Abstract
Arguably, the greatest threat to bacteria is phages. It is often assumed that those bacteria that escape phage infection have mutated or utilized phage‐defence systems; however, another possibility is that a subpopulation forms the dormant persister state in a manner similar to that demonstrated for bacterial cells undergoing nutritive, oxidative, and antibiotic stress. Persister cells do not undergo mutation and survive lethal conditions by ceasing growth transiently. Slower growth and dormancy play a key physiological role as they allow host phage defence systems more time to clear the phage infection. Here, we investigated how bacteria survive lytic phage infection by isolating surviving cells from the plaques of T2, T4, and lambda (cI mutant) virulent phages and sequencing their genomes. We found that bacteria in plaques can escape phage attack both by mutation (i.e. become resistant) and without mutation (i.e. become persistent). Specifically, whereas T4‐resistant and lambda‐resistant bacteria with over a 100,000‐fold less sensitivity were isolated from plaques with obvious genetic mutations (e.g. causing mucoidy), cells were also found after T2 infection that undergo no significant mutation, retain wild‐type phage sensitivity, and survive lethal doses of antibiotics. Corroborating this, adding T2 phage to persister cells resulted in 137,000‐fold more survival compared to that of addition to exponentially growing cells. Furthermore, our results seem general in that phage treatments with Klebsiella pneumonia and Pseudomonas aeruginosa also generated persister cells. Hence, along with resistant strains, bacteria also form persister cells during phage infection.
Bacterial infections are predicted to be the main cause of death by 2050, and persister cells often form during periods of lethal stress. Here, we show phage infections generate persister cells, and these persisters may undermine phage therapy.
INTRODUCTION
Given the lack of novel antibiotics (Van Goethem et al., 2024) and the increasing mortality due to resistant bacterial infections, ~5 M deaths/year (Murray et al., 2022), interest is surging in the use of phages to combat infections (Strathdee et al., 2023). However, bacteria can mutate rapidly to undermine some phage therapies (Little et al., 2022; Murray et al., 2022) as well as utilize effective phage‐defence systems such as toxin/antitoxins (Pecota & Wood, 1996).
Excluding phage‐defence systems, most authors have either assumed or found that cells that survive phage attack are resistant, i.e. they assume that the host survived due to genetic change. For example, a recent guide for studying plaques of lytic phages indicates only phage‐resistant bacteria are possible inside plaques, rather than transiently insensitive bacteria (Abedon, 2018), and early literature reported only the formation of phage‐resistant mutants of Pseudomonas aeruginosa spp. in the clear and confluent lysis zone (Postic & Finland, 1961). In addition, neonatal meningitis Escherichia coli colonies resistant to lytic phage EC200PP were presumed resistant, and a clearly mutant derivative (due to its different, rough colony morphology) was studied further and found to be less virulent (Pouillot et al., 2012). Furthermore, all microcolonies in plaques were assumed to be formed by phage‐resistant Klebsiella pneumoniae in a report for monitoring plaque growth with a wide‐field lensless imaging device (Perlemoine et al., 2021). Hence, the formation of persister cells as a result of phage infection has generally not been considered. A related phenotype has been reported T4 infection of stationary‐phase E. coli cells termed ‘hibernation’; however, these hibernators were not investigated for persistence (Bryan et al., 2016), and T4 resumed lytic growth in these hibernating cells once nutrients were provided (Bryan et al., 2016).
Persister cells transiently survive myriad forms of stress based their cellular inactivity (Kwan et al., 2013). The secondary messengers guanosine pentaphosphate and guanosine tetraphosphate signal the external stress and lead to ribosome dimerization, which ceases translation in persister cells (Song & Wood, 2020b). Using a single‐cell approach (Wood, 2022), it has been demonstrated that persister cells resuscitate based on the re‐activation of these dimerized ribosomes (Kim, Yamasaki et al., 2018; Song & Wood, 2020a; Yamasaki et al., 2020). Becoming persistent (i.e. dormant) is advantageous for bacteria to combat phages since slow growth/dormancy (i) increases time for phage‐defence systems to function, (ii) slows production of phage‐dependent proteins and nucleic acids, (iii) increases time for spacer acquisition for CRISPR‐Cas (van Beljouw et al., 2022), (iv) enhances genetic diversity (Schwartz et al., 2023), and (v) reduces bacteria‐phage coevolution (Schwartz et al., 2023). For example, activation of the MqsR/MqsA/MqsC tripartite toxin/antitoxin system in E. coli by phage T2 attack results in cells entering the persister state, which enables the EcoK McrBC restriction system to eliminate T2 phages (Fernández‐García et al., 2024). Moreover, expression of GTPase RsgA allows E. coli phages to better withstand T4 phage infection, likely as a result of persistence via inactivation of ribosomes (Fernández‐García et al., 2023). In a similar fashion, the Listeria spp. type VI CRISPR‐Cas system induces dormancy, which allows restriction/modification systems to eliminate phages (Williams et al., 2023). However, whether bacteria escape phage infection through persistence in general is not well studied.
Since transient resistance to phages could also undermine phage therapy by allowing pathogens to escape phage killing, we explored here whether the E. coli cells found inside plaques formed by T2, T4, and lambda cI survive by mutating or by becoming persistent. We discovered that 0.01% of the cells in suspension survive T2 infection as persister cells, rather than undergoing mutation to become resistant. Hence, our work shows for the first time that E. coli cells may become persistent to survive phage infection without cloning a phage inhibition system like MqsR/MqsA/MqsC (Fernández‐García et al., 2024) and shows for the first time that this phenomenon is general by showing persistence after phage infection with other genera.
EXPERIMENTAL PROCEDURES
Bacteria and growth conditions
Bacteria and phages are shown in Table S1, and cells were cultured at 37°C in lytic broth (Maniatis et al., 1982) (LB). The BW25113 strain was checked to ensure that it was not a lambda lysogen by PCR using tomB primers as a positive control (forward 5'‐CGATTACCTGACTTCCGCCA and reverse 5'‐TCATGGCTGGGTAAACGACC) and the cI lambda primers to check for the presence of lambda (forward 5'‐CACCCCCAAGTCTGGCTATG, and reverse‐5'ACCAAAGGTGATGCGGAGAG). The single isogenic knockouts are from the Keio Collection (Baba et al., 2006).
Kill curves
The kill curve assays to verify E. coli persister cells were formed during ampicillin and T2 phage treatment were performed by treating late exponential cells (turbidity at 600 nm of ~0.5) with 100 μg/mL ampicillin for 4 h with shaking at 250 rpm in a 15 mL flask or by treating with T2 phage at a multiplicity of infection (MOI) of 0.1 for 4 h. Samples were taken every 30 min, washed with phosphate‐buffered saline (PBS, 8 g NaC1, 0.2 g KC1, 1.15 g Na2HPO4, and 0.2 g KH2PO4 in dH2O 1000 mL), and 100 μL was serially diluted with PBS to determine the number of viable cells.
Antibiotic‐resistant bacteria
The presence of resistant bacteria was tested using 1 MIC ampicillin plates and checking for growth after 24 h.
Sensitivity assay for plaque‐derived bacteria and sequencing mutant
Escherichia coli strains were streaked on fresh LB agar plates and incubated overnight. A single colony was inoculated into 15 mL of LB broth and incubated with shaking (250 rpm) for 16 h. The overnight culture (100 μL) was used to create a double‐layer phage plaque with a 20 μL drop of T2, T4, or lambda (cI mutant) phage and incubated overnight. The next day, colonies from inside the lysis area of the phage plaque were streaked twice on fresh LB plates to purify strains and to remove phage. Single colonies were cultured overnight in LB, then diluted 100X and grown to a turbidity of 0.5 at 600 nm. T2, T4, or lambda (cI mutant) phage was added at a MOI ~ 0.1 for 3 h, then the cells were washed twice with PBS and enumerated on LB plates to measure survival in the presence of phage. Similarly, for the pinR mutant, T2 phage was added (0.1 MOI), and the number of viable cells was determined after 3 h.
To estimate the number of insensitive bacteria in plaques, 40,000 T2 phage were added via a 20 μL drop to double‐layer plates, then the number of surviving cells in the plaque was counted after overnight incubation. For nine areas approximately the size of the plaques, the number of cells was determined by resuspending in 1 mL of PBS and using the drop assay.
Persister assays
Escherichia coli persister cells were generated by 30 min rifampicin pretreatment (100 μg/mL) followed by ampicillin treatment at 10X the minimum inhibitory concentration (MIC) to lyse non‐persister cells as described previously (Kwan et al., 2013; Kim, Chowdhury et al., 2018; Kim, Yamasaki et al., 2018; Song & Wood, 2020a, 2020b; Yamasaki et al., 2020). After rifampicin treatment and washing twice with PBS, T2 phage was added for 3 h (0.1 MOI), and cell viability was tested by washing twice with PBS to remove external phage and enumerating bacteria via the drop assay.
To test for the induction of persistence during T2 treatment (but without rifampicin pretreatment), survival in antibiotics was tested by using single E. coli colonies that were cultured overnight in LB, then diluted 100X and grown to a turbidity of 0.5 at 600 nm. T2 phage was added (MOI ~ 0.01), cells were incubated for an hour, then 5 mL of cells were harvested by centrifugation at 5000 rpm for 10 min, washed with PBS, and resuspended in 5 mL of LB containing 100 μg/mL of ampicillin (10X MIC). The cultures were incubated for 3 h with shaking at 250 rpm in a 15 mL culture tube, washed twice with PBS, then 100 μL were serially diluted with PBS to determine the number of viable cells. For enumerating internal phage during the persister assay, the cell samples were washed twice with PBS to remove external phage and treated with 1% chloroform, then serially diluted with phage buffer.
Klebsiella pneumoniae and Pseudomonas aeruginosa persister assays after phage attack
Klebsiella pneumoniae obtained from a blood infection (Pacios et al., 2022) was infected with DNA lytic phage vB_KpnP‐VAC25 (Bleriot et al., 2023) at MOI 0.01, washed with PBS, and contacted with 10X MIC colistin (10 μg/mL) or 5 MIC mitomycin C (10 μg/mL). P. aeruginosa PAO1 from the Gloria Soberón collection at UNAM was infected with phage PaMx12 (DNA lytic phage, sequence https://www.ncbi.nlm.nih.gov/nuccore/386649691) at MOI 1, and surviving colonies in the centre of the plaques were isolated after 20 h and tested for phage sensitivity to determine if the cells were resistant or persistent to phage. To confirm persistence, cells were contacted with phage followed by lethal (10X MIC, gentamicin at 10 μg/mL for P. aeruginosa).
Sequencing
Genomic E. coli DNA was purified using the Qiagen DNA isolation kit following the manufacturer instructions. The quality of samples was quantified by nanodrops and Qubit. Samples were sequenced using Illumina MiSeq by the Genomics Core Facility at the Pennsylvania State University. The sequences were assembled and analysed using bv‐brc.org (Olson et al., 2023). Accession numbers for all the sequences are shown in Table S2.
RESULTS
Kill curves with T2 phage and ampicillin
We reasoned that if persister cells form and survive during phage attack, there would be a sub‐population of cells that survive in an analogous fashion to those that survive as persister cells during antibiotic killing (Kwan et al., 2013) and starvation (Kim, Chowdhury et al., 2018); a scheme showing the experimental plan for a series of experiments to explore this hypothesis is shown in Figure 1A. We used high concentrations of antibiotic since survival at these concentrations is a hallmark of persistence (Kwan et al., 2013).
FIGURE 1.
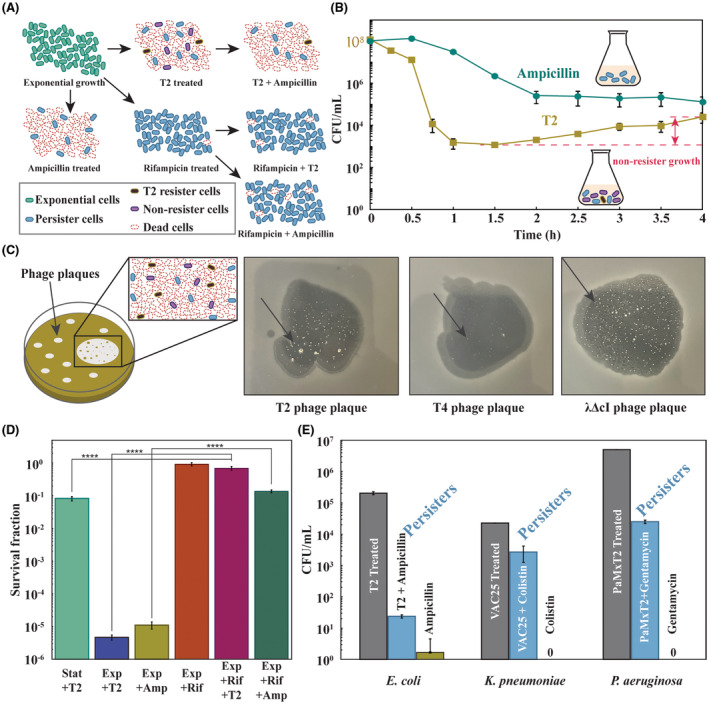
Phages produce persisters. (A) Schematic of experiments used to demonstrate that phages produce persister cells. (B) Kill curves of exponentially growing Escherichia coli BW25113 treated with ampicillin (100 μg/mL, 10X MIC, blue) or T2 phage (MOI ≈ 0.1, orange). Note the increase in cell density with T2 phage indicates that persister cells are present and have revived (with a specific growth rate of 1.2 ± 0.1 h−1). (C) Double‐layer TA plates, from left to right, of phages T2, T4, and lambda mutant cI infecting E. coli BW25113, showing surviving colonies inside the phage inhibition area (indicated with a black arrow). Colonies are visible after 1 day but allowed to grow for several days for the photo here. (D) Escherichia coli BW25113 persister cells were formed by rifampicin pre‐treatment (30 min, 100 μg/mL), and 0.1 MOI T2 phage was added for 3 h. Exponential cells (turbidity 0.5 at 600 nm) were treated with ampicillin (10X MIC) for 3 h. (E) First bar indicates initial cell density after phage attack (108 E. coli cells/mL treated with 0.01 MOI T2 phage for 1 h, 108 Klebsiella pneumoniae cells/mL treated with 0.01 MOI VAC25 phage for 1 h or 108 Pseudomonas aeruginosa cells/mL treated with 1 MOI PaMx12 phage for 1 h), second bar indicates phage and antibiotic treatment (0.01 MOI T2 phage for 1 h followed by 10X MIC of ampicillin for 3 h for E. coli, 0.01 MOI VAC25 phage for 1 h followed by 10X MIC of colistin for 3 h for K. pneumoniae or 1 MOI PaMx12 phage for 1 h followed by 10X MIC of gentamicin for 3 h), and third bar indicates antibiotic treatment alone (10X MIC of ampicillin for 3 h added to 104 cells/mL for E. coli,10X MIC of colistin for 3 h added to 104 cells/mL for K. pneumoniae or 10X MIC of gentamicin for 3 h added to 106 cells/mL for P. aeruginosa). One average deviation shown.
We found addition of ampicillin at 10X the lethal dose (10X MIC) led to 0.1% cell survival, and a clear plateau was reached in 2 h; hence, persister cells were clearly seen with ampicillin (Figure 1B). To ensure there were no antibiotic‐resistant bacteria in the exponential culture prior to ampicillin treatment, we tested for the presence of resistant cells using at 1X MIC ampicillin agar plates and found no resistant bacteria. There were also no resistant bacteria after 3 h 10X MIC ampicillin treatment. Moreover, when the cells that survived 10X MIC ampicillin treatment were regrown, they survived at the same level when retreated with 10X MIC ampicillin, indicating no heritable differences.
Similarly, the addition of T2 phage at MOI of 0.1 caused a precipitous drop in cell density that plateaued with 0.001% cell survival (Figure 1B); hence, since all cells did not die, persister cells are likely present, along with cells with successful phage‐defence systems. However, unlike the antibiotic‐treated cells, those cells with phage addition began to recover and grow after 1 h (specific growth rate of 1.2 ± 0.1 h−1), which indicates persister cells and cells with effective phage‐defence systems are resuscitating and begin growing since there was insufficient time for cells with resistance mutations to reach a population density that would affect turbidity; i.e. assuming a mutation frequency of at most 10−3/genome (Dillon Marcus et al., 2018) and given the cell density of 108 at time zero, any resistant bacteria would have at most a density of around 105 cells/mL after 1 h. Critically, no antibiotic‐resistant bacteria were present after 1 h of T2 phage treatment (at the time cell density increased, Figure 1B) as shown by plating on 1X MIC plates. Hence, the re‐growth is probably due to effective phage‐defence systems or the resuscitation of cells that survived via the persister state, rather than the growth of resistant bacteria. It is also unlikely that these cells were not initially infected since they would be susceptible to the increasing phage titre in the culture.
T2‐treated cells become persisters
To determine whether the bacteria that survive lytic phage attack are resistant or persistent, we formed plaques using T2, T4, and lambda cI phages and isolated surviving cells from inside the plaques from microcolony‐like areas (Figure 1C). The vast majority of cells die, as evidenced by the clearing of lawns when plaques forms. We then purified the cells from the phages by streaking twice on LB plates for single colonies. Some purified cells from the T4 plaque were mucoid; hence, mutation clearly occurred with some of the isolates as the original E. coli host is non‐mucoid. For both T4 and lambda cI, only resistant strains were identified with increases in survivability up to 5 × 105‐fold for T4 and 105‐fold for lambda cI (Table 1A). In contrast, with T2, all four colonies from the microcolony remained sensitive to the lytic phage (Table 1A).
TABLE 1.
Persister cells of BW25113 against phages.
| A | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| T2 | T4 | λΔcI | |||||||
| Colony | Mucoid | 3 h Survival, % | Fold‐change | Mucoid | 3 h Survival, % | Fold‐change | Mucoid | 3 h Survival, % | Fold‐change |
| WT | − | 0.003 ± 0.0002% | 1 | − | 0.002 ± 0.0002% | 1 | − | 0.027 ± 0.007% | 1 |
| 1 | − | 0.004 ± 0.001% | 1.0 | + | 720 ± 100% | 285,000 | − | 3400 ± 400% | 86,000 |
| 2 | − | 0.013 ± 0.003% | 2.8 | + | 608 ± 85% | 251,000 | − | 4000 ± 700% | 109,000 |
| 3 | − | 0.003 ± 0.001% | 1.1 | − | 121 ± 50% | 53,500 | − | 3500 ± 700% | 85,000 |
| 4 | − | 0.015 ± 0.004% | 3.6 | − | 1754 ± 400% | 516,000 | − | 31 ± 7% | 730 |
| B | ||||
|---|---|---|---|---|
| Colony | CFU/mL before infection | CFU/mL after T2 phage | CFU/mL after ampicillin | Survival % |
| WT | 1.3 × 108 ± 3 × 107 | 5.27 × 104 ± 2 × 104 | 5 ± 2 | 0.01 ± 0.006% |
| 1 | 1.0 × 108 ± 1 × 107 | 4.75 × 104 ± 2 × 104 | 34 ± 20 | 0.1 ± 0.06% |
| 2 | 1.0 × 108 ± 1 × 107 | 6.5 × 104 ± 2 × 104 | 74 ± 30 | 0.1 ± 0.06% |
| C | ||
|---|---|---|
| Colony | Internal PFU/mL after T2 phage | Internal PFU/mL after ampicillin |
| WT | 8.7 × 109 ± 2 × 109 | 1.6 × 109 ± 3 × 108 |
| 1 | 8.4 × 109 ± 2 × 109 | 1.0 × 109 ± 2 × 108 |
| 2 | 10 × 109 ± 7 × 108 | 1.2 × 109 ± 1 × 108 |
Note: (A) Survival of BW25113 against phages, mucoid phenotype of the colonies (+/−), and survival percentage of the colonies isolated from inside the phage plaque when they are retested against the original phages (MOI ~ 0.1). Survival fold‐change is based on that of the wild‐type (WT). (B) Persister cells of BW25113 (WT), colony 1 and 2 from inside the T2 phage plaque, showing CFU/mL at each step of the persister assay and survival (0.01 MOI+). (C) Phages inside cells, measured after each step of the persister assay for BW25113 (WT), colony 1 and 2 from inside the T2 phage plaque.
To enumerate the number of cells in the plaque region that survive T2 infection, we determined the percentage of surviving cells in plaques by averaging the number of cells in nine volumes equivalent to that of the plaques formed by T2 phage in 24 h and found 0.00006 ± 0.00002% survive. For example, 203 colonies formed out of roughly 2.6 × 108 ± 2 × 108 cells in one plaque. Since any reviving persister cells in the plaque would be surrounded by large numbers of T2 phage as well as contain internal phage (below), it is not surprising that persister cells were found at such a low percentage. Given that the cells that survived T2 phage were clearly not resistant; i.e. had no phage resistance (Table 1A), we focused on T2 phage for the remaining experiments with E. coli.
Sequencing T2‐surviving cells from plaques
To corroborate that the cells that survived phage attack were persistent, we sequenced the whole genome of the parental strain along with four colonies from inside each phage plate for T2, T4, and lambda cI using Illumina MiSeq (Table 2). Unlike for the resistant T4 and lambda cI colonies, which had up to 79 single mutations (Table 2) and which were shown to be resistant (Table 1A), there were far fewer mutations in the whole genome of the T2 colonies from within the plaque that were not resistant (as few as one coding change, Table 2). Moreover, for C1 from a T2 plaque, there is a single coding mutation in pinR; however, this is probably not a gain‐of‐function mutation as it is as a conservative substitution practically at the N terminus (R3Q so aa position #3 out of 196 aa's). In addition, deleting pinR did not change the growth rate in rich medium but caused a 330‐fold increase in T2 sensitivity, so PinR is necessary for host defence against T2. Furthermore, a polar mutation is unlikely since the pinR locus is monocistronic. Also, since the colonies in the plaques were sequenced after two steps of 24 h growth in plaques and 16 h of regrowth in liquid media (total 64 h growth), the mutations in the persister cells probably occurred after the persistence phase induced by T2 infection.
TABLE 2.
Summary of the SNPS and coding change mutations in the colonies isolated from inside the phage inhibition areas with T2, T4, and λΔcI phages.
| Phage | Colony | No. SNPs | aa changes | No. conserve. aa changes | No. non‐conserve. aa changes |
|---|---|---|---|---|---|
| T2 | C1 | 4 | R3Q (PinR) | 1 | 0 |
| C2 | 11 | R3Q (PinR), T51K (YdfK), R48K (NohA), R70L (NohA) | 2 | 2 | |
| C3 | 13 | R3Q (PinR), R48K (NohA), R70L (NohA), R82P (IS1‐like), H83P (IS1‐like), Y193H (IS1‐like) | 3 | 3 | |
| C4 | 7 | Q3R (PinQ), A159V (PinQ), R82P (IS1‐like), H83P (IS1‐like), Y193H (IS1‐like) | 2 | 3 | |
| T4 | C1 | 4 | Y1167C (RhsC) | 0 | 1 |
| C2 | 14 | Q3R (PinQ), R48K (NohA), R70L (NohA), T198N (YhhI), E219A (YhhI), I25M (YhhI) | 3 | 3 | |
| C3 | 22 | K48R (NohD), L70T (NohD), Q3R (PinQ), A167V (PinQ), R82P (IS1‐like), H83P (IS1‐like), Y193H (IS1‐like) | 3 | 4 | |
| C4 | 79 | L194F (DhaR), R338W (PuuP), PspD, M1V (YnbD), T333P (YnbD), E304G (FtsX), A306G (FtsX), V308G (FtsX), T11K (FtsX), M1V (YcjS), K103N (YcjS), L84F (YcjW), M225I (ISAs1), L77P (ISAs1), R82P (IS1), H83P (IS1‐like), Y193H (IS1‐like), GrcA | 4 | 12+ | |
| λΔcI | C1 | 10 | Y1167C (RhsC) | 0 | 1 |
| C2 | 13 | Y320A (MalT), H321S (MalT), P322A (MalT), L323A (MalT), F324S (MalT), F327W (MalT), L328/ (MalT), Q330R (MalT), R331S (MalT), K51T (YdfK), D83N (TfaQ), R48K (NohA), R70L (NohA), G106A (IS3‐like) | 4 | 10 | |
| C3 | 21 | Y320A (MalT), H321S (MalT), P322A (MalT), L323A (MalT), F324S (MalT), F327W (MalT), L328/ (MalT), Q330R (MalT), R331S (MalT), K51T (YdfK) | 2 | 8 | |
| C4 | 23 | R3Q (PinR) | 1 | 0 |
Note: Coding changes in italics are conservative mutations; i.e. the amino acid is substituted for another in the same group. Those proteins without a specific amino acid change indicate changes throughout the protein.
Abbreviations: conserve., conservative (i.e. similar aa substitutions); No., number; SNPs, single nucleotide polymorphisms.
Cells surviving T2 phage attacks are antibiotic persisters
To provide additional evidence that the E. coli cells isolated from plaques formed by T2 are persisters, we tested antibiotic sensitivity of the surviving cells after T2 treatment. Critically, treatment of exponentially growing cells (turbidity of 0.5 at 600 nm) with T2 phage (0.01 MOI) for 1 h (to induce persistence) followed by washing to remove external T2 phage and treatment with 10X MIC of ampicillin (to lyse non‐persister cells), revealed 0.1% of the surviving cells (colonies 1 and 2) and 0.01% of the wild‐type (Table 1B) become persistent during phage attack; i.e. these cells survived treatment with a lethal ampicillin concentration.
Since we get a small number of persisters after T2 treatment (0.01–0.1%), we hypothesized that internal T2 kills some of persister cells upon their resuscitation on the plates used to count the number of surviving cells. To investigate this, we added chloroform to washed cells after T2 treatment and found ~1010 PFU (plaque formation units)/mL from internalized phage for the wild‐type and surviving cells isolated from the inside of plaques (Table 1C) as well as found 109 PFU/mL of internalized phage after subsequent treatment with ampicillin. These results demonstrate clearly the presence of T2 phages inside the cells. Thus, we conclude that some cells probably die upon waking, and therefore the number of persister cells is even larger. Corroborating this, anomalous colony shapes (with areas of absent cells in circular colonies) were seen on the plates when quantifying the number of surviving cells.
Persister cells survive antibiotic treatment
Since T2 infection creates persister cells, we hypothesized that persister cells would withstand T2 infection better than exponentially growing cells. Hence, we used rifampicin to form E. coli persister cells and lysed non‐persister cells with ampicillin (Kwan et al., 2013; Kim, Chowdhury et al., 2018; Kim, Yamasaki et al., 2018; Song & Wood, 2020a, 2020b; Yamasaki et al., 2020). This method leads to a 105‐fold increase in persister cells and has been vetted nine ways (Kim, Yamasaki et al., 2018; Yamasaki et al., 2020) by us to show the cells generated are bona fide persister cells, and it has been used by over 33 groups to induce persistence.
We found a 212,500‐fold reduction in cell density when T2 was added to exponentially growing cells (turbidity ~0.5, MOI of 0.1, Figure 1D). This reduction in cell density was comparable to that of ampicillin treatment to exponentially growing cells (89,500‐fold, Figure 1D). However, as expected, we found that persister cells were resilient to T2 infection; i.e. there was a 137,000‐fold reduction in the ability of T2 to propagate with persister cells compared to exponentially growing cells treated with T2 (Figure 1D). We also tested the ability of stationary‐phase cells (turbidity ~ 2.5) to withstand T2 infection and found there was an 8‐fold increase in infection for stationary‐phase cells relative to persister cells (Figure 1D); therefore, persister cells are distinct from slowly growing stationary cells. Hence, persister cells are dramatically less sensitive to T2 phage infection. However, we recognize that by converting cells into the dormant persister state, they become less sensitive to myriad stresses, in addition to phage infection.
Klebsiella pneumoniae and P. aeruginosa form persister cells after phage attack that may be eradicated by mitomycin C
We also hypothesized that induction of persistence during lytic infection would be a general phenomenon. To explore whether persister cells are formed after phage attack in non‐E. coli strains, we investigated whether phages induce persistence in K. pneumoniae and P. aeruginosa. For K. pneumoniae, we found that after treatment of 108 cells/mL with VAC25 phage (0.01 MOI) for 1 h, of the remaining viable cells after phage attack (104 cells/mL), 11 ± 4% of the cells were persistent as shown by survival with 10X MIC of colistin for 3 h (Figure 1E). In comparison, starting with the same initial cell density (104 cells/mL) but omitting phage pretreatment, 0 ± 0% the cells survived 3 h 10X MIC of colistin treatment (Figure 1E). Furthermore, since mitomycin C kills persister cells (Cruz‐Muñiz et al., 2017; Kwan et al., 2015), we tested this and found that mitomycin C eradicated the K. pneumoniae persister cells that were formed after VAC25 attack.
For P. aeruginosa, we found that after treatment of 108 cells/mL with PaMx12 phage (1 MOI) for 1 h, of the remaining viable cells after phage attack (106 cells/mL), 0.5 ± 0.14% cells were persistent as shown by survival with 10X MIC of gentamicin for 3 h. In comparison, starting with the same initial cell density (106 cells/mL) but omitting phage pretreatment, 0 ± 0% the cells survived 3 h 10X MIC of gentamicin. In addition, we found that 40% of the cells from the middle of plaques formed in soft agar are persistent, whereas 60% are resistant; i.e. 60% of the cells from the plaques developed mutations that reduced plaque formation in a subsequent assay with phage PaMx12. Hence, both K. pneumoniae and P. aeruginosa form persister cells upon lytic infection.
For comparison, we also performed this set of experiments for E. coli (Figure 1E). We also found that the addition of T2 phage produces persister cells; i.e. cells that survive 3 h of lethal ampicillin treatment. Critically, no antibiotic‐resistant bacteria were present after both 10X ampicillin treatment alone and T2 + 10X ampicillin treatment, as shown by using 1X MIC agar plates. Hence, the remaining cells (Figure 1E) are persister cells.
DISCUSSION
Our results show persisters arise in phage plaques based on six lines of evidence: (i) kill curves show E. coli cells surviving phage infection are similar to cells that survive antibiotics, (ii) T2‐treated E. coli cells isolated from plaques remain sensitive to the phage to the same extent as the wild‐type, (iii) sequencing (after 64 h) shows few mutations for these sensitive E. coli cells isolated from plaques (compared to resistant cells from T4 and lambda cI), (iv) T2 addition to E. coli persister cells results in dramatically less killing compared to exponentially growing cells, (v) cells that survive T2 attack become tolerant to lethal antibiotic concentrations (10X MIC), and (vi) antibiotic‐resistant bacteria were not found prior to treating with phage and antibiotics and after phage and antibiotic treatments. Corroborating the E. coli data, we found both K. pneumoniae and P. aeruginosa formed persister cells after phage attack; hence, induction of persistence during lytic infection may be a general phenotype and should be tested on perhaps Gramme‐positive bacteria. Figure 2 summarizes our results.
FIGURE 2.
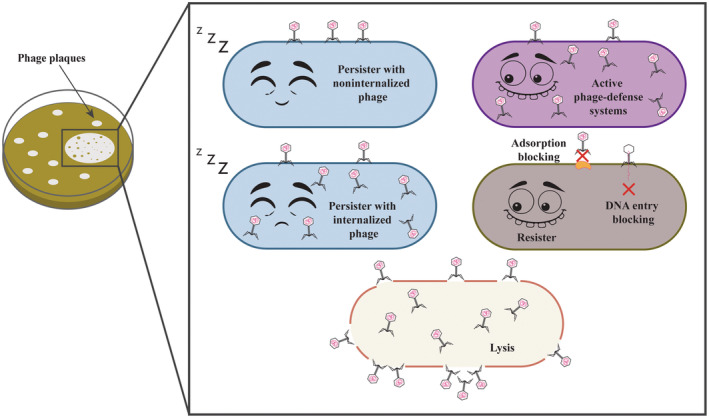
Schematic of model for cells in plaques.
The genesis of the persister cells that form during phage infection is likely a result of increased guanosine tetraphosphate that leads to ribosome dimerization (Song & Wood, 2020a, 2020b; Yamasaki et al., 2020). Moreover, the increase in guanosine tetraphosphate may be an indirect result of phage defence mechanisms like toxin/antitoxin and restriction/modification systems.
Our results with virulent lambda agree well with previous results where it was shown phages with a cI60 mutation kill persister cells (Pearl et al., 2008) since we did not isolate readily persister cells from plaques formed with virulent lambda phage. However, unlike previous results with a virulent, superinfecting lambda phage where no survivors were detected (Pearl et al., 2008), our results are in stark contrast, as we observed survival and resuscitation of persister cells after superinfection with T2 phage both from plaques, from suspension cultures, and pre‐formed persister cells. Similar survival of a small population of E. coli cells was reported previously with T4 phage although it was concluded these ‘hibernators’ wake and are lysed by T4 phage, and the hibernators were not tested for persistence (Bryan et al., 2016). Similarly, a ‘phage tolerance response’ is elicited in non‐infected Bacillus subtilis by an unknown product of cell lysis, but this response is not persistence and is, instead, is a SigX‐mediated stress response (Tzipilevich et al., 2022).
CONCLUSIONS
In prior work, it was suggested that phages could be used to target persister cells (Pearl et al., 2008), whereas we conclude it may be better to combine phages (which will generate persister cells) with anti‐persister compounds like mitomycin C, which kills persister cells by cross‐linking their DNA (Kwan et al., 2015). Mitomycin C has been shown to kill numerous pathogenic persisters (Cruz‐Muñiz et al., 2017; Kwan et al., 2015) and has been shown to work well when combined with phage therapy for Klebsiella spp. (Pacios et al., 2021). Given the internal toxicity of mitomycin C, it is best combined with phages for topical applications where it is tolerated at 40 times the effective concentration for killing persister cells (Kwan et al., 2015).
Just like resistance to phage and survival due to phage‐inhibition systems, persistence should be considered as a possible deleterious outcome of phage therapy. In addition, our results show that phage infection is similar to other stresses (e.g. antibiotics; Kwan et al., 2013), oxidative stress (Hong et al., 2012), and starvation (Kim, Chowdhury et al., 2018) that causes persistence. Moreover, since phages are envisioned for use not only in medicine but also for food preservation, disinfection of surfaces, reduction of methane for global warming, and pest control in agriculture (García‐Cruz et al., 2023), it is likely persister cells will develop in these phage applications, too.
AUTHOR CONTRIBUTIONS
Laura Fernández‐García: Methodology; investigation; writing – review and editing. Joy Kirigo: Investigation; methodology. Daniel Huelgas‐Méndez: Investigation. Michael J. Benedik: Formal analysis. María Tomás: Funding acquisition. Rodolfo García‐Contreras: Investigation; formal analysis; supervision. Thomas K. Wood: Conceptualization; funding acquisition; writing – original draft; methodology; supervision; formal analysis.
FUNDING INFORMATION
This work was supported by both a Fulbright Scholar Fellowship and a Xunta de Galicia Postdoctoral Grant for LFG. This study has been funded by the Instituto de Salud Carlos III (ISCIII) through projects PI19/00878 and PI22/00323 and co‐funded by the European Union, by a Personalized and Precision Medicine Grant from the Instituto de Salud Carlos III (MePRAM Project, PMP22/00092), and by the Study Group on Mechanisms of Action and Resistance to Antimicrobials, GEMARA (SEIMC). R.G.‐C. was supported by DGAPA, PAPIIT‐UNAM (grant number IN200224), and IN200121.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
Supporting information
Table S1.
ACKNOWLEDGEMENTS
D.H.‐M. gratefully acknowledges the Programa de Doctorado en Ciencias Bioquímicas, UNAM, and the PhD scholarship 1103451 from Consejo Nacional de Ciencia y Tecnología (CONAHCyT, México). We appreciate the assistance of Erin Essington with editing the figures.
Fernández‐García, L. , Kirigo, J. , Huelgas‐Méndez, D. , Benedik, M.J. , Tomás, M. , García‐Contreras, R. et al. (2024) Phages produce persisters. Microbial Biotechnology, 17, e14543. Available from: 10.1111/1751-7915.14543
DATA AVAILABILITY STATEMENT
All data are available in the main and supplementary materials. The sequencing data have been deposited at NCBI under the PRJNA1134382 BioProject number.
REFERENCES
- Abedon, S.T. (2018) Detection of bacteriophages: phage plaques. In: Harper, D.R. , Abedon, S.T. , Burrowes, B.H. & McConville, M.L. (Eds.) Bacteriophages: biology, technology, therapy. Cham: Springer International Publishing, pp. 1–32. [Google Scholar]
- Baba, T. , Ara, T. , Hasegawa, M. , Takai, Y. , Okumura, Y. , Baba, M. et al. (2006) Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: the Keio collection. Molecular Systems Biology, 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleriot, I. , Blasco, L. , Pacios, O. , Fernández‐García, L. , López, M. , Ortiz‐Cartagena, C. et al. (2023) Proteomic study of the interactions between phages and the bacterial host Klebsiella pneumoniae . Microbiology Spectrum, 11, e03922–e03974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, D. , El‐Shibiny, A. , Hobbs, Z. , Porter, J. & Kutter, E.M. (2016) Bacteriophage T4 infection of stationary phase E. Coli: life after log from a phage perspective. Frontiers in Microbiology, 7, 1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz‐Muñiz, M.Y. , López‐Jacome, L.E. , Hernández‐Durán, M. , Franco‐Cendejas, R. , Licona‐Limón, P. , Ramos‐Balderas, J.L. et al. (2017) Repurposing the anticancer drug mitomycin C for the treatment of persistent Acinetobacter baumannii infections. International Journal of Antimicrobial Agents, 49, 88–92. [DOI] [PubMed] [Google Scholar]
- Dillon Marcus, M. , Sung, W. , Lynch, M. & Cooper Vaughn, S. (2018) Periodic variation of mutation rates in bacterial genomes associated with replication timing. MBio, 9, 10‐1128. Available from: 10.1128/mbio.01371-01318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐García, L. , Song, S. , Kirigo, J. , Battisti, M.E. , Petersen, M.E. , Tomás, M. et al. (2024) Toxin/antitoxin systems induce persistence and work in concert with restriction/modification systems to inhibit phage. Microbiology Spectrum, 12, e03323–e03388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐García, L. , Tomás, M. & Wood, T.K. (2023) Ribosome inactivation by Escherichia coli GTPase RsgA inhibits T4 phage. Frontiers in Microbiology, 14, 1242163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García‐Cruz, J.C. , Huelgas‐Méndez, D. , Jiménez‐Zúñiga, J.S. , Rebollar‐Juárez, X. , Hernández‐Garnica, M. , Fernández‐Presas, A.M. et al. (2023) Myriad applications of bacteriophages beyond phage therapy. PeerJ, 11, e15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S.H. , Wang, X. , O'Connor, H.F. , Benedik, M.J. & Wood, T.K. (2012) Bacterial persistence increases as environmental fitness decreases. Microbial Biotechnology, 5, 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.‐S. , Chowdhury, N. , Yamasaki, R. & Wood, T.K. (2018) Viable but non‐culturable and persistence describe the same bacterial stress state. Environmental Microbiology, 20, 2038–2048. [DOI] [PubMed] [Google Scholar]
- Kim, J.‐S. , Yamasaki, R. , Song, S. , Zhang, W. & Wood, T.K. (2018) Single cell observations show persister cells wake based on ribosome content. Environmental Microbiology, 20, 2085–2098. [DOI] [PubMed] [Google Scholar]
- Kwan, B.W. , Chowdhury, N. & Wood, T.K. (2015) Combatting bacterial infections by killing persister cells with mitomycin C. Environmental Microbiology, 17, 4406–4414. [DOI] [PubMed] [Google Scholar]
- Kwan, B.W. , Valenta, J.A. , Benedik, M.J. & Wood, T.K. (2013) Arrested protein synthesis increases persister‐like cell formation. Antimicrobial Agents and Chemotherapy, 57, 1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little, J.S. , Dedrick, R.M. , Freeman, K.G. , Cristinziano, M. , Smith, B.E. , Benson, C.A. et al. (2022) Bacteriophage treatment of disseminated cutaneous Mycobacterium chelonae infection. Nature Communications, 13, 2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T. , Fritsch, E.F. & Sambrook, J. (1982) Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Murray, C.J.L. , Ikuta, K.S. , Sharara, F. , Swetschinski, L. , Robles Aguilar, G. , Gray, A. et al. (2022) Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet, 399, 629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, R.D. , Assaf, R. , Brettin, T. , Conrad, N. , Cucinell, C. , Davis, J.J. et al. (2023) Introducing the bacterial and viral bioinformatics resource center (BV‐BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Research, 51, D678–d689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios, O. , Fernández‐García, L. , Bleriot, I. , Blasco, L. , Ambroa, A. , López, M. et al. (2022) Phenotypic and genomic comparison of Klebsiella pneumoniae lytic phages: vB_KpnM‐VAC66 and vB_KpnM‐VAC13. Viruses, 14, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios, O. , Fernández‐García, L. , Bleriot, I. , Blasco, L. , González‐Bardanca, M. , López, M. et al. (2021) Enhanced antibacterial activity of repurposed mitomycin C and imipenem in combination with the lytic phage vB_KpnM‐VAC13 against clinical isolates of Klebsiella pneumoniae . Antimicrobial Agents and Chemotherapy, 65, e00900–e00921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl, S. , Gabay, C. , Kishony, R. , Oppenheim, A. & Balaban, N.Q. (2008) Nongenetic individuality in the host–phage interaction. PLoS Biology, 6, e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecota, D.C. & Wood, T.K. (1996) Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. Journal of Bacteriology, 178, 2044–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlemoine, P. , Marcoux, P.R. , Picard, E. , Hadji, E. , Zelsmann, M. , Mugnier, G. et al. (2021) Phage susceptibility testing and infectious titer determination through wide‐field lensless monitoring of phage plaque growth. PLoS One, 16, e0248917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic, B. & Finland, M. (1961) Observations on bacteriophage typing of Pseudomonas aeruginosa . The Journal of Clinical Investigation, 40, 2064–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouillot, F. , Chomton, M. , Blois, H. , Courroux, C. , Noelig, J. , Bidet, P. et al. (2012) Efficacy of bacteriophage therapy in experimental sepsis and meningitis caused by a clone O25b:H4‐ST131 Escherichia coli strain producing CTX‐M‐15. Antimicrobial Agents and Chemotherapy, 56, 3568–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D.A. , Shoemaker, W.R. , Măgălie, A. , Weitz, J.S. & Lennon, J.T. (2023) Bacteria‐phage coevolution with a seed bank. The ISME Journal, 17, 1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S. & Wood, T.K. (2020a) Persister cells resuscitate via ribosome modification by 23S rRNA pseudouridine synthase RluD. Environmental Microbiology, 22, 850–857. [DOI] [PubMed] [Google Scholar]
- Song, S. & Wood, T.K. (2020b) ppGpp ribosome dimerization model for bacterial Persister formation and resuscitation. Biochemical and Biophysical Research Communications, 523, 281–286. [DOI] [PubMed] [Google Scholar]
- Strathdee, S.A. , Hatfull, G.F. , Mutalik, V.K. & Schooley, R.T. (2023) Phage therapy: from biological mechanisms to future directions. Cell, 186, 17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzipilevich, E. , Pollak‐Fiyaksel, O. , Shraiteh, B. & Ben‐Yehuda, S. (2022) Bacteria elicit a phage tolerance response subsequent to infection of their neighbors. The EMBO Journal, 41, e109247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beljouw, S.P.B. , Sanders, J. , Rodríguez‐Molina, A. & Brouns, S.J.J. (2022) RNA‐targeting CRISPR–Cas systems. Nature Reviews. Microbiology, 21, 21–34. [DOI] [PubMed] [Google Scholar]
- Van Goethem, M.W. , Marasco, R. , Hong, P.‐Y. & Daffonchio, D. (2024) The antibiotic crisis: on the search for novel antibiotics and resistance mechanisms. Microbial Biotechnology, 17, e14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M.C. , Reker, A.E. , Margolis, S.R. , Liao, J. , Wiedmann, M. , Rojas, E.R. et al. (2023) Restriction endonuclease cleavage of phage DNA enables resuscitation from Cas13‐induced bacterial dormancy. Nature Microbiology, 8, 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, T.K. (2022) The secret lives of single cells. Microbial Biotechnology, 15, 13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki, R. , Song, S. , Benedik, M.J. & Wood, T.K. (2020) Persister cells resuscitate using membrane sensors that activate chemotaxis, lower cAMP levels, and revive ribosomes. iScience, 23, 100792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Data Availability Statement
All data are available in the main and supplementary materials. The sequencing data have been deposited at NCBI under the PRJNA1134382 BioProject number.


