Abstract
Epigenetic clocks are age predictors that use machine-learning models trained on DNA CpG methylation values to predict chronological or biological age. Increases in predicted epigenetic age relative to chronological age (epigenetic age acceleration) are connected to aging-associated pathologies, and changes in epigenetic age are linked to canonical aging hallmarks. However, epigenetic clocks rely on training data from bulk tissues whose cellular composition changes with age. Here, we found that human naive CD8+ T cells, which decrease in frequency during aging, exhibit an epigenetic age 15–20 years younger than effector memory CD8+ T cells from the same individual. Importantly, homogenous naive T cells isolated from individuals of different ages show a progressive increase in epigenetic age, indicating that current epigenetic clocks measure two independent variables, aging and immune cell composition. To isolate the age-associated cell intrinsic changes, we created an epigenetic clock, the IntrinClock, that did not change among 10 immune cell types tested. IntrinClock shows a robust predicted epigenetic age increase in a model of replicative senescence in vitro and age reversal during OSKM-mediated reprogramming.
Subject terms: Immunology, Genetics
IntrinClock is an epigenetic clock that is resistant to potential confounds arising from age-dependent changes in immune cell composition and can robustly predict epigenetic aging in multiple contexts.
Introduction
Epigenetic clocks, age predictors based on DNA methylation levels at selected CpG loci, have grown in popularity as a tool to study aging and predict health outcomes in humans. The first epigenetic clocks developed by Hannum et al.1 and Horvath2 showed remarkably high accuracy (R > 0.90) in predictions of chronological age. These “first-generation” epigenetic clocks provide unique biological insights into the aging process. For example, some but not all forms of senescence (replicative, UV-induced, etc.) accelerate epigenetic clock age predictions3. Using a later clock trained on chronological age, Kabacik et al.4 identified nutrient sensing, mitochondrial activity and stem cell composition as being associated with epigenetic aging but not telomere attrition or genomic instability. A recent report demonstrated the development of an epigenetic clock effective at predicting age across a variety of species, providing evidence for a shared mammalian aging program5.
More recently, second-generation clocks designed to predict phenotypic aging measures have been developed. These clocks, including PhenoAge6 and GrimAge7, show strong associations with diseases, such as depression8 and mortality9. DunedinPACE is a similar marker of phenotypic aging that captures the pace of aging rather than the accumulation of aging10. These clocks show promise as markers of physiological aging, but their two-step construction methodology (training a DNA methylation predictor on measures of phenotypic rather than chronological age) adds a secondary layer of complexity to interpretation.
Given the ability of epigenetic clocks to detect aging phenotypes across species and levels of organization that include cells, tissues, and organs, there is interest in understanding the underlying mechanism(s) enabling their function. Recent reports have been released on this topic, notably including one by Levine et al.11 that suggests epigenetic clocks are composites of different modules characterized by their changes during aging and reprogramming. Novel epigenetic clocks have been developed that seek to capture the aging phenomenon in more defined ways, including by identifying CpG sites predicted to be causal by Mendelian randomization12 or those capturing purely stochastic variation13. These clocks are informative about aspects of the aging process and have the potential to be particularly well-suited for certain use cases.
One major challenge in understanding the mechanism(s) underlying epigenetic clocks is the confounding effect of age-related changes in cell-type composition of many tissues. While changes in cell-type composition are an important part of aging, they can make interpreting epigenetic clocks more difficult as the relevant CpG sites may be cell-type-specific markers rather than those affecting cell-intrinsic aging. Most epigenetic clocks are trained largely on blood, which sees a drop in naïve CD8+ T cells with age and a corresponding increase in more terminally differentiated memory T-cell types14. Some clocks may be more impacted by changes in cell-type composition than others, depending on how they were constructed15. These challenges are not limited to epigenetic clocks; measures of telomere length in whole blood as predictors of age have also been shown to be linked to the proportion of naïve T-cells16. Quite recently, T-cell and NK (natural killer) cell activation have been implicated as major drivers in epigenetic clock progression17.
Other approaches have been explored to create epigenetic age predictions that are less sensitive to changes in cell type composition. Most notably, residuals from regression models that include epigenetic age and proportions of several blood cell types have been used to generate an intrinsic epigenetic age acceleration measure18. While the resulting measure is cell-type independent, it becomes challenging to biologically interpret as the underlying signal is derived from a mixture of CpG sites that can be either cell type-independent or cell type-dependent. Other modern approaches include the development of single-cell epigenetic clocks19,20, though the underlying technology will require further maturing before it can match the sensitivity and accuracy of bulk measurement-based clocks.
In this work, we report our analysis of the differences in epigenetic age predictions derived from four epigenetic clocks (Hannum1, Horvath2, Horvath Skin and Blood21, and PhenoAge6) for cytotoxic CD8+ T cells at different stages of differentiation. We found that human naïve CD8+ T cells, which decrease in humans during aging, exhibit an epigenetic age 15–20 years younger than effector memory CD8+ T cells isolated from the same individual. Interestingly, naïve T cells isolated from individuals of different ages still show a progressive increase in epigenetic age. Based on these observations, which indicate, as predicted, that current epigenetic clocks measure two independent variables, aging and immune cell composition, we created an epigenetic clock, the IntrinClock, that does not change among 10 immune cell types tested. Remarkably, this clock shows an increase in a model of replicative senescence in vitro and shows decreased aging during OSKM reprogramming. Lastly, we investigate the IntrinClock’s applicability for use in studying and detecting the effects of cell-intrinsic perturbations on aging.
Results
Existing epigenetic clock age predictions depend on CD8+ T-cell differentiation state
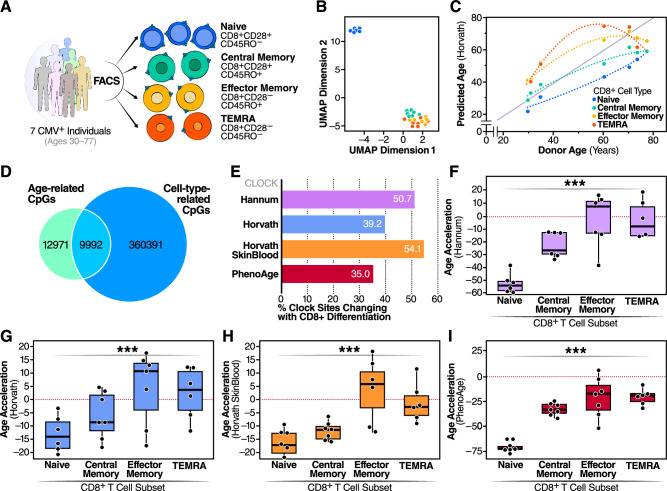
In humans, CD8+ T cells decrease in frequency, with a particularly pronounced loss of naive T cells during aging22. We used a negative bead-based selection method to isolate total T cells from seven donors (six men and one woman) of varying ages, all of whom were positive for cytomegalovirus (CMV+). We then used FACS to isolate CD8+ naive (CD8+ CD28+ CD45RO−), CD8+ central memory (CD8+ CD28+ CD45RO+), CD8+ effector memory (CD8+ CD28− CD45RO+), and CD8+ terminal effector memory RA+ (CD8+ CD28− CD45RO−) cells (Fig. 1A). After DNA isolation and profiling using the Illumina Infinium MethylationEPICTM platform, we noted a distinct clustering of CD8+ naive cells away from CD8+ central memory (CM), effector memory (EM), and terminal effector memory RA+ cells (TEMRA) (Fig. 1B) in UMAP analysis. Horvath clock epigenetic ages were measured in each of the CD8 T-cell subsets and found to correlate with age across every subset. However, strikingly, naive T cells consistently showed a significantly younger epigenetic age than other CD8+ subsets (p = .001) (Fig. 1C). This result suggests that epigenetic clock measurements are affected by CD8+ T-cell differentiation. Equally interestingly, naive CD8+ T cells from individuals of different chronological age showed an increase in epigenetic age that was parallel to chronological age but consistently lower than the chronological age (Fig. 1C). The same observation was made for CMs, EMs, and TEMRAs except that these cells’ epigenetic age appeared closer to the chronological age of the donors.
Fig. 1. CpG site changes during T-cell differentiation.
A Experimental design for determining impact of CD8+ differentiation on epigenetic clock age prediction. B UMAP dimensionality reduction of CD8+ DNA methylation profiles. C Differences between predicted epigenetic age as a function of donor age and CD8+ T-cell subset. D Comparison of shared CpG site changes between age in CD8+ T cells and CD8+ cell subset. E Percent of sites in four epigenetic clocks that are correlated with CD8+ T-cell differentiation. Comparison of the F Hannum (p = 1.1 * 10−7), G Horvath (p = 0.001), H Horvath skin and blood (p = 2.8 * 10−6), and I PhenoAge (p = 4.8 * 10−8) epigenetic age acceleration predictions for four CD8+ T-cell subsets. *** ANOVA p-value less than or equal to 0.001. Samples derived from N = 7 individuals. Boxplots are centered at median and bound one quartile on each side.
Next, using differential methylation analysis on methylation M-values, we identified 22,963 CpGs that changed with age and 370,383 CpGs that changed between naive CD8+ T cells and CD8+ CM, CD8+ EM, or CD8+ TEMRA cells. Of the 22,963 aging-related CpGs, 9,992 were also correlated with differentiation state (Fig. 1D). To understand how this could affect epigenetic clock predictions, we investigated the proportion of CpG sites used for epigenetic age prediction in the Hannum, Horvath, Horvath Skin and Blood, and PhenoAge clocks that we identified were correlated with CD8+ T-cell differentiation state. In all four clocks, more than a third of the predictive sites were correlated with differentiation state (Fig. 1E), and all four had a difference in age acceleration for CD8+ T-cell subsets. In all clocks, CD8+ TEMRA and CD8+ EM cells were predicted to be older than CD8+ CM cells, which were predicted to be older than CD8+ naive cells (Fig. 1F–I). The differences in epigenetic ages among the CD8+ T-cell subsets varied among clocks. For example, PhenoAge predicts CD8+ naive cells to be over 60 years younger than the donor chronological age, but the difference was much smaller for both Horvath clocks with an epigenetic age prediction of only approximately 12 years lower than chronological age (Fig. 1F–I).
Development of an epigenetic clock (IntrinClock) resistant to changes in immune cell composition
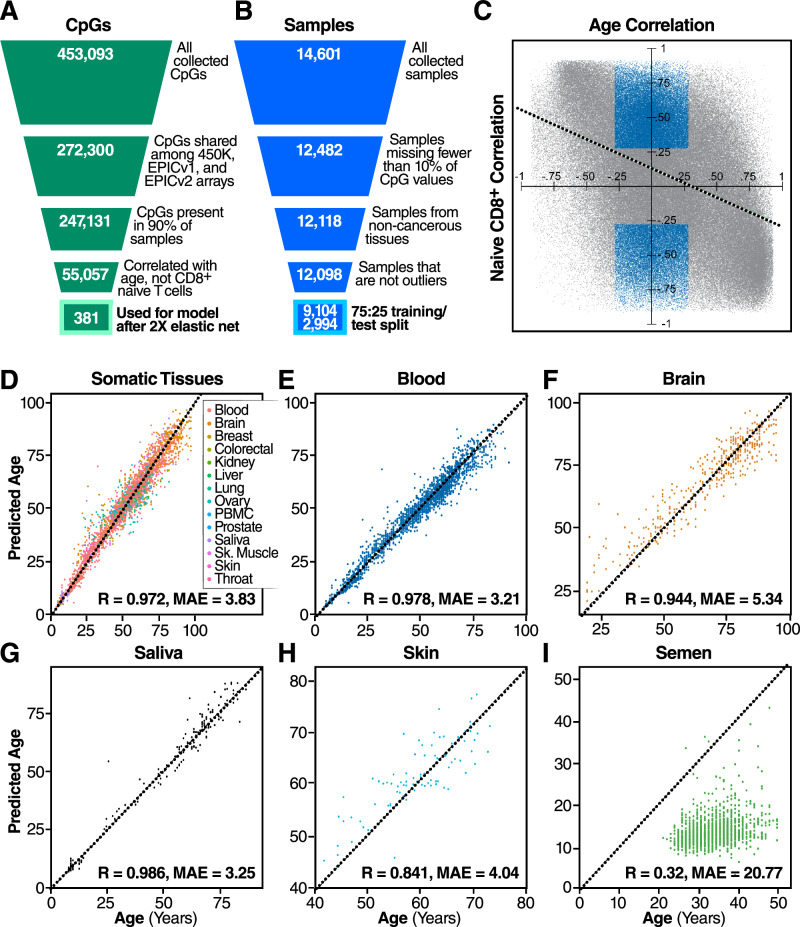
Given the overlap of DNA methylation signatures of cellular aging and CD8+ differentiation, we sought to create a new epigenetic clock that is unaffected by changes in immune cell composition. We began by generating a database of 14,601 DNA methylation samples from 71 different datasets1,22–91, generated on either the Illumina InfiniumTM HumanMethylation450 (450 K) or the Illumina InfiniumTM MethylationEPIC (EPIC) array, all sourced from the Gene Expression Omnibus (GEO) database or the Genotype-Tissue Expression project (GTEx) (Supplementary Table 1). The number of samples per dataset ranged from six to 1218, with a mean number of samples per dataset of 213 (Fig. S4A). The distribution of sexes was approximately equal (Fig. S4B). Samples were derived from a variety of tissues with the majority from blood (Fig. S4C), and the DNA methylation assay platform was split roughly evenly between the 450 K and the EPIC array. (Fig. S4D).
Once the database of samples was assembled, we performed a series of filtering and quality control steps. We filtered out all samples that were missing more than 10% of CpG sites measured by the 450 K array, those that were derived from cancerous tissue, and those that were derived from germline tissues. We then removed outliers, defining outliers as those with principal components more than two interquartile ranges away from the mean (Fig. 2B). After performing a random 75-25 training/test split, 9104 samples were used to train the model and 2994 were used to validate it.
Fig. 2. IntrinClock design strategy and performance.
A Filtering strategy for CpG sites. B Filtering strategy for samples. C Visualization of the filtering process for differentiation-independent age-related CpGs. Blue CpGs (those correlated with age but not with being a naive cell) were included in the feature set, whereas gray CpGs were not. Dashed line indicates linear least-squared regression line of relationship between CpG age correlation and CpG CD8+ naive cell correlation. D Correlation between age and IntrinClock predicted age in a variety of tissues from the test set. E–H Individual correlation plots for specific tissues in the test set. I Epigenetic age vs. chronological age correlation plot for semen samples.
Given the unique methylation pattern (Fig. 1B) and quiescent biology92 of naive CD8+ T cells, we aimed to use them as a basis on which to eliminate CpGs linked to CD8+ T-cell differentiation and performed additional filtering steps. When constructing our database of DNA methylation data, we initially collected all CpG sites measured by the 450 K array for all samples. To increase reliability, we first filtered out CpG sites that were present in fewer than 90 percent of samples. To ensure forward compatibility, we also included only CpG sites that were present on the Illumina InfiniumTM MethylationEPICv2.0 array. Next, we opted to remove any CpG sites that were correlated with a sample being a naive CD8+ sample (R > 0.3) within our CD8+ subset data (i.e., CpG sites whose methylation patterns were distinct in CD8+ naive cells as compared to CD8+ CM/EM/TEMRA cells). We also opted to include only those CpG sites correlated with age (R > 0.3) (Fig. 2C), to decrease the search space for the elastic net algorithm to identify age-predictive sites. Interestingly, we observed a negative correlation-of-correlations between the age correlation and naive CD8+ correlation of CpG sites (R = −0.45) (Fig. 2C), indicating that CpG sites that are hypermethylated with age tend to be hypomethylated in naive CD8+ cells, and vice-versa. We utilized the elastic net algorithm on the remaining 55,896 CpGs to generate a new epigenetic clock based on 410 CpG sites. To increase accuracy and reduce the number of necessary prediction sites, we used an approach whereby we employed the elastic net algorithm a second time on the training data filtered only on the 410 CpG sites used for the clock. This reduced the number of predictive CpG sites in the final model (IntrinClock) to 381, and reduced error by approximately 3 months (Fig. S5). We validated that a similar degree of improvement in predictive accuracy could not be obtained by tuning the alpha parameter (Fig. S5C).
IntrinClock is accurate across tissues, and its age predictions are not affected by adaptive immune cell compositional changes
Next, we tested the IntrinClock on a variety of tissues in the test set and observed high overall prediction accuracy (R ~ .972, mean absolute error (MAE)~3.83) (Fig. 2D). Age prediction errors on blood and saliva were particularly low (MAE~3.25, MAE ~ 3.21, respectively) (Fig. 2E, G). Tissues with less immune infiltration also had high epigenetic age correlations with chronological age (R ~ 0.944 for brain, R ~ 0.841 for skin). We were interested in discovering whether the IntrinClock would predict chronological age in semen samples, as previous epigenetic clocks have shown significant age deceleration in sperm2. We found that epigenetic age predictions of semen had only a weak correlation with chronological age (R ~ 0.32), and the predicted age of sperm samples, using a previously generated dataset93, appears to consistently be ~12 (Fig. 2I).
Importantly and as expected, IntrinClock applied to our generated CD8+ DNA methylation data showed no epigenetic age prediction differences among CD8+ T-cell subsets (p = 0.31) (Fig. 3A). As these samples were included in the training set for clock construction, we validated our approach on two external datasets94,95 with CD8+ naive and CD8+ EM DNA methylation data and found no differences in epigenetic age (paired t-test p-value = 0.17) (Fig. 3B). We also tested whether our clock could find a shift in epigenetic age between CD4+ naive and CD4+ CM cells, as the proportion of CD4+ naive cells also decreases with age96. Using two external data sets97,98, we discovered no evidence for a shift in epigenetic age between CD4+ naive and CM cells (Fig. 3C) (paired t-test p-value = 0.54), despite our filtering strategy being based only on CD8+ cells.
Fig. 3. Epigenetic age accelerations measured by different clocks.
A Differences in epigenetic age accelerations in different CD8+ subsets generated in this study. Horvath clock predictions overlaid in light gray. B Epigenetic ages of CD8+ naive cells and effector memory cells, based on data from GSE66564 and GSE83156. C Epigenetic ages of CD4+ naive cells and central memory cells, based on data from GSE121192 and GSE71825. D Epigenetic ages of PBMCs, CD8+ naive, CD8+ central memory, CD8+ combined effector and TEMRA, CD4+ naive, CD4+ central memory, B-cell naive, B-cell switched memory, CD16+CD56dim NK, and classical monocyte cells. E–I Association of percentage of e, effector memory CD8+ cells, f, central memory CD4+ cells, g, class-switched B cells, h, CD16+ CD56dim NK cells, and i, classical monocytes with epigenetic age acceleration. Samples derived from N = 9 individuals.
We also tested whether the IntrinClock would be similarly unperturbed in other immune cell types, particularly naive and memory B cells, which change in frequency with age99. We sorted CD8+ naive (CD8+CD28+CD45RO−), CD8+ CM (CD8+CD28+CD45RO+), CD8+ combined EM/TEMRA (CD8+CD28−), CD4+ naive (CD4+CD28+CD45RO−), CD4+ CM (CD4+CD28+CD45RO+), B-cell naive (CD3−CD19+CD27−IgD+), class-switched B cells (CD3−CD19+CD27+IgD−), CD16+CD56dim NK cells (CD3−CD19−CD56dimCD16+), classical monocytes (CD3−CD19−HLADR+CD14+CD16dim), and whole-peripheral blood mononuclear cell (PBMC) samples from a separate set of nine donors (five women, four men) aged 30–68 and collected DNA for methylation analysis. To increase cell recovery, we performed two sequential rounds of positive selection for CD8+ and then CD4+ cells using magnetic enrichment kits prior to flow sorting, similar to a published strategy34. Concurrently, we analyzed the PBMC samples using high-parameter spectral flow cytometry to empirically determine whether changes in immune cell composition of the PBMC samples would impact predicted epigenetic age of the whole PBMC fraction.
As predicted, we found no evidence for an association between cell subset and epigenetic age prediction (ANOVA p-value = 0.94) (Fig. 3D) or between cell subset and epigenetic age acceleration (ANOVA p-value = 0.34) (Fig. S6A). This remained consistent whether epigenetic age acceleration was defined as the difference between predicted age and chronological age or as the residual after regressing predicted epigenetic age on chronological age. In contrast, cell subset and epigenetic age acceleration were significantly correlated, according to the Hannum (p-value = 3.6 * 10−24; Fig. S6B), Horvath (p-value = 5.3 * 10−5; Fig. S6C), Horvath Skin and Blood (p-value = 6 * 10−17; Fig. S6D), and PhenoAge (p-value = 5.6 * 10−34; Fig. S6E) clocks. To further investigate how resistant IntrinClock is to the change in immune cell composition, we analyzed the correlation between the PBMC epigenetic age and percentage of several PBMC subsets. As expected, we identified no significant relationship between the PBMC epigenetic age acceleration and percentage of CD8+ EM cells (Pearson’s correlation p-value = 0.13; Fig. 3E), CD4+ CM cells (Pearson’s correlation p-value = 0.89; Fig. 3F), class-switched B cells (Pearson’s correlation p-value = 0.30; Fig. 3G), CD16+ CD56dim NK cells (Pearson’s correlation p-value = 0.63; Fig. 3H), or classical monocytes (Pearson’s correlation p-value = 0.45; Fig. 3I), relative to their parent populations. Combined with our observations of the IntrinClock’ s high accuracy across many tissues, including different cell types in the brain (Figure S6F–G), these observations indicate that shifts in immune cell composition do not impact IntrinClock age predictions.
IntrinClock is highly enriched for CpG sites upstream of transcription start sites, and its sites are enriched for motifs whose TFs are implicated in cancer
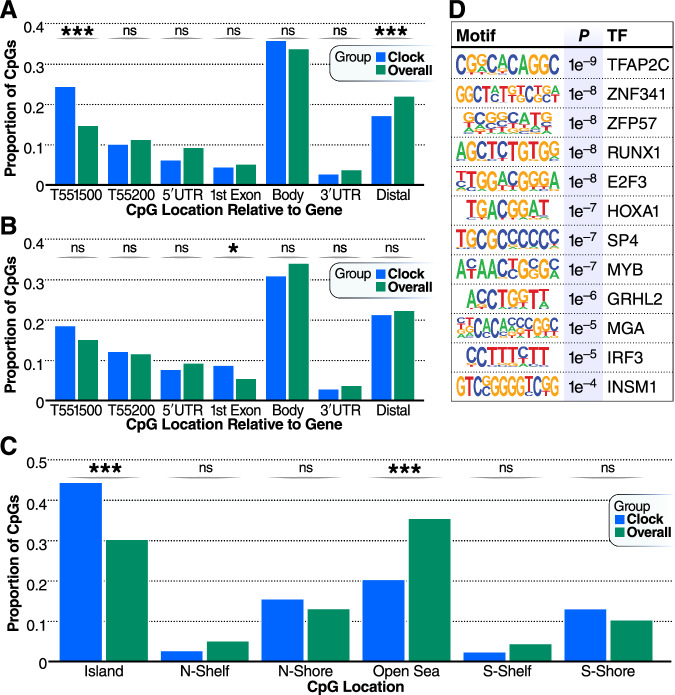
One central challenge in understanding epigenetic clocks comes from a lack of knowledge regarding to what extent epigenetic clocks are tracking a cell-autonomous or, conversely, a cell-ensemble phenomenon100. Our data provide evidence that current epigenetic clocks represent a composite of at least two variables, change in DNA methylation associated with aging in a cell intrinsic manner (IntrinClock), and a change in cell composition associated with aging. Due to the IntrinClock’s resistance to changes in immune cell composition, the CpG sites that constitute the clock may have more readily interpretable cell-autonomous biology as they are less likely to track markers of changing immune cell composition. This prediction could be particularly helpful in the context of identifying a functional or causal relationship between epigenetic clock sites and aging. We found that the sites in the IntrinClock that are hypermethylated with age are enriched within the region 200–1500 bp upstream of gene transcription start sites (25% in the clock vs 15% globally, p-value = 8.5 * 10−5), and correspondingly strongly depleted in sites distant from genes (17% in the clock vs. 25% globally, p-value = 0.008) (Fig. 4A). In sites that are hypomethylated with age, there was a significant enrichment within the first exon of genes (8% in the clock vs. 5% globally, p-value = 0.045) (Fig. 4B). DNA methylation changes within 1500 bp of the transcription start site are most closely linked to alterations in gene expression101. Similarly, IntrinClock CpGs are enriched for being located near CpG islands (45% in the clock vs. 31% globally, p-value = 1.9 * 10−9) and are depleted from open sea regions (20% in the clock vs. 36% globally, p-value = 2.3 * 10−10) (Fig. 4C).
Fig. 4. Distributions of CpG positions.
A Distributions of CpG positions relative to genes in IntrinClock sites that are hyper-methylated with age relative to background. B Distributions of CpG positions relative to genes in IntrinClock sites that are hypo-methylated with age relative to background. C Genomic distribution of IntrinClock CpG positions. D HOMER analysis of the top 12 motifs enriched within 19 bp on either side (5′ or 3′) of IntrinClock sites (40 bp total). *** one-sample proportion t-test p-value < 0.001; * < 0.05.
Transcription factor activity and DNA methylation are biologically connected both directly, as in the case of the OCT4 transcription factor preferring to bind to methylated DNA102, and indirectly, as in the case of passive methylation from lack of TF binding103,104. We investigated regions within 40 bp of IntrinClock CpG sites and used HOMER105 to identify enriched motifs associated with transcription factor-binding sites (Fig. 4D). Motifs associated with TFAP2C, ZNF341, ZFP57, RUNX1, E2F3, HOXA1, SP4, MYB, GRHL2, MGA, IRF3, and INSM1 binding were significantly enriched, compared to a 40-bp background of basepairs surrounding CpG sites that are assayed by both Illumina Infinium HumanMethylation450K and MethylationEPIC chips. Aberrant activity of each corresponding transcription factor has been associated with cancer development or worsened prognosis80,106–116. Some of these, such as E2F3117 and IRF3118, have been associated with aging-related diseases, whereas a connection for others has yet to be discovered.
We were interested in exploring general patterns of shifts in IntrinClock CpGs with age. To avoid uneven distribution of tissue samples across age groups, we focused our analysis on blood samples. Given that a linear regression model was used to build the IntrinClock, we were not surprised that the two most prevalent patterns were a linear decrease and increase, respectively, of DNA methylation with age (Fig. S7). However, we also found several CpGs (Clusters 4, 5, and 6) where the CpGs reverse their age-related direction of DNA methylation around the age of 21–30. This indicates that, for a subset of CpGs in the IntrinClock, there is a distinction between the processes of maturation and aging after sexual maturity. Interestingly, these CpGs were 2.3-fold (34% vs. 14.9%) enriched for being located 200–1500 bp upstream of a TSS, and 2-fold (19.4% vs. 10%) enriched for being located on a genomic south shore region (Fig. S8), which are more pronounced enrichments than identified for IntrinClock sites generally (Fig. 4A–C).
IntrinClock epigenetic age is accelerated in models of intrinsic hallmarks of aging and in HIV+ individuals
HIV has been associated with changes in DNA methylation state119,120, including changes in epigenetic age75. HIV infection is associated with a plethora of clinical manifestations and morbidities consistent with accelerated aging. However, HIV also causes major changes in immune cell composition121, which could skew previous versions of epigenetic clocks. As a result, it is unclear whether early results showcasing epigenetic age acceleration during HIV infection are due to changes in blood cell composition or an accelerated intrinsic rate of aging. Using the IntrinClock on previously generated data from HIV+ individuals and controls, we identified an HIV-associated increase in epigenetic age of two years (t-test p-value = 0.04), supporting the model that HIV leads to accelerated aging independently of shifts in immune cell composition (Fig. 5A). Furthermore, using a previously described cell composition prediction algorithm122 combined with a validated library123 generated on the HumanMethylation450k platform, we were able to predict changes in ten different immune cell types and their correlations with clock residuals and HIV status. We observed no association between IntrinClock residuals and immune cell proportions, including in the cases of eosinophils and neutrophils (Fig. 5B). In contrast, we observed significant associations using other epigenetic clocks, in a manner paralleling the changes seen in HIV (an increase in NK cells and a decrease in neutrophils).
Fig. 5. Impact on disease and in vitro interventions on the IntrinClock.
A IntrinClock epigenetic age in HIV+ and HIV- individuals, DNA methylation data from GSE67751. Samples derived from N = 92 individuals. B Correlation plot of HIV status, clock residuals, and predicted immune cell type proportions. C IntrinClock epigenetic age in COVID positive and COVID negative individuals, DNA methylation data from GSE167202. Samples derived from N = 525 individuals. D Correlation plot of COVID status, clock residuals, and predicted immune cell type proportions. E Epigenetic reprogramming affects fibroblast predicted IntrinClock age. DNA methylation data from GSE54848. N = 3 independent biological samples. F Induced replicative senescence in fibroblasts leads to an increase in IntrinClock predicted age. DNA methylation data from GSE91069. N = 3 independent biological samples. T-test p-values # < 0.10; * < 0.05; *** < 0.001. Boxplots are centered at median and bound one quartile on each side.
We also sought to investigate whether the IntrinClock would be accelerated by other acute immune-related diseases. Using a dataset primarily generated in 2020, we found that the IntrinClock age prediction was not affected by COVID-19 (t-test p-value = 0.88; Fig. 5C), contrary to findings in other epigenetic clocks where COVID-19 infection was associated with an increase in epigenetic age124. We utilized a library generated on the MethylationEPIC platform125 to predict changes in immune cell proportions in individuals with COVID. We did not observe an association of IntrinClock residuals with the relative proportions of any cell type (Fig. 5D). Unlike in the case of HIV infection, the associations between the epigenetic clock residuals of other epigenetic clocks and immune cell type proportions were more variable. The relationship between the residuals of several epigenetic clocks and immune cell type composition match the data we obtained on sorted populations, with naïve B and T cells generally positively correlated with epigenetic age and memory T cells associated with older epigenetic age prediction. As the data analyzed in this study were generated early in the COVID-19 pandemic, most individuals would have been acutely, rather than chronically, ill with COVID-19. It remains to be seen whether the IntrinClock will predict a higher epigenetic age in those who are infected with COVID-19 for a prolonged period (i.e., long COVID).
One application of epigenetic clocks is in tracking the effect of rejuvenating or aging interventions on cells. As the IntrinClock was developed on sites that are not shifting due to immune cell compositional changes, we reasoned it may be more sensitive to such interventions. Consistent with this idea, we used an external dataset126 to find that the IntrinClock is sensitive to Yamanaka factor–mediated reprogramming in fibroblasts. The study authors sorted cells positive for TRA-1-60+, a marker for de-differentiation, at six time points after initiation of reprogramming. We investigated IntrinClock epigenetic age predictions at each time point and found that, from an initial mean predicted epigenetic age of 31, the age prediction decreased to 20 after 11 days of OSKM-mediated reprogramming (t-test p-value = 0.03). A mean age of 0 was reached after 20 total days of reprogramming (Fig. 5E). Conversely, using publicly available data using an in vitro fibroblast model of replicative cellular senescence127, we found that the IntrinClock was progressively accelerated with cell divisions as cells become progressively more senescent. IntrinClock predicted values were greater after 14 population doublings (predicted age of 15 vs. 10), and then greater still with a predicted age of 20 after another 14 population doublings (t-test p-value = 0.038; Fig. 5F). This effect was comparable to that seen using the PhenoAge clock, and stronger relative to the Hannum, Horvath, and Horvath Skin & Blood clocks (Fig. S9).
Discussion
Epigenetic clocks hold great promise for the study of longevity due to their high correlation with age and (particularly for second-generation clocks) association with aging-related disease state. As diagnostic tools, they have the potential to serve as important predictive biomarkers for assessing biological age, determining risk for age-associated diseases, and assessing the efficacy of interventions that target the aging process128–132. Recent technical advances, such as the development of principal component clocks133 and novel techniques for cost reduction134, promise to increase reliability and usability further. However, their current status as a composite of multiple aging signals makes them difficult to interpret and to link to specific biological processes. As an example, a recent study in patients post-COVID 19 infection demonstrated a significant PhenoAge epigenetic age acceleration in individuals over the age of 50, but an epigenetic age reversal for those under the age of 50124. Further, the manner in which clocks track healthspan is not fully overlapping, as clocks can be independently predictive of mortality even when analyzed jointly135. This challenge in interpretation is equally important for cellular models of the hallmarks of aging. In models of senescence or reprogramming, the sensitivity or even direction of the perturbation on predicted epigenetic age can dramatically differ, depending on the epigenetic clock used. For example, in this study, we identified the Hannum clock as predicting an age reversal in a fibroblast model of cellular replicative senescence (Fig. S9).
The immune system changes dramatically with aging, and its decline can exacerbate or lead to many aging-related pathologies136. Clocks built solely on inflammatory markers can be used to predict age and risk of multimorbidity137. However, the presence of CpG sites that track primarily with immune cell markers makes epigenetic clocks applied to cell-intrinsic effects (e.g., cellular reprogramming in fibroblast cell culture) difficult to understand. Such sites can introduce background noise to the resulting measurement.
Here, using sorted CD8+ T-cell subsets, we observed that naive T cells consistently showed a younger epigenetic age than other CD8+ subsets (Fig. 1C), ranging from a 10-year average age under-prediction in some clocks to as high as a 60-year underprediction in others. We furthermore demonstrate that epigenetic clocks predict different ages depending on the cell type measured in subsets other than T cells, including in B cells and monocytes (Fig. S6). As many of these cell subsets are a substantial proportion of cells found in blood, it demonstrates cell composition plays a role in determining epigenetic age. Altogether, the cell subsets assessed in this study compromise between 40-60% of the PBMC blood cell fraction, and many of these subsets change in relative proportion with age138. These observations reinforce the finding that current epigenetic clocks represent the integration of at least two variables: cell intrinsic aging and changes in immune composition during aging.
To isolate these variables, we developed an epigenetic clock that is based on CpG sites that do not change with CD8+ T-cell differentiation (IntrinClock). We further observed that this clock predicts the same age in each individual across a wide variety of immune cell types. Interestingly, a filtering step based on naive CD8+ T cells can generate a clock that is not correlated with differentiation state in cells from different lineages, such as CD4+ cells or even B cells. This indicates part of a unique “CD8+ naive” signal may, in fact, be a conserved quiescence program shared by a variety of immune cells. This observation is supported by our finding that methylation patterns associated with naive CD8+ T cells have a negative correlation with those changing with aging (Fig. 2C). A connection between quiescence and aging is found in a wide variety of cell types, including neural stem cells139.
The IntrinClock’s higher proportion of sites near transcription start sites and CpG islands and its expected relationship with reprogramming and senescence suggest that it is tracking an intrinsic cellular aging program. Enrichment of IntrinClock CpG sites within motifs bound by transcription factors linked to cancer progression is consistent with a recent review investigating the connection between epigenetic clocks, global hypomethylation, cancer, and aging140. It will be important in the future to test whether acceleration of the IntrinClock is linked to disease states. This application could be a novel tool used to distinguish age-related diseases caused by aberrant cell-to-cell interactions from those caused by intrinsic cellular dysfunction. Further improvement could also be made on the IntrinClock design, such as by removing tissue-specific DNA methylation signatures from training. Although most IntrinClock CpG sites are not linked to tissue, a minority appear to be tissue-stabilizing for blood and brain tissue (Fig. S10).
The approach described here reduces the potential of cellular composition changes to be a confounder, particularly in blood or saliva samples, and will likely increase our understanding of biological aging and age-associated diseases. The IntrinClock holds the promise of being more sensitive to cell-intrinsic rejuvenation approaches, as its constituent CpG sites are not affected by immune cell composition. It may also be more closely linked to CpG sites with a functional or even causal relationship with the aging process. Overall, IntrinClock represents an instrument to add to the aging biomarker toolkit, with a potential wide variety of applications and uses.
Methods
Immune cell isolation, sorting, and DNA extraction
PBMCs were extracted from leukopheresis chambers from CMV+ donors. Donors were volunteers who donated plasma at a blood donation center in San Francisco after passing a health screening. Blood was first diluted 1:1 with PBS with 2% FBS. Diluted blood was slowly layered on top of 12 mL of Ficoll in a 50-mL Falcon conical tube. The tube was then centrifuged for 30 minutes at 2000 rpm at 21°C without applying a break. The layer containing white blood cells was removed, diluted with FBS-supplemented PBS, and centrifuged for 3 minutes at 2500 rpm. The cell pellet was re-suspended in 15 mL of ACK lysis buffer and incubated for 3 minutes. The cells were topped up with PBS with 2% FBS, centrifuged, and resuspended.
For the initial CD8+ epigenetic clock characterization experiment, an EasySepTM Human T Cell Isolation kit was used to extract T cells from the PBMC fraction. T cells were then washed, stained with 1:500 LIVE/DEADTM Fixable Near-IR Dead Cell staining kit, washed, stained with an antibody cocktail (Supplementary Table 2), and washed again. FACS was performed on a BD FACSAriaTM II instrument. DNA was isolated using a Zymo Quick-DNATM Microprep Plus kit.
For the second comprehensive immune cell-sorting experiment, 2 million PBMCs were frozen immediately after extraction. The remaining cells were then positively selected for a CD4 fraction using the EasySepTM Human CD4 Positive Selection Kit II. The CD4 cells were stained with 1:500 LIVE/DEADTM Fixable Near-IR Dead Cell staining kit, washed, and stained with CD4/CD8 antibody cocktail (Supplementary Table 2), and the remaining cells were positively selected for a CD8 fraction using the EasySepTM Human CD8 Positive Selection Kit II. Both CD8+ cells and remaining PBMCs were washed, stained with 1:500 LIVE/DEADTM Fixable Near-IR Dead Cell staining kit and washed again. CD8+ cells were stained with a CD4/CD8 antibody cocktail (Supplementary Table 2), and the remaining PBMCs were stained with a B Cell/NK Cell/Monocyte antibody cocktail (Supplementary Table 3), after blocking with human IgG. The gating strategy for T cell sorting is shown in Supplementary Fig. 1, and the gating strategy for remaining PBMCs is described in Supplementary Fig. 2. All three fractions were then subjected to FACS analysis using a BD FACSAriaTM II instrument. DNA was isolated using a Zymo Quick-DNA/RNATM Microprep Plus kit.
For both experiments, DNA was quantified using QubitTM HS dsDNA quantification reagents. Bisulfite conversion and DNA methylation assessment were performed by Diagenode. For all experiments involving FACS, post-sort validations were performed to verify cell sort purity by analyzing sorted populations via flow cytometry. The Clock Foundation assisted with facilitating DNA methylation assessment and data transfer for the initial CD8+ experiment.
High-dimensional flow cytometry
PBMCs were transferred to a 96-well V-bottom plate. Cells were re-suspended in a 1:500 dilution of LIVE/DEADTM Fixable Blue Dead Cell Stain kit in cold PBS and incubated for 30 minutes in the dark. Cells were then washed and blocked with human IgG for 30 minutes. They were then washed twice and stained with a PBMC phenotyping antibody cocktail (Supplementary Table 4). Cell phenotyping was performing on a Cytek AuroraTM instrument and analyzed using FlowJoTM. The flow cytometry analysis strategy is described in Supplementary Fig. 3.
DNA methylation analysis and pre-processing
idat files were converted into beta values by using the minfi R package141, with a functional normalization pre-processing step142. For differential methylation analyses, beta values were converted to M-values through the formula M = log2(B / (1-B)). The R package umap was used for UMAP dimensionality reduction142,143.
Dataset collection and pre-processing
All datasets used to build the novel epigenetic clock were either generated in this study or downloaded from GEO. Exact ages were obtained for GTEx data through dbGaP144, as exact chronological ages of tissues were required. For constructing the clock, the assembled database of DNA methylation data was first culled of any samples that had more than 10% of CpGs missing and of any CpGs that had more than 10% samples missing. All samples derived from cancer tissues were removed. To ensure forward compatibility, we filtered out CpGs that were not on the Infinium MethylationEPIC v2.0 array. Based on our CD8+ DNA methylation data, we tested the correlation of each CpG methylation with age and with naive CD8+ T cells. To assess whether CpGs were correlated with naive CD8+ cells, we binarized each naive sample as “ 1” and each non-naive (CM, EM, TEMRA) sample as “ 0” and then used the R cor function to compute a Spearman’s correlation between methylation and naive T-cell state. All CpGs with an absolute value correlation of .3 or greater with naive T-cell state were removed, and all CpGs with an absolute value correlation of .3 or less with age were removed.
Once CpGs and samples were filtered, the samples were split 75% for the training set and 25% for the test set. Imputation of missing was performed separately between training sets and test sets, and separately between different tissues within training sets and test sets (imputation performed using the impute R package145). Outliers were detected and removed using the outlyx function in the R watermelon package146. Untransformed beta values were used for model creation and age prediction. Prior to training the model, ages were transformed using Horvath’s formula used in his original epigenetic clock2. An elastic net model using glmnet147 was used to develop the IntrinClock, with alpha value set at .5. Once the first model was generated, the training data were a subset of only those CpGs with non-zero coefficients, which were used for training the final model. The regularization parameter for both elastic net models was generated using cross-validation (cv.glmnet() function) with ten folds.
Statistics and reproducibility
For comparisons between two measurements from one individual, as in Fig. 1B, C, paired t-tests were used for assessment of significant changes. For multiple comparisons between a group and a background reference, as in Fig. 4A, B, and C, one-sample proportional tests using the prop.test function from the R stats package were utilized with Bonferroni multiple-comparisons correction. For samples of multiple measurements, t-tests with Bonferroni multiple-comparisons corrections were used to test significance. Most graphs and figures were created with aid of the ggplot R package148.
Motif enrichment and pattern analyses
For motif enrichment analysis, the HOMER software tool was utilized105. To define sequences of interest, we investigated 40-bp windows surrounding the 381 CpG sites that compose the IntrinClock. As a background, we investigated 40-bp windows around CpG sites in our dataset immediately before removal of CpG sites associated with naive CD8+ cells and those not associated with aging. For investigating patterns of IntrinClock CpG shifts with age, beta values from blood samples were converted to M values, after which the degPatterns function from the DEGreport R package149 was utilized. Patterns with fewer than 10 CpG sites were discarded from analysis. Ages were binned into groups of 10 (0-10, 11-20, 21-30, 31-40, 41-50, 51-60, 61-70, 71-80, 81+). Each bin was confirmed to have at least 100 samples.
Epigenetic age acceleration analysis
To compute DNA methylation age for each epigenetic clock, the R methylclock package was utilized150. For experiments containing a limited number of donors or cell types, epigenetic age acceleration was defined as the difference between epigenetic age prediction and chronological age. For larger studies, epigenetic age acceleration was defined as the residual after regressing predicted epigenetic age on chronological age. To analyze associations between epigenetic clock residuals and predicted immune cell proportions, the estimateCellProp function from the ENmix151 package was utilized along with the corrplot152 package for plotting.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Materials
Acknowledgements
This research was primarily funded by the Michael Antonov Foundation, Buck institutional support, and NIH T32 training grant #AG052374. Cloud computing support was provided by the Amazon Web Services Activate program, facilitated by the On Deck Longevity Biotech fellowship. Furthermore, we extend our warm appreciation to Dr. Steve Horvath and the Clock Foundation for assisting us with sample handling and data interpretation during the early phases of this work.
Author contributions
N.A., H.M., R.T., and H.K. assisted in flow panel design and optimization. A.F. planned and conducted some of the experiments. R.R. and E.V. assisted in manuscript drafting. All authors contributed to manuscript editing and revising. A.T. designed, performed, and analyzed the experiments. A.T. drafted and wrote the manuscript.
Peer review
Peer review information
This manuscript has been previously reviewed at another Nature Portfolio journal. Communications Biology thanks Eric Latorre-Crespo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: George Inglis and Mengtan Xing. A peer review file is available.
Data availability
DNA methylation profiles generated in this study are publicly available under accession code GSE252045. Publicly available data used to generate the IntrinClock is listed in Supplementary Table 1. Source data underlying main figures are provided in Supplemental Data 1.
Code availability
Code used to generate the results in this study is available on Zenodo with 10.5281/zenodo.10426597153.
Competing interests
A.T. and E.V. are listed co-inventors on pending patents relating to work disclosed in this manuscript. Remaining authors declare no competing interests.
Ethics approval
NIH provided approval for use of phs000424/GRU (GTEx) age data via the dbGaP database approval system. Ethics approval was not required for other datasets generated. Informed consent was provided by participants in all studies analyzed for this manuscript.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06609-4.
References
- 1.Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell49, 359–367 (2013). 10.1016/j.molcel.2012.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol.14, R115 (2013). 10.1186/gb-2013-14-10-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perna, L. et al. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin. Epigenetics8, 64 (2016). 10.1186/s13148-016-0228-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kabacik, S. et al. The relationship between epigenetic age and the hallmarks of aging in human cells. Nat. Aging2, 484–493 (2022). 10.1038/s43587-022-00220-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu, A. T. et al. Universal DNA methylation age across mammalian tissues. Nat. Aging3, 1144–1166 (2023). [DOI] [PMC free article] [PubMed]
- 6.Levine, M. E. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging10, 573–591 (2018). 10.18632/aging.101414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu, A. T. et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging11, 303–327 (2019). 10.18632/aging.101684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Protsenko, E. et al. GrimAge,” an epigenetic predictor of mortality, is accelerated in major depressive disorder. Transl. Psychiatry11, 1–9 (2021). 10.1038/s41398-021-01302-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCrory, C. et al. GrimAge outperforms other epigenetic clocks in the prediction of age-related clinical phenotypes and all-cause mortality. J. Gerontol. A. Biol. Sci. Med. Sci.76, 741–749 (2021). 10.1093/gerona/glaa286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belsky, D. W. et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. eLife11, e73420 (2022). 10.7554/eLife.73420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine, M. E., Higgins-Chen, A., Thrush, K., Minteer, C. & Niimi, P. Clock Work: Deconstructing the epigenetic clock signals in aging, disease, and reprogramming. 2022.02.13.480245 Preprint at 10.1101/2022.02.13.480245 (2022).
- 12.Ying, K. et al. Causality-enriched epigenetic age uncouples damage and adaptation. Nat. Aging4, 231–246 (2024). [DOI] [PMC free article] [PubMed]
- 13.Meyer, D. H. & Schumacher, B. Aging clocks based on accumulating stochastic variation. Nat. Aging4, 871–885 (2024). [DOI] [PMC free article] [PubMed]
- 14.Goronzy, J. J., Fang, F., Cavanagh, M. M., Qi, Q. & Weyand, C. M. Naïve T cell maintenance and function in human aging. J. Immunol.194, 4073–4080 (2015). 10.4049/jimmunol.1500046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horvath, S. & Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet.19, 371–384 (2018). 10.1038/s41576-018-0004-3 [DOI] [PubMed] [Google Scholar]
- 16.Lin, Y. et al. Age-associated telomere attrition of lymphocytes in vivo is co-ordinated with changes in telomerase activity, composition of lymphocyte subsets and health conditions. Clin. Sci. Lond. Engl. 1979128, 367–377 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonkman, T. H. et al. Functional genomics analysis identifies T and NK cell activation as a driver of epigenetic clock progression. Genome Biol.23, 24 (2022). 10.1186/s13059-021-02585-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, B. H. et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging8, 1844–1859 (2016). 10.18632/aging.101020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonder, M. J. et al. Single cell DNA methylation ageing in mouse blood. 2023.01.30.526343 Preprint at 10.1101/2023.01.30.526343 (2023).
- 20.Trapp, A., Kerepesi, C. & Gladyshev, V. N. Profiling epigenetic age in single cells. Nat. Aging1, 1189–1201 (2021). 10.1038/s43587-021-00134-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath, S. et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging10, 1758–1775 (2018). 10.18632/aging.101508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazuardi, L. et al. Age-related loss of naïve T cells and dysregulation of T-cell/B-cell interactions in human lymph nodes. Immunology114, 37–43 (2005). 10.1111/j.1365-2567.2004.02006.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Policicchio, S. et al. Genome-wide DNA methylation meta-analysis in the brains of suicide completers. Transl. Psychiatry10, 1–13 (2020). 10.1038/s41398-020-0752-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pihlstrøm, L. et al. Epigenome-wide association study of human frontal cortex identifies differential methylation in Lewy body pathology. Nat. Commun.13, 4932 (2022). 10.1038/s41467-022-32619-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson, E. E. et al. Cytokine-induced molecular responses in airway smooth muscle cells inform genome-wide association studies of asthma. Genome Med12, 64 (2020). 10.1186/s13073-020-00759-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsaprouni, L. G. et al. Cigarette smoking reduces DNA methylation levels at multiple genomic loci but the effect is partially reversible upon cessation. Epigenetics9, 1382–1396 (2014). 10.4161/15592294.2014.969637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oelsner, K. T., Guo, Y., To, S. B.-C., Non, A. L. & Barkin, S. L. Maternal BMI as a predictor of methylation of obesity-related genes in saliva samples from preschool-age Hispanic children at-risk for obesity. BMC Genomics18, 57 (2017). 10.1186/s12864-016-3473-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viana, J. et al. Schizophrenia-associated methylomic variation: molecular signatures of disease and polygenic risk burden across multiple brain regions. Hum. Mol. Genet.26, 210–225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voisin, S. et al. An epigenetic clock for human skeletal muscle. J. Cachexia Sarcopenia Muscle11, 887–898 (2020). 10.1002/jcsm.12556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wockner, L. F. et al. Genome-wide DNA methylation analysis of human brain tissue from schizophrenia patients. Transl. Psychiatry4, e339–e339 (2014). 10.1038/tp.2013.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, H. et al. Sex-biased methylome and transcriptome in human prefrontal cortex. Hum. Mol. Genet.23, 1260–1270 (2014). 10.1093/hmg/ddt516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu, Z., Sandler, D. P. & Taylor, J. A. Blood DNA methylation and breast cancer: a prospective case-cohort analysis in the sister study. J. Natl Cancer Inst.112, 87–94 (2020). 10.1093/jnci/djz065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zannas, A. S. et al. Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-κB–driven inflammation and cardiovascular risk. Proc. Natl Acad. Sci.116, 11370–11379 (2019). 10.1073/pnas.1816847116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy, R. et al. DNA methylation signatures reveal that distinct combinations of transcription factors specify human immune cell epigenetic identity. Immunity54, 2465–2480.e5 (2021). 10.1016/j.immuni.2021.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Somineni, H. K. et al. Blood-derived DNA methylation signatures of Crohn’s disease and severity of intestinal inflammation. Gastroenterology156, 2254–2265.e3 (2019). 10.1053/j.gastro.2019.01.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeuchi, C. et al. Autoimmune gastritis induces aberrant DNA methylation reflecting its carcinogenic potential. J. Gastroenterol.57, 144–155 (2022). 10.1007/s00535-021-01848-2 [DOI] [PubMed] [Google Scholar]
- 37.Ringh, M. V. et al. Methylome and transcriptome signature of bronchoalveolar cells from multiple sclerosis patients in relation to smoking. Mult. Scler. J.27, 1014–1026 (2021). 10.1177/1352458520943768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roos, L. et al. Higher Nevus count exhibits a distinct DNA methylation signature in healthy human skin: implications for melanoma. J. Invest. Dermatol.137, 910–920 (2017). 10.1016/j.jid.2016.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magnaye, K. M. et al. DNA methylation signatures in airway cells from adult children of asthmatic mothers reflect subtypes of severe asthma. Proc. Natl Acad. Sci.119, e2116467119 (2022). 10.1073/pnas.2116467119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martino, D. et al. Epigenetic dysregulation of naive CD4+ T-cell activation genes in childhood food allergy. Nat. Commun.9, 3308 (2018). 10.1038/s41467-018-05608-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muse, M. E. et al. Genome-Scale DNA methylation analysis identifies repeat element alterations that modulate the genomic stability of Melanocytic Nevi. J. Invest. Dermatol.142, 1893–1902.e7 (2022). 10.1016/j.jid.2021.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicodemus-Johnson, J. et al. DNA methylation in lung cells is associated with asthma endotypes and genetic risk. JCI Insight1, (2016). [DOI] [PMC free article] [PubMed]
- 43.Nonino, C., N. Y., Noronha, C. F., Nicoletti, & M. A., Pinhel. Trait related and differential DNA Methylation in obese and normal weight Brazilian women. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166611 (2021).
- 44.Vyas, C. M. et al. Pilot study of genome-wide differences in DNA methylation among older adults with normal cognition and mild cognitive impairment, with and without neuropsychiatric symptoms. Alzheimers Dement. 17, e055497 (2021). 10.1002/alz.055497 [DOI] [Google Scholar]
- 45.Oliva, M. et al. DNA methylation QTL mapping across diverse human tissues provides molecular links between genetic variation and complex traits. Nat. Genet.55, 112–122 (2023). 10.1038/s41588-022-01248-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pai, S. et al. Differential methylation of enhancer at IGF2 is associated with abnormal dopamine synthesis in major psychosis. Nat. Commun.10, 2046 (2019). 10.1038/s41467-019-09786-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renauer, P. A., Coit, P. & Sawalha, A. H. The DNA methylation signature of human TCRαβ+CD4−CD8− double negative T cells reveals CG demethylation and a unique epigenetic architecture permissive to a broad stimulatory immune response. Clin. Immunol.156, 19–27 (2015). 10.1016/j.clim.2014.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ringh, M. V. et al. Tobacco smoking induces changes in true DNA methylation, hydroxymethylation and gene expression in bronchoalveolar lavage cells. eBioMedicine46, 290–304 (2019). 10.1016/j.ebiom.2019.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishak, M. et al. Genome-wide open chromatin methylome profiles in colorectal cancer. Biomolecules10, 719 (2020). 10.3390/biom10050719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Islam, S. A. et al. Integration of DNA methylation patterns and genetic variation in human pediatric tissues help inform EWAS design and interpretation. Epigenet. Chromat.12, 1 (2019). 10.1186/s13072-018-0245-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang, W. et al. Genome-wide identification of a methylation gene panel as a prognostic biomarker in nasopharyngeal carcinoma. Mol. Cancer Ther.14, 2864–2873 (2015). 10.1158/1535-7163.MCT-15-0260 [DOI] [PubMed] [Google Scholar]
- 52.Johnson, K. C., Houseman, E. A., King, J. E. & Christensen, B. C. Normal breast tissue DNA methylation differences at regulatory elements are associated with the cancer risk factor age. Breast Cancer Res.19, 81 (2017). 10.1186/s13058-017-0873-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kananen, L. et al. Aging-associated DNA methylation changes in middle-aged individuals: the Young Finns study. BMC Genomics17, 103 (2016). 10.1186/s12864-016-2421-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kandaswamy, R. et al. DNA methylation signatures of adolescent victimization: analysis of a longitudinal monozygotic twin sample. Epigenetics16, 1169–1186 (2021). 10.1080/15592294.2020.1853317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kasuga, Y. et al. DNA methylation analysis of cord blood samples in neonates born to gestational diabetes mothers diagnosed before 24 gestational weeks. BMJ Open Diabetes Res. Care10, e002539 (2022). 10.1136/bmjdrc-2021-002539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kho, M. et al. Epigenetic loci for blood pressure are associated with hypertensive target organ damage in older African Americans from the genetic epidemiology network of Arteriopathy (GENOA) study. BMC Med. Genomics13, 131 (2020). 10.1186/s12920-020-00791-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konigsberg, I. R. et al. Host methylation predicts SARS-CoV-2 infection and clinical outcome. Commun. Med.1, 1–10 (2021). 10.1038/s43856-021-00042-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langevin, S. M. et al. CpG island methylation profile in non-invasive oral rinse samples is predictive of oral and pharyngeal carcinoma. Clin. Epigenet.7, 125 (2015). 10.1186/s13148-015-0160-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis, S. K. et al. DNA methylation analysis validates organoids as a viable model for studying human intestinal aging. Cell. Mol. Gastroenterol. Hepatol.9, 527–541 (2020). 10.1016/j.jcmgh.2019.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kular, L. et al. DNA methylation as a mediator of HLA-DRB1*15:01 and a protective variant in multiple sclerosis. Nat. Commun.9, 2397 (2018). 10.1038/s41467-018-04732-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lunnon, K. et al. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat. Neurosci.17, 1164–1170 (2014). 10.1038/nn.3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gasparoni, G. et al. DNA methylation analysis on purified neurons and glia dissects age and Alzheimer’s disease-specific changes in the human cortex. Epigenet. Chromatin11, 41 (2018). 10.1186/s13072-018-0211-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gopalan, S. et al. Trends in DNA methylation with age replicate across diverse human populations. Genetics206, 1659–1674 (2017). 10.1534/genetics.116.195594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Witte, L.D. et al. Contribution of age, brain region, mood disorder pathology, and interindividual factors on the methylome of human microglia. Biol. Psychiatry91, 572–581 (2022). 10.1016/j.biopsych.2021.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hannon, E. et al. DNA methylation meta-analysis reveals cellular alterations in psychosis and markers of treatment-resistant schizophrenia. eLife10, e58430 (2021). 10.7554/eLife.58430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hearn, N. L., Chiu, C. L. & Lind, J. M. Comparison of DNA methylation profiles from saliva in Coeliac disease and non-coeliac disease individuals. BMC Med. Genomics13, 16 (2020). 10.1186/s12920-020-0670-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hong, S. R. et al. DNA methylation-based age prediction from saliva: High age predictability by combination of 7 CpG markers. Forensic Sci. Int. Genet.29, 118–125 (2017). 10.1016/j.fsigen.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 68.Horvath, S. et al. Obesity accelerates epigenetic aging of human liver. Proc. Natl Acad. Sci.111, 15538–15543 (2014). 10.1073/pnas.1412759111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horvath, S. et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol.17, 171 (2016). 10.1186/s13059-016-1030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao, C., Yi, S. & Huang, D. Genome-wide identification of age-related CpG sites for age estimation from blood DNA of Han Chinese individuals. Electrophoresis42, 1488–1496 (2021). 10.1002/elps.202000367 [DOI] [PubMed] [Google Scholar]
- 71.McEwen, L. M. et al. Systematic evaluation of DNA methylation age estimation with common preprocessing methods and the Infinium MethylationEPIC BeadChip array. Clin. Epigenet.10, 123 (2018). 10.1186/s13148-018-0556-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huynh, J. L. et al. Epigenome-wide differences in pathology-free regions of multiple sclerosis–affected brains. Nat. Neurosci.17, 121–130 (2014). 10.1038/nn.3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clement, J. et al. Umbilical cord plasma concentrate has beneficial effects on DNA methylation GrimAge and human clinical biomarkers. Aging Cell21, e13696 (2022). 10.1111/acel.13696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garcia-Prieto, C. A. et al. Epigenetic profiling and response to CD19 Chimeric antigen receptor T-cell therapy in B-cell malignancies. JNCI J. Natl Cancer Inst.114, 436–445 (2022). 10.1093/jnci/djab194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Horvath, S. & Levine, A. J. HIV-1 infection accelerates age according to the epigenetic clock. J. Infect. Dis.212, 1563–1573 (2015). 10.1093/infdis/jiv277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fries, G. R. et al. Accelerated hippocampal biological aging in bipolar disorder. Bipolar Disord.22, 498–507 (2020). 10.1111/bdi.12876 [DOI] [PubMed] [Google Scholar]
- 77.Estupiñán-Moreno, E. et al. Methylome and transcriptome profiling of giant cell arteritis monocytes reveals novel pathways involved in disease pathogenesis and molecular response to glucocorticoids. Ann. Rheum. Dis.81, 1290–1300 (2022). 10.1136/annrheumdis-2022-222156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davalos, V. et al. Epigenetic profiling linked to multisystem inflammatory syndrome in children (MIS-C): A multicenter, retrospective study. eClinicalMedicine50, 101515 (2022). 10.1016/j.eclinm.2022.101515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guintivano, J., Aryee, M. J. & Kaminsky, Z. A. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics8, 290–302 (2013). 10.4161/epi.23924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu, Y. et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol.31, 142–147 (2013). 10.1038/nbt.2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martino, D. et al. Longitudinal, genome-scale analysis of DNA methylation in twins from birth to 18 months of age reveals rapid epigenetic change in early life and pair-specific effects of discordance. Genome Biol.14, R42 (2013). 10.1186/gb-2013-14-5-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arpón, A. et al. Epigenome-wide association study in peripheral white blood cells involving insulin resistance. Sci. Rep.9, 2445 (2019). 10.1038/s41598-019-38980-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bacalini, M. G. et al. Identification of a DNA methylation signature in blood cells from persons with Down Syndrome. Aging7, 82–96 (2014). 10.18632/aging.100715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bartlett, T. E. et al. Antiprogestins reduce epigenetic field cancerization in breast tissue of young healthy women. Genome Med.14, 64 (2022). 10.1186/s13073-022-01063-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bauer, M. A. et al. Genome-Wide DNA methylation signatures predict the early asymptomatic doxorubicin-induced cardiotoxicity in breast cancer. Cancers13, 6291 (2021). 10.3390/cancers13246291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brennan, K. et al. NSD1 mutations deregulate transcription and DNA methylation of bivalent developmental genes in Sotos syndrome. Hum. Mol. Genet.31, 2164–2184 (2022). 10.1093/hmg/ddac026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cerapio, J. P. et al. Global DNA hypermethylation pattern and unique gene expression signature in liver cancer from patients with Indigenous American ancestry. Oncotarget12, 475–492 (2021). 10.18632/oncotarget.27890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cullell, N. et al. DNA Methylation and Ischemic Stroke Risk: An Epigenome-Wide Association Study. Thromb. Haemost. 1767–1778 10.1055/s-0042-1749328 (2022). [DOI] [PubMed]
- 89.Li, M. et al. Genomic methylation variations predict the susceptibility of six chemotherapy related adverse effects and cancer development for Chinese colorectal cancer patients. Toxicol. Appl. Pharmacol.427, 115657 (2021). 10.1016/j.taap.2021.115657 [DOI] [PubMed] [Google Scholar]
- 90.Cobben, J. M. et al. DNA methylation abundantly associates with fetal alcohol spectrum disorder and its subphenotypes. Epigenomics11, 767–785 (2019). 10.2217/epi-2018-0221 [DOI] [PubMed] [Google Scholar]
- 91.Charlton, J. et al. Methylome analysis identifies a Wilms tumor epigenetic biomarker detectable in blood. Genome Biol.15, 434 (2014). 10.1186/s13059-014-0434-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bennett, T. J., Udupa, V. A. V. & Turner, S. J. Running to Stand Still: Naive CD8+ T Cells Actively Maintain a Program of Quiescence. Int. J. Mol. Sci.21, 9773 (2020). 10.3390/ijms21249773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jenkins, T. et al. The impact of zinc and folic acid supplementation on sperm DNA methylation: results from the folic acid and zinc supplementation randomized clinical trial (FAZST). Fertil. Steril.117, 75–85 (2022). 10.1016/j.fertnstert.2021.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schlums, H. et al. Cytomegalovirus infection drives adaptive epigenetic diversification of nk cells with altered signaling and effector function. Immunity42, 443–456 (2015). 10.1016/j.immuni.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rodriguez, R. M. et al. Epigenetic networks regulate the transcriptional program in memory and terminally differentiated CD8+ T Cells. J. Immunol.198, 937–949 (2017). 10.4049/jimmunol.1601102 [DOI] [PubMed] [Google Scholar]
- 96.Li, M. et al. Age related human T cell subset evolution and senescence. Immun. Ageing16, 24 (2019). 10.1186/s12979-019-0165-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pitaksalee, R. et al. Differential CpG DNA methylation in peripheral naïve CD4+ T-cells in early rheumatoid arthritis patients. Clin. Epigenetics12, 54 (2020). 10.1186/s13148-020-00837-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Garaud, S. et al. FOXP1 is a regulator of quiescence in healthy human CD4+ T cells and is constitutively repressed in T cells from patients with lymphoproliferative disorders. Eur. J. Immunol.47, 168–179 (2017). 10.1002/eji.201646373 [DOI] [PubMed] [Google Scholar]
- 99.Chong, Y. et al. CD27+ (memory) B cell decrease and apoptosis-resistant CD27− (naive) B cell increase in aged humans: implications for age-related peripheral B cell developmental disturbances. Int. Immunol.17, 383–390 (2005). 10.1093/intimm/dxh218 [DOI] [PubMed] [Google Scholar]
- 100.Bell, C. G. et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol.20, 249 (2019). 10.1186/s13059-019-1824-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schlosberg, C. E., VanderKraats, N. D. & Edwards, J. R. Modeling complex patterns of differential DNA methylation that associate with gene expression changes. Nucleic Acids Res45, 5100–5111 (2017). 10.1093/nar/gkx078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yin, Y. et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science356, eaaj2239 (2017). 10.1126/science.aaj2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Medvedeva, Y. A. et al. Effects of cytosine methylation on transcription factor binding sites. BMC Genomics15, 119 (2014). 10.1186/1471-2164-15-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature489, 75–82 (2012). 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell38, 576–589 (2010). 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang, X. et al. TFAP2C promotes stemness and chemotherapeutic resistance in colorectal cancer via inactivating hippo signaling pathway. J. Exp. Clin. Cancer Res. CR37, 27 (2018). 10.1186/s13046-018-0683-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yan, D. et al. Developing ZNF gene signatures predicting radiosensitivity of patients with breast cancer. J. Oncol.2021, e9255494 (2021). 10.1155/2021/9255494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen, L. et al. ZFP57 suppress proliferation of breast cancer cells through down-regulation of MEST-mediated Wnt/β-catenin signalling pathway. Cell Death Dis.10, 1–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tuo, Z. et al. RUNX1 is a promising prognostic biomarker and related to immune infiltrates of cancer-associated fibroblasts in human cancers. BMC Cancer22, 523 (2022). 10.1186/s12885-022-09632-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Feng, Z. et al. E2F3 promotes cancer growth and is overexpressed through copy number variation in human melanoma. OncoTargets Ther.11, 5303–5313 (2018). 10.2147/OTT.S174103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hedrick, E., Cheng, Y., Jin, U.-H., Kim, K. & Safe, S. Specificity protein (Sp) transcription factors Sp1, Sp3 and Sp4 are non-oncogene addiction genes in cancer cells. Oncotarget7, 22245–22256 (2016). 10.18632/oncotarget.7925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cicirò, Y. & Sala, A. MYB oncoproteins: emerging players and potential therapeutic targets in human cancer. Oncogenesis10, 1–15 (2021). 10.1038/s41389-021-00309-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xiang, X. et al. Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLOS ONE7, e50781 (2012). 10.1371/journal.pone.0050781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mathsyaraja, H. et al. Loss of MGA repression mediated by an atypical polycomb complex promotes tumor progression and invasiveness. eLife10, e64212 (2021). 10.7554/eLife.64212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tian, M. et al. IRF3 prevents colorectal tumorigenesis via inhibiting the nuclear translocation of β-catenin. Nat. Commun.11, 5762 (2020). 10.1038/s41467-020-19627-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rocha, R. & Henrique, R. Insulinoma-Associated Protein 1 (INSM1): Diagnostic, prognostic, and therapeutic use in small cell lung. Cancer J. Mol. Pathol.3, 140–167 (2022). 10.3390/jmp3030013 [DOI] [Google Scholar]
- 117.Ki, S. et al. Global transcriptional profiling reveals distinct functions of thymic stromal subsets and age-related changes during thymic involution. Cell Rep.9, 402–415 (2014). 10.1016/j.celrep.2014.08.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang, X. et al. Interferon regulatory Factor 3 deficiency induces age-related alterations of the retina in young and old mice. Front. Cell. Neurosci.13, 272 (2019). 10.3389/fncel.2019.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arumugam, T., Ramphal, U., Adimulam, T., Chinniah, R. & Ramsuran, V. Deciphering DNA methylation in HIV infection. Front. Immunol.12, 795121 (2021). 10.3389/fimmu.2021.795121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mantovani, N. et al. Latency-associated DNA methylation patterns among HIV-1 infected individuals with distinct disease progression courses or antiretroviral virologic response. Sci. Rep.11, 22993 (2021). 10.1038/s41598-021-02463-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Douek, D. C. et al. Changes in thymic function with age and during the treatment of HIV infection. Nature396, 690–695 (1998). 10.1038/25374 [DOI] [PubMed] [Google Scholar]
- 122.Houseman, E. A. et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinforma.13, 86 (2012). 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Reinius, L. E. et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLOS ONE7, e41361 (2012). 10.1371/journal.pone.0041361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cao, X. et al. Accelerated biological aging in COVID-19 patients. Nat. Commun.13, 2135 (2022). 10.1038/s41467-022-29801-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Salas, L. A. et al. Enhanced cell deconvolution of peripheral blood using DNA methylation for high-resolution immune profiling. Nat. Commun.13, 761 (2022). 10.1038/s41467-021-27864-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ohnuki, M. et al. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc. Natl Acad. Sci. USA111, 12426–12431 (2014). 10.1073/pnas.1413299111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xie, W. et al. DNA methylation patterns separate senescence from transformation potential and indicate cancer risk. Cancer Cell33, 309–321.e5 (2018). 10.1016/j.ccell.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oblak, L., van der Zaag, J., Higgins-Chen, A. T., Levine, M. E. & Boks, M. P. A systematic review of biological, social and environmental factors associated with epigenetic clock acceleration. Ageing Res. Rev.69, 101348 (2021). 10.1016/j.arr.2021.101348 [DOI] [PubMed] [Google Scholar]
- 129.Duan, R., Fu, Q., Sun, Y. & Li, Q. Epigenetic clock: A promising biomarker and practical tool in aging. Ageing Res. Rev.81, 101743 (2022). 10.1016/j.arr.2022.101743 [DOI] [PubMed] [Google Scholar]
- 130.Fransquet, P. D., Wrigglesworth, J., Woods, R. L., Ernst, M. E. & Ryan, J. The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta-analysis. Clin. Epigenetics11, 62 (2019). 10.1186/s13148-019-0656-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Simpson, D. J. & Chandra, T. Epigenetic age prediction. Aging Cell20, e13452 (2021). 10.1111/acel.13452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Noroozi, R. et al. DNA methylation-based age clocks: From age prediction to age reversion. Ageing Res. Rev.68, 101314 (2021). 10.1016/j.arr.2021.101314 [DOI] [PubMed] [Google Scholar]
- 133.Higgins-Chen, A. et al. A computational solution to bolster epigenetic clock reliability for clinical trials and longitudinal tracking. Innov. Aging5, 5 (2021). 10.1093/geroni/igab046.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ultra-cheap and scalable epigenetic age predictions with TIME-Seq | bioRxiv. https://www.biorxiv.org/content/10.1101/2021.10.25.465725v1.full.
- 135.Li, X. et al. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. eLife9, e51507 (2020). 10.7554/eLife.51507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Weyand, C. M. & Goronzy, J. J. Aging of the immune system. mechanisms and therapeutic targets. Ann. Am. Thorac. Soc.13, S422–S428 (2016). 10.1513/AnnalsATS.201602-095AW [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sayed, N. et al. An inflammatory aging clock (iAge) based on deep learning tracks multimorbidity, immunosenescence, frailty and cardiovascular aging. Nat. Aging1, 598–615 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Terekhova, M. et al. Single-cell atlas of healthy human blood unveils age-related loss of NKG2C+GZMB-CD8+ memory T cells and accumulation of type 2 memory T cells. Immunity56, 2836–2854.e9 (2023). 10.1016/j.immuni.2023.10.013 [DOI] [PubMed] [Google Scholar]
- 139.Audesse, A. J. & Webb, A. E. Mechanisms of enhanced quiescence in neural stem cell aging. Mech. Ageing Dev.191, 111323 (2020). 10.1016/j.mad.2020.111323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Johnstone, S. E., Gladyshev, V. N., Aryee, M. J. & Bernstein, B. E. Epigenetic clocks, aging, and cancer. Science378, 1276–1277 (2022). 10.1126/science.abn4009 [DOI] [PubMed] [Google Scholar]
- 141.Aryee, M. J. et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics30, 1363–1369 (2014). 10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fortin, J.-P. et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol.15, 503 (2014). 10.1186/s13059-014-0503-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Konopka, T. umap: Uniform Manifold Approximation and Projection. (2023).
- 144.THE GTEX CONSORTIUM. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science369, 1318–1330 (2020). 10.1126/science.aaz1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hastie, T., Tibshirani, R., Narasimhan, B. & Chu, G. impute: Imputation for microarray data. Bioconductor version: Release (3.16) 10.18129/B9.bioc.impute (2023).
- 146.Schalkwyk, L. C. et al. wateRmelon: Illumina 450 and EPIC methylation array normalization and metrics. Bioconductor version: Release (3.16) 10.18129/B9.bioc.wateRmelon (2023).
- 147.Friedman, J., Tibshirani, R. & Hastie, T. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw., 33, 1–22 (2010). [PMC free article] [PubMed]
- 148.Wickham, H. ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag, New York, 2016).
- 149.Pantano, L. et al. DEGreport: Report of DEG analysis. Bioconductor version: Release (3.16) 10.18129/B9.bioc.DEGreport (2023).
- 150.Pelegri-Siso, D. & Gonzalez, J. R. Methylclock - DNA methylation-based clocks. Bioconductor version: Release (3.16) 10.18129/B9.bioc.methylclock (2023).
- 151.Xu, Z., Niu, L. & Taylor, J. ENmix: Quality control and analysis tools for Illumina DNA methylation BeadChip. Bioconductor version: Release (3.17) 10.18129/B9.bioc.ENmix (2023).
- 152.Wei, T. corrplot. https://www.rdocumentation.org/packages/corrplot/versions/0.92.
- 153.Tomusiak, A. IntrinClock. Zenodo 10.5281/zenodo.10426597 (2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Materials
Data Availability Statement
DNA methylation profiles generated in this study are publicly available under accession code GSE252045. Publicly available data used to generate the IntrinClock is listed in Supplementary Table 1. Source data underlying main figures are provided in Supplemental Data 1.
Code used to generate the results in this study is available on Zenodo with 10.5281/zenodo.10426597153.