Abstract
The single-stranded adeno-associated virus (AAV) genome is flanked by terminal hairpinned origins of DNA replication (terminal repeats [TRs]) that are nicked at the terminal resolution site (trs) by the AAV Rep protein in an ATP-dependent, site-specific manner. Here we determine the minimal trs sequence necessary for Rep cleavage, 3′-CCGGT/TG-5′, and show that this 7-base core sequence is required only on the nicked strand. We also identify a potential stem-loop structure at the trs. Interestingly, Rep nicking on a TR substrate that fixes this trs stem-loop in the extruded form no longer requires ATP. This suggests that ATP-dependent Rep helicase activity is necessary to unwind the duplex trs and extrude the stem-loop structure, prior to the ATP-independent Rep transesterification reaction. The extrusion of origin stem-loop structures prior to nicking appears to be a general mechanism shared by plant and animal viruses and bacterial plasmids. In the case of AAV, this mechanism of TR nicking would provide a possible regulatory function.
The small, single-stranded DNA genome of the adeno-associated virus (AAV) is flanked by terminal repeats (TRs), each folding back on itself to form terminal hairpinned structures (Fig. 1). These structures are the only cis elements required for AAV DNA replication, packaging of replicated genomes into capsids, and site-specific integration of provirus into the host genome (15). Each of these distinct activities requires interaction between the AAV nonstructural Rep proteins and the AAV TRs (12, 28, 37).
FIG. 1.
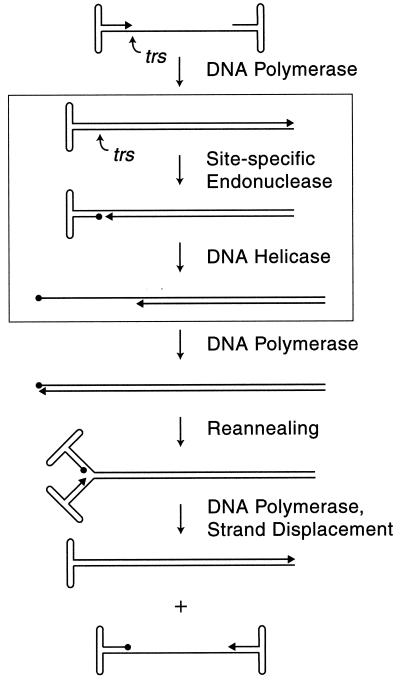
Model of AAV DNA replication. The boxed region illustrates the steps involved in the terminal resolution of AAV viral ends. In vitro, Rep68 is necessary and sufficient for both the site-specific endonuclease and helicase activities required for terminal resolution. The viral 3′ end is indicated with an arrow. Circles depict Rep covalently attached to the viral 5′ end at the trs.
The AAV nonstructural proteins, Rep78 and Rep68, contain both ATP-dependent DNA helicase and site-specific endonuclease activities. In our current model of AAV DNA replication, strand-specific nicking at the terminal resolution site (trs) and subsequent Rep-mediated unwinding within the TR generate a 3′ primer for the initiation of subsequent DNA repair of the TR, a process referred to as terminal resolution (Fig. 1) (26). During this process, the TR is nicked between the two thymidines in the sequence 3′-GGT/TGA-5′, resulting in a 5′ phosphotyrosol linkage between Rep and the nicking site (8, 24, 26). Efforts to characterize Rep trs nicking have been facilitated by the development of an in vitro trs endonuclease assay. Site-specific Rep nicking in this reaction is dependent on ATP hydrolysis and the presence of Mg2+ (or Mn2+) (8, 25, 37). The reason why the Rep transesterification reaction requires ATP hydrolysis has not been clarified.
The ATP dependence of AAV Rep-mediated nicking is a rather novel feature that is shared by the related parvovirus nonstructural protein NS-1 from the mouse minute virus (MVM) (3). However, many other rolling-circle replication initiator proteins do not seem to require ATP for the origin nicking reaction. Notably, the geminivirus Rep protein does not require ATP for in vitro nicking of single-stranded origin substrates. However, this enzyme is unable to nick biologically relevant duplex substrates under these conditions (11). Interestingly, geminivirus replication in vivo is dependent on the formation of a stem-loop structure at the nicking site (20, 27), but the relationship between geminivirus Rep ATPase activity and the formation of this origin stem-loop remains unclear. The Staphylococcus aureus plasmid pT181 also contains an origin stem-loop structure. Extrusion of this stem-loop on supercoiled substrates appears to be dependent on upstream binding of the plasmid-encoded endonuclease RepC, which nicks the origin within the loop region of this structure, but this reaction does not require ATP (9, 17).
Efficient AAV Rep nicking of the hairpinned TR appears to require three sequence recognition elements within the TR. Mutational analysis of the AAV TR has identified a core 22-bp sequence required for stable binding of Rep78 and Rep68 (23). This Rep binding element (RBE) is believed to be the primary recognition element that promotes Rep binding. Interestingly, homologues of this RBE are present at the AAV p5 promoter, in the preferential proviral integration site on human chromosome 19, and within several viral and cellular promoters. Rep78 and Rep68 bind many of these sequences in vitro and appear to regulate AAV transcription through direct interaction with promoter and TR RBEs in vivo (7, 10, 13, 21, 34, 35).
Efficient binding and nicking of hairpinned substrates also require contact between Rep and the small internal palindromes that comprise the tips of the hairpinned TR. The presence of these internal palindromes in hairpinned TRs increases nicking 5- to 50-fold (2, 8, 14, 25). Because the internal palindromes exist in two alternate configurations (flip and flop), it had previously been thought that no specific sequence within this region was recognized. However, Ryan et al. (23) have identified sequence-specific contacts between Rep and a CTTTG motif within the small internal palindromes of the TR. This internal palindrome has a constant position with respect to the trs regardless of the orientation of the TR (flip or flop).
Finally, a specific sequence at the trs itself also appears to be required for nicking. Insertion of a heterologous sequence between the nicking site and the RBE significantly reduces nicking, but nicking occurs at the correct site (25). As yet, however, the specific sequences that comprise the trs have not been identified. Curiously, high-resolution in vitro binding studies do not detect Rep contacts at the trs in the absence of ATP (23). Furthermore, ATP is not an essential cofactor for trs nicking if the trs region is present in a hairpinned TR substrate as single-stranded DNA (25). This observation led us to suggest that ATP-dependent Rep DNA helicase activity is needed to melt the duplex trs to create a single-stranded nicking intermediate. Rep DNA helicase mutants are deficient in trs nicking, supporting this conclusion (31). Additionally, studies of Rep helicase activity have shown that Rep can unwind a duplex DNA substrate provided that it contains an RBE (37).
In our previous work, we identified the sequences within the RBE and the internal palindromes that affect Rep nicking and sequence-specific DNA binding (23). Here we focus on the trs region of the TR. We identify a strand-specific trs core sequence required for Rep-catalyzed nicking through systematic mutation of sequences near the trs. Furthermore, we identify a potential stem-loop structure at the trs. Surprisingly, preferential extrusion of the nicking site stem-loop entirely removes the ATP requirement for Rep nicking. This clearly demonstrates that the AAV Rep protein requires the extrusion of a stem-loop structure containing the nicking site prior to trs cleavage. Furthermore, we conclude that the ATP-dependent Rep DNA helicase activity is necessary to unwind the duplex trs and allow formation of the stem-loop structure prior to nicking.
MATERIALS AND METHODS
Purification of baculovirus-expressed Rep68.
Rep68 was purified to homogeneity from baculovirus-infected Sf9 cells as previously described (37). Rep68 was purified by sequential chromatography on phenyl-Sepharose, single-stranded DNA-cellulose, and DEAE-cellulose. Preparations were more than 99% pure as judged by sodium dodecyl sulfate-acrylamide gel electrophoresis followed by silver staining (37).
DNA substrates.
To construct the synthetic TR substrates used in this study, 100 pmol of two annealed oligonucleotides containing the RBE and the trs sequences was ligated to 500 pmol of a third oligonucleotide containing the secondary structure element. Ligations were done at 32°C in a 100-μl reaction volume containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 25 μg of bovine serum albumin per ml, and 1,600 U of T4 DNA ligase (New England Biolabs). Complete TR constructs were purified from ethidium bromide-stained, 10% denaturing polyacrylamide gels containing 50% urea. DNA concentrations of purified substrates were determined by using the Pico Green fluorometric reagent (Molecular Probes). The entire panel of mutants was assayed together to ensure accurate relative DNA concentrations. TR substrates were 5′ labeled at 37°C in a 10-μl reaction mixture containing 200 fmol of substrate, 70 mM Tris-HCl (pH 7.6), 10 mM MgCl2, 5 mM dithiothreitol, 20 μCi of [γ-32P]ATP, and 20 U of T4 polynucleotide kinase (New England Biolabs). Final concentrations of labeled substrates were determined on the basis of specific activity and confirmed with the Pico Green fluorometric reagent.
trs endonuclease assay.
The trs endonuclease reactions were performed essentially as described previously by Im and Muzyczka (8). The 20-μl reaction mixtures contained 5 fmol of 5′-labeled substrate (average, 104 cpm/fmol) and 0.5 mM ATP. The reaction mixtures were incubated at 37°C for 1 h. Proteinase K-digested reaction products were ethanol precipitated, washed with 70% ethanol, and fractionated on 10% denaturing polyacrylamide gels containing 50% urea. The amount of product formed was determined with a phosphorimager (Fuji).
RESULTS
Determination of the minimal trs.
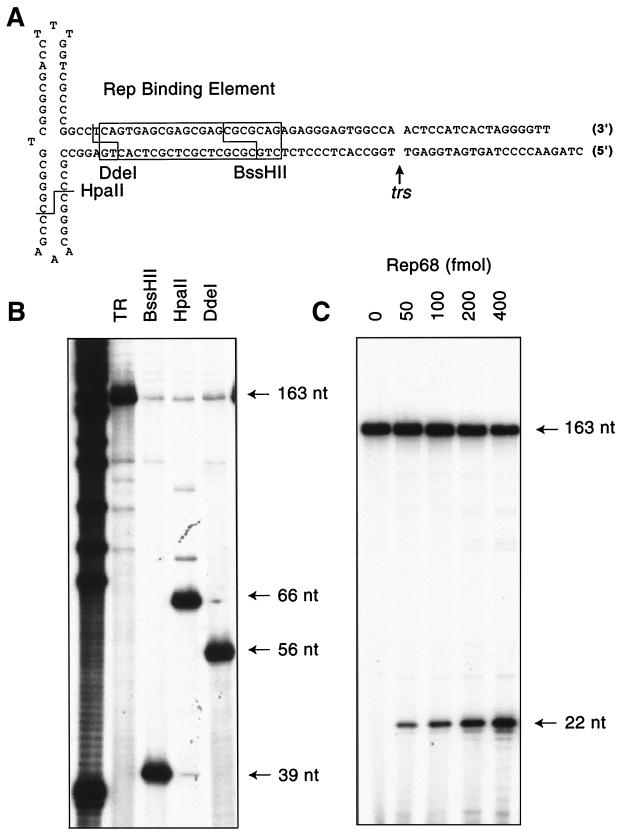
To determine the minimal trs sequences required for Rep68 nicking, a panel of mutant TR substrates was constructed. These substrates consisted of sequential single-base-pair transversions, beginning 8 bp upstream of the nicking site and extending 7 bp downstream. Mutation of these sequences was facilitated by the use of synthetic oligonucleotides. To construct TR substrates, two annealed oligonucleotides containing the RBE and the trs sequences were ligated to a third oligonucleotide containing the terminal palindromes (Fig. 2A and B). Complete TR constructs were purified from ethidium bromide-stained, denaturing polyacrylamide gels, and DNA concentrations of the purified substrates were determined as described in Materials and Methods. The mutants were then 5′-end-labeled with [γ-32P]ATP and T4 polynucleotide kinase, and the final concentrations of labeled substrates were determined on the basis of specific activity and confirmed with fluorometry. Rep trs endonuclease activity on these substrates was assayed in vitro by using homogeneously pure Rep68. All substrates were assayed together, and each substrate was assayed at several Rep concentrations. Typically (Fig. 2C), Rep titrations contained 0.05 to 1 pmol of enzyme in reaction mixtures containing 5 fmol of a given substrate. The results of the nicking reactions with each mutant are shown in Fig. 3.
FIG. 2.
Synthetic AAV TR substrates. (A) The sequence of wt AAV TR substrates used in this study is shown. The boxed region denotes the canonical RBE. The positions of the trs and relevant restriction enzyme sites are also indicated. (B) Restriction enzyme analysis was performed on 5′-labeled wt TR substrate. The products were fractionated on a 10% denaturing polyacrylamide gel. TR indicates undigested substrate. (C) Rep68 endonuclease assays were performed on 5′-labeled wt TR substrate (see Materials and Methods for details). Products were resolved on a 10% denaturing polyacrylamide gel. The positions of the 163-nucleotide (nt) substrate and 22-nucleotide product are indicated. The total amount of Rep68 used in the reaction is indicated above each lane.
FIG. 3.
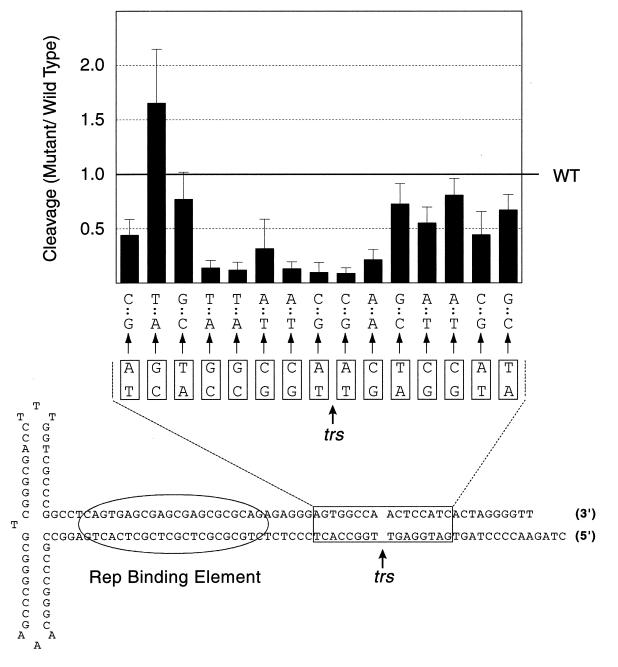
Rep68 endonuclease assays on 1-bp transversion mutants. Individual base pairs were mutated to transversions and assayed for Rep68 endonuclease activity. Boxed regions denote wt AAV sequence. Arrows point to mutant sequence. Mutants were assayed together as a panel that included wt substrate (see Materials and Methods for details). Rep68 nicking activity on wt substrate is indicated with a horizontal line across the graph. (n = 11 for mutations within the sequence 3′-CCGGTTGAGG-5′, and n = 6 for mutations flanking this sequence. Error bars indicate standard deviations of the means.)
Analysis of the mutants revealed a 7-bp sequence that appeared to be the core recognition element required for trs activity. Mutations in this sequence, 3′-CCGGT/TG-5′, reduced Rep68 nicking 3- to 10-fold compared to that with the wild-type substrate. Since this core sequence included the actual nicking site, we assumed that this significant inhibition reflected direct Rep68 interaction with these nucleotides during trs nicking. One of the mutated bases in this sequence, the first G, was not as deleterious as other core mutations, implying that the Rep68 sequence requirements at this position were less stringent.
Most of the substrates with mutations flanking this 7-bp core sequence were also slightly defective for nicking, 20 to 50% lower than wild type, and the results from these 1-bp transversions were consistent with data obtained from overlapping 2-bp transversions (data not shown). This was surprising because we assumed that the inclusion of numerous flanking mutations would define a region of the trs where transversions would not affect Rep68 nicking activity. However, we were unable to define such a region, suggesting that the entirety of the mutated sequences somehow contributed to Rep68 trs nicking. Additionally, the panel of 1-bp transversions included a mutant that was nicked an average of 1.7-fold more efficiently than the wild type (wt). Rep68 nicking assays were performed on this mutant several times, and this substrate was consistently nicked at elevated levels compared to the wt.
Although we felt that our mutation analysis had defined the core trs sequence, 5′-CCGGT/TG-3′, the significance of mutations outside this core remained unclear. There seemed to be at least two classes of mutations, those within the core trs sequence that greatly reduced Rep68 nicking and a second set of mutations that had a more modest effect.
Strand specificity of Rep68 trs nicking.
Though we had identified a core duplex sequence required for Rep68 nicking, we had not determined the strand specificity of Rep contacts within this sequence. Previous work indicated that insertion of heterologous sequences directly opposite the nick site reduced Rep68 cleavage, yet deletion of these same sequences, resulting in a single-stranded trs, did not reduce nicking. However, the single-stranded trs substrates were nicked at two sites with approximately the same efficiency, the trs and a site 11 nucleotides downstream, indicating a possible role for the opposite strand in Rep68 nicking specificity (25). Hence, the contribution of the strand opposite the trs during Rep68 nicking remained unclear. To clarify these inconsistencies and to determine the strand specificity of Rep68 contacts during nicking, a new panel of trs mutants was constructed. These substrates contained a single-nucleotide transversion either on the trs-containing strand or on the opposite strand (Fig. 4). In this way, each nucleotide of a given base pair was independently mutated within the core trs sequence.
FIG. 4.
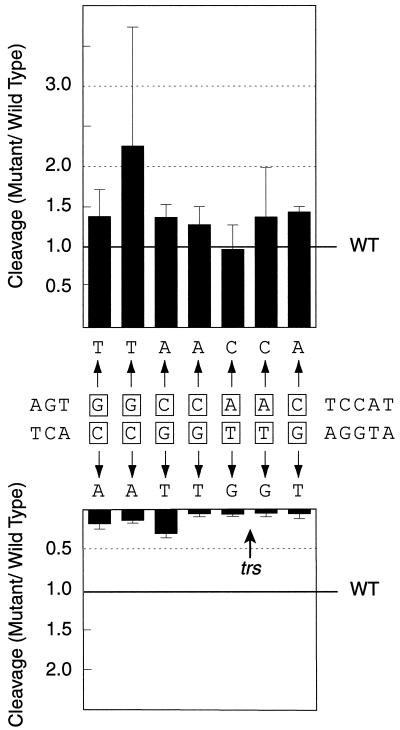
Rep68 endonuclease assays on 1-nucleotide transversion mutants. Individual nucleotides within the 7-bp core sequence were mutated to transversions and assayed for Rep68 endonuclease activity. The sequence mutated in Fig. 3 is shown. Boxed regions denote wt AAV sequence. Arrows point to mutant sequence. Mutants were assayed together as a panel that included wt substrate (see Materials and Methods for details). Relative Rep68 nicking activity on wt substrate is indicated with a horizontal line across the graph. (n = 4 for all mutants. Error bars indicate standard deviations of the means.)
As expected, mutations on the trs-containing strand reduced Rep68 cleavage by 3- to 10-fold. These data were consistent with the results from the single base-pair transversions and confirmed the importance of the 7-base core sequence 3′-CCGGT/TG-5′ in Rep nicking. Surprisingly, transversions on the strand opposite the nicking site did not inhibit nicking. Indeed, Rep68 cleaved the mutants on the trs+ strand at elevated levels compared to wt. Because the standard deviations in the trs mutant experiments were comparable to the increase in nicking, the significance of the improved nicking was not clear. Nevertheless, the elevated Rep68 nicking activity observed on virtually all of the trs+ mutants suggested a general phenomenon. One possible explanation was that these mutations slightly disrupted the duplex trs+, allowing enhanced Rep68 accessibility to the nicking site.
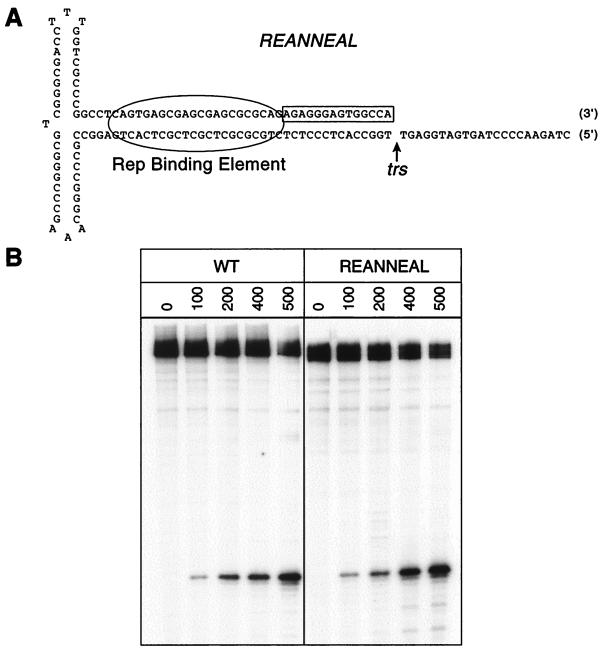
The nicking data for these single-nucleotide transversion mutants indicated that Rep was making strand-specific contacts during nicking. Though inconsistent with the insertion mutants mentioned above, this finding was consistent with Rep68 nicking on substrates containing an extensively single-stranded region extending through the trs (25). This substrate was nicked by Rep68 at wt levels despite the absence of sequences opposite the nicking site. Our data (Fig. 4) were also consistent with results obtained from another partially single-stranded mutant that we constructed. Unlike the extensively single-stranded mutant used in previous studies, our substrate was designed to mimic the 3′ viral terminus (Fig. 5). As such, this substrate was double stranded through the RBE to the trs, but the nicked strand was single-stranded directly distal of the nicking site. Rep68 nicked this mutant an average of 1.6-fold more efficiently than it did the wt (standard deviation, 0.45-fold [Fig. 5]). Yet, unlike the extensively single-stranded substrates, nicking on our substrate mostly occurred at the trs. Hence, it appeared that the strand opposite the nicking site somehow contributed to the specificity of Rep68 nicking, but no particular base within this strand was essential.
FIG. 5.
Rep68 endonuclease reactions on single-stranded TR mutants. (A) The sequence of our substrate designed to mimic the 3′ viral end (REANNEAL) is given. The boxed region indicates additional sequences deleted opposite the trs in the extensively single-stranded substrate described by Snyder et al. (25). (B) Rep68 endonuclease reactions were performed on wt and REANNEAL substrates as described in Materials and Methods. Products were resolved on a 10% denaturing polyacrylamide gel. Numbers above lanes indicate total amounts of Rep68 in the reactions expressed in femtomoles.
Secondary structure at the trs.
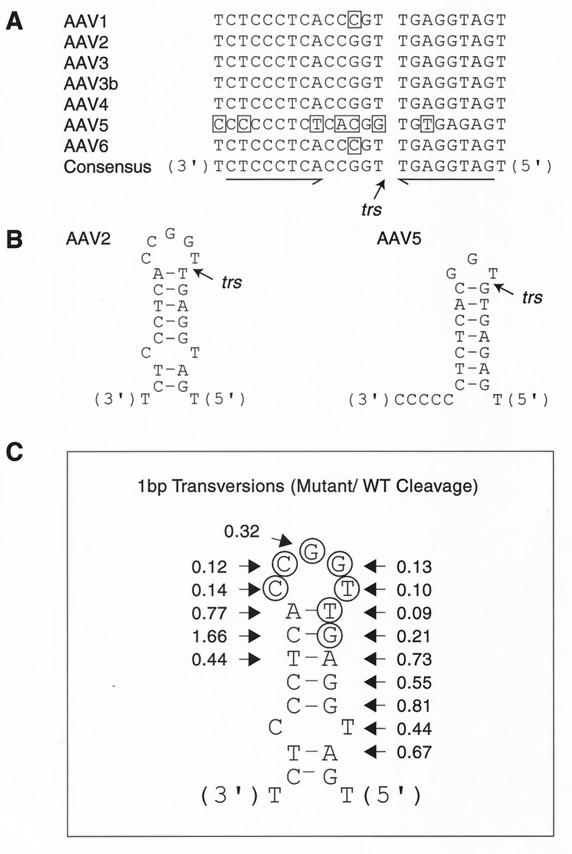
In an attempt to better understand our data, we aligned TR sequences from the seven AAV serotypes. With the exception of AAV5, the region near the trs showed stark conservation among all serotypes. Curiously, this conservation included an inverted repeat flanking the first 5 nucleotides of the trs core sequence (Fig. 6). Additionally, although AAV5 trs sequences were quite disparate with respect to the other serotypes, these sequences also contained an inverted repeat flanking the nick site.
FIG. 6.
Secondary structure at the AAV trs. (A) Sequences near the trs from the various AAV serotypes were downloaded from GenBank and aligned. Boxed nucleotides denote sequence changes from the consensus. Palindromic sequences are underlined. (B) Palindromic sequences near the AAV2 and AAV5 trs depicted as stem-loop structures. Location of AAV-5 trs is from the work of Chiorini et al. (1). (C) Rep68 nicking data from 1-bp transversion mutants have been superimposed on the predicted AAV2 trs stem-loop structure. Circles denote minimal trs as determined by 1-nucleotide mutants.
We, therefore, wondered if these inverted repeats were involved in Rep trs nicking. We predicted that these inverted repeats would form an 8-bp stem structure flanking a 5-nucleotide loop (Fig. 6). This structure seemed energetically improbable, but other origins of DNA replication also contained such improbable structures that appear necessary for in vivo origin function. The geminivirus origin of replication is an 11-bp stem structure flanking an 11-nucleotide loop (20, 27), and the plasmid pT181 origin is a 9-bp stem flanking a 6-nucleotide loop (17). In both examples, the single-stranded loop regions include the origin-nicking site. Extrusion of these structures seems to be dependent on other factors. In the case of pT181, origin stem-loop formation appears to require binding of the RepC endonuclease, which uses the free energy of the circular DNA superhelix to melt the loop region and promote cruciform extrusion (17). By analogy, we hypothesized that the Rep DNA helicase activity might facilitate the extrusion of this energetically unfavorable structure at the trs and that such unwinding on a linear DNA molecule would require ATP.
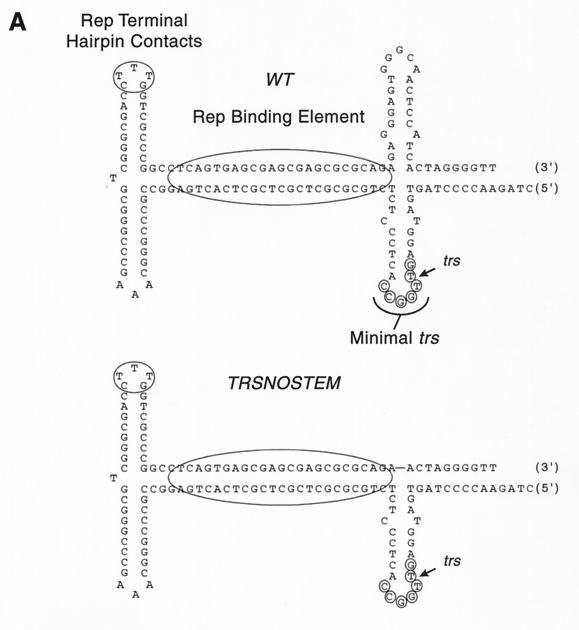
To determine the role of this predicted stem-loop in Rep68 trs nicking, a new mutant wherein the predicted hairpin on the strand opposite the nick site was deleted was constructed (Fig. 7). We reasoned that this mutant would force the sequences on the trs-containing strand to adopt the predicted stem-loop structure (Fig. 7). Surprisingly, Rep not only nicked this mutant, but it nicked this mutant an average of 2.8-fold more efficiently than it did the wt (standard deviation, 0.92-fold). This result confirmed our previous conclusion (Fig. 4) that Rep made contacts with only the trs+ strand (the strand that is cut) during nicking. Furthermore, the enhanced Rep nicking activity on this mutant compared to that on the wt also indicated that the extruded stem-loop was a reaction intermediate. In fact, the significant difference in Rep68 nicking activity between the two substrates implied that the extrusion of the trs stem-loop was probably a rate-limiting step in the trs endonuclease reaction.
FIG. 7.
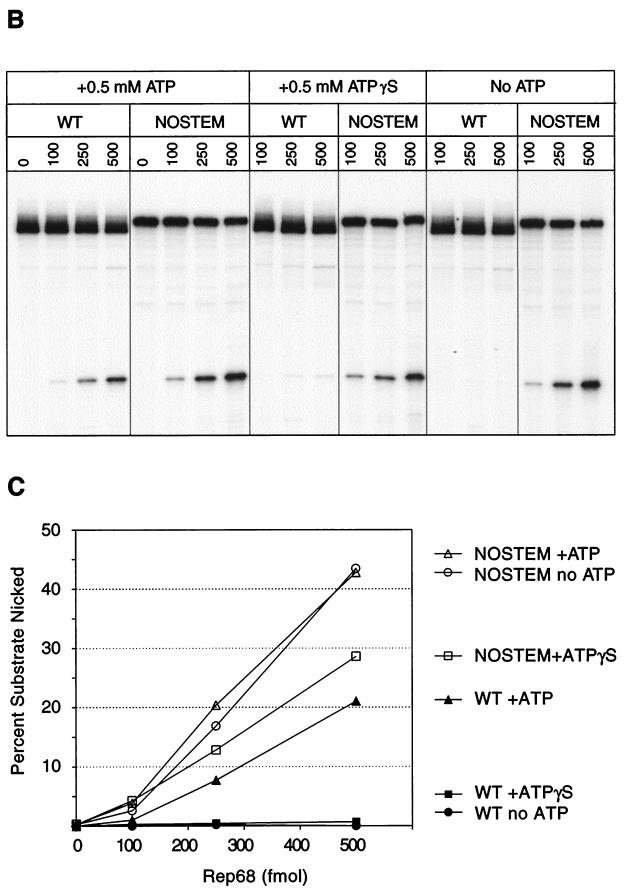
Role of trs stem-loop in Rep68 endonuclease reaction. (A) wt and nostem substrates are depicted after the formation of trs stem-loop structure. The RBE and Rep contacts with the small internal palindromes of the terminal hairpin are indicated with ovals. The minimal trs as determined by 1-nucleotide mutants is also indicated. (B) Rep68 endonuclease reactions were performed on wt and nostem substrates in the presence of 0.5 mM ATP, 0.5 mM ATPγS, or no ATP as described in Materials and Methods. Products were resolved on a 10% denaturing polyacrylamide gel. Numbers above lanes indicate the total amount of Rep68 in the reactions expressed in femtomoles. (C) The gel from panel B was phosphorimaged, and the amounts of substrate and product were determined. These data were graphed to show relative specific activities of Rep68 on the two substrates under the different ATP conditions.
Elimination of the ATP requirement for Rep nicking.
If extrusion of the trs stem-loop is a necessary step prior to nicking, and if the Rep-associated DNA helicase is necessary for extrusion to occur, then we would expect that the trs nostem substrate (Fig. 7) would be ATP independent for nicking. Indeed, trs endonuclease reactions with the stem-loop mutant were no longer dependent on ATP. Rep cleavage of the mutant was essentially the same in both the presence and the absence of ATP, indicating that ATP was not required for the Rep68 transesterification reaction at the trs (Fig. 7). These data were consistent with Rep68 trs assays using the extensively single-stranded substrate that was nicked in the absence of ATP (25). The single-stranded region of this mutant included the inverted repeat necessary for the predicted trs stem-loop formation (Fig. 5). Hence, it is probable that this partially single-stranded substrate could form the trs stem-loop in solution. However, unlike this single-stranded substrate, the nostem mutant used here was nicked only at the correct site.
Since Rep68 did not nick wt substrates in the absence of ATP (Fig. 7), the predicted trs stem-loop did not seem to be energetically favorable. Rather, our data suggested that this structure must be formed and stabilized prior to Rep68-mediated trs cleavage through an active mechanism. The unwinding of the duplex trs and formation of the stem-loop would be facilitated by Rep DNA helicase activity. We have shown previously that Rep can unwind a duplex DNA molecule with no single-stranded regions, provided that the duplex contains an internal RBE (37). Thus, Rep unwinding of the upstream RBE present in our wt substrates would melt the adjacent trs region and extrude the trs stem loop. The requirement for RBE-dependent DNA helicase activity would then account for the ATP requirement during trs nicking.
Interestingly, addition of ATPγS to in vitro trs assays inhibited Rep68 nicking of the nostem mutant compared to that with addition of ATP or no cofactor (Fig. 7). The level of nicking activity in the presence of ATPγS was comparable to what was seen with the wt substrate in the presence of ATP. Since ATPγS is a nonhydrolyzable analogue, it is possible that it binds to Rep and remains associated with the enzyme for extended periods, in contrast to ATP. This would suggest that the Rep complex involved in the actual transesterification reaction at the trs may be inhibited by the presence of ATP bound to the enzyme. Thus, this result suggested the existence of at least two functional Rep68 conformations, the native enzyme conformation and one resulting from ATP binding. Of these, the native form appeared to be the more efficient for nicking, supporting the conclusion that the Rep68 endonuclease activity is ATP independent.
DISCUSSION
Mapping the minimal trs.
During the course of this study, we have analyzed the effect of mutations near the AAV trs on Rep68-catalyzed origin nicking. Our results indicate that a 7-base core nucleotide sequence, 3′-CCGGT/TG-5′, is required for Rep nicking at the trs. This core sequence is strand specific. It is required only on the nicked strand, and it does not matter if the complementary sequence on the opposite strand is intact. Interestingly, the first 5 nucleotides of this core trs sequence are flanked by an 8-bp inverted repeat that appears to form a stem-loop intermediate prior to nicking. Formation of this stem-loop seems to be ATP dependent and is presumably facilitated by Rep DNA helicase activity acting on the nearby RBE. Conversely, Rep does not require ATP to nick the trs once this stem-loop structure is formed, indicating that the endonuclease reaction is ATP independent.
Since the actual nicking site is contained within the single-stranded loop region of this structure, it is possible that a single-stranded nicking site is the only requirement for Rep-mediated trs cleavage in the absence of ATP. However, Snyder et al. (25) did not observe Rep cleavage in the absence of ATP with a mismatched substrate that created a single-stranded bubble within the sequence 3′-GGT/TGA-5′, suggesting that the stem-loop structure itself is important. Moreover, our substrate that fixes the predicted stem-loop in the extruded form is nicked 2.8-fold more efficiently than is the wt, and this reaction results in a singular, authentic reaction product. Finally, this predicted stem-loop structure is conserved among all AAV serotypes despite differences in actual sequences. Thus, our results suggest that Rep trs nicking is a two-step reaction, requiring both a specific sequence and a structure at the trs.
Though we believe we have identified the minimal trs sequence required for Rep68 nicking, the nature of the Rep68 interaction with this core sequence still remains unclear. We expect some of these core nucleotides to directly interact with the Rep68 active site while others may be involved only in the extrusion and stabilization of the trs stem-loop. Although Rep68 cleavage was significantly reduced on all single-nucleotide transversions in the nicked strand, endonuclease activity with these mutants was variable. Individual mutation of nucleotides within the nicked strand sequence 3′-GT/TG-5′ resulted in an average two- to fivefold reduction in Rep68 cleavage activity compared to that with the other single-nucleotide mutations in the core trs sequence. Perhaps this indicates that these four nucleotides interact with the Rep68 active site during nicking and that the other nucleotides within the core sequence are involved in stem-loop formation.
Initially, the data obtained from single-base-pair mutants with mutations flanking the core trs sequence were difficult to interpret. We did not understand why most of the flanking mutations reduced Rep68 trs nicking. However, it now seems likely that these flanking mutations interfered with trs stem-loop formation. Such interference would arise from at least two possible mechanisms. First, mutations within the stem sequence would reduce the number of base pairs within this stem-loop structure and effectively reduce its thermodynamic stability. Second, mutations may also affect Rep helicase interactions with these sequences, preventing the extrusion of the stem-loop. Most of our mutations flanking the core trs sequence would reduce the number of base pairs within the predicted stem region. However, the underlined thymidine in the sequence 3′-GT/TGAGGT-5′ would not form a Watson-Crick base pair after stem-loop extrusion (Fig. 6), and yet Rep68 nicked the 1-bp transversion of this position less than half as efficiently as it did wt. Perhaps this indicates direct Rep interaction with this nucleotide during stem-loop extrusion.
In general, our conclusions are consistent with results from other studies. Wang et al. (33) concluded that the two thymidines flanking the nicking site in the context of uncharacterized upstream sequences were not sufficient for wt Rep68 trs endonuclease activity. Additionally, Snyder et al. (25) concluded that sequences directly across from the nicking site contributed to Rep68 trs endonuclease activity. Yet, the 7-nucleotide insertion mutant used in the Snyder study would have disrupted stem-loop formation opposite the nicking site, reducing the probability of trs stem-loop formation. However, our data do not explain results obtained from another mutant used in this same study. Rep68 nicking activity was reduced when a 2-nucleotide transversion was made directly across from the two thymidines at the nicking site (25). Neither of our single-nucleotide transversions at these two positions had a deleterious effect on Rep68 nicking, suggesting a cumulative phenomenon or experimental error.
Architecture of the AAV TR.
The finding that specific sequences at the trs are involved in Rep-catalyzed nicking is consistent with our working model of Rep interactions with the AAV TR. In this model, site-specific Rep cleavage at the trs requires sequence-specific interactions with the RBE, the trs, and the small internal palindromes that comprise the tips of the hairpinned TR. In addition to these elements, our data indicate that a stem-loop structure near the trs is also involved in Rep-mediated nicking.
The formation of this nicking site stem-loop structure may facilitate several requirements for Rep trs nicking. First, formation of the stem-loop would present the Rep active site with a single-stranded trs. This single-stranded trs appears to be the relevant nicking substrate, since Rep nicks extensively single-stranded TRs and our stem-loop mutant in the absence of ATP. Second, the stem-loop structure would effectively reposition the trs closer to the RBE. This seems important since binding studies do not detect Rep contacts near the trs. It appears that the trs must be brought inward toward the RBE for the Rep active site to associate with the nicking site. Third, the stem-loop itself would help to stabilize trs sequences in the proper conformation and position for nicking.
It appears that two stem-loop structures are present within the TR during Rep nicking, the thermodynamically favorable terminal hairpin and the trs hairpin, resulting in a saddle structure (Fig. 7A). We assume that Rep is making contacts with loop regions in both of these hairpins during nicking since binding assays detect terminal hairpin contacts (23), and our nicking assays detect trs contacts. The involvement of AAV terminal hairpin and trs stem-loop structures in Rep-catalyzed nicking may have ramifications for Rep activity on other substrates. Most notably, Rep78 cleaves human chromosome 19 substrates that include the region of site-specific, AAV proviral integration (29, 30). This site is referred to as AAVS1 and includes an RBE homologue and the trs homologue 3′-GTTG-5′. Urcelay et al. (30) have demonstrated Rep-dependent nicking at the AAVS1 trs homologue and have proposed that the initiation of DNA replication from this site is involved in AAV proviral integration.
Interestingly, AAVS1 does not contain sequences near the RBE and trs homologues capable of forming secondary structures. However, the spacing between the RBE and trs homologues is decreased in AAVS1 compared to that in the AAV TR. This decreased spacing between the AAVS1 RBE and trs may allow RBE-bound Rep to interact with the trs on this substrate in the absence of other structural features. Yet, we assume that the formation of a single-stranded nicking site is required prior to nicking, and so the mechanism of Rep AAVS1 nicking remains unclear. Perhaps this site contains other long-range sequence elements that contribute to Rep nicking by stabilizing the nicking complex or by facilitating a single-stranded nicking site through heterologous protein interaction.
Why is ATP required for Rep68 nicking?
Our data indicate that ATP is required during Rep trs nicking to melt the duplex trs and allow formation of a stem-loop structure at the nicking site. Once the trs stem-loop has been formed, the actual Rep nicking reaction does not require ATP. In fact, the Rep68 transesterification reaction appears to be inhibited by bound ATP. Although Rep68 nicking activity on the stem-loop mutant is the same in the absence or in the presence of ATP, the addition of ATPγS to the reaction reduced nicking activity to the level observed on wt substrates. Since ATPγS is a nonhydrolyzable analogue, it would presumably remain bound to Rep, unlike ATP, which would be hydrolyzed. Thus, it appears that Rep adopts at least two functional conformations during trs nicking, ATP bound and native. The ATP-bound form appears to facilitate Rep helicase activity, and the native form appears to be the more efficient endonuclease.
The modulation of these two Rep conformations through ATP binding suggests a model for Rep interaction with the AAV TR. During nicking, Rep first binds the TR through the RBE. Subsequent ATP binding results in a conformation that allows Rep to reach out from the RBE and make downstream sequence contacts necessary for trs unwinding. After hydrolysis of ATP, Rep would relax into the native conformation. It is unclear if Rep maintains its original contacts and pulls unwound downstream sequences toward the RBE, allowing them to self-anneal into the trs stem-loop, or if stem-loop formation is more passive in nature. In either case, the net result of Rep helicase activity would be the formation of the trs stem-loop. Once formed, this structure presents the single-stranded trs to the native Rep active site.
The Rep proteins of the autonomously replicating parvoviruses also require ATP hydrolysis for in vitro origin nicking (3). The MVM NS-1 replication protein appears to be functionally homologous to AAV Rep (3–5), suggesting that the mechanism of origin nicking is similar in the two systems. Thus, we assume that NS-1 also requires a single-stranded nicking site for cleavage. Yet, the MVM origin sequences do not appear to include secondary structures at the nicking site. Although NS-1 is a helicase (36) and could presumably unwind the duplex MVM origin, the lack of secondary structure at the nicking site implies that MVM origin nicking is distinct from that in AAV.
Unlike AAV, MVM origin nicking requires the accessory DNA binding proteins HMG (at the right origin) and parvoviral initiation factor (at the left origin) (3, 5). Although the exact nature of the interaction between these accessory proteins and the MVM origins is unknown, Cotmore and Tattersall (5) have suggested that these accessory proteins facilitate nicking through origin binding and direct interaction with NS-1. Considering our results, the role of these accessory proteins may be similar to that of the AAV trs stem-loop, i.e., stabilizing a single-stranded nicking intermediate.
General mechanisms of origin nicking.
The similarity between AAV Rep and other viral replication proteins is not limited to the parvoviruses. Geminivirus origin sequences include a predicted stem-loop structure at the nicking site similar to that in AAV. This structure is required for in vivo replication (20, 27), but the mechanism of stem-loop extrusion is unknown. Although it is tempting to draw mechanistic homologies between AAV Rep and geminivirus Rep, it is worth noting that the geminivirus Rep does not exhibit helicase activity (6). This observation raises questions about the functional significance of the geminivirus Rep ATPase activity. This activity is required for viral DNA replication, but the basis of this requirement is unclear (6). Interestingly, the ATPase activities of both AAV and geminivirus Rep proteins are not DNA dependent (6, 37). This rather novel shared characteristic suggests a functional role for ATP-dependent conformational changes outside the context of DNA unwinding.
In the geminivirus system, this conformational switching may be required for interactions between Rep and other proteins. Similar to AAV Rep, geminivirus Rep protein nicks single-stranded origin substrates in vitro. However, geminivirus Rep is unable to nick biologically relevant double-stranded origin substrates (11). Without evidence of geminivirus Rep helicase activity, others have suggested that unidentified accessory proteins are necessary for origin nicking in vivo (6). Such hypothetical proteins would provide the helicase activity for origin unwinding and nicking site stem-loop formation. The geminivirus Rep ATPase activity may facilitate conformation-dependent interactions with these hypothetical proteins.
In contrast to geminivirus, the extrusion of stem-loop structures at other origins of DNA replication appears to require only direct interaction with initiator proteins. The S. aureus plasmid pT181 contains an energetically improbable stem-loop structure required for plasmid DNA replication (32). Like those in AAV and geminivirus, this stem-loop contains the origin nicking site. However, pT181 origin stem-loop extrusion seems to involve both the plasmid-encoded endonuclease RepC and plasmid DNA topology. Upstream binding of RepC increases S1 nuclease sensitivity of origin stem-loop sequences on supercoiled substrates, indicating conversion of these duplex sequences to the stem-loop structure. This S1 nuclease sensitivity is not observed on RepC-bound linear substrates, suggesting that superhelical twisting is necessary to drive the formation of the origin stem-loop (17).
Thus, it appears that AAV, geminivirus, and pT181 share a general mechanism of origin nicking. The sequences of each origin are capable of forming a stem-loop with the actual nicking site located within the single-stranded loop of this structure. Formation of this stem-loop appears necessary for origin nicking in all three systems, indicating that the preferred nicking substrate is single-stranded DNA. In the case of AAV and pT181, origin stem-loop extrusion clearly involves interaction with cognate origin recognition proteins. However, the mechanism of extrusion differs between the two systems. pT181 appears to use the energy associated with superhelical coiling to drive formation of the stem-loop (17). In contrast, the linear AAV genome would require an active mechanism of origin stem-loop extrusion. Our data indicate that this mechanism involves endogenous Rep helicase activity.
Regulation of AAV DNA replication.
The extrusion of the AAV trs stem-loop structure appears to be rate-limiting in Rep68 trs nicking. Zhou et al. (37) recently described the reaction kinetics of Rep68 nicking on wt TR substrates as sigmoidal with respect to enzyme concentration. They concluded that at least a dimer was required for Rep68 endonuclease activity on these substrates. However, these studies would not have measured the reaction kinetics of the endonuclease reaction, per se, but would have measured the kinetics of the rate-limiting step of the reaction. Thus, we conclude that the extrusion and stabilization of the stem-loop structure require at least a dimer of Rep68.
During viral DNA replication, AAV Rep must discriminate between 3′ viral termini and internally replicated TRs (Fig. 1). Nicking of 3′ viral termini would result in dead-end replication products whereas nicking of internal TRs would create a 3′ hydroxyl primer for continued DNA synthesis. Our nicking results from a partially single-stranded TR substrate designed to mimic these viral termini (Fig. 4) indicate that Rep endonuclease is active on 3′ viral termini in vitro. However, analysis of AAV DNA replication by two-dimensional gel electrophoresis indicates that very little nicking of 3′ viral termini occurs in vivo (16). Thus, Rep trs nicking appears to be regulated in vivo, ensuring efficient viral replication.
The multistep Rep trs nicking reaction would allow regulation of trs nicking through modulation of origin stem-loop formation. Since Rep helicase activity appears to facilitate extrusion of this structure, one possible model of trs nicking regulation would involve modulation of Rep helicase activity by phosphorylation. This type of regulation has been observed with the MVM NS-1 protein. Phosphorylation seems to stimulate NS-1 DNA helicase, ATPase, and nicking activities in vitro and to increase viral replication in vivo (18, 19).
Rep trs nicking could also be regulated through direct inhibition of origin stem-loop extrusion. Cellular factors may regulate Rep68 nicking through such a mechanism. Another group has recently identified a cellular protein that binds sequences within the AAV trs stem-loop. This D-stem binding protein (ssD-BP) shows strand-specific binding activity (22). The core binding sequence appears to be 3′-AGTGA-5′, and this sequence appears to be preferentially bound as single-stranded DNA (33). Thus, the binding site of this cellular factor overlaps several nucleotides of the predicted trs stem-loop at the 3′ viral termini. ssD-BP appears to regulate AAV DNA replication by preventing initiation through interaction with its cognate binding site (22). Perhaps binding of ssD-BP also prevents Rep trs nicking of 3′ viral termini by preventing extrusion of the trs stem-loop structure. Such a regulatory mechanism would not prevent Rep nicking of replicated, duplex trs sites since ssD-BP binding shows a preference for a single-stranded binding site.
ACKNOWLEDGMENT
This work was supported by a grant from the National Institutes of Health (RO1 GM35723).
REFERENCES
- 1.Chiorini J A, Afione S, Kotin R M. Adeno-associated virus (AAV) type 5 Rep protein cleaves a unique terminal resolution site compared with other AAV serotypes. J Virol. 1999;73:4293–4298. doi: 10.1128/jvi.73.5.4293-4298.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen J, Cotmore S F, Tattersall P. Parvovirus initiation factor PIF: a novel human DNA-binding factor which coordinately recognizes two ACGT motifs. J Virol. 1997;71:5733–5741. doi: 10.1128/jvi.71.8.5733-5741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotmore S F, Christensen J, Nuesch J P, Tattersall P. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2–3. J Virol. 1995;69:1652–1660. doi: 10.1128/jvi.69.3.1652-1660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotmore S F, Tattersall P. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J Virol. 1998;72:8477–8484. doi: 10.1128/jvi.72.11.8477-8484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desbiez C, David C, Mettouchi A, Laufs J, Gronenborn B. Rep protein of tomato yellow leaf curl geminivirus has an ATPase activity required for viral DNA replication. Proc Natl Acad Sci USA. 1995;92:5640–5644. doi: 10.1073/pnas.92.12.5640. . (Erratum, 92:11322.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermonat P L. Down-regulation of the human c-fos and c-myc proto-oncogene promoters by adeno-associated virus Rep78. Cancer Lett. 1994;81:129–136. doi: 10.1016/0304-3835(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 8.Im D S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 9.Jin R, Zhou X, Novick R P. The inactive pT181 initiator heterodimer, RepC/C, binds but fails to induce melting of the plasmid replication origin. J Biol Chem. 1996;271:31086–31091. doi: 10.1074/jbc.271.49.31086. [DOI] [PubMed] [Google Scholar]
- 10.Kokorina N A, Santin A D, Li C, Hermonat P L. Involvement of protein-DNA interaction in adeno-associated virus Rep78-mediated inhibition of HIV-1. J Hum Virol. 1998;1:441–450. [PubMed] [Google Scholar]
- 11.Laufs J, Traut W, Heyraud F, Matzeit V, Rogers S G, Schell J, Gronenborn B. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc Natl Acad Sci USA. 1995;92:3879–3883. doi: 10.1073/pnas.92.9.3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarty D M, Ni T H, Muzyczka N. Analysis of mutations in adeno-associated virus Rep protein in vivo and in vitro. J Virol. 1992;66:4050–4057. doi: 10.1128/jvi.66.7.4050-4057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarty D M, Pereira D J, Zolotukhin I, Zhou X, Ryan J H, Muzyczka N. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J Virol. 1994;68:4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarty D M, Ryan J H, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaughlin S K, Collis P, Hermonat P L, Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni T H, McDonald W F, Zolotukhin I, Melendy T, Waga S, Stillman B, Muzyczka N. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol. 1998;72:2777–2787. doi: 10.1128/jvi.72.4.2777-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noirot P, Bargonetti J, Novick R P. Initiation of rolling-circle replication in pT181 plasmid: initiator protein enhances cruciform extrusion at the origin. Proc Natl Acad Sci USA. 1990;87:8560–8564. doi: 10.1073/pnas.87.21.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nuesch J P, Corbau R, Tattersall P, Rommelaere J. Biochemical activities of minute virus of mice nonstructural protein NS1 are modulated in vitro by the phosphorylation state of the polypeptide. J Virol. 1998;72:8002–8012. doi: 10.1128/jvi.72.10.8002-8012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuesch J P, Dettwiler S, Corbau R, Rommelaere J. Replicative functions of minute virus of mice NS1 protein are regulated in vitro by phosphorylation through protein kinase C. J Virol. 1998;72:9966–9977. doi: 10.1128/jvi.72.12.9966-9977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orozco B M, Hanley-Bowdoin L. A DNA structure is required for geminivirus replication origin function. J Virol. 1996;70:148–158. doi: 10.1128/jvi.70.1.148-158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereira D J, McCarty D M, Muzyczka N. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J Virol. 1997;71:1079–1088. doi: 10.1128/jvi.71.2.1079-1088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qing K, Wang X S, Kube D M, Ponnazhagan S, Bajpai A, Srivastava A. Role of tyrosine phosphorylation of a cellular protein in adeno-associated virus 2-mediated transgene expression. Proc Natl Acad Sci USA. 1997;94:10879–10884. doi: 10.1073/pnas.94.20.10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan J H, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snyder R O, Im D S, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) Rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder R O, Im D S, Ni T, Xiao X, Samulski R J, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snyder R O, Samulski R J, Muzyczka N. In vitro resolution of covalently joined AAV chromosome ends. Cell. 1990;60:105–113. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- 27.Stanley J. Analysis of African cassava mosaic virus recombinants suggests strand nicking occurs within the conserved nonanucleotide motif during the initiation of rolling circle DNA replication. Virology. 1995;206:707–712. doi: 10.1016/s0042-6822(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 28.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. Adeno-associated virus Rep proteins target DNA sequences to a unique locus in the human genome. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urabe M, Hasumi Y, Kume A, Surosky R T, Kurtzman G J, Tobita K, Ozawa K. Charged-to-alanine scanning mutagenesis of the N-terminal half of adeno-associated virus type 2 Rep78 protein. J Virol. 1999;73:2682–2693. doi: 10.1128/jvi.73.4.2682-2693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Urcelay E, Ward P, Wiener S M, Safer B, Kotin R M. Asymmetric replication in vitro from a human sequence element is dependent on adeno-associated virus Rep protein. J Virol. 1995;69:2038–2046. doi: 10.1128/jvi.69.4.2038-2046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker S L, Wonderling R S, Owens R A. Mutational analysis of the adeno-associated virus type 2 Rep68 protein helicase motifs. J Virol. 1997;71:6996–7004. doi: 10.1128/jvi.71.9.6996-7004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P Z, Projan S J, Henriquez V, Novick R P. Origin recognition specificity in pT181 plasmids is determined by a functionally asymmetric palindromic DNA element. EMBO J. 1993;12:45–52. doi: 10.1002/j.1460-2075.1993.tb05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X S, Qing K, Ponnazhagan S, Srivastava A. Adeno-associated virus type 2 DNA replication in vivo: mutation analyses of the D sequence in viral inverted terminal repeats. J Virol. 1997;71:3077–3082. doi: 10.1128/jvi.71.4.3077-3082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weger S, Wistuba A, Grimm D, Kleinschmidt J A. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep proteins. J Virol. 1997;71:8437–8447. doi: 10.1128/jvi.71.11.8437-8447.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weitzman M D, Kyostio S R, Kotin R M, Owens R A. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson G M, Jindal H K, Yeung D E, Chen W, Astell C R. Expression of minute virus of mice major nonstructural protein in insect cells: purification and identification of ATPase and helicase activities. Virology. 1991;185:90–98. doi: 10.1016/0042-6822(91)90757-3. [DOI] [PubMed] [Google Scholar]
- 37.Zhou X, Zolotukhin I, Im D S, Muzyczka N. Biochemical characterization of adeno-associated virus Rep68 DNA helicase and ATPase activities. J Virol. 1999;73:1580–1590. doi: 10.1128/jvi.73.2.1580-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]