Abstract
The human genome harbors 25 to 50 proviral copies of the endogenous retrovirus type K (HERV-K), some of which code for the characteristic retroviral proteins Gag, Pol, and Env. For a genome-wide cloning approach of full-length and intact HERV-K proviruses, a human P1 gene library was screened with a gag-specific probe. Both HERV-K type 1 and 2 clones were isolated. Sixteen HERV-K type 2 proviral genomes were characterized by direct coupled in vitro transcription-in vitro translation assays to analyze the coding potential of isolated gag, pol, and env amplicons from individual P1 clones. After determination of long terminal repeat (LTR) sequences and adjacent chromosomal integration sites by inverse PCR techniques, two HERV-K type 2 proviruses displaying long retroviral open reading frames (ORFs) were assigned to chromosomes 7 (C7) and 19 (C19) by using a human-rodent monochromosomal cell hybrid mapping panel. HERV-K(C7) shows an altered (YIDD-to-CIDD) motif in the reverse transcriptase domain. HERV-K(C19) is truncated in the 5′ LTR and harbors a defective protease gene due to a point mutation. Direct amplification of proviral structures from single chromosomes by using chromosomal flanking primers was performed by long PCR for HERV-K(C7) and HERV-K(C19) and for type 1 proviruses HERV-K10 and HERV-K18 from chromosomes 5 and 1, respectively. HERV-K18, in contrast to HERV-K10, bears no intact gag ORF and shows close homology to HERV-K/IDDMK1,222. In transfection experiments, HERV-K(C7) and HERV-K cDNA-based expression vectors yielded the proteins Gag and cORF whereas HERV-K10 vectors yielded Gag alone. The data suggest that the human genome does not contain an entire, intact proviral copy of HERV-K.
Although the HERV-K group comes closest of all known human endogenous retroviruses (HERVs) to containing infectious virus, no corresponding replication-competent virus has so far been described (3, 15). The prototype full-length sequence, HERV-K10, was described as being defective in the gag and env genes (30). However, other HERV-K genes with long open reading frames (ORFs), which potentially express the Gag, Pol, and Env proteins, have been isolated (14, 16). In addition, a doubly spliced HERV-K mRNA encoding a 12-kDa protein, cORF, with homology to lentivirus Rev proteins was identified; it is expressed by HERV-K type 2 proviruses, which differ from type 1 genomes by the presence of a 292-bp 5′-terminal env gene segment (17).
Some human teratocarcinoma cell (TC) lines constitutively produce core proteins which are assembled and form apparently noninfectious virus-like particles (VLP) formerly named human teratocarcinoma derived virus (HTDV) (5, 11). Particles resembling such HERV-K/HTDV VLP could be produced in a heterologous expression system by using recombinant baculoviruses carrying HERV-K cDNA-based proviral cassettes (36). Like the VLP produced in TC lines (35), those particles package HERV-K RNA (36). Reverse transcriptase (RT) activity was detected in both TC- and insect cell-derived VLPs by ultrasensitive PCR-based assays (36, 38).
At present, the possibility that HERV-K VLP, like their closest relatives, mouse mammary tumor virus and jaagsiekte sheep retrovirus, have a narrow host range cannot be excluded (3). On the other hand a processed form of expressed HERV-K Env proteins, a prerequisite for infection, could not be found in TC or in highly expressing transformed mammalian or insect cells (37). It is therefore possible that, in terms of infectious virion production, HERV-K is defective at multiple levels, including the observed arrest during budding, inefficient RT enzyme activity, and incomplete Env expression (15).
It has recently been reported that only a small subset of human chromosomes carries ORFs for both HERV-K Gag and Env proteins; these are chromosomes 7, 19, and Y (20, 21). One provirus bearing all ORFs, called HERV-K(HML-2.HOM), was assigned to human chromosome 7p22 (22). However, this HERV-K element is expected to express a nonfunctional reverse transcriptase.
The aim of this study was to isolate, based on a genome-wide search using a P1 genomic library, full-length HERV-K proviruses in the human genome which have the capacity to encode all retroviral proteins. Here we show that only two proviruses with these properties could be found; one HERV-K element is located on human chromosome 7 and appears to be an allelic variant to HERV-K(HML-2.HOM), and a second provirus resides on chromosome 19, has a single point mutation in the protease gene, and lacks most of the 5′ long terminal repeat (LTR). Different appropriate expression vectors generated HERV-K core proteins and cORF but no apparent Env upon transfection into heterologous mammalian cells. These results suggest that HERV-K virions with replicative capacities might be formed only by complementation of different expressed proviruses.
MATERIALS AND METHODS
DNA sources and probes.
Screening of human P1 genomic library high-density filters (FP1-2285; Genome Systems, St. Louis, Mo.), which represented the genome approximately twice, was performed with a HERV-K gag-specific probe (EcoRV-PstI 813-bp fragment) (Fig. 1) derived from plasmid pcG3gag (36). The fragment was labelled for hybridizations by random priming (8) with [α-32P]dCTP (3,000 Ci/mmol; Amersham Pharmacia Biotech, Freiburg, Germany). Isolated P1 clones were subjected to a second screening with oligonucleotide YB23.3 (GTTGCCATCCACCAAG; nucleotides [nt] 6564 to 6576) (Fig. 1) specific for the 292-bp segment present in type 2 proviruses (17). For a third hybridization analysis, oligonucleotide 23S10/Ins (CGTTTACGGCGTTTACTGCCCTTA, nt 4158 to 4144) bearing a 9-bp insertion at positions 4149 to 4150 (shown in italics) was used. Both oligonucleotides were 5′-end labelled with T4 polynucleotide kinase by using [γ-32P]ATP (>5,000 Ci/mmol) (Amersham) or were labelled with a nonradioactive 3′-end-labelling system (Amersham). For 32P-labelled probes, 1 × 106 to 2 × 106 cpm of probe/ml was used in the hybridizations. P1 DNA was prepared by standard phenol-chloroform extractions as specified by the manufacturer (Genome Systems).
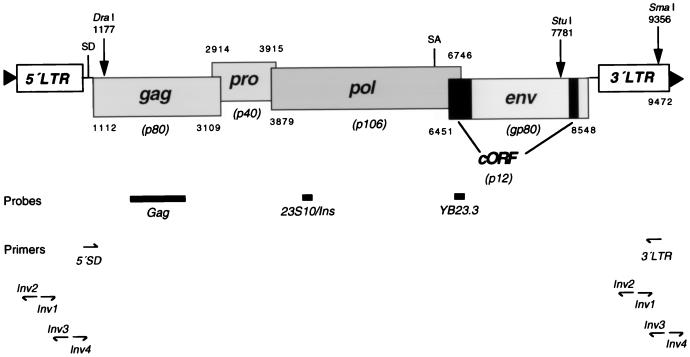
FIG. 1.
Map of HERV-K type 2 proviral structure. Genes and corresponding ORFs with theoretical molecular masses of proteins are shown. The protein cORF is derived from a doubly spliced mRNA (17). Locations of probes and primers used for clone isolations and of restriction enzyme sites and primers instrumental for inverse PCR techniques are indicated. SD, splice donor; SA, splice acceptor. Numbers denote first and last nucleotides of ORFs. Triangles represent direct repeats in chromosomal DNA.
An SphI restriction fragment (∼15 kb) of a λ bacteriophage harboring a HERV-K proviral sequence (ΦP23/HERV-K) isolated from a human genomic placenta DNA library was cloned into pGem-3Zf (Promega), yielding pHERV-K(gP23-C19). A 1.9-kb EcoRI restriction fragment bearing part of the HERV-K pol and env genes from this provirus (nt 6079 to 7991) has been recently described (37). Numbering of sequences is based on the HERV-K type 2 sequence as described previously (17).
PCR amplification of HERV-K proviral sequences from human chromosomes.
For identification of full-length HERV-K proviruses, a long PCR technique was used with the Expand Long System (Boehringer, Mannheim, Germany). PCR primers designed from the 5′ untranslated region upstream of the gag gene (HERV-K/5′SD; GCTTGCGCGCTCGGAAGAAGCTAGGGTGA, nt 1081 to 1109) and from the U5 region of the 3′ LTR (HERV-K/3′LTR; AGAGAAAGAAAGAAGGGGACCCGGGGAACCAGC, nt 9380–9348) (Fig. 1) were used on human monochromosomal mapping panel DNA as templates.
Full-length proviruses HERV-K10 and HERV-K18 (29) were generated from human monochromosomal mapping panel DNA by using primers specific for adjacent chromosomal DNA sequences (29), i.e. 5′-HERV-K10 (AGTGGCATGATCTCAACTCACTGCTG), 3′-HERV-K10 (GAACTAGGAGGCGGAGGTTGCAGTTG), 5′-HERV-K18 (TACAACATAAGCGGAATCTGAGACTG), and 3′-HERV-K18 (CCCAAACCTTTAAATATTGTCTCATG).
Products were cloned into pGEM-T or pGEM-T Easy (Promega), yielding plasmids pHERV-K(SD-C7-3′LTR), pHERV-K(SD-C19-3′LTR), pHERV-K10, and pHERV-K18, respectively.
The human-rodent somatic cell hybrid mapping panel (repository no. MBP0002) was obtained from Coriell Cell Repositories, Camden, N.J. For PCR amplifications, 100-ng samples of hybrid DNA containing single human chromosomes were used. Oligonucleotide primers were purchased from ARK Scientific (Darmstadt, Germany).
Generation of adjacent chromosomal sequences.
Genomic sequences adjacent to HERV-K proviral LTRs were cloned by inverse PCR techniques. Whole genomic DNA isolated from P1 clones was digested with restriction endonucleases (NEB), generating blunt ends (Fig. 1). After heat inactivation and alcohol precipitation, 1 to 5 μg of DNA fragments was resuspended in water and self-ligated overnight at 16°C with 4 U of T4 DNA ligase (Life Technologies) in a 300-μl volume. Circularized DNA fragments were precipitated and resuspended in 50 μl of water, of which 2.5 μl served as the template for PCR with inversely orientated oligonucleotides Inv1 (TCTGAAATATGGCCTCGTGG, nt 361 to 380/nt 8865 to 8884) and Inv2 (TATCACATGGGGAGAAACCT, nt 359 to 340/nt 8863 to 8844) or Inv3 (GTATAGAGAAAGAAATAAGGGGAC, nt 880 to 867/nt 9394 to 9371) and Inv4 (TCCAAATCTCTCGTCCCACCTTAC, nt 904 to 927/nt 9408 to 9431), respectively (Fig. 1). Amplifications based on a cycle scheme of initial denaturation for 10 min at 94°C, 35 cycles of 45 s at 94°C, 45 s at 54°C, and 6 min at 72°C, an additional 6 cycles of 45 s at 94°C, 45 s at 51°C, and 6 min at 72°C, and a final extension for 10 min at 72°C were performed with 2.5 U of AmpliTaq Gold (Perkin-Elmer Cetus) in a 50-μl reaction volume (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2, 0.01% [wt/vol] gelatin, 0.2 mM each deoxynucleoside triphosphate) with 20 pmol of each primer. Gel-purified amplification products were cloned into pGEM-T Easy (Promega) or pCRII-TOPO (Invitrogen). Plasmid DNA was prepared with Qiagen (Hilden, Germany) systems.
Chromosomal assignment of HERV-K sequences.
Chromosomal locations of selected clones showing multiple ORFs were determined by PCR analyses with oligonucleotide HERV-K/SD-inv (CTAGCTTCTTCCGAGCGCGCAAGC, nt 1104 to 1081) combined with a P1 clone-specific 5′-flanking primer (5′ Fl-51C12/69L1, ATTTAGATTCAGGGGGTTCATGTGTAGTG, nt −324 to −295) and with a HERV-K-specific oligonucleotide (HERV-K/Env-for, AGAGACAGCGACCATCGAGAACGG, nt 8437 to 8460) combined with specific 3′-flanking primers (3′ Fl-51C12/69L1, GAATTAGGCTTTCGGGACTTGAACATTGGA, nt +130 to +101; and 3′ Fl-214M19/221L7/359N6/367M9, ATTATGCAACCTGGGGCTGGTCAG, nt +47 to +24). The plus and minus signs indicate primer locations relative to the first and last nucleotides of HERV-K, respectively. HERV-K10 and HERV-K18 were chromosomally assigned by using primers 5′-HERV-K10 and 5′-HERV-K18 in combination with HERV-K/SD-inv and primers 3′-HERV-K10 and 3′-HERV-K18 combined with HERV-K/Env-for, respectively.
Sequence analysis of HERV-K elements.
Both strands of PCR-amplified proviral sequences and of suitable subclones of P1 and bacteriophage isolates in plasmid vectors were cycle sequenced on 373A and 377 DNA sequencing systems (Applied Biosystems, Weiterstadt, Germany). Sequencing reactions were performed in a Vistra DNA Labstation 625 (Molecular Dynamics, Krefeld, Germany) by using the Thermo Sequenase fluorescent dye-terminator cycle sequencing and precipitation kits as specified by the manufacturer (Amersham).
Identification of ORFs.
The remaining 16 P1 clones were subjected to a direct coupled transcription-translation assay allowing the rapid identification of ORFs by the protein truncation test (PTT) (32). The gag, pol, and env sequences were amplified in conjunction with a T7 promoter by using oligonucleotides T7-HERV-K gag-for (GGATCCTAATACGACTCACTATAGGAACAGACCATAATGGGGCAAACTAAAAGT, nt 1109 to 1129), HERV-K gag-rev (AGGCAGTGGGCCATATACCCCTG, nt 3194 to 3172), and T7-HERV-K pol-for (TAATACGACTCACTATAGGGAACAGGGCTGTAAACGCCGTAATTC, nt 4148 to 4166) and HERV-K pol-rev (GACTGCCCGAATTAAGGGCGG, nt 6797 to 6776), T7-HERV-K env-for (TAATACGACTCACTATAGGAACAGACCACCATGAACCCATCAGAGATGCA, nt 6448 to 6469) and HERV-K env-rev (AACAGAATCTCAAGG CAGAAGA, nt 8620 to 8598). T7 promoter and spacer sequences are shown in italics.
Amplicons were in vitro transcribed and in vitro translated by using the TNT T7 quick coupled transcription-translation system (Promega) as specified by the manufacturer. Products with incorporated [35S]methionine were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by autoradiography. HERV-K10 Gag was produced accordingly with primers T7-HERV-K gag-for and HERV-K gag-rev for amplification and subsequent transcription-translation of pHERV-K10 gag.
Generation of mammalian expression vectors.
The HERV-K expression vector pJW/HERV-K(SD-C7-LTR) harboring a retroviral gag-pro-pol-env gene cassette was generated by cloning a blunted NotI restriction fragment derived from pHERV-K(SD-C7-3′LTR) into expression plasmid pJW4303 downstream of the cytomegalovirus-intron A promoter (19, 37). Accordingly, the HERV-K expression vector pJW/HERV-K10, harboring a retroviral gag-pro-pol gene cassette, was generated by cloning a blunted BamHI restriction fragment (nt 825 to 7751) derived from pHERV-K10 into pJW4303.
A fully cDNA-based HERV-K type 2 expression plasmid including LTRs was produced by cloning the LTR sequence (nt 1 to 825) from pcK30 (17) into the BamHI site of pcG31 (36) at position 825, generating pcG31-LTR. An LTR containing the SphI-HindIII fragment (nt 238 to 1080) from pcG31-LTR was cloned into pcPK23/30 (36) with the partial LTR sequence deleted downstream of the SphI site at position 8737, producing pcPK23/30-LTR. An ∼630-bp SstI (pBS)-SstI (nt 4411) fragment was excised from pcPK23/30-LTR and replaced by an ∼4,415-bp SstI (pBS)-SstI fragment isolated from pcG31-LTR. The resulting plasmid, termed pcHERV-K, harbors a 9,472-bp HERV-K cDNA. A derivative called pcHERV-K/Ires-βGeo was generated by cloning an indicator and resistance gene cassette bearing a β-galactosidase gene fused in frame with a neo gene downstream of an internal ribosomal entry site (25) into the SphI restriction enzyme site of pcHERV-K.
Monkey kidney (COS7), dog osteosarcoma (D17), and human teratocarcinoma (GH) cell lines were transfected with Effecten (Qiagen) or Lipofectamine (Life Technologies) and 2 to 5 μg of recombinant plasmid DNA, which was prepared with the EndoFree system (Qiagen). At 24 to 72 h posttransfection, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (Sigma) staining and/or indirect immunofluorescence for the analysis of HERV-K Gag, Env, and cORF protein expression was performed with a laser-scanning microscope as described previously (36, 37).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been submitted to the EMBL nucleotide sequence database and have been assigned the accession no. Y17832 [HERV-K(C7)], Y17833 [HERV-K(SD-C7-3′LTR)], Y17834 [HERV-K(gP23-C19)], and Y18890 (HERV-K18).
RESULTS
Isolation of HERV-K proviral sequences from genomic libraries.
A human genome-wide screening for intact full-length proviral structures of HERV-K members was performed with a genomic library cloned in the P1 vector. This approach was chosen because PCR-based strategies for complete amplification of HERV-K sequences including LTR sequences are less appropriate due to the presence of multimeric proviral templates (30) and of 10,000 to 25,000 solitary LTR sequences (13). A HERV-K gag specific probe was used (Fig. 1) to retrieve proviruses with coding capacity for retroviral core proteins. Screening revealed 77 positive signals, of which 62 P1 clones harboring inserts of 75 to 105 kb were further characterized. Isolated clones were used for a second hybridization with a probe consisting of oligonucleotide YB23.3 (Fig. 1), which is specific for the 292-bp segment present in type 2 proviruses and which is a prerequisite for expression of HERV-K Env and cORF proteins (17, 37). Sixteen P1 clones harboring HERV-K type 2 proviral structures were identified, and DNA was prepared from them. The HERV-K recombinant P1 clones were designated 21L23, 50M12, 51C12, 63G16, 69L1, 82H14, 104G12, 146L5, 214M19, 221L7, 247N15, 271F7, 300A7, 323N1, 359N6, and 367M9 and were further analyzed for their coding capacities (see below). A third hybridization analysis with 23S10/Ins as a probe was performed and demonstrated that P1 clones 214M19, 221L7, 359N6, and 367M9 carry a 9-bp insertion (see below).
A HERV-K type 2 proviral sequence located on a λ bacteriophage (ΦP23/HERV-K), which was isolated previously from a human genomic placenta DNA library, was further characterized. Part of the HERV-K pol and env genes from this provirus (nt 6079 to 7991) showed an ORF (37). Complete sequence analysis of the HERV-K element located on subclone pHERV-K(gP23-C19) revealed ORFs for gag (nt 1112 to 3109), pol (nt 3879 to 6746) and env (nt 6451 to 8548). The protease ORF (nt 2914 to 3915) is disrupted by a single nucleotide insertion (A) at nt 3377. The pol gene bears the highly conserved YXDD motif of RT at nt 4461 to 4472 and exhibits a 9-bp in-frame insertion (nt 4149 to 4150) compared with the standard sequence found in the four P1 clones 214M19, 221L7, 359N6, and 367M9 (see above). The 5′ LTR element is incomplete since it starts at position 946 and includes the primer binding site for tRNALys (nt 971 to 988). A hexanucleotide sequence (AGGTAT) upstream of the truncated 5′ LTR is also found adjacent to the 3′ LTR (nt 8505 to 9472), probably representing a direct repeat sequence. In the U3 region of the 3′ LTR element, an identical duplication of 26 bp containing the glucocorticoid response element was identified. A GenBank database search using the 5′-flanking chromosomal sequence as the query retrieved a cosmid (R26450; accession no. AC004164) with 100% overlap homology. The insert of 36,798 bp at one end (nt 1 to 104) exhibited a partial HERV-K 5′LTR (nt 1049 to 946) in reverse orientation. Furthermore, this cosmid harbored a second reversely oriented partial HERV-K LTR at nt 32421 to 32138. The cosmid clone was isolated from a chromosome 19-specific library and is located near the centromeric boundary of 19q12.
Long-PCR-mediated cloning of proviral HERV-K from single chromosomes.
Direct amplification of HERV-K proviruses excluding the 5′ LTR and part of the 3′ LTR sequences was performed with monochromosomal human DNA and oligonucleotides HERV-K/5′SD and HERV-K/3′ LTR as primers (Fig. 1). PCR experiments revealed signals in the range of the expected size (∼8.3 kb) and in most cases smaller bands on human chromosomes (data not shown). One single amplification product derived from chromosome 7 was isolated, cloned into a T/A vector, designated pHERV-K(SD-C7-3′LTR), and analyzed by sequencing. Likewise, long PCR with chromosome 19 as a template revealed a band of ∼8.3 kb and two smaller amplification products. No product was obtained from human chromosome Y. Sequence analysis revealed that the provirus derived from chromosome 7 bears ORFs for gag (nt 1112 to 3109), pro (nt 2914 to 3915), pol (3879 to 6746), and env (nt 6451 to 8548). In the pol gene, a single nucleotide mutation (A to G, nt 4462) converts the highly conserved motif YIDD of the RT into CIDD. The env gene in pHERV-K(SD-C7-3′LTR) showed 99.9% homology to the cDNA sequences of pcK30env and pcE12env reported previously (17, 37) and exhibited a C-to-A nucleotide change at position 7298, suggesting that this provirus is transcribed in teratocarcinoma cells.
The HERV-K proviral sequence isolated from chromosome 19 proved to be identical to pHERV-K(gP23-C19) (see above). Notably, the pol gene exhibited the YIDD motif and contained a 9-bp in frame insertion (at nt 4149 to 4150).
Determination of HERV-K ORFs in P1 clones.
For analysis of coding potential, individual HERV-K genes amplified by PCR from 16 P1 clones were subjected to direct coupled transcription-translation assays. These protein truncation tests (PTT) for gag, pol, and env putatively located on P1 clones demonstrated that six HERV-K recombinants in vitro encode full-length Gag, Pol, and Env proteins (Fig. 2). These clones are 51C12 (Fig. 2, lane 3), 69L1 (lane 5), 214M19 (lane 9), 221L7 (lane 10), 359N6 (lane 15), and 367M9 (lane 16). A fully cDNA-based HERV-K construct (pcHERV-K) (lane 17) and plasmid pHERV-K(SD-C7-3′LTR) (lane 18) served as controls. Completely synthesized Gag of 80 kDa in all cases was prominent compared to truncated products (Fig. 2A). For Pol and Env, in vitro translation yielded products of ∼106 kDa (Fig. 2B) and ∼80 kDa (Fig. 2C), respectively. However, the translation efficiency was not as high as for gag sequences. In clones 214M19, 221L7, 359N6, 367M9, and pHERV-K(SD-C7-3′LTR), Pol protein was produced at barely detectable levels (Fig. 2B). In vitro translation of env sequences yielded more or less prominent readthrough products besides the cognate Env protein of ∼80 kDa (Fig. 2C). P1 clones 51C12, 69L1, 214M19, 221L7, 359N6, and 367M9 were selected for further molecular analyses.
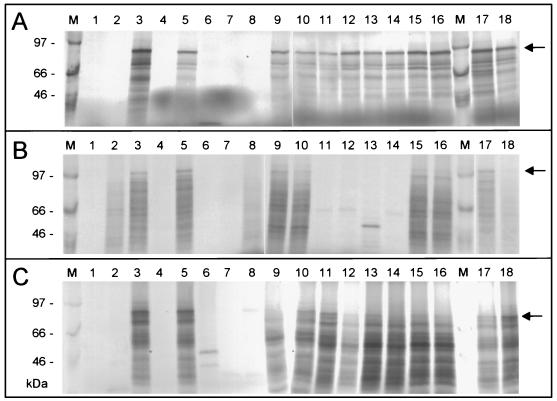
FIG. 2.
Direct coupled in vitro transcription-in vitro translation assays for HERV-K genes located on P1 clones. (A) HERV-K Gag proteins; (B) HERV-K Pol proteins, (C) HERV-K Env proteins. P1 clones used: lane 1, 21L23; lane 2, 50M12; lane 3, 51C12; lane 4, 63G16; lane 5, 69L1; lane 6, 82H14; lane 7, 104G12; lane 8, 146L5; lane 9, 214M19; lane 10, 221L7; lane 11, 247N15; lane 12, 271F7; lane 13, 300A7; lane 14, 323N1; lane 15, 359N6; lane 16, 367M9; lane 17, pcHERV-K; lane 18, pHERV-K (SD-C7-LTR); lane M, 14C-labeled methylated ladder (in kilodaltons) (Amersham). Arrows denote full-length translation products (sizes are indicated in Fig. 1).
Allocation of full-length HERV proviruses to human chromosomes.
A comparison of restriction fragment patterns and Southern blot analyses suggested close relationships or identities of P1 clones 51C12 and 69L1 and of clones 214M19, 221L7, 359N6, and 367M9 (data not shown). Since unique genome specific sequences are required for cloning of complete proviruses including both LTRs, inverse PCR experiments were performed. For clones 51C12 and 69L1, 5′ integration sites were determined by cloning amplification products derived from DraI-religated restriction products with primers Inv1 and Inv2. An amplicon derived from StuI-religated fragments revealed 3′ integration sites (data not shown). For characterization of 3′-flanking sequences of P1 clones 214M19, 221L7, 359N6, and 367M9, primers Inv3 and Inv4 were used on SmaI-religated restriction fragments. No 5′-flanking sequences could be generated by this approach for these clones (data not shown; see below). All PCR products were analyzed by sequencing, which demonstrated that clones 51C12 and 69L1 and clones 214M19, 221L7, 359N6, and 367M9 have identical flanking sequences.
Chromosomal assignment for clones 51C12 and 69L1 based on the integration site sequence information gained was performed using one flanking primer and one HERV-K specific primer in PCR experiments (Fig. 3). With oligonucleotides HERV-K/Env-for and 3′Fl-51C12/69L1 used on the monochromosomal mapping panel as the template, a product of ∼1,200 bp was exclusively obtained for human chromosome 7 (Fig. 3A). Likewise, a specific ∼1,400-bp PCR product was generated on chromosome 7 with 5′Fl-51C12/69L1 combined with HERV-K/SD-inv (data not shown). The entire proviral sequence including flanking sequences was PCR amplified with primers 5′Fl-51C12/69L1 and 3′Fl-51C12/69L1, yielding an amplification product of ∼9,930 bp for P1 clones 51C12 and 69L1 (Fig. 3B) and demonstrating that these P1 clones are derived from chromosome 7. Analysis of the sequence adjacent to the provirus on chromosome 7 showed that this HERV-K element has integrated into an Alu sequence at position 110 (Fig. 3C). The Alu element is truncated at its 5′ end and starts at position 55. Sequences adjacent to the split Alu element are unique as revealed by database searches. A second Alu sequence in reverse orientation was found 159 bp downstream of the first one, i.e., upstream relative to HERV-K (Fig. 3C). The proviral structure is flanked by two hexanucleotide sequences (GGTTTC) representing the direct repeats. The 5′ and 3′ LTR sequences differ by 10 nt.
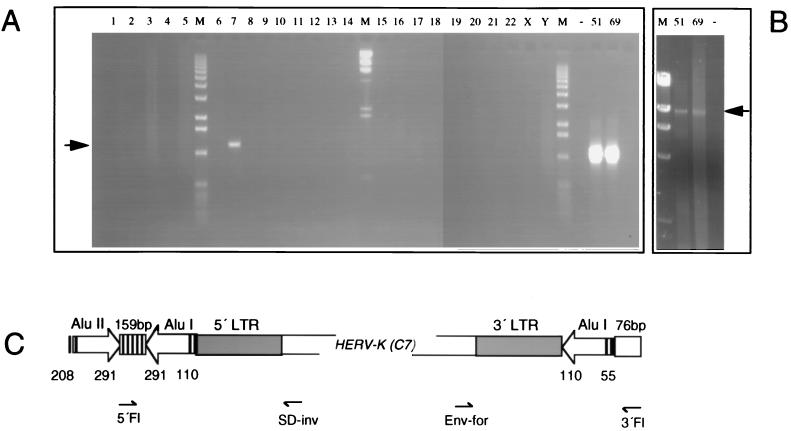
FIG. 3.
Chromosomal assignment of P1 clone HERV-K(C7). (A) An env-specific primer (Env-for) in combination with a 3′-flanking primer (3′Fl) of the P1 clone 51C12 revealed an amplification product of ca. 1,200 bp on chromosome 7 DNA. Complementary analysis was performed with a 5′-flanking primer (5′Fl) and a splice donor site specific primer (SD-inv) (not shown). Lanes 1 to 22, X, and Y, single human chromosomes in rodent background; lanes 51 and 69, P1 clones 51C12 and 69L1; lane −, water control; lanes M, DNA molecular weight markers. (B) Long PCR amplification of HERV-K(C7) on P1 clones 51C12 and 69L1 with primers 5′Fl and 3′Fl. For abbreviations, see above. (C) Graph summarizing the chromosome 7 integration site of provirus HERV-K(C7) (clone 51C12). Two Alu elements (I and II), separated by 159 bp of unique sequence, and their orientations (open arrows) are indicated. Numbers denote nucleotide positions according to the reference Alu elements (Alu I, AluSx [U14574]; Alu II, AluSp [U14572]) which gave highest homologies in GenBank searches. Note that the Alu I element is split by the HERV-K provirus and that its 5′ end starts with nt 55. The Alu II element was not sequenced beyond nt 208. Proviral genes are omitted for clarity. Diagnostic primers for chromosomal allocations are indicated.
A database search revealed that provirus HERV-K(HML-2.HOM) (accession no. AF074086 [22]) has a sequence highly homologous to HERV-K(C7). HERV-K(HML-2.HOM) is located at 7p22 and in total shows six exchanges compared to the sequence reported here (at nt 833, 1468, 1469, 2672, 6331, and 7298). In the 3′-flanking segment of HERV-K(HML-2.HOM) deposited in GenBank, the overlapping sequence differs at nt 110 and 131, indicating that HERV-K(HML-2.HOM) resides at the same location as HERV-K(C7). The chromosomal location of P1 clones 214M19, 221L7, 359N6, and 367M9 was determined by using a unique 3′-flanking oligonucleotide in combination with primer HERV-K/Env-for. A PCR product of ∼1,100 bp was generated exclusively with DNA from human chromosome 19 (data not shown). The 3′-flanking sequence obtained by inverse PCR was identical to pHERV-K(gP23-C19) (data not shown), suggesting that these four P1 clones are derived from the same chromosomal location as the bacteriophage ΦP23/HERV-K. The fact that inverse PCR for 5′-flanking sequences based on LTR primers was not feasible for these P1 clones was due to the almost complete absence of a 5′ LTR in this HERV-K provirus located on chromosome 19.
Cloning of type 1 proviruses HERV-K10 and HERV-K18.
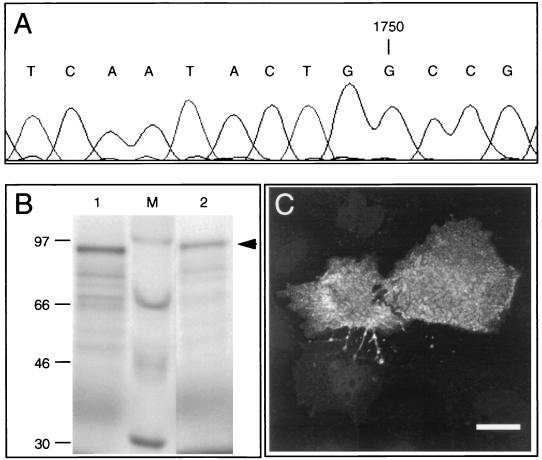
Similar to the amplification of full-length HERV-K type 2 proviruses from chromosomes 7 and 19, direct amplification of previously reported HERV-K10 (30) and of partially described HERV-K18 (29) was carried out. First, chromosomal assignments were performed by using specific 5′-flanking primers in combination with HERV-K SD-inv. HERV-K10 and HERV-K18 turned out to be located on chromosome 5 (Fig. 4A) and on chromosome 1 (Fig. 4C), respectively. An additional LTR amplification product on chromosome 6 with HERV-K10 primers (Fig. 4A) was obtained. However, this result was not confirmed when the entire provirus was generated with 5′- and 3′-flanking primers, yielding a product of 9,180 bp exclusively on chromosome 5 DNA (Fig. 4B), possibly indicating the existence of a partially duplicated HERV-K sequence. Likewise, PCR with flanking primers specific for HERV-K18 revealed a product of 9,234 bp on DNA from chromosome 1 (Fig. 4D). In contrast to the published sequence of HERV-K10, the allele amplified from chromosome 5 exhibits a single nucleotide insertion (G) at position 1750 in the gag gene (Fig. 5A), providing a contiguous ORF as opposed to the split gene structure shown previously (30) (see below).
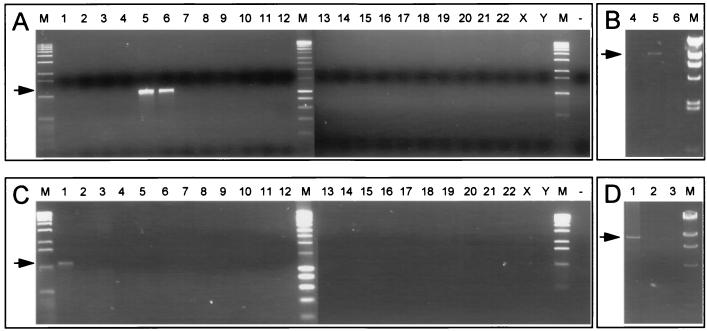
FIG. 4.
Chromosomal assignment and PCR amplification of HERV-K10 and HERV-K18 proviruses. (A) Allocation of HERV-K10. Chromosome-specific DNA primer 5′-HERV-K10 was used in combination with HERV-K splice donor site-specific primer (SD-inv). The arrow indicates the 1,100-bp amplification product. (B) Long PCR amplification of the HERV-K10 provirus with primers 5′-HERV-K10 and 3′-HERV-K10. The arrow denotes the 9,180-bp product. (C) Allocation of HERV-K18. The chromosome-specific DNA primer 5′-HERV-K18 was used in combination with the HERV-K splice donor site-specific primer (SD-inv). The arrow indicates the 1,100-bp amplification product. (D) Long PCR amplification of HERV-K18 provirus with primers 5′-HERV-K18 and 3′-HERV-K18. The arrow denotes the 9,234-bp product. Complementary analysis was performed with an env-specific primer (Env-for) combined with chromosome-specific DNA primers 3′-HERV-K10 and 3′-HERV-K18, respectively (not shown). Lanes: 1 to 22, X, and Y, single human chromosomes in rodent background; −, water control; M, DNA molecular weight markers.
FIG. 5.
Partial sequence of HERV-K10 gag and Gag protein expression. (A) Part of a chromatogram file of the gag sequence showing an additional nucleotide (G) at position 1750. (B) Direct coupled in vitro transcription-in vitro translation assay for the HERV-K10 gag gene. Lane 1, pHERV-K10 Gag; lane 2, pHERV-K (SD-C7-LTR) Gag; lane M, molecular mass markers (in kilodaltons). The arrow denotes the Gag protein of 80 kDa. (C) Indirect immunofluorescence analysis of HERV-K10 Gag expression in COS7 cells transfected with pJW/HERV-K10. Cells were incubated with goat anti-Gag antisera (4). Bar, 25 μm.
Sequence analysis of HERV-K18 demonstrated only a partial gag ORF (nt 1113 to 1872) due to multiple mutations, a pro ORF (nt 2971 to 3972), a pol ORF (nt 3935 to 6371), and an env ORF (nt 6543 to 8231). The latter demonstrates extensive homology to the superantigen sequence of denominated provirus IDDMK1,222 reported by Conrad et al. (6). However, the superantigen sequence at amino acid 97 shows a tyrosine instead of a cysteine and the stop codon is located at amino acid 561. The 5′- and 3′-LTR sequences of HERV-K18 differ by 38 nt whereas HERV-K10 LTRs show only two differences (29).
Expression of recombinant HERV-K sequences in mammalian cells.
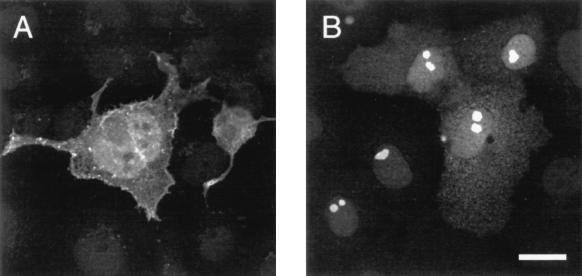
Isolated proviral HERV-K sequences were investigated for their potential to encode proteins in tissue culture. Prior to expression in mammalian cells, the intactness of the HERV-K10 gag ORF was demonstrated in PTT experiments where the full-length 80-kDa protein was produced, similar to HERV-K(C7)-derived Gag (Fig. 5B). Upon transfection into COS-7 cells, HERV-K10 Gag was found at the membrane of the cell membrane, as revealed by laser-scanning microscopy analysis (Fig. 5C). Since the K10 provirus is a member of the type 1 subfamily of HERV-K, most of the first exon of cORF is missing and the protein can therefore not be expressed from this genome. By contrast, a HERV-K(C7)-derived expression vector [pJW/HERV-K(SD-C7-LTR)] efficiently generated Gag and cORF proteins after transfection into COS-7 cells (Fig. 6). Rarely, Env expression was observed in a lower percentage of transfected cells than the percentage in which Gag and cORF expression was observed (data not shown).
FIG. 6.
Indirect immunofluorescence analysis of HERV-K(C7) Gag and cORF expression. COS7 cells were transiently transfected with the HERV-K expression vector pJW/HERV-K (SD-C7-LTR) and were fixed and stained at 48 h posttransfection. (A) Cells incubated with goat anti-Gag antisera (4). (B) Cells incubated with rabbit anti-cORF antisera (17). Bar, 25 μm.
When HERV-K LTR sequences were used as promoter sequences instead of the heterologous cytomegalovirus promoter, expression of construct pcHERV-K in COS-7 cells dropped to negligible levels (data not shown). D17 dog osteosarcoma cells were chosen after a variety of mammalian cells were screened for their potential to transcribe HERV-K LTRs by using an internal β-galactosidase indicator gene attached to an Ires sequence downstream of HERV-K env in a complex expression cassette (pcHERV-K/Ires-βGeo). As a positive control for transfection and for LTR activity, human teratocarcinoma cells were used, which efficiently expressed β-galactosidase (data not shown). Immunofluorescence analyses showed that D17 cells expressed Gag and cORF from constructs pcHERV-K and pcHERV-K/Ires-βGeo at lower levels than those expressed from pJW/HERV-K(SD-C7-LTR) in COS-7 cells (data not shown). Stable transfection experiments with pcHERV-K/Ires-βGeo were performed, and clones were established. However, increased expression in selected stable clones was found neither in the short term nor over longer periods of cultivation. Upon cotransfection of pcHERV-K/Ires-βGeo and of pJW-Ex30 or pJW-tPA-SP/EOP (36), the production of HERV-K particles as visualized by colocalization of Gag and Env with appropriate antibodies (4, 36) was not detected (not shown).
DISCUSSION
Replication-competent HERV-K has not yet been identified. Even though HTDV particles in teratocarcinoma cells (5, 11) have been linked to complex mRNA expression of HERV-K (14, 16) no transmission of these VLPs to animal or human cells has been achieved, suggesting that no intact full-length provirus exists in the human genome and/or that dominant-negative phenotypes prevent HERV-K particles from being infectious, as has been shown for HIV virions (39). The only known full-length provirus, HERV-K10, is 9.2 kb long and shows a pol ORF but disrupted gag and env (30). Enzyme activities for recombinant protease (34) and integrase (10) but not RT (38) derived from homologous HERV-K sequences have been reported. Recently, however, RT activities for in vitro-expressed cDNA clones of different HERV-K pol sequences have been described (2).
Characterization of two full-length HERV-K type 2 proviruses.
We have cloned and chromosomally assigned HERV-K proviral sequences by a genome-wide screening approach. Of 16 isolated P1 clones harboring HERV-K type 2 sequences, 2 proviruses located on chromosomes 7 and 19 displayed ORFs for gag, pol, and env. In addition HERV-K(C7) has a protease ORF and contains two LTRs, whereas HERV-K(C19) displays a frameshift mutation in the protease gene and is almost completely devoid of the 5′ LTR. The direct repeats adjacent to provirus HERV-K(C19) probably indicate that this element was originally integrated without the 5′LTR rather than having lost this sequence as a secondary event. A partially overlapping sequence with a cosmid clone deposited in GenBank suggests that HERV-K(C19) is located near the centromeric boundary of 19q12. In addition, both HERV-K(C7) and HERV-K(C19) proviral sequences were independently generated by long PCR from a monochromosomal mapping panel. No other HERV-K showing the expected size for intact proviral organization and the complete set of proviral ORF could be identified by these two approaches.
PTT analyses of individually cloned HERV-K gag and env genes had shown that only human chromosomes 7, 19, and Y might bear proviruses encompassing both ORFs (20, 21). The experiments presented here clearly confirm and extend these data by demonstrating entire proviral structures on C7 and C19. However, our approaches did not reveal the presence of a full-length HERV-K on chromosome Y, either by genomic methods or by PCR-mediated cloning.
HERV-K(C7) is probably an allelic variant of HERV-K(HML-2.HOM), a proviral sequence recently reported which also bears all ORFs and which was assigned to human chromosome 7p22 by FISH analysis (22). The identity of the 3′-flanking portions indicates the same chromosomal locations of these proviruses. Like the pol gene in HERV-K(C7), provirus HERV-K(HML-2.HOM) displays a YIDD-to-CIDD mutation in the highly conserved motif of RT and supposedly expresses a nonfunctional RT.
This assumption is confirmed by a report on in vitro RT activities of different recombinant HERV-K pol gene segments, where clone 11.2 displays the CIDD motif but no RT activity, in contrast to three clones (7.1, 10.1, and 10.9) bearing the YIDD sequence (2). Since HERV-K(C19) has no 5′LTR and thus misses the cognate promoter and since the authors used expressed pol sequences from bone marrow for their studies, this type of pol sequence showing an in-frame insertion of 3 amino acids (between nt 4149 and 4150) was not tested (2).
Characterization of HERV-K type 1 proviruses.
Cloning and characterization of type 1 proviruses, K10 and K18, demonstrated localization on human chromosomes 5 and 1, respectively. The gag gene of the HERV-K10 provirus presented here, in contrast to the published sequence (30), bears a long ORF which encodes an 80-kDa protein as demonstrated in vitro (Fig. 5B). Recombinant expression of HERV-K10 gag and protease genes in COS-7 cells produces Gag at the cellular membrane (Fig. 5C) and generates particles with an HTDV-like phenotype as in teratocarcinoma cells, where budding of particles occurs (5). These results are in agreement with the demonstration of in vitro protease activity of HERV-K10 (34) and, in particular, with our previously reported data that recombinant baculoviruses bearing HERV-K gag and protease genes produce VLP in insect cells (36). Hence, it is likely that HERV-K10 as a type 1 provirus has the potential to express VLP in vivo.
In contrast, HERV-K18 has no intact gag ORF but displays protease and pol ORFs, preventing this provirus from generating VLP. Most prominently, the ORF of the env segment of HERV-K18, which differs from the HERV-K(C7) Env sequence, shows striking homology to the SAG sequence of a HERV-K proviral sequence, designated IDDMK1,222. This retroviral sequence has been implicated in the etiology of type I diabetes (6). However, HERV-K18 shows one amino acid difference and the ORF encoding 560 amino acids extends into the 3′ LTR, suggesting that this provirus is closely related but distinct from IDDMK1,222, which so far has not been fully characterized.
Expression of HERV-K and biological functions.
The high homology scores of HERV-K proviral genes displaying intact but also defective ORFs calls into question the significance of specific alleles being expressed in human tissues. The association of IDDMK1,222 expression with disease, postulated by Conrad et al. (6), could not be confirmed (1, 9, 12, 18, 26, 27), probably since ubiquitous expression of HERV-K mRNA occurs in human tissues (18, 23). On the other hand, tumor-associated specific antibody responses against HERV-K proteins have been described (4, 33), suggesting that expression of these retroviral proteins has been impaired. Our data demonstrate that in addition to HERV-K type 1 proviruses, type 2 proviral genomes not only have the capacity to form VLP but also efficiently express proteins like cORF from spliced HERV-K mRNA (Fig. 6). However, no replicating HERV-K could be established with such permissive heterologous recombinant expression systems, although individual structural proteins and enzymes can be produced. One reason for this phenomenon probably involves the inefficiently cleaved Env precursor protein (37). Second, even though in vitro RT activity has been demonstrated (2), RT in purified particles isolated from human tissues and cell lines can be detected only by ultrasensitive RT assays (31, 35, 36, 38), indicating that HERV-K RT is a relatively weak enzyme. It is therefore conceivable that different proviruses contribute to a replication-competent virion by complementation.
It has been suggested that phylogenetically more recent integrations of HERV-K sequences have arisen from retrotransposition events triggered by HERV-K. For instance, HERV-K(HML-2.HOM) (22), a distinct cluster of HERV-K LTRs (24), and a HERV-K LTR in the DQ region of the human MHC (7) have been found exclusively in the genomes of humans but not of great apes.
Since HERV-K found on human chromosome 7 appears to be the most intact proviral structure of this endogenous retrovirus family, which, however, most probably lacks RT activity, it remains unclear which entity initiated retrotransposition and where HERV-K(C7) originates. We have attempted to quantify retrotransposition frequencies of appropriately designed HERV-K expression vectors in mammalian cells. The data suggest that HERV-K indeed has the capacity to trigger retrotransposition but at extremely low levels in comparison to those of exogenous retroviruses (28).
The possibility that due to our specific screening strategies, other (intact) members of this large family of human endogenous retroviruses were missed cannot be excluded. Additionally and alternatively, unidentified exogenous HERV-K homologues may still exist. The failure to identify such clones, however, provides good evidence for their absence from the human genome. In fact, the number of clones isolated for HERV-K(C7) (four clones) and HERV-K(C19) (two clones) is in the range of the twofold redundancy of the P1 genomic library. Our data, taken together with the results of the distinct strategy leading to the isolation of HERV-K(HML-2.HOM) (22), make it very unlikely that infectious HERV-K proviruses exist.
ACKNOWLEDGMENTS
This study was supported by a grant from the European Union (GENE-CT 93-0019) and by a donation from the Heinz Kuthe de Mouson Foundation, Basel, Switzerland.
The technical assistance of Gundula Braun and Nicole Fischer is gratefully acknowledged. We thank Peter Mountford, Victoria, Australia, for providing plasmid pIresβgeo.
REFERENCES
- 1.Badenhoop K, Donner H, Neumann J, Herwig J, Kurth R, Usadel K H, Tönjes R R. IDDM patients neither show humoral reactivities against endogenous retroviral envelope protein nor do they differ in retroviral mRNA expression from healthy relatives or normal individuals. Diabetes. 1999;48:215–218. doi: 10.2337/diabetes.48.1.215. [DOI] [PubMed] [Google Scholar]
- 2.Berkhout B, Jebbink M, Zsíros J. Identification of an active reverse transcriptase enzyme encoded by a human endogenous HERV-K retrovirus. J Virol. 1999;73:2365–2375. doi: 10.1128/jvi.73.3.2365-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeke J D, Stoye J P. Retroposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 4.Boller K, Janssen O, Schuldes H, Tönjes R R, Kurth R. Characterization of the antibody response specific for the human endogenous retrovirus HTDV/HERV-K. J Virol. 1997;71:4581–4588. doi: 10.1128/jvi.71.6.4581-4588.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boller K, König H, Sauter M, Mueller-Lantzsch N, Löwer R, Löwer J, Kurth R. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma-derived retrovirus HTDV. Virology. 1993;196:349–353. doi: 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 6.Conrad B, Weissmahr R N, Böni J, Arcari R, Schüpbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–313. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 7.Donner H, Tönjes R R, Bontrop R E, Kurth R, Usadel K H, Badenhoop K. Intronic sequence motifs of HLA-DQB1 are shared between humans, apes and old world monkeys, but a retroviral LTR element (DQLTR3) is human specific. Tissue Antigens. 1999;53:551–558. doi: 10.1034/j.1399-0039.1999.530605.x. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg A P, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 9.Jaeckel E, Heringlake S, Berger D, Brabant G, Hunsmann G, Manns M P. No evidence for association between IDDMK1,222, a novel isolated retrovirus, and IDDM. Diabetes. 1999;48:209–214. doi: 10.2337/diabetes.48.1.209. [DOI] [PubMed] [Google Scholar]
- 10.Kitamura Y, Ayukawa T, Ishikawa T, Kanda T, Yoshiike K. Human endogenous retrovirus K10 encodes a functional integrase. J Virol. 1996;70:3302–3306. doi: 10.1128/jvi.70.5.3302-3306.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurth R, Löwer R, Löwer J, Harzmann R, Pfeiffer R, Schmidt C G, Fogh J, Frank H. Oncovirus synthesis in human teratocarcinoma cultures and an increased anti-viral immune reactivity in corresponding patients. In: Essex M, Todaro G J, zur Hausen H, editors. Viruses in naturally occurring cancers. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1980. pp. 835–846. [Google Scholar]
- 12.Lan M S, Mason A, Coutant R, Chen Q-Y, Vargas A, Rao J, Gomez R, Chalew S, Garry R, Maclaren N K. HERV-K10s and immune-mediated (type 1) diabetes. Cell. 1998;95:11–14. doi: 10.1016/s0092-8674(00)81777-8. [DOI] [PubMed] [Google Scholar]
- 13.Leib-Mösch C, Haltmeier M, Werner T, Geigl E M, Brack-Werner R, Francke U, Erfle V, Hehlmann R. Genomic distribution and transcription of solitary HERV-K LTRs. Genomics. 1993;18:261–269. doi: 10.1006/geno.1993.1464. [DOI] [PubMed] [Google Scholar]
- 14.Löwer R, Boller K, Hasenmaier B, Korbmacher C, Mueller-Lantzsch N, Löwer J, Kurth R. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Löwer R, Löwer J, Tondera-Koch C, Kurth R. A general method for the identification of transcribed retrovirus sequences (R-U5 PCR) reveals the expression of the human endogenous retrovirus loci HERV-H and HERV-K in teratocarcinoma cells. Virology. 1993;192:501–511. doi: 10.1006/viro.1993.1066. [DOI] [PubMed] [Google Scholar]
- 17.Löwer R, Tönjes R R, Korbmacher C, Kurth R, Löwer J. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J Virol. 1995;69:141–149. doi: 10.1128/jvi.69.1.141-149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Löwer R, Tönjes R R, Boller K, Denner J, Kaiser B, Phelps R C, Löwer J, Kurth R, Badenhoop K, Donner H, Usadel K H, Miethke T, Lapatschek M, Wagner H. Development of insulin-dependent diabetes mellitus does not depend on specific expression of the human endogenous retrovirus HERV-K. Cell. 1998;95:11–14. doi: 10.1016/s0092-8674(00)81776-6. [DOI] [PubMed] [Google Scholar]
- 19.Lu S, Arthos J, Montefiori D C, Yasutomi Y, Manson K, Mustafa F, Johnson E, Santor J C, Wissink J, Mullins J I, Haynes J R, Letvin N L, Wyand M, Robinson H L. Simian immunodeficiency virus DNA vaccine trial in macaques. J Virol. 1996;70:3978–3991. doi: 10.1128/jvi.70.6.3978-3991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer J, Meese E U, Mueller-Lantzsch N. Multiple human endogenous retrovirus K (HERV-K) loci with Gag open reading frames in the human genome. Cytogenet Cell Genet. 1997;78:1–5. doi: 10.1159/000134614. [DOI] [PubMed] [Google Scholar]
- 21.Mayer J, Meese E U, Mueller-Lantzsch N. Chromosomal assignment of human endogenous retrovirus K (HERV-K) Env open reading frames. Cytogenet Cell Genet. 1997;79:157–161. doi: 10.1159/000134709. [DOI] [PubMed] [Google Scholar]
- 22.Mayer J, Sauter M, Rácz A, Scherer D, Mueller-Lantzsch N, Meese E U. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 1999;21:257–258. doi: 10.1038/6766. [DOI] [PubMed] [Google Scholar]
- 23.Medstrand P, Blomberg J. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential expression in normal human tissues. J Virol. 1993;67:6778–6787. doi: 10.1128/jvi.67.11.6778-6787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medstrand P, Mager D L. Human-specific integrations of the HERV-K endogenous retrovirus family. J Virol. 1998;72:9782–9787. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mountford P, Zevnik B, Düwel A, Nichols J, Li M, Dani C, Robertson M, Chambers I, Smith A. Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc Natl Acad Sci USA. 1994;91:4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muir A, Ruan Q-G, Marron M P, She J-X. The IDDMK1,222 retrovirus is not detectable in either mRNA or genomic DNA from patients with type 1 diabetes. Diabetes. 1999;48:219–222. doi: 10.2337/diabetes.48.1.219. [DOI] [PubMed] [Google Scholar]
- 27.Murphy V J, Harrison L C, Rudert W A, Luppi P, Trucco M, Fierabracci A, Biro P A, Bottazzo G F. Retroviral superantigens and type 1 diabetes mellitus. Cell. 1998;95:9–11. doi: 10.1016/s0092-8674(00)81775-4. [DOI] [PubMed] [Google Scholar]
- 28.Nan, X., R. R. Tönjes, and R. Kurth. Unpublished data.
- 29.Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol. 1986;58:937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589–598. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patience C, Simpson G R, Coletta A A, Welch H M, Weiss R A, Boyd M T. Human endogenous retrovirus expression and reverse transcriptase activity in the T47D mammary carcinoma cell line. J Virol. 1996;70:2654–2657. doi: 10.1128/jvi.70.4.2654-2657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roest P A M, Roberts R G, Sugino S, van Ommen G J B. Protein truncation test (PTT) for rapid detection of translation-terminating mutations. Hum Mol Genet. 1993;2:1719–1721. doi: 10.1093/hmg/2.10.1719. [DOI] [PubMed] [Google Scholar]
- 33.Sauter M, Schommer S, Kremmer E, Remberger K, Dölken G, Lemm I, Buck M, Best B, Neumann-Haefelin D, Mueller-Lantzsch N. Human endogenous retrovirus K10: expression of Gag proteins and detection of antibodies in patients with seminomas. J Virol. 1995;69:414–421. doi: 10.1128/jvi.69.1.414-421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schommer S, Sauter M, Kräusslich H G, Best B, Mueller-Lantzsch N. Characterization of the human endogenous retrovirus K (HERV-K) proteinase. J Gen Virol. 1996;77:375–379. doi: 10.1099/0022-1317-77-2-375. [DOI] [PubMed] [Google Scholar]
- 35.Simpson G R, Patience C, Löwer R, Tönjes R R, Moore H D, Weiss R A, Boyd M T. Endogenous D-type (HERV-K) related sequences are packaged into retroviral particles in the placenta and possess open reading frames for reverse transcriptase. Virology. 1996;222:451–456. doi: 10.1006/viro.1996.0443. [DOI] [PubMed] [Google Scholar]
- 36.Tönjes R R, Boller K, Limbach C, Lugert R, Kurth R. Characterization of human endogenous retrovirus type K virus-like particles generated from recombinant baculoviruses. Virology. 1997;233:280–291. doi: 10.1006/viro.1997.8614. [DOI] [PubMed] [Google Scholar]
- 37.Tönjes R R, Limbach C, Löwer R, Kurth R. Expression of human endogenous retrovirus type K envelope glycoprotein in insect and mammalian cells. J Virol. 1997;71:2747–2756. doi: 10.1128/jvi.71.4.2747-2756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tönjes R R, Löwer R, Boller K, Denner J, Hasenmaier B, Kirsch H, König H, Korbmacher C, Limbach C, Lugert R, Phelps R C, Scherer J, Thelen K, Löwer J, Kurth R. HERV-K: the biologically most active human endogenous retrovirus family. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13(Suppl. 1):S261–S267. doi: 10.1097/00042560-199600001-00039. [DOI] [PubMed] [Google Scholar]
- 39.Zybarth G, Kräusslich H G, Partin K, Carter C. Proteolytic activity of novel human immunodeficiency virus type 1 proteinase proteins from a precursor with a blocking mutation at the N terminus of the PR domain. J Virol. 1994;68:240–250. doi: 10.1128/jvi.68.1.240-250.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]