Abstract
The differential susceptibilities of mouse strains to xenotropic and polytropic murine leukemia viruses (X-MLVs and P-MLVs, respectively) are poorly understood but may involve multiple mechanisms. Recent evidence has demonstrated that these viruses use a common cell surface receptor (the X-receptor) for infection of human cells. We describe the properties of X-receptor cDNAs with distinct sequences cloned from five laboratory and wild strains of mice and from hamsters and minks. Expression of these cDNAs in resistant cells conferred susceptibilities to the same viruses that naturally infect the animals from which the cDNAs were derived. Thus, a laboratory mouse (NIH Swiss) X-receptor conferred susceptibility to P-MLVs but not to X-MLVs, whereas those from humans, minks, and several wild mice (Mus dunni, SC-1 cells, and Mus spretus) mediated infections by both X-MLVs and P-MLVs. In contrast, X-receptors from the resistant mouse strain Mus castaneus and from hamsters were inactive as viral receptors. These results suggest that X-receptor polymorphisms are a primary cause of resistances of mice to members of the X-MLV/P-MLV family of retroviruses and are responsible for the xenotropism of X-MLVs in laboratory mice. By site-directed mutagenesis, we substituted sequences between the X-receptors of M. dunni and NIH Swiss mice. The NIH Swiss protein contains two key differences (K500E in presumptive extracellular loop 3 [ECL 3] and a T582 deletion in ECL 4) that are both required to block X-MLV infections. Accordingly, a single inverse mutation in the NIH Swiss protein conferred X-MLV susceptibility. Furthermore, expression of an X-MLV envelope glycoprotein in Chinese hamster ovary cells interfered efficiently with X-MLV and P-MLV infections mediated by X-receptors that contained K500 and/or T582 but had no effect on P-MLV infections mediated by X-receptors that lacked these amino acids. In contrast, moderate expression of a P-MLV (MCF247) envelope glycoprotein did not cause substantial interference, suggesting that X-MLV and P-MLV glycoproteins interfere nonreciprocally with X-receptor-mediated infections. We conclude that P-MLVs have become adapted to utilize X-receptors that lack K500 and T582. A penalty for this adaptation is a reduced ability to interfere with superinfection. Because failure of interference is a hallmark of several exceptionally pathogenic retroviruses, we propose that it contributes to P-MLV-induced diseases.
Recent evidence has demonstrated that a single cell surface receptor (the X-receptor) from humans mediates infections by both xenotropic and polytropic host range groups of murine leukemia viruses (X-MLVs and P-MLVs, respectively) (1, 37, 40), consistent with previous evidence that these classes of MLVs might use a common receptor. For example, laboratory mice including NIH Swiss are highly susceptible to P-MLVs (also called mink cell focus-forming viruses [MCFs]) (5, 11) but are resistant to X-MLVs (21), whereas most wild mice, including Mus spretus, are variably susceptible to both X-MLVs and P-MLVs (19). Crosses of NIH Swiss mice with M. spretus implied that susceptibility to X-MLVs is caused by a dominant allele of the P-MLV receptor gene Rmc1 (18, 19). Moreover, expression of a P-MLV-related envelope glycoprotein in M. spretus caused weak interference to infections by both groups of virus (19). Cross-interference between X-MLVs and P-MLVs has also been reported for Mus dunni fibroblasts (2, 27). In contrast, Mus castaneus is resistant to both X-MLVs and P-MLVs (24, 25). In crosses of M. castaneus with NIH Swiss or DBA/2 laboratory mice, resistance was inherited as a recessive allele of the Rmc1 gene (24). Surprisingly, however, recent evidence has demonstrated that resistance to X-MLVs and P-MLVs is dominant in crosses of M. castaneus with mice that contain the M. spretus Rmc1 allele (25). Apparently, an X-MLV-related envelope glycoprotein that is endogenously inherited in M. castaneus interferes with the M. spretus X-receptor but not with the NIH Swiss X-receptor (25). Together, these results imply that the X-receptors of mice must be polymorphic and that susceptibilities to infections by X-MLVs and P-MLVs are regulated by these polymorphisms and also by inherited interference factors that differentially interact with X-receptors of distinct mouse strains. In addition, a lipoprotein factor in the sera of most mouse strains specifically inactivates X-MLVs and P-MLVs but not other host range classes of MLVs (15, 21–23). X-MLV gene expression is also partially silenced in certain mouse cells (13, 19a). This evidence has implied that multiple mechanisms of defense have evolved to control diseases caused by the X-MLV/P-MLV family of retroviruses. Indeed, P-MLVs (MCFs) are exceptionally pathogenic and have been implicated as critical causal factors in murine retroviral leukemogenesis and lymphomagenesis (4, 8, 11, 12, 14, 32, 34, 36).
Although it was recently shown that the human X-receptor mediates infections by both X-MLVs and P-MLVs (1, 37, 40) and that an X-receptor from NIH Swiss mice functions as a P-MLV receptor when expressed in hamster cells (40), the latter receptor was not tested for mediation of X-MLV infections. Such testing is necessary because susceptibility of mice to X-MLVs and P-MLVs can be regulated not only by receptors but also, as described above, by many other host factors. Moreover, X-receptor cDNAs from other mouse strains have not been previously described. To address these issues, we cloned and functionally analyzed X-receptor cDNAs from a representative group of mouse strains that are differentially susceptible to X-MLVs and P-MLVs, as well as from mink cells that are morphologically altered by P-MLVs and from Chinese hamster ovary (CHO) cells. Moreover, we constructed and tested a series of mutations in the X-receptors of M. dunni and NIH Swiss mice in order to identify the amino acid sequence differences that are responsible for resistance of laboratory mice to X-MLVs.
MATERIALS AND METHODS
Mice, cell lines, and viruses.
NIH Swiss inbred NFS/N, M. spretus, and M. castaneus wild-derived mice were obtained from Jackson Laboratory, Bar Harbor, Maine. M. dunni tail fibroblasts and SC-1, BALB/3T3, and mink Mv-1-Lu (CCL64) cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS). Chinese hamster ovary (CHO) cells were grown in α-modified minimal essential medium supplemented with 10% FBS.
LacZ(X-MLV), LacZ(MCF13), and LacZ(MCF247) pseudotype viruses were generated as previously described (37).
Receptor cDNA cloning.
X-receptor cDNAs were isolated by reverse transcription-PCR amplification with total RNAs. Total RNAs were prepared from M. dunni, SC-1, mink Mv-1-Lu, and hamster (CHO) cells with the RNeasy Midi kit (Qiagen, Valencia, Calif.), whereas total RNAs from mouse kidney tissue, isolated from NIH Swiss, M. spretus, and M. castaneus, were prepared by the cesium chloride method (3). The 2.1-kb X-receptor cDNAs were amplified by using primers complementary to the 5′ and 3′ ends of the human X-receptor coding region (upstream primer, 5′-GGGGGATCCATGAAGTTCGCCGAGCACCTC-3′, containing a BamHI restriction site [underlined sequence]; downstream primer, 5′-GGTCGAGGAAAGGATTGTAG-3′). PCR was run for 30 cycles with a 60°C annealing temperature for 1 min and an extension temperature of 72°C for 3 min. The amplified X-receptor DNAs were cloned into the mammalian expression vector pcDNA3.1.V5His-TOPO (Invitrogen, Carlsbad, Calif.). The DNA sequences were determined at the Microbiology and Molecular Immunology Core Facility on a PE/ABD 377 sequencer with dye terminator cycle sequencing chemistry (Applied Biosystems, Foster City, Calif.).
Transfection and infection assays.
Mouse BALB/3T3 and CHO cells expressing NIH Swiss, M. dunni, SC-1, M. castaneus, mink, and hamster X-receptors were generated by transient or stable transfection of the corresponding cDNAs with SuperFect transfection reagent (Qiagen). Transient transfectants were challenged with either LacZ(X-MLV) or LacZ(MCF247) pseudotype virus, with overnight incubations with virus beginning 24 h after transfection. Infected cells were stained by treating cells with 0.25% glutaraldehyde and assayed for β-galactosidase activity with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as a substrate (26). Blue CFU were counted, and the titer of infection was expressed as the number of CFU per milliliter of virus supernatant. Stable transfectants were generated by selection with G418 (1 mg/ml). G418-resistant clones were then pooled and tested for sensitivity to LacZ(X-MLV), LacZ(MCF13), and LacZ(MCF247), as outlined above.
Mutagenesis of mouse xenotropic receptor cDNAs.
NIH Swiss and M. dunni X-receptor residues were mutated by PCR mutagenesis with two complementary mutagenic primers containing the targeted point mutation and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). Plasmid DNA from three independent clones was sequenced to confirm the mutations. The DNA sequence was determined as described above. The mutants were designated by the parental X-receptor name followed by the mutated amino acid followed by the residue number and the new amino acid.
Cross-interference assay.
CHO cells were transfected with either FBXsalf (xenotropic envelope gene NZB expression vector [37]) or phMCF (MCF247 envelope gene cloned into the FBsalf expression vector, kindly provided by Jean-Michel Heard, Pasteur Institute, Paris, France) DNAs by the calcium phosphate-DNA precipitation method (9). Transfectants were selected with phleomycin (50 μg/ml), and resistant clones were screened for envelope expression by immunofluorescence assay using goat anti-Bv2 polyclonal antibody (for xenotropic envelope glycoprotein) and goat anti-gp70 antibody (for MCF247 envelope glycoprotein) (20). The highest envelope-expressing clones were used in the cross-interference assay.
Interference assays were performed as follows. Control CHO cells, xenotropic (CHO.Xenv) cells, and MCF247 (CHO.MCF247env) envelope-expressing cells were transfected with X-receptor cDNAs by using SuperFect transfection reagent (Qiagen). After 24 h, the transfected cells were tested for susceptibility to infections with LacZ(X-MLV) and LacZ(MCF247), as outlined above.
Nucleotide sequence accession numbers.
The X-receptor cDNA sequences have been assigned GenBank accession numbers as follows: NIH Swiss, AF131096; M. dunni, AF131097; SC-1, AF131098; CHO, AF131099; CCL64, AF131100; M. spretus, AF13101; M. castaneus, AF131102.
RESULTS
Sequences of X-receptors.
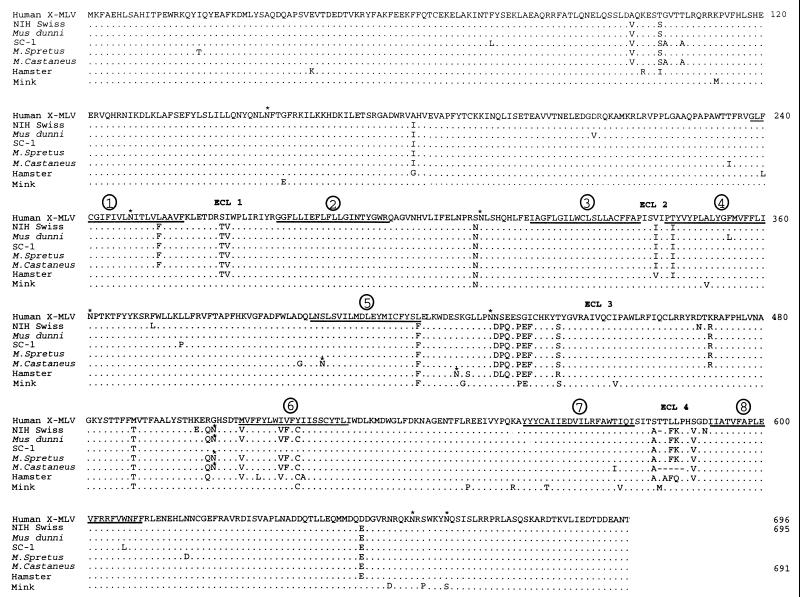
Seven new X-receptor cDNAs were cloned by reverse transcriptase PCR, as described in Materials and Methods. A comparison of the predicted amino acid sequences of these X-receptors with the human X-receptor is shown in Fig. 1, which also indicates the eight hydrophobic potential membrane-spanning sequences and seven NX(S/T) sites for potential N-linked glycosylation. Based on the absence of a recognizable signal sequence in these proteins and on evidence for a cytosolic localization of the hydrophilic amino-terminal region (35, 37) we have inferred a topology for the X-receptor that includes the presumptive extracellular loop (ECL) regions indicated (Fig. 1). Interestingly, the mouse X-receptor sequences are polymorphic, and each contains multiple nonconservative substitutions, deletions, and/or novel glycosylation sites. All of the seven NX(S/T) sites in the human protein are conserved in the other X-receptors except for the unique NNS sequence at position 432. However, additional potential glycosylation sites occur in the M. castaneus protein at position 402, in the Chinese hamster protein at position 426, and in the NIH Swiss, M. dunni, M. spretus, and M. castaneus X-receptors at position 503. Although many of the distinguishing sequences are scattered throughout these X-receptors, almost all of the nonconservative changes likely to be functionally important for virus infections occur between positions 430 and 590, with particular clustering in the hydrophilic regions labeled ECL 3 and ECL 4. For example, the laboratory mouse (NIH Swiss), M. castaneus, and hamster X-receptors all have nonconservative substitutions and/or deletions in ECL 4. Moreover, the NIH Swiss X-receptor has additional nonconservative substitutions in ECL 3.
FIG. 1.
Amino acid sequence comparison of human, mouse, mink, and hamster X-receptors. The mouse sequences were derived from an NIH Swiss laboratory strain and from the wild mouse strains M. dunni, SC-1, M. spretus, and M. castaneus. The hamster sequence was from CHO cells, and the mink sequence was from CCL64 lung fibroblasts. Positions of identity to the human sequence are indicated by dots, whereas deletions in the NIH Swiss and M. castaneus sequences are indicated by dashes. The eight hydrophobic potential membrane-spanning sequences are indicated by bars underlying the human sequence and are identified by numbers 1 to 8. The putative ECL regions are indicated. Numbers at the right of the sequences correspond to the position of the last amino acid shown. Potential N-linked glycosylation sites are indicated by asterisks.
Properties of X-receptor cDNAs expressed in resistant cells.
We assayed the viral receptor functions of these X-receptors by expressing them in naturally resistant cells and assaying for susceptibilities to β-galactosidase-encoding X-MLV (NZB) (29), MCF13 (5), and MCF247 (16) pseudotype viruses, as previously described (37). For this purpose, we used CHO cells, which are fully resistant to P-MLVs (but weakly susceptible to X-MLVs) and mouse BALB/3T3 fibroblasts, which are fully resistant to X-MLVs (Table 1). All of the X-receptor cDNAs, including the 2.1-kbp human coding region, were expressed with the pcDNA3.1.V5His-TOPO vector. These results clearly established that the X-receptors from humans, minks, the wild mice M. dunni and M. spretus, and SC-1 cells all conferred significant but variably efficient susceptibilities to the MCFs and X-MLV used in these assays. In contrast, the X-receptor from NIH Swiss mice mediated infections of MCFs but not of X-MLV, whereas the X-receptors from M. castaneus and Chinese hamsters were inactive as viral receptors in these assays. Interestingly, these patterns of infectivity mimic the susceptibilities of the animals from which the cDNAs were derived (see Discussion). Consequently, we infer that the resistances of these animals to infections by X-MLVs and P-MLVs are principally determined by the specific sequences of their X-receptors, with additional effects possibly caused by other mechanisms, such as interfering glycoproteins (19, 25), serum lipoprotein factors (15, 21, 23), or transcriptional silencing (13, 19a). Thus, xenotropism in this system (i.e., the resistance of inbred laboratory mice to X-MLVs) is caused, at least partly, by an inherent property of the X-receptor protein in these mice.
TABLE 1.
Mediation of infections by X-receptors from different mammalian species and mouse strains
| Expt | X-receptor assayed | Titer of LacZ pseudotype (CFU/ml)a
|
||
|---|---|---|---|---|
| CHO cells
|
BALB/3T3 cells
|
|||
| MCF13 | MCF247 | X-MLV (NZB) | ||
| 1b | None | 0 | 0 | 0 |
| Human | 1.2 × 102 | 2.2 × 103 | 2.3 × 103 | |
| NIH Swiss | 4.7 × 103 | 1.5 × 103 | 0 | |
| M. dunni | 8.8 × 104 | 2.4 × 104 | 1.9 × 103 | |
| SC-1 | 6.0 × 101 | 2.8 × 102 | 4.7 × 103 | |
| M. castaneus | 0 | 0 | 0 | |
| 2c | None | ND | 0 | 0 |
| Human | ND | 4.6 × 103 | 1.0 × 104 | |
| M. spretus | ND | 1.3 × 102 | 2.2 × 102 | |
| Mink | ND | 2.6 × 102 | 1.0 × 102 | |
| Hamster | ND | 0 | 0 | |
Infectivity assays for MCF viruses were done with CHO cells, whereas those for X-MLV were done with BALB/3T3 fibroblasts. This was done because these untransfected cells have zero backgrounds for the viruses. CHO cells have a weak susceptibility to X-MLVs that complicates their use for study of these viruses (Fig. 2B). The titers indicated are all averages of three independent experiments. ND, not done.
Cells were stably transfected with X-receptor expression vectors, and pooled populations of transfected clones were employed (see Materials and Methods). Similar results were obtained when the cells were transiently transfected (data not shown).
These X-receptors were assayed after transient transfection of expression vectors into the cells (see Materials and Methods).
Interestingly, the M. castaneus X-receptor is inactive in mediating infections by X-MLVs and MCFs in heterologous cells (Table 1), in agreement with the resistance of M. castaneus to these viruses (24, 25). As described above, M. castaneus contains an X-MLV-related envelope glycoprotein that can block the viral receptor function of the M. spretus X-receptor but not of the NIH Swiss X-receptor (25). This previous evidence did not establish whether the M. castaneus X-receptor was inherently inactive or whether it was also masked by the endogenous interfering glycoprotein. Our results strongly suggest that the M. castaneus X-receptor is inherently inactive as a receptor for X-MLVs and P-MLVs. Similarly, CHO cells are resistant to P-MLVs and only slightly susceptible to X-MLVs, consistent with the results in Table 1.
At least some human cell lines, including HeLa cells, are resistant to P-MLV infections despite their susceptibility to X-MLVs and despite the ability of the human X-receptor to mediate P-MLV infections in heterologous cells (5) (Table 1). Surprisingly, expression of the M. dunni and NIH Swiss X-receptors in HeLa cells did not confer susceptibilities to P-MLVs (data not shown). Consequently, certain cell lines, including HeLa cells, may have a dominant resistance to P-MLVs.
Identification of two amino acids within the presumptive ECL 3 and ECL 4 that specifically control X-MLV infections.
To determine which region(s) of the M. dunni and NIH Swiss X-receptors is responsible for their different abilities to mediate X-MLV infections, we used PCR site-directed mutagenesis to substitute sequences between their presumptive extracellular regions. The NIH Swiss X-receptor differs from that of M. dunni at seven positions, including five nonconservative differences. Of these, one (V210D) is in the intracellular amino-terminal region, two (D469N and K500E) are in ECL 3, and two (T582Δ and D590N) are in ECL 4 (Fig. 1). Furthermore, we considered the ECL 4 region potentially important because the M. castaneus X-receptor is inactive in mediating infections and contains a deletion of five amino acids that overlaps the single T582Δ deletion in the NIH Swiss protein (Fig. 1).
Based on this information, we first made the M. dunni mutant T582Δ by deleting T582 and the corresponding inverse NIH Swiss mutant Δ582T by inserting T at this position. Both mutants as well as the wild-type X-receptors were then transiently expressed in resistant cells and tested for mediation of X-MLV and P-MLV infections. As shown in Table 2, the NIH Swiss Δ582T mutant X-receptor was able to mediate X-MLV infection, whereas the reciprocal M. dunni T582Δ mutation did not eliminate X-MLV infectivity. These results implied that a second sequence difference must account for the different activities of the M. dunni T582Δ and NIH Swiss X-receptors.
TABLE 2.
Specific effects of a T582 deletion mutation on X-MLV infections differ for NIH Swiss and M. dunni X-receptors
| X-receptor assayedb | Titer of LacZ pseudotype (CFU/ml)a
|
|
|---|---|---|
| MCF247 | X-MLV (NZB) | |
| None | 0 | 0 |
| M. dunni | 5.3 × 103 | 1.1 × 103 |
| M. dunni T582Δ | 5.3 × 103 | 7.6 × 103 |
| NIH Swiss | 1.4 × 103 | 0 |
| NIH Swiss Δ582T | 2.8 × 102 | 8.0 × 102 |
Infectivity assays for the LacZ(MCF247) virus were done in CHO cells, whereas those for LacZ(X-MLV) were done in BALB/3T3 fibroblasts. This was done because these untransfected or mock transfected control cells have zero backgrounds for the viruses (Table 1). The results are averages of three independent assays.
Cells were transiently transfected with X-receptor expression vectors prior to infections with the viruses indicated (see Materials and Methods).
Consequently, we constructed a second series of mutations in the M. dunni X-receptor. These included three single-residue substitutions (D469N, K500E, and D590N), three double-residue substitutions (D469N-T582Δ, K500E-T582Δ, and D590N-T582Δ), and two triple-residue substitutions (D469N-T582Δ-D590N and K500E-T582Δ-D590N). These mutants as well as the wild-type M. dunni X-receptor were then tested for mediation of X-MLV and P-MLV infections. As shown in Table 3, all of these mutants mediated P-MLV (MCF247) infections with the same efficiency as the wild-type X-receptor. Interestingly, complete abrogation of X-MLV infections required a combination of two mutations, K500E in presumptive ECL 3 and T582Δ in ECL 4.
TABLE 3.
Redundant roles of K500 and T582 in mediation of X-MLV infections by the M. dunni X-receptor
| X-receptor assayedb | Titer of LacZ pseudotype (CFU/ml)a
|
|
|---|---|---|
| MCF247 | X-MLV (NZB) | |
| None | 0 | 0 |
| M. dunni | 6.1 × 102 | 1.5 × 102 |
| Single substitutions | ||
| D590N | 1.7 × 102 | 3.8 × 102 |
| K500E | 1.1 × 103 | 4.0 × 101 |
| D469N | 1.2 × 103 | 1.4 × 102 |
| Double mutations | ||
| D590N-T582Δ | 7.4 × 102 | 2.0 × 102 |
| K500E-T582Δ | 1.8 × 103 | 0 |
| D469N-T582Δ | 2.0 × 102 | 3.2 × 102 |
| Triple mutations | ||
| K500E-T582Δ-D590N | 4.6 × 102 | 0 |
| D469N-T582Δ-D590N | 4.2 × 102 | 7.7 × 102 |
Infectivity assays for the LacZ(MCF) virus were done in CHO cells, whereas those for LacZ(X-MLV) were done in BALB/3T3 fibroblasts because of the zero backgrounds of infections in these control untransfected or mock-transfected cells (Tables 1 and 2). The results are averages of three independent assays.
Cross-interference assay.
Previous interference analyses of X-MLV and P-MLV viruses used various cells that expressed only their endogenous receptors (2, 5, 27). For example, the susceptibility of laboratory mouse cells to P-MLVs was unaffected by expression of an X-MLV (NZB) envelope glycoprotein (2). This is not surprising, because the X-receptors of laboratory mice presumably cannot interact with the X-MLV glycoprotein. In contrast, varying degrees of nonreciprocal cross-interference between X-MLVs and P-MLVs have been observed in cells that are susceptible to both groups of virus, with X-MLVs generally interfering strongly with P-MLVs and P-MLVs only interfering weakly with X-MLVs (2, 27). In several assays it was also reported that P-MLV-related envelope glycoproteins only weakly interfered with P-MLVs (2, 19). We have also observed only weak interference with P-MLV in M. dunni, NIH 3T3, and mink CCL64 cells expressing P-MLV envelope glycoproteins (data not shown). Based on this information, it was proposed that X-MLVs and P-MLVs may share a common receptor and that X-MLVs may in addition have a unique receptor (27). However, an alternative interpretation of this evidence, as discussed elsewhere, is that X-MLVs and P-MLVs may compete unequally for a common receptor (37).
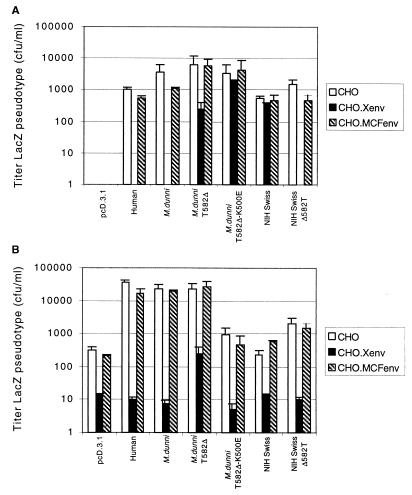
To study this issue more conclusively, we made CHO cell derivatives that stably expressed either a xenotropic (NZB) or a P-MLV (MCF247) envelope glycoprotein (see Materials and Methods). These cells as well as control CHO cells were transiently transfected with X-receptor expression vectors, and the cells were subsequently assayed for susceptibilities to infections by LacZ(X-MLV) and LacZ(MCF) viruses. Figure 2 summarizes the results of three experiments in which we used human, M. dunni, M. dunni T582Δ, M. dunni T582Δ-K500E, NIH Swiss, and NIH Swiss Δ582T X-receptors. As shown in Fig. 2A, control CHO cells that lack exogenous X-receptors are completely resistant to MCF infections, whereas all of the tested X-receptors conferred MCF susceptibilities. Compatible with previous reports (2, 31), expression of the X-MLV (NZB) envelope glycoprotein in CHO.Xenv cells resulted in complete interference with P-MLV infections mediated by the human and M. dunni X-receptors but not with P-MLV infections mediated by the X-receptor of the NIH Swiss laboratory mouse. Interestingly, however, P-MLV infection mediated by the NIH Swiss Δ582T mutant X-receptor was completely blocked by expression of the X-MLV envelope glycoprotein, suggesting that interference occurs only with X-receptors that strongly interact with the glycoprotein. Similarly, mediation of P-MLV infections by the M. dunni T582Δ-K500E double mutant, which is inactive in X-MLV infections, was also unaffected by expression of the X-MLV glycoprotein. Interestingly, P-MLV and X-MLV infections mediated by the M. dunni T582Δ X-receptor mutant were only partially blocked by expression of the X-MLV glycoprotein, despite the fact that this mutant functions as a strong X-MLV receptor (Table 2). These results suggest that a T at position 582 strongly interacts with the X-MLV envelope glycoprotein and makes a critical contribution to interference. These results also show that expression of the MCF247 envelope glycoprotein in CHO.MCFenv cells caused only slight or negligible interference with infections by either X-MLVs or P-MLVs. This is not entirely surprising, because P-MLVs apparently do not interact significantly with K500 or T582, which are critical for infection and interference of X-MLVs (see Discussion). Thus, in all cases interference is strong only if the glycoprotein functionally interacts with K500 and/or T582 of the receptor.
FIG. 2.
Studies of interference with wild-type and mutant human, M. dunni, and NIH Swiss mouse X-receptors. The assays were done with control CHO cells or CHO derivative clones that stably express either the X-MLV (NZB) envelope glycoprotein or the MCF247 envelope glycoprotein. These cell clones are called CHO.Xenv and CHO.MCFenv, respectively (see Materials and Methods). These cells were transiently transfected with expression vectors for the human, M. dunni, M. dunni T582Δ, M. dunni K500E-T582Δ, NIH Swiss, and NIH Swiss Δ582T X-receptors. Infections with LacZ pseudotype viruses were initiated 24 h after beginning the transfections. (A) Infections of LacZ(MCF247) virus; (B) infections of LacZ(X-MLV) virus.
Similarly, Fig. 2B shows data for X-MLV infections of the same control and envelope glycoprotein-expressing CHO cells. These results are slightly more complex, because control CHO cells are weakly susceptible to X-MLVs. Moreover, their endogenous susceptibility to X-MLVs is partially inhibited (ca. 10-fold) by expression of the X-MLV envelope glycoprotein. Despite this background of infectivity, the results basically support our other evidence. Specifically, these data are compatible with our conclusions that the human, M. dunni, M. dunni T582Δ, and NIH Swiss Δ582T X-receptors can mediate X-MLV infections (Tables 1 and 2); that wild-type NIH Swiss and M. dunni T582Δ-K500E double mutant X-receptors are not significantly active in X-MLV infection compared with control CHO cells (see also Table 3); that the X-receptor residues T582 and K500 are important for the interference caused by X-MLVs; and that the MCF envelope glycoprotein does not significantly interfere with X-MLV infections. The background of X-MLV infections in CHO cells makes them less useful than mouse cells for accurately measuring low titers of this virus.
DISCUSSION
Polymorphisms in X-receptors of mice control susceptibilities to X-MLV and P-MLV infections.
In this investigation we have cloned and functionally characterized X-receptor cDNAs from five representative strains of mice that differ in their susceptibilities to X-MLV and P-MLV infections, from mink CCL64 cells that form foci after infection with P-MLVs (MCFs), and from Chinese hamster cells that are completely resistant to P-MLVs and only slightly susceptible to X-MLVs. Importantly, our results substantiate previous genetic evidence, which implied that mouse X-receptors must be polymorphic in their sequences and that these polymorphisms help to control infections by the P-MLV/X-MLV family of retroviruses (18, 19, 24, 25). In particular, we have found that these cloned X-receptors mediate the same patterns of viral susceptibility as the cells from which the cDNAs were derived (Table 1). In addition, the differential activities of these X-receptors in X-MLV infections were closely similar when the assays were done with BALB/3T3 and CHO cells (e.g., see Table 1 and Fig. 2). Thus, the X-receptor from NIH Swiss mice conferred susceptibility to P-MLVs but resistance to X-MLVs; the X-receptors from M. dunni, SC-1 cells, M. spretus, and minks conferred susceptibilities with different efficiencies to both of these host range groups of MLVs; and the corresponding proteins from M. castaneus and CHO cells were inactive as virus receptors in our transient expression and infection assays. The only minor discrepancy in these results was the inactivity of the CHO X-receptor in our assays, despite the weak susceptibility of CHO cells to X-MLVs (compare Table 1 and Fig. 2). This could reflect a relative insensitivity of our transient transfection-infection assays or perhaps an inactivity of this receptor in BALB/3T3 fibroblasts. There is some cell type specificity in assays for X-receptor activities, as illustrated by our observation that resistance of HeLa cells to P-MLVs is dominant (see Results).
It is well established that additional factors also contribute to host resistances to these viruses. In particular, X-MLVs appear to be transcriptionally repressed in some mouse cells (13, 19a), and a lipoprotein factor(s) in many mouse sera can specifically inactivate X-MLVs and P-MLVs (15, 21, 23). In addition, endogenously inherited envelope glycoproteins can cause resistance to infection by an interference mechanism (e.g., see references 19, 25, and 34). In some cases these glycoprotein factors act differentially to block infections mediated by X-receptors of only certain mice. For example, a glycoprotein encoded by M. castaneus can apparently interfere with the X-receptor of M. spretus but not with the X-receptor of NIH Swiss laboratory mice (25). Our results provide an example of this specificity because the X-MLV (NZB) envelope glycoprotein also interferes with the X-receptor of wild mice but not with the X-receptor of laboratory mice (Fig. 2). Thus, the specificity of interfering glycoproteins for X-receptors of particular mouse strains is a secondary consequence of X-receptor polymorphisms. These considerations support our conclusion that X-receptor polymorphisms are an important primary cause for the differential susceptibilities of mice to the P-MLV/X-MLV family of retroviruses.
Molecular basis for X-MLV xenotropism.
By site-directed mutagenesis, we have compared the X-receptors of M. dunni and NIH Swiss mice, which differ in their abilities to mediate X-MLV infections but are both capable of mediating P-MLV infections. Thus, testing for P-MLV infections provided a positive control to demonstrate cell surface expression of the mutant X-receptors used in this investigation. In essence, these studies strongly implied that K500 in presumptive ECL 3 and T582 in ECL 4 make important but somewhat redundant contributions to infections mediated by the M. dunni X-receptor (Tables 2 and 3). Thus, in the context of the M. dunni X-receptor, either K500 or T582 was sufficient to allow X-MLV infections. However, none of the mutations had any effect on P-MLV infections. Consistent with this evidence, a single Δ582T insertion mutation in the NIH Swiss X-receptor was sufficient to confer activity in X-MLV infections (Table 2). These results imply that NIH Swiss mice contain two mutations, K500E and T582Δ, that are necessary to prevent X-MLV infections and that either K500 or T582 is sufficient for X-MLV susceptibility (Tables 2 and 3 and Fig. 2).
In this context, it is notable that the M. castaneus X-receptor contains K500 although it is completely inactive in both X-MLV and P-MLV infections. Thus, K500 is sufficient to allow X-MLV infections only in the context of certain X-receptors. The M. castaneus X-receptor contains a five-amino-acid deletion in ECL 4 that overlaps the deletion at position 582 that occurs in NIH Swiss mice (Fig. 1). Moreover, the hamster X-receptor also contains K500, although CHO cells are inactive in P-MLV infections and only weakly active in X-MLV infections. These X-receptors apparently contain structural features that prevent their utilization by either X-MLVs or P-MLVs.
Interference between X-MLVs and P-MLVs.
It is striking that X-MLVs and P-MLVs interact so differently with the same X-receptor proteins, as indicated by our infectivity and interference assays. Most significantly, X-MLVs rely heavily on K500 and T582 in the M. dunni X-receptor, and the double mutant K500E-T582Δ is completely inactive (Tables 2 and 3 and Fig. 2). In contrast, these mutations have no effect on P-MLV infections. Moreover, the same amino acids are critical for interference by the X-MLV (NZB) envelope glycoprotein (Fig. 2), suggesting that they may be necessary for strong glycoprotein-receptor binding. Thus, the amino acids in the X-receptor that are most critical for X-MLVs are ignored by P-MLVs. However, X-receptors may lack alternative sites for strong virus attachments because P-MLVs are very weak in their interference properties (e.g., see Fig. 2).
Based on these considerations, we propose that P-MLVs evolved from X-MLVs in response to X-receptor mutations (for example, K500E and T582Δ) that enabled many European mice to evade X-MLV infections. These European mice that were resistant to X-MLVs were later used to generate common inbred laboratory strains. Presumably, it was difficult for the viruses to overcome this resistance barrier because K500 and T582 are essential for strong functional interactions with the X-MLV envelope glycoprotein and because alternative sites for strong interactions may not occur in exposed positions on the X-receptor surface. According to this idea, the P-MLV adaptations were sufficient for infections but were inadequate for the strong competitive binding needed to establish efficient interference to superinfections. As a consequence, P-MLV glycoproteins may be able to cause significant interference only if they are highly overexpressed. Such overexpression was not an issue in our assays but would be expected to distort interference studies with cells that are chronically infected and therefore heavily superinfected with P-MLVs. Evidence with mice supports our conclusion that P-MLVs may cause only weak interferences to superinfection (12, 36).
Potential relevance to P-MLV pathogenesis.
It is surprising that P-MLV formation in mice has been highly correlated with the onset of severe pathogenesis whereas X-MLVs have not been associated with disease even in susceptible strains of wild mice (8, 11, 21, 32). Moreover, the only common feature of P-MLVs that distinguishes them from ecotropic and xenotropic MLVs is the amino-terminal receptor-determining region of their envelope glycoproteins (7, 17, 30, 38). Thus, class I P-MLVs typically are recombinants that contain long terminal repeat sequences derived from endogenous X-MLVs, MCF-specific env gene regions derived from endogenously inherited sequences, and a remainder that is derived from an ecotropic MLV (7, 17, 30, 38). These results have implied that P-MLV pathogenesis may involve interaction of the viral glycoprotein with its cell surface receptor. Consequently, the recent demonstration that X-MLVs and P-MLVs use a common cell surface receptor despite their distinct pathogenic effects implies that a difference in their interactions with the X-receptor could have a pathogenic consequence. From this perspective, our observation that X-MLVs and P-MLVs interact in a distinct manner with X-receptors is intriguing. Moreover, several other exceptionally pathogenic retroviruses, in addition to P-MLVs, cause only weak interference with superinfection. For example, the feline leukemia virus FeLV-FAIDS-EECC causes rapid immunodeficiency and T-cell destruction compared to closely related nonpathogenic isolates (28). The most distinguishing characteristic of FeLV-FAIDS-EECC is its inability to cause interference with superinfection (6, 28). Similar results occur with some cytopathic isolates of avian leukosis virus (39), ovine visna lentivirus (10), and human immunodeficiency virus type 1 (33). In agreement with our proposal that there may be a failure of interference in P-MLV-induced diseases, the process of thymic lymphomagenesis in AKR mice has been associated with massive superinfections by MCFs within single cells (12, 36). Hallmarks of this superinfection include numerous MCF proviruses integrated into the DNA of single cells and large quantities of unintegrated MCF-specific proviral DNA (12, 36). Additional investigations will be required to evaluate this hypothesis concerning the mechanism of P-MLV-induced diseases.
ACKNOWLEDGMENTS
This research was supported by NIH grants CA25810 and CA54149 from the National Cancer Institute.
We are grateful to our colleagues Emily Platt, Navid Madani, and Shawn Kuhmann for suggestions and encouragement.
REFERENCES
- 1.Battini J L, Rasko J E, Miller A D. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc Natl Acad Sci USA. 1999;96:1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chesebro B, Wehrly K. Different murine cell lines manifest unique patterns of interference to superinfection by murine leukemia viruses. Virology. 1985;141:119–129. doi: 10.1016/0042-6822(85)90188-6. [DOI] [PubMed] [Google Scholar]
- 3.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 4.Cloyd M W, Hartley J W, Rowe W P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1980;151:542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cloyd M W, Thompson M M, Hartley J W. Host range of mink cell focus-inducing viruses. Virology. 1985;140:239–248. doi: 10.1016/0042-6822(85)90362-9. [DOI] [PubMed] [Google Scholar]
- 6.Donahue P R, Quackenbush S L, Gallo M V, deNoronha C M, Overbaugh J, Hoover E A, Mullins J I. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol. 1991;65:4461–4469. doi: 10.1128/jvi.65.8.4461-4469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans L H, Malik F G. Class II polytropic murine leukemia viruses (MuLVs) of AKR/J mice: possible role in the generation of class I oncogenic polytropic MuLVs. J Virol. 1987;61:1882–1892. doi: 10.1128/jvi.61.6.1882-1892.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischinger P J, Nomura S, Bolognesi D P. A novel murine oncornavirus with dual eco- and xenotropic properties. Proc Natl Acad Sci USA. 1975;72:5150–5155. doi: 10.1073/pnas.72.12.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorman C. High efficiency gene transfer into mammalian cells. In: Glover D M, editor. DNA cloning. Vol. 2. Oxford, United Kingdom: IRL Press; 1985. pp. 143–190. [Google Scholar]
- 10.Harris J D, Blum H, Scott J, Traynor B, Ventura P, Haase A. Slow virus visna: reproduction in vitro of virus from extrachromosomal DNA. Proc Natl Acad Sci USA. 1984;81:7212–7215. doi: 10.1073/pnas.81.22.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartley J W, Wolford N K, Old L J, Rowe W P. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci USA. 1977;74:789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herr W, Gilbert W. Free and integrated recombinant murine leukemia virus DNAs appear in preleukemic thymuses of ARK/J mice. J Virol. 1984;50:155–162. doi: 10.1128/jvi.50.1.155-162.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishimoto A, Hartley J W, Rowe W P. Detection and quantitation of phenotypically mixed viruses: mixing of ecotropic and xenotropic murine leukemia viruses. Virology. 1977;81:263–269. doi: 10.1016/0042-6822(77)90143-x. [DOI] [PubMed] [Google Scholar]
- 14.Kabat D. Molecular biology of Friend viral erythroleukemia. Curr Top Microbiol Immunol. 1989;148:1–42. doi: 10.1007/978-3-642-74700-7_1. [DOI] [PubMed] [Google Scholar]
- 15.Kane J P, Hardman D A, Dimpfl J C, Levy J A. Apolipoprotein is responsible for neutralization of xenotropic type C virus by mouse serum. Proc Natl Acad Sci USA. 1979;76:5957–5961. doi: 10.1073/pnas.76.11.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan A S. Nucleotide sequence analysis establishes the role of endogenous murine leukemia virus DNA segments in formation of recombinant mink cell focus-forming murine leukemia viruses. J Virol. 1984;50:864–871. doi: 10.1128/jvi.50.3.864-871.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch W, Zimmermann W, Oliff A, Friedrich R. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J Virol. 1984;49:828–840. doi: 10.1128/jvi.49.3.828-840.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak C A. Genetic mapping of a mouse chromosomal locus required for mink cell focus-forming virus replication. J Virol. 1983;48:300–303. doi: 10.1128/jvi.48.1.300-303.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak C A. Susceptibility of wild mouse cells to exogenous infection with xenotropic leukemia viruses: control by a single dominant locus on chromosome 1. J Virol. 1985;55:690–695. doi: 10.1128/jvi.55.3.690-695.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Kozak, S., and D. Kabat. Unpublished results.
- 20.Kozak S L, Kabat D. Ping-pong amplification of a retroviral vector achieves high-level gene expression: human growth hormone production. J Virol. 1990;64:3500–3508. doi: 10.1128/jvi.64.7.3500-3508.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy J A. Xenotropic type C viruses. Curr Top Microbiol Immunol. 1978;79:111–213. doi: 10.1007/978-3-642-66853-1_4. [DOI] [PubMed] [Google Scholar]
- 22.Levy J A, Dimpfl J, Hardman D, Kane J P. Transfer of mouse anti-xenotropic virus neutralizing factor to human lipoproteins. J Virol. 1982;42:365–371. doi: 10.1128/jvi.42.2.365-371.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy J A, Ihle J N, Oleszko O, Barnes R D. Virus-specific neutralization by a soluble non-immunoglobulin factor found naturally in normal mouse sera. Proc Natl Acad Sci USA. 1975;72:5071–5075. doi: 10.1073/pnas.72.12.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyu M S, Kozak C A. Genetic basis for resistance to polytropic murine leukemia viruses in the wild mouse species Mus castaneus. J Virol. 1996;70:830–833. doi: 10.1128/jvi.70.2.830-833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyu M S, Nihrane A, Kozak C A. Receptor-mediated interference mechanism responsible for resistance to polytropic leukemia viruses in Mus castaneus. J Virol. 1999;73:3733–3736. doi: 10.1128/jvi.73.5.3733-3736.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacGregor G R, Mogg A E, Burke J F, Caskey C T. Histochemical staining of clonal mammalian cell lines expressing E. coli beta galactosidase indicates heterogeneous expression of the bacterial gene. Somatic Cell Mol Genet. 1987;13:253–265. doi: 10.1007/BF01535207. [DOI] [PubMed] [Google Scholar]
- 27.Miller A D, Wolgamot G. Murine retroviruses use at least six different receptors for entry into Mus dunni cells. J Virol. 1997;71:4531–4535. doi: 10.1128/jvi.71.6.4531-4535.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mullins J I, Chen C S, Hoover E A. Disease-specific and tissue-specific production of unintegrated feline leukaemia virus variant DNA in feline AIDS. Nature. 1986;319:333–336. doi: 10.1038/319333a0. [DOI] [PubMed] [Google Scholar]
- 29.O’Neill R R, Buckler C E, Theodore T S, Martin M A, Repaske R. Envelope and long terminal repeat sequences of a cloned infectious NZB xenotropic murine leukemia virus. J Virol. 1985;53:100–106. doi: 10.1128/jvi.53.1.100-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quint W, Boelens W, van Wezenbeek P, Cuypers T, Maandag E R, Selten G, Berns A. Generation of AKR mink cell focus-forming viruses: a conserved single-copy xenotrope-like provirus provides recombinant long terminal repeat sequences. J Virol. 1984;50:432–438. doi: 10.1128/jvi.50.2.432-438.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rein A, Schultz A. Different recombinant murine leukemia viruses use different cell surface receptors. Virology. 1984;136:144–152. doi: 10.1016/0042-6822(84)90255-1. [DOI] [PubMed] [Google Scholar]
- 32.Rosenberg N, Jolicoeur P. Retroviral pathogenesis. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 475–585. [PubMed] [Google Scholar]
- 33.Shaw G M, Hahn B H, Arya S K, Groopman J E, Gallo R C, Wong-Staal F. Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in the acquired immune deficiency syndrome. Science. 1984;226:1165–1171. doi: 10.1126/science.6095449. [DOI] [PubMed] [Google Scholar]
- 34.Silver J. Role of mink cell focus-inducing virus in leukemias induced by Friend ecotropic virus. J Virol. 1984;50:872–877. doi: 10.1128/jvi.50.3.872-877.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spain B H, Koo D, Ramakrishnan M, Dzudzor B, Colicelli J. Truncated forms of a novel yeast protein suppress the lethality of a G protein alpha subunit deficiency by interacting with the beta subunit. J Biol Chem. 1995;270:25435–25444. doi: 10.1074/jbc.270.43.25435. [DOI] [PubMed] [Google Scholar]
- 36.Stoye J P, Moroni C, Coffin J M. Virological events leading to spontaneous AKR thymomas. J Virol. 1991;65:1273–1285. doi: 10.1128/jvi.65.3.1273-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tailor C S, Nouri A, Lee C G, Kozak C, Kabat D. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc Natl Acad Sci USA. 1999;96:927–932. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas C Y, Coffin J M. Genetic alterations of RNA leukemia viruses associated with the development of spontaneous thymic leukemia in AKR/J mice. J Virol. 1982;43:416–426. doi: 10.1128/jvi.43.2.416-426.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weller S K, Joy A E, Temin H M. Correlation between cell killing and massive second-round superinfection by members of some subgroups of avian leukosis virus. J Virol. 1980;33:494–506. doi: 10.1128/jvi.33.1.494-506.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y L, Guo L, Xu S, Holland C A, Kitamura T, Hunter K, Cunningham J M. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat Genet. 1999;21:216–219. doi: 10.1038/6005. [DOI] [PubMed] [Google Scholar]