Abstract
Infection by the neuropathogenic murine leukemia virus (MLV) TR1.3 results in hemorrhagic disease that correlates directly to in vivo syncytium formation of brain capillary endothelial cells (BCEC). This phenotype maps to amino acid 102 in the envelope (Env) protein of TR1.3. Substitution of glycine (G) for tryptophan (W) at this position (W102G Env) in the nonpathogenic MLV FB29 induces both syncytium formation and neurologic disease in vivo. Using an in vitro gene reporter cell fusion assay, we showed that fusion either with murine NIH 3T3 cells or with nonmurine target cells that expressed receptors at or below endogenous murine levels mirrored that seen in BCEC in vivo. In these instances only TR1.3 and W102G Env induced cell fusion. In contrast, when receptor levels on nonmurine cells were raised above endogenous murine levels, FB29 Env was as fusogenic as the neuropathogenic TR1.3 and W102G Env. These results indicate that TR1.3 Env and W102G Env are intrinsically more fusogenic than FB29 Env, that the induction of fusion requires a threshold number of receptors that is greater for FB29 Env than for TR1.3 or W102G Env, and that receptor density on murine NIH 3T3 cells and BCEC is below the threshold for FB29-dependent fusion. Surprisingly, receptor density on NIH 3T3 cells could not be increased by stable expression of exogenous receptors, and FB29-dependent fusion was not observed in NIH 3T3 cells that transiently expressed elevated receptor numbers. These results suggest that an additional undefined host cell factor(s) may limit both receptor expression and fusion potential in murine cells.
Endothelial cell damage is thought to be an important initiating event in the development of a number of diseases, including atherosclerosis, stroke, and cerebral hemorrhage (4, 14, 40). Evidence from several systems now suggests that infectious agents may participate in these processes. Both herpes simplex virus type 1 and cytomegalovirus infect endothelial cells (16, 22), induce the up-regulation of adhesion molecules (2, 13, 18, 41, 45), and are associated with the formation of atherosclerotic plaques (4, 28, 53). There is also evidence that patients infected with varicella-zoster virus (36, 42) or with human immunodeficiency virus type 1 (8, 12) suffer a high frequency of stroke. However, the precise mechanisms whereby infectious agents cause acute or chronic endothelial cell damage and disease have not been defined.
Infection of endothelial cells is characteristic of several murine leukemia viruses (MLV) (10, 23, 34). MLV infection may lead to neuropathy, which has been linked to the extent of virus replication in brain capillary endothelial cells (BCEC) (10, 26, 27, 46). The Friend MLV clone TR1.3 replicates aggressively in neonatal BCEC and induces syncytium formation of BCEC in vivo (30–32). Syncytium formation of BCEC triggers a cascade of pathologic changes that include breakdown of the blood-brain barrier, stroke formation, neurological disease, and death of infected mice (31, 32). The demonstration of a direct relationship between neurological disease and cytopathic syncytium formation of BCEC during in vivo infection by TR1.3 provides a unique and powerful model to investigate the molecular and cellular mechanisms of both endothelial damage and retroviral pathology within the central nervous system.
Sequence comparison indicates that TR1.3 is closely related to FB29, a leukemogenic Friend MLV that is endothelial-cell tropic but neither pathogenic nor syncytium inducing in BALB/c mice (33). The primary determinant of in vivo pathogenesis was mapped by using recombinant viruses to position 102, which lies within variable region A of the external (surface [SU]) domain of Env (33). The introduction of a tryptophan-to-glycine substitution at position 102 (W102G) in FB29 Env is sufficient to produce BCEC fusion, stroke formation, and mortality that are indistinguishable from the progression seen with TR1.3. The mechanism whereby this change in Env mediates cell and organismal pathology is a central question of ongoing studies in our laboratories. In this regard, early comparisons of the cytopathic effects that accompany TR1.3 infection in vivo and in vitro brought a striking paradox to light. TR1.3 infects endothelium throughout the body, but cell fusion and pathology are detected only within BCEC (30). In contrast, TR1.3 induces widespread infection and fusion of murine fibroblast lines in vitro. Taken together, these observations suggest that unique features of both viruses and host cells regulate the susceptibility to cell fusion.
We attempted to understand TR1.3 pathology by comparing syncytium induction by TR1.3 and W102G Env with that induced by FB29 Env in murine, human, and quail cells that expressed various levels of ecotropic virus receptors. Here we show that the W102G substitution renders Env protein more intrinsically fusogenic, that induction of syncytium formation requires a threshold level of virus receptors, and that the intrinsic difference in fusion ability results in a greater threshold receptor level for FB29 Env than for the TR1.3 or W102G Env protein. We also show that the endogenous level of receptors on murine cells is below the threshold for FB29 Env but above that for the TR1.3 and W102G Env proteins. Augmentation of receptor expression on NIH 3T3 cells increased the overall level of fusion but failed to elevate fusion with FB29 Env to that seen with either TR1.3 or W102G Env. In contrast, FB29 was as fusogenic as TR1.3 and W102G when receptor levels were increased in human 293 cells. These observations suggest that a host cell factor(s) may limit receptor density in NIH 3T3 cells but not in transformed quail or human cells. A model for the mechanism of TR1.3 and W102G neuropathogenesis is discussed.
MATERIALS AND METHODS
Constructs.
Molecular clones of FB29, TR1.3, or W102G in pUC19B (51) were first digested with AscI and BsaAI enzymes to prepare MLV env constructs. env gene fragments were agarose gel purified, and overhanging ends were filled in with the Klenow fragment of Escherichia coli DNA polymerase I (Promega) and then ligated into pcDNA I (Invitrogen) at the EcoRV site. The orientation of env was confirmed by restriction endonuclease analysis. The plasmid pJET, which encodes the ecotropic receptor murine cationic amino acid transporter (MCAT-1), was the kind gift of James Cunningham (Harvard Medical School). Construction of the Mcat-1 expression plasmid utilized pcDNA3 (Invitrogen) under the control of the cytomegalovirus promoter as previously reported (25). Robert Doms (University of Pennsylvania) kindly provided vTF1.1, a recombinant vaccinia virus encoding the T7 RNA polymerase (5). The luciferase-T7 plasmid was purchased from Promega.
Cells.
The Japanese quail fibrosarcoma cell line QT6 (ATCC CRL-1708) was provided by Paul Bates (University of Pennsylvania). The following cell lines were obtained from the American Type Culture Collection: NIH 3T3, murine embryo cells (CRL-1658), and 293T, a transformed primary human embryonal kidney cell line expressing the simian virus 40 T antigen. Cell lines 387, 1475, and 427, which express various levels of MCAT-1, were derived by transfection of human 293 cells (ATCC CRL-1573) with pJET or with the pcDNA3 MCAT-1 plasmid as previously described (1, 25). Tissue culture media and supplements were purchased from Life Technologies, Inc., unless otherwise noted. All cell lines were maintained in Dulbecco’s modified Eagle medium (DMEM) with a high glucose concentration and supplemented with 10% fetal bovine serum (FBS) (HyClone), 2 mM glutamine, and penicillin-streptomycin. Cells were grown at 37°C in 5% CO2.
Viruses.
Infectious MLV were generated according to previously described methods (33). Virions were purified by centrifugation at 70,000 × g over a 20% sucrose cushion for 2 h (7). Purified virions were quantified by immunoblotting or by reverse transcriptase (RT) assay as described previously (33). For quantification by immunoblotting, goat anti-SU gp70 and goat anti-capsid p30 (Quality Biotech, Camden, N.J.) were reacted with viral proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Reactive antibody was detected with 125I-protein G (Dupont NEN) and then quantified on a STORM image analysis system (Molecular Dynamics).
Gene reporter cell fusion assay.
Cell fusion was quantified by the gene reporter fusion assay described by Nussbaum et al. (29). Effector cells expressed T7 polymerase and Env protein. Target cells expressed luciferase under the control of the T7 promoter in addition to the ecotropic receptor (MCAT-1) under the control of the simian virus 40 promoter. Effector cells were produced by exposing QT6 cells to the recombinant vaccinia virus vTF1.1 (multiplicity of infection of 10) in DMEM–2% FBS at 37°C for 5 h. After 1 h of incubation, a calcium phosphate precipitate of the appropriate Env constructs was added to this mixture. Cells were then washed once with phosphate-buffered saline (PBS) and incubated at 32°C overnight in the presence of rifampin. Target cells were produced by calcium phosphate cotransfection of the pJET plasmid encoding the MLV receptor and the luciferase-T7 plasmid, followed by one wash in PBS and overnight incubation in fresh medium at 37°C. To initiate fusion, 2 × 105 effector cells were added to 1 × 105 target cells in 24-well plates at 37°C in the presence of 1-β-d-arabinofuranosylcytosine to inhibit further vaccinia virus replication. In some experiments, 2 × 105 target cells were used. To quantitate fusion at different times after initiation, lysates were prepared in 0.5% Triton X-100. The Promega luciferase assay was performed on 50-μl aliquots of lysates according to the manufacturer’s instructions, and the luciferase activity was measured as relative light units (RLU) on a Wallac luminometer. Data were analyzed by calculating the ratio of RLU observed in Env-positive effectors mixed with the specified target cells to the RLU observed in Env-negative effectors mixed with the same target cells (defined as the luciferase activity ratio). Statistical analyses were performed on these data by using a Student two-tailed t test and probability calculations (39).
Virus binding assay.
Surface expression of MLV receptors was quantified by determining the relative number of virus binding sites per cell by using a flow cytometry assay with the following modifications (21, 52). Briefly, a total of 2 × 105 cells were mixed with 200 μl of concentrated FB29 virus over a dilution range in the presence of 8 μg Polybrene per ml and incubated at 37°C for 20 min with gentle agitation. The cells were washed twice with wash buffer (5% FBS and 0.1% sodium azide in PBS) and then incubated with 100 μl of a 1:50 dilution of polyclonal goat anti-Rauscher SU (anti-gp70; Quality Biotech) in wash buffer for 1 h at 4°C. Following two washes, the cells were incubated for 1 h at 4°C in a 1:50 dilution of phycoerythrin-conjugated rabbit anti-goat immunoglobulin G (Sigma). After two final washes, the cells were resuspended in 4% paraformaldehyde in PBS on ice and analyzed by flow cytometry (FACS Star Plus; Becton Dickinson, San Jose, Calif.). The mean channel fluorescence (MCF) intensity of cells incubated with different dilutions of concentrated virus was normalized to the amount of RT activity. The saturating MCF intensities were determined and averaged for statistical analysis of the standard error of the mean. The background MCF was quantified on 293 cells lacking MLV receptors or on NIH 3T3 cells incubated with normal goat serum (isotype control) and secondary antibodies after binding virus. Data presented represent the MCF minus the background MCF. The MCF was linear and saturated within the range of FB29 particle dilutions used for all lines tested.
Cell fusion assay from without.
Cells were plated in 24-well plates at 1 × 105 to 2 × 105 cells per well and treated with 50 μM zidovudine (AZT) (Sigma) in DMEM overnight to inhibit retroviral RT activity and virus infection (9). Cells were then incubated overnight with equivalent amounts of purified viruses in the presence of AZT and 8 μg of Polybrene per ml. Virus concentrations were normalized by RT activity. Cells were then fixed in methanol and stained with methylene blue. Syncytia were quantified by light microscopy, counting nuclei in 10 random fields under a magnification of ×200.
RESULTS
The FB29, TR1.3, and W102G Env proteins mediate comparable levels of cell fusion in quail and human cells expressing MLV receptor MCAT-1.
Syncytium formation of murine BCEC in vivo and SC-1 cells in vitro is induced by TR1.3 and W102G Env but not FB29 Env (33). In order to isolate and then analyze the individual contributions of Env, receptor, and host cell type to fusion, we employed an in vitro cell fusion assay that utilizes a vaccinia virus-based gene reporter (29). This assay measures fusion mediated by Env expressed within cells, termed fusion from within, which is believed to be the primary pathway of in vivo syncytium formation (20). Briefly, effector cells that expressed MLV Env and T7 RNA polymerase were mixed with target cells that expressed ecotropic receptor MCAT-1 and a T7 promoter-driven luciferase reporter gene. Membrane fusion between effector and target cells results in transcription of the luciferase gene by T7 RNA polymerase. Luciferase activity thus provides an indirect, but proportional, measurement of cell fusion.
We first quantified fusion between quail (QT6) cells that transiently expressed different env constructs, shown in Fig. 1, and human fetal kidney (293T) cells that transiently expressed Mcat-1 cDNA (293T-MCAT-1). In striking contrast to the differential fusion of murine BCEC and SC-1 cells by TR1.3 and FB29, equivalent levels of fusion were observed in effector QT6 cells that expressed either TR1.3, W102G, or FB29 Env (Fig. 2A). Each MLV Env protein stimulated an approximately 10-fold increase in cell fusion relative to fusion controls. Env controls were QT6 effectors transfected with vector alone (QT6-pcDNA I), while receptor controls were 293T targets transfected with vector alone (293T-pcDNA I).
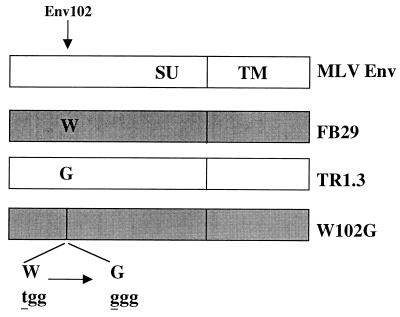
FIG. 1.
Diagram of expression constructs of MLV envelope proteins. The amino acid at position 102 is indicated for each virus. The nucleotide (lowercase) and amino acid (capitals) sequences for the region of W102G Env appear at the bottom.
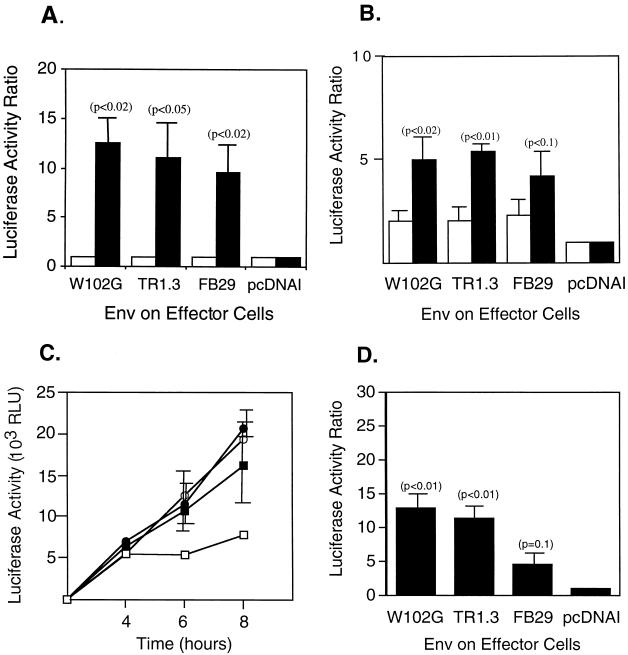
FIG. 2.
Cell fusion directed by MLV Env and MCAT-1 in a fusion-from-within assay. QT6 effector cells transiently expressing Env and T7 RNA polymerase were mixed with QT6, 293T, or NIH 3T3 target cells containing a luciferase reporter gene under the control of the T7 promoter and, in the instance of QT6 and 293T cells, transiently expressing the ecotropic MLV receptor MCAT-1. Data are representative of three independent experiments, each performed in triplicate. Error bars represent standard deviations. Significance was determined by the Student t test. Probability values of Env-positive compared to Env-negative (pcDNA I) controls are presented in parentheses. (A) Fusion between QT6 Env-positive effector cells and human 293T target cells transfected with either Mcat-1 (solid bars) or the vector control (open bars) at 8 h. (B) Fusion between QT6 Env-positive effector cells and QT6 target cells transfected with either Mcat-1 (solid bars) or the vector control (open bars) at 8 h. (C) Kinetics of fusion between QT6 receptor-positive cells and QT6 effector cells transfected with either FB29 Env (filled circles), TR1.3 Env (open circles), W102G Env (filled squares), or the vector control (open squares). (D) Fusion between NIH 3T3 target cells expressing endogenous MCAT-1 and QT6 effector cells transfected with either MLV Env or the vector control.
Similar analyses were conducted with QT6 cells as targets (QT6-MCAT-1). As shown in Fig. 2B, the overall level of fusion with QT6 targets was comparable to that seen with 293T targets. More importantly, fusion of effector cells expressing either TR1.3 or W102G Env was indistinguishable from fusion of effector cells expressing FB29 Env. We also used this system to examine the time course of cell fusion with different MLV Env proteins. As shown in Fig. 2C, the magnitude of fusion was the same for each Env protein for up to 8 h after cell mixing.
TR1.3 and W102G Env proteins mediate greater fusion than FB29 Env with murine NIH 3T3 target cells.
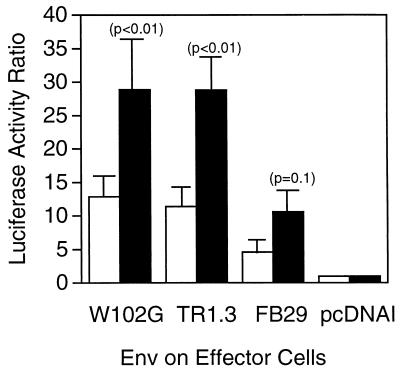
The results of studies which utilized nonmurine cells as fusion targets were at odds with previous in vivo and in vitro observations on murine targets (30, 33). Therefore, we examined the effectiveness of murine NIH 3T3 cells that expressed endogenous MCAT-1 to serve as fusion targets with QT6 effectors that expressed either TR1.3, W102G, or FB29 Env. As shown in Fig. 2D, both TR1.3 and W102G Env stimulated a >10-fold increase above the background in cell fusion with NIH 3T3 target cells. In contrast, fusion with FB29 Env effectors was not significantly above the background and was statistically less than seen with either TR1.3 (P < 0.02) or W102G (P < 0.02) Env.
Differential syncytium formation of murine and nonmurine cells in a fusion-from-without assay.
In view of these observations, we compared Env-mediated syncytium formation by another fusion pathway, termed fusion from without. In this instance MLV Env was provided directly by virus particles, in high concentrations, instead of expressed on the cell surface. Virus particles were first purified by separation through a sucrose cushion and then analyzed for particle integrity both by electron microscopy and by immunoblotting with anti-Env and anticapsid antiserum (data not shown). The relative level of virus was quantified by RT activity. Fusion of nonmurine cells was determined with 293T-MCAT-1 cells as targets, and fusion with murine cells utilized NIH 3T3 cells as targets. Cells were pretreated with and maintained in AZT during incubation with virus particles to inhibit virus replication and Env expression. The analysis of fusion with nonmurine 293T-MCAT-1 cells is shown in Fig. 3. In agreement with the results described above for the fusion-from-within assay, identical levels of syncytia were observed with each virus, TR1.3 (Fig. 3B), W102G (Fig. 3C), and FB29 (Fig. 3D), at all concentrations tested. The threshold at which cell fusion was observed above the background (Fig. 3A) was 800 cpm of RT activity, and 100% of cells in culture formed syncytia at virus concentrations in excess of 4,000 cpm. Virus-induced cell fusion was blocked by the addition of goat anti-Env antiserum but not preimmune goat serum (data not shown). Syncytium formation was not observed in incubations of cells with concentrated mouse mammary tumor virus, a non-syncytium-inducing retrovirus, or in incubations of MLV with 293 cells that lacked MCAT-1 (data not shown).
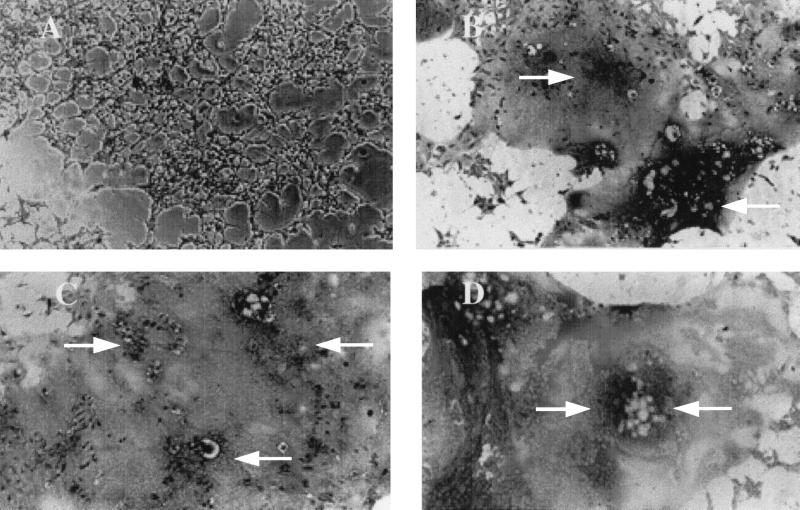
FIG. 3.
MLV-dependent syncytium formation in a nonmurine line by a fusion-from-without assay. AZT-treated human 293 cells that transiently expressed MCAT-1 were incubated overnight with medium (A), purified FB29 virus (B), purified TR1.3 virus (C), or purified W102G virus (D). Cells were photographed under a light microscope (magnification, ×200) after staining with methylene blue. Data are representative of results obtained in three independent experiments. Arrows demarcate areas of cell fusion.
The relative potential of MLV to induce syncytium formation in the fusion-from-without assay when incubated with murine NIH 3T3 cells that express endogenous receptors is presented in Fig. 4. In this instance, and as described for fusion from within, syncytium formation was observed with TR1.3 (Fig. 4C) and W102G (Fig. 4D) but not FB29 (Fig. 4B). Approximately 10 to 30% of cells in culture formed syncytia at 4,000 cpm of RT activity, and this increased to 100% syncytium formation at >10,000 cpm of RT activity. In contrast to these results, syncytia were never observed upon addition of FB29 virus, even when added to cultures in a 10-fold excess relative to the concentration of TR1.3 (data not shown).
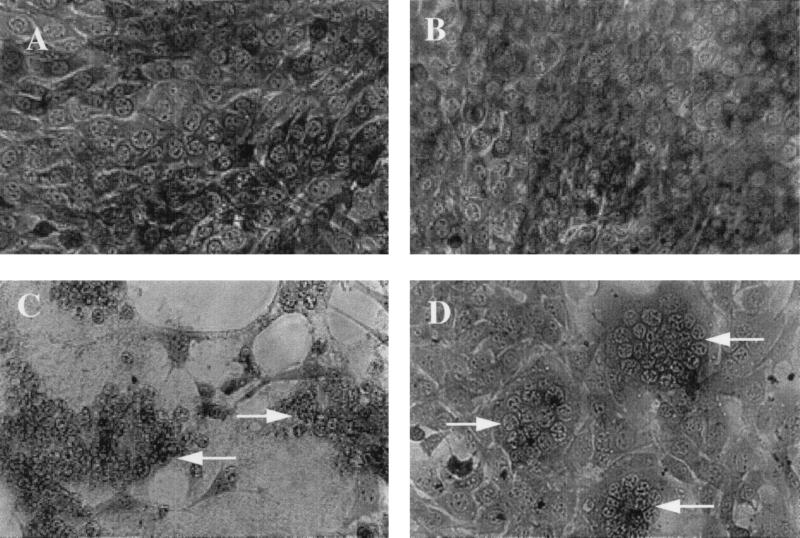
FIG. 4.
MLV-dependent syncytium formation in murine NIH 3T3 cells by a fusion-from-without assay. AZT-treated NIH 3T3 cells were incubated overnight with medium (A), purified FB29 virus (B), purified TR1.3 virus (C), or purified W102G virus (D). Cells were photographed under a light microscope (magnification, ×200) after staining with methylene blue. Data are representative of results obtained in three independent experiments. Arrows demarcate areas of cell fusion.
Augmenting the number of receptors on NIH 3T3 cells has no effect on MLV-dependent cell fusion.
In considering why FB29 Env was able to fuse nonmurine but not murine target cells, we noted that in the in vitro fusion assay effector and target cells were mixed at the time of peak receptor expression, shortly after transfection of Mcat-1 cDNA. If the fusion capacity is variably regulated by a threshold of available receptors, then an exceptionally high number of receptors might enhance FB29 Env-mediated fusion to levels comparable to that seen with TR1.3 or W102G Env. This hypothesis implies that the endogenous level of ecotropic MLV receptors on NIH 3T3 cells is above the threshold for W102G- and TR1.3-induced fusion but below that for FB29-induced fusion. If this were true, then increasing the number of receptors on murine targets should enhance the fusion potential of FB29 Env, perhaps to that seen with TR1.3 or W102G Env.
Augmentation of endogenous MCAT-1 levels in NIH 3T3 cells was achieved by transient transfection with exogenous receptor cDNA (0.5 to 5.0 μg/incubation). Cells were then assayed for fusion (Fig. 5), using the T7-luciferase gene reporter system. Although different in magnitude, the results of these studies were similar to those reported for naïve NIH 3T3 cells. Fusion of NIH 3T3-MCAT-1 targets with QT6 effector cells that expressed either TR1.3, W102G, or FB29 Env was significantly greater than that with cDNA controls (see P values in Fig. 5) and was amplified approximately twofold over levels seen with naïve NIH 3T3 targets. Nevertheless, fusion with TR1.3 (P < 0.02) or W102G (P < 0.05) Env effectors remained significantly greater than fusion with FB29 Env effectors.
FIG. 5.
Impact of enhanced MCAT-1 expression on cell fusion in NIH 3T3 cells. Env-positive QT6 effector cells were mixed with NIH 3T3 target cells that either expressed endogenous receptors (open bars) or were transfected with Mcat-1 cDNA (2 μg/incubation) (solid bars). Data are expressed as the luciferase activity ratio after 8 h and are representative of three independent experiments, each performed in triplicate. Error bars represent standard deviations. Significance was determined by the Student t test. Probability values of Env-positive compared to Env-negative (pcDNAI) controls are presented in parentheses.
This method of analysis was limited by the efficiency of cDNA transfection, which did not exceed 20% as determined by cotransfection markers. Analysis of ecotropic receptor expression on these cells by flow cytometry confirmed expression on a minor proportion of the NIH 3T3 cell population. As it was not possible to directly compare the magnitude of increase in receptor binding sites to increased cell fusion on a per-cell basis, we also attempted to generate stable NIH 3T3-derived cell lines that expressed high levels of exogenous receptors in addition to endogenous receptors. To this end, NIH 3T3 cells were transfected with the pcDNA3 MCAT-1 expression plasmid containing a linked bacterial neomycin resistance gene. Numerous stable G418-resistant colonies were obtained in six independent experiments, but none exhibited more than a 15% increase in receptor expression as determined by cationic amino acid transport (data not shown). Moreover, transfected NIH 3T3 cells showed a progressive decrease over time in exogenous receptor expression to below detectable levels (21a).
Decreasing the number of receptors to levels below that of NIH 3T3 cells results in a differential fusion pattern in nonmurine cell lines.
In view of the difficulties in assessing the impact of augmented MCAT-1 receptor expression on NIH 3T3 cells, we separately addressed the effects of reduced MCAT-1 receptor expression on human 293 cells. If the capacity for virus fusion is regulated by a threshold of available receptors, then there should be a receptor density on nonmurine cells at or below which fusion with FB29 Env effector cells is significantly less than fusion with TR1.3 or W102G Env effectors.
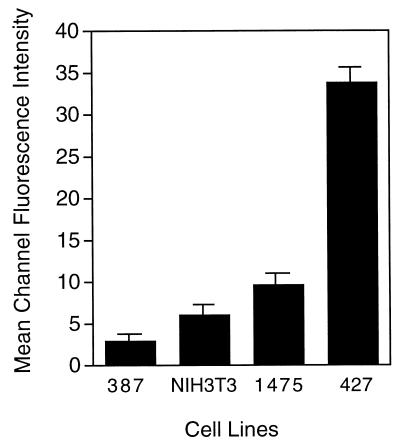
We first established the relative numbers of MLV binding sites on three human 293-derived cell lines that displayed stable expression of exogenous MCAT-1. For comparison we also measured the binding sites on murine NIH 3T3 cells which were previously reported to express 5 × 105 MLV Env binding sites per cell (11). Virus binding sites were quantified by flow cytometry following incubation with a saturating concentration of purified FB29 virus, followed by detection with anti-Env antiserum and phycoerythrin-conjugated secondary antibody. The MCF per cell, minus the MCF for receptor-negative 293 cells, was used as a relative measure of the number of binding sites. Results presented in Fig. 6 illustrate that the 427 cell line expressed the greatest number of receptors (MCF = 33.8 ± 1.35 [mean ± standard deviation]), followed by the 1475 line (MCF = 9.6 ± 0.7), the NIH 3T3 line (MCF = 6.0 ± 1.7), and the 387 line (MCF = 2.9 ± 0.8).
FIG. 6.
Comparison of ecotropic receptor levels on human 293 and NIH 3T3 cells. Cell suspensions were incubated with purified FB29 virions at 4°C to allow virus binding but not internalization. Bound virus was then detected with goat anti-SU antiserum and rabbit anti-goat immunoglobulin G conjugated to phycoerythrin. The relative number of receptors per cells was determined by measuring the MCF intensity of bound phycoerythrin by flow cytometry. Data are presented as the MCF of cell lines 387, 1475, and 427 minus the MCF of human 293 cells that lack receptor or, in the instance of NIH 3T3 cells, minus the MCF of NIH 3T3 cells incubated with virus and then normal goat serum and secondary antiserum. Error bars represent standard deviations of data from three independent experiments.
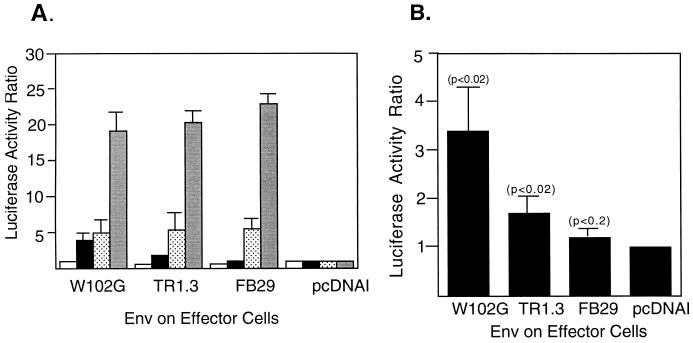
The impact of altered receptor levels on cell fusion by FB29, TR1.3, and W102G was then determined by using the T7-luciferase reporter assay described above. Human 293 cell lines that expressed high (line 427), intermediate (line 1475), or low (line 387) receptor levels and the T7 promoter-driven luciferase gene served as fusion targets. QT6 cells that expressed either TR1.3, W102G, or FB29 Env and T7 RNA polymerase served as effectors. As shown in Fig. 7A, several interesting observations emerged from this analysis. The first was that the relative degree of cell fusion was directly proportional to the level of receptors on the target cell line. Line 427 displayed approximately fourfold greater fusion potential than line 1475, which in turn showed potential approximately twofold greater than line 387. The second observation was that fusion was indistinguishable with FB29, TR1.3, and W102G Env in the 427 and 1475 cell lines, both of which expressed a greater number of virus binding sites than NIH 3T3 cells. Lastly, and as shown in Fig. 7B, differential Env fusion of TR1.3 and W102G relative to FB29 was observed with line 387, which expressed fewer virus binding sites than NIH 3T3 cells. Although the magnitude of fusion observed with line 387 was low, presumably because of the low MCAT-1 receptor number, statistical analysis conducted on data from multiple independent experiments indicated that the fusion responses of both TR1.3 Env and W102G Env were significantly greater (P < 0.02) than that of FB29 Env. In addition, the fusion response of FB29 Env was not significantly different from that of the vector control, although W102G Env was more fusogenic than TR1.3 Env (P < 0.02).
FIG. 7.
Comparison of cell fusion directed by MLV Env in human 293 lines that express various levels of receptors. (A) QT6 effector cells expressing Env and T7 RNA polymerase were mixed for 8 h with 293 target cell lines that displayed stable expression of ecotropic MLV receptors and transiently expressed a luciferase reporter gene under the control of the T7 promoter. Grey bars, line 427; stippled bars, line 1475; black bars, line 387; white bars, receptor-negative 293 cells. Data are representative of three independent experiments, each performed in triplicate. Error bars represent standard deviations. (B) Amplified scale of fusion with line 387 from panel A. Significance was determined by the Student t test. Probability values of Env-positive compared to Env-negative (pcDNAI) controls are presented in parentheses.
The evaluation of fusion from without in lines 427, 1475, and 387 yielded similar but not identical results. As seen in Fig. 3, uniformly high levels of syncytium formation were observed in both 427 and 1475 cells when incubated with either TR1.3, W102G, or FB29 virus (data not shown). Analysis of fusion in 387 cells was less conclusive. As expected, line 387 displayed a much lower level of syncytium formation (10 to 30% of maximum) with TR1.3 or W102G virus than either the 427 or 1475 cell line. Unfortunately, the variability in fusion at these levels did not permit statistical distinction of fusion between either TR1.3 or W102G and FB29.
DISCUSSION
Infection of neonatal Fv-1b mice with TR1.3 MLV induces widespread syncytium formation of BCEC that uniformly results in destruction of the blood-brain barrier, stroke formation, and death. This model of acute disease provides an important tool for dissecting the fundamental mechanisms of both virus-dependent cell fusion and endothelial cell pathology. The genetic basis of TR1.3-induced syncytium formation and disease is a W102G substitution in Env within the SU domain. However, the process whereby this change in Env mediates cellular and organismal pathology is unclear. One hypothesis is that these genetic changes directly alter the inherent fusion capacity of Env. Alternatively, these changes might affect the expression and/or availability of Env or ecotropic receptors, or they might affect the premature cleavage of transmembrane protein (TM) to the fusogenic form. In this study we utilized quantitative cell fusion assays to evaluate the inherent fusion capacity of TR1.3, W102G, and FB29 Env, as well as the role of ecotropic receptor expression in syncytium formation induced by MLV. Our results indicated that TR1.3 Env and W102G Env are inherently more fusogenic than FB29 Env. Nevertheless, we also observed that syncytium formation was regulated by the level of ecotropic receptors, as well as other host cell determinants. Taken together, these observations indicate that syncytium formation is dependent upon both the fusion capacity of individual MLV and the capacity of target cells to achieve the necessary threshold level of receptors to initiate fusion. We propose that BCEC fusion and cytopathology observed in TR1.3 and W102G, but not in FB29, infection in vivo result from a reduced threshold needed for MLV fusion in BCEC compared to other small-vessel endothelial cells.
There are several means by which mutations in MLV Env may alter fusion potential. For example, MLV, Mason-Pfizer monkey virus, and equine infectious anemia virus control envelope fusion activity through the cleavage of the TM subunit pre15ETM to release p15E and a short C-terminal fragment termed the R peptide (6, 17, 38, 44). Normally, the R peptide is cleaved by the viral protease after the virus particle is released from infected cells, preventing the premature activation of envelope protein fusion (35, 37). Mutations in TM that abrogate R-peptide cleavage inhibit virus-mediated cell fusion even though these Env proteins are properly processed and transported to the cell surface (50). In contrast, truncation of TM before the R peptide renders the “R-less” p15E protein hyperfusogenic and results in reduced production of infectious particles (19) and a loss of virus infectivity (37). Therefore, one explanation for the extraordinary fusion capability of TR1.3 and W102G Env might be that the W102G change either enhances virus-protease cleavage or otherwise enables premature cleavage of the R peptide by a host cell protease. Premature cleavage of R peptide within cells might explain enhanced fusion from within observed with TR1.3 and W102G MLV. However, it is less clear whether this mechanism could account for enhanced fusogenicity of mature virus particles as seen in Fig. 3 and 4. Lastly, there was no difference in the production of infectious virus among TR1.3, W102G, and FB29 in either murine or nonmurine lines. Nevertheless, the hypothesis that a single W102G point mutation in SU could induce fusion by increased R-peptide cleavage is currently being investigated.
An alternative mechanism is that the W102G change in Env results in structural changes that affect events in the fusion process itself. In the envelope protein of the highly homologous Friend 57 MLV, the planar aromatic rings on the side chain of W102 lie directly under the side chain of aspartate 86 (D86) (15), a residue shown to be essential for Env binding to receptors (24). Perhaps the absence of the bulky tryptophan side chain and/or the peptide chain flexibility added by the glycine replacement allows for more rapid movement of D86 upon receptor binding. This change might alter the avidity or kinetics of Env binding to receptors and thereby enhance fusion potential. Alternatively, this structural change might affect the conformation of SU and/or TM, resulting in enhancement of subsequent events in the fusion process, such as exposure of the fusion peptide. In related studies, two of us (8a) observed that TR1.3 and W102G viruses have a lower binding avidity for receptors than does FB29, consistent with the presence of structural changes in Env. This preliminary observation highlights the complexity of the fusion process, as a decrease in binding avidity might be anticipated to proportionally decrease cell fusion. A major goal of our ongoing studies is to utilize TR1.3, W102G, and related viruses as tools to define the parameters of Env binding and the biophysics of MLV fusion. These analyses are an essential component of our ability to predict and combat virus pathology of MLV and other retroviruses.
Although the W102G substitution accurately reconstitutes the cell and organismal pathology of TR1.3, there may be subtle aspects of fusion which are not be fully reproduced. An example of this was provided in the analysis of fusion in line 387, which expressed low levels of ecotropic receptors (Fig. 7B). While both W102G Env and TR1.3 Env were more fusogenic than FB29 Env, W102G Env was in turn more fusogenic (P < 0.02) than TR1.3 Env. It is also interesting that in previous studies members of our group were unable to generate infectious virus from the reciprocal substitution, G102W, within the backbone of TR1.3 (33). These results suggest that other regions within Env may interact with or influence residue 102 in the regulation of cell fusion and pathology. We have identified and analyzed two additional amino acid substitutions within the first 200 residues of the N terminus of TR1.3 Env. Replacement of these residues within FB29 had no effect on syncytium formation or disease (33). We have not yet investigated the role of more distal C-terminal substitutions on fusion.
One of the most important questions raised by these studies is why TR1.3-dependent syncytium formation is specific to BCEC in vivo. The answer may lie in part with the physiology of BCEC and the cellular distribution of ecotropic receptors on these cells. It is well established that BCEC are a unique cell type as evidenced by the formation of tight junctions that delineate the blood-brain barrier (3). Unfortunately, there is no analysis of ecotropic receptor or cationic amino acid transporter distribution in these cells. Related studies on kidney epithelial cells demonstrated that cationic amino acid transporters were localized in the basolateral membrane created by tight junctions in vivo (48). A similar distribution of receptors was also observed in confluent in vitro cultures of Madin-Darby canine kidney cells and human 293 kidney epithelial cells that express exogenous ecotropic receptors (21a). Moreover, MLV assemble and bud exclusively from the basolateral membranes of infected polarized cells (47). These observations suggest that receptors and virus envelope proteins may be concentrated on directly apposing surfaces below tight junctions, thereby facilitating the fusion process.
The hypothesis that receptor concentration serves to regulate the magnitude of cell fusion is also reinforced by our in vitro analysis on human 293T lines that expressed various levels of ecotropic receptors. Lines 1475 and 427, which expressed approximately 50 and 400% more receptors, respectively, than NIH 3T3 cells, displayed robust fusion with all MLV Env proteins tested. In contrast, the 387 cell line, which expressed lower receptor numbers than NIH 3T3 cells, fused only in the presence of TR1.3 or W102G Env. These results indicate that induction of cell fusion requires a threshold level of virus receptors that is greater for FB29 Env than for TR1.3 and W102G Env.
These findings also led to the expectation that increasing receptor expression in NIH 3T3 cells should render them equally amenable to FB29 Env-mediated fusion. We attempted to increase receptor expression in NIH 3T3 cells by transient transfection of receptor cDNA but found that FB29 Env still did not mediate fusion comparably to TR1.3 and W102G Env. It is difficult to draw concrete conclusions from this analysis, as we detected only 20% transfection efficiency based on cotransfection markers. Therefore, it is possible that we have not sufficiently amplified ecotropic receptors on NIH 3T3 cells to reach the threshold of FB29 Env fusion. In view of this we also attempted to generate NIH 3T3 cells with stable, high-level expression of exogenous receptors. None of these lines displayed more than a 15% increase in receptor expression, and this increase was lost upon passage. In contrast, receptor expression could be increased by up to 400% in transformed human 293T cells. The inability to force a substantial increase in receptor expression in NIH 3T3 cells suggests either that this limitation is due to a host cell factor(s) available in limited amounts in NIH 3T3 cells or that this phenomenon is related to the transformed phenotype of 293T and QT6 cells. Evidence of an accessory factor for MLV-dependent fusion in Chinese hamster cells has been previously reported, but such a factor has not been characterized for murine cells (43). Transformed cells may also display greater susceptibility to MLV fusion, perhaps as a result of alterations in the cytoskeletal network, expression of cell surface proteases, and/or membrane glycolipid composition (49). Studies to resolve these issues with regard to TR1.3-dependent fusion are ongoing in our laboratories.
We propose the following model for the mechanism of TR1.3 virus syncytium induction and neurovirulence in vivo. TR1.3 first infects small-vessel endothelial cells throughout the body. In the majority of infected cells, available receptors are distributed throughout the plasma membrane, while in BCEC, receptors are concentrated at the basolateral membrane in association with tight junctions. This distribution comprises a unique feature that increases the local receptor density in BCEC such that it exceeds the threshold receptor level for TR1.3- and W102G-mediated fusion but not FB29-mediated fusion. Virus Env is also directed to the basolateral membranes of infected BCEC during virus replication, concentrating the two principal participants of cell fusion in the region where adjacent cell membranes are in intimate contact. This process may be augmented by a putative decrease in TR1.3 Env avidity, which might serve to limit the down-regulation of receptor expression by Env. Thus, syncytium formation occurs in BCEC, but not in other cell types, as a consequence of viral and cellular events, yielding the neuropathology characteristic of TR1.3 infection.
ACKNOWLEDGMENTS
This work was supported by NIH grants NS30606 (G.N.G.) and AI33410 (L.M.A.) and by the Robert D. and Alma W. Moreton Oncology Research Endowment Fund (L.M.A.).
We are grateful to Robert Doms, Paul Bates, Rachel Damico, and David Kabat for both technical support and intellectual guidance of this project and to Maureen Hynes for administrative assistance.
REFERENCES
- 1.Albritton L M, Kim J W, Tseng L, Cunningham J M. Envelope binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altieri D C, Etingin O R, Fair D S, Brunck T K, Geltosky J E, Hajjar D P, Edgington T S. Structurally homologous ligand binding of integrin Mac-1 and viral glycoprotein C receptors. Science. 1991;254:1200–1202. doi: 10.1126/science.1957171. [DOI] [PubMed] [Google Scholar]
- 3.Beilke M A. Vascular endothelium in immunology and infectious disease. Rev Infect Dis. 1989;11:273–283. doi: 10.1093/clinids/11.2.273. [DOI] [PubMed] [Google Scholar]
- 4.Benditt E P, Barrett T, McDougall J K. Viruses in the etiology of atherosclerosis. Proc Natl Acad Sci USA. 1983;80:6386–6389. doi: 10.1073/pnas.80.20.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brody B A, Rhee S S, Hunter E. Postassembly cleavage of a retroviral glycoprotein cytoplasmic domain removes a necessary incorporation signal and activates fusion activity. J Virol. 1994;68:4620–4627. doi: 10.1128/jvi.68.7.4620-4627.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carneiro A V, Ferro J, Figueiredo C, Costa L, Campos J, dePauda F. Herpes zoster and contralateral hemiplegia in one African patient infected with HIV-1. Acta Med Port. 1994;4:91–92. [PubMed] [Google Scholar]
- 8a.Chung, M., and G. N. Gaulton. Unpublished data.
- 9.Clavel F, Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol. 1994;68:1179–1185. doi: 10.1128/jvi.68.2.1179-1185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czub S, Lynch W P, Czub M, Portis J L. Kinetic analysis of spongiform neurodegenerative disease induced by a highly virulent murine retrovirus. Lab Investig. 1994;70:711–723. [PubMed] [Google Scholar]
- 11.DeLarco J, Todaro G J. Membrane receptors for murine leukemia virus: characterization using the purified viral envelope glycoprotein, gp71. Cell. 1976;8:365–371. doi: 10.1016/0092-8674(76)90148-3. [DOI] [PubMed] [Google Scholar]
- 12.Engstrom J W, Lowenstein D H, Bredesen D E. Cerebal infarctions and transient neurologic deficits associated with acquired immunodeficiency syndrome. Am J Med. 1989;86:528–532. doi: 10.1016/0002-9343(89)90379-3. [DOI] [PubMed] [Google Scholar]
- 13.Etingin O R, Silverstein R L, Haggar D P. Identification of a monocyte receptor on herpesvirus-infected endothelial cells. Proc Natl Acad Sci USA. 1991;88:7200–7203. doi: 10.1073/pnas.88.16.7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fabricant C G. Atherosclerosis: the consequence of infection with a herpes virus. Adv Vet Sci Comp Med. 1985;30:39–66. [PubMed] [Google Scholar]
- 15.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 16.Fish K, Soderberg-Naucler C, Mills L, Stenglein S, Nelson J. Human cytomegalovirus persistently infects aortic endothelial cells. J Virol. 1998;72:5661–5668. doi: 10.1128/jvi.72.7.5661-5668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajjar D P. Viral pathogenesis of atherosclerosis. Am J Pathol. 1991;139:1195–1211. [PMC free article] [PubMed] [Google Scholar]
- 19.Januszeski M M, Cannon P M, Chen D, Rozenberg Y, Anderson W F. Functional analysis of the cytoplasmic tail of Moloney murine leukemia virus envelope protein. J Virol. 1997;71:3613–3619. doi: 10.1128/jvi.71.5.3613-3619.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones J S, Risser R. Cell fusion induced by the murine leukemia virus envelope glycoprotein. J Virol. 1993;67:67–74. doi: 10.1128/jvi.67.1.67-74.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaden M J, Sturm S, Anderson W F, Eglitis M A. Detection of receptor-specific murine leukemia virus binding to cells by immunofluorescence analysis. J Virol. 1992;66:2281–2287. doi: 10.1128/jvi.66.4.2281-2287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Kizhatil, K., and L. M. Albritton. Unpublished data.
- 22.Lathey J, Wiley C, Verity M, Nelson J. Cultured human brain capillary endothelial cells are permissive for infection by human cytomegalovirus. Virology. 1990;176:266–273. doi: 10.1016/0042-6822(90)90252-m. [DOI] [PubMed] [Google Scholar]
- 23.Lynch W P, Czub S, McAtee F J, Hayes S F, Portis J L. Murine retrovirus-induced spongiform encephalopathy: productive infection of microglia and cerebellar neurons in accelerated CNS disease. Neuron. 1991;7:365. doi: 10.1016/0896-6273(91)90289-c. [DOI] [PubMed] [Google Scholar]
- 24.MacKrell A J, Soong N W, Carmel C M, Anderson W F. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J Virol. 1996;70:1768–1774. doi: 10.1128/jvi.70.3.1768-1774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malhotra S, Scott A G, Zavorotinskaya T, Albritton L M. Analysis of the murine ecotropic leukemia virus receptor reveals a common biochemical determinant on diverse cell surface receptors that is essential to retrovirus entry. J Virol. 1996;70:321–326. doi: 10.1128/jvi.70.1.321-326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda M, Hanson C A, Dugger N V, Robbins D S, Wilt S G, Ruscetti S K, Hoffman P M. Capillary endothelial cell tropism of PVC-211 murine leukemia virus and its application for gene transduction. J Virol. 1997;71:6168–6173. doi: 10.1128/jvi.71.8.6168-6173.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda M, Hoffman P M, Ruscetti S K. Viral determinants that control the neuropathogenicity of PVC-211 murine leukemia virus in vivo determine brain capillary endothelial cell tropism of the virus in vitro. J Virol. 1993;67:4580–4587. doi: 10.1128/jvi.67.8.4580-4587.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melnick J, Petrie B, Dreesman G, Burek J, McCollum C, DeBakey M. Cytomegalovirus antigen within human arterial smooth muscle cells. Lancet. 1983;ii:644–647. doi: 10.1016/s0140-6736(83)92529-1. [DOI] [PubMed] [Google Scholar]
- 29.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park B H, Lavi E, Blank K J, Gaulton G N. Intracerebral hemorrhages and syncytium formation induced by endothelial cell infection with a murine leukemia virus. J Virol. 1993;67:6015–6024. doi: 10.1128/jvi.67.10.6015-6024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park B H, Lavi E, Gaulton G N. Intracerebral hemorrhages and infarction induced by a murine leukemia virus is influenced by host determinants within endothelial cells. Virology. 1994;203:393–396. doi: 10.1006/viro.1994.1500. [DOI] [PubMed] [Google Scholar]
- 32.Park B H, Lavi E, Stieber A, Gaulton G N. Pathogenesis of cerebral infarction and hemorrhages induced by a murine leukemia virus. Lab Investig. 1994;71:78–85. [PubMed] [Google Scholar]
- 33.Park B H, Matuschke B, Lavi E, Gaulton G N. A point mutation in the env gene of a murine leukemia virus induces syncytium formation and neurologic disease. J Virol. 1994;68:7516–7524. doi: 10.1128/jvi.68.11.7516-7524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pitts O M, Powers J M, Bilello J A, Hoffman P M. Ultrastructural changes associated with retroviral replication in central nervous system capillary endothelial cells. Lab Investig. 1987;56:401–409. [PubMed] [Google Scholar]
- 35.Ragheb J A, Anderson W F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawlinsin W D, Cunningham A L. Contralateral hemiplegia following thoracic herpes zoster. Med J Aust. 1991;155:344–346. doi: 10.5694/j.1326-5377.1991.tb142297.x. [DOI] [PubMed] [Google Scholar]
- 37.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice N R, Henderson L E, Sowder R C, Copeland T D, Oroszlan S, Edwards J F. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J Virol. 1990;64:3770–3778. doi: 10.1128/jvi.64.8.3770-3778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosen B. Fundamentals of biostatistics. Boston, Mass: PWS-Kent Publishing Co.; 1990. [Google Scholar]
- 40.Ross R. The pathogenesis of atherosclerosis—an update. N Engl J Med. 1986;314:488–500. doi: 10.1056/NEJM198602203140806. [DOI] [PubMed] [Google Scholar]
- 41.Sedmak D, Knight D, Vook N, Waldman W. Divergent patterns of ELAM-1, ICAM-1, and VCAM-1 expression on cytomegalovirus-infected endothelial cells. Transplantation. 1994;58:1379–1385. [PubMed] [Google Scholar]
- 42.Shuper A, Vining E P, Freeman J M. Central nervous system vasculitis after chicken pox: cause or coincidence? Arch Dis Child. 1990;65:1245–1248. doi: 10.1136/adc.65.11.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siess D C, Kozak S L, Kabat D. Exceptional fusogenicity of Chinese hamster ovary cells with murine retroviruses suggests roles for cellular factor(s) and receptor clusters in the membrane fusion process. J Virol. 1996;70:3432–3439. doi: 10.1128/jvi.70.6.3432-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sommerfelt M A, Petteway S R, Dreyer G B, Hunter E. Effect of retroviral protease inhibitors on Mason-Pfizer monkey virus maturation and transmembrane glycoprotein cleavage. J Virol. 1992;66:4220–4227. doi: 10.1128/jvi.66.7.4220-4227.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Span A, van Dam-Mieras M, Mullers W, Endert J, Muller A, Bruggeman C. The effect of virus infection on the adherence of leukocytes or platelets to endothelial cells. Clin Investig. 1991;21:331–338. doi: 10.1111/j.1365-2362.1991.tb01378.x. [DOI] [PubMed] [Google Scholar]
- 46.Stoica G, Illanes O, Tasca S I, Wong P K. Temporal central and peripheral nervous system changes induced by a paralytogenic mutant of Moloney murine leukemia virus TB. Lab Investig. 1993;69:724–735. [PubMed] [Google Scholar]
- 47.Tucker S P, Compans R W. Virus infection of polarized epithelial cells. Adv Virus Res. 1993;42:187–247. doi: 10.1016/S0065-3527(08)60086-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White M F. The transport of cationic amino acids across mammalian cells. Biochim Biophys Acta. 1985;822:355–364. doi: 10.1016/0304-4157(85)90015-2. [DOI] [PubMed] [Google Scholar]
- 49.Wilson C A, Marsh J W, Eiden M V. The requirements for viral entry differ from those for virally induced syncytium formation in NIH 3T3/DTras cells exposed to Moloney murine leukemia virus. J Virol. 1992;66:7262–7269. doi: 10.1128/jvi.66.12.7262-7269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang C, Compans R W. Analysis of the cell fusion activities of chimeric simian immunodeficiency virus-murine leukemia virus envelope proteins: inhibitory effects of the R peptide. J Virol. 1996;70:248–254. doi: 10.1128/jvi.70.1.248-254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 52.Yu H, Soong N, Anderson W F. Binding kinetics of ecotropic (Moloney) murine leukemia retrovirus with NIH 3T3 cells. J Virol. 1995;69:6557–6562. doi: 10.1128/jvi.69.10.6557-6562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y, Leon M, Waclwiw M, Popma J, Finkel T, Epstein S. Association between prior cytomegalovirus infection and the risk of restenosis after coronary atherectomy. N Engl J Med. 1996;335:624–630. doi: 10.1056/NEJM199608293350903. [DOI] [PubMed] [Google Scholar]