Summary
Human brain organoid models have emerged as a promising tool for studying human brain development and function. These models preserve human genetics and recapitulate some aspects of human brain development, while facilitating manipulation in an in vitro setting. Despite their potential to transform biology and medicine, concerns persist about their fidelity. To fully harness their potential, it is imperative to establish reliable analytic methods, ensuring rigor and reproducibility. Here, we review current analytical platforms used to characterize human forebrain cortical organoids, highlight challenges, and propose recommendations for future studies to achieve greater precision and uniformity across laboratories.
Graphical abstract
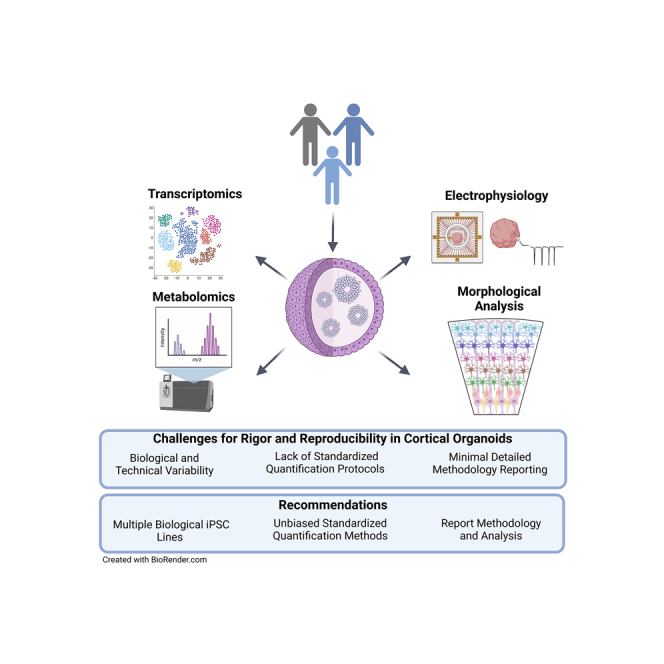
Sandoval et al. reviewed the current quantitative methods used for cellular, molecular, and functional analyses of brain organoid models, with a focus on cortical organoids. The authors identified the challenges posed by this powerful and innovative technology and proposed recommendations to improve the reliability and reproducibility of data generated across laboratories.
Introduction
Human brain development precisely orchestrates the formation of numerous functionally distinct cell types, regions, and connections, underpinning our unique social, cognitive, and sensory-motor abilities. This complex process is governed by intricate molecular mechanisms, the understanding of which is essential for developing effective treatments for various brain disorders. The challenge, however, lies in the inaccessibility of human brain tissues, especially from prenatal stages, and the limitations of direct experimentation, thus necessitating reliance on animal models. Yet, mounting evidence underscores substantial differences in developmental processes between humans, mice, and other model organisms (Sousa et al., 2017). Furthermore, human neuronal and glial populations exhibit greater cellular diversity than their rodent counterparts, and the uniqueness of many aspects of human metabolism, including drug metabolism, adds another layer of complexity. Consequently, gaining insights into the intricacies of the developing human brain necessitates the use of human models.
Human brain organoid models have surfaced as an appealing platform for studying human brain development and function. Derived from human pluripotent stem cells (hPSCs), these organoids can self-organize or be patterned to form highly enriched neural cell aggregates that resemble diverse regions of the central nervous system, including the cerebral cortex (Kadoshima et al., 2013; Lancaster et al., 2013, 2017; Renner et al., 2017), midbrain (Jo et al., 2016; Smits et al., 2019), striatum (Chen et al., 2022), hippocampus (Sakaguchi et al., 2015), cerebellum (Atamian et al., 2024; Chen et al., 2023; Muguruma et al., 2015), spinal cord (Lee et al., 2022), thalamus (Kiral et al., 2023), hypothalamus (Huang et al., 2021), brain-stem (Eura et al., 2020), choroid plexus (Pellegrini et al., 2020), and retina (Zhong et al., 2014). In addition, various research groups have pioneered the fusion of region-specific organoids to create assembloids, which emulate the functional relationships between distinct brain regions as observed in the developing human brain. Regional combinations include dorsal-ventral forebrain, cortex-striatum, cortex-thalamus, hypothalamus-pituitary, and cortico-motor assembloids (Andersen et al., 2020; Bagley et al., 2017; Birey et al., 2017; Kasai et al., 2020; Miura et al., 2020; Xiang et al., 2017, 2019), among others. Moreover, brain organoids can also be generated to include cell types with diverse embryonic lineages generated through separate differentiation protocols, such as microglia (Abud et al., 2017; Lin et al., 2018; Nzou et al., 2018; Ormel et al., 2018; Shi et al., 2020; Song et al., 2019) and endothelial cells (Ham et al., 2020; Pham et al., 2018). They exhibit network activities akin to in vivo multi-frequency oscillations (Trujillo et al., 2019), further confirming that brain organoids preserve aspects of human development both spatially and temporally in an in vitro setting (Dolmetsch and Geschwind, 2011). Thus, brain organoids have become invaluable for investigating both normal development (Bagley et al., 2017) and pathological conditions (Marton and Pasca, 2020; Sloan et al., 2018), including autism spectrum disorders (Villa et al., 2022), Down syndrome (Xu et al., 2019), Rett Syndrome (Gomes et al., 2020), and fragile X syndrome (Kang et al., 2021), showcasing their potential for disease modeling.
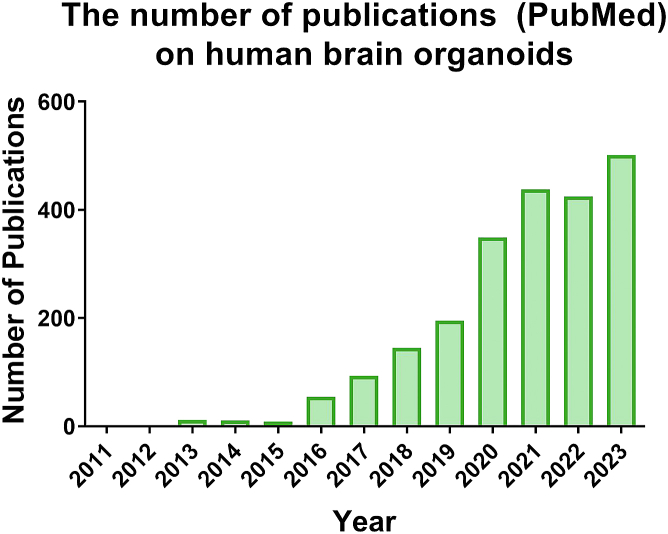
Despite their transformative potential, as evidenced by the proliferation of published data on organoids since their inception (Figure 1), major challenges remain in terms of fidelity and reproducibility of these models. As a starting point, the quality of hPSC lines used to generate the organoids and their cultivation conditions significantly impact the quality of organoids and the outcome of organoid models (Glass et al., 2023; Watanabe et al., 2022). Adherence to stringent standards for hPSC maintenance, including regular testing for pluripotency, the presence of pathogens, and genetic integrity, as recommended by the International Society for Stem Cell Research (ISSCR) (Ludwig et al., 2023), is imperative. Once generated, organoids must be quantitatively analyzed using precise methods and reported in sufficient detail for rigor and reproducibility. However, quantitative analysis of cellular phenotypes within organoids is highly challenging. This difficulty is underscored by the lack of standardized and clear guidelines for distinguishing between technical and biological replicates, leading to misleading statistical analyses if these variables are not factored into the analysis.
Figure 1.
The number of publications on human brain organoids in PubMed
To fully harness the potential of brain organoids as experimental models that bridge animal models and human brain development, it is thus critical to develop reliable and standardized methods for phenotypic analysis, ensuring rigor and reproducibility. This review provides a comprehensive overview of the current quantitative analyses of cortical organoids, the most commonly used organoid type, from cellular to molecular and functional analyses, offering guidelines to bolster the reliability and reproducibility of research within this burgeoning field.
Cellular analyses of organoids
Brain organoids serve as pivotal models for understanding the three-dimensional development of the human brain, necessitating precise evaluation of both their architectural features and the spatial distribution of different cell types. Although histological analyses of cell lineage markers indicate that organoids roughly resemble the complex human brain cytoarchitecture (Kadoshima et al., 2013; Lancaster et al., 2013; Pasca et al., 2015; Qian et al., 2016, 2020), the availability and consistency of quantitative data on these aspects remain limited. Here, we provide an overview of the current methodologies employed for quantifying cell types in cortical organoids generated using directed differentiation protocols (e.g., Birey et al., 2017; Kadoshima et al., 2013; Qian et al., 2016). Furthermore, recognizing the critical need for standardization in this emerging field, we propose a unified reporting framework to enhance rigor and reproducibility of brain organoid measurements.
Quantitative analysis of three-dimensional (3D) architecture and cell types
Several cell type-specific markers developed for in vivo brain analyses can be used for cellular and architectural comparisons between the developing human brain and brain organoid models (Table S1). These gene markers, including PAX6 (Thakurela et al., 2016) and SOX2 (Zhang and Cui, 2014) for the ventricular zone (VZ), EOMES and PPP1R17 for the apical subventricular zone (SVZ), HOPX for the outer subventricular zone (oSVZ), and RBFOX3/NeuN, TUBB3, and MAP2 for neurons in the cortical plate (CP) (Anastasaki et al., 2022), enable researchers to map organoid development with precision. Additional markers such as TBR1 (layer 6); BCL11B/CTIP2 (layer 5); SATB2 (layer 4); POU3F2/BRN2, CUX1, CUX2, and SATB2 (layers 2 and 3); and RELN (layer 1) are used to identify specific neuronal layers, while ALDH1L1, S100B, OLIG2, and MBP distinguish non-neuronal cells such as astrocytes and oligodendrocytes (Fleming and Coutts, 1988; Voulgaris et al., 2022). Notably, each cortical layer contains multiple cell types (Ma et al., 2022), most of which are best defined by a combination of markers rather than a single one. These markers, together with cell counting platforms such as CellProfiler, Imaris, and ImageJ (Tables S2 and S4), have been useful for qualitative comparisons of brain organoids and the human brain (Bhaduri et al., 2020; Gordon et al., 2021; Pollen et al., 2019; Zhou et al., 2024).
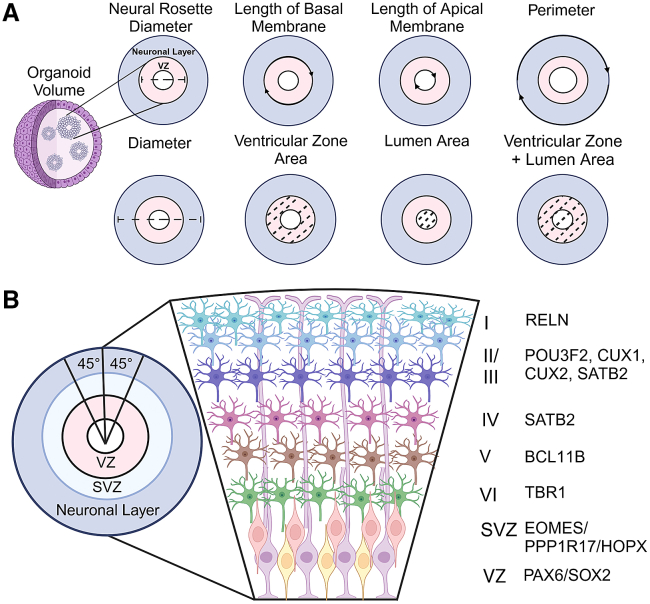
In addition to identifying cell types within the organoid, morphological analyses such as measuring the diameter, perimeter, area, and volume of whole organoids (Figure 2A) provide basic structural insights. During the early stages of differentiation, cortical organoids contain one or multiple neural rosettes. The cross-sections of neural rosettes resemble the cross-sections of neural tube which has a central lumen and several layers of cells comprising neural progenitor cells and sometimes immature neurons. To expand on these measures, Qian and colleagues developed a method to quantify the neuroepithelium of rosettes using SOX2 and BCL11B to differentiate between the ventricular zone (SOX2-positive progenitors), subventricular zone (SOX2-positive progenitors and BCL11B-positive neurons), and cortical plate (BCL11B-positive neurons) (Qian et al., 2016). After defining these layers, the authors performed three radial measurements, separated by a 45-degree angle, to quantify ventricular and cortical plate thicknesses (Figure 2B), important measures of organoid maturation. Another common technique to evaluate cellular distribution without the need to differentiate between specific layers based on cell markers is “cell binning” (Kang et al., 2021; Qian et al., 2020). By dividing the region of interest into discrete sections or “bins,” researchers can quantitatively assess cell types within each segment. Cell distribution along the layers can then be easily compared across experimental groups or conditions (Kang et al., 2021; Qian et al., 2016, 2020), offering a reliable view of organoid composition and development. Such measurements have proven instrumental in validating organoids as models of developmental disorders characterized by significant variations in cell numbers, such as microcephaly and macrocephaly (Brighi et al., 2021; Tang et al., 2021; Zhang et al., 2019).
Figure 2.
Morphological analyses that have been used for organoids
(A) Morphological analyses of organoids include organoid volume, organoid neural rosette number, diameter, perimeter, lumen area, ventricular zone area, ventricular zone + lumen area, length of basal membrane, and length of apical membrane.
(B) Cellular architectural analyses of organoids include the measurements of thickness of cellular layers resembling cortical layers in the brain as identified by common developmental markers. The thicknesses of cell layers were obtained by three measurements equally distanced by 45°.
Challenges
Quantitative assessment of brain organoids presents a multifaceted challenge, primarily due to the variability in organoid development and the lack of standardized reporting guidelines. For example, early differentiation of cortical organoids features neural rosettes, structures that resemble the neural tube (Zhang et al., 2001). However, unlike the developing brain that has a single neural tube, cortical organoids have more than one neural tube-like rosette structure and exhibit significant variability even among organoids differentiated from the same hPSC line (Glass et al., 2023). This variability is further complicated by the lack of clearly defined cortical layers in later developmental stages, diverging from the typically developing human brain in vivo. Furthermore, markers commonly used to identify certain cell populations within organoids may not be specific for a given cell type, necessitating coexpression analyses of multiple markers for accurate cell identification. Moreover, additional markers are needed to better define neuronal layers and cell types. Single-cell gene expression analysis is of particular value in confronting this issue, having identified new cell types and cell type-specific markers (Ma et al., 2022; Pollen et al., 2014). Compounding this issue, there is also a lack of specific and well-characterized antibodies for individual cell types. Techniques like single-molecule fluorescence in situ hybridization (FISH) (e.g., RNAscope) have partially addressed this by detecting RNA rather than protein, but the RNA expression does not always linearly correlate with the protein expression (Ma et al., 2022). Therefore, significant efforts should be dedicated to generating specific, reproducible, and open-source antibodies for human cell lineage analysis, improving the precision of organoid characterization. Finally, cell types, distributions, and architecture can vary widely, depending on the organoid generation protocol used (Kelava and Lancaster, 2016; Poli et al., 2019), highlighting the need for a more unified approach to organoid cultivation and analysis.
To date, there are no standardized reporting guidelines for quantitative analyses of organoids, in stark contrast to human and animal brain studies, which benefit from published atlases and well-established measurement methods (Hawrylycz et al., 2012; Poli et al., 2019). This lack of standardization is evident in the reporting practices of organoid studies, where less than half of published brain organoid studies include quantitative analyses of cell types and architecture (Bremond Martin et al., 2021). Furthermore, the details provided in these studies often lack the detail required for reproducibility, such as the number of organoids analyzed per condition, the number of regions quantified within each organoid, and the methods employed for data analysis. Often, single cryosections from a single organoid are quantified, without consideration of the entire organoid or differentiation batch, introducing potential sample bias and further complicating comparative analyses across studies.
Optical clearing for morphological analysis of organoids
Various imaging techniques are used for analyzing organoids, including confocal and light sheet microscopies (Dekkers et al., 2019; Ntziachristos and Razansky, 2010; Shnaider and Pristyazhnyuk, 2021). Confocal microscopies require thin-section preparations (4–20 μm), which makes 3D reconstruction challenging. On the other hand, light sheet microscopy, when combined with optical clearing methods (Table S3), enables sufficient in-plane resolution and deep light penetration for both qualitative and quantitative analyses of neuroanatomical structures in intact whole organoids (Susaki and Takasato, 2021). Optical clearing removes the bias of choosing individual cryosections and eliminates sectioning-induced artifacts, such as tissue distortion (Albanese et al., 2020). Nevertheless, it is important to optimize antibody concentration, permeabilization, and incubation times, since the concentrations may be higher and incubations longer when using optical clearing compared to conventional staining practices. For further detail, Smrek and Stelzer offer recommendations on immunostaining whole mounts based on different fixation and permeabilization criteria (Smyrek and Stelzer, 2017). Once an optimized framework has been established, tissue clearing for organoids allows for fast and high-throughput characterization.
A range of clearing techniques (Kolesova et al., 2021; Susaki and Takasato, 2021) have been adapted for organoids, including methods based on organic solvents, hydrophilic reagents, and hydrogels. The choice of the optimal clearing method depends upon the experimental goals and the method of organoid generation. For example, a comparison of three clearing methods on neurospheres found that the ClearT2 protocol preserved structural integrity more effectively, whereas ScaleSQ caused tissue expansion, and SeeDB resulted in tissue shrinkage (Boutin and Hoffman-Kim, 2015). In contrast, different studies showed tissue shrinkage with ClearT2 in spheroids (Diosdi et al., 2021; Nurnberg et al., 2020), and another comparison showed that, for on-chip clearing, SeeDB and ScaleSQ outperformed ClearT2 (Grist et al., 2016). However, SeeDB and ScaleSQ had drawbacks, such as induced autofluorescence and tissue expansion, respectively. These data underscore the importance of conducting comparative analysis of clearing methods to ascertain the optimal approach for specific projects.
Cleared organoids allow for extraction of multiple measurements. These range from large features such as the organoid volume and shape to finer details such as the volume of VZ-like zones, which can be generated by creating surfaces around cells positive for PAX6 and NCAD (Glass et al., 2023). In addition, quantification of cell types based on markers and neuronal migration is possible across the 3D image (Meng et al., 2023).
Challenges
Cleared organoids generate large amounts of data with greater complexity than two-dimensional (2D) immunohistochemical sections; therefore, data analysis and management plans are paramount. However, data analysis poses a significant challenge to image-based quantitation using cleared organoids, especially when stitching multiple z stacks of images into a cohesive 3D structure and aligning multiple channel images to correct chromatic aberration (Krupa et al., 2021). Stitching usually involves intentionally overlapping image portions and thus is prone to errors, including doubling of image features (like nuclei) that can alter counts along the stitching seams. Stitched stacks must therefore be visually checked for accuracy. Moreover, despite the existence of numerous tools for 2D nuclear segmentation (Greenwald et al., 2022), the options for 3D segmentation are limited (Diosdi et al., 2021; Stringer et al., 2021), making it difficult to accurately separate nuclei among densely packed cells even in high-resolution images (Borland et al., 2021).
Lack of standardized methods for labeling can lead to significant variability across studies. For example, one of the issues in the analysis of organoid morphology is whether necrotic cores—regions of dying or dead cells at the center of larger organoids, typically resulting from insufficient nutrient perfusion—should be included in volume measurements. This practice is contentious because these cores lack healthy or viable cells, raising questions about their relevance to meaningful morphological assessments. Consequently, new analytical tools are needed to automatically count nuclei within regions densely packed with cells, identify distinct zones within the organoid such as VZ- or CP-like areas and necrotic cores, and trace axonal connections.
Molecular analyses of organoids
Transcriptome analyses
Almost every aspect of cellular physiology is dependent on and regulated by the transcriptome. Advances in next-generation sequencing, such as bulk and single-cell RNA sequencing (scRNA-seq), have unlocked the potential to delineate transcriptomic signatures of multiple types of cells across cell development, disease states, and specific brain regions. However, despite its promise, the application of human brain transcriptomics is constrained by the limited availability of primary human tissue. Thus, human brain organoids emerge as a powerful surrogate, facilitating the exploration of the transcriptome at the single-cell level. Indeed, scRNA-seq data from cortical and retinal organoids are concordant with scRNA-seq data from the developing human cerebral cortex and retina, respectively (Fernando et al., 2022), showcasing organoid transcriptomic fidelity. In addition, different organoid models—cortical vs. thalamic vs. medial ganglionic eminence organoids—have distinct transcriptome profiles (Xiang et al., 2017, 2019), further supporting their fidelity as model systems. One recent study built an integrated human neural organoid cell atlas that has harmonized a number of scRNA-seq transcriptomic data from organoids and mapped developing human brain data onto the atlas for comparison to primary cell types (He et al., 2023b).
Organoid transcriptomics not only illuminate development in health and disease but also reveal the maturation of cellular composition over time (Quadrato et al., 2017; Sun et al., 2021). For instance, analysis of 6-month-old organoids unveiled gene expression profiles indicative of cell types absent in 3-month-old organoids (Gordon et al., 2021), demonstrating their evolving complexity. Most recently, trajectory inference approaches elucidated the differentiation sequence of cell types within cortical organoids (He et al., 2023a; Qiu et al., 2017), such as radial glia differentiation into mature excitatory or inhibitory neurons (Velasco et al., 2019).
While scRNA-seq has predominantly been used to study mRNA, its versatility extends to studies of cell type-specific long non-coding RNAs (lncRNAs) (Field et al., 2019), expanding the relevance of this method for mechanistic insights of human brain development and pathology. These methods can reveal new insights into numerous types of disorders (Klaus et al., 2019; Leong et al., 2022; Sawada et al., 2020), marking a transformative era in our understanding of cellular dynamics in the context of neurodevelopmental health and pathology.
Challenges
Organoid variability poses a challenge for transcriptomics studies, largely due to differing growth conditions and extracellular matrix components across labs (Chiaradia et al., 2023; Martins-Costa et al., 2023; Pagliaro et al., 2023). Certain cell types such as neuroprogenitors, excitatory neurons, and astrocytes, are consistently detected across models, while others, such as inhibitory neurons, appear sporadically. It is thus imperative that culture conditions are standardized to enhance comparisons across studies and models.
In addition, single-cell transcriptomics analyses of organoids are mostly done using large-scale platforms such as droplet-seq (10× Genomics) (Yang et al., 2024) or split-seq (Parse Biosciences) (Glass et al., 2023). Although many cells can be sampled, the depth of sequencing per single cell is low, which undermines in-depth cell type or differential gene expression analyses requiring the presence of genes with low expression levels.
Furthermore, data analysis pipelines such as Cell Ranger, while constantly evolving, present challenges in experimental application and dataset comparisons. A recently published machine learning tool, Brain and Organoid Manifold Alignment (BOMA), represents a step forward in integrating scRNA-seq datasets to identify common or diverse developmental trajectories between human organoids and brains (He et al., 2023a).
Finally, scRNA-seq alone is limited in defining cell types and cell lineages. The integration of multiomics approaches offers a more nuanced view of gene regulatory networks critical for cellular functions. For example, combining transcriptomic data with chromatin accessibility measurements (e.g., single-cell assay for transposase-accessible chromatin with sequencing or scATAC-seq) has revealed aspects of gene regulatory mechanisms such as linking transcription factors and distant regulatory elements to target genes, which transcriptomics alone might miss (Zhang et al., 2022). Despite their promise to enrich our understanding of molecular mechanisms governing early human brain development, single-cell multiomic technologies face unique challenges such as high noise and dropout rates, batch effects, and the complexity of data integration. The field still awaits the development of computational methods capable of effectively integrating single-cell multiomics data and comparing brains and organoids, which, despite the current high costs, would significantly advance our comprehension of the genetic and epigenetic underpinnings of brain organoid development and function.
Metabolic analyses
The brain, as the most energy-demanding organ in the body, demands a continuous and substantial supply of adenosine triphosphate (ATP) to maintain its complex functions. This energy requirement underpins not only the basic cellular activities but also the higher-order processes such as neuronal firing, essential for cognitive and sensory-motor functions (Rangaraju et al., 2014). Moreover, brain diseases often manifest with alterations in diverse cellular processes, including metabolism, which is highly dynamic and specific to cell type (Chong et al., 2022). Notably, metabolism is profoundly influenced by cellular environment. For organoid cultures, this environment depends on external conditions such as culture media composition and the levels of oxygen and carbon dioxide. Thus, to understand—and potentially target—the effects of metabolism on brain development and disease, we must ensure that organoid culture conditions are as close to physiological conditions as possible.
Various methods are available to assess metabolic activity and metabolic stress, as detailed in Table S4. Some of these methods represent endpoint analyses, while others can be conducted on live cells. Widely utilized methods include colorimetric assays and biochemical measures of mitochondrial activity. Additionally, emerging technologies for comprehensive metabolome and lipidome assessments encompass targeted and untargeted metabolomics, and imaging mass spectrometry for examining spatial distribution of metabolites.
Microscopy assays
Among live assays, fluorescence microscopy and fluorescence lifetime imaging microscopy (FLIM) stand out as valuable tools for non-invasive real-time studies of metabolic activity (Okkelman et al., 2020). For example, specific fluorescent probes allow estimation of the optical redox ratio in a given cell. This method can indicate the mitochondrial nicotinamide adenine dinucleotide (NAD+/NADH) redox potential (Blacker and Duchen, 2016), where higher redox ratio suggests prevalence of oxidative phosphorylation (OXPHOS) (Meleshina et al., 2017), as well as oxidized glutathione (Jiang et al., 2017, 2019), which can reflect reactive oxygen species (ROS) production in a cell. In turn, FLIM measures the contribution of exogenous and endogenous fluorophores over time (Meleshina et al., 2016) and has been employed to estimate the relative amounts of free or enzyme-bound NAD(P)H in brain spheroids (Kashirina et al., 2021; Stringari et al., 2012).
Plate-based assays
Specific glucose and lactate assays offer the capability to measure the respective metabolite concentration in both tissue and culture medium (Cho et al., 2021). Simultaneously, genetically encoded pH sensors, designed to selectively target distinct cellular compartments (Martynov et al., 2018), assess tissue pH (Kashirina et al., 2021). Collectively, these assays provide information on tissue acidity, indirectly indicating anaerobic conditions and cellular stress. Furthermore, Seahorse analysis can provide information on mitochondrial respiration by measuring oxygen consumption rate and glycolysis by measuring extracellular acidification rate. Although Seahorse is primarily designed for monolayer cultures, new protocols for measuring bioenergetics in intestine-derived and brain-derived organoids are emerging. Methods involve dissociating organoids to monolayer culture before Seahorse analysis and sequentially administrating mitochondrial inhibitors (the ATP synthase inhibitor oligomycin, the proton ionophore carbonyl cyanide-p-trifluoromethoxyphenyl hydrazone [FCCP], the mitochondrial complex I inhibitor rotenone, and the mitochondrial respiratory chain inhibitor antimycin A) and blocking mitochondrial membrane potential and electron transport (Le et al., 2021; Ludikhuize et al., 2021). These complex experiments can tease out specific mitochondrial processes that collectively occur in each organoid sample.
Nanoparticle-based assays
Intra-organoid oxygen levels can be measured by oxygen-sensitive phosphorus nanoparticles, which emit phosphorescence based on oxygen levels. To implement this technique, nanoparticles are diffused into brain organoids incubated in medium containing Pt(II) meso-tetra(pentafluorophenyl)porphine (PtTFPP)-poly(urethane acrylate nonionomer) (PUAN) (Choi et al., 2012). Higher oxygen levels result in lower phosphorescence of the nanoparticles, which is attributed to collisional quenching between oxygen molecules and PtTFPP (Kashirina et al., 2021).
Metabolomics analyses
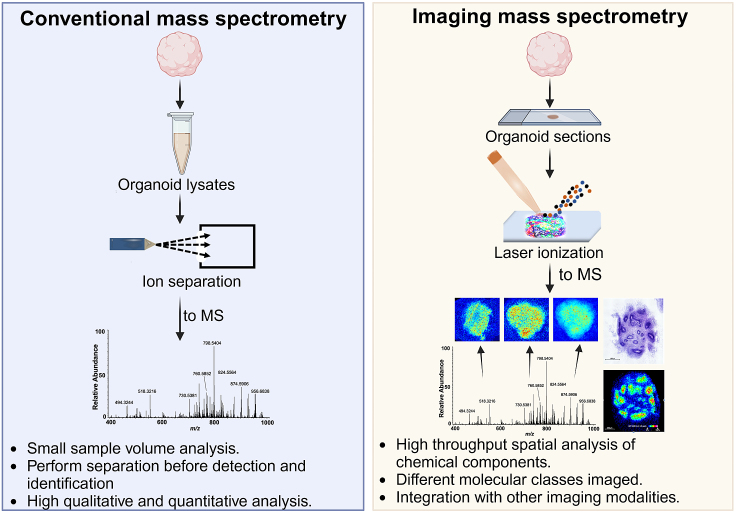
To comprehensively assess metabolism in organoids, targeted and untargeted metabolomics are powerful approaches. Although extensively employed in cancer models, the utilization of these methods in human brain organoid models has been relatively limited (Gomez-Giro et al., 2019; Notaras et al., 2021; Sun et al., 2021). Targeted analysis of the glycolytic and fatty acid pathways, OXPHOS, and other metabolites would provide specific metabolome insights that, coupled with transcriptome analyses, would generate the precise metabolic landscape in organoid models. Most recently, imaging mass spectrometry, which provides spatial localization of hundreds of metabolites in a given sample at the same time, without the need for special probes or labeling, generated the first map of lipids in a human brain organoid model (Cappuccio et al., 2023) (Figure 3). Imaging mass spectrometry has been envisioned as a spatial metabolomics tool, paving the way for future studies of the metabolic/lipidomic status of diverse organoid models over time. Despite technical challenges, these mass spectrometry-based methodologies offer the most comprehensive insights into organoid metabolomes, particularly when coupled with transcriptome analyses of enzymes. Embracing these advanced techniques holds promise for unraveling the complex metabolic signatures and organoid stress responses in human brain organoid models.
Figure 3.
Distinct mass spectrometry technologies offer cell type-specific analyses of the metabolome
Conventional mass spectrometry (liquid chromatography-mass spectrometry [LC-MS]) detects the metabolome content in a low-volume sample lysate, while imaging mass spectrometry (IMS) maps the distribution of metabolites in tissue sections, thus providing spatial information on thousands of label-free metabolites.
Challenges
Metabolic studies in organoids are challenging and intrinsically linked to the complexity of metabolic processes and the constraints of current methodologies for organoid growth and analysis. These challenges influence the precision, consistency, and depth of our understanding of metabolism in these complex models.
One of the most significant challenges in organoid culture is the diffusional limitation of oxygen and nutrients (Bhaduri et al., 2020; Qian et al., 2016). As organoids grow in size, their innermost cells may experience hypoxia and nutrient deprivation, leading to a cascade of cellular stress responses. Several studies have reported markers of oxidative stress in organoids, such as DNA damage, lipid peroxidation, protein oxidation products, and unfolded protein response as well as increased markers of glycolysis (such as the glycolysis PGK1 gene), endoplasmic reticulum (ER) stress (such as ARCN1 and GORASP2 genes), and electron transport pathways (Bhaduri et al., 2020; Huang et al., 2022b; Pollen et al., 2019). Even though this process starts in the inner parts of the organoid, it alters the metabolic landscape throughout, as dying cells release their acidic content into the tissue. This process can skew metabolic assessments and has necessitated development of strategies to enhance nutrient and oxygen penetration (Giandomenico et al., 2019; Qian et al., 2020).
Creating physiological metabolic conditions is a challenge of its own because metabolism—and particularly generation of ATP for energy demands—is cell type dependent. Achieving a cell’s ATP demands hinges on the delicate balance between glycolysis and OXPHOS, the former distinctly favored by proliferating cells, such as neuroprogenitors, and the latter by postmitotic cells, such as neurons (Vander Heiden et al., 2009). This metabolism is reflected in mitochondrial adaptations: proliferating cells have smaller, shorter mitochondria favoring glycolysis and lipid metabolism, while postmitotic cells contain complex, elongated mitochondria, multiplied in number to accommodate the increased demand for OXPHOS. The coexistence of proliferating and non-proliferating cell types in the organoids—albeit at varying degrees—presents a challenge in ensuring optimal nutrient supplies to meet their respective metabolic demands across developmental stages, as well as assessing metabolism of specific cell types.
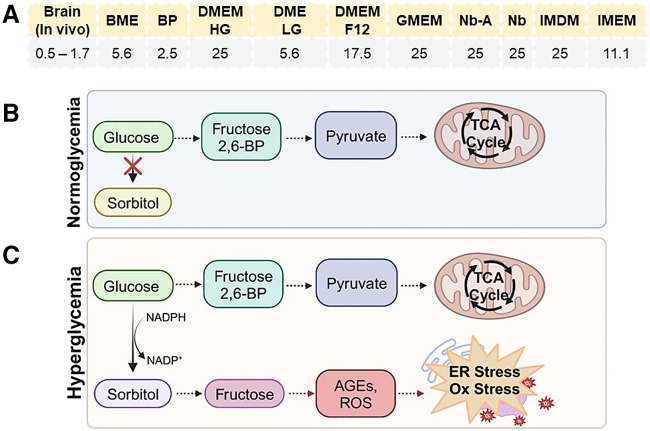
Indeed, the standard culture conditions often do not accurately mimic the in vivo environment of human tissues, inadvertently contributing to stress responses in cells. For example, current standard brain organoid culture media are hyperglycemic (Figure 4A), which may heighten neuronal stress as cells increase oxygen consumption to metabolize the high glucose content, consequently generating more ROS (Yu et al., 2006) (Figures 4B and 4C). Further, glucose concentration influences the balance between glycolysis and OXPHOS, which in turn affects neuronal differentiation rates (Chen et al., 2018) and function (Bardy et al., 2015). The variable glucose culture conditions across organoid protocols might contribute to some observed variability between studies. In turn, oxygen concentration also plays a key role in cellular metabolism. Cortical organoids grown in hypoxic (<1% O2) or hyperoxic (40% O2) conditions, regardless of the glucose concentration in the media, both demonstrate oxidative stress, disruption of protein homeostasis, and cell death (Pasca et al., 2019).
Figure 4.
Hyperglycemia causes cellular stress via the polyol pathway
(A) Glucose concentrations (mM) of commonly used basal media are significantly higher compared to those in the brain (Brain, in vivo). BME, basal medium Eagle. BP, BrainPhys medium. DMEM, Dulbecco’s modified Eagle medium. HG, high glucose. LG, low glucose. GMEM, Glasgow minimum essential medium. Nb, Neurobasal medium. IMDM, Isocove’s modified Dulbecco’s medium. IMEM, improved minimum essential medium.
(B) Under normoglycemia, glucose is phosphorylated to produce glucose 6-phosphate to initiate glycolysis, which feeds pyruvate into the tricarboxylic acid (TCA) cycle.
(C) Under chronic hyperglycemia, the enzyme that converts glucose into glucose 6-phosphate, hexokinase, becomes saturated. As glucose builds up, aldose reductase converts glucose into sorbitol, which is then converted into fructose. Fructose accumulation increases advanced glycation end products (AGEs) which cause ER stress. In addition, the conversion of glucose to sorbitol depletes the nicotinamide adenine dinucleotide phosphate (NADPH) required for the antioxidant system, thereby increasing oxidative stress. Hyperglycemia also leads to an increase in the overall rate of glycolysis with a concurrent decrease in the rate of oxidative phosphorylation, termed a glycolytic shift, which is associated with numerous disease phenotypes across tissue types.
The variability in organoid generation protocols, culture conditions, and analytical methods can lead to inconsistencies in metabolic data across studies. Indeed, reports on metabolic stress in organoids are quite diverse, with some highlighting substantial stress and questioning the validity of organoid models (Bhaduri et al., 2020; Huang et al., 2022b; Tanaka et al., 2020) and others not reporting significant changes in their models (Gordon et al., 2021; Samarasinghe et al., 2021). The issue is compounded by reliance on transcriptomic data, which provide valuable insights but do not capture enzyme activity, a key aspect of metabolic pathways. Enzymes are activated when the substrate is available and all other conditions for an enzymatic reaction are met, and this information can only be gleaned from metabolomic analyses. Further, in contemporary transcriptome analyses, stressed/apoptotic cells are filtered out (Vertesy et al., 2022), but the reported number of filtered cells is rarely provided, even though it could serve as a metric for organoid stress.
Finally, studies of metabolism in organoid models face technical limitations of current assays as many of them have been designed for monolayer cell cultures and may not be directly applicable or fully informative for three-dimensional organoid cultures.
Overall, cellular stress, including mitochondrial stress, emerges as a key player in organoid development, growth and maturation, cell specification, and fidelity with respect to fetal tissue counterparts. This intricate relationship underscores the importance of understanding the interplay between environmental factors, cellular metabolism, and stress responses in organoid models and integrating metabolic assessments into their analysis. Addressing these complexities requires concerted efforts to refine organoid culture techniques, develop standardized metabolic assessment protocols and data integration analyses, and foster interdisciplinary collaborations to enhance the fidelity of brain organoids for studies of human brain development and diseases.
Functional measurements
Brain organoids can be maintained in vitro for months, which enables analysis of functional maturation, a critical aspect when modeling neurodevelopmental disorders. Multiple technologies have been used to measure both single-cell and network activities in neurons in brain organoids. The most common approaches include traditional patch-clamp methods, calcium imaging, and multielectrode arrays (MEAs) (Table 1).
Table 1.
Methods for analyzing electrophysiology of organoids
| Ephys method | Organoid preparation | Age | References |
|---|---|---|---|
| Calcium imaging: fluo-4 | Dissociated and sliced | 64 days | Lancaster et al. (2013); Lancaster et al. (2017) |
| Voltage and current clamp, calcium imaging: fura-2 and fluo-4 | Sliced and dissociated | 90–130 days | Pasca et al. (2015) |
| Whole-cell patch clamp and calcium imaging: GCAMP6f | Sliced culture | 77–84 days | Watanabe et al. (2017) |
| Calcium imaging: GCaMP6f | Whole-mount and in vivo | 78–141 days | Mansour et al. (2018) |
| Calcium imaging: fluo-4 | Whole-mount | 76–104 days | Sakaguchi et al. (2019) |
| Current clamp | Whole-mount | 120–150 days | Sun et al. (2019) |
| Whole-cell patch clamp | Whole-mount | 76–91 days | Wu et al. (2022) |
| Voltage and current clamp and MEA | Whole mount | 42–224 days | Trujillo et al. (2019) |
| Whole-cell patch clamp, current clamp, and MEA | Whole-mount and sliced | 150 days | Giandomenico et al. (2019) |
| MEA | Whole-mount | 7–21 days | Hartmann et al. (2023) |
| Kirigami electronics (KiriE) | Whole-mount | 75–179 days | Yang et al. (2024) |
Patch-clamp recording
Patch clamp is the most widely used electrophysiology technique, offering unparalleled insights into individual cell activities with high temporal and spatial resolution (Cummins et al., 2009). Patch clamp adapts to various organoid preparations—whole-mount, sliced, or dissociated—with each method presenting unique benefits and drawbacks (Table 1). Whole-mount organoid recordings offer a holistic view of organoid activity but are limited to surface neurons. Organoid sectioning or cutting partially conserves the 3D organoid cytoarchitecture, but it risks severing some neuronal connections and potentially damaging neurons, which may alter electrophysiological properties. Finally, organoid dissociation allows access to the innermost neurons at the cost of disrupting the 3D structure and neuronal networks.
Functional maturation of neurons within organoids has been documented through patch-clamp studies. Trujillo et al. (2019) and Giandomenico et al. (2019) independently provided initial proof that neurons in organoids exhibit spontaneous and evoked electrophysiological activities indicative of developmental progression, mirroring human cortical development. Extending beyond developmental maturation and circuit organization insights, whole-cell recordings can measure intrinsic biophysical properties of human neurons, revealing critical differences between human and rodent models such as membrane capacitance (Eyal et al., 2016), dendritic input resistance (Beaulieu-Laroche et al., 2021), and dendritic compartmentalization (Beaulieu-Laroche et al., 2018). These functional differences are critical for understanding human-specific neuronal properties and their implications in neurological conditions such as epilepsy (Rich et al., 2022; Wu et al., 2022) and Angelman syndrome (Sun et al., 2019), showcasing organoids as a relevant model for human brain disorders.
Finally, patch-seq, a novel advancement, merges patch clamp with single-cell sequencing (Cadwell et al., 2016), broadening our understanding of the molecular underpinnings of neuronal function. Patch-seq has elucidated gene expression signatures underlying neuronal activity across various brain slice models, from mouse (Foldy et al., 2016) to non-human primate (Gao et al., 2023) and human (Berg et al., 2021), as well as in hPSC-derived neurons (van den Hurk et al., 2018). By correlating electrical activity with transcriptome profiles at the level of individual neurons, patch-seq offers a comprehensive view of neuronal diversity and function in organoids. Rigorous design based on statistical power and transparent reporting are essential for robust patch-seq and patch-clamp studies, ensuring meaningful and replicable results.
Challenges
Patch-clamp recording requires specific expertise, is laborious, and has low throughput. For organoids, patching neurons located in the inner part of organoids has been challenging. In addition, patch-clamp recording is not suitable for studying neural network activities among neurons within the 3D structure of organoids.
Calcium imaging
Traditional patch-clamp electrophysiology, known for its high temporal resolution, faces limitations in throughput and broad network activity measurements. Calcium imaging overcomes both challenges, albeit with lower temporal resolution. Fluorescent calcium indicators detect changes in intracellular calcium concentration, indicative of action potentials as shown by simultaneous patch-clamp and calcium imaging methods (Wei et al., 2020). By selecting the correct calcium indicator and imaging platform, calcium imaging can be effectively used to detect neuronal activity and synaptic plasticity (Grienberger and Magee, 2022). The rapidly expanding catalog of calcium indicators, alongside advancements in cell type-specific targeting, has made calcium imaging highly accessible to many neuroscientists.
Calcium indicators fall into two categories: chemical indicators and genetically encoded indicators (GECIs) (de Melo Reis et al., 2020), each of which have been successfully used in cortical organoids (Table 1). However, chemical indicators diffuse very slowly and inefficiently through the organoid and are predominantly absorbed by glia, limiting their use for long-term imaging essential for neurodevelopmental studies. In turn, GECIs—particularly the genetically encoded calcium indicator ((GCaMP) family—offer advantages over chemical indicators by allowing specific, long-term neuronal expression and imaging (Zhang et al., 2023). Viral delivery methods enhance the applicability of GECIs, enabling targeted cell type analysis and deep organoid imaging using multi-photon microscopy (Grienberger and Konnerth, 2012). This approach has elucidated functional neuronal networks within organoids (Sakaguchi et al., 2019; Watanabe et al., 2017) and synchronous calcium oscillations in cortical-medial ganglionic eminence assembloids (Xiang et al., 2017). Calcium imaging reports synaptic maturity and integration of human organoids engrafted into animal models, including aberrancies in disease states, which cannot be observed in in vitro models (Mansour et al., 2018). Calcium imaging can be paired with optogenetics (Andersen et al., 2020), expanding the scope of complex research questions.
Challenges
Although calcium imaging provides many advantages, calcium indicators are too slow to distinguish fast neuronal spikes, and they also sequester intracellular calcium, potentially altering signal intensities (McMahon and Jackson, 2018). Voltage imaging, offering faster kinetics capable of capturing individual action potentials, has recently emerged as an alternative (Kiral et al., 2023; Puppo et al., 2021), despite its lower signal-to-noise ratio compared to calcium imaging and the need for specialized microscopy equipment.
MEAs
MEA platforms offer a novel alternative to studying functional neuronal networks, bridging the gap between traditional in vitro and in vivo neural analyses. Comprising arrays of electrodes—embedded in culture plates or implanted into animals—MEA records extracellular voltage changes, providing insights into both single-cell and collective neuronal network activity. This high-throughput technology allows non-invasive monitoring of neural activity over time, eliminating the need for cultures to undergo disruptive procedures during data collection.
MEA platforms vary in their electrode density, catering to different research needs. MEAs range from the widely used systems such as the Axion Biosystems Maestro Pro—accommodating multiple well formats (6-, 24-, 48-, or 96-well tissue culture plates) and recording from 8 to 64 electrodes per well—to high-density options such as the MaxWell MaxOne, boasting up to 26,400 electrodes per well to enable increased spatial and temporal resolution, quantification of action potential propagation speeds, single-cell functional resolution, and even quantification of neurite arborization. High-density MEA can also be paired with optogenetic techniques, stimulations, and axonal tracking for examining 2D and 3D neuronal cultures derived from hPSCs and conducting drug screening or neurotoxicity tests (Hartmann et al., 2023; Shafer, 2019; Shen et al., 2023; Guo et al., 2023; Yokoi et al., 2022), thanks to their ability to capture intricate details of neural activity and network organization over extended periods.
The utilization of MEA for organoid research presents both unique opportunities and challenges. Studies utilizing in vitro MEA have demonstrated a progressive increase in organoid electrical activity from 2 to 8 months in culture, mirroring brain development in vivo (Trujillo et al., 2019). Advanced MEA setups have enabled detection of not only network activity but also long-distance spatial connections within organoid slices, rather than just neighbor-to-neighbor connections among neurons. Low-pass filtering of electrical activity in organoids, particularly from penetrating electrodes that span several layers of the cortical sheet, may reveal whether local field potentials (LFPs), a hallmark of complex network organization in animal studies, occur in organoids. For instance, a recent study identified spatial and temporal correlations in the theta frequency (4–12 Hz) in brain organoid slices (Sharf et al., 2022), suggesting that organoid neurons may encode information. Further, MEA has been used to examine the activity of engrafted human organoids in a rodent model in vivo, providing single-cell spike data (Mansour et al., 2018).
Challenges
Commercially available MEA systems have several limitations, particularly when applied to 3D structures like organoids. Primarily, these systems are designed for 2D cultures, limiting their effectiveness in capturing the full scope of activity within a 3D organoid. As a result, MEAs provide measures of the organoid surface or cells that have migrated outward over time. While it is possible to dissociate or slice organoids, doing so disrupts potentially important neuronal connections, presenting a significant drawback. Furthermore, MEA plates are coated with an extracellular matrix to secure the organoid, requiring optimization for long-term recordings. To circumvent these challenges, innovative 3D MEA configurations have been developed to enable more comprehensive activity measurements within organoids (Choi et al., 2021; Shin et al., 2021). Comparative studies of organoids on 2D low-density (8x8), 3D low-density (8x8), and 2D high-density (64x64) MEA have shown that 2D high-density MEA are more adept at detecting activity within organoids (Muzzi et al., 2023), suggesting that higher electrode density may offer better suitability for organoid recordings. Emerging MEA platforms, such as mesh nanoelectronics and organoid-integrating scaffolds, promise to facilitate 3D and high-resolution recordings within cortical organoids (Huang et al., 2022a; Liu et al., 2020; Passaro and Stice, 2020; Yang et al., 2024). However, the application of these novel approaches is limited.
Recommendations for comprehensive evaluation of organoids
Human brain organoids have emerged as important models for studying early human development and pathology, paving the way toward personalized precision medicine. While organoids may not recapitulate all aspects of brain development and function, their value in probing human development and developmental disorders is clear. Challenges such as reproducibility across labs and the absence of standardized characterization approaches persist. The recent ISSCR standards for stem cell research (Ludwig et al., 2023) are a step forward, offering guidelines to elevate the quality of organoid research by establishing clear criteria for stem cell quality, sample sizes, and reporting standards. In addition, new technologies, such as high-throughput and high-resolution microscopy; single-cell spatial genomics, proteomics, and metabolomics; and 3D MEA, offer excellent analytical tools with high resolution needed for characterization of these complex models. However, clear standardized guidelines for organoid analyses are still lacking, and here we propose a structured approach to organoid analysis to address variability and improve reproducibility (Table 2).
Table 2.
Recommended standards for quantitative analyses of organoids
| Methods of analysis | Minimum recommendation | Ideal recommendation |
|---|---|---|
| All methods | When possible use hPSC lines derived from multiple individuals for each condition; if hPSC lines from multiple individuals are not available, differentiate organoids from multiple clones of the same hPSC line. Alternatively, perform independent differentiations from the available hPSC lines and ensure the use of multiple organoids from each differentiation. | As recommended by ISSCR, power analysis should be used to determine sample size. |
| Report number of organoids used from each differentiation batch and from each hPSC line. Specify criteria for any excluded organoids. | Report number of organoids used from each differentiation batch and from each hPSC line in figure legends and dedicated supplemental tables. Specify criteria for any excluded organoids. | |
| Report codes, packages, and analysis software | Use or develop open-source methods; Report codes, packages, and analysis software. | |
| Morphological and cell lineage | Random sampling of 3–5 serial sections | Perform organoid clearing to analyze 3D reconstructions |
| Sample from 3 to 5 organoids in each differentiation batch | Sample size pre-determined by trial experiments and power analysis | |
| Provide quantification of cell lineage markers used. | Use unbiased automated or semi-automated methods | |
| Transcriptomics | Report number of organoids used from each differentiation batch and hPSC line. | Combine multiomic approaches (e.g., scATAC-seq, spatial genomics) |
| Functional | When appropriate, report how many neurons from each organoid are recorded and how many organoids were assessed | Use at least two complimentary methods to confirm results (e.g., MEA and patch clamp) |
| Report media composition before and during recordings | ||
| Metabolic | Analysis of lactate in culture media (e.g., Abcam kit) | Targeted qPCR or targeted glycolysis assay |
| Fluorescent reporters that use ratiometric analysis of redox state | Analysis of lactate in organoid tissues (ten organoids per batch if using proton nuclear magnetic resonance [NMR]). | |
| Transcriptome-metabolome coupling analysis |
First, to address the variability inherent to organoid models and improve reproducibility, we emphasize the importance of distinguishing between technical and biological replicates. Technical replicates should stem from a single differentiation batch (one hPSC clone), whereas biological replicates should originate from differentiation batches of separate hPSC lines. Incorporating multiple hPSC lines in a single study is critical to account for genetic variability and ensure robust conclusions, especially in instances when isogenic controls are unavailable. A strong rationale should be provided for the chosen sample size, which ideally includes power analysis. Finally, we recommend using power analyses to determine the number of hPSC lines and the number of organoids needed.
Next, data documentation is of the absolute essence. All methods and criteria used in a study must be meticulously documented, including the quantitative assessments of specific structures such as rosettes, cortical layers, and organoids as a whole. Next, it is critical to specify how measurements are obtained, including the software packages and features or markers assessed. Clarifying the boundaries for inclusion and exclusion will also aid in standardizing data collection and analysis. Comprehensive sample documentation should detail the number of hPSC lines and clones used, the number of independent differentiations performed for each line, and the number of organoids analyzed from each differentiation batch. Additionally, describing the sampling methods and the number of cryosections evaluated per organoid will enhance the reproducibility of findings. This practice will facilitate cross-laboratory comparisons and improve the field’s overall rigor.
For histological analyses of cell types and organoid structure, efforts should be made to develop and use specific antibodies and markers for cell type analysis. Where antibodies fall short, single-molecule FISH techniques such as RNAscope may provide a valuable alternative, despite the caveats regarding RNA and protein expression correlation. We recommend analyzing multiple sections per organoid to capture its 3D architecture accurately. Random sampling of serial sections can be used for quantitative stereology, but sections should be sufficiently distanced to prevent repeated measurements of the same cell. The number of sections selected needs to be determined using statistical methods. Finally, to minimize bias and increase the accuracy of quantitative measurements, we recommend using semi-automatic and automatic cell counting platforms such as CellProfiler or Imaris, and ImageJ (Table S3). Promoting the use of open-source software for data analysis and encouraging sharing of raw data and code will facilitate methodological standardization and independent validation of findings.
Clearing methods for organoid characterization should include consistent protocols and develop open-source tools to advance organoid research. Data documentation is essential, mandating transparent reporting such as whether necrotic cores are included or excluded from morphological analyses. To mitigate sampling bias and ensure comprehensive analysis, sufficient sampling of organoid sections is recommended, advocating for the analysis of at least 10 discrete areas of sufficient separation. Ideally, these efforts will lead to deeper understanding of organoid structure and the development of organoid atlases across methodologies.
For single-cell gene expression analysis of organoids, multiple organoids from multiple hPSC lines should be used to draw conclusions. Use of multiomics data and integrative approaches that combine transcriptomics with chromatin accessibility, chromatin interaction, and spatial omics may help to further deconvolute the data. Comparing transcriptome data with embryonic and fetal human brain gene expression data and revealing shared cell developmental trajectories using new bioinformatics tools such as BOMA (He et al., 2023a) will help validate the organoid model and disease mechanisms. Predicting gene regulatory networks, especially involving epigenomics, driving gene expression patterns can also help us understand developmental mechanisms across brains and organoids.
For monitoring the metabolic status in live organoids, methods such as lactate analysis in culture media and organoids and utilizing fluorescent reporters for ratiometric assessments of the redox states are relatively straightforward. These approaches offer a glimpse into the organoid bioenergetic state, providing valuable insights into their metabolic dynamics. For a more comprehensive endpoint analysis, targeted qPCR and glycolysis assays that measure pyruvate/lactate levels would be instrumental in elucidating the first-tier bioenergetics across both proliferating and postmitotic cell types. Moreover, multiomic strategies that integrate transcriptomic and metabolomic data stand out as particularly potent tools. These methods facilitate a deep dive into the metabolic machinery in organoids, enabling identification of specific metabolic pathways, including enzymes and metabolites, involved in particular experimental conditions. Such analyses are crucial for a thorough assessment and documentation of organoid metabolism and stress responses. Employing these assays is thus recommended to enhance the depth and breadth of our understanding of metabolic processes in organoid models.
Functional assays should leverage the strengths of multiple complementary methods to validate phenotypes robustly, thereby strengthening data interpretation and drawn conclusions. As patch clamping is straightforward, our recommendation here centers on statistical analysis of these data. For example, power analysis determining the number of cells needed and the number of cells to be recorded when combining patch clamp with patch-seq is highly recommended, as this information is lacking in most publications. It is also important to report the inclusion and exclusion criteria when analyzing these data and any thresholds that have been considered. If using calcium indicators, we strongly recommend that detailed methodological descriptions—from microscopy technique and imaging parameters to pre-processing steps (e.g., low and high pass filters before inserting the data into an analysis pipeline) and analysis software—are critical for enhancing reproducibility and facilitating cross-study comparisons. We also recommend targeting calcium indicators to specific cell types to refine data interpretation, underscoring the importance of methodological precision in advancing the field of organoid research. Finally, when considering MEA, we recommend two strategies for examining neural activity within organoids: 1) employing high-density MEA for detailed but lower-throughput analysis or 2) opting for low-density MEA to maximize sample sizes and conditions. Another key consideration in utilizing MEA is the ability to accurately identify the source of recorded activity on a single channel and across multiple channels, also known as spike sorting. Spike sorting is essential for discerning whether changes in channel activity are attributable to alterations in a single neuron’s firing rate or involvement of multiple neurons. Currently, many computational methods exist for spike sorting (Lee et al., 2020; Pachitariu et al., 2023) that can distinguish the neurons that contribute to the electrical activity within and across channels. Accurate spike sorting is critical for assessing pairwise correlations that can unravel functional coupling between neurons within neural networks (Ventura and Gerkin, 2012). Thus, for comprehensive neuronal activity analysis, it is advisable to supplement MEA data with post hoc spike sorting and complement these findings with other techniques such as calcium imaging or patch-clamp electrophysiology. This multifaceted approach will enable a deeper understanding of specific cell types and the intricate dynamics of neuronal networks within organoids.
Finally, for data analysis, including omics and electrophysiological function assays, the code, packages, and methodologies (not only the raw data) should be shared to enhance standardization of approaches across labs. To further enhance reproducibility, it is important for investigators to use and publish open-source packages that can be readily tested by others as our common goal is to better understand organoid structure and function.
By adhering to these recommendations and embracing a comprehensive and standardized approach to quantitative analysis of cortical organoids, we can significantly enhance the reliability and reproducibility of research in this rapidly evolving field. Although these recommendations are tailored to cortical organoids—the most commonly used organoid type—they are broadly applicable to other organoid models as well. Ultimately, this concerted effort aims not only to refine existing techniques but also to cultivate a collaborative ethos to propel our comprehension of the human brain to new heights.
Consortia
Cross-IDDRC Human Stem Cell Consortium: Aislinn Williams (University of Iowa), Mark Niciu (University of Iowa), Chris Proschel (University of Rochester), Krishnan Padmanabhan (University of Rochester), Harley Kornblum (UCLA), Bennett Novitch (UCLA), Jason Stein (UNC), Andre Sousa (University of Wisconsin-Madison), Anita, Bhattacharyya (University of Wisconsin-Madison), Xinyu Zhao (University of Wisconsin-Madison), Daifeng Wang (University of Wisconsin-Madison), Qiang Chang (University of Wisconsin-Madison), Mustafa Sahin (Boston Children’s Hospital), Elizabeth Buttermore (Boston Children’s Hospital), Dosh Whye (Boston Children’s Hospital), Mirjana Maletic-Savatic (Baylor College of Medicine), Deborah French (Children’s Hospital of Philadelphia), Elisa Waxman (Children’s Hospital of Philadelphia), Stewart Anderson (Children’s Hospital of Philadelphia), Kazue Hashimoto-Torii (Children National Medical Center), Kristin Kroll (Washington University), Herbert M Lachman (Albert Einstein College of Medicine). For more information, please see Document S1 of the supplemental information.
Acknowledgments
We thank Dr. Tracy King (NICHD) for input and support. This study was made possible by the following IDDRC grants: P50HD105353 to the Waisman Center of University of Wisconsin-Madison, P50HD103556 to the University of Iowa, P50HD105351 to Boston Children's Hospital, P50HD103536 to the University of Rochester, P50HD103573 to University of North Carolina at Chapel Hill, P50HD103555 to Baylor College of Medicine.
In addition, this work was supported by grants from the National Institutes of Health (R01MH118827, R01NS105200, and R01MH116582 to X.Z.; a diversity supplement to R01MH118827 and R36MH136790 for S.O.S.; RF1MH128695, R01NS064025, R01AG067025, and U01MH116492 to D. Wang; KL2TR002536 to A.W.; R01MH130356 to M.M.-S.; R01MH130441 and R01MH121433 to J.L.S.; R01HD106197 to A.B.); DOD IIRA grant W81XWH-22-1-0621 (to X.Z. and A.B.); the Translational Research Institute for Space Health through NASA Cooperative Agreement NNX16AO69A grant RAD01013 and NASA grant 21-3DTMPS 2-0020 (M.M.-S.); UW Vilas Mid-Career Award, Kellett Mid-Career Award, Wisconsin Alumni Research Foundation, and Jenni and Kyle Professorship (to X.Z.); Simons Foundation Autism Research Initiative pilot grants (to X.Z. A.M.M.S., D. Wang, and M.M.-S.); NSF Career Award 2144475 (to D. Wang); Brain Research Foundation Seed Grant (BRFSG-2023-11 to A.M.M.S.); NARSAD (30056, to K.K.); SciMed scholarships (to N.M.M.-A. and S.O.S.), T32 GM141013 Molecular Pharmacology training grant and predoctoral fellowship from the Wisconsin Stem Cell and Regeneration Medicine Center (to N.M.M.-A.); Autism Speaks (to G.C.); and postdoctoral fellowships from FRAXA and from the Autism Science Foundation (to C.L.S.).
Author contributions
X.Z., M.M.-S., and A.W. conceptualized and supervised this project. S.O. and M.M.-S. wrote the introduction; S.O.S. and X.Z. wrote the cellular and functional measurement sections and created Tables 2 and S1–S3. S.O. and D. Wang wrote transcriptomics; K.P. wrote a portion of MEA in the functional measurement section; G.C., S.M.K., K.K., A.W., and M.M.-S. wrote the metabolic section. K.K., G.C., A.W., and M.M.-S. created Table S4; X.Z. created Figure 1; S.O.S. created Figure 2; G.C. and S.M.K. created Figure 3; K.K. and A.W. created Figure 4. N.M.M.-A. finalized all the figures. S.O.S. added references. K.K., E.A.W., J.L.S., A.B., E.D.B., and D. Whye performed detailed editing. All authors have edited and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.stemcr.2024.04.008.
Contributor Information
Aislinn Williams, Email: aislinn-williams@uiowa.edu.
Mirjana Maletic-Savatic, Email: mirjana.maletic-savatic@bcm.edu.
Xinyu Zhao, Email: xinyu.zhao@wisc.edu.
Cross-IDDRC Human Stem Cell Consortium:
Stewart Anderson, Anita, Bhattacharyya, Elizabeth Buttermore, Qiang Chang, Deborah French, Kazue Hashimoto-Torii, Harley Kornblum, Kristin Kroll, Herbert M. Lachman, Mirjana Maletic-Savatic, Mark Niciu, Bennett Novitch, Krishnan Padmanabhan, Chris Proschel, Mustafa Sahin, Andre Sousa, Jason Stein, Daifeng Wang, Elisa Waxman, Dosh Whye, Aislinn Williams, and Xinyu Zhao
Supplemental information
References
- Abud E.M., Ramirez R.N., Martinez E.S., Healy L.M., Nguyen C.H.H., Newman S.A., Yeromin A.V., Scarfone V.M., Marsh S.E., Fimbres C., et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron. 2017;94:278–293.e9. doi: 10.1016/j.neuron.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese A., Swaney J.M., Yun D.H., Evans N.B., Antonucci J.M., Velasco S., Sohn C.H., Arlotta P., Gehrke L., Chung K. Multiscale 3D phenotyping of human cerebral organoids. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-78130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasaki C., Wilson A.F., Chen A.S., Wegscheid M.L., Gutmann D.H. Generation of human induced pluripotent stem cell-derived cerebral organoids for cellular and molecular characterization. STAR Protoc. 2022;3 doi: 10.1016/j.xpro.2022.101173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J., Revah O., Miura Y., Thom N., Amin N.D., Kelley K.W., Singh M., Chen X., Thete M.V., Walczak E.M., et al. Generation of Functional Human 3D Cortico-Motor Assembloids. Cell. 2020;183:1913–1929.e26. doi: 10.1016/j.cell.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamian A., Birtele M., Hosseini N., Nguyen T., Seth A., Del Dosso A., Paul S., Tedeschi N., Taylor R., Coba M.P., et al. Human cerebellar organoids with functional Purkinje cells. Cell Stem Cell. 2024;31:39–51.e6. doi: 10.1016/j.stem.2023.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley J.A., Reumann D., Bian S., Lévi-Strauss J., Knoblich J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy C., van den Hurk M., Eames T., Marchand C., Hernandez R.V., Kellogg M., Gorris M., Galet B., Palomares V., Brown J., et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc. Natl. Acad. Sci. USA. 2015;112:E2725–E2734. doi: 10.1073/pnas.1504393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu-Laroche L., Brown N.J., Hansen M., Toloza E.H.S., Sharma J., Williams Z.M., Frosch M.P., Cosgrove G.R., Cash S.S., Harnett M.T. Allometric rules for mammalian cortical layer 5 neuron biophysics. Nature. 2021;600:274–278. doi: 10.1038/s41586-021-04072-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu-Laroche L., Toloza E.H.S., van der Goes M.S., Lafourcade M., Barnagian D., Williams Z.M., Eskandar E.N., Frosch M.P., Cash S.S., Harnett M.T. Enhanced Dendritic Compartmentalization in Human Cortical Neurons. Cell. 2018;175:643–651.e14. doi: 10.1016/j.cell.2018.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J., Sorensen S.A., Ting J.T., Miller J.A., Chartrand T., Buchin A., Bakken T.E., Budzillo A., Dee N., Ding S.L., et al. Human neocortical expansion involves glutamatergic neuron diversification. Nature. 2021;598:151–158. doi: 10.1038/s41586-021-03813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaduri A., Andrews M.G., Mancia Leon W., Jung D., Shin D., Allen D., Jung D., Schmunk G., Haeussler M., Salma J., et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578:142–148. doi: 10.1038/s41586-020-1962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F., Andersen J., Makinson C.D., Islam S., Wei W., Huber N., Fan H.C., Metzler K.R.C., Panagiotakos G., Thom N., et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacker T.S., Duchen M.R. Investigating mitochondrial redox state using NADH and NADPH autofluorescence. Free Radic. Biol. Med. 2016;100:53–65. doi: 10.1016/j.freeradbiomed.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland D., McCormick C.M., Patel N.K., Krupa O., Mory J.T., Beltran A.A., Farah T.M., Escobar-Tomlienovich C.F., Olson S.S., Kim M., et al. Segmentor: a tool for manual refinement of 3D microscopy annotations. BMC Bioinf. 2021;22:260. doi: 10.1186/s12859-021-04202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin M.E., Hoffman-Kim D. Application and assessment of optical clearing methods for imaging of tissue-engineered neural stem cell spheres. Tissue Eng. C Methods. 2015;21:292–302. doi: 10.1089/ten.TEC.2014.0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brémond Martin C., Simon Chane C., Clouchoux C., Histace A. Recent Trends and Perspectives in Cerebral Organoids Imaging and Analysis. Front. Neurosci. 2021;15 doi: 10.3389/fnins.2021.629067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brighi C., Salaris F., Soloperto A., Cordella F., Ghirga S., de Turris V., Rosito M., Porceddu P.F., D'Antoni C., Reggiani A., et al. Novel fragile X syndrome 2D and 3D brain models based on human isogenic FMRP-KO iPSCs. Cell Death Dis. 2021;12:498. doi: 10.1038/s41419-021-03776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell C.R., Palasantza A., Jiang X., Berens P., Deng Q., Yilmaz M., Reimer J., Shen S., Bethge M., Tolias K.F., et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 2016;34:199–203. doi: 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuccio G., Khalil S.M., Osenberg S., Li F., Maletic-Savatic M. Mass spectrometry imaging as an emerging tool for studying metabolism in human brain organoids. Front. Mol. Biosci. 2023;10 doi: 10.3389/fmolb.2023.1181965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Saiyin H., Liu Y., Wang Y., Li X., Ji R., Ma L. Human striatal organoids derived from pluripotent stem cells recapitulate striatal development and compartments. PLoS Biol. 2022;20 doi: 10.1371/journal.pbio.3001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Shen W.B., Yang P., Dong D., Sun W., Yang P. High Glucose Inhibits Neural Stem Cell Differentiation Through Oxidative Stress and Endoplasmic Reticulum Stress. Stem Cell. Dev. 2018;27:745–755. doi: 10.1089/scd.2017.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Bury L.A., Chen F., Aldinger K.A., Miranda H.C., Wynshaw-Boris A. Generation of advanced cerebellar organoids for neurogenesis and neuronal network development. Hum. Mol. Genet. 2023;32:2832–2841. doi: 10.1093/hmg/ddad110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaradia I., Imaz-Rosshandler I., Nilges B.S., Boulanger J., Pellegrini L., Das R., Kashikar N.D., Lancaster M.A. Tissue morphology influences the temporal program of human brain organoid development. Cell Stem Cell. 2023;30:1351–1367.e10. doi: 10.1016/j.stem.2023.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A.N., Jin Y., An Y., Kim J., Choi Y.S., Lee J.S., Kim J., Choi W.Y., Koo D.J., Yu W., et al. Microfluidic device with brain extracellular matrix promotes structural and functional maturation of human brain organoids. Nat. Commun. 2021;12:4730. doi: 10.1038/s41467-021-24775-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J.S., Lee H.J., Rajaraman S., Kim D.H. Recent advances in three-dimensional microelectrode array technologies for in vitro and in vivo cardiac and neuronal interfaces. Biosens. Bioelectron. 2021;171 doi: 10.1016/j.bios.2020.112687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N.W., Verbridge S.S., Williams R.M., Chen J., Kim J.Y., Schmehl R., Farnum C.E., Zipfel W.R., Fischbach C., Stroock A.D. Phosphorescent nanoparticles for quantitative measurements of oxygen profiles in vitro and in vivo. Biomaterials. 2012;33:2710–2722. doi: 10.1016/j.biomaterials.2011.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Z.S., Khong Z.J., Tay S.H., Ng S.Y. Metabolic contributions to neuronal deficits caused by genomic disruption of schizophrenia risk gene SETD1A. Schizophrenia (Heidelb) 2022;8:115. doi: 10.1038/s41537-022-00326-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins T.R., Rush A.M., Estacion M., Dib-Hajj S.D., Waxman S.G. Voltage-clamp and current-clamp recordings from mammalian DRG neurons. Nat. Protoc. 2009;4:1103–1112. doi: 10.1038/nprot.2009.91. [DOI] [PubMed] [Google Scholar]
- de Melo Reis R.A., Freitas H.R., de Mello F.G. Cell Calcium Imaging as a Reliable Method to Study Neuron-Glial Circuits. Front. Neurosci. 2020;14 doi: 10.3389/fnins.2020.569361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers J.F., Alieva M., Wellens L.M., Ariese H.C.R., Jamieson P.R., Vonk A.M., Amatngalim G.D., Hu H., Oost K.C., Snippert H.J.G., et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 2019;14:1756–1771. doi: 10.1038/s41596-019-0160-8. [DOI] [PubMed] [Google Scholar]
- Diosdi A., Hirling D., Kovacs M., Toth T., Harmati M., Koos K., Buzas K., Piccinini F., Horvath P. A quantitative metric for the comparative evaluation of optical clearing protocols for 3D multicellular spheroids. Comput. Struct. Biotechnol. J. 2021;19:1233–1243. doi: 10.1016/j.csbj.2021.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch R., Geschwind D.H. The human brain in a dish: the promise of iPSC-derived neurons. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eura N., Matsui T.K., Luginbühl J., Matsubayashi M., Nanaura H., Shiota T., Kinugawa K., Iguchi N., Kiriyama T., Zheng C., et al. Brainstem Organoids From Human Pluripotent Stem Cells. Front. Neurosci. 2020;14:538. doi: 10.3389/fnins.2020.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal G., Verhoog M.B., Testa-Silva G., Deitcher Y., Lodder J.C., Benavides-Piccione R., Morales J., DeFelipe J., de Kock C.P., Mansvelder H.D., Segev I. Unique membrane properties and enhanced signal processing in human neocortical neurons. Elife. 2016;5 doi: 10.7554/eLife.16553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando M., Lee S., Wark J.R., Xiao D., Lim B.Y., O'Hara-Wright M., Kim H.J., Smith G.C., Wong T., Teber E.T., et al. Differentiation of brain and retinal organoids from confluent cultures of pluripotent stem cells connected by nerve-like axonal projections of optic origin. Stem Cell Rep. 2022;17:1476–1492. doi: 10.1016/j.stemcr.2022.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field A.R., Jacobs F.M.J., Fiddes I.T., Phillips A.P.R., Reyes-Ortiz A.M., LaMontagne E., Whitehead L., Meng V., Rosenkrantz J.L., Olsen M., et al. Structurally Conserved Primate LncRNAs Are Transiently Expressed during Human Cortical Differentiation and Influence Cell-Type-Specific Genes. Stem Cell Rep. 2019;12:245–257. doi: 10.1016/j.stemcr.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming R., Coutts J.R. LHRH analogues for ovulation induction, with particular reference to polycystic ovary syndrome. Baillieres Clin. Obstet. Gynaecol. 1988;2:677–687. doi: 10.1016/s0950-3552(88)80052-x. [DOI] [PubMed] [Google Scholar]
- Földy C., Darmanis S., Aoto J., Malenka R.C., Quake S.R., Südhof T.C. Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc. Natl. Acad. Sci. USA. 2016;113:E5222–E5231. doi: 10.1073/pnas.1610155113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Dong Q., Arachchilage K.H., Risgaard R., Sheng J., Syed M., Schmidt D.K., Jin T., Liu S., Knaack S.A., et al. Multimodal analysis reveals genes driving neuronal maturation in the primate prefrontal cortex. bioRxiv. 2023 doi: 10.1101/2023.06.02.543460. Preprint at. [DOI] [Google Scholar]
- Giandomenico S.L., Mierau S.B., Gibbons G.M., Wenger L.M.D., Masullo L., Sit T., Sutcliffe M., Boulanger J., Tripodi M., Derivery E., et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 2019;22:669–679. doi: 10.1038/s41593-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass M.R., Waxman E.A., Yamashita S., Lafferty M., Beltran A., Farah T., Patel N.K., Matoba N., Ahmed S., Srivastava M., et al. Cross-site reproducibility of human cortical organoids reveals consistent cell type composition and architecture. bioRxiv. 2023 doi: 10.1101/2023.07.28.550873. Preprint at. [DOI] [Google Scholar]
- Gomes A.R., Fernandes T.G., Vaz S.H., Silva T.P., Bekman E.P., Xapelli S., Duarte S., Ghazvini M., Gribnau J., Muotri A.R., et al. Modeling Rett Syndrome With Human Patient-Specific Forebrain Organoids. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.610427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Giro G., Arias-Fuenzalida J., Jarazo J., Zeuschner D., Ali M., Possemis N., Bolognin S., Halder R., Jäger C., Kuper W.F.E., et al. Synapse alterations precede neuronal damage and storage pathology in a human cerebral organoid model of CLN3-juvenile neuronal ceroid lipofuscinosis. Acta Neuropathol. Commun. 2019;7:222. doi: 10.1186/s40478-019-0871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A., Yoon S.J., Tran S.S., Makinson C.D., Park J.Y., Andersen J., Valencia A.M., Horvath S., Xiao X., Huguenard J.R., et al. Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat. Neurosci. 2021;24:331–342. doi: 10.1038/s41593-021-00802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald N.F., Miller G., Moen E., Kong A., Kagel A., Dougherty T., Fullaway C.C., McIntosh B.J., Leow K.X., Schwartz M.S., et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 2022;40:555–565. doi: 10.1038/s41587-021-01094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienberger C., Konnerth A. Imaging calcium in neurons. Neuron. 2012;73:862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Grienberger C., Magee J.C. Entorhinal cortex directs learning-related changes in CA1 representations. Nature. 2022;611:554–562. doi: 10.1038/s41586-022-05378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grist S.M., Nasseri S.S., Poon T., Roskelley C., Cheung K.C. On-chip clearing of arrays of 3-D cell cultures and micro-tissues. Biomicrofluidics. 2016;10 doi: 10.1063/1.4959031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Shen M., Dong Q., Méndez-Albelo N.M., Huang S.X., Sirois C.L., Le J., Li M., Jarzembowski E.D., Schoeller K.A., et al. Elevated levels of FMRP-target MAP1B impair human and mouse neuronal development and mouse social behaviors via autophagy pathway. Nat. Commun. 2023;14:3801. doi: 10.1038/s41467-023-39337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham O., Jin Y.B., Kim J., Lee M.O. Blood vessel formation in cerebral organoids formed from human embryonic stem cells. Biochem. Biophys. Res. Commun. 2020;521:84–90. doi: 10.1016/j.bbrc.2019.10.079. [DOI] [PubMed] [Google Scholar]
- Hartmann J., Henschel N., Bartmann K., Dönmez A., Brockerhoff G., Koch K., Fritsche E. Molecular and Functional Characterization of Different BrainSphere Models for Use in Neurotoxicity Testing on Microelectrode Arrays. Cells. 2023;12 doi: 10.3390/cells12091270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L., et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Kalafut N.C., Sandoval S.O., Risgaard R., Sirois C.L., Yang C., Khullar S., Suzuki M., Huang X., Chang Q., et al. BOMA, a machine-learning framework for comparative gene expression analysis across brains and organoids. Cell Rep. Methods. 2023;3 doi: 10.1016/j.crmeth.2023.100409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Dony L., Fleck J.S., Szałata A., Li K.X., Slišković I., Lin H.-C., Santel M., Atamian A., Quadrato G., et al. An integrated transcriptomic cell atlas of human neural organoids. bioRxiv. 2023 doi: 10.1101/2023.10.05.561097. Preprint at. [DOI] [Google Scholar]
- Huang Q., Tang B., Romero J.C., Yang Y., Elsayed S.K., Pahapale G., Lee T.J., Morales Pantoja I.E., Han F., Berlinicke C., et al. Shell microelectrode arrays (MEAs) for brain organoids. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abq5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Huang F., Zhang H., Yang Y., Lu J., Chen J., Shen L., Pei G. In vivo development and single-cell transcriptome profiling of human brain organoids. Cell Prolif. 2022;55 doi: 10.1111/cpr.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.K., Wong S.Z.H., Pather S.R., Nguyen P.T.T., Zhang F., Zhang D.Y., Zhang Z., Lu L., Fang W., Chen L., et al. Generation of hypothalamic arcuate organoids from human induced pluripotent stem cells. Cell Stem Cell. 2021;28:1657–1670.e10. doi: 10.1016/j.stem.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Chen J., Bajić A., Zhang C., Song X., Carroll S.L., Cai Z.L., Tang M., Xue M., Cheng N., et al. Quantitative real-time imaging of glutathione. Nat. Commun. 2017;8 doi: 10.1038/ncomms16087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Zhang C., Chen J., Choi S., Zhou Y., Zhao M., Song X., Chen X., Maletić-Savatić M., Palzkill T., et al. Quantitative Real-Time Imaging of Glutathione with Subcellular Resolution. Antioxidants Redox Signal. 2019;30:1900–1910. doi: 10.1089/ars.2018.7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J., Xiao Y., Sun A.X., Cukuroglu E., Tran H.D., Göke J., Tan Z.Y., Saw T.Y., Tan C.P., Lokman H., et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell. 2016;19:248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoshima T., Sakaguchi H., Nakano T., Soen M., Ando S., Eiraku M., Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Zhou Y., Li Y., Han Y., Xu J., Niu W., Li Z., Liu S., Feng H., Huang W., et al. A human forebrain organoid model of fragile X syndrome exhibits altered neurogenesis and highlights new treatment strategies. Nat. Neurosci. 2021;24:1377–1391. doi: 10.1038/s41593-021-00913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai T., Suga H., Sakakibara M., Ozone C., Matsumoto R., Kano M., Mitsumoto K., Ogawa K., Kodani Y., Nagasaki H., et al. Hypothalamic Contribution to Pituitary Functions Is Recapitulated In Vitro Using 3D-Cultured Human iPS Cells. Cell Rep. 2020;30:18–24.e5. doi: 10.1016/j.celrep.2019.12.009. [DOI] [PubMed] [Google Scholar]
- Kashirina A., Gavrina A., Kryukov E., Elagin V., Kolesova Y., Artyuhov A., Momotyuk E., Abdyyev V., Meshcheryakova N., Zagaynova E., et al. Energy Metabolism and Intracellular pH Alteration in Neural Spheroids Carrying Down Syndrome. Biomedicines. 2021;9 doi: 10.3390/biomedicines9111741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelava I., Lancaster M.A. Stem Cell Models of Human Brain Development. Cell Stem Cell. 2016;18:736–748. doi: 10.1016/j.stem.2016.05.022. [DOI] [PubMed] [Google Scholar]
- Kiral F.R., Cakir B., Tanaka Y., Kim J., Yang W.S., Wehbe F., Kang Y.J., Zhong M., Sancer G., Lee S.H., et al. Generation of ventralized human thalamic organoids with thalamic reticular nucleus. Cell Stem Cell. 2023;30:677–688.e5. doi: 10.1016/j.stem.2023.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus J., Kanton S., Kyrousi C., Ayo-Martin A.C., Di Giaimo R., Riesenberg S., O'Neill A.C., Camp J.G., Tocco C., Santel M., et al. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat. Med. 2019;25:561–568. doi: 10.1038/s41591-019-0371-0. [DOI] [PubMed] [Google Scholar]
- Kolesová H., Olejníčková V., Kvasilová A., Gregorovičová M., Sedmera D. Tissue clearing and imaging methods for cardiovascular development. iScience. 2021;24 doi: 10.1016/j.isci.2021.102387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupa O., Fragola G., Hadden-Ford E., Mory J.T., Liu T., Humphrey Z., Rees B.W., Krishnamurthy A., Snider W.D., Zylka M.J., et al. NuMorph: Tools for cortical cellular phenotyping in tissue-cleared whole-brain images. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Corsini N.S., Wolfinger S., Gustafson E.H., Phillips A.W., Burkard T.R., Otani T., Livesey F.J., Knoblich J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S., Petersilie L., Inak G., Menacho-Pando C., Kafitz K.W., Rybak-Wolf A., Rajewsky N., Rose C.R., Prigione A. Generation of Human Brain Organoids for Mitochondrial Disease Modeling. J. Vis. Exp. 2021:e62756. doi: 10.3791/62756. [DOI] [PubMed] [Google Scholar]
- Lee J., Mitelut C., Shokri H., Kinsella I., Dethe N., Wu S., Li K., Reyes E.B., Turcu D., Batty E., et al. YASS: Yet Another Spike Sorter applied to large-scale multi-electrode array recordings in primate retina. bioRxiv. 2020 doi: 10.1101/2020.03.18.997924. Preprint at. [DOI] [Google Scholar]
- Lee J.H., Shin H., Shaker M.R., Kim H.J., Park S.H., Kim J.H., Lee N., Kang M., Cho S., Kwak T.H., et al. Production of human spinal-cord organoids recapitulating neural-tube morphogenesis. Nat. Biomed. Eng. 2022;6:435–448. doi: 10.1038/s41551-022-00868-4. [DOI] [PubMed] [Google Scholar]
- Leong Y.C., Di Foggia V., Pramod H., Bitner-Glindzicz M., Patel A., Sowden J.C. Molecular pathology of Usher 1B patient-derived retinal organoids at single cell resolution. Stem Cell Rep. 2022;17:2421–2437. doi: 10.1016/j.stemcr.2022.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.T., Seo J., Gao F., Feldman H.M., Wen H.L., Penney J., Cam H.P., Gjoneska E., Raja W.K., Cheng J., et al. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer's Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron. 2018;98:1141–1154.e7. doi: 10.1016/j.neuron.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li X., Chen J., Yuan C. Micro/Nano Electrode Array Sensors: Advances in Fabrication and Emerging Applications in Bioanalysis. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.573865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludikhuize M.C., Meerlo M., Burgering B.M.T., Rodríguez Colman M.J. Protocol to profile the bioenergetics of organoids using Seahorse. STAR Protoc. 2021;2 doi: 10.1016/j.xpro.2021.100386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig T.E., Andrews P.W., Barbaric I., Benvenisty N., Bhattacharyya A., Crook J.M., Daheron L.M., Draper J.S., Healy L.E., Huch M., et al. ISSCR standards for the use of human stem cells in basic research. Stem Cell Rep. 2023;18:1744–1752. doi: 10.1016/j.stemcr.2023.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Skarica M., Li Q., Xu C., Risgaard R.D., Tebbenkamp A.T.N., Mato-Blanco X., Kovner R., Krsnik Ž., de Martin X., et al. Molecular and cellular evolution of the primate dorsolateral prefrontal cortex. Science. 2022;377 doi: 10.1126/science.abo7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour A.A., Gonçalves J.T., Bloyd C.W., Li H., Fernandes S., Quang D., Johnston S., Parylak S.L., Jin X., Gage F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018;36:432–441. doi: 10.1038/nbt.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Costa C., Pham V.A., Sidhaye J., Novatchkova M., Wiegers A., Peer A., Möseneder P., Corsini N.S., Knoblich J.A. Morphogenesis and development of human telencephalic organoids in the absence and presence of exogenous extracellular matrix. EMBO J. 2023;42 doi: 10.15252/embj.2022113213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton R.M., Pașca S.P. Organoid and Assembloid Technologies for Investigating Cellular Crosstalk in Human Brain Development and Disease. Trends Cell Biol. 2020;30:133–143. doi: 10.1016/j.tcb.2019.11.004. [DOI] [PubMed] [Google Scholar]
- Martynov V.I., Pakhomov A.A., Deyev I.E., Petrenko A.G. Genetically encoded fluorescent indicators for live cell pH imaging. Biochim. Biophys. Acta Gen. Subj. 2018;1862:2924–2939. doi: 10.1016/j.bbagen.2018.09.013. [DOI] [PubMed] [Google Scholar]
- McMahon S.M., Jackson M.B. An Inconvenient Truth: Calcium Sensors Are Calcium Buffers. Trends Neurosci. 2018;41:880–884. doi: 10.1016/j.tins.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meleshina A.V., Dudenkova V.V., Bystrova A.S., Kuznetsova D.S., Shirmanova M.V., Zagaynova E.V. Two-photon FLIM of NAD(P)H and FAD in mesenchymal stem cells undergoing either osteogenic or chondrogenic differentiation. Stem Cell Res. Ther. 2017;8:15. doi: 10.1186/s13287-017-0484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meleshina A.V., Dudenkova V.V., Shirmanova M.V., Shcheslavskiy V.I., Becker W., Bystrova A.S., Cherkasova E.I., Zagaynova E.V. Probing metabolic states of differentiating stem cells using two-photon FLIM. Sci. Rep. 2016;6 doi: 10.1038/srep21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Yao D., Imaizumi K., Chen X., Kelley K.W., Reis N., Thete M.V., Arjun McKinney A., Kulkarni S., Panagiotakos G., et al. Assembloid CRISPR screens reveal impact of disease genes in human neurodevelopment. Nature. 2023;622:359–366. doi: 10.1038/s41586-023-06564-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura Y., Li M.Y., Birey F., Ikeda K., Revah O., Thete M.V., Park J.Y., Puno A., Lee S.H., Porteus M.H., Pașca S.P. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. Biotechnol. 2020;38:1421–1430. doi: 10.1038/s41587-020-00763-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muguruma K., Nishiyama A., Kawakami H., Hashimoto K., Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015;10:537–550. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- Muzzi L., Di Lisa D., Falappa M., Pepe S., Maccione A., Pastorino L., Martinoia S., Frega M. Human-Derived Cortical Neurospheroids Coupled to Passive, High-Density and 3D MEAs: A Valid Platform for Functional Tests. Bioengineering. 2023;10 doi: 10.3390/bioengineering10040449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaras M., Lodhi A., Barrio-Alonso E., Foord C., Rodrick T., Jones D., Fang H., Greening D., Colak D. Neurodevelopmental signatures of narcotic and neuropsychiatric risk factors in 3D human-derived forebrain organoids. Mol. Psychiatr. 2021;26:7760–7783. doi: 10.1038/s41380-021-01189-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos V., Razansky D. Molecular imaging by means of multispectral optoacoustic tomography (MSOT) Chem. Rev. 2010;110:2783–2794. doi: 10.1021/cr9002566. [DOI] [PubMed] [Google Scholar]
- Nürnberg E., Vitacolonna M., Klicks J., von Molitor E., Cesetti T., Keller F., Bruch R., Ertongur-Fauth T., Riedel K., Scholz P., et al. Routine Optical Clearing of 3D-Cell Cultures: Simplicity Forward. Front. Mol. Biosci. 2020;7:20. doi: 10.3389/fmolb.2020.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzou G., Wicks R.T., Wicks E.E., Seale S.A., Sane C.H., Chen A., Murphy S.V., Jackson J.D., Atala A.J. Human Cortex Spheroid with a Functional Blood Brain Barrier for High-Throughput Neurotoxicity Screening and Disease Modeling. Sci. Rep. 2018;8:7413. doi: 10.1038/s41598-018-25603-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkelman I.A., Neto N., Papkovsky D.B., Monaghan M.G., Dmitriev R.I. A deeper understanding of intestinal organoid metabolism revealed by combining fluorescence lifetime imaging microscopy (FLIM) and extracellular flux analyses. Redox Biol. 2020;30 doi: 10.1016/j.redox.2019.101420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormel P.R., Vieira de Sá R., van Bodegraven E.J., Karst H., Harschnitz O., Sneeboer M.A.M., Johansen L.E., van Dijk R.E., Scheefhals N., Berdenis van Berlekom A., et al. Microglia innately develop within cerebral organoids. Nat. Commun. 2018;9:4167. doi: 10.1038/s41467-018-06684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachitariu M., Sridhar S., Stringer C. Solving the spike sorting problem with Kilosort. bioRxiv. 2023 doi: 10.1101/2023.01.07.523036. Preprint at. [DOI] [Google Scholar]
- Pagliaro A., Finger R., Zoutendijk I., Bunschuh S., Clevers H., Hendriks D., Artegiani B. Temporal morphogen gradient-driven neural induction shapes single expanded neuroepithelium brain organoids with enhanced cortical identity. Nat. Commun. 2023;14:7361. doi: 10.1038/s41467-023-43141-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pașca A.M., Park J.Y., Shin H.W., Qi Q., Revah O., Krasnoff R., O'Hara R., Willsey A.J., Palmer T.D., Pașca S.P. Human 3D cellular model of hypoxic brain injury of prematurity. Nat. Med. 2019;25:784–791. doi: 10.1038/s41591-019-0436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paşca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.Y., O'Rourke N.A., Nguyen K.D., et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passaro A.P., Stice S.L. Electrophysiological Analysis of Brain Organoids: Current Approaches and Advancements. Front. Neurosci. 2020;14 doi: 10.3389/fnins.2020.622137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Bonfio C., Chadwick J., Begum F., Skehel M., Lancaster M.A. Human CNS barrier-forming organoids with cerebrospinal fluid production. Science. 2020;369 doi: 10.1126/science.aaz5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham M.T., Pollock K.M., Rose M.D., Cary W.A., Stewart H.R., Zhou P., Nolta J.A., Waldau B. Generation of human vascularized brain organoids. Neuroreport. 2018;29:588–593. doi: 10.1097/WNR.0000000000001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli D., Magliaro C., Ahluwalia A. Experimental and Computational Methods for the Study of Cerebral Organoids: A Review. Front. Neurosci. 2019;13:162. doi: 10.3389/fnins.2019.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen A.A., Bhaduri A., Andrews M.G., Nowakowski T.J., Meyerson O.S., Mostajo-Radji M.A., Di Lullo E., Alvarado B., Bedolli M., Dougherty M.L., et al. Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell. 2019;176:743–756.e17. doi: 10.1016/j.cell.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen A.A., Nowakowski T.J., Shuga J., Wang X., Leyrat A.A., Lui J.H., Li N., Szpankowski L., Fowler B., Chen P., et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppo F., Sadegh S., Trujillo C.A., Thunemann M., Campbell E.P., Vandenberghe M., Shan X., Akkouh I.A., Miller E.W., Bloodgood B.L., et al. All-Optical Electrophysiology in hiPSC-Derived Neurons With Synthetic Voltage Sensors. Front. Cell. Neurosci. 2021;15 doi: 10.3389/fncel.2021.671549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Su Y., Adam C.D., Deutschmann A.U., Pather S.R., Goldberg E.M., Su K., Li S., Lu L., Jacob F., et al. Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell. 2020;26:766–781.e9. doi: 10.1016/j.stem.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Min Yang S., Berger D.R., Maria N., Scholvin J., Goldman M., Kinney J.P., et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju V., Calloway N., Ryan T.A. Activity-driven local ATP synthesis is required for synaptic function. Cell. 2014;156:825–835. doi: 10.1016/j.cell.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner M., Lancaster M.A., Bian S., Choi H., Ku T., Peer A., Chung K., Knoblich J.A. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36:1316–1329. doi: 10.15252/embj.201694700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich S., Moradi Chameh H., Lefebvre J., Valiante T.A. Loss of neuronal heterogeneity in epileptogenic human tissue impairs network resilience to sudden changes in synchrony. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110863. [DOI] [PubMed] [Google Scholar]
- Sakaguchi H., Kadoshima T., Soen M., Narii N., Ishida Y., Ohgushi M., Takahashi J., Eiraku M., Sasai Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi H., Ozaki Y., Ashida T., Matsubara T., Oishi N., Kihara S., Takahashi J. Self-Organized Synchronous Calcium Transients in a Cultured Human Neural Network Derived from Cerebral Organoids. Stem Cell Rep. 2019;13:458–473. doi: 10.1016/j.stemcr.2019.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarasinghe R.A., Miranda O.A., Buth J.E., Mitchell S., Ferando I., Watanabe M., Allison T.F., Kurdian A., Fotion N.N., Gandal M.J., et al. Identification of neural oscillations and epileptiform changes in human brain organoids. Nat. Neurosci. 2021;24:1488–1500. doi: 10.1038/s41593-021-00906-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada T., Chater T.E., Sasagawa Y., Yoshimura M., Fujimori-Tonou N., Tanaka K., Benjamin K.J.M., Paquola A.C.M., Erwin J.A., Goda Y., et al. Developmental excitation-inhibition imbalance underlying psychoses revealed by single-cell analyses of discordant twins-derived cerebral organoids. Mol. Psychiatr. 2020;25:2695–2711. doi: 10.1038/s41380-020-0844-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer T.J. Application of Microelectrode Array Approaches to Neurotoxicity Testing and Screening. Adv. Neurobiol. 2019;22:275–297. doi: 10.1007/978-3-030-11135-9_12. [DOI] [PubMed] [Google Scholar]
- Sharf T., van der Molen T., Glasauer S.M.K., Guzman E., Buccino A.P., Luna G., Cheng Z., Audouard M., Ranasinghe K.G., Kudo K., et al. Functional neuronal circuitry and oscillatory dynamics in human brain organoids. Nat. Commun. 2022;13:4403. doi: 10.1038/s41467-022-32115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Sirois C.L., Guo Y., Li M., Dong Q., Méndez-Albelo N.M., Gao Y., Khullar S., Kissel L., Sandoval S.O., et al. Species-specific FMRP regulation of RACK1 is critical for prenatal cortical development. Neuron. 2023;111:3988–4005.e11. doi: 10.1016/j.neuron.2023.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Sun L., Wang M., Liu J., Zhong S., Li R., Li P., Guo L., Fang A., Chen R., et al. Vascularized human cortical organoids (vOrganoids) model cortical development in vivo. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Jeong S., Lee J.H., Sun W., Choi N., Cho I.J. 3D high-density microelectrode array with optical stimulation and drug delivery for investigating neural circuit dynamics. Nat. Commun. 2021;12:492. doi: 10.1038/s41467-020-20763-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnaider T.A., Pristyazhnyuk I.E. CLARITY and Light-Sheet microscopy sample preparation in application to human cerebral organoids. Vavilovskii Zhurnal Genet. Selektsii. 2021;25:889–895. doi: 10.18699/VJ21.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan S.A., Andersen J., Pașca A.M., Birey F., Pașca S.P. Generation and assembly of human brain region-specific three-dimensional cultures. Nat. Protoc. 2018;13:2062–2085. doi: 10.1038/s41596-018-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits L.M., Reinhardt L., Reinhardt P., Glatza M., Monzel A.S., Stanslowsky N., Rosato-Siri M.D., Zanon A., Antony P.M., Bellmann J., et al. Modeling Parkinson's disease in midbrain-like organoids. NPJ Parkinsons Dis. 2019;5:5. doi: 10.1038/s41531-019-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrek I., Stelzer E.H.K. Quantitative three-dimensional evaluation of immunofluorescence staining for large whole mount spheroids with light sheet microscopy. Biomed. Opt Express. 2017;8:484–499. doi: 10.1364/BOE.8.000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Yuan X., Jones Z., Vied C., Miao Y., Marzano M., Hua T., Sang Q.X.A., Guan J., Ma T., et al. Functionalization of Brain Region-specific Spheroids with Isogenic Microglia-like Cells. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-47444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A.M.M., Meyer K.A., Santpere G., Gulden F.O., Sestan N. Evolution of the Human Nervous System Function, Structure, and Development. Cell. 2017;170:226–247. doi: 10.1016/j.cell.2017.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringari C., Edwards R.A., Pate K.T., Waterman M.L., Donovan P.J., Gratton E. Metabolic trajectory of cellular differentiation in small intestine by Phasor Fluorescence Lifetime Microscopy of NADH. Sci. Rep. 2012;2:568. doi: 10.1038/srep00568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer C., Wang T., Michaelos M., Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods. 2021;18:100–106. doi: 10.1038/s41592-020-01018-x. [DOI] [PubMed] [Google Scholar]
- Sun A.X., Yuan Q., Fukuda M., Yu W., Yan H., Lim G.G.Y., Nai M.H., D'Agostino G.A., Tran H.D., Itahana Y., et al. Potassium channel dysfunction in human neuronal models of Angelman syndrome. Science. 2019;366:1486–1492. doi: 10.1126/science.aav5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Meng X., Liu Y., Song D., Jiang C., Cai J. Applications of brain organoids in neurodevelopment and neurological diseases. J. Biomed. Sci. 2021;28:30. doi: 10.1186/s12929-021-00728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki E.A., Takasato M. Perspective: Extending the Utility of Three-Dimensional Organoids by Tissue Clearing Technologies. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.679226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Cakir B., Xiang Y., Sullivan G.J., Park I.H. Synthetic Analyses of Single-Cell Transcriptomes from Multiple Brain Organoids and Fetal Brain. Cell Rep. 2020;30:1682–1689.e3. doi: 10.1016/j.celrep.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X.Y., Xu L., Wang J., Hong Y., Wang Y., Zhu Q., Wang D., Zhang X.Y., Liu C.Y., Fang K.H., et al. DSCAM/PAK1 pathway suppression reverses neurogenesis deficits in iPSC-derived cerebral organoids from patients with Down syndrome. J. Clin. Invest. 2021;131 doi: 10.1172/JCI135763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakurela S., Tiwari N., Schick S., Garding A., Ivanek R., Berninger B., Tiwari V.K. Mapping gene regulatory circuitry of Pax6 during neurogenesis. Cell Discov. 2016;2 doi: 10.1038/celldisc.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo C.A., Gao R., Negraes P.D., Gu J., Buchanan J., Preissl S., Wang A., Wu W., Haddad G.G., Chaim I.A., et al. Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell. 2019;25:558–569.e7. doi: 10.1016/j.stem.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk M., Erwin J.A., Yeo G.W., Gage F.H., Bardy C. Patch-Seq Protocol to Analyze the Electrophysiology, Morphology and Transcriptome of Whole Single Neurons Derived From Human Pluripotent Stem Cells. Front Mol Neurosci. 2018;11:261. doi: 10.3389/fnmol.2018.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A., et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura V., Gerkin R.C. Accurately estimating neuronal correlation requires a new spike-sorting paradigm. Proc. Natl. Acad. Sci. USA. 2012;109:7230–7235. doi: 10.1073/pnas.1115236109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vértesy Á., Eichmüller O.L., Naas J., Novatchkova M., Esk C., Balmaña M., Ladstaetter S., Bock C., von Haeseler A., Knoblich J.A. Gruffi: an algorithm for computational removal of stressed cells from brain organoid transcriptomic datasets. EMBO J. 2022;41 doi: 10.15252/embj.2022111118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa C.E., Cheroni C., Dotter C.P., López-Tóbon A., Oliveira B., Sacco R., Yahya A.Ç., Morandell J., Gabriele M., Tavakoli M.R., et al. CHD8 haploinsufficiency links autism to transient alterations in excitatory and inhibitory trajectories. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110615. [DOI] [PubMed] [Google Scholar]
- Voulgaris D., Nikolakopoulou P., Herland A. Generation of Human iPSC-Derived Astrocytes with a mature star-shaped phenotype for CNS modeling. Stem Cell Rev. Rep. 2022;18:2494–2512. doi: 10.1007/s12015-022-10376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Buth J.E., Haney J.R., Vishlaghi N., Turcios F., Elahi L.S., Gu W., Pearson C.A., Kurdian A., Baliaouri N.V., et al. TGFbeta superfamily signaling regulates the state of human stem cell pluripotency and capacity to create well-structured telencephalic organoids. Stem Cell Rep. 2022;17:2220–2238. doi: 10.1016/j.stemcr.2022.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Buth J.E., Vishlaghi N., de la Torre-Ubieta L., Taxidis J., Khakh B.S., Coppola G., Pearson C.A., Yamauchi K., Gong D., et al. Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. Cell Rep. 2017;21:517–532. doi: 10.1016/j.celrep.2017.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Lin B.J., Chen T.W., Daie K., Svoboda K., Druckmann S. A comparison of neuronal population dynamics measured with calcium imaging and electrophysiology. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Yao H., Negraes P.D., Wang J., Trujillo C.A., de Souza J.S., Muotri A.R., Haddad G.G. Neuronal hyperexcitability and ion channel dysfunction in CDKL5-deficiency patient iPSC-derived cortical organoids. Neurobiol. Dis. 2022;174 doi: 10.1016/j.nbd.2022.105882. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Tanaka Y., Cakir B., Patterson B., Kim K.Y., Sun P., Kang Y.J., Zhong M., Liu X., Patra P., et al. hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell. 2019;24:487–497.e7. doi: 10.1016/j.stem.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Tanaka Y., Patterson B., Kang Y.J., Govindaiah G., Roselaar N., Cakir B., Kim K.Y., Lombroso A.P., Hwang S.M., et al. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell. 2017;21:383–398.e7. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Brawner A.T., Li S., Liu J.J., Kim H., Xue H., Pang Z.P., Kim W.Y., Hart R.P., Liu Y., Jiang P. OLIG2 Drives Abnormal Neurodevelopmental Phenotypes in Human iPSC-Based Organoid and Chimeric Mouse Models of Down Syndrome. Cell Stem Cell. 2019;24:908–926.e8. doi: 10.1016/j.stem.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Forró C., Li T.L., Miura Y., Zaluska T.J., Tsai C.T., Kanton S., McQueen J.P., Chen X., Mollo V., et al. Kirigami electronics for long-term electrophysiological recording of human neural organoids and assembloids. Nat. Biotechnol. 2024 doi: 10.1038/s41587-023-02081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi R., Shigemoto-Kuroda T., Matsuda N., Odawara A., Suzuki I. Electrophysiological responses to seizurogenic compounds dependent on E/I balance in human iPSC-derived cortical neural networks. J. Pharmacol. Sci. 2022;148:267–278. doi: 10.1016/j.jphs.2021.12.006. [DOI] [PubMed] [Google Scholar]
- Yu T., Robotham J.L., Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc. Natl. Acad. Sci. USA. 2006;103:2653–2658. doi: 10.1073/pnas.0511154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Cui W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cell. 2014;6:305–311. doi: 10.4252/wjsc.v6.i3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.C., Wernig M., Duncan I.D., Brüstle O., Thomson J.A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001;19:1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- Zhang W., Yang S.L., Yang M., Herrlinger S., Shao Q., Collar J.L., Fierro E., Shi Y., Liu A., Lu H., et al. Modeling microcephaly with cerebral organoids reveals a WDR62-CEP170-KIF2A pathway promoting cilium disassembly in neural progenitors. Nat. Commun. 2019;10:2612. doi: 10.1038/s41467-019-10497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Rózsa M., Liang Y., Bushey D., Wei Z., Zheng J., Reep D., Broussard G.J., Tsang A., Tsegaye G., et al. Fast and sensitive GCaMP calcium indicators for imaging neural populations. Nature. 2023;615:884–891. doi: 10.1038/s41586-023-05828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yang C., Zhang X. scDART: integrating unmatched scRNA-seq and scATAC-seq data and learning cross-modality relationship simultaneously. Genome Biol. 2022;23:139. doi: 10.1186/s13059-022-02706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Gutierrez C., Xue T., Hampton C., Vergara M.N., Cao L.H., Peters A., Park T.S., Zambidis E.T., Meyer J.S., et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Song H., Ming G.L. Genetics of human brain development. Nat. Rev. Genet. 2024;25:26–45. doi: 10.1038/s41576-023-00626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.