Abstract
Purpose
To construct a prognostic model for pancreatic cancer based on glycolytic and hypoxic metabolic subtypes. To analyze the biological characteristics of these subtypes and explore potential therapeutic options.
Methods
We obtained mRNA, simple nucleotide variation (SNP), and clinical data for pancreatic cancer from The Cancer Genome Atlas (TCGA). Patients were classified into four metabolic subtypes. We focused on glycolysis and hypoxia subtypes. Single-sample gene set enrichment analysis (ssGSEA) assessed immune cell infiltration. We evaluated the effects of immunotherapy and chemotherapy on these subtypes. Cox regression and random survival forest algorithms were used to build a prognostic model. Validation was performed using data from the International Cancer Genome Consortium (ICGC) and ArrayExpress database.
Results
We identified four subtypes. Kaplan-Meier survival analysis showed the glycolytic subtype had the longest survival, while the hypoxic subtype had the shortest. The glycolytic subtype exhibited higher immune cell infiltration. Immunotherapy and chemotherapy appeared more beneficial for the glycolytic subtype. KRAS mutations were more frequent in the hypoxic subtype. Our prognostic model indicated a worse prognosis for high-risk groups, validated by external data.
Conclusion
The glycolytic metabolic subtype of pancreatic cancer is associated with longer survival and better response to chemotherapy and immunotherapy compared to the hypoxic subtype.
Keywords: Pancreatic cancer, Biomarkers, Glycolysis, Prognosis, Hypoxia
Highlights
-
•
Prognostic Model: Developed and validated a prognostic model for pancreatic cancer based on glycolytic and hypoxic subtypes.
-
•
Survival Analysis: Found that the glycolytic subtype had the longest survival, while the hypoxic subtype had the shortest.
-
•
Immune Response: Glycolytic subtype showed higher immune cell infiltration and better response to anti-PD-1 therapy and certain chemotherapeutic agents.
-
•
KRAS Mutations: Identified higher frequency of KRAS mutations in the hypoxic subtype, correlating with poorer prognosis.
-
•
External Validation: The model was validated with significant prognostic value using external datasets from ICGC and ArrayExpress.
1. Introduction
Pancreatic cancer is one of the most fatal malignancies, with a one-year survival rate of only 20 % and an overall survival rate of just 8 % [1]. Studies predict that by 2030, pancreatic cancer will become the second leading cause of cancer-related mortality [2]. The absence of obvious clinical symptoms and the lack of effective biomarkers in early stages make early diagnosis challenging [3].
Unlike other solid tumors, pancreatic cancer is characterized by abnormal stromal proliferation, which can account for up to 90 % of the tumor volume. This results in poor vascularity and the formation of a hypoxic microenvironment [4]. Hypoxia inhibits aerobic glucose oxidation and promotes glycolysis. Both hypoxia [5] and glycolysis [6] are significant factors contributing to poor cancer prognosis. Metabolic subtypes exhibit varying sensitivities to chemotherapeutic agents [7]. Studies have confirmed that glycolysis and hypoxia impact the prognosis of pancreatic cancer. Hypoxia leads to more aggressive tumor cells [8,9], and hypoxia-based metabolic staging can predict patient survival [10]. Lower immune infiltration in glycolytic subtypes is linked to shorter survival times [11]. However, no study has yet analyzed both metabolic subtypes in tandem.
In our study, we identified four subtypes based on hypoxia and glycolysis genes. Patients with the glycolytic subtype had longer overall survival compared to those with the hypoxic subtype. The glycolytic subtype also showed higher immune cell infiltration. Immunotherapy and chemotherapy were more beneficial for the glycolytic subtype. We constructed a prognostic model using the random survival forest algorithm and COX regression analysis, which was validated with data from TCGA and two external cohorts.
2. Materials and methods
2.1. Patient data
To obtain a sufficient number of pancreatic cancer samples, we conducted a comprehensive search using TCGA (https://portal.gdc.cancer.gov/), ICGC (PACA-CA) (https://daco.icgc.org/), and ArrayExpress (E-MTAB-6134) (https://www.ebi.ac.uk/arrayexpress/). From TCGA, we downloaded clinical data, mRNA data, and SNP data for pancreatic cancer. Additionally, we obtained mRNA and clinical data from ICGC and ArrayExpress.
2.2. Defining the subtypes
To identify glycolytic and hypoxic genes, we utilized the Gene Set Enrichment Analysis (GSEA) tool (https://www.gsea-msigdb.org/gsea/msigdb/search.jsp). We retrieved the hypoxic gene set “BUFFA_HYPOXIA_METAGEN” (n = 50) and the glycolytic gene set “REACTOME_GLYCOLYSIS” (n = 72). We then performed consensus clustering of these genes using ConsensusClusterPlus (parameters: reps = 100, pItem = 0.8, pFeature = 1). Ward.D2 and Euclidean distances were employed as the clustering algorithm and distance metric, respectively.
Samples and genes were grouped after reanalyzing the clusters. Based on the gene expression profiles of these grouped samples, we calculated the median expression of glycolytic and hypoxic genes, setting the median to 0 as a reference value. Subtypes were defined using the median expression levels as follows: quiescent (hypoxia ≤0, glycolysis ≤0), mixed (hypoxia >0, glycolysis >0), glycolysis (hypoxia <0, glycolysis ≥0), and hypoxia (hypoxia >0, glycolysis ≤0).
2.3. Immune cells infiltration and immune checkpoint genes
Two different algorithms, ssGSEA and MCPcounter, were used to calculate the infiltration of immune cells in TCGA datasets. To predict immune cell infiltration, we referred to the methodology described by Bindea et al. [12]. In their study, 24 immune cell gene sets were identified. Accordingly, we evaluated the infiltration of 24 different immune cell types using ssGSEA.
Since MCPcounter does not require additional gene sets, we utilized it to evaluate the infiltration of 10 immune cell types. Additionally, we investigated the expression of 22 immune checkpoint genes within the TCGA dataset. This comprehensive analysis allowed us to assess the immune landscape of pancreatic cancer accurately.
2.4. The prediction of immunotherapy and chemotherapy response
Using subclass mapping, we predicted the response of different pancreatic cancer subtypes to immunotherapy through the online tool GenePattern (https://cloud.genepattern.org/gp) [[13], [14], [15]]. To determine the responsiveness of different subtypes to chemotherapy, we utilized the largest publicly available pharmacogenomics database, the Genomics of Drug Sensitivity in Cancer (GDSC) (https://www.cancerrxgene.org/).
The prediction process involved using the R package “pRRophetic” to estimate the half-maximal inhibitory concentration (IC50) of the samples via ridge regression. Prediction accuracy was assessed using 10-fold cross-validation based on the GDSC training set. All parameters were set to their default values, and duplicate gene expressions were averaged. This approach allowed for a robust evaluation of drug sensitivity across different metabolic subtypes of pancreatic cancer.
2.5. Real-time fluorescence quantitative PCR to detect mRNA expression levels of prognostic genes in pancreatic cancer cell lines cultured in hypoxia
The human PAAD cell line, PaTu8988, was maintained at the Central Laboratory of Shanghai Fengxian District Central Hospital. Cells were cultured in DMEM supplemented with 10 % fetal bovine serum (FBS) and 1 % penicillin-streptomycin (both from Gibco, Thermo Fisher Scientific, Inc.), and kept in a humidified incubator with 5 % CO2 at 37 °C. RNA extraction and quantitative fluorescence PCR were performed at 24, 48, and 72 h.
Additionally, the PaTu8988 cell line was cultured under a cobalt chloride-induced hypoxic environment, and RNA extraction and quantitative fluorescence PCR were conducted at the same time points: 24, 48, and 72 h. This setup allowed for the comparison of gene expression under normoxic and hypoxic conditions.
2.6. Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from tissues and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The extracted RNA was then reverse transcribed into cDNA using the PrimeScript™ RT Reagent Kit (Takara Bio, Inc.) under the following conditions: 15 min at 37 °C and 5 s at 85 °C. Quantitative PCR (qPCR) was subsequently performed on an ABI 7300 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the SYBR® Premix Ex Taq™ II kit (Takara Bio, Inc.).
The thermocycling conditions for qPCR were as follows.
-
1.
Initial denaturation at 95 °C for 30 s;
-
2
40 cycles of denaturation at 95 °C for 5 s;
-
3.
Annealing at 58 °C for 30 s;
-
4.
Extension at 72 °C for 30 s.
The following primer pairs were used for qPCR.
-
1.
POM121C: forward 5′-CCCTTGCCTGCTCCCTTCTTC-3′ and reverse 5′-TACTCACACTGCTGCTGCTCAC-3′;
-
2.
LDHA: forward 5′-TCAGCCCGATTCCGTTACCTAATG-3′ and reverse 5′-CACCAGCAACATTCATTCCACTCC-3′;
-
3.
GAPDH: forward 5′-TTGGTATCGTGGAAGGACTCA-3′ and reverse 5′-TGTCATCATATTTGGCAGGTT-3′.
All primers were purchased from Sangon Biotech Co., Ltd. The relative mRNA expression levels were quantified using the 2−ΔΔCq method, with GAPDH serving as the internal control.
2.7. Statistical analysis
The Wilcoxon rank sum test was used to compare continuous variables between two groups. For variables with more than two categories, the Kruskal-Wallis test was chosen. Kaplan-Meier survival analysis was conducted using the “survival” package and evaluated with the log-rank test. Heatmaps were generated using the “pheatmap” package, and mutations were analyzed with “GenVisR.” All statistical analyses were performed using R software (version 3.6.2). All P-values were two-sided, and P < 0.05 was considered statistically significant.
3. Results
3.1. Four subtypes were identified by clustering analysis
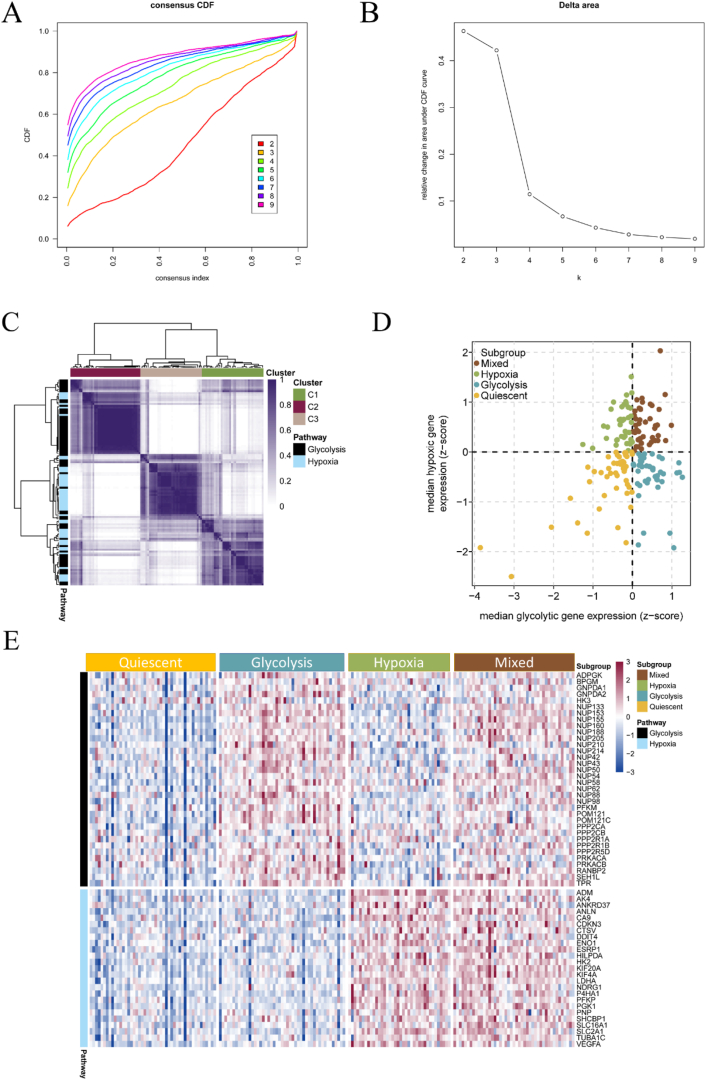
We used the collected 50 hypoxic and 72 glycolytic genes to perform subtype analysis on 178 pancreatic cancer samples from TCGA. Through unsupervised clustering analysis, all pancreatic cancers were classified into k (k = 2–9) different subtypes. By plotting the cumulative distribution function curves of the consensus score, we determined that k = 3 was optimal (Fig. 1A and B). Ultimately, 34 glycolysis-related genes and 25 hypoxia-related genes were successfully clustered using ConsensusClusterPlus (Fig. 1C).
Fig. 1.
Molecular subtypes based on glycolytic and hypoxic genes. (A) The cumulative distribution function (CDF) curves in consensus cluster analysis. (B) The delta area score displayed the relative growth in cluster stability. (C) Heatmap showing the consensus score matrix of all samples when K = 3. (D) The samples were divided into four different subtypes based on the median values of glycolytic and hypoxic genes. (E) Heatmap showing the expression of glycolytic and hypoxic genes in the four subtypes.
Next, we calculated the median expression levels of glycolytic and hypoxic genes for each sample, which allowed us to classify 177 samples into one of four subtypes based on their gene expression levels. Of these, 47 samples were identified as quiescent, 47 as glycolytic, 38 as hypoxic, and 45 as mixed (Fig. 1D; Supplementary Tables 1 and 2).
A heatmap was used to visualize the expression of glycolysis and hypoxia-related genes across the four subtypes. In this heatmap, columns represent samples and rows represent gene expression. In the quiescent group, both glycolysis- and hypoxia-related genes showed low expression. In the glycolytic group, glycolysis-related genes were highly expressed while hypoxia-related genes were lowly expressed. Conversely, in the hypoxic group, glycolysis-related genes were lowly expressed while hypoxia-related genes were highly expressed. In the mixed group, both glycolytic and hypoxic genes were highly expressed (Fig. 1E).
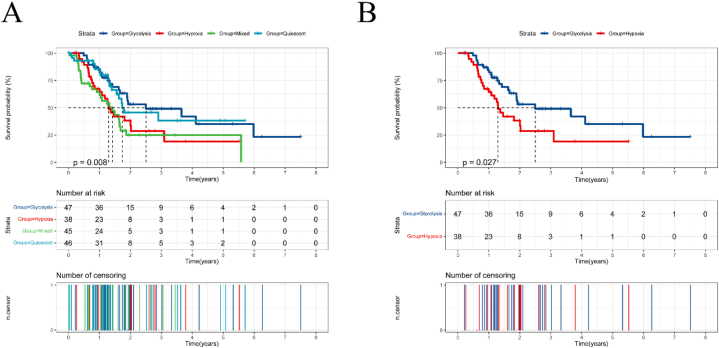
To compare the prognoses of different subtypes, we plotted Kaplan-Meier survival curves. The analysis revealed significant differences in overall survival between subtypes (Fig. 2A). Specifically, the hypoxic subtype had shorter overall survival times compared to the glycolytic subtype. In the survival curves, the blue line represents the glycolytic group, and the red line represents the hypoxic group. These lines did not intersect, indicating that the glycolytic group had both a longer median survival time and a longer overall survival time (Fig. 2B).
Fig. 2.
Survival analysis of the four subtypes. (A) Overall survival analysis of the four subtypes. (B) Overall survival of glycolytic and hypoxic subtypes.
3.2. Higher immune cells infiltration/immune checkpoint genes in glycolytic subtype
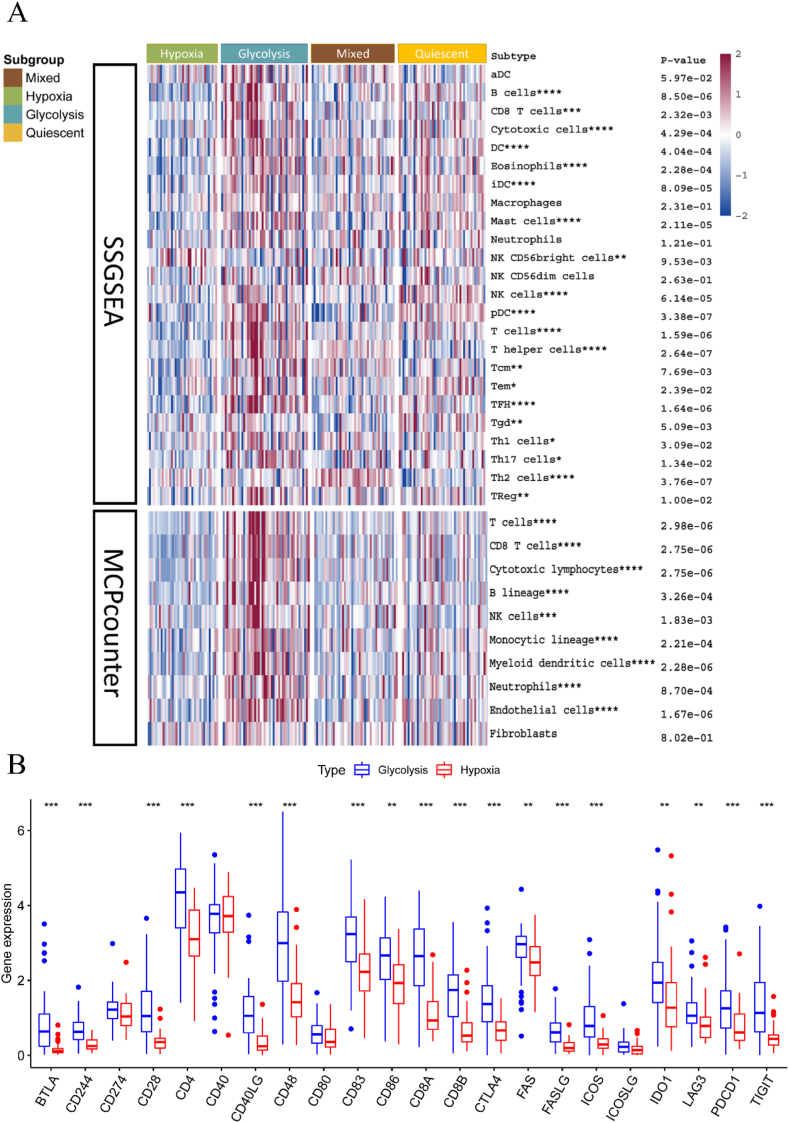
To explore variations in immune cell infiltration across different subtypes, we used two algorithms: ssGSEA and MCPcounter. The heatmap illustrates the infiltration of immune cells in various subtypes (Fig. 3A). Using the Kruskal-Wallis test for comparison, we observed that the glycolytic subtype exhibits more immune cell infiltration than the hypoxic subtype.
Fig. 3.
Subtypes with immune cells infiltration and immune checkpoint genes. (A) Infiltration of immune cells in different subtypes was inferred using two different methods, ssGSEA and MCPcounter. (B) Violin plot showing immune checkpoint genes in glycolytic subtype and hypoxic subtype.
We also compared the expression of 22 immune checkpoint genes between glycolytic and hypoxic subtypes. Our results show that immune checkpoint genes have significantly higher expression in the glycolytic subtype compared to the hypoxic subtype (Supplementary Fig. 1). Classical immune checkpoint genes, including PDCD1 (PD-1), were markedly more expressed in the glycolytic subtype than in the hypoxic subtype, as depicted in the violin plot (Fig. 3B). Thus, the glycolytic group had a higher inflammatory infiltrate, which is associated with a worse prognosis.
3.3. Glycolytic subtypes are more sensitive to immunotherapy/chemotherapy
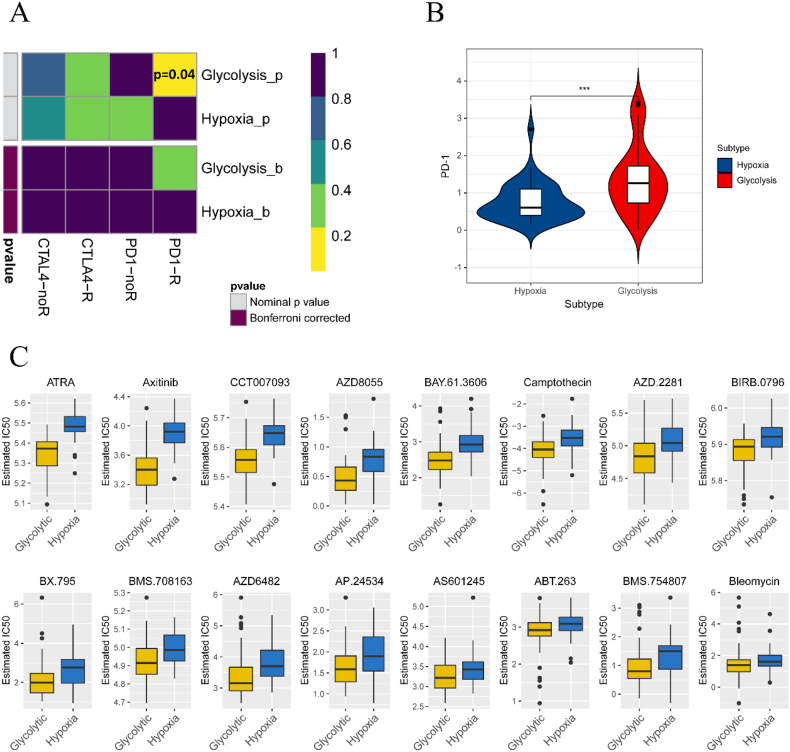
Subclass mapping was used to forecast the response of subtypes to immunotherapy. We compared the expression profiles of glycolytic and hypoxic subtypes with a published dataset containing 47 melanoma patients who responded to immunotherapy [14,15]. Our analysis indicated that the glycolytic subtype may be more promising for anti-PD-1 therapy (p = 0.04) (Fig. 4A). Additionally, we observed a difference in PD-1 expression between the two subtypes (Fig. 4B). This suggests that the glycolytic group is more likely to benefit from immunotherapy.
Fig. 4.
Subtypes response to immunotherapy and chemotherapy. (A) Using subclass mapping to infer subtypes response to immunotherapy. (B) PD-1 expression in glycolytic and hypoxic subtypes. (C) Effect of different chemotherapeutic agents on subtypes.
We further explored the sensitivity of subtypes to chemotherapeutic drugs. A prediction model was trained using the GDSC cell line dataset with ridge regression. The prediction accuracy was satisfactory, as confirmed by 10-fold cross-validation. After estimating the IC50 of samples in the TCGA dataset, we found that certain chemotherapeutic drugs, such as Axitinib and all-trans-retinoic acid (ATRA), were more effective in the glycolytic subtype, as evidenced by lower IC50 values (Fig. 4C). This implies that the glycolytic group receiving chemotherapy may achieve longer survival times and lower recurrence rates.
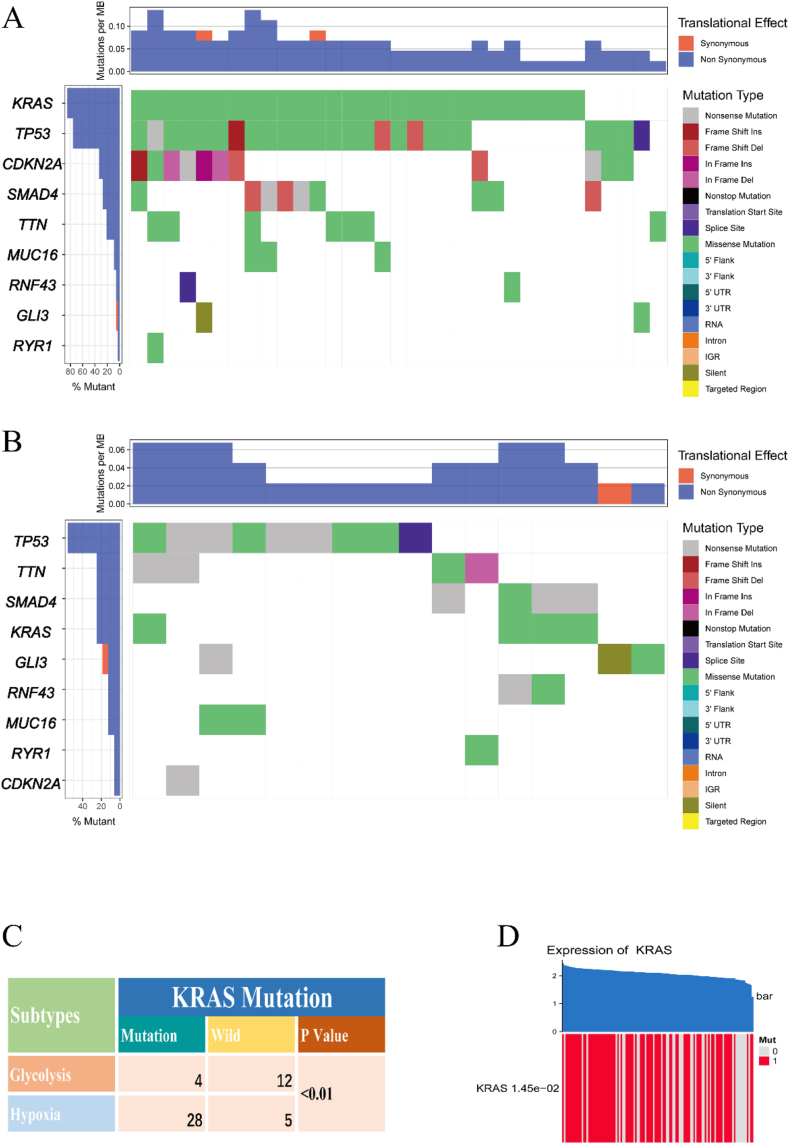
3.4. KRAS mutations are more frequent in the hypoxic subtype
To investigate mutations in subtypes, we downloaded SNP data from TCGA. We plotted the mutation landscape of glycolytic and hypoxic subtypes, respectively (Fig. 5A and B). Using the chi-square test to analyze these nine genes, we found that KRAS mutations were more frequent in the hypoxic subtype compared to the glycolytic subtype (Fig. 5C). Additionally, we examined the relationship between KRAS mutations and their expression. The permutation test indicated that samples with KRAS mutations had higher KRAS expression (Fig. 5D).
Fig. 5.
Subtypes and mutations. (A) Mutation landscape in the hypoxic subtype. (B) Mutation landscape in glycolytic subtype. (C) KRAS mutations in hypoxic and glycolytic subtypes. (D) Relationship between KRAS expression and mutation.
3.5. Construction of prognostic model and stratified analysis
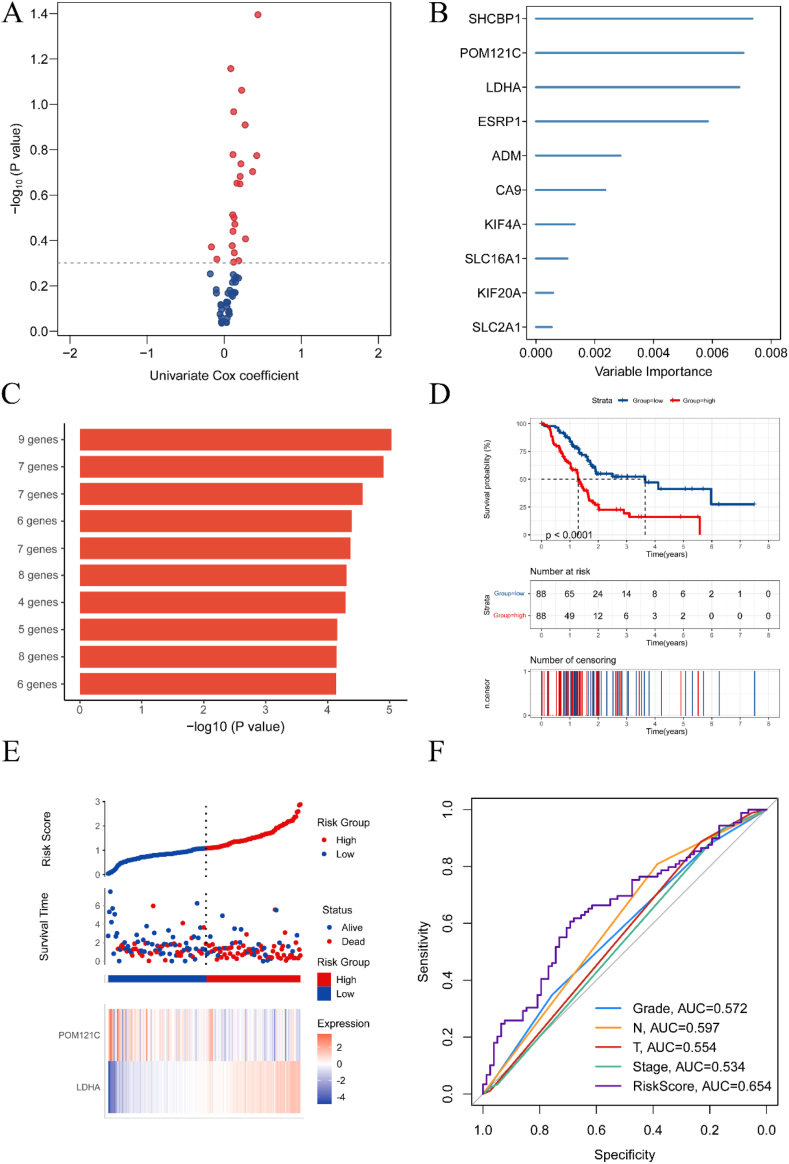
To construct a prognostic model based on glycolytic and hypoxic genes, we used 59 genes successfully clustered by ConsensusClusterPlus, including 34 glycolysis-related and 25 hypoxia-related genes. We initially included 22 genes with p < 0.5 in the subsequent analysis using univariate COX regression (Fig. 6A). We then screened 10 genes using the random survival forest algorithm (Fig. 6B). Since these 10 genes could form a total of 1023 risk models, we performed Kaplan-Meier analysis to identify the best risk models. The optimal module, which ranked first by comparing the -log10 P log-rank values of these 1023 models, included nine genes. We selected these nine genes for subsequent multivariate COX analysis (Fig. 6C). As some of these nine genes were not present in the two separate validation groups, we retained only the five genes present in all three datasets.
Fig. 6.
Establishment of a prognostic model. (A) Volcano plot showing the genes analyzed by univariate COX regression with P < 0.5. (B) The 10 candidate genes defined using random survival forest analysis. (C) The top ten combinations with the smallest P-values were selected using Kaplan-Meier analysis. (D) Survival analysis of the high-risk group and the low-risk group. (E) Distribution of risk scores and overall outcomes in TCGA. (F) ROC curves demonstrating the predictive power of the model.
The multivariate COX analysis ultimately identified two genes: LDHA and POM121C. We constructed a prognostic model based on these two genes. The prognostic risk score formula is: LDHA × 0.64 - POM121C × 0.29 (Table 1). Using the median risk score as the cut-off value, we classified the training group (TCGA) into high-risk and low-risk groups. The high-risk group, due to typically skewed risk score distribution, showed a worse prognosis (p < 0.0001) (Fig. 6D). Poor outcomes were more frequent in the high-risk group (Fig. 6E). We plotted receiver operating characteristic (ROC) curves for RiskScore, T-stage, N-stage, stage, and grade, with area under the curve (AUC) values of 0.654, 0.554, 0.597, 0.534, and 0.572, respectively (Fig. 6F). This demonstrated our model's higher predictive ability.
Table 1.
Multivariate Cox regression analysis of in the TCGA.
| coef | HR (95%CI) | P | |
| LDHA | 0.64 | 1.91 (1.41–2.57) | <0.01 |
| POM121C | −0.29 | 0.75 (0.50–1.11) | 0.15 |
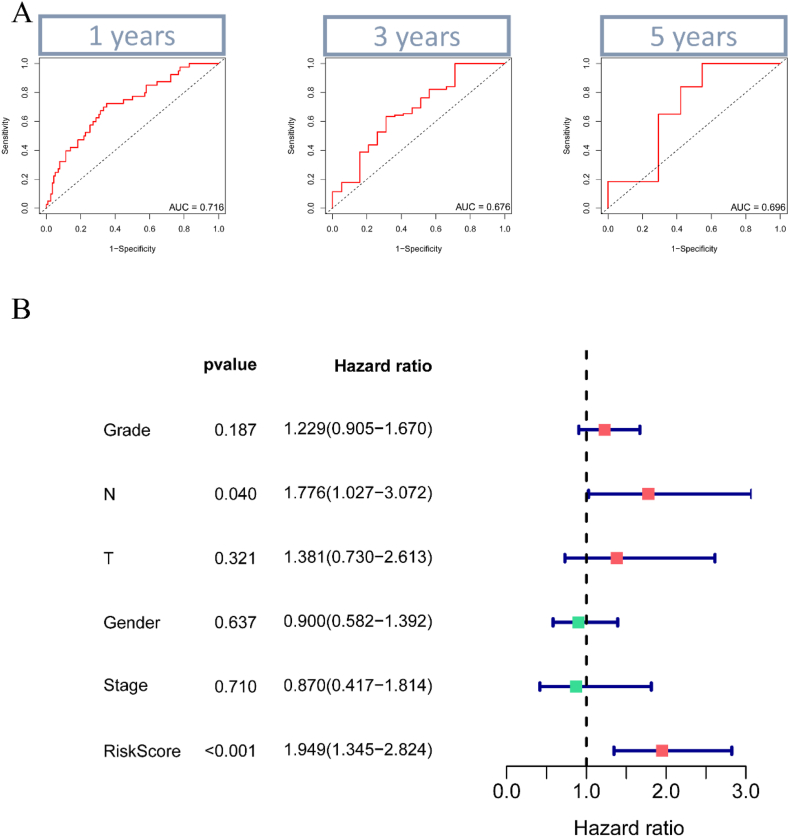
To test our model's predictive power, we plotted ROC curves at 1, 3, and 5 years, showing AUC values of 0.716, 0.676, and 0.696, respectively (Fig. 7A). Additionally, multivariate COX analysis confirmed that our model is an independent prognostic factor (Fig. 7B). Correlation analysis suggested a positive correlation between LDHA and T-stage (Supplementary Fig. 2).
Fig. 7.
ROC curves and multivariate COX analysis. (A) ROC curves were plotted at 1, 3, and 5 years, and the area under the curve (AUC) values were 0.716, 0.676, and 0.696 at 1, 3, and 5 years. (B) Multivariate COX analysis showed that our model was an independent prognostic factor.
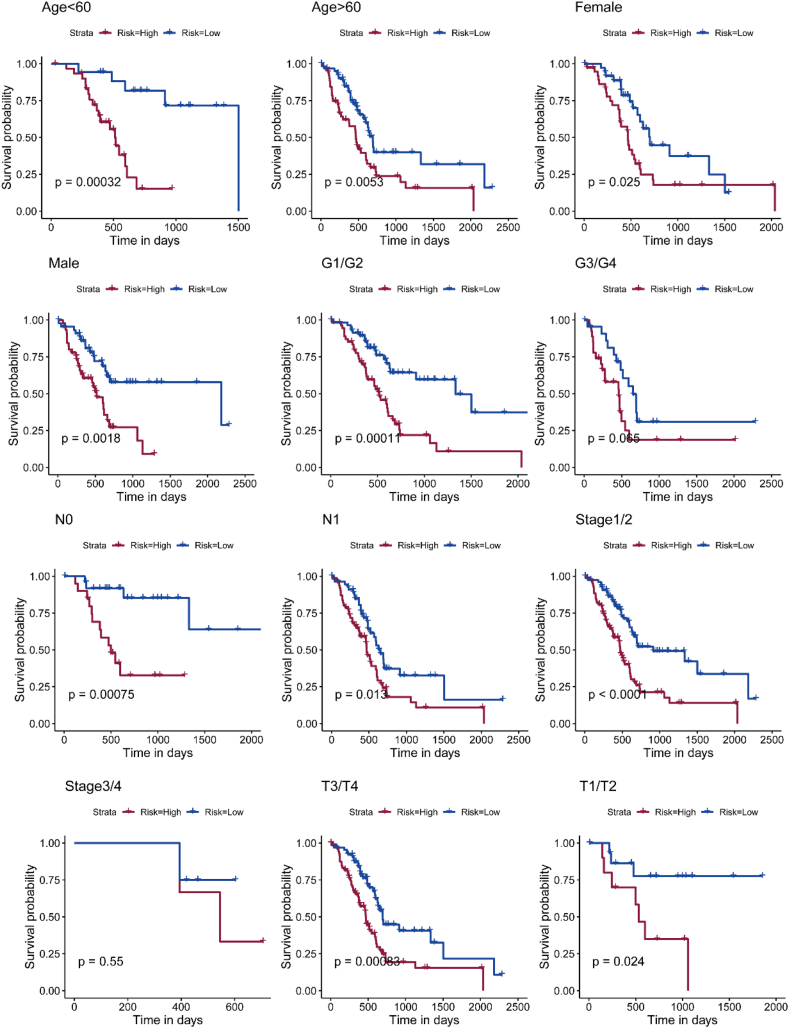
We also conducted a stratification analysis, revealing that our model remained clinically and statistically significant in predicting overall survival (OS) when stratified by age, stage, and tumor histological grade (Fig. 8). Compared to traditional tumor staging, our metabolism-related prognostic model predicts patient survival more accurately.
Fig. 8.
Prognostic model and stratification analysis. Stratification was performed according to age, stage and tumor histological grade.
3.6. POM121C and LDHA belong to the genes related to hypoxia and glycolysis in PAAD
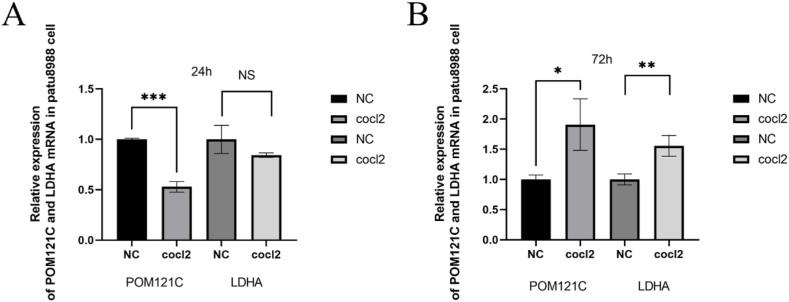
In the human PAAD cell line PaTu8988, RT-qPCR was used to analyze the differences in mRNA expression levels of LDHA and POM121C in normal and hypoxic environments. Compared to the normally cultured pancreatic cancer cell line, there was no significant increase in POM121C or LDHA gene expression at the 24th hour of hypoxic culture (Fig. 9A). However, the mRNA expression of LDHA and POM121C was significantly elevated in PaTu8988 at the 72 nd h of hypoxic culture (Fig. 9B).
Fig. 9.
Relative expression of LDHA and POM121C at molecular level detected by qPCR in pancreatic cancer cell line patu8988. (A) 24th hour in culture. (B) 72 nd h in culture, the mRNA levels of both genes, LDHA and POM121C, were significantly elevated.
3.7. External validation of the prediction model
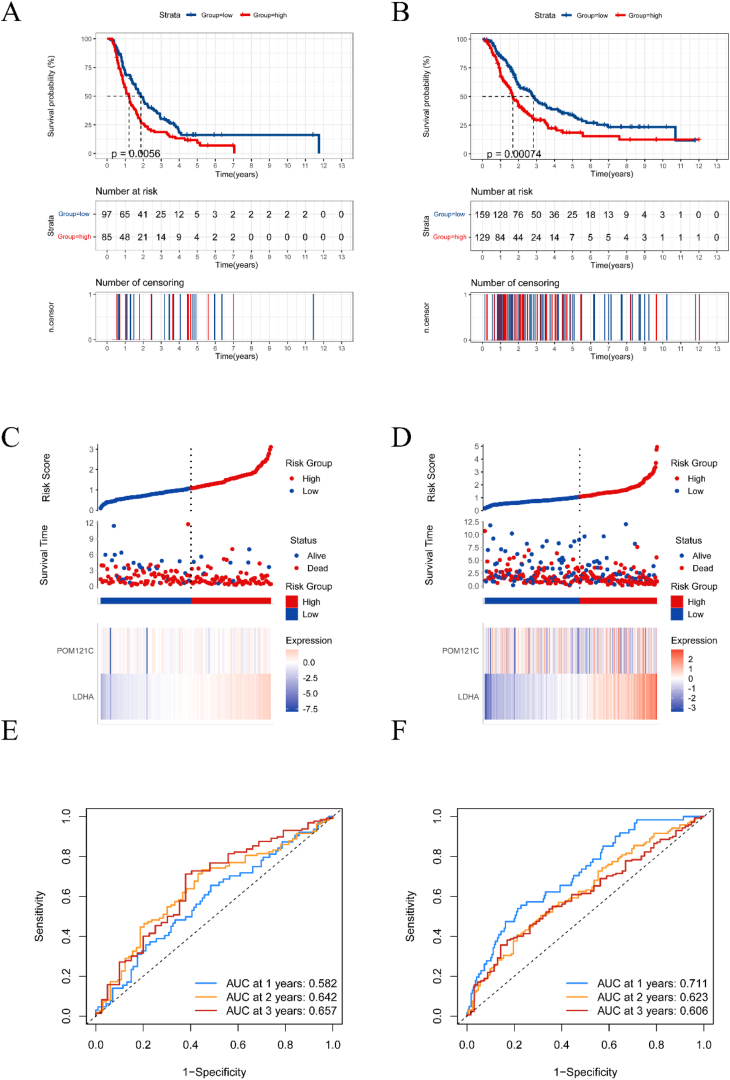
To further validate our model, we used two external validation datasets: ICGC and ArrayExpress. We first excluded patients with missing clinical data and then calculated risk scores using the previously established formula. Kaplan-Meier curves demonstrated that our model has significant prognostic value in both ICGC and ArrayExpress datasets (Fig. 10A and B). Similar to the TCGA cohort, we observed that patients in the high-risk group had a worse prognosis more frequently in both external validation groups (Fig. 10C and D).
Fig. 10.
External validation of the prognostic model. (A, B) Kaplan-Meier curves showing survival in the high-risk and low-risk groups in the ICGC and ArrayExpress cohorts, respectively. (C, D) Distribution of risk scores and overall outcomes distribution in the ICGC and ArrayExpress cohorts. (E, F) the receiver operating characteristic (ROC) curves demonstrating the predictive power of the model in the ICGC and ArrayExpress cohorts.
We also plotted the corresponding ROC curves. The AUC values were 0.582, 0.642, and 0.657 at 1, 2, and 3 years for the ICGC group (Figs. 10E), 0.711 and 0.623, and 0.606 at 1, 2, and 3 years for the ArrayExpress group (Fig. 10F). These results indicate that our risk score accurately stratified patients by their survival outcomes in both datasets. Therefore, our prognostic model is a meaningful and valuable tool for predicting survival in patients with pancreatic cancer.
4. Discussion
In the histology, abnormal proliferation of the stroma, as the most striking features of PDAC, can occupy 90 % of tumor tissue volume. This results in extensive fibrosis, lack of vascularity, hypoxia and immune cells infiltration [4,16,17]. Previous studies have shown that pancreatic cancer has the lowest oxygen levels of all solid tumors [18]. In addition, unlike normal cellular metabolism, most cancer cells rely primarily on glycolytic metabolism for energy, a phenomenon known as the Warburg effect [19]. Thus, glycolysis and hypoxia constitute an active loop, on the one hand, the hypoxic environment in pancreatic cancer prevents aerobic oxidation of sugar and promotes glycolysis, and on the other hand, glycolysis allows massive growth of tumor cells, and the large number of tumor cells leads to abnormal proliferation of stroma, thus creating a hypoxic microenvironment [20]. A large amount of experiments have pointed out that both hypoxia and glycolysis are both causes to the worse prognosis of pancreatic cancer [21,22].
Our study explored the relationship between glycolytic and hypoxic genes based on their association with pancreatic cancer. The glycolytic subtype had a better prognosis compared to the hypoxic subtype. Previous studies have indicated that in pancreatic cancer, higher glycolysis scores tend to predict a worse prognosis, so we hypothesized that hypoxia may be more likely to lead to a poor prognosis than glycolysis in pancreatic cancer [[23], [24], [25]].
We also evaluated the immune cells infiltration in pancreatic cancer, and we found that the immune cells infiltration was much more abundant in the glycolytic subtype, especially some anti-tumor immune cells such as NK cells, CD8+ T cells, and the infiltration of these immune cells may be one of the reasons for the relatively better prognosis of the glycolytic subtype [26].
In the analysis of immune checkpoint genes, we found that PD-1 (PDCD1), CTLA4 and other immune checkpoint genes were significantly higher in glycolytic subtype than in hypoxic subtype. Nowadays, immunotherapy has been used as a novel therapy in many tumors, and immune checkpoint blockade (ICB) using PD-1/PD-L1 antibodies has revolutionized the treatment of some cancers, especially non-small cell lung cancer, melanoma, uroepithelial and renal cancers [27]. Although single PD-1/PD-L1 blockade is not very effective in treating pancreatic cancer, it has been suggested that PD-1/PD-L1 blockade combined with CSF1R blockade is more effective in tumor suppression, and in addition, PD-1/PD-L1 blockade combined with GM-CSF cell-based vaccines (GVAX) can also improve prognosis of patients [28,29]. Notably, when we analyzed glycolytic subtype and hypoxic subtype using subclass mapping, we found an interesting phenomenon that glycolytic subtype seemed to be more effective for anti-PD-1 treatment. In addition, we also performed chemotherapy drug prediction using GDSC, and we found that two chemotherapy drugs, all-trans-retinoicacid (ATRA) and Axitinib, had smaller IC50 in the glycolytic subtype, implying that the glycolytic subtype is more sensitive to these two drugs. Previous studies have shown that ATRA inhibits tumor growth by restoring retinoic acid (RA) stores in pancreatic stellate cells, and in addition, ATRA inhibits aberrantly activated pathways in pancreatic cancer [[30], [31], [32]]. Axitinib is an inhibitor of the vascular endothelial growth factor receptor. Although a previous randomized phase 2 trial of gemcitabine combined with or without axitinib for advanced pancreatic cancer showed a better outcome in patients treated with the combination of axitinib, the results of a subsequent phase 3 trial were discouraging: axitinib combined with gemcitabine could not refine the outcome of patients with advanced pancreatic cancer [33]. Hypoxia decreases the responsiveness of tumors to most anti-cancer drugs. This is because during prolonged hypoxia, most cells end their lives by programmed death, however, some cells adapt to their environment and survive by avoiding necrosis and apoptosis. Some tumor cells may even become more aggressive, and in a hypoxic environment, tumors adapt through activation of hypoxia-inducible factor (hif). However, hif in turn increases the expression of genes related to angiogenesis, metabolic regulation, pH homeostasis and apoptosis, thus enhancing the viability of tumor cells. The presence of the Pasteur effect in tumor cells gives them an innate growth and replication advantage over somatic cells. Animal and cellular experiments by Milane L et al. demonstrated that tumors derived from hypoxic cells had a higher MDR (the development of multi-drug resistance) than tumors derived from normoxic cells in a mouse model. This was especially true at the margins of the tumors. Tumor cells formed by hypoxia exhibited more aggressive growth [[34], [35], [36]].
Besides, when we studied SNP, we found that KRAS mutations were more frequent in the hypoxic subtype, and KRAS mutations are one of the strongest oncogenic drivers in all cancers [37]. In animal models of pancreatic cancer, excision of KRAS mutations resulted in complete or significant tumor regression, making KRAS mutations an important therapeutic target [38,39]. In pancreatic cancer, the relationship between KRAS mutations and hypoxia involves complex interactions that enhance tumor survival and proliferation. Specifically, the interaction between the Receptor for Advanced Glycation End Products (RAGE) and mutant KRAS increases under hypoxic conditions, sustaining KRAS signaling pathways. These pathways include RAF-MEK-ERK and PI3K-AKT, both of which are instrumental in promoting cellular proliferation and survival under hypoxic stress [40].
Furthermore, mutant KRAS enhances the stabilization and activation of HIF1α by increasing its interaction with RAGE, leading to a survival advantage in hypoxic tumor environments. This interaction particularly benefits pancreatic tumor cells by sustaining cellular metabolism and supporting invasive characteristics through enhanced angiogenesis and anaerobic metabolism. This mechanistic insight highlights the pivotal role of mutant KRAS in not only driving pancreatic tumorigenesis but also in adapting to the hypoxic microenvironment, thereby supporting tumor growth and resistance to therapy [40].
We also constructed a prognostic model using glycolytic and hypoxic genes, demonstrating robust predictive power in both the TCGA cohort and an external validation cohort. Lactate dehydrogenase A (LDHA), which converts pyruvate to lactate, is frequently overexpressed and activated in various cancers. Numerous studies have highlighted the role of LDHA in promoting tumor progression and invasion, making it a potential target for cancer prevention and treatment [41]. Conversely, research on POM121C in the context of pancreatic cancer remains limited, necessitating further investigation.
We examined the relationship between different subtypes of pancreatic cancer and prognosis using glycolytic and hypoxic genes. Our findings indicate that the glycolytic subtype is associated with a better prognosis compared to the hypoxic subtype. Additionally, both immunotherapy and chemotherapy appear to be more effective for the glycolytic subtype. Our prognostic model, incorporating glycolytic and hypoxic genes, demonstrated high accuracy across all three datasets.
5. Conclusion
Through clustering analysis, we identified distinct metabolic subtypes of glycolysis and hypoxia in pancreatic cancer and developed a metabolic subtype-based survival prediction model. The hypoxic metabolic subtype was associated with a poorer prognosis, a finding that was corroborated by subsequent tumor microenvironment and SNP analyses. TIDE analysis revealed that enhanced glycolytic metabolism increases the sensitivity of pancreatic cancer to antitumor drugs and immunotherapy.
However, this study has notable limitations. Due to constraints, we were unable to perform protein-level validation experiments. Additionally, we could not integrate our findings with the gene sequencing data from patients at our center, which may affect the reliability of our conclusions.
Data availability statement
The data used in our study are available in TCGA, ICGC and ArrayExpress.
Funding information
This research was funded by Suzhou Science and Technology Bureau (SSTB) (SKJY2021053)
CRediT authorship contribution statement
Yujie Huang: Conceptualization, Data curation, Investigation, Methodology. Qilu Zhu: Project administration, Resources. Yizhang Sun: Investigation, Software. Weigang Zhang: Formal analysis, Software, Visualization. Jiayue Zou: Software, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Weigang Zhang reports financial support was provided by Suzhou Science and Technology Bureau (SSTB). If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e34104.
Contributor Information
Yujie Huang, Email: 1017693747@qq.com.
Qilu Zhu, Email: 453832409@qq.com.
Yizhang Sun, Email: a18896777080@163.com.
Weigang Zhang, Email: zhangwg1116@hotmail.com.
Jiayue Zou, Email: jiayuezou56@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA A Cancer J. Clin. 2019 Jan;69(1):7–34. doi: 10.3322/caac.21551. Epub 2019 Jan 8. PMID: 30620402. [DOI] [PubMed] [Google Scholar]
- 2.Bekkali N.L.H., Oppong K.W. Pancreatic ductal adenocarcinoma epidemiology and risk assessment: could we prevent? Possibility for an early diagnosis. Endosc Ultrasound. 2017 Dec;6(Suppl 3):S58–S61. doi: 10.4103/eus.eus_60_17. PMID: 29387690; PMCID: PMC5774073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kakkar A., Choudhuri J., Mukherjee I. The subtypes of pancreatic ductal adenocarcinomas. Sanamed. 2016;11(3):239–242. doi: 10.24125/sanamed.v11i3.151. [DOI] [Google Scholar]
- 4.Kamphorst J.J., Cross J.R., Fan J., de Stanchina E., Mathew R., White E.P., Thompson C.B., Rabinowitz J.D. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. U. S. A. 2013 May 28;110(22):8882–8887. doi: 10.1073/pnas.1307237110. Epub 2013 May 13. PMID: 23671091; PMCID: PMC3670379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu J., Lin J., Dai Z., Lin B., Zhang J. Hypoxia-associated autophagy flux dysregulation in human cancers. Cancer Lett. 2024 May 28;590 doi: 10.1016/j.canlet.2024.216823. Epub 2024 Mar 21. PMID: 38521197. [DOI] [PubMed] [Google Scholar]
- 6.Yang K., Zhong Z., Zou J., Liao J.Y., Chen S., Zhou S., Zhao Y., Li J., Yin D., Huang K., Li Y. Glycolysis and tumor progression promoted by the m6A writer VIRMA via m6A-dependent upregulation of STRA6 in pancreatic ductal adenocarcinoma. Cancer Lett. 2024 May 28;590 doi: 10.1016/j.canlet.2024.216840. Epub 2024 Apr 9. PMID: 38604311. [DOI] [PubMed] [Google Scholar]
- 7.Daemen A., Peterson D., Sahu N., McCord R., Du X., Liu B., Kowanetz K., Hong R., Moffat J., Gao M., Boudreau A., Mroue R., Corson L., O'Brien T., Qing J., Sampath D., Merchant M., Yauch R., Manning G., Settleman J., Hatzivassiliou G., Evangelista M. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2015 Aug 11;112(32):E4410–E4417. doi: 10.1073/pnas.1501605112. Epub 2015 Jul 27. PMID: 26216984; PMCID: PMC4538616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J.J., Shao C., Yin Y.X., Sun Q., Li Y.N., Zha Y.W., Li M.Y., Hu B.L. Hypoxia-related signature is a prognostic biomarker of pancreatic cancer. Dis. Markers. 2022 Jun 25;2022 doi: 10.1155/2022/6449997. PMID: 35789607; PMCID: PMC9250441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Integrated Analysis Revealed Hypoxia Signatures and LDHA Related to Tumor Cell Dedifferentiation and Unfavorable Prognosis in Pancreatic Adenocarcinoma: Hypoxia in PDAC. [DOI] [PMC free article] [PubMed]
- 10.Chen D., Huang H., Zang L., Gao W., Zhu H., Yu X. Development and verification of the hypoxia- and immune-associated prognostic signature for pancreatic ductal adenocarcinoma. Front. Immunol. 2021 Oct 6;12 doi: 10.3389/fimmu.2021.728062. PMID: 34691034; PMCID: PMC8526937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Follia L., Ferrero G., Mandili G., Beccuti M., Giordano D., Spadi R., Satolli M.A., Evangelista A., Katayama H., Hong W., Momin A.A., Capello M., Hanash S.M., Novelli F., Cordero F. Integrative analysis of novel metabolic subtypes in pancreatic cancer fosters new prognostic biomarkers. Front. Oncol. 2019 Feb 27;9:115. doi: 10.3389/fonc.2019.00115. PMID: 30873387; PMCID: PMC6400843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roh W., Chen P.L., Reuben A., Spencer C.N., Prieto P.A., Miller J.P., Gopalakrishnan V., Wang F., Cooper Z.A., Reddy S.M., Gumbs C., Little L., Chang Q., Chen W.S., Wani K., De Macedo M.P., Chen E., Austin-Breneman J.L., Jiang H., Roszik J., Tetzlaff M.T., Davies M.A., Gershenwald J.E., Tawbi H., Lazar A.J., Hwu P., Hwu W.J., Diab A., Glitza I.C., Patel S.P., Woodman S.E., Amaria R.N., Prieto V.G., Hu J., Sharma P., Allison J.P., Chin L., Zhang J., Wargo J.A., Futreal P.A. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 2017 Mar 1;9(379) doi: 10.1126/scitranslmed.aah3560. Erratum in: Sci Transl Med. 2017 Apr 12;9(385): PMID: 28251903; PMCID: PMC5819607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B., Liu J., Freeman G.J., Brown M.A., Wucherpfennig K.W., Liu X.S. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018 Oct;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. Epub 2018 Aug 20. PMID: 30127393; PMCID: PMC6487502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoshida Y., Brunet J.P., Tamayo P., Golub T.R., Mesirov J.P. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS One. 2007 Nov 21;2(11) doi: 10.1371/journal.pone.0001195. PMID: 18030330; PMCID: PMC2065909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma X., Zhao X., Ouyang H., Sun F., Zhang H., Zhou C., Shen H. The metabolic features of normal pancreas and pancreatic adenocarcinoma: preliminary result of in vivo proton magnetic resonance spectroscopy at 3.0 T. J. Comput. Assist. Tomogr. 2011 Sep-Oct;35(5):539–543. doi: 10.1097/RCT.0b013e318227a545. PMID: 21926845. [DOI] [PubMed] [Google Scholar]
- 16.Hingorani S.R. Cellular and molecular conspirators in pancreas cancer. Carcinogenesis. 2014 Jul;35(7):1435. doi: 10.1093/carcin/bgu138. PMID: 24987024. [DOI] [PubMed] [Google Scholar]
- 17.Koong A.C., Mehta V.K., Le Q.T., Fisher G.A., Terris D.J., Brown J.M., Bastidas A.J., Vierra M. Pancreatic tumors show high levels of hypoxia. Int. J. Radiat. Oncol. Biol. Phys. 2000 Nov 1;48(4):919–922. doi: 10.1016/s0360-3016(00)00803-8. PMID: 11072146. [DOI] [PubMed] [Google Scholar]
- 18.Warburg O., Wind F., Negelein E. The metabolism of tumors in the body. J. Gen. Physiol. 1927 Mar 7;8(6):519–530. doi: 10.1085/jgp.8.6.519. PMID: 19872213; PMCID: PMC2140820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spivak-Kroizman T.R., Hostetter G., Posner R., Aziz M., Hu C., Demeure M.J., Von Hoff D., Hingorani S.R., Palculict T.B., Izzo J., Kiriakova G.M., Abdelmelek M., Bartholomeusz G., James B.P., Powis G. Hypoxia triggers hedgehog-mediated tumor-stromal interactions in pancreatic cancer. Cancer Res. 2013 Jun 1;73(11):3235–3247. doi: 10.1158/0008-5472.CAN-11-1433. Epub 2013 Apr 30. PMID: 23633488; PMCID: PMC3782107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011 Mar 4;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. PMID: 21376230. [DOI] [PubMed] [Google Scholar]
- 21.Li J., Zhu C., Yue P., Zheng T., Li Y., Wang B., Meng X., Zhang Y. Identification of glycolysis related pathways in pancreatic adenocarcinoma and liver hepatocellular carcinoma based on TCGA and GEO datasets. Cancer Cell Int. 2021 Feb 19;21(1):128. doi: 10.1186/s12935-021-01809-y. PMID: 33607990; PMCID: PMC7893943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamasaki A., Yanai K., Onishi H. Hypoxia and pancreatic ductal adenocarcinoma. Cancer Lett. 2020 Aug 1;484:9–15. doi: 10.1016/j.canlet.2020.04.018. Epub 2020 May 5. PMID: 32380129. [DOI] [PubMed] [Google Scholar]
- 23.Sun J.S., Zhang X.L., Yang Y.J., Nie Z.G., Zhang Y. Hypoxia promotes C-X-C chemokine receptor type 4 expression through microRNA-150 in pancreatic cancer cells. Oncol. Lett. 2015;10(2):835–840. doi: 10.3892/ol.2015.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y., Yu X., Zhang F., Dai J. Propofol inhibits pancreatic cancer progress under hypoxia via ADAM8. J Hepatobiliary Pancreat Sci. 2019 Jun;26(6):219–226. doi: 10.1002/jhbp.624. Epub 2019 May 11. PMID: 30945470. [DOI] [PubMed] [Google Scholar]
- 25.Ino Y., Yamazaki-Itoh R., Shimada K., Iwasaki M., Kosuge T., Kanai Y., Hiraoka N. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br. J. Cancer. 2013 Mar 5;108(4):914–923. doi: 10.1038/bjc.2013.32. Epub 2013 Feb 5. PMID: 23385730; PMCID: PMC3590668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macherla S., Laks S., Naqash A.R., Bulumulle A., Zervos E., Muzaffar M. Emerging role of immune checkpoint blockade in pancreatic cancer. Int. J. Mol. Sci. 2018 Nov 7;19(11):3505. doi: 10.3390/ijms19113505. PMID: 30405053; PMCID: PMC6274962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu Y., Knolhoff B.L., Meyer M.A., Nywening T.M., West B.L., Luo J., Wang-Gillam A., Goedegebuure S.P., Linehan D.C., DeNardo D.G. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014 Sep 15;74(18):5057–5069. doi: 10.1158/0008-5472.CAN-13-3723. Epub 2014 Jul 31. PMID: 25082815; PMCID: PMC4182950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le D.T., Lutz E., Uram J.N., Sugar E.A., Onners B., Solt S., Zheng L., Diaz L.A., Jr., Donehower R.C., Jaffee E.M., Laheru D.A. Evaluation of ipilimumab in combination with allogeneic pancreatic tumor cells transfected with a GM-CSF gene in previously treated pancreatic cancer. J. Immunother. 2013 Sep;36(7):382–389. doi: 10.1097/CJI.0b013e31829fb7a2. PMID: 23924790; PMCID: PMC3779664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Froeling F.E., Feig C., Chelala C., Dobson R., Mein C.E., Tuveson D.A., Clevers H., Hart I.R., Kocher H.M. Retinoic acid-induced pancreatic stellate cell quiescence reduces paracrine Wnt-β-catenin signaling to slow tumor progression. Gastroenterology. 2011 Oct;141(4):1486–1497. doi: 10.1053/j.gastro.2011.06.047. 1497.e1-1497. Epub 2011 Jun 24. PMID: 21704588. [DOI] [PubMed] [Google Scholar]
- 30.Ene-Obong A., Clear A.J., Watt J., Wang J., Fatah R., Riches J.C., Marshall J.F., Chin-Aleong J., Chelala C., Gribben J.G., Ramsay A.G., Kocher H.M. Activated pancreatic stellate cells sequester CD8+ T cells to reduce their infiltration of the juxtatumoral compartment of pancreatic ductal adenocarcinoma. Gastroenterology. 2013 Nov;145(5):1121–1132. doi: 10.1053/j.gastro.2013.07.025. Epub 2013 Jul 25. PMID: 23891972; PMCID: PMC3896919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Maggio F., Arumugam P., Delvecchio F.R., Batista S., Lechertier T., Hodivala-Dilke K., Kocher H.M. Pancreatic stellate cells regulate blood vessel density in the stroma of pancreatic ductal adenocarcinoma. Pancreatology. 2016 Nov-Dec;16(6):995–1004. doi: 10.1016/j.pan.2016.05.393. Epub 2016 Jun 1. PMID: 27288147; PMCID: PMC5123629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kindler H.L., Ioka T., Richel D.J., Bennouna J., Létourneau R., Okusaka T., Funakoshi A., Furuse J., Park Y.S., Ohkawa S., Springett G.M., Wasan H.S., Trask P.C., Bycott P., Ricart A.D., Kim S., Van Cutsem E. Axitinib plus gemcitabine versus placebo plus gemcitabine in patients with advanced pancreatic adenocarcinoma: a double-blind randomised phase 3 study. Lancet Oncol. 2011 Mar;12(3):256–262. doi: 10.1016/S1470-2045(11)70004-3. PMID: 21306953. [DOI] [PubMed] [Google Scholar]
- 33.Koritzinsky M., Magagnin M.G., van den Beucken T., et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25(5):1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milane L., Duan Z., Amiji M. Role of hypoxia and glycolysis in the development of multi-drug resistance in human tumor cells and the establishment of an orthotopic multi-drug resistant tumor model in nude mice using hypoxic pre-conditioning. Cancer Cell Int. 2011 Feb 14;11:3. doi: 10.1186/1475-2867-11-3. PMID: 21320311; PMCID: PMC3045873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graham K., Unger E. Overcoming tumor hypoxia as a barrier to radiotherapy, chemotherapy and immunotherapy in cancer treatment. Int. J. Nanomed. 2018 Oct 4;13:6049–6058. doi: 10.2147/IJN.S140462. PMID: 30323592; PMCID: PMC6177375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Almoguera C., Shibata D., Forrester K., Martin J., Arnheim N., Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988 May 20;53(4):549–554. doi: 10.1016/0092-8674(88)90571-5. PMID: 2453289. [DOI] [PubMed] [Google Scholar]
- 37.Collins M.A., Bednar F., Zhang Y., Brisset J.C., Galbán S., Galbán C.J., Rakshit S., Flannagan K.S., Adsay N.V., Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Invest. 2012 Feb;122(2):639–653. doi: 10.1172/JCI59227. Epub 2012 Jan 9. PMID: 22232209; PMCID: PMC3266788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying H., Kimmelman A.C., Lyssiotis C.A., Hua S., Chu G.C., Fletcher-Sananikone E., Locasale J.W., Son J., Zhang H., Coloff J.L., Yan H., Wang W., Chen S., Viale A., Zheng H., Paik J.H., Lim C., Guimaraes A.R., Martin E.S., Chang J., Hezel A.F., Perry S.R., Hu J., Gan B., Xiao Y., Asara J.M., Weissleder R., Wang Y.A., Chin L., Cantley L.C., DePinho R.A. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012 Apr 27;149(3):656–670. doi: 10.1016/j.cell.2012.01.058. PMID: 22541435; PMCID: PMC3472002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sheng S.L., Liu J.J., Dai Y.H., Sun X.G., Xiong X.P., Huang G. Knockdown of lactate dehydrogenase A suppresses tumor growth and metastasis of human hepatocellular carcinoma. FEBS J. 2012 Oct;279(20):3898–3910. doi: 10.1111/j.1742-4658.2012.08748.x. Epub 2012 Sep 13. PMID: 22897481. [DOI] [PubMed] [Google Scholar]
- 40.Kang R., Hou W., Zhang Q., Chen R., Lee Y.J., Bartlett D.L., Lotze M.T., Tang D., Zeh H.J. RAGE is essential for oncogenic KRAS-mediated hypoxic signaling in pancreatic cancer. Cell Death Dis. 2014 Oct 23;5(10) doi: 10.1038/cddis.2014.445. PMID: 25341034; PMCID: PMC4237264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fantin V.R., St-Pierre J., Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006 Jun;9(6):425–434. doi: 10.1016/j.ccr.2006.04.023. Erratum in: Cancer Cell. 2006 Aug;10(2):172. PMID: 16766262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in our study are available in TCGA, ICGC and ArrayExpress.