Abstract
The 86-kDa IE2 protein of human cytomegalovirus (HCMV) is an important regulator of viral and host cell gene expression. Still, besides its function as a transcription factor, little is known about the biological activities of IE2. Here, we show that IE2 can induce a G1 arrest in several different cell lines, including HCMV-permissive U-373 cells. The known transcriptional activation domains of IE2 are dispensable for G1 arrest, favoring a posttranscriptional mechanism mediating this cell cycle effect. We show that like human primary fibroblasts U-373 cells arrest in G1 upon infection with HCMV. This G1 arrest occurs within 24 h after infection and in proliferating cells depends on viral gene expression. Our data therefore suggest that IE2 is at least partially responsible for blocking the transition from G1 to S phase, which is induced when cells are infected with HCMV.
Following entry into the cell, human cytomegalovirus (HCMV) gene expression is temporally regulated, giving rise to immediate-early (IE), early, and late gene products. IE genes together with virion factors pave the way for the following stages of the viral replicative cycle. Driven by the strong CMV promoter-enhancer, the so-called major IE gene is the most intensively transcribed region during IE times of infection. As a result of different splicing events, several gene products originate from this IE gene, among them, as the first and most abundant, the 72-kD IE protein (IE1) and the 86-kD IE protein (IE2). These two nuclear phosphoproteins have been extensively studied with respect to their ability to regulate transcription of numerous viral and cellular promoters (reviewed in reference 33). IE2, especially, is a strong, somewhat promiscuous transcriptional activator. In this capacity, it can interact with a variety of basal (e.g., TBP [18], TFIIB [6], and TAF130 [29]) as well as promoter-specific transcription factors (e.g., Sp1 [30], CREB [26], and Egr-1 [51]).
Both IE1 and IE2 have also been connected with cell cycle-regulated transcription. IE1 has been reported to activate the dihydrofolate reductase (32) and DNA polymerase α promoters (21), both of which are physiologically induced during G1-S cell cycle transition. IE2 stimulates transcription from the cyclin E promoter (3), which gives rise to a gene product which itself actively regulates cell cycle transition at the G1-S boundary. Furthermore, it has been shown that in HCMV-infected cells IE1 is associated with p107 (40) and IE2 is associated with the retinoblastoma protein (pRb) (17), two members of the family of pocket proteins (reviewed in reference 34). These interactions seem to be of functional relevance for cell cycle-dependent transcription because the suppression effects of these pocket proteins on an E2F-responsive promoter can be reversed by IE1 or IE2 (17, 40). In addition, IE2 can counteract a flat-cell phenotype resulting from overexpression of wild-type pRb in SAOS-2 cells which lack endogenous pRb (13). IE1 has even been described to have kinase activity able to phosphorylate the pocket proteins p107 and p130, as well as the E2Fs 1, 2, and 3 in vitro (35). However, the functional consequences of these phosphorylation events are unknown. IE2 can also bind p53, another potent regulator of cell cycle progression, abolishing its ability to activate transcription of an artificial reporter gene (46). The biological function of the p53-IE2 interaction is still unknown: the DNA damage checkpoint function of p53 is not impaired by IE2 (2), and the antiapoptotic activities reported for IE2 (and IE1) do not seem to depend on p53 (53).
Given that IE1 and IE2 can influence the activity of several key regulators of cell cycle progression and growth control, it is somewhat surprising that nothing is known about their effects on cell cycle progression per se. One reason might be that the above-mentioned S phase-promoting activities of IE1 and IE2 appear to contradict the current knowledge of HCMV-induced cell cycle regulations. HCMV has been shown to arrest the host cell cycle at various stages—predominantly the G1-S transition (4, 12, 23, 27). These data together with those of an earlier study (10) seem to suggest that cellular DNA replication, hence S phase, may counteract efficient viral DNA replication, which is consistent with more recent findings showing that HCMV infection during S phase leads to a significant delay in the onset of viral replication when compared to viral infection of G1 cells (42).
However, in contrast to its cell cycle arrest function, HCMV infection also stimulates growth-regulated pathways. Like a growth factor stimulus, HCMV infection rapidly activates the expression of the proto-oncogenes c-fos, c-jun, and c-myc in quiescent cells (1). Moreover, a number of enzymes involved in nucleotide metabolism and DNA replication are upregulated during HCMV replication (reviewed in reference 33). These findings favor a model in which HCMV mediates a cell cycle state that supports viral DNA replication by upregulating replication factors and increasing nucleotide pools but specifically inhibits competitive cellular DNA synthesis and mitosis.
As outlined above, IE1 and IE2 fit into this model in that they upregulate several gene products involved in cell cycle progression and DNA synthesis. Here we present data demonstrating that, in addition, IE2 can block cell cycle progression in G1, thereby mimicking an important cell cycle regulatory function of HCMV. This newly discovered biological function of IE2 seems to be separable from its function as a transcriptional activator. Thus, IE2 is a multifunctional protein reflecting at least partially the complex interplay between HCMV and the host cell cycle.
MATERIALS AND METHODS
Plasmids.
The IE2 expression vector pHM121 (38) and the IE1 expression vector pSV2IE1 (18), both containing full-length cDNAs under the control of the simian virus 40 early promoter-enhancer, have been described. The p16INK4a expression vector pXmycp16 was a generous gift from Jiri Lukas (Danish Cancer Society). To generate pSG5CD20, the CD20 cDNA was removed from pCMVCD20 (54) by BamHI digestion and inserted into the BamHI cloning site of pSG5 (16). This was necessary to avoid the repressive effects exerted on the CMV promoter by IE2. For cloning of pSG5-3HA, a triple hemagglutinin (HA) tag consisting of three tandem copies of the HA1 epitope was PCR amplified from pGTEpi (50) and inserted between the EcoRI and BglII sites of pSG5. To create IE2 mutants with N-terminal or C-terminal deletions, the corresponding cDNA fragments were PCR amplified from pHM121, thereby introducing flanking BamHI/BglII sites. For C-terminal deletion constructs, translation stop codons were added to the new 3′ end of the cDNA clone. The PCR products were cloned into the BglII site of pSG5-3HA, in frame with the HA tag. The internal deletion mutant was created by cutting out the coding region for amino acids 136 to 289 by SmaI and XhoI from pSG5-3HA-IE2 and in frame religation. The c-fosCAT construct (19) and the β-galactosidase expression plasmid p97b (11) have been described elsewhere. All plasmids were purified by CsCl ethidium bromide equilibrium centrifugation.
Cell culture.
All cell lines were maintained as monolayers in Dulbecco’s modified Eagle medium supplemented with 5% newborn and 5% fetal calf serum (FCS) and 100 U of penicillin-streptomycin per ml. U-373-MG cells were purchased from Cell Lines Service (Heidelberg, Germany). Human embryonic primary lung fibroblasts had passage numbers of 13 to 15 and were kindly provided by S. Proesch (Institute of Virology, Charité, Germany).
DNA transfections.
Approximately 106 cells per 100-mm-diameter dish were seeded on the day prior to transfection. By using the calcium phosphate coprecipitation method (7), 16 μg of effector plasmid and 4 μg of pSG5CD20 or reporter plasmid (c-fosCAT) were applied to the cells. To control for transfection efficiency, a β-galactosidase expression plasmid was also included. At 48 h posttransfection, cells were harvested and aliquots were processed for flow cytometry, immunoblotting, or chloramphenicol acetyltransferase (CAT) assays. Where indicated, nocodazole at a final concentration of 50 ng/ml was added to the cells 22 h before harvesting. Growth factor starvation was achieved by completely omitting serum for 48 h, followed by restimulation with 10% FCS and harvesting at the indicated time points.
Virus infections.
Two days before infection, cells were plated at a density of 1.5 × 104 cells/cm2. For synchronizing, cells in G0 were kept in serum-free medium for 72 h prior to infection. Stocks of the HCMV laboratory strain AD169 (a generous gift of S. Proesch [Institute of Virology]) were used to infect these cells at a multiplicity of infection (MOI) of 5 PFU per cell, unless stated differently. Virus adsorption was allowed for 1 h. Mock-infected control cultures were exposed to an equal volume of medium containing the same serum concentration as the viral stocks. At 24 h postinfection, cells were harvested by trypsinization and washed once with phosphate-buffered saline (PBS) and aliquots were processed for flow cytometry or immunoblotting. For UV inactivation, viral stocks were irradiated with 1200 J/m2 in a UV Stratalinker 2400 (Stratagene) as described elsewhere (28).
Flow cytometry.
In order to determine the infection rate with HCMV, cells were fixed with PBS–2% paraformaldehyde for 30 min at 0°C. After cells were washed with PBS, two successive antibody binding reactions were carried out, each of them with 5 μg of antibody per ml in PBS–0.1% saponin–20% FCS for 40 min at 0°C, followed by two washing steps with PBS–0.1% saponin. The primary antibody (clone 9121; NEN) recognizes the common N terminus of IE1 and IE2. The secondary antibody was a fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody (Becton Dickinson). To determine the cell cycle profile of virus-infected cultures, cells were fixed with 70% ethanol and stained with propidium iodide, according to standard procedures (8). Flow cytometry was performed on a FACScan (Becton Dickinson) equipped with CellQuest software. Cell cycle analysis of transfected cells was done by a previously described double-staining procedure by using FITC-conjugated anti-CD20 antibodies (Pharmingen) and propidium iodide (54). Single cells with an FITC staining at least 20 times stronger than that in the untransfected subpopulation were gated on the FACScan and analyzed for DNA content as previously described (22).
Immunoblotting and CAT assays.
Cells were lysed in 0.25 M Tris (pH 7.5) by freezing and thawing. Extracts were clarified by centrifugation, and the protein concentration was determined with the Bio-Rad protein assay. For immunoblot analysis, standard techniques were employed (8). Briefly, equal amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 8% acrylamide and transferred to polyvinylidene difluoride membranes. After being blocked with 6% dry milk in 0.1% Tween 20–0.1 M Tris (pH 7.6)–0.15 M NaCl (TTBS), the membranes were probed with anti-HCMV antibody (9121; NEN) or with anti-HA antibody (12CA5; Boehringer) and developed with peroxidase-conjugated anti-mouse immunoglobulin antibody (Sigma) and by enhanced chemiluminescense as recommended by the manufacturer (Amersham). CAT assays were performed essentially as previously described (16). They were quantified by scanning with a GS-250 Molecular Imager (Bio-Rad). The CAT conversion data were normalized for plate-to-plate variations in transfection efficiency by using the cotransfected β-galactosidase gene (driven by the cell cycle-independent Rous sarcoma virus promoter) as an internal reference. The β-galactosidase activity was assayed as previously reported (20).
RESULTS
IE2 arrests U-373 cells in G1.
The observation that the CMV 72-kDa IE1 and 86-kDa IE2 proteins can functionally interact with cell cycle regulatory factors prompted us to ask whether IE1 and IE2 can directly influence cell cycle progression. Since it has previously been shown that upon HCMV infection host cells arrest (predominantly) in the G1 phase of the cell cycle, we decided to investigate whether IE1 and IE2 could specifically contribute to those effects.
In order to address the question of a possible role of IE1 and/or IE2 in regulating cell cycle progression, we employed a transient transfection system in which the viral IE and control DNAs were transfected together with a CD20 cDNA, enabling us to select transiently transfected cells for DNA content analysis by flow cytometry (54). Transfections were done in the HCMV-permissive cell line U-373, which can be transfected with high efficiency. Together, these aspects make this system a particularly useful tool for addressing the experimental question outlined above.
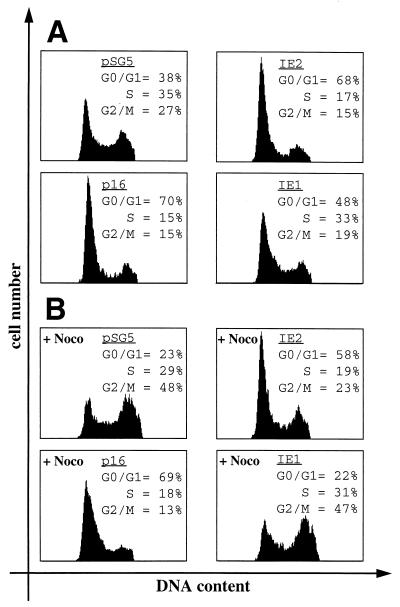
The normal cell cycle distribution pattern of control transfected proliferating U-373 cells is depicted in Fig. 1A. IE2 but not IE1 expression in these cells induced a dramatic change in this pattern (Fig. 1A). Upon IE2 expression, the G1 population increased from 38 to 68% and both the S phase and G2/M cell populations were reduced by approximately 50% (35 to 17% for S phase and 27 to 15% for G2/M). This IE2-mediated distribution pattern is similar to that induced by cyclin-dependent kinase inhibitors like p16INK4a (Fig. 1A), which is known to block cells in G1.
FIG. 1.
Expression of IE2 causes a G1 arrest in HCMV permissive cells. U-373 cells were transiently transfected with plasmids expressing IE1, IE2, or p16INK4a (p16) together with the CD20 expression vector pSG5CD20 in a 4:1 molar ratio. As a control, pSG5CD20 was cotransfected with pSG5. Cells were harvested 48 h after transfection and stained for CD20 expression and DNA content prior to analysis by flow cytometry. DNA histograms show CD20-positive cell populations in which the relative DNA content was plotted against cell number. (A) Proteins were expressed in asynchronously cycling cells. (B) The G2/M blocker nocodazole (Noco) was added to the cells 24 h before harvest. Data shown in panels A and B are from a single experiment representative of multiple experiments with similar results.
To differentiate whether the IE2-induced increase of cells in G1 is also due to a G1 arrest or, alternatively, to a change in the relative length of time cells spent in G1 versus S and G2 phases, cells were treated with nocodazole after transfection. Nocodazole blocks spindle formation, and consequently, cell cycle progression is blocked in G2, where cells finally accumulate. Therefore, after 24 h of nocodazole treatment, only 23% of control transfected cells had a G1 DNA content, whereas 48% of the cells had accumulated in G2 (Fig. 1B). In contrast, the majority of cells expressing IE2 were found to have a G1 DNA content (58%) unable to enter G2. This distribution pattern closely resembles that of cells expressing p16INK4a (69% of cells in G1) and is characteristic for proteins inducing a G1 arrest. Under these conditions, IE1 did not induce significant changes to the cell cycle profile. Thus, IE2 appears to induce a G1 arrest when transiently expressed in proliferating U-373 cells.
IE2 blocks S phase entry after growth stimulation of serum-starved U-373 cells.
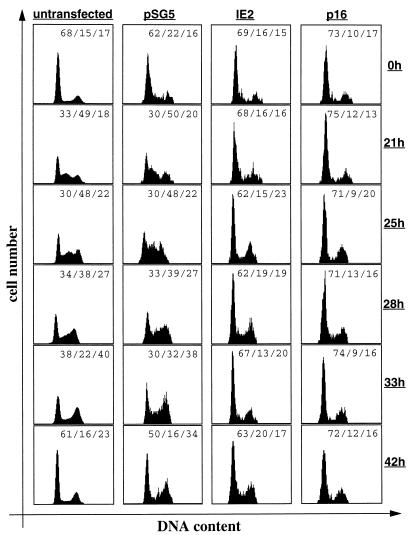
Unlike some transformed cell lines (e.g., HeLa), U-373 cells can withdraw from the cell cycle upon growth factor (i.e., serum) starvation. After readdition of serum, a subpopulation of about one-third of the entire culture of U-373 cells synchronously reenters the cell cycle, and 21 h later, this subpopulation of cells can be visualized in mid-S phase. Subsequently, these cells can be monitored to passage through the cell cycle, and between 33 and 42 h after serum readdition, one round of the cell cycle has been completed and cells reappear in G1 (Fig. 2). pSG5 control transfected cells follow the same kinetics as nontransfected cells, although with a slight delay of about 2 to 3 h (Fig. 2). To further characterize the cell cycle function of IE2, we next asked whether IE2 could overcome a cell growth stimulus employed on serum-starved, and hence quiescent, U-373 cells and whether IE2 could truly arrest cells in G1 rather than only slowing down cell cycle progression. To address this issue, U-373 cells were transfected with IE2 or p16INK4a expression vectors, serum deprived, and restimulated as described above. As expected, both IE2- and p16INK4a-expressing cells accumulate in G0 and G1 after serum starvation. However, in contrast to untransfected and pSG5 control transfected cells, IE2-expressing cells failed to enter S phase 21 h after growth factor readdition, indicating that IE2 indeed interferes with serum-induced S phase entry of quiescent cells. Importantly, IE2 not only delays S phase entry but also appears to block cells in G0 and G1, since even 33 to 42 h after growth factor addition (when control cells had completed one entire round of the cell cycle) IE2-transfected cells remained in G1 (Fig. 2). Only a small fraction of cells escaped the IE2-mediated G1 block. Between 21 and 25 h, 6% of cells appeared to have entered G2 from G1. However, this is also true for p16INK4a-transfected cells (4% during this time period). Therefore, these minor subpopulations most likely reflect cells which express CD20 but are not sufficiently transfected with IE2 or p16 expression plasmids and consequently contaminate the G2 population. In the entire experiment, IE2 again behaved like p16INK4a, one of the most powerful known inducers of a G1 arrest. In addition, the same results were obtained when NIH 3T3 fibroblasts were used instead of U-373 cells (see below and data not shown). These experiments demonstrate that IE2 can truly arrest cells prior to S phase induction and that a growth factor stimulus employed on quiescent cells cannot overcome this cell cycle arrest function of IE2. Therefore, these data support the conclusions drawn from the experiments shown in Fig. 1.
FIG. 2.
IE2 inhibits quiescent cells to enter S phase after serum restimulation. U-373 cells were transfected as described in Fig. 1. At 12 h after removal of DNA precipitates, cells were serum starved for 48 h followed by serum restimulation. At the end of the starvation period (0 h) and at the indicated time points after restimulation, cells were harvested and stained and cell cycle distribution was analyzed by flow cytometry. DNA histograms of IE2, p16, or control transfected CD20-positive cell populations and of a CD20-negative (untransfected) cell population are shown. The quantitation of cell cycle phases is given as percent G0-G1/percent S/percent G2/M.
IE2 induces a G1 arrest in many different cell lines.
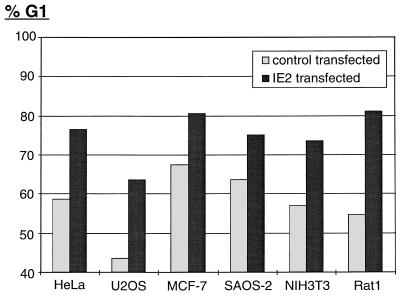
To exclude the possibility that the IE2-induced cell cycle arrest function observed in U-373 cells is a cell type-specific event, we analyzed the cell cycle distribution pattern of various cell lines after IE2 transfection. The panel of human cell lines tested included the cervical carcinoma cell line HeLa, osteosarcoma cell lines U2OS and SAOS-2, and breast carcinoma cell line MCF-7. In addition, immortalized fibroblasts from mouse (NIH 3T3) and rat (Rat1) were analyzed. Even in the absence of nocodazole, the ability of IE2 to induce a G1 arrest in every cell line tested could readily be shown (Fig. 3). The degree of the IE2-induced G1 block seems to vary slightly between different cell lines. However, this primarily depended on the percentage of G1 cells in the control population: the lower the G1 population in control transfected cells, the higher the degree of the IE2-mediated G1 arrest (e.g., compare U2OS and SAOS-2 cells in Fig. 3). More interestingly, the different cell lines tested have various genetic lesions (overview in reference 48), some of which occur in genes of known cell cycle regulatory proteins: MCF-7, NIH 3T3, and U2OS cells lack functional p16INK4a. U-373 cells lack functional p53, and Rat1 cells lack functional p21CIP1. SAOS-2 cells lack functional p53 and pRb, and in HeLa cells, these two tumor suppressor genes are inactivated by papillomavirus gene products E6 and E7. In view of these known genetic defects, the IE2-mediated G1 arrest does not appear to be strictly dependent on any of the aforementioned cell cycle regulators. In addition, the IE2 effect is neither cell type nor species specific.
FIG. 3.
IE2 blocks G1-S progression in many different cell lines. IE2 was coexpressed with CD20 in different asynchronously growing cell lines, as indicated. Transfections and subsequent cell cycle analyses were performed exactly as described for Fig. 1A. The bar chart compares the percentages of IE2 and control transfected cells in G1. Mean values of results from at least three independent experiments are shown.
IE2 mimics an HCMV-induced cell cycle arrest in U-373 cells.
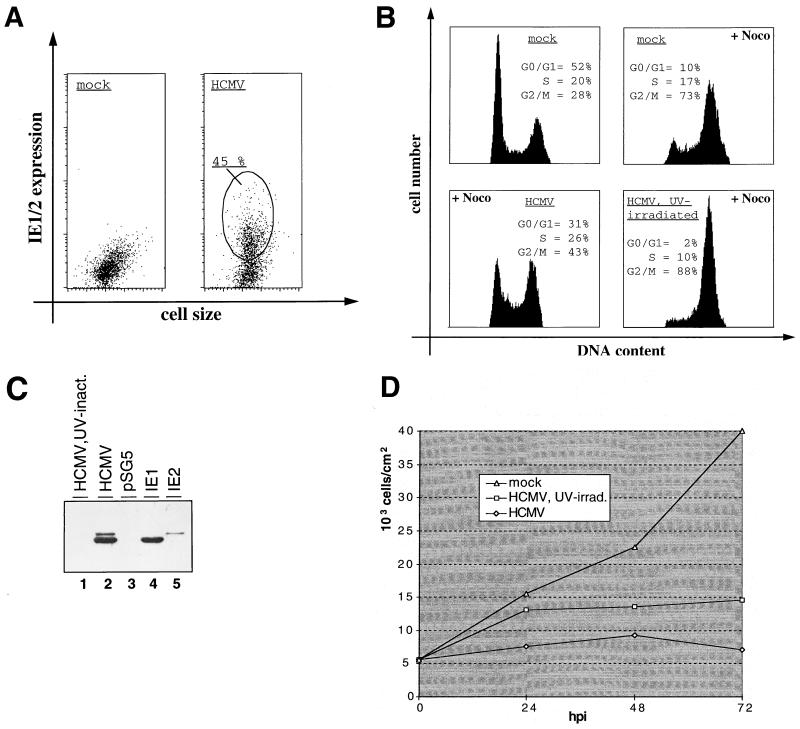
Recently, several groups have shown that upon infection with HCMV human fibroblasts arrest at various stages of the cell cycle—predominantly in G1 (4, 12, 23, 27). To start to answer the question of whether the G1 arrest function of IE2 defined under transient transfection conditions in U-373 cells could be of physiological significance for HCMV infection, we asked whether HCMV infection would also cause a G1 arrest in U-373 cells. To this end, proliferating U-373 cells were either HCMV or mock infected and treated with nocodazole 8 h after infection. Even at an MOI of 5, U-373 cells could be infected only up to about 60%, which is consistent with other reports (5, 40). In the experiment described here, 45% of U-373 cells expressed IE1 and IE2 after viral infection (MOI = 1) of the culture, indicating that approximately every second cell of the culture was infected with HCMV (Fig. 4A). This result was confirmed by immunofluorescence microscopy, which showed a clear nuclear staining with the IE1- and IE2-specific antibody (data not shown).
FIG. 4.
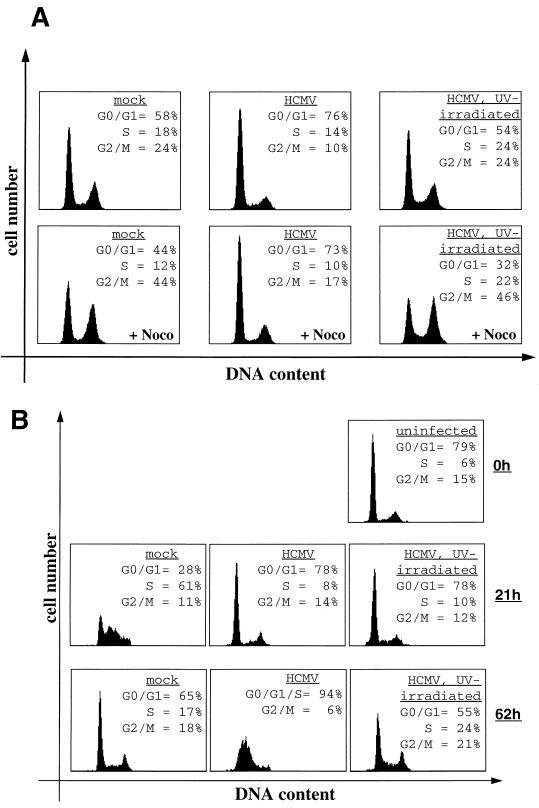
HCMV arrests U-373 cells in G1 depending on viral gene expression. U-373 cells were either mock infected or infected with HCMV or UV-irradiated HCMV, as indicated. At 24 h postinfection, cells were analyzed for infection rate, cell cycle distribution, and IE1 and IE2 expression levels. (A) To determine the infection rate, cells were immunostained for IE1 and IE2 expression and analyzed by flow cytometry. In the diagram, IE1 and IE2 expression (measured as FITC fluorescence) is plotted against cell size (measured as forward scatter). HCMV-infected cells (encircled) with an FITC signal over background fluorescence of mock-infected cells were considered HCMV infected and subsequently quantitated. (B) Analysis of cell cycle distribution patterns of infected and noninfected cells. U-373 cells were stained for DNA content and subjected to flow cytometry. Where indicated, cells were treated with nocodazole (+Noco) 8 h postinfection. DNA histograms of cultures including infected as well as uninfected cells according to panel A are shown. (C) Comparison of IE1 and IE2 protein levels in HCMV-infected versus IE1- or IE2-transfected cells. Equal amounts of extracts from U-373 cells either infected with UV-irradiated HCMV or HCMV or from U-373 cells transiently transfected with control plasmid (pSG5) or IE1 and IE2 expression vectors were separated by SDS-PAGE and analyzed by immunoblotting with an antibody for specifically recognizing the common N termini of IE1 and IE2 (9121; NEN). inact., inactivated. (D) Growth curve of infected primary fibroblasts. Every 24 h after infection, cells were harvested and counted with a hemacytometer. The calculated cell density is depicted against the time point of harvest. hpi, hours postinfection.
As expected, mock-infected cells accumulated in G2 under nocodazole treatment (G1 = 10%; G2 = 73%) (Fig. 4B). In contrast, 24 h after HCMV infection more than three times as many cells (31%) remained in G1, whereas only 43% of cells were found to have a G2 DNA content. Given the number of infected cells in the culture, this result correlates well with HCMV-infected U-373 cells being arrested in G1. In this respect, U-373 cells seem to behave like human primary fibroblasts. Next, we investigated whether the HCMV-induced G1 arrest in U-373 cells depends on viral gene expression. Therefore, an additional culture was infected with UV-irradiated virus. UV irradiation was done under conditions which were previously reported to shut off viral gene expression but to leave virion proteins intact (52). In an immunoblot analysis, IE1 and IE2 proteins were readily detectable in extracts from HCMV-infected U-373 cells. However, the antibodies failed to detect IE1 and IE2 in extracts from cells infected with UV-irradiated virus, indicating that gene expression was successfully eliminated under UV treatment (Fig. 4C, lanes 1 and 2). In good agreement with previously published work (28), UV-irradiated HCMV, like normal HCMV, imposes a growth inhibitory effect onto human fibroblasts in our hands (Fig. 4D), which assigns biological activity to the UV-irradiated virus used here, indicating a functional virion after UV treatment. As can be seen from Fig. 4B, cells infected with UV-inactivated virus were not arrested in G1. Thus, HCMV induces a G1 arrest in U-373 cells, and viral gene expression within the first 24 h of the infection is necessary for this cell cycle regulatory step to occur. Indeed, we noticed that the G1 population of this culture was even further diminished (from 10 to 2%), indicating that the UV-irradiated virus had led to an accelerated G1-S transition.
Furthermore, using corresponding amounts of extracts from IE1- and IE2-transfected U-373 cells (aliquots from the experiments shown in Fig. 1), we could show that the expression levels of the two IE proteins correlated well between infected and transfected U-373 cells (Fig. 4C, compare lanes 2, 4, and 5). Given the estimated transfection rate of 10% (data not shown), this result demonstrates that the concentration of IE1 and IE2 in infected cells is very similar to that in transfected cells. Therefore, in our experimental system, the amount of IE2 protein found in infected U-373 cells reflects that which is sufficient to cause a G1 arrest after transient transfection of IE2. Furthermore, a dose-response analysis demonstrated that the IE2-mediated G1 arrest was quantitatively unchanged, even when the level of the IE2 protein was reduced by a factor of 5 to 10 (data not shown). Together, these data show that concentrations of IE2 which readily cause transfected cells to arrest in G1 were routinely reached in HCMV-infected U-373 cells, indicating that the transient transfection assays used here are a valid system for studying the cell cycle effects of the IE2 protein.
Taken together, our findings that (i) the HCMV-induced G1 arrest in U-373 cells depends on viral gene products expressed during the initial 24 h of an infection and (ii) physiological concentrations of IE2 can mimic the G1 arrest function of HCMV suggest that IE2 is at least partially responsible for the HCMV-induced G1 arrest in U-373 cells.
U-373 cells behave like human fibroblasts when infected with HCMV.
Since the cell cycle distribution pattern of HCMV-infected U-373 cells has not been looked at before, we wanted to analyze whether in this respect U-373 cells behave like human fibroblasts, which, with an MOI of 5, were readily infected to nearly 100% (as determined by immunofluorescence microscopy; data not shown). As can be seen from Fig. 5A, the results obtained with proliferating human fibroblasts infected with normal and UV-inactivated HCMV correlate well with the ones from U-373 cells. Both in the presence and absence of nocodazole, HCMV but not UV-irradiated HCMV infection causes human primary fibroblasts to arrest in G1. As in U-373 cells, infection with UV-inactivated virus also leads to a decrease of G1 phase cells, which is particularly noticeable under nocodazole treatment. In human fibroblasts, however, this effect is less pronounced than in U-373 cells, which is likely due to the higher proliferation rate of U-373 cells.
FIG. 5.
Cell cycle analysis of HCMV-infected human primary fibroblasts. (A) Subconfluent, cycling cells were either mock infected or infected with HCMV or UV-irradiated HCMV. Cells in the lower panels were additionally treated with nocodazole (+Noco) 8 h postinfection. (B) Cells were serum starved for 72 h prior to infection. At the time of infection, cells were refed with serum and harvested at the indicated time points after infection, and cultures were then analyzed for cell cycle distribution.
We also looked at HCMV infection of human fibroblasts synchronized in G0 by serum deprivation for 72 h (Fig. 5B). At the time of infection, again using an MOI of 5, cells were refed with serum, and the cell cycle distribution was analyzed 21 h later, when more than 50% of the mock-infected cells had synchronously entered S phase, and 62 h later, when mock-infected cells were showing the normal cell cycle distribution pattern of proliferating cells (Fig. 5B). HCMV-infected cells were found to be arrested in G1 21 h after infection, and another 41 h later the DNA distribution pattern showed a single broad peak, which is characteristic of viral DNA replication and consistent with other reports (12, 23, 28). We also included infections with UV-irradiated virus in this set of experiments. As in the experiment described above, the UV irradiation was monitored by loss of IE1 and IE2 expression and a growth inhibition curve (data not shown). Also, UV-irradiated virus and the normal virus used in one experimental setup originated from the same stock. Under these conditions, UV-irradiated virus was able to sufficiently block serum-starved, quiescent cells from entering S phase after 21 h (Fig. 5B). After 62 h, the G0-G1 arrest was less obvious, which might be due to the fading presence of virion factors in the absence of viral gene expression. These results are in contrast to the inability of UV-irradiated virus to block either proliferating human fibroblasts (Fig. 5A) or U-373 cells (Fig. 4B) in G1. The main difference between these experimental approaches is that proliferating cells enter S phase from the preceding mitosis, while serum-deprived cells need to exit from G0 in order to enter S phase. Therefore, these results may suggest that UV-irradiated virus has retained the ability to block G0-G1 progression but has lost the ability to block G1-S transition, which in turn requires viral gene expression.
IE2 rescues the cell cycle arrest phenotype in U-373 cells infected with UV-irradiated virus.
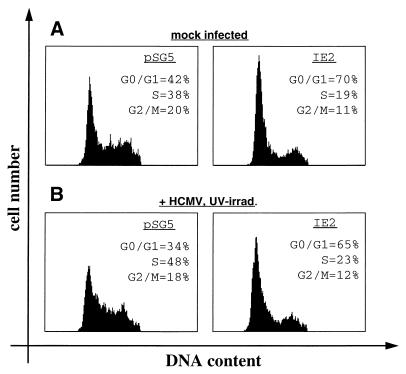
We next wanted to analyze whether IE2 could reconstruct the G1 arrest phenotype of U-373 cells infected with UV-irradiated HCMV. To test this hypothesis, we cotransfected IE2-expressing and control vectors together with a CD20 expression plasmid into U-373 cells. At 12 h after transfection, cells were infected with UV-irradiated HCMV, and they were analyzed for DNA content by flow cytometry another 24 h later. Again, the UV-irradiated virus used in this experiment conforms to the criteria presented in Fig. 4. In order to minimize the artificial character of the experimental setup, we omitted nocodazole from these experiments. Even in the absence of nocodazole, both the IE2-mediated G1 arrest and the S phase-promoting activity of UV-irradiated HCMV can be observed (Fig. 1 and 6 and data not shown), although the latter effect appears less pronounced in the absence of nocodazole, since cells can reenter G1. As seen before, IE2 causes a G1 arrest in the absence of virus with the G1 phase population increasing from 42 to 70% and the S phase population falling from 38 to 19% (Fig. 6A). When control transfected cells were infected with UV-irradiated virus, no G1 arrest could be observed. Instead, G1 cells were diminished, resulting in an increase of S phase cells from 38 to 48% (Fig. 6B). This observation is consistent with the results presented in Fig. 4B and 5A. In contrast, IE2-transfected cells superinfected with UV-irradiated virus had a dominant IE2 phenotype. The G1 phase population increased from 34 to 65%, and the S phase population declined from 48 to 23%, compared to the populations in cells which were control transfected and superinfected (Fig. 6B). These results show that IE2 expression is sufficient to arrest cells in G1, even in the presence of UV-irradiated virus, which by itself leads to a slight increase of the S phase population.
FIG. 6.
IE2 reconstructs the G1 arrest in cells infected with UV-irradiated HCMV. Control plasmid pSG5 or IE2 expression plasmid was cotransfected with pSG5CD20 into U-373 cells as described for Fig. 1. At 12 h after removal of DNA precipitates, cells were mock infected (A) or infected with UV-irradiated HCMV (B). At 24 h postinfection, cells were harvested, stained, and analyzed by flow cytometry. Shown are DNA histograms of the CD20-positive cell populations.
Deletion analysis favors a posttranscriptional mechanism for the IE2-mediated G1 arrest.
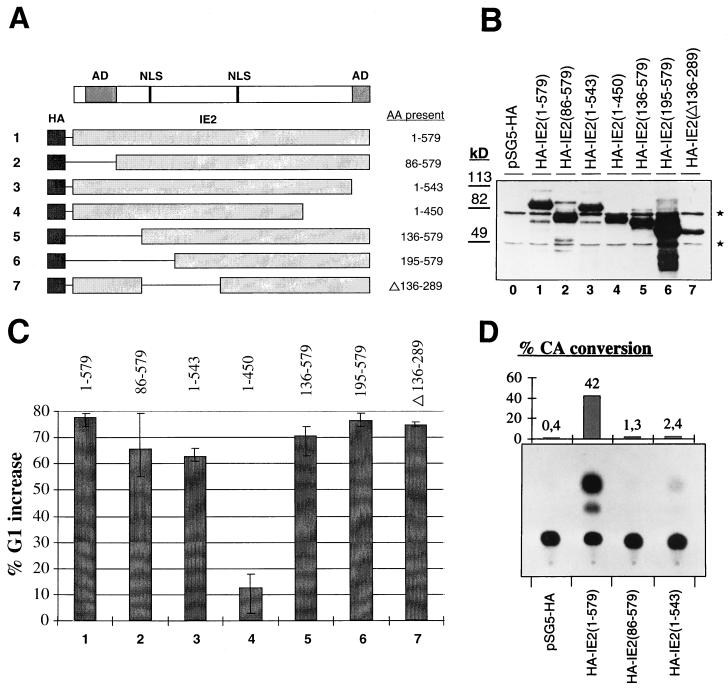
The IE2 protein contains several domains, some of whose functions have been characterized. For instance, large portions of IE2 contribute to its role as a transcription factor and the N and C termini contain independent transactivation domains necessary for transcriptional activation (31, 36, 47). Other domains of IE2 were primarily defined as interaction sites for cellular proteins, like p53 (49) or pRb (13, 17). In order to define regions of IE2 involved in mediating the observed G1 arrest and to ask whether known protein functions of IE2 may contribute to its cell cycle regulatory activity, we generated a panel of deletion constructs (Fig. 7A). All constructs encode IE2 mutants which contain at least one of the two known nuclear localization signals. This has been shown to be sufficient for nuclear localization of IE2 (36). Furthermore, all mutants were linked to the HA epitope tag, which allowed us to directly compare expression levels. In all assays employed, IE2 behaved exactly like HA-IE2, indicating that the epitope tag did not interfere with IE2 function (data not shown). IE2 and all deletion mutants were expressed to similar levels (Fig. 7B). Consequently, any changes observed in the cell cycle regulatory function of IE2 mutants would not simply be attributable to differences in their protein concentrations.
FIG. 7.
Cell cycle and transcriptional activities of IE2 deletion mutants. All results shown were obtained with aliquots of the same extracts. (A) Schematic of the IE2 deletion constructs used. The amino acids (AA) present in each construct are indicated. AD, activation domain; NLS, nuclear localization signal. (B) Equal amounts of extracts from U-373 cells transiently transfected with either control plasmid (pSG5-HA) or vectors expressing the HA-tagged forms of full-length IE2 or the indicated IE2 deletion mutants were separated by SDS-PAGE and analyzed by immunoblotting with an antibody against the HA epitope tag. Size markers are given on the left. Nonspecific background bands occurring in all lanes indicate equal gel loadings and are marked by asterisks. (C) Cell cycle analysis of transfected cells was performed as described for Fig. 1. The bar chart gives the relative increase in G1 populations found in cells transfected with IE2 or the indicated IE2 mutants versus the G1 population of cells transfected with the control plasmid. The highest and lowest values of three independent experiments are indicated by error bars. (D) Extracts from U-373 cells expressing the control plasmid (pSG5-HA), full-length HA-IE2 [HA-IE2(1-579)], or IE2 mutants lacking the independent transcriptional activation domains [HA-IE2(86-579) and HA-IE2(1-543)] were analyzed for their transcriptional activity on a c-fos reporter plasmid (18) in CAT assays. Results of a representative CAT assay are shown in the bottom panel. The bar chart in the top panel shows for the same experiment the ratios of acetylated to total chloramphenicol as percent conversions corrected for variations in transfection efficiency.
In order to assay for their ability to induce a G1 arrest (in the absence of nocodazole), IE2 mutants were transiently transfected into U-373 cells, together with CD20. In this assay, HA-IE2 consistently led to a nearly 80% (e.g., from a 40 to 72% G1 content) increase in the G1 population of asynchronously growing cells (Fig. 7C). Interestingly, neither the N-terminus [HA-IE2(86-579)]- nor C-terminus [HA-IE2(1-543)]-independent transcriptional activation domains of IE2 were required for the G1 arrest (Fig. 7C). Even a deletion of the N terminus up to amino acid 194 [HA-IE2(195-579)] or an internal deletion of amino acids 136 to 289 [HA-IE2(Δ136-289)] impinged on the ability of IE2 to induce a G1 arrest (Fig. 7C). However, a further C-terminal deletion of residues 451 to 543 [HA-IE2(1-450)] virtually abolished the G1 arrest function of IE2 (Fig. 7C). This analysis narrows the region important for the cell cycle regulatory function of IE2 to residues 289 to 543.
The data further suggest that the mechanism employed by IE2 to induce the G1 arrest may not be transcriptional, since the two mutants lacking the independent activation domains of IE2 were still capable of arresting cells in G1. To address this point more directly, we cotransfected full-length IE2 next to HA-IE2(86-579) and HA-IE2(1-543), together with a reporter CAT construct which had previously been shown to be IE2 responsive (19). Only full-length IE2 was able to significantly transactivate this reporter construct in U-373 cells. In contrast, both the N- and C-terminal deletion mutants of IE2 were transcriptionally inactive but fully capable of causing a G1 arrest (Fig. 7C and D). All other IE2 mutants were equally unable to transactivate transcription, with the exception of HA-IE2(Δ136-289) (data not shown) (45). This mutant showed a severely reduced capacity to transactivate (data not shown) (43) but retained its ability to induce a G1 arrest (Fig. 7C). These data show that domains of IE2 absolutely required for transactivation are dispensable for the G1 arrest function. Therefore, the mechanism by which IE2 mediates the cell cycle block in G1 may well be posttranscriptional.
DISCUSSION
Here we present data identifying HCMV IE2 as an inducer of a G1 arrest. This observation fits in well with HCMV physiology, since during viral infection cell cycle progression of human fibroblasts is blocked at multiple points—predominantly in G1 (4, 12, 23, 27). Consistent with that, we show that the HCMV permissive astrocytoma cell line U-373 is also blocked in G1 upon HCMV infection. In a direct comparison, we show that U-373 cells behave like human fibroblasts in that respect, implying that U-373 cells are a valid model system to study HCMV-induced cell cycle regulatory effects. The view that the IE2-mediated G1 arrest is of physiological significance during HCMV infection is supported by the following observations. (i) The HCMV-induced G1 arrest of cycling cells (both U-373 cells and human fibroblasts) requires viral gene expression, since UV-inactivated virus did not induce cell cycle blockage in these cells. (ii) HCMV gene products expressed during the initial 24 h of infection were sufficient for mediating this G1 block. (iii) Cotransfected IE2 can rescue the G1 arrest phenotype of U-373 cells infected with UV-inactivated HCMV. (iv) The concentration of IE2 sufficient to induce a G1 arrest after transient transfection is also achieved during viral infection. (v) The IE2-mediated cell cycle effect is a true block of G1-S transition lasting for days, rather than a consequence of a slowed cell cycle progression. (vi) The IE2-mediated cell cycle arrest was found to be a potent effect. Throughout this work, we have included p16INK4a as a positive control, one of the most efficient inducers of a G1 arrest known to date, and the G1 arrest function of IE2 was comparable in strength to that of p16INK4a.
Importantly, IE2 not only mimics the HCMV-mediated G1 arrest but also reflects, more generally, seemingly contradictory findings during HCMV infection. As outlined in the introduction, HCMV can arrest cells in G1 (4, 12, 27) but at the same time induces S phase-promoting activities like hyperphosphorylated forms of pRb (23) or increased cyclin E-associated kinase activity (4, 23) and also activates the expression of proliferating cell nuclear antigen (12), as well as other proliferation-coupled factors required for DNA synthesis (33). Normally, these findings characterize a cell cycle state which lies beyond the restriction point (39). The restriction point is the most important stage of growth control in mammalian cells and is typically activated during the G1 arrest imposed on cells by overexpression of cyclin kinase inhibitors, such as p16INK4a, p21CIP, or p27KIP (44). Upon passage through the restriction point, a cell is usually committed to replicate its DNA (39). However, HCMV-infected cells, although showing postrestriction point characteristics, do not start cellular DNA replication, suggesting that the virus may at least partially dissociate cell cycle progression from cellular DNA synthesis.
There are several lines of evidence suggesting that IE2 is at least partially responsible for this exceptional state of HCMV-infected cells—possibly by exerting some S phase-promoting activities characteristic of oncogenes from small DNA tumor viruses coupled with the ability to finally block cell cycle progression in G1. First, although IE2, like E1A, E7, and large T antigen, binds pRb, compromising its transcriptional repressor activity (17), IE2-expressing cells do not proceed into S phase but rather are blocked in G1. Second, IE2, like large T antigen, can reverse the retinoblastoma-induced flat-cell phenotype of SAOS-2 cells (13) but, unlike large T antigen, fails to reverse the G1 arrest of those cells (13). Third, again as with large T antigen (9), IE2 overexpression can reverse the cell cycle-specific transcriptional defects of cells containing a temperature-sensitive mutant of the TAFII250 gene (29), but unlike large T antigen, IE2 is unable to abrogate the G1 arrest of these cells (29). These findings, together with the observation that IE2 activates the cyclin E promoter (3), are consistent with the view that IE2 might be part of an HCMV activity which guides infected cells past the restriction point but at the same time disallows cells to enter S phase. The finding that IE2 blocks S phase entry of both proliferating cells (Fig. 1) and cells being stimulated to leave the quiescent state (Fig. 2) would also suggest that the IE2-induced cell cycle block occurs later in G1 after the routes of the M-G1-S and G0-G1-S transition have merged (this notion does not exclude the possibility that IE2 also employs distinct mechanisms to block S phase entry of cells coming from mitosis and G0). Interestingly, a recent publication suggested the existence of a late G1 checkpoint after the restriction point (55). Such a late checkpoint could, for instance, be activated by IE2. Alternatively, IE2 may even dissociate aspects of cell cycle progression from cellular DNA synthesis as in principle has been shown for the papillomavirus E7 protein (14, 24).
In addition to IE2, other viral factors seem to contribute to the regulation of G1-S transition in infected cells. Lu and Shenk have recently shown in transient transfection experiments that the virion protein UL69 can also arrest cells in G1 (28), and our own analysis of UL69 is consistent with that (unpublished data). At the same time, however, a second virion factor, pp71, was found to accelerate G1-S transition when transiently overexpressed (reference 25 and unpublished observation). Therefore, the virion appears to contain at least two factors with opposing activities on G1-S transition. Under different conditions, these factors may to a lesser or greater extent contribute to the net cell cycle effect of the virion. Our cell cycle analysis after infection with UV-irradiated virus shows that cycling cells were not arrested in G1, indicating that viral gene expression is necessary for blocking the G1-S transition step, and IE2 seems to play an important role here. However, we also found that UV-irradiated virus can block S phase entry when cells exit from G0 rather than from mitosis. This might be an indication that virion factors are sufficient to block the G0-G1 transition. In addition, infection of cycling cells with UV-irradiated virus leads to a decrease of cells in G1, which would be consistent with the described pp71 function. However, the reconstruction experiment (Fig. 6) demonstrates that the G1-S transition induced by UV-irradiated virus can be counteracted sufficiently by ectopic expression of IE2. Taken together, these findings suggest that HCMV has evolved several factors which contribute to the complex cell cycle state of infected cells and that IE2 appears to be at least partially responsible for the G1-S block, assigning a new biological activity to IE2.
The demonstration that IE2 causes a G1 arrest in many established cell lines may also help to explain why relatively few cell lines stably expressing IE2 have been reported. Still, due to genetic instabilities of established cell lines and the strong selection pressure applied during the procedure of generating stable cell lines, it might be possible to select single clones able to proliferate in the presence of IE2. However, at least in our hands, forced expression of IE2 generally appears to select against growth.
What might be the mechanism of the IE2-mediated cell cycle arrest? As noted above, IE2 can induce a G1 arrest in several different cell lines with distinct genetic defects in genes coding for known cell cycle regulatory proteins, suggesting that the IE2-mediated G1 arrest does not strictly depend on the cyclin kinase inhibitors p16INK4a and p21CIP or the tumor suppressor proteins pRb and p53. Somewhat surprisingly, mutants of IE2 defective in activating the c-fos promoter were still sufficient to induce a G1 arrest (Fig. 7). In particular, the transactivation domains of IE2 were found to be dispensable for mediating the cell cycle block. Despite numerous studies analyzing IE2 regions necessary for transcriptional activation (31, 36, 37, 43, 45, 47), to our knowledge there has been no report demonstrating transcriptional activity of IE2 mutants lacking the N- or C-terminal activation domains. It is conceivable, therefore, that IE2 employs (so far unrecognized) posttranscriptional mechanisms to block cells in G1. Work in progress aims to define these mechanisms. It will be important to examine whether interfering with these mechanisms may hamper the ability of HCMV to replicate its DNA.
ACKNOWLEDGMENTS
We thank S. Proesch, T. Stamminger, Jiri Lukas, and T. Kouzarides for reagents and M. Truss and N. Robinson for their opinions on the manuscript. We thank I. Gruska for excellent technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Klinische Forschergruppe Paediatrische Molekularbiologie, GA167/6-1 to G. Gaedicke and C.H. and SFB 421-A3 to C.H.).
REFERENCES
- 1.Boldogh I, AbuBakar S, Albrecht T. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science. 1990;247:561–564. doi: 10.1126/science.1689075. [DOI] [PubMed] [Google Scholar]
- 2.Bonin L R, McDougall J K. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J Virol. 1997;71:5861–5870. doi: 10.1128/jvi.71.8.5861-5870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bresnahan W A, Albrecht T, Thompson E A. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J Biol Chem. 1998;273:22075–22082. doi: 10.1074/jbc.273.34.22075. [DOI] [PubMed] [Google Scholar]
- 4.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 5.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Inhibition of cellular cdk2 activity blocks human cytomegalovirus replication. Virology. 1997;231:239–247. doi: 10.1006/viro.1997.8489. [DOI] [PubMed] [Google Scholar]
- 6.Caswell R, Hagemeier C, Chiou C-J, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 7.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, editors. Current protocols in immunology. New York, N.Y: John Wiley & Sons; 1991. [Google Scholar]
- 9.Damania B, Alwine J C. TAF-like function of SV40 large T antigen. Genes Dev. 1996;10:1369–1381. doi: 10.1101/gad.10.11.1369. [DOI] [PubMed] [Google Scholar]
- 10.DeMarchi J M. Correlation between stimulation of host cell DNA synthesis by human cytomegalovirus and lack of expression of a subset of early virus genes. Virology. 1983;129:274–286. doi: 10.1016/0042-6822(83)90167-8. [DOI] [PubMed] [Google Scholar]
- 11.Dennig J, Beato M, Suske G. An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J. 1996;15:5659–5667. [PMC free article] [PubMed] [Google Scholar]
- 12.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortunato E A, Sommer M H, Yoder K, Spector D H. Identification of domains within the human cytomegalovirus major immediate-early 86-kilodalton protein and the retinoblastoma protein required for physical and functional interaction with each other. J Virol. 1997;71:8176–8185. doi: 10.1128/jvi.71.11.8176-8185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funk J O, Waga S, Harry J B, Espling E, Stillman B, Galloway D. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green S, Issemann I, Sheer E. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeier C, Walker S M, Sissons P J G, Sinclair J H. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J Gen Virol. 1992;73:2385–2393. doi: 10.1099/0022-1317-73-9-2385. [DOI] [PubMed] [Google Scholar]
- 20.Hall C V, Jacob P E, Ringold G M, Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2:101–109. [PubMed] [Google Scholar]
- 21.Hayhurst G P, Bryant L A, Caswell R C, Walker S M, Sinclair J H. CCAAT box-dependent activation of the TATA-less human DNA polymerase α promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J Virol. 1995;69:182–188. doi: 10.1128/jvi.69.1.182-188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heuvel S V D, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 23.Jault F M, Jault J-M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones D L, Alani R M, Münger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaleita R F, Bechtel J T, Shenk T. Abstracts of the 23rd International Herpesvirus Workshop. 1998. Human cytomegalovirus protein pp71 (UL82) stimulates cell cycle progression, abstr. 79. [Google Scholar]
- 26.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu M, Shenk T. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J Virol. 1999;73:676–683. doi: 10.1128/jvi.73.1.676-683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukac D M, Harel N Y, Tanese N, Alwine J C. TAF-like functions of human cytomegalovirus immediate-early proteins. J Virol. 1997;71:7227–7239. doi: 10.1128/jvi.71.10.7227-7239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukac D M, Manuppello J R, Alwine J C. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;65:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margolis M J, Pajovic S, Wong E L, Wade M, Jupp R, Nelson J A, Azizkhan J C. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J Virol. 1995;69:7759–7767. doi: 10.1128/jvi.69.12.7759-7767.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2448–2492. [Google Scholar]
- 34.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 35.Pajovic S, Wong E L, Black A R, Azizkhan J C. Identification of a viral kinase that phosphorylates specific E2Fs and pocket proteins. Mol Cell Biol. 1997;17:6459–6464. doi: 10.1128/mcb.17.11.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pizzorno M C, Mullen M-A, Chang Y-N, Hayward G S. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J Virol. 1991;65:3839–3852. doi: 10.1128/jvi.65.7.3839-3852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pizzorno M C, O’Hare P, Sha L, LaFemina R L, Hayward G S. trans-Activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J Virol. 1988;62:1167–1179. doi: 10.1128/jvi.62.4.1167-1179.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plachter B, Vornhagen R, Britt B, Stamminger T, Jahn G. Analysis of proteins encoded by IE-regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology. 1993;193:642–652. doi: 10.1006/viro.1993.1172. [DOI] [PubMed] [Google Scholar]
- 39.Planas-Silva M D, Weinberg R A. The restriction point and control of cell proliferation. Curr Opin Cell Biol. 1997;9:768–772. doi: 10.1016/s0955-0674(97)80076-2. [DOI] [PubMed] [Google Scholar]
- 40.Poma E E, Kowalik T F, Zhu L, Sinclair J H, Huang E-S. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J Virol. 1996;70:7867–7877. doi: 10.1128/jvi.70.11.7867-7877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ripalti A, Boccuni M C, Campani F, Landini M P. Cytomegalovirus-mediated induction of antisense mRNA expression to UL44 inhibits virus replication in an astrocytoma cell line: identification of an essential gene. J Virol. 1995;69:2047–2057. doi: 10.1128/jvi.69.4.2047-2057.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salvant B S, Fortunato E A, Spector D H. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J Virol. 1998;72:3729–3741. doi: 10.1128/jvi.72.5.3729-3741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scully A L, Sommer M H, Schwartz R, Spector D H. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein interactions. J Virol. 1995;69:6533–6540. doi: 10.1128/jvi.69.10.6533-6540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 45.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Speir E, Modali R, Huang E-S, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 47.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stott F J, Bates S, James M C, McConnell B B, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden K H, Peters G. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai H-L, Kou G-H, Chen S-C, Wu C-W, Lin Y-S. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J Biol Chem. 1996;271:3534–3540. [PubMed] [Google Scholar]
- 50.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoo Y D, Chiou C J, Choi K S, Yi Y, Michelson S, Kim S, Hayward G S, Kim S J. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor beta1 gene through an Egr-1 binding site. J Virol. 1996;70:7062–7070. doi: 10.1128/jvi.70.10.7062-7070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu H, Cong J-P, Shenk T. Use of a differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu L, Heuvel S V D, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 55.Zhu X, Ohtsubo M, Böhmer R M, Roberts J M, Assoian R K. Adhesion-dependent cell cycle progression linked to the expression of cyclin D1, activation of cyclin E-cdk2, and phosphorylation of the retinoblastoma protein. J Cell Biol. 1996;133:391–403. doi: 10.1083/jcb.133.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]