Summary
Background
In the United States (U.S.), hantavirus pulmonary syndrome (HPS) and non-HPS hantavirus infection are nationally notifiable diseases. Criteria for identifying human cases are based on clinical symptoms (HPS or non-HPS) and acute diagnostic results (IgM+, rising IgG+ titers, RT-PCR+, or immunohistochemistry (IHC)+). Here we provide an overview of diagnostic testing and summarize human Hantavirus disease occurrence and genotype distribution in the U.S. from 2008 to 2020.
Methods
Epidemiological data from the national hantavirus registry was merged with laboratory diagnostic testing results performed at the CDC. Residual hantavirus-positive specimens were sequenced, and the available epidemiological and genetic data sets were linked to conduct a genomic epidemiological study of hantavirus disease in the U.S.
Findings
From 1993 to 2020, 833 human hantavirus cases have been identified, and from 2008 to 2020, 335 human cases have occurred. Among New World (NW) hantavirus cases detected at the CDC diagnostic laboratory (representing 29.2% of total cases), most (85.0%) were detected during acute disease, however, some convalescent cases were detected in states not traditionally associated with hantavirus infections (Connecticut, Missouri, New Jersey, Pennsylvania, Tennessee, and Vermont). From 1993 to 2020, 94.9% (745/785) of U.S. hantaviruses cases were detected west of the Mississippi with 45.7% (359/785) in the Four Corners region of the U.S. From 2008 to 2020, 67.7% of NW hantavirus cases were detected between the months of March and August. Sequencing of RT-PCR-positive cases demonstrates a geographic separation of Orthohantavirus sinnombreense species [Sin Nombre virus (SNV), New York virus, and Monongahela virus]; however, there is a large gap in viral sequence data from the Northwestern and Central U.S. Finally, these data indicate that commercial IgM assays are not concordant with CDC-developed assays, and that “concordant positive” (i.e., commercial IgM+ and CDC IgM+ results) specimens exhibit clinical characteristics of hantavirus disease.
Interpretation
Hantaviral disease is broadly distributed in the contiguous U.S, viral variants are localised to specific geographic regions, and hantaviral disease infrequently detected in most Southeastern states. Discordant results between two diagnostic detection methods highlight the need for an improved standardised testing plan in the U.S. Hantavirus surveillance and detection will continue to improve with clearly defined, systematic reporting methods, as well as explicit guidelines for clinical characterization and diagnostic criteria.
Funding
This work was funded by core funds provided to the Viral Special Pathogens Branch at CDC.
Keywords: Orthohantavirus, Rodent, Bunyavirus, Disease surveillance, Genetics, HPS, HCPS, HFRS, Hantavirus pulmonary syndrome, Hantavirus cardiopulmonary syndrome, Hantavirus haemorrhagic fever with renal syndrome
Research in context.
Evidence before this study
Hantaviruses are rodent-borne viruses with worldwide distribution that cause human disease following exposure to rodent excreta (in homes, recreational or occupational settings) or from rodent bites (less frequent); limited human-to-human transmission has only been documented for Andes virus in South America. Rodents are the natural reservoir for hantaviruses in the U.S., but little is known about the distribution of U.S. cases and the hantavirus strains that cause human disease. We conducted a literature review in PubMed for all studies examining previous human hantavirus cases published between 1990 and 2023. We used the search terms “hantavirus disease” and “hantavirus disease U.S.” Previous publications have reviewed US human hantavirus cases from 1993 to 2008, 1993–2009, and 1993–2013, and reported the epidemiological details associated with hantaviral disease, or reported details about individual human cases, but no existing studies summarise the epidemiologic and genetic data associated with human hantavirus cases in the U.S.
Added value of this study
Here, an updated view of hantavirus surveillance in the U.S. is presented. We report the spatial and temporal details of human hantaviral disease and summarize the human serological response. Most [94.9% (745/785)] U.S. hantaviruses cases were detected west of the Mississippi between the months of March–August. Only 45.7% (359/785) of human cases were detected in the Southwest Four Corners region of the U.S., an area traditionally associated with hantaviral prevalence, and cases were infrequently detected in Southeastern states. New World hantaviral species localised to distinct geographic regions and serological evidence of Old World hantavirus infections were detected in states with major ports. We also highlight discordance between two diagnostic detection methods.
Implications of all the available evidence
Our findings demonstrate the temporal and spatial distribution of human hantaviral disease and hantaviral strain prevalence in the continental U.S. The research demonstrates the geographic gaps in hantaviral disease detection and the need for an improved detection algorithm.
Introduction
Hantaviruses are negative-sense multi-segmented viruses in the Bunyavirales order with worldwide distribution that persistently infect rodents.1 Infection in rodents is predominantly asymptomatic,2 while spillover hantaviral infection in humans typically results in disease.1,3 Humans acquire disease following exposure to rodent excreta (in homes, recreational or occupational settings) or from rodent bites (less frequent)4; human-to-human transmission has only been documented for a single viral species (Orthohantavirus andesense).5,6 Human disease can be broadly categorised into hantavirus haemorrhagic fever with renal syndrome (HFRS)—primarily affecting the kidneys caused by Old World (OW) hantaviruses predominantly located in Europe and Asia, and hantavirus cardiopulmonary syndrome (HPS)—primarily affecting the lungs and caused by New World (NW) hantaviruses predominantly located in North and South America.7 However, human disease can range from asymptomatic to mild or severe, and pulmonary and renal involvement have been documented in both HFRS and HPS cases, respectively.8, 9, 10, 11
Currently in the United States, hantavirus infection is a nationally notifiable disease, and state, tribal, local, and territorial (STLT) health departments report case surveillance data from acutely-infected hantavirus cases via case report forms to the national hantavirus surveillance registry hosted at the U.S. Centers for Disease Control and Prevention (CDC).12,13 The criteria to report positive cases are based on the national case definition, which includes clinical symptoms (HPS or non-HPS) and acute laboratory diagnostic results, such as: 1) IgM positive; 2) IgG positive with rising titers; 3) immunohistochemistry positive; or 4) PCR positive.8,9 Specimens meeting this case definition should be reported by STLT jurisdictions via the National Notifiable Diseases Surveillance System (NNDSS) and a case report form submitted to CDC. There is no standardised hantavirus serological or RT-PCR test used in the U.S. Hantavirus cases are identified diagnostically at the CDC, state public health labs using CDC-developed assays, state public health labs using other diagnostic assays, or at commercial labs. While state health departments report acute hantavirus-positive cases to the hantavirus registry, this surveillance tool misses the detection of convalescent (IgG-only positive, without rising titers) hantavirus cases in the U.S., because STLT jurisdictions are not asked to report convalescent case results.
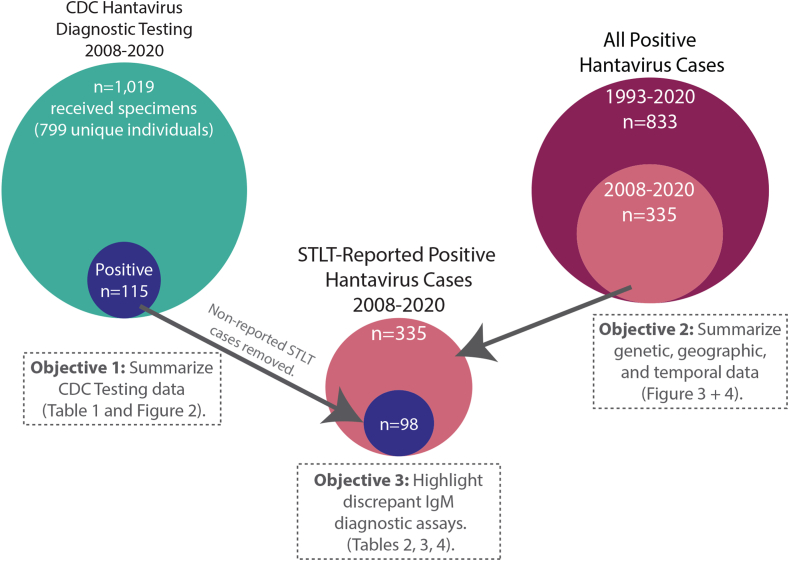
To present a more complete picture of hantavirus surveillance in the U.S., three datasets were utilised: 1) acute hantavirus cases reported by STLT health departments to the hantavirus registry from 1993 to 2020 (n = 833), 2) acute hantavirus cases reported by STLT health departments to the hantavirus registry from 2008 to 2020 (n = 335), and 3) hantavirus acute and convalescent diagnostic testing performed at the CDC/Viral Special Pathogens Branch (VSPB) from 2008 to 2020, representing 29.2% (98/335) of total acute hantavirus cases (Fig. 1). Previous publications have reviewed U.S. human hantavirus cases reported to the registry from 1993 to 2008,14 1993–2009,12 and 1993–2013,13 but none have reported any details about convalescent case detection in the U.S., nor about the duration of antibody responses. By combining the registry dataset with detailed information from CDC diagnostic testing and sequencing over the same time period, an updated view of hantavirus surveillance in the U.S. is presented, which includes the geographic distribution of human exposures and Orthohantavirus genotypes causing human disease. Currently, no standardised hantavirus serological or RT-PCR test is used in the US, and reference testing performed at the CDC has demonstrated that IgM false-positive results have been observed from commercial diagnostic assays, suggesting that a possible overestimation of acute hantavirus cases may be reported from states that do not perform confirmatory testing at a reference lab. A suggested testing algorithm for hantavirus diagnostic testing is provided for state public health departments to be considered for adoption.
Fig. 1.
Overview of datasets used to evaluate human Orthohantavirus disease prevalence and genotype distribution in the U.S. Overall 11,019 unique specimens were received at VSPB from 2008 to 2020 for diagnostic testing and 115 unique individuals were identified as hantavirus positive. Seventeen individuals tested by CDC were not reported as hantavirus-positive cases by STLT (n = 98). From 2008 to 2020, 335 unique people were reported as hantavirus-positive cases by STLT and of those VSPB performed diagnostic testing for 98 unique people. Altogether, 833 unique individuals were reported as Hantavirus-positive cases by STLT health departments. The relationships between these datasets, objectives, and figures/tables are highlighted.
Methods
Human subject
The project was reviewed by the CDC Human Subject advisors and determined to meet the requirements of a public health surveillance activity that involved the use of residual human specimens. Additionally, the use of domestic surveillance data for this project was also reviewed by the CDC Human Subject advisors and determined to meet the requirements of public health surveillance activities. Therefore, IRB approval was not required for this work.
Study design
This project was a retrospective analysis of multiple datasets (Fig. 1). 1) All acute hantavirus cases reported by STLT health departments to the NNDSS and hantavirus registry from 1993 to 2020 (n = 833), 2) acute hantavirus cases reported by STLT health departments to the NNDSS and hantavirus registry from 2008 to 2020 (n = 335), and 3) hantavirus acute and convalescent diagnostic testing performed at the CDC/Viral Special Pathogens Branch (VSPB) from 2008 to 2020, representing 29.2% (98/335) of total acute hantavirus cases (Fig. 1).
Data agglomeration and distribution analysis
The summary of hantavirus laboratory testing performed at VSPB from 2008 to 2020 was generated by parsing the reports sent by VSPB to submitters (state public health labs and requesting doctors) using a pdf parser (https://github.com/evk3/hantavirus_US_distribution). Health departments review the laboratory and clinical data associated with suspect hantavirus cases and submit confirmed positive cases to the NNDSS. A summary of HPS- and non-HPS symptoms can be found in Supplemental Table S1. A summary of confirmed hantavirus cases from the NNDSS is provided to VSPB epidemiologists, who request additional case data from STLT jurisdictions using a hantavirus disease report form.4,12 These data are reconciled annually with the case surveillance data reported in the NNDSS to ensure all reported cases were included in the registry. Location information captured by the case report form included city, county, state, and zip code of residence and city, county, and state of potential rodent exposures within the 6 weeks prior to illness. As a caveat, the exposure location captured on the case report form may be the best estimate and/or may not represent the true exposure location -- which could be unknown. Sex (but not gender) and ethnicity data are collected on hantavirus case report forms and have been summarised by St. Maurice et al.4 Additionally, patient information for suspected hantavirus cases tested at a commercial laboratory prior to testing at the CDC was utilised to investigate the concordance of diagnostic results. For patients whose samples were tested at the CDC but did not have a corresponding case report form available (i.e., tested negative), information on signs and symptoms, exposure, and general clinical course were abstracted from clinical consultation summaries when available. Data from the national hantavirus registry, and from hantavirus testing performed at CDC was compared, summarised, and manipulated using Python3 (version 3.7). Serological and PCR results versus time of symptom onset were visualised using Python3 with pandas plot.kde (bw_method = 0.3). Geographic and temporal distributions were visualised using QGIS (3.22.3) and R (version 4).
Laboratory testing of suspect hantaviral specimens
Whole blood (WB) or serum specimens submitted to CDC for hantavirus diagnostics were tested for the presence of New World (NW) Orthohantavirus reactive immunoglobulin G (IgG) (using recombinant SNV nucleocapsid protein as antigens) and immunoglobulin M (IgM) antibodies (using native SNV antigens),15 as well as for the presence of Old World (OW) Orthohantavirus-reactive IgG and IgM (using native SEO antigens).16 Hantavirus RNA positive specimens were assessed using the hantavirus conventional PCR assay from Woods et al.17 Acute specimens were defined by the presence of Orthohantaviral reactive IgM, IgM/IgG or PCR positivity and presence of clinical (HPS and non-HPS symptoms) and epidemiological factors (rodent exposure). Convalescent specimens were defined by the presence of only Orthohantaviral reactive IgG.
Next generation sequencing
Residual whole blood (WB) or serum specimens submitted for diagnostic testing at CDC were inactivated with Tripure (Roche, USA) or 5× Magmax 96 Viral Isolation kit (Applied Biosystems Inc., USA). RNA was extracted from Tripure-inactivated WB or serum by phase-separation using 1-bromo-3-chloropropane (Sigma–Aldrich, St. Louis, MO) and applied to Clean and Concentrate-25 columns (Zymo Research, Irvine, CA) for further purification and concentration. RNA was extracted from residual FFPE tissue specimens by cutting one 16 μm scroll and extracting RNA according to Bhatnagar et al.18 To evaluate the level of fragmentation and presence of PCR inhibitors, RNA extracted from FFPE tissue specimens was also tested by housekeeping gene 18S rRNA RT-PCR using QuantumRNA Classic 18S Internal Standard (Life Technologies, Carlsbad, CA, U.S.) following the manufacturer's instructions. To perform next-generation sequencing (NGS), extracted RNA was treated with RNase-free DNase (Roche, Basel, Switzerland) and prepared for unbiased NGS using a TruSeq RNA Access Library preparation kit (Illumina, San Diego, CA) with pan-hantavirus-specific enrichment oligos. Specimens were enriched using single-plex enrichment with 0.625 pmol probes per enrichment and reducing the reaction volumes by a quarter. The pan-hantavirus probe set was designed by downloading all available hantavirus genomes from Genbank, removing duplicate virus entries and designing 80bp 5′ biotinylated probes offset every 200bp (https://github.com/evk3/Nipah_phylogenetics), resulting in 5325 hantavirus-specific probes (Twist Biosciences). Libraries were sequenced using either an Illumina MiSeq or MiniSeq (High Output 2 × 150 cycles).
Bioinformatics and phylogenetics
Genomes were constructed using a guided de novo assembly approach and scripts are available on https://github.com/evk3/hantavirus_US_distribution. First, reads were trimmed for quality (prinseq-lite -min_qual_mean 25 -trim_qual_right 20 -min_len 50) and de novo assembled to make contigs (spades.py -k auto). Contigs were blasted to identify the closest reference sequence on GenBank. Genomes were iteratively assembled by mapping reads and contigs to the closest refseq sequence (SNV, Monongahela, New York or Seoul viruses). Consensus genomes were called from bam files using iVar (v1.3, -m X -n N), and threshold setting (-m) was set above the level of reference-mapping reads observed in the negative extraction control or sequencing control (lowest threshold was 10-fold, or 10-fold if no negative extraction control was available). For 4/66 genomes (200605818_L, 200605818_L, 2020030197_M, 201802465_L), mapping of reads and contigs introduced indels or frameshifts, in these cases, consensus genomes were re-built using Geneious (0% majority, assign quality total). Patient metadata and a summary of genetic coverage and accession numbers can be found in Supplemental Table S3.
To build phylogenetic trees, sequences generated in this study were analysed with all available full-length SNV, Monongahela, New York, Black Creek Canal, and Choclo segments downloaded from GenBank and aligned using MAFFT (v7.471). Trees were constructed with RAxML (v7.3.0, -m GTRGAMMA -p $RANDOM -f a -x $RANDOM -N 1000); and visualised with ggtree (v1.11.3). Genomes were submitted to GenBank, accession numbers OQ999105-OQ999170.
Statistical analysis, endpoints and outcomes
In Table 1 and Fig. 2 (“Objective 1: Summarize CDC Testing data”) the summary of hantaviral diagnostics performed in the Viral Special Pathogens Branch at CDC (2008–2020) was calculated using an iPython notebook (Python 3.9). Mean and Mode were calculated using the pandas and numpy libraries. Geographic and temporal distributions in Fig. 2 were calculated from the same data set. In Figs. 2 and 3 (“Objective 2: Summarize genetic, geographic, and temporal data”), we summarize the genetic, geographic and temporal characteristics of human hantaviral cases reported to the NNDSS by STLT health departments between 1993–2020 and 2008–2020. In Tables 2 and 3 (“Objective 4: Highlight discrepant IgM diagnostic assays”) we compare the results between different diagnostic assays tested on the same sample sets. Statistical calculations for agreement between datasets were generated using 2 × 2 contingency tables to calculate percent agreement [also known as “diagnostic accuracy” ((TP + TN)/(TP + TN + FP + FN)) ], and Cohen's Kappa (Tables 2 and 3) using [k = [(Po—Pe)/(1- Pe)]]. Clinical symptoms (eg, fever, thrombocytopenia, elevated hematocrit, etc) and diagnostic results (eg, CDC IgG+) of persons with Commerical IgM+ results were compared between those with CDC IgM+ or CDC IgM-results using Barnard's exact test. All analyses were performed using Python (3.9) and p-values <0.05 were considered statistically significant. Spearman'r ρ and Pearson's r in Supplemental Figure S2 were calculated using the ∗.corr function from pandas.
Table 1.
Summary of hantavirus diagnostics performed in the viral special pathogens branch, US CDC, 2008–2020.
| New world Orthohantavirus |
Old world Orthohantavirus |
Pan-hanta |
|||||
|---|---|---|---|---|---|---|---|
| IgM+ (Acute) | IgM+ and IgG+ (Acute/Recovery) | IgG+ (Recovery) | IgM+ (Acute) | IgM+ and IgG+ (Acute/Recovery) | IgG+ (Recovery) | RT-PCR+ (L segment) | |
| Total specimens percent positive | 5.9% (61/1019) | 5.9% (61/1019) | 1.9% (19/1019) | 1.6% (17/1019) | 1.0% (11/1019) | 0.3% (4/1019) | 9.0% (21/231) |
| Total unique individuals percent positive | 5.8% (47/799) | 6.6% (53/799) | 2.2% (18/799) | 1.0% (8/799) | 0.6% (5/799) | 0.2% (2/799) | 10.3% (21/202) |
| Pan-Hanta RT-PCR+ Individualsa % (positive/total) | 50.0% (7/14) | 55.5% (10/18) | 0% (0/0) | 0% (0/4) | 0% (0/2) | 0% (0/0) | (0/164)b |
| Median time frame from symptom onset to specimen collection [range in Days] | 5 [1, 159] | 7 [0, 32] | N/A | 16 [11, 21] | 8 [6, 10] | N/A | 5.0 [1, 48] |
| Modal time frame from symptom onset to specimen collection (Days) | 4 | 5 | N/A | 16 | 8 | N/A | 4 |
| Number of specimens with symptom onset dates | 51 | 59 | 0 | 2 | 4 | 0 | 19 |
| Percent of positive specimens with symptom onset dates | 83.6% (51/61) | 96.7% (59/61) | N/A | 11.7% (2/17) | 36.3% (4/11) | N/A | 90.4% (19/21) |
4 additional individuals were PCR positive: 1 serologically negative; 1 serologically indeterminate, IHC+; 2 individuals with tissues received and serology was not performed, IHC+.
164 PCR tests were performed on serologically-negative individuals; all were PCR negative.
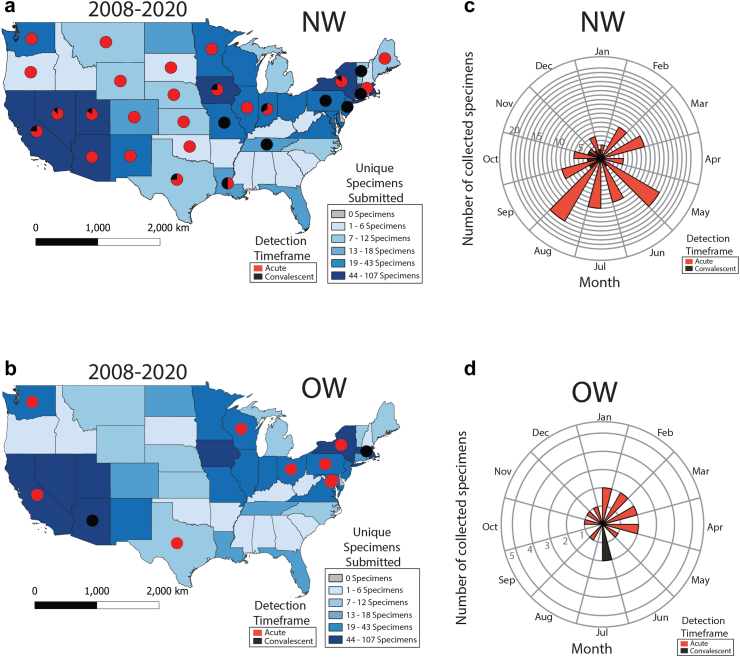
Fig. 2.
Initial detection of acute or convalescent human hantavirus cases by VSPB-CDC presented by state and time, 2008–2020. a and b) Map of initial detection of acute or convalescent NW (a) or OW (b) human hantavirus cases. Pie charts indicate ratio of initial acute or convalescent cases detected per state. State color indicates number of unique specimens received per state for hantavirus diagnostic testing. A single specimen was received from Alaska, Puerto Rico, and the U.S. Virgin Islands, and two specimens from Hawaii, but all were hantavirus negative. No other specimens were received from other U.S. territories. c and d) Seasonality of initial detection of acute (red) or convalescent (black) NW (c) or OW (d) human hantavirus cases by VSPB-CDC from 2008 to 2020.
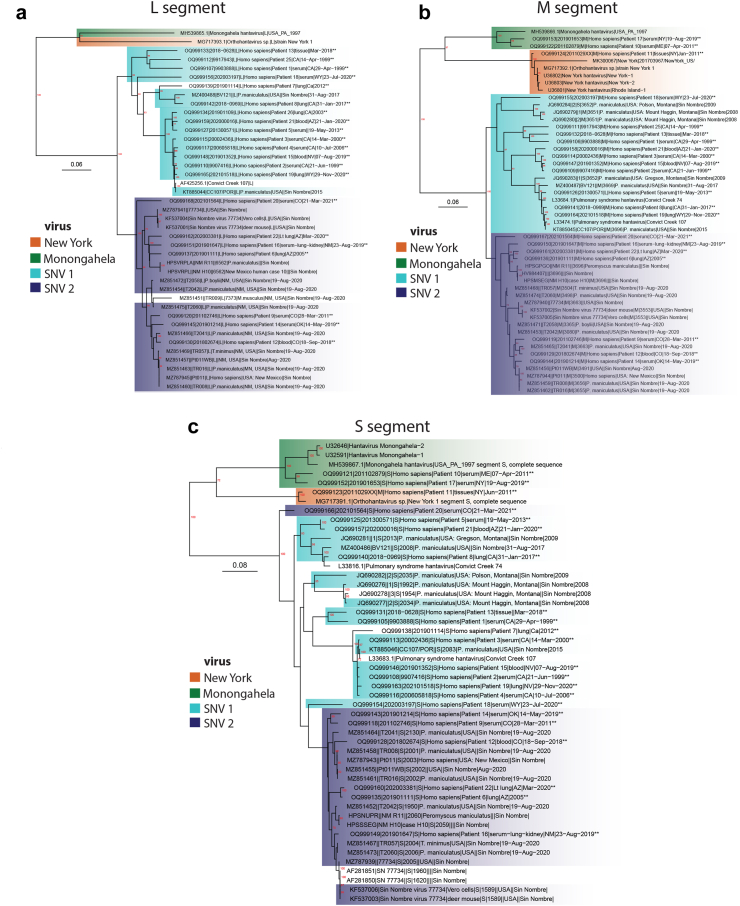
Fig. 3.
Inferred relationships of domestic human Orthohantavirus sinnombreense cases. Viral genotypes, New York virus, Monongahela virus and Sin Nombre virus are highlighted orange, green or blue, respectively (similar to Fig. 4). The two major SNV clades are colored light or dark blue according to the SNV M segment to highlight possible reassortment between segments and sequences without highlight did not match with a specific M segment. New sequences are indicated with “∗∗” at the end of sequence names. Bootstrap support (n = 1000) greater than 70% is labeled in red at internal nodes and scale bars are in units of substitutions/site. a) Phylogenetic tree of all available full length Orthohantavirus sinnombreense species L segments. b) Phylogenetic tree of all available full length Orthohantavirus sinnombreense species M segments. c) Phylogenetic tree of all available full length Orthohantavirus sinnombreense species S segments.
Table 2.
Concordance of NW serological diagnostic results from VSPB- and commercial lab-based testing, 2018–2020.
| Samples received with commercial hantavirus DXa | Number of commercial lab positive tests | Number of paired positive commercial lab and VSPB total tests | Number of paired positive commercial lab and positive VSPB tests | Percent agreement | Cohen's kappa | |
|---|---|---|---|---|---|---|
| IgM+ Results | 92 | 78 | 76 | 13 | 29% | 0.09 |
| IgG+ Results | 77 | 25 | 24 | 5 | 70% | 0.83 |
| IgM+ & IgG+ Results | 76 | 16 | 10 | 4 | 40% | 0.11 |
Percent agreement and Cohen's Kappa values were calculated using the 2 × 2 contingency tables included in Supplemental Table S2.
Data is from all individuals, including duplicate testing.
Table 3.
Concordance of OW serological diagnostic results from VSPB- and commercial lab-based testing, 2018–2020.
| Samples received with commercial hantavirus DXa | Number of commercial lab positive tests | Number of paired positive commercial lab and VSPB total tests | Number of paired positive commercial lab and positive VSPB tests | Percent agreement | Cohen's kappa | |
|---|---|---|---|---|---|---|
| IgM+ Results | 18 | 14 | 13 | 12 | 82% | 0.48 |
| IgG+ Results | 18 | 15 | 14 | 10 | 76% | 0.45 |
| IgM+ & IgG+ Results | 18 | 11 | 9 | 8 | 89% | 0.29 |
Percent agreement and Cohen's Kappa values were calculated using the 2 × 2 contingency tables included in Supplemental Table S2.
Data is from all individuals, including duplicate testing.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Summary of VSPB Orthohantavirus diagnostic testing, 2008–2020
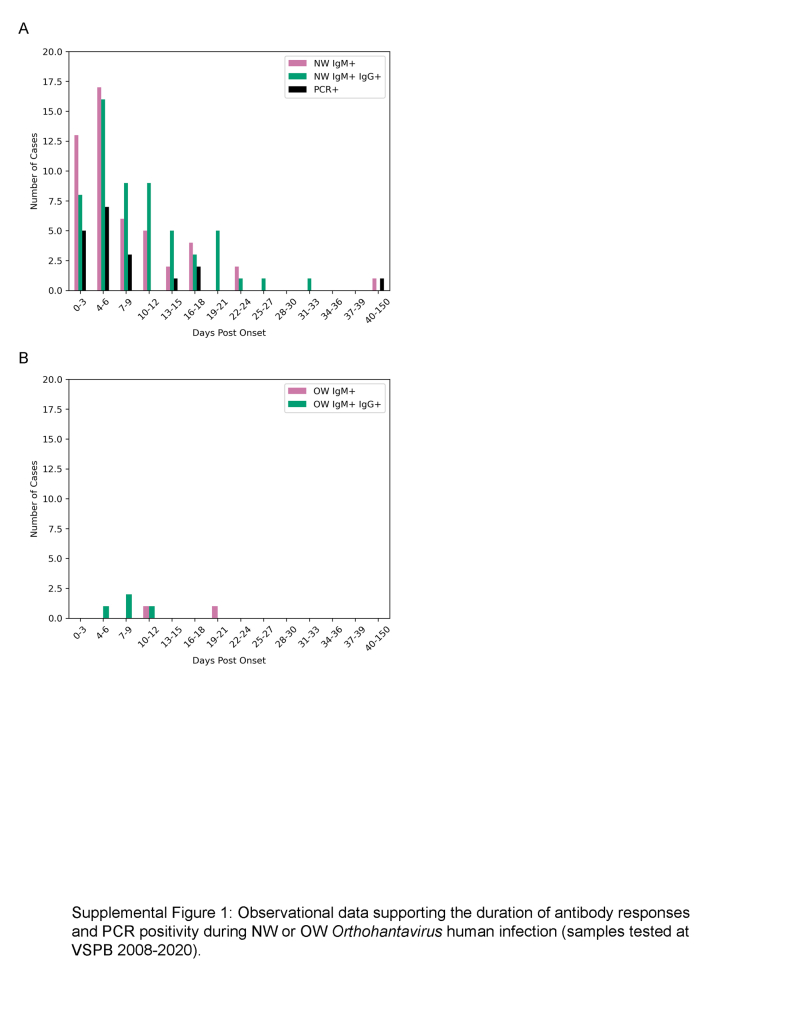
Between 1993 and 2020, 833 human hantavirus cases have been identified, and from 2008 to 2020, 335 human cases have occurred (Fig. 1). These results were reported by STLT health departments as a result of diagnostic testing performed at CDC, state public health labs using CDC-developed assays, state public health labs using other diagnostic assays, or at commercial labs. There is no standardised hantavirus serological or RT-PCR test used in the U.S. From 2008 to 2020, 1019 domestic samples were received by CDC-VSPB for hantavirus diagnostic testing and all specimens were tested for the presence of NW-specific and OW-specific IgM and IgG (Table 1). 115 specimens were identified as positive by CDC, and by matching these cases with the same 98 individuals reported as hantavirus-positive cases by STLT (representing 29.2% (98/335) of total acute confirmed cases reported by health departments) we identified 17 non-STLT reported cases (Fig. 1). Most positive samples tested at CDC were both NW IgM and IgG seropositive (5.9%, 61/1019), and NW IgM-only seropositive (5.9%, 61/1019), followed by NW IgG-only seropositive samples (1.9%, 19/1019) (Table 1). A smaller proportion were identified as OW seropositive (0.3–1.6% IgM-only, IgM/IgG, or IgG-only positive). When these results were filtered to remove duplicate samples tested from the same individual, similar seropositive rates were observed between the total specimens and unique individual NW-reactive and OW-reactive datasets (Table 1). Specimens that were collected less than 14 days after symptom onset were also evaluated for the presence of hantaviral RNA (n = 231 total specimens and n = 202 unique individuals tested) and only 21 unique individuals (10.3%, 21/202) were identified as PCR-positive (Table 1). PCR positive individuals were either NW-reactive IgM or IgM and IgG positive (Table 1). Patient-reported symptom onset dates were available for 83.6% of IgM-only and 96.7% of IgM/IgG seropositive NW specimens and 11.7% of IgM-only and 36.3% of IgM/IgG seropositive OW specimens (Table 1). The median time from symptom onset to specimen collection for patients with detectable NW IgM was 5 [1, 159] days and OW IgM was 16 [11, 21] days. In contrast, specimens from individuals with hantavirus RNA detected by RT-PCR were within 5.0 [1, 48] days post onset (Table 1). When these serological and PCR responses were plotted temporally, concurrent IgM and IgG responses for NW and OW were observed (Supplemental Figure S1). This observational data suggests that NW and OW IgM were most frequently detected at 4 and 16 days post onset (dpo), respectively.NW IgM and IgG responses were most frequently detected at 5 dpo (Table 1 and Supplemental Figure S1). Hantaviral RNA was most frequently detected at 4 dpo (Table 1 and Supplemental Figure S1).
Geographic and temporal detection of U.S. hantavirus cases
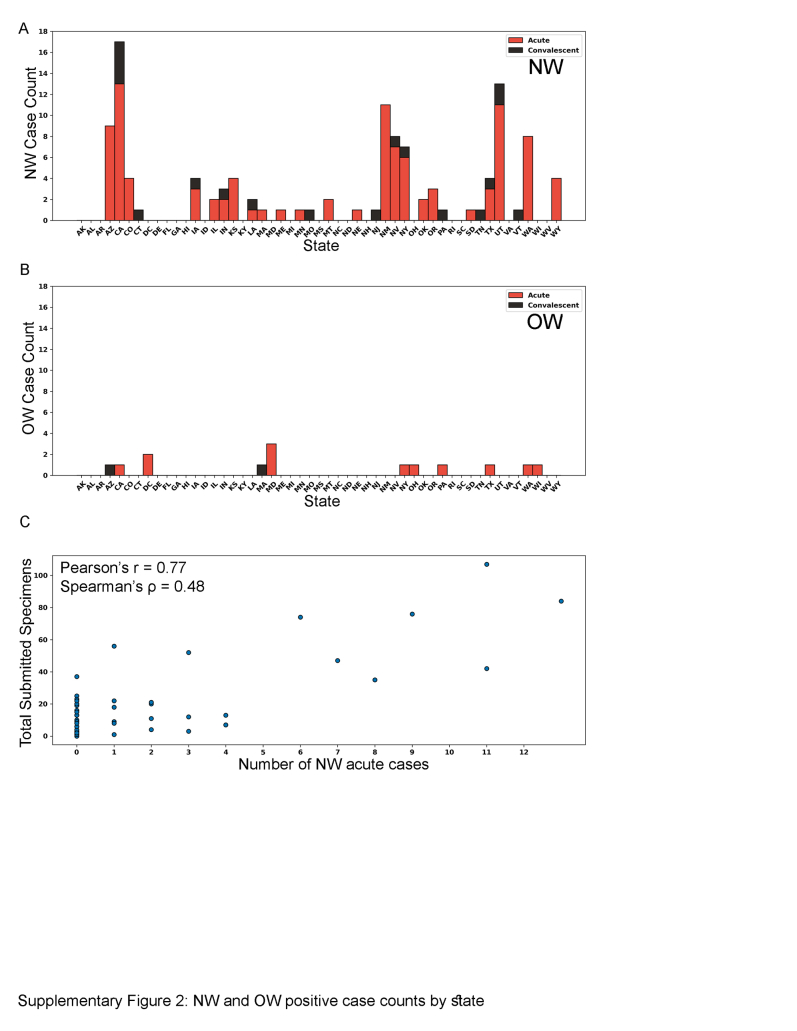
Specimens submitted to CDC for diagnostic testing were evaluated by exploring when and where individuals were first detected as positive cases, using the same acute or convalescent criteria used by jurisdictions [i.e.,—acute = IgM+, RT-PCR+ or convalescent = IgG+ -only; no IHC+ or IgG+ cases with rising titers were identified]. State location was assigned based on reported exposure state [65.2% (88/135); the 135 denominator represents 115 acute and 20 convalescent individuals], and when exposure state was not available, residence state (7.4%, 10/135) or state lab report was sent to (27.4%, 37/135) were used, respectively (only three individuals reported multiple exposure states and are not included on the map). While diagnostic testing at CDC only represents a fraction of total acute hantavirus positive cases reported by health departments in the U.S., this dataset identifies some convalescent cases (IgG+) that are not currently reported to the NNDSS or the hantavirus registry. NW-specific cases (n = 102 acute, 18 convalescent) were observed to have wide geographic distribution (29/50 states), while OW-specific cases (n = 13 acute, 2 convalescent; not associated with the 2017 SEO rattery outbreak16) were observed in only 19/50 states and Washington, DC (Fig. 2a and b and Supplemental Figure S2A and B). Overall, 86.6% (13/15) of OW- and 85.0% (102/120) of NW-specific cases were detected during acute disease (Fig. 2A–B); and most positive NW cases were from California, Utah, and New Mexico (Supplemental Figure S2A and B). Individuals were detected only during the NW convalescent phase in Connecticut, Missouri, New Jersey, Pennsylvania, Tennessee, and Vermont and OW convalescent phase in Massachusetts and Arizona—most of these states are outside of the Four Corners region (AZ, CO, UT, and NM) that are not traditionally associated with hantaviral cases. Furthermore, there was a high positive correlation between the number of specimens sent by a state to CDC for hantaviral testing and the number of positive NW cases detected in that state (Pearson's r = 0.77, Spearman's ρ = 0.48) (Supplemental Figure S2C). When exploring the temporal distribution of domestic hantaviral cases, we observed that NW-specific acute cases were detected most frequently during May, June, July, and August, whereas OW-specific acute cases were detected most frequently during January–April (Fig. 2c and d).
Phylogenetic relatedness of U.S. Orthohantavirus genotypes causing human disease
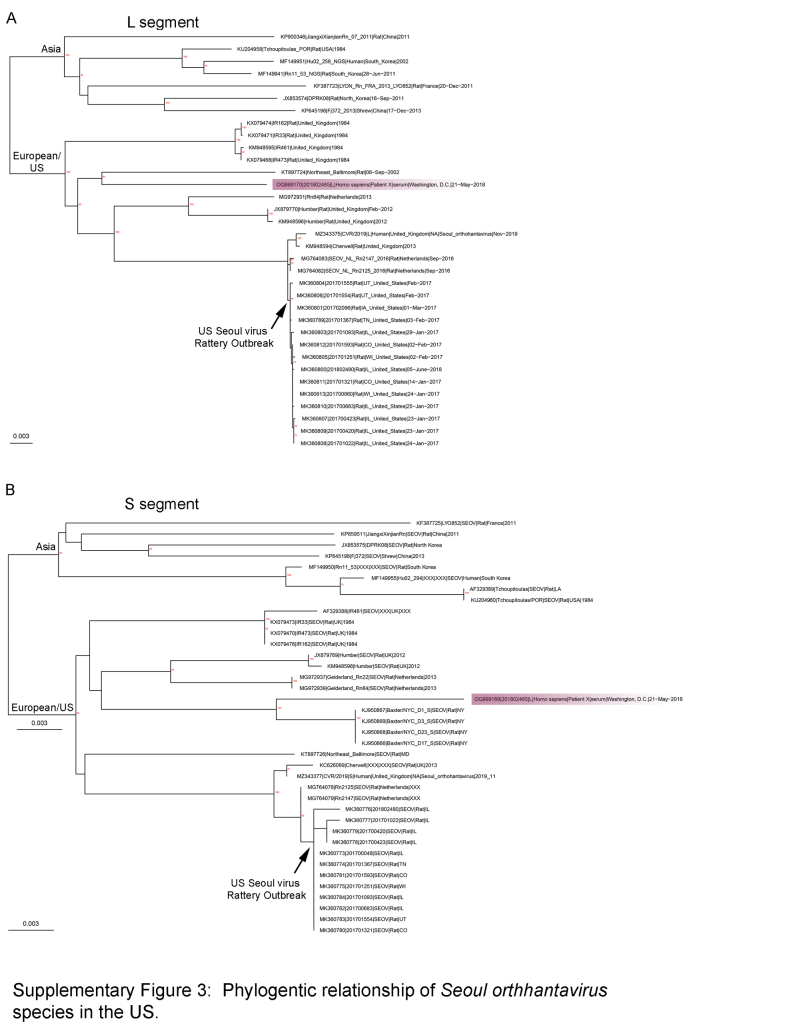
To better understand the inferred relatedness of domestic Orthohantavirus sinnombreense genotypes causing human disease, all available PCR-positive human specimens collected between 2008 and 2020 (n = 21 specimens), as well as a subset of historic human hantaviral cases from 1999 to 2008 (representing 60 blood, serum, or tissue specimens from 40 unique individuals from 1999 to 2020) were deep sequenced (Supplemental Table S3). From 23 unique individuals, 20, 19, and 21 L, M and S segments, respectively, with greater than 69% coverage (at >10× -fold depth) were generated. Within the O. sinnombreense species, two new Monongahela sequences and one New York sequence were generated and these cluster with related reference sequences (Fig. 3a–c). The SNV clade separated into two distinct well supported sub-clades (SNV1 and SNV2) for the L and the M segments (Fig. 3a and b). In contrast, the separation between these clades was not strongly supported for the S segment, but the current S topology mimics the separation observed for the L and M segments (Fig. 3c). Except for S segments from two sequences (OQ999166 and OQ999154), there was no strong evidence for segment reassortment between the SNV1 and SNV2 subclades. In the Orthohantavirus seoulense species, a single Seoul virus (SEO) sequence collected from an individual in Washington, D.C. was generated, and it clustered with SEO sequences from Northeastern Baltimore and New York City (Supplemental Figure S3).
Geographic and temporal distribution of STLT-reported acute hantaviral disease
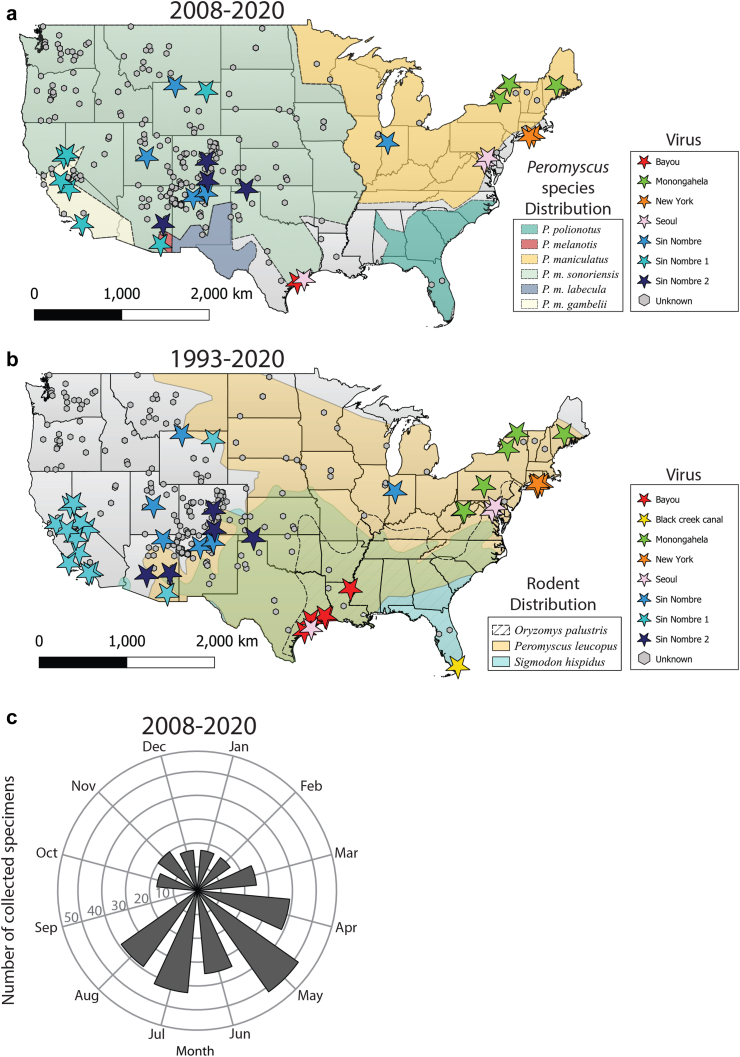
Since 1993, 833 human hantaviral disease cases have been identified by STLT jurisdictions and 31 of these cases were identified retrospectively as occurring before 1993. Combining the available genetic information with known exposure locations for all acute human hantavirus cases demonstrates the domestic U.S. Orthohantavirus prevalence, and genotype distribution (Fig. 4a and b) between 2008–2020 (n = 335) and 1993–2020 (n = 802). Since 1993, twelve cases reported multiple exposure locations, five cases were exposed outside of the U.S, and seven Seoul rattery cases are not included on the map. Exposure locations were reported for a majority of cases, and when unavailable, residence location was used (Supplemental Figure S4A and B) (n = 785). From 1993 to 2020, the highest local concentration of hantavirus cases in the continental US cluster in the Four Corners region of the US 45.7% (359/785) and 94.9% (745/785) of U.S. hantaviruses cases occured west of the Mississippi (Fig. 4b). Hantavirus cases also occur in the upper Midwest (IL, WI, IN, MI), mid-Atlantic (VA, WV, MD, PA, DE) and Northeastern (NY, CT, RI, MA, VT, ME, NH) regions. In contrast, fewer than 3 hantavirus cases were observed in Southeastern states (TN, NC, SC, MS, AL, GA, FL). Viral genotype was confirmed either by sequencing of a subset of PCR-positive cases (n = 44) from 1993 to 2020 [either through WGS (n = 27) or partial Sanger sequencing (n = 16) directly from human specimens or a rodent collected from the exposure site19] or serology (n = 2) (SEO20 and Bayou21 viruses in TX). Monongahela virus was localized to the Northeastern and the Mid-Atlantic regions, whereas SNV was primarily found west of the Mississippi and Ohio Rivers (Fig. 4a and b). A single SNV case was identified in Indiana, but sequence data is otherwise lacking from the Central U.S. Bayou and Black Creek Canal viruses exhibited a local distribution in TX/LA and FL, respectively. The phylogenetically distinct SNV1 and SNV2 sub-clades were not associated with localized geographic clustering. Consistent with seasonality observed from VSPB-specific diagnostic testing (Fig. 2b), we observed that most STLT-reported hantavirus cases occurred in May through August (Fig. 4c).
Fig. 4.
Geographic and temporal distribution of STLT-reported human hantaviral cases and viral genotypes from 2008 to 2020 and a comprehensive geographic distribution of all available human hantavirus cases from 1993 to 2020. a) Geographic distribution of human hantaviral cases from 2008 to 2020 (points) and Peromyscus species distributions (shading) from Greenbaum et al.28 Viral genotypes are indicated by star color and cases without sequence data are labeled as “Unknown.” No human hantavirus cases have been detected in Alaska, Hawaii, or U.S. territories. b) Geographic distribution of human hantaviral cases from 1993 to 2020 (points) and hantavirus rodent vector distributions (shading). Map is colored according to panel A. No human hantavirus cases have been detected in Alaska, Hawaii, or U.S. territories. c) Seasonality of state-reported human hantaviral cases from 2008 to 2020.
Discordance between VSPB and commercial diagnostic results
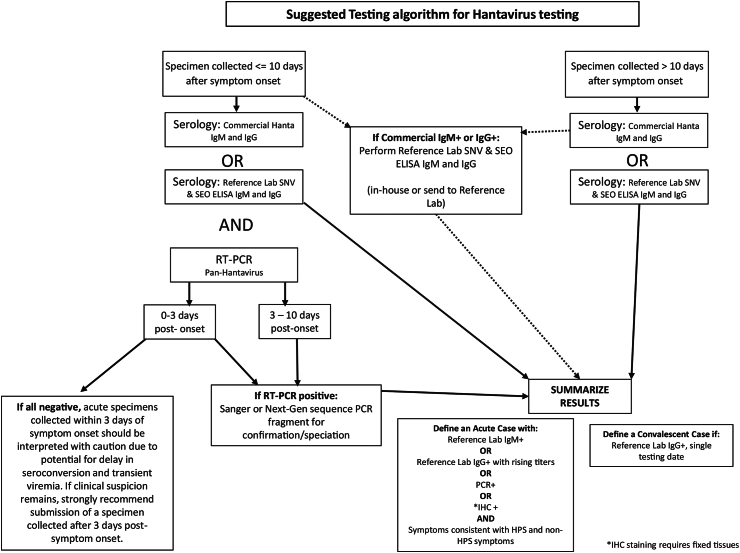
From May 2018 to December 2020, discordant NW IgM results were identified when duplicate specimens were tested using the VSPB laboratory-derived NW ELISA assays and commercial hantavirus NW IgM assays; this change was noted to have occurred while commercial labs changed hantavirus diagnostic assays. The percent agreement between NW IgM+ positive samples was 29% (n = 76), whereas the percent agreement between NW IgG+ samples was 70% (n = 24) and NW IgG+/IgM+ was 40% (n = 10) (Table 2 and Supplemental Table S2). Cohen's κ (a measure of inter-rater reliability relative to agreement by chance) from this dataset supported a minimal relationship for NW IgM+ and NW IgM+/IgG+ results, whereas NW IgG+ results were strongly supported by percent agreement and Cohen's κ. In contrast, a high percent agreement of 76–89% was observed for commercial and VSPB OW IgM (n = 13), IgM/IgG (n = 14), and IgG serological testing (n = 9). For OW results, Cohen's κ supported poor agreement, but the magnitude of Cohen's κ is likely reduced due to a small number of paired results from IgM and IgG specimens (n = 13, 14, and 9, respectively) (Table 3 and Supplemental Table S2). An analysis of symptoms associated with dual positive (VSPB and commercial) IgM+/IgM+ patients versus single positive (only commercial test positive) IgM−/IgM+ patients finds that a subset of clinical information significantly associates with these “true” hantavirus cases (ie–IgM+/IgM+ patients). Hospitalisation, thrombocytopenia, elevated haematocrit, elevated creatinine, IgG+ seroconversion and PCR-positive results were significantly associated with IgM+/IgM+ patients, but not IgM−/IgM+ patients (Table 4). Based on the commercial IgM false-positive results observed here, a revised testing algorithm is suggested that includes reflex testing with a different serological assay for all commercial IgM-positive samples (Fig. 5). The remainder of the testing algorithm and diagnoses criteria remain unchanged.
Table 4.
Symptomology associated with VSPB IgM+/Commercial IgM+ versus VSPB IgM−/Commercial IgM+ Acute hantaviral cases, 2018–2020.
| Clinical information | Commercial IgM+ |
p-value | |||||
|---|---|---|---|---|---|---|---|
| CDC IgM+ (n = 14)a |
CDC IgM− (n = 54) |
||||||
| n | denom | % | n | denom | % | ||
| CRF or other source available? | 14 | 14 | 100.0% | 52 | 54 | 96.3% | |
| Hospitalizedd | 13 | 14 | 92.9% | 28 | 48 | 58.3% | 0.012 |
| Known rodent exposured | 12 | 14 | 85.7% | 36 | 46 | 78.3% | 0.381 |
| Symptoms | |||||||
| Feverd | 7 | 14 | 50.0% | 33 | 48 | 68.8% | 1.000 |
| Thrombocytopeniad | 10 | 13 | 76.9% | 12 | 38 | 31.6% | 0.003 |
| Elevated hematocritd | 6 | 12 | 50.0% | 5 | 38 | 13.2% | 0.015 |
| Elevated creatinined | 8 | 13 | 61.5% | 7 | 32 | 21.9% | 0.007 |
| Supplemental O2d | 7 | 13 | 53.8% | 11 | 29 | 37.9% | 0.211 |
| Intubationd | 2 | 13 | 15.4% | 6 | 34 | 17.6% | 1.000 |
| CXR suggestive ARDSd | 7 | 11 | 63.6% | 14 | 37 | 37.8% | 0.085 |
| Any resp signsb | 10 | 13 | 76.9% | 31 | 39 | 79.5% | 1.000 |
| Any flu-like signsb | 9 | 10 | 90.0% | 26 | 31 | 83.9% | 0.476 |
| Any gastrointestinal signsb | 6 | 8 | 75.0% | 14 | 26 | 53.8% | 0.199 |
| Alternate Dxb | 0 | 14 | 0.0% | 9 | 52 | 17.3% | 1.000 |
| Underlying conditionb | 3 | 14 | 21.4% | 10 | 52 | 19.2% | 0.546 |
| Other test results | |||||||
| Commercial IgG+ | 10 | 13 | 76.9% | 5 | 44 | 11.4% | <0.001 |
| CDC IgG+ | 6 | 14 | 42.9% | 0 | 54 | 0.0% | <0.001 |
| CDC PCR+ | 6 | 13 | 46.2% | 0 | 29 | 0.0% | <0.001 |
| State reporting | |||||||
| Reported as HPS | 9 | 14 | 64.3% | 0 | 54 | 0.0% | <0.001 |
| Reported as non-HPSc | 3 | 14 | 21.4% | 0 | 54 | 0.0% | 0.017 |
| Not reported as case | 2 | 14 | 14.3% | 54 | 54 | 100.0% | 1.000 |
Probability values are calculated as the likelihood of having greater hantavirus-specific symptoms in the IgM+/IgM+ group using Barnard's test. Bolded text denotes p values below the significance cutoff of 0.05.
Includes 3 patients that tested IgM+ for Seoul virus at the CDC and one patient that tested IgM+ for Puumala virus at the CDC.
Information not collected by standardized case report form.
Includes 2 confirmed Seoul virus cases and 1 confirmed Puumala case with exposure outside of the United States.
Systematically collected on case report forms.
Fig. 5.
Suggested testing algorithm for Orthohantavirus diagnostic testing.
Discussion
Here, a broad and updated view of human Orthohantavirus prevalence, genotype distribution, and diagnostics between 2008 and 2020 are presented. The goal of this work is to extend earlier observations by Mills et al. (U.S. hantavirus exposure locations from 1993 to 2008),14 MacNeill et al. (hantavirus seasonality 1993–2009),12 Knust et al. (U.S. hantavirus case counts from 1993 to 2013),13 while also including detailed information about Orthohantavirus diagnostic testing, and viral genotype information combined with human exposure locations.
Mills et al. (2010)14 summarised the state of knowledge of hantavirus disease ecology in the US and presented a geographic mapping of U.S. hantavirus cases and rodent vectors, but these visualisations did not include any viral sequence data, and thus, were not able to plot hantaviral genotype distribution in the US. Here, a geographic separation of the Orthohantavirus sinnombreense species (SNV—primarily west of the Mississippi, New York virus—Long Island, NY; and Monongahela virus—Northeastern/Mid-Atlantic) is presented. However, hantavirus sequence data is limited, and large gaps in viral sequence data from the Pacific Northwest and Central U.S. remain, suggesting that additional viral genotypes causing human disease could exist in the U.S.
It has been postulated that hantaviruses co-speciate with their rodent hosts,22, 23, 24 however, widespread molecular surveillance and phylogenetic analyses of hantaviruses have demonstrated that spillover or host switching to unrelated taxa (e.g., Chiroptera, Soricidae, Talpidae) has occurred14,25, 26, 27 challenging this hypothesis. On smaller taxonomic scales, however, patterns of co-speciation may be upheld. The geographic distribution of specific SNV viruses observed here is suggestive that Orthohantaviruses may have co-evolved within the Peromyscus maniculatus species complex (Peromyscus maniculatus, P. m. sonoriensis, and P. m. gambelii); however, elements of Peromyscus systematics and taxonomy, such as recognising cryptic species, remain unresolved,28 and, likely, the diversity of hantaviruses in the U.S. is still largely unexplored. Nonetheless, phylogeographic patterns of U.S. hantaviruses may shed light on potential species boundaries within the P. maniculatus complex or within other potential rodent, shrew, or bat vector species (e.g., multiple hantaviruses are associated with Sigmodon hispidus which is also thought be comprised of multiple species).14
Of particular interest to public health are the U.S. regions, such as the southeastern and lower mid-Atlantic U.S., where human hantavirus cases have not occurred at high frequency or are undetected. These regions are within the home range of known hantavirus rodent vectors (Peromyscus sonorensis, Oryzomyz palustris, Peromyscus leucopis, and Sigmodon hispidus),28 suggesting that hantavirus spillover is still a public health threat. However, one possibility is that a “Southeastern” U.S. hantavirus remains infrequently detected, either due to a lack of spillover human cases or due to under-reporting/under-detection of human cases as a result of mild or undetected disease. Improving the clinical characterisation of hantavirus infection, as well as clinician education to recognise non-pulmonary and/or mild disease could improve case detection.
Using the results of CDC/VSPB-specific human diagnostic testing (representing 29.2% of total U.S. hantavirus cases), we quantified whether suspect hantavirus cases were detected during acute or convalescent disease. Most NW hantavirus cases 85.0% (102/120) were detected during acute disease, however, some cases were identified in the convalescent phase in states outside of the Four Corners region of the U.S. not usually associated with hantavirus infections (Connecticut, Missouri, New Jersey, Pennsylvania, Tennessee, and Vermont). These data are subject to bias due to patient-reported and incomplete exposure locations, the number of specimens and likelihood of laboratories submitting to CDC for diagnostic testing; however, it provides the only available snapshot of convalescent hantavirus detection in the U.S. (which is not reported nationally), and better case-controlled data collection is needed to quantify gaps in hantavirus surveillance. As demonstrated here, there was a positive correlation between the number of suspect hantavirus cases submitted to CDC for diagnosis and the number of hantavirus positive cases detected per state (Supplemental Figure S2C). Overall, enhancing clinician education and outreach to consider hantavirus infection as a differential diagnosis, improving access and quality of hantavirus acute diagnostic assays, and re-evaluating the hantavirus case definition could improve rates of hantavirus detection. For example, Armien et al. demonstrated that a subset (48%, 25/52) of Choclo virus-infected patients only exhibited mild disease consisting of fever with mild or no pulmonary oedema.29 An additional component to better understand disease prevalence is improving rodent hantavirus surveillance, especially in non-traditional hantavirus-positive states. Limited rodent surveillance in national parks, and in connection with human hantavirus cases has begun to elucidate new geographic areas containing hantavirus seropositive rodents.27,30, 31, 32 Increased clinician vigilance combined with improved rodent surveillance should yield improved detection of acute human cases.
Crucial for detecting positive hantavirus cases is the need for a more standardised testing algorithm. Currently, no standardised hantavirus serological or PCR test is used in the U.S., and CDC does not systematically track what hantavirus assays are run in jurisdictional public health laboratories; furthermore, jurisdictions may change assay usage over time and are not required to inform CDC. The data presented here demonstrate that discordance exists between CDC-specific and commercial NW IgM diagnostic assays; thus false-positive hantavirus cases confirmed only by commercial NW IgM-positive results may be reported by health departments. Hantavirus-negative specimens identified by commercial labs are not routinely sent for confirmatory testing with the CDC assay, thus, the false-negative rate of the commercial assays is currently unknown. Previous commercial testing during the 2012 Yosemite hantavirus outbreak identified a similar false-positive issue that required an increase in the assay threshold to eliminate the false-positive results.33 Public health jurisdictions may consider free confirmatory testing at the CDC for samples with positive serological results from commercial assays. Concordant positive (i.e., commercial IgM+ and CDC IgM+ results) individuals clinically exhibited characteristics of traditional hantavirus-associated disease (thrombocytopenia, elevated haematocrit, elevated creatinine, IgG + seroconversion, and/or PCR positive results). The five-point screening tool of peripheral blood smears identified a similar set of laboratory findings (thrombocytopenia, elevated haemoglobin/haematocrit, a left shift on neutrophils, absence of significant toxic granulation of the neutrophils, and immunoblasts and plasma cells more than 10% of lymphoid cells) that are hallmarks of hantavirus infection.34 These clinical symptoms have also been predictive of hantaviral disease during the COVID-19 pandemic and as a rapid screening tool during early and indistinguishable disease presentation.35 Development of current and future diagnostic assays will need to demonstrate concordance with known symptomology, as well as sensitivity and specificity to detect diverse hantavirus genotypes. As sequencing gains a stronger foothold in public health laboratories, a better characterization of hantaviruses along with important metadata, such as clinical symptoms (mild versus severe versus pulmonary), is needed to evaluate whether the Orthohantavirus sinnombreense species (New York, SNV, Monongahela) are associated with specific symptoms. Sequencing of RT-PCR positive hantavirus specimens can also be performed at CDC free of charge to the submitting public health laboratory.
Looking forward, the goal of hantavirus surveillance and detection over the next twenty years should aim for improved coordination between STLT health departments, CDC, commercial and academic partners. As demonstrated by Armien et al., accurate diagnostic testing is needed to better define the spectrum of hantavirus disease, refine the case definition, and yield improved case detection. However, without standardised diagnostic methods and coordination between partners, this sequential refinement of hantavirus surveillance is not supported. Ongoing efforts by PAHO to strengthen regional Hantavirus capacity in South America by aligning diagnostic and surveillance protocols, developing regional guidelines and training workshops is an example of ongoing efforts to refine hantavirus surveillance.36 Hantavirus surveillance and detection will continue to improve with clearly defined, systematic reporting methods, improved detection methods and assays, as well as explicit guidelines for clinical characterisation and diagnostic criteria.
Contributors
Conceptualization, S.L.M.W., A.W., J.K.
Methodology, S.L.M.W., A.W., M.M., E.T., S.Z., D.C., C-F.C., A.G., I.K., M.M-B., B.K., B.A.
Software, S.L.W.M., A.W., M.M., E.T.
Data Verification, Raw data access, and validation, S.L.M.W., A.W., M.M., E.T.
Decision to submit for publication: S.L.M.W., A.W., J.M.M., J.D.K.
Formal Analysis, S.L.M.W, A.W., M.M., E.T., E.S.
Investigation, S.L.M.W., A.W., M.M., E.T., E.S., S.M., M.D., J.B., L.E., S.Z., D.C., C-F.C., A.G., I.K., M.M-B., M.C., B.K., B.A.
Resources, S.M., M.D., J.B., L.E., J.M, T.S., J.K.
Data Curation, Verification and Raw Data Access, S.L.M.W., A.W., M.M., E.T., D.C., C-F.C., A.G., I.K., M.M-B., M.C., B.K., T.S., J.K.
Writing—Original Draft, S.L.M.W., J.K.
Writing—Review & Editing, S.L.M.W., A.W., C.C., S.M., M.C., B.K., B.A., J.K.
Visualization, S.L.M.W., A.W., M.M., E.T.
Supervision, J.M, T.S., J.K.
Project Administration, J.M, T.S., J.K.
Funding Administration, J.M, T.S., J.K.
All authors contributed important intellectual content during manuscript drafting or revision and accept accountability for the overall work by ensuring that questions pertaining to the accuracy or integrity of any portion of the work are appropriately investigated and resolved. All authors had final responsibility for the decision to submit for publication.
Data sharing statement
An anonymised, deidentified version of the dataset can be made available upon reasonable request to the corresponding author to allow all results to be reproduced.
Editor note
The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Declaration of interests
The authors declare no competing interests.
Acknowledgements
We acknowledge the previous and current Viral Special Pathogens Branch Diagnostic team members that performed hantaviral diagnostic testing. We also acknowledge state, tribal, local, and territorial public jurisdictions who completed case report forms and assisted with national hantavirus surveillance. The views expressed in this Article are those of the authors and do not necessarily represent the official position of the U.S. Centers for Disease Control and Prevention, the California Department of Public Health, nor the California Health and Human Services Agency.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lana.2024.100836.
Appendix A. Supplementary data
Supplemental Figure 1.
Observational data supporting the duration of antibody responses and PCR positivity during NW or OW Orthohantavirus human infection (samples tested at VSPB 2008–2020), including duplicate specimens. A. Serological and PCR positivity during NW infection. Bars represent the number of cases for each 3-day binned time point. B. Serological and PCR positivity during OW infection. Bars represent the number of cases for each 3-day binned time point.
Supplemental Figure S2.
Summary of human hantavirus-positive cases initially detected by VSPB as acute or convalescent infection by state. A) NW-positive human cases. B) OW-positive human cases. C) Relationship between the total number of submitted specimens per state compared to total number of positive NW acute cases.
Supplemental Figure S3.
Inferred relationship of domestic human Orthohantavirus seoulense case (highlighted purple) relative to sequences from the Seoul virus Rattery outbreak (UK, Netherlands, U.S.), historic domestic U.S. sequences (lower clade) and a subset of Seoul sequences from Asia (upper clade).
National Notifiable Diseases Surveillance System (NNDSS) Hantavirus Infection Clinical Descriptions.
Summary of Patient Metadata and Hantaviral Genetic Coverage Results. Hantaviral sequences were generated directly from patient clinical material and the metadata associated with host, specimen collection date, residence state and viral strain are included. L, M, and S segment Accession numbers were assigned by Genbank. L, M, and S segment percent coverage was calculated as the amount of genetic sequence coverage for each segment relative to the Genbank SNV hantaviral refseq (NC_077669-71).
2 x 2 Contingency Tables Comparing VSPB/CDC and Commercial Hantavirus Serological Results.
References
- 1.Viruses ICotTo. Bunyaviridae. https://ictv.global/report_9th/RNAneg/Bunyaviridae2023
- 2.Lee H.W., French G.R., Lee P.W., Baek L.J., Tsuchiya K., Foulke R.S. Observations on natural and laboratory infection of rodents with the etiologic agent of Korean hemorrhagic fever. Am J Trop Med Hyg. 1981;30(2):477–482. doi: 10.4269/ajtmh.1981.30.477. [DOI] [PubMed] [Google Scholar]
- 3.Nunez J.J., Fritz C.L., Knust B., et al. Hantavirus infections among overnight visitors to Yosemite national park, California, USA, 2012. Emerg Infect Dis. 2014;20(3):386–393. doi: 10.3201/eid2003.131581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de St Maurice A., Ervin E., Schumacher M., et al. Exposure characteristics of hantavirus pulmonary syndrome patients, United States, 1993-2015. Emerg Infect Dis. 2017;23(5):733–739. doi: 10.3201/eid2305.161770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enria D., Padula P., Segura E.L., et al. Hantavirus pulmonary syndrome in Argentina. Possibility of person to person transmission. Medicina (B Aires) 1996;56(6):709–711. [PubMed] [Google Scholar]
- 6.Martinez V.P., Di Paola N., Alonso D.O., et al. "Super-spreaders" and person-to-person transmission of Andes virus in Argentina. N Engl J Med. 2020;383(23):2230–2241. doi: 10.1056/NEJMoa2009040. [DOI] [PubMed] [Google Scholar]
- 7.Jonsson C.B., Figueiredo L.T., Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev. 2010;23(2):412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(NNDSS) NNDSS . CDC; 2015. Hantavirus infection, non-hantavirus pulmonary syndrome 2015 case definition.https://ndc.services.cdc.gov/case-definitions/hantavirus-infection-non-hantavirus-pulmonary-syndrome-2015/ [Google Scholar]
- 9.(NNDSS) NNDSS . CDC; 2015. Hantavirus pulmonary syndrome (HPS) 2015 case definition.https://ndc.services.cdc.gov/case-definitions/hantavirus-pulmonary-syndrome-2015/ [Google Scholar]
- 10.Rasmuson J., Andersson C., Norrman E., Haney M., Evander M., Ahlm C. Time to revise the paradigm of hantavirus syndromes? Hantavirus pulmonary syndrome caused by European hantavirus. Eur J Clin Microbiol Infect Dis. 2011;30(5):685–690. doi: 10.1007/s10096-010-1141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Passaro D.J., Shieh W.J., Hacker J.K., et al. Predominant kidney involvement in a fatal case of hantavirus pulmonary syndrome caused by Sin Nombre virus. Clin Infect Dis. 2001;33(2):263–264. doi: 10.1086/321832. [DOI] [PubMed] [Google Scholar]
- 12.MacNeil A., Ksiazek T.G., Rollin P.E. Hantavirus pulmonary syndrome, United States, 1993-2009. Emerg Infect Dis. 2011;17(7):1195–1201. doi: 10.3201/eid1707.101306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knust B., Rollin P.E. Twenty-year summary of surveillance for human hantavirus infections, United States. Emerg Infect Dis. 2013;19(12):1934–1937. doi: 10.3201/eid1912.131217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mills J.N., Amman B.R., Glass G.E. Ecology of hantaviruses and their hosts in North America. Vector Borne Zoonotic Dis. 2010;10(6):563–574. doi: 10.1089/vbz.2009.0018. [DOI] [PubMed] [Google Scholar]
- 15.Khan A.S., Gaviria M., Rollin P.E., et al. Hantavirus pulmonary syndrome in Florida: association with the newly identified Black Creek Canal virus. Am J Med. 1996;100(1):46–48. doi: 10.1016/s0002-9343(96)90010-8. [DOI] [PubMed] [Google Scholar]
- 16.Knust B., Brown S., de St Maurice A., et al. Seoul virus infection and spread in United States home-based ratteries: rat and human testing results from a multistate outbreak investigation. J Infect Dis. 2020;222(8):1311–1319. doi: 10.1093/infdis/jiaa307. [DOI] [PubMed] [Google Scholar]
- 17.Woods C., Palekar R., Kim P., et al. Domestically acquired seoul virus causing hemorrhagic fever with renal syndrome-Maryland, 2008. Clin Infect Dis. 2009;49(10):e109–e112. doi: 10.1086/644742. [DOI] [PubMed] [Google Scholar]
- 18.Bhatnagar J., Blau D.M., Shieh W.J., et al. Molecular detection and typing of dengue viruses from archived tissues of fatal cases by rt-PCR and sequencing: diagnostic and epidemiologic implications. Am J Trop Med Hyg. 2012;86(2):335–340. doi: 10.4269/ajtmh.2012.11-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes L.V., 3rd, Huang C., Sanchez A.J., et al. Hantavirus pulmonary syndrome associated with Monongahela virus, Pennsylvania. Emerg Infect Dis. 2000;6(6):616–621. doi: 10.3201/eid0606.000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roig I.L., Musher D.M., Tweardy D.J. Severe pulmonary involvement in a case attributed to domestically acquired Seoul hantavirus in the United States. Clin Infect Dis. 2012;54(1):91–94. doi: 10.1093/cid/cir748. [DOI] [PubMed] [Google Scholar]
- 21.Rivers M.N., Alexander J.L., Rohde R.E., Pierce J.R., Jr. Hantavirus pulmonary syndrome in Texas: 1993-2006. South Med J. 2009;102(1):36–41. doi: 10.1097/SMJ.0b013e318187d06f. [DOI] [PubMed] [Google Scholar]
- 22.Morzunov S.P., Rowe J.E., Ksiazek T.G., Peters C.J., St Jeor S.C., Nichol S.T. Genetic analysis of the diversity and origin of hantaviruses in Peromyscus leucopus mice in North America. J Virol. 1998;72(1):57–64. doi: 10.1128/jvi.72.1.57-64.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plyusnin AaM S.P. In: Schmaljohn CSaN S.T., editor. Vol. 256. Springer-Verlag; 2001. Virus evolution and genetic diversity of hantaviruses and their rodent hosts; pp. 47–76. (Current topics in microbiology). [DOI] [PubMed] [Google Scholar]
- 24.Yates T.L., Mills J.N., Parmenter C.A., et al. The ecology and evolutionary history of an emergent disease: hantavirus pulmonary syndrome. Bioscience. 2002;52(11):989–998. [Google Scholar]
- 25.Guo W.P., Lin X.D., Wang W., et al. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 2013;9(2) doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes E.C., Zhang Y.Z. The evolution and emergence of hantaviruses. Curr Opin Virol. 2015;10:27–33. doi: 10.1016/j.coviro.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Goodfellow S.M., Nofchissey R.A., Schwalm K.C., et al. Tracing transmission of Sin Nombre virus and discovery of infection in multiple rodent species. J Virol. 2021;95(23) doi: 10.1128/JVI.01534-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenbaum I.F., Honeycutt R.L., Chirhart S.E. From field to laboratory: a memorial volume in honor of Robert J Baker. Museum of Texas Tech University; 2019. Taxonomy and phylogenetics of the Peromyscus maniculatus species group; pp. 559–575. [Google Scholar]
- 29.Armien B., Pascale J.M., Munoz C., et al. Hantavirus fever without pulmonary syndrome in Panama. Am J Trop Med Hyg. 2013;89(3):489–494. doi: 10.4269/ajtmh.12-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mills J.N., Johnson J.M., Ksiazek T.G., et al. A survey of hantavirus antibody in small-mammal populations in selected United States National Parks. Am J Trop Med Hyg. 1998;58(4):525–532. doi: 10.4269/ajtmh.1998.58.525. [DOI] [PubMed] [Google Scholar]
- 31.Liphardt S.W., Kang H.J., Dizney L.J., Ruedas L.A., Cook J.A., Yanagihara R. Complex history of codiversification and host switching of a newfound soricid-borne Orthohantavirus in North America. Viruses. 2019;11(7):637. doi: 10.3390/v11070637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kjemtrup A.M., Messenger S., Meza A.M., et al. New exposure location for hantavirus pulmonary syndrome case, California, USA, 2018. Emerg Infect Dis. 2019;25(10):1962–1964. doi: 10.3201/eid2510.190058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prince H.E., Lieberman J.M. Impact of the Yosemite hantavirus outbreak on hantavirus antibody testing at a national reference laboratory. Clin Vaccine Immunol. 2013;20(8):1213–1216. doi: 10.1128/CVI.00326-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koster F., Foucar K., Hjelle B., et al. Rapid presumptive diagnosis of hantavirus cardiopulmonary syndrome by peripheral blood smear review. Am J Clin Pathol. 2001;116(5):665–672. doi: 10.1309/CNWF-DC72-QYMR-M8DA. [DOI] [PubMed] [Google Scholar]
- 35.Oliver T.T., Dyal J.W., Talker D.L., et al. Successful implementation of a rapid screening tool for hantavirus cardiopulmonary syndrome: 5 years of experience from a community hospital in an endemic region. Am J Clin Pathol. 2022;157(4):498–501. doi: 10.1093/ajcp/aqab170. [DOI] [PubMed] [Google Scholar]
- 36.PAHO . PAHO; 2024. PAHO strengthens capacities for hantavirus and arenavirus surveillance in the Americas.https://www.paho.org/en/news/13-3-2024-paho-strengthens-capacities-hantavirus-and-arenavirus-surveillance-americas Available from: [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
National Notifiable Diseases Surveillance System (NNDSS) Hantavirus Infection Clinical Descriptions.
Summary of Patient Metadata and Hantaviral Genetic Coverage Results. Hantaviral sequences were generated directly from patient clinical material and the metadata associated with host, specimen collection date, residence state and viral strain are included. L, M, and S segment Accession numbers were assigned by Genbank. L, M, and S segment percent coverage was calculated as the amount of genetic sequence coverage for each segment relative to the Genbank SNV hantaviral refseq (NC_077669-71).
2 x 2 Contingency Tables Comparing VSPB/CDC and Commercial Hantavirus Serological Results.