Abstract
The adeno-associated virus type 2 (AAV) replication (Rep) proteins Rep78 and 68 (Rep78/68) exhibit a number of biochemical activities required for AAV replication, including specific binding to a 22-bp region of the terminal repeat, site-specific endonuclease activity, and helicase activity. Individual and clusters of charged amino acids were converted to alanines in an effort to generate a collection of conditionally defective Rep78/68 proteins. Rep78 variants were expressed in human 293 cells and analyzed for their ability to mediate replication of recombinant AAV vectors at various temperatures. The biochemical activities of Rep variants were further characterized in vitro by using Rep68 His-tagged proteins purified from bacteria. The results of these analyses identified a temperature-sensitive (ts) Rep protein (D40,42,44A-78) that exhibited a delayed replication phenotype at 32°C, which exceeded wild-type activity by 48 h. Replication activity was reduced by more than threefold at 37°C and was undetectable at 39°C. Stability of the Rep78 protein paralleled replication levels at each temperature, further supporting a ts phenotype. Replication differences resulted in a 3-log-unit difference in virus yields between the permissive and nonpermissive temperatures (2.2 × 106 and 3 × 103, respectively), demonstrating that this is a relatively tight mutant. In addition to the ts Rep mutant, we identified a nonconditional mutant with a reduced ability to support viral replication in vivo. Additional characterization of this mutant demonstrated an Mg2+-dependent phenotype that was specific to Rep endonuclease activity and did not affect helicase activity. The two mutants described here are unique, in that Rep ts mutants have not previously been described and the D412A Rep mutant represents the first mutant in which the helicase and endonuclease functions can be distinguished biochemically. Further understanding of these mutants should facilitate our understanding of AAV replication and integration, as well as provide novel strategies for production of viral vectors.
Adeno-associated virus type 2 (AAV) is a nonpathogenic human parvovirus that generally depends on coinfection with a helper virus (adenovirus or herpesvirus) for efficient replication (reviewed in reference 10). The linear, single-stranded DNA genome of AAV contains two open reading frames (rep and cap) flanked by 145-bp inverted terminal repeats (ITRs) (62). Replication of the AAV genome requires two viral components, the ITR that serves as the origin of replication (26, 55, 57, 63) and the rep gene products (27, 57, 65). The rep gene encodes four multifunctional proteins (27, 45, 65, 67) that are expressed from two promoters at map units 5 (p5) and 19 (p19). The larger Rep proteins transcribed from the p5 promoter (Rep78 and Rep68 [Rep78/68]), are essentially identical except for a unique carboxy terminus generated from unspliced (Rep78) and spliced (Rep68) transcripts (62). Two smaller Rep proteins (Rep52 and Rep40), transcribed from the p19 promoter, are amino-terminal truncations of Rep78 and Rep68, respectively.
Several biochemical activities of Rep78/68 have been characterized as being necessary for AAV replication. These include specific binding to the AAV ITR (4, 30, 59) and site-specific endonuclease cleavage at the terminal resolution site (trs) (31, 32, 58, 60). Rep78/68 also possess ATP-dependent DNA-DNA helicase (31, 32) and DNA-RNA helicase (73) as well as ATPase (73) activities. In addition to these activities, which are required for replication, Rep78/68 also regulate transcription from the viral promoters (8, 38, 39, 50, 66) and have been shown to mediate viral targeted integration (6, 40, 51, 74). How Rep78/68 mediate these diverse activities in vivo is still under investigation.
Mutant studies of the Rep proteins have indicated that the activities of Rep can be divided into partially distinct functional domains (Fig. 1A) that are spread throughout the protein (14, 21, 43, 48, 68–71, 77). These include regions required for binding to the ITR, a putative nucleoside triphosphate (NTP) binding/ATPase domain, a nuclear localization domain, and residues putatively required for nicking and helicase functions. Rep proteins with mutations within the NTP binding/ATPase domain that lacked trs endonuclease and viral replication were also defective for transactivation functions, suggesting a need for further mutant analysis (43). Since most mutants disrupt multiple Rep-mediated functions for the AAV life cycle, the detailed characterization of distinct functions has been difficult (21, 43, 48, 68–71, 77).
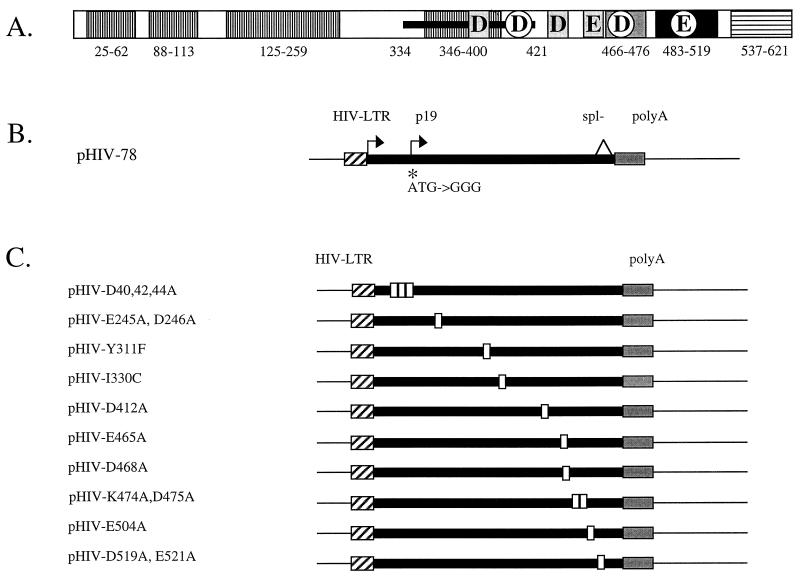
FIG. 1.
AAV Rep78. (A) Schematic diagram of previously described functional domains (see text) within Rep78/68. Regions described as required for binding (vertical stripes), nuclear localization (black box) (amino acids 483 to 519), oligomerization (gray box) (amino acids 466 to 476), and ATPase and helicase activities (black line) (amino acids 334 to 421) and the unique region between Rep78 and Rep68 (horizontal stripes) (amino acids 537 to 621) are indicated. Overlapping DD35E motifs are indicated as boxed residues corresponding to one motif at D368, D429, and E465 and as circled residues corresponding to a second motif at D412, D468, and E504. (B) Eucaryotic expression cassette (pHIV-78) for generating Rep78 proteins. Protein expression is under control of the HIV LTR promoter and the poly(A) signal from simian virus 40. The initiation codon (ATG) at p19 was changed to GGG to eliminate expression of Rep52 and Rep40, and the splice donor site was modified (spl−) as described previously (74) to eliminate expression of Rep68. (C) Positions of point and clustered alanine mutations (open boxes) within the pHIV-78 expression plasmid. Amino acid changes are indicated by the nomenclature for each construct.
The use of temperature-sensitive (ts) mutations has proven to be an effective method for elucidating the essential functions of viral proteins (11, 17, 46, 53). One approach for generating ts mutants has been to utilize the charge-to-alanine mutagenesis strategy (7, 9, 18, 22, 49, 72). This approach has been utilized by others to characterized AAV Rep functions (21, 68); however, none of these mutants were assayed for temperature sensitivity. The rationale of this approach is that since most charged residues are found on the protein surface, they are expected to exert little effect on protein folding or stability (18, 19, 72), but could feasibly make a protein more thermosensitive by disrupting electrostatic and H-bonding interactions (22).
In an attempt to generate ts Rep mutants, we used alanine substitution to target a number of the functional domains critical for Rep-mediated activities. We describe the effects of these mutations on Rep78-mediated replication of an ITR-containing vector in adenovirus-infected human cells. We also assayed several biochemical activities in vitro (i.e., ITR binding, trs endonuclease, and helicase) by using Rep68 His-tagged fusion proteins expressed and purified from bacteria. From these analyses, we identified three classes of mutants (Table 1). Class I mutants were normal by in vivo replication and in vitro biochemical analyses. A class II mutant (D40,42,44A-78) was ts for expression and intracellular replication of AAV, resulting in a 3-log-unit difference in viral titer between the permissive and nonpermissive temperatures. In addition, a class III mutant demonstrated a defect for replication, but protein stability was wild type (wt) in nature. Further analysis of this mutant demonstrated Mg2+-dependent trs endonuclease activity, which was unrelated to either DNA helicase or DNA binding activity. This mutant Rep suggests that a specific interaction between residue D412 and Mg2+ is critical for endonuclease activity. These data represent the first description of a ts AAV Rep protein as well as the putative identification of an Mg2+ binding pocket required for Rep-specific DNA nicking activity. Further characterization of these mutants should prove useful in studying AAV replication and integration and should provide new strategies for AAV vector-packaging cell lines.
TABLE 1.
Summary of Rep protein mutations
| Repa | Activity
|
||||
|---|---|---|---|---|---|
| Expression | Replication | Binding | Nicking | Helicase | |
| Class I (wt) | |||||
| Rep78 (wt) | + | + | + | + | + |
| E245A,D246A | + | + | NDb | ND | ND |
| K474A,D475A | + | + | ND | ND | ND |
| D519A,E521A | + | + | ND | ND | ND |
| I330C | + | + | ND | ND | ND |
| Y311F | + | + | + | + | + |
| E465A | + | + | + | + | + |
| D468A | + | + | + | + | + |
| E504A | + | + | +c | +c | +c |
| Class II (ts), D40,42,44A | +/−de | +/−e | +f | +f | ND |
| Class III, D412A | + | +/− | + | +g | + |
Mutant proteins are designated by the site(s) of their amino acid change(s).
ND, not determined.
Activity required 50-fold more protein than for the wt.
+/−, activity was less than 50% of wt.
Activity was ts.
Activity reduced relative to wt.
Activity was sensitive to magnesium concentration.
MATERIALS AND METHODS
Plasmids and site-directed mutagenesis.
All recombinant DNA manipulations were performed by standard protocols (5). Unless otherwise noted, all enzymes were purchased from New England Biolabs and used according to supplier recommendations. The mutants used in this study were originally generated in the plasmid pHIV-78 (74). pHIV-78 (Fig. 1B) is a derivative of pHIV-Rep (2). pHIV-Rep contains a wt rep gene and expresses all four Rep proteins from the human immunodeficiency virus (HIV) long terminal repeat (LTR). To express Rep78 in the absence of the other rep gene products, the splice site was modified to prevent splicing and subsequent generation of the Rep68 and Rep40 proteins (74). In addition, the initiation codon following the p19 promoter was changed to GGG (13) to abrogate expression of Rep52. For simplicity, we have designated the Rep78 protein expressed from pHIV-78 the wt Rep78 control, with the recognition that the rep gene has been modified as described above.
Mutations were generated in the pHIV-78 construct by using a Vent polymerase site-directed mutagenesis procedure essentially as described previously (12), with an additional DpnI digestion step to remove the template DNA. DNA sequencing (done at the University of North Carolina sequencing facility) confirmed point mutations. Regions immediately adjacent to the mutations were subcloned back into the original pHIV-78 vector backbone and sequenced to eliminate the possibility that additional mutations were introduced during PCR mutagenesis.
Several of these mutations were also subcloned from pHIV-78 into an inducible bacterial expression vector, pStump68 (78a), that expresses the Rep68 protein. The Rep68 proteins contain a C-terminal His6-tag (Rep68H6) for purification of the recombinant proteins over nickel columns. Subcloned mutations were confirmed by DNA sequencing, and inducible expression in Escherichia coli was confirmed by Western blot analysis (data not shown).
The mutant D40,42,44A was also generated in the plasmid pIM45, which contains the wt AAV genome without the terminal repeats (TRs) as previously described (42). For clarity, these mutant Rep proteins, which are under the control of the p5 promoter, will be designated mutD40,42,44A to distinguish them from D40,42,44A-78 described in Fig. 1C. The mutD40,42,44A construct was generated by site-directed mutagenesis as previously described (37) with the oligonucleotide 5′ CCG CCA GCT TCT GCC ATG GCT CTG AAT 3′. The pIM45 (wt) and mutD40,42,44A plasmids were used to produce virus from the plasmid pTRUF5, which contains the green florescent protein (GFP) flanked by the AAV TRs (79), to generate the virus rAAV-UF5 (GFP). An additional recombinant AAV (rAAV) plasmid, pAB-11, that was used in these studies has previously been described (24). Briefly, pAB-11 contains the β-galactosidase gene under the control of the cytomegalovirus immediate-early promoter flanked by the AAV ITRs.
Intracellular replication assays and viral replication time course.
Subconfluent 293 cells were cotransfected with mutant or wt pHIV-78 helper constructs and the rAAV plasmid (pAB-11) at a 3:1 molar ratio, respectively, via Lipofectin (Gibco/BRL) essentially as described in the supplier protocol. After 14 h, the DNA-Lipofectin complexes were replaced with medium containing enough adenovirus type 5 (dl309 [33]) to infect cells at a multiplicity of infection (MOI) of 5. When transfections were performed to evaluate ts activity of the mutants, all reagents for transfection, including the cells and medium, were prewarmed and incubated at the appropriate temperatures (32, 37, or 39°C) following transfection. At 48 h postinfection, low-molecular-weight DNA was extracted (28), treated with RNase A, and digested extensively with DpnI to remove input plasmid DNA. Southern blots were performed on digested DNA (5) and probed with 32P-labeled β-galactosidase. Viral replication was determined by coinfecting subconfluent human embryonic kidney (HEK) 293 cells with rAAV-UF5 (MOI = 5) and adenovirus type 5 (MOI = 5) and simultaneously transfecting with the mutD40A,D42A,D44A or pIM45 plasmid. Hirt DNA was analyzed by Southern blotting with a vector GFP-specific probe.
Production of rAAV GFP virus by using ts Rep plasmids.
In order to produce rAAV-UF5 virus, HEK 293 cells were cotransfected by the CaPO4 method as previously described (61) with pTRUF5 (79), helper plasmid pXX6 to supply adenovirus functions (75), and either wt pIM45 or mutD40,42,44A to supply Rep functions. Following transfection, the cells were incubated for 48 h, harvested, and subjected to three successive freeze-thaw cycles to liberate the virus. Incubations were performed at 32, 37, and 39.5°C. The crude viral preparations containing rAAV-UF5 were then titrated by counting GFP single-cell fluorescence.
ECL-Western blotting.
ECL-Western blots analyses were performed on transfected-cell lysates essentially as recommended by the supplier (Amersham), and blots were probed with a monoclonal antibody raised against all four of the AAV Rep proteins (29).
Purification of mutant and wt Rep68 His-tagged fusion proteins.
His-tagged Rep68 fusion proteins (Rep68H6) were purified from SG13009 cells (Qiagen) by passage over nickel columns as previously described (78a) with minor modifications. Briefly, cleared cell extracts were incubated with Ni-nitrilotriacetic acid agarose (Qiagen) for 3 to 4 h at 4°C, followed by elution with a 0.1 to 0.5 M imidazole linear gradient. Eluted proteins were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and silver staining (see Fig. 4) according to the manufacturer’s protocol (Bio-Rad Silver Stain Plus). Protein concentrations were determined with a bicinchoninic acid kit (Pierce) with bovine serum albumin (BSA) as a standard and by direct comparison to known amounts of BSA on a silver-stained SDS-polyacrylamide gel.
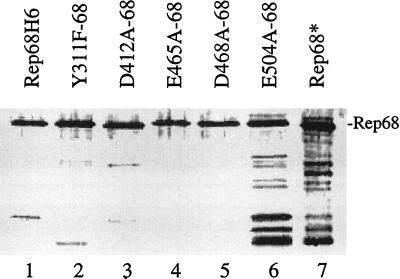
FIG. 4.
SDS-polyacrylamide gel electrophoresis–silver stain analysis of purified mutant and wt Rep68H6 proteins overexpressed in E. coli. Proteins were expressed from the inducible expression cassette pStump68 and purified via a His6 tag by passage over nickel-agarose columns (Qiagen) as described in Materials and Methods. The position of full-length Rep68H6 is indicated. The same amount of protein was loaded in each lane, except for E504A-68, which required 50-fold more protein to achieve the same level of full-length protein. The lower-molecular-weight products visible in the gel (especially in lanes 6 and 7) were mostly degradation products as indicated by Western blot analysis (data not shown). Rep68* was expressed in a baculovirus system and purified as described previously (30).
DNA substrates for in vitro biochemical assays.
The AAV TR hairpin DNA used in the majority of the biochemical assays was prepared by EcoRI digestion of pDD (76), generating a 171-bp DNA fragment with D sequences flanking either side of the TR (D′ A C′ C B′ B A′ D). The substrate was boiled and snap cooled to form a hairpin with a double-stranded trs and then labeled with [γ-32P]ATP by using T4 polynucleotide kinase to generate the substrate, 32P-TR.
An AAV TR substrate with a single-stranded trs region (32P-TRss) was generated by digestion of psub201 (55) with XbaI and PuvII as previously described (30) and labeled at the 5′ end with T4 polynucleotide kinase.
The DNA substrate for the helicase assays was generated by annealing a 24-base primer (catalog no. 1224; New England Biolabs) to an M13 single-stranded phage (catalog no. 70704; United States Biochemical) essentially as described previously (31). The substrate (M13/24) was labeled with [α-32P]dATP by using the Klenow fragment of DNA polymerase in the presence of 0.5 mM dTTP and dGTP. For all substrates, the unincorporated nucleotides were removed by passage over G25 spin columns (Boehringer Mannheim).
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays were performed essentially as previously described (44). Briefly, 0.01 to 0.02 pmol of 32P-TR was incubated with up to 9 ng (∼0.15 pmol) of mutant or wt Rep68H6 protein in 20-μl binding reaction mixtures (40 mM KCl, 10 mM HEPES-KOH [pH 7.5], 0.2 mM dithiothreitol, 5% [vol/vol] glycerol, 0.5 μg of BSA, and 1 μg of poly[dI-dC]) at 25°C for 30 min. Protein-DNA complexes were resolved on 5% nondenaturing polyacrylamide gels and quantitated with a phosphorimager. Assays were performed in triplicate, and standard errors are indicated in the figures.
trs endonuclease assays.
Site-specific endonuclease activity was screened by trs endonuclease assays as previously described (32), except where indicated otherwise. Briefly, 20-μl reaction mixtures (25 mM HEPES-KOH [pH 7.5], 5 mM MgCl2, 1 mM dithiothreitol, 0.4 mM ATP, 10 μg of BSA per ml) containing 0.005 to 0.02 pmol of 32P-TR (or 32P-TRss) and up to 9 ng (∼0.15 pmol) of mutant or wt Rep68H6 were incubated at 37°C for 60 min. Reaction products were treated with protease K for 60 min, extracted with phenol-chloroform, precipitated, and resolved on denaturing sequencing gels. The amount of product formed was determined with a phosphorimager. Assays were performed in triplicate, and standard errors are indicated in the figures.
Helicase assays.
The helicase assays (31) were performed in the same reaction buffer as for the trs endonuclease assay except where indicated otherwise. Briefly, 20-μl reaction mixtures containing 0.01 to 0.02 pmol of M13/24 and up to 9 ng of mutant or wt Rep68H6 protein were incubated for 30 min at 37°C. Reactions were terminated by addition of 10 μl of stop solution (0.5% SDS, 50 mM EDTA [pH 7.5], 50% glycerol, 0.1% bromphenol blue, 0.1% xylene cyanole), resolved by electrophoresis on a 5% nondenaturing polyacrylamide gel, and quantitated with a phosphorimager. Assays were repeated a minimum of three times, and standard errors are indicated in the figures.
Structural modeling.
Computational results were obtained by using software programs from Molecular Simulations Inc. Dynamics calculations were performed with the Discover Æ program, using the CFF91 forcefield; ab initio calculations were done with the DMol program; and graphical displays were printed out from the Cerius2 molecular modeling system.
RESULTS
Generation of mutant and “wt” Rep78 constructs.
In an attempt to generate ts Rep mutants, we used a charge-to-alanine substitution strategy (7, 9, 18) to target a number of the putative functional domains critical for Rep-mediated activities (Fig. 1A). Site-directed mutations were generated in a eucaryotic expression construct, pHIV-78 (74), that expresses only the 78-kDa AAV Rep protein (Fig. 1B). In this construct, the rep gene has been altered to prevent p19 protein expression (ATG converted to GGG), as previously described (13). In addition, the 5′ splice site was inactivated so that only the Rep78 protein initiated from the p5 promoter was expressed (as described in Materials and Methods) (74). For simplicity, we have designated the Rep78 protein expressed from pHIV-78 the wt Rep78 control. By using intracellular expression, AAV-mediated replication, and in vitro biochemical assays, the mutants described in Fig. 1C were grouped into three classes (class I, non-ts and normal; class II, ts for replication; and class III, non-ts and defective for replication) (Table 1).
Intracellular expression of mutant and wt Rep78 proteins.
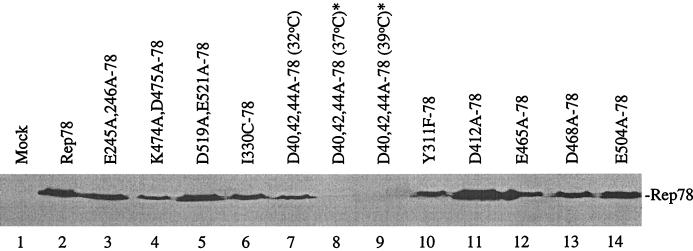
All of the mutant plasmids were sequenced and characterized for the ability to express full-length Rep protein by transfection in 293 cells. ts expression was determined by transfecting mutant and wt pHIV-78 constructs into 293 cells at 32, 37, and 39°C, followed by Western blot analysis. A representative blot of protein expression is shown in Fig. 2. Based on protein expression at different temperatures, we designated two classes of Rep mutants. The class I mutant proteins were expressed at normal levels from the HIV LTR promoter, whereas the class II mutant D40,42,44A-78 was present at levels below the detection sensitivity of the ECL-Western blot analysis at 37 and 39°C (Fig. 2, lanes 8 and 9). Expression of this class II mutant was confirmed at 37°C by overexposing Western blots (data not shown) and by comparing lysates prepared from cells transfected at 32°C (Fig. 2, lane 7). Detection of the class II mutant protein at 32°C but not at 37°C suggested a ts Rep phenotype (22).
FIG. 2.
Immunoblot analysis of the various Rep78 proteins expressed from the HIV LTR in transiently transfected 293 cells. Mutant and wt pHIV-78 constructs were transfected into 293 cells at 32, 37, and 39°C as described in Materials and Methods. A representative blot of protein expression is shown. With the exception of D40,42,44A, all lysates were prepared from cells transfected at 37°C. Blots were hybridized with a monoclonal antibody that recognizes all four Rep proteins. All lanes were loaded with 15 μl of cellular extract. ∗, proteins were detected after long-term exposure of blots.
Replication activities of mutant Rep78 proteins.
The intracellular functional activities of the Rep mutants (Fig. 3A) and wt Rep78, wt Rep68, and a construct (wt Rep) expressing all four Rep proteins (Fig. 3A, compare lanes 1, 14, and 15) were determined by assaying the ability to mediate replication of an rAAV plasmid (pAB-11) in the presence of adenovirus. The replication activities of control plasmids expressing either wt Rep78 or Rep68 appeared to be equivalent (Fig. 3A, compare lanes 1 and 14). As expected, the construct expressing all four Rep proteins (wt Rep) generated slightly more replicated DNA in this assay (Fig. 3A, compare lanes 1 and 14 to 15). Mutants that had less than 50% of wt Rep78 activity at 37°C were considered to be defective for replication. Intracellular replication assays were also performed at 32 and 39°C to determine if the Rep mutants were ts in activity.
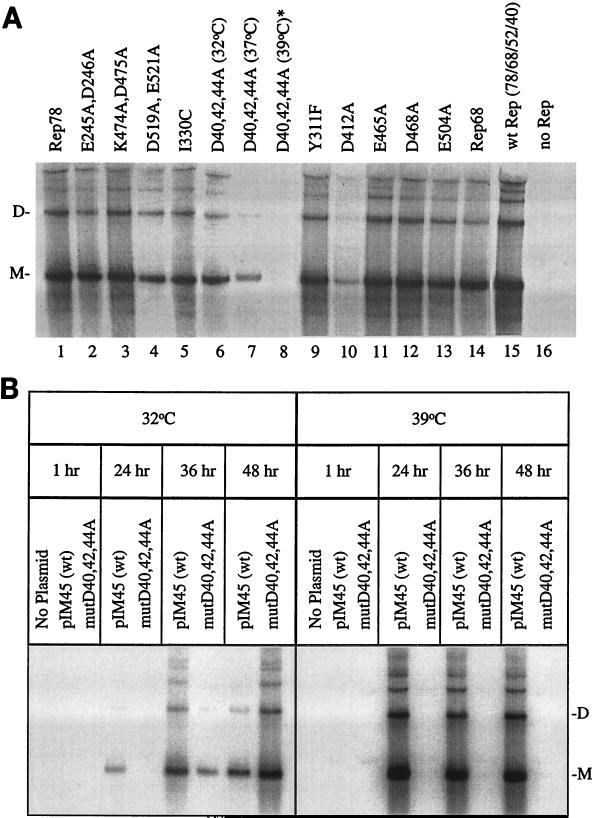
FIG. 3.
Southern blot analysis of intracellular replication mediated by various Rep78 proteins. (A) Representative Southern blot of rAAV-LacZ (pAB-11) replication mediated by mutant and wt Rep78 proteins expressed from the HIV LTR in adenovirus type 5-infected 293 cells. The replication assays shown were performed at 37°C, except as indicated for D40,42,44A (lanes 6 to 8). The pHIV-78 and pAB-11 constructs were transfected at a 3:1 molar ratio as described in Materials and Methods. Following transfection, the cells were incubated for 48 h with adenovirus type 5 (MOI of 5). At 48 h postinfection, Hirt DNA was extracted, digested with DpnI, and analyzed by Southern blotting with a 32P-labeled probe specific for β-galactosidase sequences. Replicative monomer (M) and dimer (D) DNA forms are indicated. ∗, replicated forms of pAB-11 were detected only after long-term exposure of blots. Rep68 was expressed from pHIV-68, a derivative of the pHIV-78 construct that has the rep gene intron removed as described previously (74). wt Rep is expressed from the pHIV-Rep construct, which expresses all four rep gene products, as described in Materials and Methods. (B) Southern blot analysis of replication of an rAAV-UF5 virus (GFP) mediated by transfected wt pIM45 or mutD40A,D42A,D44A pIM45 Rep expression plasmids. The pIM45 constructs express all four rep gene products (see Materials and Methods). Transfection infections were performed at permissive (32°C) and nonpermissive (39.5°C) temperatures in adenovirus type 5-infected HEK 293 cells. Rep-containing plasmids (wt pIM45 or mutD40,D42,D44A pIM45) were transfected into subconfluent HEK 293 cells and simultaneously coinfected with adenovirus type 5 (MOI of 5) and rAAV-UF5 (MOI of 5). At various time points, Hirt DNA was extracted and analyzed by Southern blotting as described above with a 32P-labeled probe specific for GFP sequences. No-plasmid lanes, cells were infected with adenovirus type 5 only; pIM45 (wt) lanes, cells were coinfected with adenovirus type 5 and rAAV-UF5 and transfected with wt pIM45; mutD40,42,44A lanes, cells were coinfected with adenovirus type 5 and rAAV-UF5 and transfected with mutD40,D42,D44A,pIM45.
All of the class I mutants except D412A possessed replication activities that were similar to that of wt Rep78 under physiological (Fig. 3A, lanes 2 to 5, 9, and 11 to 13) and nonphysiological (data not shown) conditions. The class II mutant, D40,42,44A-78, exceeded wt Rep78 replication at 32°C, but its activity was reduced over threefold at 37°C and diminished to almost nondetectable levels at 39°C (Fig. 3A, lanes 6 to 8). Both the temperature-dependent expression and replication profiles for D40,42,44A-78 (Fig. 2, lanes 7 to 9, and 3A, lanes 6 to 8) indicate that this variant had a ts phenotype.
In addition to assaying AAV replication with the ts mutant expressed from the HIV promoter cassette, the mutant was also assayed in the context of all of the other AAV Rep proteins (pIM45 helper construct, see Materials and Methods). In these assays, replication was determined by Southern blotting of Hirt extracts with an rAAV GFP vector (pTRUF5) for replication and GFP transduction for virus yield (Fig. 3B and Table 2, respectively). At 32°C, wt Rep demonstrated highest levels of replication at 36 h, which diminished by 48 h (Fig. 3B). At 32°C the ts mutant was delayed at 24 h, but replication increased steadily and surpassed that of the wt at the 48-h time point. At the nonpermissive temperature of 39°C, the ts mutant was nonviable, while wt Rep demonstrated normal levels of AAV replication at all time points (Fig. 3B). Virus yields for the ts mutant (mutD40,42,44A) at 32, 37, and 39°C were 2.2 × 106, 3 × 104, and 3 × 103 transducing units/ml, respectively, thus displaying over a 3-log-unit difference in titer between the permissive and nonpermissive temperatures (Table 2). From these analyses, charge-to-alanine mutant D40,42,44A-78 displayed all of the characteristics of a ts Rep protein.
TABLE 2.
Yield of rAAV-UF5 (GFP) virus
| Rep | Yield (transducing units/ml)a at:
|
||
|---|---|---|---|
| 32°C | 37°C | 39.5°C | |
| pIM45 (wtb) | 9.4 × 105 | 1.7 × 106 | 6.8 × 105 |
| mutD40,42,44Ac | 2.2 × 106 | 3.0 × 104 | 3.0 × 103 |
See text.
All four rep gene products are expressed.
Mutations were generated in the pIM45 vector.
After characterization of this ts mutant Rep protein, we turned our analysis to other variants that displayed a replication phenotype but did not appear to be temperature sensitive. The mutant D412A-78, which was originally identified as having a class I mutant Rep phenotype based on protein expression (Fig. 2, lane 11), had AAV replication activity 5- to 10-fold lower than that of wt Rep78 at all temperatures (Fig. 3A, lane 10, and data not shown). For this reason, D412A-78 was defined separately as a class III mutant (normal protein levels but decreased replication activity and not ts) and was further analyzed.
In addition to the class II and III mutants characterized in this replication assay, a high level of replication was mediated by three class I mutant proteins, K474,D475A-78, Y311F-78, and E465A-78 (Fig. 3A, lanes 3, 9, and 11, respectively). These results are noteworthy considering that mutants with similar mutations at these positions have previously been shown to be negative for trs endonuclease activity when measured in vitro in the context of the maltose-binding protein (MBP)–Rep68Δ fusion protein (21, 69).
In vitro expression and purification of mutant Rep proteins.
In an effort to further characterize the non-ts Rep mutants initially evaluated in the intracellular replication study, several mutant constructs (Y311F, D412A, E465A, D468A, and E504A) were subcloned into the bacterial expression vector pStump68 (78a) for purification and characterized of Rep-specific biochemical activities. The pStump68 construct allowed for high-level expression and purification of Rep68 proteins from E. coli via a His6-tag fusion. Overexpression and purification from this vector enabled us to compare the activities of the mutant Rep proteins to well-established biochemical activities previously assigned for wt Rep68 (30–32). Silver stained SDS-polyacrylamide gels (Fig. 4) and Western blot analysis (data not shown) demonstrated that the His6-tagged proteins were purified to greater than 95% homogeneity. With the exception of mutant E504A-68, the Rep mutants expressed similar amounts of stable Rep68 protein (Fig. 4). Mutant E504A-68 required 50 times more total protein when assayed under these conditions and appeared to have a pronounced profile of breakdown products that were similar to those of Rep68 purified from baculovirus (Fig. 4, lanes 6 and 7). This was in contrast to its expression in 293 cells, which appeared to be normal (Fig. 2, lane 14). These two contradicting data suggest that this mutant may be unstable in the context of the His-tagged fusion protein. While this result exemplifies the concern of modifying Rep for E. coli expression, all other constructs generated identical protein profiles both in vivo and in vitro, supporting further analysis of their biochemical activities.
Interaction of Rep68H6 proteins with the AAV TR.
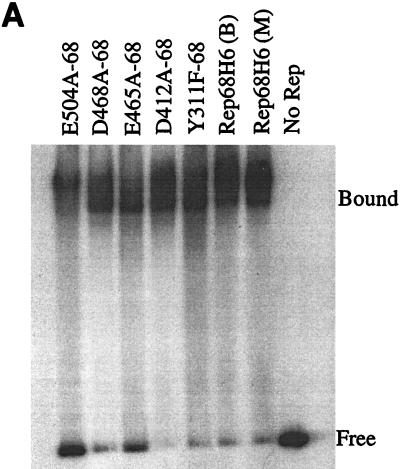
Standard mobility shift assays were used to compare the specific binding of mutant and wt Rep68H6 proteins to the hairpin TR substrate (32P-TR). Binding titration profiles performed with each class of mutant protein were similar to that for wt Rep68H6 (data not shown). A representative binding assay with a protein-to-DNA molar ratio of ∼10:1 demonstrated that all of the His-tagged E. coli-produced proteins bound specifically to 32P-TR (Fig. 5A). Specific binding to the TR suggested that neither the point mutations nor the addition of the His6-tag significantly altered the DNA binding activities of these Rep68 proteins. However, as observed in the silver stain analysis, approximately 50-fold more total protein was used in the binding assays for E504A-68 relative to wt Rep68H6.
FIG. 5.
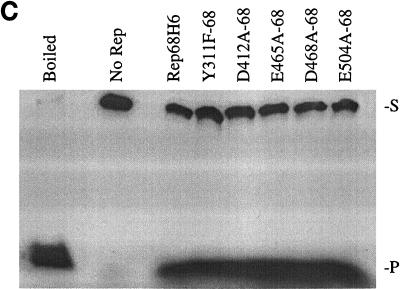
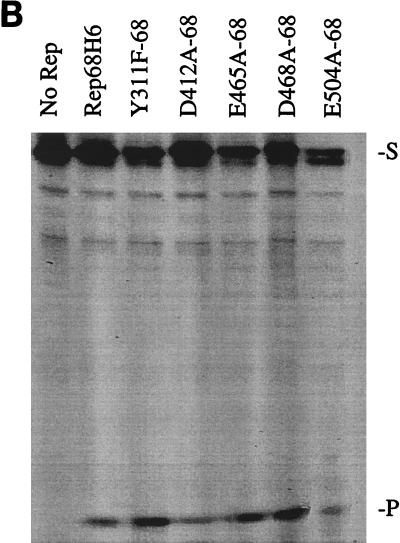
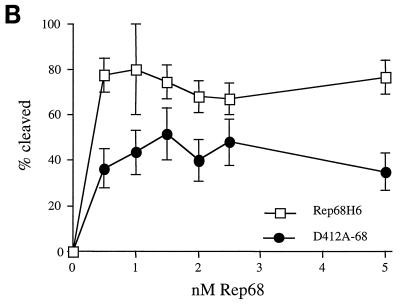
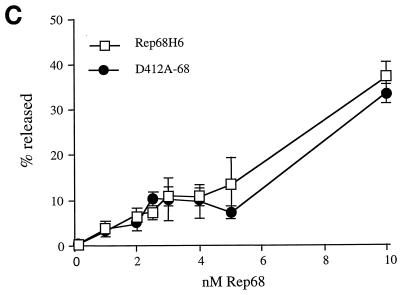
In vitro biochemical analyses of His-tagged Rep68 proteins. (A) Gel mobility shift assay of mutant and wt Rep68H6 proteins expressed in E. coli. Standard binding reaction mixtures contained 0.5 nM 32P-labeled AAV TR hairpin DNA (32P-TR) and 6 nM purified Rep68 protein containing the indicated mutation. Reactions were carried out in standard binding reaction buffer (see Materials and Methods) at 25°C for 30 min, and products were resolved on nondenaturing 5% polyacrylamide gels. Mutant and Rep68H6 (B) proteins were purified by one passage over nickel columns (batch) while Rep68H6 (M) was further purified by passage over a Mono Q column. (B) Terminal resolution by His-tagged Rep68 proteins. Standard trs endonuclease assays contained 0.5 nM 32P-TR substrate and 5 nM His-tagged protein. Nicking reactions were performed in standard nicking buffer (see Materials and Methods) for 60 min at 37°C, followed by protease K digestion and phenol-chloroform extraction, and products were resolved on denaturing polyacrylamide sequencing gels. Substrate (S) and cleavage products (P) are indicated. (C) Standard helicase reactions were performed at 37°C for 60 min in nicking buffer containing 0.5 nM substrate (M13/24) and 6 nM protein, and products were resolved on 5% nondenaturing polyacrylamide gels. Substrate (S) and released product (P) are indicated. Boiled, positive control. In all of the assays described for Fig. 5, E504A-68 required 50-fold more protein to achieve the level of activity shown.
Effect of mutations on Rep68H6 trs endonuclease activity.
Site-specific and strand-specific endonuclease cleavage of the AAV TR by Rep78/68 is an essential part of AAV replication (47). All of the mutants examined specifically nicked the TR at the trs, generating the proper-sized product (Fig. 5B). In the nicking assay shown (Fig. 5B), a protein-to-DNA molar ratio of ∼10:1 was used for all of the mutants except E504A-68, which required ∼50-fold more total protein to obtain the same level of cleaved product.
Also of interest was the wt level of cleavage observed with the mutants Y311F-68 and D465A-68 (Fig. 5B). From previously published studies, Rep proteins with mutations at these positions were not expected to display nicking activity (69). We suspect that the discrepancy between our analysis and others concerning in vitro biochemical activities (69) are related to the MBP-Rep68 fusion protein. With the exception of mutant E504A-68, we did not observe an influence on Rep activity in the context of the Rep68H6 protein. In addition, these same mutants when expressed from the HIV LTR promoter cassette in vivo without the His modification all displayed AAV replication activity, supporting the in vitro biochemical analyses.
Effect of mutations on Rep68H6 helicase activity.
Under the reaction conditions used in these assays (∼10:1 protein-to-DNA molar ratio), all of the Rep68H6 mutants examined exhibited helicase activity (Fig. 5C).
The retention of nicking and helicase activities by D412A-68 was somewhat surprising, considering that this mutant was notably defective in the intracellular replication assay (Fig. 3, lane 10). In an effort to assign a phenotype to the D412A mutation, more comprehensive biochemical analyses comparing the activity of D412A-68 to that of Rep68H6 were conducted.
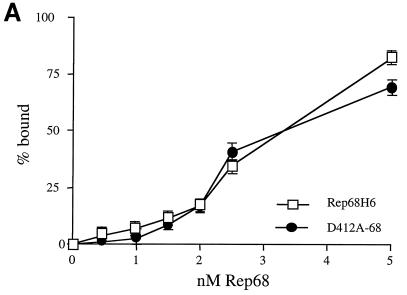
Affinity of D412A-68 versus Rep68H6 for the hairpin TR.
In the assays described above, a difference in binding between D412A-68 and wt Rep68H6 was not detected. This suggested that the replication defect observed with D412A-78 in the intracellular assays was not due simply to an inability to interact efficiently with the viral TR. While we used 500-fold less Rep protein than used in other studies (69), we still had excess protein relative to the TR concentration, potentially masking any subtle differences in binding efficiency. In order to determine the concentrations of D412A-68 and wt Rep68H6 required to bind 50% of the substrate (KD), binding assays were performed as a function of protein concentration. The apparent KD values were determined from plots of the percentage of TR bound versus protein concentration (Fig. 6A). The apparent KD values for wt Rep68H6 and D412A-68 for the TR were essentially the same, at about ∼3 nM (Fig. 6A). These values, which were derived from assays performed in triplicate, were also within the nanomolar range previously described for baculovirus-purified Rep68 binding to a hairpin TR substrate (44).
FIG. 6.
Binding, trs endonuclease, and helicase activities of D412A-68 and Rep68H6 as a function of protein concentration. (A) Standard binding reactions were performed as described for Fig. 5A and carried out in triplicate with 1 nM substrate (32P-TR) and the indicated concentration of each protein. (B) Standard nicking reactions were performed as described for Fig. 5B and carried out in triplicate with 1 nM 32P-TR substrate and the indicated concentration of each protein. (C) Standard helicase reactions were performed as described for Fig. 5C and carried out four times in nicking buffer containing 1 nM substrate (M13/24) and the indicated concentration of protein.
Titration of trs endonuclease and helicase activities.
In an effort to identify gross biological differences between the mutant and wt Rep (Fig. 5B and C), protein levels were in excess relative to the respective DNA substrates. In the following assays, the efficiency of D412A-68 compared to that of wt Rep68H6 in the trs endonuclease and helicase activities was determined as a function of protein concentration (Fig. 6B and C, respectively). In the nicking assay comparisons, nearly threefold more D412A-68 protein was required to nick 50% of the substrate, and about 25% less substrate was cleaved overall relative to that with wt Rep68H6 (Fig. 6B). In contrast, no significant differences were observed between the efficiencies of D412A and wt Rep68H6 in the helicase assay (Fig. 6C).
Effect of magnesium concentration on Rep68 functions.
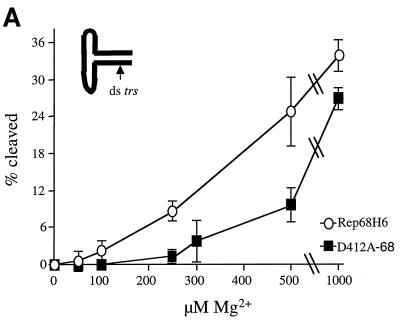
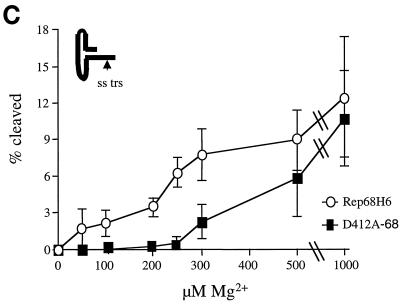
Previous studies have shown that the endonuclease activity of Rep68 requires magnesium (Mg2+) (31). In the nicking reactions described above, the Mg2+ concentration [Mg2+] was held constant at 5 mM, which is approximately 10-fold greater than would be present in the intracellular environment (1). Under such conditions, if an interaction between Mg2+ and Rep68 was necessary to form an active transition state for nicking, a difference in binding affinity for Mg2+ might go unnoticed. Excess Mg2+ in the nicking reactions should shift the equilibrium in favor of binding, therefore potentially masking a mutant protein phenotype. For this reason, nicking reactions were performed under conditions of low [Mg2+] (Fig. 7A). Neither of the proteins was very efficient at nicking the TR when the [Mg2+] was <500 μM. However, wt Rep68H6 activity increased linearly with [Mg2+] and cleaved about 14-fold more substrate at 250 μM Mg2+. In contrast, we could not detect any cleaved substrate with D412A-68 at <200 μM Mg2+. The sensitivity to [Mg2+] with D412A-68 suggested that this mutant had a diminished ability to bind Mg2+ and that an interaction between Mg2+ and D412 was necessary for efficient trs endonuclease activity in vitro.
FIG. 7.
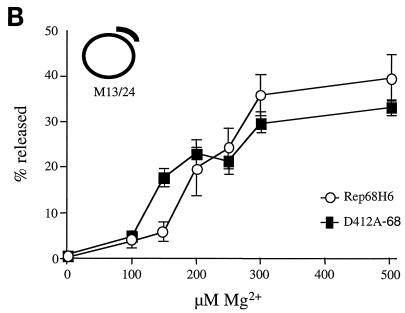
trs endonuclease and helicase activities of D412A-68 and Rep68H6 as a function of magnesium concentration. (A) Endonuclease reactions were performed as described for Fig. 5B and carried out in triplicate. Nicking buffer contained 1 nM substrate (32P-TR) with a double-stranded (ds) nicking site (trs), 3 nM protein, and the indicated [Mg2+]. (B) Helicase assays were performed as described for Fig. 5C and carried out in triplicate in nicking buffer that contained 1 nM helicase substrate (M13/24), 3 nM protein, and the indicated [Mg2+]. (C) Nicking reactions were performed as described for Fig. 5B and carried out in triplicate. Nicking buffer contained 1 nM substrate (32P-TRss) with a single-stranded (ss) nicking site (trs), 3 nM protein, and the indicated [Mg2+].
In addition to requiring Mg2+ for endonuclease activity, Rep also requires the ATP-dependent helicase activity to nick the viral TR (59). The Rep helicase activity was suggested to be necessary to unwind the double-stranded TR in order to expose the trs (59). This function also requires Mg2+ (31). Thus, the sensitivity to [Mg2+] in the above-described nicking assays could also reflect an impairment of helicase activity. To address this possibility, we titrated [Mg2+] levels in a Rep-dependent helicase assay previously described (31). From this analysis, D412A-68 and wt Rep68H6 had similar Mg2+ requirements in the helicase assay (Fig. 7B).
To further demonstrate that the Mg2+ dependence of the class III mutant D412A was related to Rep’s trs endonuclease activity and not related to helicase function, we performed TR nicking assays with a modified TR substrate. Previously, Snyder et al. (59) generated a TR substrate with the nicking site (trs) existing as a single-stranded DNA. Those authors were able to demonstrate wt Rep nicking activity in the absence of ATP, suggesting that the helicase function to expose the nicking site was not required. We performed identical nicking assays with the D412A mutant Rep and demonstrated that the amount of cleaved product with this single-stranded substrate (32P-TRss) was also sensitive to Mg2+ levels (Fig. 7C). This data supports the role of Rep amino acid D412 in Mg2+ binding and suggests that this interaction is important in AAV trs endonuclease activity.
DISCUSSION
In the absence of a crystal structure, assignment of structure-function activities to the AAV Rep proteins has relied on mutational analysis studies. Mutational analysis of the AAV Rep proteins has indicated that Rep-mediated activities can be attributed to somewhat distinct functional domains (Fig. 1A) (14, 21, 43, 48, 68–71, 77). However, in these mutational studies, multiple effects were observed due to the disruption of more than one Rep biological activity. In this study we used a charge-to-alanine mutagenesis strategy (7, 9, 18) to target a number of Rep78/68 functional domains in an attempt to generate a collection of ts mutants (22, 49, 72). We describe the effects of these mutations on Rep78-mediated replication of an rAAV vector in adenovirus-infected 293 cells and on in vitro biochemical assays with purified Rep68 His-tagged fusion proteins expressed in bacteria. Using this alanine substitution strategy, we generated mutants that were grouped into three classes based on their functional activities (Table 1). These included class I mutants (E245A,D246A, Y311F, I330C, E465A, D468A, K474A,D475A, and E504A), which retained normal activity; a class II mutant (D40,42,44A), which was ts for replication; and a class III mutant (D412A), which was not ts but was defective for replication and more [Mg2+] dependent for in vitro endonuclease activity.
The expression and replication activity of the mutant Rep proteins relative to those of a wt Rep78 protein in 293 cells at permissive and nonpermissive temperatures were characterized. Similar to results of previous alanine mutagenesis studies (9), the majority of the mutants (class I and III) were stably expressed at levels similar to those of wt protein (wt Rep78) (Fig. 2). The class I mutants also exhibited wt activities in the intracellular replication assays (Fig. 3A) and in the biochemical assays in vitro (Fig. 5). Interestingly, we determined that three of the mutants (Y311F, E465A, and K474,D475A) grouped within this class were active, whereas similar mutants have previously been shown to lack endonuclease activity in the context of the MBP-Rep68 fusion protein (21, 69). The discrepancy between these observations suggests a need for concern when evaluating only in vitro Rep activity by using fusion proteins. For this reason, we first assayed all of the mutants in intracellular replication assays in the context of the Rep78 protein (Fig. 3A) and supported these analyses by in vitro studies. Of the mutant Rep proteins that we analyzed in vitro, only mutant E504A displayed protein stability characteristics contrary to those seen in in vivo analysis (compare Fig. 2, lane 14, to Fig. 4, lane 6). Since completion of our study, Davis et al. (21) generated an MBP Rep fusion protein with mutations identical to that of the ts mutant we describe here. In these in vitro studies, D40,42,44–MBP-68 does not bind or nick the TR, contrary to our in vitro and in vivo results.
While steady-state protein levels for the class II ts mutant (D40,42,44A) at 37°C were generally below the sensitivity limits of the ECL-Western blot analyses (Fig. 2), expression was shown to be at wt levels at 32°C. This class II mutant was also notably defective for replication under physiological conditions (Fig. 3A). D40,42,44A-78 mediated replication much more efficiently at 32°C (∼3-fold) than at 37°C and was essentially inactive at 39°C. The instability of the class II mutant Rep protein (D40,42,44A) corroborated ts replication data (Fig. 3), allowing us to assign a ts replication phenotype. When this mutant was assayed in the context of all four Rep proteins (pIM45 plasmid), its activity was similar to that seen with the HIV construct (Fig. 3B), although the stability of the pIM45 mutant Rep was not characterized. In addition, a 3-log-unit difference in titer was observed when permissive and nonpermissive temperatures were compared (Table 2). All of these analyses support Rep mutant D40,42,44A-78 having a ts phenotype. The 3-log-unit difference in Rep function demonstrates that this is a tight mutant and should be useful in further analysis of Rep function.
Mg2+ binding mutant.
The mutant D412A was categorized in a separate class (class III), since it was expressed at wt levels (class I) but did not exhibit a temperature-dependent phenotype (class II). However, this mutant was markedly reduced in its ability to mediate replication in the intracellular assays (Fig. 3A). When the biochemical activities of D412A were analyzed in the context of the Rep68 His-tagged fusion protein, this mutant bound efficiently to the TR (Fig. 6A) but had a reduced level of endonuclease activity relative to wt Rep68H6 (Fig. 6B). The reduced nicking activity of D412A-68 was shown with both single-stranded and double-stranded substrates and appeared to be more Mg2+ dependent for activity (Fig. 7A and C). While Mg2+ is also required for Rep ATP-dependent helicase activity, we did not see an [Mg2+]-dependent phenotype for the D412A mutation in helicase assays (Fig. 7B). The biochemical assays that we have described support that the Rep mutant D412A has diminished ability to interact with Mg2+ and that an interaction between D412 and Mg2+ is necessary for efficient trs endonuclease activity. These results were reproducible with multiple Rep protein extracts, further supporting that the active DNA cleavage site of Rep78/68 consists, at least in part, of D412 and Mg2+.
Nearly all endonucleases employ Mg2+ or some divalent cation as part of their catalytic mechanism. Since the nicking activity of Rep78/68 is required for replication of AAV, a decreased affinity for Mg2+ could effectively explain the intracellular replication defect observed with D412A-78. An inability to efficiently interact with Mg2+ would especially inhibit intracellular activity, where limited Mg2+ reserves (0.5 mM) must be utilized by all other proteins present in the cellular milieu. For instance, the binding affinity of the restriction endonuclease EcoRV for Mg2+ is at physiological levels, and thus any disruption in Mg2+ binding results in loss of function (56). Supporting this conclusion, we have shown that increasing the intracellular Mg2+ levels (23a) can increase the replication activity of D412A-78 in an Mg2+-dependent manner. While the data presented in this study implies a role for Mg2+, the overall phenotype of this mutant may be in part related to other defects (e.g., an inability to associate with host factors, form stable Rep-Rep complexes, migrate efficiently to the nucleus, or be sequestered in AAV replication centers). Further analysis will help elucidate the complete role of the Mg2+-dependent mutant phenotype and AAV replication in vivo.
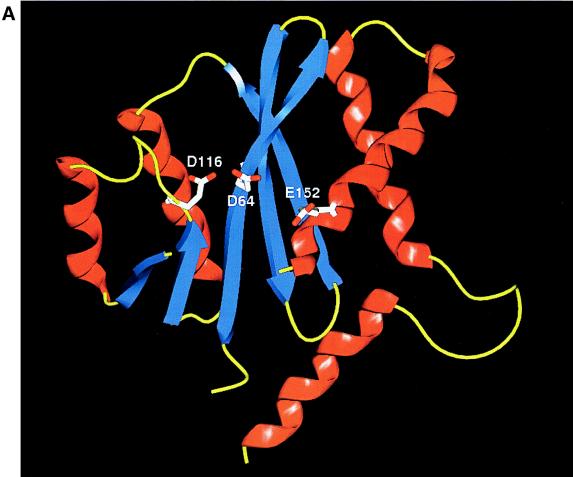
DD35E motif.
The D412 mutation maps to a region of Rep78/68 that contains a cluster of acidic amino acid residues that have the potential to form overlapping DD35E motifs (Fig. 1). The acidic amino acids of the DD35E motif have been shown to form an Mg2+ binding pocket that is critical for the nicking and strand transfer reactions of bacterial transposases and retroviral integrases (16, 36, 52). One might speculate that the decreased affinity for Mg2+ with D412A was consistent with the partial disruption of a similar putative Mg2+ binding domain. Based on this assumption, mutations at any of the conserved DD35E motif residues should result in the loss of some in vitro function as previously described (35, 36). Rep78/68 mutants generated within this putative DD35E motif (D468A and E504A) were identified as class I mutants with wt phenotypes, suggesting that these residues are not critical for catalytic function. However, this does not rule out the possibility that the second putative DD35E motif within Rep78/68 was involved in the coordination of the active-site Mg2+ ion(s). We also introduced a mutation at E465A within this second putative DD35E motif (at residues D368, D429, and E465) (71) to test the relevance of this DD35E cluster. The activity of this mutant was also similar to that of wt Rep78/68.
Based on this analysis, these two distinct motifs may be unrelated to any function in AAV Rep. However, the presence of overlapping DD35E motifs raised the interesting question of whether these acidic acid clusters were capable of cooperating to coordinate Mg2+, thus compensating for mutations at E465A, D468A, and/or E504A. To test this possibility we have subsequently generated a double mutant (D412A,E465A-78) that contains a mutation in each of the two putative DD35E Mg2+ binding pockets described above (23a). The phenotype of this double mutant was negative for AAV replication (23a). Continued studies are under way to determine the role, if any, of the two overlapping DD35E motifs in the AAV Rep proteins (23a).
Mg2+ binding pocket.
One of the reasons that we were originally intrigued by the DD35E motif was that Rep mediates targeted integration by an as-yet-unknown mechanism. Also, the DD35E motif is highly conserved among enzymes that mediate transposition and integration (16). A number of crystal structures have been solved for various Mg2+-dependent nucleases, such as HIV integrase (23, 41) and Mu transposase (54), which utilize the DD35E motif, as well as E. coli RNase H (34), HIV RNase H (20), and the E. coli resolvase RuvC (3). Genetic and crystallography studies have demonstrated that the catalytic cores of these enzymes consist of a cluster of acidic residues that interact directly with Mg. While the primary sequences of these proteins are quite diverse, the overall secondary and tertiary structures of these enzymes are strikingly similar (25, 78). As shown with HIV integrase (Fig. 8A), The active domains of these structures consist of five central beta strands flanked by a various number of alpha helices (23, 41).
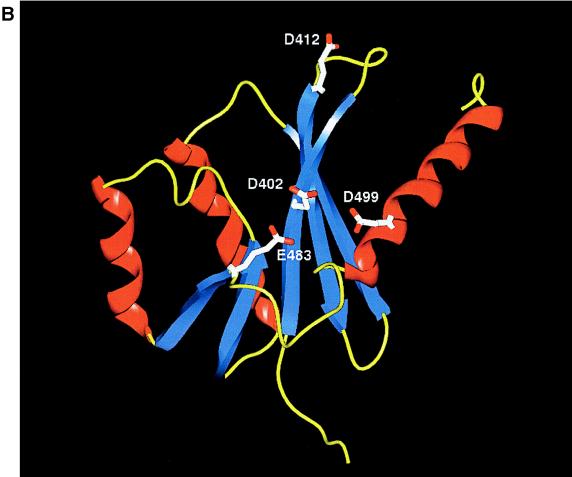
FIG. 8.
Structural analysis of the magnesium binding pocket and catalytic active site for nuclease activity. (A) Crystal structure of the active site for the HIV integrase, INT (23). Residues critical for magnesium binding and nuclease activity are indicated; beta strands are colored blue, and helices are colored red. (B) Theoretical structural model of the Rep78/68 active site for nicking. Residues 391 to 534 were used to generate a theoretical model of the magnesium binding domain of Rep by using secondary structural predictions (64, 79a) and the computer software programs from Molecular Simulations Inc. (see Materials and Methods). Residues potentially critical for magnesium binding and Rep nuclease activity based on this model are shown.
Using the data from secondary structural predictions (64, 64a), and homology-modeling software programs from Molecular Simulations Inc., we generated a theoretical structure for the active site of Rep78/68 (Fig. 8B) that was similar to those of HIV integrase and RuvC. In this theoretical model, residue D412 is positioned within the central beta strands of the putative catalytic core (Fig. 8B). In addition, other amino acids that would potentially be critical in this model are identified (Fig. 8B). These amino acids are also highly conserved among the AAV serotypes (15). Studies to support or dismiss the theoretical model described here for a Rep-dependent Mg2+ binding pocket are under way. However, it should be noted that in addition to this motif, AAV has a number of domains that are similar to published structural motifs (i.e., Walker A and B domain for NTP binding and helicase activities, DD35E and HxH metal binding domains, and the YXXXY motif utilized by φX174 for rolling-circle replication). All of these predictions are supported by sequence homology or genetic studies and still await crystal analysis of Rep for final confirmation.
The results of this study in conjunction with those of studies examining the frequency at which D412A and the other mutants, especially tsD40,42,44A-78, mediate targeted integration should also help clarify the relationship between the replication and integration activities mediate by Rep. An Mg2+ dependence for efficient targeted integration with the D412A mutant would also provide valuable insight into the mechanism of this Rep-mediated activity. Finally, the availability of the ts and non-ts AAV Rep proteins described in this study should provide novel approaches for generating rAAV packaging systems, including packaging cell lines, adenovirus and/or herpes simplex virus helper viruses, and in vitro packaging systems.
ACKNOWLEDGMENTS
We thank lab members of the Gene Therapy Center (specifically, Doug McCarty) for helpful discussions and Terry Van Dyke for technical contributions to the manuscript.
This work was supported by NIH grant HL48347, and D.K.G. is supported by NIH postdoctoral fellowship GM19342.
REFERENCES
- 1.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular biology of the cell. 2nd ed. New York, N.Y: Garland Publishing, Inc.; 1989. [Google Scholar]
- 2.Antoni B A, Rabson A B, Miller I L, Trempe J P, Chejanovsky N, Cater B J. Adeno-associated virus Rep protein inhibits human immunodeficiency virus type 1 production in human cells. J Virol. 1991;65:396–404. doi: 10.1128/jvi.65.1.396-404.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariyoshi M, Vassylyev D G, Iwasaki H, Nakamura H, Shinagawa H, Morikawa K. Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli. Cell. 1994;78:1063–1072. doi: 10.1016/0092-8674(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 4.Ashktorab H, Srivastava A. Identification of nuclear proteins that specifically interact with the adeno-associated virus 2 inverted terminal repeat hairpin DNA. J Virol. 1989;63:3034–3039. doi: 10.1128/jvi.63.7.3034-3039.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 3. New York, N.Y: John Wiley & Sons; 1996. [Google Scholar]
- 6.Balague C, Kalla M, Zhang W W. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bass S H, Mulkerrin M G, Wells J A. A systematic mutational analysis of hormone-binding determinants in the human growth factor receptor. Proc Natl Acad Sci USA. 1991;88:4498–4502. doi: 10.1073/pnas.88.10.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beaton A, Palumbo P, Berns K I. Expression from the adeno-associated virus p5 and p19 promoters is negatively regulated in trans by the Rep protein. J Virol. 1989;63:4450–4454. doi: 10.1128/jvi.63.10.4450-4454.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett W F, Paoni N F, Keyt B A, Botstein D, Jones A J S, Presta L, Wurm F M, Zoller M J. High resolution analysis of functional determinants on human tissue-type plasminogen activator. J Biol Chem. 1991;266:5191–5201. [PubMed] [Google Scholar]
- 10.Berns K I. Parvoviridae: the viruses and their replication. In: Fields B N, editor. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Raven Press; 1996. pp. 2173–2197. [Google Scholar]
- 11.Burns C C, Richards O C, Ehrenfeld E. Temperature-sensitive polioviruses containing mutations in RNA polymerase. Virology. 1992;189:568–582. doi: 10.1016/0042-6822(92)90580-i. [DOI] [PubMed] [Google Scholar]
- 12.Byrappa S, Gavin D K, Gupta K C. A highly efficient procedure for site-directed mutagenesis of full length plasmids using Vent DNA polymerase. Genome Res. 1995;5:404–407. doi: 10.1101/gr.5.4.404. [DOI] [PubMed] [Google Scholar]
- 13.Chejanovsky N, Carter B J. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology. 1989;173:120–128. doi: 10.1016/0042-6822(89)90227-4. [DOI] [PubMed] [Google Scholar]
- 14.Chejanovsky N, Carter B J. Mutation of a consensus purine nucleotide binding site in the adeno-associated virus rep gene generates a dominant negative phenotype for DNA replication. J Virol. 1990;64:1764–1770. doi: 10.1128/jvi.64.4.1764-1770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiorini J A, Kin F, Yang L, Kotin R M. Cloning and characterization of adeno-associated virus type 5. J Virol. 1999;73:1309–1319. doi: 10.1128/jvi.73.2.1309-1319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craig N L. Unity in transposition reactions. Science. 1995;270:253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- 17.Crowe J E, Firestone C Y, Whitehead S S, Collins P L, Murphy B R. Acquisition of the ts phenotype by a chemically mutagenized cold-passaged human respiratory syncytial virus vaccine candidate results from the acquisition of a single mutation in the polymerase (L) gene. Virus Genes. 1996;13:269–273. doi: 10.1007/BF00366988. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham B C, Wells J A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 19.Dao-pin S, Anderson D E, Baase W A, Dahlquist F W, Matthews B W. Structural and thermodynamic consequences of burying a charged residue within the hydrophobic core of T4 lysozyme. Biochemistry. 1991;30:11521–11529. doi: 10.1021/bi00113a006. [DOI] [PubMed] [Google Scholar]
- 20.Davis J F, Hostomska Z, Hostomsky Z, Jordan S R, Matthews D A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 21.Davis M D, Wonderling R S, Walker S L, Owens R A. Analysis of the effects of charge cluster mutations in adeno-associated virus Rep68 protein in vitro. J Virol. 1999;73:2084–2093. doi: 10.1128/jvi.73.3.2084-2093.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diamond S E, Kirkegaard K. Clustered charged-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J Virol. 1994;68:863–876. doi: 10.1128/jvi.68.2.863-876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyda F, Hickman A B, Jenkins T M, Engleman A, Craige R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotide transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 23a.Gavin, D. K., and R. J. Samulski. Unpublished data.
- 24.Goodman S, Xiao X, Donahue R E, Moulton A, Miller J, Walsh C, Young N S, Samulski R J, Nienhuis A W. Recombinant adeno-associated virus-mediated gene transfer into hematopoietic progenitor cells. Blood. 1994;84:1492–1500. [PubMed] [Google Scholar]
- 25.Grindley N F, Leschziner A E. DNA transposition: from black box to a color monitor. Cell. 1995;83:1063–1066. doi: 10.1016/0092-8674(95)90132-9. [DOI] [PubMed] [Google Scholar]
- 26.Hauswirth W W, Berns K I. Origin and termination of adeno-associated virus DNA replication. Virology. 1977;78:488–499. doi: 10.1016/0042-6822(77)90125-8. [DOI] [PubMed] [Google Scholar]
- 27.Hermonat P L, Labow M A, Wright R, Berns K I, Muzyczka N. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J Virol. 1984;51:329–333. doi: 10.1128/jvi.51.2.329-339.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 29.Hunter L A, Samulski R J. Colocalization of adeno-associated virus Rep and capsid proteins in the nuclei of infected cells. J Virol. 1992;66:317–324. doi: 10.1128/jvi.66.1.317-324.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Im D-S, Muzyczka N. Factors that bind to the AAV terminal repeats. J Virol. 1989;63:3095–3104. doi: 10.1128/jvi.63.7.3095-3104.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Im D-S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 32.Im D S, Muzyczka N. Partial purification of adeno-associated virus Rep78, Rep52, and Rep40 and their biochemical characterization. J Virol. 1992;66:1119–1128. doi: 10.1128/jvi.66.2.1119-1128.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones N C, Shenk T. An adenovirus type 5 early region gene function regulates expression of other early viral genes. Proc Natl Acad Sci USA. 1979;76:3665–3669. doi: 10.1073/pnas.76.8.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Ikehara M, Matsuzaki T, Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature. 1990;347:306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- 35.Kim K, Namgoog S, Jayaram M, Harshey R. Step-arrest mutant of phage Mu transposase. J Biochem. 1995;270:1472–1479. doi: 10.1074/jbc.270.3.1472. [DOI] [PubMed] [Google Scholar]
- 36.Kulkosky J, Jones K S, Katz R A, Mack J P G, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retrovial/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kyostio S R, Owens R A, Weitzman M D, Antoni B A, Chejanovsky N, Carter B J. Analysis of adeno-associated virus (AAV) wild-type and mutant Rep proteins for their abilities to negatively regulate AAV p5 and p19 mRNA levels. J Virol. 1994;68:2947–2957. doi: 10.1128/jvi.68.5.2947-2957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labow M A, Hermonat P L, Berns K I. Positive and negative autoregulation or the adeno-associated virus type 2 genome. J Virol. 1986;60:251–258. doi: 10.1128/jvi.60.1.251-258.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaMartina S, Roscilli G, Rinaudo D, DelMastro P, Toniatti C. Lipofection of purified adeno-associated virus Rep68 protein: toward a chromosome-targeting nonviral particle. J Virol. 1998;72:7653–7658. doi: 10.1128/jvi.72.9.7653-7658.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maignan S, Guilloteau J P, Zhou-Liu Q, Clement-Mella C, Mikol V. Crystal structure of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J Mol Biol. 1998;282:359–368. doi: 10.1006/jmbi.1998.2002. [DOI] [PubMed] [Google Scholar]
- 42.McCarty D M, Christensen M, Muzyczka N. Sequences required for the coordinate induction of the adeno-associated virus p19 and p40 promoters by the Rep protein. J Virol. 1991;65:2936–2945. doi: 10.1128/jvi.65.6.2936-2945.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCarty D M, Ni T H, Muzyczka N. Analysis of mutations in adeno-associated virus Rep protein in vivo and in vitro. J Virol. 1992;66:4050–4057. doi: 10.1128/jvi.66.7.4050-4057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarty D M, Ryan J H, Zolotukhin S, Zhou X, Muzyczka N. Interaction of the adeno-associated virus Rep protein with a sequence within the A palindrome of the viral terminal repeat. J Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendelson E, Trempe J P, Carter B J. Identification of the trans-active Rep proteins of adeno-associated virus by antibodies to a synthetic oligopeptide. J Virol. 1986;60:823–832. doi: 10.1128/jvi.60.3.823-832.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy B R, Prince G A, Collins P L, Van Wyke C K, Olmstead R A, Spriggs M K, Parrott R H, Kim H W, Brandt C D, Chonock R M. Current approaches to vaccines effective against parainfluenza and respiratory syncytial viruses. Virus Res. 1988;11:1–15. doi: 10.1016/0168-1702(88)90063-9. [DOI] [PubMed] [Google Scholar]
- 47.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 48.Owens R A, Weitzman M D, Kyostio S R, Carter B J. Identification of a DNA-binding domain in the amino terminus of adeno-associated virus Rep protein. J Virol. 1993;67:997–1005. doi: 10.1128/jvi.67.2.997-1005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parkin N T, Chiu P, Coelingh K L. Temperature sensitive mutants of influenza A virus generated by reverse genetics and clustered charged to alanine mutagenesis. Virus Res. 1996;46:31–44. doi: 10.1016/s0168-1702(96)01373-1. [DOI] [PubMed] [Google Scholar]
- 50.Pereira D J, McCarty D M, Muzyczka N. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J Virol. 1997;71:1079–1088. doi: 10.1128/jvi.71.2.1079-1088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pieroni L, Fipaldini C, Moncotti A, Cimini D, Sgura A, Fattori E, Epifano O, Cortese R, Palombo F, La Monica N. Targeted integration of adeno-associated virus-derived plasmids in transfected human cells. Virology. 1998;249:249–259. doi: 10.1006/viro.1998.9332. [DOI] [PubMed] [Google Scholar]
- 52.Polard P, Chandler M. Bacterial transposases and retroviral integrases. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 53.Rhode S L. Replication process of the parvovirus H-1. X. Isolation of a mutant defective in replication-form DNA replication. J Virol. 1978;25:215–223. doi: 10.1128/jvi.25.1.215-223.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice P A, Mizuuchi K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell. 1995;82:209–220. doi: 10.1016/0092-8674(95)90308-9. [DOI] [PubMed] [Google Scholar]
- 55.Samulski R J, Srivastava A, Berns K I, Muzyczka N. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell. 1983;33:135–143. doi: 10.1016/0092-8674(83)90342-2. [DOI] [PubMed] [Google Scholar]
- 56.Selent U, Ruter T, Kohler E, Liedtke M, Thielking V, Alves J, Oelgeschlager T, Wolfes H, Peters F, Pingoud A. A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry. 1992;31:4808–4815. doi: 10.1021/bi00135a010. [DOI] [PubMed] [Google Scholar]
- 57.Senepathy P, Tratschin J D, Carter B J. Replication of adeno-associated virus DNA. Complementation of naturally occurring rep mutants by a wild-type genome or an ori− mutant and correction of terminal deletions. J Mol Biol. 1984;179:1–20. doi: 10.1016/0022-2836(84)90303-6. [DOI] [PubMed] [Google Scholar]
- 58.Snyder R O, Im D-S, Muzyczka N. Evidence for covalent attachment of the adeno-associated virus (AAV) Rep protein to the ends of the AAV genome. J Virol. 1990;64:6204–6213. doi: 10.1128/jvi.64.12.6204-6213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Snyder R O, Im D S, Ni T, Xiao X, Samulski R J, Muzyczka N. Features of the adeno-associated virus origin involved in substrate recognition by the viral Rep protein. J Virol. 1993;67:6096–6104. doi: 10.1128/jvi.67.10.6096-6104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Snyder R O, Samulski R J, Muzyczka N. In vitro resolution of covalently joined AAV chromosome ends. Cell. 1990;60:105–133. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- 61.Snyder R O, Xiao X, Samulski R J. Current protocols in human genetics. Vol. 2. New York, N.Y: John Wiley & Sons; 1996. [Google Scholar]
- 62.Srivastava A, Lusby E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Straus S E, Sebring E D, Rose J A. Concatemers of alternating plus and minus strands are intermediates in adenovirus-associated virus DNA synthesis. Proc Natl Acad Sci USA. 1976;73:742–746. doi: 10.1073/pnas.73.3.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stultz C M, Nambudripad R, Lathrop R H, White J V. Predicting protein structure with probabilistic models. In: Allewell N, Woodward C, editors. Protein structural biology in bio-medical research. Greenwich, Conn: JAI Press; 1997. [Google Scholar]
- 64a.Stultz C M, Nambudripad R, Lathrop R H, White J V. revision date. Predicting protein structure with probabilistic models. In N. Allewell and C. Woodward (ed.), Protein structural biology in bio-medical research. [Online.] JAI Press, Greenwich, Conn. http://bmerc-www.bu.edu/psa/. [30 August 1999, last date accessed.] 9 July 1999. [Google Scholar]
- 65.Tratschin J-D, Miller I L, Carter B J. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J Virol. 1984;51:611–619. doi: 10.1128/jvi.51.3.611-619.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tratschin J-D, Tal J, Carter B J. Negative and positive regulation in trans of gene expression from adeno-associated virus vectors in mammalian cells by a viral Rep gene product. Mol Cell Biol. 1986;6:2884–2894. doi: 10.1128/mcb.6.8.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trempe J P, Mendelson E, Carter B J. Characterization of adeno-associated virus rep proteins in human cells by antibodies raised against rep expressed in Escherechia coli. Virology. 1987;161:18–28. doi: 10.1016/0042-6822(87)90166-8. [DOI] [PubMed] [Google Scholar]
- 68.Urabe M, Hasumi Y, Kume A, Surosky R T, Kurtzman G J, Tobita K, Ozawa K. Charged-to-alanine scanning mutagenesis of the N-terminal half of adeno-associated type 2 Rep78 protein. J Virol. 1999;73:2682–2693. doi: 10.1128/jvi.73.4.2682-2693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walker S L, Wonderling R S, Owens R A. Mutational analysis of the adeno-associated virus Rep68 protein: identification of critical residues necessary for site-specific endonuclease activity. J Virol. 1997;71:2722–2730. doi: 10.1128/jvi.71.4.2722-2730.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walker S L, Wonderling R S, Owens R A. Mutational analysis of the adeno-associated virus type 2 Rep68 protein helicase motifs. J Virol. 1997;71:6996–7004. doi: 10.1128/jvi.71.9.6996-7004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weitzman M D, Kyostio S R M, Carter B J, Owens R A. Interaction of wild-type and mutant adeno-associated virus (AAV) Rep proteins on AAV hairpin DNA. J Virol. 1996;70:2440–2448. doi: 10.1128/jvi.70.4.2440-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wertman K F, Drubin D G, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wonderling R S K, Sirkka R, Owens R A. A maltose-binding protein/adeno-associated virus Rep68 fusion protein has DNA-RNA helicase and ATPase activities. J Virol. 1995;69:3542–3548. doi: 10.1128/jvi.69.6.3542-3548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao W. PhD. thesis. Chapel Hill: University of North Carolina; 1996. [Google Scholar]
- 75.Xiao X, Li J, Samulski R J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao X, Xiao W, Juan L, Samulski R J. A novel 165-base-pair terminal repeat system is the sole cis requirement for adeno-associated virus life cycle. J Virol. 1997;71:941–948. doi: 10.1128/jvi.71.2.941-948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang Q, Kadam A, Trempe J P. Mutational analysis of the adeno-associated virus rep gene. J Virol. 1992;66:6058–6069. doi: 10.1128/jvi.66.10.6058-6069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang W, Steitz T A. Recombining the structures of HIV integrase, RuvC and RNase H. Structure. 1995;3:131–134. doi: 10.1016/s0969-2126(01)00142-3. [DOI] [PubMed] [Google Scholar]
- 78a.Young, S., and R. J. Samulski. Unpublished data.
- 79.Zolotukhin S, Potter M, Hauswirth W W, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]