Abstract
The study of species groups in which the presence of interspecific hybridization or introgression phenomena is known or suspected involves analysing shared bi-parentally inherited molecular markers. Current methods are based on different categories of markers among which the classical microsatellites or the more recent genome wide approaches for the analyses of thousands of SNPs or hundreds of microhaplotypes through high throughput sequencing. Our approach utilizes intron-targeted amplicon sequencing to characterise multi-locus intron polymorphisms (MIPs) and assess genetic diversity. These highly variable intron regions, combined with inter-specific transferable loci, serve as powerful multiple-SNP markers potentially suitable for various applications, from species and hybrid identification to population comparisons, without prior species knowledge. We developed the first panel of MIPs highly transferable across fish genomes, effectively distinguishing between species, even those closely related, and populations with different structures. MIPs offer versatile, hypervariable nuclear markers and promise to be especially useful when multiple nuclear loci must be genotyped across different species, such as for the monitoring of interspecific hybridization. Moreover, the relatively long sequences obtained ease the development of single-locus PCR-based diagnostic markers. This method, here demonstrated in teleost fishes, can be readily applied to other taxa, unlocking a new source of genetic variation.
Keywords: Forensic identification, High-throughput DNA sequencing, Molecular markers, Multiple-SNP haplotypes, Non-model organisms, Teleost fishes
Subject terms: Biodiversity, Molecular ecology
Introduction
Species identification, population genomics, and molecular ecology studies rely on the availability of highly polymorphic independent markers. Decreasing isolation and characterisation costs and increasing transferability across species are major challenges in improving efficiency and applicability of markers1,2. Special attention is paid to non-model organisms usually involved in molecular ecology studies, for which genetic and genomic information is very scarce or incomplete. Mitochondrial DNA has been for decades, and still is, the most widely used tool for species identification and cataloguing. However, its maternal inheritance does not allow for the investigation of hybridization and introgression processes in most animal species. For this reason, with the aim of stressing the aspects related to interspecific hybridization, we have intentionally decided to focus on nuclear markers only. Long before the establishment of Next Generation Sequencing (NGS) technology, traditional genetic approaches preferentially involved the use of highly variable microsatellites to study patterns of distribution of genetic diversity, parental relationships, and species and hybrid identification1. Despite their high power in diversity assessment, microsatellites present several problems mostly associated with low transferability across species and reproducibility across laboratories, homoplasy, as well as high costs for marker isolation and genotyping. Advances in NGS have driven the costs of sequencing down, shifting the attention to the genotyping of single nucleotide polymorphism (SNP) markers1,3. SNPs are mostly bi-allelic loci less variable than microsatellites, but more abundant within genomes and therefore preferable in order to obtain information at different evolutionary scales1. Typically, methods relying on reduced representation sequencing (RRS) techniques, such as different variants of restriction site-associated DNA sequencing (RADseq4) and Genotyping by Sequencing (GBS5), are employed to reduce genome complexity and to ease the genotyping of stand-alone SNPs in many individuals simultaneously. Due to their high abundance, wide distribution, and codominant Mendelian inheritance, SNPs became preferentially used to study genetic differentiation patterns, intraspecific demographic processes, and interspecies relationships with high resolution power6–9. Once identified, a panel of SNPs can be genotyped in several ways. In the case of routine analyses conducted on a large number of samples, it may be convenient to develop species-specific SNP chips10,11. On the contrary, when it is necessary to analyse a few samples at a time, it may be convenient to use alternative methods. Even when all SNPs of a specific region must be considered in their different combinations, with the possibility of having more than two of alleles at the same locus, the SNP chips cannot be used as they detect a single polymorphism per time. These closely linked single nucleotide polymorphisms are called Microhaplotypes. They capture genetic variation across multiple adjacent SNPs providing improved resolution power6,12. An interesting application of this approach is represented by the genotyping-in-thousands by sequencing (GT-seq) in which several targeted sequences are simultaneously characterized13. In the present study, we adapted this last approach to the characterization of intronic regions which, besides having a high level of variability are flanked by conserved regions often shared by related species. The proposed approach is called Multi-locus Intron Polymorphisms (MIPs).
Introns are non-coding regions of DNA found within genes of all eukaryotic genomes. They can contain regulatory elements that control the expression of genes (including enhancers and silencers) and the splicing process14. In spite of these diverse roles, introns can exhibit a significant level of variability in terms of length and sequence composition. In fact, unlike exons, which often contain protein-coding sequences that are under selective pressure, introns are not subject to the same constraints and can accumulate mutations more freely leading to higher sequence diversity15. Accordingly, even if intron features cannot be simply associated to a random mutational model, their high sequence variability16–19 makes them good candidates as sources of multi-alleles DNA markers.
So far, introns have been considered as a potential alternative to commonly used nuclear markers. The pioneer study on introns involved the use of exon-primed intron-crossing PCR (EPIC-PCR20) to amplify these regions with primer pairs designed on the more conserved exon-flanking regions. However, before the advent of NGS, the application of introns as molecular markers was nontrivial and the exploitation of their full potential has been hindered by demanding laboratory procedures (e.g., the need of cloning amplicons to reliably genotype each locus). For this reason, studies focused almost exclusively on loci showing length polymorphisms, thus avoiding direct sequencing and cloning procedures21–24. Analysis of intron sequence variability with single-locus approaches was applied only to a limited number of species25,26, mainly for species identification and forensic purposes27–29. Only recently, with the increase of the number of available genomes, the approaches shifted towards an in silico isolation of predicted introns to analyse length polymorphisms or sequence variability on a multi-locus, genome-wide scale30–32. However, the limited number of species used in the development and definition of intron loci has not promoted the horizontal applicability to multiple species that these markers might have.
In this study, we present a new method for assessing genetic diversity by genotyping a predefined panel of MIP markers using a targeted amplicon high-throughput sequencing approach. The power of the method was first evaluated for cross-species/cross-genera transferability and species identification by testing 65 teleost species.
Then the method was corroborated using three case studies whose sample sets were already characterized by microsatellites or SNPs33–35. The first case study (case study 1) evaluated power of MIPs to differentiate Antarctic species of suborder Notothenioidei. Some notothenioid species, particularly the genus Chionodraco (family Channichthyiadae), are morphologically very similar and the lack of diagnostic traits limits the potential for precise assessment of fish diversity by morphological identification only. Moreover, the occurrence of interspecific hybridisation was recently suggested for the genus Chionodraco33,36. The second case study (case study 2) explored the effectiveness of MIPs to differentiate cryptic species of sympatric sole species: the Common Sole (Solea solea, SS) and Egyptian Sole (Solea aegyptiaca, SA). These two species are valuable fishery resources, and their meaningful management would benefit from the availability of diagnostic molecular markers for fast species identification tools37. The third case study (case study 3) examined the power of MIPs to identify population genetic structure among geographical samples of SS and SA from the Mediterranean Sea. Case study 3 represents the first application of MIPs to detect subtler levels of intraspecies differentiation.
We demonstrated how MIPs substantially provide a good power in identifying species, even phylogenetically closely related, and how these markers are also suitable to investigate relationships among populations in genetic studies as alternative source of variability. We also discussed limits and potentials of the MIPs panel here proposed for teleost fishes, and suggested actions that could promote the implementation of the approach in other taxonomic groups.
Results
Sequencing performance
The two MiSeq v3 runs with a total of 384 individuals (including 33 technical replicates) genotyped at 121 intron loci yielded 32,455,511 paired-end raw sequences, with a number per individual ranging from 706 to 199,362 (mean 84,520). Almost all individuals with less than 10,000 raw sequences (N = 23) were old samples of Atherina spp. stored for more than 20 years at −20 °C, indicating that the low quality of DNA can strongly affect the results. After the first filtering phase by Cutadapt, 24.4% (6,370,386) of the raw reads did not exceed the established thresholds and were removed from further bioinformatic steps. The 26,085,125 demultiplexed sequences that passed the above step were distributed across samples with a mean of 67,930, while the number of sequences retained per locus ranged from 10 (Locus_76) to 969,783 (Locus_15) with a mean of 215,580 pointing to high heterogeneity in locus amplification performances.
The measure used to decrease artefact formation in the library amplification steps successfully limited the formation of chimeric sequences that were detected by bioinformatics pipelines at very low frequencies and easily removed from further analysis. Of 46,464 expected single-locus genotypes (384 individuals genotyped at 121 loci), 24.4% (11,340) were discarded during merging of paired-ends. Most frequently, this owed to sequencing problems in one direction or, more rarely, the excessive intron length (3,021 cases) that prevented paired-ends from merging reliably. Of the remaining 35,124 single-locus genotyping that successfully passed the merging phase, 32,499 (92.5%) showed for-rev overlapping over 90% of the total sequences per individual per locus. After the final filtering phase by SeekDeep, which allowed the generation of alleles and genotypes, a total of 25,643,808 high-quality filtered and merged sequences were retained for further analysis, with a mean of 67,130 sequences per individual and 215,494 sequences per locus. The mean coverage associated with each allele per locus per individual was 1,762 reads.
Exploring the transferability of MIP loci
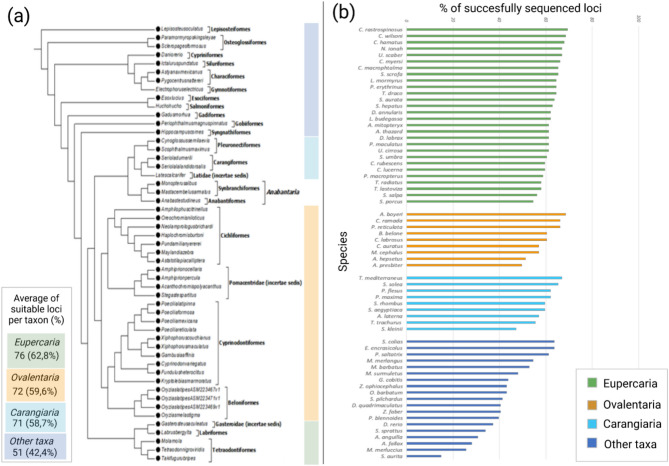
On average, from each species, 56% of the 121 MIP loci were successfully amplified and sequenced. These loci were different among species. Based on the classification reported in Betancur-R et al. (2017)38 and Nelson et al. (2016)39, for species included within Eupercaria, Ovalentaria, and Carangiaria a greater number of loci were successfully sequenced with a mean of 76 loci per species (62.8%), 72 (59.6%), and 71 (58.7%) respectively for the three clades (Fig. 1a,b). For species included in other taxa such as the orders of Clupeiformes, Gadiformes, Cypriniformes (with a single species tested, Danio rerio) and Anguilliformes (with a single species tested, Anguilla anguilla), genotyping yielded lower performance with genotypes obtained on average at 51 loci (42.4%) per species.
Figure 1.
Transferability of loci across tested species and reproducibility of the approach. (a) Modified version of the Ensembl phylogeny and taxon nomenclature reported by Betancur-R et al.38 and Nelson et al.39 with the mean number of loci successfully genotyped per taxon. We reported in the phylogeny only the species for which we used the genome in the first part of the isolation of intron loci. (b) Distribution of the percentage of loci successfully amplified and sequenced in 65 species of fishes tested. Species are grouped based on taxa reported in (Fig. 1a). Figure refined with Biorender.
Replicates showed a percentage of loci with identical genotypes on average of 87.2% and 88.1%, respectively, considering high-quality filtered data and further filtering the output excluding alleles with a coverage lower than 30 sequences. Comparisons included replicates analysed and sequenced within the same MiSeq run or in different runs. The replicates within or between runs were comparable with a mean genotype identity between replicates of 89.65% ± 6.05 intra-run and 84.18% ± 7.42 inter-run, indicating good reproducibility of the approach (see Supplementary Material—Figure S1). Discrepancies between replicates were primarily observed at loci that had amplification difficulties for the species in question (alleles with low coverage and substantial missing data) possibly due to the presence of mismatches at the primer binding sites. This is somehow expected as the panel of loci is applied to species for which no a priori information about the analysed loci is available. Therefore, once it was established that the discrepancies between replicated were probably not due to technical causes but rather to suboptimal loci for the species in question, the issue was resolved by removing them from the analyses and considering only the ones for which reproducibility between replicates was nearly total.
In addition to the good observed transferability of MIP markers across teleost species, these loci also revealed a strong potential for species identification. Pairwise allele sharing between samples (within and among species) was graphically represented in Figure S2 (Supplementary Material) including species with at least three individuals genotyped with high coverage at almost all loci. The clustering of individuals perfectly reflected the species to which they belong, suggesting the presence of many private alleles per species at different loci (Figure S2, Supplementary Material).
Case study 1: Exploring species identification power in antarctic notothenioids
Of the 121 MIP loci tested, 54 (44%) passed all filters (described in Methods) and were selected for the identification of notothenioid species in this case study. Most loci were polymorphic with many alleles per species. Most of the alleles were private and therefore informative for species discrimination (Table 1). The species A. mitopterix showed the highest Allelic richness and private Allelic richness, suggesting a clear differentiation from the other notothenioid species also according to its phylogenetic position [340, 41] (Table 1). Some inbreeding coefficients were negative reflecting an apparent excess of heterozygotes intraspecies. Out of 424 probability tests for HWE, 41 showed a significant departure from HWE, with a nominal threshold of 0.05. However, it is noteworthy that 90 tests failed because loci were monomorphic within the species sample. Loci showing HW disequilibrium are different across species. After correction for multiple tests, only 5 probability tests for HWE are statistically significant (loci 7 and 71 in C. hamatus, locus 24 in C. myersi and loci 7 and 31 in C. rastrospinosus) (Table 1).
Table 1.
Descriptive statistics calculated at 54 loci within icefish species and on the total dataset without the 11 individuals with dubious identification (N = 69). The individual originally classified as C. hamatus but resulted as a mislabelling of the sample tube has been removed from this calculation. For each species, samples size (N), proportion of polymorphic loci (PN), missing data percentage across individuals per species (MVs), unbiased expected heterozygosity (HE), observed heterozygosity (HO), inbreeding coefficient (FIS), mean number of alleles per locus (Na), Allelic richness (AR) and private Allelic richness (pAr) are reported. Ar and pAr values are calculated on a minimum sample size of six diploid individuals. Some inbreeding coefficients were negative reflecting an apparent excess of heterozygotes intraspecies.
| Species | N | PN (%) | MVS (%) | HE | HO | FIS | Na | Ar | pAr |
|---|---|---|---|---|---|---|---|---|---|
| C. hamatus | 11 | 88.68 | 11.49 | 0.425 | 0.353 | 0.168 | 4.189 | 3.320 | 1.140 |
| C. myersi | 12 | 86.79 | 9.74 | 0.425 | 0.429 | − 0.011 | 4.359 | 3.380 | 1.020 |
| C. rastrospinosus | 14 | 88.68 | 5.12 | 0.428 | 0.409 | 0.043 | 4.566 | 3.450 | 1.140 |
| P. macropterus | 6 | 69.81 | 11.32 | 0.316 | 0.337 | − 0.067 | 2.674 | 5.490 | 1.460 |
| P. maculatus | 6 | 50.94 | 9.11 | 0.201 | 0.213 | − 0.058 | 1.755 | 4.640 | 0.870 |
| N. ionah | 6 | 79.25 | 9.11 | 0.376 | 0.390 | − 0.038 | 2.830 | 2.830 | 1.980 |
| C. wilsoni | 8 | 84.91 | 2.12 | 0.441 | 0.424 | 0.039 | 4.509 | 3.880 | 2.210 |
| A. mitopteryx | 6 | 81.13 | 8.80 | 0.448 | 0.500 | − 0.116 | 4.000 | 9.060 | 3.490 |
The differentiation between specimens within and between population samples based on 54 intron loci was represented by PCAs (Fig. 2). Without considering a priori sample classification based on morphology, the first, the second, and the third principal components (PC1, PC2, and PC3) explained, respectively, 16.14, 10.19 and 8.75% of the variance in the dataset, and almost all samples clustered according to the original morphological identification of species. The same clustering was obtained by the Structure analysis (see Supplementary Material—Figure S3). The only exception was a single individual originally labelled C. hamatus (Cham_PS82_2711), which was completely different from all other individuals in the data set including all C. hamatus specimens. Sequencing of the cytochrome oxidase I (COI) mitochondrial gene confirmed the above evidence, assigning the sample to the notothenioid species Trematomus scotti. This species was not meant to be included in tested species and it was a clear case of sample mislabelling, since C. hamatus and T. scotti are morphologically very different and easy to distinguish (they belong to two well separated notothenioid families, Channichthyidae and Notothenioodea, respectively).
Figure 2.
Plot of the principal component analysis of variation based on 54 intron loci genotyped at a total of 81 icefish individuals (including 8 species defined based on morphological features and 11 individuals with unknown species origin marked as Chionodraco spp.). Colours indicate species. The eigenvalues of the first three axes explain 16.14, 10.19 and 8.75% of the variation. The individual genetically assigned to the species Trematomus scotti was originally mis-assigned to C. hamatus due to sample mislabelling and is highlighted by a red circle. (a) PC1 versus PC2. (b) PC1 vs PC3. Figure refined with Biorender.
The eleven animals identified to the genus level Chionodraco spp. resulted to be pure ancestry and undoubtedly assigned to one of the three Chionodraco species, as shown by PCA-based analysis (Fig. 2) and by Structure-based analysis (see Supplementary Material—Figure S3).
Case study 2: species and population differentiation within the genus Solea
Of the 121 MIP markers tested, 43 (35%) passed all filters and were selected for analysis. All selected loci were polymorphic in S. solea and 93.1% in S. aegyptiaca. The S. solea species showed higher genetic diversity with an overall Allelic richness (Ar) overall of 10.91 while S. aegyptiaca ranked lower with an overall Ar of 4.32 (Table 2). Furthermore, many private and therefore diagnostic alleles were identified at almost all loci (pAr = 10.8 and pAr = 4.24, respectively; Table 2) suggesting a high power to differentiate the two species as confirmed by cluster analysis (Fig. 3a). Of 43 MIP markers, 39 showed alternative alleles for the two species and each one of these, taken alone, could be used for future development of rapid single locus tools to differentiate the two species. No interspecific hybrids were observed, suggesting complete reproductive isolation between the two species.
Table 2.
Descriptive statistics calculated at 43 loci within the genus Solea. Statistics were calculated per species based on the total dataset (of 68 and 38 individuals for Solea solea and Solea aegyptiaca, respectively) and per populations. Individuals processed in replicate were considered only once. The three outlier individuals, morphologically misclassified as S. solea and resulted instead S. aegyptiaca based on genetic identification, were considered as S. aegyptiaca for the total dataset but were a priori removed for the analysis population-based. For each samples, size (N), proportion of polymorphic loci (PN), missing data percentage across individuals per species/populations (MVs), unbiased expected heterozygosity (HE), observed heterozygosity (HO), inbreeding coefficient (FIS), mean number of alleles per locus (Na), Allelic richness (Ar) and private Allelic richness (pAr) are reported. Ar and pAr values are calculated on a minimum sample size of 12 and 5 diploid individuals respectively for S. solea and S. aegyptiaca.
| Species | N | PN (%) | MVS (%) | HE | HO | FIS | Na | Ar | pAr |
|---|---|---|---|---|---|---|---|---|---|
| Solea solea | 68 | 100 | 2.599 | 0.592 | 0.493 | 0.167 | 14.14 | 10.91 | 10.82 |
| Solea aegyptiaca | 38 | 93.02 | 1.101 | 0.364 | 0.260 | 0.286 | 4.32 | 4.32 | 4.24 |
| Populations of Solea solea | |||||||||
| SS-GLI | 14 | 95.3 | 2.66 | 0.604 | 0.551 | 0.089 | 6.19 | 6.75 | 2.21 |
| SS-TYR | 15 | 97.7 | 5.12 | 0.613 | 0.567 | 0.074 | 7.40 | 6.78 | 2.38 |
| SS-ADR | 14 | 97.7 | 1.83 | 0.565 | 0.512 | 0.094 | 5.72 | 5.41 | 1.06 |
| SS-GRE | 12 | 95.4 | 1.36 | 0.493 | 0.449 | 0.089 | 4.51 | 4.51 | 1.05 |
| SS-TUR | 13 | 93.0 | 1.61 | 0.436 | 0.371 | 0.149 | 3.86 | 3.76 | 0.79 |
| Populations of Solea aegyptiaca | |||||||||
| SA-TYR | 13 | 48.8 | 1.97 | 0.168 | 0.153 | 0.090 | 1.72 | 1.54 | 0.15 |
| SA-ADR | 5 | 62.8 | 0.93 | 0.287 | 0.321 | − 0.118 | 2.09 | 2.09 | 0.42 |
| SA-GRE | 6 | 72.1 | 0 | 0.364 | 0.407 | − 0.118 | 2.67 | 2.54 | 0.78 |
| SA-TUR | 6 | 53.5 | 0 | 0.220 | 0.248 | − 0.126 | 1.76 | 1.72 | 0.21 |
| SA-ALE | 5 | 58.1 | 0.47 | 0.258 | 0.287 | − 0.115 | 2.00 | 2.00 | 0.24 |
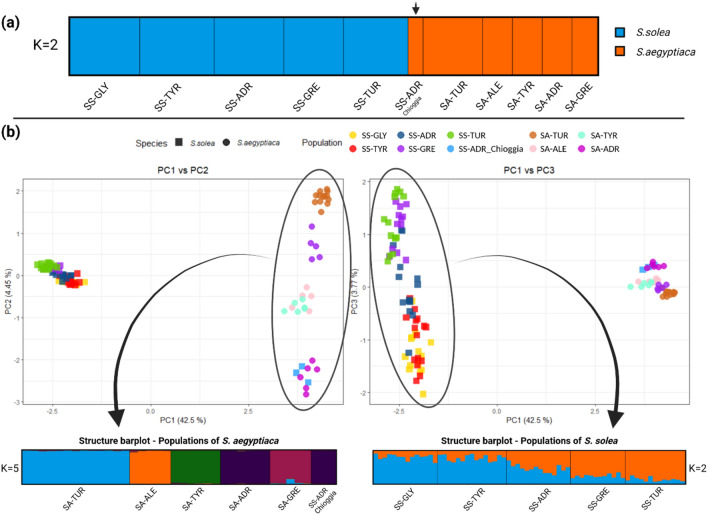
Figure 3.
Distribution pattern of genetic differentiation between species and populations of the Solea genus based on genotype information at 43 intron loci. (a) Structure bar-plot showing the membership probabilities of cluster assignment for each of the 106 individuals of the two species (71 S. solea and 35 S. aegyptiaca). The best number of clusters (K = 2) that overlaps the species subdivision is reported. The black arrow indicates the three individuals morphologically classified as S. solea, which cluster with S. aegyptiaca according to their mitochondrial barcode information. (b) Plot of the principal component analysis of variation (PC1 vs PC2 on the left, PC1 vs PC3 on the right). Eigenvalues of the first three axes explain 42.5, 4.45 and 3.77% of the variation. Shapes indicate species, while colours indicate populations. Below each PCA, the best cluster subdivision obtained by structure analysis performed within species is also reported as bar-graph showing individual membership probabilities. Figure refined with Biorender.
In the PCA analysis without a priori information of species identity, the first, the second, and the third principal components (PC1, PC2, and PC3) explained 42.5, 4.45 and 3.77% of the variance in the dataset, respectively. Most of the variation (PC1) was explained by species divergence (Fig. 3b) while PC2 and PC3 explained subtler subdivisions. The total FST values for the two population samples of the two sole species were 0.061 (p < 0.0001) and 0.342 (p < 0.0001) respectively for S. solea and S. aegyptiaca. In S. aegyptiaca, all analyses were consistent in identifying the more supported clustering as congruent with the geographical population subdivision (Fig. 3b on the left; Supplementary Material—Table S1C). Concerning the species S. solea, the MIPs analysis substantially overlapped what was already identified based on previous studies42,43, showing a gradient of divergence from the west to east side of the Mediterranean basin, with the Adriatic Sea hosting an admixed population of the two Mediterranean clusters (Fig. 3b on the right). A recently published paper on population genetics of S. solea analysed at 380 SNP loci44, included the specimens here analysed with 43 MIPs. We estimated Fst values among populations considering the same individuals analysed by the two approaches and obtained comparable results, with overall Fst values of 0.075 and 0.061 for SNPs and MIPs respectively. The Fst values between pairwise populations are reported in the Supplementary Material—(Table S1A,B).
The only exception was represented by three individuals fished in the North Adriatic Sea, sampled at the Chioggia Fish Market, and morphologically identified as S. solea which clustered with the Adriatic population of S. aegyptiaca in all the analyses. This discrepancy was confirmed by COI sequencing that assigned all samples to the species S. aegyptiaca, thus confirming the original misclassification.
Discussion
In the last decades, the analysis of genetic diversity was increasingly based on SNPs analysed by whole genome approaches, such as GBS45 and RAD methods4, while the development and application of methods for the analysis of intron polymorphisms struggled so far to take off due to demanding laboratory procedures and the need of optimisation of single-locus sequencing.
Here, we leveraged on the major advances in next-generation technologies and developed a protocol for the simultaneous analyses of multiple, highly transferable selected introns, using teleost fishes as test group. The results obtained go beyond our expectations, and the approach based on MIPs showed not only a high transferability but also a flexible resolving power, being successfully applied both at the species and population levels.
Transferability across teleost species
Transferability across species is one of the strength of the approach here proposed. It must be considered that 30 (46%) and 12 (18%) of the species analysed in this work were represented by one and two individuals, respectively, and that the origin of the different samples was very heterogeneous, as well as their state of sample preservation. Therefore, it is possible that some species were represented by a few animals with too low DNA quality. This led to an underestimation of the degree of transferability because loci for which amplification failed due to poor DNA quality may perform better with higher DNA quality. Despite this, MIP markers proved to be highly transferable and, in most species, more than 50% of loci were successfully genotyped. Given the wide taxonomic range of our sample dataset, we consider this to be a very good result. Even by reversing the perspective and evaluating the percentage of species on which each locus is amplifiable, the results are very satisfactory, and more than 60% of the loci were amplified in more than 50% of the species. In addition, in this case, these percentages are underestimated due to DNA quality, as previously described.
The panel of MIP loci proposed here was the result of a pilot experiment carried out on 65 species and 121 loci. Both the number of species analysed and the number of tested loci may be increased in the future, allowing the preparation of a reference data set that will enable the identification of a priori effective MIP loci to be genotyped for the different species or groups of species.
Limits and potential of using MIPs: comparative considerations among different approaches
MIPs fit into a heterogeneous scenario in which a variety of molecular markers (where the most used are microsatellites and SNPs scored with different approaches) can already provide the necessary information in many analytical contexts. Features of MIPs such as high transferability, codominance, bi-parental Mendelian inheritance, high number of alleles per locus, high mutation rate and applicability at both species and population level, taken individually, can also be found in other markers. For example, a high number of alleles per locus is usually common for microsatellites or GT-seq, or the possibility to simultaneously genotype hundreds of individuals at thousands of loci in a single sequencing is typical of bi-allelic SNPs used in RAD approaches. What makes MIPs unique is that they exhibit these characteristics together, which makes them an option to consider when initially selecting a marker to be used in a genetic diversity study. The approach conceptually most similar to MIPs is certainly Genotyping-in-Thousands by sequencing (GT-seq), in which multiple amplifications on target loci result in sequence information simultaneously from hundreds of loci. Both of these methods share, compared to RAD approaches, the advantage of being directly based on an initial amplification reaction, without necessarily starting from a restriction reaction, which imposes certain constraints in terms of the quantity and quality of the starting DNA. However, compared to GT-seq, MIPs offer high transferability between species, making them particularly versatile and very useful for the analysis of complexes of related species.
In our opinion, the most interesting feature of MIPs is the high horizontal transferability across large systematic groups such as teleost fishes. Indeed, the universal nature of MIPs primers makes their application to any other fish species and the characterisation of homologous loci even with a high degree of differentiation straightforward. This makes MIPs very attractive for all applications where different species need to be studied simultaneously, as for instance detection and monitoring interspecific hybridization events.
Given the several analogies that readers may glimpse between MIP and RAD markers as well as with genotyping by sequencing approaches in general, it is appropriate to engage in a focused discussion regarding their similarities and differences. Both approaches share similarities in terms of the experimental effort, involving the preparation of genomic libraries and subsequent Illumina platform sequencing. However, significant differences also exist. The first is that RAD approaches aim to characterize a very high number of regions selected by the restriction process, without any a-priori information about the loci, whereas MIPs concentrate on a smaller number of loci that are pre-selected for their high transferability across species. Another difference is that despite RAD methods provide microhaplotypes for which the diversity of the entire region can potentially be utilized, typically only the information from a single SNP per locus is used, excluding all other polymorphisms in linkage from the analyses, probably for the more complex downstream analytical process that the use of microhaplotype information would require. Consequently, each locus is represented by a single bi-allelic polymorphism and the variability represented by the entire sequence (haplotypes) is not fully considered. Finally, as better described in the following paragraph the intrinsic transferability of MIPs makes them more suitable for studies on species complexes of interspecific hybridization.
MIP loci for species and hybrid identification
The efficiency at the species level, potentially very useful for the identification of species and hybrids, was clearly demonstrated in the present study by several evidence. First, in case studies 1 and 2 on Antarctic fishes and Solea spp., all species were perfectly distinguished. Second, the perfect clustering of the individuals of other 22 taxa was concordant with the original morphological identification of species (simply considering presence/absence of information of alleles at different loci, Figure S2 Supplementary Material) suggesting a high potential of this panel for species identification in teleost species. The only two cases of discrepancy between a priori classification and genetic analysis turned out to be the mislabelling of one individual of T. scotti (case study 1) and the identification error of three individuals of S. solea (case study 2). Unfortunately, no certain interspecific hybrids were available in our case studies. However, the significant number of private alleles detected in both Icefish and in Solea species suggest that MIPs might be particularly well-suited for hybrid identification. It is worth noting that an ideal marker for hybrid identification should be codominant, exhibit biparental Mendelian inheritance and preferably span multiple loci. For identifying first-generation interspecific hybrids multi-locus markers are not essential since diagnostic alleles from both parental species can always be observed at each diagnostic locus. In such instances, having multiple loci available can simply provide stronger support to the identification. Conversely, when dealing with backcrosses or multispecies hybrids resulting from mating hybrids with a third species, having multiple loci available significantly increases the likelihood of detecting the contributions of all species involved. In our laboratory, we have already started the application of MIPs for hybrid identification on sturgeons and pikes (Esox lucius and E. flaviae), yielding promising preliminary results. In case of pure species identification, the analysis with MIPs (as well as with any other nuclear marker) does not bring advantages compared to sequencing mtDNA (e.g. DNA barcode, metabarcoding).
MIP loci at the population level
MIPs were not developed for population genetics studies but rather as highly transferable markers across a wide range of species. However, results obtained with solefish have unexpectedly shown that, in the case of geographically structured populations, MIPs can also successfully detect intraspecific differentiation patterns. In fact, the analysis on various populations of Egyptian sole showed a clear geographical pattern of differentiation. Also in the Common sole, the differentiation gradient detected by MIPs from the east to west in the Mediterranean basin confirms the results previously obtained with microsatellites42,43. For studies of this nature, other approaches are naturally more suitable, such as RAD sequencing, that doesn’t need a priori information about the species but in which the availability of numerous loci empowers the analyses and allows the identification of even less clear patterns. Nevertheless, the fact that MIPs are multiallelic loci allows them to partially offset the considerably smaller number of loci compared to RAD sequencing, making them conceptually closer in terms of their applicability to microsatellite loci. Once the scoring of MIP loci is completed, and the genotype of each individual is determined, the dataset can be analysed just as if it was a microsatellite dataset. Unlike microsatellites, moreover, MIPs also provide sequence information, which can be especially valuable, for tasks like the development of rapid diagnostic markers. One notable advantage of MIPs over microsatellites, in addition to their greater marker transferability across species, is the lower expected rate of homoplasy. In microsatellites, homoplasy is inherently high because variability arises from the addition or removal of repeated units, allowing the same allele to be easily generated independently multiple times. On the other hand, the mutation rate of microsatellites is probably higher than that of MIPs, for which the maximum number of observed alleles in species rarely exceeds 10 (Table S2). However, this consideration should be taken with caution because of the small number of animals analysed for each species in the present study, with the exception of solefish.
Future developments
Among the future developments of the MIPs, the more promising ones are presented below. Firstly, exploring the effectiveness of MIPs in paternity tests, relatedness analysis and genetic tagging could help provide highly informative markers for the management of genetic diversity in species/populations at risk of extinction. Since introns have a Mendelian pattern of inheritance, it is likely expected that they will be very efficient. Microsatellites are currently the markers of choice for this purpose, but for some species they can be difficult to isolate, not enough polymorphic, or difficult to interpret as in the case of polyploid species in which inferring the allelic dosage based on the intensity of the peaks is challenging. When dealing with MIPs, the exact genotype can be inferred directly from the coverage of the various alleles, but dedicated tests must be performed to confirm this.
Secondly, investigating the informative potential of MIPs in phylogenetic and phylogeographic studies based on intron sequence information. In this context, the high transferability of loci across species may simplify the characterisation of homologous loci between different species or populations.
Thirdly, developing and making available an automated and user-friendly bioinformatics pipeline for the processing of sequencing output will facilitate workflow management and data analysis.
Fourthly, increasing efficiency and decreasing costs will be achieved by developing an interactive reference database dedicated to MIP markers where all available intron information will be collected. This would help researchers in consulting, downloading, selecting a priori the best loci for each case study and, in a longer-temporal perspective, integrating data resulting thereof.
Finally, another important possible development is the transfer of this approach to other taxonomic groups. Indeed, the identification of conserved intron regions across species to develop a panel of MIP markers would be theoretically possible for Once the panel of loci is identified for a given taxonomic group, it would become the reference panel for all species in that group, further speeding up the entire process.
Conclusions
MIPs markers, developed and tested on teleosts, are potentially suitable for any other taxon with available genomic information. This wide applicability, combined with their intermediate characteristics compared to other types of markers, positions MIPs as a valuable alternative genetic tool for exploring genetic diversity at different levels of complexity. By providing both allelic frequency and sequence information, they are suitable for a wide range of applications. However, in our opinion, the most interesting aspect is their high transferability between species, making these markers particularly useful for monitoring hybridization processes.
In conclusion, the introduction of MIPs as a new genetic tool offers a promising approach for advancing genetic studies across diverse taxa, with significant potential for applications in conservation, evolutionary biology, and biodiversity monitoring.
Methods
Samples
A total of 65 species belonging to 20 different orders of teleost fishes were used to evaluate cross-species/cross-genera transferability of MIPs. A total of 361 individuals were used: 350 classified at the species level based on morphology and 11 samples identified at the genus level (Chionodraco spp.) due to morphological ambiguities (Table 3). The number of individuals analysed per species ranged from one to five except for the species involved in the three case studies for which a higher number of individuals from different populations were used (see below and Table 1).
Table 3.
Species and relative sample sizes used for library preparation and sequencing. The species further involved in the proposed case studies are in bold.
| Order | Species | N | Order | Species | N |
|---|---|---|---|---|---|
| Acanthuriformes | Sciaena umbra | 1 | Perciformes | Chionodraco spp.†‡ | 11 |
| Umbrina cirrosa | 1 | Dicentrarchus labrax | 2 | ||
| Anguilliformes | Anguilla anguilla | 3 | Neopagetopsis ionah† | 6 | |
| Atheriniformes | Atherina boyeri | 60 | Pagetopsis macropterus† | 6 | |
| Atherina hepsetus | 2 | Pagetopsis maculatus† | 6 | ||
| Atherina presbiter | 1 | Platichthys flesus | 4 | ||
| Beloniformes | Belone belone | 1 | Scorpaena porcus | 1 | |
| Carangiformes | Arnoglossus laterna | 3 | Scorpaena scrofa | 2 | |
| Clupeiformes | Alosa fallax | 1 | Serranus hepatus | 2 | |
| Engralius encrasicolus | 5 | Trachinus draco | 3 | ||
| Sardina pilchardus | 4 | Trachinus radiates | 1 | ||
| Sprattus sprattus | 2 | Trigloporus lastoviza | 1 | ||
| Danio rerio | 1 | Pleuronectiformes | Psetta maxima | 1 | |
| Cypriniformers | Sardinella aurita | 1 | Solea aegyptiaca | 35 | |
| Cyprinodontiformes | Poecilia reticulata | 6 | Synapturichthys kleinii | 1 | |
| Gadiformes | Merlangius merlangus | 2 | Scophothalmus rhombus | 1 | |
| Phycis blennoides | 1 | Solea solea | 71 | ||
| Merluccius merluccius | 1 | Trachurus mediterraneus | 6 | ||
| Deltentosteus quadrimaculatus | 2 | Trachurus trachurus | 1 | ||
| Gobius cobitis | 1 | Priacanthiformes | Cepola macrophtalma | 1 | |
| Gobiiformes | Zosterisessor ophiocephalus | 4 | Cepola rubescens | 1 | |
| Mugiliformes | Chelon auratus | 1 | Scombriformes | Pomatomus saltatrix | 1 |
| Chelon labrosus | 2 | Scomber colias | 3 | ||
| Chelon ramada | 1 | Spariformes | Diplodus annularis | 1 | |
| Mugil cephalus | 1 | Lophius budegassa | 2 | ||
| Ophidiiformes | Ophidion barbatum | 1 | Lithognathus mormyrus | 5 | |
| Perciformes | Auxis thazard | 1 | Pagellus erythrinus | 2 | |
| Aethotaxis mitopteryx† | 6 | Sparus aurata | 5 | ||
| Chaenodraco wilsoni† | 8 | Sarpa salpa | 1 | ||
| Chelidonichthys lucerna | 1 | Syngnathiformes | Mullus barbatus | 2 | |
| Chionodraco hamatus† | 17 | Mullus surmuletus | 1 | ||
| Chionodraco myersi† | 16 | Uranoscopiformes | Uranoscopus scaber | 1 | |
| Chionodraco rastrospinosus† | 14 | Zeiformes | Zeus faber | 2 |
†Samples obtained during multiple R/V Polarstern cruises. Sampling was approved by the competent national authority for Antarctic research (Umweltbundesamt, UBA, Germany).
‡Chionodraco individuals with ambiguous morphological features which prevented a clear species classification based on morphology or putative hybrids based on previous studies (Schiavon et al.33).
For case study 1, a total of 81 individuals of 8 notothenioid (Aethotaxis mytopterix, Chaenodarco wilsoni, the congeneric Chionodraco hamatus, C. myersi, and C. rastrospinosus, Neopagetopsis ionah, and the congeneric Pagetopsis macropterus and P. maculatus) were analysed (Table 1). Further 11 specimens of Chionodraco spp. (Table 1), for which the morphological and mitochondrial characterisation were discordant or the analysis of microsatellite genotypes indicated some degrees of interspecific hybridisation (admixed individuals,33), were also included.
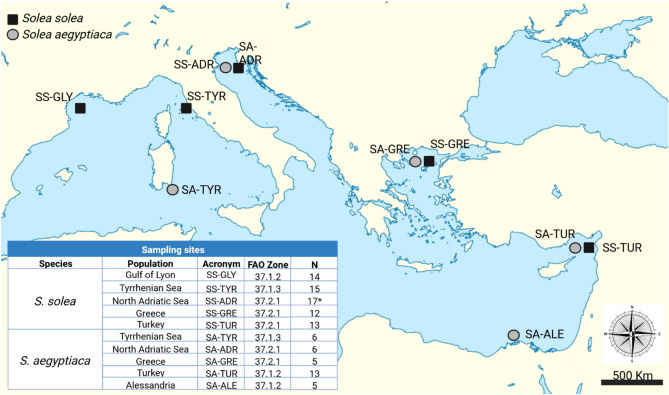
For case studies 2 and 3, a total of 71 Solea solea (SS) and a total of 35 S. aegyptiaca (SS) were analysed. Samples of SS were obtained at five locations across the Mediterranean Sea (Gulf of Lyon, Tyrrhenian Sea, North Adriatic Sea, Greece, and Turkey). Samples of SA were obtained at the same locations except for Alessandria (which replace Gulf of Lyon) (Fig. 4). Samples were part of the FishPop Trace Consortium except three individuals obtained locally at the Chioggia Fish Market, which were morphologically confined as SS and added to the sample from Adriatic Sea.
Figure 4.
Sampling sites and population details of the Solea solea and S. aegyptiaca species involved in case studies 2 and 3. Almost all samples are legacy FishPopTrace Consortium (Nielsen et al., 2012). The only exception is represented by three individuals of the North Adriatic population of S. solea (marked with * in the table inset) that were sampled at the Chioggia Fish Market and morphologically classified as S. solea. Wikimedia Commons map refined with Biorender (https://www.biorender.com/).
All experiments were performed in accordance with relevant guidelines and regulations. The majority of samples used in the present study were collected from the fish market and no live animal was used during the study. For what concerns Chionodraco sp., specimens were sampled for a previous study33 in accordance to and within laws, guidelines and policies of the German and European Animal Welfare legislations. Sample collection conducted during the cruises with R/V Polarstern was approved by the competent German authority for Antarctic research, the UBA (Umweltbundesamt).
Additionally, the present study was carried out in compliance with the ARRIVE guidelines.
For all samples, genomic DNA was purified from fin clips using DNeasy Blood and Tissue kit (Qiagen) with an elution volume of 100 µl; obtained extracts were stored at −20 °C until analysis.
In silico isolation of candidate intron markers and primer design and validation
Putative intron candidate markers, with high potential to be transversally applicable for ecological studies on teleost fishes, were isolated by aligning 48 annotated fish genomes distributed cross the phylogeny (see Supplementary Material—Table S2). FASTA files and GTF files, which contain the coordinates of different annotated genes on the reference genomes, were downloaded from Ensembl database and intron positions embedded into well-conserved automatically generated exonic alignment were considered.
Firstly, according to the GTF files, the exon-flanking regions of each intron were extracted and 100 bp before and after the intron junction were retained for further analysis. On average, 135,488 sequences were retrieved from each genome, for a total of 6,503,466 introns. Secondly, an intra-locus sequencing clustering was performed using CD-HIT program version 4.8.146–48 and setting (i) sequence identity to 0.9, (ii) the ‘-g’ option to 1 allowing for a slower but more accurate clustering, and (iii) the word length to 5. The clustering step produced a total of 4,846,172 different clusters. The CD-HIT clusters were further processed to retain only those who were composed by sequences belonging to at least 15 different genomes with the aim to identify candidate intron markers whose exon-flanking regions are potentially highly conserved across teleost genomes. A total of 2,441 clusters passed the above filter and were considered for further analysis, for a total of 43,744 sequences.
With the final aim to further maximise the species representation within filtered clusters, the last step included a screening for potential non predicted genes. For each cluster, the representative sequence provided by CD-HIT was aligned on the reference genomes of all species excluded during the sequencing clustering step. The alignment was implemented in BLAST49 by using highly stringent criteria. Only sequences aligned with a percentage of identity of at least 90% and a sequence coverage of 100% were retained as previously undetected homologous sequences. As a result of this process, a total of 16,593 sequences were retrieved and added to the previously filtered clusters.
The 2,441 filtered clusters were examined for primer construction and introns potentially usable as highly transferable markers were selected to be tested on 65 species based on the following criteria: (i) selection of clusters showing an average intron length between 200 and 800 bp calculated on the genomes successfully aligned for each cluster, (ii) the highest possible number of genomes matching cluster, and iii) the more conserved exon-flanking regions. These criteria allowed to concentrate efforts on loci with predicted dimension compatible with Illumina sequencing, easily amplifiable with PCR and potentially more conserved across species for which genome was not yet available.
Based on these criteria, a panel of 121 candidate intron markers was selected for the present study, and locus specific primer pairs were designed on exon-flanking regions, close to intron junctions, based on consensus sequences for each cluster (table S3).
All primer pairs were validated by single-locus PCR on a few individuals belonging to 15 different species (of which only two, Danio rerio and Psetta maxima, had available genomic information), thus confirming their cross-applicability.
Library preparation and amplicon sequencing
The DNA concentration was normalized across individuals, and the samples were processed with an optimized method to obtain individual intron libraries to be simultaneously sequenced on Illumina platform. The proposed method includes a two-steps PCR protocol (see Supplementary Material—Figure S4). The first step consisted in multiplexing intron loci using locus-specific primer-pairs opportunely modified by adding a tail at 5’ position (5’CTACACGACGCTCTTCCGATCTTCAGA3’ and 5’CAGACGTGTGCTCTTCCGATCT3’ respectively for forward and reverse primer) and which will act as the binding site for primers used in the second step. Six multiplex reactions (M1-6) have been performed with the Multiplex PCR Master Mix (Qiagen) following the manufactures protocol. The final PCR volume of 50 µl for each individual was divided into three wells during amplification to reduce biases introduced by PCR. Cycling parameters were the following: denaturation at 95 °C for 15’, 35 cycles with denaturation at 94 °C for 30’’, annealing for 1′30’’at 58 and 60 °C respectively for M1-2 and M3-6, and extension at 72 °C for 1’, followed by a final extension at 60 °C for 30’. Detailed information on multiplex composition are reported in Supporting Information (Table S3).
After pooling the three reactions for each multiplex and all multiplex PCRs with different loci for each sample, the second step consisted of indexing-PCRs with primer pairs usually used in the 2bRAD protocol50,51 with the aim of adding individual-specific 7 bp barcodes for demultiplexing and adapters compatible with the Illumina platform (P5-P7). For all indexing PCRs, the following reagents were used in a final volume of 50 µl (always divided into three reactions for amplification as described above): 1X HF Buffer (5X), 0.31 mM of dNTPs (20 mM), 0.5 µM of primer F and Barcoded-Primer R (10 µM), 0.2 µM of primer 2bRAD_amp_F and 2bRAD_amp_R (10 µM), 0.04 U/µl of Taq Phusion (New England Biolabs) and the purified multiplexed PCR products opportunely diluted (1:200). Cycling parameters were the following: denaturation at 95 °C for 3’, 10 cycles with denaturation at 94 °C for 30’’, annealing at 60 °C for 30’’, and extension at 72 °C for 45’’, and a final extension at 72 °C for 5’.
Size selection and purification were performed for each individual library after the first amplification step, and for the final library after indexing-PCRs and pooling amplicons from different samples. The size selection involved the use of SPRISelect magnetic beads (Beckman Coulter, 0.5X and 0.56X are the ratios used for the first and second purification steps, respectively) and allowed removing primer dimers and amplicons larger than 800 bp, which might negatively affect sequencing performances. The final libraries of targeted intron amplicons were visualized on agarose gel and quantified by TapeStation 4150 (Agilent).
A total of 384 samples (351 individuals of which 33 were processed as replicates) were successfully amplified and sequenced on two 300 bp paired-end Illumina MiSeq v3 runs performed at the Norwegian Sequencing Centre (Oslo, Norway; https://www.sequencing.uio.no/).
Bioinformatic data processing for haplotype generation
The raw reads in fastq format were firstly processed with Cutadapt 2.852 to demultiplex reads based on the individual-specific barcode and to search for the presence of the intron-specific primer pairs used to amplify each locus. Cutadapt was also used to discard low quality bases and remove any possible adapters that remain inside by setting (i) the quality cutoff ‘-q’ at 15, (ii) the minimum read length after trimming to 100 pb, and (iii) the minimum overlap length with the primer sequence at 15 bp. During the processing, when one of the paired-end sequences was eliminated, the entire for-rev pair was removed from the entire data set. Secondly, the FLASH v 2.2.00 program53 was used to merge paired-ends intra-sample for each locus. Given the heterogeneous dimensional range expected for different intron loci in different species genomes, an effective merging of paired-ends was expected only for shorter introns. Consequently, whenever the number of merged for-rev reads at a locus was less than 20% of the total, the locus was discarded at the species level as too long to generate reliable haplotypes. At this step, loci showing a coverage lower than 30 sequences were also removed at the species level.
Finally, the SeekDeep pipeline (v 3.0.0)54 was implemented to genotype MIP markers by generating a de novo clustering of allelic variants at each locus based on an intra- and inter-individual analyses. The qluster package was implemented to predict and estimate the frequencies and relative abundance of trailing locus-specific alleles at the individual level. Qluster was run using default parameters to collapse errors, group reads, and identify chimeric sequences. After that, the processClusters package was used to compare alleles from each locus across samples. ProcessClusters applies a filter to the final results of qcluster to remove low frequency alleles, singlets and chimeric alleles getting rid of artefacts. To compare alleles across samples, the option ‘noErrors’ was used.
The final output of the SeekDeep pipeline was further filtered by retaining for each locus and each sample only the first ni alleles (sorting them by decreasing frequency) whose cumulative abundance was greater than 75% of the total count.
Selection of loci for the analysis of case studies and data analysis
The detected alleles for different loci were grouped per species and formatted in FASTA format using BioPhyton tools55,56. The FASTA files for each locus per species were aligned and analysed with MEGAX57 and all sequences were manually checked.
Loci were screened separately per species target of each case study. Each locus was retained in accordance with the following criteria: (i) the presence of the genotype in at least the 75% of individuals per species or population, (ii) an allele coverage greater than 30 high-quality assembled reads, (iii) concordant genotypes between replicates, (iv) no more than two called alleles per genotype, and (v) the absence of a homopolymer stretch or microsatellites in the merging region of the for-rev reads.
The data from the final panel of 121 MIPs were converted into genepop format, as outlined in the Supplementary Material (SF1). Subsequently, basic descriptive statistics and structure analyses were performed on the data from the two case studies following details and statistics described in Supplementary Material (SF1).
Supplementary Information
Acknowledgements
We thank “Ittico Sostenibile” (Dr. Andrea Chinellato) which contributed to sample collection or sharing and Dr. Leonardo Girlanda that provided support to laboratory procedures. The authors are grateful to Dr. Magnus Lucassen and Nils Koschnick (Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research, AWI, Bremerhaven, Germany) and the ship crew for their help in collecting samples on board the R/V Polarstern. We thank EuroFishMarket for future support in dissemination of present MIP markers applicability.
Author contributions
E.B. and L.C. designed the study; E.B., S.D.P. and A.S. performed research; N.V. and A.S. developed the bioinformatics pipeline and assembled obtained reads; E.B., L.C., and L.Z. contributed new reagents or analytical tools and obtained funding; E.B., A.S. and L.S. analysed data; E.B. and L.C. interpreted the results and co-wrote the main manuscript; A.C. and C.P. provided samples; all authors reviewed the manuscript.
Funding
The work was partially funded by the FSBI Small Research Grants (n. 256475). LZ and LC acknowledge support under the National Recovery and Resilience Plan (NRRP), Mission 4. Component 2 Investment 1.4—Call for tender No. 3138 of 16 December 2021, rectified by Decree n.3175 of 18 December 2021 of Italian Ministry of University and Research funded by the European Union—NextGenerationEU; Award Number: Project code CN_00000033, Concession Decree No. 1034 of 17 June 2022. Adopted by the Italian Ministry of University and Research,C93C22002810006, Project title “National Biodiversity Future Center—NBFC”. C.P. acknowledges financial support of the European Marie Curie Project “Polarexpress” Grant No. 622320 and the University of Padova BIRD Grant No.164793. L.Z. acknowledges support by the Italian National Programme of Antarctic Research (PNRA) Project 2016_00307.
Data availability
Raw fastq sequencing reads are deposited in NCBI SRA (BioProject PRJNA1044401).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-68065-8.
References
- 1.Camacho-Sanchez, M. et al. Comparative assessment of range-wide patterns of genetic diversity and structure with SNPs and microsatellites: A case study with Iberian amphibians. Ecol. Evol.10, 10353–10363. 10.1002/ece3.6670 (2020). 10.1002/ece3.6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toews, D. P. L. & Brelsford, A. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol.21(16), 3907–3930. 10.1111/j.1365-294X.2012.05664.x) (2012). 10.1111/j.1365-294X.2012.05664.x) [DOI] [PubMed] [Google Scholar]
- 3.Allendorf, F. W. Genetics and the conservation of natural populations: Allozymes to genomes. Mol. Ecol.26(2), 420–430. 10.1111/mec.13948 (2017). 10.1111/mec.13948 [DOI] [PubMed] [Google Scholar]
- 4.Baird, N. A. et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE3(10), e3376. 10.1371/journal.pone.0003376 (2008). 10.1371/journal.pone.0003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elshire, R. J. et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE6(5), e19379. 10.1371/journal.pone.0019379 (2011). 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baetscher, D. D., Clemento, A. J., Ng, T. C., Anderson, E. C. & Garza, J. C. Microhaplotypes provide increased power from short-read DNA sequences for relationship inference. Mol. Ecol. Res.18, 296–305. 10.1111/1755-0998.12737 (2018). 10.1111/1755-0998.12737 [DOI] [PubMed] [Google Scholar]
- 7.Christiansen, H. et al. Facilitating population genomics of non-model organisms through optimized experimental design for reduced representation sequencing. BMC Genom.22, 625. 10.1186/s12864-021-07917-3 (2021). 10.1186/s12864-021-07917-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodel, R. G. J. et al. The report of my death was an exaggeration: A review for researchers using microsatellites in the 21st century. Appl. Plant Sci.4(6), 1600025. 10.3732/apps.1600025 (2016). 10.3732/apps.1600025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puckett, E. E. Variability in total project and per sample genotyping costs under varying study designs including with microsatellites or SNPs to answer conservation genetic questions. Conserv. Genet. Res.9(2), 289–304. 10.1007/s12686-016-0643-7 (2017). 10.1007/s12686-016-0643-7 [DOI] [Google Scholar]
- 10.LaFramboise, T. Single nucleotide polymorphism array: A decade of biological, computational and technological advances. Nucleic Acids Res.37(13), 4181–4193. 10.1093/nar/gkp552 (2009). 10.1093/nar/gkp552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pujolar, J. M., Limborg, M. T., Ehrlich, M. & Jaspers, C. High throughput SNP chip as cost effective new monitoring tool for assessing invasion dynamics in the comb jelly Mnemiopsis leidyi. Front. Mar. Sci.9, 1019001. 10.3389/fmars.2022.1019001 (2022). 10.3389/fmars.2022.1019001 [DOI] [Google Scholar]
- 12.Pakstis, A. J. et al. The population genetics characteristics of a 90 locus panel of microhaplotypes. Hum. Genet.140, 1753–1773. 10.1007/s00439-021-02382-0 (2021). 10.1007/s00439-021-02382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell, N. R., Harmon, S. A. & Narum, S. R. Genotyping-in-thousands by sequencing (GT-seq): A cost effective SNP genotyping method based on custom amplicon sequencing. Mol. Ecol. Res.15, 855–867. 10.1111/1755-0998.12357 (2014). 10.1111/1755-0998.12357 [DOI] [PubMed] [Google Scholar]
- 14.Ressayre, A. et al. Introns structure patterns of variation in nucleotide composition in arabidopsis thaliana and rice protein-coding genes. Genome Bio. Evol.7(10), 2913–2928. 10.1093/gbe/evv189 (2015). 10.1093/gbe/evv189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forcina, G., Camacho-Sanchez, M., Tuh, F. Y. Y., Moreno, S. K. & Leonard, J. A. Markers for genetic change. Helyion7(1), e05583. 10.1016/j.helyion.2020.e05583 (2021). 10.1016/j.helyion.2020.e05583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bong-Seok, J. & Sun Shim, C. Introns: The functional benefits of introns in genomes. Genom. Inform.13(4), 112–118. 10.5808/GI.2015.13.4.112 (2015). 10.5808/GI.2015.13.4.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chorev, M. & Carmel, L. The function of introns. Front. Genet.3, 55. 10.3389/fgene.2012.00055 (2012). 10.3389/fgene.2012.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irimia, M. & Roy, S. W. Spliceosomal introns as tools for genomic and evolutionary analysis. Nucleic Acids Res.36(5), 1703–1712. 10.1093/nar/gkn012 (2008). 10.1093/nar/gkn012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeo, G., Hoon, S., Venkatesh, B. & Burge, C. B. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. PNAS101(44), 15700–15705. 10.1073/pnas.0404901101 (2004). 10.1073/pnas.0404901101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lessa, E. P. Rapid surveying of DNA sequence variation in natural populations. Mol. Biol. Evol.9, 323–330 (1992). [DOI] [PubMed] [Google Scholar]
- 21.Daguin, C. & Borsa, P. Genetic characterisation of Mytilus galloprovincialis Lmk. In North West Africa using nuclear DNA markers. J. Exp. Mar. Biol. Ecol.235(1), 55–65. 10.1016/S0022-0981(98)00163-4 (1999). 10.1016/S0022-0981(98)00163-4 [DOI] [Google Scholar]
- 22.Corte-Real, H. B. S., Dizon, D. R. & Holland, P. W. H. Intron-targeted PCR: a new approach to survey neutral DNA polymorphism in bivalve populations. Mar. Biol.120(3), 407–413. 10.1007/BF00680214 (1994). 10.1007/BF00680214 [DOI] [Google Scholar]
- 23.Daguin, C., Bonhomme, F. & Borsa, P. The zone of sympatry and hybridization of Mytilus edulis and M. galloprovincialis, as described by intron length polymorphism at locus mac-1. Heredity86(3), 342–354. 10.1046/j.1365-2540.2001.00832.x (2001). 10.1046/j.1365-2540.2001.00832.x [DOI] [PubMed] [Google Scholar]
- 24.Villablanca, F. X., Roderick, G. K. & Palumbi, S. R. Invasion genetics of the mediterranean fruit fly: Variation in multiple nuclear introns. Mol. Ecol.7(5), 547–560. 10.1046/j.1365.294x.1998.00351.x (1998). 10.1046/j.1365.294x.1998.00351.x [DOI] [PubMed] [Google Scholar]
- 25.Chow, S. & Hazama, K. Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol. Ecol.7, 1247–1263 (1998). [PubMed] [Google Scholar]
- 26.Palumbi, S. R. & Baker, C. S. Contrasting population structure from nuclear intron sequences and mtDNA of humpback whales. Mol. Biol. Evol.11(3), 426–435. 10.1093/oxfordjournals.molbev.a040115 (1994). 10.1093/oxfordjournals.molbev.a040115 [DOI] [PubMed] [Google Scholar]
- 27.Boscari, E. et al. Species and hybrid identification of sturgeon caviar: A new molecular approach to detect illegal trade. Mol. Ecol. Res.14(3), 489–498. 10.1111/1755-0998.12203 (2014). 10.1111/1755-0998.12203 [DOI] [PubMed] [Google Scholar]
- 28.Boscari, E. et al. Fast genetic identification of the Beluga sturgeon and its sought-after caviar to stem illegal trade. Food Control75, 145–152. 10.1016/j.foodcont.2016.11.039 (2017). 10.1016/j.foodcont.2016.11.039 [DOI] [Google Scholar]
- 29.Boscari, E. et al. Genetic identification of the caviar-producing Amur and Kaluga sturgeons revealed a high level of concealed hybridization. Food Control82, 243–250. 10.1016/j.foodcont.2017.07.001 (2017). 10.1016/j.foodcont.2017.07.001 [DOI] [Google Scholar]
- 30.Igea, J., Juste, J. & Castresana, J. Novel intron markers to study the phylogeny of closely related mammalian species. BMC Evol. Biol.10(1), 369. 10.1186/1471-2148-10-369 (2010). 10.1186/1471-2148-10-369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, C., Riethoven, J. J. M. & Ma, L. Exon-primed intron-crossing (EPIC) markers for non-model teleost fishes. BMC Evol. Biol.10, 90. 10.1186/1471-2148-10-90 (2010). 10.1186/1471-2148-10-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ströher, P. R., Li, C. & Pie, M. R. Exon-primed intron-crossing (EPIC) markers as a tool for ant phylogeography. Revista Brasileira Entomologia10.1590/S0085-56262013005000039 (2013). 10.1590/S0085-56262013005000039 [DOI] [Google Scholar]
- 33.Schiavon, L. et al. Species distribution, hybridization and connectivity in the genus Chionodraco: Unveiling unknown icefish diversity in Antarctica. Divers. Distrib.27(5), 766–789. 10.1111/ddi.13249 (2021). 10.1111/ddi.13249 [DOI] [Google Scholar]
- 34.Garoia, F., Guarniero, I., Grifoni, D., Marzola, S. & Tinti, F. Comparative analysis of AFLPs and SSRs efficiency in resolving population genetic structure of mediterranean Solea vulgaris. Mol. Ecol.16(7), 1377–1387. 10.1111/j.1365-294X.2007.03247.x (2007). 10.1111/j.1365-294X.2007.03247.x [DOI] [PubMed] [Google Scholar]
- 35.Rolland, J. L. et al. Population structure of the common sole (Solea solea) in the Northeastern atlantic and the mediterranean sea: Revisiting the divide with epic markers. Mar. Biol.151, 327–341. 10.1007/s00227-006-0484-0 (2007). 10.1007/s00227-006-0484-0 [DOI] [Google Scholar]
- 36.Marino, I. A. M. et al. Evidence for past and presence hybridization in three Antarctic icefish species provides new perspectives on an evolutionary radiation. Mol. Ecol.22(20), 5148–5161. 10.1111/mec.12458 (2013). 10.1111/mec.12458 [DOI] [PubMed] [Google Scholar]
- 37.Sabatini, L. et al. Good practices for common sole assessment in the Adriatic sea: Genetic and morphological differentiation of Solea solea (Linnaeus, 1758) from S. aegyptiaca (Chabanaud, 1927) and stock identification. J. Sea Res.137, 57–64. 10.1016/j.seares.2018.04.004 (2018). 10.1016/j.seares.2018.04.004 [DOI] [Google Scholar]
- 38.Betancur-R, R. et al. Phylogenetic classification of bony fishes. BMC Evol. Biol.17(1), 162. 10.1186/s12862-017-0958-3 (2017). 10.1186/s12862-017-0958-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson, J. S., Grande, T. C. & Wilson, M. V. Fishes of the World (Wiley, 2016). [Google Scholar]
- 40.Near, T. J. et al. Ancient climate change, antifreeze, and evolutionary diversification of Antarctic fishes. PNAS109(9), 3434–3439. 10.1073/pnas.1115169109 (2012). 10.1073/pnas.1115169109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Near, T. J. et al. Phylogenetic analysis of Antarctic notothenioids illuminates the utility of RADseq for resolving cenozoic adaptive radiations. Mol. Phylogenet. Evol.129, 268–279. 10.1016/j.ympev.2018.09.001 (2018). 10.1016/j.ympev.2018.09.001 [DOI] [PubMed] [Google Scholar]
- 42.Corti, R. Moving toward stock units identification based on spatial population structure of marine species in the Mediterranean Sea and adjacent waters through multidisciplinary and holistic approaches for the sustainability of fisheries resources. Universitty of Bologna. PhD thesis. (2022).
- 43.Corti, R., et al. Seascape genomics approach to describe population structure of two marine species: Solea solea and Merluccius merluccius case studies. Proc. 9th Congress of the Italian Society for Evolutionary Biology (SIBE), 4–7, Ancona (Italy). https://www.sibe-iseb.it/_files/ugd/744a74_4775ee0936dd424181479d6f63ef0a13.pdf (2022).
- 44.Corti, R. et al. A multidisciplinary approach to describe population structure of Solea solea in the mediterranean sea. Front. Mar. Sci.11, 1372743. 10.3389/fmars.2024.1372743 (2024). 10.3389/fmars.2024.1372743 [DOI] [Google Scholar]
- 45.He, J. et al. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci.10.3389/fpls.2014.00484 (2014). 10.3389/fpls.2014.00484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: Accelerated for clustering the next generation sequencing data. Bioinformatics28, 3150–3152. 10.1093/bioinformatics/bts565 (2012). 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Limin, F., Beifang, N., Zhengwei, Z., Sitao, W. & Weizhong, L. CD-HIT: Accelerated for clustering the next generation sequencing data. Bioinformatics28(23), 3150–3152. 10.1093/bioinformatics/bts565 (2012). 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weizhong, L. & Godzik, A. CD-HIT: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics22, 1658–1659 (2006). 10.1093/bioinformatics/btl158 [DOI] [PubMed] [Google Scholar]
- 49.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol.215, 403–410 (1990). 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 50.Boscari, E. et al. A population genomics insight by 2b-RAD reveals populations’ uniqueness along the Italian coastline in Leptopsammia pruvoti(Scleractinia, Dendrophylliidae). Divers. Distrib.25, 1101–1117. 10.1111/ddi.12918 (2019). 10.1111/ddi.12918 [DOI] [Google Scholar]
- 51.Wang, S., Meyer, E., McKay, J., McKay, J. K. & Matz, M. V. 2b-RAD: A simple and flexible method for genome-wide genotyping. Nat. Methods9, 808–810. 10.1038/nmeth.2023 (2012). 10.1038/nmeth.2023 [DOI] [PubMed] [Google Scholar]
- 52.Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J.17, 10–12. 10.14806/ej.17.1.200 (2011). 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 53.Magoc, T. & Salzberg, S. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics27(21), 2957–2963 (2011). 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hathaway, N. J., Parobek, C. M., Juliano, J. J. & Bailey, J. A. SeekDeep: single-base resolution de novo clustering for amplicon deep sequencing. Nucleic Acids Res.46(4), e21. 10.1093/nar/gkx1201 (2018). 10.1093/nar/gkx1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cock, P. J. et al. Biopython: Freely available python tools for computational molecular biology and bioinformatics. Bioinformatics25(11), 1422–1423. 10.1093/bioinformatics/btp163 (2009). 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Rossum, G., Drake, J. R. & Fred, L. Python Reference Manual (Centrum voor Wiskunde en Informatica Amsterdam, 1995). [Google Scholar]
- 57.Kumar, S., Stecher, G., Li, M., Knyazm, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.35(6), 1547–1549. 10.1093/molbev/msy096 (2018). 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw fastq sequencing reads are deposited in NCBI SRA (BioProject PRJNA1044401).