeptide of HSV-1 (strain KOS) is depicted by the open rectangle, with the portion (amino acids 208 to 243) containing site-directed mutations indicated by hatching. The structures of the UL41 polypeptides encoded by HSV-1(KOS) and the K/3/K(208;243) chimera are shown, with only those amino acids that differ indicated. The UL41 polypeptides encoded by a number of mutant alleles are shown, with only those amino acids at which the mutants differ from KOS indicated. Amino acid coordinates are those for the KOS allele. The [DNA]0.3 for each allele is shown at the right of the figure, expressed as a fraction of the [DNA]0.3 for KOS. The error bars represent standard errors of the means.
Interestingly, Rev (240;242), which contains the T229;G230 pair along with two other strain 333-specific amino acids, was less active than the parental KOS allele (Fig. 6). This was even more the case for the mutant H210, which is identical to KOS except for a histidine at position 210 (Fig. 7). In fact, the activity of this mutant was so low that a value of [DNA]0.3 could not be determined because it failed to inhibit lacZ expression to 30% of the control value for every DNA concentration that was tested. Inclusion of the pair of strain 333-specific residues T229 and G230 along with histidine 210 increased the inhibitory activity of the allele somewhat (H210;T229;G230), but not to the level of KOS (Fig. 7). A similar observation was made for Rev 230, which is identical to the K/3/K(208;243) chimera except at amino acid 230 yet was not as active as KOS (Fig. 7). In sum, the strain 333-specific residue histidine 210 inhibited vhs activity when introduced into an otherwise KOS background, and most or all of the other five 333-specific residues were required to restore activity to that of the K/3/K(208;243) chimera.
Spontaneous mutations.
During mutant construction, several UL41 alleles were isolated that had lost all ability to inhibit reporter gene expression in the transient-expression assay. Upon sequencing, these alleles were found to contain several spontaneous point mutations and deletions. The allele Rev 423; FrSh 425 contains a deletion of the first base of codon 426 in a background of the Rev 423 allele. As was seen in Fig. 7, Rev 423 has a vhs activity that is indistinguishable from that of KOS. The deletion in Rev 423; FrSh 425 causes a frameshift, resulting in production of a 432-amino-acid UL41 polypeptide containing the first 425 amino acids of Rev 423 fused to the sequence SGDPGLF. As can be seen in Fig. 8, this allele lacked detectable vhs activity, suggesting that sequences beyond amino acid 425 are critical to vhs activity.
The allele Q11;RSE19;H435 contains a point mutation that changes arginine 435 to histidine, in a background that is identical to KOS, except for the amino acids Q11, R19, S22, and E25. This allele is identical to Q11;RSE19, except for the presence of a histidine instead of arginine at position 435. Nevertheless, Q11;RSE19;H435 failed to inhibit reporter gene expression, even though Q11;RSE19 was almost as active as the RSE19 allele and considerably more active than KOS (Fig. 8). Thus, a change of arginine to histidine at position 435 abolished detectable vhs activity.
Similarly, the allele RSE19;F131;RLQ200 is identical to RSE19 except for the four amino acids F131, R200, L201, and Q200. Nevertheless, it failed to inhibit reporter gene expression in the transient-expression assay (Fig. 8). This loss of activity probably was not due to the change of serine to phenylalanine at position 131, since the same mutation did not adversely affect the activity of the SMR65;ET86 allele (compare the SMR65;ET86 and SMR65;ET86;F131 alleles in Fig. 2). Thus, the change of leucine 200, tyrosine 201, and histidine 202 to arginine, leucine, and glutamine, respectively, appears to abrogate UL41 activity.
DISCUSSION
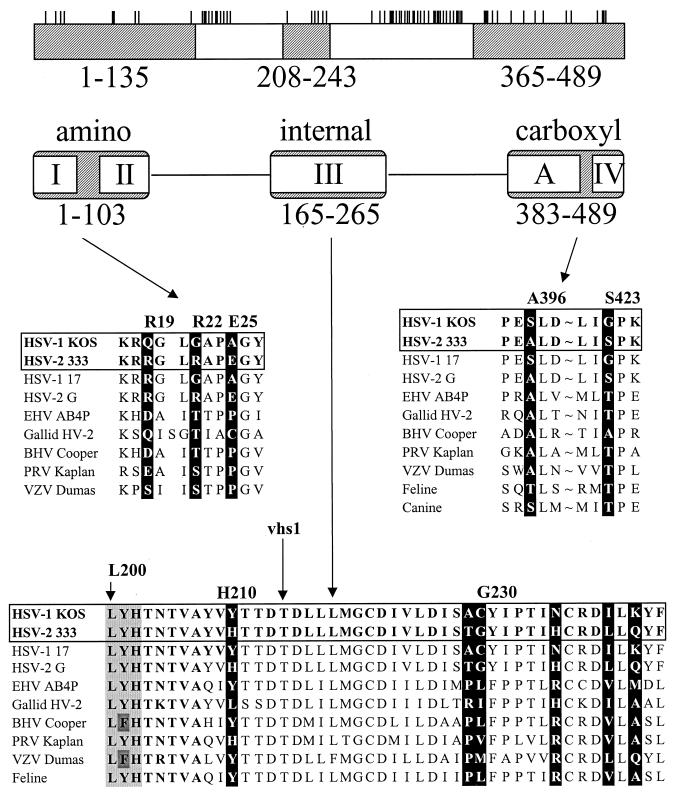
This study identifies several type-specific amino acids that are largely responsible for the difference in mRNA degradative activities of the UL41 homologues of HSV-1(KOS) and HSV-2(333). In an earlier report (9), we identified three regions of the strain 333 polypeptide that significantly enhance the activity of KOS when substituted for the corresponding portions of the HSV-1 protein, either singly or in combination. As is shown in Fig. 9, these regions overlap stretches of amino acids that are conserved in the UL41 homologues of the other alphaherpesviruses (9, 19). These results have now been extended through the use of site-directed mutagenesis to introduce selected strain 333-specific amino acids into an HSV-1 background and to revert strain 333-specific amino acids to their HSV-1 counterparts.
FIG. 9.
Summary of the residues important to type-specific differences in UL41 activity. The UL41 polypeptide of HSV-1 (strain KOS) is depicted by the open rectangle at the top of the figure. The short vertical lines above the rectangle indicate the sites at which the strain KOS and 333 polypeptides differ (9). Replacement of the shaded regions of the KOS polypeptide (amino acids 1 to 135, 208 to 243, and 365 to 489) with the corresponding regions of the HSV-2 (strain 333) polypeptide has been shown to significantly enhance UL41 activity (9). The regions of the KOS polypeptide that are conserved in the UL41 homologues of the alphaherpesviruses are summarized, with roman numerals I through IV referring to the conserved regions identified by Berthomme and coworkers (3), while the region labeled A was identified by Jones and colleagues (19). The sequences of UL41 homologues corresponding to the portions of the HSV-1 and HSV-2 polypeptides that are responsible for type-specific differences are shown in the middle and lower portions of the figure. The sequences of the various UL41 homologues are taken from references cited in the text. The amino acids identified in this study as being important for the difference in UL41 activity of HSV-1(KOS) and HSV-2(333) are highlighted by white type in black boxes. Amino acid numbers refer to the positions of the KOS residues. Sequences between amino acids 398 and 421 were omitted (indicated by ∼). Threonine 214 that is mutated to isoleucine in the mutant vhs 1 is indicated by an arrow. The triplet of amino acids (leucine 200, tyrosine 201, and histidine 202) that is altered in one of the spontaneous mutants is indicated by the light shading in the bottom portion of the figure. Abbreviations: EHV, equine herpesvirus; HV-2, herpesvirus 2; BHV, bovine herpesvirus; PRV, pseudorabies virus; VZV, varicella-zoster virus.
The key amino acids responsible for the type-specific difference in UL41 activity are summarized in Fig. 9. The most dramatic effect was seen for an allele containing just three strain 333-specific amino acids (arginine 19, arginine 22, and glutamic acid 25) in an otherwise KOS background. This allele inhibited reporter gene expression just as efficiently as an intertypic chimera [3/K(135)] containing 10 HSV-2 specific residues scattered between positions 11 and 131, and almost as well as the parental HSV-2 allele. Comparison of alleles containing various combinations of these three amino acids suggested that the enhancement was due primarily to arginine 22 and glutamic acid 25 and that, although by itself either residue enhanced activity somewhat, the full effect required the pair of strain 333-specific amino acids. Similarly, an allele with just two strain 333-specific amino acids (alanine 396 and serine 423) inhibited lacZ expression even more efficiently than the chimera K/3(365), which has 14 HSV-2 amino acids scattered between positions 374 and 472. While this allele did not inhibit the reporter quite as well as the parental strain 333 allele, it was substantially more active than KOS, requiring 10 times less UL41 DNA to achieve the same level of inhibition. Taken together, the data indicate that four HSV-2-specific amino acids (R22, E25, A396, and S423) can account for much of the difference in activities of the UL41 alleles from strains KOS and 333.
Although not as dramatic in their effect as changes at positions 22, 25, 396, and 423, the introduction of six strain 333-specific amino acids between residues 210 and 242 had a modest but reproducible effect upon UL41 activity. Thus, the chimera K/3/K(208;243) required two and one half times less UL41 DNA to yield the same level of inhibition as KOS (Fig. 1 and 6) (9). Some enhancement of KOS activity could be obtained by introducing the strain 333-specific residues, threonine 229 and glycine 230. However, the full effect required the substitution of all six strain 333-specific amino acids for their KOS counterparts. Interestingly, in several instances, the substitution of fewer than all six strain 333-specific amino acids resulted in UL41 alleles that inhibited lacZ expression less efficiently than the parental KOS allele. The most striking example was the substitution of a strain 333-specific histidine for tyrosine at position 210, which almost abolished UL41 activity. The data are consistent with the possibilities that these six amino acids are part of a single functional domain and that they must be altered in a coordinated fashion to maintain activity.
Comparison of the UL41 alleles of alphaherpesviruses reveals a preference for certain amino acids at some of the key positions identified above (Fig. 9). Thus, 8 of 11 alleles have a serine or threonine at position 423, while all 11 have alanine, serine, or threonine at position 396. Although both of the sequenced HSV-1 alleles have a glycine at position 22, and both HSV-2 alleles have an arginine, the UL41 alleles of five other alphaherpesviruses have a serine or threonine at this position. Interestingly, alteration of the KOS allele to introduce a serine at position 22, along with a glutamic acid at position 25, significantly increased its activity.
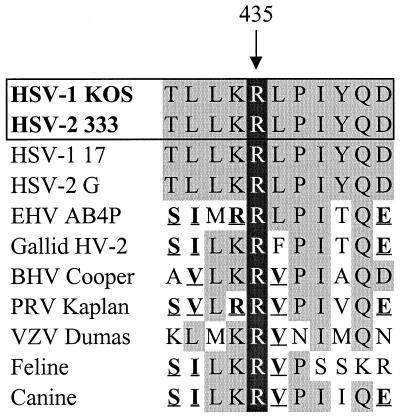
In all likelihood, UL41 polypeptides contain certain amino acids or motifs that are absolutely required for activity, as well as others that, while not required, modulate the amplitude of the activity. Amino acids that vary between the HSV-1 and HSV-2 polypeptides and are responsible for the difference in their activities probably fall into the second category. Many of the required amino acids may be invariant between UL41 alleles or undergo only conservative changes. These may include amino acids that were altered in the spontaneous mutants that had lost all UL41 activity. In this study, a point mutation that changed arginine 435 to histidine resulted in a UL41 allele that lacked activity. This effect could not be explained simply by an alteration in the charge of the protein, since both arginine and histidine are positively charged at neutral pH. Interestingly, arginine 435 is invariant in all UL41 alleles that have been sequenced (Fig. 10), suggesting that it may be a required residue for vhs activity. Similarly, a cluster of point mutations that changed leucine 200, tyrosine 201, and histidine 202 to arginine, leucine, and glutamine, respectively, abolished detectable UL41 activity. This triplet of amino acids is highly conserved among the currently sequenced UL41 alleles (Fig. 9), with the only variations being the conservative change of tyrosine 201 to phenylalanine in two of the alleles. Leucine 200 is also homologous to a leucine that is conserved in a number of cellular nucleases with homology to UL41 (9, 10), further supporting the idea that it may be a required amino acid for UL41 activity.
FIG. 10.
Sequences of UL41 homologues surrounding arginine 435. Arginine 435, which is invariant in all of the sequenced UL41 homologues of alphaherpesviruses, is highlighted by white type in black boxes. Alteration of this residue to histidine completely abolished UL41 activity. Amino acids that are identical to residues in HSV-1 (strain KOS) are shown in lightly shaded boxes, while conservative changes are shown in boldface and underlined. Abbreviations: EHV, equine herpesvirus; HV-2, herpesvirus 2; BHV, bovine herpesvirus; PRV, pseudorabies virus; VZV, varicella-zoster virus.
In addition to these point mutations, a UL41 allele containing a frameshift after codon 425 was inactive in the transient-expression assay, suggesting that some amino acids between positions 426 and 489 are required for mRNA degradative activity. This is consistent with the earlier finding that a nonsense mutant allele encoding a 382-amino-acid UL41 polypeptide was inactive in the transient-expression assay and produced vhs-deficient virions after introduction into virus (30). This was true even though the truncated UL41 polypeptide was incorporated into virions. Similarly, Strelow and Leib reported that virions carrying a nonsense mutation that truncated the UL41 polypeptide at 460 amino acids lacked virion host shutoff activity, although they did not show that the mutant polypeptide was incorporated into virus (45).
All of the experiments in this study utilized a transient-expression assay of vhs activity that measures the ability of a transfected UL41 allele to inhibit expression of a cotransfected lacZ reporter gene. During virus infections, the vhs activity of HSV strains may differ for a variety of reasons. The UL41 polypeptides may differ in mRNA degradative activity, with regard to how much of the protein is incorporated into virions, or how rapidly and efficiently it is released from incoming virus particles. While all these factors ultimately may be important, a useful first step in deciphering strain-specific differences in vhs activity would be to compare the mRNA degradative activities of UL41 polypeptides in the absence of potential complications due to interactions with other viral gene products.
The transient-expression assay of UL41 activity offers this advantage. Nevertheless, the results from these experiments should be interpreted with the caveat that, because different transfected UL41 alleles may express different amounts of the UL41 polypeptide, the assay does not permit a truly quantitative measurement of the specific mRNA degradative activities of different UL41 proteins. For example, some mutations might result in UL41 proteins with increased stability, leading to increased amounts of the polypeptide within transfected cells. These alleles might be perceived as more active in the transient-expression assay even if the UL41 polypeptides do not have enhanced mRNA degradative activity. This does not appear to be the explanation for the increased activity of the 3/K(135) allele, the most active of the intertypic chimeras. Examination of transfected cells by indirect immunofluorescence and Western blotting indicates that there is significantly less of the UL41 polypeptide in cells transfected with the 3/K(135) chimera than in those transfected with the parental KOS allele (31). Conversely, for a number of mutant UL41 alleles that have decreased activity in the transient-expression assay, the vhs polypeptides are expressed at higher levels than is the KOS protein (10, 30). Thus, the transient-expression assay appears to underestimate the difference between the activities of many of the most active and least active UL41 alleles, since the polypeptides encoded by the more active alleles are present in lower amounts than the proteins encoded by the less active alleles. In sum, the results of the current study indicate that the UL41 polypeptides of strains KOS and 333 differ in their intrinsic mRNA degradative activities. However, one cannot exclude the possibilities that, within infected cells, they also differ in their interactions with other viral proteins, and that this contributes to the difference in the host shutoff activities of HSV-1 and HSV-2. Efforts are under way to introduce some of the mutant UL41 alleles into virus to examine their activities in the context of a virus infection.
While these studies identify several amino acids that modulate UL41 activity, they do not indicate why these residues should have such a major effect upon vhs function. This is because the precise activity of the UL41 protein remains to be determined. Specifically, it is unclear whether UL41 is itself a RNase or somehow activates a cellular enzyme. Data suggesting that the vhs protein is a RNase include the observation that it shares regions of sequence homology with a number of nucleases from mammalian cells, Saccharomyces cerevisiae, and bacteria (7, 9, 10). Site-directed mutagenesis of several UL41 residues corresponding to amino acids critical to the nuclease activity of cellular homologues shows that they are critical to vhs activity as well (10). In addition, extracts of partially purified virions exhibit a RNase activity that appears to be vhs dependent since it is present in extracts of wild-type but not vhs mutant virions and can be blocked by UL41-specific antisera (53). If UL41 indeed is a RNase, a number of questions remain concerning its specificity or targeting. First, while vhs degrades mRNAs, a number of its putative cellular homologues have DNase activity. It is unclear what features of the UL41 protein restrict it to RNA. Second, although vhs does not appear to discriminate between different kinds of mRNA, it exhibits a strong preference for mRNAs over non-mRNAs. This is true both in vivo (29, 38, 46) and in in vitro reactions involving cytoplasmic extracts from infected cells (22, 42). In addition, recent studies indicate that the in vivo degradation of at least one target mRNA initiates at or near the 5′ end of the mRNA (20). Whether this was due to targeting of the vhs protein to 5′ ends or to regions of translation initiation is unclear. In contrast, the RNase activity observed in virion extracts was not restricted to mRNAs and cleaved target RNAs at multiple internal sites (53). The data are consistent with the possibility that the purified UL41 polypeptide will turn out to be a RNase with significantly less specificity than that which is observed in vivo. The precise nature of the UL41 activity and how it is targeted are questions that will be answered by further genetic and biochemical characterization of the vhs polypeptide.
ACKNOWLEDGMENTS
We thank Mary Patterson and Pinghui Feng for helpful discussions concerning all aspects of these experiments. We are indebted to Alfred Esser for allowing us to use his 96-well plate reader and to Krys Morris of the UMKC Molecular Biology Core Facility for sequencing the mutant UL41 alleles. Our colleague Lindsey Hutt-Fletcher had helpful suggestions at all stages of this work.
This work was supported in part by grant AI21501 from the National Institute of Allergy and Infectious Diseases and by a grant from the University of Missouri Research Board.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker Y, Tavor E, Asher Y, Berkowiltz C, Moyal M. Effect of herpes simplex virus type-1 UL41 gene on the stability of mRNA from the cellular genes: beta-actin, fibronectin, glucose transporter-1, and docking protein, and on virus intraperitoneal pathogenicity of newborn mice. Virus Genes. 1993;7:133–143. doi: 10.1007/BF01702393. [DOI] [PubMed] [Google Scholar]
- 3.Berthomme H, Jacquemont B, Epstein A. The pseudorabies virus host-shutoff homolog gene: nucleotide sequence and comparison with alphaherpesvirus protein counterparts. Virology. 1993;193:1028–1032. doi: 10.1006/viro.1993.1221. [DOI] [PubMed] [Google Scholar]
- 4.Caponigro G, Parker R. Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darteil R, Bublot M, Laplace E, Bouquet J F, Audonnet J C, Riviere M. Herpesvirus of turkey recombinant viruses expressing infectious bursal disease virus (IBDV) VP2 immunogen induce protection against an IBDV virulent challenge in chickens. Virology. 1995;211:481–490. doi: 10.1006/viro.1995.1430. [DOI] [PubMed] [Google Scholar]
- 6.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 7.Doherty A J, Serpell L C, Pointing C P. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everett R D, Fenwick M L. Comparative DNA sequence analysis of the host shutoff genes of different strains of herpes simplex virus: type 2 strain HG52 encodes a truncated UL41 product. J Gen Virol. 1990;71:1387–1390. doi: 10.1099/0022-1317-71-6-1387. [DOI] [PubMed] [Google Scholar]
- 9.Everly D N, Jr, Read G S. Mutational analysis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): characterization of HSV type 1 (HSV-1)/HSV-2 chimeras. J Virol. 1997;71:7157–7166. doi: 10.1128/jvi.71.10.7157-7166.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everly, D. N., Jr., and G. S. Read. Unpublished data.
- 11.Feng X, Thompson Y G, Lewis J B, Caughman G B. Expression and function of the equine herpesvirus 1 virion-associated host shutoff homolog. J Virol. 1996;70:8710–8718. doi: 10.1128/jvi.70.12.8710-8718.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenwick M L, Everett R D. Inactivation of the shutoff gene (UL41) of herpes simplex virus types 1 and 2. J Gen Virol. 1990;71:2961–2967. doi: 10.1099/0022-1317-71-12-2961. [DOI] [PubMed] [Google Scholar]
- 13.Fenwick M L, Everett R D. Transfer of UL41, the gene controlling virion-associated cell shutoff, between strains of herpes simplex virus. J Gen Virol. 1990;71:411–418. doi: 10.1099/0022-1317-71-2-411. [DOI] [PubMed] [Google Scholar]
- 14.Fenwick M L, McMenamin M M. Early virion-associated suppression of cellular protein synthesis by herpes simplex virus is accompanied by inactivation of mRNA. J Gen Virol. 1984;65:1225–1228. doi: 10.1099/0022-1317-65-7-1225. [DOI] [PubMed] [Google Scholar]
- 15.Fenwick M L, Morse L S, Roizman B. Anatomy of herpes simplex virus DNA. XI. Apparent clustering of functions effecting rapid inhibition of host DNA and protein synthesis. J Virol. 1979;29:825–827. doi: 10.1128/jvi.29.2.825-827.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwicke M A, Sandri-Goldin R M. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and the regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jennings S R, Rice P L, Kloszewski E D, Anderson R W, Thompson D L, Tevethia S S. Effect of herpes simplex virus types 1 and 2 on surface expression of class I major histocompatibility antigens on infected cells. J Virol. 1985;56:757–766. doi: 10.1128/jvi.56.3.757-766.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones F E, Smibert C A, Smiley J R. Mutational analysis of the herpes simplex virus virion host shutoff protein: evidence that vhs functions in the absence of other viral proteins. J Virol. 1995;69:4863–4871. doi: 10.1128/jvi.69.8.4863-4871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karr, B. M., and G. S. Read. The virion host shutoff function of herpes simplex virus degrades the 5′ end of a target mRNA prior to the 3′ end. Virology, in press. [DOI] [PubMed]
- 21.Koelle D M, Tigges M A, Burke R L, Symington F M, Riddell S R, Abbo H, Corey L. Herpes simplex virus infection of human fibroblasts and keratinocytes inhibits recognition by cloned CD8+ cytotoxic T lymphocytes. J Clin Invest. 1993;91:961–966. doi: 10.1172/JCI116317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krikorian C R, Read G S. An in vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J Virol. 1991;65:112–122. doi: 10.1128/jvi.65.1.112-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwong A D, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc Natl Acad Sci USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwong A D, Kruper J A, Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam Q, Smibert C A, Koop K E, Lavery C, Capone J P, Weinheimer S P, Smiley J R. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 1996;15:2575–2581. [PMC free article] [PubMed] [Google Scholar]
- 26.Leib D A, Harrison T E, Laslo K M, Machalek M A, Moorman N J, Virgin H W. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J Exp Med. 1999;189:663–672. doi: 10.1084/jem.189.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 28.Oroskar A A, Read G S. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate-early (alpha) mRNAs. J Virol. 1987;61:604–606. doi: 10.1128/jvi.61.2.604-606.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oroskar A A, Read G S. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pak A S, Everly D N, Knight K, Read G S. The virion host shutoff protein of herpes simplex virus inhibits reporter gene expression in the absence of other viral gene products. Virology. 1995;211:491–506. doi: 10.1006/viro.1995.1431. [DOI] [PubMed] [Google Scholar]
- 31.Patterson, M., D. N. Everly, Jr., and G. S. Read. Unpublished data.
- 32.Posavad C M, Rosenthal K L. Herpes simplex virus-infected human fibroblasts are resistant to and inhibit cytotoxic T-lymphocyte activity. J Virol. 1992;66:6264–6272. doi: 10.1128/jvi.66.11.6264-6272.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Read G S. Control of mRNA stability during herpes simplex virus infections. In: Harford J B, Morris D R, editors. mRNA metabolism and posttranscriptional gene regulation. New York, N.Y: Wiley-Liss, Inc.; 1997. pp. 311–321. [Google Scholar]
- 34.Read G S, Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate-early) polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read G S, Karr B M, Knight K. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J Virol. 1993;67:7149–7160. doi: 10.1128/jvi.67.12.7149-7160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Remond M, Sheldrick P, Lebreton F, Nardeux P, Foulon T. Gene organization in the UL region and inverted repeats of the canine herpesvirus genome. J Gen Virol. 1999;77:37–48. doi: 10.1099/0022-1317-77-1-37. [DOI] [PubMed] [Google Scholar]
- 37.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schek N, Bachenheimer S L. Degradation of cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of Vero cells. J Virol. 1985;55:601–610. doi: 10.1128/jvi.55.3.601-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmelter J, Knez J, Smiley J R, Capone J P. Identification and characterization of a small modular domain in the herpes simplex virus host shutoff protein sufficient for interaction with VP16. J Virol. 1996;70:2124–2131. doi: 10.1128/jvi.70.4.2124-2131.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simard C, Langlois I, Styger D, Vogt B, Vicek C, Chalifour A, Trudel M, Schwyzer M. Sequence analysis of the UL39, UL38, and UL37 homologues of bovine herpesvirus 1 and expression studies of UL40 and UL39, the subunits of ribonucleotide reductase. Virology. 1995;212:734–740. doi: 10.1006/viro.1995.1533. [DOI] [PubMed] [Google Scholar]
- 41.Smibert C A, Papova B, Xiao P, Capone J P, Smiley J R. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J Virol. 1994;68:2339–2346. doi: 10.1128/jvi.68.4.2339-2346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorenson C M, Hart P A, Ross J. Analysis of herpes simplex virus-induced mRNA destabilizing activity using an in vitro mRNA decay system. Nucleic Acids Res. 1991;19:4459–4465. doi: 10.1093/nar/19.16.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strelow L, Smith T, Leib D. The virion host shutoff function of herpes simplex virus type 1 plays a role in corneal invasion and functions independently of the cell cycle. Virology. 1997;231:28–34. doi: 10.1006/viro.1997.8497. [DOI] [PubMed] [Google Scholar]
- 44.Strelow L I, Leib D A. Role of the virion host shutoff of herpes simplex virus type 1 in latency and pathogenesis. J Virol. 1995;69:6779–6786. doi: 10.1128/jvi.69.11.6779-6786.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strelow L I, Leib D A. Analysis of conserved domains of UL41 of herpes simplex virus type 1 in virion host shutoff and pathogenesis. J Virol. 1996;70:5665–5667. doi: 10.1128/jvi.70.8.5665-5667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strom T, Frenkel N. Effects of herpes simplex virus on mRNA stability. J Virol. 1987;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Telford E A R, Watson M S, McBride K, Davison A J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:304–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 48.Tharun S, Parker R. Mechanisms of mRNA turnover in eukaryotic cells. In: Harford J B, Morris D R, editors. mRNA metabolism and posttranscriptional gene regulation. New York, N.Y: Wiley-Liss, Inc.; 1997. pp. 181–199. [Google Scholar]
- 49.Tigges M A, Leng S, Johnson D C, Burke R L. Human herpes simplex virus (HSV)-specific CD8+ CTL clones recognize HSV-2-infected fibroblasts after treatment with IFN-gamma or when virion host shutoff functions are disabled. J Immunol. 1996;156:3901–3910. [PubMed] [Google Scholar]
- 50.Tomazin R, Hill A B, Jugovic P, York I, Van Endert P, Ploegh H L, Andrews D W, Johnson D C. Stable binding of the herpes simplex virus ICP47 protein to the peptide binding site of TAP. EMBO J. 1996;15:3256–3266. [PMC free article] [PubMed] [Google Scholar]
- 51.Willoughby K, Bennett M, Williams R A, McCracken C, Gaskell R M. Sequences of the ribonucleotide reductase-encoding genes of felid herpesvirus 1 and molecular phylogenetic analysis. Virus Genes. 1997;15:203–218. doi: 10.1023/a:1007924419113. [DOI] [PubMed] [Google Scholar]
- 52.York I A, Roop C, Andrews D W, Riddell S R, Graham F L, Johnson D C. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 53.Zelus B D, Stewart R S, Ross J. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J Virol. 1996;70:2411–2419. doi: 10.1128/jvi.70.4.2411-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]