Abstract
Huntington’s disease (HD) causes selective degeneration of striatal and cortical neurons, resulting in cell mosaicism of coexisting still functional and dysfunctional cells. The impact of non-cell autonomous mechanisms between these cellular states is poorly understood. Here we generated telencephalic organoids with healthy or HD cells, grown separately or as mosaics of the two genotypes. Single-cell RNA sequencing revealed neurodevelopmental abnormalities in the ventral fate acquisition of HD organoids, confirmed by cytoarchitectural and transcriptional defects leading to fewer GABAergic neurons, while dorsal populations showed milder phenotypes mainly in maturation trajectory. Healthy cells in mosaic organoids restored HD cell identity, trajectories, synaptic density, and communication pathways upon cell-cell contact, while showing no significant alterations when grown with HD cells. These findings highlight cell-type-specific alterations in HD and beneficial non-cell autonomous effects of healthy cells, emphasizing the therapeutic potential of modulating cell-cell communication in disease progression and treatment.
Subject terms: Cellular neuroscience, Huntington's disease, Stem-cell differentiation
Mosaic organoids where pathological and healthy cells are grown together, reveal the rescue of phenotypes in pathological cells due to communication with healthy cells without harming them, as demonstrated by single-cell RNA-sequencing data.
Introduction
Huntington’s disease (HD) is a neurodegenerative disorder caused by the expansion of a CAG repeat region in the HTT gene leading to the selective loss of striatal medium spiny neurons (MSNs) and, subsequently, of deep layer cortical neurons. Although HD typically presents in adulthood, some studies suggest that abnormalities may already be present during brain development, with an impact later in life1–5. In the adult brain, selective vulnerability has been associated with striatum-specific transcriptional dysregulation6–8, increased somatic CAG instability9 and formation of mutant HTT (mHTT) aggregates10. Dysfunction in other brain regions may also contribute to HD pathophysiology. Studies have shown that HD is associated with altered cortico-striatal connectivity with a reduction of synaptic contacts11–13, leading to the loss of neurotrophic support for striatal neurons14,15 and the presence of glutamatergic abnormalities16. Additionally, an altered dopaminergic neurotransmission has been observed to exacerbate HD pathophysiology17 highlighting the role of cell-cell communication in driving dysfunction in the disease. The interplay between cell types in HD has been primarily investigated using adult conditional mouse models with restricted expression of mHTT in specific brain regions11,18–20 or by investigating the interaction between neuronal and glial cells21,22. However, there is a lack of a more systematic approach to understand both cell autonomous and non-cell autonomous mechanisms in the context of human HD. As possible future therapies involving cell replacement and gene silencing techniques will inevitably create mosaicism within the brain, with healthy cells co-existing with dysfunctional cell types, a more precise understanding of their interactions becomes crucial.
In this study, we established long-term culture of control (CTRL) and HD telencephalic organoids both in monoculture or mosaic co-culture configurations. Using single cell RNA sequencing (scRNAseq) we then investigated the cell type-specific alterations of HD organoids grown alone or co-cultured with healthy cells. These organoids consisted of intermixed populations of progenitors and neurons with ventral and dorsal identities. HD organoids exhibited abnormal transcriptional profile particularly in the ventral rather than dorsal component, revealing cytoarchitectural and neurodevelopmental alterations. Importantly, cell-type composition, differentially expressed genes (DEGs), maturation trajectories, synaptic density, and communication pathways in HD cells were rescued when grown together with CTRL cells in the same mosaic organoid. In contrast, the presence of HD cells did not significantly alter the CTRL counterparts.
These results collectively point to the presence of cell type-specific alterations during early development, underscoring the importance of investigating aspects of neurodevelopment in HD. Significantly, our mosaic organoids approach demonstrated the disease-modifying effect of healthy cells in restoring ventral developmental programs. This highlights and amplifies the therapeutic potential of cell replacement and gene silencing strategies in HD, underscoring the beneficial non-cell autonomous effects of healthy cells on HD cells.
Results
Telencephalic organoids show ventral and dorsal fate acquisition with altered differentiation pattern in HD
To investigate the cell type-specific role of mHTT, we developed an organoid protocol that promotes ventral telencephalic identity first, followed by the appearance of dorsal identity. Ventral sub-populations are generated within a short time window of rostro-ventral patterning obtained by exposure to the WNT-βCatenin inhibitor DKK and the Sonic HedgeHog SHH while intermixed dorsal identities emerge by default23,24 from later progenitors, upon removal of the morphogens (Fig. 1A). Telencephalic organoids were generated from the isogenic hESC RUES2 cell series25 including two CTRL lines (parental and edited line carrying 20CAG) and three HD lines (edited with 48, 56 and 72CAG repeats). These isogenic lines offer “mutation corrected” cell lines that share the same genetic background, i.e. resulting from Cas9-assisted HTT targeting of the same parental RUES2 cell line25.
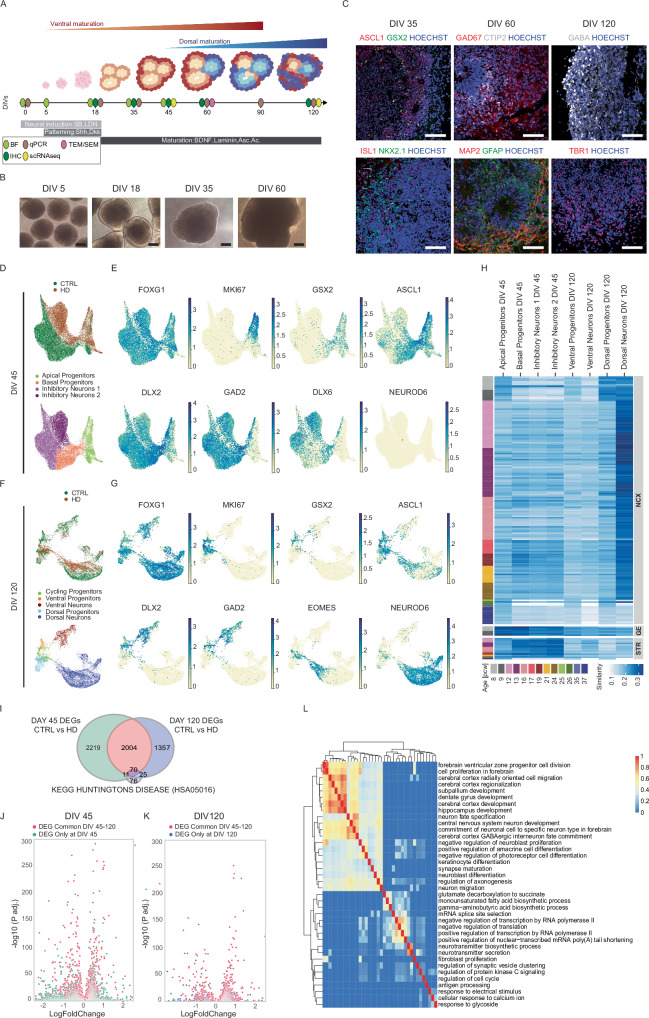
Fig. 1. Telencephalic organoids present ventral and dorsal identities with impaired neuronal transcriptional signatures in HD.
A Scheme of the protocol for telencephalic organoids. Created with BioRender.com. released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Abbreviations: BF=brightfield, IHC=immunohistochemistry, qPCR=quantitative PCR, scRNAseq=single cell RNA sequencing, TEM/SEM=transmission and scanning electron microscopy. B Brightfield images during differentiation. Scale bars = 250 µm C Immunohistochemistry analyses of telencephalic organoids during differentiation. At DIV 35 for ASCL1, GSX2, ISLT1 and NKX2.1; at DIV 60 for GAD67, CTIP2, GFAP, MAP2; at DIV 120 for GABA and TBR1. Scale bars = 100 µm D UMAP plots of scRNAseq analyses on CTRL (20CAG) and HD (56CAG) organoids at DIV 45 showing genotypes and clusters of subpopulations E UMAP plots for specific markers of telencephalic development (FOXG1) with progenitors (MKI67) and ventral identity (GSX2, ASCL1, DLX2, GAD2, DLX6) or dorsal identity (NEUROD6) F UMAP plots of scRNAseq analyses on CTRL (20CAG) and HD (56CAG) organoids at DIV 120 showing the genotypes and clusters of subpopulations G UMAP plots for specific markers of telencephalic development (FOXG1) with progenitors (MKI67) and ventral identity (GSX2, ASCL1, DLX2, GAD2) or dorsal identity (EOMES, NEUROD6) H Voxhunt heatmap of similarity between individual subpopulations of our organoids and human fetal brain by mapping onto BrainSpan human transcriptomic dataset I Venn diagram of DEGs between CTRLs and HD organoids showing which are in common between DIV 45, DIV 120, and KEGG pathway associated to HD (Wilcoxon test, two-sided, p < 0.05) J, K Volcano plot of p-value adjusted (p.adj.) for DEGs at DIV 45 or DIV 120, Wilcoxon test two-sided L Heatmap showing the results of gene ontology analysis of the common DEGs where the enriched GO terms are grouped according to their semantic similarity. (Biological Replicates for scRNAseq: At DIV 45 N=3 individual organoids per genotype and at DIV 120 N=a pool of 10 organoids from 2 cell lines per genotype. See fig. S1).
The telencephalic organoids show initial neuroepithelial features with progressive growth and maturation over time (Fig. 1B and Supplementary Fig 1A). At early time points, they presented organized structures of progenitors (SOX2, KI67, NESTIN, BLBP) and neurons (βIII-TUBULIN) in a cell sorting out fashion, as identified by SOX2/βIII-TUBULIN (Supplementary Fig 1A, B), with early ventral specification (GSX2/ASCL1, NKX2.1/ISLT1, GAD67, EBF1, Fig. 1C and Supplementary Fig 1B) followed by later maturation of dorsal identities (TBR1, SATB2) intermixed with ventral neurons (GABA) (Fig. 1C and Supplementary Fig 1B). Gene expression data confirmed the progressive differentiation (MAP2, SNAP25) with initial ventral (ASCL1, SIX3) and subsequent dorsal (TBR2, SATB2; Supplementary Fig 1C) maturation. Starting from DIV 60 the organoids presented neuronal features displaying complex neuronal networks (Supplementary Fig 1D) and synapses visible by TEM (Supplementary Fig 1E).
scRNAseq performed at DIV 45 and 120 on CTRL (20CAG) and HD (56CAG) organoids (Supplementary Fig 1F, G) confirmed their telencephalic identity (Fig. 1D–G). At DIV 45 we found four clusters corresponding to subpopulations of apical progenitors (AP), basal progenitors (BP) and inhibitory neurons (IN) (Fig. 1D) displaying markers typical of forebrain (FOXG1), cycling cells (MKI67), and ventral inhibitory lineage (GSX2, ASCL1, DLX2, GAD2, DLX6) with almost no cortical populations (NEUROD6) (Fig. 1E). At DIV 120, we identified cycling progenitors (CP), ventral progenitors (VP), ventral inhibitory neurons (VN), dorsal progenitors (DP) and dorsal excitatory neurons (DN), confirming the duality of the differentiation (Fig. 1F, G). The distinction between progenitors and neurons was also verified by the progressive expression of neuronal maturation genes26,27 (Supplementary Fig 1H, I). Lack of IRX2, GBX2, and EN2 confirmed that neither CTRL nor HD organoids differentiated towards more caudal brain lineages (Supplementary Fig 1J, K).
To validate the regional identity of our organoids, we exploited the VoxHunt algorithm28 mapping our datasets to the BrainSpan human transcriptomic dataset representing neocortex (NCX), ganglionic eminences (GE) and striatum (STR) from 8 to 37 post-conception weeks (pcw). Our analysis revealed that subpopulations of DIV 45 exhibited a higher resemblance to the GE of human samples at 8-9 pcw compared to DIV 120 (Fig. 1H). The ventral identities of basal progenitors, inhibitory neurons, and ventral neurons were also confirmed by their similarity to the STR observed in samples at 12 to 19 pcw. Consistently, the dorsal populations displayed a stronger correlation with the NCX, with dorsal progenitors resembling samples from 8-9 pcw and dorsal neurons showing the highest similarity with samples from 12–19 pcw (Fig. 1H). Further comparison with the dataset of human fetal brain from Braun and colleagues29 revealed that our cell type populations exhibit similarities with forebrain Intermediate Progenitor Cells (IPC), forebrain neurons, and striatum radial glia at both DIV 45 and 120 time points, along with the ventral midbrain neurons (likely due to common ventral genes regulated by SHH). Additionally, similarities with cortex IPC and cortex radial glia were found only at DIV 120, which is not unexpected since cortical maturation in our organoids occurs subsequently and happens when SHH is no longer added to the cultures (Supplementary Fig 2A, B). Lastly, we compared our organoids scRNAseq dataset with the dataset retrieved from a recent cell atlas of human neural organoids30, which integrates 26 different organoid protocols. This analysis confirmed that our progenitors are most similar to radial glia, progenitors, or neuroblasts populations from many different protocols (Supplementary Fig 2C), and that our ventral and inhibitory subpopulations resemble GABAergic neurons and interneurons, while dorsal neurons in our organoids resemble cerebral cortex neurons (Supplementary Fig 2C).
These findings provide evidence of the telencephalic dorso-ventral specification of our organoids, supporting their ability to recapitulate human fetal brain regional identity. Our protocol therefore enables us to interrogate the impact of the HD mutation on both ventral and dorsal neuronal maturation within the same experimental model.
In the datasets from the two time points, we identified a total of 5686 DEGs between CTRL and HD organoids, with 2074 of these genes overlapping between DIV 45 and DIV 120 (Fig. 1I). When comparing these DEGS with the list of genes known to be associated to HD from the specific KEGG pathway, we observed that many genes belonging to this pathway were also identified as DEGs between our CTRL and HD organoids (Fig. 1I). Additionally, we conducted a comparison of the dysregulated genes in our HD organoids with those found in caudate/putamen (striatal) cell types of grades 2–4 human HD post-mortem samples using snRNA-seq7. Despite the differences between the two paradigms, we discovered that many DEGs found in postmortem samples overlapped with the DEGs between CTRL and HD at both DIV 45 and DIV 120, for both upregulated and downregulated genes (Supplementary Fig 2D). This comparison strengthens the validity of our model for investigating HD in vitro, with the advantage of maintaining species-specific genes.
Volcano plots for p-values at each individual time point show how the DEGs common to both timepoints are the most significant ones while the DEGs present at only one timepoint are usually less significative (Fig. 1J, K). These common DEGs show a strong enrichment in terms related to brain development, neuron migration, and synapse maturation (Fig. 1L), suggesting that these processes remain highly altered during forebrain differentiation and maturation in HD organoids.
Compromised ventral composition and maturation in HD organoids
To understand the biological significance of terms altered in HD, we separated GO analyses of upregulated and downregulated DEGs (Supplementary Fig 3A, B). Top upregulated terms include “cortical cytoskeleton organization” in multiple cell populations (Fig. 2A). Consistent with this, HD neurons exhibit enlarged processes (Fig. 2B, C), confirming the axonal abnormalities observed in an HD mouse model31. Another upregulated term in HD is “regulation of the cell cycle”, which is enriched in progenitor populations (Supplementary Fig 3C). Analysis of mitotic spindle orientation by discrimination of horizontal (0°–30°), oblique (30°–60°), or vertical (60°–90°) divisions (Supplementary Fig 3D) showed that HD organoids with longer CAG repeats exhibit more horizontal asymmetric divisions compared to CTRL (Supplementary Fig 3E), in line with previous studies4,25,32. HD organoids also showed a significantly lower number of KI67+ cells, a higher fraction of mitotic cells (PH3+/KI67+; Supplementary Fig 3F, G), and a higher cell cycle exit ratio (Supplementary Fig 3H–I), resulting in a smaller organoid size at DIV 35 (Supplementary Fig 3J).
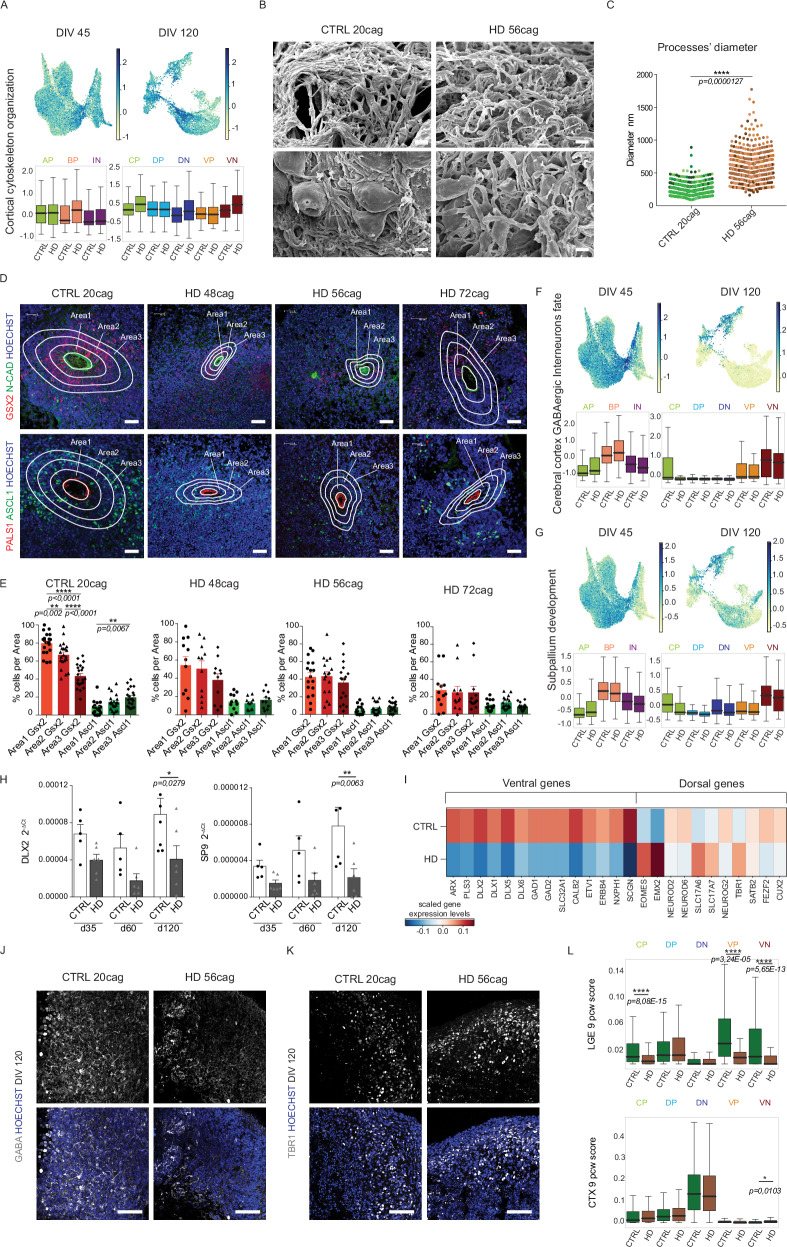
Fig. 2. Self-organization and ventral maturation are altered in HD organoids.
A UMAP and boxplot of score of DEGs of term “cortical cytoskeleton organization” upregulated in HD organoids at both time points. Abbreviations: AP=Apical Progenitors; BP=Basal Progenitors; IN=Inhibitory Neurons; CP=Cycling Progenitors; DP=Dorsal Progenitors; DN=Dorsal Neurons; VP=Ventral Progenitors; VN=Ventral Neurons B Scanning electron microscopy (SEM) images of CTRL and HD organoids at DIV 60. Scale bars = 5 µm C Diameter of neuronal processes (Dots shown are all the multiple processes measured in N = 6 organoids of independent biological replicates, but statistics is performed on the mean of processes per organoid. Dots coloured based on the 6 replicates). Unpaired two-tailed t-test, ****p<0,0001) D Immunohistochemistry analyses of CTRL (parental and 20CAG) and HD (48, 56 and 72 CAG) organoids at DIV 45-60 for GSX2, ASCL1, N-CADHERIN and PALS1. Scale bar 30 µm E Numbers of GSX2+ or ASCL1+ cells in each area (Manual counting. Anova One Way, Bonferroni post test, **p < 0,01, ****p < 0,0001. N ≥ 11 VZ-like structures, derived from 10 organoids of 2/3 biological replicates) F, G UMAP and boxplot of score of DEGs of terms “cerebral cortex GABAergic interneurons” and “subpallium development” downregulated in HD organoids H q-PCR for DLX2 and SP9 on CTRL and HD organoids at DIV 35, 60, and 120. (N = 5 independent biological replicates where each one is a pool of 4 organoids; error bars represent ± SEM; Anova One Way, Bonferroni post test, *p < 0,05; **p < 0,01) I Heatmap of gene expression of key dorso-ventral telencephalic markers in CTRL and HD organoids at DIV 120 J, K Immunohistochemistry analyses of CTRL and HD organoids at DIV 120 for GABA and TBR1. Scale bar 100 μm L Boxplots of the signature score of each subpopulation of organoids at DIV 120 and the bulk RNAseq from human fetal brain LGE and CTX (cortex) at 9pcw39 (Wilcoxon test, two-sided, Bonferroni post test, ****p<0,0001). For A, F, G, and L the box plots show the median (centre line), upper and lower quartiles (box limits), and the highest and lowest values within 1.5× the IQR of the nearest hinge (whiskers). Source data are provided as a Source Data file.
The analysis of downregulated GO terms in HD organoids (Supplementary Fig 3B) revealed terms such as “forebrain neuron differentiation” and “positive regulation of organ growth”, which include genes crucial for telencephalic development and cell adhesion. Based on these findings, we investigated the cell-adhesion and cytoarchitectural properties by examining the ability of cells to self-organize into VZ-like structures, which recapitulate features of brain development and polarity through multilaminar radial organization33. In the HD allelic series, all organoids exhibited a reduced number of VZ-like structures (Supplementary Fig 4A, B) along with smaller lumen size (Supplementary Fig 4C, D), suggesting a decreased pool of polarized progenitors. This is consistent with a recent model of HD striatal organoids34. Additionally, since neural stem cell maintenance and polarity are regulated by primary cilia35,36, we stained all organoids for Arl13b and observed longer cilia (Supplementary Fig 4E, F) and increased cilium density (Supplementary Fig 4G) in HD organoids. These observations align with reports of enlarged cilia in HD rodent striatal cells37 and HD human fetal brain4. Taken together, these data demonstrate that HD organoids exhibit developmental impairments.
The adult striatum originates from the ventral telencephalon and is the first and most affected region in HD. Our CTRL organoids exhibit a resemblance to the fetal VZ and SVZ organization of the ventral telencephalon38–40, with GSX2+ progenitors predominantly located in apical area 1 and ASCL1+ cells positioned more externally (Fig. 2D, E). This radial distribution is peculiar of organoids41,42 as demonstrated by the validation of our quantification on organoids generated following a previous protocol43 (Supplementary Fig 4H, I). In contrast, this organization of ventral progenitors is disrupted in the organoids from the entire HD allelic series (Fig. 2D, E).
Consistent with these findings, HD organoids exhibited downregulation of GO terms associated with ventral populations, such as “subpallium development” and “cerebral cortex GABAergic interneuron fate commitment” (Supplementary Fig 3B and Fig. 2F, G). q-PCR analysis of DLX2 and SP9 genes in independent biological replicates further confirmed impaired ventral maturation in DIV 120 HD organoids (Fig. 2H). Importantly, analysis of DIV120 organoids revealed that the ventral fate was more significantly affected compared to dorsal specification. Indeed, pivotal genes for subpallial identity, such as the DLX family (DLX2, DLX1, DLX5, DLX6), GABA production and uptake (GAD1, GAD2, SLC32A1), and interneuron markers (SCGN, CALB2, ERBB4), were strongly repressed in HD organoids (Fig. 2I). In contrast, genes associated with the dorsal lineage showed milder alterations with different trends among genes. These included the upregulation of early markers (EOMES and EMX2) and mature markers (TBR1 and SLC17A6), along with modest downregulation of NEUROD2, NEUROD6, FEZF2, and CUX2 (Fig. 2I). The specific impairment of the ventral lineage was confirmed by IHC, which revealed fewer GABAergic neurons in HD organoids compared to controls at DIV 120. However, an equal amount of TBR1+ dorsal neurons was observed in both HD and control organoids at the same time point (Fig. 2J, K).
Finally, by comparing our subpopulations at DIV 120 with bulk RNAseq data from human fetal LGE and cortex at 9 pcw39, we found that HD organoids exhibited lower scores specifically for ventral identities (Fig. 2L). Together, these data confirm a cell-type-specific ventral alteration in HD.
Non-cell autonomous rescue of HD ventral defects in mosaic CTRL-HD organoids
During the early to mid-stages of HD, the brain consists of cells exhibiting diverse levels of pathology, also as a consequence of the different somatic CAG expansions exhibited by various brain regions44. This cellular mosaicism will likewise be replicated in future interventions involving cell replacement45,46 or gene silencing47, where healthy cells will coexist with dysfunctional and dying HD cells. This phenomenon emphasizes the importance of comprehending the interactions between these distinct cell populations. Cellular mosaicism can be reproduced in organoids by co-culturing CTRL and HD cells, providing a platform to investigate cell autonomous and non-cell autonomous mechanisms within the brain microenvironment in vitro.
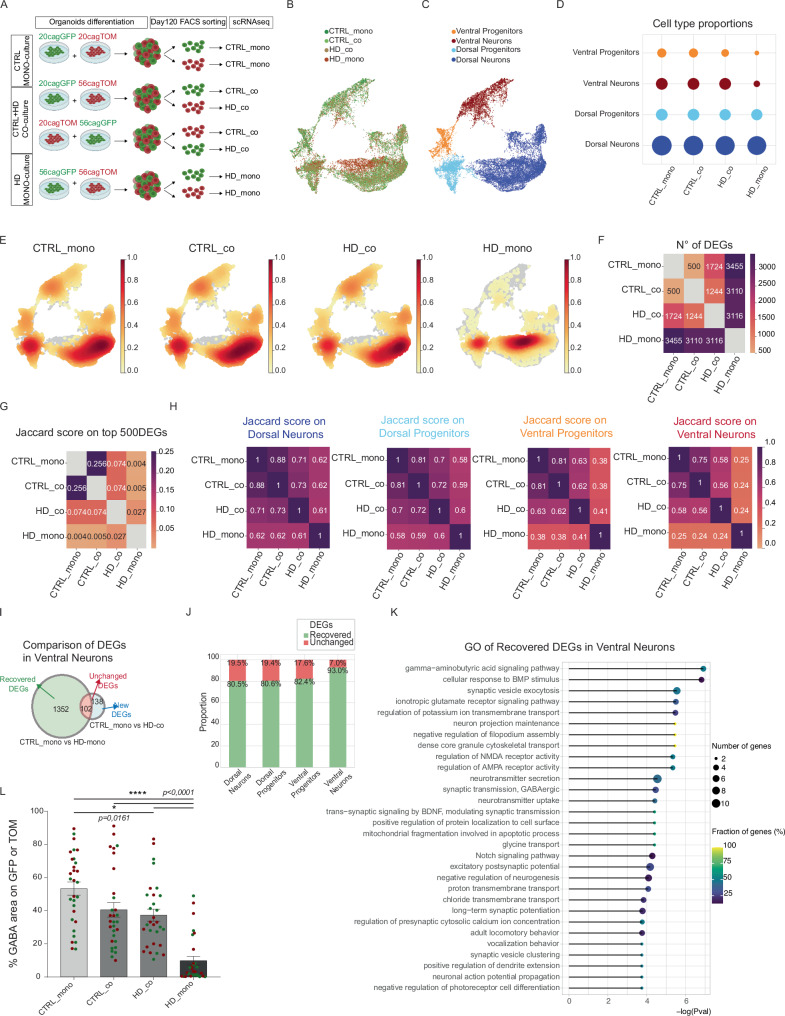
To achieve this, we initially generated sub-clones of CTRL 20CAG and HD 56CAG cells that express either ubiquitous GFP or TOMATO (TOM) (Supplementary Fig 5A). Following quality controls for pluripotency, karyotype, and differentiation capacity (Supplementary Fig 5B–D), we produced mono-culture organoids where GFP and TOM cells of the same genotype were grown together (20CAG-GFP with 20CAG-TOM and 56CAG-GFP with 56CAG-TOM). Additionally, we generated mosaic organoids by co-culturing cells of different genotypes together (20CAG-GFP with 56CAG-TOM and 20CAG-TOM with 56CAG-GFP, Fig. 3A and Supplementary Fig 5E). During differentiation the organoids were checked for progressive formation of VZ-like structures and neurons (Supplementary Fig 5F, G). Finally, they were dissociated and FACS sorted at DIV 120 to dissect four biological conditions: “CTRL_mono” (CTRL cells grown alone); “CTRL_co” (CTRL cells that were grown in the presence of HD cells and then isolated); “HD_co” (HD cells that were grown in the presence of CTRL cells and then isolated); and “HD_mono” (HD cells grown alone, as for CTRL_mono). Each of the eight conditions (considering both GFP and TOM lines) were individually subjected to scRNAseq (Fig. 3A, B). In all samples, scRNAseq analyses using known markers for neuronal maturation and regional identity (Supplementary Fig 6A, B), revealed four subpopulations of ventral progenitors (VP), ventral neurons (VN), dorsal progenitors (DP), and dorsal neurons (DN) (Fig. 3C).
Fig. 3. Mosaic organoids reveal non-cell autonomous recovery of HD ventral identities.
A Experimental scheme for the CTRL-HD mosaic organoids. Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license B, C UMAP plots of scRNAseq analyses on mosaic organoids at DIV 120 for culture conditions (CTRL-mono, CTRL_co, HD_co, and HD_mono) and clusters of subpopulations identified with the Louvain algorithm (Ventral Progenitors, Ventral Neurons, Dorsal Progenitors, Dorsal Neurons) D Bubble plot of cell type proportions per condition E Cell density plots showing how the cells are concentrated in each condition. F Heatmap of number of DEGs (Wilcoxon test, two-sided, p < 0.05) between samples in a pairwise comparison G Heatmap of Jaccard similarity score, based on the proportion of the top 500 DEGS (defined by Wilcoxon rank-sum test) specifically identifying each condition (CTRL_mono, CTRL_co, HD_co, HD_mono) that are in common between two distinct conditions H Heatmap of Jaccard similarity score divided per subpopulations (Dorsal Neurons, Dorsal Progenitors, Ventral Progenitors, Ventral Neurons). I Venn diagram of DEGs in Ventral neurons revealing recovered genes by comparing the list of DEGs between CTRL organoids (CTRL_mono) and HD organoids (HD_mono) with a second list of DEGs between CTRL_mono and HD_co: the genes missing in this second list are the Recovered ones (in green), while genes included in both lists are the Unchanged one (in red) and genes that compare in the co-culture condition are new DEGs (in light blue) J Proportion between Recovered DEGs (in green) and Unchanged DEGs (in red) in each subpopulation K Top 30 GO terms associated to the recovered DEGs in ventral neurons, resulting from GO analysis performed with R package topGO (Fisher test, one-sided). L Automatic quantification of the area positive for GABA over GFP or TOM area (N=8 organoids from 2 independent biological replicates. error bars represent ± SEM. Anova One Way, Bonferroni post test. ****p < 0.0001, *p < 0.05). Source data are provided as a Source Data file.
Analysis of cell composition showed that HD organoids grown alone (HD_mono) displayed fewer ventral progenitors and ventral neurons compared to other conditions (Fig. 3D). In contrast, HD cells grown with CTRLs (HD_co) showed compositions similar to CTRL organoids grown alone (CTRL_mono), indicating recovery in ventral sub-populations. Furthermore, the profile of CTRL cells grown with HD (CTRL_co) showed similar proportions to CTRL_mono (Fig. 3D), suggesting that HD cells are not detrimental to CTRL cells for this parameter. The difference in ventral subpopulations density between HD_mono and HD_co is confirmed in each sub-clone (Supplementary Fig 6C). Moreover, the UMAP cell density plots (Fig. 3E), the number of DEGs in pairwise comparisons between culture conditions (Fig. 3F), and the Jaccard similarity scores (JS), based on the proportion of DEGs specific for each condition that are in common between two distinct conditions (Fig. 3G), confirmed the beneficial effect of healthy cells on HD cells.
Furthermore, the average expression scores of DEGs between CTRL and HD organoids which overlapped with the DEGs from the KEGG pathway associated with HD (already shown in Fig. 1I), indicated a downregulation in HD organoids. This downregulation is rescued in HD_co at both bulk and single sub-populations levels (Supplementary Fig 6D, E). These results suggest a non-cell autonomous transcriptional recovery in HD cells grown together with CTRL cells. The fact that CTRL_co (CTRL cells grown with HD cells) are similar to CTRL_mono suggests the absence of a major transfer of toxicity from HD cells to CTRL cells.
To determine which sub-population of HD organoids is most affected and highly recovered in mosaic cultures, we performed JS analysis also focusing on single subpopulations. JS comparison across different culture conditions revealed that ventral neurons in HD_mono (HD cells cultured alone) are the most affected population (Fig. 3H), with a stronger non-cell autonomous recovery in HD_co for ventral neurons compared to other subpopulations (Fig. 3H).
This is also confirmed at the level of differentially expressed genes (DEGs) in ventral neurons, which were highest in HD_mono compared to all other cultures and confirmed restored expression levels in HD_co (Supplementary Fig 7A), consistent with the previous result at a bulk level (Fig. 3F). To assess the impact of this rescue, we compared the list of DEGs between CTRL and HD cells grown alone (CTRL_mono vs HD_mono), with a second list of DEGs between CTRL cells grown alone and HD cells grown with CTRLs (CTRL_mono vs HD_co). We found that most genes were recovered (Fig. 3I), with only a few unchanged or new DEGs. Recovered DEGs represented the majority in all subpopulations (Supplementary Fig 7B), but the ratio was higher in ventral neurons (Fig. 3J). This is further confirmed at the level of DEGs that were uniquely expressed in each subpopulation (Supplementary Fig 7C), indicating a cell-type-specific recovery.
Recovered DEGs in HD_co ventral neurons were enriched in terms related to neuronal identity and activity (Fig. 3K). This category included GABA signaling, glutamate receptor activity, and BDNF synaptic transmission (Fig. 3K), which are known to be critical for subpallial neurons during both development and HD pathogenesis14,48. Moreover, among the genes enriched in ventral neurons recovered in HD_co we found pivotal genes for striatal neurons development, including PBX3, SP9, and SYNPR39 (Supplementary Fig 7D, E). The cell type specific recovery was further validated by qPCR analyses conducted on independent biological replicates (Supplementary Fig 7F) and by the quantification of GABAergic neurons which were rescued in both GFP and TOM HD_co cells grown with CTRL (Fig. 3L and Supplementary Fig 8A).
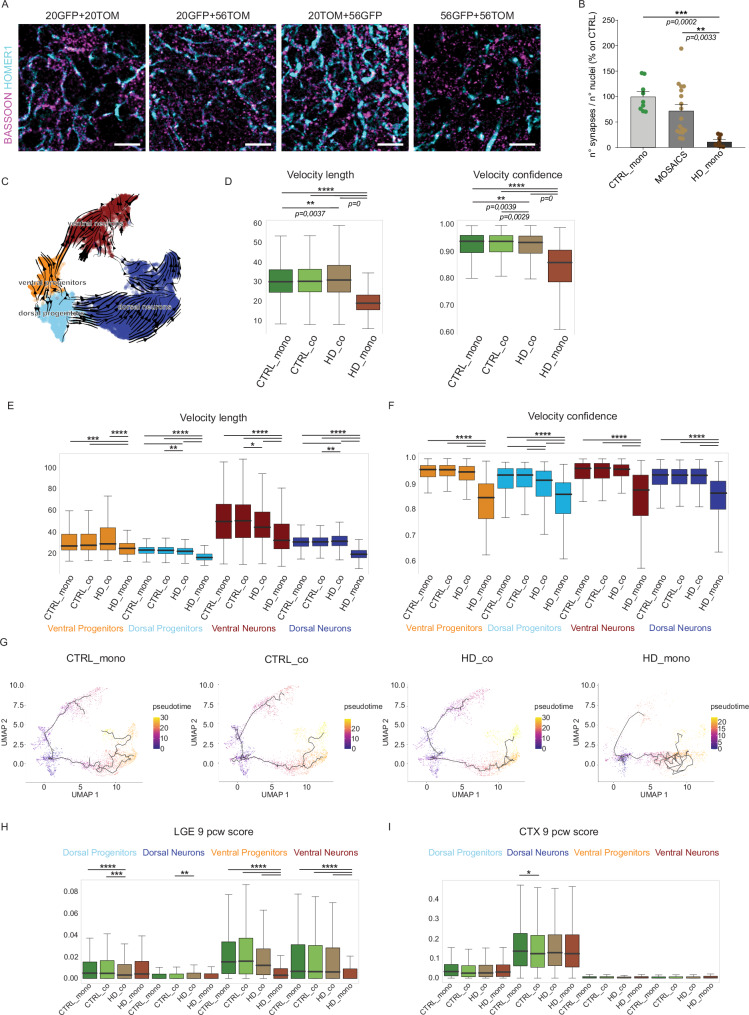
Additionally, since the GO of rescued genes in ventral neurons (Fig. 3K) revealed terms related to synaptic activity and neurotransmitters regulation, we evaluated the Gene Set Enrichment Analysis (GSEA) for those and other synapse-related terms (Supplementary Fig 8B). CTRL_mono ventral neurons compared to HD_mono showed positive enrichment for synaptic terms like neurotransmitter uptake, GABAergic synaptic transmission, and synaptic vesicle membrane (Supplementary Fig 8B). This suggests lower synaptic density or activity in HD organoids. The extent of synapses presence was further evaluated by assessing the puncta of colocalized Bassoon and Homer1 signals49. We found that synaptic density was reduced in HD_mono compared to CTRL_mono organoids, highlighting a phenotype typically associated with more advanced, pre-symptomatic stages of the disease in mice50–52 (Fig. 4A, B and Supplementary Fig 8C). Strikingly, this impairment was rescued in co-culture mosaic organoids (defined MOSAICS in Fig. 4I). We also verified the specificity of this primary synapse pathology by normalizing the Homer1 density to the Map2 positive area, revealing that HD organoids have similar neuronal content but fewer synapses, which are rescued in HD_co organoids (Supplementary Fig 8D, E).
Fig. 4. HD cells recover maturation and fate determination when grown with CTRL cells.
A Immunohistochemistry analyses of CTRL_mono (20GFP+20TOM) co-culture mosaic organoids (20GFP+56TOM and 20TOM+56GFP) and HD_mono (56GFP+56TOM) organoids at DIV 120 of differentiation for BASSOON and HOMER1. Scale bar 20 µm B Automatic quantification of numbers of synapses as BASSOON/HOMER1 co-localizing puncta in each condition. As it was not possible to quantify single GFP or TOM cells overlapping with the puncta staining, the quantification reports the total number of synapses in the co-culture mosaic condition (N = 8 organoids from 2 independent biological replicates. Data are normalized over the mean of CTRL values, error bars represent ± SEM. Anova One Way, Bonferroni post test. ***p < 0.001, **p < 0.01). C Differentiation trajectories of all mosaic organoids together, inferred with the method of RNA velocity D Length and confidence of the velocity vectors in each culture sample (Wilcoxon test, two-sided, Bonferroni post test, **p < 0,01; ****p < 0,0001) E, F Length and confidence of the velocity vectors in each individual subpopulation (Wilcoxon test, two-sided, Bonferroni post test, *p < 0,05; **p < 0,01; ***p < 0,001; ****p < 0,0001) G Pseudotime analysis in each culture sample performed using Monocle3 H, I Boxplots for the signature score (as described in methods) of comparison between our mosaic organoids over the bulk RNAseq from human fetal LGE and CTX (cortex) at 9pcw39, focusing on individual subpopulation of each sample (Wilcoxon test, two-sided, Bonferroni post test, *p < 0,05; **p < 0,01; ***p < 0,001; ****p < 0,0001). For D, E, F, H, and I the box plots show the median (centre line), upper and lower quartiles (box limits), and the highest and lowest values within 1.5× the IQR of the nearest hinge (whiskers). For E, F and H, I exact p-values can be found in Supplementary Table 2 inside the Supplementary Information. Source data are provided as a Source Data file.
In parallel to the recovery, we wondered what the few unchanged DEGs indicate by looking at their GO terms. We found that the unchanged DEGs reflected genes with broader functions, ranging from metabolism to translation (Supplementary Fig 9A). This indicates that a small core set of altered basal programs could not be recovered even in the presence of CTRL cells. These include some polarity and cell migration genes such as NNAT, CNTNAP2, EPS8L2, and ROBO1 which are not recovered (Supplementary Fig 9B). Accordingly, the organization of progenitors in VZ-like structures was only partially restored in mosaic organoids, as shown by PALS1 staining (Supplementary Fig 9C, D). To have a broader view of unchanged genes and networks, we looked at transcription factor (TF) activity inference in HD_mono and HD_co both compared to CTRL_mono. Among the TFs more active in HD_mono and not rescued in HD_co, we found SP1 (Supplementary Fig 9E), known to bind HTT and already described as dysregulated in HD in in-vitro derived neurons53, further suggesting the relevance of our model. Moreover, in HD_mono, many TFs related to stress and cell death emerged, such as XBP1, ATF4, and FOXO3, which are partially rescued in HD_co (Supplementary Fig 9E). We confirmed higher cell apoptosis in HD_mono organoids compared to CTRLs by cleaved Caspase-3 immunohistochemistry, which is not fully prevented in co-culture mosaic organoids (Supplementary Fig 9F–G). From these data, we can exclude mechanisms of polarity or cell survival as drivers of the recovery observed in HD_co grown with CTRL cells.
To understand how cell fate determination occurs in each condition, we evaluated maturation trajectories. Velocyto analysis54 highlighted the presence of dual branching lineages of ventral and dorsal trajectories (Fig. 4C), with reduced length and confidence of the velocity vectors in HD cells grown alone (HD_mono) compared to all other conditions (Fig. 4D). This reduction is consistent across each single subpopulation (Figs. 4E, F). Monocle analysis55 confirmed a significant defect in ventral lineage commitment in HD cells grown alone, as evidenced by the obstruction of their progression towards ventral neurons (Fig. 4G). Additionally, it revealed an irregular trajectory toward dorsal neurons (Fig. 4G), suggesting an overall slower and more heterogeneous rate of maturation compared to CTRL organoids, consistent with the results from Velocyto analysis. Importantly, all these defects were recovered in HD_co and were not altered in CTRL_co (Fig. 4D–G), supporting the notion that CTRL cells rescue abnormal trajectories of HD cells, while HD cells do not alter developmental programs in CTRLs.
Rescue in ventral fate determination was further confirmed by comparing our dataset with the bulk RNA-seq signature scores of human fetal LGE and cortex at 9 pcw39. The lower similarity score in ventral subpopulations of HD_mono was recovered in HD cells grown with CTRL cells (HD_co), while the similarity with the cortex was generally comparable between CTRL and HD (Fig. 4H, I). These results on trajectories and synapses suggest that the presence of CTRL cells in mosaic organoids promotes the recovery of aspects associated with neuronal maturation and synapse density in HD cells.
Overall, our approach using mosaic organoids allowed us to identify a non-cell autonomous beneficial effect in cell compositions and maturation.
Recovery of the HD ventral phenotypes occurs through cell-cell interactions
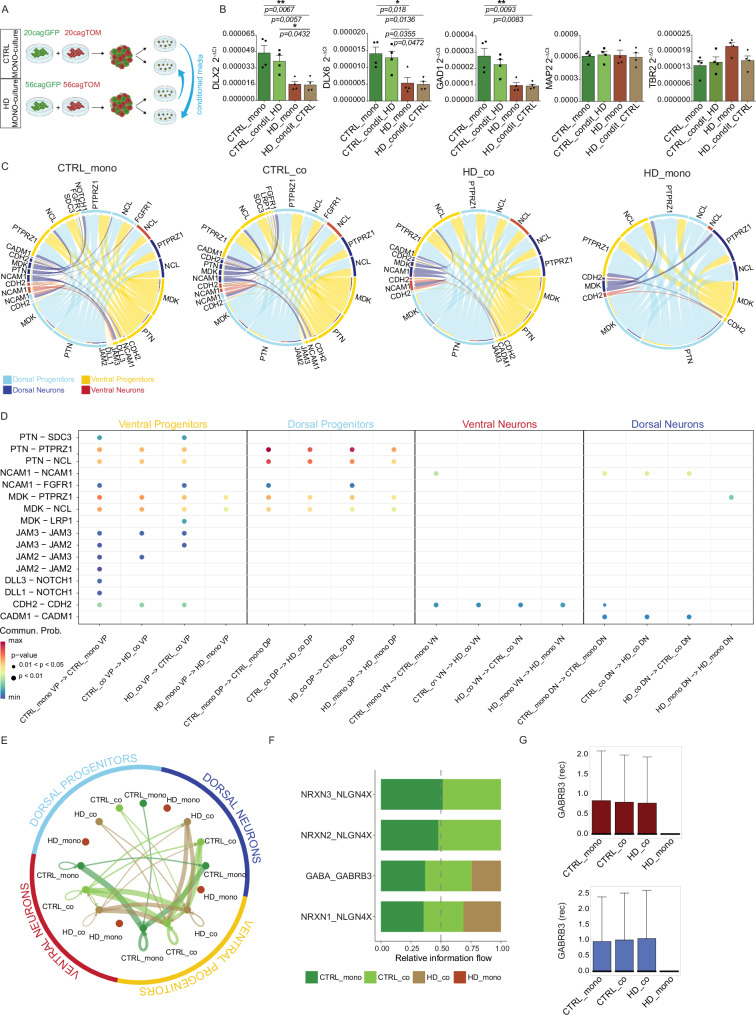
To analyze the nature of the non-cell autonomous rescue in HD_co, we first tested the potential contribution of factors released in the media (Fig. 5A). However, no rescue of gene expression was detected in HD organoids upon incubation with medium conditioned from CTRL organoids (HD condit CTRL) (Fig. 5B), indicating that cell-cell contact is necessary for the recovery. We then explored our mosaic organoids for CTRL-HD cell communications using CellChat, which relies on ligand-receptor expression and interactions56. This analysis revealed reduced communication pathways in HD_mono, especially in ventral HD progenitors (yellow connections), which were largely recovered in HD_co (Fig. 5C). In contrast, the communications in dorsal progenitors (light blue connections) are less affected, consistent with the less compromised dorsal populations of HD organoids. The communications in CTRL_co organoids were overall similar to CTRL_mono (Fig. 5C), confirming that CTRL cells were minimally affected by the presence of HD cells.
Fig. 5. Cell-cell communications show cell type specific changes in mosaic organoids.
A Experimental scheme for conditioned medium organoids with CTRL receiving media from HD and vice-versa. Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license B q-PCR for DLX2, DLX6, GAD1, MAP2, and TBR2 on CTRL and HD canonical (mono) and organoids conditioned from HD (condit HD) or from CTRL (condit CTRL) at DIV 120. (N = 4 pools of organoids from 2 independent biological replicates; error bars represent ± SEM; Anova One Way, Bonferroni post test, *p < 0,05; **p < 0,01) C CellChat circle plot communications between all subpopulations in each individual condition D CellChat bubble plot of gene expression for couples of ligands and receptors between cells grown together in the same organoid per each subpopulation. Abbreviations: VP=Ventral Progenitors; DP=Dorsal Progenitors; VN=Ventral Neurons; DN=Dorsal Neurons. Wilcoxon test, two-sided, Bonferroni adjustments for multiple comparisons E NeuronChat circle plot showing total communications between all subpopulations and conditions F NeuronChat stacked bar plot of gene expression for couples of ligands and receptors in each condition G Box plots of gene expression for GABRB3 receptor in ventral neurons and dorsal neurons for each condition. For G the box plots show the median (centre line), upper and lower quartiles (box limits), and the highest and lowest values within 1.5× the IQR of the nearest hinge (whiskers). Source data are provided as a Source Data file.
CellChat found more communication pathways within the subpopulations of progenitors than in neurons (Fig. 5D), reflecting what happens in the developing human LGE and in 2D striatal neurons in vitro57. In particular, the Pleiotrophin (PTN) pathway communications displayed a specific rescue in number of connections intra the ventral progenitors and between this and other subpopulations (Supplementary Fig 10A), as also indicated by the levels of the PTN ligand and its receptors PTPRZ1 and NCL (Supplementary Fig 10B), which are critical for the development of outer radial glial cells in the human brain26. The Junctional Adhesion Molecule (JAM) transmembrane receptors and ligands, which are physiologically localized at tight junctions, showed both highly specific expression (Supplementary Fig 10C) and recovery (Supplementary Fig 10D) in ventral progenitors. HD_co ventral progenitors also recovered N-cadherin (CDH2) expression (Supplementary Fig 10E, F), whose distribution is altered in human HD fetal cortical tissues4 and whose proteolysis is affected in the HD adult brain58.
Together with the other rescued pathways, such as NCAM1 and CADM1 in dorsal neurons (Fig. 5D), the majority of rescued ligands and receptors were transmembrane proteins or cell adhesion molecules, suggesting the importance of cell contacts and interactions.
Further investigation on cell-cell communication for neuronal specific signaling using NeuronChat59 corroborated the downregulated expression of ligands and receptors in HD organoids compared to CTRL ones and, importantly, the rescue observed in HD_co mosaic organoids (Fig. 5E). Specifically, this tool highlighted neuronal cell communication, where we observed the rescue of Neurexin1 (NRXN1), a neuronal cell surface protein involved in cell adhesion and synaptic junctions, along with GABRB3 receptor (Fig. 5F). These findings demonstrated both cell-type-specific expression and rescue in dorsal and ventral neurons (Fig. 5G).
Consistent with synapse loss in our model (Fig. 4A, B) and as a predominant feature in the brain of adult HD mice51,52, also our results from CellChat and NeuronChat (Fig. 5D–F) indicate a loss of function for ligands and receptors that are expressed in the synaptic compartment. In particular, we found that the expression of N-Cadherin (CADH2) and Neurexin1 (NRXN1) is reduced in HD organoids. This reduction is rescued in HD_co that were grown with CTRL cells.
Given that progenitors are commonly transplanted in adult HD hosts45 and we have shown the majority of beneficial communications in those types of cells, we hypothesized that the same underlying biological interactions driving the reported rescue could be beneficial also when progenitor cells are transplanted and in contact with the host adult cells in vivo. Indeed, in the mosaic organoids the communications between a CTRL progenitor (CTRL_co DP and CTRL_co VP) and a HD neuron (HD_co DN and HD_co VN) were rescued and equal to the same type of interactions in CTRL_mono (Supplementary Fig 10G), suggesting that CDH2 and NCL could drive beneficial interactions between those cell types.
Concurrently, our data indicate that communication among HD cells is weaker rather than abnormal (Fig. 5C–F). However, as cell communication is bilateral, we also examined transcriptomic changes in CTRL_co cells. We observed that only the DLL1/3-NOTCH1 (Fig. 5C, D) pathway is expressed in CTRL_mono but not in CTRL_co suggesting a potential loss of function of this specific pathway in CTRL cells grown with HD. While this effect is minor effect compared to the different pathways rescued in HD_co, we concluded that healthy cells are beneficial for HD cells without being hardly affected by them in return.
In conclusion, these data indicate a reduced capability of HD cells per se to communicate. We suggest that the presence of CTRL cells in co-culture mosaic organoids restores a threshold of cell interactions in HD cells by rescuing the expression of receptors and ligands for communication pathways in a non-cell autonomous manner.
Discussion
Three-dimensional organoids offer a significant advantageous opportunity for studying cell-cell interactions, making them a superior cell model for investigating both cell autonomous and non-autonomous mechanisms in HD compared to traditional two-dimensional cultures. To address this, we developed a telencephalic organoid system to model HD phenotypes by growing non-pathologic (CTRL) and CAG-expanded (HD) isogenic cell lines either alone or in mosaic organoids. We characterized the cell type-specific transcriptional profile of HD organoids using scRNAseq for both mono-culture and mosaic organoids. These telencephalic organoids demonstrated progressive maturation over time, forming organized structures composed of progenitors and mature neurons. Immunolabeling and gene expression analyses revealed distinct sub-populations of progenitors and neurons with early specification of ventral identities followed by the emergence of dorsal subpopulations. We identified neurodevelopmental and cytoarchitectural alterations in HD organoids, including reduced multilaminar radial organization, altered polarity, mitotic spindle, cell cycle, and primary cilium. These findings align with data on HD human cortical fetal tissue4 and underscore the importance of deeply investigating neurodevelopmental aspects of the disease in a human context. In addition to modeling these neurodevelopmental defects, our organoids also exhibited certain mature phenotypes of the adult brain. We observed a specific defect in ventral neurons, characterized by the downregulation of key subpallial genes and fewer GABAergic neurons in HD organoids. HD organoids also exhibited dysregulation of genes belonging to the HD KEGG pathway, and many of our DEGs also overlapped with DEGs identified in striatal cell types of grades 2–4 human HD post-mortem samples7. Additionally, HD organoids displayed fewer synapses and transcriptional dysregulation, phenotypes in line with studies on HD mice and postmortem human HD material60–62 and described to correlate with clinical progression12.
HD organoids did not show mHTT aggregates, as expected, since aggregates are typically not found in cell culture systems, both in 2D and 3D, ranging from human PSCs to primary neurons from HD mice15,63–65. More studies are needed to understand the complexities surrounding the presence and significance of mHTT aggregates and how closely they provide a measure of pathological disease stage66,67, given their variable occurrence in human postmortem brains68,69 and their presence in mouse models that do not exhibit behavioral abnormalities nor neurodegeneration70.
In conclusion, the phenotypes observed in our HD organoids underscore the relevance of this model for studying various facets of HD pathology, from early developmental stages to more advanced disease states. Considering future therapeutic approaches for HD, such as cell replacement45,46,71 and gene silencing47, which will create mosaicism with coexisting healthy and dysfunctional cells within the brain microenvironment, it becomes crucial to unravel how cells in different states can interact and influence each other72. Importantly, in our mosaic organoids where CTRL and HD cells are co-cultured, we observed a significant beneficial non-cell autonomous effect of healthy CTRL cells on HD progenitors and neurons, particularly within the ventral identities. The presence of CTRL cells enabled the HD cells to successfully re-establish cell composition, DEGs, trajectories, ventral maturation, synapses, and communication pathways. We found that the communication of HD cells was not altered with abnormal pathways but was weaker, suggesting a reduced capability of HD cells per se to communicate that is restored by CTRL cells in mosaic organoids. Moreover, since the treatment of HD organoids with medium conditioned from CTRL cells alone failed to mitigate HD transcriptional alterations, these data further suggest that cell-to-cell interactions play a pivotal role in reversing the abnormal HD transcriptional state.
Although we observed a near-full transcriptional recovery in HD cells grown with CTRL ones in mosaic organoids, some phenotypes, such as cell polarity in progenitors’ organization and apoptosis, were not fully rescued. While further investigations are required, we speculate that the mechanism driving these defects involves genes with cell autonomous regulation, likely linked to the intrinsic toxicity of mHTT, and therefore unaffected and not rescued by interactions with control cells.
One remaining limitation of this study lies in the model. Organoids are complex multicellular cultures that, compared to other culture systems, can produce a broader spectrum of cell types better reflecting the diversity of the endogenous brain. However, they are still not heterogeneous and oriented enough to fully mimic the brain. This limitation could impede investigations of more advance phenotypes due to the lack of specific cell types or connections between areas. Despite this, in the absence of faithful experimental models for human neuronal biology, organoids remain one of the most valuable alternatives.
In mouse chimeras, the transplantation of human HD glial progenitors cells into the wild-type mouse brain resulted in HD phenotypes, whereas CTRL glial progenitors cells could rescue these effects22. We did not observe harmful effects on CTRL cells from HD ones in our study and this discrepancy could be due to differences between the in vivo mouse glia-based chimerism and our in vitro human specific chimerism, which is based only on neuronal populations without glial cells. Nonetheless, both studies indicate the involvement of non-cell autonomous mechanisms in HD22. Our data emphasize the disease-modifying effect of healthy human cells, highlighting the potential therapeutic role of cell-cell communication in HD progression and therapy. In line with our study, a recent work by Pipicelli and colleagues73 demonstrated non-cell autonomous regulation of interneuron fate specification using an organoid model with mutations in the extracellular matrix gene LGALS3BP, which is associated with cortical malformations.
In conclusion, our study provides valuable insights to the investigation of neurodevelopmental aspects in HD, cell type-specific defects, and synaptic abnormalities associated with the pre-symptomatic stage, while at the same time offering evidence of beneficial non-cell autonomous mechanisms in HD pathogenesis. This work provides an ideal platform for disease modeling application, offering invaluable inspiration to open new avenues for investigating therapeutic interventions.
Methods
Culture of human pluripotent stem cells
The use of RUES2 human embryonic stem cell lines and derivatives in the laboratory has been approved by the ethical committee of the University of Milano (Neurostemcell Repair on 18/04/2016, and CHDI on 16/05/2016).
The RUES2 lines were derived from Prof. Ali Brivanlou’s Laboratory (Rockefeller University) and provided by the CHDI Foundation (New York, NY, USA). The lines were regularly tested and maintained mycoplasma-free (Eurofins). Karyotype for each cell line was monitored every 3 months during the passages by Q-banding analyses (by ISENET group). Cells were kept in monolayer on 120–180 µg/ml Geltrex (Thermo Fisher Scientific, A14133-02) coated cell culture dishes and in feeder-free conditions in mTeSR1 medium (StemCell Technology, 85850) added with Penicillin (100 U/ml) and Streptomycin (100 μg/ml) (Euroclone, EC3001D). Once a week, hESC colonies were split with a ratio of 1:10 by ReleSR (StemCell Technology, 100-0484) dissociation. Cells were kept in culture for 3 months and then a new batch was thawed. Cells were frozen in cold KSR (Knockout serum replacement, Thermo Fisher Scientific, 10828028) containing 10% DMSO (Sigma Aldrich, D2650), 0,1% RI Y-27632 (Rock Inhibitor, 10 µM, provided by CHDI Foundation, CHDI-00197406-0001-007) at −150 °C and thawed in mTeSR1 with RI.
Cell lines used in this work are reported in Table 1A inside the Supplementary Information.
Generation of telencephalic organoids
hESC were kept in a 6 cm2 dish until they were 70–80% confluent and ready for aggregation (day0). The cells were detached with 1 ml of Accutase (Millipore, SF006) for 4-5’ and resuspended in mTeSR1 with RI (10 µM, CHDI-00197406-0001-007). Using a multichannel the cells were seeded at a 2 x 104 cells/well concentration in a V-bottom 96well plate with 150 µl/well and centrifugated at 150 g for 1’. After 2 days the medium was replaced with the Neural Induction Medium that contains DMEM/F12 (21331, Thermo Fisher Scientific) with Penicillin (100 U/ml) and Streptomycin (100 μg/ml) (Euroclone, EC3001D), L-Glutamine (2 mM, Thermo Fisher Scientific, 25030081) N2 supplement (1:100, Thermo Fisher Scientific, 17502-048), B27-RA supplement (1:50, Thermo Fisher Scientific, 12587-010), NEAA (1:100, Thermo Fisher Scientific, 11140-050), Heparin (1mg/ml, Sigma Aldrich, H3393), Insulin (5 µg/ml, Sigma Aldrich, I9278), SB431542 (10 µM, CHDI Foundation, CHDI-00447536), LDN (500 nm, CHDI Foundation, CHDI-00396388). This neural induction determined the day 0.
Every two days the aggregates receive a partial medium change by taking out 50 µl of old medium and adding 50 µl of fresh medium. From day 6 the medium is replaced with Patterning Medium which add Shh (200 ng/ml, R&D Systems, 464-SH) and Dkk1 (100 ng/ml, Peprotech, 120–30) to the previous one.
At around day 18 the EBs were transferred to a 6 cm2 dish on orbital shaker, with fresh medium (6 ml) and then embedded in 25 µl Geltrex (Thermo Fisher Scientific, A14133-02), one by one using a cut 200 µl pipette tip switching into Neuronal Differentiation Medium: DMEM/F12 (21331, Thermo Fisher Scientific) and Neurobasal (21103049, Thermo Fisher Scientific) in ratio 1:1 with N2 supplement (1:100, Thermo Fisher Scientific, 17502-048), B27 complete supplement (1:50, Thermo Fisher Scientific, 17504-044), NEAA (1:100, Thermo Fisher Scientific, 11140-050), Insulin (5µg/ml, Sigma Aldrich, I9278), Laminin (200 ng/ml, Thermo Fisher Scientific, 23017-015), BDNF (10 ng/ml, Peprotech, 450-02), 2-Mercaptoethanol (50 µM, Thermo Fisher Scientific, 31350-010), Ascorbic acid (100µM, Sigma Aldrich, A4544). From now to the end of differentiation, half of the medium was changed two days a week and the dish was kept on the orbital shaker at 37 °C.
Generation of cortical organoids
This protocol was used only as comparison for the assay of VZ-like structures concentric areas. It was an adaptation from Lancaster’s original one43,74. The major differences consisted in using hanging drop plates for the initial aggregation, putting the aggregates on rotation on the orbital shaker since day5 and maintaining them in rotation after Matrigel embedding instead of bioreactors, while the media were the same from the published protocols.
hESC were kept in a 6 cm dish until they are 70–80% confluent. The cells were detached with 1 ml of Accutase for 4-5’, centrifugated at 200 g for 3’ at room temperature, resuspended in Aggregation medium: DMEM/F12 (21331, Thermo Fisher Scientific); KSR (20%, Knockout serum, Thermo Fisher Scientific, 10828028); NEAA (1:100, Thermo Fisher Scientific, 11140-050); 2-Mercaptoethanol (100nM, Thermo Fisher Scientific, 31350-010); Rock Inhibitor (50 µM, CHDI Foundation, only for 48 h) and plated as 1 x 104 cells/well concentration in a Perfecta3D hanging drop 96well plate (HDP1096, 3D Biomatrix) in 35 µl/well. The day after 5 µl of Aggregation medium were added to each well of the plate. At day 3 the EBs were transferred in a 6 cm dish with 6ml of Aggregation medium. Around day 6–8 when the EBs were about 500–600 µm in diameter the media was changed with Neural Induction Medium: DMEM/F12 (21331, Thermo Fisher Scientific); N2 supplement (1:100, Thermo Fisher Scientific, 17502-048); NEAA (1:100, Thermo Fisher Scientific, 11140-050); Heparin (1 µg/ml, Sigma Aldrich, H3393) which was partially changed every other day. Between day 13–16 the EBs were embedded into Matrigel droplets. Aggregates were embedded in a drop of 25 µl of liquid Cultrex (Cultrex BME Pathclean, Trevigen 343201001) similarly to telencephalic organoids and kept with Neuronal Differentiation medium: DMEM/F12 (21331, Thermo Fisher Scientific); N2 supplement (1:100, Thermo Fisher Scientific, 17502-048); NEAA (1:100, Thermo Fisher Scientific, 11140-050); Insulin (2,5 ng/ml, Sigma Aldrich, I9278); B27-RA supplement (1:100, Thermo Fisher Scientific, 12587-010); 2-Mercaptoethanol (100nM, Thermo Fisher Scientific, 31350-010). From day 30 until the end of differentiation, the Neuronal Differentiation medium was changed in Final Differentiation medium by substitution of B27-RA with B27 compete (1:50, Thermo Fisher Scientific, 17504-044) to support neuronal differentiation and maturation and partially changed twice a week.
Mosaic organoids composed of WT and HD telencephalic cells
Lentivirus production
To achieve stable expression of GFP and TOM, the Rues2 hESC line were infected with lentivirus particles containing the fluorescent genes under the control of chicken β-actin promoter and the puromycin selection cassette75. The lentivirus particles were generated through HEK293 cultured in DMEM (Thermo Fisher Scientific, 11965092) supplemented with 10% Fetal Bovine Serum (Thermo Fisher Scientific, 16000044), 0,5% Penicillin (100 U/ml) and Streptomycin (100 μg/ml) (Euroclone, EC3001D), 2 mM Glutamine (Thermo Fisher Scientific, 25030081), 1 mM Sodium Pyruvate (Thermo Fisher Scientific, 11360070) and 1% NEAA (Thermo Fisher Scientific, 11140-050). The cells were transfected in OptiMEM (Thermo Fisher Scientific, 31985070) medium with helpers pVSVG (envelope plasmid) and pPAX2 (packaging plasmid). The lentiviral particles were then collected by centrifugation and concentrated by Amicon column.
Generation of GFP and TOM stable cell lines
hES RUES2 cell lines were infected with GFP and TOM lentiviruses to generate: WT-Rues2-20CAG-GFP; WT-Rues2-20CAGT-OM; HD-Rues2-48CAG-GFP; HD-Rues2-48CAG-TOM; HD-Rues2-56CAG-GFP and HD-Rues2-56CAG-TOM. Cells in self renewal were detached with Accutase (Millipore, SF006) and resuspended at the concentration of 100,000 cells/ml in mTeSR (StemCell Technology, 85850) containing RI Y-27632 (10 µM, CHDI Foundation, CHDI-00197406-0001-007). The viruses (50 µl for each ml of cells) were added at this suspension and placed in tubes at 37 °C in the incubator for 30 min. The infected cells were then plated on Geltrex (Thermo Fisher Scientific, A14133-02) coated plates at 37 °C and after 2, 5 h the media was changed with fresh mTeSR containing RI. The following day the medium was changed without RI and after 3 days Puromycin (300 ng/ml) was added for selection. After one week in selection with daily medium change the colonies were picked and plated in presence of RI. After one week in Puromycin selection the cells were frozen and fixed for IF to check levels of fluorescent proteins (with AbI anti-GFP and anti-RFP) and pluripotency (with AbI anti-Oct4). The selected clones were thawed, amplified, and used for organoids experiment.
Co-aggregation of CTRL and HD cells to generate mosaic organoids
We followed the procedures described before to generate telencephalic organoids. The different cell lines (20CAG-GFP, 20CAG-TOM, 56CAG-GFP and 56CAG-TOM) were detached, counted and resuspended in parallel. At day0 the cells were plated in 96well V bottom plate with a mixture of two lines in a ratio 1:1 in mTeSR added with RI (10 µM, CHDI Foundation). The following steps were the same of telencephalic organoids protocols. Around day 8 and 18 the EBs were checked at the microscope for endogenous levels of fluorescent proteins in live. During differentiation, the mosaic organoids were routinely tested for GFP and TOM with Immunohistochemistry analyses.
Dissociation ad FACS sorting of co-culture organoids for scRNAseq
The mosaic co-culture organoids were differentiated until day 120 and dissociated in single cells suspension, following the same procedure described for scRNAseq. These suspensions were FACS sorted to isolate CTRL and HD populations inside the co-culture by their GFP or TOM signal. The sorting was performed with the BD FACSAria cell sorter and analyzed by the FACS Diva v. 6.1.3 software. Pools of 10 organoids per genotype were used because there was not sufficient material from one individual organoid to conduct scRNAseq analyses after FACS sorting.
EdU Click-iT assay
EdU (5-ethynyl-2’-deoxyuridine), a nucleoside analogous of thymidine, is incorporated into DNA during active DNA synthesis and is fluorescently labelled with a bright, photostable Alexa Fluor dye. We used the Click-iT™ EdU Cell Proliferation Kit for Imaging (Thermo Fisher Scientific, C10340). At day 35 of differentiation EdU (10 uM) was added through a partial change of the medium for organoids. EdU medium was kept for 90 min in rotation on orbital shaker, after which organoids were immediately collected and fixed, or washed with PBS and fed with fresh medium until later collection for other time points. EdU click-it assay was then performed on fixed organoids sections following manufacture’s instruction and immuno-staining.
RNA extraction and Real-time q-PCR
At specific time points during differentiation, organoids were collected and processed for total RNA extraction using TRIzolTM Reagent (Thermo Fisher Scientific, 15596018) according to the manufacturer’s instructions. The integrity of the purified RNA and the absence of genomic DNA contamination were assessed by non-denaturing agarose gel electrophoresis. In presence of genomic DNA contamination, Ambion® DNA-free™ Dnase Treatment and Removal Reagents (Invitrogen, Cat. No. AM1906) was used. 500 ng of total RNA was retrotranscribed using iScript cDNA Synthesis Kit (BioRad, Cat. No. 1708891) following the manufacturer’s instructions. Quantitative RT-PCR (RT-qPCR) was performed using a CFX96TM Real-Time System (Bio-Rad) and analyzed with the CFX Manager Software (Bio-Rad). All reactions were performed in 15 µL containing 50ng cDNA and SsoFast EvaGreen Supermix Kit (BioRad, Cat. No. 1725204). Primer pairs used are reported in Table 1B inside the Supplementary Information.
Immunohistochemistry and imaging
Organoids were fixed in 4% (vol/vol) paraformaldehyde (Thermo Fisher Scientific, J19943-K2) for 1 h at 4 °C. They were washed 3 times in PBS (Euroclone, ECB40041) and transferred in a 15% sucrose (Thermofisher, A15583) solution at 4 °C for 24 h and 30% sucrose solution for the following 24 h. Next, organoids were transferred into Tissue-Tek OCT compound (Sakura, 4583), frozen immediately on dry ice and stored at −80 °C. Sections of 12/15 μm thickness were then generated using a cryostat (Leica CM1520). Cryo-sections were left overnight at room temperature to dry and then washed with PBS and permeabilized with 0.5% Triton X-100 (Euroclone, EMR237500) in PBS for 15 min. For antigen retrieval, cryo-sections were treated with Sodium Citrate 10 mM in a water bath at 90° for 30 minutes and then blocked with 10% NGS (Vector, S-1000), 0.1% Triton X-100 in PBS at room temperature for 1 h. Primary antibodies used were diluted in solution containing 3% NGS and 0.1% Triton X-100 in PBS at 4 °C overnight. The following day sections were washed three times in PBS at room temperature. Secondary antibodies conjugated to Alexa fluorophores 488, 514, 568 or 647 and produced in goat (Molecular Probe, Life Technologies) were used 1:500 in solution containing 3% NGS and 0.1% Triton X-100 in PBS at room temperature for 1 h mixed with Hoechst 33258 (5 μg/ml; Thermo Fisher Scientific) to visualize nuclei. Then the sections were washed three times in PBS and finally mounted with Dako Glycergel (Aqueous Mounting Medium, Agilent, C056330-2) at room temperature overnight. The following day the sections were dry enough to be visualized at the microscope and then conserved at 4 °C. Images were acquired with either fluorescent widefield microscope (Leica AF6000) or confocal microscope (Leica SP5) and analyzed with software for imaging (Fiji, CellProfiler v.2,2 and NIS-Elements AR v5.11).
Primary antibodies and concentrations were as follows
ARL13B (Rabbit, 1:500, Abcam, cat. n ab83879), ASCL1 (mouse, 1:500; Becton Dickinson, cat. n. 556604), BASSOON (Mouse, 1:500, Enzo Life Sciences, cat. n. ADI-VAM-PS003-F), BLBP (Rabbit, 1:500; Millipore, cat. n. ABN14), βIII-TUBULIN (rabbit, 1:1,000; Becton Dickinson, cat. n. 802001), CLEAVED CASPASE-3 (Rabbit, 1:400, Cell Signaling, cat. n. 9661), CTIP2 (rat, 1:500; Abcam, cat. n. ab18465), EBF1 (Mouse, 1:1000; Santa Cruz, cat. n. sc-137065), GABA (Rabbit, 1:500; Sigma, cat. n. A2052), GAD67 (Mouse, 1:1000, Millipore, cat. n. MAB5406), GFP (chicken, 1:1000, Abcam, cat. n. ab13970), GSX2 (rabbit, 1:200, GeneTex, cat. n. gtx129390), HOMER1 (Rabbit, 1:300, Genetex, cat. n. GTX103278), ISLT1/2 (mouse, 1:1,000; Hybridoma Bank, cat. n. 39.405), KI67 (rabbit, 1:500; Abcam, cat. n. ab16667), MAP2 (mouse, 1:500; Becton Dickinson, cat. n. 556320), N-CADHERIN (mouse, 1:800; Becton Dickinson, cat. n. 610921), NESTIN (mouse, 1:300; Millipore, cat. n. MAB5326), NKX2.1/TTF1 (rabbit, 1:200; Abcam, cat. n. ab76013), OCT3/4 (Mouse, 1:100, Santa Cruz, cat. n. sc-5279), PALS1 (rabbit, 1:500; Protein tech, cat. n. 17710-1-AP), PAX6 (rabbit, 1:300; Biolegend, cat. n. 901302), PERICENTRIN (Rabbit, 1:1000, Abcam, cat. n. ab4448), PH3 (Mouse, 1:1000, Cell Signaling, cat. n. #9706), RFP (Rabbit, 1:500, MBL, cat. n. PM005), SATB2 (Mouse, 1:500, Abcam, cat. n. ab51502), SOX2 (rabbit, 1:200; Millipore, AB5603), TBR1 (rabbit, 1:1,000; Abcam, cat. n. ab31940), TBR2 (rabbit, 1:100; Abcam, ab23345), VIMENTIN (mouse, 1:100; Becton Dickinson, cat. n. 550513), p-VIMENTIN (Mouse, 1:1000, MBL, D076-3), ZO-1 (Mouse, 1:100, Thermo Scientific, cat. n. 33-9100).
Quantitative assessment of IHC on organoids
The masks of concentric areas for VZ-like structures (Fig. 2, Supplementary Fig 4) were drawn with ImageJ in a semi-automatic approach through a manual drawing of the lumen ‘s perimeter. Then the automatic enlargement of the lumen’s mask draws three or four concentric areas with similar paths sharing the same central core and the same distance from the lumen. To quantify nuclear markers (GSX2, ASCL1, PAX6, TBR2, CTIP2, and TBR1) the cells were counted manually with Fiji ImageJ inside each area and the percentage quantification was based on positive cells over total nuclei marked by Hoechst staining.
Mitotic spindle (Supplementary Fig 3) was manually drawn and quantified with Fiji ImageJ software in VZ-like structures by p-VIMENTIN (to identify dividing cells) and PERICENTRIN (to visualize the plane of dividing cells). The orientation of the mitotic spindle was investigated in relation to the prospective ventricular surface in at least 40–50 VZ like structures per cell lines.
For quantification of cell cycle (Supplementary Fig 3) nuclear markers (KI67, PH3; EdU) the positive cells were counted with CellProfiler platform.
Quantification of primary cilia (Supplementary Fig 4), based on ARL13B staining, was done with NIS element. The confocal images taken at x63 with 4zoom factor were deconvolved with the Richardson-Lucy algorithm (10 iterations) to increase the resolution and then automatically quantified by the general analyses tool following a pipeline set on parameters such as background, dimensions and circularity.
Quantification of GABA area (Fig. 3L and Supplementary Fig 7), based on the area double positive for GABA and GFP or TOM staining, was performed with NIS-element. The confocal images acquired at x20 magnification were deconvolved with the Richardson-Lucy algorithm (10 iterations) to increase the resolution. Subsequently, automatic quantification was carried out by a general analysis tool employing a custom pipeline set on parameters such as background and dimension.
For the quantification of synapse density (Fig. 4H and Supplementary Fig 8), based on BASSOON/HOMER1/MAP2 staining, NIS-elements was also used. The confocal images taken at 63x with 1.7 zoom factor were deconvolved with the Richardson-Lucy algorithm (20 iterations) to increase the resolution. Synapses were automatically quantified using a custom pipeline developed in NIS-elements to recognize the number of BASSOON/HOMER1 co-localizing puncta. The same pipeline was used for all the conditions. The number of BASSOON+/HOMER1+ puncta was then normalized to the number of nuclei based on the Hoechst staining for each field of view. Additionally, the area of MAP2 detected in µm was normalized to the number of nuclei, while the number of HOMER1 puncta was normalized to the area of MAP2.
For Caspase-3 quantification (Supplementary Fig 9) the organoids were analyzed on DIV 120. Images were acquired by IN Cell Analyzer 6000 (GE Healthcare Life Sciences) instrument and processed with the software ImageJ (version 2.0.0). Automatic quantification was performed with CellProfiler software (version 4.2.1) as Caspase-3 positive signal area normalized on total Hoechst positive signal area.
Electron microscopy
TEM sample preparation
Organoids were fixed using 2.5% glutaraldehyde (Electron Microscopy Sciences, catalog 16220), and 2% paraformaldehyde (Electron Microscopy Sciences, catalog 19200) as fixatives, both in sodium cacodylate buffer 0.15 M (pH 7.4) (Electron Microscopy Sciences, catalog 12300). organoids were manually cut in small pieces (1 mm³) and post-fixed for an additional 24 h at 4 °C. Samples were then washed with 0.1 M cold sodium cacodylate buffer and then postfixed in a reduced osmium solution (i.e., 1.5% potassium ferrocyanide, Electron Microscopy Sciences, catalog 20150) with 2% osmium tetroxide (Electron Microscopy Sciences, catalog 19170) in 0.15 M cacodylate buffer for 1 h on ice. After the first heavy metal incubation, the samples were washed with ddH2O at room temperature and then placed in the 0.22 μm Millipore- filtered 1% thiocarbohydrazide (TCH) (Electron Microscopy Sciences, catalog 21900) in ddH2O solution for 20 min at room temperature. Organoids were then rinsed again in ddH2O and incubated in 2% osmium tetroxide in ddH2O for 30 min at room temperature. After several washings at room temperature in ddH2O, they were then placed in 1% uranyl acetate (aqueous) and left overnight at 4 °C. Samples were washed once again and then immersed en bloc in Walton’s lead aspartate solution (0.066 gr lead nitrate, Electron Microscopy Sciences, catalog 17900) dissolved in 10 ml of 0.003 M aspartic acid solution, pH 5.5) at 60 °C for 30 min. The organoids were washed and then dehydrated stepwise through an ethanol series and finally placed in anhydrous ice-cold acetone for 10 min. Infiltration was performed with an acetone (Sigma-Aldrich, catalog 179124)/Epon812 (Electron Microscopy Sciences, catalog 14120) mixture with 3:1 volume proportion for 2 h, then 1:1 overnight. The samples were left for 2 h in pure resin and then embedded in Epon812 resin and placed in a 60 °C oven for 48 h for polymerization. For TEM imaging, 70 nm thick ultrathin sections were prepared using an UltraCut E Ultramicrotome (Reichert) and placed on TEM copper grids. These grids were then observed using a Tecnai G2 Spirit transmission electron microscope (FEI, Eindhoven, the Netherlands). The microscope operated at an acceleration voltage of 120 kV and was equipped with a lanthanum hexaboride thermionic source, a twin objective lens, and a bottom-mount 11MP Gatan Orius SC1000 CCD camera (Gatan, Pleasanton, USA).
SEM sample preparation
The organoids were removed from the differentiation medium, washed with NaCacodylate 0.1 M and fixed with glutaraldehyde 2% in NaCacodylate 0.1 M for 24 h at 4 °C. After the fixation they were washed three times with NaCacodylate 0.1 M for 10 min, and post-fixed with osmium tetroxide (OsO4) 2% in NaCacodylate 0.1 M for one hour at room temperature. After removing the OsO4 solution and rinsing twice with bi-distilled water, the samples were gradually dehydrated by consecutive 10 min’ incubations 20%, 30%, 40%, 50%, 70%, 80% and 90% ethanol-H2O, and 100% ethanol, followed by 50% (v/v) ethanol–hexamethyldisilazane (HMDS) and 100% HMDS that was air-dried overnight at room temperature. All reagents were purchased from Electron Microscopy Sciences (Hatfield, PA, USA). Organoids were opened to reveal the internal structure with 0.5 mm tungsten needles using a Leica stereomicroscope (Wien, Austria) and mounted onto 12 mm specimen stubs. Before SEM imaging, the samples were coated with 20 nm-thick film of Pt-Pd using a Cressington 208HR sputter coater (Watford, UK) operated at a current of 40 mA. The scanning electron microscopy (SEM) imaging was performed by a Zeiss Merlin field emission gun (FEG)-SEM (Oberkochen, Germany), working at an acceleration voltage of 5 kV, a beam current of 200 pA and a working distance of 10 mm, and acquiring the secondary electron (SE) signal by an Everhart-Thornley (ET) in-chamber detector. 4.7.1 SEM sample preparation.
Single-cell RNA-seq on telencephalic organoids
Single-cell dissociation of telencephalic organoids for scRNAseq
The organoids were transferred in a dish to cut away the excess of Geltrex with a blade and then they were processed by dissociation using Papain Dissociation System with DNase (Worthington, LK003150) following the manufacturer’s recommendations. The incubation time was adjusted, based on size, at 20 min in the orbital shaker at 37 °C. Then the samples were centrifugated for 1 min at 200 g and resuspended in DMEM/F12 (21331, Thermo Fisher Scientific) and Neurobasal (21103049, Thermo Fisher Scientific) in ratio 1:1 with Penicillin (100 U/ml) and Streptomycin (100 μg/ml) (Euroclone, EC3001D), L-Glutamine (2mM, Thermo Fisher Scientific, 25030081), N2 supplement (1:100, Thermo Fisher Scientific, 17502-048), B27 complete supplement (1:50, Thermo Fisher Scientific, 17504-044) and Rock Inhibitor Y-27632 (10µM, CHDI Foundation). For the dissociation into single cell suspension, the samples were pipetted up and down first with 1000 µl pipettes and following with glass pipettes of increasingly smaller tip diameter (fire-polished). The suspension was filtered with disposable filter of 20 µm (Celltrics Sysmex, 04-004-2325). The cells were counted with Countess Automated Cell Counter to check viability (higher than 90%) and concentration. The suspension was centrifugated for 1 min at 200 g and resuspended at the concentration of 1 x 104 cells /µl in PBS containing 0.04% weight/volume BSA (400 µg/ml).
scRNA-seq library preparation and sequencing
Single cell sequencing libraries were prepared using the Chromium Single Cell 3′ Library & Gel Bead Kit v2 (PN- 120237) and v3 for the rest of the batches, Chromium Single Cell 3′ Chip kit v2 (PN-120236) and Chromium i7 Multiplex Kit (PN-120262) according to the manufacturer’s instructions. Libraries were generated and were sequenced by GATC Eurofins company with the Illumina NovaSeq platform (2 x 100 bp), using paired end sequencing with single indexing.
Single-cell RNA-seq analysis
For all samples, the sequencing data was pre-processed using a Nextflow pipeline which made use of the Cell Ranger Single-Cell Software Suite76 (v.2.1.1, 10x Genomics). The Cellranger count command was used to perform alignment, filtering, barcode counting and UMI counting. Cellranger uses STAR77 to map reads on the reference genome (version Hg38). It was used a custom gene annotation called Gencode HFB (introduced by ref. 39) consisting in a modified version of GENCODE release 25, integrated with both already known lincRNAs (obtained from Cabili and FANTOM dataset) and a list of 1116 de novo identified lincRNAs. The generated count matrix was then analyzed using Scanpy78 version ≥1.7.1. The quality control was performed removing from the dataset all genes expressed in less than three cells and keeping only cells with more than two hundred detected genes and with unique gene counts that is between five hundred and four thousand, in order to eliminate doublets and empty droplets. This step was followed by the removal of dead or damaged cells, keeping only cells with a proportion of mitochondrial genes lower than 0.1 for the DIV45 dataset and 0.24 for the DIV120 datasets. The remaining counts were subjected to normalization for library size and log-transformation. The datasets were restricted to only highly variable genes (HVGs), defined as those genes having an average normalized expression between 0.0125 and 3 and a dispersion greater than 0.5. Variation in gene expression given by the number of detected molecules and mitochondrial gene expression was regressed out to avoid any possible bias in the following analysis. Dimensionality reduction was achieved using principal component analysis (PCA), the number of principal components retained was selected following the Elbow method, keeping only the ones incorporating the majority of variance, which corresponded to the first eight components for DIV45 and fifteen for both DIV120 dataset. A K-nearest neighbors (KNN) graph was constructed linking each cell to the K most similar cells, selecting a K value of two hundred for DIV45 and ten for the DIV120 datasets. Successively, the cells have been visualized in a two-dimensional space using the UMAP algorithm. The UMAP of the DIV120 mono-cultures and of DIV120 including also the co-cultures naturally exhibited visible similarities, but the similar structures appear as mirrored images. In order to facilitate comparisons, we have reversed the coordinates of UMAP1 for the DIV120 mono-culture plot, while the UMAP2 remained unaltered, allowing for clearer visual continuity between the figures. In the next step of the analysis, cells have been grouped into communities using the Louvain algorithm for DIV45 and the Leiden algorithm for DIV120 datasets. The resolution parameter was set to 0.3 for DIV45 and DIV120-mono and 0.2 for DIV120-co. Lastly, cell communities have been detected performing a Wilcoxon rank sum test to identify genes specific of each subpopulation and plotting the expression of canonical ventral and dorsal marker genes. Differential gene expression analysis between CTRL and HD at DIV45 and DIV120 was performed applying Wilcoxon rank sum test and all genes exhibiting a p.value lower that 0.05 were represented in a Volcano plot to show the log fold change and adjusted p-value of DEGs common at both timepoints or timepoint-specific. Among all differentially expressed genes, some genes of particular interest were selected and their gene expression value in each subpopulation and/or culture was represented using a heatmap. We compared our DEGs with publicly accessible human HD data by identifying the overlap between our DEGs lists and those included in the supplementary data of Lee and colleagues7. The results of this comparison were visualized as a Venn diagram.
Gene Ontology analysis
Gene Ontology terms related to genes dysregulated at both time points have been identified using the R package topGO79 v. 2.44.0, terms with p-value < 0.05 were then grouped together according to semantic similarity using the R package rrvigo80 1.4.4. For all the other go analysis, the top thirty significative terms among those with p-value < 0.05 were visualized in a lollipop plot showing the number of genes in each term and the fraction of genes associated with the GO term. For all the analysis, the gene set of interest was selected considering the DEGs with a p-value < 0.05, the background gene list consisted in the final number of genes of the processed Anndata object (HVGs) and the GO terms were retrieved from the BioMart database system using biomaRt81 v.2.48.3. After selecting relevant go terms, their score was computed using the Scanpy function scanpy.tl.score_genes, calculating the mean expression of the genes inside those GO term and subtracting the mean expression of random genes in the dataset (the random seed was set to 0). The differences in score between the genotypes have been assessed using the Wilcoxon test with Bonferroni p-value correction.
Trajectory analysis
RNA velocity analysis was performed with the DIV120 co-culture dataset using Velocyto54 v.0.17.17, counting spliced and unspliced reads on top of the Cellranger output to generate the loom files. The run velocyto command was incorporated to the nextflow pipeline with default parameters and allowing the masking of confounding factors given by repetitive elements. The post-processing was completed importing the loom file into the Jupyter notebook using Loompy 3.0.6 and then using the scvelo82 python package (v.0.2.3), selecting the generalized stochastic model. After computing the RNA velocities, they were embedded into a UMAP space. The length of the velocity vector, representing the speed of differentiation and the coherence of the vector field, representing how a velocity vector correlates with its neighboring velocities, were computed using the built-in function scv.tl.velocity_confidence. Differences among populations and genotypes in this two metrics were assessed using the Wilcoxon Test and then adjusting the p-value with Bonferroni correction.
Trajectory was further investigated with a pseudotime analysis, the dataset was converted into a Seurat object using SeuratDisk v0.0.0.9015, and the analysis was performed separately for each culture, using the Monocle355,83 v.0.1.0 R package with default parameters. The root or starting point for the trajectory was chosen manually by locating an endpoint of the tree within one cell among the progenitors in similar positions for each condition. We considered the starting point of the trajectory to be the most immature cell type.
Similarity analysis
We first validated our results through similarity with external in-vivo dataset using the R package VoxHunt28 v1.0.1 that allows the comparison with RNA-seq data of microdissected human brain regions belonging to stages between 10 and 24 pcw from BrainSpan Atlas of the Developing Human Brain (https://www.brainspan.org). The similarity was computed on top of the expression of the default set of marker genes, the regions selected for comparison were the neocortex (NCx), ganglionic eminences (GE) and striatum (STR).
Then, we further compared our results with fetal samples using 9pcw bulk RNA-seq data from ref. 39. Gene expression from each culture and cell type was compared to in vivo LGE and cortex data. Firstly, a list of top one hundred differentially expressed genes in LGE and in cortex was selected and then the similarity was assessed through a signature score84 that consists in the product of two values, the fraction of genes of the list expressed for each cell and the expression level of each gene, each score is then scaled between 0 and 1 by dividing by the raw score by the maximum value. In this way, the score increases both when the number of genes of interest is higher and when the expression of these genes increases. For the other comparison with human fetal data from Braun et al. 29, we used the built-in model of CellTypist59 v1.6.2., which was trained on the Braun et al. 29 dataset, to compare the cell types of different tissues in this atlas with our cell types at DIV 45 and 120. Predicted labels from CellTypist were calculated using the ‘best-match’ mode.
To compare the cell types in our study across different time points with those in established organoid protocols from the literature, we utilized transcriptomic data from the Human Neural Organoid Cell Atlas (HNOCA)30. We accessed the publicly available section of HNOCA, which encompasses all integrated protocols except for three that remain unpublished. We employed the CellHint tool85 v0.1.1 to harmonize cell types across various organoid protocols and to compute a cross-dataset distance matrix. This matrix included 1665880 cells and 161 cell types from 26 different protocols, as indicated in the ‘assay_differentiation’ section of the HNOCA metadata. During the harmonization process with CellHint, each cell was assigned to the most similar cell type from each dataset, resulting in a comprehensive assignment matrix. For ease of analysis, we focused on specific comparisons: excluding all cell types labeled as ‘unknown’ and retaining only those comparisons that involved our datasets and had a score greater than 0.5 for at least one cell type. Finally, we visualized the inferred similarities among cell populations using a heatmap, which displayed unsupervised hierarchical clustering.
For the comparison between different cultures and different cell populations, the Jaccard similarity index was used. The Wilcoxon rank-sum test was applied to generate a list of the top five hundred differentially expressed genes in each culture with respect to the others, then the Jaccard similarity index was used to assess similarity between the list of the top DEGs for each culture according to the number of genes in common or not. This value was expected to be small as we were looking for similarities among the set of most different genes. Then, considering each culture separately, we computed the top five hundred genes differentially expressed in each cell population with respect to the others using the Wilcoxon rank-sum test. The Jaccard similarity score was used again in order to assess similarity among the various populations of each culture.
Cell communication analysis
The cell-cell communication analysis between control and HD cells at DIV120 was performed using the R package CellChat56 v.1.1.1. The analysis was completed using the human ligand-receptor interaction database to identify overexpressed ligands and receptor in each cell type. This tool has been used to identify biologically significant cell-cell communications, associating ligands-receptors pairs with a signaling pathway to compute a cell-cell communication network. The communication networks were first visualized separately per each culture using a chord diagram, then all the significant interactions (L-R pairs) from one group to another were shown using netVisual_bubble function. Interaction between groups not actually in contract during the experiment (e.g., from one group belonging to a mono-culture to one of the co-culture) were manually excluded from the analysis. Furthermore, the package NeuronChat57 v.1.0.0 was employed to infer and visualized neural-specific L-R interactions, performing a comparative analysis to confront the weights of the communication for individual interaction pairs in each cultures. Finally, inferred communication networks of relevant signaling pathways identified with CellChat and NeuronChat have been considered by visualizing them using a circle plot using the built-in function netVisual_aggregate and by plotting the gene expression levels of ligand and receptor belonging to those pathways.
GSEA
The Gene Set Enrichment Analysis was performed using GSEApy v1.1.1 on the ventral neuron subset of the dataset, utilizing the pre-rank module. The ranked gene list was obtained as the output of differential expression analysis in pseudo-bulk, using the package Decoupler v1.6.0.
TF activity inference
Transcription factor activity inference was computed using Decoupler’s Univariate Linear Model method, with CollecTRI as the reference network.
Statistics and reproducibility
GraphPad Software was used for statistical analysis of immunohistochemistry, scanning electron microscopy, and q-PCR. In all analysis comparison of mean values was conducted with unpaired t-test or ANOVA One Way. The tests used are described in the figure legends and the threshold of statistical significance was set at ≤ 0.05 (****p < 0.0001, ***p < 0.001; **p < 0.01; *p < 0.05; in the graphs multiple lines with only one symbol mean they have the same significance). Statistics and graph were done with PRISM software v. 6. Data derived from different technical replicates (individual organoids) of independent biological replicates (differentiation experiments) are specified in each legend and are represented with mean and SEM.
For single-cell RNA sequencing we sequenced 6 individual organoid (3 per genotype) at DIV 45 and 8 samples (4 pools of 10 organoids that are FACS sorted, with 2 cell lines per genotype) at DIV 120.
Immunohistochemistry and brightfield images (such as Figs. 1B, C, 2B and D, 2J–K, and 4A) derived from independent experiments repeated three times with similar results, collecting at least 12 individual organoids per time point.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank A. Fasciani of the Imaging Facility and M. Crosti of the Cytofluorometry Facility from INGM for scientific and technical assistance. We acknowledge Ernest Arenas (Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden) for discussions and comments. This study was supported by the European Union’s funded Consortia “Nsc-Reconstruct: Novel Strategies for Cell based Neural Reconstruction” (Horizon 2020, GA No. 874758) and by JPND (EU) ModelPolyQ (Grant No. 643417) and, partially, by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 742436). Single cell RNA sequencing was partially supported by CHDI Foundation (No. JSC A11103). All the grants were referred to E.C.
Author contributions
M.G. and E.C. conceived the study; M.G. performed the organoids experiments, the immunohistochemistry analyses, the images quantifications, and the qPCR data with the contributions from M.Cr.; M.R.N. performed computational analysis with the support of V.D.B.; P.C. performed the single-cell experiments with contributions from M.G.; E.V. and A.Fal. performed the electron microscopy analysis; M.Ce. C.M., R.I., A.S., and C.Z. performed additional image analyses and helped for revisions; C.C. supported the work through the INGM imaging facility; I.E.C. generated the GFP and TOM cell lines; A.Fae. contributed to the initial organoids protocol; V.R. supported the work through the INGM bioinformatic platforms; M.G., V.D.B., P.C., C.Z., D.B. and E.C. contributed to assays identification, data interpretation and presentation. D.F and T.V. gave further experimental suggestions on the mosaics approach; M.G. assembled the figures and wrote the first draft of the paper which was then revised by E.C., D.B., M.R.N., V.D.B., P.C. and C.Z., edited and proofread by all authors. E.C. coordinated the study, secured the funding and established the collaborations.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
All scRNA-seq data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE272271, and also in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress/) under accession no E-MTAB-12924. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/21/2025
A Correction to this paper has been published: 10.1038/s41467-025-60108-6
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-50877-x.
References
- 1.Nopoulos, P. C. et al. Smaller intracranial volume in prodromal Huntington’s disease: evidence for abnormal neurodevelopment. Brain134, 137–142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee, J. K. et al. Measures of growth in children at risk for Huntington disease. Neurology79, 668–674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molero, A. E. et al. Selective expression of mutant huntingtin during development recapitulates characteristic features of Huntington’s disease. Proc. Natl. Acad. Sci.113, 5736–5741 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnat, M. et al. Huntington’s Disease Alters Human Neurodevelopment. Science369, 787–793 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hickman, R. A. et al. Developmental malformations in Huntington disease: neuropathologic evidence of focal neuronal migration defects in a subset of adult brains. Acta Neuropathol.141, 399–413 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desplats, P. A. et al. Selective deficits in the expression of striatal-enriched mRNAs in Huntington’s disease. J. Neurochem96, 743–757 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Lee, H. et al. Cell Type-Specific Transcriptomics Reveals that Mutant Huntingtin Leads to Mitochondrial RNA Release and Neuronal Innate Immune Activation. Neuron107, 891–908 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu, X. et al. Uninterrupted CAG repeat drives striatum-selective transcriptionopathy and nuclear pathogenesis in human Huntingtin BAC mice. Neuron110, 1173–1192 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goula, A. V. et al. Transcription Elongation and Tissue-Specific Somatic CAG Instability. PLoS Genet8, e1003051 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menalled, L. B. et al. Early motor dysfunction and striosomal distribution of huntingtin microaggregates in Huntington’s disease knock-in mice. J. Neurosci.22, 8266–8276 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu, X. et al. Pathological cell-cell interactions are necessary for striatal pathogenesis in a conditional mouse model of Huntington’s disease. Mol. Neurodegener.2, 1–11 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veldman, M. B. & Yang, X. W. Molecular insights into cortico-striatal miscommunications in Huntington’s disease. Curr. Opin. Neurobiol.48, 79–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plotkin, J. L. & Surmeier, D. J. Corticostriatal synaptic adaptationns in Huntington’s disease. Curr. Opin. Neurobiol.33, 53–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuccato, C. & Cattaneo, E. Huntington’s disease. Handb. Exp. Pharm.220, 357–409 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Virlogeux, A. et al. Reconstituting Corticostriatal Network on-a-Chip Reveals the Contribution of the Presynaptic Compartment to Huntington’s Disease. Cell Rep.22, 110–122 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Sepers, M. D. & Raymond, L. A. Mechanisms of synaptic dysfunction and excitotoxicity in Huntington’s disease. Drug Discov. Today19, 990–996 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Cepeda, C., Murphy, K. P. S., Parent, M. & Levine, M. S. The role of dopamine in Huntington’s disease. in. Prog. Brain Res.211, 235–254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown, T. B., Bogush, A. I. & Ehrlich, M. E. Neocortical expression of mutant huntingtin is not required for alterations in striatal gene expression or motor dysfunction in a transgenic mouse. Hum. Mol. Genet17, 3095–3104 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas, E. A. et al. In vivo cell-autonomous transcriptional abnormalities revealed in mice expressing mutant huntingtin in striatal but not cortical neurons. Hum. Mol. Genet20, 1049–1060 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu, X. et al. Pathological cell-cell interactions elicited by a neuropathogenic form of mutant huntingtin contribute to cortical pathogenesis in HD mice. Neuron46, 433–444 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Bradford, J. et al. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. Proc. Natl. Acad. Sci. USA106, 22480–22485 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benraiss, A. et al. Human glia can both induce and rescue aspects of disease phenotype in Huntington disease. Nat. Commun.7, 1–13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaspard, N. et al. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature455, 351–357 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Muñoz-Sanjuán, I. & Brivanlou, A. H. Neural induction, the default model and embryonic stem cells. Nat. Rev. Neurosci.3, 271–280 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Ruzo, A. et al. Chromosomal instability during neurogenesis in Huntington’s disease. Development145, dev156844 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Pollen, A. A. et al. Molecular identity of human outer radial glia during cortical development. Cell163, 55–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, Y. et al. Interneuron origin and molecular diversity in the human fetal brain. Nat. Neurosci.24, 1745–1756 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Fleck, J. S. et al. Inferring and perturbing cell fate regulomes in human brain organoids. Nature621, 365–372 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Braun, E. et al. Comprehensive cell atlas of the first-trimester developing human brain. Science382, eadf1226 (2023). [DOI] [PubMed] [Google Scholar]
- 30.He, Z. et al. An integrated transcriptomic cell atlas of human neural organoids. Biorxiv, 1–23 (2023). [DOI] [PMC free article] [PubMed]
- 31.Marangoni, M. et al. Age-related axonal swellings precede other neuropathological hallmarks in a knock-in mouse model of Huntington’s disease. Neurobiol. Aging35, 2382–2393 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Molina-Calavita, M. et al. Mutant Huntingtin Affects Cortical Progenitor Cell Division and Development of the Mouse Neocortex. J. Neurosci.34, 10034–10040 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Lullo, E. & Kriegstein, A. R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci.18, 573–584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, X. et al. Human striatal organoids derived from pluripotent stem cells recapitulate striatal development and compartments. PLoS Biol.20, e3001868 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bisgrove, B. W. & Yost, H. J. The roles of cilia in developmental disorders and disease. Development133, 4131–43 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Higginbotham, H. et al. Arl13b-regulated cilia activities are essential for polarized radial glial scaffold formation. Nat. Neurosci.16, 1000–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keryer, G. et al. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J. Clin. Invest121, 4372–4382 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onorati, M. et al. Molecular and functional definition of the developing human striatum. Nat. Neurosci.17, 1804–1815 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Bocchi, V. D. et al. The coding and long noncoding single-cell atlas of the developing human fetal striatum. Science372, eabf5759 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Shi, Y. et al. Mouse and human share conserved transcriptional programs for interneuron development. Science374, eabj6641 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, Y. et al. Modeling human telencephalic development and autism-associated SHANK3 deficiency using organoids generated from single neural rosettes. Nat. Commun.13, 5688 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian, X. et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell165, 1238–1254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinto, R. M. et al. Patterns of CAG repeat instability in the central nervous system and periphery in Huntington’s disease and in spinocerebellar ataxia type 1. Hum. Mol. Genet29, 2551–2567 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Besusso, D. et al. Stem Cell-Derived Human Striatal Progenitors Innervate Striatal Targets and Alleviate Sensorimotor Deficit in a Rat Model of Huntington Disease. Stem Cell Rep.14, 876–891 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosser, A. E. et al. Translating cell therapies for neurodegenerative diseases: Huntington’s disease as a model disorder. Brain145, 1584–1597 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wild, E. J. & Tabrizi, S. J. Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol.16, 837–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garret, M., Du, Z., Chazalon, M., Cho, Y. H. & Baufreton, J. Alteration of GABAergic neurotransmission in Huntington’s disease. CNS Neurosci. Therapeutics24, 292–300 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verstraelen, P. et al. Systematic Quantification of Synapses in Primary Neuronal Culture. iScience23, 101542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barry, J., Bui, M. T. N., Levine, M. S. & Cepeda, C. Synaptic pathology in Huntington’s disease: Beyond the corticostriatal pathway. Neurobiol. Dis.162, 105574 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng, Y. P., Wong, T., Bricker-Anthony, C., Deng, B. & Reiner, A. Loss of corticostriatal and thalamostriatal synaptic terminals precedes striatal projection neuron pathology in heterozygous Q140 Huntington’s disease mice. Neurobiol. Dis.60, 89–107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murmu, R. P., Li, W., Szepesi, Z. & Li, J. Y. Altered sensory experience exacerbates stable dendritic spine and synapse loss in a mouse model of huntington’s disease. J. Neurosci.35, 287–298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conforti, P. et al. RUES2 hESCs exhibit MGE-biased neuronal differentiation and muHTT-dependent defective specification hinting at SP1. Neurobiol. Dis.146, 105140 (2020). [DOI] [PubMed] [Google Scholar]
- 54.La Manno, G. et al. RNA velocity of single cells. Nature560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol.32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun.12, 1–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conforti, P. et al. In vitro-derived medium spiny neurons recapitulate human striatal development and complexity at single-cell resolution. Cell Rep. Methods2, 100367 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vezzoli, E. et al. Inhibiting pathologically active ADAM10 rescues synaptic and cognitive decline in Huntington’s disease. J. Clin. Investig.129, 2390–2403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Domínguez Conde, C. et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science376, eabl5197 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mätlik, K. et al. Cell-type-specific CAG repeat expansions and toxicity of mutant Huntingtin in human striatum and cerebellum. Nat. Genet56, 383–394 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kacher, R. et al. CYP46A1 gene therapy deciphers the role of brain cholesterol metabolism in Huntington’s disease. Brain142, 2432–2450 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Wilton, D. K. et al. Microglia and complement mediate early corticostriatal synapse loss and cognitive dysfunction in Huntington’s disease. Nat. Med. 29, 2866–2884 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.iPSC, H. D. Consortium. Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat. Neurosci.20, 648–660 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haremaki, T. et al. Self-organizing neuruloids model developmental aspects of Huntington’s disease in the ectodermal compartment. Nat. Biotechnol.37, 1198–1208 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Chen, S. et al. Altered synaptic vesicle release and ca2+ influx at single presynaptic terminals of cortical neurons in a knock-in mouse model of huntington’s disease. Front Mol. Neurosci.11, 478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arrasate, M. & Finkbeiner, S. Protein aggregates in Huntington’s disease. Exp. Neurol.238, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hickman, R. A., Faust, P. L., Marder, K., Yamamoto, A. & Vonsattel, J. P. The distribution and density of Huntingtin inclusions across the Huntington disease neocortex: regional correlations with Huntingtin repeat expansion independent of pathologic grade. Acta Neuropathol. Commun.10, 55 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gutekunst, C.-A. et al. Nuclear and Neuropil Aggregates in Huntington’s Disease: Relationship to Neuropathology. J. Neurosci.19, 2522–2534 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Di Figlia et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science277, 1990–1993 (1997). [DOI] [PubMed] [Google Scholar]
- 70.Slow, E. J. et al. Absence of Behavioral Abnormalities and Neurodegeneration in Vivo despite Widespread Neuronal Huntingtin Inclusions. Proc. Natl. Acad. Sci. USA102, 11402–11407 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reidling, J. C. et al. Human Neural Stem Cell Transplantation Rescues Functional Deficits in R6/2 and Q140 Huntington’s Disease Mice. Stem Cell Rep.10, 58–72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paquola, A. C. M., Erwin, J. A. & Gage, F. H. Insights into the role of somatic mosaicism in the brain. Curr. Opin. Syst. Biol.1, 90–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pipicelli, F. et al. Non-cell-autonomous regulation of interneuron specification mediated by extracellular vesicles. Sci. Adv.9, eadd8164 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lancaster, M. A. & Knoblich, J. A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science (1979)345, 1247125–1247125 (2014). [DOI] [PubMed] [Google Scholar]
- 75.Espuny-Camacho, I. et al. Pyramidal Neurons Derived from Human Pluripotent Stem Cells Integrate Efficiently into Mouse Brain Circuits In Vivo. Neuron77, 440–456 (2013). [DOI] [PubMed] [Google Scholar]
- 76.Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun.8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol.20, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alexa, A. & Rahnenführer, J. Gene set enrichment analysis with topGO. Bioconductor Improvments27, 1–26 (2007).
- 80.Sayols, S. rrvgo: a Bioconductor package to reduce and visualize Gene Ontology terms. MicroPubl Biol.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10155054/ (2023). [DOI] [PMC free article] [PubMed]
- 81.Smedley, D. et al. BioMart - Biological queries made easy. BMC Genomics10, 1–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol.38, 1408–1414 (2020). [DOI] [PubMed] [Google Scholar]
- 83.Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Della Chiara, G. et al. Epigenomic landscape of human colorectal cancer unveils an aberrant core of pan-cancer enhancers orchestrated by YAP/TAZ. Nat. Commun.12, 2340 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu, C. et al. Automatic cell-type harmonization and integration across Human Cell Atlas datasets. Cell186, 5876–5891 (2023). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scRNA-seq data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE272271, and also in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress/) under accession no E-MTAB-12924. Source data are provided with this paper.