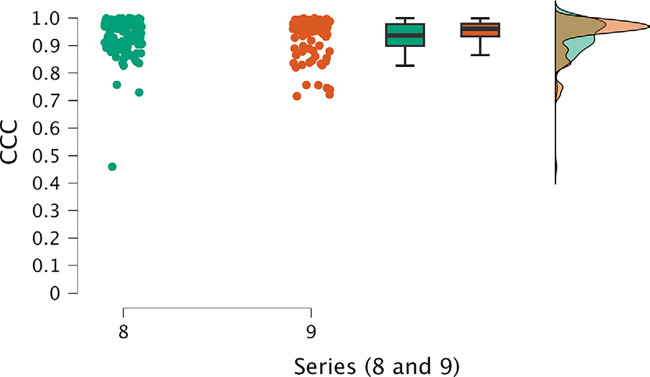Abstract
Purpose
To evaluate the reproducibility of radiomics features extracted from T2-weighted MR images in patients with neuroblastoma.
Materials and Methods
A retrospective study included 419 patients (mean age, 29 months ± 34 [SD]; 220 male, 199 female) with neuroblastic tumors diagnosed between 2002 and 2023, within the scope of the PRedictive In-silico Multiscale Analytics to support cancer personalized diaGnosis and prognosis, Empowered by imaging biomarkers (ie, PRIMAGE) project, involving 746 T2/T2*-weighted MRI sequences at diagnosis and/or after initial chemotherapy. Images underwent processing steps (denoising, inhomogeneity bias field correction, normalization, and resampling). Tumors were automatically segmented, and 107 shape, first-order, and second-order radiomics features were extracted, considered as the reference standard. Subsequently, the previous image processing settings were modified, and volumetric masks were applied. New radiomics features were extracted and compared with the reference standard. Reproducibility was assessed using the concordance correlation coefficient (CCC); intrasubject repeatability was measured using the coefficient of variation (CoV).
Results
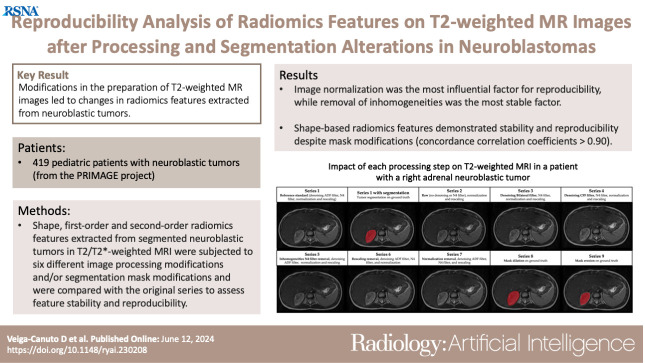
When normalization was omitted, only 5% of the radiomics features demonstrated high reproducibility. Statistical analysis revealed significant changes in the normalization and resampling processes (P < .001). Inhomogeneities removal had the least impact on radiomics (83% of parameters remained stable). Shape features remained stable after mask modifications, with a CCC greater than 0.90. Mask modifications were the most favorable changes for achieving high CCC values, with a radiomics features stability of 70%. Only 7% of second-order radiomics features showed an excellent CoV of less than 0.10.
Conclusion
Modifications in the T2-weighted MRI preparation process in patients with neuroblastoma resulted in changes in radiomics features, with normalization identified as the most influential factor for reproducibility. Inhomogeneities removal had the least impact on radiomics features.
Keywords: Pediatrics, MR Imaging, Oncology, Radiomics, Reproducibility, Repeatability, Neuroblastic Tumors
Supplemental material is available for this article.
© RSNA, 2024
See also the commentary by Safdar and Galaria in this issue.
Keywords: Pediatrics, MR Imaging, Oncology, Radiomics, Reproducibility, Repeatability, Neuroblastic Tumors
Summary
Modifications in the preparation of T2-weighted MR images led to changes in radiomics features extracted from neuroblastic tumors.
Key Points
■ Shape, first-order, and second-order radiomics features extracted from segmented neuroblastic tumors in T2/T2*-weighted MR images were subjected to six different image processing modifications and/or segmentation mask modifications and were compared with the original series to assess feature stability and reproducibility.
■ Image normalization was identified as the most influential factor for reproducibility, with a radiomics features stability of only 5%, while removal of inhomogeneities was the most stable factor, with a parameters stability of 83%.
■ Shape-based radiomics features demonstrated stability and reproducibility despite mask modifications, with a concordance correlation coefficient greater than 0.90.
Introduction
Radiomics involves the mathematical extraction of quantitative features from regions of interest in images, providing shape, textural, and voxel interrelationships information (1,2). Radiomics analysis aims to obtain clinically meaningful information through prediction models. Several studies have demonstrated that radiomics signatures can aid in characterizing tumors (1,3,4).
However, feature stability and reproducibility are key limitations in the generalization of these types of studies. The stability of radiomics analyses is a major challenge because of the inherent variations in image acquisition and reconstruction parameters, as well as modifications in the radiomics pipeline. Radiomics analyses involve several preliminary image preparation steps, such as image denoising, signal normalization, and voxel size resampling, and throughout this workflow, multiple factors may influence the results (2,5,6). Several attempts to homogenize the radiomics extraction process and empower the quality of radiomics processing and reporting have been made, but a compromise on the standardization process of radiomics analysis has not been reached (7,8). However, numerous studies have been conducted to evaluate the repeatability (ie, scan-rescan approach) (9–11) and reproducibility (ie, by studying the impact of imaging perturbations such as filters [12–14] and the changes induced by different segmentation masks performed by different readers [15–17]) of radiomics features. The robustness of radiomics extraction is crucial for its successful translation to the clinical setting.
Neuroblastic tumors are the most common solid extracranial tumors in children. They are heterogeneous in appearance and location and display diverse behavior, which is influenced by various biologic, clinical, and prognostic factors. While some tumors undergo spontaneous regression, others advance and produce fatal outcomes despite therapeutic interventions (18). Radiomics has been linked to pathologic differentiation of peripheral neuroblastic tumors (9) and MYCN amplification prediction (19), but the heterogeneous behavior of these tumors emphasizes the importance of studying the reproducibility of radiomics, as it can have a substantial impact on the reproducibility and robustness of predictive models, which may impact the current state of the art for tumor predictions.
This study aimed to evaluate the reproducibility of radiomics features extracted from T2/T2*-weighted MR images from a large dataset of patients with neuroblastic tumors in the scope of the PRedictive In-silico Multiscale Analytics to support cancer personalized diaGnosis and prognosis, Empowered by imaging biomarkers (PRIMAGE) project (20). Tumors were automatically segmented (21,22) after image preparation (denoising, inhomogeneity bias field correction, signal normalization, and resampling). Radiomics features were extracted as the reference standard and compared with those obtained after modifying these steps to assess stability.
Materials and Methods
Patients
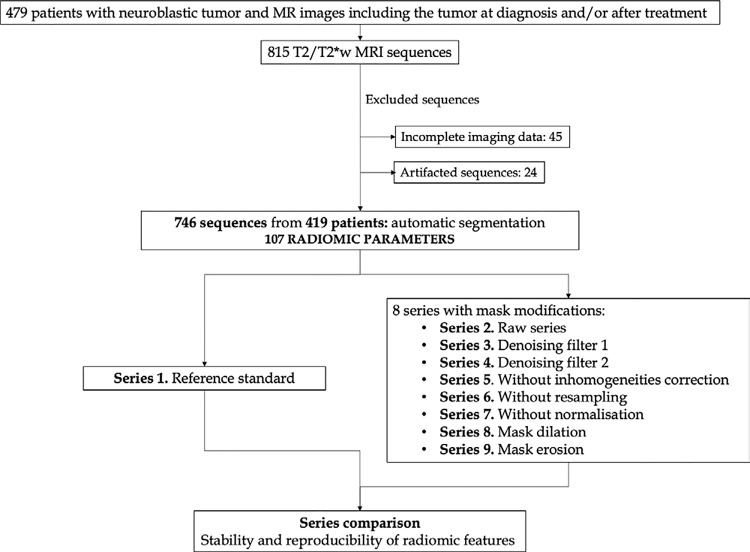
This retrospective international multicenter exploratory study included 419 pediatric patients with pathologically proven neuroblastic tumors between 2002 and 2023 (23) from the PRIMAGE European Union Horizon 408 2020 research and innovation act project (topic SC1-DTH-07–2018, grant agreement no. 826494). Additionally, most of the patients participated in two clinical trials led by the Society of Pediatric Oncology European Neuroblastoma Network (SIOPEN): (a) High Risk Neuroblastoma Study (HR-126 NBL1/SIOPEN) with patients from 12 countries, led by St. Anna Children’s Cancer Research Institute (Vienna, Austria) and (b) the SIOPEN European Low and Intermediate Risk Neuroblastoma Protocol clinical trial (ie, LINES/SIOPEN), led by La Fe University and Polytechnic Hospital (Valencia, Spain). Thus, this is a multi-institutional study with a largely heterogeneous dataset. Three hundred of the 419 patients have been previously reported (22). This prior article dealt with independent validation of a previously developed automatic segmentation tool, whereas in this article, we report the reproducibility of extracted radiomics features. The study received institutional ethics committee approvals from all involved institutions; it was approved by the Ethics Committee for Investigation with Medicinal Products of the University and Polytechnic La Fe Hospital, ethics code 2018/0228. Signed informed consent was waived due to the observational and retrospective study design. All patients underwent an MRI examination of the anatomic region where the tumor was located (328 in the abdominopelvic region, 91 in the cervicothoracic region) either at diagnosis (625 sequences) and/or after initial chemotherapy treatment (121 sequences). Of the original 815 T2/ T2*-weighted MR images, 69 were excluded due to incomplete or degraded images; thus, 746 T2/T2*-weighted MR images were included in the final analysis (Fig 1, Table 1). The analysis was performed on T2/T2*-weighted images, as they yield the maximum contrast between the tumor and the surrounding structures. To avoid losing sample size and to increase generalizability by analyzing different T2 sequences, all T2-weighted spin-echo, T2-weighted spin-echo fat-suppression, T2-weighted short inversion time inversion recovery, and T2*-weighted gradient-echo fat-saturation images that met the inclusion criteria were incorporated. Subsequently, in 92 patients, only one image was included per patient, while in 327 patients, two images were included per patient. All sequences had been performed separately and independently. The PRIMAGE data repository and platform are currently being integrated into the European Federation for Cancer Images (ie, EUCAIM), which is the largest European cancer imaging research infrastructure created to date.
Figure 1:
Flowchart of selected MR images and series. There were 419 selected patients with 815 T2/T2*-weighted MR images. After exclusion, 746 images remained. A total of 107 radiomics parameters were extracted (series 1). Eight different modifications were performed and compared with the original series to assess the stability and reproducibility of radiomics parameters.
Table 1:
Characteristics of Study Dataset
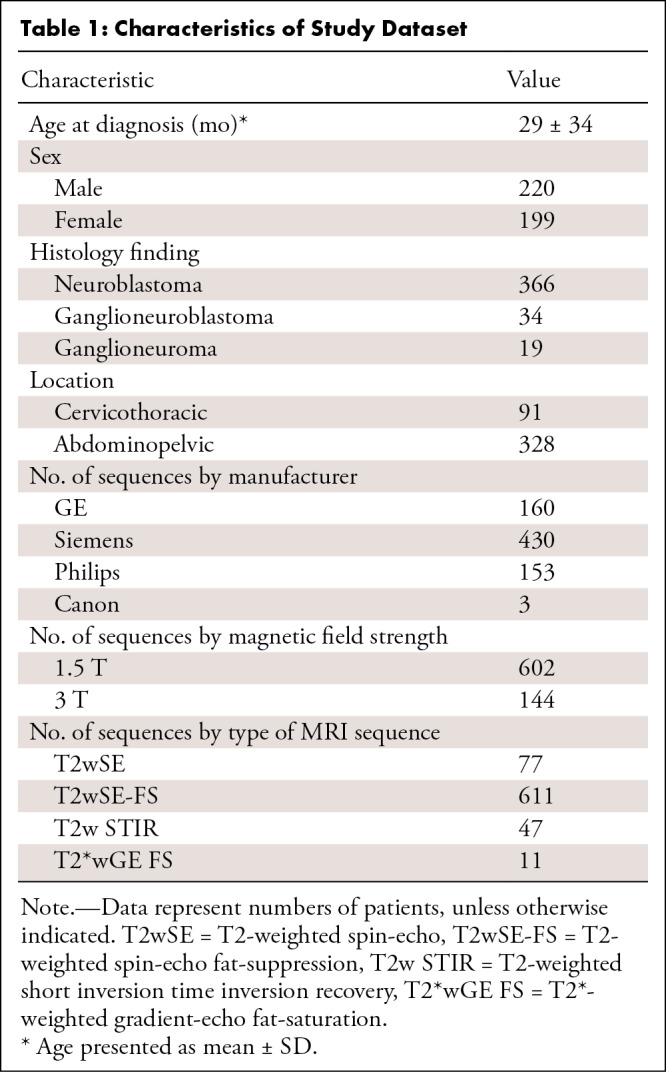
Image Processing
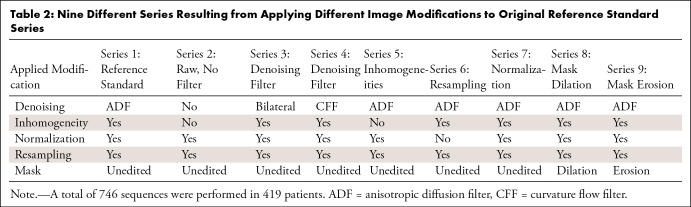
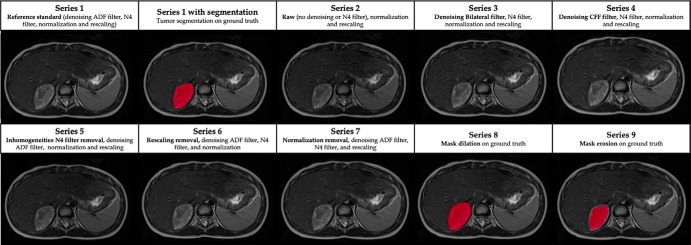
Images were prepared to ensure a common framework before tumor segmentation (20). This preparatory phase consisted of applying an anisotropic diffusion filter (ADF) for denoising, N4 bias field correction to correct signal inhomogeneity, z score signal normalization for standardization, and resampling for spatial harmonization. These image modification steps were applied and considered as the reference standard for radiomics extraction, referred to as series 1 (Table 2). Segmentation was performed after the image was improved with these modifications, which represents a real-world scenario in which images are harmonized.
Table 2:
Nine Different Series Resulting from Applying Different Image Modifications to Original Reference Standard Series
Tumor Segmentation
An nnU-Net convolutional neural network automatic tool (21,22,24) for tumor detection and segmentation was implemented to delineate the tumor as a region of interest on all MR images, which saved time for completing this process. This tool was previously trained with 132 T2-weighted manually segmented cases (21) and independently validated with 535 T2-weighted studies, which were automatically segmented and visually validated by a radiologist (D.V.C.), who performed manual corrections when necessary (22). All 746 tumors were successfully identified and segmented automatically in the T2- and T2*-weighted MR images of patients with a primary neuroblastic tumor at diagnosis or after the first line of treatment with chemotherapy. The segmentation masks were visually validated and manually edited, if needed, by a pediatric radiologist with 7 years of experience (D.V.C.). The in-house automatic segmentation tool is publicly available at https://github.com/lcerdaal/MRNeuroblastomaSegmentation/tree/main.
Image and Mask Modifications
For comparison, eight additional sets of images were generated after modifying the processing and segmentation steps. The different filters and modifications, which were open-source codes with an affordable computational cost and widely used in previous studies, are detailed below.
-
Denoising: Three commonly used “edge-preserving” filters were applied. These filters are integrated into the SimpleITK library (25):
- (a) ADF, which acts as a high-pass filter, removing high-frequency noise. In a previous study performed in the scope of the PRIMAGE project (26), this filter showed the best results regarding reproducibility of radiomics features.
- (b) Curvature flow filter (CFF), which smooths perpendicular to the isointensity contours (27).
- (c) Bilateral filter, which replaces the intensity of each pixel with a weighted average of intensity values from nearby pixels (28).
Inhomogeneities: N4 bias field correction was used to correct low-frequency intensity nonuniformity present in MRI data, known as a bias or gain field. This filter has been widely used and is a state-of-the-art method for correcting the bias field to optimize MRI-based quantification (29,30). This filter was applied using ANTS software (31).
Resampling: All MR image series were resampled to a voxel size of 1 × 1 × 6 mm for spatial harmonization to allow image comparison. This step was employed directly in the PyRadiomics environment (32).
Normalization: z score methods were applied as a harmonization process to reduce systematic variations due to image acquisition, reconstruction, and postprocessing (2,33). This filter is widely used to standardize data across images and has previously shown strong results for radiomics analysis (34). This step was employed directly in the PyRadiomics environment.
Mask editing: Erosion and dilation of the masks were performed as if tumors had been segmented by different readers. A disc-shaped structuring element with a diameter of 2 voxels was used for this purpose. A modification with a fixed thickness was performed to maintain consistent variability in the masks. These modifications were performed using SimpleITK library (25).
A scheme of the modifications for each series is presented in Table 2 and depicted in Figure 2. Series 2–5 underwent normalization and resampling similar to the reference standard and were used for evaluation of denoising and inhomogeneity filters. Series 2 did not have any denoising or inhomogeneity filters. In series 3 and 4, different denoising filters were evaluated (bilateral and CFF). Series 5 evaluated the effect of removing the inhomogeneity filter, with the ADF denoising filter being the same as the reference standard. Series 6 and 7 were used for evaluation of resampling and normalization: They had the same denoising and inhomogeneity filters as the reference standard, but the modifications related to resampling (series 6) or normalization (series 7) were removed. Series 8 and 9 had the same parameters as the reference standard, but a mask modification was performed either with dilation (series 8) or erosion (series 9) to evaluate if minor shape variations affect radiomics.
Figure 2:
Impact of each processing step on T2-weighted MR images in a 6-year-old female patient with a right adrenal neuroblastic tumor. ADF = anisotropic diffusion filter, CFF = curvature flow filter.
All the modifications were applied in the PRIMAGE project’s environment, where a cloud-based platform has been established with an international repository of neuroblastic tumors (20). The analysis modules were tested, integrated, and deployed on the platform.
Segmentation was performed on the reference standard and then extrapolated to the rest of the series (except for 8 and 9, mask modifications).
Image Feature Extraction
A total of 107 radiomics features were extracted from the 746 T2/T2*-weighted MR images and from all of the nine series datasets. Extracted features were classified as shape (n = 14), original intensity-based histogram or first order (n = 18), and original texture or second order (n = 75), including gray-level co-occurrence matrix (n = 24), gray-level dependence matrix (n = 14), gray-level run-length matrix (GLRLM, n = 16), gray-level size-zone matrix (GLSZM, n = 16), and neighboring gray-tone difference matrix (n = 5). Analyses were performed using PyRadiomics (32). A complete list of the obtained features is included in Table S1.
For the series in which the shape was not altered (series 2 to 7), shape features were not analyzed, as the tumor shape and mask remained consistent across all transformations. In these cases, 93 first- and second-order radiomics features were considered. Conversely, in the series in which the mask was modified, shape, first-order, and second-order radiomics features were evaluated.
Performance and Statistical Analysis
According to recommended terminology for the technical performance of quantitative imaging biomarkers, reproducibility is a measure of precision when repeating measurements under varying conditions (35). For assessing reproducibility, the concordance correlation coefficient (CCC) was used. This is a commonly used method to evaluate agreement between paired data (36), calculated as follows:
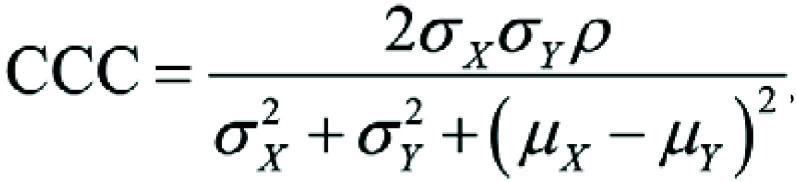 |
where μX and μY are the mean values, 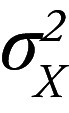 and
and 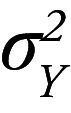 are the corresponding variances, and ρ is the correlation coefficient as assessed for each radiomics feature over all individuals within different acquisitions. CCC values close to 1 are indicative of high reproducibility. CCC was classified as excellent (if ≥ 0.90), good (0.75–0.89), moderate (0.50 to <0.75), or poor (<0.50) (6).
are the corresponding variances, and ρ is the correlation coefficient as assessed for each radiomics feature over all individuals within different acquisitions. CCC values close to 1 are indicative of high reproducibility. CCC was classified as excellent (if ≥ 0.90), good (0.75–0.89), moderate (0.50 to <0.75), or poor (<0.50) (6).
As this calculation depends on the natural variance of the underlying data (2), the coefficient of variation (CoV) was also calculated for each radiomics parameter. This precision repeatability test is frequently used for reproducibility investigations based on intrasubject variability, defined as follows:
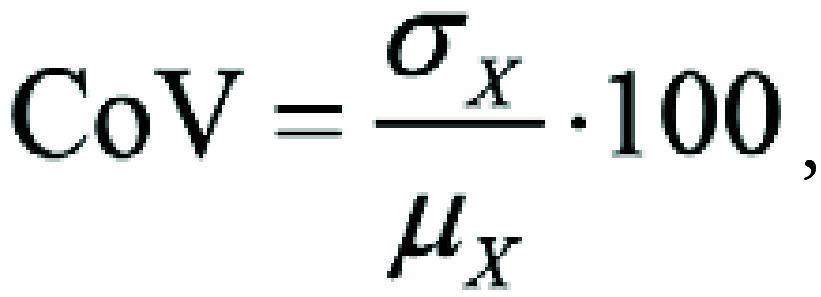 |
where σX is the SD and μX is the mean of the absolute difference of feature values between two MRI scans. A low CoV indicates that the data points are closely clustered around the mean, suggesting less variability and greater consistency. The classification of CoV results was as follows: excellent (CoV ≤ 10%), good (11–20%), moderate (21–30%), and poor (>30%) (5).
Additionally, to assess the differences between series, an initial exploratory descriptive analysis was performed to assess the differences between series, and to confirm the findings, an analysis of variance (ANOVA) was conducted. A post hoc multiple comparisons test was applied, employing the Bonferroni method. Only P values less than .001 were considered as statistically significant. Statistical analysis was performed with JASP software (version 0.18.1).
Results
Patient Characteristics
Characteristics of the 419 included patients (mean age, 29 months ± 34 [SD]; 220 male, 199 female) are presented in Table 1.
Interseries Radiomics Reproducibility
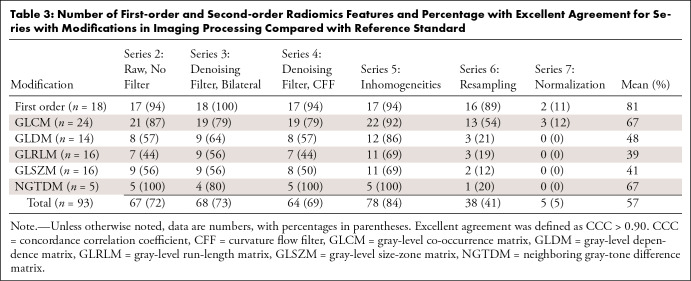
The number of radiomics features and the percentage of those within each level of agreement (excellent, good, moderate, poor) were analyzed per series and are summarized in Table S2. As the study aim was to evaluate the radiomics features that showed the highest reproducibility, only those variables that demonstrated excellent agreement (CCC > 0.90) were selected as stable for each series (Tables 3, 4).
Table 3:
Number of First-order and Second-order Radiomics Features and Percentage with Excellent Agreement for Series with Modifications in Imaging Processing Compared with Reference Standard
Table 4:
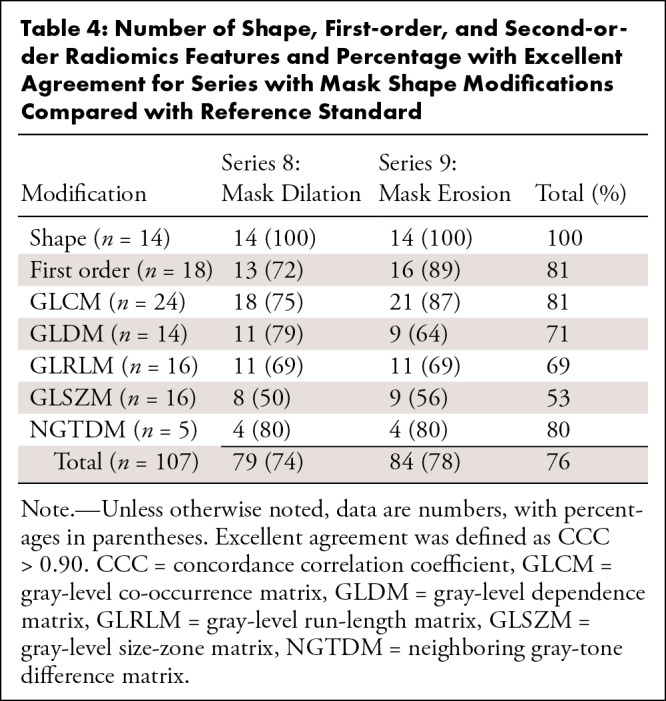
Number of Shape, First-order, and Second-order Radiomics Features and Percentage with Excellent Agreement for Series with Mask Shape Modifications Compared with Reference Standard
Reproducibility after Filter Modification
Upon analysis of the radiomics variables with a CCC greater than 0.90 after filter modifications (series 2 to 5: raw, denoising, and inhomogeneities filters), series 5 without the N4 inhomogeneity filter displayed the highest number of reproducible variables (78 of 93 radiomics features, 84%) (Table 3). This indicates that inhomogeneities removal had the least impact on radiomics. Of the 93 radiomics features, series 3 (bilateral denoising filter) and series 4 (CFF denoising filter) showed a CCC greater than 0.90 in 68 (73%) and 64 (69%) features, respectively. Series 2 (without denoising or inhomogeneity filter) had a total of 67 reproducible features (72%).
When series 2–5 were combined and the influence of filtering was analyzed, 57 of 93 (61%) radiomics features demonstrated high reproducibility (CCC > 0.90) (Table S3).
Reproducibility after Removing Resampling and Normalization
The impact of normalization and resampling was analyzed as part of the curation and harmonization algorithms applied to images in real-world data before radiomics feature extraction. Among the radiomics features with a CCC greater than 0.90 after resampling or normalization removal, 38 of 93 features maintained a high level of agreement when resampling was not performed (41%), while only five of 93 (5%) exhibited a CCC greater than 0.90 if z score normalization was not applied (Table 3). When series 6 and 7 were put together, there were 41 of 93 (44%) radiomics features that had high reproducibility (Table S4).
Reproducibility with Mask Modification
In these analyses, radiomics shape features were also included. There were some radiomics variables that showed a CCC greater than 0.90 after mask size modification (series 8 and 9). In series 8 (mask dilation), 79 of 107 (74%) features showed excellent agreement compared with the reference standard. In series 9 (mask erosion), a higher number of reproducible variables with a CCC greater than 0.90 was observed (n = 84, 78%) (Table 4). All the shape variables remained stable and reproducible in both series 8 and 9 (14 of 14 radiomics features, 100% in each series). When the morphology modifications were combined, 75 of 107 features (70%) remained with a CCC greater than 0.90. Table 4 lists the radiomics features that had high reproducibility after mask size modification. The radiomics features with an excellent level of agreement regarding erosions and dilations are listed in Table S5.
Lower Reproducibility after Image Perturbations
Second-order radiomics features GLRLM and GLSZM were the group of parameters that had the lowest mean percentage with an excellent level of agreement (Tables 3, 4): After processing modification (series 2 to 7), a mean of 39% ([44 + 56 + 44 + 69 + 19 + 0]/6) of GLRLM features and 41% ([56 + 56 + 50 + 69 + 12 + 0]/6) of GLSZM features reached an excellent level of agreement. After mask modifications, these values were 69% for GLRLM ([69 + 69]/2) and 53% ([50 + 56]/2) for GLSZM. There were 15 second-order radiomics features that showed a poor level of agreement in all the series (Table S6).
Additional Analyses
To assess the differences between series, two exploratory analyses were initially conducted for series 2 to 7 and for series 8 and 9 separately.
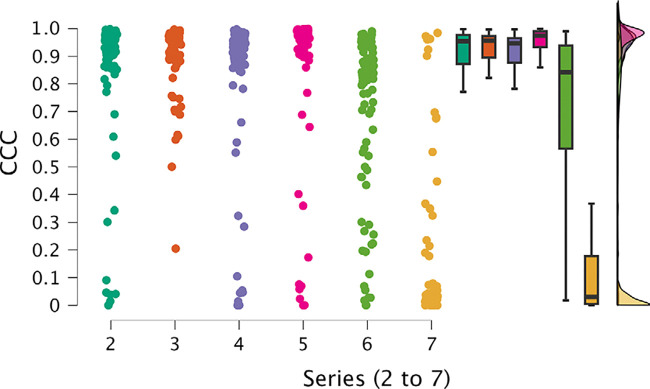
The first analysis, for series 2 to 7, revealed significant differences among series. Notably, series 6 and 7 exhibited remarkably poorer performance compared with the other series, as evident in their lower mean and median values and the proportion of cases falling below the 0.9 threshold for CCC (Fig 3). Additionally, these series displayed higher dispersion values. Conversely, series 2–5 displayed comparable results. The most noteworthy results were observed in the analysis of series 8 and 9 (Fig 4), which emerged as the top-performing series, with higher mean and median CCC values and lower SDs.
Figure 3:
Individual value, box and whisker, and raincloud plots of CCC show notable differences among series 2–7. Series 6 and 7 exhibited poorer performance, with lower mean and median values and a higher proportion of cases falling below the 0.9 threshold for CCC, along with higher dispersion values. Series 2–5 displayed comparable results with one another. Box plots represent the median and first and third quartiles, and the whiskers represent the lines extending from the box in both directions to the minimum and maximum values. Raincloud plots allow for visualization of the distribution of the data. CCC = concordance correlation coefficient.
Figure 4:
Individual value, box and whisker, and raincloud plots of CCC of series 8 and 9, which were the top-performing series, exhibit higher mean and median CCC values and lower SDs. Box plots represent the median and first and third quartiles, and the whiskers represent the lines extending from the box in both directions to the minimum and maximum values. Raincloud plots allow for visualization of the distribution of the data. CCC = concordance correlation coefficient.
To further confirm these findings, we conducted two ANOVA analyses (for each group of series, 2 to 7 and 8 and 9), treating the series as a factor while considering the data’s organizational structure as blocks, due to the calculation of 93 parameters for the first group of series and 107 parameters for the second group. For series 2 to 7, the ANOVA results confirmed statistically significant differences among the series (Table S7, Fig S1). The analysis revealed no evidence of differences among series 2–5. Nevertheless, series 6 and 7 displayed significant differences compared with the rest of the series and also compared with one another (Table S8). A detailed analysis of the residuals showed no indication of any potential influence on the results from factors other than the series.
Series 8 and 9, characterized by lower SDs, achieved higher average CCC values. The sole distinction between these two high-performing series was the utilization of either dilation or erosion in the mask. The analysis focusing on these two series, considering their paired data, concluded that there was no evidence of differences between them (Table S9).
Intrasubject Variability
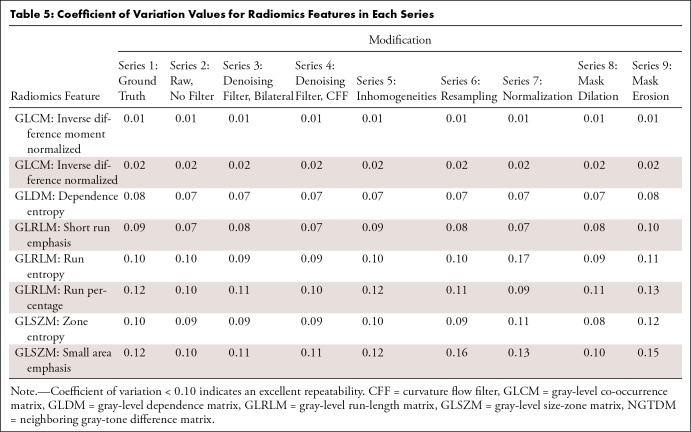
When analyzing intrasubject variability through the CoV, only eight of 107 second-order radiomics features showed an excellent result of less than 0.10 in at least two series (7%) (Table 5).
Table 5:
Coefficient of Variation Values for Radiomics Features in Each Series
Clinical Impact
Within the PRIMAGE project, a model for overall survival prediction was developed (summary available at: https://cordis.europa.eu/project/id/826494/results/es, DOI: 10.3030/826494). Among the variables with a significant impact on this overall survival model, two main radiomics features stand out: shape feature maximum two-dimensional diameter (x-z plane) and first-order feature skewness. In our study, the first variable demonstrated reproducibility and stability after mask modifications, achieving a CCC of 0.99 for erosions and dilations. Skewness was also among the more stable variables, showing a high level of agreement (CCC > 0.9) for all the modifications applied, except for dilation.
Discussion
To effectively translate radiomics to clinical practice, it is essential to evaluate the robustness of radiomics features. Radiomics repeatability can be evaluated by testing the consistency of performance by scan-rescan approach, but our real-world data study does not allow this analysis. Reproducibility can be evaluated by applying image perturbations, such as filters or normalization, and different segmentation masks, as has been demonstrated in this study. To our knowledge, this is the first study to analyze the reproducibility of radiomics in pediatric tumors, although previous works have analyzed the precision of radiomics features in other tumors, such as hepatocellular carcinoma (5) or glioblastoma (13,14). One of the main strengths of our study is the large multicentric cohort of patients with neuroblastic tumors recruited during a long period of time (21-year data collection), which are heterogeneous in location and behavior.
In this analysis, different changes were applied to a reference standard, including an ADF algorithm for denoising, N4 bias field correction for inhomogeneities removal, normalization with z score, and resampling. Images were improved with these modifications to represent a real-world scenario in which images are harmonized.
Among the different imaging modifications, inhomogeneities removal (filter N4, series 5) had the lowest impact on radiomics, showing more reproducible features compared with other series. A previous study analyzing the influence of bias field correction and denoising in radiomics for glioblastoma multiforme showed that bias field correction followed by noise filtering introduced more stable and reproducible features than noise filtering followed by bias field correction (13).
Resampling and normalization, crucial steps before comparison, altered the radiomics results, with statistically significant differences between normalization and resampling compared with the other modifications. When removing normalization, only five of 93 (5%) of the radiomics features remained stable (two first-order radiomics features and three second-order gray-level co-occurrence matrix features). Normalization has a great impact on first- and second-order radiomics, mainly due to the fact that the value of the intensities is modified by applying a z score, with an average intensity value of 0. Normalization should be considered carefully to avoid biasing the results and not evaluating the effect of the tumor but the effect of modifications in image preparation. Our results support the conclusions extracted in previous works (14). One previous study analyzed the effects of MR image normalization in prostate cancer radiomics (12) and revealed that normalization had a huge impact on the majority of radiomics features, which could have a remarkable impact on the results of radiomics models. These results demonstrate that comparing radiomics research may not be reliable if normalization has not been done with the same standardized methods.
The stability of shape radiomics features was anticipated in the series where the shape was not altered, as the tumor shape and mask remained consistent across all transformations. Consequently, shape radiomics features were not included in series 2–7 analysis, as the segmentation mask remained unchanged. Similar results have been previously reported, with high robustness and reproducibility of shape features in radiomics in glioblastoma multiforme (13). For series in which the mask’s shape was modified (erosion or dilation of the mask), the shape variables remained stable and reproducible, with a CCC above the cutoff point of 0.9. In these cases, the modifications of the shape were relatively small, so these radiomics variables did not change significantly. This implies that small modifications in the shape of the segmentations that may be due to discrepancies in manual segmentation and interobserver variability do not affect the reproducibility of this radiomics feature but do affect first- and second-order radiomics. Mask dilation and erosion were found to impact the radiomics results. When applying erosion, radiomics features remained more stable. This implies that the reduction of the area of the tumor still encases only tumoral voxels and that radiomics remains more stable compared with dilation, where voxels that do not belong to the tumor are added to the radiomics analyses. Segmentation mask–related changes have also been reported. A study comparing radiomics results in segmentations performed by independent professionals of different disciplines showed that variability in segmentation affects radiomics feature stability for CT-based radiomics studies in pancreatic cancer (15). Additionally, a study including manual segmentations performed by different individuals in three different tumor types on CT images showed that interobserver delineation variability had a relevant influence on radiomics analysis and was strongly influenced by tumor type (37).
Regarding intrasubject reproducibility, many of the radiomics features did not show high variability over multiple measurements. However, some second-order radiomics features presented excellent reproducibility in all series, which could be considered as stable biomarkers and could have potential clinical value. Previous studies addressing the CoV after gray-level resampling have shown that this modification improves second and higher order radiomics features (38).
Our study had several limitations. It was conducted on a heterogeneous dataset with varying acquisition parameters, time points (at diagnosis or after treatment), and locations, which could have influenced the results. However, all these variations reflect the real-world nature of the large retrospective datasets used for tumor phenotyping by radiomics. Further investigation should consider the differences by stratifying the results based on variables such as sex, location, previous treatment, or MR manufacturer. Another potential limitation was that this study was performed only in neuroblastic tumors, and future opportunities would include studying different cancer types. Furthermore, the analysis included only T2-weighted images, so future work could explore the reproducibility in other MRI sequences. Future work should focus on the construction of prognostic models integrating clinical variables to assess the impact of radiomics on clinical outcomes of interest.
In summary, it is essential to report each step of image processing to ensure reproducible radiomics feature extraction. We propose the use of an ADF for denoising, N4 bias field correction to correct signal inhomogeneity, z score signal normalization for standardization, and resampling for spatial harmonization. Any modification of these preparation steps can lead to changes in radiomics features, with normalization being the most influential step.
This project has received funding from the European Union Horizon 2020 research and innovation program under grant agreement number 826494. This study was funded by PRIMAGE (PRedictive In-silico Multiscale Analytics to support cancer personalized diaGnosis and prognosis, Empowered by imaging biomarkers), a Horizon 408 2020 research and innovation action project (topic SC1-DTH-07-2018), grant agreement number 826494.
Disclosures of conflicts of interest: D.V.C. European Union Horizon 2020 Research and Innovation Programme under grant agreement number 826494, project coordinator Luis Martí-Bonmatí, PhD, and Adela Cañete, PhD. M.F.P. No relevant relationships. L.C.A. No relevant relationships. A.J.P. No relevant relationships. A.G.M. Support from La Fe Research Institute. J.M.C.S. No relevant relationships. C.S.N. No relevant relationships. B.M.d.l.H. Work was developed within the PRedictive In-silico Multiscale Analytics to support cancer personalized diaGnosis and prognosis, Empowered by imaging biomarkers (PRIMAGE) Project (H2020 funding programme of the European Commission), author was employed by this funding to work in the project as pediatric oncologist. U.P. No relevant relationships. S.T.M. No relevant relationships. E.N. President of the European Society of Oncologic Imaging; Quality Safety and Standards Committee chair of the European Society of Radiology; International Affairs Delegate, Italian Society of Medical and Interventional Radiology; consultant to the editor for Radiology: Artificial Intelligence. A.C. No relevant relationships. R.L. No relevant relationships. B.H. European Funding within the project PRIMAGE. A.A.B. Treasurer of the European Society of Medical Imaging Informatics; major shareholder and CEO at Quibim. L.M.B. No relevant relationships.
Abbreviations:
- ADF
- anisotropic diffusion filter
- ANOVA
- analysis of variance
- CCC
- concordance correlation coefficient
- CFF
- curvature flow filter
- CoV
- coefficient of variation
- GLRLM
- gray-level run-length matrix
- GLSZM
- gray-level size-zone matrix
- PRIMAGE
- PRedictive In-silico Multiscale Analytics to support cancer personalized diaGnosis and prognosis Empowered by imaging biomarkers
- SIOPEN
- Society of Pediatric Oncology European Neuroblastoma Network
References
- 1. Zhang X , Zhang Y , Zhang G , et al . Deep Learning With Radiomics for Disease Diagnosis and Treatment: Challenges and Potential . Front Oncol 2022. ; 12 : 773840 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Timmeren JE , Cester D , Tanadini-Lang S , Alkadhi H , Baessler B . Radiomics in medical imaging-“how-to” guide and critical reflection . Insights Imaging 2020. ; 11 ( 1 ): 91 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lambin P , Leijenaar RTH , Deist TM , et al . Radiomics: the bridge between medical imaging and personalized medicine . Nat Rev Clin Oncol 2017. ; 14 ( 12 ): 749 – 762 . [DOI] [PubMed] [Google Scholar]
- 4. Tomaszewski MR , Gillies RJ . The Biological Meaning of Radiomic Features . Radiology 2021. ; 298 ( 3 ): 505 – 516 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carbonell G , Kennedy P , Bane O , et al . Precision of MRI radiomics features in the liver and hepatocellular carcinoma . Eur Radiol 2022. ; 32 ( 3 ): 2030 – 2040 . [DOI] [PubMed] [Google Scholar]
- 6. Sun M , Baiyasi A , Liu X , et al . Robustness and reproducibility of radiomics in T2 weighted images from magnetic resonance image guided linear accelerator in a phantom study . Phys Med 2022. ; 96 : 130 – 139 . [DOI] [PubMed] [Google Scholar]
- 7. Zwanenburg A , Vallières M , Abdalah MA , et al . The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping . Radiology 2020 ; 295 ( 2 ): 328 – 338 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Park JE , Kim D , Kim HS , et al . Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement . Eur Radiol 2020. ; 30 ( 1 ): 523 – 536 . [DOI] [PubMed] [Google Scholar]
- 9. Wang H , Zhou Y , Wang X , et al . Reproducibility and Repeatability of CBCT-Derived Radiomics Features . Front Oncol 2021. ; 11 : 773512 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berenguer R , Pastor-Juan MdR , Canales-Vázquez J , et al . Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters . Radiology 2018. ; 288 ( 2 ): 407 – 415 . [DOI] [PubMed] [Google Scholar]
- 11. Euler A , Laqua FC , Cester D , et al . Virtual Monoenergetic Images of Dual-Energy CT-Impact on Repeatability, Reproducibility, and Classification in Radiomics . Cancers (Basel) 2021. ; 13 ( 18 ): 4710 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Isaksson LJ , Raimondi S , Botta F , et al . Effects of MRI image normalization techniques in prostate cancer radiomics . Phys Med 2020. ; 71 : 7 – 13 . [DOI] [PubMed] [Google Scholar]
- 13. Moradmand H , Aghamiri SMR , Ghaderi R . Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma . J Appl Clin Med Phys 2020. ; 21 ( 1 ): 179 – 190 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoebel KV , Patel JB , Beers AL , et al . Radiomics Repeatability Pitfalls in a Scan-Rescan MRI Study of Glioblastoma . Radiol Artif Intell 2020. ; 3 ( 1 ): e190199 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wong J , Baine M , Wisnoskie S , et al . Effects of interobserver and interdisciplinary segmentation variabilities on CT-based radiomics for pancreatic cancer . Sci Rep 2021. ; 11 ( 1 ): 16328 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jensen LJ , Kim D , Elgeti T , Steffen IG , Hamm B , Nagel SN . Stability of Liver Radiomics across Different 3D ROI Sizes-An MRI In Vivo Study . Tomography 2021. ; 7 ( 4 ): 866 – 876 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ramli Z , Karim MKA , Effendy N , et al . Stability and Reproducibility of Radiomic Features Based on Various Segmentation Techniques on Cervical Cancer DWI-MRI . Diagnostics (Basel) 2022. ; 12 ( 12 ): 3125 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cohn SL , Pearson AD , London WB , et al . The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report . J Clin Oncol 2009. ; 27 ( 2 ): 289 – 297 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen X , Wang H , Huang K , et al . CT-Based Radiomics Signature With Machine Learning Predicts MYCN Amplification in Pediatric Abdominal Neuroblastoma . Front Oncol 2021. ; 11 : 687884 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martí-Bonmatí L , Alberich-Bayarri Á , Ladenstein R , et al . PRIMAGE project: predictive in silico multiscale analytics to support childhood cancer personalised evaluation empowered by imaging biomarkers . Eur Radiol Exp 2020. ; 4 ( 1 ): 22 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Veiga-Canuto D , Cerdà-Alberich L , Sangüesa Nebot C , et al . Comparative Multicentric Evaluation of Inter-Observer Variability in Manual and Automatic Segmentation of Neuroblastic Tumors in Magnetic Resonance Images . Cancers (Basel) 2022. ; 14 ( 15 ): 3648 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Veiga-Canuto D , Cerdà-Alberich L , Jiménez-Pastor A , et al . Independent Validation of a Deep Learning nnU-Net Tool for Neuroblastoma Detection and Segmentation in MR Images . Cancers (Basel) 2023. ; 15 ( 5 ): 1622 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Damián Segrelles Quilis J , López-Huguet S , Lozano P , Blanquer I . A federated cloud architecture for processing of cancer images on a distributed storage . Future Gener Comput Syst 2023. ; 139 : 38 – 52 . [Google Scholar]
- 24. Isensee F , Jaeger PF , Kohl SAA , Petersen J , Maier-Hein KH . nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation . Nat Methods 2021. ; 18 ( 2 ): 203 – 211 . [DOI] [PubMed] [Google Scholar]
- 25. Lowekamp BC , Chen DT , Ibáñez L , Blezek D . The Design of SimpleITK . Front Neuroinform 2013. ; 7 : 45 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fernández Patón M , Cerdá Alberich L , Sangüesa Nebot C , et al . MR Denoising Increases Radiomic Biomarker Precision and Reproducibility in Oncologic Imaging . J Digit Imaging 2021. ; 34 ( 5 ): 1134 – 1145 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sethian JA . Level set methods and fast marching methods: evolving interfaces in computational geometry, fluid mechanics, computer vision, and materials science . Cambridge University Press; , 1999. . [Google Scholar]
- 28. Banterle F , Corsini M , Cignoni P , Scopigno R . A Low-Memory, Straight-forward and Fast Bilateral Filter Through Subsampling in Spatial Domain . Comput Graph Forum 2012. ; 31 ( 1 ): 19 – 32 . [Google Scholar]
- 29. Tustison NJ , Avants BB , Cook PA , et al . N4ITK: improved N3 bias correction . IEEE Trans Med Imaging 2010. ; 29 ( 6 ): 1310 – 1320 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dovrou A , Nikiforaki K , Zaridis D , et al . A segmentation-based method improving the performance of N4 bias field correction on T2weighted MR imaging data of the prostate . Magn Reson Imaging 2023. ; 101 : 1 – 12 . [DOI] [PubMed] [Google Scholar]
- 31. Avants BB , Tustison NJ , Song G , Duda JT , Johnson HJ . Advanced Normalization Tools (ANTS) . https://antsx.github.io/ANTs/ .
- 32. van Griethuysen JJM , Fedorov A , Parmar C , et al . Computational Radiomics System to Decode the Radiographic Phenotype . Cancer Res 2017. ; 77 ( 21 ): e104 – e107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hadjiiski L , Cha K , Chan HP , et al . AAPM task group report 273: Recommendations on best practices for AI and machine learning for computer-aided diagnosis in medical imaging . Med Phys 2023. ; 50 ( 2 ): e1 – e24 . [DOI] [PubMed] [Google Scholar]
- 34. Haga A , Takahashi W , Aoki S , et al . Standardization of imaging features for radiomics analysis . J Med Invest 2019. ; 66 ( 1.2 ): 35 – 37 . [DOI] [PubMed] [Google Scholar]
- 35. Sullivan DC , Obuchowski NA , Kessler LG , et al . Metrology Standards for Quantitative Imaging Biomarkers . Radiology 2015. ; 277 ( 3 ): 813 – 825 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin LI . A concordance correlation coefficient to evaluate reproducibility . Biometrics 1989. ; 45 ( 1 ): 255 – 268 . [PubMed] [Google Scholar]
- 37. Pavic M , Bogowicz M , Würms X , et al . Influence of inter-observer delineation variability on radiomics stability in different tumor sites . Acta Oncol 2018. ; 57 ( 8 ): 1070 – 1074 . [DOI] [PubMed] [Google Scholar]
- 38. Shafiq-Ul-Hassan M , Zhang GG , Latifi K , et al . Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels . Med Phys 2017. ; 44 ( 3 ): 1050 – 1062 . [DOI] [PMC free article] [PubMed] [Google Scholar]