Abstract
Kaposi’s sarcoma (KS)-associated herpesvirus (KSHV) is a lymphotropic virus strongly linked to the development of KS, an endothelial cell neoplasm frequent in persons with AIDS. Reactivation from latency in B cells is thought to be an important antecedent to viral spread to endothelial cells during KS pathogenesis. Earlier experiments have posited a role for the transcriptional activator encoded by KSHV open reading frame 50 (ORF50) in such reactivation, since ectopic overexpression of this protein induces reactivation in latently infected B cells. Here we have explored several aspects of the expression, structure, and function of this protein bearing on this role. The ORF50 gene is expressed very early in lytic reactivation, before several other genes implicated as candidate regulatory genes in related viruses, and its expression can upregulate their promoters in transient assays. The protein is extensively phosphorylated in vivo and bears numerous sites for phosphorylation by protein kinase C, activators of which are potent stimulators of lytic induction. The C terminus of the ORF50 protein contains a domain that can strongly activate transcription when targeted to DNA; deletion of this domain generates an allele that expresses a truncated protein which retains the ability to form multimers with full-length ORF50 and functions as a dominant-negative protein. Expression of this allele in latently infected cells ablates spontaneous reactivation from latency and strikingly suppresses viral replication induced by multiple stimuli, including phorbol ester, ionomycin, and sodium butyrate. These results indicate that the ORF50 gene product plays an essential role in KSHV lytic replication and are consistent with its action as a putative molecular switch controlling the induction of virus from latency.
Kaposi’s sarcoma (KS)-associated herpesvirus (KSHV; also called human herpesvirus 8) has been established as a key factor in the pathogenesis of both AIDS-related and classical KS (10, 49). KS is a complex neoplasm characterized by marked hyperplasia of spindle cells of endothelial lineage and striking concomitant neoangiogenesis (45). Taxonomically, KSHV belongs to the gamma, or lymphotropic, subfamily of herpesviruses (46). Consistent with this fact, in the peripheral blood of KS patients, viral DNA is found principally in CD19+ B cells (1, 56) (and possibly other mononuclear cells) (7); KSHV is also intimately associated with diseases of abnormal lymphoproliferation, including multicentric Castleman’s disease and primary effusion lymphoma (12, 52). Infection by KSHV precedes KS development, with latent infection being established well before progression to full-blown disease (37, 39). In KS tumors, viral DNA resides primarily in endothelial lineage-derived spindle cells (9, 53).
Although KSHV infection of spindle cells is predominantly latent, two lines of evidence support the inference that active, lytic viral replication is also important in KS development. First, treatment of AIDS patients at risk for KS with ganciclovir, a drug which blocks lytic but not latent KSHV replication, strikingly decreases the incidence of KS development (36). Second, the viral load in peripheral blood mononuclear cells increases with progression to clinical KS (1, 56). Taken together, these findings suggest that reactivation from latency and subsequent lytic replication are important events in KS pathogenesis. Such events would be expected to be augmented by AIDS and likely represent an important part of the mechanism underlying the association between human immunodeficiency virus infection and KS.
Lytic reactivation can be envisioned to relate to KS pathogenesis in at least two ways. First, since KSHV is a lymphotropic virus but KS is an endothelial cell neoplasm, viral reactivation within and dissemination from the lymphoid reservoir must occur in order to recruit endothelial cell targets. Second, the KSHV genome encodes numerous homologues of cellular signaling molecules; many of these genes are expressed as lytic cycle genes (28, 40, 46, 48, 54). Several such genes (e.g., those encoding viral macrophage inflammatory protein II and viral G-protein-coupled receptor) have been shown to promote neoangiogenesis (2, 4, 8)—one of the signature features of KS histology—in a paracrine fashion. Since a small percentage of KS spindle cells have been shown to support the lytic cycle (53, 54), local reservoirs of such paracrine factors exist in the tumor and could contribute directly to the angiogenic and inflammatory components of the lesion. Thus, understanding the mechanisms involved in the reactivation of KSHV from latency is important for a full understanding of the natural history of KSHV infection.
Although the molecular events in the reactivation of latent KSHV are just beginning to be understood, considerable insight has been gained from the study of similar issues for the distantly related Epstein-Barr virus (EBV). A critical event in the reactivation of EBV is the expression of the Zta (also called Z or Zebra) protein, a known transcriptional activator. The central role of Zta in reactivation was established by a number of experimental findings: (i) ectopic expression of Zta triggers lytic reactivation in B cells (15); (ii) Zta mRNA is one of the first viral mRNAs expressed after reactivation in latently infected B cells (reviewed in reference 38); (iii) Zta can activate expression from delayed-early (DE) EBV promoters required for viral DNA replication (14, 26, 27, 50); and (iv) a dominant-negative variant of Zta called RAZ inhibits lytic reactivation (20).
Significantly, EBV also expresses a second immediate-early (IE) protein, termed Rta (or R), but Rta had until recently not been considered a likely switch protein in B cells for the following reasons: (i) overexpression of Rta with a weak promoter does not stimulate reactivation of latent EBV (16); and (ii) Rta appears to be a downstream target of Zta transactivation (19, 29, 30, 51). However, Rta overexpression can reactivate EBV in an epithelial cell model of latency (57), and a recent report demonstrated that forced expression of Rta from a stronger promoter can reactivate latent EBV in B cells (43). These results indicate that even in B cells, EBV Rta can occasionally function as a switch protein or that it is a key downstream effector of switch protein action.
KSHV expresses proteins with distant amino acid homology to the EBV lytic transactivators Zta, Rta, and M; these proteins are encoded by the K8, open reading frame 50 (ORF50), and ORF57 loci, respectively (46). One or more of these are likely candidates for switch proteins involved in the control of lytic reactivation. We (34) and others (55) have recently shown that the KSHV Rta homologue (ORF50) can induce markers of lytic reactivation in B-cell models of KSHV latency. Our model system includes the PEL cell line BCBL-1, which harbors the viral genome in a latent state. Upon treatment with inducing agents, such as 12-O-tetradecanoylphorbol-13-acetate (TPA) or sodium butyrate, the lytic gene cascade is induced, culminating in viral replication, cell lysis, and release of virions (44). Ectopic expression of ORF50 in BCBL-1 cells (34) (or in BC-1 cells [55]) induces the expression of DE (ORF59) and late (K8.1) lytic cycle proteins and cytopathic effects in a manner quantitatively similar to that of phorbol ester treatment. We have also demonstrated that ORF50 is a nuclear protein that can directly transactivate KSHV DE but not late promoters (34).
These findings suggest that this gene product may be a key switch protein in KSHV lytic reactivation. However, because the key experiments implicating ORF50 involve artificial overexpression of the ORF50 gene, we sought additional evidence that would support a role for the protein in this process. Here we demonstrate that the expression of the ORF50 mRNA is significantly upregulated before the onset of expression of the major Zta (K-bZIP) and M (ORF57) homologues. In accordance with these induction kinetics, we demonstrate that ORF50 can strongly activate the promoters of K-bZIP and ORF57 in a dose-dependent manner. The ORF50 protein is heavily phosphorylated and bears numerous consensus sites for phosphorylation by protein kinase C, activators of which are potent inducers of KSHV lytic replication. By identifying a strong carboxy-terminal activation domain in the protein, we have been able to construct a dominant-negative ORF50 allele. Expression of this allele in BCBL-1 cells ablates spontaneous lytic reactivation and strongly inhibits the induction of KSHV replication by other known inducers (e.g., TPA, ionomycin, and sodium butyrate). These data provide conclusive evidence that the ORF50 protein is essential for the reactivation of KSHV in B cells and are consistent with the suggestion that it is a molecular switch.
MATERIALS AND METHODS
Plasmids.
All plasmids were propagated as described previously (34).
pcDNA3-gORF50, described elsewhere (34), is a KSHV genomic clone which expresses the full-length ORF50 protein.
pBS-1.3 contains the 5′ ORF50 1.3-kb SacI fragment cloned into the SacI site of pBluescript II KS(+). pBS-2.3 contains the 2.3-kb KpnI/EcoRI fragment containing 3′ ORF50, K8, and K8.1 sequences cloned into the KpnI/EcoRI sites in pBluescript II KS(+). pBS-3.9 contains the 3.9-kb SacI fragment containing 3′ ORF50, K8, K8.1, ORF52, and ORF53 and 5′ ORF54 sequences cloned into the SacI site of pBluescript II KS(+).
pTA-Adv-N50 contains the product of 5′ rapid amplification of cDNA ends (RACE) analysis of the ORF50 transcript (see below).
pGem3-FLc50 was constructed by first subcloning the EcoRI fragment from pTA-Adv-N50 into pGem3 (Promega). The remainder of ORF50 was then added by ligation of the EagI/XbaI fragment from pBS-Sal7 (34). pcDNA3-FLc50 was constructed by subcloning the EcoRI fragment from pGem3-FLc50 into pcDNA3 (Invitrogen).
pCMV-myc-nuc-50ΔSTAD expresses amino acids (aa) 1 to 530 of ORF50 fused C terminally to three copies of the simian virus 40 (SV40) large T antigen nuclear localization signal (NLS) and a myc epitope tag. A PCR product spanning nucleotides (nts) 1 to 1590 of ORF50 was digested with EcoRV and XhoI (which had been introduced by the primers) and cloned into pCMV-myc-nuc (Invitrogen) that had been digested with NcoI, Klenow fragment blunted, and then digested with XhoI. pV5-50ΔSTAD expresses the same truncation of ORF50 fused C terminally to the V5 epitope and a histidine tag. The plasmid was generated by cloning the same PCR product as that used above into the EcoRV/XhoI sites of pcDNA3.1/V5-His A (Invitrogen). pGem3-50ΔSTAD expresses aa 1 to 514 of ORF50 in rabbit reticulocyte lysates (RRL). A PCR product spanning nts 1 to 1552 of ORF50 was digested with EcoRV and SalI (which had been introduced by the primers) and cloned into pGem3 that had been digested with the same enzymes. The 3′ primer introduced an in-frame stop codon as well.
N-terminal fusions to the Gal4 DNA binding domain were made with plasmid pSG0 (18) (see Fig. 5A for diagrams of polypeptides expressed from each). pSG-C50 was constructed by subcloning the 3′ ORF50 XmnI/SalI fragment into pSG0 that had been digested with SmaI and SalI. pSG-FL50ΔBam was constructed by inserting the BamHI/XbaI fragment of pcDNA3-FLc50 into pSG0 that had been digested with the same enzymes. pSG-FL50ΔSTAD was constructed by generating a PCR product spanning nts 1 to 1590 of the ORF50 cDNA; the primers introduced 5′ EcoRV and 3′ SalI sites. The fragment resulting from digestion with these enzymes was cloned into pSG0 that had been digested with SmaI and SalI. pSG-FL50 was constructed by inserting the EcoRV/BamHI fragment from the above-described PCR product into pSG-FL50ΔBam that had been digested with SmaI and BamHI.
FIG. 5.
Analysis of the transcriptional activation domain of ORF50. (A) Gal4-ORF50 fusions. The top of the figure shows a schematic of the ORF50 polypeptide. Putative domains are indicated as follows: LZ, leucine zipper; ST, serine/threonine-rich region; AD, activation domain. Below this schematic are representations of the four ORF50 variants fused N-terminally to the DNA binding domain of Gal4. Amino acid numbers indicate the end of each truncation. (B) ORF50 contains a potent carboxy-terminal activation domain. CV-1 cells were transfected with 3 μg of pGal4-tk-luc (see the text) and 1 or 3 μg of each of the indicated plasmids. Cell extracts were analyzed as described in the legend to Fig. 4. All activation levels were normalized to that of pSG0 (Gal4 alone), here given as 1×. Error bars show standard deviations. (C) The ORF50 activation domain functions in B cells and spindle cells. BJAB and SLK cells were transfected and extracts were analyzed as described in the legend to Fig. 4. Error bars show standard deviations. (D) Expression of Gal4 fusion proteins in CV-1 cells. CV-1 cells were transfected with the indicated vectors, and cells were harvested and extracts were analyzed as described in the legend to Fig. 3. Positions of molecular weight markers are indicated.
pcDNA3-gZ was cloned by digesting pBS-3.9 with SacI, blunting with T4 DNA polymerase, and digesting with KpnI. The resulting 3.7-kb fragment was inserted into pcDNA3 that had been digested with KpnI and EcoRV. pBS-Z-LZ was cloned by removing the HindIII/PstI fragment (containing exon 3 of K-bZIP) and cloning it into pBluescript II KS(+) that had been digested with the same enzymes.
KSHV DE reporter plasmids pGL3-TK, pGL3-pol, pGL3-DBP, and pGL3-nut1 were all described elsewhere (34). pORF57-GL3 contains the BamHI/ApaLI fragment from upstream of ORF57 cloned into pGL3-basic (Promega). pK-bZIP-GL3 contains the 0.7-kb SacI/AflIII fragment from upstream of K8 cloned into the SacI/MluI sites of pGL3-basic.
pGal4-tk-luc contains five binding sites for Gal4 linked to the herpes simplex virus thymidine kinase (TK) TATA box in pGL3-basic.
pnut24 and pcDNA3.1lacZ were described elsewhere (34).
All PCR-generated plasmids were sequenced to confirm that no mutations had been introduced.
Cell lines and transfections.
The cell line BCBL-1 was propagated and maintained as described previously (44). Electroporations were performed as described previously (34) but with the following modifications. After one wash in phosphate-buffered saline (PBS), cells were resuspended in unsupplemented minimal RPMI 1640 medium at a density of 2.2 × 107 cells/ml. Cell volumes of 0.45 ml were placed in electroporation cuvettes (0.42 cm), and DNA was added. Cells were electroporated at 960 μF and 200 mV and then transferred to 20 ml of complete RPMI medium. For induction experiments, 20 μg of the putative inducing plasmid and 4 μg of pnut24 were electroporated.
The B-cell lymphoma cell line BJAB was propagated, maintained, and transfected in a manner similar to that used for BCBL-1 cells, except for the use of 960 μF and 250 mV.
The human diploid endothelial cell line SLK (25) was propagated, maintained, and transfected as described previously (47). For each transfection, 1 μg of reporter plasmid and 1 or 2 μg of transactivator were used.
CV-1 cells were propagated, maintained, and transfected as described previously (34). Cos-7 cells were propagated and maintained similarly.
In all transient transfection experiments, pcDNA3 was used as a filler plasmid to normalize total DNA per transfection. pcDNA3.1lacZ, expressing β-galactosidase, was included in each transfection as an internal control. Data reported are representative of more than one experiment with each transfection performed in triplicate.
Luciferase and β-galactosidase assays.
For the BJAB cell line, cells were collected by centrifugation, washed once with 10 ml of PBS, and washed once with 1.4 ml of PBS. The pellet was resuspended in 0.2 ml of reporter lysis buffer (Promega), debris was spun out, and the extracts were transferred to new tubes. Extracts for SLK and CV-1 cells were prepared as described previously (34). BJAB, CV-1, and SLK extracts were analyzed as described previously (34).
Northern blotting.
Total RNA from BCBL-1 cells was purified as described previously (28). The Northern blot data reported here represent RNA from a single induction, separated on one of three gels prepared in triplicate. A 15-μg quantity of total RNA per time point was blotted and hybridized as described previously (28). Blots were sequentially probed for transcripts, and representative results are reported.
Specific information concerning the double-stranded (ds) probes can be obtained by contacting the authors.
Single-stranded probes were generated and hybridized as described previously (28). Sense K-bZIP transcripts were detected with an antisense probe transcribed from the NarI/HindIII fragment of exon 3. Sense ORF50 transcripts were detected with an antisense probe transcribed from the 5′ exon 2 SacI fragment; antisense ORF50 transcripts were detected with a sense probe transcribed from the same fragment.
cDNA cloning.
A cDNA library prepared from lytically induced BCBL-1 cells (58) was probed with four ORF50 probes (three 5′ probes and one 3′ probe; contact the authors for specific information); probe synthesis and hybridization were performed as for ds probe Northern hybridization (see above). Three rounds of plaque purification were performed before sequencing of rescued phagemids.
RACE.
5′ RACE was performed with a 5′/3′ RACE kit (Boehringer Mannheim Biochemicals) in accordance with manufacturer recommendations. Briefly, total cellular RNA was isolated from BCBL-1 cells which had been stimulated with TPA for 12 or 48 h. Reverse transcription was performed with a 25-base oligonucleotide proceeding antisense beginning at nt 73030. The resulting cDNAs were purified with QiaQuick columns (Qiagen). Two successive rounds of PCR were performed with an oligo(dT) anchor primer with nested ORF50-specific primers. Products were cloned by using an AdvanTAge PCR cloning kit (Clontech), and 31 products were sequenced.
S1 nuclease assays.
Total RNA was isolated from BCBL-1 cells stimulated with TPA for 48 h or from BJAB cells. An S1 assay kit (Ambion) was used in accordance with manufacturer recommendations. RNA (10 μg) was hybridized to 105 cpm of a 50-base oligonucleotide which spanned either the ORF50 start site at nt 71560 or the splice acceptor at nt 72572. Probes were end labelled with [γ-32P]ATP and T4 polynucleotide kinase, and hybridization was carried out overnight at room temperature. Digestions were performed with 1:200 dilutions of the supplied S1 nuclease. Products were visualized by separation by 10% denaturing gel electrophoresis. Sequencing ladders for sizing were generated with a Thermo-Sequenase kit (Amersham).
Primer extension.
Total RNA was prepared as described above, and primer extension reactions were performed as described previously (3). The antisense primer used was a 35-mer oligonucleotide starting at nt 72797 or a 40-mer oligonucleotide starting at nt 72830. RNA (10 μg) was hybridized with the oligonucleotides at 56°C for 5 min. Reverse transcription was carried out for 1 h at 42°C with 5 U of avian myeloblastosis virus reverse transcriptase (RT; Boehringer). Products were separated by 10% denaturing polyacrylamide gel electrophoresis (PAGE). Sequencing ladders were prepared as described above.
RT PCR.
Total RNA was isolated from BCBL-1 cells (28). After treatment with RNase-free DNase, RNA was precipitated, and 2 μg was annealed to 25 ng of an antisense 21-nt oligonucleotide (whose 5′ end was at nt 74675). Annealing was performed as in the primer extension protocol (see above), except for a 15-min incubation at 65°C. The RT reaction was performed for 1 h at 55°C with 16 U of avian myeloblastosis virus RT. A control reaction was performed in tandem under the same conditions but without the RT. Products were purified with QiaQuick columns. PCRs were carried out with each of three templates and four different primer pairs. The templates were pcDNA3-FLg50, total RNA with RT, and total RNA without RT. The primer pairs were as follows (with the 5′ nucleotide of the 5′ primer first, followed by the 5′ nucleotide of the 3′ primer; specific sequences can be obtained from the authors): pair 1, 71560-72886; pair 2, 72686-73339; pair 3, 73315-73958; and pair 4, 73936-74675. Each template (1 μl) was mixed with each primer pair (50 μM), and PCR was performed for 40 cycles of 94°C for 1 min, 47°C for 1 min, and 72°C for 2 min. One microliter of each genomic product and 6 μl of each total RNA product were visualized by electrophoresis on a 2% agarose gel with ethidium bromide staining. Products were TA cloned and sequenced to confirm sequences.
SDS-PAGE analyses of cellular extracts.
Cos-7 cells were transfected with various plasmids by use of Lipofectamine (Gibco/BRL) in accordance with manufacturer suggestions. At 48 h posttransfection, cells were harvested in 10s buffer (11) supplemented with protease inhibitor cocktail set III (Calbiochem). Extracts were diluted 1:5 in 5× Laemmli sample buffer, boiled for 5 min, and then separated by sodium dodecyl sulfate (SDS)-PAGE.
BCBL-1 or BJAB cells were treated with TPA (34) for 24 h or left untreated, and extracts were prepared and analyzed as described above.
Alkaline phosphatase treatment of cellular extracts.
Cellular extracts were prepared as described above, dialyzed, and treated with 40 U of alkaline phosphatase (Boehringer) in a buffer supplied by the manufacturer (5). Control reactions received only the buffer. After 2 h at 37°C, extracts were diluted 1:5 in 5× Laemmli sample buffer and separated on SDS–8% polyacrylamide gels. RRL-generated ORF50 was also electrophoresed on the same gels. ORF50 protein was detected by Western blotting.
Coimmunoprecipitations.
For coimmunoprecipitations, pcDNA3, pcDNA3FLg50, and pV5-50ΔSTAD were cotransfected into Cos-7 cells in various combinations (all transfections contained a total of 10 μg of DNA), and cells were harvested as described above. One eighth of each extract was removed and used to identify input proteins. Equal volumes of extracts were incubated with anti-V5 antibody (Invitrogen) for 2 h at 4°C with rotation. Forty microliters of protein A-Sepharose (Sigma; 50% solution preequilibrated in 10s buffer) was added, and incubation was continued for 1 h. Beads were washed three times with 0.5 ml of 10s buffer and boiled in Laemmli sample buffer, and immunoprecipitated proteins were detected by Western blotting of SDS-polyacrylamide gels with anti-ORF50 serum (34) or horseradish peroxidase-conjugated anti-V5 antibody (V5 is a 14-amino-acid epitope from P/V proteins of paramyxovirus SV4).
Alternatively, pGem3, pGem3-FLc50, and pGem3-50ΔSTAD were transcribed and translated with a TNT T7 Quick coupled transcription-translation kit (Promega) in various combinations. All reactions contained a total of 2 μg of DNA, and l-[35S]methionine was included to label the translated products. Following incubation, 1/15th of each lysate was removed and used to identify input polypeptides. The remaining lysates were each brought to 0.15 ml in 1× NETN (20 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) supplemented with protease inhibitors. Four microliters of anti-ORF50 serum (34) was added, lysates were rotated at 4°C for 2 h, and protein A beads (equilibrated in NETN) were added. Beads were washed five times with 0.5 ml of NETN and boiled in Laemmli sample buffer, and immunoprecipitated proteins were displayed on SDS–8% polyacrylamide gels. Gels were fixed for 30 min, treated with Amplify (Amersham) for 1 h, dried, and then analyzed by autoradiography.
IVT-T.
For in vitro transcription-translation (IVT-T) of unlabelled proteins, we used the TNT T7 Quick coupled transcription-translation kit with unlabelled methionine substituted for labelled methionine.
Western blotting.
Following electrophoresis, proteins were transferred to nitrocellulose (Schleicher & Schuell) in an electroblotter (Bio-Rad) containing 25 mM Tris base–190 mM glycine–20% methanol. Following transfer, the membrane was probed with anti-ORF50 serum (diluted 1:5,000) (34), anti-Gal4 DNA binding domain antibody (diluted 1:100; antibody RK5C1; Santa Cruz), or anti-c-myc antibody (diluted 1:1,000; Covance); the secondary antibody was detected with an ECL kit in accordance with manufacturer recommendations (Amersham). Washes were performed with PBS–0.05% Tween 20.
Transfection and induction of BCBL-1 cells.
Experiments with TPA- or ionomycin-stimulated cells were analyzed by immunofluorescence as described previously (34) at 72 h postelectroporation. Experiments with sodium butyrate-stimulated cells were analyzed at 36 h postelectroporation.
RESULTS
Multiple, divergent transcripts are expressed from the ORF50 locus.
To begin our comparative analysis of the kinetics of the expression of potential regulators of KSHV reactivation, we first analyzed transcripts of the ORF50 locus (Fig. 1A). In an initial Northern blot analysis of total RNA isolated from BCBL-1 or BC-1 cells (data not shown), we used a ds (non-strand-specific) probe derived from a segment of the ORF50 coding region. This analysis revealed a doublet of mRNAs in the 3.4- to 3.8-kb range, as well as several minor transcripts. All species were upregulated with identical kinetics by TPA and sodium butyrate (data not shown). To identify which transcripts had coding potential for ORF50, we performed Northern blotting with single-stranded probes and total RNA from uninduced or TPA-induced BCBL-1 cells. The right panel of Fig. 1A shows that only one of the transcripts produced from this locus corresponds to sense RNA; this transcript is approximately 3.6 kb long and is upregulated by TPA. The remainder of the transcripts, including a very abundant transcript of ca. 3.4 kb and three minor RNAs of ca. 7.5, 2.0, and 1.3 kb, are transcribed from the antisense DNA template (Fig. 1A, left panel). Both the sense transcript and the major antisense transcript are weakly expressed in uninduced cells, likely because of the small percentage of BCBL-1 cells which spontaneously undergo lytic reactivation (1 to 3%). Currently, we do not know if the antisense transcripts have a coding function or serve in another (e.g., regulatory) capacity.
FIG. 1.
Analysis of transcription of potential lytic cycle regulatory genes in KSHV. (A) Multiple divergent transcripts are expressed in the ORF50 locus. Total RNA from untreated BCBL-1 cells (lane 0) or BCBL-1 cells induced with TPA for 12 h (lane 12) was Northern blotted and probed with single-stranded probes detecting antisense (left) or sense (right) ORF50 transcripts. Sizes of RNA molecular weight markers are shown at left. (B) Kinetics of transcript expression of potential lytic regulatory genes. Total RNA was isolated from uninduced and induced BCBL-1 cells as in panel A at the indicated times after TPA addition. Single-stranded probes were used to detect the sense ORF50 and K-bZIP transcripts, and ds probes were used to detect the other transcripts (see Materials and Methods). DBP, DNA binding protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Kinetics of ORF50 expression.
An important predicted property of any putative switch protein is an onset of expression extremely early in the lytic cycle. Most such proteins are encoded by IE genes, classically defined by mRNA expression in the absence of protein synthesis. Two recent analyses of KSHV expression in induced BC-1 cells in the presence of cycloheximide (CHX) concluded that ORF50 mRNA accumulation was resistant to CHX inhibition (54, 59) and assigned the gene to the IE class on that basis. Using BCBL-1 cells, we (43a) and others (59) have been unable to find any KSHV mRNA that stably accumulates after treatment with CHX, even at low drug concentrations, perhaps owing to the pronounced cytotoxicity of the drug in this cell line. Because of this finding, we opted to simply examine the kinetics of ORF50 expression in the absence of protein synthesis inhibitors.
Accordingly, we prepared Northern blots of total RNAs from untreated or TPA-induced BCBL-1 cells at 1, 4, 8, 12, and 24 h postinduction and hybridized these RNAs to probes for the viral genes shown in Fig. 1B. This experiment revealed that the sense ORF50 transcript is easily detected within 1 h of TPA treatment and is abundantly expressed prior to the detectable expression of transcripts corresponding to ORF57, K-bZIP, K3, K5, and the known DE gene for DNA binding protein (DBP). Cellular glyceraldehyde-3-phosphate dehydrogenase served as a loading control and was unchanged at all time points. This very early expression of ORF50 RNA is consistent with its assignment as an IE gene; together with the known ability of ORF50 to directly transactivate the DBP promoter and other DE promoters (34), these data can account for the kinetics of DE gene expression. The order of expression of the K-bZIP, ORF57, K3, and K5 genes (which are the KSHV homologues of known IE genes of other herpesviruses) also raises the possibility that the prior expression of ORF50 may augment the expression of these genes as well (see Discussion). Sun and colleagues (54) have found a similar temporal relationship between ORF50 and K-bZIP upregulation in BC-1 cells; taken together with our data, these data suggest that the order of expression of these two genes in KSHV infection is opposite that in EBV infection.
Fine structure of ORF50 mRNA.
In our initial attempts to isolate a cDNA clone for ORF50, we screened an oligo(dT)-primed cDNA library prepared from TPA-induced BCBL-1 cells. From this experiment, we obtained no full-length clones: the longest cDNA (Fig. 2A) extended from a canonical poly(A) site at nt 76714 to nt 74025. This cDNA represented a transcript with four exons; the splice donor and acceptor sites are identical to the splice signals recently described as part of the K-bZIP transcript (22, 33). This finding confirms an earlier report that K-bZIP RNA sequences can be found in a bicistronic arrangement with ORF50 RNA sequences as well as in a monocistronic mRNA (22, 33).
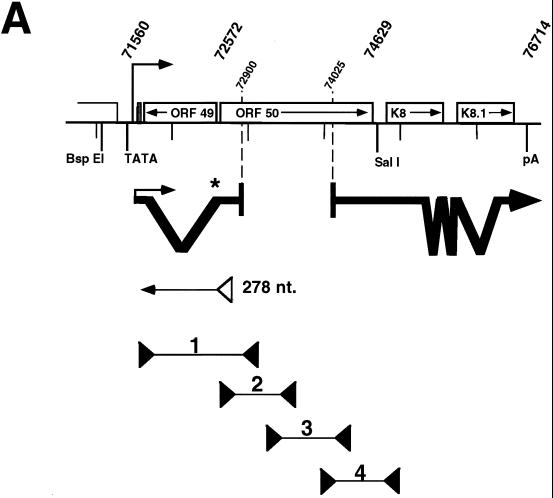
FIG. 2.
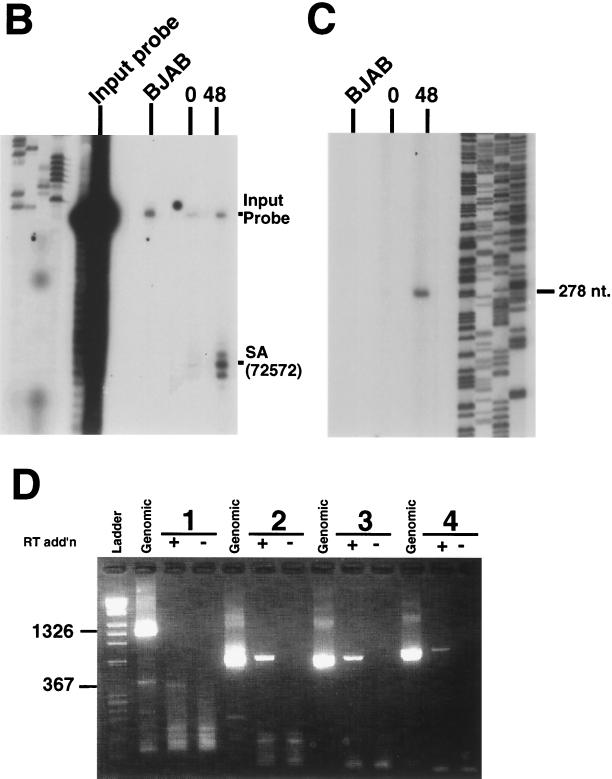
Detailed analysis of transcription in the ORF50 locus. (A) Summary of methods used to analyze ORF50 transcripts. The top of the figure displays a schematic of the genomic features of the ORF50 locus. Transcripts detected by 5′ RACE and cDNA library screening are indicated below the locus, to the left and right, respectively. The asterisk shows the location of the oligonucleotide probe used for S1 nuclease analysis. The open triangle immediately below the 5′ RACE product depicts the primer used in the primer extension reaction. The four lines below that represent the PCR amplification products expected from PCR performed on KSHV genomic DNA with the indicated primers (solid triangles). The rightmost primer was used as the primer for the RT reaction. (B) S1 nuclease analysis of the splice acceptor site at nt 72572. Total RNA from unstimulated BCBL-1 cells, BCBL-1 cells treated with TPA for 48 h, or BJAB cells was analyzed by S1 nuclease digestion with the oligonucleotide indicated in panel and described in Materials and Methods. Products were separated on an 8% denaturing polyacrylamide gel. The locations of the input probe and the digestion product (splice acceptor [SA]) are indicated. (C) Primer extension analysis of the start site of the ORF50 transcript. Total BCBL-1 and BJAB RNA was isolated as described above, and primer extension was performed with the primer indicated in panel A and described in Materials and Methods. Products were visualized as for panel B. A sequencing ladder at the right provides a size reference. (D) RT PCR analysis of ORF50 cDNA. Total RNA was isolated as described above, half was reverse transcribed, and the other half was incubated with the RT primer in the absence of RT. The resulting reactions were divided among four PCRs with the four primer pairs depicted in panel A. A genomic ORF50 clone was used as a control in similar PCRs (Genomic). Products were visualized by agarose gel electrophoresis and ethidium bromide staining. Numbers at the top indicate the primer pairs; +, RT addition; −, RT omission. The ladder is a mixture of phage lambda DNA HindIII and phage φX174 DNA HaeIII fragments. The molecular sizes given at left refer to the genomic and cDNA amplification products generated with primer pair 1.
To locate the 5′ end of the nascent ORF50 transcript, we performed 5′ RACE by using an RT primer lying within the contiguous open reading frame located downstream of the initiator AUG identified in the initial genomic sequencing of virus. Total RNA was harvested from induced BCBL-1 cells at 12 and 48 h after TPA addition, and 31 5′ RACE clones were sequenced. Of these, 10 corresponded to the clone shown in Fig. 2A [18 others were produced by mispriming of the poly(dT) primer, and 3 were the result of strong stops in the 5′ end of the transcript]. These experiments revealed that the 5′ end of the RNA contained a single splice, with a splice donor at position 71613 and a splice acceptor at position 72572. The transcript initiated at position 71560, 23 nts downstream of a potential TATA box; its first AUG was located at position 71596. Thus, the 5′ exon of ORF50 is 53 nts long and adds 60 amino acids to the N terminus of the ORF50 protein predicted by the genomic KSHV sequence.
In order to confirm the splice acceptor site, we performed S1 nuclease analysis with BCBL-1 cell total RNA by using a 50-base oligonucleotide straddling nt 72572. Figure 2B shows that a 25-nt band corresponding to the correct splice site resulted; no degradation products were evident in similar analyses of total RNA from BJAB cells. Moreover, there was no protection of the full-length probe above the background level; this result suggests the absence of stable ORF50 transcripts that are unspliced in this region.
To confirm the start site found by 5′ RACE, we performed primer extension analyses with the same RNA samples as those shown in Fig. 2B. Figure 2C shows a single, strong band only in lane 3, corresponding to total RNA from cells induced for 48 h. By comparison with a sequencing ladder generated from clones of this genomic DNA region, the RT product was determined to be 278 nts long, corresponding precisely to the start site identified in the 5′ RACE analyses. This start site was confirmed by primer extension analyses with a second (more downstream) primer, as well as by S1 nuclease analysis (data not shown). The start site identified for BCBL-1 cell RNA differs from that found for ORF50 in BC-1 cells (55). However, the first AUG of both transcripts is identical.
The 3′ end of the 5′ RACE clones, at position 72900, and the 5′ end of the longest library-derived cDNA, at position 74025 (Fig. 2A), resulted in a gap of 1,125 nts for which we had no information concerning transcript architecture. Therefore, we performed RT PCR across the entire ORF50 coding sequence to search for any additional splices. Four primer pairs were designed to generate overlapping PCR products following reverse transcription of BCBL-1 cell RNA with the 3′-most oligonucleotide as the RT primer (Fig. 2A; see also Materials and Methods). Figure 2D shows the results of this analysis. Primer pairs 2, 3, and 4 each yielded amplification products indistinguishable in size from the corresponding products of the genomic template amplification, indicating that there were no additional splices within the coding sequence of ORF50. (Primer pair 1 yielded a 367-bp band, consistent with the 959-bp intron identified by 5′ RACE).
Taken together, these data reveal that the sense ORF50 transcript derived from BCBL-1 cells is 3,402 nts long [without the poly(A) tail] and has a single 5′ intron and three 3′ introns. Accounting for the addition of the nontemplated poly(A) tail, this size corresponds well with the sense transcript size predicted by the Northern blots shown in Fig. 1. The transcript is capable of encoding an ORF50 polypeptide of 691 aa.
The ORF50 polypeptide is highly phosphorylated in mammalian cells.
To characterize the polypeptide produced by ORF50, we transfected CV-1 and Cos-7 cells with either an empty expression vector or vectors expressing ORF50 from either a cDNA clone or a genomic fragment. A comparison of the proteins expressed from the ORF50 vectors revealed no obvious difference in mobility analyzed by Western blotting after SDS-PAGE (data not shown). However, Fig. 3A shows that the apparent molecular mass of the ORF50 polypeptide in these analyses is approximately 110 kDa, about 36 kDa higher than the predicted molecular mass of 73.7 kDa. In addition, when the cDNA is transcribed and translated in RRL, the apparent molecular mass is approximately 90 kDa (Fig. 3A). These observations suggest that the ORF50 polypeptide may be altered posttranslationally, resulting in its reduced apparent mobility in denaturing electrophoresis.
FIG. 3.
The ORF50 polypeptide is highly phosphorylated in mammalian cells. (A) Transfected cells. Cos-7 cells were transfected with pcDNA3 (Vector) or pcDNA3-FLg50 (ORF50), and whole-cell extracts were isolated with 10s buffer at 48 h posttransfection (see Materials and Methods). Extracts were incubated with (+) or without (−) CIP (see Materials and Methods). The resulting products were separated on SDS–8% polyacrylamide gels and detected by Western blotting as described in Materials and Methods. The leftmost lane contains the product of IVT-T of ORF50 in RRL. The migration of protein molecular weight standards is indicated at the left. (B) BCBL-1 cells. BCBL-1 or BJAB cells were treated with TPA for 24 h or left untreated and harvested in 10s buffer as described for panel A. Total protein in each extract was quantitated by Bradford analysis, and equal amounts of protein were analyzed as described for panel A. The ORF50 RRL product and extracts from vector- or pcDNA3-FLg50-transfected Cos-7 cells were coelectrophoresed as size references. The migration of protein molecular weight markers is indicated at the left.
Analysis of the ORF50 primary amino acid sequence reveals multiple potential sites of phosphorylation, including a C-terminal region rich in serines and threonines (see Fig. 6), and 20 other consensus sites for phosphorylation by the serine/threonine kinases casein kinase II and protein kinase C (PKC) (data not shown). To determine whether or not the reduced mobility of the ORF50 protein could be explained, at least in part, by phosphorylation, we transfected Cos-7 cells with either empty expression vectors or vectors expressing ORF50, prepared whole-cell extracts, and treated them with calf intestinal phosphatase (CIP) (see Materials and Methods). The ORF50 protein treated with CIP runs at an apparent molecular mass similar to that of the ORF50 protein produced by RRL (compare the first and last lanes in Fig. 3A). (The ORF50 protein treated with phosphatase buffer alone [Fig. 3A, fourth lane] shows no mobility shift relative to that seen with untreated extracts [Fig. 3B].) Since the background band visible in all four lanes of transfected Cos-7 cells shows no difference in mobility upon CIP treatment, we can conclude that the mobility change of the ORF50 protein is not due to a nonspecific proteolytic event occurring under the reaction conditions. Assuming that all phosphates in the ORF50 protein are accessible to CIP action, these data also suggest that RRL are incapable of phosphorylating the ORF50 protein. Therefore, phosphorylation accounts for most but not all of the abnormal migration of this protein in SDS-polyacrylamide gels.
FIG. 6.
The ORF50 activation domain shows sequence conservation with numerous cellular and viral transcription factors. (A) Sequence of aa 486 to 691 of ORF50, representing the peptide fused to the Gal4 DNA binding domain in plasmid pSG0-C50 (Fig. 5A). The putative NLS, a serine/threonine-rich region (S/T-rich), and four conserved repeats (AD-1 to AD-4) are indicated. Underlining represent regions of overlap between adjacent conserved repeats. The asterisk indicates the location of the stop codon. (B) Alignment of the four conserved repeats of the ORF50 activation domain with similar domains found in cellular (Sp1 and CTF) and viral (VP-16, HSV VP16; E1A, adenovirus E1A; HVS, herpesvirus saimiri ORF50) transcription factors. R, genetic designation for EBV ORF50 homolog. Conserved bulky hydrophobic amino acids are boxed. A, B, 2A, 2B, and 2C designate independent domains of each protein (24). The asterisk indicates F442 in VP16 domain A, discussed in the text. (Adapted from reference 24 with permission.)
To examine whether similar phosphorylation events occur in authentic KSHV infection, we examined the electrophoretic mobility of the ORF50 protein in TPA-induced BCBL-1 cells. As shown in Fig. 3B, the ORF50 protein produced in such cells was indistinguishable from the recombinant protein produced in Cos-7 cells.
The ORF50 protein can function in many cell types and can directly transactivate the promoters of ORF57 and K-bZIP.
Since ORF50 expression precedes that of the other putative IE activators of gene expression (ORF57 and K-bZIP) in lytic induction (Fig. 1), we were interested in examining whether its product could directly activate the promoters of those genes. Figure 4A demonstrates that ORF50 strikingly upregulates the promoters of both ORF57 and K-bZIP in transient cotransfections of CV-1 cells. This effect is dose dependent, with the magnitude of the activation reaching 150- to 250-fold—much greater than the 10- to 40-fold that we typically observe for classical KSHV DE genes (e.g., DBP, TK and nut-1 [34]). This ability to directly upregulate the promoters of these regulatory viral genes is highly consistent with the observed kinetics of appearance of their transcripts after TPA stimulation of BCBL-1 cells.
FIG. 4.
Transient transcriptional activation of KSHV promoters by ORF50. (A) ORF50 directly transactivates the promoters of ORF57 and K-bZIP in a dose-dependent fashion. CV-1 cells were transfected with the indicated reporter vectors and increasing amounts of pcDNA3-FLgORF50 (see Materials and Methods). Empty pcDNA3 was used to normalize total DNA. Cell extracts were analyzed for luciferase and β-galactosidase activities 42 to 48 h posttransfection (see Materials and Methods). (B) ORF50 transactivates DE promoters in the human cell lines BJAB and SLK. The indicated promoter constructs were electroporated or transfected into BJAB or SLK cells, together with various amounts of ORF50, as described in Materials and Methods. Cell extracts were analyzed as described for panel A. The maximal amount of activation of each promoter is shown. Error bars show standard deviations.
Since KSHV reactivates from latency both in B lymphocytes and in endothelial (spindle) cells in KS lesions, we analyzed the ability of ORF50 to transactivate its target promoters in two human cell lines corresponding to these sites of tropism. These were the EBV-negative B-cell lymphoma line BJAB and the SLK cell line, a diploid endothelial (spindle) cell line established from a KS tumor biopsy (25). The target promoters tested were those for the TK, DBP, nut-1, and DNA polymerase DE genes. We transfected filler plasmid or increasing amounts of an ORF50 expression vector, together with constant amounts of the respective reporter plasmids, into both cell lines and assayed for reporter gene expression. Figure 4B shows the maximal fold activation by ORF50 for each promoter tested. Although the maximal levels of activation for each promoter are consistently ca. 1.5-fold lower in these human cells than in CV-1 cells, the rank order of activation of the promoters was identical in all three cell lines, and in none of these cell lines was the DNA polymerase promoter upregulated by ORF50 (see also reference 34). We have confirmed that the promoters of ORF57 and K-bZIP are also activated by ORF50 in BJAB cells (data not shown). These data indicate that activation by ORF50 displays little cell type specificity, suggesting that ORF50 function may not be an important determinant of cellular tropism in KSHV infection.
The ORF50 protein contains a potent carboxy-terminal activation domain.
To gain insight into the mechanism of transactivation of ORF50 and the function of ORF50 as a lytic switch in KSHV reactivation, we mapped the activation domain of the protein by fusing various regions of the ORF50 polypeptide downstream of the DNA binding domain of the yeast protein Gal4. The regions of the ORF50 polypeptide that were fused are shown in Fig. 5A. These ORF50 variants included full-length (FL) ORF50, two amino-terminal truncations (FL50 ΔBam and C50), and a carboxy-terminal truncation (FL50 ΔSTAD). Plasmids expressing these constructs were transfected into CV-1 cells together with a luciferase reporter driven by a promoter consisting of five Gal4 elements upstream of the herpes simplex virus TK TATA box; 48 h later, cell extracts were assayed for luciferase expression (Fig. 5B). Both full-length ORF50 and the two amino-terminal truncations of ORF50 significantly activated transcription, while a deletion of the C-terminal 161 aa of ORF50 resulted in a protein incapable of transcriptional activation. (As shown in Fig. 5D, all constructs produced stable immunoreactive fusion proteins.) These results suggest that the principal activation domain of ORF50 lies at its extreme carboxy terminus between aa 531 and 691. This conclusion was confirmed by showing that the C50 construct, containing only 205 carboxy-terminal aa of ORF50, retained the full transactivation function of full-length ORF50 when targeted to DNA as a Gal4 fusion. As expected from the broad host range of wild-type ORF50 activation (Fig. 4B), the Gal4-C50 fusion was fully active in both the BJAB and the SLK cell lines (Fig. 5C).
The primary amino acid sequence of the C50 construct (aa 486 to 691) is shown in Fig. 6A. This region of ORF50 contains numerous charged amino acids interspersed with regularly repeating bulky hydrophobic residues. In fact, this region of the protein contains four overlapping domains, which we have termed AD-1 to AD-4, in which the bulky hydrophobic repeats can be aligned with similar repeats found in the activation domains of a variety of cellular and viral transcription factors (17, 24). These repeats are shown in Fig. 6A, and their alignments with the other transcription factors are shown in Fig. 6B. Phenylalanine 442 in VP16 activation domain A, a critical amino acid required for transcriptional activation by Gal4-VP16, also is indicated in Fig. 6B (17). This position is conserved as a bulky hydrophobic side chain in all of the aligned sequences shown—mutations in F442 create a dominant-negative mutant of Gal4-VP16 (17).
Deletion of the ORF50 protein activation domain creates a dominant-negative mutant.
To activate the transcription of some EBV promoters, the EBV Rta protein must dimerize and bind DNA (21, 23, 35, 41); these functions of EBV Rta localize to its amino terminus (35). Since this region is detectably conserved in KSHV ORF50, we hypothesized that the N terminus of ORF50 would have a similar function. If so, this domain organization would suggest that removal of the carboxy-terminal activation domain from the KSHV ORF50 protein might generate a dominant-negative allele capable of inhibiting wild-type ORF50 function.
Using Gal4-FL50ΔSTAD as a guide, we truncated wild-type ORF50 C terminally at aa 530, and this coding region was fused to an epitope tag from the c-myc gene and three repeats of the SV40 large T antigen NLS (see Materials and Methods). This procedure generated the clone pCMV-myc-nuc-50ΔSTAD. When this plasmid was transfected into Cos-7 cells, the fusion protein (ORF50ΔSTAD) was stably and detectably expressed (Fig. 7B). As expected (Fig. 7A), this mutant was unable to transactivate a cotransfected reporter gene driven by the ORF57 promoter, a promoter known to be strongly activated by wild-type ORF50 (Fig. 4). We then examined whether this mutant could interfere with the activity of wild-type ORF50 by cotransfecting increasing amounts of the mutant together with a constant amount (1 μg) of wild-type ORF50 and the aforementioned reporter. (All transfections were brought to a fixed DNA concentration with vector DNA to control for nonspecific effects of the input DNA.) Figure 7A shows that increasing amounts of ORF50ΔSTAD severely inhibited the ability of wild-type ORF50 to transactivate the ORF57 promoter in a dose-dependent fashion; maximal inhibition approached 95%. ORF50ΔSTAD also comparably inhibited wild-type ORF50 transactivation of the nut-1 promoter (data not shown). As a specificity control, ORF50ΔSTAD had no significant effect on the expression of an irrelevant reporter, pCMV-lacZ, in the presence (data not shown) or absence (Fig. 7A) of wild-type ORF50; this result confirms the finding that the suppression of luciferase observed in Fig. 7 is not the trivial result of cellular toxicity or transcriptional squelching.
FIG. 7.
Deletion of the ORF50 activation domain creates a dominant-negative transcriptional mutant. (A) ORF50ΔSTAD inhibits transactivation of the ORF57 promoter by wild-type ORF50. CV-1 cells were transfected with ORF57-GL3 (P57) and 0 to 12 μg of pCMV-myc-nuc-50ΔSTAD alone (left ordinate; ○) or with 1 μg of pcDNA3-FLg50 (left ordinate; □). Alternatively, 5 μg of pCMV-βgal was transfected with increasing amounts of ORF50ΔSTAD (right ordinate; ×). Empty pcDNA3 was included to normalize total DNA in each transfection. Extracts were prepared and analyzed as described in the legend to Fig. 4. Error bars show standard deviations. (B) ORF50ΔSTAD is expressed in Cos-7 cells. Cos-7 cells were transfected with empty pcDNA3, pCMV-myc-nuc-ΔSTAD, or pcDNA3-FLg50, harvested, and analyzed as described in the legend to Fig. 5D. The numbers at the left indicate the positions of protein molecular weight markers. (C) ORF50ΔSTAD heterodimerizes with wild-type ORF50 in transfected Cos-7 cells. Cos-7 cells were transfected with the indicated plasmids; total DNA was normalized to 10 μg with vector plasmid in all transfections. Extracts were prepared and incubated with anti-V5 antibody (see Materials and Methods). Immunoprecipitated proteins were analyzed by SDS-PAGE, Western blotted, and probed with anti-ORF50 serum. HC, migration of the heavy chain of the antibody in each lane. Input lanes contained one-eighth the volume of total extract before immunoprecipitation. Molecular weight markers are shown at the left. (D) ORF50ΔSTAD heterodimerizes with wild-type ORF50 in RRL. RRL were programmed with the indicated plasmids, and translated products were labelled by including l-[35S]methionine in the reactions. All reactions were programmed with equal amounts of DNA. Lysates were incubated with anti-ORF50 serum (see Materials and Methods), and immunoprecipitated proteins were analyzed by SDS-PAGE and autoradiography of the fixed, amplified, and dried gel. Input lanes contained 1/15th the volume of total lysate before immunoprecipitation. Molecular weight markers are shown at the left.
To explore a possible mechanism for the dominant-negative mutation, we examined whether C-terminally truncated ORF50 mutants could form heterodimers with wild-type ORF50. Cos-7 cells were transfected with an empty vector, the plasmid expressing wild-type ORF50, or plasmid pV5-50 ΔSTAD (see Materials and Methods) alone or in combination, and total DNA was normalized to 10 μg in all transfections by the addition of the empty vector. Wild-type ORF50 was immunoprecipitated only when cotransfected with pV5-ΔSTAD (Fig. 7C, fourth lane); wild-type ORF50 was not immunoprecipitated when expressed in the presence of the filler plasmid. Thus, wild-type ORF50 and ORF50ΔSTAD form heterodimers in Cos-7 cell extracts.
As further evidence for heterodimer formation between the wild-type and dominant-negative proteins, we also generated a clone (pGem3-50ΔSTAD; see Materials and Methods) which expressed a C-terminally truncated protein similar to ORF50ΔSTAD in RRL. In vitro transcripts for the wild-type and mutant chains were translated in RRL, either individually or together, precipitated with an anti-ORF50 antibody, and examined by SDS-PAGE. Figure 7D shows that while the antibody recognized only wild-type ORF50, when the chains were cotranslated, the truncated protein could be efficiently coprecipitated, indicating heterodimer formation. Importantly, the truncated protein was not immunoprecipitated when translated alone. If homodimerization is required for the function of wild-type ORF50, heterodimer formation with the mutant protein could be the basis for the dominant-negative effect of the mutation. However, these results do not exclude other models for the mechanism of this class of dominant-negative mutation (see Discussion).
Transactivation by ORF50 is necessary for lytic reactivation.
Next, the requirement for transcriptional activation by ORF50 in inducing lytic reactivation was tested by ectopically expressing ORF50ΔSTAD in BCBL-1 cells. We have previously established that the ectopic expression of wild-type ORF50 in these cells induces the lytic reactivation of KSHV with an efficiency similar to that of TPA (34). In these assays, we transfect the ORF50 expression vector into BCBL-1 cells together with an expression vector for hepatitis delta antigen, whose expression can be monitored by immunofluorescence to mark successfully transfected cells. At least 1,000 transfected cells are then scored by immunofluorescence for the expression of two markers of KSHV lytic reactivation: (i) ORF59 protein, a DNA polymerase-associated processivity factor expressed in a DE manner (13, 32), and (ii) K8.1 protein, a viral envelope glycoprotein expressed with late kinetics (31, 42). Each of these two proteins was detected with monoclonal antibodies specific for the corresponding marker (34).
Both wild-type ORF50 and pCMV-myc-nuc-50ΔSTAD were transfected into BCBL-1 cells, as was the empty expression vector for each protein (pcDNA3 and pCMV-myc-nuc). The cells were either left untreated or stimulated with TPA at 15 to 18 h posttransfection. Lytic reactivation was scored at 72 h posttransfection by the methods described above. Figure 8A shows the results of this analysis. In the absence of TPA, the characteristic 0.3 to 0.8% of BCBL-1 cells underwent spontaneous reactivation in both of the empty vector-transfected populations. This value for the pcDNA3-transfected cells was set as the background level of reactivation against which the other transfections were compared. After the addition of TPA to similarly transfected cultures, ORF59 protein was induced 10- to 12-fold and K8.1 protein was induced about 7-fold, consistent with our previous observations (34). These results indicate that the expression of the short SV40 NLS-containing peptide from pCMV-myc-nuc does not alter the reactivation characteristics of pcDNA3-transfected cells.
FIG. 8.
Transactivation by ORF50 is necessary for lytic reactivation. BCBL-1 cells were electroporated with 20 μg of activator or empty vector and 4 μg of a vector expressing hepatitis delta antigen. Cells either were left untreated or were treated with TPA (A) or sodium butyrate (nBA) (B) 15 to 18 h postelectroporation. Successfully transfected cells, identified by positive staining for hepatitis delta antigen, were scored 72 h (A) or 36 h (B) postelectroporation for expression of ORF59 and K8.1. At least 1,000 transfected cells were counted for each electroporation. The percentage of transfected cells expressing the respective KSHV lytic marker is indicated above each bar. The bars indicate the fold induction of the lytic markers relative to that in cells electroporated with the control pcDNA3 vector in the absence of TPA.
Wild-type ORF50-transfected cells also showed the expected result—ectopic expression of ORF50 induced ORF59 and K8.1 in a manner indistinguishable from that of TPA. The addition of TPA to cells expressing wild-type ORF50 resulted in an even greater enhancement of reactivation—the percentage of transfected cells expressing both ORF59 and K8.1 was approximately doubled relative to the value obtained with transfection of ORF50 alone or TPA treatment alone. However, electroporation of the ORF50ΔSTAD expression vector resulted in a complete inhibition of detectable spontaneous reactivation in the uninduced cultures. Moreover, ectopically expressed ORF50ΔSTAD potently suppressed the induction of lytic cycle markers by TPA (Fig. 8A) and sodium butyrate (Fig. 8B), inducers with different biochemical mechanisms of action (ORF50ΔSTAD also suppressed induction by ionomycin [data not shown]). Taken together, these results indicate that, under conditions in which ORF50 is produced at natural levels in the context of the intact viral genome, its product is required for the lytic induction of KSHV by all known inducing stimuli.
DISCUSSION
The data presented here support a central role for the ORF50 protein in the lytic reactivation of KSHV in BCBL-1 cells. We have demonstrated that the 3.6-kb ORF50 transcript is one of the first to be expressed after the induction of KSHV replication (Fig. 1B). Its onset of expression precedes even that of the transcripts of other candidate viral IE genes (e.g., ORF57, K3, and K5). This finding suggests that the expression of the KSHV ORF50 protein might augment the expression of these other potential regulatory proteins—an inference supported by data (Fig. 4) showing that the promoters of both ORF57 and K-bZIP are potently upregulated by ORF50 coexpression. However, we do not know if these genes are strictly dependent upon ORF50 for their expression and do not believe that they should be assigned to the DE class on this basis. We note that the availability of a dominant-negative version of ORF50 may provide a useful reagent for investigation of this question, since classically defined IE genes should be expressed in its presence. This approach has the additional virtue of not requiring the use of toxic inhibitors of protein synthesis which, in BCBL-1 cells at least, produce unacceptable nonspecific cytotoxicity.
Our analysis of the physical properties of the ORF50 protein demonstrates that a substantial fraction of its abnormal electrophoretic mobility can be attributed to phosphorylation (Fig. 3). In fact, the sequence analysis of the ORF50 polypeptide reveals numerous consensus elements for two cellular serine/threonine kinases, casein kinase II and PKC. Interestingly, the EBV switch protein Zta has been shown to be regulated posttranslationally through serine/threonine phosphorylation by PKC, a potent stimulator of Zta transactivation (6). We note that lytic reactivation induced by the ORF50 protein is further enhanced by the addition of TPA (Fig. 8A). One potential explanation for this effect of TPA may be upregulation of the activity of ORF50 by modulation of its phosphorylation state. However, many other models could also explain this observation.
Our finding that ORF50 transactivates DE promoters in the human cell lines BJAB and SLK (Fig. 4B) indicates that the protein is active in the major cell types in which viral latency is known to be established and from which reactivation must occur in vivo. Together with its activity in the simian fibroblast cell line CV-1, the data suggest that the transcriptional activity of the protein displays little cell type specificity. This conclusion is consistent with the simplest model for its action—namely, that it binds to DNA in a site-specific fashion and recruits or activates ubiquitous components of the basal transcription machinery. However, models in which the protein might be targeted to DNA via interactions with ubiquitous DNA binding proteins are not excluded; indeed, it is possible that the protein uses both modes of action, for example, under different physiological circumstances or on different target promoters. We are currently defining the target sites for ORF50 action both genetically and biochemically.
The identification of an activation domain in the C terminus of the protein allowed us to construct a potent and specific dominant-negative allele. Several possible models can be proposed for the mechanism by which the mutant blocks ORF50 transactivation. Since the mutant can form heterodimers with wild-type ORF50, one model might be that such mixed oligomers would be inactive for transactivation. Alternatively, mutant homodimers might compete for promoter binding sites with the wild-type protein. Regardless of the details of the mechanism, however, the effects of ORF50ΔSTAD expression on lytic reactivation—either spontaneous or induced—demonstrate the necessity of ORF50 for all known reactivation events and strongly implicate its transcriptional activity as the basis for this role. While not in itself diagnostic of a role for ORF50 as the molecular switch governing the latent-lytic transformation, this finding, together with the extremely early kinetics of ORF50 expression and the inductive effects of its forced overexpression (34, 55), strongly supports such a role. Given the importance of lytic reactivation in the natural history of KSHV infection, the enumeration of the targets of ORF50 action and the analysis of its biochemical mechanism of action are now critical subjects for investigation.
ACKNOWLEDGMENTS
We thank the Ganem laboratory members for helpful advice and discussion.
We thank the Howard Hughes Medical Institute for generous support. D.M.L. is a postdoctoral fellow of the Irvington Institute for Immunological Research.
REFERENCES
- 1.Ambroziak J, Blackbourn D, Herndier B, Glogan R, Gullet J, McDonald A, Lennette E, Levy J. Herpesvirus-like sequences in HIV-infected and uninfected Kaposi’s sarcoma patients. Science. 1995;268:582–583. doi: 10.1126/science.7725108. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 4.Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka E G, Gutkind J S, Asch A S, Cesarman E, Gershengorn M C, Mesri E A. G-protein-coupled receptor of Kaposi’s sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. [DOI] [PubMed] [Google Scholar]
- 5.Barber J R, Verma I M. Modification of fos proteins: phosphorylation of c-fos, but not v-fos, is stimulated by 12-tetradecanoyl-phorbol-13-acetate and serum. Mol Cell Biol. 1987;7:2201–2211. doi: 10.1128/mcb.7.6.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumann M, Mischak H, Dammeier S, Kolch W, Gires O, Pich D, Zeidler R, Delecluse H-J, Hammerschmidt W. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-tetradecanoylphorbol-13-acetate-induced phosphorylation. J Virol. 1998;72:8105–8114. doi: 10.1128/jvi.72.10.8105-8114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasig C, Zietz C, Haar B, Neipel F, Esser S, Brockmeyer N H, Tschachler E, Colombini S, Ensoli B, Sturzl M. Monocytes in Kaposi’s sarcoma lesions are productively infected by human herpesvirus 8. J Virol. 1997;71:7963–7968. doi: 10.1128/jvi.71.10.7963-7968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, Moore P S, Chang Y, Weiss R A. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 9.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O’Leary J J. Kaposi’s sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 10.Boshoff C, Weiss R. Kaposi’s sarcoma-associated herpesvirus. Adv Cancer Res. 1998;75:57–86. doi: 10.1016/s0065-230x(08)60739-3. [DOI] [PubMed] [Google Scholar]
- 11.Boyer T G, Berk A J. Functional interaction of adenovirus E1A with holo-TFIID. Genes Dev. 1993;7:1810–1823. doi: 10.1101/gad.7.9.1810. [DOI] [PubMed] [Google Scholar]
- 12.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 13.Chan S R, Bloomer C, Chandran B. Identification and characterization of human herpesvirus-8 lytic cycle-associated ORF 59 protein and the encoding cDNA by monoclonal antibody. Virology. 1998;240:118–126. doi: 10.1006/viro.1997.8911. [DOI] [PubMed] [Google Scholar]
- 14.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox M A, Leahy J, Hardwick J M. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J Virol. 1990;64:313–321. doi: 10.1128/jvi.64.1.313-321.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cress W D, Triezenberg S J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 18.Fearon E R, Finkel T, Gillison M L, Kennedy S P, Casella J F, Tomaselli G F, Morrow J S, Van Dang C. Karyoplasmic interaction selection strategy: a general strategy to detect protein-protein interactions in mammalian cells. Proc Natl Acad Sci USA. 1992;89:7958–7962. doi: 10.1073/pnas.89.17.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flemington E, Speck S H. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1227–1232. doi: 10.1128/jvi.64.3.1227-1232.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furnari F, Zacny V, Quinlivan E B, Kenney S, Pagano J S. RAZ, an Epstein-Barr virus transdominant repressor that modulates the viral reactivation mechanism. J Virol. 1994;68:1827–1836. doi: 10.1128/jvi.68.3.1827-1836.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruffat H, Duran N, Buisson M, Wild F, Buckland R, Sergeant A. Characterization of an R-binding site mediating the R-induced activation of the Epstein-Barr virus BMLF1 promoter. J Virol. 1992;66:46–52. doi: 10.1128/jvi.66.1.46-52.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruffat H, Portes-Sentis S, Sergeant A, Manet E. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) encodes a homologue of the Epstein-Barr virus bZip protein EB1. J Gen Virol. 1999;80:557–561. doi: 10.1099/0022-1317-80-3-557. [DOI] [PubMed] [Google Scholar]
- 23.Gruffat H, Sergeant A. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 1994;22:1172–1178. doi: 10.1093/nar/22.7.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardwick J M, Tse L, Applegren N, Nicholas J, Veliuona M A. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herndier B G, Werner A, Arnstein P, Abbey N W, Demartis F, Cohen R L, Shuman M A, Levy J A. Characterization of a human Kaposi’s sarcoma cell line that induces angiogenic tumors in animals. AIDS. 1994;8:575–581. doi: 10.1097/00002030-199405000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Holley-Guthrie E A, Quinlivan E B, Mar E-C, Kenney S. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J Virol. 1990;64:3753–3759. doi: 10.1128/jvi.64.8.3753-3759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenney S, Holley-Guthrie E, Mar E-C, Smith M. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J Virol. 1989;63:3878–3883. doi: 10.1128/jvi.63.9.3878-3883.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirshner J R, Staskus K, Haase A, Lagunoff M, Ganem D. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi’s sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J Virol. 1999;73:6006–6014. doi: 10.1128/jvi.73.7.6006-6014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolman J L, Taylor N, Gradoville L, Countryman J, Miller G. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J Virol. 1996;70:1493–1504. doi: 10.1128/jvi.70.3.1493-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeRoux F, Sergeant A, Corbo L. Epstein-Barr virus (EBV) EB1/Zta protein provided in trans and competent for the activation of productive cycle genes does not activate the BZLF1 gene in the EBV genome. J Gen Virol. 1996;77:501–509. doi: 10.1099/0022-1317-77-3-501. [DOI] [PubMed] [Google Scholar]
- 31.Li M, McKey J, Czajak S C, Desrosiers R C, Lackner A A, Jung J U. Identification and characterization of Kaposi’s sarcoma-associated herpesvirus K8.1 glycoprotein. J Virol. 1999;73:1341–1349. doi: 10.1128/jvi.73.2.1341-1349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin K, Dai C Y, Ricciardi R P. Cloning and functional analysis of Kaposi’s sarcoma-associated herpesvirus DNA polymerase and its processivity factor. J Virol. 1998;72:6228–6232. doi: 10.1128/jvi.72.7.6228-6232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin S-F, Robinson D R, Miller G, Kung H-J. Kaposi’s sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol. 1999;73:1909–1917. doi: 10.1128/jvi.73.3.1909-1917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 35.Manet E, Rigolet A, Gruffat H, Giot J-F, Sergeant A. Domains of the Epstein-Barr virus (EBV) transcription factor R required for dimerization, DNA binding and activation. Nucleic Acids Res. 1991;19:2661–2667. doi: 10.1093/nar/19.10.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin D F, Kuppermann B D, Wolitz R A, Palestine A G, Li H, Robinson C A. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N Engl J Med. 1999;340:1063–1070. doi: 10.1056/NEJM199904083401402. [DOI] [PubMed] [Google Scholar]
- 37.Martin J N, Ganem D E, Osmond D H, Page-Shafer K A, Macrae D, Kedes D H. Sexual transmission and the natural history of human herpesvirus 8 infection. N Engl J Med. 1998;338:948–954. doi: 10.1056/NEJM199804023381403. [DOI] [PubMed] [Google Scholar]
- 38.Miller G. The switch between latency and replication of Epstein-Barr virus. J Infect Dis. 1990;161:833–844. doi: 10.1093/infdis/161.5.833. [DOI] [PubMed] [Google Scholar]
- 39.Moore P S, Kingsley L A, Holmberg S D, Spira T, Gupta P, Hoover D R, Parry J P, Conley L J, Jaffe H W, Chang Y. Kaposi’s sarcoma-associated herpesvirus infection prior to onset of Kaposi’s sarcoma. AIDS. 1996;10:175–180. doi: 10.1097/00002030-199602000-00007. [DOI] [PubMed] [Google Scholar]
- 40.Neipel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi’s sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinlivan E B, Holley-Guthrie E A, Norris M, Gutsch D, Bachenheimer S L, Kenney S C. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 1993;21:1999–2007. doi: 10.1093/nar/21.8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raab M S, Albrecht J C, Birkmann A, Yaguboglu S, Lang D, Fleckenstein B, Neipel F. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J Virol. 1998;72:6725–6731. doi: 10.1128/jvi.72.8.6725-6731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72:7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Renne, R., and D. Lukac. Unpublished data.
- 44.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 45.Roth W K, Brandstetter H, Sturzl M. Cellular and molecular features of HIV-associated Kaposi’s sarcoma. AIDS. 1992;6:895–913. doi: 10.1097/00002030-199209000-00001. [DOI] [PubMed] [Google Scholar]
- 46.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sadler R, Wu L, Forghani B, Renne R, Zhong W, Herndier B, Ganem D. A complex translational program generates multiple novel proteins from the latently expressed Kaposin (K12) locus of Kaposi’s sarcoma-associated herpesvirus. J Virol. 1999;73:5722–5730. doi: 10.1128/jvi.73.7.5722-5730.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarid R, Flore O, Bohenzky R A, Chang Y, Moore P S. Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol. 1998;72:1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schulz T. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 50.Shimizu N, Sakuma S, Ono Y, Takada K. Identification of an enhancer-type sequence that is responsive to Z and R trans-activators of Epstein-Barr virus. Virology. 1989;172:655–658. doi: 10.1016/0042-6822(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 51.Sinclair A J, Brimmell M, Shanahan F, Farrell P J. Pathways of activation of the Epstein-Barr virus productive cycle. J Virol. 1991;65:2237–2244. doi: 10.1128/jvi.65.5.2237-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 53.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun R, Lin S-F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi’s sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun R, Lin S F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Boshoff C, Hatzioannou T, Suggett F E, Aldam D M, Denton A S, et al. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi’s sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 57.Zalani S, Holley-Guthrie E, Kenney S. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93:9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu F, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi’s sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]