Abstract
Walleye epidermal hyperplasia virus types 1 and 2 (WEHV1 and WEHV2, respectively) are associated with a hyperproliferative skin lesion on walleyes that appears and regresses seasonally. We have determined the complete nucleotide sequences and transcriptional profiles of these viruses. WEHV1 and WEHV2 are large, complex retroviruses of 12,999 and 13,125 kb in length, respectively, that are closely related to one another and to walleye dermal sarcoma virus (WDSV). These walleye retroviruses contain three open reading frames, orfA, orfB, and orfC, in addition to gag, pol, and env. orfA and orfB are adjacent to one another and located downstream of env. The OrfA proteins were previously identified as cyclin D homologs that may contribute to the induction of cell proliferation leading to epidermal hyperplasia and dermal sarcoma. The sequence analysis of WEHV1 and WEHV2 revealed that the OrfB proteins are distantly related to the OrfA proteins, suggesting that orfB arose by gene duplication. Presuming that the precursor of orfA and orfB was derived from a cellular cyclin, these genes are the first accessory genes of complex retroviruses that can be traced to a cellular origin. WEHV1, WEHV2, and WDSV are the only retroviruses that have an open reading frame, orfC, of considerable size (ca. 130 amino acids) in the leader region preceding gag. While we were unable to predict a function for the OrfC proteins, they are more conserved than OrfA and OrfB, suggesting that they may be biologically important to the viruses. The transcriptional profiles of WEHV1 and WEHV2 were also similar to that of WDSV; Northern blot analyses detected only low levels of the orfA transcripts in developing lesions, whereas abundant levels of genomic, env, orfA, and orfB transcripts were detected in regressing lesions. The splice donors and acceptors of individual transcripts were identified by reverse transcriptase PCR. The similarities of WEHV1, WEHV2, and WDSV suggest that these viruses use similar strategies of viral replication and induce cell proliferation by a similar mechanism.
The family Retroviridae consists of seven genera of avian and mammalian viruses that are grouped by common morphology, genomic structure, and host range (14). Two piscine retroviruses that are not congruous with the classification of these genera have been cloned and analyzed: walleye dermal sarcoma virus (WDSV) and snakehead fish retrovirus (SnRV) (20, 22), both of which are complex viruses that have type C morphology (18, 37). WDSV is etiologically associated with a nodular skin lesion, walleye dermal sarcoma (WDS) (36, 52), whereas the pathogenicity of SnRV is unknown (20). Based on pol sequences and genomic organization, WDSV and SnRV are only distantly related, making it difficult to judge if either is representative of fish retroviruses in general.
Recently, we identified two novel retroviruses associated with walleye epidermal hyperplasia (WEH) (52, 57), WEH virus types 1 and 2 (WEHV1 and WEHV2, respectively) (31). WEHV1 and WEHV2 are type C retroviruses that are closely related to one another (76% amino acid [aa] identity in pol) and to WDSV (ca. 65% aa identity in pol) (22, 31). Interestingly, like WDS, epidermal hyperplasia appears and regresses on a seasonal basis. Lesions develop in the fall, regress in the spring, and are absent in the summer (7, 8). Evidence suggesting that WEHV1 and WEHV2 are the causative agents of WEH includes the following: (i) WEHV1 and WEHV2 pol sequences were identified and cloned by reverse transcriptase PCR (RT-PCR) from virion preparations derived from hyperplastic tissue (31); (ii) WEHV1 and WEHV2 genomic DNA is found in diseased tissue, but not in surrounding normal tissue (31); and (iii) WEH has been experimentally transmitted to walleye fingerlings with cell-free inocula from hyperplasias, and WEHV1 and/or WEHV2 viral DNAs were detected in the lesions by PCR (6). While the two are commonly found together in lesions, WEHV2 has been detected in the absence of WEHV1, suggesting that WEHV2 alone can cause disease (6, 31). However, the relative capacities of WEHV1 and WEHV2 to cause disease have not been investigated.
Members of our group recently found that WEHV1, WEHV2, and WDSV encode cyclin D homologs (30). Since cyclin D1 is an oncogene that has been implicated in many types of human tumors, the retroviral cyclins (rv-cyclins) may induce the cell proliferation that ultimately leads to WEH and WDS (1, 30). As part of our efforts to develop this unique model of tumor induction and tumor regression, we have completed the DNA sequences of WEHV1 and WEHV2 and have identified their transcripts in developing and regressing tumors. The comparisons of the DNA sequences and transcriptional patterns of WEHV1, WEHV2, and WDSV emphasize the relatedness of these viruses and suggest that they use similar replication strategies that likely play a role in pathogenesis. In addition, the sequence analyses suggest that two of their accessory genes, orfA (rv-cyclin) and orfB, arose by gene duplication. This represents the first example where the origin of complex retrovirus accessory genes can be traced to a cellular gene.
MATERIALS AND METHODS
Characterization of WEHV1 and WEHV2 genomic clones.
All probes were labeled with 32P by using a random priming kit (Boehringer). λ DNAs were prepared, digested with restriction enzymes, electrophoresed on 1% agarose gels, and blotted onto nitrocellulose by standard procedures (46). All blots were hybridized with probes in 50% formamide buffer at 37°C for 24 h and washed in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 55°C. Fragments for subcloning were gel purified using a Qiaex II gel extraction kit (Qiagen) and were cloned into Bluescript II SK(−) (Stratagene).
WEHV1 and WEHV2 partial genomic clones were identified from a λ library derived from a pool of WEHs by using pol probes as previously described (31). The WEHV1 and WEHV2 long terminal repeat (LTR) probes were generated by PCR with primers that amplified positions −203 to +116 and positions −210 to +120, respectively. The WEHV1 and WEHV2 orfC probes, encompassing the entire gene from positions 494 to 895 and 452 to 853, respectively, were also generated by PCR.
Eight λ plaques hybridized with the WEHV1 pol probe and six hybridized with the WEHV2 pol probe. Each plaque was purified and used to isolate λ DNA (46). Since the WEHV pol regions are closely related to WDSV, we reasoned that the WDSV 1.05 probe (36), which contains WDSV LTR sequences, the primer binding site (PBS), and orfC, might cross-hybridize with WEHV sequences. The WEHV1 and WEHV2 λ DNAs were digested and hybridized with the WDSV 1.05 probe or the appropriate WEHV pol probe. As expected, DNA isolated from all λ clones hybridized with their cognate pol probes. In addition, several of the clones hybridized with the WDSV 1.05 probe, whereas others did not. Since DNA cloned into λ must be approximately 20 kb in size, each pol-containing viral clone should contain at least one LTR. The simplest explanation for the lack of 1.05 hybridization with some clones is that the hybridizing sequences were not in the LTR but in adjacent sequences, i.e., the PBS, leader sequences, or orfC. An EcoRI fragment from a WEHV1 λ clone and a BamHI fragment from a WEHV2 λ clone that hybridized with both 1.05 and their specific pol probes were gel purified and cloned. Since the PBS is often conserved among related retroviruses, we used primers complementary to the PBS to sequence the 5′ LTR and downstream sequences in these subclones. The PBS and surrounding sequences were identical in WEHV1, WEHV2, and WDSV (see Fig. 2), whereas LTR and orfC sequences were divergent, suggesting that the WDSV 1.05 probe hybridized with the PBS region. To identify full-length WEHV1 and WEHV2 genomic clones, λ DNAs were digested with various enzymes and hybridized with WEHV1 and WEHV2 LTR-specific probes. In every case, only one restriction fragment hybridized with the probes, indicating that only one LTR was present in each clone, and therefore no full-length viral clones were isolated.
FIG. 2.
WEHV1 (A) and WEHV2 (B) LTRs. The predicted boundaries of U3, R, and U5, along with the start of transcription (+1), are indicated. Potential transcription factor binding sites are shown. A 7-bp repeat sequence, which is identical to the core sequence of the growth hormone factor 1 binding site, is shown. pA, polyadenylation signal sequence; PEA3, polyoma enhancer activator; GRE, glucocorticoid response element. The lowercase letters represent nucleotides in the PBS region that are conserved in WEHV1, WEHV2, and WDSV.
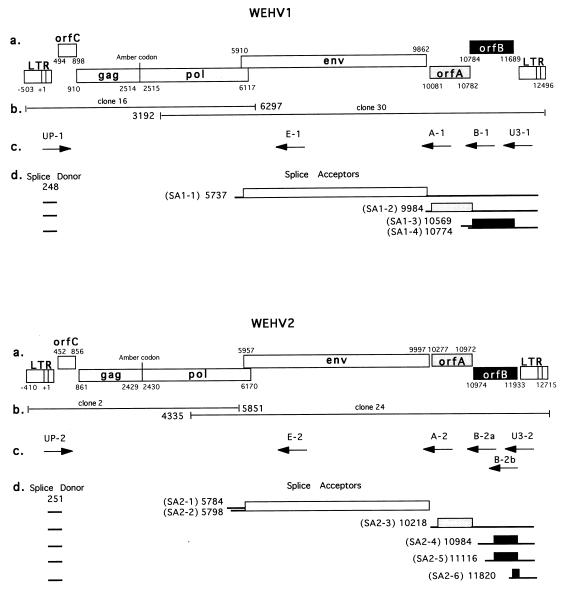
To identify WEHV1 and WEHV2 partial genomic clones that contained the 5′ or 3′ LTR, we hybridized λ DNAs with specific LTR, pol, and orfC probes. Clones that hybridized with LTR, pol, and orfC probes were assumed to contain a 5′ segment of the virus, while those that did not hybridize with orfC were assumed to contain the 3′ segment of the virus. A SmaI fragment from a WEHV1 λ clone and a XbaI-SalI fragment from a WEHV2 λ clone containing the 3′ regions were subcloned. In summary, subclones 16 and 30 represent the 5′ LTR-to-pol and pol-to-3′ LTR segments of WEHV1, respectively, and subclones 2 and 24 represent the 5′ LTR-to-pol and pol-to-3′ LTR segments of WEHV2, respectively (Fig. 1, panels b).
FIG. 1.
WEHV1 and WEHV2. For each, panels a to d are as follows. (a) Genomic organization. The numbers mark the boundaries of the open reading frames and of the LTRs relative to the start of transcription (+1). (b) Schematic of the subclones used for DNA sequence analysis. The numbers mark the ends of the overlapping subclones. (c) Primers used for RT-PCR. (d) Transcriptional profiles. The positions of the major splice donor and SAs relative to the start of transcription are indicated.
DNA sequencing.
Plasmids for sequencing were purified with a Qiagen Maxi Prep kit. Both DNA strands of the viral clones were sequenced by walking using automated fluorescent sequencing with rhodamine dye terminator chemistry at the Cornell University Bioresource Center DNA Sequencing Facility. cDNA clones from transcriptional mapping were sequenced with the Sequenase sequencing kit, version 2.0 (U.S. Biochemicals).
Database searches and amino acid alignments.
Database searches were done by the BlastX algorithm (National Center for Biotechnology Information). Amino acid alignments were done with Megalign (DNAstar) and Geneworks (IntelliGenetics, Inc.). The amino acid substitutions allowed for determination of sequence similarities were as follows: F = Y = W, M = L = V = I, S = A, S = T, D = E, N = Q, and R = K = H.
Northern blotting.
Total RNA was isolated from WEH samples collected from individual fish in the spring (0.2 g) with RNAzol B (Tel-test, Inc.). Fall hyperplasias were difficult to obtain, and therefore lesions from 2 to 3 fish were pooled to produce enough material (1 g of tissue) for each of two independent poly(A)+ RNA isolations. poly(A)+ RNA was enriched from fall lesions with the PolyATract IV mRNA Isolation System (Promega). Spring and fall lesions were not derived from the same fish. Ten micrograms of total RNA or 1 μg of poly(A)+ RNA was electrophoresed in formaldehyde gels and blotted onto nitrocellulose (46). The blots were hybridized by using WEHV1 or WEHV2 virus-specific LTR probes to detect all viral transcripts. WEHV1 or WEHV2 LTR probes were made and hybridized for 48 h as described above. Blots were washed in 0.2× SSC at 55°C and exposed to film for 24 h (spring lesions) or 1 week (fall lesions).
Transcriptional mapping.
Transcriptional mapping was done by RT-PCR; the oligonucleotides used are listed in Table 1 and shown schematically in Fig. 1, panels c. Total RNA was isolated from three or more spring WEH lesions derived from individual fish for the analysis of WEHV1 and WEHV2 transcripts. cDNA synthesis (50-μl reaction mixture) was done with 1 μg of total RNA, 10 pmol of antisense primer (U3-1 RT or U3-2 RT), and 4 U of murine leukemia virus (MLV) reverse transcriptase (New England Biolabs). The cDNA (2 μl) was amplified by PCR (50-μl reaction mixture) with Taq polymerase (Gibco-BRL). All samples were denatured at 96°C for 5 min, and 2.5 U of Taq polymerase was added to the reaction mixture. The amplification program consisted of 30 s at 94°C, 30 s at 50°C, and 2 min at 72°C for 35 cycles and 10 min at 72°C for 1 cycle. The PCR products were extracted with phenol-chloroform and precipitated with ethanol. WEHV1 products digested with XhoI and NotI and WEHV2 products digested with XhoI and SacI were cloned into Bluescript II SK(−). Some products were gel purified prior to cloning.
TABLE 1.
Oligonucleotides used for WEHV1 and WEHV2 transcriptional mapping
| Virus and designation | Sequencea | Positionb |
|---|---|---|
| WEHV1 | ||
| UP-1 | CTACTCGAGTTCCATCATCGGATCCTGCT | 179 |
| E-1 | ATTAGCGGCCGCGTAACTGTTCCATCTGAGTGCTT | 6517 |
| A-1 | ATTAGCGGCCGCGATCCATGTAGCTCAGGAACT | 10595 |
| B-1 | ATTAGCGGCCGCACTACCAAAGCTGCGCTTGCA | 11175 |
| U3-1 | ATTAGCGGCCGCCATGTTAAGCCTAACATTTCGTA | 11899 |
| U3-1 RT | CTACTCGAGCATACTCAACAGGTTCCCAAT | 12008 |
| WEHV2 | ||
| UP-2 | CTACTCGAGCTTGGATACCATCGACACCT | 177 |
| E-2 | CTAGAGCTCTGAATTACCCAACTGATAGAGCA | 6498 |
| A-2 | CTAGAGCTCTGGTGATGTAGCTGAGTCAGA | 10689 |
| B-2a | CTAGAAGCTCTGATTCGCAGCTACAGCTGATA | 11392 |
| B-2b | CTAGAGCTCAACGACCCACTGTCACT | 11878 |
| U3-2 | CTAGAGCTCAGAGTATCCTTGATTCTTAGTTCCCTCTT | 12222 |
| U3-2 RT | CTACTCGAGTGTTCAAATTGTGCAACACAG | 12491 |
Oligonucleotide sequences are written in the 5′-to-3′ direction. Added restriction sites are underlined.
UP-1 and UP-2 are forward primers; all the others are reverse primers.
Start of transcription.
5′ rapid amplification of cDNA ends (RACE) (Life Technologies) was used to identify viral transcriptional start sites. cDNA was synthesized (50-μl reaction mixture) with 1 μg of total RNA, 2 pmol of gag-specific downstream primers for WEHV1 (5′-AGATCCAGGCCATCCTGCTAA-3′) and WEHV2 (5′-ATGTTCTGCTATCTGATCTC-3′), and 4 U of MLV RT. The cDNA was purified as previously described (31). Two microliters of the cDNA was amplified by PCR with an upstream GI anchor primer (Gibco-BRL) and downstream primers in orfC for WEHV1 (5′-ATTAGCGGCCGCACCATGGCCTGGAACAAAAAGCAT-3′) and WEHV2 (5′-CTCTCTAGATTACTAAAAGGTATTTCCGGTTTG-3′) (engineered restriction sites are underlined). The samples were denatured for 5 min at 96°C prior to the addition of 2.5 U of Taq polymerase and amplified as follows: 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min. The WEHV1 and WEHV2 PCR products were sufficiently abundant to be gel purified and cloned into Bluescript II SK(−) with SalI-NotI and SalI-XbaI, respectively.
Nucleotide sequence accession numbers.
The DNA sequences of WEHV1 and WEHV2 were deposited in GenBank under accession no. AF133051 and AF133052, respectively.
RESULTS
Sequence analysis of WEHV1 and WEHV2.
The complete DNA sequence for each virus was obtained from a single pair of 5′ and 3′ overlapping subclones (Fig. 1, panels b). The sequences of the 5′ and 3′ LTRs and overlapping pol regions showed little variation. Therefore, we assume that the sequence analyses reported here are representative of the complete WEHV1 and WEHV2 genomes. The sequence analyses showed that the genomic organizations of WEHV1 and WEHV2 are similar to that of WDSV; each virus contains three open reading frames in addition to the genes found in all retroviruses, gag, pol, and env (Fig. 1, panels a). The sizes of the WEHV1 and WEHV2 proviruses are 12,999 bp and 13,125 bp, respectively, and the analyses of their genomes are described below.
LTRs.
The boundaries of the 5′ LTRs were identified by comparison with their 3′ LTR sequences. The WEHV1 and WEHV2 LTRs are shown in Fig. 2. The boundary of the U3 promoter region and R is defined as the start site for transcription. The WDSV transcriptional start site begins with the sequence GTCTCA (22). This sequence is conserved in WEHV1 and WEHV2, and by 5′ RACE we confirmed that the G of this sequence is the transcriptional start site (position +1) for WEHV1 and WEHV2 (data not shown). TATA boxes are located at positions −31 and −30 in the WEHV1 and WEHV2 U3 regions, respectively. The boundaries of U3, R, and U5 for WEHV1 and WEHV2 were predicted as was done previously with the WDSV LTR (22). The WEHV1 LTR is 643 bp in length, with U3, R, and U5 regions of 503, 81, and 59 bp, respectively (Fig. 2A). The WEHV2 LTR is 550 bp in length, with U3, R, and U5 regions of 410, 79, and 61 bp, respectively (Fig. 2B). The consensus polyadenylation signal sequence, AATAAA, occurs at position 61 in both viruses, and the putative 3′ ends of R for WEHV1 and WEHV2 are located at positions 81 and 79, respectively.
Several potential transcription factor binding sites that may be involved in the regulation of viral gene expression were identified in the WEHV1 and WEHV2 promoters (Fig. 2). Sites that are common to the WEHV1, WEHV2, and WDSV LTRs include AP-1, polyoma enhancer activator (PEA3), and NF-IL6, but their spatial orientation and number are not conserved. The WEHV1 promoter contains two glucocorticoid response elements and the WDSV promoter contains one (22), but the WEHV2 promoter does not contain an obvious glucocorticoid response element sequence. Additionally, there is a repeat sequence (ATAAATG) present in the WEHV1 and WEHV2 LTRs that is identical to the core sequence of the binding site for growth hormone factor 1 (17).
PBSs for synthesis of proviral DNA.
Like WDSV, WEHV1 and WEHV2 are predicted to use a histidyl-tRNA (tRNAHis) primer for initiation of minus-strand DNA synthesis (22). The nucleic acid sequence flanking the PBS is conserved in WEHV1, WEHV2, and WDSV (Fig. 2). A polypurine tract, AAAAGGGG, presumably used for priming plus-strand DNA synthesis, is located upstream of the 3′ LTR, and its sequence is conserved in WEHV1 (position 11842), WEHV2 (position 12154), and WDSV (22). Like spumaviruses and lentiviruses, WDSV, WEHV1, and WEHV2 have two additional polypurine tracts in the 3′ portion of pol (13, 22, 28). The sequence CAAAGGGGG is found in WEHV1 pol at positions 4427 and 5028 and in WEHV2 pol at positions 4345 and 5081. One of the polypurine tracts in WDSV pol has this sequence, and the other varies in two positions (22).
Leader region and orfC.
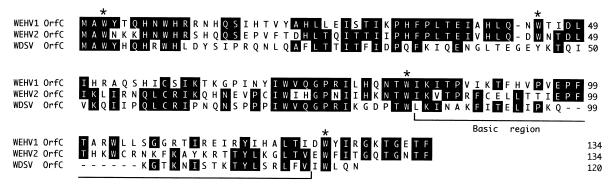
The leader sequence of retroviruses is usually defined as extending from position +1 to the initiation codon of gag. By this definition, the predicted lengths of the leader sequences for WEHV1 and WEHV2 are 910 and 861 bp, respectively. Like WDSV, WEHV1 (position 494) and WEHV2 (position 452) each have a large open reading frame (orfC) found in the leader region (22). The predicted OrfC proteins of WEHV1 and WEHV2 are 134 aa in length, share 46% aa identity, and have 25 and 30% aa identity with the WDSV OrfC, respectively (Table 2). All three OrfC proteins are very basic. Notably, several tryptophan (W) residues are conserved in the OrfC proteins of WEHV1, WEHV2, and WDSV (Fig. 3). Four of the Trp residues have a striking periodicity (WX41WX36WX41W) that is more evident for the WEHV1 and WEHV2 OrfC proteins than for the WDSV OrfC, which has a tyrosine in place of the second conserved Trp (WX42YX36WX33W). Additionally, many of the hydrophobic residues in the X regions are also conserved. Interestingly, the spacing of the Trp residues is reminiscent of the WW domain that is found in some signaling, regulatory, and cytoskeletal proteins (4, 5). The WW domain is believed to play a role in mediating protein-protein interactions with proteins containing proline-rich regions (4, 5). Recently, it was shown that the Rous sarcoma virus Gag protein late domain interacts at the plasma membrane with the WW protein, Yes-associated protein, in a process that may be important to viral budding (19). However, since we were unable to detect any obvious homology to proteins in the databases, we cannot assign a putative function to the OrfC proteins.
TABLE 2.
Comparison of the predicted proteins encoded by WEHV1, WEHV2, and WDSV
| Comparison | Amino acid sequence identity (%)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OrfC | Gag
|
Pol | Env
|
OrfA | OrfB | |||||
| MA | p10/20a | CA | NC | SU | TM | |||||
| WEHV1/WEHV2 | 46 | 58 | 22 | 66 | 52 | 79 | 36 | 59 | 36 | 26 |
| WEHV1/WDSV | 25 | 23 | 17 | 50 | 26 | 64 | 19 | 36 | 25 | 14 |
| WEHV2/WDSV | 30 | 27 | 17 | 51 | 22 | 65 | 19 | 38 | 20 | 14 |
p10/20 was identified in virions for WDSV. It is assumed that an analogous protein is proteolytically cleaved from the WEHV1 and WEHV2 Gag precursor.
FIG. 3.
Alignment of the WEHV1, WEHV2, and WDSV predicted OrfC proteins. The conserved tryptophan (W) residues comprising a possible WW domain are marked with asterisks. The basic region near the C terminus of these proteins is bracketed.
gag.
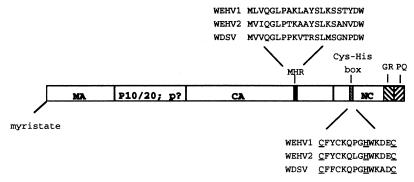
The amino acid sequences of the WEHV1 and WEHV2 gag open reading frames were aligned with that of WDSV Gag to identify putative cleavage products. The N termini of the WDSV virion proteins MA, CA, p10/p20, and NC have been determined (22). The MA proteins of WEHV1 (position 910), WEHV2 (position 861), and WDSV begin with the amino acid sequence MGN and are predicted to be myristylated, as shown schematically in Fig. 4. WEHV1 and WEHV2 share 58% aa identity in MA and they share 23 and 27% aa identity, respectively, with WDSV MA (Table 2).
FIG. 4.
Schematic of the WEHV1, WEHV2, and WDSV Gag polyproteins. MA, matrix; CA, capsid; NC, nucleocapsid; MHR, major homology region. The Gag proteins are predicted to be myristylated at a glycine residue located at the penultimate amino acid residue. p10/p20 is the WDSV protein located between the MA and CA proteins, and p? represents the hypothetical proteins for WEHV1 and WEHV2 in the same region. Underlines denote signature residues. The GR-rich and proline-glutamine (PQ)-rich regions are shown with slashed boxes.
In most retroviruses other than lentiviruses, the region between MA and CA encodes one or two small proline-rich proteins. This protein, or the protein adjacent to CA if two proteins are made, contains an assembly domain [i.e., PPP(W/Y)V] that is required late in the budding process (55). Two hydrophilic, glutamine-rich virion proteins, p10 and p20, having the same N terminus are encoded in this region of WDSV Gag (22). As also found in other retroviruses, this is the least conserved segment of Gag among the three viruses. WEHV1 (positions 1189 to 1503) and WEHV2 (positions 1140 to 1442) share only 22% aa identity in this region, and each shares only 17% aa identity with WDSV (Table 2). In addition, this region of WDSV (156 aa) is substantially longer than those encoded by WEHV1 (105 aa) and WEHV2 (101 aa). WEHV1, WEHV2, and WDSV do not contain sequences resembling known late assembly domains in this region.
The CA proteins of WEHV1 (position 1504), WEHV2 (position 1443), and WDSV comprise 206 aa that are highly conserved (Table 2); the WEHV1 and WEHV2 CAs share 66% aa identity, and each shares about 50% aa identity with the WDSV CA. The major homology region is highly conserved in WEHV1 (position 1954), WEHV2 (position 1893), and WDSV, but the C termini of these sequences are quite different from those of other retroviruses (Fig. 4) (56).
The predicted sizes of the NC proteins of WEHV1 (position 2122) and WEHV2 (position 2061) (122 and 130 aa, respectively) are similar to that of WDSV (127 aa). The WEHV1 and WEHV2 NCs share 56% aa identity and exhibit 26 and 22% aa identity, respectively, with WDSV NC (Table 2). Like WDSV and members of the MLV group, the NC proteins of WEHV1 and WEHV2 contain one Cys-His box at positions 2188 and 2121, respectively (Fig. 4). Just downstream of the Cys-His box, the WEHV NC proteins contain a glycine-arginine (GR)-rich region followed by a proline-glutamine-rich region. WDSV NC has similar motifs, but they are much smaller. The Gly-Arg-rich sequences near the C termini are reminiscent of the GR boxes found in the NC proteins of spumaviruses and other RNA binding proteins (33). However, unlike the moderately conserved GR boxes in spumaviruses (21), the amino acid sequences in the GR-rich regions are not conserved within the walleye viruses.
pol.
The WEHV1 and WEHV2 genomic 5′ and 3′ subclones overlap in the pol region. The WEHV1 pol overlap is 3,105 bp and the WEHV2 pol overlap is 1,516 bp (Fig. 1, panels b). Sequence analysis showed that the pol overlap regions in the WEHV1 5′ and 3′ subclones are identical. The WEHV2 pol overlap regions in the 5′ and 3′ subclones differ by 18 nucleotides, resulting in three amino acid substitutions (31). Like WDSV and MLV, the gag and pro-pol genes of WEHV1 and WEHV2 are in the same translational frame, and suppression of the amber stop codon at the end of gag is likely responsible for the translation of pro-pol (22, 59). pro-pol is the most conserved gene among WEHV1 (position 2515), WEHV2 (position 2430), and WDSV, and the alignment of the three pol genes has been previously reported (31). The protease signature sequence (LVTGA) for WEHV1 and WEHV2 is located at positions 2599 and 2514, respectively. By analogy with other retroviruses, the N terminus for RT is expected to begin 10 to 20 aa downstream of the GRD (position 2809 for WEHV1 and position 2724 for WEHV2) sequence in the protease (45). As in other retroviruses, a putative zinc finger motif is present in the integrase proteins (24): HGVSH at positions 5008 and 5061, followed by CX2C (underlines denote signature residues) at positions 5110 and 5163 for WEHV1 and WEHV2, respectively. The RT-RNase H and RNase H-integrase boundaries cannot be predicted from the amino acid sequence alignments.
env.
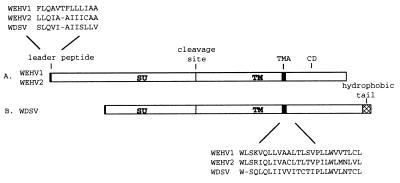
The N terminus of the WDSV transmembrane protein (TM) and, therefore, the C terminus of the extracellular surface protein (SU) were determined previously by amino acid sequencing (22). The sequences of the WEHV and WDSV Env proteins are sufficiently similar to allow the WEHV1 and WEHV2 SU and TM components be identified. The SUs of WEHV1 (position 5910), WEHV2 (position 5957), and WDSV are not highly conserved; the WEHV1 and WEHV2 SUs share 36% aa identity, and each shares 19% aa identity with WDSV (Table 2). The SUs of WEHV1 (711 aa) and WEHV2 (739 aa) are substantially larger than that of WDSV (470 aa) (Fig. 5). The N termini of the WEHV1, WEHV2, and WDSV SUs contain a similar hydrophobic sequence of 14 aa that may be the hydrophobic core of the secretion signal peptide (Fig. 5). Analogous to the WDSV SU-TM boundary, the putative WEHV1 and WEHV2 SU-TM boundaries are marked by a consensus proteolytic cleavage site, RX(R/K)R.
FIG. 5.
Schematic of WEHV1 and WEHV2 (A) and WDSV (B) envelope proteins. SU, extracellular surface protein; TM, transmembrane protein; TMA, transmembrane anchor; CD, cytoplasmic domain of the TM. A possible hydrophobic leader peptide is shown at the N terminus of the SU. The cleavage site between the WEHV SU and TM proteins was predicted based on the N-terminal amino acid sequencing of the WDSV TM. The sequences of the TMAs as predicted by hydrophilicity plots are shown. The hydrophobic tail of the WDSV TM is shown by a hatched box.
The predicted N termini of the WEHV1 and WEHV2 TMs are located at positions 8043 and 8174, respectively. The TMs of WEHV1 and WEHV2 are more conserved than is the SU region; WEHV1 and WEHV2 share 59% aa identity, and they exhibit 36 and 38% aa identity, respectively, with WDSV (Table 2). Hydrophilicity profiles of the three TMs (data not shown) showed a 26-aa hydrophobic region that is likely to be the single spanning transmembrane anchor of WEHV1 (position 9165), WEHV2 (position 9296), and WDSV (position 8514) (Fig. 5). The regions downstream of the transmembrane anchors encode cytoplasmic domains that are proline rich and contain several charged residues. The cytoplasmic domains of WEHV1 and WEHV2 are about 200 aa long. The cytoplasmic domain of WDSV Env is quite different from those of the WEHVs; it is much longer (∼350 aa), and the last 65 aa are hydrophobic (Fig. 5) (22). Since WEHV1, WEHV2, and WDSV have analogous predictable and comparable transmembrane spanning regions, the previous assignment of the C-terminal hydrophobic stretch of amino acids as the transmembrane anchor of WDSV Env was likely in error (22).
3′ orfs.
By analogy with other complex retroviruses, we presume that the open reading frames located downstream of env encode proteins that may contribute to the regulation of viral gene expression and/or replication. The orfA genes of WEHV1 (position 10081), WEHV2 (position 10277), and WDSV were found to encode cyclin D homologs (233, 231, and 297 aa in length, respectively), and their relationships to each other and to cellular cyclins have been discussed (30). The similarity of the rv-cyclins and known cyclins is restricted to the conserved cyclin box motif (3). Analogous to cellular cyclins, the rv-cyclins may promote cell cycle progression (47), thereby stimulating the cell division needed for viral replication. It is also likely that the rv-cyclins induce the cell proliferation that ultimately leads to WEH and WDS (30).
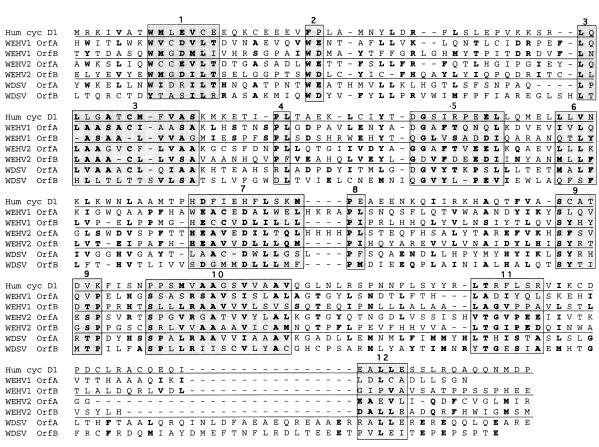
The orfB genes of WEHV1 (position 10784) and WEHV2 (position 10974) are predicted to encode proteins of 301 and 319 aa, respectively. WEHV1 and WEHV2 BlastX searches with WEHV1 and WDSV orfB sequences did not identify proteins with obvious similarity, but the same search with WEHV2 orfB showed a weak homology to WDSV OrfA protein. By visual inspection, we were able to align conserved regions in the OrfA and OrfB proteins of the three walleye retroviruses. The homology between the OrfA and OrfB proteins is limited to the cyclin box region (Fig. 6). Human cyclin D1 was included in the alignment as a reference because of its similarity to the walleye OrfA rv-cyclins (30, 32). Regions where the amino acid sequences of the OrfA and OrfB proteins, within the same virus and between viruses, could be aligned are shaded (Fig. 6). The overall amino acid similarity for any of the OrfA-OrfB pairs is low (Table 3), with WEHV2 OrfA and WEHV2 OrfB being most similar, sharing 24% aa identity and 39% aa similarity. The similarity of WEHV2 OrfA and OrfB is most evident in block 7, where the sequence HEAV.D.L is conserved (Fig. 6). The sequence S..LRAAVV in block 10 shows the similarity of WDSV OrfA and WEHV1 OrfB. Additionally, the sequence ALLE is present in cyclin D1, WEHV2 OrfB, and WDSV OrfA in block 12. These data strongly suggest that orfA and orfB arose by a gene duplication following capture of a cellular cyclin.
FIG. 6.
Alignment of the WEHV1, WEHV2, and WDSV OrfA and OrfB proteins within the cyclin box region. Shaded regions 1 through 12 represent areas of greatest similarity between the OrfA and OrfB proteins. The amino acid substitutions allowed are listed in Materials and Methods. The amino acids shown in bold represent the following: (i) those that are similar in four or more of the OrfA or OrfB proteins (i.e., excluding human cyclin D1) or (ii) those that are similar in an OrfA or OrfB pair within a single virus (between solid lines). Both conditions may be shown in a single column. Human cyclin D1 is included to show the relationship of the OrfAs and OrfBs to cyclin proteins.
TABLE 3.
Amino acid comparisons of OrfA and OrfB proteins within the cyclin box regiona
| Protein | % Amino acid similarity/identity with OrfB of:
|
||
|---|---|---|---|
| WEHV1 | WEHV2 | WDSV | |
| OrfA of: | |||
| WEHV1 | 18/29 | 20/33 | 11/19 |
| WEHV2 | 17/31 | 24/39 | 14/27 |
| WDSV | 18/28 | 20/33 | 18/27 |
| Human cyclin D1 | 13/28 | 18/31 | 11/18 |
OrfA represents the rv-cyclins.
The N and C termini of the WEHV1, WEHV2, and WDSV OrfB proteins have motifs found in regulatory proteins and/or transcriptional activators. The N termini of WEHV1 and WEHV2 OrfB contain polyproline tracts analogous to those of other proteins that have been implicated in protein-protein interactions (data not shown) (54). The C termini of the three OrfB proteins contain acidic domains analogous to those found in many transcriptional activators (e.g., VP-16 of herpes simplex virus) (data not shown) (43).
Analysis of viral gene expression by Northern blotting and transcriptional mapping.
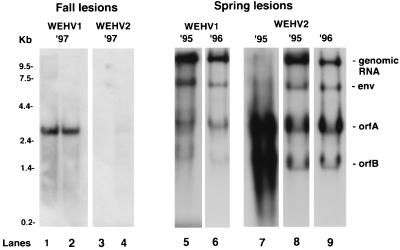
Previous Northern blot analyses have shown that abundant levels of WDSV viral RNA (full-length and subgenomic transcripts) are present in regressing dermal sarcomas (spring) and that low levels of WDSV viral RNA (predominantly orfA and orfB) are present in developing sarcomas (fall) (11, 44). Northern blot analysis with LTR-specific probes showed a similar expression pattern for WEHV1 and WEHV2 (Fig. 7). WEHV2 RNA was present at levels much lower than WEHV1 RNA in the fall samples and was not detected in one of the samples (Fig. 7, lane 3). A low level of a 2.6-kb transcript, which was shown earlier to hybridize with an orfA probe (30), was observed in WEHV fall lesions (Fig. 7, lanes 1, 2, and 4). In contrast, abundant levels of WEHV1 and WEHV2 genomic and subgenomic transcripts (∼13, 7.0, 2.6, and 1.8 kb) were detected in spring hyperplasias collected in different years (Fig. 7, lanes 5 to 9). The sizes of the transcripts corresponded to those predicted for genomic RNA, env, orfA, and orfB transcripts, respectively. Notably, one sample collected in the spring contained abundant levels of the orfA and orfB transcripts but only low levels of full-length and genomic RNA (Fig. 7, lane 7). Qualitatively, this pattern is similar to that observed for developing hyperplasias and dermal sarcomas. This sample was prepared from a fish that was 80% covered with hyperplasias (data not shown). The significance of this observation is not known. Based on the intensity of hybridization, amount of RNA loaded, and exposure time, we estimate that there is 10- to 50-fold more of the 2.6-kb transcript (orfA) in the spring lesions than in the fall lesions (Fig. 7, compare lanes 1 and 2 with lanes 5 and 6). The differences in the patterns and levels of RNA expression between spring and fall lesions are similar to those observed for WDSV, for which there is at least 100-fold more of the 2.8-kb message (orfA) present in spring than is present in fall lesions (44).
FIG. 7.
Northern blot analysis of WEHV1 and WEHV2 viral transcripts in fall and spring lesions collected in different years, using LTR-specific probes. poly(A)+ RNA (1 μg) from fall lesions was hybridized with WEHV1 (lanes 1 and 2) or WEHV2 (lanes 3 and 4) LTR probes. Lesions from two to three fish were pooled to generate each of the two RNA samples represented in lanes 1 to 4. Total RNA (10 μg) from spring lesions isolated from individual fish was hybridized with WEHV1 (lanes 5 and 6) and WEHV2 (lanes 7 to 9) LTR probes; RNA shown in lane 7 was isolated from lesions obtained from a fish with a severe case of epidermal hyperplasia; i.e., 80% of its body was covered with the disease.
Individual WEHV1 and WEHV2 RNA transcripts were characterized by RT-PCR with upstream primers in R (UP-1 and UP-2, respectively) and downstream primers located in individual genes (env, orfA, and orfB) or in U3 (U3-1 and U3-2, respectively), as shown in Table 1 and Fig. 1, panels c. WEHV1 and WEHV2 cDNAs were cloned from three spring hyperplastic tissue samples from different fish to identify transcripts that were generated from replication-competent viruses. The results of these analyses are shown in Fig. 1, panels d, and Table 4. Sequence analyses of the cDNA clones showed that the major splice donor sequence used by WDSV, CTGGTGAGTAC, is also used by WEHV1 (position 250) and WEHV2 (position 251) (44). In contrast to WDSV, where two additional splice donors were identified, only the major splice donor was identified in WEHV1 and WEHV2.
TABLE 4.
WEHV1 and WEHV2 splice donors and acceptors
| Splice sitea | Position (aa) | Sequenceb |
|---|---|---|
| WEHV1 | ||
| Donor | ||
| MSD | 250 | CTG/GTGAGT |
| Acceptors | ||
| SA1-1 | 5737 | ACATGCTCTCTCAG/A |
| SA1-2 | 9984 | TCTCCTCCCCAAAG/A |
| SA1-3 | 10569 | AATATTCCCTCCAG/G |
| SA1-4 | 10774 | TGATCTACTCTCAG/G |
| Consensus | NNNYYTYCYCYCAG/R | |
| WEHV2 | ||
| Donor | ||
| MSD | 251 | CTG/GTGAGT |
| Acceptors | ||
| SA2-1 | 5784 | ACATGATCTTTCAG/C |
| SA2-2 | 5798 | CTTATCACACACAG/G |
| SA2-3 | 10218 | TTTTGTTAAAACAG/G |
| SA2-4 | 10984 | AAAATGGCCTCTAG/G |
| SA2-5 | 11116 | TCCCCTTCCTATAG/G |
| SA2-6 | 11820 | ATTTCATTGGATAG/G |
| Consensus | NYYYYNYYNYRYAG/G |
MSD, major splice donor.
/, position of the splice site.
As in all retroviruses, WEHV1 and WEHV2 appear to express Env from a singly spliced message (Fig. 1, panels d). The splice acceptor (SA) for WEHV1 env is located at position 5737 (SA1-1) (Table 4). The predicted AUG initiation codon for env is located 173 bases downstream of the SA (position 5910) and is in a good translational context (26). Two WEHV2 env transcripts were identified that used SAs for env at positions 5784 (SA2-1) and 5798 (SA2-2). Both WEHV2 env transcripts are predicted to use the AUG located at position 5957 for initiation of Env translation. The sizes of the WEHV1 and WEHV2 env transcripts are consistent with the 6.8-kb bands observed by Northern blotting (Fig. 7).
Sequencing of the first 200 bases of WEHV2 env cDNAs identified clones that had eight nucleotide differences from the sequence of the WEHV2 partial genomic clone 24. A thymidine-to-cytosine change at position 5963 generated a BamHI site and changed a serine to a proline at amino acid position 3 in Env, whereas the other mutations were silent. All of the mutations were silent in Pol (pol and env overlap in this region). The BamHI site-containing cDNA was isolated from three independent tissue samples, whereas the genomic clone-like transcript was cloned from one sample. These data suggest that the latter sequence may be more representative of env in WEHV2 and, along with the variation in the overlapping pol sequences discussed above, that there is heterogeneity in the WEHV2 population that we did not observe among isolates of the WEHV1 genomic and cDNA sequences.
The WEHV1 and WEHV2 OrfA proteins are predicted to be expressed from a singly spliced message (Fig. 1, panels d). WEHV1 orfA uses an SA at position 9984 (SA1-2), and WEHV2 orfA uses an SA at position 10218 (SA2-3) (Table 4). The predicted initiation codons for WEHV1 and WEHV2 OrfA proteins are located at positions 10081 and 10277, respectively, and they are in a suitable translational context. The generation of the WEHV1 and WEHV2 orfA transcripts appears to be less complex than that of WDSV, in which singly and multiply spliced transcripts containing full-length and truncated versions of orfA were identified (44). The sizes of the predicted WEHV1 and WEHV2 orfA transcripts are consistent with the 2.6-kb bands observed on the Northern gel (Fig. 7).
The WEHV1 OrfB protein is predicted to be expressed from two singly spliced orfB transcripts. One transcript uses a splice acceptor within orfA at position 10569 (SA1-3) (Fig. 1 and Table 4), with the predicted translational initiation codon at position 10784. In an analogous fashion, the orfB transcript observed for WDSV has a leader region that comprises ∼300 bases of the 3′ end of orfA (44). The other WEHV1 orfB transcript uses an SA at position 10774 (SA1-4), with the predicted initiation codon only 11 bp downstream. In contrast, a WEHV2 orfB transcript that would encode a full-length OrfB protein was not isolated with any of three different downstream primers, B2-a, B2-b, or U3-2 (Table 1 and Fig. 1). However, three truncated orfB transcripts were identified from several independent tissue samples. The largest orfB transcript uses a consensus SA at position 10984 (SA2-4) (Fig. 1 and Table 4). This transcript lacked the first 12 bases of orfB, and the first AUG codon that is not closely followed by a stop codon is located at position 11232. If translated, this transcript would encode a protein lacking the first 86 aa (the proline-rich N terminus) of OrfB. The second orfB transcript uses a consensus SA at position 11116 (SA2-5). SA2-5 is located just downstream of a long stretch of cytosines (21 cytosines of 24 bases) that encodes the polyproline tract found in the N terminus of the predicted WEHV2 OrfB protein. There are several AUGs in the leader sequence of this transcript, but they are shortly followed by stop codons. An AUG at position 11232 could serve as the orfB initiation codon, but analogous to the transcript above, the predicted protein would lack the first 86 aa of OrfB. An analogous WEHV1 orfB transcript was not identified, but WEHV1 does have an SA site just downstream of the region encoding the WEHV1 polyproline tract at position 10887, leaving open the possibility that it exists. The third WEHV2 orfB transcript uses an SA at position 11820 (SA2-6) (Fig. 1 and Table 4) that is followed by two AUGs at positions 11823 and 11829, but neither is in a favorable translational context. If translated, this transcript would encode a peptide of 36 aa. The significance of these truncated WEHV2 transcripts is unknown, since analogous transcripts were not identified for WEHV1 or WDSV. Despite our inability to clone a cDNA that would encode the full-length OrfB protein, we infer that the 1.8-kb band on the Northern blot represents the full-length transcript by analogy with WEHV1 and WDSV (Fig. 7). However, we have not ruled out the possibility that the transcript lacking the first 12 bases is not present in the 1.8-kb band. Neither of the two smaller transcripts were detected by Northern blotting (Fig. 7).
Together, the Northern blot and transcriptional analyses suggest that WEHV1, WEHV2, and WDSV use similar strategies to regulate viral gene expression and replication and that the expression of specific viral genes correlates with different stages of their cognate diseases.
DISCUSSION
The sequence analyses of WEHV1, WEHV2, and WDSV have shown them to be closely related, with characteristics in common with several retrovirus genera: spumaviruses (large size, location and structure of 3′ orfs, and multiple polypurine tracts); lentiviruses (long cytoplasmic domain in Env and multiple polypurine tracts); and mammalian type C simple retroviruses (type C particle morphology, a single Cys-His box in NC, and predicted amber suppression for translation of the Gag-Pol polyprotein). Additionally, these viruses are unique among the Retroviridae: they use a histidyl-tRNA as the primer for first-strand synthesis; they encode a large open reading frame in the leader region upstream of gag (orfC); they encode cyclin homologs (orfA) that may be involved in the induction of cell proliferation (30); and they encode accessory genes, orfA and orfB, that likely arose by gene duplication and whose origin can be traced to a cellular gene.
The leader sequence at the 5′ end of genomic retroviral RNA is required for RNA dimerization, RNA encapsidation, and initiation of reverse transcription (34). In some cases, this region encodes peptides that may be required for viral replication. For example, the avian leukemia and sarcoma viruses encode three short peptides in their leader sequences that are important for packaging genomic RNA (16). By comparison, each of the leader sequences of the walleye retroviruses harbors an open reading frame, orfC, that encodes a protein of approximately 14 kDa. Although we cannot infer a function based on sequence analysis, the conservation of the OrfC proteins strongly suggests that they are biologically significant to the viruses. Alternatively, or possibly in parallel, we cannot rule out the possibility that an underlying RNA secondary structure in this region is responsible for conservation of orfC. Interestingly, the OrfC proteins contain tryptophans at regularly spaced intervals, similar to those found in the protein-binding module, the WW domain (48). WW domain-containing proteins are a diverse group of cellular proteins that bind ligands containing a PPPPY (PY motif) or PY-like motifs (42). The WW domain of the signaling protein, Yes-associated protein, binds to the PPPPY late assembly domain of Rous sarcoma virus in vitro, suggesting that cellular WW domain-containing proteins may be involved in the budding process (19). It is tempting to speculate that the tryptophan repeats found in the OrfCs form a WW domain that could mediate protein interactions with Gag at the plasma membrane necessary for viral budding, but we have no data suggesting that this is the case.
If orfC is translated, it is unclear how the Gag proteins are made by the walleye retroviruses. Simple stop codon suppression or frameshifting mechanisms are not likely to produce an OrfC-Gag polyprotein because different stop codons and spacing between orfC and gag exist in the three viruses and these mechanisms seem too inefficient to produce a sufficient amount of Gag for virion assembly. It is possible that Gag is translated from a spliced mRNA from which orfC has been removed, but we have failed to identify such a spliced transcript by RT-PCR (29). It has been shown that a cap-independent internal ribosomal entry mechanism can be used by MLV for Gag and Gag-Pol polyprotein translation (4, 27). It is possible that a similar mechanism could be used by the walleye retroviruses to produce Gag, but this remains to be tested.
The envelope proteins of the walleye retroviruses are quite different from those of other retroviruses. In the most general view, the envelope proteins are much larger, with the TM proteins of WEHV1 and WEHV2 (ca. 600 aa) and WDSV (755 aa) being two to four times larger than the TMs of prototypical retroviruses like MLV and human immunodeficiency virus (HIV) (ca. 170 aa and 345 aa, respectively). Previously, it was suggested that the long hydrophobic tail at the C terminus of the WDSV TM protein was the transmembrane anchor and that WDSV Env lacked a cytoplasmic domain (22). We have identified a small hydrophobic region (26 aa) in the WEHV1, WEHV2, and WDSV TM proteins that is similar in sequence and position in all three viruses. Since the WEHVs lack the hydrophobic tail at the C terminus of Env, we predict that this is the transmembrane anchor for the three TM proteins. The cytoplasmic domains for the WEHVs (ca. 200 aa) and WDSV (ca. 350 aa, including the 65-aa hydrophobic tail) are longer than those observed for any other retrovirus (40). Additionally, WDSV is the only retrovirus whose cytoplasmic domain ends in a long hydrophobic tail that may be buried within the protein or imbedded in the cell membrane (22). The biological significance of the unusual envelope proteins encoded by these viruses is not known, but it is speculated that they play some role in stabilizing the virus in an aquatic environment. It is likely that WDS and WEH are horizontally transmitted during the walleye spring spawning run when viral particles are shed from the lesions (11). Experimental transmission of WDS to walleye fingerlings has been achieved by injection of cell-free tumor filtrates, topical application to fingerlings, and oral lavage, suggesting that contact and/or the oral ingestion of tumors is a route of natural transmission (9, 37). Recently, efficient experimental transmission of WDS was observed when tumor-negative fish were housed in flowing stream water that harbored tumor-positive fish upstream, indicating that virions remain infectious in water (10).
By analogy with other complex retroviruses, we suggest that the proteins encoded by the orfA and orfB transcripts provide accessory or regulatory functions for the walleye viruses (15). The orfA genes of WEHV1, WEHV2, and WDSV encode cyclin D homologs that are likely to play a central role in the induction of cell proliferation observed in developing lesions and may be necessary for viral replication (30). While inducing cell proliferation may be the central function of the rv-cyclins, it is also possible that the rv-cyclins contribute to other aspects of viral replication, e.g., gene regulation. Independent of cell cycle functions, cyclins have been shown to be important in transcription (35, 60). A potentially relevant example of a cyclin regulating viral gene expression is the novel Cdk9-cyclin T complex that is believed to activate HIV transcription by acting as a cellular cofactor for binding of the HIV Tat protein to the TAR sequence (53, 58).
While the OrfB proteins have sequence similarities with the D cyclins and the rv-cyclins within the cyclin box, we can only speculate about their biochemical properties. It seems unlikely that they interact with cellular cyclin-dependent kinases because they lack the critical lysine and glutamate residues in α-helices C and E that are necessary for cyclin-dependent kinase binding (12, 23, 31). However, because the amino acid sequences of the cyclin box can vary significantly between cyclin subfamilies (3), it is possible that all or some of the cyclin box tertiary structure has been retained in OrfB. Notably, two important cellular proteins have structural similarity with cyclins: the general transcription factor TFIIB contains a partial cyclin box and its crystal structure is remarkably similar to that of cyclin A (2), and the retinoblastoma tumor suppressor protein (Rb) has two cyclin box motifs that form the Rb pocket, which contains several potential binding sites for cellular and viral proteins (25). The N termini of the OrfB proteins contain polyproline tracts that may be involved in protein-protein interactions, and their C termini contain clusters of acidic residues analogous to those found in VP-16 of herpes simplex virus and Taf of simian foamy virus (38, 43, 54).
The similarity, albeit limited, of the OrfA and OrfB proteins and the adjacent positions of orfA and orfB in the genomes of the walleye viruses suggest that these genes arose by duplication (50). This is the second example of inferred gene duplication in a retrovirus, the first being that the vpx gene of the HIV type 2/simian immunodeficiency virus group arose by gene duplication of the vpr gene (50). The additional observations that orfA and orfB are distantly related to cellular cyclins and that the WDSV OrfA protein can rescue cyclin-deficient yeast from growth arrest suggest that they were derived from the host genome. It has been proposed that complex retroviruses evolved from simple retroviruses that acquired additional sequences from the host by transduction (39, 49), but sequence and functional data supporting capture and evolution of accessory genes from cellular genes in complex retroviruses have not been forthcoming (39). HIV/SIV Nef is the only example of an accessory protein that has sequence similarities to cellular proteins, including Ras and Src kinases, mammalian G proteins, and other cellular proteins, but the significance of this is unknown (41). In contrast, the finding that the WDSV OrfA protein complements cyclin deficiency in yeast clearly suggests that it was derived from a cellular gene with a similar function (30). Also, unlike the genes acquired by acutely transforming simple retroviruses, the sequences of orfA and orfB are very divergent from cellular cyclin sequences, suggesting that the capture of the cellular cyclin event occurred early in the emergence of the walleye retroviruses (30). Although convergent evolution cannot be excluded, we suggest that orfA and orfB are the first accessory genes of complex retroviruses whose origin is unambiguously cellular.
The pattern of gene expression observed for WEHV1, WEHV2, and WDSV in fall and spring tumors parallels the early and late phases of viral replication of complex retroviruses in cell culture; low levels of predominantly subgenomic transcripts (orfA and orfB) are observed in developing lesions collected in the fall, and high levels of genomic and subgenomic transcripts are observed in lesions collected in the spring. The transcriptional profiles of WEHV1 and WEHV2 are similar to that observed for WDSV, suggesting that they use similar strategies for replication and that the expression of specific viral genes correlates with different stages of their cognate diseases. The major difference between the profiles of the WEHVs and WDSV is that only one singly spliced transcript is predicted to encode each of the WEHV OrfA proteins, whereas several multiply spliced transcripts are predicted to encode the WDSV OrfA protein (44). Although a transcript of predicted size for WEHV2 orfB was identified by Northern blotting, we were unable to analyze this cDNA by RT-PCR and clone it. At present, we do not know why we were unable to isolate this cDNA; possibly the gene is unstable in Escherichia coli or the cDNA was underrepresented in the population of amplified products.
Previous phylogenetic analyses based on the conserved region of RT suggested that these retroviruses were most closely related to the mammalian type C (MLV-related) and spumavirus clades, but they are clearly divergent from both groups (31, 51). In addition, WEHV1, WEHV2, and WDSV are very different from SnRV, the only other fish retrovirus for which sequence is available (20). Based on the data presented herein, we suggest that WEHV1, WEHV2, and WDSV warrant consideration as a new genus in the family Retroviridae. Further characterization of these viruses will likely contribute to our understanding of retroviral biology, including viral replication, pathogenesis, and evolution.
ACKNOWLEDGMENTS
We thank V. Vogt and A. Eaglesham for critical reading of the manuscript.
The work was supported by a grant from the American Cancer Society (RPG-96-040-03). L.A.L. was supported by an NIH training grant (CA09682).
REFERENCES
- 1.Arnold A. The cyclin D1-PRAD1 oncogene in human neoplasia. J Investig Med. 1995;43:543–549. [PubMed] [Google Scholar]
- 2.Bagby S, Kim S, Maldonado E, Tong K I, Reinberg D, Ikura M. Solution structure of the C-terminal core domain of human TFIIB: similarity to cyclin A and interaction with TATA-binding protein. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- 3.Bazan J F. Helical fold prediction for the cyclin box. Proteins. 1996;24:1–17. doi: 10.1002/(SICI)1097-0134(199601)24:1<1::AID-PROT1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 4.Berlioz C, Darlix J-L. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 6.Bowser P R, Earnest-Koons K, Wooster G A, LaPierre L A, Holzschu D L, Casey J W. Experimental transmission of discrete epidermal hyperplasia in walleyes. J Aquat Anim Health. 1998;10:282–286. [Google Scholar]
- 7.Bowser P R, Wolfe M J, Forney J L, Wooster G A. Seasonal prevalence of skin tumors from walleye (Stizostedion vitreum) from Oneida Lake, New York. J Wildl Dis. 1988;24:292–298. doi: 10.7589/0090-3558-24.2.292. [DOI] [PubMed] [Google Scholar]
- 8.Bowser P R, Wooster G A. Regression of dermal sarcoma in adult walleyes (Stizostedion vitreum) J Aquat Anim Health. 1991;3:147–150. [Google Scholar]
- 9.Bowser P R, Wooster G A, Earnest-Koons K. Effects of fish age and challenge route in experimental transmission of walleye dermal sarcoma in walleye by cell-free tumor filtrates. J Aquat Anim Health. 1997;9:274–278. [Google Scholar]
- 10.Bowser P R, Wooster G A, Getchell R G. Transmission of walleye dermal sarcoma and lymphocystis via water-borne exposure. J Aquat Anim Health. 1999;11:158–161. [Google Scholar]
- 11.Bowser P R, Wooster G A, Quackenbush S L, Casey R N, Casey J W. Comparison of fall and spring tumors as inocula for experimental transmission of walleye dermal sarcoma. J Aquat Anim Health. 1996;8:78–81. [Google Scholar]
- 12.Brown N R, Noble M E, Endicott J A, Garman E F, Wakatsuki S, Mitchell E, Rasmussen B, Hunt T, Johnson L N. The crystal structure of cyclin A. Structure. 1995;3:1235–1247. doi: 10.1016/s0969-2126(01)00259-3. [DOI] [PubMed] [Google Scholar]
- 13.Charneau P, Clavel F. A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J Virol. 1991;65:2415–2421. doi: 10.1128/jvi.65.5.2415-2421.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin J M. Structure and classification of retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 1. New York, N.Y: Plenum Press; 1992. pp. 19–49. [Google Scholar]
- 15.Cullen B. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol Rev. 1992;56:375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donze O, Spahr P F. Role of the open reading frames of Rouse sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faisst S, Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992;20:3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frerichs G N, Morgan D, Hart D, Skerrow C, Roberts R J, Onions D E. Spontaneously productive C-type retrovirus infection of fish cell lines. J Gen Virol. 1991;72:2537–2539. doi: 10.1099/0022-1317-72-10-2537. [DOI] [PubMed] [Google Scholar]
- 19.Garnier L, Wills J W, Verderame M F, Sudol M. WW domains and retrovirus budding. Nature. 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 20.Hart D, Frerichs G N, Rambaut A, Onions D E. Complete nucleotide sequence and transcriptional analysis of the snakehead fish retrovirus. J Virol. 1996;70:3606–3616. doi: 10.1128/jvi.70.6.3606-3616.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holzschu D L, Delaney M A, Renshaw R W, Casey J W. The nucleotide sequence and spliced pol mRNA levels of the nonprimate spumavirus bovine foamy virus. J Virol. 1998;72:2177–2182. doi: 10.1128/jvi.72.3.2177-2182.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holzschu D L, Martineau D, Fodor S K, Vogt V M, Bowser P R, Casey J W. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J Virol. 1995;69:5320–5331. doi: 10.1128/jvi.69.9.5320-5331.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeffrey P D, Russo A A, Poluak K, Gibbs E, Hurwitz J, Massague J, Pavletich N. Mechanisms of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 24.Katz R A, Skalka A M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 25.Kim H Y, Cho Y. Structural similarity between the pocket region of retinoblastoma tumour suppressor and the cyclin-box. Nat Struct Biol. 1997;4:390–395. doi: 10.1038/nsb0597-390. [DOI] [PubMed] [Google Scholar]
- 26.Kozak M. An analysis of 5′-noncoding regions from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eukaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kupiec J, Tobaly-Tapiero J, Canivet M, Santillana-Hayat M, Flugel R M, Peries J, Emanoil-Ravier R. Evidence for a gapped linear duplex DNA intermediate in the replicative cycle of human and simian spumaviruses. Nucleic Acids Res. 1988;16:9557–9565. doi: 10.1093/nar/16.20.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LaPierre, L. A., and J. W. Casey. Unpublished data.
- 30.LaPierre L A, Casey J W, Holzschu D L. Walleye retroviruses associated with skin tumors and hyperplasias encode cyclin D homologs. J Virol. 1998;72:8765–8771. doi: 10.1128/jvi.72.11.8765-8771.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaPierre L A, Holzschu D L, Wooster G A, Bowser P R, Casey J W. Two closely related but distinct retroviruses are associated with walleye discrete epidermal hyperplasia. J Virol. 1998;72:3484–3490. doi: 10.1128/jvi.72.4.3484-3490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lew D J, Dulic V, Reed S I. Isolation of three novel human cyclins by rescue of G1 cyclin Cln function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 33.Lochelt M, Flugel R M. The molecular biology of human and primate spuma retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 4. New York, N.Y: Plenum Press; 1995. pp. 239–292. [Google Scholar]
- 34.Luciw P A, Leung N J. Mechanisms in retroviral replication. In: Levy J, editor. The Retroviridae. Vol. 1. New York, N.Y: Plenum Press; 1992. pp. 159–298. [Google Scholar]
- 35.Makela T P, Parvin J D, Kim J, Huber L J, Sharp P A, Weinberg R A. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:5174–5178. doi: 10.1073/pnas.92.11.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martineau D, Bowser P R, Renshaw R R, Casey J W. Molecular characterization of a unique retrovirus associated with a fish tumor. J Virol. 1992;66:596–599. doi: 10.1128/jvi.66.1.596-599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martineau D, Bowser P R, Wooster G A, Armstrong G A. Experimental transmission of a dermal sarcoma in fingerling walleyes (Stizostedion vitreum vitreum) Vet Pathol. 1990;27:230–234. doi: 10.1177/030098589002700403. [DOI] [PubMed] [Google Scholar]
- 38.Mergia A, Renshaw-Gegg L W, Stout M W, Renne R, Herchenröeder O. Functional domains of the simian foamy virus type 1 transcriptional activator (Taf) J Virol. 1993;67:4598–4604. doi: 10.1128/jvi.67.8.4598-4604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myers G, Pavlakis G N. Evolutionary potential of complex retroviruses. In: Levy J, editor. The Retroviridae. Vol. 1. New York, N.Y: Plenum Press; 1992. pp. 51–105. [Google Scholar]
- 40.Pancino G, Ellerbrok H, Sitbon M, Sonigo P. Conserved framework of envelope glycoproteins among lentiviruses. Curr Top Microbiol Immunol. 1994;188:77–105. doi: 10.1007/978-3-642-78536-8_5. [DOI] [PubMed] [Google Scholar]
- 41.Peterlin M. Molecular biology of HIV. In: Levy J, editor. The Retroviridae. Vol. 4. New York, N.Y: Plenum Press; 1995. pp. 185–238. [Google Scholar]
- 42.Pirozzi G, McConnell S J, Uveges A J, Carter J M, Sparks A B, Kay B K, Fowlkes D M. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- 43.Ptashne M, Gann A A F. Activators and targets. Nature. 1990;335:329–331. doi: 10.1038/346329a0. [DOI] [PubMed] [Google Scholar]
- 44.Quackenbush S L, Holzschu D L, Bowser P R, Casey J W. Transcriptional analysis of walleye dermal sarcoma virus (WDSV) Virology. 1997;237:107–112. doi: 10.1006/viro.1997.8755. [DOI] [PubMed] [Google Scholar]
- 45.Rao J K M, Erikson J W, Wlodawer A. Structural and evolutionary relationships between retroviral and eucaryotic aspartic proteinases. Biochemistry. 1991;30:4663–4671. doi: 10.1021/bi00233a005. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 47.Sherr C J. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 48.Sudol M, Chen H I, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module—the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 49.Swain A, Coffin J F. Mechanism of transduction by retroviruses. Science. 1992;255:841–845. doi: 10.1126/science.1371365. [DOI] [PubMed] [Google Scholar]
- 50.Tristem M, Marshall C, Karpas A, Petrik J, Hill F. Origin of vpx in lentiviruses. Nature. 1990;347:341–342. doi: 10.1038/347341b0. [DOI] [PubMed] [Google Scholar]
- 51.Tristem M, Myles T, Hill F. A highly divergent retroviral sequence in the tuatara (Sphendon) Virology. 1995;210:206–211. doi: 10.1006/viro.1995.1333. [DOI] [PubMed] [Google Scholar]
- 52.Walker R. Virus associated with epidermal hyperplasia in fish. Natl Cancer Inst Monogr. 1969;31:195–207. [PubMed] [Google Scholar]
- 53.Wei P, Garber M E, Fang S, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 tat and mediates its high-affinity, loop specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 54.Williamson M P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto T, Kelly R K, Nielson O. Epidermal hyperplasia of walleye, Stizostedion vitreum vitreum (Mitchell), associated with retrovirus-like type-C particles: prevalence, histologic and electron microscopic observations. J Fish Dis. 1985;8:425–436. [Google Scholar]
- 58.Yang X, Herrmann C, Rice A. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxy-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshinaka Y, Katoh I, Copeland T D, Oroszlan S J. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc Natl Acad Sci USA. 1985;82:1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zwijsen R M L, Wientjens E, Klompmaker R, Van Der Sman J, Bernards R, Michalides R J A M. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]