Abstract
The herpes simplex virus (HSV) virion host shutoff (vhs) protein (UL41 gene product) is a component of the HSV virion tegument that triggers shutoff of host protein synthesis and accelerated mRNA degradation during the early stages of HSV infection. vhs displays weak amino acid sequence similarity to the fen-1 family of nucleases and suffices to induce accelerated RNA turnover through endoribonucleolytic cleavage events when it is expressed as the only HSV protein in a rabbit reticulocyte in vitro translation system. Although vhs selectively targets mRNAs in vivo, the basis for this selectivity remains obscure, since in vitro activity is not influenced by the presence of a 5′ cap or 3′ poly(A) tail. Here we show that vhs activity is greatly altered by placing an internal ribosome entry site (IRES) from encephalomyocarditis virus or poliovirus in the RNA substrate. Transcripts bearing the IRES were preferentially cleaved by the vhs-dependent endoribonuclease at multiple sites clustered in a narrow zone located immediately downstream of the element in a reaction that did not require ribosomes. Targeting was observed when the IRES was located at the 5′ end or placed at internal sites in the substrate, indicating that it is independent of position or sequence context. These data indicate that the vhs-dependent nuclease can be selectively targeted by specific cis-acting elements in the RNA substrate, possibly through secondary structure or a component of the translational machinery.
Many viruses selectively inhibit host cell protein synthesis as a key element of their strategy of reprogramming the cellular biosynthetic machinery to support efficient virus replication. In the best-understood cases, picornaviruses employ multiple mechanisms to inactivate the cap-binding translation initiation factor eIF4F, thereby preventing translation of most cellular mRNAs. The eIF4F complex is composed of three proteins: eIF4E (the cap-binding protein), eIF4A (an RNA helicase), and eIF4G (which serves as a scaffold for assembly of the complex). Once bound to the mRNA through the 5′ cap structure, eIF4F recruits eIF3, which in turn serves as bridge between eIF4F and the incoming 40S ribosomal subunit (for a review, see reference 24). Members of the Enterovirus, Rhinovirus, and Aphthovirus genera of the Picornaviridae family encode proteases (e.g., the poliovirus 2Apro protease) that cleave eIF4G into two fragments (3, 5, 8, 21, 23, 41). The N-terminal fragment contains the eIF4E binding site, while the C-terminal fragment contains the binding sites for eIF4A and eIF3 (27, 40). Thus, these viral proteases uncouple the cap recognition and ribosome recruitment functions of eIF4F, thereby inhibiting cap-dependent translation. Picornavirus mRNAs escape shutoff by utilizing a cap-independent mode of translation initiation mediated by the C-terminal fragment of eIF4G (48, 55), which binds to the highly structured internal ribosome entry site (IRES) elements found in the 5′ untranslated regions (UTRs) of picornavirus RNAs (28, 29, 53, 54). Recently, Gradi and colleagues have identified an additional mammalian homologue of eIF4G, termed eIF4GII. These authors showed that both eIF4GI (the original eIF4G) and eIF4GII are cleaved in poliovirus- or rhinovirus-infected cells and concluded that cleavage of both proteins is required for shutoff of host cell protein synthesis (20, 21, 68).
Picornavirus infection also leads to inactivation of the cap-binding protein (eIF4E) component of eIF4F by two distinct mechanisms. First, the active, phosphorylated form of eIF4E (reviewed in reference 63) is dephosphorylated, resulting in the loss of activity (33). Second, the eIF4E inhibitor 4E-BP1 is also dephosphorylated (18, 19), leading to its activation. Active 4E-BP1 binds to eIF4E and prevents it from interacting with eIF4G, thereby inhibiting cap-dependent translation (22, 43, 45). Inactivation of eIF4E seems to be the major mechanism contributing to the host shutoff induced by cardioviruses (exemplified by encephalomyocarditis virus [EMCV]), which do not induce cleavage of eIF4G (18, 19, 33). More recently, poliovirus and coxsackievirus have been shown to induce cleavage of the poly(A)-binding protein (PABP) (30, 32). This observation is significant because PABP directly interacts with components of eIF4F to synergistically stimulate cap-dependent translation (26) (for a review, see reference 16). Moreover, PABP has been shown to compensate for a partial loss of eIF4E function and stimulate translation initiation of uncapped mRNAs in the yeast Saccharomyces cerevisiae (56, 71). Therefore, by inactivating eIF4G, eIF4E, and PABP, picornaviruses ensure the complete shutoff of cap-dependent translation.
Adenovirus and influenza virus also target eIF4E for inactivation as a component of their host shutoff strategies (10, 25, 76). Adenovirus late mRNAs escape this shutoff by utilizing a mode of translation that is less dependent on intact eIF4F. This is mediated by the tripartite leader (found in the 5′ UTRs of late mRNAs), which directs efficient translation initiation in the presence of limiting amounts of functional eIF4F by ribosome jumping (74). Similarly, the 5′ UTRs of influenza virus mRNAs have been shown to possess cis-acting sequence elements that direct efficient translation initiation in the presence of limiting amounts of functional eIF4F (17, 52).
Herpes simplex virus (HSV) also causes a dramatic reduction of host cell protein synthesis. Shutoff occurs in two distinct phases that occur at early and intermediate times postinfection, respectively (for a review, see reference 11). The early host shutoff induced by HSV provides a striking contrast to the strategies employed by picornaviruses and adenovirus, since it involves rapid degradation of preexisting cellular mRNAs rather than alterations to the translational apparatus per se. This effect is triggered by the infecting virus particle and requires the virion host shutoff protein (vhs) product of the HSV gene UL41 (12–15, 31, 34–37, 46, 47, 49, 50, 57, 59, 64, 67, 69, 70). vhs is a 58-kDa phosphoprotein that is produced late during infection and is packaged into the virion tegument (the space between the nucleocapsid and the viral envelope) (58, 60). vhs inhibits reporter gene expression in transiently transfected mammalian cells (31, 51) and triggers translational arrest and accelerated degradation of reporter RNAs when it is produced in a rabbit reticulocyte lysate (RRL) in vitro system (7, 75). Thus, vhs suffices to induce shutoff in the absence of other HSV proteins.
Although vhs is not essential for virus replication, vhs mutants display a ca. 10-fold reduction in virus yield in tissue culture (57, 62) and cause severe defects in the nervous system of the mouse (66), indicating that this protein plays an important role during the viral life cycle. vhs presumably helps viral mRNAs compete for the cellular translation machinery by reducing the level of cellular mRNAs in the cytoplasm. vhs also significantly destabilizes HSV mRNAs belonging to all three temporal classes (14, 36, 37, 49, 50, 57, 67), an effect that sharpens the transitions between the successive phases of viral protein synthesis by tightly coupling changes in the rate of mRNA synthesis to altered mRNA levels. Although HSV mRNAs are susceptible to vhs-induced decay, Fenwick and Owen demonstrated that the onset of viral protein synthesis leads to a significant increase in the half-lives of viral mRNAs (14). This observation suggests that a newly synthesized HSV protein(s) partially downregulates the vhs activity of the infecting virion, allowing viral mRNAs to accumulate after host mRNAs have been degraded (14). vhs binds to the virion transactivator VP16 (61), and VP16 null mutants undergo a severe vhs-induced translational arrest at intermediate times postinfection (39). These data strongly suggest that VP16 plays an important role in the downregulation of vhs activity.
The mechanism of action of vhs remains to be precisely defined. vhs displays weak but significant amino acid sequence similarity to the fen-1 family of nucleases that are involved in DNA replication and repair in eukaryotes and archaebacteria (6), and recent studies have shown that human fen-1 cleaves both RNA and DNA substrates (65). These data suggest that vhs may be a ribonuclease. Consistent with this hypothesis, Zelus and colleagues showed that extracts of partially purified HSV virions contain a vhs-dependent ribonuclease activity that is inhibited by anti-vhs antiserum (75). Although these data do not exclude the possibility that this ribonuclease contains one or more cellular subunits, they strongly suggest that vhs is an integral and required component of the enzyme.
We and others have shown that vhs induces translational arrest and degradation of reporter mRNA in vitro when it is expressed as the only HSV product in RRLs (7, 75). We further demonstrated that decay occurs through an endoribonucleolytic mechanism that does not depend on a 5′ cap or a 3′ poly(A) tail in the RNA substrate (7). During a survey of the mode of decay of a variety of RNAs in this system, we discovered that picornavirus IRES elements profoundly alter the degradation profile of substrate RNAs. In this communication, we show that the IRES elements of EMCV and poliovirus strongly direct vhs-induced endoribonucleolytic cleavage to sequences located immediately 3′ to the IRES. This targeting activity was observed in several distinct sequence contexts, demonstrating that these IRESs serve as movable targeting elements for vhs-dependent cleavage.
MATERIALS AND METHODS
Plasmids.
The vhs in vitro transcription vector pSP6vhs has been previously described (7). pCITE-1 (Novagen) contains residues 255 to 836 of the EMCV 5′ UTR (the IRES element) 9 bp downstream of the T7 RNA polymerase promoter start site. The IRES element is followed by a 1,166-bp fragment corresponding to the extreme 5′ end of the EMCV open reading frame. pCITE RI/AvrII (lacking the 5′-most 159 bp of the IRES) was constructed by self-ligating EcoRI- and AvrII-digested pCITE-1 DNA after filling in the ends with the Klenow fragment of DNA polymerase I. pCITE Msc/RI (lacking all of the IRES) was generated in the same way, using MscI- and EcoRI-cleaved pCITE-1 DNA. pSexAI IRES was constructed by ligating a 600-nucleotide (nt) EcoRI-MscI fragment of pCITE-1 (bearing the IRES) into the SexAI site of pCITE Msc/RI (after filling in all of the ends with the Klenow fragment). The resulting plasmid contains the EMCV IRES 729 bp downstream of the T7 start site. pSPSR19N contains a cDNA encoding the canine signal recognition particle α subunit (SRPα), initiating at an engineered NcoI site, inserted into pSPUTK (9, 73). pSP19StuI IRES was constructed by inserting the 600-nt EcoRI-MscI fragment of pCITE-1 into the unique StuI site of pSPSR19N (after all ends were made flush). The resulting plasmid bears the EMCV IRES element 1,721 bp downstream of the SP6 RNA polymerase start site.
The plasmid pCITE Msc/RI P2 is a pCITE-1 derivative in which the EMCV IRES is replaced with the IRES of poliovirus type 2. It was constructed by exchanging the ca. 600-bp EcoRI-MscI fragment of pCITE-1 with a ca. 600-bp HindIII-MscI fragment from the plasmid pP2CAT (after making all ends flush with Klenow fragment). P2CAT was a generous gift from N. Sonenberg, McGill University. pSexAI P2 was generated by ligating the ca. 600-bp HindIII-MscI fragment from pP2CAT into the SexAI site of pCITE Msc/RI after repairing the ends with Klenow fragment. The resulting plasmid lacks the EMCV IRES and contains the poliovirus IRES ca. 729 nt from the 5′ end of the T7 RNA polymerase transcript.
In vitro transcription and RNA labeling.
Transcription reactions were carried out with the Riboprobe in vitro transcription system (Promega) according to the vendor’s protocol. vhs mRNA destined for in vitro translation was generated by transcribing 3 to 5 μg of supercoiled pSP6vhs plasmid DNA in a 50-μl reaction mixture for 30 min at 30°C, using 40 U of SP6 RNA polymerase in the presence of 0.5 mM cap primer 7mG(5′)ppp(5′)G (Pharmacia), 12.5 μM GTP, and 0.25 mM each CTP, ATP, and UTP. Following digestion of plasmid DNA with 5 U of RQ1 DNase (Promega), the reaction mixture was extracted once with phenol-chloroform-isoamyl alcohol and once with chloroform. The resulting solution was brought to 2.5 M ammonium acetate, and the mRNA was precipitated with 95% ethanol. The mRNA pellet was then washed with 70% ethanol, dried, and resuspended in RNase-free water.
Capped, internally labeled reporter RNAs were generated as described above, except that the template was linearized at an appropriate site (see below) prior to transcription and 1 μCi of [α-32P]GTP was added to the transcription reaction. Uncapped, internally labeled reporter RNAs were produced in a similar fashion, except that the cap primer was omitted and the GTP concentration was increased to 0.25 mM. Transcription of reporter mRNAs was terminated by adding 20% RNA loading buffer (50% glycerol, 1 mM EDTA, 10 mg of xylene cyanol/ml, and 10 mg of bromophenol blue/ml) and immediately loading the sample on a 1% agarose gel in 1× TBE (90 mM Tris-borate, 2 mM EDTA) buffer. Following electrophoresis for 2 h at approximately 7 V/cm, gel slices containing full-length transcripts (detected with UV light after ethidium bromide staining) were excised and equilibrated in 0.5× TBE for 10 min. The RNA was then electroeluted from gel slices into a 100-μl 7.5 M ammonium acetate trap in a six-well v-channel electrolutor (IBI) at 100 V for 30 min. The RNA was then recovered from the salt solution by ethanol precipitation. The RNA pellets were washed two times with 70% ethanol, dried, and resuspended in RNase-free water.
Reporter RNAs transcribed from pCITE, pCITE RI/AvrII, pCITE Msc/RI, pCITE Msc/RI P2, pSexAI IRES, and pSexAI P2 were generated by using T7 RNA polymerase and EcoNI-linearized plasmid DNAs as templates to yield runoff transcripts of ca. 2.3, 2.1, 1.7, 2.3, 2.3, and 2.3 kb, respectively. A shorter transcript of pCITE RI/AvrII (437 nt) was generated by using T7 RNA polymerase and MscI-linearized pCITE RI/AvrII plasmid DNA as a template. SRPα and SRPα StuI IRES reporter mRNAs were generated by using SP6 RNA polymerase and EcoRV-linearized pSPSR19N and pSP19StuI IRES plasmid DNAs as templates to yield runoff transcripts of 2.4 and 3 kb, respectively.
Cap-labeled reporter RNAs were generated from gel-purified uncapped, unlabeled runoff transcripts by using vaccinia virus guanylyltransferase in the presence of [α-32P]GTP. Approximately 500 ng of RNA in a solution containing 50 mM Tris-HCl (pH 7.9), 1.25 mM MgCl2, 6 mM KCl, 2.5 mM dithiothreitol, 0.1 mg of RNase-free bovine serum albumin, 1 U of RNasin/μl, and 0.1 mM S-adenosyl-l-methionine was combined with 1 to 3 U of guanylyltransferase (Gibco-BRL) and 50 μCi of [α-32P]GTP in a total reaction volume of 30 μl. Following a 45-min reaction at 37°C, the reaction mixture was extracted once with phenol-chloroform-isoamyl alcohol and once with chloroform and the RNA was recovered by ethanol precipitation.
In vitro translation and vhs activity assay.
Approximately 5 μg of vhs mRNA was translated in a 50-μl RRL (Promega or Novagen) reaction mixture containing 40 μCi of [35S]methionine, in accordance with the vendor’s protocol. Translation reactions were carried out for 1 h at 30°C. Blank RRL controls were generated as described above except that mRNA was omitted from the translation reactions. Samples of the translation reaction products were assessed for [35S]methionine incorporation by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis (38).
To assay for vhs activity, reporter RNA substrates were added to RRL containing pretranslated vhs and the reaction mixtures were incubated at 30°C. Aliquots (5 μl) of reaction mixtures were removed at various times and immediately added to 200 μl of Trizol (Gibco-BRL) containing 20 μg of carrier Escherichia coli tRNA (Sigma). The samples were extracted by the addition of 40 μl of chloroform, and the resulting aqueous phase was reextracted with chloroform. RNA was recovered by isopropanol precipitation, resuspended in 100 μl of RNase-free water, and reprecipitated with 95% ethanol. Following a 70% ethanol wash, the RNA pellet was dried and resuspended in RNase-free water. The RNA samples were then analyzed by electrophoresis through agarose-formaldehyde or polyacrylamide sequencing gels or by primer extension.
Agarose gel electrophoresis and Northern blot analysis.
RNA samples were resuspended in 4.5 μl of RNase-free water and then combined with 2 μl of 10× MOPS buffer (200 mM 3-n-morpholinopropanesulfonic acid [pH 7.0], 50 mM sodium acetate, and 5 mM EDTA), 10 μl of deionized formamide, and 3.5 μl of a 37% formaldehyde solution. Following a 10-min incubation at 75 to 80°C, the solution was combined with 6 μl of RNA loading buffer and subjected to electrophoresis through a 1% agarose gel containing 6% formaldehyde. Electrophoresis was carried out in 1× MOPS buffer containing 6% formaldehyde at approximately 5 V/cm for 3 to 4 h. The gel was then washed in water for 10 min, treated with 50 mM NaOH–10 mM NaCl (20 min), and neutralized with 100 mM Tris-HCl (20 min). RNA was then transferred to a Nytran Plus membrane in 20× SSC (3 M sodium chloride, 0.3 M sodium citrate). Following UV cross-linking (Stratalinker 2400; Stratagene), 32P-labeled RNA fragments were detected by exposure to Kodak X-OMAT AR film at −70°C.
Unlabeled RNA fragments cross-liked to Nytran Plus membranes were detected by Northern blot analysis (4). Briefly, the membranes were prehybridized in Church buffer (250 mM sodium phosphate buffer [pH 7.2], 7% SDS, 1% bovine serum albumin, 1 mM EDTA) at 62°C for 1 h. The membrane shown in Fig. 1C was hybridized to a 5′-32P-labeled oligonucleotide (AB9899; 5′-CATCATCCTCTCCATCAG-3′) complementary to residues 729 to 746 of the pCITE transcript. The membranes in Fig. 5C and D were hybridized to a 400-nt EcoRV-EcoRI fragment of pSPSR19N corresponding to the 3′-most portion of the SRPα transcript (32P labeled by random priming). Rabbit 18S rRNA was detected with a 5′-32P-labeled oligonucleotide complementary to residues 269 to 299 of that rRNA (AB12084; 5′-TATCTAGAGTCACCAGAGCCGCCGAGCCCA-3′). Hybridization was carried out in Church buffer at 62°C for 13 to 17 h. The membrane was then washed twice (10 min each) in 2× SSC–0.1% SDS and twice (10 min each) in 0.1× SSC–0.1 SDS prior to being subjected to autoradiography.
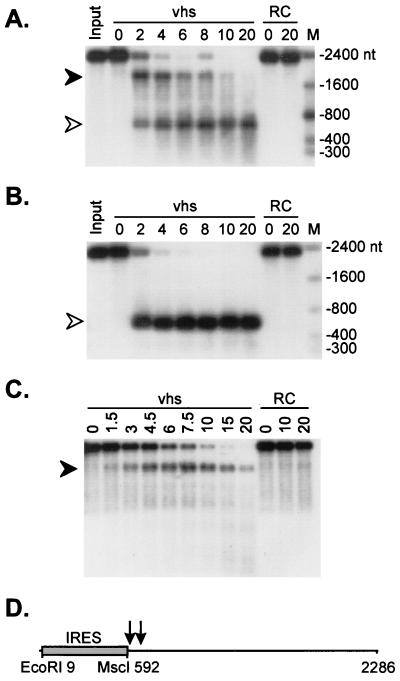
FIG. 1.
vhs induces preferential endonucleolytic cleavage in the vicinity of the 3′ boundary of the EMCV IRES. A 2.3-kb runoff transcript of pCITE-1 was added to RRLs containing pretranslated vhs (lanes marked vhs) and to control RRL (lanes marked RC), and samples were removed at the indicated times (in minutes). RNA was extracted from each sample, resolved on a 1% agarose–6% formaldehyde gel, and transferred to a Nytran Plus membrane. (A) Capped, internally labeled RNA. The closed and open arrowheads indicate the ca. 1,800- and 600-nt fragments, respectively. (B) 5′-Cap-labeled RNA. (C) Capped, unlabeled RNA. The radioactive signals in panels A and B were detected by autoradiography. The RNA products in panel C were detected by hybridization to a 5′-labeled oligonucleotide probe complementary to residues 729 to 746 of the pCITE transcript, followed by autoradiography. The numbers to the right of panels A and B indicate the sizes of RNA markers (lanes marked M) in nucleotides. (D) Diagram of the pCITE transcript indicating the approximate position(s) of the cleavage site(s) (arrows).
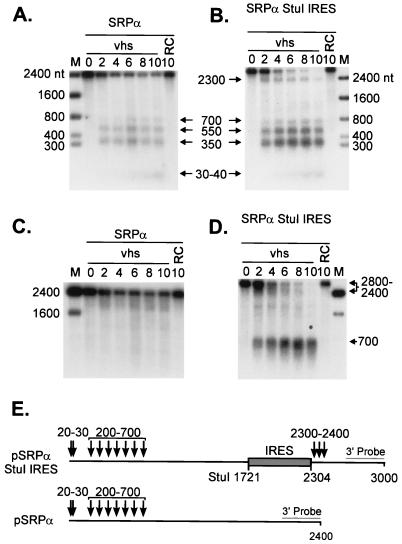
FIG. 5.
The EMCV IRES targets vhs-induced cleavage of SRPα mRNA. Cap-labeled SRPα (A) and SRPα StuI IRES (B) RNAs were added to RRLs containing vhs (diluted 1:4 in naive RRL) (lanes marked vhs) and to control RRL (lanes marked RC), and RNA extracted from samples removed at the indicated times (in minutes) was analyzed by agarose-formaldehyde gel electrophoresis as for Fig. 1. (C D) The membranes displayed in panels A and B, respectively, were hybridized to a probe corresponding to the 3′-most 400 nt of SRPα RNA (after the radioactive signal from the cap label had been allowed to decay for more than 6 half-lives). Arrows and associated numbers indicate the positions and sizes of the vhs-induced cleavage intermediates. The numbers to the left of panels A and C and to the right of panels B and D indicate the sizes of RNA markers (lanes M) in nucleotides. (E) Diagram showing the structures of both transcripts, with the approximate locations of vhs-induced cleavages indicated by arrows. The shaded rectangle represents the EMCV IRES.
Primer extension.
RNA samples were suspended in 10 μl of annealing buffer (10 mM Tris-HCl [pH 7.9], 1 mM EDTA, 250 mM KCl) containing 50,000 Cerenkov cpm of an appropriate 5′-32P-labeled oligonucleotide. The following oligonucleotide primers were used to detect vhs-induced cleavages downstream of the IRES elements: AB11388 (5′-CATTCTTCATCATACTTTAGCAGGT-3′), complementary to residues 685 to 710 of pCITE-1 RNA (see Fig. 2C and 7B); AB11259 (5′-CCATTAGGCAGGTTATCCTTGGACC-3′), complementary to residues 1447 to 1471 of pSexAI IRES RNA (see Fig. 4); and AB11260 (5′-GCAGCTCCCACCTTGTCATCAATGG-3′), complementary to residues 2424 to 2448 of SRPα StuI IRES RNA (see Fig. 6). After annealing for 1 h at 65°C, the samples were combined with 25 μl of PE buffer (20 mM Tris-HCl [pH 8.7], 10 mM MgCl2, 5 mM dithiothreitol, 330 μM each deoxynucleoside triphosphate 10 μg of actinomycin D/ml, and 10 U of SuperScript II [Gibco-BRL] reverse transcriptase) and the extension reaction was carried out for 1 h at 42°C. Nucleic acids were precipitated with 95% ethanol, washed with 70% ethanol, dried, and resuspended in water. The samples were then combined with equal volumes of sequencing-gel loading buffer, heated to 80°C for 2 to 3 min, and resolved on 8% polyacrylamide sequencing gels. The radioactive signal was detected by autoradiography.
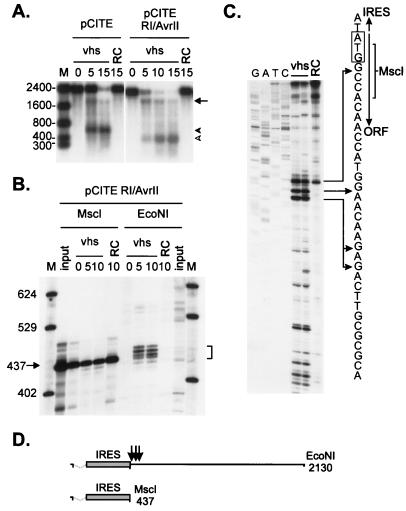
FIG. 2.
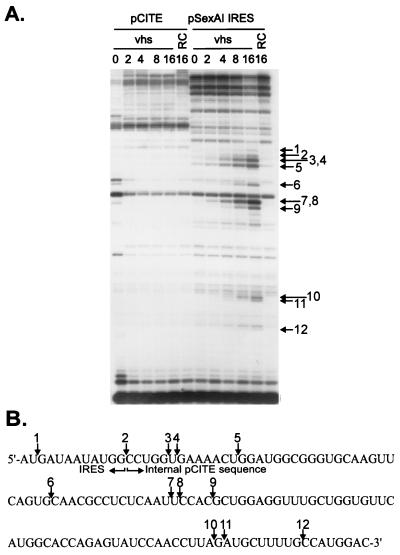
The pCITE transcript is cleaved immediately downstream of the IRES. (A) Uncapped, internally labeled transcripts of pCITE and pCITE RI/AvrII (lacking the 5′-most 159 nt of the IRES) were added to RRLs containing vhs, and the reaction products were analyzed as for Fig. 1A. The arrow and arrowheads indicate the 3′ and 5′ degradation products, respectively. The numbers to the left indicate the sizes of RNA markers (lane M) in nucleotides. RC, control RRL. (B) 5′-Cap-labeled runoff transcripts of pCITE RI/AvrII were added to RRL containing vhs, and reaction products extracted at the indicated times (in minutes) were resolved on an 8% polyacrylamide sequencing gel. The template DNA was linearized with MscI or EcoNI prior to transcription, generating runoff transcripts of 437 and 2,127 nt, respectively. The numbers on the left indicate the sizes of DNA markers (lane M) in nucleotides. The bracket indicates the four prominent 5′ products. (C) A capped, unlabeled EcoNI runoff transcript of pCITE was incubated for 10 min in RRL containing pretranslated vhs or in control RRL (RC). The RNA was then extracted and analyzed by primer extension, using a 5′-labeled oligonucleotide complementary to residues 685 to 710 of the transcript. The primer extension products were resolved on an 8% polyacrylamide sequencing gel along with a DNA sequencing ladder generated from pCITE-1 DNA by using the same primer. The diagram accompanying panel C shows the nucleotide sequence at the IRES/open reading frame boundary. The EMCV translational initiation codon is indicated by a rectangle, and arrows indicate the MscI cleavage site and the positions of some of the 5′ ends generated by vhs-induced cleavage. (D) Structures of the pCITE RI/AvrII runoff transcripts. Dashed lines indicate the extent of the IRES deletion, and arrows indicate the approximate locations of the vhs-dependent cleavage sites.
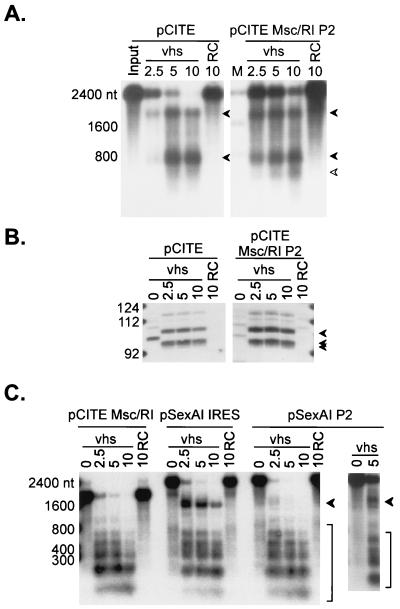
FIG. 7.
The poliovirus IRES serves as a movable targeting element for vhs-induced RNA cleavage. (A) Uncapped, internally labeled pCITE and pCITE Msc/RI P2 RNAs were reacted with RRLs containing vhs and with control RRL (RC), and samples removed at the indicated times (in minutes) were analyzed by agarose-formaldehyde gel electrophoresis as for Fig. 1. pCITE Msc/RI P2 is a derivative of pCITE in which the EMCV IRES has been replaced by the IRES of poliovirus type 2. (B) Uncapped, unlabeled pCITE and pCITE Msc/RI P2 RNAs were added to RRLs containing vhs and to control RRL. RNAs extracted from samples removed at the indicated times (in minutes) were analyzed by primer extension, using a 5′-labeled oligonucleotide that anneals to residues 685 to 710 of pCITE RNA (and residues ca. 685 to 710 of pCITE Msc/RI P2 RNA). The arrowheads indicate the positions of the novel 5′ ends generated by vhs-induced cleavage. These correspond to the three primer extension products indicated in Fig. 2C. The fainter band running just above the 112-nt marker is the reverse transcriptase pause site at the MscI site of pCITE (Fig. 2C). The numbers to the left indicate the sizes of DNA markers in nucleotides. (C) Cap-labeled runoff transcripts of the indicated plasmids were combined with RRLs containing vhs (lanes marked vhs) and with control RRL (lanes marked RC), and samples removed at the indicated times (in minutes) were analyzed by agarose-formaldehyde gel electrophoresis as for Fig. 1. pCITE Msc/RI lacks an IRES, while pSexAI IRES and pSexAI P2 bear the EMCV and poliovirus type 2 IRES elements, respectively, inserted at the SexAI site of pCITE Msc/RI. Arrowheads in panels A and C indicate vhs-induced cleavage intermediates. The numbers to the left of panels A and C indicate the sizes of RNA markers (lanes M) in nucleotides.
FIG. 4.
vhs induces cleavage downstream of an internally located EMCV IRES. (A) Uncapped, unlabeled transcripts of pCITE and pSexAI IRES were added to RRLs containing vhs (lanes marked vhs) and to control RRL (lanes marked RC), and samples were removed at the indicted times (in minutes). Following extraction, the reaction products were analyzed by primer extension, using a 5′-labeled oligonucleotide complementary to residues 1447 to 1471 of the pSexAI IRES RNA (and residues 1438 to 1462 of the pCITE transcript). Primer extension products were analyzed on an 8% polyacrylamide sequencing gel. Numbered arrows indicate primer extension products resulting from vhs-induced cleavage. (B) Diagram showing the nucleotide sequence at the 3′ boundary of the IRES in pSexAI IRES RNA. Numbered arrows correspond to those in panel A and indicate the locations of the novel 5′ ends produced by vhs-induced cleavage.
FIG. 6.
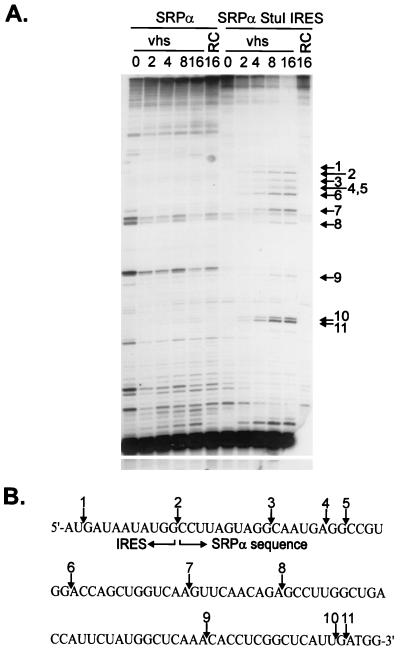
The EMCV IRES targets cleavage to 3′-flanking SRPα sequences. (A) Uncapped, unlabeled SRPα and SRPα StuI IRES RNAs were combined with RRLs containing vhs (lanes marked vhs) and with control RRL (lanes marked RC), and samples were withdrawn at the indicated times (in minutes). RNAs were then extracted and analyzed by primer extension, using a 5′-labeled oligonucleotide that anneals to residues 1841 to 1865 of SRPα RNA (and residues 2424 to 2448 of the SRPα StuI IRES transcript). Numbered arrows indicate the positions of novel 5′ ends resulting from vhs-induced RNA cleavage. (B) Diagram showing the nucleotide sequence at the 3′ boundary of the IRES in SRPα StuI IRES RNA. Numbered arrows correspond to those in panel A and indicate the locations of the vhs-induced cleavage sites.
RESULTS
Preferential vhs-induced cleavage near the 3′ boundary of the EMCV IRES.
We and others have previously shown that HSV type 1 (HSV-1) vhs triggers endoribonucleolytic cleavage of exogenous RNA substrates when it is produced as the only HSV protein in an in vitro translation system derived from RRL (7, 75). During a survey of the mode of decay of a variety of substrate RNAs, we discovered that a 2.3-kb transcript of pCITE-1 (bearing the EMCV IRES at its 5′ end) gave rise to a strikingly simple pattern of early degradation intermediates. As shown in Fig. 1A, capped, internally labeled pCITE-1 RNA yielded two prominent products of ca. 1,800 and 600 nt when it was incubated in RRL containing pretranslated vhs. The ca. 600-nt fragment was stable throughout the course of the reaction, while the ca. 1,800-nt fragment was subject to further decay. Only the ca. 600-nt fragment was detected when 5′-cap-labeled RNA was used as the substrate (Fig. 1B), indicating that it is derived from the 5′ end of the RNA. Inasmuch as the two fragments roughly sum to yield the size of the intact transcript (ca. 2,300 nt), these data suggested that vhs-induced endoribonucleolytic cleavage occurs at one or several closely spaced sites located approximately 600 nt from the 5′ end of the RNA. Consistent with this interpretation, an oligonucleotide complementary to residues 729 to 746 of the transcript hybridized to only the larger (3′) fragment (Fig. 1C). Similarly, only the larger fragment was detected with a probe corresponding to the 3′-most 253 nt of the RNA (data not shown).
As shown schematically in Fig. 1D, the prominent cleavage site(s) detected above maps to the vicinity of the 3′ boundary of the IRES. These data raised the possibility that the IRES element plays a role in targeting vhs-induced cleavage and indicated that the majority of the IRES sequence is highly resistant to degradation.
Cleavage occurs immediately downstream of the EMCV IRES element.
We mapped the location(s) of the prominent early cleavage site(s) more precisely, through high-resolution analysis of the cleavage products. To increase the resolution of the experiment, we used a pCITE derivative that lacks the 5′-most 156 nt of the EMCV IRES (pCITE RI/AvrII) to characterize the 5′ products. This mutant IRES element retains the ability to promote cap-independent initiation of translation (72). We first established that this transcript is cleaved in the vicinity of the 3′ boundary of the IRES in the same fashion as pCITE-1 RNA. Figure 2A shows that internally labeled pCITE RI/AvrII RNA gave rise to products of ca. 1,800 and 450 nt. The larger fragment comigrated with the 3′ fragment of pCITE-1 RNA (Fig. 2A), and the estimated size of the smaller product (ca. 450 nt) approximately corresponds to that of the deleted IRES (437 nt) (Fig. 2D). Thus, these data strongly suggest that pCITE RI/AvrII RNA is cleaved at or near the 3′ boundary of the IRES. We then used 5′-cap-labeled pCITE RI/AvrII RNA as a substrate and analyzed the products on an 8% polyacrylamide sequencing gel. As shown in Fig. 2B, a 2,130-nt EcoNI runoff transcript (Fig. 2D) gave rise to four prominent 5′ products, ranging in size from approximately 437 to 460 nt, as well as a fainter, ca. 430-nt band. In contrast, a 437-nt MscI runoff transcript was essentially immune to vhs-induced cleavage. The MscI cleavage site is located 5 nt downstream of the EMCV initiation codon (Fig. 2C) and is operationally considered to mark the 3′ boundary of the IRES. Thus, these data indicate that vhs-induced cleavage occurs primarily at several sites located just downstream of the IRES. This conclusion was confirmed by primer extension analysis of the cleavage products of pCITE RNA, using a primer complementary to sequences located 91 to 115 nt downstream of the MscI site (Fig. 2C). The results indicate that the majority of the novel vhs-induced 5′ ends detected in this region are located 3′ to the MscI site. Three of the most prominent of these 5′ ends have been mapped at the nucleotide level and are indicated in Fig. 2C. The strong primer extension signal at the MscI site was also observed in RNA samples incubated in a control reticulocyte lysate (Fig. 2C) and likely represents pausing of reverse transcriptase due to the extensive secondary structure of the IRES.
The EMCV IRES serves as a movable targeting element for vhs-induced cleavage.
The data described above demonstrate that prominent sites of initial vhs-induced cleavage are located just downstream of the EMCV IRES in the pCITE transcript. In principle, this narrow clustering of preferred cleavage sites might depend on the nucleotide sequence of the cleavage sites, the structure or function of the adjacent IRES, or the fact that this region corresponds to the 5′-most section of the transcript that is accessible to the vhs-induced endoribonuclease (the IRES itself is highly resistant to cleavage).
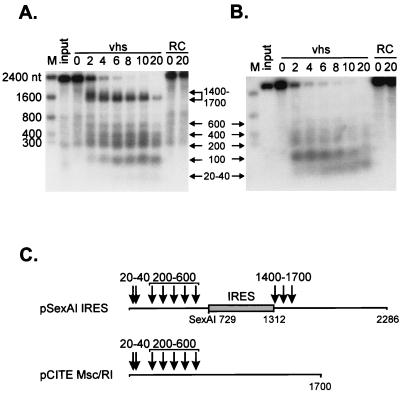
As one approach to distinguish between these possibilities, we asked whether the EMCV IRES element would induce novel vhs-dependent cleavages if it were transplanted to the middle of the pCITE transcript. To this end, we deleted the IRES element from the 5′ end to yield construct pCITE RI/MscI, then reinserted the IRES at a SexAI site located 729 nt into the pCITE RI/McsI RNA (construct pSexAI IRES [Fig. 3C]). 5′-Cap-labeled pCITE RI/MscI RNA (lacking an IRES) gave rise to a heterogeneous set of 5′ products ranging in size from ca. 20 to 600 nt (Fig. 3B). Although we have not yet examined the mode of degradation of this transcript in detail, the pattern of 5′ fragments produced is similar to that previously observed with SRPα RNA, which is preferentially cleaved at a variety of sites distributed over the 5′ quadrant of the transcript early during the reaction (7) (see Fig. 5 below). pSexAI IRES RNA displayed a prominent novel band of ca. 1,400 nt, in addition to these smaller 5′ products (Fig. 3A). This novel band was quite broad at early times (extending from ca. 1,400 to 1,700 nt) (Fig. 3A), then sharpened into a more discrete signal at ca. 1,400 nt as the reaction proceeded. These data indicate that the IRES present in the pSexAI IRES transcript provokes novel cleavage events in the region extending from ca. 1,400 to 1,700 nt from the 5′ end of the RNA. As diagrammed in Fig. 3C, these cleavage sites map at or close to the 3′ boundary of the IRES. The intensity of the 1,400- to 1,700-nt signal declined as the reaction proceeded, while the 20- to 600-nt products accumulated, suggesting that the 5′ products of the IRES-directed cleavages may be substrates for additional cleavage events. Inasmuch as we were able to easily detect the cleavage sites located 1,400 to 1,700 nt from the 5′ end of the pSexAI IRES transcript by using 5′-cap-labeled RNA, the data displayed in Fig. 3B argue that the most prominent sites of initial cleavage of the pCITE RI/MscI RNA are confined to the 5′ quadrant of this transcript.
FIG. 3.
The EMCV IRES serves as a movable targeting element for vhs-induced RNA cleavage. 5′-Cap-labeled EcoNI runoff transcripts of pSexAI IRES (A) and parental pCITE Msc/RI (B) were added to RRLs containing vhs (lanes marked vhs) and to control RRL (lanes marked RC), and samples removed at the indicated times (in minutes) were analyzed by agarose-formaldehyde gel electrophoresis as for Fig. 1. Arrows and associated numbers indicate the positions and sizes (in nucleotides) of the vhs-dependent RNA cleavage products. The numbers to the left indicate the sizes of RNA markers (lanes M) in nucleotides. (C) Diagram showing the structures of the two transcripts and the approximate locations of the vhs-induced cleavages. The shaded box represents the EMCV IRES.
As shown in Fig. 2, pCITE RNA is cleaved just downstream of the IRES, within the 3′-flanking sequences. To determine if this is also the case with the pSexAI IRES transcript, we mapped some of the cleavage sites by primer extension, using a primer complementary to residues 1,447 to 1,471 of that RNA (Fig. 4). pSexAI IRES RNA displayed a variety of vhs-dependent novel 5′ ends in the region examined (Fig. 4A). The precise positions of some of these 5′ ends were determined by resolving the primer extension products beside a DNA sequencing ladder generated from pSexAI IRES DNA, using the same primer (data not shown) (diagrammed in Fig. 4B). The majority of the new 5′ ends were located downstream of the 3′ boundary of the IRES. Only one very faint band (labeled 1) mapped within the IRES itself. Although pCITE RNA has exactly the same nucleotide sequence as the pSexAI IRES transcript downstream of site 2, prominent vhs-induced cleavages were not observed in this region of the pCITE RNA.
Taken in combination, these data indicated that the IRES present in the pSexAI IRES RNA provoked novel vhs-induced RNA cleavage events in the 3′-flanking sequences. In this sense, the IRES behaved as a movable targeting element for vhs-induced cleavage.
We tested the generality of this finding by asking whether the EMCV IRES would similarly alter the degradation pattern of the entirely unrelated RNA encoding SRPα (Fig. 5). We inserted the IRES at a StuI site located at residue 1721 of the transcript, generating construct pSRPα StuI IRES (Fig. 5E). SRPα and SRPα StuI IRES RNAs were then 5′-cap labeled and added to RRL containing pretranslated vhs. As previously reported, SRPα RNA gave rise to 5′ fragments ranging in size from ca. 30 to 700 nt (Fig. 5A), reflecting the clustering of the preferred sites of initial cleavage over the 5′ quadrant of this RNA (7). SRPα StuI IRES RNA displayed these same products, as well as an additional ca. 2,300-nt fragment that was not observed with SRPα RNA (Fig. 5B). The size of this novel 5′ fragment agreed well with that predicted to arise from cleavage at the 3′ boundary of the inserted IRES (Fig. 5E). Such cleavage events would also generate novel 3′ fragments of ca. 700 nt. We tested for these by hybridizing the membranes shown in Fig. 5A and B to a probe for the extreme 3′ end of the RNA (after allowing the 5′-cap label to decay for 6 half-lives). The results clearly indicated that SRPα StuI IRES RNA gave rise to the predicted set of novel ca. 700-nt 3′ fragments (Fig. 5D). As previously reported, SRPα RNA generated a heterogeneous set of early 3′ products ranging in size from 1,800 to 2,200 nt (Fig. 5C), reflecting the 5′ clustering of initial cleavage events on this transcript (7). Analogous high-molecular-weight 3′ products were also observed with SRPα StuI IRES RNA (ca. 2,400 to 2,700 nt) (Fig. 5D). Taken in combination, these data indicate that SRPα StuI IRES RNA is cleaved over its 5′-most 600 nt in the same fashion as SRPα RNA and is additionally cleaved downstream of the inserted IRES. The two sets of cleavage events appear to occur independently.
In this case as well, primer extension analysis, using a primer complementary to residues 2424 to 2448 of the SRPα StuI IRES transcript, revealed that the cleavage events induced by the IRES occurred in the 3′-flanking sequences (Fig. 6A). Only one weak cleavage site was detected within the IRES element (site 1). Interestingly, both this site and the site at the junction between the IRES and SRPα sequences (site 2) are the same as those observed with pSexAI IRES (compare Fig. 6B and 4B).
Taken together, these data clearly show that the EMCV IRES targets vhs-dependent endoribonucleolytic cleavage events to 3′-flanking sequences. This activity is independent of sequence context and operates when the IRES is placed at a variety of locations in the substrate RNA.
The poliovirus IRES also serves as a movable vhs-targeting element.
We asked whether the unrelated poliovirus IRES also acts to target vhs-dependent cleavage events. To this end, we constructed pCITE derivatives in which the EMCV IRES element in pCITE and pSexAI IRES was replaced with a 600-nt fragment corresponding to the poliovirus type 2 IRES, yielding pCITE Msc/RI P2 and pSexAI P2, respectively.
Internally labeled pCITE Msc/RI P2 RNA gave rise to ca. 1,800- and 600-nt products at early times (Fig. 7A). These fragments comigrated with those generated from pCITE RNA and are therefore likely correspond to the 3′ and 5′ fragments, respectively, produced by cleavage near the 3′ end of the poliovirus IRES. pCITE Msc/RI P2 RNA also produced a ca. 450-nt fragment (Fig. 7A) which we have yet to definitively identify. Primer extension revealed that the IRES-directed cleavages occurred at exactly the same sites downstream of the IRES element in both RNAs (Fig. 7B), strongly suggesting that both elements function in the same way to target vhs-dependent cleavage events.
Taken in combination, these data indicate that the poliovirus IRES targets vhs-dependent cleavage to 3′-flanking sequences when it is placed at the 5′ end of the pCITE transcript. To determine if the poliovirus IRES retains this activity when it is placed at an internal site, we compared the degradation profile of cap-labeled pSexAI P2 RNA to that of the pSexAI IRES transcript (Fig. 7C). As described above, pSexAI IRES RNA gives rise to a novel 1,400-nt 5′ fragment representing cleavage downstream of the internal IRES, in addition to the 20- to 600-nt products observed with the parental pCITE Msc/RI construct (Fig. 7C). pSexAI P2 RNA also gave rise to the ca. 1,400-nt fragments, albeit at much lower levels than pSexAI IRES (Fig. 7C). These data therefore indicate that the poliovirus IRES displays weak but detectable targeting activity when it is located at an internal site.
The IRES targeting function does not require ribosomes.
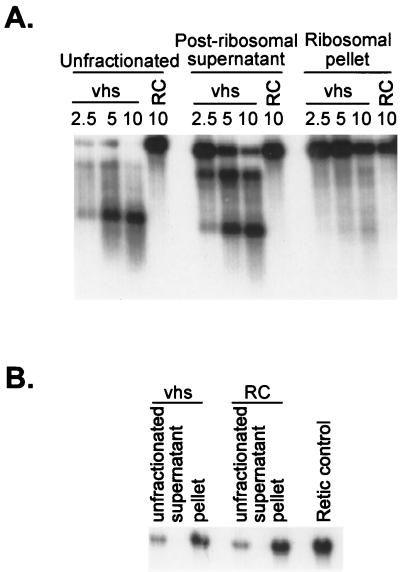
vhs-induced RNA degradation occurs in the presence of agents that block translational initiation and elongation in HSV-1-infected cells (13, 59, 67), and the RNA-destabilizing activity present in extracts of infected cells partitions with the postribosomal fraction (64). These data indicate that an RNA need not be actively translated in order to be degraded. Consistent with this conclusion, we have shown that vhs-dependent degradation of SRPα does not require ribosomes in the RRL in vitro system (7). As described above, the IRES-dependent cleavage events described herein appear to take place independently of those that occur over the 5′ quadrants of SRPα and pCITE RI/MscI RNAs (Fig. 3 and 5). This observation, and the fact that IRES elements serve to recruit translation initiation components and ribosomes to mRNAs, prompted us to ask whether ribosomes are required for the IRES to target vhs-dependent cleavage events to 3′-flanking sequences (Fig. 8). To answer this question, vhs was first generated in RRL and then the ribosomes were removed from the lysate by ultracentrifugation. The extent of ribosomal clearance was verified by assaying the postribosomal supernatant for rabbit 18S rRNA by Northern blot hybridization (Fig. 8B). We found that the postribosomal supernatant retained the ability to cleave internally labeled pCITE RNA downstream of the EMCV IRES, while the ribosomal pellet displayed substantially less activity. Inasmuch as the postribosomal fraction was devoid of detectable 18S rRNA, these data indicate that ribosomes are not required for the targeting activity of the IRES.
FIG. 8.
IRES-targeted cleavage occurs in the absence of ribosomes. vhs was translated in RRL, and then ribosomes were removed by ultracentrifugation as previously described (7). (A) The postribosomal supernatant and ribosomal pellets were combined with uncapped, internally labeled pCITE RNA, and samples recovered at various times (in minutes) were analyzed by agarose-formaldehyde gel electrophoresis as for Fig. 1. RC, blank RRL control. (B) Northern blot analysis of the postribosomal fractions and controls, using a 5′-32P-labeled oligonucleotide probe complementary to rabbit 18S rRNA.
DISCUSSION
Presently available data indicate that vhs selectively targets mRNA in vivo and in vitro (7, 34, 49, 50, 75). However, the basis for this apparent selectivity remains obscure. We and others have previously demonstrated that the in vitro vhs-induced RNA cleavage activity is independent of the 5′ cap structure and 3′ poly(A) tail (7, 75), thereby excluding the two most obvious structural features that distinguish mRNAs from other cytoplasmic RNA species. Thus, uncovering the basis for substrate recognition by the vhs-dependent endoribonuclease is of considerable interest. The experiments described in this report demonstrate that RNA substrates bearing the EMCV IRES are preferentially cleaved by the vhs-dependent endoribonuclease at multiple sites clustered in a narrow zone located immediately downstream of the IRES. This selective targeting of cleavage events was observed when the IRES was placed at the 5′ end of the pCITE-1 substrate or moved to internal sites in this and one other, unrelated RNA (Fig. 1 to 6, and additional data not shown). Taken together, these observations indicate that the targeting phenomenon does not depend on the sequence context or the location of the IRES within the transcript. The cleavages provoked by the inserted IRES occur at sites that are not preferentially used in substrates lacking the element (Fig. 3 to 6), demonstrating that the IRES behaves as a movable element that targets cleavage to 3′-flanking sequences. The unrelated poliovirus IRES displayed detectable but weaker activity, demonstrating that the EMCV IRES is not unique in this regard and raising the possibility that a variety of IRES elements are capable of similarly targeting vhs-dependent cleavage events.
How do the EMCV and poliovirus IRES elements target vhs-dependent cleavage events to 3′-flanking sequences? One intriguing possibility is that this activity reflects the translational initiation function of these elements. As reviewed in the introduction, picornavirus IRES elements bind translational initiation factors that recruit the 40S ribosomal subunit to the RNA, thereby promoting cap-independent initiation of translation. Although our data demonstrate that ribosomes are not required for IRES-directed cleavage, it is possible that the vhs-dependent endoribonuclease activity is delivered to the RNA substrate through interactions with one or more of the translation initiation factors that act upstream of ribosome loading (e.g., eIF3, eIF4A, eIF4B, and eIF4G). This is an attractive hypothesis, since it could provide a functional link between vhs and the translational apparatus and thus potentially explain how mRNAs (including those that lack an IRES) are selectively targeted for degradation in vivo. Moreover, this mechanism would preferentially target those mRNAs that are translated at the highest rate. Two additional features of our data are consistent with this model. First, the cluster of vhs-dependent cleavages provoked by the IRES in the pCITE-1 RNA is located around the site where the 40S ribosomal subunit loads (i.e., the initiation codon). Second, the EMCV IRES is substantially more active than the poliovirus IRES in targeting vhs-dependent cleavage, particularly when it is placed at an internal site in the RNA substrate (Fig. 7). This difference correlates with the relative translational initiation efficiency of these elements, since the EMCV IRES is significantly more active in promoting translation than the poliovirus IRES in RRL in vitro and in some cell lines in vivo (1, 2). The hypothesis that translation initiation factors serve to selectively target vhs activity to mRNAs is also generally consistent with our previous observation that vhs-induced degradation of SRPα mRNA (lacking an IRES) appears to initiate by endoribonucleolytic cleavage events clustered over the 5′ quadrant of the RNA (7).
Several lines of evidence could be interpreted to argue against the aforementioned hypothesis. First, in vivo experiments using translational inhibitors have demonstrated that mRNAs need not be engaged in ongoing translation to be targeted by vhs (59, 67). However, all of the drugs used in these studies act at, or downstream of, ribosome loading onto the mRNA. Therefore, these data do not necessarily exclude a role for initiation factors that function to recruit ribosomes to the mRNA. Second, the hypothesis that vhs is recruited to mRNAs through interactions with translation initiation factors predicts that the 5′ cap should markedly stimulate degradation of mRNAs that lack an IRES. However, we have previously shown that the presence of a 5′ cap does not detectably alter the rate or mode of degradation of SRPα mRNA in the rabbit reticulocyte in vitro system (7). However, it is worth noting that translation initiation in RRLs is relatively cap independent (44). Therefore, this line of evidence does not definitively exclude the hypothesis. Third, Zelus et al. (75) have shown that extracts of partially purified HSV-1 virions contain vhs-dependent ribonuclease activity. These data suggest, but do not prove, that vhs displays detectable activity in the absence of translation initiation factors. If so, then it would be highly informative to determine whether IRES elements retain their ability to target vhs-induced cleavage in such virion extracts.
Alternatively, it is possible that targeting is mechanistically unrelated to translation and instead directly depends on one or more structural features of the IRES element. We can envision at least three distinct structure-based mechanisms that could give rise to the observed cleavage pattern.
(i) The IRES might serve as a preferred loading site for the vhs-dependent endoribonuclease. In this scenario, the nuclease directly recognizes and binds to the IRES (likely through one or more features of the extensive secondary structure adopted by the element), then tracks along the RNA into flanking sequences until it encounters relatively unstructured regions that are susceptible to cleavage. Our observation that cleavage occurs only in 3′-flanking sequences could be readily explained under this scenario if vhs tracks exclusively in a 5′-to-3′ direction. In this context, it is interesting that vhs displays weak but significant amino acid sequence similarity to the fen-1 family of nucleases that are involved in DNA replication and repair (6). fen-1 loads onto the 5′ end of DNA substrates, then tracks in a 3′ direction until it encounters structural features that trigger cleavage (for a review, see reference 42). Moreover, recent evidence suggests that fen-1 interacts with RNA substrates in a similar fashion (65).
The hypothesis that vhs tracks along the RNA in the 5′-to-3′ direction in search of cleavage sites is interesting, since it also potentially explains our observation that two transcripts lacking an IRES are initially cleaved at multiple sites located over the 5′ quadrant of the RNA (7) (Fig. 3). Specifically, if one assumes that the 5′ end of the RNA also serves as a preferred loading site for the vhs-dependent nuclease (as is the case for fen-1), then 5′ loading and tracking would give rise to the observed pattern.
(ii) The highly structured IRES might serve as a barrier to the movement of the nuclease along the RNA, resulting in the accumulation of the enzyme and clustering of cleavage events at the boundary of the IRES. To accommodate the observation that only 3′-flanking sequences are targeted, one would likely have to propose that the nuclease tracks in a 3′-to-5′ direction. This requirement is not obviously consistent with our observations that transcripts containing an internal IRES are also cleaved over their 5′ quadrant and that these 5′ cleavage events appear to occur independently of those provoked by the IRES. Indeed, these observations are more compatible with the first scenario as described above. Moreover, our preliminary results indicate that the highly structured human immunodeficiency virus TAR element does not serve to target vhs-dependent cleavage, arguing that targeting requires specific structural features rather than secondary structure per se (53a).
(iii) The vhs-dependent nuclease might directly recognize and cleave the 3′ junction between highly structured and relatively unstructured regions. Although we cannot exclude this possibility, we note that it does not directly predict that the IRES-induced cleavages would be distributed over a fairly broad zone (which in the case of the pSexAI IRES transcript extends over ca. 300 nt [Fig. 3]). This observation seems more compatible with the first two scenarios.
The possibility that the vhs-dependent nuclease directly recognizes a structural feature of the IRES raises questions about the possible biological significance of this activity. Perhaps vhs is designed to selectively target cellular or viral transcripts that contain IRES elements or other highly structured regions. Alternatively, the activity might mirror another function of vhs. As noted above, vhs displays limited but significant amino acid sequence similarity to the fen-1 family of nucleases (6). These enzymes are structure-specific endonucleases involved in DNA replication and repair (for a review, see reference 42). fen-1 specifically recognizes and cleaves 5′ flap structures in DNA substrates by loading onto the 5′ end of the unpaired strand and tracking in the 3′ direction until it reaches the base of the flap. fen-1 also performs structure-specific cleavage of RNA substrates, at the 5′ base of stem-loops (65). The RNase activity of fen-1 appears to mirror its DNase activity in that fen-1 likely loads at the 5′ end of the RNA and then tracks 5′ to 3′ until it encounters the secondary-structure elements that trigger cleavage. By analogy with fen-1, it is conceivable that vhs is capable of structure-specific cleavage of both DNA and RNA substrates.
We have previously reported that vhs-dependent cleavage events at the extreme 5′ end of SRPα RNA tend to occur between purine residues (7). In the present study, we have mapped 26 additional cleavage sites at a high resolution (Fig. 2, 4, and 6). In addition, Zelus and colleagues mapped six cleavage sites in β-globin RNA (75). The combined data from these three studies indicate that of 37 sites analyzed, 19 occur between purine residues. The others are GC (six), UG (five), AC (two), GU, CU, UU, UC, and CG (one each). In combination, these data suggest that the vhs-induced endoribonuclease displays a relaxed sequence specificity.
The experiments outlined in this report describe a novel and unanticipated effect of picornavirus IRES elements in targeting vhs-dependent endoribonucleolytic cleavage events. We suspect that further analysis of this activity will lead to increased understanding of how vhs targets mRNAs in vivo.
ACKNOWLEDGMENTS
We thank Joanne Duncan, Carol Lavery, and Rob Maranchuk for superb technical assistance and David Andrews and Nahum Sonenberg for gifts of plasmids.
This work was supported by a grant from the National Cancer Institute of Canada [NCI(C)]. J.R.S. was a Terry Fox Senior Scientist of the NCI(C).
REFERENCES
- 1.Borman A M, Bailly J L, Girard M, Kean K M. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 1995;23:3656–3663. doi: 10.1093/nar/23.18.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borman A M, Le Mercier P, Girard M, Kean K M. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckley B, Ehrenfeld E. The cap-binding protein complex in uninfected and poliovirus-infected HeLa cells. J Biol Chem. 1987;262:13599–13606. [PubMed] [Google Scholar]
- 4.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devaney M A, Vakharia V N, Lloyd R E, Ehrenfeld E, Grubman M J. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J Virol. 1988;62:4407–4409. doi: 10.1128/jvi.62.11.4407-4409.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doherty A J, Serpell L C, Ponting C P. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elgadi M M, Hayes C E, Smiley J R. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J Virol. 1999;73:7153–7164. doi: 10.1128/jvi.73.9.7153-7164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Etchison D, Fout S. Human rhinovirus 14 infection of HeLa cells results in the proteolytic cleavage of the p220 cap-binding complex subunit and inactivates globin mRNA translation in vitro. J Virol. 1985;54:634–638. doi: 10.1128/jvi.54.2.634-638.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falcone D, Andrews D W. Both the 5′ untranslated region and the sequence surrounding the start site contribute to efficient initiation of translation in vitro. Mol Cell Biol. 1991;11:2656–2664. doi: 10.1128/mcb.11.5.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feigenblum D, Schneider R J. Modification of eukaryotic initiation factor 4F during infection by influenza virus. J Virol. 1993;67:3027–3035. doi: 10.1128/jvi.67.6.3027-3035.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenwick M L. The effect of herpesviruses on cellular macromolecule synthesis. In: Fraenkel-Conrat H, Wagner R K, editors. Comprehensive virology. Vol. 19. New York, N.Y: Plenum Press; 1984. pp. 359–390. [Google Scholar]
- 12.Fenwick M L, Clark J. Early and delayed shut-off of host protein synthesis in cells infected with herpes simplex virus. J Gen Virol. 1982;61:121–125. doi: 10.1099/0022-1317-61-1-121. [DOI] [PubMed] [Google Scholar]
- 13.Fenwick M L, McMenamin M M. Early virion-associated suppression of cellular protein synthesis by herpes simplex virus is accompanied by inactivation of mRNA. J Gen Virol. 1984;65:1225–1228. doi: 10.1099/0022-1317-65-7-1225. [DOI] [PubMed] [Google Scholar]
- 14.Fenwick M L, Owen S A. On the control of immediate early (alpha) mRNA survival in cells infected with herpes simplex virus. J Gen Virol. 1988;69:2869–2877. doi: 10.1099/0022-1317-69-11-2869. [DOI] [PubMed] [Google Scholar]
- 15.Fenwick M L, Walker M J. Suppression of synthesis of cellular macromolecules by herpes simplex virus. J Gen Virol. 1978;41:37–51. doi: 10.1099/0022-1317-41-1-37. [DOI] [PubMed] [Google Scholar]
- 16.Gallie D R. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 17.Garfinkel M S, Katze M G. Translational control by influenza virus. Selective translation is mediated by sequences within the viral mRNA 5′-untranslated region. J Biol Chem. 1993;268:22223–22226. [PubMed] [Google Scholar]
- 18.Gingras A C, Sonenberg N. Adenovirus infection inactivates the translational inhibitors 4E-BP1 and 4E-BP2. Virology. 1997;237:182–186. doi: 10.1006/viro.1997.8757. [DOI] [PubMed] [Google Scholar]
- 19.Gingras A C, Svitkin Y, Belsham G J, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci USA. 1996;93:5578–5583. doi: 10.1073/pnas.93.11.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gradi A, Imataka H, Svitkin Y V, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gradi A, Svitkin Y V, Imataka H, Sonenberg N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci USA. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–5709. doi: 10.1002/j.1460-2075.1995.tb00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haghighat A, Svitkin Y, Novoa I, Kuechler E, Skern T, Sonenberg N. The eIF4G-eIF4E complex is the target for direct cleavage by the rhinovirus 2A proteinase. J Virol. 1996;70:8444–8450. doi: 10.1128/jvi.70.12.8444-8450.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hentze M W. eIF4G: a multipurpose ribosome adapter? Science. 1997;275:500–501. doi: 10.1126/science.275.5299.500. . (Erratum, 275:1553.) [DOI] [PubMed] [Google Scholar]
- 25.Huang J T, Schneider R J. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell. 1991;65:271–280. doi: 10.1016/0092-8674(91)90161-q. [DOI] [PubMed] [Google Scholar]
- 26.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol Cell Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joachims M, Van Breugel P C, Lloyd R E. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J Virol. 1999;73:718–727. doi: 10.1128/jvi.73.1.718-727.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones F E, Smibert C A, Smiley J R. Mutational analysis of the herpes simplex virus virion host shutoff protein: evidence that vhs functions in the absence of other viral proteins. J Virol. 1995;69:4863–4871. doi: 10.1128/jvi.69.8.4863-4871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerekatte V, Keiper B D, Badorff C, Cai A, Knowlton K U, Rhoads R E. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J Virol. 1999;73:709–717. doi: 10.1128/jvi.73.1.709-717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleijn M, Vrins C L, Voorma H O, Thomas A A. Phosphorylation state of the cap-binding protein eIF4E during viral infection. Virology. 1996;217:486–494. doi: 10.1006/viro.1996.0143. [DOI] [PubMed] [Google Scholar]
- 34.Krikorian C R, Read G S. In vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J Virol. 1991;65:112–122. doi: 10.1128/jvi.65.1.112-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwong A D, Frenkel N. The herpes simplex virus virion host shutoff function. J Virol. 1989;63:4834–4839. doi: 10.1128/jvi.63.11.4834-4839.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwong A D, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc Natl Acad Sci USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kwong A D, Kruper J A, Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Lam Q, Smibert C A, Koop K E, Lavery C, Capone J P, Weinheimer S P, Smiley J R. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 1996;15:2575–2581. [PMC free article] [PubMed] [Google Scholar]
- 40.Lamphear B J, Kirchweger R, Skern T, Rhoads R E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- 41.Lamphear B J, Yan R, Yang F, Waters D, Liebig H D, Klump H, Kuechler E, Skern T, Rhoads R E. Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human coxsackievirus and rhinovirus. J Biol Chem. 1993;268:19200–19203. [PubMed] [Google Scholar]
- 42.Lieber M R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 43.Lin T A, Kong X, Haystead T A, Pause A, Belsham G, Sonenberg N, Lawrence J C., Jr PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- 44.Lodish H F, Rose J K. Relative importance of 7-methylguanosine in ribosome binding and translation of vesicular stomatitis virus mRNA in wheat germ and reticulocyte cell-free systems. J Biol Chem. 1977;252:1181–1188. [PubMed] [Google Scholar]
- 45.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishioka Y, Silverstein S. Degradation of cellular mRNAs during infection by herpes simplex virus. Proc Natl Acad Sci USA. 1977;74:2370–2374. doi: 10.1073/pnas.74.6.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishioka Y, Silverstein S. Requirement of protein synthesis for the degradation of host mRNA in Friend erythroleukemia cells infected with herpes simplex virus type 1. J Virol. 1978;27:619–627. doi: 10.1128/jvi.27.3.619-627.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohlmann T, Rau M, Pain V M, Morley S J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- 49.Oroskar A A, Read G S. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oroskar A A, Read G S. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate-early (alpha) mRNAs. J Virol. 1987;61:604–606. doi: 10.1128/jvi.61.2.604-606.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pak A S, Everly D N, Knight K, Read G S. The virion host shutoff protein of herpes simplex virus inhibits reporter gene expression in the absence of other viral gene products. Virology. 1995;211:491–506. doi: 10.1006/viro.1995.1431. [DOI] [PubMed] [Google Scholar]
- 52.Park Y W, Katze M G. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J Biol Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 53.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 53a.Perez-Parada, J., M. Elgadi, and J. R. Smiley. Unpublished data.
- 54.Pestova T V, Maslova S V, Potapov V K, Agol V I. Distinct modes of poliovirus polyprotein initiation in vitro. Virus Res. 1989;14:107–118. doi: 10.1016/0168-1702(89)90032-4. [DOI] [PubMed] [Google Scholar]
- 55.Pestova T V, Shatsky I N, Hellen C U. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Preiss T, Hentze M W. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 57.Read G S, Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of α (immediate early) viral polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Read G S, Karr B M, Knight K. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J Virol. 1993;67:7149–7160. doi: 10.1128/jvi.67.12.7149-7160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schek N, Bachenheimer S L. Degradation of cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of Vero cells. J Virol. 1985;55:601–610. doi: 10.1128/jvi.55.3.601-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smibert C A, Johnson D C, Smiley J R. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. J Gen Virol. 1992;73:467–470. doi: 10.1099/0022-1317-73-2-467. [DOI] [PubMed] [Google Scholar]
- 61.Smibert C A, Popova B, Xiao P, Capone J P, Smiley J R. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J Virol. 1994;68:2339–2346. doi: 10.1128/jvi.68.4.2339-2346.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smibert C A, Smiley J R. Differential regulation of endogenous and transduced β-globin genes during infection of erythroid cells with a herpes simplex virus type 1 recombinant. J Virol. 1990;64:3882–3894. doi: 10.1128/jvi.64.8.3882-3894.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonenberg N. Regulation of translation and cell growth by eIF4E. Biochimie. 1994;76:839–846. doi: 10.1016/0300-9084(94)90185-6. [DOI] [PubMed] [Google Scholar]
- 64.Sorenson C M, Hart P A, Ross J. Analysis of herpes simplex virus-induced mRNA destabilizing activity using an in vitro mRNA decay system. Nucleic Acids Res. 1991;19:4459–4465. doi: 10.1093/nar/19.16.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stevens A. Endonucleolytic cleavage of RNA at 5′ endogenous stem structures by human flap endonuclease 1. Biochem Biophys Res Commun. 1998;251:501–508. doi: 10.1006/bbrc.1998.9499. [DOI] [PubMed] [Google Scholar]
- 66.Strelow L I, Leib D A. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J Virol. 1995;69:6779–6786. doi: 10.1128/jvi.69.11.6779-6786.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strom T, Frenkel N. Effect of herpes simplex virus on mRNA stability. J Virol. 1987;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Svitkin Y V, Gradi A, Imataka H, Morino S, Sonenberg N. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J Virol. 1999;73:3467–3472. doi: 10.1128/jvi.73.4.3467-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sydiskis R J, Roizman B. The disaggregation of host polyribosomes in productive and abortive infection with herpes simplex virus. Virology. 1967;32:678–686. doi: 10.1016/0042-6822(67)90043-8. [DOI] [PubMed] [Google Scholar]
- 70.Sydiskis R J, Roizman B. Polysomes and protein synthesis in cells infected with a DNA virus. Science. 1966;153:76–78. doi: 10.1126/science.153.3731.76. [DOI] [PubMed] [Google Scholar]
- 71.Tarun S Z, Jr, Wells S E, Deardorff J A, Sachs A B. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Witherell G W, Wimmer E. Encephalomyocarditis virus internal ribosomal entry site RNA-protein interaction. J Virol. 1994;68:3183–3192. doi: 10.1128/jvi.68.5.3183-3192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Young J C, Ursini J, Legate K R, Miller J D, Walter P, Andrews D W. An amino terminal domain containing hydrophobic and hydrophilic sequences binds the signal recognition particle receptor alpha subunit to the beta subunit on the endoplasmic reticulum membrane. J Biol Chem. 1995;270:15650–15657. doi: 10.1074/jbc.270.26.15650. [DOI] [PubMed] [Google Scholar]
- 74.Yueh A, Schneider R J. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev. 1996;10:1557–1567. doi: 10.1101/gad.10.12.1557. [DOI] [PubMed] [Google Scholar]
- 75.Zelus B D, Stewart R S, Ross J. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J Virol. 1996;70:2411–2419. doi: 10.1128/jvi.70.4.2411-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Y, Feigenblum D, Schneider R J. A late adenovirus factor induces eIF-4E dephosphorylation and inhibition of cell protein synthesis. J Virol. 1994;68:7040–7050. doi: 10.1128/jvi.68.11.7040-7050.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]