Abstract
In paramyxovirus transcription, viral RNA polymerase synthesizes each monocistronic mRNA by recognizing the gene start (S) and end (E) signals flanking each gene. These signal sequences are well conserved in the virus family; nevertheless, they do exhibit some variations even within a virus species. In Sendai virus (SeV) Z strain, the E signals are identical for all six genes but there are four (N, P/M/HN, F, and L) different S signals with one or two nucleotide variations. The significance of these variations for in vitro and in vivo replication has been unknown. We addressed this issue by SeV reverse genetics. The luciferase gene was placed between the N and P gene so that recombinant SeVs expressed luciferase under the control of each of the four different S signals. The S signal for the F gene was found to drive a lower level of transcription than that of the other three, which exhibited comparable reinitiation capacities. The polar attenuation of SeV transcription thus appeared to be not linear but biphasic. Then, a mutant SeV whose F gene S signal was replaced with that used for the P, M, and HN genes was created, and its replication capability was examined. The mutant produced a larger amount of F protein and downstream gene-encoded proteins and replicated faster than wild-type SeV in cultured cells and in embryonated eggs. Compared with the wild type, the mutant virus also replicated faster in mice and was more virulent, requiring a dose 20 times lower to kill 50% of mice. On the other hand, the unique F start sequence as well as the other start sequences are perfectly conserved in all SeV isolates sequenced to date, including highly virulent fresh isolates as well as egg-adapted strains, with a virulence several magnitudes lower than that of the fresh isolates. This moderation of transcription at the F gene may therefore be relevant to viral fitness in nature.
Sendai virus (SeV) is an enveloped virus, with a nonsegmented negative-strand RNA genome of 15,384 bases, and belongs to the genus Respirovirus in the family Paramyxoviridae. The SeV genome is organized starting with the short 3′ leader region, followed by six genes encoding the N (nucleocapsid), P (phospho-), M (matrix), F (fusion), HN (hemagglutinin-neuraminidase), and L (large) proteins, and ending with a short 5′ trailer region. In addition to the P protein, the second gene expresses the accessory V and C proteins by a process known as cotranscriptional editing to insert a single nontemplated G residue (34, 35, 41, 42) and by alternative translational initiations, respectively (10, 28). The genome is tightly associated with the N protein, forming the helical ribonucleoprotein complex (RNP). This RNP, but not the naked RNA, is the template for both transcription and replication (30). There is only a single promoter at the 3′ end of the viral RNA polymerase, comprising the P and L proteins (11). By recognizing the short conserved end (E) and restart (S) signals at each gene boundary, the polymerase gives rise to leader RNA and each mRNA (7). There is a trinucleotide intergenic sequence between the E and S signals, which is not transcribed (9, 31). Since the reinitiation efficiency of transcription at each gene boundary is high but not perfect, the transcripts from the downstream genes are less abundant than those from the upstream genes. Therefore, each mRNA is not synthesized in equimolar quantities in infected cells but there is polar attenuation of transcription toward the 5′ end (7, 17, 30).
After translation of the mRNAs and accumulation of translation products, genome replication takes place. Here, the same viral RNA polymerase replicates the same RNP template, but now it somehow ignores the successive E and S signals for mRNAs and generates a full-length antigenomic positive-sense RNP (30). The polymerase enters the promoter at the 3′ end of the positive-sense RNP to generate the genomic negative-sense RNP, which serves as the template for the next round of transcription and replication.
The E sequence (3′-AUUCUUUUUU-5′ in the negative-sense genome) is completely conserved among the six genes in the SeV genome. Five U residues in the latter half allow polymerase slippage, generating poly(A). In contrast, the S signals are variable and are generalized as 3′-UCCCWVUUWC-5′ (9). They are UCCCACUUUC for the P, M, and HN genes, UCCCAgUUUC for the N gene, UCCCuaUUUC for the F gene, and UCCCACUUaC for the L gene. Identical differences are seen in all SeV strains sequenced to date, regardless of differences in isolation procedure, passage history, and virulence for mice, the natural host, suggesting that the variations are locus specific (Table 1). Thus, it has to be defined whether these variations have any significance for SeV replication and pathogenesis. To address this issue, we took advantage of SeV reverse genetics, which has been previously employed for identifying the gene functions and their contributions to viral pathogenesis (21, 22, 26, 32, 33, 37).
TABLE 1.
Gene start sequences of various SeV strains with database accession numbers
| Strain | Passage and propagation host | Mouse virulencea | N UCCCAgUUUC | P UCCCACUUUC | M UCCCACUUUC | F UCCCuaUUUC | HN UCCCACUUUC | L UCCCACUUaC |
|---|---|---|---|---|---|---|---|---|
| Z | Egg | Low | M30202 | M30202 | M30202 | M30202 | M30202 | M30202 |
| Fushimi | Egg | Low | X17218 | X17008 | X53050 | D00152 | X56131 | X58886 |
| Enders | Egg | Low | X00583 | X00583 | X00584 | X00585 | X00586 | X00587 |
| Harris | Egg | Low | M29347 | NDb | K02742 | X02131 | M12397 | ND |
| F1-R | Cell line | Low | M30203 | M30203 | M30203 | M30203 | M30203 | M30203 |
| ts-F1 | Cell line | Low | M30204 | M30204 | M30204 | M30204 | M30204 | M30204 |
| T5 | Cell line | Low | M69040 | M69040 | M69040 | M69040 | M69040 | M69040 |
| Oh-MVC11 | Cell line | Low | AB005796 | AB005796 | AB005796 | AB005796 | AB005796 | AB005796 |
| Oh-M1 | Mouse | High | AB005795 | AB005795 | AB005795 | AB005795 | AB005795 | AB005795 |
| Hamamatsu | Mouse | High | ND | ND | D11446 | D11446 | X57213 | ND |
Low, LD50 values higher than 104 PFU/mouse; high, those less than 102 PFU/mouse.
ND, not determined.
In this study, the firefly luciferase gene fused with the novel upstream E and S signals was inserted in the downstream noncoding region of the N gene. The S signals were changed exactly according to the four naturally occurring variations described above. In the recombinant viruses, the N mRNA transcription starts by its own S signal and terminates by the synthetic E signal within the inserted reporter sequence. Reporter gene expression, which was driven by each of the different S signals, was quantitated and compared each to the other. Thus, assessed reinitiation activity was remarkably lower for the natural S signal of the F gene than for those of the other three. Furthermore, when the natural S signal for the F gene was converted to that with a higher reinitiation activity, the recombinant virus was found to replicate faster, be more cytopathic in cell culture, and be more virulent for mice. SeV replication capability thus appeared to be moderated by a modification of the F gene S signal, and the significance of such moderation is discussed.
MATERIALS AND METHODS
Cell cultures and virus infection.
The monkey kidney-derived cell lines LLCMK2 and CV1, were grown in minimal essential medium (MEM) supplemented with 10% fetal bovine serum at 37°C. Monolayer cultures of these cells were infected with the mutant viruses recovered from cDNAs at an input multiplicity of infection (MOI) of 10 PFU/cell unless otherwise noted and maintained in serum-free MEM. The wild-type SeV (Z strain) recovered from the cDNA (20) was used as a control.
Creation of an insertion site after the N ORF.
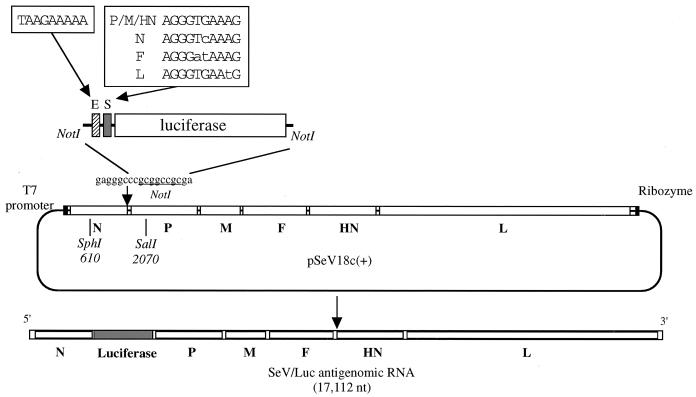
The plasmid pSeV(+) contained a cDNA copy of the full-length SeV antigenome (20) and was used as the starting material for plasmid construction. Eighteen nucleotides (gagggcccgcggccgcga) containing the NotI restriction site were inserted between nucleotides 1698 and 1699 at the 3′ end of SeV genome which was located within the 5′ noncoding (in the negative sense) region of the N gene as shown in Fig. 1 (38). For the insertion, we used site-directed mutagenesis by a PCR-mediated overlap primer extension method (16) essentially according to our previous report (12). Briefly, two primers (NmF, 5′-gagggcccgcggccgcga1699TACGAGGCTTCAAGGTACTT1718-3′, and NmR, 5′-tcgcggccgcgggccctc1698TGATCCTAGATTCCTCCTAC1670-3′) with overlapping 18-nucleotide ends and two outer primers (OP1, 5′-61CAAAGTATCCACCACCCTGAGGAGCAGGTTCCAGACCCTTTGCTTTGC105-3′, and OP2, 5′-2467TTAAGTTGGTVAGTGACTC2449-3′) were synthesized. The first PCRs were performed with the OP1/NmF and the OP2/NmF primer pairs, using pSeV(+) as a template to gave rise to 1.6- and 0.8-kDa fragments, respectively. The second PCR was then performed with the OP1/OP2 primer pair, using the purified 1.6- and 0.8-kDa fragments to generate a single 2.4-kDa fragment with the 18 nucleotides. The 2.4-kDa fragment was purified and digested with SphI and SalI. Plasmid pSeV(+) was cut at positions 610 and 2070 on the SeV genome by these enzymes. The sequence of the resulting 1.47-kDa fragment was verified by sequencing on an AFLII automated DNA sequencer (Pharmacia, Uppsala, Sweden) and replaced with the same fragment of parental pSeV(+), thus generating pSeV18c(+), containing a unique restriction site after the N open reading frame (ORF) (Fig. 1).
FIG. 1.
Construction of plasmid pSeV18c(+) and insertion of the luciferase gene into the downstream region of the N ORF. An 18-nucleotide-fragment designed to contain a NotI site was inserted between nucleotides 1698 and 1699 from the 3′ end (38) of the SeV genome in pSeV(+) by site-directed mutagenesis. The resulting plasmid encoding the SeV antigenome with the 18-nucleotide insertion was named pSeV18c(+). The ORF of the luciferase gene was PCR amplified with four sets of NotI-tagged primers (ESn/NotLr, ESp/NotLr, ESf/NotLr, and ESl/NotLr) from the template plasmid, pHvLuc-RT4 (20), to generate the fragments containing the conserved E signal and each of the different natural S signals placed at the head of the luciferase gene. These amplified fragments were digested with NotI and introduced into the same site of pSeV18c(+). The resulting plasmids, named pSeV(+)SnLuc, pSeV(+)SpLuc, pSeV(+)SfLuc, and pSeV(+)SlLuc, were used to recover the recombinants SeV/SnLuc, SeV/SpLuc, SeV/SfLuc, and SeV/SlLuc, respectively.
Insertion of the luciferase gene with various S elements into pSeV18c(+).
The luciferase gene from the firefly Photinus pyralis derived from pHVlucRT4(−) (20) was amplified by PCR with the following four primer pairs corresponding to the four different S sequences: ESp, 5′-TTgcggccgcGTAAGAAAAACTTAGGGTGAAAGTTCACTTCACGATGGAAGACGGCAAAAACAT-3′; ESn, 5′-TTgcggccgcGTAAGAAAAACTTAGGGTCAAAGTTCACTTCACGATGGAAGACGGCAAAAACAT-3′; ESf, 5′-TTgcggccgcGTAAGAAAAACTTAGGGATAAAGTTCACTTCACGATGGAAGACGGCAAAAACAT-3′; and ESl, 5′-TTgcggccgcGTAAGAAAAACTTAGGGTGAATGTTCACTTCACGATGGAAGACGGCAAAAACAT-3′ and one common reverse primer NotLr, 5′-TCgcggccgcTATTACAATTTGGACTTTCCG-3′. Underlined are a new set of SeV E and S signals connected to the conserved intergenic trinucleotide; the lowercase letters represent the NotI restriction site. The boldface letters represent each unique nucleotide in the S signals. The 1.7-kDa fragments amplified with these primer pairs were purified, digested with NotI, and directly introduced into the NotI site of pSeV18c(+) (Fig. 1). The final constructs were named pSeV(+)SpLuc, pSeV(+)SnLuc, pSeV(+)SfLuc, and pSeV(+)SlLuc, respectively.
Mutagenesis to modify the S signal of the F gene in pSeV(+).
A two-nucleotide exchange was performed on the S sequence of the F gene by two successive steps. At the first step, pSeV(+) was cleaved by BanIII at SeV positions 2088 and 5333 in pSeV(+) and the resulting 3.4-kDa fragment was recloned into the same restriction site of pBluescrit KS(+) (Stratagene, La Jolla, Calif.) to make pB/BanIII. Then, site-directed mutagenesis by a PCR-mediated overlap primer extension method (16) was performed as described above, using two inner primers (mGS1F, 5′-4810CTTAGGGTGAAAGTCCCTTGT4830-3′, and mGS1R, 5′-4830ACAAGGGACTTTCACCCTAAG4810-3′) and two outer primers (M1F, 5′-3931TACCCATAGGTGTGGCCAAAT3951-3′, and T7, 5′-TAATACGACTCACTATAGGGC-3′). Underlined are the mutagenesis points. The first PCR was performed with the primer pairs MF1/mGS1R and T7/mGS1F, using pB/BanIII as a template, and yielded 0.9- and 0.6-kDa fragments, respectively. The second PCR was then performed with the M1F/T7 primer pair using these two purified fragments, generating a single 1.5-kDa fragment with the two nucleotide mutations. This fragment was purified and digested with BanIII and recloned into the same restriction site of pSeV(+) to make pSeV(+)mGSf. The authenticity of sequences to be cloned was verified by nucleotide sequencing.
Virus recovery from cDNAs.
Viruses were recovered from cDNAs essentially according to the previously described procedures (20). Briefly, 2 × 106 LLCMK2 cells in 6-cm-diameter plates were infected with vaccinia virus (VV), vTF7-3, a gift of B. Moss (5), at an MOI of 2 PFU/cell. Then, 10 μg of the parental or mutated pSeV(+) and the plasmids encoding trans-acting proteins, pGEM-N (4 μg), pGEM-P or the mutated pGEM-P (see above) (2 μg), and pGEM-L (4 μg) were transfected simultaneously with the aid of Lipofectin reagent (DOTAP; Boehringer-Mannheim, Mannheim, Germany). The cells were maintained in serum-free MEM in the presence of 40 μg/ml of araC (1-β-d-arabinofuranosylcytosine) and 100 μg/ml of rifampin to minimize VV cytopathogenicity and thereby maximize the recovery rate (20). Forty hours after transfection, cells were harvested, disrupted by three cycles of freezing and thawing, and inoculated into 10-day-old embryonated hen eggs. After 3 days of incubation, the allantoic fluid was harvested. The titers of recovered viruses were expressed in hemagglutination units and PFU/milliliter as described previously (20). The helper VV contaminating the allantoic fluid of the eggs containing 108 to 109 PFU/ml of the recovered SeVs was eliminated by the second propagation in eggs at a dilution of 10−7. This second passage of fluids, stored at −80°C, was used as the seed virus for all experiments.
Luciferase assay.
The expression of luciferase activity from SeV was studied in 5 × 105 CV1 cells/well in 6-well plates at input multiplicities of from 1 to 300 PFU per cell. Under the single-cycle growth conditions, cells were harvested at 0, 6, 14, 20, and 26 h postinfection. Primary virus transcription was studied by incubating infected cells with 100 μg/ml of cycloheximide (Sigma, St. Louis, Mo.) for 12 h, followed by incubation without cycloheximide for an additional 0, 2, and 4 h. The luciferase activity of harvested cells was measured by a luciferase assay kit (Promega, Madison, Wis.) with a luminometer (Luminos CT-9000D, Dia-Iatron, Tokyo, Japan) as described before (12, 20).
RNA extraction and Northern hybridization.
RNAs were extracted from the cells using TRIzol (Gibco BRL) or from culture supernatants and egg allantoic fluids using TRIzol/LS (Gibco BRL). For Northern hybridization, the RNAs were ethanol precipitated, dissolved in formamide-formaldehyde solution, electrophoresed in 0.9% agarose-formamide–MOPS gels, and capillary transferred onto Hibond-N filters (Amersham, Buckinghamshire, United Kingdom). They were probed with 32P-labeled probes made by the multiprime labeling kit (Amersham). For the luciferase probe, the NarI/HincII (1,270-bp) fragment was purified from pHvlucRT4 (20). For the SeV N probe, the PstI/PvuI (1,189-bp) fragment was purified from pGEM-N. For the P probe, 792 bp of the SmaI/SmaI fragment was purified from pGEM-P. For the M, F, HN, and L probes, the NdeI/NdeI (878-bp), BamHI/BamHI (902-bp), ScaI/ScaI (1,108-bp), and BamHI/BamHI (1,654-bp) fragments, respectively, were purified from pSeV(+).
Western blotting.
CV1 cells (2 × 105) grown in 6-well plates were infected at an MOI of 10 PFU per cell with the wild type or with SeV/mSf and harvested at various times postinfection. The cell pellets were lysed and run in sodium dodecyl sulfate–12.5% polyacrylamide gels (29) and immunoprobed with anti-SeV rabbit serum as described previously (19, 20).
Virus passages in eggs and virus detection.
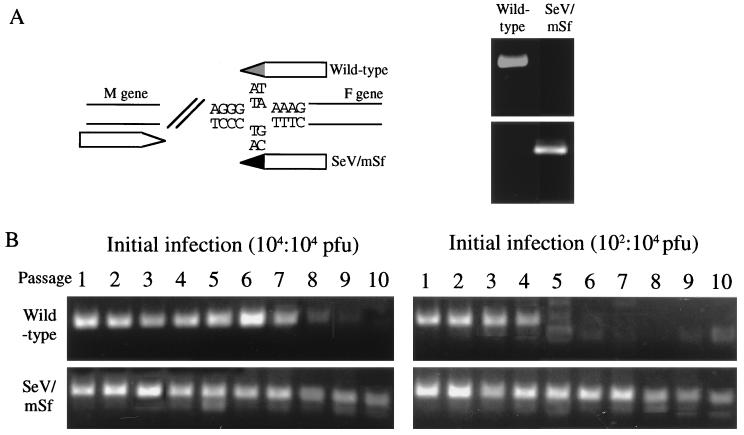
The wild-type SRV and SeV/mSf, which was a mutant virus recovered from the plasmid pSeV(+)mGSf, were coinoculated into two embryonated hen eggs with the respective doses of 104 PFU/egg or both 104 and 102 PFU/egg. Every 3 days postinoculation, the allantoic fluids were harvested and after dilution of 10−6, reinoculated into new eggs. These reinoculations were successively repeated 10 times. The viruses grown in the allantoic fluids were semiquantitatively measured by reverse transcription-PCR (RT-PCR) with specific primer pairs. The RNA was extracted from 25 μl of each allantoic fluid, reverse transcribed with primer HvM (5′-4448TTTTCTCACTTGGGTTAATC4467-3′) at 50°C for 30 min, using Superscript II (Gibco BRL), and heat denatured at 94°C for 2 min. The cDNAs were amplified with primers HvM and GS2WR (5′-4836GCACTCACAAGGGACTTTCA4817-3′) for wild-type SeV and with primers HvM and GS2MR (5′-4836GCACTCACAAGGGACTTTat4817-3′) for SeV/mSf as described previously (21, 28). The lowercase letters represent the mutated dinucleotides. The specific products were analyzed by electrophoresis in agarose gels as described above.
Infection of mice.
Specific-pathogen-free, 3-week-old BALB/c and 4-week-old BALB/c (nu/nu) male mice were purchased from Charles River Laboratories. These mice were infected intranasally with 104, 105, 106, 107, or 108 PFU/mouse of the wild type or SeV/mSf under mild anesthetization with ether (23). Their body weights were individually measured every day up to 14 days. At 0, 1, 3, 5, 7, and 9 days postinfection, three mice in each group were sacrificed and the virus titers in the lungs were measured for BALB/c and BALB/c (nu/nu) mice inoculated with 104 PFU. Consolidation in the lungs was scored at the same time. The consolidation scores are expressed as follows: 0, no visible lesions or atrophy; 1, less than 25% of follicles affected; 2, 25 to 50% of follicles affected; 3, 50 to 75% of follicles affected; 4, more than 75% of follicles affected. When the mice died, one point was added for a score of 5.
RESULTS
Recovery of recombinant viruses expressing the luciferase gene under control of different S signals and their characterization.
To insert the luciferase gene with a synthetic set of E and S signals, a unique NotI site was created downstream of the N ORF within the N gene essentially according to our previous work for the insertion of the same gene in the upstream noncoding region of the N ORF (12). In the present case, insertion of an 18-nucleotide (GAGGGCCCGCGGCCGCGA) stretch containing a NotI restriction site was not deleterious for virus rescue, with the recovery of a recombinant virus possessing full infectivity and replication capability similar to those recovered from the parental pSeV(+) (data not shown). Then, the luciferase genes fused to each of the four S sequences (Sn, Sp, Sf, and Sl) were inserted into the NotI site of the cDNA plasmid, pSeV18c(+) (Fig. 1). In all attempts, recombinant viruses were recovered. They were named SeV/SpLuc, SeV/SnLuc, SeV/SfLuc, and SeV/SlLuc, respectively, according to the S signals used.
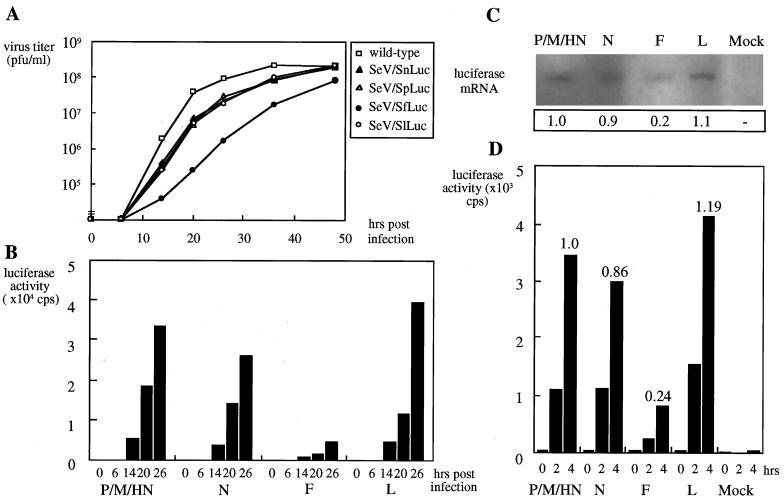
The recombinant viruses were found to replicate more slowly than the wild type in CV1 cells (Fig. 2A), probably because of accommodation of an extra gene as long as 1,728 nucleotides (12). Among the four recombinants, SeV/SfLuc was still more attenuated because of reduced reinitiation activity at this particular S sequence (see below). Luciferase activities expressed from the recombinant SeVs were compared with each other. In all cases, the activities increased as infection proceeded (Fig. 2B) and their levels correlated well with those of luciferase mRNAs, which were identified as monocistronic transcripts by Northern hybridization (data not shown). These data unequivocally demonstrated that the synthetic E and S signals inserted just before the luciferase ORF were correctly recognized by the viral RNA polymerase. Remarkably, there were striking differences in luciferase activities, which appeared to be brought about by the S signal variations. The highest activity was obtained with SeV/SlLuc, and the lowest activity was obtained with SeV/SfLuc at 26 h postinfection (Fig. 2B). SeV/SpLuc and SeV/SnLuc had slightly lower activities than SeV/SlLuc at 26 h postinfection. However, this was not seen at 14 and 20 h postinfection. Thus, the reinitiation capacities of Sp, Sn, and Sl were regarded as comparable.
FIG. 2.
Luciferase expression of SeV/SnLuc, SeV/SpLuc, SeV/SfLuc, and SeV/SlLuc. The recombinant viruses were inoculated onto CV1 cells at an MOI of 10 PFU/cell. Virus titers in the culture supernatants (A) and the luciferase activity in the cells (B) were measured at the times indicated. The recombinant viruses were inoculated onto CV1 cells at an MOI of 100 PFU/cell. The cells were cultured in the presence of cycloheximide for 12 h. Portions of the cells were harvested to prepare RNA and probed with the luciferase probe (C). The intensities relative to that of P/M/HN are also shown. The remainder of the cells was additionally incubated for 0, 2, and 4 h without cycloheximide to allow protein synthesis and lysed to measure the luciferase activity (D).
To see whether the above differences were primarily brought about at the level of transcription but not as part of the replication process, cells infected with the recombinants were incubated in the presence of cycloheximide, which inhibits protein synthesis and hence blocks viral replication requiring de novo viral protein synthesis. Under these conditions, only primary virus transcription catalyzed by the virion-associated RNA polymerase is allowed. After primary transcription and accumulation of viral mRNAs, cycloheximide was washed out from the culture. Cells were either lysed immediately to prepare the viral RNA (Fig. 2C) or incubated for an additional 0, 2, or 4 h to allow protein synthesis for measuring luciferase activities (Fig. 2D). The activities increased as the incubation period after cycloheximide removal was prolonged; SeV/SfLuc was again significantly lower than the other three in luciferase expression. The amounts of luciferase mRNA in each of the virus-infected cell groups correlated well with the activities of luciferase (Fig. 2C). The luciferase activities at 4 h of incubation were normalized to that of SeV/SpLuc, as this type of S signal is shared with three of the six genes. SeV/SnLuc and SeV/SlLuc activities were 0.86 and 1.19, respectively, and thus nearly comparable to SeV/SpLuc. In contrast, SeV/SfLuc reached only 0.24 of the control (Fig. 2D). These results strongly suggested that the signal used for F gene expression possessed a lower reinitiation potential than the other S signals.
Creation of an SeV mutant with an altered S signal for the F gene.
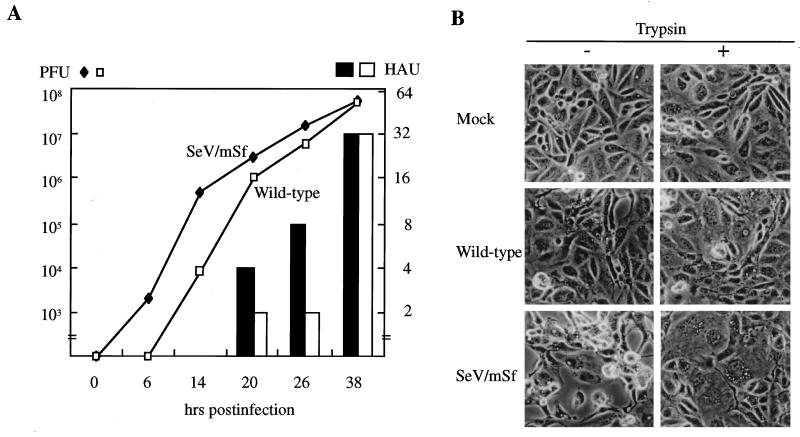
The results described above suggested that there was a down-regulation of transcription at the F gene in the natural genome context of SeV. To substantiate this, we next created the mutant SeV/mSf, whose S signal for the F gene was replaced with that for the P/M/HN gene, and compared its replication with that of the wild type. SeV/mSf was found to grow faster than the wild type in CV1 cells (Fig. 3A). Cytopathogenicity, as manifested by cell rounding and cell detaching in the absence of trypsin and by cell fusion in the presence of exogenous trypsin to proteolytically activate the F glycoprotein, was greater for the mutant than for the wild type (Fig. 3B).
FIG. 3.
Growth kinetics and cytopathogenicity of SeV/mSf. (A) The titers of the wild type and mutant SeV/mSf were measured at the times indicated, under single-cycle conditions. Open bars and filled bars represent hemagglutination units (HAU) of the wild type and mutant viruses, respectively. Lines with open and filled symbols represent PFU per milliliter of the wild-type and mutant viruses, respectively. (B) CV1 cells were infected with the wild type or SeV/mSf at an MOI of 20 PFU/cell in the presence (+) or absence (−) of trypsin. The pictures were taken at 48 h postinfection.
Expression of SeV/mSf genes.
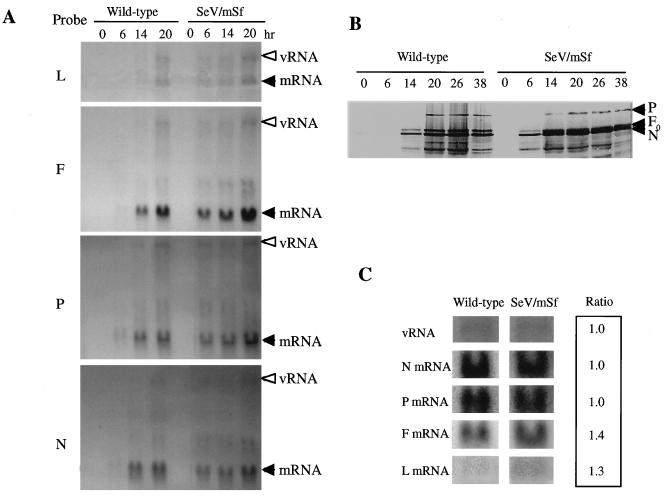
The mRNA levels in CV1 cells infected with the wild-type and mutant viruses were compared by Northern blotting at various times postinfection. As shown in Fig. 4A, the F and L transcripts from SeV/mSf were detected earlier and reached remarkably higher levels than those from the wild-type infection. The P and N transcripts were also detected earlier in SeV/mSf infection, although the peak levels were comparable. Accordingly, the levels of F0 protein in the former were significantly higher than in the latter (Fig. 4B) at any time point throughout infection. The downstream gene products, HN and L, were not well resolved in this experiment. Enhanced expression of the F and L genes, but not of the N and P genes, was also clearly seen under the conditions of blocking de novo protein synthesis by cycloheximide (Fig. 4C). These results again unequivocally demonstrated that the S signal naturally occurring in F gene transcription possesses a lower reinitiation/promoter activity and hence down-regulates expression of F and downstream genes. Probably because of enhanced L gene expression, the virion RNA levels were higher for the mutant than for the wild type throughout infection (Fig. 4A). Earlier detection of mRNAs in mutant-infected cells, as demonstrated in Fig. 4A, might be also due to increased L gene expression.
FIG. 4.
Intracellular expression of viral genes. (A) CV1 cells infected with the wild type or SeV/mSf virus were analyzed by Northern hybridization with the viral N, P, F, or L gene probes at various times postinfection. The positions of mRNAs and genomic/antigenomic RNA (vRNA) are marked. (B) Intracellular expression of viral genes in CV1 cells was analyzed by Western blotting with anti-SeV antibody at the times (hours) indicated at the top of each lane. (C) CV1 cells were infected with either virus at an MOI of 100 PFU in the presence of cycloheximide. RNAs were extracted after 12 h of inoculation and analyzed by Northern hybridization. Approximate ratios of SeV/mSf to wild-type mRNA were determined with the BAS 2000 Image Analyzer.
Successive copassages of the wild type and SeV/mSf in embryonated hen eggs.
Although wild-type SeV replicated more slowly than SeV/mSf in CV1 cells as shown in Fig. 2A, the possibility still remained that the naturally occurring down-regulation of transcription for the F and downstream genes would be advantageous for the persistence of SeV in the host, chicken embryos, than the artificially introduced up-regulation. We thus examined whether either the wild type or SeV/mSf would compete each other out during copassages of the two viruses in eggs. Coinfection was initiated with 104 PFU/egg of the two viruses or with 102 PFU/egg of the wild type and 104 PFU/egg of the mutant. At each inoculation up to the 10th passage, specific RT-PCR was performed to amplify either of the two viral genomes isolated from virions in the allantoic fluids, as shown in Fig. 5A. It was found that the wild-type genome was competed out by the eighth passage in the case of a 104-104 initial inoculation and by the fifth passage following a 102-106 initial inoculation (Fig. 5B). In control experiments, each virus was individually passaged and the genome sequences were determined. The results indicated that both viral genomes were stably maintained during 10 successive passages without any nucleotide change in the regions sequenced. These data indicated that the naturally occurring F gene S signal conferred no replication advantage on SeV at least in ovo.
FIG. 5.
Competition assays of the wild type and SeV/mSf in serial copassages. (A) Specific primer sets (left) detect either of the viral RNAs (right). (B) Each passage was initiated with input doses of 104 (SeV/mSf) and 104 (wild-type virus) PFU/egg or 104 (SeV/mSf) and 102 (wild-type virus) PFU/egg. The allantoic fluids were harvested every 3 days, diluted to 10−6, and coinoculated into new eggs serially up to 10 passages. Viral RNAs were extracted and analyzed by a one-step RT-PCR method, using the specific primer sets.
Pathogenicity of wild-type SeV and SeV/mSf for mice.
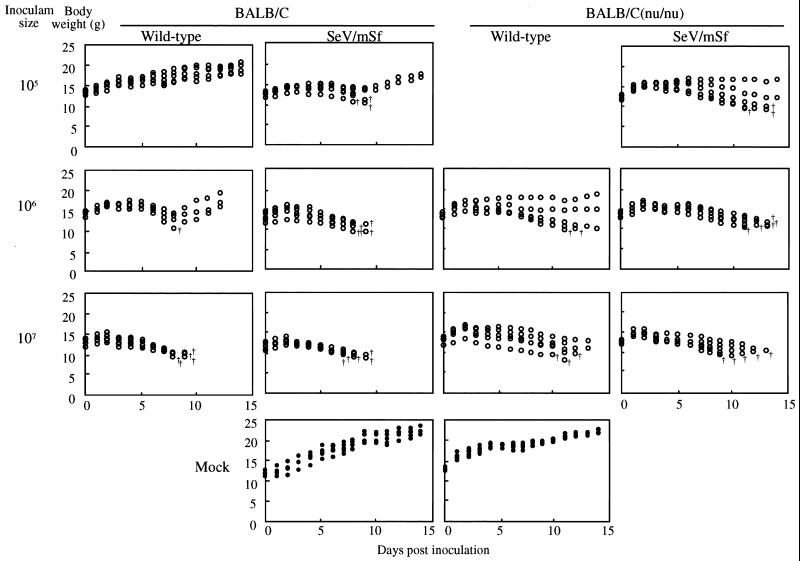
The final issue addressed in this work was whether the mutant SeV/mSf would replicate faster and be more pathogenic than the wild type in mice, the natural host, for which much more complex conditions exist than for cultured cells or eggs. Infections of BALB/c mice with the wild-type and mutant viruses were initiated intranasally at doses of 104, 105, 106, 107, and 108 PFU/mouse (Fig. 6). The mouse body weight gain was strongly disturbed by inoculations of 107 PFU of both viruses. All mice were killed by either virus at similar days postinfection. At 106 PFU, significant differences were found between the two viruses. Infection with SeV/mSf more strongly affected body weight gain than infection with the wild type. The former killed all mice, while the latter killed only one and allowed the remaining to regain the weight. At 105 PFU, all mice infected with the wild type showed a pattern of weight gain nearly comparable to that of mock-infected mice and survived, while those infected with the mutant did not. Thus, SeV/mSf was clearly more virulent than the wild type. The difference in virulence was quantitated by determining the 50% lethal dose (LD50); the LD50 was 1.78 × 106 PFU for the wild type and 7.94 × 104 PFU for the mutant (Table 2). The mutant virus was thus 22 times more virulent than the wild type for the BALB/c strain. These results suggested that the naturally occurring F gene S signal attenuated SeV to some extent so that infected mice survive longer.
FIG. 6.
Body weight gain of normal BALB/c and thymus-deficient BALB/c (nu/nu) mice infected with the wild-type and SeV/mSf viruses. Five mice were inoculated intranasally with various doses of virus (104 to 107 PFU per mouse). The weight gain of mice was measured in grams (top) every day up to 14 days postinoculation. Dead mice are marked by †.
TABLE 2.
LD50 values of the wild-type and mutant viruses for BALB/c and nude mice
| Virus and inoculum | BALB/cb | LD50 | BALB/c (nu/nu)b | LD50 |
|---|---|---|---|---|
| Wild-type | ||||
| 108 | 5/5 | NTb | ||
| 107 | 5/5 | 3/5 | ||
| 106 | 1/4 | 1.78 × 106 | 2/5 | 3.16 × 106 |
| 105 | 0/5 | NT | ||
| 104 | 0/5 | 0/5 | ||
| SeVmSf | ||||
| 108 | 5/5 | NT | ||
| 107 | 5/5 | 5/5 | ||
| 106 | 5/5 | 7.94 × 104 | 5/5 | 7.94 × 104 |
| 105 | 3/5 | 3/5 | ||
| 104 | 0/5 | 0/5 |
No. dead/no. inoculated.
NT, not tested.
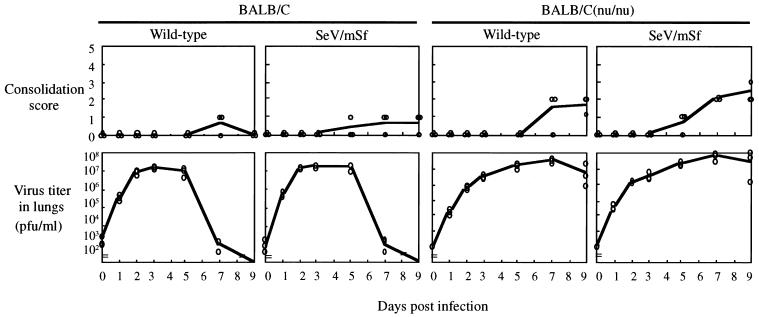
Cytotoxic T lymphocytes (CTL) modulate SeV pathogenesis in two different ways. They contribute to eliminating or clearing the virus from body on the one hand, and on the other, they accelerate disease progression by immunopathological processes. We also examined the pathogenicity of the wild-type and mutant viruses for thymus-deficient nude mice. The LD50 values of each virus were comparable for nude mice and for the parental normal mice, and a similar difference (∼40-fold) between the two viruses was found for the nude mice (Table 2). This result suggested that CTL and other thymus-dependent defenses did not play a major role in the pathogenesis of both wild-type and mutant viruses during the observation period (14 days), at least on the basis of LD50. However, nude mice infected with both the wild-type and mutant viruses survived longer (Fig. 6). They nevertheless supported extended virus replication and increased lung consolidation (Fig. 7). These data suggested that CTL and other thymus-dependent responses played at least a part in viral pathogenesis (immunopathogenesis).
FIG. 7.
Consolidation and viral loads in the lungs of BALB/c and BALB/c (nu/nu) mice. Each mouse was intranasally inoculated with 104 PFU of the indicated viruses. Three mice were sacrificed at 0, 1, 2, 3, 5, 7, and 9 days postinoculation to grade consolidation scores (top) and to determine virus titers in the lungs (bottom). All these values are individually shown for each mouse.
DISCUSSION
All nonsegmented negative-strand RNA genomes possess semiconserved, similarly sized S and E signals. Conservation of the sequences of these signals is extensively high within a viral genus and within a family and is extremely high among genes of a given virus species (4). However, there are some variations even within a virus species. In the case of the SeV S signal, there are four variations (9). Remarkably, these variations are fixed perfectly at the same respective genes of all strains, including fresh isolates, which are highly virulent for the natural host, rodents, and those isolated decades ago, passaged under different laboratory conditions and attenuated to various extents (Table 1). This fact suggested the possibility that the regulation of transcription, which presumably depends on the S motifs, is important for SeV life cycle and ecology.
Previously, several studies with model template systems of various nonsegmented negative-strand RNA viruses indicated that the S signals were indeed critically required for transcriptional initiation but able to tolerate variations in sequence to some extent (1, 2, 18, 25, 36, 40). Certain nucleotide exchanges in their S signals were shown to initiate less transcription, suggesting that gene expression was also modulated by naturally occurring variations in the viral life cycle (26, 27, 39). However, in the model template systems, any event required early in the natural life cycle, like primary transcription, is bypassed by the successive and constant supply of trans-acting proteins (32). The transcription and replication of minigenomes are uncoupled in these systems. T7 polymerase-expressing VV often used to produce the trans-acting proteins might mask the subtle effects of mutations by, for example, the posttranscriptional modifications by VV encoding capping enzyme. In addition, the transfection efficiencies might not be equal throughout the experiment. Thus, to address the roles of S and E signals comprehensively, it has been necessary to introduce relevant mutations into a full-length viral genome (3, 15).
The results obtained here by SeV reverse genetics clearly showed that the S sequence for the F gene was remarkably less potent in initiation than the other three S sequences. That the reduced luciferase gene expression by the F-specific signal was indeed caused primarily at the transcriptional level but was not a secondary result of replication was confirmed by blocking de novo protein synthesis and hence eliminating genome replication (Fig. 2). The primary transcription experiment further assessed that the reinitiation activity driven by the S sequence for the F gene was approximately one fourth those of the other three. These observations are in good agreement with the previous observation that the amounts of N, P, and M mRNAs in SeV-infected cells were almost equally high, but F and HN mRNAs were present in about threefold-lower amounts (17). The results taken together may argue against the view of simple polar attenuation of transcription for SeV, which is essentially linear toward the 5′ end of template. Rather, the transcriptional attenuation between the N and P genes as well as that between the P and M genes may be small or negligible. Although the main issue was the steep transcriptional attenuation at the subsequent F gene, the possibility that the low level of mRNA might be the result of the shorter life of the transcripts due to the differences at the S signal could not be ruled out. There is no plausible explanation for the extremely low copy numbers of L mRNA (about 1/30 those of N, P, and M mRNAs) (17). The reinitiation activity of the S sequence for the L gene studied by recombinant simian virus 5 with the reporter green fluorescent protein gene was considerably low (15). The same signal for respiratory syncytial virus (RSV), studied by a model template system, appeared to be as active as the other RSV signals (27). The same was true for SeV as shown here. The contribution of the S signal to L gene expression thus appeared to be variable among the viruses, or the copy number of the extremely long L gene transcripts may be influenced by some other factors, such as processivity of the polymerase.
The reinitiation capacity of different S sequences was then assessed by replacing the natural S sequence of the F gene with that of the P/M/HN genes and by examining the replication capability of the recovered virus (SeV/mSf) in cultured cells, in ovo, and in mice. It was unequivocally demonstrated that the inserted S sequence enhanced F and downstream gene expression, again at the transcriptional level (Fig. 4). The mutant virus with the new S sequence for the F gene replicated faster and was more cytopathic in cultured cells (Fig. 3) and competed out the wild-type virus in eggs (Fig. 5). Thus, the naturally occurring S signal for the F gene appears to moderate the initiation of F gene transcription and viral replication. Since polymerase enters the 3′ end of the genome in transcription as well as in replication, once moderated at the F gene, transcription of the downstream genes including the L gene encoding the catalytic subunit of polymerase was also moderated. Then, the next round of both replication and transcription could be affected, ultimately leading to reduced virion RNA synthesis and progeny production.
The in vivo study showed that SeV/mSf had 20 times lower LD50 values and hence was more virulent than the wild type for BALB/c mice (Table 2). CTL generally play a major role in virus clearance by eliminating virus-infected cells. They are also important for immunopathological processes leading to damage of infected cells and tissues. The increased level of F protein expression from the mutant virus would lead to a stronger CTL response. These aspects of CTL appeared to be involved not only in wild-type SeV pathogenesis but also in increased pathogenicity of SeV/mSf as determined by LD50. Thus, the increased pathogenicity of SeV/mSf may simply be attributable to its increased replication capacity.
So far, our data suggest that the F gene S signal imposes a restriction of productive infection on SeV. Its presence in the natural SeV genome did not appear to be advantageous at least in ovo in competition assays with a signal with a higher promoter capacity (Fig. 5). It is therefore quite difficult to conceptualize the basis for the strong conservation of an S sequence with a lower initiation activity at the same position in SeV genomes sequenced to date, including those of highly virulent fresh isolates. One estimate suggested that the LD50 of a fresh isolate is as low as 10 PFU/mouse, whereas that of the egg-adapted strain used here was as high as 105 to 106 PFU/mouse (21, 22, 28). If such a virulent field strain is further potentiated by incorporating an S signal with a higher reinitiation activity into the F gene, infected rodents would be killed so rapidly that they would have less opportunity to transmit virus to new hosts. Indeed, it was recently shown that some newly emerging influenza virus isolates of the H5N1 subtype were too virulent to be transmitted to a neighbor host in a mouse colony (6, 8). Moderation of replication by transcriptional attenuation at the F gene may thus be advantageous for SeV to persist in nature.
RNA-dependent RNA polymerase is error prone (13, 24). The natural S signal for the F gene was here shown to be less advantageous than that with a higher reinitiation activity in hen eggs. Thus, one can predict that SeV with an F start of higher reinitiation capacity would evolve during serial passages in eggs. However, this has not happened over the years, as indicated by strict conservation of the natural F start sequence in all strains sequenced to date. A similar situation is seen for another SeV cis-acting element, the editing motif in the P gene, as it is totally dispensable or even restrictive for viral replication under laboratory conditions but strictly conserved in SeV isolates, including egg-adapted strains (14, 22). These sequence conservations may be somehow required for optimizing replication capacity even in cultured cells and eggs. Alternatively, nucleotide misincorporation may not be equal throughout the genome but is somehow restricted in these regions. Based on this hypothesis, S signal mutation that down-regulates F expression could be beneficial for SeV to persist in nature, since it was fixed early in the evolutionary process and then has been maintained in laboratory strains.
ACKNOWLEDGMENTS
We thank D. Kolakofsky and B. Moss for the gifts of pGEM-N, pGEM-P, and pGEM-L and vTF7-3, respectively. The Genome Net services and databases were provided by the Human Genome Center (HGC) at the University of Tokyo.
This work was supported by research grants from the Ministry of Education, Science, Sports and Culture, Japan, and from the Bio-Oriented Technology Research Advancement Institution (BRAIN), Japan.
REFERENCES
- 1.Barr J N, Whelan S P J, Wertz G W. Role of the intergenic dinucleotide in vesicular stomatitis virus RNA transcription. J Virol. 1997;71:1794–1801. doi: 10.1128/jvi.71.3.1794-1801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr J N, Whelan S P, Wertz G W. cis-acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J Virol. 1997;71:8718–8725. doi: 10.1128/jvi.71.11.8718-8725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bukreyev A, Camargo E, Collins P L. Recovery of infectious respiratory syncytial virus expressing an additional, foreign gene. J Virol. 1996;70:6634–6641. doi: 10.1128/jvi.70.10.6634-6641.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldmann H, Muhlberger E, Randolf A, Will C, Kiley M, Sanchez A, Klenk H D. Marburg virus, a filovirus: messenger RNAs, gene order, and regulatory elements of the replication cycle. Virus Res. 1992;24:1–19. doi: 10.1016/0168-1702(92)90027-7. [DOI] [PubMed] [Google Scholar]
- 5.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley A J, Kawaoka Y. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol. 1999;73:3184–3189. doi: 10.1128/jvi.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glazier K, Raghow R, Kingsbury D W. Regulation of Sendai virus transcription: evidence for a single promoter in vivo. J Virol. 1997;21:863–871. doi: 10.1128/jvi.21.3.863-871.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gubareva L V, McCullers J A, Bethell R C, Webster R G. Characterization of influenza A/HongKong/156/97 (H5N1) virus in a mouse model and protective effect of zanamivir on H5N1 infection in mice. J Infect Dis. 1998;178:1592–1596. doi: 10.1086/314515. [DOI] [PubMed] [Google Scholar]
- 9.Gupta K C, Kingsbury D W. Complete sequences of the intergenic and mRNA start signals in the Sendai virus genome: homologies with the genome of vesicular stomatitis virus. Nucleic Acids Res. 1984;12:3829–3841. doi: 10.1093/nar/12.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta K C, Ono E. Stimulation of Sendai virus C′ protein synthesis by cycloheximide. Biochem J. 1997;321:811–818. doi: 10.1042/bj3210811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamaguchi M, Yoshida T, Nishikawa K, Naruse H, Nagai Y. Transcriptive complex of Newcastle disease virus. I. Both L and P proteins are required to constitute an active complex. Virology. 1983;128:105–117. doi: 10.1016/0042-6822(83)90322-7. [DOI] [PubMed] [Google Scholar]
- 12.Hasan M K, Kato A, Shioda T, Sakai Y, Yu D, Nagai Y. Creation of an infectious recombinant Sendai virus expressing the firefly luciferase gene from the 3′ proximal first locus. J Gen Virol. 1997;78:2813–2820. doi: 10.1099/0022-1317-78-11-2813. [DOI] [PubMed] [Google Scholar]
- 13.Hausmann S, Jacques J P, Kolakofsky D. Paramyxovirus RNA editing and the requirement for hexamer genome length. RNA. 1996;2:1033–1045. [PMC free article] [PubMed] [Google Scholar]
- 14.Hausmann S, Garcin D, Morel A-S, Kolakofsky D. Two nucleotides immediately upstream of the essential A6G3 slippery sequence modulate the pattern of G insertions during Sendai virus mRNA editing. J Virol. 1999;73:343–351. doi: 10.1128/jvi.73.1.343-351.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He B, Paterson R G, Ward C D, Lamb R A. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- 16.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 17.Homann H E, Hofschneider P H, Neubert W J. Sendai virus gene expression in lytically and persistently infected cells. Virology. 1990;177:131–140. doi: 10.1016/0042-6822(90)90467-6. [DOI] [PubMed] [Google Scholar]
- 18.Hwang L N, Englund N, Pattnaik A K. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J Virol. 1998;72:1805–1813. doi: 10.1128/jvi.72.3.1805-1813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato A, Fujino M, Nakamura T, Ishihama A, Otaki Y. Gene organization of chicken anemia virus. Virology. 1995;209:480–488. doi: 10.1006/viro.1995.1280. [DOI] [PubMed] [Google Scholar]
- 20.Kato A, Sakai Y, Shioda T, Kondo T, Nakanishi M, Nagai Y. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells. 1996;1:569–579. doi: 10.1046/j.1365-2443.1996.d01-261.x. [DOI] [PubMed] [Google Scholar]
- 21.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 1997;16:578–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato A, Kiyotani K, Sakai Y, Yoshida T, Shioda T, Nagai Y. Importance of the cysteine-rich carboxyl-terminal half of V protein for Sendai virus pathogenesis. J Virol. 1997;71:7266–7272. doi: 10.1128/jvi.71.10.7266-7272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiyotani K, Takao S, Sakaguchi T, Yoshida T. Immediate protection of mice from lethal wild-type Sendai virus (HVJ) infections by a temperature-sensitive mutant, HVJpi, possessing homologous interfering capacity. Virology. 1990;177:65–74. doi: 10.1016/0042-6822(90)90460-9. [DOI] [PubMed] [Google Scholar]
- 24.Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo L, Fearns R, Collins P L. The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome. J Virol. 1996;70:6143–6150. doi: 10.1128/jvi.70.9.6143-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo L, Grosfeld H, Cristina J, Hill M G, Collins P L. Effect of mutation in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J Virol. 1996;70:6892–6901. doi: 10.1128/jvi.70.10.6892-6901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuo L, Fearns R, Collins P L. Analysis of gene start and gene end signals of human respiratory syncytial virus; quasi-templated initiation at position 1 of the encoded mRNA. J Virol. 1997;71:4944–4953. doi: 10.1128/jvi.71.7.4944-4953.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuronati A, Kiyotani K, Kato A, Shioda T, Sakai Y, Mizumoto K, Yoshida T, Nagai Y. The paramyxovirus, Sendai virus, C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells. 1998;3:111–124. doi: 10.1046/j.1365-2443.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. New York, N.Y: Lippincott-Raven; 1996. pp. 1177–1204. [Google Scholar]
- 31.Luk D, Masters P S, Gill D S, Banerjee A K. Intergenic sequences of the vesicular stomatitis virus genome (New Jersey serotype): evidence for two transcription initiation sites within the L gene. Virology. 1987;160:88–94. doi: 10.1016/0042-6822(87)90048-1. [DOI] [PubMed] [Google Scholar]
- 32.Nagai Y. Paramyxovirus replication and pathogenesis. Reverse genetics transforms understanding. Rev Med Virol. 1999;9:83–99. doi: 10.1002/(sici)1099-1654(199904/06)9:2<83::aid-rmv244>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 33.Nagai Y, Kato A. Paramyxovirus reverse genetics is coming of age. Microbiol Immunol. 1999;43:613–624. doi: 10.1111/j.1348-0421.1999.tb02448.x. [DOI] [PubMed] [Google Scholar]
- 34.Park K H, Krystal M. In vivo model for pseudo-templated transcription in Sendai virus. J Virol. 1992;66:7033–7039. doi: 10.1128/jvi.66.12.7033-7039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paterson R G, Lamb R A. RNA editing by G-nucleotide insertion in mumps virus P gene mRNA transcripts. J Virol. 1990;64:4137–4145. doi: 10.1128/jvi.64.9.4137-4145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rassa J C, Parks G D. Molecular basis for naturally occurring elevated readthrough transcription across the M-F junction of the paramyxovirus SV5. Virology. 1998;247:274–286. doi: 10.1006/viro.1998.9266. [DOI] [PubMed] [Google Scholar]
- 37.Sakaguchi T, Kiyotani K, Kato A, Asakawa M, Fujii Y, Nagai Y, Yoshida T. Phosphorylation of the Sendai virus M protein is not essential for virus replication either in vitro or in vivo. Virology. 1997;235:360–366. doi: 10.1006/viro.1997.8701. [DOI] [PubMed] [Google Scholar]
- 38.Shioda T, Hidaka Y, Kanda T, Shibuta H, Nomota A, Iwasaki K. Sequence of 3687 nt from the 3′ end of Sendai virus genome RNA and the predicted amino acid sequences of viral NP, P and C proteins. Nucleic Acids Res. 1983;11:7317–7330. doi: 10.1093/nar/11.21.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stillman E A, Whitt M A. Mutational analyses of the intergenic dinucleotide and the transcriptional start sequence of vesicular stomatitis virus (VSV) define sequences required for efficient termination and initiation of VSV transcripts. J Virol. 1997;71:2127–2137. doi: 10.1128/jvi.71.3.2127-2137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stillman E A, Whitt M A. The length and sequence composition of vesicular stomatitis virus intergenic regions affect mRNA levels and the site of transcript initiation. J Virol. 1998;72:5565–5572. doi: 10.1128/jvi.72.7.5565-5572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas S M, Lamb R A, Paterson R G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell. 1988;54:891–902. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vidal S, Curran J, Kolakofsky D. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J Virol. 1990;64:239–246. doi: 10.1128/jvi.64.1.239-246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]