Abstract
Enhancing cardiomyocyte proliferation is essential to reverse or slow down the heart failure progression in many cardiovascular diseases such as myocardial infarction (MI). Long non-coding RNAs (lncRNAs) have been reported to regulate cardiomyocyte proliferation. In particular, lncRNA urothelial carcinoma-associated 1 (lncUCA1) played multiple roles in regulating cell cycle progression and cardiovascular diseases, making lncUCA1 a potential target for promoting cardiomyocyte proliferation. However, the role of lncUCA1 in cardiomyocyte proliferation remains unknown. This study aimed at exploring the function and underlying molecular mechanism of lncUCA1 in cardiomyocyte proliferation. Quantitative RT-PCR showed that lncUCA1 expression decreased in postnatal hearts. Gain-and-loss-of-function experiments showed that lncUCA1 positively regulated cardiomyocyte proliferation in vitro and in vivo. The bioinformatics program identified miR-128 as a potential target of lncUCA1, and loss of miR-128 was reported to promote cardiomyocyte proliferation by inhibiting the SUZ12/P27 pathway. Luciferase reporter assay, qRT-PCR, western blotting, and immunostaining experiments further revealed that lncUCA1 acted as a ceRNA of miR-128 to upregulate its target SUZ12 and downregulate P27, thereby increasing cyclin B1, cyclin E, CDK1 and CDK2 expression to promote cardiomyocyte proliferation. In conclusion, upregulation of lncRNA UCA1 promoted cardiomyocyte proliferation by inhibiting the miR-128/SUZ12/P27 pathway. Our results indicated that lncUCA1 might be a new therapeutic target for stimulating cardiomyocyte proliferation.
Keywords: lncRNA UCA1, Cardiomyocyte proliferation, miR-128, suz12, P27
Abbreviations
- AAV9
(adeno-associated virus 9)
- BSA
(bovine serum albumin)
- CMs
(cardiomyocytes)
- CFs
(cardiac fibroblasts)
- DMEM
(Dulbecco's modified Eagle medium)
- E15.5
(embryonic day 15.5)
- ECs
(endothelial cells)
- EdU
(5-ethynyl-2′-deoxyuridine)
- FBS
(fetal bovine serum)
- FISH
(fluorescent in situ hybridization)
- LAD
(left anterior descending coronary artery)
- LncRNAs
(long non-coding RNAs)
- LncUCA1
(long non-coding RNA urothelial carcinoma-associated 1)
- MI
(myocardial infarction)
- P1
(postnatal day 1)
- P7
(postnatal day 7)
- pH3
(phosphorylated histone H3)
- qRT-PCR
(quantitative RT-PCR)
- SD
(Sprague Dawley)
- siRNA
(small interfering RNA)
- WGA
(wheat germ agglutinin)
1. Introduction
Cardiovascular diseases continue to be a leading cause of mortality worldwide [1]. Due to the irreversible loss of cardiomyocytes (CMs) and excessive cardiac fibrosis, many cardiovascular diseases such as myocardial infarction (MI) would progress to heart failure [2]. After myocardial infarction, the limited CM self-renewal rate in the adult mammalian heart is insufficient to compensate for CM loss [3]. However, myocardial injury within 1 week after birth in neonatal mice could trigger cardiac regeneration and result in complete functional recovery within 1 month [3]. Thus, reactivation of cardiomyocyte proliferation in adult hearts is essential to reverse or slow down the progression of heart failure progression in cardiovascular diseases [4].
Long non-coding RNAs are a class of transcripts longer than 200 nt in length and have no or limited ability to encode proteins [5]. LncRNAs have been reported to participate in many biological and pathological processes, such as cell proliferation [5] and cell pyroptosis [6]. Increasing evidence has revealed that lncRNA expression is closely related to the occurrence and development of various cardiovascular diseases such as hypertension, myocardial ischemia/reperfusion injury and cardiac hypertrophy [7,8]. A larger number of studies have confirmed that lncRNAs could awake postnatal CMs to re-enter the cell cycle and trigger cardiac regeneration [9,10]. LncRNA AZIN2-sv was the first lncRNA identified as a regulator of cardiomyocyte proliferation. AZIN-sv upregulated during postnatal heart development and regulated CM proliferation through the miR-214/PTEN/Akt pathway [11]. Other lncRNAs such as CPR [12] and ECRAR [13] were also reported to stimulate sufficient cardiac regeneration. These studies indicated that lncRNAs are ideal targets for regulating cardiomyocyte proliferation. Further explorating the roles of lncRNAs in cardiomyocyte proliferation might result in new therapeutic targets for treating human IHD and its related diseases.
LncRNA Urothelial carcinoma-associated 1 (lncUCA1) was first reported to be a biomarker in bladder transitional cell carcinoma and was of great importance in regulating cell proliferation [14]. To date, lncUCA1 has been reported to play multiple roles in regulating cardiovascular diseases [[15], [16], [17]]. Recent studies showed that hypoxia-conditioned hMSC-derived lncUCA1 plays a cardioprotective role in rat MI models by inhibiting the miR-873-5p/XIAP axis, and circulating exosomal lncUCA1 could be a marker of AMI [15]. Another study reported that lncUCA1 protected rat cardiomyocytes from hypoxia/reoxygenation-induced apoptosis by inhibiting the miR-143/MDM2/p53 axis [16]. Moreover, upregulation of lncUCA1 by morphine post-conditioning could alleviate autophagy in ischemia-reperfusion induced cardiac injury [17]. Although lncUCA1 has been reported to play important roles in regulating cell cycle progression and several cardiovascular diseases, it is still unknown whether lncUCA1 could be a novel target for regulating cardiomyocyte proliferation.
The integrated bioinformatic analysis has shown that lncUCA1 harbors predictive binding sites for miR-12817. MiR-128 was reported to be a negative regulator of cardiomyocyte proliferation and loss of miR-128 induced cardiac regeneration by inhibiting the SUZ12/P27 pathway [18]. Based on the relationship between lncUCA1 and miR-128, we hypothesized that lncUCA1 might act as a ceRNA of miR-128 to abrogate its effect, thereby controlling the SUZ12/P27 pathway to regulate cardiomyocyte proliferation. To address our hypothesis, we first detected lncUCA1 expression in rat hearts from different ages and the effect of lncUCA1 on cardiomyocyte proliferation. Moreover, we investigated the interaction between lncUCA1 and miR-128, and then explored whether lncUCA1 regulated cardiomyocyte proliferation through the miR-128/SUZ12/P27 axis.
2. Materials and methods
2.1. Experimental animals
Sprague Dawley (SD) rats were purchased from Beijing Vital River Laboratory Animal Technology Co t. For the in vitro experiments, CMs were isolated from 1-day and 7-day-old SD neonatal rats. For the in vivo experiments, 1-day-old SD neonatal rats and 8-week-old adult rats were used to perform MI surgery. The experimental protocol in this study conformed to the NIH Guidelines for the Care and Use of Laboratory Animals and were approved by the Biomedical Ethics Committee of Haikou People's Hospital.
2.2. qRT-PCR
The rats were deeply anesthetized with 2 % inhaled isoflurane and euthanized by cervical dislocation. Then, the hearts were dissected out and washed with PBS. The total RNA Kit II (Omega) was used to extract total RNA from isolated CMs, and ventricular neonatal or adult rat heart tissue. NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) were used to extract the nuclear and cytoplasmic fractions of CMs. Then RNA was transcribed to cDNA by reverse transcription using PrimeScript™ RT reagent Kit (TaKaRa Bio). QRT-PCR was performed with SYBR Green PCR Master Mix (TaKaRa, China) using a LightCycler480 (Roche). GAPDH was used as the internal reference and relative gene expression was calculated with the ΔΔCt method. The primer pair sequences used for qRT-PCR are listed in Supplementary Table 1.
2.3. Bioinformatics analysis
Gene expression profiles of three embryonic hearts and three adult hearts were obtained from the Gene Expression Omnibus (GEO) database (GSE184905 and GSE150661). These datasets were merged, normalized after removing batch effects by using the R package SVA. Differentially expressed gene (DEG) analysis was conducted by the “limma” R package. Genes with a p value < 0.05 and |log2fc| (log2 fold change) > 1 were assigned as differentially expressed. Enriched GO analysis of differentially expressed genes and Gene Set Enrichment Analysis (GSEA) were performed by using clusterProfiler package. Single sample Gene Set Enrichment Analysis (ssGSEA) was performed on each sample by using the GSVA package. The correlation between the lncRNA expression level and ssGSEA scores of each sample, and the correlation between the expression level of lncRNA and mRNA were analysed by using Pearson's correlation coefficient. The correlation network between lncRNA and mRNA was visualized using Cytoscape (v3.9).
2.4. Ventricular CMs isolation
The ventricular CMs were isolated with a modified protocol as previously described [18]. Briefly, after washing and mincing the neonatal rat hearts with PBS (without Ca2+, Mg2+) supplemented with 20 mM BDM (Sigma-Aldrich), tissue fragments were incubated in the isolation medium (PBS supplemented with 20 mM BDM and 0.0125 % trypsin) with gentle agitation at 4 °C for 14 h. Predigested tissue fragments were then transferred into a freshly made digestion solution [1.5 mg/ml collagenase type II (Roche) and 5 mg/ml bovine serum albumin (BSA) from Sigma-Aldrich were digested in PBS] and incubated for 20 min at 37 °C. Cell suspension was collected and centrifuged to yield the isolated cell pellets. Cells were plated and incubated for 2 h in a cell culture incubator. The adherent fibroblasts were further cultured and harvested. The non-adherent CMs were resuspended and cultured on slides coated with 10 μg/ml fibronectin (Sigma-Aldrich) in DMEM/F12 containing 10 % FBS and 1 % penicillin/streptomycin at 37 °C with 5 % CO2.
2.5. RNA fluorescence in Situ hybridization
Specific probes targeting lncUCA1 were synthesized by GenePharma (Shanghai, China) and the RNA FISH assay was performed using a FISH kit (Shanghai GenePharma Co., Ltd.) according to the manufacturer's protocols. Briefly, CMs were incubated with the lncUCA1 probe at 37 °C overnight after fixation and permeabilization. Cells were then counterstained with DAPI (1 μg/ml) for 10 min at room temperature before visualization using a Nikon TE2000 epifluorescent microscope with deconvolution (Volocity; PerkinElmer).
2.6. Cell transfection
The siRNA against lncUCA1 and the negative control, as well as the mimics, were synthesized by Ribobio (Guangzhou, China). The target sequence of si-LncUCA1 was 5′-CCATCAGATCCTT GCCCAT-3'. P1 CMs were allowed to adhere for 24 h at a density of 5 × 104 cells/cm2, and then the si-lncUCA1 (200 nmol/l), miR-128 mimic (100 nmol/l), as well as their negative control were transfected into CMs using LipoRNA fit (Hanbio, Shanghai, China) according to the manufacturer's instructions. After 48 h, cells were harvested for analysis.
The adv vector overexpressing lncUCA1 was synthesized by OBiO (Shanghai, China) and the transduction was performed according to the manufacturer's instructions. Briefly, P7 CMs at a density of 5 × 104 cells/cm2 were transduced with the adv vector (100 MOI) for 12 h and then cultured with DMEM/F12 containing 10 % FBS and 1 % penicillin/streptomycin at 37 °C with 5 % CO2. After 36 h, cells were harvested for analysis.
2.7. MI model establishment and AAV9 vector injection
Myocardial infarction (MI) in neonatal (P1) or adult (8 weeks) rats were induced by ligation of the left anterior descending coronary artery (LAD) as previously described [3]. AAV9 containing the CM-specific cTNT promoter to overexpress or knock down lncUCA1 were synthesized by Kidan (Guangzhou, China) and the target sequence of AAV9-shlncUCA1 was 5ʹ-CAGTTCCCTAACTTGAGCGTTA-3ʹ. Briefly, P1 rats were anesthetized by cooling on an ice bed for 3–4 min, whereas P56 rats were sedated with 1.2 % Avertin and were artificially ventilated following tracheal intubation. Lateral thoracotomy at the fourth intercostal space was performed by blunt dissection of the intercostal muscles following a skin incision. Following ligation of the LAD, AAV9-NC or AAV9-lncUCA1 vectors were immediately injected into the myocardium of adult rat at 5–6 sites with a dose of 1 × 1012 viral genome particles per animal (approximately 50 μl), whereas AAV9-shNC or AAV9-shlncUCA1 vectors were immediately injected into the myocardium of neonatal rat at 3–4 sites with a dose of 5 × 1011 viral genome particles per animal (approximately 25 μl). The thoracic wall and skin incision were then sutured with 6.0 non-absorbable silk sutures. All rats were then placed under a heat lamp until they recovered from anesthesia. After that, neonatal rats were timely returned to their mother.
2.8. Immunofluorescence staining
The cultured cells and heart sections (5–8 μm) were fixed with 4 % paraformaldehyde for 15 min, permeabilized with 0.5 % Triton X-100 in PBS for 5 min, blocked with 1 % BSA in PBS for 1 h at room temperature, and incubated with primary antibodies at 4 °C overnight. Anti-cTnT antibody (Santa Cruz, sc-20025, 1:100) was used to identify CM. Anti-Ki67 (Abcam, ab15580, 1:200), anti-pH3 (Abcam, ab170904), and Aurora B (MilliporeSigma, A5102, 1:100) antibodies were used to analyze cell cycle activity, karyokinesis, and cytokinesis, respectively. After triple washing in PBS, slides were incubated for 60 min at 37 °C with fluorescence conjugated secondary antibodies (Biosynthesis). DAPI was used for nuclear counterstaining. EdU staining was performed with the YF®594 Click-iT EdU Imaging Kits (C6017,UE) according to the manufacturer's instructions. Cyclin B1 (Santa Cruz, sc-245, 1:100) and CDK1 (Proteintech, 19532-1-AP, 1:100) antibodies were used to perform the immunostaing experiments. Fluorescent imaging was performed on a Nikon TE2000 epifluorescent microscope with deconvolution (Volocity; PerkinElmer).
2.9. WGA staining
For wheat germ agglutinin (WGA) staining, rat heart slides were deparaffinized, rehydrated, and subjected to citrate-based heat-mediated antigen retrieval. Slides were incubated with Alexa Fluor 647–preconjugated WGA (W32466, Invitrogen) overnight at 4 °C and mounted using Prolong Gold mounting medium (P36934, Thermo fisher). The cardiomyocyte cross-sectional area was determined using an automated algorithm with Image J.
2.10. Quantification of cardiomyocyte numbers
CM numbers were quantified by calculating the cTnT-positive cells. Briefly, unfiltered cells resultant from a whole heart were resuspended in 2 ml of PBS. After fully resuspended, a 20 μl aliquot was diluted 1:50 in PBS and cultured in the confocal dishes. For the gain-of-function experiment, cells were transduced with Adv-NC or Adv-lncUCA1; For the loss-of-function experiment, cells were transfected with si-NC or si-lncUCA1. All groups were blinded until data analysis was completed. Cardiomyocytes were distinguished from non-cardiomyocytes by cTnT immuno-staining. To determine the best possible estimation of CM numbers, Ten 4X fields of each confocal dish were randomly selected for calculation and average.
2.11. Luciferase reporter assay
The pGL3-based luciferase reporter plasmids (Invitrogen; Thermo Fisher Scientific, Inc.) containing the 3′-UTR region of lncUCA1-WT or lncUCA1-MUT mutated with altered miR-128 binding sites were designed and constructed. A total of 5 × 104 cells were co-transfected with miRNA mimics and luciferase using Lipofectamine 3000 (Invitrogen). After incubation for 48 h, the luciferase activities were measured using a Dual-Luciferase Reporter Assay System (Promega, Wisconsin, USA). Relative luciferase activity was normalized to Renilla luciferase.
2.12. Western blot
Cells were lysed with ice-cold cell lysis buffer plus protease inhibitor (Sigma-Aldrich, P8340). Total cell protein concentration was determined using a BCA Protein Assay Kit. Western blotting was performed as described previously with the following antibodies: SUZ12 (Abcam, ab12073, 1:1000), p27 (Cell Signaling Technology, 3686, 1:1000), Cyclin B1 (Santa Cruz, sc-245, 1:500), Cyclin E (Santa Cruz, sc-481, 1:500), CDK1 (Proteintech, 19532-1-AP, 1:1000), CDK2 (Abcam, ab32147, 1:1000), and GAPDH (Proteintech, 10494-1-AP, 1:10000).
2.13. Statistical analysis
The data were presented as the mean ± standard deviation (SD) and all analysis were performed using SPSS 17.0 statistical packages (SPSS, Inc.). Student's t-test was used to compare two groups. Comparisons among multiple groups were performed by one-way ANOVA analysis, followed by Tukey's post hoc test. Spearman's correlation analysis was used to detect the correlation between gene expression. A P-value less than 0.05 was considered statistically significant.
3. Results
3.1. LncRNA UCA1 expression decreased in postnatal hearts
To clarify whether lncUCA1 is one of the candidate lncRNAs involved incardiomyocyte proliferation, we first conducted an analysis using lncRNA expression profiles obtained from fetal and adult hearts (GSE184905 [19] and GSE150661 [20]) and found that lncRNAs are highly correlated within groups (Fig. 1A and B). A total of 1819 differentially expressed mRNAs and 127 differentially expressed lncRNAs were identified (Fig. 1C). GO enrichment analysis revealed that differentially expressed mRNAs were mainly involved in the cell cycle, mitotic nuclear division, and DNA replication (Fig. 1D). The Gene set enrichment analysis (GSEA) further indicated that fetal hearts have a stronger cell cycle activity compared with adult hearts (Fig. 1E and F). The venn diagram and heatmap further showed that lncUCA1 was one of the 30 common genes that were positively correlated with the cell cycle and upregulated in fetal hearts (Fig. 1G and H). Moreover, correlation and co-expression network analysis revealed that lncUCA1 might be involved in the positive regulation of cardiac cell cycle activity (Fig. 1I and J). We next detected lncUCA1 expression in E15.5, P1, P7, and P56 rat hearts, and found that lncUCA1 expression decreased in postnatal hearts (Fig. 1K). Additionally, lncUCA1 was highly expressed in the heart compared with the lung, spleen, or muscle (Fig. 1L). At the cellular level, lncUCA1 expression was higher in CMs than in cardiac fibroblasts and endothelial cells (Fig. 1M). Both qRT-PCR and FISH assay results further showed that lncUCA1 was primarily located in the cytoplasm of CMs (Fig. 1N-O). Therefore, these results indicated that lncUCA1 might be involved in cardiomyocyte proliferation.
Fig. 1.
LncRNA UCA1 expression decreased in postnatal hearts. (A) The analysis of normalized expression levels in fetal and adult hearts. (B) Correlation analysis between fetal and adult hearts. (C) Differential gene expression analysis showing up- and down-regulated mRNAs and LncRNAs between fetal and adult hearts. A |log2FC|>1 and a p value < 0.05 were considered to be statistically significant. (D) GO enrichment analysis of up- and down-regulated mRNAs. The x-axis indicated gene counts and the y-axis specified GO terms. (E) GSEA of cell cycle DNA replication, positive regulation of cell cycle, and positive regulation of cytokinesis in fetal hearts. (F) Single-sample GSEA score of cell cycle DNA replication, positive regulation of cell cycle, and positive regulation of cytokinesis in fetal and adult hearts. (G) Venn diagram analysis showed the number of lncRNAs that positively correlated with the cell cycle and were upregulated in fetal hearts. (H) Heatmap displaying the expression of 30 shared lncRNAs in fetal and adult hearts. (I) Correlation between single sample GSEA score of the cell cycle and lncUCA1 expression in fetal and adult hearts. (J) Co-expression network analysis of lncUCA1 and cell cycle-related genes. Orange represents genes most closely related to lncUCA1. (K) QRT-PCR analysis of lncUCA1 expression in SD rat hearts at different ages (n = 5). (L) QRT-PCR analysis of lncUCA1 expression in different organs of 1-day-old SD rats (n = 5). (M) QRT-PCR analysis of lncUCA1 expression in cardiomyocytes (CMs), cardiac fibroblasts (CFs), and endothelial cells (ECs) (n = 5). (N) The nuclear and cytoplasmic distribution of lncUCA1 in P1 SD rat CMs (n = 5). (O) Representative image of RNA FISH to confirm lncUCA1 location in P1 CMs isolated from SD rats. Statistical significance was calculated using one-way ANOVA analysis in K-M and Student's t-test in N. Data are represented as means ± SD. *P < 0.05. Bar = 20 μm.
3.2. LncRNA UCA1 overexpression promoted P7 CM proliferation
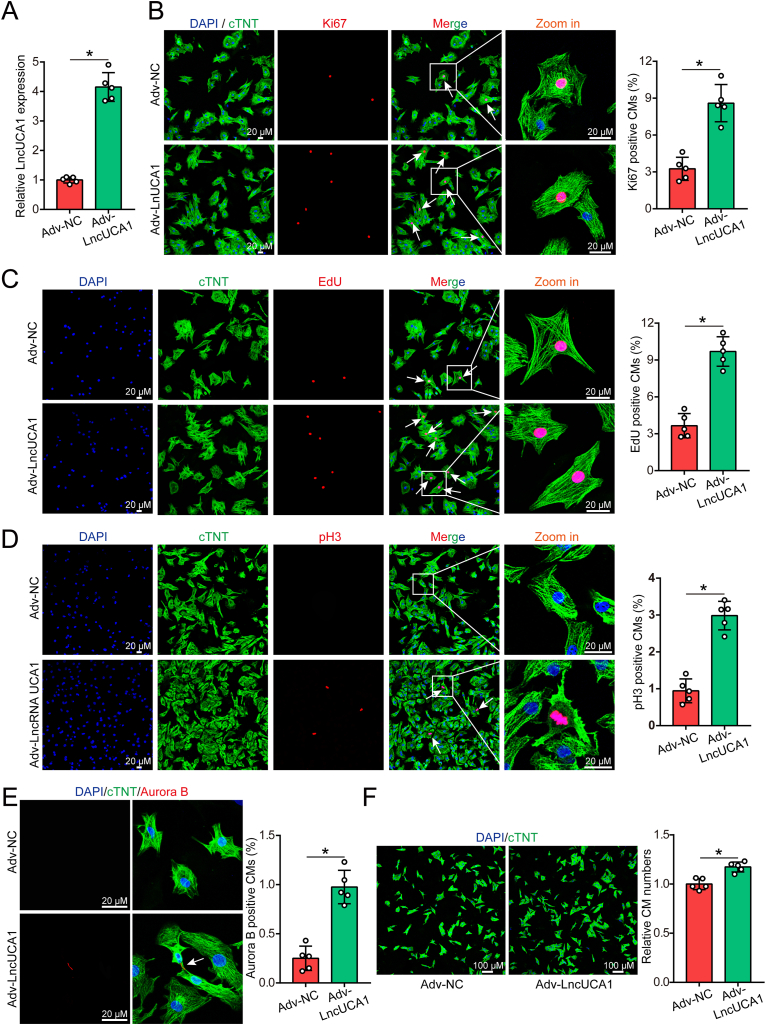
We next investigated whether lncUCA1 overexpression could promote P7 CM proliferation during which most CMs normally exit the cell cycle. After transducing the adv virus overexpressing lncUCA1 (Adv-LncRNA UCA1) and its negative control (Adv-NC) into P7 CMs, we observed that lncUCA1 expression was upregulated in the Adv-LncRNA UCA1 group (Fig. 2A). The effect of lncUCA1 on P7 CM proliferation was evaluated by using the cell cycle activity marker Ki67, DNA synthesis marker EdU, mitosis marker pH3 and cytokinesis marker aurora-B. LncUCA1 overexpression significantly increased the ratio of P7 CMs expressing Ki67 (from 3.26 ± 0.94 % to 8.60 ± 1.51 %; Fig. 2B), EdU (from 3.66 ± 0.99 % to 9.69 ± 1.20 %; Fig. 2C), pH3 (from 0.94 ± 0.33 % to 2.99 ± 0.39 %; Fig. 2D) and aurora B (from 0.25 ± 0.12 % to 0.98 ± 0.17 %; Fig. 2E). Furthermore, we observed a 17 % increase in CM numbers in the Adv-LncRNA UCA1 group compared to the control group (Fig. 2F). In general, these results indicated that lncRNA UCA1 overexpression promoted P7 CM proliferation in vitro.
Fig. 2.
LncRNA UCA1 overexpression promoted P7 cardiomyocyte proliferation. (A) QRT-PCR analysis of lncRNA UCA1 expression in P7 SD rat CMs after lncUCA1 overexpression (n = 5). (B–E) Ki67, EdU, pH3 and Aurora B immunostaining of P7 SD rat CMs transduced with Adv-NC or Adv-LncUCA1 (n = 5), bar = 20 μm. (F) Quantification of cardiomyocyte numbers after lncUCA1 overexpression (n = 5), bar = 100 μm. Statistical significance was calculated using Student's t-test in A-F. Data are represented as means ± SD. *P < 0.05.
3.3. LncRNA UCA1 overexpression promoted adult rat cardiomyocyte proliferation after myocardial infarction
To examine whether lncUCA1 promotes adult rat cardiomyocyte proliferation after myocardial infarction, we used AAV9 vectors carrying the lncRNA UCA1 with CM-specific cTnT promoter to overexpress lncUCA1 specifically in the CMs of adult rat MI models and observed an increase in lncUCA1 expression in the AAV9-LncRNA UCA1 group (Fig. 3A). Analysis of wheat germ agglutinin staining revealed that lncUCA1 overexpression caused little effect on the cross-sectional area of cardiomyocytes (Fig. 3B). Moreover, lncUCA1 overexpression increased the proportion of ki67-positive CMs (from 1.41 ± 0.33 % to 5.05 ± 0.66 %; Fig. 3C), pH3-positive CMs (from 0.55 ± 0.14 % to 1.93 ± 0.23 %; Fig. 3D) and aurora B-positive CMs (from 0.11 ± 0.02 % to 0.76 ± 0.15 %; Fig. 3E) in the adult rat MI models. Taken together, lncRNA UCA1 overexpression promoted adult rat cardiomyocyte proliferation after MI.
Fig. 3.
LncRNA UCA1 overexpression promoted adult rat cardiomyocyte proliferation after myocardial infarction. (A) QRT-PCR analysis of lncRNA UCA1 expression in adult SD rat MI model hearts after lncUCA1 overexpression (n = 5). (B) WGA staining of left ventricular (LV) heart section transduced with AAV9-NC or AAV9-LncUCA1 at 14 days post-MI (n = 5), bar = 50 μm. (C–E) Ki67, pH3 and Aurora B immunostaining of adult SD rat hearts transduced with AAV9-NC or AAV9-LncUCA1 at 14 days post-MI (n = 5), bars = 20 μm (left) and 10 μm (right). Statistical significance was calculated using Student's t-test in A-E. Data are represented as means ± SD. *P < 0.05.
3.4. Downregulation of lncRNA UCA1 inhibited neonatal rat cardiomyocyte proliferation
Since that the lncUCA1 expression decreased in postnatal hearts, we next evaluated whether downregulation of lncUCA1 could inhibit neonatal rat cardiomyocyte proliferation. We used siRNA to knock down lncUCA1 in vitro and observed that lncUCA1 expression was decreased in the si-LncRNA UCA1 group (Fig. 4A). Downregulation of lncUCA1 significantly decreased the ratio of P1 CMs expressing Ki67 (from 7.74 ± 0.85 % to 2.67 ± 0.40 %; Fig. 4B) and aurora B (1.16 ± 0.14 % to 0.42 ± 0.09 %; Fig. 4C). We observed a 22 % decrease in CM numbers in the si-LncRNA UCA1 group compared to the control group (Fig. 4D). To assess the in vivo effect of lncUCA1 downregulation, we used AAV9 vectors carrying the lncUCA1 shRNA with CM-specific cTnT promoter to knock down lncUCA1 specifically in the CMs of neonatal rat MI models. QRT-PCR results showed that lncUCA1 expression was decreased in the AAV9-shLncUCA1 group (Fig. 4E). Downregulation of lncUCA1 decreased the proportion of ki67-positive CMs (from 3.10 ± 0.43 % to 1.44 ± 0.17 %; Fig. 4F), pH3-positive CMs (from 0.86 ± 0.12 % to 0.40 ± 0.08 %; Fig. 4G), and aurora B-positive CMs (from 0.23 ± 0.04 % to 0.10 ± 0.01 %; Fig. 4H) in the neonatal rat MI models. Therefore, these data suggested that downregulation of lncRNA UCA1 inhibited neonatal rat cardiomyocyte proliferation.
Fig. 4.
Downregulation of lncRNA UCA1 inhibited neonatal rat cardiomyocyte proliferation. (A) QRT-PCR analysis of lncRNA UCA1 expression in P1 SD rat CMs after lncUCA1 downregulation (n = 5). (B–C) Ki67 and Aurora B immunostaining of P1 SD rat CMs transfectd with si-NC or si-LncUCA1 (n = 5), bar = 20 μm. (D) Quantification of cardiomyocyte numbers after lncUCA1 downregulation (n = 5), bar = 100 μm. (E) QRT-PCR analysis of lncRNA UCA1 expression in neonatal SD rat MI model hearts after lncUCA1 downregulation (n = 5). (F–G) Ki67 and pH3 immunostaining of neonatal SD rat hearts transduced with AAV9-shNC or AAV9-shLncUCA1 at 12 days post-MI (n = 5), bars = 20 μm (left) and 10 μm (right). (H) Aurora B immunostaining of neonatal SD rat hearts at 12 days post-MI (n = 5), bar = 20 μm. Statistical significance was calculated using Student's t-test in A-H. Data are represented as means ± SD. *P < 0.05.
3.5. LncRNA UCA1 directly interacted with miR-128 and regulated its expression
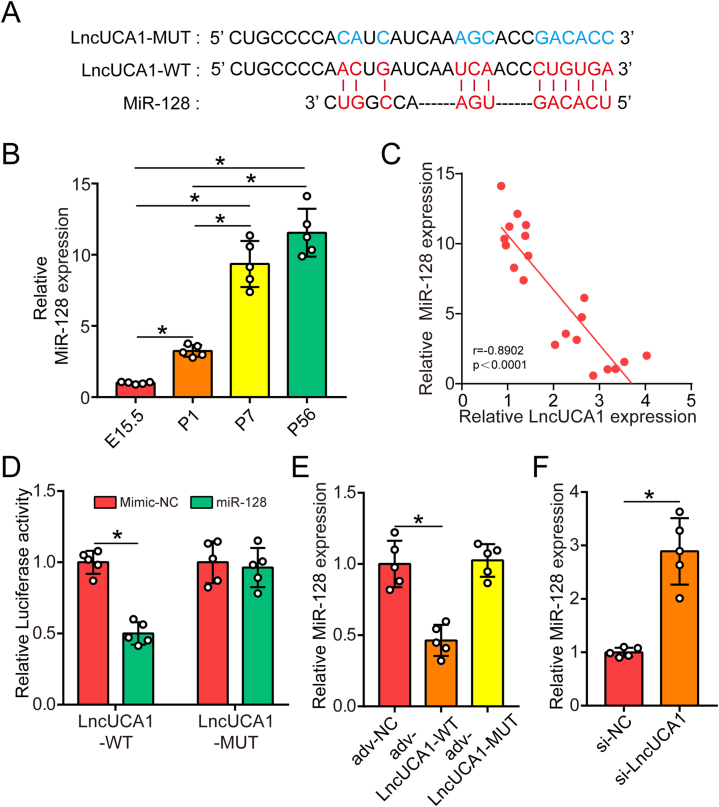
To further investigate the underlying molecular mechanism of lncUCA1 in regulating cardiac regeneration, we used a bioinformatics program to screen the miRNAs that might bind to lncUCA1. We found that lncUCA1 might bind with miR-128 which has been reported to inhibit cardiomyocyte proliferation (Fig. 5A). We next detected miR-128 expression in rat hearts at different ages and found that miR-128 expression significantly increased during heart development (Fig. 5B). We also found a negative correlation between lncUCA1 and miR-128 expression in ventricular heart samples from different aged rats (Fig. 5C). We then produced a luciferase construction of lncUCA1 (Luc-LncUCA1-WT) and a mutated form with a derivative devoid of the miR-128 binding site (Luc-LncUCA1-MUT). The luciferase assay showed that miR-128 could suppress the luciferase activity of Luc-LncUCA1-WT, but had little effect on the Luc-LncUCA1-MUT (Fig. 5D), indicating that lncUCA1 could directly bind to miR-128. Next, we examined whether lncUCA1 could regulate miR-128 expression. QRT-PCR results showed that lncUCA1-WT overexpression decreased the expression of miR-128 while lncUCA1-MUT overexpression caused no effect on the expression of miR-128 (Fig. 5E). Additionally, downregulation of lncUCA1 increased the expression of miR-128 (Fig. 5F). Collectively, these findings indicated that lncUCA1 directly interacted with miR-128 and regulated its expression.
Fig. 5.
LncRNA UCA1 interacted with miR-128 and regulated its expression. (A) Online prediction revealed the binding site between lncUCA1 and miR-128, and the information of lncUCA1-WT and lncUCA1-MUT. (B) QRT-PCR analysis of miR-128 expression in rat hearts at different ages (n = 5). (C) Correlation between miR-128 and lncUCA1 expressions in 20 ventricular heart samples from different aged rats. (D) Luciferase activity of isolated P1 SD rat CMs transfected with Luc-LncUCA1-WT or Luc-LncUCA1-MUT (n = 3). (E) QRT-PCR analysis of miR-128 expression in P7 SD rat CMs transduced with adv-NC, adv-LncUCA1-WT or adv-LncUCA1-MUT (n = 5). (F) QRT-PCR analysis of miR-128 expression in P7 SD rat CMs transfected with si-NC and si-LncUCA1 (n = 5). Statistical significance was calculated using one-way ANOVA analysis in B and E, spearman's test in C, and Student's t-test in D and F. Data are represented as means ± SD. *P < 0.05.
3.6. LncRNA UCA1 promoted cardiomyocyte proliferation by inhibiting the miR-128/SUZ12/P27 pathway
A previous study reported that loss of miR-128 could promote cardiomyocyte proliferation through the SUZ12/P27 pathway [18], so we next determined whether lncUCA1/miR-128 axis could regulate the SUZ12/P27 pathway and cell proliferative effect in CMs. QRT-PCR and western blotting experiments showed that lncUCA1 could act as a ceRNA of miR-128 to upregulate its target suz12 and downregulate P27, thereby increasing the expression of cell cycle regutors such as cyclin B1, cyclin E, CDK1 and CDK2 (Fig. 6A and B). In addition, immunostaining showed that lncUCA1 overexpression upregulated the expression of cell cycle regulators such as cyclin B1 and CDK1, which was counteracted by miR-128 (Fig. 6C and D). Moreover, Ki67 and aurora-B immunostaining showed that miR-128 overexpression counteracted the proliferative effect of cardiomyocytes induced by lncUCA1 (Fig. 6E and F). Taken together, these findings indicated that lncUCA1 acted as a ceRNA of miR-128 to upregulate its target suz12 and downregulate P27, thereby increasing the expression of cyclin B1, cyclin E, CDK1 and CDK2 to promote cardiomyocyte proliferation.
Fig. 6.
LncRNA UCA1 regulated cardiomyocyte proliferation through the miR-128/SUZ12/P27 pathway. (A) The mRNA levels of SUZ12 and cell cycle regulators in P7 SD rat CMs after miR-128 and lncUCA1 interference (n = 5). (B) Western blotting analysis of SUZ12, P27, Cyclin B1, Cyclin E, CDK1, and CDK2 protein levels in P7 SD rat CMs after miR-128 and lncUCA1 interference (n = 4). (C–D) Cyclin B1 and CDK1 immunostaining in P7 SD rat CMs after miR-128 and lncUCA1 interference (n = 5). (E–F) Ki67 and aurora-B immunostaining in P7 SD rat CMs after miR-128 and lncUCA1 interference (n = 5). Statistical significance was calculated using one-way ANOVA in A-F. Data are represented as means ± SD. *P < 0.05. Bar = 20 μm.
4. Discussion
In the present study, we found that lncUCA1 expression decreased in postnatal hearts. Overexpression of lncUCA1 promoted P7 cardiomyocyte proliferation and adult rat cardiomyocyte proliferation after MI, while downregulation of lncUCA1 inhibited P1 cardiomyocyte proliferation and neonatal rat cardiomyocyte proliferation after injury. Furthermore, lncUCA1 acted as a ceRNA of miR-128 to upregulate its target SUZ12 and downregulate P27, thereby increasing the expression of cyclin B1, cyclin E, CDK1, and CDK2 to promote cardiomyocyte proliferation.
LncUCA1 was a long non-coding RNA originally identified in bladder cancer [14]. LncUCA1 plays an important role in regulating cell cycle progression [21,22]. Several studies have demonstrated that lncUCA1 has an oncogenic effect in many cancers, such as breast cancer [23], colorectal cancer [24] and gastric cancer [25]. A recent study also reported that lncUCA1 alleviated As-induced G2/M phase arrest in human liver cells by destabilizing EZH2 and facilitating NFATc2 expression [26]. Regulation of cell cycle progression has been an attractive strategy to stimulate cardiomyocyte proliferation [27]. Hence, after finding that lncUCA1 has different expression levels in embryonic and adult hearts, we used the proliferative markers Ki67, EdU, pH3, and aurora B to investigate whether lncUCA1 could regulate cardiomyocyte proliferation. Upregulation of lncUCA1 increased the proportion of ki67, EdU, pH3 and aurora B-positive CMs, thereby rising P7 CM numbers. Downregulation of lncUCA1 significantly decreased the proportion of ki67-or aurora B-positive CMs, leading to the reduction of P1 CM numbers. Moreover, lncUCA1 overexpression promoted cardiomyocyte proliferation in the adult rat MI model while lncUCA1 deficiency inhibited the cardiac regenerative response in the neonatal rat MI model. Therefore, our findings demonstrated that lncUCA1 could regulate the cell cycle progression of cardiomyocytes. Recent studies showed that lncUCA1 overexpression protected cardiomyocytes from apoptosis [16,28] and autophagy [17]. These studies and our results indicated that lncUCA1 has multiple beneficial effects in CMs, and lncUCA1 would act as an attractive therapeutic target to slow down the progression of ischemic heart diseases.
By using the bioinformatics program and performing rescue experiments, we found that lncUCA1 promoted cardiomyocyte proliferation by inhibiting the miR-128/SUZ12/P27 pathway. MiR-128 has been reported to be a negative regulator of cell cycle progression in CMs [18]. Loss of miR-128 could upregulate SUZ12 protein level, downregulate negative cell cycle regulators p27 protein level and upregulate cyclin E and CDK2 protein levels [18]. In this study, the bioinformatics program, correlation analysis, luciferase assay, and qRT-PCR experiments indicated that lncUCA1 could directly bind to miR-128 and negatively regulate its expression. Moreover, we found that lncUCA1 and miR-128 interference could regulate the mRNA and protein levels of SUZ12 and p27, indicating that lncUCA1 might act as a ceRNA of miR-128 to primarily posttranscriptionally regulate the expression of SUZ12 and p27. Among the downstream targets of miR-128, p27 was widely regulated by lncUCA1 in different diseases. In pancreatic cancer, p27 protein level was negatively correlated with lncUCA1 expression [29]. Besides, lncUCA1 could sustain acute myeloid leukemia cell proliferation through inhibiting p27kip1 [30], and HBx-upregulated lncUCA1 could promote hepatic LO2 cell growth and tumorigenesis by recruiting EZH2 and repressing p27Kip1/CDK2 signaling [31]. A recent study also showed that upregulation of the lncUCA1/mTOR axis in cancer-associated fibroblasts could suppress miR-143 and p27, thereby inducing cell proliferation and metastasis of colorectal cancer SW480 cells [32]. In line with these studies, our study showed that the lncUCA1/miR-128 axis upregulated SUZ12 protein levels to suppress p27, thereby increasing the expression of P27-related cell cycle regulators such as cyclin B1, cyclin E, CDK1, and CDK2 to promote cardiomyocyte proliferation. Taken together, p27 protein and its related cell cycle regulators (cyclin B1, cyclin E, CDK1, and CDK2) were essential for the lncUCA1/miR-128/SUZ12 axis to regulate cardiomyocyte proliferation.
Our study has several limitations. Firstly, since that lncUCA1 could enhance cardiomyocyte proliferation and inhibit cardiomyocyte apoptosis and autophagy, future studies should clarify the interaction among lncUCA1-induced multiple protective effects in CMs. Secondly, in addition to mir-128, lncUCA1 has been reported to act as a ceRNA of several miRNAs involved in cardiomyocyte proliferation, such as mir-214 [11,33], mir-143 [16, 34, 35] and mir-495[36, 37]. It is worthy investigating the roles of mir-214, mir-143 and mir-495 in lncUCA1-mediated CM proliferation. Thirdly, previous studies reported that lncUCA1 also played important roles in regulating cardiac regeneration-related factors and pathways including EZH2 [38], Hippo [39], and PI3K-AKT [40] pathways. It would be better to perform RNA-seq or proteomics to explore the comprehensive mechanism of lncUCA1-mediated CM proliferation in the future.
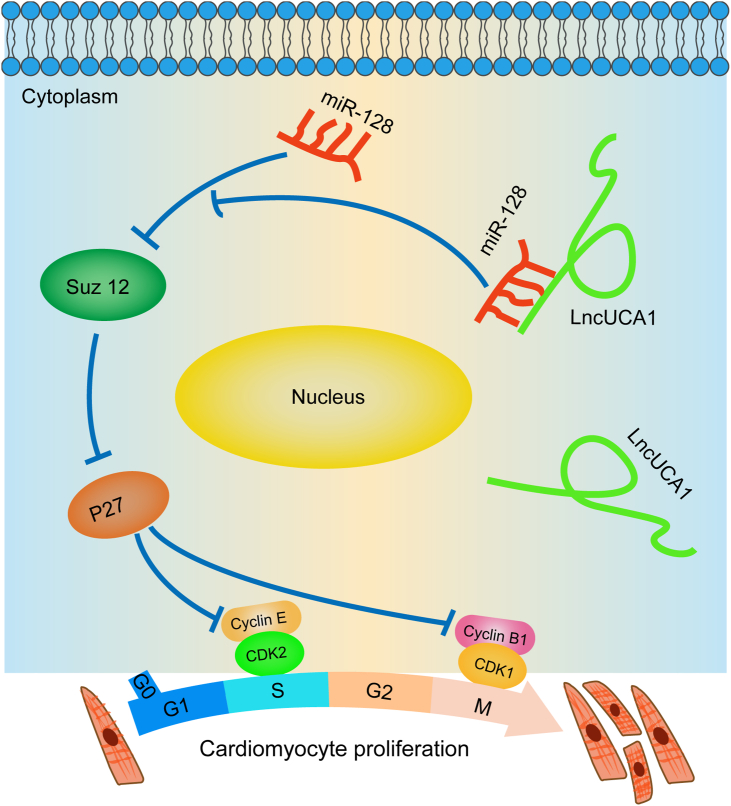
In conclusion, our study showed that lncUCA1 promoted cardiomyocyte prolferation by inhibiting the miR-128/SUZ12/P27 pathway (Fig. 7), indicating that lncUCA1 could be a new therapeutic target to promote cardiac regenerative response after MI.
Fig. 7.
Schematic diagram illustrating the mechanisms of lncRNA UCA1 promoting cardiomyocyte proliferation. LncRNA UCA1 directly bound to miR-128 and inhibited the miR-128/SUZ12/P27 pathway, thereby increasing Cyclin B1, Cyclin E, CDK1, and CDK2 expression to promote cardiomyocyte proliferation.
Ethics approval and consent to participate
All experimental procedures conformed to the guidelines of Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes and the NIH Guidelines for the Care and Use of Laboratory Animals, and were approved by the Biomedical Ethics Committee of Haikou People's Hospital.
Consent for publication
All authors agree to publish this article.
Availability of data and materials
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
Funding
This work was supported by a grant to Kang Huang and Denggao Huang from the Scientific Research Project of Hainan Provincial Health Commission (20A200122&21A200149) and Finance science and technology project of Hainan Province (822MS200).
Statement
This study is reported in accordance with ARRIVE guidelines.
CRediT authorship contribution statement
Kang Huang: Writing – original draft, Investigation, Funding acquisition, Data curation. Denggao Huang: Writing – review & editing, Writing – original draft, Supervision, Funding acquisition, Conceptualization. Qiang Li: Investigation, Data curation. Jianghua Zhong: Validation, Methodology, Data curation. Yilei Zhou: Investigation, Data curation. Zanrui Zhong: Methodology, Investigation. Shilin Tang: Investigation, Data curation. Wei Zhang: Methodology, Investigation, Formal analysis. Zibin Chen: Investigation, Formal analysis. Shijuan Lu: Writing – review & editing, Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e34181.
Contributor Information
Denggao Huang, Email: hdg_qx@163.com.
Shijuan Lu, Email: lushijuan0927@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Martin S.S., Aday A.W., Almarzooq Z.I., Anderson C.A.M., Arora P., Avery C.L., Baker-Smith C.M., Barone Gibbs B., Beaton A.Z., Boehme A.K., Commodore-Mensah Y., Currie M.E., Elkind M.S.V., Evenson K.R., Generoso G., Heard D.G., Hiremath S., Johansen M.C., Kalani R., Kazi D.S., Ko D., Liu J., Magnani J.W., Michos E.D., Mussolino M.E., Navaneethan S.D., Parikh N.I., Perman S.M., Poudel R., Rezk-Hanna M., Roth G.A., Shah N.S., St-Onge M.P., Thacker E.L., Tsao C.W., Urbut S.M., Van Spall H.G.C., Voeks J.H., Wang N.Y., Wong N.D., Wong S.S., Yaffe K., Palaniappan L.P. 2024 heart disease and stroke statistics: a report of us and global data from the american heart association. Circulation. 2024;149:e347–e913. doi: 10.1161/CIR.0000000000001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maruyama K., Imanaka-Yoshida K. The pathogenesis of cardiac fibrosis: a review of recent progress. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23052617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan Y., Cheng Y., Li Y., Chen B., Wang Z., Wei T., Zhang H., Guo Y., Wang Q., Wei Y., Chen F., Sha J., Guo X., Wang L. Phosphoproteomic analysis of neonatal regenerative myocardium revealed important roles of checkpoint kinase 1 via activating mammalian target of rapamycin c1/ribosomal protein s6 kinase b-1 pathway. Circulation. 2020;141:1554–1569. doi: 10.1161/CIRCULATIONAHA.119.040747. [DOI] [PubMed] [Google Scholar]
- 4.Lin Z., Pu W.T. Strategies for cardiac regeneration and repair. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3006681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meng X., Lou Q.Y., Yang W.Y., Wang Y.R., Chen R., Wang L., Xu T., Zhang L. The role of non-coding rnas in drug resistance of oral squamous cell carcinoma and therapeutic potential. Cancer Commun. 2021 Oct;41(10):981–1006. doi: 10.1002/cac2.12194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao J., Chen X., Wei P., Wang Y., Li P., Shao K. Regulation of pyroptosis in cardiovascular pathologies: role of noncoding rnas. Mol. Ther. Nucleic Acids. 2021;25:220–236. doi: 10.1016/j.omtn.2021.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan Z., Huang W. New developments in exosomal lncrnas in cardiovascular diseases. Frontiers in cardiovascular medicine. 2021;8 doi: 10.3389/fcvm.2021.709169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu L., Li N., Sun L., Zheng D., Shao G. Non-coding rnas: the key detectors and regulators in cardiovascular disease. Genomics. 2021;113:1233–1246. doi: 10.1016/j.ygeno.2020.10.024. [DOI] [PubMed] [Google Scholar]
- 9.Yuan T., Krishnan J. Non-coding rnas in cardiac regeneration. Front. Physiol. 2021;12 doi: 10.3389/fphys.2021.650566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abbas N., Perbellini F. Non-coding rnas: emerging players in cardiomyocyte proliferation and cardiac regeneration. Basic Res. Cardiol. 2020;115(5):52–115. doi: 10.1007/s00395-020-0816-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X., He X., Wang H., Li M., Huang S., Chen G., Jing Y., Wang S., Chen Y., Liao W., Liao Y., Bin J. Loss of azin2 splice variant facilitates endogenous cardiac regeneration. Cardiovasc. Res. 2018;114:1642–1655. doi: 10.1093/cvr/cvy075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponnusamy M., Liu F., Zhang Y.H., Li R.B., Zhai M., Liu F., Zhou L.Y., Liu C.Y., Yan K.W., Dong Y.H., Wang M., Qian L.L., Shan C., Xu S., Wang Q., Zhang Y.H., Li P.F., Zhang J., Wang K. Long noncoding rna cpr (cardiomyocyte proliferation regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation. 2019;139:2668–2684. doi: 10.1161/CIRCULATIONAHA.118.035832. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y., Li X., Li B., Wang H., Li M., Huang S., Sun Y., Chen G., Si X., Huang C., Liao W., Liao Y., Bin J. Long non-coding rna ecrar triggers post-natal myocardial regeneration by activating erk1/2 signaling. Mol. Ther. : the journal of the American Society of Gene Therapy. 2019;27:29–45. doi: 10.1016/j.ymthe.2018.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghafouri-Fard S., Taheri M. Uca1 long non-coding rna: an update on its roles in malignant behavior of cancers. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;120 doi: 10.1016/j.biopha.2019.109459. [DOI] [PubMed] [Google Scholar]
- 15.Sun L., Zhu W., Zhao P., Wang Q., Fan B., Zhu Y., Lu Y., Chen Q., Zhang J., Zhang F. Long noncoding rna uca1 from hypoxia-conditioned hmsc-derived exosomes: a novel molecular target for cardioprotection through mir-873-5p/xiap axis. Cell Death Dis. 2020;11:696. doi: 10.1038/s41419-020-02783-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q.S., Zhou J., Li X. Lncrna uca1 protects cardiomyocytes against hypoxia/reoxygenation induced apoptosis through inhibiting mir-143/mdm2/p53 axis. Genomics. 2020;112:574–580. doi: 10.1016/j.ygeno.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Chen Z., Liu R., Niu Q., Wang H., Yang Z., Bao Y. Morphine postconditioning alleviates autophage in ischemia-reperfusion induced cardiac injury through up-regulating lncrna uca1. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;108:1357–1364. doi: 10.1016/j.biopha.2018.09.119. [DOI] [PubMed] [Google Scholar]
- 18.Huang W., Feng Y., Liang J., Yu H., Wang C., Wang B., Wang M., Jiang L., Meng W., Cai W., Medvedovic M., Chen J., Paul C., Davidson W.S., Sadayappan S. Loss of microrna-128 promotes cardiomyocyte proliferation and heart regeneration. Nat. Commun. 2018 Feb 16;9(1):700. doi: 10.1038/s41467-018-03019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gou Z., Zhou Y., Jia H., Yang Z., Zhang Q., Yan X. Prenatal diagnosis and mrna profiles of fetal tetralogy of fallot. BMC Pregnancy Childbirth. 2022;22:853. doi: 10.1186/s12884-022-05190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai F.C., Chang G.J. Ubiquitin pathway is associated with worsening left ventricle function after mitral valve repair: a global gene expression study. Int. J. Mol. Sci. 2020 Jul 18;21(14):5073. doi: 10.3390/ijms21145073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apicella C., Ruano C.S.M., Jacques S., Gascoin G., Méhats C., Vaiman D., Miralles F. Urothelial cancer associated 1 (uca1) and mir-193 are two non-coding rnas involved in trophoblast fusion and placental diseases. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.633937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang D., Wang Y., Zheng Y., Dai F., Liu S., Yuan M., Deng Z., Bao A., Cheng Y. Silencing of lncrna uca1 inhibited the pathological progression in pcos mice through the regulation of pi3k/akt signaling pathway. J. Ovarian Res. 2021;14:48. doi: 10.1186/s13048-021-00792-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choudhry H. Uca1 overexpression promotes hypoxic breast cancer cell proliferation and inhibits apoptosis via hif-1α activation. Journal of oncology. 2021;2021 doi: 10.1155/2021/5512156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S.J., Li Z.Q., Wang X.Y., Liu F., Xiao Z.M., Zhang D.C. Lncrna uca1 induced by sp1 and sp3 forms a positive feedback loop to facilitate malignant phenotypes of colorectal cancer via targeting mir-495. Life Sci. 2021;277 doi: 10.1016/j.lfs.2021.119569. [DOI] [PubMed] [Google Scholar]
- 25.Cheng H., Sharen G., Wang Z., Zhou J. Lncrna uca1 enhances cisplatin resistance by regulating cyp1b1-mediated apoptosis via mir-513a-3p in human gastric cancer. Cancer Manag. Res. 2021;13:367–377. doi: 10.2147/CMAR.S277399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong Z., Gao M., Li C., Xu M., Liu S. Lncrna uca1 antagonizes arsenic-induced cell cycle arrest through destabilizing ezh2 and facilitating nfatc2 expression. Adv. Sci. 2020;7(11):1903630. doi: 10.1002/advs.201903630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohamed TMA., Ang YS., Radzinsky E., Zhou P., Huang Y., Elfenbein A., Foley A., Magnitsky S., Srivastava D. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell. 2018;173:104–116.e112. doi: 10.1016/j.cell.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J., Hu Q., Zhang B.F., Liu X.P., Yang S., Jiang H. Long noncoding rna uca1 inhibits ischaemia/reperfusion injury induced cardiomyocytes apoptosis via suppression of endoplasmic reticulum stress. Genes Genomics. 2019;41:803–810. doi: 10.1007/s13258-019-00806-w. [DOI] [PubMed] [Google Scholar]
- 29.Chen P., Wan D., Zheng D., Zheng Q., Wu F., Zhi Q. Long non-coding rna uca1 promotes the tumorigenesis in pancreatic cancer. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2016;83:1220–1226. doi: 10.1016/j.biopha.2016.08.041. [DOI] [PubMed] [Google Scholar]
- 30.Liang Y., Li E. Silencing of lncrna uca1 curbs proliferation and accelerates apoptosis by repressing sirt1 signals by targeting mir-204 in pediatric aml. J. Biochem. Mol. Toxicol. 2020 Mar;34(3) doi: 10.1002/jbt.22435. [DOI] [PubMed] [Google Scholar]
- 31.Hu J.J., Song W., Zhang S.D., Shen X.H., Qiu X.M., Wu H.Z., Gong P.H., Lu S., Zhao Z.J., He M.L., Fan H. Hbx-upregulated lncrna uca1 promotes cell growth and tumorigenesis by recruiting ezh2 and repressing p27kip1/cdk2 signaling. Sci. Rep. 2016;6 doi: 10.1038/srep23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jahangiri B., Khalaj-Kondori M., Asadollahi E., Sadeghizadeh M. Cancer-associated fibroblasts enhance cell proliferation and metastasis of colorectal cancer sw480 cells by provoking long noncoding rna uca1. Journal of cell communication and signaling. 2019;13:53–64. doi: 10.1007/s12079-018-0471-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang H., Tu B., Luo M., Hou P., Wang J., Zhang R., Wu L. Knockdown of uca1 attenuated the progression of alcoholic fatty disease by sponging mir-214. Mamm. Genome : official journal of the International Mammalian Genome Society. 2022;33:534–542. doi: 10.1007/s00335-022-09953-0. [DOI] [PubMed] [Google Scholar]
- 34.Ren Z., Liu Y., Cai A., Yu Y., Wang X., Lan L., Guo X., Yan H., Gao X., Li H., Tian Y., Ji H., Chen H., Ding F., Ma W., Wang N., Cai B., Yang B. Cannabidiol represses mir-143 to promote cardiomyocyte proliferation and heart regeneration after myocardial infarction. Eur. J. Pharmacol. 2024;963 doi: 10.1016/j.ejphar.2023.176245. [DOI] [PubMed] [Google Scholar]
- 35.Gong R., Wang X., Li H., Liu S., Jiang Z., Zhao Y., Yu Y., Han Z., Yu Y., Dong C., Li S., Xu B., Zhang W., Wang N., Li X., Gao X., Yang F., Bamba D., Ma W., Liu Y., Cai B. Loss of m(6)a methyltransferase mettl3 promotes heart regeneration and repair after myocardial injury. Pharmacol. Res. 2021;174 doi: 10.1016/j.phrs.2021.105845. [DOI] [PubMed] [Google Scholar]
- 36.Yuan H.H., Zhang X.C., Wei X.L., Zhang W.J., Du X.X., Huang P., Chen H., Bai L., Zhang H.F., Han Y. Lncrna uca1 mediates cetuximab resistance in colorectal cancer via the mir-495 and hgf/c-met pathways. J. Cancer. 2022;13:253–267. doi: 10.7150/jca.65687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark A.L., Naya F.J. Micrornas in the myocyte enhancer factor 2 (mef2)-regulated gtl2-dio3 noncoding rna locus promote cardiomyocyte proliferation by targeting the transcriptional coactivator cited2. J. Biol. Chem. 2015;290:23162–23172. doi: 10.1074/jbc.M115.672659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yue Z., Chen J., Lian H., Pei J., Li Y., Chen X., Song S., Xia J., Zhou B., Feng J., Zhang X., Hu S., Nie Y. Pdgfr-β signaling regulates cardiomyocyte proliferation and myocardial regeneration. Cell Rep. 2019;28:966–978.e964. doi: 10.1016/j.celrep.2019.06.065. [DOI] [PubMed] [Google Scholar]
- 39.Liu S., Li R.G., Martin J.F. The cell-autonomous and non-cell-autonomous roles of the hippo pathway in heart regeneration. J. Mol. Cell. Cardiol. 2022;168:98–106. doi: 10.1016/j.yjmcc.2022.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y., Xu T., Li M., Li C., Ma Y., Chen G., Sun Y., Zheng H., Wu G., Liao W., Liao Y., Chen Y., Bin J. Inhibition of senp2-mediated akt desumoylation promotes cardiac regeneration via activating akt pathway. Clin. Sci. 2021;135:811–828. doi: 10.1042/CS20201408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request.