Abstract
There is serological evidence that bovine group C rotaviruses exist in the United States, but there are no reports of their isolation. Ninety fecal samples from calves with diarrhea, 81 samples from adult cows with diarrhea (winter dysentery), and 20 fecal samples from healthy adult cows were tested for group C rotaviruses by polyacrylamide gel electrophoresis, immune electron microscopy, and reverse transcription-PCR (RT-PCR). Three samples from adult cow diarrhea cases were positive only by RT-PCR, and a group C rotavirus was isolated from a positive sample in monkey kidney (MA104) cells (WD534tc/C). Genetically and serologically, the WD534tc/C strain was more closely related to the Cowden porcine group C strain than to the Shintoku bovine strain. Because the original cow feces also contained a group A rotavirus (detected after passage in cell culture), we hypothesized that such dual-rotavirus infections might play a role in the pathogenesis and host adaptation of rotaviruses. Thus, we examined the pathogenesis of WD534tc/C alone or combined with virulent (IND/A) or attenuated (NCDV/A) bovine group A rotaviruses in gnotobiotic calves. WD534tc/C alone induced diarrhea without (or with limited) virus shedding in inoculated calves (n = 3). In contrast, all calves coinfected with WD534tc/C and IND/A (n = 2) developed diarrhea and shed both viruses, whereas calves coinfected with WD534tc/C and NCDV/A (n = 3) developed diarrhea but did not shed either virus. Infection with WD534tc/C or NCDV/A alone caused only mild villous atrophy (jejunum and/or ileum), whereas dual infection with both viruses induced lesions throughout the small intestine. Although IND/A alone caused villous atrophy, more-widespread small intestinal lesions occurred in calves coinfected with WD534tc/C and IND/A. In conclusion, coinfection of calves with group A rotaviruses enhanced fecal shedding of a bovine group C rotavirus and the extent of histopathological lesions in the small intestines. Thus, our findings suggest a potential novel hypothesis involving dual infections for the adaptation of heterologous rotaviruses to new host species.
Rotaviruses are a major cause of diarrhea in young children and animals including cattle (21, 35) and belong to seven distinct antigenic groups (A to G) (7, 21, 24). Although group C rotaviruses are antigenically distinct from group A rotaviruses in serological tests, they are genetically related and they share a minor nonneutralizing epitope on VP6 detected with monoclonal antibodies (47). The VP6 genes of group C rotaviruses share 88.4 to 90.6% homology (porcine Cowden, bovine Shintoku, and human Bristol) and 41.3 and 16.3% homology with the VP6 gene of group A (bovine RF) and group B (human ADRV) rotaviruses, respectively (19). There is antigenic and genetic variation within group C rotaviruses. There are at least three G types (VP7) identified by two-way cross-neutralization tests and sequence analysis of the VP7 genes (70 to 75% homology among the serotypically distinct strains), and the Cowden porcine and Shintoku bovine strains are different serotypes (45, 46).
Group A rotaviruses are endemic, and group B rotaviruses cause sporadic cases of diarrhea in calves and cows in the United States (5, 31, 34, 35). However, although a moderate prevalence of antibodies to group C rotaviruses in sera of cattle (47 to 56%) in the United States (44) was reported, to date, the Shintoku group C bovine rotavirus, which was isolated from an adult cow in Japan, is the only bovine isolate of group C rotavirus (31, 48). The first group C rotavirus of any species was isolated in 1980 from nursing pigs (33) and more recently identified as a cause of enzootic diarrhea in neonates (27) and older finishing (fattening) pigs (22). Serological studies suggest that group C rotaviruses are endemic in U.S. swine herds (40, 44). In humans, group C rotaviruses are potential emerging enteric pathogens for all ages including adults (4, 17, 31). Since their first detection in humans in 1983 (28), group C rotaviruses have been associated with large outbreaks of diarrhea in Japan (4, 16), and with family or sporadic cases of diarrhea in children and adults worldwide including the United States (4, 17).
Assays to detect group C rotaviruses, including polyacrylamide gel electrophoresis (PAGE), immune electron microscopy (IEM), enzyme-linked immunosorbent assay (ELISA), and cell culture immunofluorescence (CCIF) tests have been described elsewhere (3, 12, 18, 40, 44). However, because of the limited shedding or instability of group C rotaviruses in feces (4, 17, 31), highly sensitive assays are needed for their efficient detection (17). In this study, we surveyed fecal samples from calves and adult cows with diarrhea for the presence of group C rotaviruses by reverse transcription-PCR (RT-PCR). A group C rotavirus (WD534tc/C strain) was isolated from a diarrheic adult cow fecal sample, which also contained group A rotavirus. The WD534tc/C strain showed a higher relatedness to Cowden porcine group C rotavirus by serological and genetic analysis than to the Shintoku bovine group C rotavirus and was pathogenic in gnotobiotic pigs. During our preliminary study of the pathogenicity of the WD534tc/C infection, gnotobiotic calves inoculated with this strain developed diarrhea without or with limited (1-day) virus shedding. Because the original field sample was a mixed infection with group A rotavirus, and the isolate was potentially a porcine group C strain, we hypothesized that such dual-rotavirus infections might play a role in the pathogenesis and adaptation of heterologous group C rotaviruses to cattle. Thus, we investigated the coinfection of gnotobiotic calves with group A bovine rotaviruses and the WD534tc/C bovine group C rotavirus isolate to determine clinical signs, virus shedding patterns, and the extent of histopathological lesions in the intestines.
MATERIALS AND METHODS
Viruses and cells.
Reference group C rotaviruses included the Shintoku bovine strain (48) and the Cowden porcine strain (41). Group A rotaviruses included a virulent serotype G6 IND (P[5]G6) strain (6) and an attenuated vaccine NCDV (P[1]G6) strain from the American Tissue Culture Collection (Atlanta, Ga.). The Shintoku/C, attenuated Cowden/C, and NCDV/A strains were grown in monkey kidney cells (MA104 cells) in the presence of pancreatin (50 μg/ml) (36, 37, 41, 48). The virulent IND/A and Cowden/C strains were prepared by virus passage in gnotobiotic calves or piglets, respectively, and infected feces or intestinal contents were collected as a source of virus. Mock-infected cell culture samples and mock-infected gnotobiotic calf feces or pig intestinal contents were used as negative controls.
Field samples.
Analysis for bovine group C rotaviruses was performed on the following samples: 90 diarrheic calf fecal samples from Ohio, California, Wyoming, South Dakota, and Nebraska; 81 adult cow diarrheic fecal samples (winter dysentery cases) from Ohio, New York, and California; and 20 healthy adult cow fecal samples (from two Ohio herds that were matched case controls for the winter dysentery cases) (39).
Extraction and electrophoresis of rotavirus dsRNA.
Rotavirus double-stranded RNA (dsRNA) was extracted from cell culture-propagated viruses and fecal or intestinal viruses by previously described procedures (13). Rotavirus dsRNA in extracted samples was analyzed by PAGE to confirm the presence of dsRNA and to examine the genomic electropherotypes as previously described (6, 13, 31). The discontinuous buffer system of Laemmli was utilized, and dsRNA was resolved on 10% polyacrylamide slab gels (23). Electrophoresis was conducted at 12 mA for 14 to 16 h. The dsRNA bands were visualized by silver staining (13).
IEM and CCIF assay.
Fifty-three fecal samples from diarrheic adult cows (field samples) and fecal samples after single or dual inoculation of gnotobiotic calves and pigs and cell culture isolates were processed for IEM as previously described (32). The processed samples were incubated separately with group A-, B-, C-, or bovine coronavirus-specific hyperimmune antisera, and then the mixtures were pelleted, negatively stained with phosphotungstic acid, applied to grids, and examined by electron microscopy (29, 32). A CCIF assay was used to confirm if group A or C rotaviruses were present in virus isolates and to detect virus shedding after challenge of gnotobiotic calves and pigs with group A and/or C rotaviruses as described previously (20). Briefly, the fecal or cell-passaged rotavirus samples were serially diluted with minimal essential medium (MEM) from 1:10 to 1:107 and were inoculated onto monolayers of MA104 cells grown in 96-well plates. After adsorption at 37°C for 1 h, the mixtures were discarded and 200 μl (per well) of MEM containing 50 μg of pancreatin per ml was added to the cells and incubated at 37°C for 20 h. The medium was discarded, and the cells were fixed with 80% acetone and stained with fluorescein isothiocyanate (FITC)-conjugated anti-OSU porcine group A or anti-Cowden porcine group C rotavirus serum (20, 40). The numbers of fluorescent focus units (FFU) in each well were counted, and titers were expressed as FFU per milliliter.
RT-PCR assay.
Field fecal samples and fecal samples collected after virus challenge of gnotobiotic calves and pigs were tested for group A and C rotaviruses by RT-PCR (6). The RT-PCR primers were designed to produce partial-length group C rotavirus VP6 based on Cowden and Shintoku VP6 genes and a full-length group A rotavirus VP7 gene. cDNA was synthesized and amplified in an RT-PCR mixture containing 10× RT-PCR buffer (200 mM Tris-HCl [pH 8.3], 2.5 mM MgCl2, KCl, 0.05% gelatin), deoxynucleoside triphosphates (10 mM [each]), 200 ng of primer A (sense strands for group C VP6 [5′-GAAGCTGTATGTGATGATGA-3′] and group A VP7 [5′-GGCCGGATTTAAAAGCGACAATTT-3′]), 200 ng of primer B (antisense strands for group C VP6 [5′-CATAGCAGCTGGTCTAATCA-3′] and for group A VP7 [5′-GGTCACATCATACAACTCTA-3′]), (6), 5 U of RNasin (Boehringer Mannheim Biochemicals, Indianapolis, Ind.), 10 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim Biochemicals), and 2.5 U of Taq polymerase (Boehringer Mannheim Biochemicals). First-strand cDNA synthesis was accomplished by incubating the above mixture for 90 min at 42°C. Thirty amplification cycles were conducted, with each cycle consisting of 94°C for 1 min (denaturation), 52°C for 1 min (annealing), and 72°C for 2 min (extension), followed by a 7-min extension step at 72°C. The PCR products were analyzed on 1% agarose gels by standard procedures (38).
Isolation of a group C rotavirus from fecal samples.
Two adult cow diarrheic fecal samples (from the same Ohio farm) which were group C positive by RT-PCR were inoculated into MA104 cells in roller tubes for virus isolation (36, 37). They were blind passaged until cytopathic effects were seen and monitored for group A and C rotaviruses by CCIF (20, 40).
Cloning and sequence analysis of the full-length VP6 gene of WD534tc.
The full-length VP6 gene of WD534tc was produced by RT-PCR with 5′- and 3′-end primers and cloned in the pCRII vector (Invitrogen, San Diego, Calif.). The sequence analysis was performed by the primer extension method (Sequenase version 2.0; Amersham, Arlington Height, Ill.), and the data were analyzed with the DNASTAR program (DNASTAR Inc., Madison, Wis.).
Antigenic analyses of WD534tc, Cowden, Shintoku group C rotaviruses, and IND group A rotavirus by two-way FFN tests.
The isolate WD534tc, Cowden and Shintoku group C rotaviruses, and IND group A rotavirus were tested in two-way fluorescent focus neutralization (FFN) tests using antisera to each virus, which were prepared by hyperimmunization of a gnotobiotic piglet with WD534tc/C and of guinea pigs with Cowden, Shintoku, or IND strain, as described previously (20, 40). Briefly, the antisera were serially diluted with MEM from 1:10 to 1:20,480, mixed with the homologous or heterologous virus strains (104 FFU/ml), and reacted at 37°C for 1 h. The mixtures (100 μl) were inoculated onto monolayers of MA104 cells grown in 96-well plates. After adsorption at 37°C for 1 h, the mixtures were discarded and 200 μl (per well) of MEM containing 50 μg of pancreatin per ml was added to the cells and incubated at 37°C for 20 h. The medium was discarded, and the cells were fixed with 80% acetone and stained with FITC-conjugated pig anti-OSU group A or anti-Cowden group C rotavirus serum. Neutralizing antibody titers were expressed as the reciprocal of the highest dilution that resulted in 80% or greater reduction in the numbers of FFU. Serum samples from unexposed gnotobiotic pigs were used for negative controls.
Virus challenge of gnotobiotic calves and piglets.
The challenge was either single or dual inoculation (simultaneous) of group A and group C rotavirus strains into 16 gnotobiotic calves and 6 gnotobiotic piglets (Table 1). All calves and piglets were from 1 to 24 days old and were derived, fed, inoculated, and maintained as previously described (25, 29). Briefly, the gnotobiotic calves and piglets were oronasally inoculated with diluted (1:10) fecal filtrates or virus-infected cell culture lysates. Viruses were inoculated at similar doses (WD534/C, 107 FFU for calves and 106 FFU for piglets; IND/A, 107 FFU; NCDV/A, 105 FFU for single inoculation and 105 or 107 FFU for dual inoculation; virulent and attenuated Cowden/C, 107 FFU for calves and 106 FFU for piglets) in a total of 50 ml of calves and 10 ml for pigs. Dual inoculation (WD534tc/C and IND/A, WD534tc/C and NCDV/A or virulent Cowden/C and IND/A) of gnotobiotic calves was done with the same virus titers (except for WD534tc/C and NCDV/A, when either 105 or 107 FFU for NCDV/A was used) and volumes as single inoculations. A titer of 105 FFU for single inoculation of calves with NCDV/A was used because it is the recommended dose of commercial rotavirus vaccines for use in calves (42). After inoculation, calves and piglets were examined twice daily for clinical signs and development of diarrhea, and the fecal consistency was scored as follows: 0, normal; 1, pasty; 2, semiliquid; 3, liquid. For the data summarized in the tables, diarrhea was noted as a fecal consistency of ≥2 and questionable or mild diarrhea was noted as fecal consistency of 1. Feces and serum samples were collected daily and weekly, respectively, and stored at −20°C until tested. Each fecal sample was tested for the presence of groups A and C rotaviruses by CCIF, IEM, and RT-PCR, and each serum sample was tested for antibodies to group A and C rotaviruses by the FFN test as described previously (20, 40). Seroconversion was indicated as serum antibody titer increases from <50 for acute serum to ≥50 for convalescent serum.
TABLE 1.
Experimental design for single and dual inoculation of gnotobiotic calves and piglets with group C and group A rotaviruses
| Group | Virus inoculuma | No. of animals in group |
|---|---|---|
| Calvesb | ||
| 1 | IND/A | 2 |
| 2 | NCDV/A | 3 |
| 3 | WD534tc/C | 4 |
| 4 | Virulent Cowden/C | 1 |
| 5 | WD534tc/C + IND/A | 2 |
| 6 | Cowden/C (virulent) + IND/A | 1 |
| 7 | WD534tc/C + NCDV/A | 2 |
| 8 | None (buffer) (control) | 1 |
| Pigletsc | ||
| 9 | WD534tc/C | 3 |
| 10 | Virulent Cowden/C | 1 |
| 11 | Attenuated Cowden/C | 2 |
Viruses were inoculated at the following doses (WD534tc/C, 107 FFU for calves and 106 FFU for piglets; IND/A, 107 FFU; NCDV/A, 105 or 107 FFU; virulent and attenuated Cowden, 107 FFU for the calf and 106 FFU for piglets).
Calves were challenged at 3 to 8 days of age, except for one older calf (24 days old) in group 6.
Piglets were challenged at 4 days of age, except for one piglet (24 days old) in group 11.
Histopathological evaluation.
Single or dual inoculated calves were euthanized within 12 h of diarrhea onset (postinoculation days [PID] 2 to 4) and the duodenum, jejunum, and ileum from the small intestines and the mesenteric lymph nodes were collected and placed in fixative (Prefer; Anatech Ltd., Battle Creek, Mich.). The tissues were processed, embedded in paraffin, and stained with hematoxylin and eosin (50). Histological evaluation was done on coded samples, and a comparison was made with tissues from age-matched controls (50).
Nucleotide sequence accession number.
The GenBank accession number of the VP6 gene from WD534tc/C is AF162434.
RESULTS
IEM, PAGE, and RT-PCR of field samples.
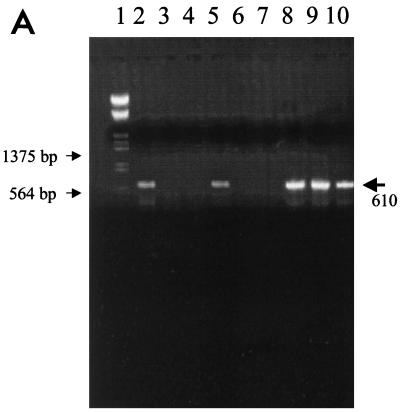
None of 53 fecal field samples from diarrheic adult cows tested were group C rotavirus positive by IEM. By PAGE, none of the fecal samples from calves and adult cows with diarrhea and from healthy adult cows were group C rotavirus positive. Fifty-four calf fecal samples (of 90), one adult cow diarrheic fecal sample (of 81), and none of the healthy adult cow fecal samples (of 20) were group A rotavirus positive by PAGE. The partial VP6 gene (expected length, 610 bp) and full-length VP7 gene (expected length, 1,062 bp) were generated for reference group C and group A rotavirus strains, respectively, using the selected primer pairs (Fig. 1). The RT-PCR assay was rotavirus group specific, and no PCR product was seen when the RT-PCR assay was applied to the heterologous rotaviruses and negative-control samples. No group C rotaviruses were detected in 90 diarrheic calf samples, whereas 66 samples were group A positive by RT-PCR. For diarrheic adult cow fecal samples, three samples (3 of 81, 2 from the same farm) and 1 sample were group C and group A rotavirus positive by RT-PCR, respectively. None of the fecal samples from healthy case-matched control cows were positive for group A or group C rotaviruses.
FIG. 1.
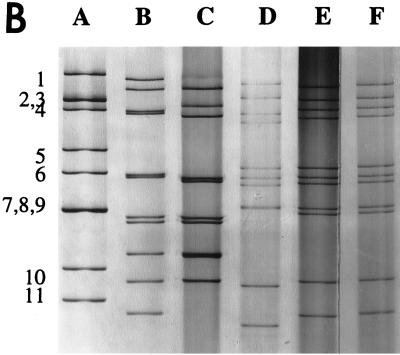
(A) The RT-PCR products of reference and field samples detected with primers specific for the partial VP6 gene of group C rotavirus. Lane 1, molecular size markers; lanes 2 to 5, field fecal samples; lane 6, IND/A; lane 7, NCDV/A; lane 8, Cowden/C; lane 9, Shintoku/C; lane 10, WD534tc/C. (B) dsRNA electropherotypes of the reference and WD534tc strains. Lane A, IND/A; lane B, ATI/B; lane C, WD653/B; lane D, Shintoku/C; lane E, Cowden/C; lane F, WD534tc/C. Numbers to the left of lane A designate group A rotavirus dsRNA segments.
Isolation of a group C rotavirus-positive sample in MA104 cells.
One adult cow diarrheic fecal sample showed cytopathic effects after six blind passages in MA104 cells (WD534tc/C). However, both group C and A rotaviruses were subsequently detected in the inoculated cell culture lysates by CCIF. After the G and P types of the group A rotavirus were determined, the group A rotavirus (P[5]G6) was eliminated by using hyperimmune antiserum to group A rotavirus (polyclonal and monoclonal antibodies to the G6 serotype) to neutralize the group A rotavirus. The group C rotavirus was cloned by plaque isolation twice to further eliminate the group A rotavirus.
Characterization of WD534tc/C.
By IEM, the WD534tc/C isolate showed typical rotavirus morphology and was aggregated by anti-group C rotavirus (Cowden) serum, but not by anti-bovine group A rotavirus serum, by IEM (data not shown). By PAGE, WD534tc/C had a typical group C rotavirus electropherotype by PAGE (4-3-2-2 band pattern) (Fig. 1). By sequence analysis, the full-length VP6 gene of the WD534tc isolate showed higher homologies (over 98%) with the Cowden porcine group C rotavirus than with the Shintoku bovine group C rotavirus (81.0%). Compared to the group A IND rotavirus, the similarity of the VP6 gene was 55.8% (Table 2). By antigenic analysis of WD534tc by the FFN test, the WD534tc/C rotavirus was more closely related to the Cowden (FFN antibody titers were identical [5,120] or only twofold different [5,120 versus 10,240]) porcine group C rotavirus than to the Shintoku (64-fold different [1,280 versus 20]) bovine group C rotavirus in two-way FFN tests (Table 2). The WD534tc/C rotavirus was antigenically unrelated to the IND group A rotavirus in the FFN test (FFN titers of <10) (Table 2).
TABLE 2.
Characterization of the WD534tc strain of bovine group C rotavirus in comparison with other group C rotaviruses and group A rotavirus
| Rotavirus strain | VP6 nucleotide identity (%) to WD534tc | Virus neutralization titera of hyperimmune antiserum to:
|
|||
|---|---|---|---|---|---|
| Shintoku | Cowden | WD534tc | IND/A | ||
| Group C | |||||
| Shintoku (bovine) | 81.0 | 1,280 | 40 | 20 | <10 |
| Cowden (porcine) | 98.7 | 40 | 5,120 | 10,240 | <10 |
| WD534tc (bovine) | 100 | 20 | 5,120 | 5,120 | <10 |
| Group A | |||||
| IND (bovine) | 55.8 | <10 | <10 | <10 | 2,560 |
Neutralizing antibody titers were expressed as the reciprocal of the highest dilution that resulted in 80% or greater reduction in the number of FFU.
Single inoculation of gnotobiotic calves with IND/A, NCDV/A, WD534tc/C, or Cowden/C rotavirus.
The virulent IND/A rotavirus caused diarrhea (from PID 2 or 3) and virus shedding (from PID 2; n = 2) detected by RT-PCR, IEM, and CCIF (Table 3). The CCIF titers of IND/A shed were about 105 to 106 FFU/ml (Table 3). With the attenuated NCDV/A, all calves (three calves but two had mild diarrhea before inoculation) developed mild diarrhea (Table 3) without virus shedding and seroconverted to the homologous virus (n = 2) (calf 3 was euthanized at PID 3 for histopathology) 3 weeks after inoculation. All calves (n = 4) inoculated with WD534tc/C developed diarrhea from PID 1 to 3 with limited virus shedding detected by only RT-PCR or RT-PCR and IEM (PID 1 only for two calves [calves 7 and 9]) or without virus shedding (n = 2) and seroconverted to WD534tc/C (n = 3; calf 9 was euthanized at PID 3 for histopathology) 3 weeks after inoculation (Table 3). The calf inoculated with virulent Cowden/C porcine rotavirus had no diarrhea and limited virus shedding detected by only IEM (PID 1 only) and seroconverted to homologous Cowden/C virus 3 weeks after inoculation (Table 3). The control calf (n = 1) inoculated with diluent showed no diarrhea or virus shedding (data not shown).
TABLE 3.
Pathogenesis of virulent IND/A, attenuated NCDV/A, and WD534tc/C bovine rotaviruses and Cowden/C porcine rotavirus in gnotobiotic calves
| Group, virus, and calf no. | Age (days) at inoculation | Clinical sign | Diarrheaa or virus sheddingb at PID:
|
Seroconversionc | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||||
| Group 1, IND/A | ||||||||||
| 1 | 5 | Diarrhea | − | + | + | EUT | ND | |||
| Virus shedding | −/−/− | +/+/+ | +/+/+ | |||||||
| 2 | 4 | Diarrhea | − | + | + | + | EUT | ND | ||
| Virus shedding | −/−/− | +/+/+ | +/+/+ | +/+/+ | ||||||
| Group 2,d NCDV/A | ||||||||||
| 3 | 5 | Diarrhea | + | + | EUT | ND | ||||
| Virus shedding | −/−/− | −/−/− | ||||||||
| 4e | 6 | Diarrhea | +/− | +/− | +/− | +/− | +/− | +/− | +/− | + |
| Virus shedding | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
| 5e | 6 | Diarrhea | +/− | +/− | +/− | +/− | +/− | +/− | +/− | + |
| Virus shedding | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
| Group 3, WD534tc/C | ||||||||||
| 6 | 6 | Diarrhea | + | + | + | + | + | + | +f | + |
| Virus shedding | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
| 7 | 21 | Diarrhea | + | + | + | + | + | + | +f | + |
| Virus shedding | +/−/− | +/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
| 8 | 2 | Diarrhea | − | − | + | + | + | + | +f | + |
| Virus shedding | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
| 9 | 6 | Diarrhea | − | − | + | EUT | ND | |||
| Virus shedding | +/+/− | −/−/− | −/−/− | |||||||
| Group 4, Cowden/C | ||||||||||
| 10 | 8 | Diarrhea | − | − | − | − | − | − | − | + |
| Virus shedding | −/+/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
+, fecal consistency of ≥2; +/−, questionable or mild diarrhea (fecal consistency of 1); −, no diarrhea (normal feces). EUT, euthanized for histopathology.
Virus shedding determined by three assays. Virus detected (+) or not detected (−) by RT-PCR, IEM, and CCIF assays (results shown in this order separated by slashes in table). The CCIF titers were 105 to 106 FFU/ml.
Seroconversion to NCDV/A, WD534tc/C, or Cowden/C rotavirus after PID 21 determined by FFN test (antibody titers of ≥50 compared to <50 for acute sera). +, seroconversion detected; ND, not done.
All calves in this group were inoculated with 105 FFU.
Calf had mild diarrhea before challenge.
Diarrhea persisted from PID 8 to 11.
Dual inoculation of calves with WD534tc/C or Cowden/C and virulent or attenuated group A rotavirus.
Dual inoculation of calves with WD534tc/C and IND/A caused diarrhea (from PID 1 or 3) with both group A and C virus shedding detected by CCIF, IEM, and RT-PCR (n = 2) (Table 4). The CCIF titers of WD534tc/C and IND/A shed were about 102 and 105 to 106 FFU/ml, respectively (Table 4). Cowden/C and IND/A dual inoculation of one calf caused diarrhea after PID 2 with both group A and C rotavirus shedding detected by RT-PCR, IEM, and CCIF (calf 13 [Table 4]). The CCIF titers of Cowden/C and IND/A shed were about 102 and 105 to 106 FFU/ml, respectively (Table 4).
TABLE 4.
Pathogenesis of WD534tc/C in gnotobiotic calves coinfected with either virulent IND/A or attenuated NCDV/A bovine rotaviruses or virulent Cowden/C porcine rotaviruses
| Group, viruses, and calf no. | Age (days) at inoculation | Clinical signa | Diarrheab or virus sheddingc at PID:
|
Seroconversiond | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||||
| Group 5, WD534tc/C + IND/A | ||||||||||
| 11 | 6 | Diarrhea | − | − | + | + | EUT | ND | ||
| gpA shedding | +/+/+ | +/+/+ | +/+/+ | +/+/+ | ||||||
| gpC shedding | −/−/− | +/+/+ | +/+/+ | +/+/− | ||||||
| 12 | 24 | Diarrhea | + | + | EUT | ND | ||||
| gpA shedding | +/+/+ | +/+/+ | ||||||||
| gpC shedding | +/+/− | +/+/+ | ||||||||
| Group 6, Cowden/C + IND/A | ||||||||||
| 13 | 6 | Diarrhea | − | + | + | + | + | + | +e | + |
| gpA shedding | +/+/+ | +/+/+ | +/+/+ | +/+/+ | +/+/+ | +/+/+ | +/+/+ | |||
| gpC shedding | +/+/+ | +/+/− | +/+/− | +/−/− | +/−/− | +/−/− | −/−/− | |||
| Group 7, WD534tc/C + NCDV/A | ||||||||||
| 14f | 5 | Diarrhea | + | + | EUT | ND | ||||
| gpA shedding | −/−/− | −/−/− | ||||||||
| gpC shedding | −/−/− | −/−/− | ||||||||
| 15f | 6 | Diarrhea | − | + | + | + | + | + | +g | + |
| gpA shedding | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
| gpC shedding | +/−/− | +/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
| 16h | 6 | Diarrhea | − | + | − | + | + | + | − | + |
| gpA shedding | +/+/− | +/+/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
| gpC shedding | +/−/− | +/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
gpA shedding, group A rotavirus shedding; gpC shedding, group C rotavirus shedding.
+, fecal consistency of ≥2; +/−, questionable or mild diarrhea (fecal consistency of 1); −, no diarrhea (normal feces). EUT, euthanized for histopathology.
Virus shedding determined by three assays. Virus detected (+) or not detected (−) by RT-PCR, IEM, and CCIF assays (results shown in this order separated by slashes in table). The CCIF titers of group A and group C rotavirus were 105 to 106 FFU/ml and about 102 FFU/ml, respectively.
Seroconversion to NCDV/A, WD534tc/C, or Cowden/C rotavirus after PID 21 determined by FFN test (antibody titers of ≥50 compared to <50 for acute sera). +, seroconversion detected; ND, not done.
Diarrhea lasted until PID 9.
Challenge dose of NCDV/A was 105 FFU.
Diarrhea lasted until PID 7.
Challenge dose of NCDV/A was 107 FFU.
Dual inoculation of calves with WD534tc/C and attenuated NCDV/A rotavirus induced diarrhea in all calves (n = 3) with variable, transient virus shedding depending on the inoculation titer of the attenuated NCDV/A rotavirus. Using a low titer (105 FFU) of attenuated NCDV/A (calves 14 and 15), the effects of dual inoculation were similar to those of single inoculation of calves with only WD534tc/C: group C shedding was detected by only RT-PCR (calf 15) or there was no virus shedding (calf 14) (Table 4). However, the use of a higher titer (107 FFU) of NCDV/A rotavirus for dual inoculation induced both group A and group C virus shedding of limited amounts (detected by only RT-PCR and IEM for NCDV/A and by only RT-PCR for WD534tc/C) and for a limited period (PID 1 and 2, calf 16 [Table 4]).
Inoculation of gnotobiotic piglets with WD534tc/C and virulent or attenuated Cowden/C rotavirus.
WD534tc/C and virulent Cowden/C caused diarrhea and virus shedding in gnotobiotic piglets (n = 3 and n = 1, respectively), whereas the attenuated Cowden/C strain (n = 2) induced no diarrhea, and virus shedding was detected only by RT-PCR (Table 5).
TABLE 5.
Pathogenesis of WD534tc/C in gnotobiotic piglets compared to that of virulent or attenuated Cowden/C porcine strains
| Virus and pig no. | Age (days) at inoculation | Clinical sign | Diarrheaa or virus sheddingb at PID:
|
Seroconversionc | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||||
| WD534tc/C | ||||||||||
| 1 | 4 | Diarrhea | − | − | + | + | + | + | +d | + |
| Virus shedding | −/−/− | +/+/− | +/+/+ | +/+/− | +/+/− | +/+/− | +/−/− | |||
| 2 | 4 | Diarrhea | − | − | + | + | EUT | ND | ||
| Virus shedding | −/−/− | +/+/+ | +/+/+ | +/+/+ | ||||||
| 3 | 4 | Diarrhea | − | − | + | EUT | ND | |||
| Virus shedding | −/−/− | +/+/+ | +/+/+ | |||||||
| Virulent Cowden/C | ||||||||||
| 4 | 4 | Diarrhea | − | − | + | + | EUT | ND | ||
| Virus shedding | +/+/− | +/+/+ | +/+/+ | +/+/+ | ||||||
| Attenuated Cowden/C | ||||||||||
| 5 | 26 | Diarrhea | − | − | − | − | − | − | − | + |
| Virus shedding | +/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | −/−/− | |||
| 6 | 4 | Diarrhea | − | − | − | − | − | − | − | + |
| Virus shedding | −/−/− | −/−/− | +/−/− | +/−/− | +/−/− | −/−/− | −/−/− | |||
+, fecal consistency of ≥2; +/−, questionable or mild diarrhea (fecal consistency of 1); −, no diarrhea (normal feces). EUT, euthanized for histopathology.
Virus shedding determined by three assays. Virus detected (+) or not detected (−) by RT-PCR, IEM, and CCIF assays (results shown in this order separated by slashes in table). The CCIF titers were about 104 FFU/ml.
Seroconversion to NCDV/A, WD534tc/C, or Cowden/C rotavirus after PID 21 determined by FFN test (antibody titers of ≥50 compared to <50 for acute sera). +, seroconversion detected; ND, not done.
Diarrhea lasted until PID 7.
Histopathology studies.
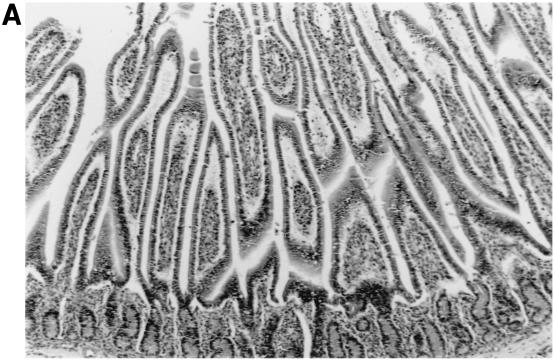
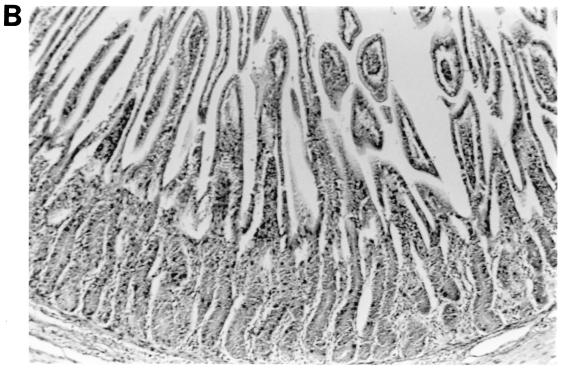
The morphologic changes observed in the small intestinal tissues of calves euthanized at PID 3 to 4 following single or dual rotavirus inoculation are summarized (Table 6 and Fig. 2). Virulent IND/A induced lesions typical of group A rotavirus infection characterized by loss of normal absorptive cells, detachment of absorptive cells from their basement membranes (especially over villous tips), blunting and fusion of villi, crypt hyperplasia, and lymphoreticular hyperplasia within villous tip stroma, small intestinal submucosal lymphoid aggregates, Peyer’s patches, and mesenteric lymph nodes. Inoculation with IND/A alone caused mild to moderate intestinal lymphoid hyperplasia and scattered foci of atrophied villi throughout all regions of the small intestine examined. Inoculation with NCDV/A alone elicited mild intestinal lymphoid hyperplasia and sparse villous atrophy in the jejunum and ileum. Inoculation with WD534tc/C alone elicited little to no lymphoid changes and only slight villous changes in the jejunum. Mild to moderate intestinal lymphoid hyperplasia was observed in the intestinal tissues in calves coinfected with WD534tc/C and either IND/A or NCDV/A, and villous atrophy was more pronounced and more frequently observed throughout all regions of the small intestine (duodenum, jejunum, and ileum) at PID 2 and 4 (Table 6 and Fig. 2).
TABLE 6.
Intestinal morphologic changes in individual calves following single or dual inoculation with group A and C bovine rotaviruses
| Virus inoculuma | PIDEb | Lymphoid hyperplasiac
|
Villous atrophyc
|
|||
|---|---|---|---|---|---|---|
| Mesenteric lymph nodes | Small intestined | Duodenum | Jejunum | Ileum | ||
| IND/A | 3 | + | ++ | + | + | + |
| NCDV/A | 4 | + | +/− | − | + | +/− |
| WD534tc/C | 4 | − | +/− | − | +/− | − |
| WD534tc/C + IND/A | 2 or 4 | + | ++ | ++ | ++ | ++ |
| WD534tc/C + NCDV/A | 4 | + | + | + | + | + |
Calves were inoculated at 6 to 8 days of age.
PIDE, postinoculation day of euthanasia.
Degree of histologic change (lymphoid hyperplasia and villous atrophy) from age-matched control: −, none (same as control); +/−, minimal, focal; +, mild, scattered; ++, moderate, widespread.
Small intestinal lamina propria, submucosa, and Peyer’s patch lymphoid tissues evaluated.
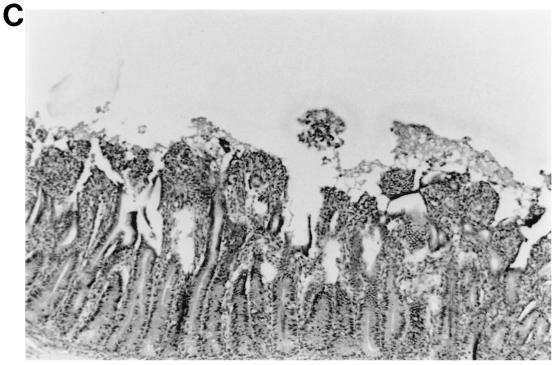
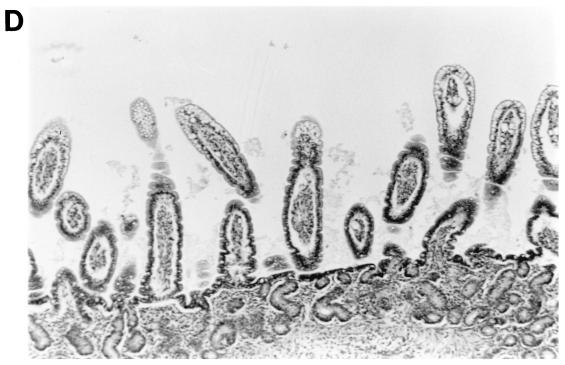
FIG. 2.
Intestinal lesions in the jejuna of gnotobiotic calves following oronasal inoculation with virus. The calves were mock infected (2 PID) (A) or infected with WD534tc/C (4 PID) (B) or IND/A (3 PID) (C) or coinfected with IND/A and WD534tc/C (2 PID) (D). The slides were stained with hematoxylin and eosin. Note that WD534tc alone caused minimal changes (B), whereas IND/A and coinfection with IND/A and WD534tc/C caused villous atrophy (C and D), vacuolation of absorptive cells (D), and hyperplasia of cryptic cells (C and D) in the jejunum. Magnification, ×50.
DISCUSSION
Group C rotaviruses have been reported in pigs, humans, and cattle. Unlike group A rotaviruses, group C rotaviruses are associated with diarrhea in humans of all ages (infants, children, and adults) (4, 16, 17, 28, 31). They have been reported worldwide since the early 1980s and are regarded as a potential emerging pathogen (4, 31). Although there is serological evidence for group C rotaviruses in cattle, so far there are no reports of their detection or isolation from cattle in the United States (31, 44). When we screened adult cow diarrheal samples from winter dysentery outbreaks for group C rotaviruses by RT-PCR (n = 81), we found only three positive samples which were negative by IEM and PAGE. These results suggest that the prevalence of bovine group C rotaviruses in the field may be low, the viruses may be unstable in feces, or virus shedding from infected animals may be limited. Therefore, to survey for bovine group C rotaviruses in the field, highly sensitive assays like RT-PCR are required as used in our study. Jiang et al. (17) also reported difficulty in detecting group C rotaviruses in samples from diarrheic children because of the low titers of virus in fecal samples and noted that the use of conventional methods such as PAGE may underestimate the true detection rate of human group C rotaviruses.
From one diarrheic adult cow fecal sample, we isolated a group C rotavirus (WD534tc/C strain) in MA104 cells. It was interesting that genetically and serologically, the WD534tc/C strain was more closely related to the Cowden/C porcine strain than to the Shintoku/C bovine strain. Challenge of three gnotobiotic calves with the WD534tc/C strain induced diarrhea without virus shedding or with limited virus shedding, and calves seroconverted to this virus, which indicates that the WD534tc/C strain is capable of limited replication in the intestine of the calves. The more-extensive diarrhea and shedding of WD534tc/C in piglets, comparable to that seen with virulent Cowden porcine group C rotavirus, suggests that the natural host for the WD534tc/C strain is more likely pigs, rather than cattle.
Because the original sample contained both group A and C rotaviruses, we were interested in the effects of their dual infection on gnotobiotic calves. Single inoculation of calves with the virulent IND/A strain caused diarrhea and virus shedding in the inoculated calves. Theil and McCloskey (42) reported that a titer of 105 FFU of NCDV/A in a commercial modified live bovine rotavirus-coronavirus vaccine induced mild diarrhea at PID 3 to 6 without virus shedding (except for 1 rotavirus-positive sample of 41 fecal samples examined) by IEM in all three gnotobiotic calves, and all calves seroconverted to NCDV/A rotavirus. In this study, although single inoculation of calves with 105 FFU of the attenuated NCDV/A strain caused mild diarrhea in one calf (the other two calves had mild diarrhea preexposure) and seroconversion to homologous virus in all calves tested 3 weeks after inoculation, no virus shedding was detected in fecal samples, suggesting the limited replication of this attenuated virus in the intestines of calves.
Using the virulent IND/A strain, dual infection with WD534tc/C consistently enhanced shedding of the WD534tc/C strain, as was seen also for porcine Cowden/C after dual inoculation of a calf with Cowden/C and IND/A. Using a low titer (105 FFU) of attenuated NCDV/A, the effect of dual inoculation was similar to that seen after inoculation with only WD534tc (calves 14 and 15 [Table 4]): no synergistic effect was seen by coinfection with both viruses. However, the use of a higher titer (107 FFU) of NCDV/A for dual infection induced limited amounts of both group A and group C virus shedding (detected by RT-PCR and IEM for NCDV/A and by RT-PCR for WD534tc/C) for a limited period (PID 1 and 2, calf 16 [Table 4]). Differences between the virulent and attenuated group A rotaviruses and different titers of attenuated group A rotavirus in their effects on dual infection could be due to differences in their replication ability in the intestine and/or the extent of cytopathology (villous atrophy) induced.
In the histopathological study, the intestinal lesions (Fig. 2 and Table 6) were generally more severe in calves with dual infection than with single infection of group A and C rotaviruses. Infection with WD534tc/C or NCDV/A alone caused only mild villous atrophy in the jejunum or jejunum and ileum, respectively, whereas dual infection with both viruses induced lesions throughout the small intestine. Although IND/A alone caused villous atrophy throughout the small intestine, more-widespread small intestinal lesions occurred in calves coinfected with WD534tc/C and IND/A.
Interspecies transmission of group A rotaviruses has been suggested, especially between humans and cattle. Gerna et al. (9) reported G6 serotype strains from humans (PA151 and PA169) (9, 10, 15), which were recognized by G6 monoclonal antibodies (bovine G6) and showed some genetic variation (about 10% in amino acid identity) with typical bovine G6 strains (UK and NCDV strains). By RNA-RNA hybridization, most of the gene segments of PA151 G6 strains were closely related to AU-1 human or NCDV bovine strains (15). Urasawa et al. (49) reported human G10 group A rotaviruses and suggested that this strain originated from a bovine host. Blackhall et al. (2) confirmed a bovine G1 strain analogous to human G1 serotypes in Argentina by serological assays and sequence analysis of the VP7 genes. In addition, rotavirus strain 116E, isolated from the fecal specimen of a newborn asymptomatic infant from India was identified as a P[11] type which was previously reported only in cattle (8).
Coinfection with group C and group A rotaviruses were detected in diarrheic fecal samples from children and finishing pigs (17, 22). Kim et al. (22) reported a diarrhea outbreak associated with group C rotaviruses in finishing (fattening) pigs and found these fecal samples also had group A rotaviruses detectable only by a highly sensitive second-round PCR assay. Ishimaru et al. (16) studied the epidemiology of a gastroenteritis outbreak associated with group C rotaviruses in the Matsuyama district of Japan and noted that group C rotavirus gastroenteritis occurred following an epidemic of group A rotavirus infections, primarily in children aged 4 to 7 years but rarely in those aged 0 to 1 years.
Coinfections by group A rotaviruses and other enteropathogens including Escherichia coli in calves are common in nature (26, 30). The effects of coinfection of calves with group A rotavirus and E. coli have been studied (11, 14, 30). Generally coinfected calves had more severe diarrhea, lesions, and shedding of both agents than infection by either agent alone, but little information is available regarding the detailed mechanisms for these synergistic effects. When rotaviruses and E. coli were coinoculated into calves, Runnels et al. (30) observed that there was more severe villous atrophy in the ilea of calves inoculated with rotavirus and E. coli than in calves inoculated with only rotavirus. They also found more-intensive small intestinal colonization by E. coli after coinfection with rotavirus. Gouet et al. (11) reported that experimental inoculation of calves with rotavirus, although not lethal in itself, followed by inoculation with a nonlethal dose of E. coli, led to dehydration and death in newborn colostrum-deprived calves. Hess et al. (14) reported that coinfection of newborn colostrum-deprived calves with rotavirus and E. coli resulted in not only more severe histological small intestinal lesions but also increased shedding of both pathogens.
The mechanism for the enhancement of virus shedding of the WD534tc/C strain by coinfection with the virulent group A rotavirus is unclear. We hypothesize that the WD534tc/C strain is likely highly cell associated in the calf intestine as seen in tissue culture (40), and following coinfection by a cytolytic group A rotavirus, the group C virus may be more readily released by cytolysis from the infected cells. Alternatively, structural or nonstructural proteins of group A rotavirus could assist in the replication or release of the WD534tc/C strain in coinfected cells or could have an indirect bystander effect on group C rotavirus-infected cells. A potential candidate is the nonstructural group A rotavirus protein NSP4, which has recently been shown to function as a viral enterotoxin in mouse studies and has membrane destabilization activity (1, 43).
Based on our results, which showed only limited or no virus shedding of the WD534tc/C strain in singly inoculated calves and the genetic and antigenic similarity of this strain to porcine group C rotavirus, we propose that the WD534tc/C strain may have originated from a porcine host and thus replicates only to a limited extent in the calf intestine. However, if dual infections with both group A and C rotaviruses occur in the bovine host, synergic effects may cause the release of the WD534tc/C strain into the feces, leading to intraspecies (bovine) transmission and further host adaptation after serial passage in the new host. Interestingly, calves dually infected with WD534tc/C and attenuated group A rotavirus (analogous to currently used oral attenuated vaccine strain) also briefly shed both viruses. Thus, our findings suggest a potential novel hypothesis for the adaptation of heterologous rotaviruses to new host species.
To explore potential mechanisms for the observed enhanced shedding of WD534tc/C rotavirus by coinfection with virulent group A rotavirus, in future studies, we plan to examine the effects of group A and C rotavirus coinfection and of cotransfection and infection of a group A rotavirus NSP4 gene and group C rotavirus in an vitro cell culture system.
ACKNOWLEDGMENTS
We thank D. C. Hodgins for help with gnotobiotic calf work and C. Nielsen for help with histopathological studies.
Salaries and research support were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center (OARDC), The Ohio State University. This work was partially supported by USDA NRICGP competitive grant 97-35204-4682 and grant RO1 AI 3356-04 from the National Institute of Allergy and Infectious Diseases, NIH.
REFERENCES
- 1.Ball J M, Tian P, Zeng C Q, Morris A P, Estes M K. Age dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science. 1996;272:101–104. doi: 10.1126/science.272.5258.101. [DOI] [PubMed] [Google Scholar]
- 2.Blackhall J, Bellinzoni R, Mattion N, Estes M K, LaTorre J L, Magnusson G. A bovine rotavirus serotype 1: serological characterization of the virus and nucleotide sequence determination of the structural glycoprotein VP7 gene. Virology. 1992;189:833–837. doi: 10.1016/0042-6822(92)90617-x. [DOI] [PubMed] [Google Scholar]
- 3.Bohl E H, Saif L J, Theil K W, Agnes A G, Cross R F. Porcine pararotavirus: detection, differentiation from rotavirus, and pathogenesis in gnotobiotic pigs. J Clin Microbiol. 1982;15:312–319. doi: 10.1128/jcm.15.2.312-319.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridger J C. Non-group A rotaviruses. In: Kapikian A Z, editor. Virus infections of the gastrontestinal tract. New York, N.Y: Marcel Dekker; 1994. pp. 369–407. [Google Scholar]
- 5.Chang K O, Parwani A V, Smith D, Saif L J. Detection of group B rotaviruses in fecal samples from diarrheic calves and adult cows and characterization of their VP7 genes. J Clin Microbiol. 1997;35:2107–2110. doi: 10.1128/jcm.35.8.2107-2110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang K O, Parwani A V, Saif L J. The characterization of VP7 (G type) and VP4 (P type) genes of bovine group A rotaviruses from field samples using RT-PCR and RFLP analysis. Arch Virol. 1996;141:1727–1739. doi: 10.1007/BF01718295. [DOI] [PubMed] [Google Scholar]
- 7.Estes M K. Replication of rotaviruses. In: Fields B N, Knipe D M, editors. Fields Virology. 2nd ed. New York, N.Y: Raven Press; 1990. pp. 1353–1404. [Google Scholar]
- 8.Gentsch J R, Das B K, Jiang B, Bhan M K, Glass R I. Similarity of the VP4 protein of human rotavirus strain 116E to that of the bovine B223 strain. Virology. 1993;194:424–430. doi: 10.1006/viro.1993.1280. [DOI] [PubMed] [Google Scholar]
- 9.Gerna G, Sarasini A, Parea M, Arista S, Miranda P, Brussow H, Hoshino Y, Flores J. Isolation and characterization of two distinct human rotavirus strains with G6 specificity. J Clin Microbiol. 1994;30:9–16. doi: 10.1128/jcm.30.1.9-16.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerna G, Sears J, Hoshino Y, Steele A D, Nakagomi O, Sarasini A, Flores J. Identification of a new VP4 serotype of human rotaviruses. Virology. 1994;200:66–71. doi: 10.1006/viro.1994.1163. [DOI] [PubMed] [Google Scholar]
- 11.Gouet P, Contrepois M, Dubourguier H C, Riou Y, Scherrer R, Laporte J, Vautherot J F, Cohen J, L’Haridon R. The experimental production of diarrhoea in colostrum deprived axenic and gnotoxenic calves with enteropathogenic Escherichia coli, rotavirus, coronavirus and in a combined infection of rotavirus and E. coli. Ann Rech Vet. 1978;9:433–440. [PubMed] [Google Scholar]
- 12.Gouvea V, Allen J R, Glass R I, Fang Z, Bremont M, Cohen J, McCrae M A, Saif L J, Sinarachaatanant P, Caul E O. Detection of group B and C rotaviruses by polymerase chain reaction. J Clin Microbiol. 1991;29:519–523. doi: 10.1128/jcm.29.3.519-523.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herring A J, Inglis N F, Ojeh C K, Snodgrass D R, Menzies J D. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J Clin Microbiol. 1982;16:473–477. doi: 10.1128/jcm.16.3.473-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hess R G, Bachmann P A, Baljer G, Mayr A, Pospischil A, Schmid G. Synergism in experimental mixed infections of newborn colostrum-deprived calves with bovine rotavirus and enterotoxigenic Escherichia coli (ETEC) Zentbl Vetmed Reihe B. 1984;31:585–596. doi: 10.1111/j.1439-0450.1984.tb01339.x. [DOI] [PubMed] [Google Scholar]
- 15.Iizuka M, Chiba M, Masamune O, Gerna G, Nakagomi O. Molecular characterization of human rotavirus VP4 genes by polymerase chain reaction and restriction fragment length polymorphism assay. Microbiol Immunol. 1992;37:729–735. doi: 10.1111/j.1348-0421.1993.tb01698.x. [DOI] [PubMed] [Google Scholar]
- 16.Ishimaru Y, Nakano S, Nakano H, Oseto M, Yamashita Y. Epidemiology of group C rotavirus gastroenteritis in Matsuyama, Japan. Acta Paediatr Jpn. 1991;33:50–56. doi: 10.1111/j.1442-200x.1991.tb01519.x. [DOI] [PubMed] [Google Scholar]
- 17.Jiang B, Dennehy P H, Spangenberger S, Gentsch J R, Glass R I. First detection of group C rotavirus in fecal specimens of children with diarrhea in the United States. J Infect Dis. 1995;172:45–50. doi: 10.1093/infdis/172.1.45. [DOI] [PubMed] [Google Scholar]
- 18.Jiang B, Qian Y, Tsunemitsu H, Green K Y, Saif L J. Analysis of the gene encoding the outer capsid glycoprotein (VP7) of group C rotaviruses by Northern and dot blot hybridization. Virology. 1991;184:433–436. doi: 10.1016/0042-6822(91)90864-8. [DOI] [PubMed] [Google Scholar]
- 19.Jiang B, Tsunemitsu H, Gentsch J R, Glass R I, Green K Y, Qian Y, Saif L J. Nucleotide sequence of gene 5 encoding the inner capsid protein (VP6) of bovine group C rotavirus: comparison with corresponding genes of group C, A, and B rotaviruses. Virology. 1992;190:542–547. doi: 10.1016/0042-6822(92)91250-x. [DOI] [PubMed] [Google Scholar]
- 20.Kang S Y, Saif L J, Miller K L. Reactivity of VP4-specific monoclonal antibodies to a serotype 4 porcine rotavirus with distinct serotypes of human (symptomatic and asymptomatic) and animal rotaviruses. J Clin Microbiol. 1989;27:2744–2750. doi: 10.1128/jcm.27.12.2744-2750.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapikian A Z, Chanock R M. Rotaviruses. In: Fields B N, Knipe D M, editors. Fields virology. 2nd ed. New York, N.Y: Raven Press; 1990. pp. 1353–1404. [Google Scholar]
- 22.Kim Y, Chang K O, Straw B, Saif L J. Characterization of group C rotaviruses associated with diarrhea outbreaks in feeder pigs. J Clin Microbiol. 1999;37:1484–1488. doi: 10.1128/jcm.37.5.1484-1488.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;27:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Mattion N M, Cohen J, Estes M K. The rotavirus proteins. In: Kapikian A Z, editor. Virus infections of the gastrontestinal tract. New York, N.Y: Marcel Dekker; 1994. pp. 169–249. [Google Scholar]
- 25.Meyer R C, Bohl E H, Kohler E M. Procurement and maintenance of germ-free swine for microbiological investigations. Appl Microbiol. 1964;12:295–300. doi: 10.1128/am.12.4.295-300.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon H W, McClurkin A W, Isaacson R E, Pohlenz J, Skartvedt S M, Gillette K G, Baetz A L. Pathogenic relationships of rotavirus, Escherichia coli, and other agents in mixed infections in calves. J Am Vet Med Assoc. 1978;173:577–583. [PubMed] [Google Scholar]
- 27.Morin M, Magar R, Robinson Y. Porcine group C rotavirus as a cause of neonatal diarrhea in a Quebec swine heard. Can J Vet Res. 1990;54:385–389. [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolas J C, Cohen J, Fortier B, Lourenco M H, Bricout F. Isolation of a human pararotavirus. Virology. 1983;124:181–184. doi: 10.1016/0042-6822(83)90302-1. [DOI] [PubMed] [Google Scholar]
- 29.Parwani A V, Lucchelli A, Saif L J. Identification of group B rotaviruses with short genome electropherotypes from adult cows with diarrhea. J Clin Microbiol. 1996;34:1303–1305. doi: 10.1128/jcm.34.5.1303-1305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Runnels P L, Moon H W, Matthews P J, Whipp S C, Woode G N. Effects of microbial and host variables on the interaction of rotavirus and Escherichia coli infections in gnotobiotic calves. Am J Vet Res. 1986;47:1542–1550. [PubMed] [Google Scholar]
- 31.Saif L J, Jiang B. Nongroup A rotaviruses of humans and animals. In: Ramig R F, editor. Rotaviruses. Berlin, Germany: Springer-Verlag; 1994. pp. 339–371. [DOI] [PubMed] [Google Scholar]
- 32.Saif L J, Bohl E H, Kohler E M, Hughes J H. Immune electron microscopy of transmissible gastroenteritis virus and rotavirus (reovirus-like agent) of swine. Am J Vet Res. 1977;38:13–20. [PubMed] [Google Scholar]
- 33.Saif L J, Bohl E H, Theil K W, Cross R F, House J A. Rotavirus-like, calicivirus-like and 23-nm virus-like particles associated with diarrhea in young pigs. J Clin Microbiol. 1980;12:105–111. doi: 10.1128/jcm.12.1.105-111.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saif L J, Brock K V, Redman D R, Kohler E M. Winter dysentery in dairy herds: electron microscopic and serological evidence for an association with coronavirus infection. Vet Rec. 1991;128:447–449. doi: 10.1136/vr.128.19.447. [DOI] [PubMed] [Google Scholar]
- 35.Saif L J, Rosen B I, Parwani A V. Animal rotavirus. In: Kapikian A Z, editor. Virus infections of the gastrontestinal tract. New York, N.Y: Marcel Dekker; 1994. pp. 289–314. [Google Scholar]
- 36.Saif L J, Rosen B I, Kang K Y, Miller K L. Cell culture propagation of rotaviruses. J Tissue Cult Methods. 1988;11:147–156. doi: 10.1007/BF01404267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saif L J, Terrett L A, Miller K L, Cross R F. Serial propagation of porcine group C rotavirus (pararotavirus) in a continuous cell line and characterization of the passaged virus. J Clin Microbiol. 1988;26:1277–1282. doi: 10.1128/jcm.26.7.1277-1282.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Smith D R, Fedorka-Cray P J, Mohan R, Brock K V, Wittum T E, Morley P S, Hoblet K H, Saif L J. Epidemiologic herd-level assessment of causative agents and risk factors for winter dysentery in dairy cattle. Am J Vet Res. 1998;59:994–1001. [PubMed] [Google Scholar]
- 40.Terrett L A, Saif L J, Theil K W, Kohler E M. Physicochemical characterization of porcine pararotavirus and detection of virus and viral antibodies using cell culture immunofluorescence. J Clin Microbiol. 1987;25:268–272. doi: 10.1128/jcm.25.2.268-272.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terrett L A, Saif L J. Serial propagation of porcine group C rotavirus (pararotavirus) in primary porcine kidney cell cultures. J Clin Microbiol. 1987;25:1316–1319. doi: 10.1128/jcm.25.7.1316-1319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Theil K W, McCloskey C M. Rotavirus shedding in feces of gnotobiotic calves orally inoculated with a commercial rotavirus-coronavirus vaccine. J Vet Diagn Invest. 1995;7:427–432. doi: 10.1177/104063879500700401. [DOI] [PubMed] [Google Scholar]
- 43.Tian P, Ball J M, Zeng C Q, Estes M K. The rotavirus nonstructural glycoprotein NSP4 possesses membrane destabilization activity. J Virol. 1996;70:6973–6981. doi: 10.1128/jvi.70.10.6973-6981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsunemitsu H, Jiang B, Saif L J. Detection of group C rotavirus antigens and antibodies in animals and humans by enzyme-linked immunosorbent assays. J Clin Microbiol. 1992;30:2129–2134. doi: 10.1128/jcm.30.8.2129-2134.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsunemitsu H, Jiang B, Saif L J. Sequence comparison of the VP7 gene encoding the outer capsid glycoprotein among animal and human group C rotaviruses. Arch Virol. 1996;141:705–713. doi: 10.1007/BF01718328. [DOI] [PubMed] [Google Scholar]
- 46.Tsunemitsu H, Jiang B, Yamashita Y, Oseto M, Ushijima H, Saif L J. Evidence of serologic diversity within group C rotaviruses. J Clin Microbiol. 1992;30:3009–3012. doi: 10.1128/jcm.30.11.3009-3012.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsunemitsu H, Ojeh C K, Jiang B, Simkins R A, Weilnau P A, Saif L J. Production and characterization of monoclonal antibodies to porcine group C rotaviruses cross-reactive with group A rotaviruses. Virology. 1992;91:272–281. doi: 10.1016/0042-6822(92)90189-v. [DOI] [PubMed] [Google Scholar]
- 48.Tsunemitsu H, Saif L J, Jiang B M, Shimizu M, Hiro M, Yamaguchi H, Ishiyama T, Hirai T. Isolation, characterization, and serial propagation of a bovine group C rotavirus in a monkey kidney cell line (MA104) J Clin Microbiol. 1991;29:2609–2613. doi: 10.1128/jcm.29.11.2609-2613.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urasawa S, Hasegawa A, Urasawa T, Taniguchi K, Wakasugi F, Suzuki H, Inouye S, Pongprot B, Supawadee J, Suprasert S. Antigenic and genetic analyses of human rotaviruses in Chiang Mai, Thailand: evidence for a close relationship between human and animal rotaviruses. J Infect Dis. 1992;166:227–234. doi: 10.1093/infdis/166.2.227. [DOI] [PubMed] [Google Scholar]
- 50.Ward L A, Rosen B I, Yuan L, Saif L J. Pathogenesis of an attenuated and a virulent strain of group A human rotavirus in neonatal gnotobiotic pigs. J Gen Virol. 1996;77:1431–1441. doi: 10.1099/0022-1317-77-7-1431. [DOI] [PubMed] [Google Scholar]