Abstract
Recently we described rescue of defective Kunjin virus (KUN) RNAs with small deletions in the methyltransferase and RNA polymerase motifs of the ns5 gene, using BHK cells stably expressing KUN replicon RNA (repBHK cells) as helper (A. A. Khromykh et al., J. Virol. 72:7270–7279, 1998). We have now extended our previous observations and report successful trans-complementation of defective KUN RNAs with most of the ns5 gene deleted or substituted with a heterologous (dengue virus) ns5 sequence. Replication of full-length KUN RNAs with 3′-terminal deletions of 136 (5%), 933 (34%), and 1526 (56%) nucleotides in the ns5 gene was complemented efficiently in transfected repBHK cells. RNA with a larger deletion of 2,042 nucleotides (75%) was complemented less efficiently, and RNA with an even larger deletion of 2,279 nucleotides (84%) was not complemented at all. Chimeric KUN genomic RNA containing 87% of the KUN ns5 gene replaced by the corresponding sequence of the dengue virus type 2 ns5 gene was unable to replicate in normal BHK cells but was complemented in repBHK cells. These results demonstrate for the first time complementation of flavivirus RNAs with large deletions (as much as 75%) in the RNA polymerase gene and establish that translation of most of the N-terminal half of NS5 is essential for complementation in trans. A model of formation of the flavivirus replication complex implicating a possible role in RNA replication of conserved coding sequences in the N-terminal half of NS5 is proposed based on the complementation and earlier results with KUN and on reported data with other flaviviruses.
Kunjin virus (KUN) is an Australian member of the Flavivirus genus within the family Flaviviridae and is closely related to other members of the Japanese encephalitis virus subgroup (23). The KUN genome consists of single-stranded RNA of positive polarity comprising 11,022 nucleotides (13) with one long open reading frame coding for 3,433 amino acids in three structural proteins (C, prM, and E) and seven nonstructural (NS) proteins (NS1 to NS5) for which the boundaries of all flavivirus genes were first defined (6). KUN has long been a very useful model for studying the events of flavivirus replication and in particular the composition and functions of the RNA replication complex (RC). Earlier we proposed a model for flavivirus RNA replication demonstrating the role of double-stranded RNA (dsRNA) as the template for RNA synthesis late in infection (4). Our later studies on partial purification of the RC (5), coprecipitations of NS proteins and dsRNA in a radioimmunoprecipitation reaction, and colocalizations of NS proteins and dsRNA defined by electron microscopy using immunogold labelling of cryosections (22, 31, 32) demonstrated involvement of nearly all of the NS proteins in the flavivirus RC. Of particular interest is the role in RNA replication of NS5 protein due to the presence in its sequence of several domains associated with conserved RNA-dependent RNA polymerase (RdRp) motifs (16, 27) and the demonstrated activity for dengue virus type 1 (DEN1) NS5 protein of nonspecific copying of RNA in an in vitro RdRp assay (30). There are currently eight identified conserved RdRp motifs (16, 26, 27) which in the KUN ns5 gene are situated in the 3′-terminal half extending approximately from nucleotides 1370 to 2214 (Fig. 1) (6). These motifs are thought to be involved in substrate binding, RNA binding, catalytic activity, and formation of the proper secondary structure of RdRps (16, 26). The N-terminal half of the flavivirus NS5 protein contains two conserved domains characteristic for methyltransferases (MT motif [Fig. 1]) (17).
FIG. 1.
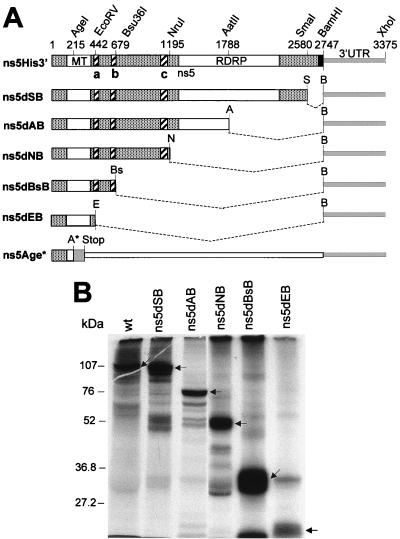
KUN ns5 deletion mutants. (A) Fragments of pBS-based intermediate plasmids representing ns5 and 3′UTR regions. ns5His3′ represents part of the pBSns5His3′UTR sequence that codes for KUN NS5 comprising 905 amino acids (large box), an additional six histidines and BamHI site (filled box), followed by the complete KUN 3′UTR sequence (thick line), which was used in the construction of deletion mutants as described in Materials and Methods. Open boxes within the ns5 gene represent regions containing MT motifs (17) and RdRp motifs (16). Striped boxes show three amino acid sequences, a, b, and c, strongly conserved among flaviviruses (6). AgeI, EcoRV (E), Bsu36I (Bs), NruI (N), AatII (A), SmaI (S), BamHI (B), and XhoI denote recognition sites for corresponding restrictases, respectively, with the numbers indicating their nucleotide positions starting in the KUN RNA sequence commencing from the first nucleotide of the ns5 gene. A four-nucleotide insertion at the AgeI site (ns5Age* construct; see Materials and Methods) produced a frameshift in the subsequent coding sequence. (B) In vitro translation products of RNAs prepared from the pBS-based intermediate plasmids shown in panel A, performed as described in Materials and Methods. Arrowheads show positions of the corresponding deleted NS5 proteins. The expected number of amino acids in wild-type (wt) NS5 is 905 and in the deleted proteins, including the number of additional amino acids derived from the vector during construction (indicated in parentheses), are as follows: ns5dSB, 867 (5); ns5dAB, 600 (5); ns5dNB, 407 (9); ns5dBsB, 236 (9); and ns5dEB, 156 (9). Numbers on the left show positions of proteins from low-range prestained protein molecular weight standards (Bio-Rad).
One way of identifying the role of a particular motif in the functional activity of a viral protein is to delete it and examine the effect of the deletion on normal function of the protein as well as to examine whether this defective function can be complemented by addition of the native undeleted protein. The development of the stable KUN full-length infectious cDNA clone (13) and of a functional cDNA clone for production of subgenomic KUN replicon RNA with deleted structural genes (14) allows us to study the effects of introduced deletions and mutations in NS proteins on their functions in RNA replication as well as to devise a system for trans-complementation of these defective proteins. Thus we showed that deletion of the active site GDD in one of the RdRp motifs and the S-adenosylmethionine-binding site in one of the MT motifs of KUN ns5 gene in the full-length RNA (FLdGDD and FLdSAM constructs, respectively) resulted in a total loss of the replication ability of these RNAs (15), hence demonstrating an essential role of these motifs in the NS5 protein for RNA replication. We were also able to complement replication of these defective full-length RNAs by providing functional NS5 protein in trans from the KUN replicon RNA persistently replicating in repBHK cells (15). repBHK cells were prepared by antibiotic G418 selection of BHK cells transfected with KUN replicon RNA C20DXrepNeo deleted of most of the KUN structural region and containing an insertion of encephalomyocarditis virus internal ribosomal entry site-neomycin resistance gene (Neo) cassette in the KUN 3′ untranslated region (3′UTR) (15). In this paper, we show that deletions of the carboxy-terminal coding region comprising up to 56% of NS5 (including the entire RdRp region) can be efficiently complemented in trans by NS5 expressed in repBHK cells. In contrast, coding deletions extending toward the amino-terminal half of NS5 were complemented either much less efficiently or not at all. These results as well as our failure to complement a frameshift mutation at the beginning of NS5 allowed us to postulate a role for the translated N-terminal part of NS5 protein as a cis-acting element for RNA replication and to propose a model for formation of the flavivirus RC.
MATERIALS AND METHODS
Cells.
BHK-21 cells were maintained in the Dulbecco’s modification of minimal essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS). repBHK cells containing stably replicating KUN replicon RNA (15) were maintained in DMEM–10% FBS supplemented with 1 mg of G418 (Geneticin; Gibco BRL) per ml.
Construction of plasmids.
C-terminal deletions in the ns5 gene were prepared initially using plasmid pBSns5His3′UTR (ns5His3′ [Fig. 1A]) (12) by digestion first with BamHI and then with SmaI, AatII, NruI, Bsu36I, or EcoRV, then blunt ending and religating the resulting purified vectors to obtain intermediate plasmid pBSns5dSB3′UTR, pBSns5dAB3′UTR, pBSns5dNB3′UTR, pBSns5dBsB3′UTR, or pBSns5dEB3′UTR, respectively. Full-length KUN plasmids ns5dSB, ns5dAB, ns5dNB, ns5dBsB, and ns5dEB were then obtained by transferring the AgeI-XhoI fragments containing deleted ns5 sequences followed by the KUN 3′UTR sequence from the corresponding intermediate pBS plasmids (see above) into the KUN full-length FLSDX vector (15) digested with AgeI and XhoI (Fig. 1A). The ns5Age* (frameshift) construct (Fig. 1A) was prepared by digesting FLSDX plasmid (15) with AgeI restrictase, filling in with Klenow DNA polymerase, and religating the resulting vector. These manipulations introduced a four-nucleotide insertion at the former AgeI site (AgeI* in Fig. 1A), leading to a frameshift in the subsequent coding sequence (grey box in ns5Age* construct in Fig. 1A) and introduction of an artificial translation termination codon 39 codons downstream of the mutated AgeI site (Fig. 1A, Stop).
Plasmid DENns5 containing an in-frame replacement of most of the KUN ns5 gene by the corresponding region of the DEN ns5 gene was constructed by preparing an AgeI-XmaI PCR fragment amplified from the DEN2 cDNA clone pMK8.5 (10) (kindly provided by R. Padmanabhan, University of Kansas Medical Center), using primers DENns5F (5′-CACACCgGtAGGGAAAGTA-3′) and DENns5R (5′-GGTGGCCCgGgTTGTTAG-3′) with incorporated AgeI and XmaI sites (underlined nucleotides; Fig. 2A) and high-fidelity Pfu DNA polymerase. Lowercase nucleotides in the primer sequences denote the mutated nucleotides in the DEN2 sequence that were incorporated to create restriction sites. This fragment was cloned into the KUN full-length FLSDX clone digested with AgeI and XmaI (Fig. 2A). To confirm an open reading frame of the chimeric ns5 gene by in vitro translation, the AgeI-XhoI fragment from the DENns5 clone (Fig. 2A) was transferred into plasmid pBSNS5wt (15) digested with AgeI and XhoI to obtain plasmid pBSDENns5.
FIG. 2.
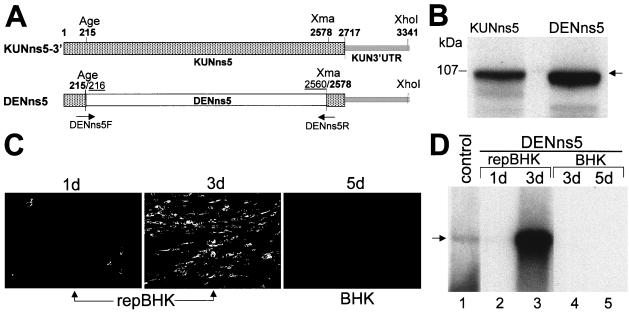
Complementation of chimeric KUN-DEN2 RNA. (A) Fragments from the chimeric full-length cDNA constructs KUNns5-3′ and DENns5, representing the ns5 and 3′UTR regions. Hatched boxes and thick lines represent KUN ns5 and KUN 3′UTR sequences, respectively, as in Fig. 1A except that the additional sequence coding for six histidine residues and BamHI site was not present in these constructs. The open box represents the DEN ns5 sequence PCR amplified from the DEN2 cDNA clone pMK8.5 (10). Age, Xma, and Xho denote positions of recognition sites for AgeI, XmaI, and XhoI restrictases in the KUN cDNA (13) used in construction of chimeric clone as described in Materials and Methods. Numbers in bold indicate positions of KUN nucleotides, and underlined numbers mark the positions of DEN2 nucleotides, both commencing from the first nucleotide of the corresponding ns5 gene. (B) In vitro translations of chimeric RNAs transcribed from an intermediate plasmid DNA, pBSNS5wt (KUNns5 lane) or pBSDENns5 (DENns5 lane), performed as described in Materials and Methods. The position of a 107-kDa protein from prestained low-range molecular weight standards (Bio-Rad) is shown on the left. (C) IF analysis with KUN anti-E antibodies of DENns5-transfected repBHK cells at day 1 (1d panel) and day 3 (3d panel) after transfection and of DENns5-transfected BHK cells at day 5 after transfection (BHK 5d panel). (D) Northern blot analysis of total RNA isolated from repBHK cells and from normal BHK cells at 1 day and 3 days after transfection with DENns5 RNA. An arrowhead indicates the position of RNA of about 11 kb, determined as described for Fig. 3B. The control lane contains ∼5 ng of in vitro-transcribed full-length KUN RNA.
RNA transcription and transfection.
All full-length RNA transcripts were prepared with SP6 RNA polymerase from XhoI-linearized plasmid DNAs and electroporated into BHK-21 or repBHK cells as described previously (14, 15).
In vitro translation.
All intermediate pBS-based plasmid DNAs with deleted and substituted ns5 mutants were linearized with XhoI (Fig. 1A and 2A), and corresponding RNAs were transcribed in a standard in vitro transcription reaction with T7 RNA polymerase (Promega). Approximately 1 μg of purified RNA was used in 10-μl in vitro translation reactions with rabbit reticulocyte lysate (Promega) essentially as described by the manufacturer. Aliquots of 2 μl of the radioactive translation reaction mixture were subjected to electrophoresis in a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel, and labelled proteins were detected by exposure of the dried gel to an X-ray film.
Immunofluorescence and Northern blot analyses.
Detection of replication of complemented KUN full-length RNA in transfected cells was performed by indirect immunofluorescence (IF) analysis of acetone-fixed cells with KUN anti-E antibodies and by Northern blot hybridization of total cell RNA with a 32P-labelled AatII-ClaI cDNA fragment representing 568 nucleotides of the KUN virus prM-E region (KUN nucleotides 522 to 1089 [6, 13]) as described previously (15).
RESULTS
Complementation of defective KUN RNAs with large deletions in the ns5 gene.
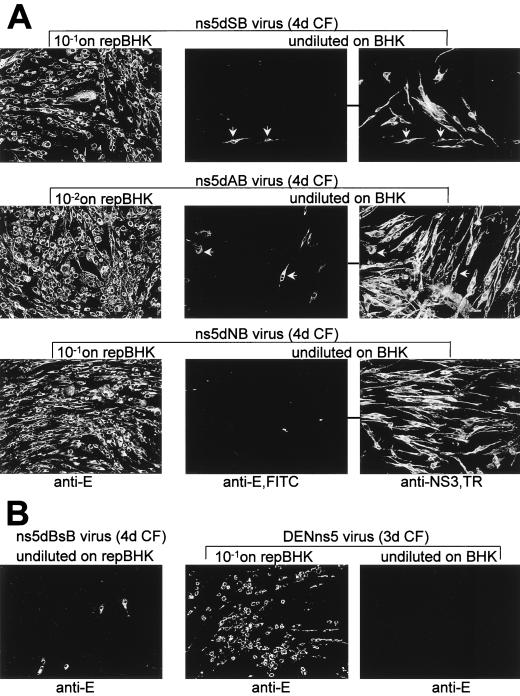
Encouraged by our recent success in the trans-complementation of defective KUN RNAs with a small deletion in either the RNA polymerase or the MT motifs of the ns5 gene (15), we decided to define the maximum extent of the deletion in the ns5 gene which can be complemented in trans. To rescue replication of deleted RNAs, we used BHK cells persistently expressing KUN replicon RNA (repBHK cells [15]). Use of repBHK cells as a helper for complementation of full-length KUN RNAs with introduced deletions allows quick evaluation of the replication of complemented RNAs by IF analysis with KUN anti-E antibodies, which was shown to correlate well with the accumulation of complemented RNA detected by Northern blot analysis (15).
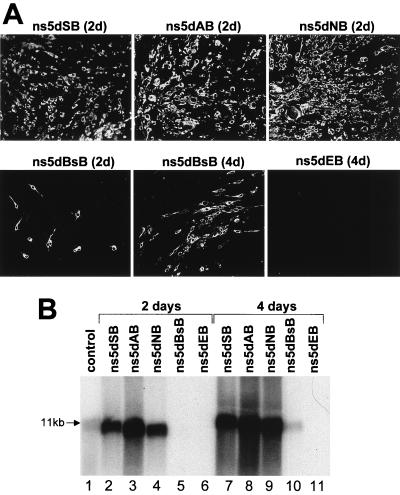
The NS5 mutants containing progressive 3′-terminal deletions were initially prepared in intermediate pBS-based plasmids (Materials and Methods; Fig. 1A). In vitro translation of these pBS-based plasmids produced NS5 protein products of expected sizes (Fig. 1B), thus demonstrating correct translation of the deleted RNAs. We then used these intermediate plasmids to prepare full-length KUN cDNA plasmids with corresponding deletions in the ns5 gene (Materials and Methods; Fig. 1A). The RNA transcripts prepared from these NS5-deleted full-length cDNA plasmids were electroporated into repBHK cells for complementation. IF analysis showed that 100% of repBHK cells were anti-E positive by 2 days after electroporation of ns5dSB, ns5dAB, and ns5dNB RNAs, thus demonstrating efficient complementation of these RNAs (Fig. 3A). Noticeably, the largest efficiently complemented deletion was 1,526 nucleotides (ns5dNB), which represents more than half of the ns5 gene including all the RNA polymerase motifs (Fig. 1A) (16). Further deletion of 516 nucleotides (ns5dBsB [Fig. 1A]) produced significantly less complementation of the corresponding deleted RNA in repBHK cells, with only a few single anti-E-positive cells by day 2 after electroporation, shown in the ns5dBsB (2d) panel in Fig. 3A. However, foci of anti-E-positive cells were observed at day 4 after electroporation, as shown in the ns5dBsB (4d) panel in Fig. 3A, indicating a slow spread of the complemented virus in repBHK cells. No anti-E-positive cells were detected even at 5 to 7 days after transfection of any of these deleted RNAs into normal BHK cells (data not shown). Further deletion of another 237 nucleotides (ns5dEB in Fig. 1A) resulted in complete inability of the ns5dEB RNA to be complemented in repBHK cells, as judged by the absence of anti-E-positive cells at day 4, as shown in the ns5dEB (4d) panel in Fig. 3A, and at day 6 (results not shown) after electroporation.
FIG. 3.
Complementation of KUN RNAs with large deletions in the ns5 gene. (A) IF analysis with KUN anti-E antibodies of repBHK cells transfected with deleted RNAs as shown. (B) Northern blot analysis with a radioactive prM-E cDNA probe of the total RNA isolated from repBHK cells transfected with deleted RNAs. Designations for RNAs used for transfections and the time in days after transfections when the analyses were performed are as indicated. The arrow in panel B indicates the position in the gel of RNA of about 11 kb, determined relative to migration in the same gel of an ethidium bromide-stained 1 Kb Plus DNA Ladder (GibcoBRL). The control lane in panel B contains ∼5 ng of in vitro-transcribed full-length KUN RNA.
We next confirmed the IF results on complementation of defective RNAs by Northern blot analysis of the parallel samples of total cell RNA by using a radioactive cDNA probe representing part of the KUN prM-E region which is absent in the KUN replicon RNA (Fig. 3B). Appropriate differences in size of complemented RNAs were observed in the blots, as expected from the differences in size of the corresponding deletions (Fig. 3B, lanes 1 to 4 and 7 to 10). These results demonstrated that replication of defective KUN RNAs with lethal C-terminal coding deletions in the KUN ns5 gene ranging from as few as 137 nucleotides up to as many as ∼2,070 nucleotides, representing more than two-thirds of the gene, could be complemented in trans by the functional ns5 gene expressed from the KUN replicon RNA. The results also showed that the KUN ns5 sequence between nucleotides 442 and 679 (present in ns5dBsB but not in ns5dEB) is absolutely essential for maintaining the ability of the corresponding RNA to be complemented in trans and thus probably represents part of a cis-acting element. In a separate experiment, we could not complement KUN full-length RNA ns5Age* with a frameshift mutation at the AgeI site (construct shown in Fig. 1A) (results not shown), demonstrating that translation of the amino acid sequence of the first 679 nucleotides of ns5 gene rather than the RNA sequence per se in this region is required for trans-complementation to occur.
Complementation of KUN RNA containing the chimeric DEN2-KUN ns5 gene.
Having established an essential role for translation of the N-terminal half of the KUN ns5 gene in complementation, we then investigated the stringency of these amino acid sequence requirements by substituting in the KUN sequence a heterologous ns5 sequence from a distantly related flavivirus. For this purpose, we prepared a full-length KUN cDNA construct containing a chimeric DEN2-KUN ns5 gene. The sequence from AgeI to XmaI sites representing approximately 87% of the KUN ns5 gene (including the essential region for complementation in the N-terminal half shown above) was replaced by the corresponding sequence of the DEN2 ns5 gene that has about 29% difference in amino acid sequence (Fig. 2A) (2, 6). Retention of the open reading frame in the chimeric ns5 gene was confirmed by in vitro translation of the RNA transcribed from the corresponding pBSDENns5 plasmid (Fig. 2B). We then addressed the question of whether this chimeric RNA can replicate in transfected cells by itself and, if it cannot, whether the function of this chimeric NS5 protein can be complemented in trans by the functional KUN replication complex. No replication was detected even by 5 days after transfection of DENns5 RNA into normal BHK cells (Fig. 2D, lanes 4 and 5), suggesting that chimeric DEN-KUN NS5 protein could not amplify KUN RNA. However, replication of DENns5 RNA was complemented in transfected repBHK cells, as judged by IF and Northern blot analyses (Fig. 2C and D). The noticeable increase in the amount of anti-E-positive cells (Fig. 2C) and in the amount of accumulated RNA (Fig. 2D) from day 1 to day 3 after transfection indicates spread of complemented virus. These results demonstrate that a chimeric NS5 protein comprising most of the DEN2 NS5 could not recognize the KUN RNA molecule for amplification, but this chimeric RNA molecule with an authentic KUN 3′UTR was apparently recognized and amplified by the functional KUN RC present in repBHK cells. Therefore, we concluded that the RNA polymerase function of chimeric DEN-KUN NS5 protein was efficiently complemented by the native KUN NS5 protein produced as a part of functional KUN RC in repBHK cells.
Characterization of secreted complemented viruses.
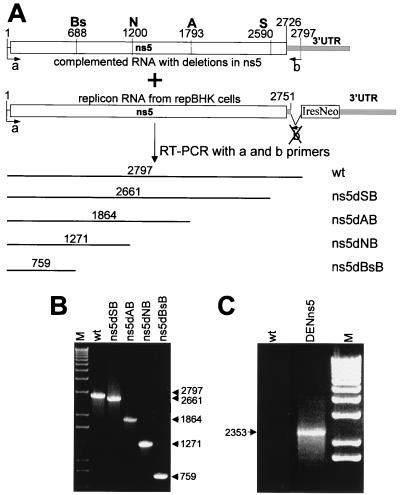
The presence of deleted or substituted RNAs in the recovered defective viruses was confirmed by RT-PCR analysis of RNA isolated from the secreted defective virus particles immunoprecipitated with anti-E antibodies (Fig. 4). The primers used for reverse transcription (RT)-PCR analysis were designed so that they could amplify packaged defective full-length RNA but not packaged helper replicon RNA (Fig. 4A). To ensure that RT-PCR amplification occurred only from RNA purified from recovered complemented virus particles, culture fluids (CFs) from transfected repBHK cells were exhaustively treated with RNase A and DNase before and during the precipitation with anti-E antibodies (see the legend to Fig. 4). The gel migrations of the RT-PCR-amplified fragments correlated well with the predicted sizes, which were 2,797 nucleotides for purified full-length KUN virion RNA, 2,661 nucleotides for ns5dSB virus RNA, 1,864 nucleotides for ns5dAB virus RNA, 1,271 nucleotides for ns5dNB virus RNA, 759 nucleotides for ns5dBSB virus RNA (Fig. 4A and B), and 2,353 nucleotides for DENns5 virus RNA (Fig. 2A and 4C). Although a fragment of the correct size was detected in the RT-PCR product from RNA purified from 6-day CF of ns5dBsB virus (Fig. 5B), no RT-PCR product was detected in the RNA sample isolated from 4-day CF of ns5dBsB virus or in a control reaction with RNA isolated from 4-day CF collected from BHK cells transfected with FLdGDD RNA (results not shown). The latter represents full-length KUN RNA with a coding deletion of the RNA polymerase motif GDD described previously (15). These negative RT-PCR results confirmed the very inefficient complementation of nd5dBsB RNA and the slow secretion of complemented virus observed in transfection experiments (Fig. 3), as well as demonstrating the specificity of the RT-PCRs.
FIG. 4.
Characterization of secreted complemented viruses by RT-PCR analysis. (A) Schematic representation of the KUN full-length and replicon genomes in the ns5-3′UTR sequence. The numbers represent nucleotide positions in the KUN RNA sequence in the ns5-3′UTR sequence including an additional nine nucleotides incorporated in primer a described below. These numbers were used to calculate the size of RT-PCR fragments shown above the lines. Bs, N, A, and S show positions of the restriction sites used for generation of the deletions as in Fig. 1A, with addition of nine nucleotides present in primer a. Primers used in RT-PCRs, indicated by a and b, were as follows: (a) 5′-ggtcatatgGGTGGGGCAAAAGGA-3′ (nucleotides in capital letters correspond to nucleotides 1 to 15 of the KUN ns5 gene [6]; nucleotides in lowercase show an additional nine nucleotides, not present in the KUN sequence), and (b) 5′-CACACTAAACACTATTATAAAGCTAAA-3′ (minus sense, complementary to nucleotides 2771 to 2797 of the KUN ns5-3′UTR sequence, which contains an additional nine nucleotides derived from the 5′ end of primer a). Note that primer b cannot bind to the 3′UTR sequence in the replicon RNA due to a deletion introduced during its construction (see Materials and Methods and reference 14). (B) RT-PCR analysis of recovered defective viral RNAs. Aliquots of 630 μl of 1-ml CFs collected at day 3 (DENns5), day 4 (ns5dSB, ns5dAB, and ns5dNB), and day 6 (ns5dBsB) after transfection of repBHK cells with corresponding defective RNAs were treated by addition of 50 μg of RNase A (Sigma) per ml and 5 U of RQ DNase (Promega) per ml for 30 min at 37°C to ensure the absence of any possible DNA and RNA contaminations from transfected in vitro transcription mixtures. CFs still containing RNase A and DNase were then incubated overnight at 4°C with 70 μl of anti-E monoclonal antibodies followed by a further 2-h incubation with 100 μl of a 10% slurry of protein A-Sepharose beads (Pharmacia). The precipitates on the washed beads were treated with proteinase K in the presence of 0.5% SDS, followed by phenol-chloroform extraction and ethanol precipitation of the RNA. Precipitated RNA was dissolved in 6 μl of diethyl pyrocarbonate-treated H2O, and 1 μl of this RNA was used in a 10-μl RT-PCR performed with the SuperScript One-Step RT-PCR system (GibcoBRL) essentially as described by the manufacturer and with primers a and b, defined above. In panel C, primers for amplifying DENns5 virus RNA by RT-PCR were DENns5F and DENns5R (see Materials and Methods). KUN lanes shown as wt in panels B and C represent RT-PCRs with ∼10 ng of KUN virion RNA purified as described previously (13). M lanes in panels A and B show the 1 Kb Plus DNA Ladder (GibcoBRL).
FIG. 5.
Characterization of the recovered viruses by IF analysis. (A) IF analysis of repBHK and BHK cells which were infected with diluted or undiluted CFs (as indicated) collected at 4 days after transfection of ns5dSB, ns5dAB, and ns5dNB RNAs. repBHK panels show IF analysis at day 2 after infection of repBHK cells using anti-E antibodies (anti-E panels). Pairs of joined BHK panels representing the same selected field were dual labelled with anti-E (anti-E,FITC panels) and anti-NS3 antibodies (anti-NS3,TR panels). White arrows indicate selected cells positive in IF with both anti-E and anti-NS3 antibodies. (B) IF analysis using anti-E antibodies of repBHK and BHK cells infected with CFs containing complemented ns5dBsB and DENns5 viruses. Designations are as in panel A. FITC-conjugated secondary antibodies were used in all experiments with repBHK and BHK cells for IF detection of E protein, and TR-conjugated secondary antibodies were used for IF detection of NS3 protein in experiments with dual labelling in BHK cells.
We then investigated replication of the recovered defective viruses in infected repBHK and normal BHK cells by dual-label (fluorescein isothiocyanate [FITC] and Texas red [TR]) IF analysis. Infection of repBHK cells with 1/10 dilution of ns5dSB or ns5dNB or 1/100 dilution of ns5dAB defective viruses secreted in the 4-day CF resulted in detection of 100% cells positive in IF analysis with anti-E antibodies by 2 days after infection (repBHK panels in Fig. 5A), indicating transmission and efficient replication of the complemented defective viruses in repBHK cells. Infection of repBHK cells with 1/10 dilution of DENns5 virus (3-day CF) produced ∼60 to 70% anti-E-positive cells by 2 days after infection (Fig. 5B), also showing efficient replication of DENns5 virus. As expected from inefficient complementation of ns5dBsB RNA (Fig. 3), infection of repBHK cells with undiluted ns5BsB virus (4-day CF) resulted in detection of only a few anti-E-positive cells at day 2 after infection (Fig. 5B), and a slight increase in number of positive cells was observed at day 5 after infection (results not shown). To compare relative efficiencies of complementation, we titrated repBHK CFs harvested at 4 days after transfection of ns5dSB, ns5dAB, ns5dNB, and DENns5 RNAs by infection of repBHK cells. Infectious titers were calculated by counting foci of anti-E IF-positive cells at 2 days after the infection. The number of IF foci decreased linearly with the dilutions of CFs, and the titers were ∼3 × 105 to 5 × 105 infectious units (IU) per ml for ns5dSB, ns5dNB, and DENns5 viruses and ∼5 × 106 IU per ml for ns5dAB virus. These results with secreted viruses correlated well with our observations that ns5dAB RNA was complemented more efficiently in transfected repBHK cells in the same experiment than were ns5dSB and ns5dNB RNAs (compare, for example, lane 3 with lanes 2 and 4 in Fig. 3B). The titers of 4-day and 6-day CFs of ns5dBSB virus were ∼102 and ∼5 × 102 IU per ml, respectively, thus confirming the very low efficiency of ns5dBsB RNA complementation observed in transfection experiments (see above).
We showed previously in complementation experiments with FLdGDD and FLdSAM RNAs performed with repBHK cells (15) that infection of normal BHK cells with recovered defective complemented viruses may lead to detection of rare anti-E-positive cells due to simultaneous coinfection of these cells with two types of particles, one containing packaged replicon RNA from repBHK cells (replicon RNA particles) and the other containing packaged complemented defective full-length RNA (defective RNA particles). This coinfection event occurred rarely and these two types of particles could be distinguished by dual IF analysis using anti-NS3 antibodies (able to detect replication of both replicon RNA and deleted full-length RNA) and anti-E antibodies (able to detect replication of only deleted full-length RNA). The results of such dual IF analyses performed at 2 days after infection of normal BHK cells with undiluted CFs containing ns5dSB, ns5dAB, and ns5dNB complemented viruses showed that while a significant number of these normal BHK cells were anti-NS3 positive (BHK, anti-NS3 panels in Fig. 5A), only very few of these anti-NS3 positive cells were also positive in IF with anti-E antibodies (BHK, anti-E panels in Fig. 5A), as shown previously for complemented FLdGDD and FLdSAM viruses (15). No anti-E-positive cells were detected after infection of BHK cells with undiluted CF containing complemented DENns5 virus (Fig. 5B). Interestingly, longer incubation of normal BHK cells infected with ns5dSB, ns5dAB, and ns5dNB viruses resulted in detection of a small number of slowly spreading anti-E-positive foci by day 5 after infection (results not shown) which may have arisen either from the spread of secreted complemented virus in the neighboring cells infected with replicon RNA particles or from self-replicating recombinant virus. Further passaging of the CFs collected from these infected BHK cells with spreading anti-E-positive foci on fresh BHK or repBHK cells did not produce anti-E-positive BHK cells, but the majority of repBHK cells were anti-E positive (results not shown), indicating the presence of only defective virus particles in the repBHK-complemented virus material passaged once on BHK cells. These passaging results for BHK-infected CFs as well as results for initial infections of repBHK and BHK cells with recovered complemented viruses (Fig. 5) and results of RT-PCR analysis of secreted complemented viral particles (Fig. 4) clearly demonstrate that the majority of secreted complemented virus particles recovered in repBHK transfection experiments contained defective full-length KUN RNAs able to replicate in repBHK cells but deficient in the ability to independently replicate in normal BHK cells. Thus, possible recombinant self-replicating viruses were either absent or present in negligible or undetectable amounts.
DISCUSSION
We constructed several KUN mutants whose genomes contained large deletions, a frameshift mutation, and a heterologous (DEN) substitution retaining only 13% of the KUN NS5 coding region, all in the RNA polymerase gene ns5. None of the mutant RNAs was able to replicate independently in transfected BHK cells, thus demonstrating an essential role of the deleted or substituted amino acid sequences in the NS5 protein for RNA replication. We then used repBHK cells stably expressing KUN replicon RNA as a helper to trans-complement replication of these mutant RNAs. The RNAs containing deletions representing the C-terminal half of NS5 were complemented efficiently in repBHK cells, while RNAs with deletions extending into the N-terminal half of NS5 were complemented either much less efficiently or not at all (Table 1). This is the first report on successful trans-complementation of large deletions (as much as 75%) in the flavivirus RNA polymerase gene. In other experiments, RNA with a frameshift mutation at codon 71 in the KUN ns5 gene (ns5Age* [Fig. 1A]) was not complemented (see Results). Interestingly, KUN RNA with a 3′-terminal deletion of 509 codons including the entire NS5 RdRp region was still complemented efficiently (ns5dNB [Table 1]). Thus, our results clearly demonstrate that despite the absence of translation in cis of all of the amino acid sequence representing the enzymatically functional region of the RdRp gene, the polymerase functions can be complemented by another (native) RdRp supplied in trans.
TABLE 1.
Complementation of KUN RNAs with deletions or a frameshift mutation in the ns5 gene
| Construct | Size of C-terminal deletionsa | N-terminal conserved motifs retainedb | Complementation by IFc | Titersd (IU/ml) |
|---|---|---|---|---|
| ns5dSB | 45 | a, b, c | ++++ | (∼3–5) × 105 |
| ns5dAB | 312 | a, b, c | ++++ | ∼5 × 106 |
| ns5dNB | 509 | a, b, c | ++++ | (∼3–5) × 105 |
| ns5dBsB | 680 | a, b | + | ∼102 |
| ns5dEB | 760 | − | ND | |
| ns5Age* | 832e | − | ND |
Number of amino acids deleted from NS5 proteins as deduced from the nucleotide sequence of corresponding cDNA constructs.
a, b, and c represent three amino acid sequences conserved among flaviviruses (see text, Fig. 1A and Fig. 6).
Relative efficiency of complementation, determined as the approximate percentage of repBHK cells positive in IF with anti-E antibodies at 2 days after transfection with mutated RNAs (see text and Fig. 3A): ++++, 100% positive cells; +, 0.5 to 1% (few single positive cells); −, no positive cells.
Titer of complemented virus in 4-day CFs from repBHK cells transfected with the corresponding RNAs and determined by infection of repBHK cells with serial dilutions followed by counting of anti-E IF-positive foci at 2 days after infection. ND, not determined.
Number of amino acids unable to be translated in the correct reading frame due to a frameshift mutation (see Fig. 1A and Materials and Methods).
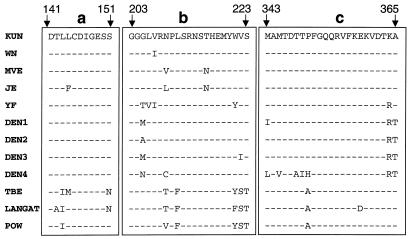
Comparative amino acid analysis of the N-terminal half of the flavivirus NS5 protein revealed the presence of three highly conserved regions comprising 11, 21, and 23 amino acids situated within the first 365 amino acids of NS5 (motifs a, b, and c, respectively, in Fig. 6; references 2 and 6). The presence of one of these conserved regions (c) was previously noted by Coia et al. (6), but as yet no functions have been assigned to any of these three homology motifs. The striking difference between the efficient complementation of ns5dNB RNA (retaining all three motifs) and the very inefficient complementation of ns5dBsB RNA (retaining only the first two motifs, a and b) (Table 1) suggests an important role of at least motif c in retaining the ability of deleted RNA to be efficiently complemented in trans. Removal of all three conserved regions resulted in total loss of the ability of ns5dEB RNA to be complemented (Table 1). Interestingly, defective chimeric RNA in which 87% of the KUN ns5 gene (including the N-terminal coding region for motifs a, b, and c) was replaced by a corresponding DEN2 ns5 sequence was efficiently complemented in repBHK cells (Results and Fig. 2). Notably, all these three motifs are highly conserved in both KUN and DEN2 NS5 proteins (Fig. 6), while the remainder of the replaced amino acid sequence differed by approximately 29% (see Results). Importantly, this chimeric RNA was not able to establish self-replication in normal BHK cells, which demonstrates that chimeric NS5 protein could not form a functional RC with KUN RNA. The lack of complementation of RNA with a frameshift mutation at codon 71 (ns5Age* results; see above) and the successful complementation of RNAs with the deleted and substituted NS5 mutants clearly demonstrate that only when the N-terminal half of the ns5 gene (encoding conserved motifs a, b, and c) was translated in cis could the RNA be recognized and amplified efficiently by a functional RC.
FIG. 6.
Conserved motifs in the N-terminal half of the flavivirus ns5 gene. a, b, and c represent alignment of NS5 amino acid sequences (KUN NS5 amino acids 141 to 151, 203 to 223, and 343 to 365, respectively, as indicated) conserved with trivial variations only for all flaviviruses (2, 6).
Although NS5 of DEN1 was shown to copy RNA in an in vitro reaction, there was no specificity in regard to template RNA (30). Our analyses of the native KUN RC late in infection both in vivo and in vitro have shown that it has a consensus composition of NS1, NS3, NS5, NS2A, and NS4A. This was established by cryoimmunoelectron microscopy of infected cells showing colocalizations with the putative dsRNA template (22, 31, 32), by purification of the native RdRp after detergent treatment of the semipurified active membrane fraction (5), and/or by radioimmunoprecipitation of detergent-treated cytoplasmic extracts with antibodies to dsRNA (32). We also showed in binding assays that NS2A and NS4A bind to the same constellation of proteins as above, that NS4A binds very strongly to itself, and that NS2A binds to KUN 3′UTR (22) as does NS5 (12). Others have shown that flavivirus NS5 and NS3 in Japanese encephalitis virus-infected cell lysates can be cross-linked by UV irradiation specifically to the putative stem-loop within the RNA positive-strand 3′UTR (3) or be coprecipitated in DEN2-infected cells by antibodies to DEN2 NS3 or NS5 and also to bind to each other in vitro (11). DEN2 NS3 was recently shown to have intrinsic RNA helicase activity as well as the RNA-stimulated nucleoside triphosphatase activity (18) reported for several other flaviviruses (for references, see reference 18), implying strong affinity of this protein for RNA. Significantly, substitutions of homologous sequences of West Nile virus RNA in the bottom half of the DEN2 3′ stem-loop were lethal (36).
Like flavivirus RNAs, picornavirus RNA is translated from one long open reading frame with the gene order of structural genes followed by NS genes, and after the genomic RNA is copied by the viral replicase into an RNA minus strand, a double-stranded template is formed for initiation of synthesis of progeny RNA in association with membranes (35). Some comparisons with replication of the well-studied poliovirus therefore have some relevance. For example, Novak and Kirkegaard (24) reported successful complementation by helper replicon RNA of poliovirus full-length RNA with a frameshift mutation at the beginning of the 3Dpol RNA polymerase gene (codon 28), thus eliminating translation of virtually all of the 3Dpol. Similarly, deletion of the entire KUN NS5 RdRp region was also efficiently complemented in trans (ns5dNB [Table 1]). Interestingly, the N-terminal region of one of the poliovirus 3Dpol subunits was proposed to be involved in interaction with the C-terminal region of another 3Dpol subunit during oligomerization (8), and the N-terminal region (amino acids 40 to 140) of brome mosaic virus RdRp (2a protein) was shown to interact with the helicase-like domain of 1a protein (9, 25). However, there are no reports of specific interactions of the N-terminal region of flavivirus NS5.
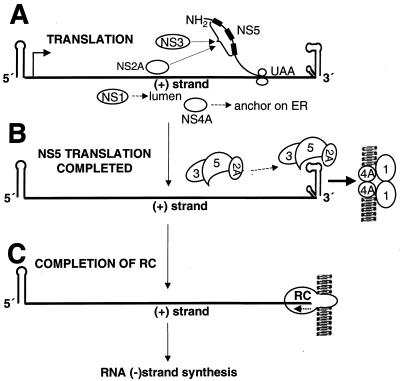
In view of the considerations above and assuming that the compositions of the RC are similar early and late in infection, it is tempting to speculate that the N-terminal region of the flavivirus NS5 protein interacts via conserved motifs (a, b, and c) with NS3 and possibly other NS proteins such as NS2A during translation to initiate formation of the RC, most likely on the 3′UTR of genomic RNA (Fig. 7). We propose that the RNA template with the partially formed RC is then transported to an anchor region on virus-induced membranes to complete the RC where initiation of synthesis of RNA negative strand can then occur (Fig. 7). The mode of transport and anchoring of the RC to membranes is speculative. Possible roles for NS2A arising from our KUN data include targeting the viral RNA and RC to cytoplasmic membranes, and we suggested that NS4A may play a targeting or membrane-anchoring role within the RC (22, 31, 32). NS1 dimerizes after synthesis becoming membrane bound, with increased hydrophobicity (34). Proposals for the role of NS1 include interaction with the RC by directing it to membranes (20, 32) or a role prior to or at initial RNA negative-strand synthesis involving assembly of the components of the RC directly via protein-protein interactions or via the relationship between NS1 and cellular membranes (19). The relationship of NS4A to NS1 shown in the model (Fig. 7B) is supported by a recent report from Lindenbach and Rice (20) showing genetic interaction between these products as a determinant of replicase function. Once formed, the RC appears to be stable because it remains active for several hours after complete inhibition of all protein synthesis by cycloheximide treatment (33). Hence, the RC requires no supplementation by recently translated NS proteins or by polyprotein precursors which are cleaved rapidly during KUN virus RNA translation in cells synchronized in translation (29).
FIG. 7.
Proposed model for formation of the flavivirus RC and initiation of RNA negative-strand [(−) strand] synthesis. (A) NS3 is assumed to bind to NS5 (3, 11) at one or more of the conserved regions a, b, and c (each indicated by a thickened line within the bracket) in the N-terminal region (Fig. 6), possibly associated also with NS2A. ER, endoplasmic reticulum. (B) On completion of translation, the complex attaches to the 3′UTR via binding of NS2A (21) probably at the 3′-terminal stem-loop at which the NS3 and NS5 components also bind (3, 7). The location of the complex shown on the 3′ stem-loop is arbitrary. The complex attached to the RNA positive strand [(+) strand] is transported (indicated by a thick arrow) to the membrane site of replication by affinity of the hydrophobic regions of NS2A interacting with those of NS4A (shown as dimers), which in turn is bound by hydrophilic extensions in the lumen between transmembrane domains (6) to dimeric NS1 in the lumen (20). (C) The RC is now complete and may undergo a rearrangement as the RdRp domains of NS5 bind to the template plus strand, allowing copying to proceed. The RC is represented as an extended circle on the 3′UTR, and a short dashed arrow indicates the direction of synthesis (5′ to 3′) of the initiating RNA negative strand. The association with membranes and the consensus composition of the RC (NS1, NS3, NS5, NS2A, and NS4A) are described in the text. A role for NS1 in synthesis of the RNA negative strand early in infection has been proposed by Lindenbach and Rice (19, 20) based on complementation experiments with mutated yellow fever virus RNA.
According to the proposed model (Fig. 7), deletions in the C-terminal half of the NS5 protein (as in ns5dSB, ns5dAB, and ns5dNB) should still allow interaction of other NS proteins with motifs a, b, and c in the N-terminal half of NS5 and thus eventual formation of a defective RC, probably consisting of a number of NS proteins (presumably NS3 and NS2A) bound to truncated NS5 protein and to the terminal region of the 3′UTR of genomic RNA (see the legend to Fig. 7). In complementation experiments, this defective RC would still be able to transport the defective RNA (possibly directed by NS2A) to the site of helper (replicon) RNA synthesis anchored on membranes in repBHK cells, thus allowing initiation of synthesis of a deleted RNA negative strand by exchanging the helper RC or its components with the defective RC. Translation of RNA with deletion of c, one of the proposed protein-binding motifs, as in ns5dBsB may result in the loss of binding of truncated NS5 to one or more NS proteins, leading to formation of an unstable or incomplete defective RC, unable to efficiently exchange components with helper RC. Finally, loss of all proposed protein-protein binding motifs as in ns5dEB and ns5Age* would not allow any interaction between truncated NS5 and other NS proteins and thus would result in an inability to form any defective RC capable of transporting RNA to the location of the required helper RC. This model is speculative at present but is based on our results obtained in complementation experiments with KUN NS5-deleted RNAs and on the extensive results summarized above describing the composition of the KUN RC, as well as on the results of binding studies and cited data of other groups. Further detailed experiments on protein-protein and RNA-protein binding with deleted NS5 proteins as well as with other NS proteins are required to determine whether this model accurately represents the events in flavivirus RNA replication. There is a need to explore also the role in replication of cellular proteins such as EF-1α shown to bind to the 3′-terminal stem-loop of several flaviviruses by Blackwell and Brinton (1), who suggested that EF-1α may be involved in targeting flavivirus RNA to intracellular membranes or in assembly of the viral RC. The proposed model is the first attempt to provide a comprehensive interpretation of a wide range of reported observations possibly relevant to the formation of the flavivirus RC for initiation of RNA negative-strand synthesis and should facilitate further studies on unraveling the events of flavivirus replication.
ACKNOWLEDGMENTS
We are grateful to R. Padmanabhan for supplying plasmid pMK8.5 with DEN2 cDNA and R. Hall for supplying KUN anti-E monoclonal antibodies.
This work was supported by grant N981442 from the National Health and Medical Research Council of Australia.
Footnotes
Publication no. 93 from the Sir Albert Sakzewski Virus Research Centre.
REFERENCES
- 1.Blackwell J L, Brinton M A. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang G-J. Molecular biology of dengue viruses. In: Gubler D J, Kuno G, editors. Dengue and dengue hemorrhagic fever. Wallingford, United Kingdom: CAB International; 1997. pp. 175–198. [Google Scholar]
- 3.Chen C-J, Kulo M-D, Chien L-J, Hsu S-L, Wang Y-M, Lin J-H. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu P W, Westaway E G. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology. 1985;140:68–79. doi: 10.1016/0042-6822(85)90446-5. [DOI] [PubMed] [Google Scholar]
- 5.Chu P W, Westaway E G. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch Virol. 1992;125:177–191. doi: 10.1007/BF01309636. [DOI] [PubMed] [Google Scholar]
- 6.Coia G, Parker M D, Speight G, Byrne M E, Westaway E G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988;69:1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- 7.Cui T, Sugrue R J, Xu Q, Lee A K W, Chan Y-C, Fu J. Recombinant dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by NS5. Virology. 1998;246:409–417. doi: 10.1006/viro.1998.9213. [DOI] [PubMed] [Google Scholar]
- 8.Hansen J L, Long A M, Schultz S C. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure. 1997;5:1109–1122. doi: 10.1016/s0969-2126(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 9.Kao C C, Ahlquist P. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J Virol. 1992;66:7293–7302. doi: 10.1128/jvi.66.12.7293-7302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapoor M, Zhang L, Mohan P M, Padmanabhan R. Synthesis and characterization of an infectious dengue virus type-2 RNA genome (New Guinea C strain) Gene. 1995;162:175–180. doi: 10.1016/0378-1119(95)00332-z. [DOI] [PubMed] [Google Scholar]
- 11.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner K E, Padmanbhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 12.Khromykh, A. A. Unpublished data.
- 13.Khromykh A A, Westaway E G. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J Virol. 1994;68:4580–4588. doi: 10.1128/jvi.68.7.4580-4588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khromykh A A, Westaway E G. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khromykh A A, Kenney M T, Westaway E G. trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J Virol. 1998;72:7270–7279. doi: 10.1128/jvi.72.9.7270-7279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koonin E V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 17.Koonin E V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J Gen Virol. 1993;74:733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- 18.Li H, Clum S, You S, Ebner K E, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol. 1999;73:3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindenbach B D, Rice C M. trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindenbach B D, Rice C M. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999;73:4611–4621. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackenzie J M, Jones M K, Young P R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 22.Mackenzie J M, Khromykh A A, Jones M K, Westaway E G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 23.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D. Virus taxonomy: classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Arch Virol Suppl. 1995;10:415–427. [Google Scholar]
- 24.Novak J E, Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 25.O’Reilly E K, Paul J D, Kao C C. Analysis of the interaction of viral RNA replication proteins by using two-hybrid assay. J Virol. 1997;71:7526–7532. doi: 10.1128/jvi.71.10.7526-7532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Reilly E K, Kao C C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 27.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Racaniello V R, Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci USA. 1981;78:4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrader A P, Westaway E G. Translation mapping with the flavivirus Kunjin: gene order and anomalities in translation of NS5. Virus Res. 1988;9:323–334. doi: 10.1016/0168-1702(88)90091-3. [DOI] [PubMed] [Google Scholar]
- 30.Tan B-H, Fu J, Sugrue R J, Yap E-H, Chan Y-C, Tan Y H. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996;216:317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- 31.Westaway E G, Khromykh A A, Kenney M T, Mackenzie J M, Jones M K. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology. 1997;234:31–41. doi: 10.1006/viro.1997.8629. [DOI] [PubMed] [Google Scholar]
- 32.Westaway E G, Mackenzie J M, Kenney M T, Jones M K, Khromykh A A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westaway E G, Khromykh A A, Mackenzie J M. Nascent flavivirus RNA co-localized in situ with double-stranded RNA in stable replication complexes. Virology. 1999;258:108–117. doi: 10.1006/viro.1999.9683. [DOI] [PubMed] [Google Scholar]
- 34.Winkler G, Maxwell S E, Ruemmler C, Stollar V. Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology. 1989;171:302–305. doi: 10.1016/0042-6822(89)90544-8. [DOI] [PubMed] [Google Scholar]
- 35.Xiang W, Paul A V, Wimmer E. RNA signals in entero- and rhinovirus genome replication. Semin Virol. 1997;8:256–273. [Google Scholar]
- 36.Zheng L, Falgout B, Markoff L. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J Virol. 1998;72:7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]