Abstract
Extracellular vesicles (EVs) are membrane-enclosed bio-nanoparticles secreted by cells and naturally evolved to transport various bioactive molecules between cells and even organisms. These cellular objects are considered one of the most promising bio-nanovehicles for the delivery of native and exogenous molecular cargo. However, many challenges with state-of-the-art EV-based candidates as drug carriers still exist, including issues with scalability, batch-to-batch reproducibility, and cost-sustainability of the final therapeutic formulation. Microalgal extracellular vesicles, which we named nanoalgosomes, are naturally released by various microalgal species. Here, we evaluate the innate biological properties of nanoalgosomes derived from cultures of the marine microalgae Tetraselmis chuii, using an optimized manufacturing protocol. Our investigation of nanoalgosome biocompatibility in preclinical models includes toxicological analyses, using the invertebrate model organism Caenorhabditis elegans, hematological and immunological evaluations ex vivo and in mice. We evaluate nanoalgosome cellular uptake mechanisms in C. elegans at cellular and subcellular levels, and study their biodistribution in mice with accurate space-time resolution. Further examination highlights the antioxidant and anti-inflammatory bioactivities of nanoalgosomes. This holistic approach to nanoalgosome functional characterization demonstrates that they are biocompatible and innate bioactive effectors with unique bone tropism. These findings suggest that nanoalgosomes have significant potential for future therapeutic applications.
Subject terms: Bioinspired materials, Cell delivery
This study highlights the biocompatibility, tropism, antioxidant and anti-inflammatory properties of nanoalgosomes, extracellular vesicles (EVs) derived from the marine microalga Tetraselmis chuii, suggesting their potential for EV-based therapies.
Introduction
Cell-free therapy has emerged as a promising approach in the field of biomedical research, offering innovative strategies for the treatment of various diseases1–3. Among the major players in this field, extracellular vesicles (EVs) stand out as promising cell-derived therapeutic effectors1,3–5. Within naturally occurring nanoparticles, EVs have shown immense potential as intercellular mediators due to their ability to transfer bioactive cargo molecules to recipient cells and thereby influence their behavior1,3,6. This cargo includes growth factors, cytokines, microRNAs and other signaling molecules that can modulate cellular processes and promote cell survival, proliferation, differentiation, tissue repair and regeneration7. The therapeutic potential of EVs is influenced by their cellular origin, resulting in different changes in the target cells. For example, EVs derived from human mesenchymal stem cells (MSCs) have shown regenerative properties, while EVs derived from cancer cells promote tumor progression4,7. Another important aspect to consider when using EVs as therapeutic effectors is their immunomodulatory properties8–10. Milk-derived EVs, for instance, have remarkable regenerative properties due to their high content of growth factors and immunomodulatory molecules9. Some plant-derived EVs also show therapeutic potential due to their bioactive cargo, including phytochemicals and secondary metabolites that can influence immune responses and promote an anti-inflammatory environment11,12. In addition, EVs derived from microorganisms such as bacteria offer an interesting opportunity for therapeutic applications, particularly as vaccines, as they can carry a wide range of functional molecules and have unique bioactivity derived from their microbial origin that promotes enhanced immune activation13. Furthermore, EVs can be loaded with exogenous therapeutic entities (such as small molecule drugs, nucleic acid, proteins, and CRISPR/Cas9) and their endogenous cargo can intensify the effect of the encapsulated drugs, creating a combinatorial effect14–16. In recent years, interest in EVs derived from human MSCs has increased due to their therapeutic potential in preclinical studies and clinical trials across regenerative medicine in diverse tissues, such as lung, kidney, liver, central nervous system, cartilage, bone, and heart17. However, the therapeutic potential of MSC-EVs is still debated due to the complexity and variability of the donor MSCs, including their tissue origin and cell culture conditions, and the manufacturing scalability, maintaining consistent batch-to-batch quality, and ensuring cost-effectiveness. Thus, the academic and industrial communities have been investigating alternative approaches to obtain EVs from other biological sources12,18–22. In this context, we are showcasing and further exploring a new specific type of EVs that we call “nanoalgosomes” or “algosomes”23. Nanoalgosomes are small EVs (sEVs) isolated from microalgae conditioned media, that are surrounded by a lipid bilayer membrane, contain EV biomarkers, have a typical EV size distribution, morphology and density, are highly stable in human blood plasma, non-cytotoxic in vitro and can be taken up by various cellular systems23,24. We focused on small EVs isolated from the photosynthetic marine chlorophyte Tetraselmis chuii, which is rich in vitamin E, carotenoids and chlorophyll with anti-inflammatory and antioxidant properties23–25. Through our patented platform, we have achieved high-yield production of quality-controlled EVs from microalgae, a bioresource that can be grown in scalable, renewable and environmentally sustainable photo-bioreactors26. The current study aims to fully elucidate the biological properties of nanoalgosomes and to validate their safe use as innate bioactive effectors in EV-based therapy. Our analyses show that nanoalgosomes are biocompatible and exert anti-inflammatory and antioxidant effects both in vitro as well as in vivo, using mouse models and the invertebrate model organism Caenorhabditis elegans, a faster and less expensive alternative to mammalian models, whose body transparency is advantageous for studying nanoparticle toxicity, distribution and uptake in living organisms25.
Results and discussion
Biological features of nanoalgosomes
Production and quality checking of nanoalgosomes
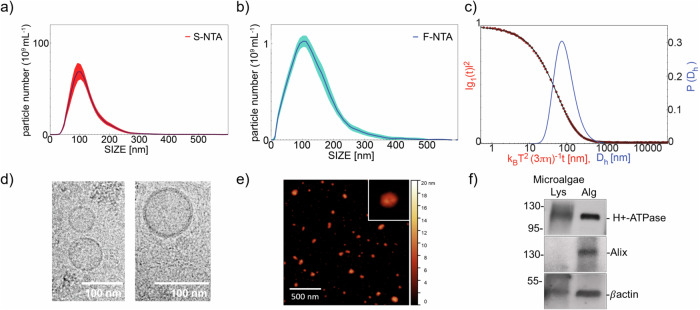
The extracellular vesicles derived from microalgae (e.g., nanoalgosomes) presented here are isolated from T. chuii-conditioned media consisting of ultrafiltered seawater supplemented with salts and vitamins (e.g., F/2 medium)23. Compared to extracellular vesicles isolated from other sources, such as those from plasma or in vitro cell cultures, nanoalgosome batches have the significant advantage of exhibiting very low to negligible presence of co-isolates (i.e., proteins and lipoproteins). All nanoalgosome samples have been characterized and described as required by the MISEV2018 reference guidelines more recently encouraged by MISEV202327,28. Specifically, the nanoalgosome samples are tested and validated according to a specific reference checklist adapted for microalgal EVs23. Multiple orthogonal physical and molecular techniques have been applied, including Nanoparticle Tracking Analyses (NTA), fluorescent-NTA (F-NTA) following membrane-labeling of nanoalgosomes, Dynamic Light Scattering (DLS), cryo-Transmission Electron Microscopy (cryo-TEM), Atomic Force Microscopy (AFM), and immunoblotting analyses of nanoalgosome markers (Fig. 1a–f). This approach allowed to demonstrate the presence of small EVs with expected characteristics: (i) the size distributions measured by NTA, F-NTA and DLS and shown in Fig. 1a–c, peak at 100 nm, 100 nm and 70 nm respectively (mode), and extend down to 60 nm, 40 nm and 40 nm, respectively (half width at half maximum); therefore, considering that the distribution sampled by scattering techniques is weighted by mass, the larger EV number is in the range 40–100 nm, coherent with results obtained by AFM and cryo-TEM; (ii) the EV morphology and the presence of a bilayered membrane is clearly assessed by cryo-TEM and AFM images in Fig. 1c, d; (iii) the presence of lipid membrane and lack of large number of non-vesicle contaminants are also assessed by F-NTA (Fig. 1b) measured on EV labeled by Di-8-ANEPPS; (iv) the presence of EV markers (e.g., Alix, H+-ATPase and β-actin positivity) is assessed by immunoblotting (Fig. 1f); (v) moreover the EV particle number/protein ratio (e.g., 1 µg of total EV protein corresponds to a range of 5–10 × 109 in all nanoalgosome batches, n = 6) is coherent with the estimate of EV particles/μg of EV proteins, as reported by Sverdlov (2012)29. All these features were evaluated in different nanoalgosome batches (n = 3) and showed repeatedly yielding vesicles in several (n = 6) preparations (~1012 nanoalgosomes/L of microalgal conditioned-medium, corresponding to ~104 nanoalgosomes/microalgal cell). These results on nanoalgosomes are consistent with our previous studies, which extensively have described the experimental approach and evaluation of the nanoalgosome characterization study23.
Fig. 1. Quality control of nanoalgosome preparations.
a Nanoalgosome size distribution and concentration measured by NTA, the red deviation is relative to the five measurements analysed per sample. b Size distribution of nanoalgosomes stained with Di-8-ANEPPS measured by F-NTA (using laser wavelength of 488 nm), the blue deviation is relative to five measurements analysed per sample, and (c) DLS. Morphology of nanoalgosomes was analyzed by (d) cryo-TEM and (e) AFM. f Immunoblot analyses were performed on T.chuii lysate (Lys) and nanoalgosomes (Alg) to detect the markers H+-ATPase, Alix and β-actin. Representative results of three independent biological replicates (n = 3 biologically independent samples) are presented.
Nanoalgosome internalization mechanism in vitro and in vivo
In our previous study, we set up and validated the labeling of nanoalgosomes with different fluorescent dyes (i.e., Di-8-ANEPPS, PKH26, or DiR), and demonstrated that labeled nanoalgosomes are specifically taken up and localized in vitro in the cytoplasm of cells and in vivo in the cytoplasm of C. elegans intestinal cells24,25. In the present study, following our validated labeling protocol, we used fluorescently labeled nanoalgosomes along with the relative unbound dye negative controls to exclude any artifacts due to free/self-aggregate dye not incorporated into EVs.
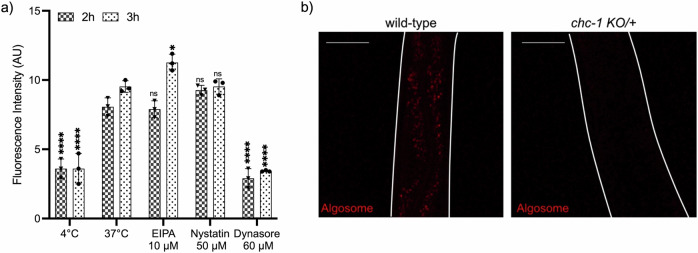
Since we proved that nanoalgosomes are internalized within human cells in a dose- and time-dependent manner through an energy-dependent mechanism, we hypothesized active endocytic pathways, including macropinocytosis, clathrin- and caveolae-mediated endocytosis, as possible mechanisms of nanoalgosome internalization that were reported for other EVs30. Therefore, here we investigated the molecular mechanisms involved in nanoalgosome internalization in vitro and in vivo. To test among alternative hypotheses, we used three specific blocking agents in 1-7 HB2 cells: dynasore to inhibit clathrin-mediated endocytosis, nystatin to interfere with caveolae-dependent uptake, and 5-[N-ethyl-N-isopropyl] amiloride (EIPA) to inhibit macropinocytosis31–34. We labeled and quality checked nanoalgosome with Di-8-ANEPPS and monitored intracellular nanoalgosome uptake in cells by measuring the fluorescence intensity of Di-8-ANEPPS-labeled nanoalgosomes after 2 and 3 h of incubation, with or without different inhibitor treatments (Fig. 2a, Supplementary Fig. 1a, b). The results showed that cells treated with dynasore (60 µM) had a nanoalgosome internalization trend similar to the negative control (i.e., cells incubated at 4 °C, which are inhibited for all energy-dependent processes), thus indicating that dynasore inhibited nanoalgosome cellular uptake. In contrast, no significant endocytosis inhibition was observed in cells treated with EIPA (10 µM) or nystatin (50 µM), which showed a nanoalgosome internalization level similar to the positive control (i.e., cells incubated at 37 °C). Cells are viable after each treatment, demonstrating that none of the inhibitors used, at any concentration, were toxic to the cells (Supplementary Fig. 1a). These results indicate that clathrin-dependent endocytosis plays a role in the cellular uptake of nanoalgosomes.
Fig. 2. Molecular mechanism of nanoalgosome internalization in human cell lines and in vivo.
a Effect of metabolic inhibitors of endocytosis on nanoalgosome uptake in 1–7 HB2 cells treated with EIPA, nystatin and dynasore. Results are presented as arbitrary unit of nanoalgosome fluorescence intensity inside cells after 2 h and 3 h of incubation. Each value represents the mean ± SD from three independent experiments (n = 3 biologically independent experiments). One-way ANOVA statistical test was used to assess the statistical significance of the differences: 37 °C (2 h) (3 h) vs 4 °C (2 h) (3 h), EIPA 10 µM (2 h) (3 h), nystatin 50 µM (2 h) (3 h) and dynasore 60 µM (2 h) (3 h), ****p < 0.0001, *p < 0.01, ns=not significant (p > 0.5). b Representative confocal images of wild-type and chc-1 KO heterozygote C. elegans animals after 24 h of treatment with Di-8-ANEPPS labeled nanoalgosomes (red). Fluorescent signal is observed only in the intestinal cells of wild-type animals, while in chc-1 KO heterozygote animals the fluorescent signal is strongly reduced. Animal body is outlined with white lines. Scale bar is 50 µm.
To confirm the results obtained in vitro, we used the model system C. elegans, taking advantage of the ease to use genetic mutants and dissect molecular pathways in this model. In particular, when the clathrin heavy chain gene is mutated in C. elegans, the uptake of synthetic nanoparticles is impaired35. Thus, we used a KO mutant in the clathrin heavy chain, chc-1(ok2369), and since chc-1 is an essential gene and its depletion causes animal lethality, we analyzed heterozygous balanced animals (chc-1 KO/+). We treated for 24 h animals with nanoalgosome fluorescently labeled with Di-8-ANEPPS and observed, in wild-type animals, a fluorescent signal in the intestinal cells (Fig. 2b). On the contrary, in all chc-1 KO/+ animals analyzed, the fluorescent signal in the intestinal cells was strongly reduced (Fig. 2b). The identification of clathrin-mediated endocytosis as the primary mechanism responsible for the cellular uptake of nanoalgosomes is aligned to previous studies, which investigated the routes and mechanisms of mammalian cell-derived EV internalization, highlighting the conservation of this endocytic mechanism towards EVs from different species3,30. The clathrin-mediated endocytosis relies on receptor-mediated, hydrophobic or electrostatic interactions in areas of clathrin expression on the cell membrane, and is also the most common route for synthetic nanoparticles and virus uptake in non-specialized mammalian cells36,37.
Nanoalgosome intracellular localization in vitro and in vivo
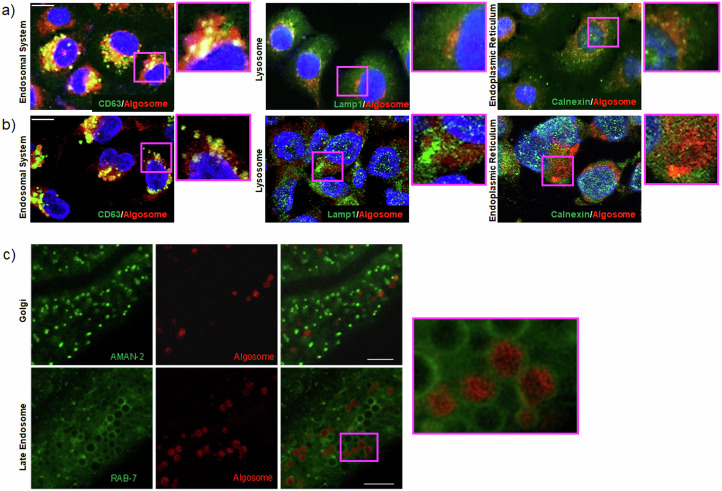
We further investigated the subcellular fate of labeled nanoalgosomes once internalized, both in vitro and in vivo. The subcellular localization of quality checked PKH26-labeled nanoalgosomes in 1-7 HB2 and MDA-MB 231 cell lines was determined using immunofluorescence and fluorescent and confocal microscopy analyses (Fig. 3a, b, respectively). Three distinct subcellular compartments were evaluated for in vitro study: the endosomal compartment, the lysosomal system, and the endoplasmic reticulum (ER), using established biomarkers (CD63, LAMP1, and calnexin, respectively).
Fig. 3. Intracellular nanoalgosome localization.
a Representative fluorescence microscopy images, showing the cellular uptake of PKH26-fluorescent nanoalgosomes (red) in 1-7 HB2 cells (nuclei in blue) incubated at 37 °C for 24 h. b Representative confocal microscopy images, showing the cellular uptake of PKH26-fluorescent nanoalgosomes (red) in MDA-MB 231 cells (nuclei in blue) incubated at 37 °C for 24 h. In green are the endosomal system (CD63), the endoplasmic reticulum (calnexin) and lysosomes (Lamp1). Scale Bar are 50 µm. Experiments were repeated >3 times, with similar results. c Confocal microscopy analysis of C. elegans intestinal cells. In green the Golgi marker AMAN-2 and late endosomes marker RAB-7. In red Di-8-ANEPPs labeled nanoalgosomes. Scale bar 25 µm. An enlargement of the pink box is visible on the right.
After 24 h of incubation, we found a co-localization of nanoalgosomes (red signal) with the endosomal protein CD63 (green signal). Indeed, fluorescence images showed this co-localization in a large portion of the cells (Fig. 3a, Supplementary Fig. 2), suggesting nanoalgosome localization in endosomal compartments. Confocal images (Fig. 3b) further confirmed this direct relation with the endosomal system in the area where the fluorescence appears yellow. Furthermore, the localization of PKH26-labeled nanoalgosomes was evaluated in relation to two other intracellular markers, LAMP-1 and calnexin (Fig. 3a, b). Both fluorescence and confocal images showed that intracellular LAMP-1- and calnexin-positive compartments (green) did not co-localize with internalized PKH26-labeled vesicles, suggesting that lysosomes and ER were not involved in their intracellular trafficking.
To confirm these observations in vivo, we took advantage of C. elegans’ transparency and transgenics expressing fluorescent proteins in the Golgi (GFP fused to AMAN-2 protein) or in late endosomes (GFP fused to RAB-7 protein). AMAN-2 is a membrane protein of the Golgi, and we did not observe any co-localization with Di-8-ANEPPS-labeled nanoalgosomes, as shown in Fig. 3c (upper panels). On the other hand, when we used a marker for late endosome membranes (RAB-7), we observed the nanoalgosome fluorescent signal inside the endosomes (Fig. 3c, lower panels).
Taken together, our results show that after nanoalgosomes are internalized by clathrin-mediated endocytosis, the fluorescent signal of nanoalgosome membrane co-localizes only with the endosome and not with the lysosomes, thus demonstrating that nanoalgosomes are not destined to lysosomal degradation.
Nanoalgosome biocompatibility in vitro and in vivo
In vitro and in vivo genotoxicity
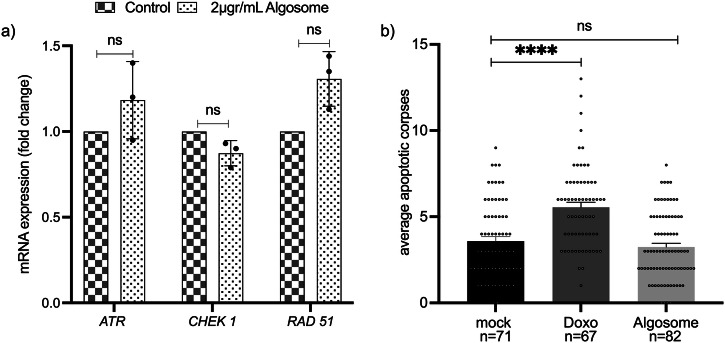
Our group recently demonstrated that nanoalgosomes did not elicit cytotoxicity, hepatotoxicity or genotoxicity in different cell lines23,24. Here, we have examined whether nanoalgosome treatment could trigger the activation of DNA-damage pathways by conducting gene expression analysis in 1–7 HB2 normal mammary epithelial cells. Typically, DNA damage leads to cell cycle arrest, regulation of DNA replication, and activation of the repair pathway. DNA damage triggers the activation of DNA-damage sensors such as ATR (ATM and Rad3-related serine/threonine kinase) and their recruitment to DNA damage sites38,39. In addition, checkpoint kinase 1 (CHEK1) is a key downstream molecule of DNA-damage response signaling; CHEK1 phosphorylates various intracellular substrate proteins, including the RAD51 recombinase which is central to the homologous recombination pathway, and binds single-stranded DNA at damage sites, forming filaments observed microscopically as nuclear foci. Therefore, these genes are considered to be involved in the response to DNA damage maintaining genome integrity, with ATR initiating a signaling cascade that activates CHEK1 and RAD5140. Gene expression analysis of 1-7 HB2 cells treated with 2 µg/mL of nanoalgosomes for 24 h showed not significant changes in the expression level of these selected DNA-damage sensors (ATR, CHECK1, RAD51), thus suggesting that the outcomes related to nanoalgosomes are negligible on the activation of the DNA damage signaling pathway (Fig. 4a).
Fig. 4. Genotoxicity assessment in vitro and in vivo.
a Real-time PCR for quantification of DNA-damage sensors expression levels following exposure to nanoalgosomes (2 µg/mL for 24 h in 1-7 HB2 cells). Fold-changes were normalized with β-actin expression and given as relative to control. Each experiment was performed in triplicate (n = 3 biologically independent samples). By One-way ANOVA differences of treated cells were determined not statistically significant (ns) when compared with the control (p > 0.05). b Quantification of SYTO12 positive apoptotic corpses in the C. elegans germline after treatment with mock (PBS), Doxo (doxorubicin 2.4 μM), and nanoalgosomes (20 µg/mL). Bars represent means and dots the single animals; error bar is the SEM. n is the number of animals analyzed. Kruskal-Wallis one-way ANOVA was used to assess the statistical significance of the differences: mock vs nanoalgosomes ns=not significant (p > 0.7); mock vs Doxo****, p < 0.0001.
For in vivo analyses of nanoalgosome genotoxicity, we used C. elegans that allows the evaluation of the putative toxic effect of nanoparticles on a whole living animal, with fewer ethical concerns, lower costs, and an important reduction in the number of vertebrate animals used. Specifically, the C. elegans germline can be used as a tool to study genotoxicity in vivo41. Physiological germline apoptosis occurs in wild-type animals in absence of any stress and can be visualized as an average of three apoptotic corpses per gonadal arm using SYTO12, an apoptotic-DNA fluorescent marker42,43. Genotoxic agents, such as doxorubicin, are capable of increasing germline apoptosis as consequence of DNA damage44. We performed a chronic treatment for 72 h of animals with nanoalgosomes at 20 µg/mL and we evaluated the germline apoptosis to assess nanoalgosomes genotoxicity in vivo. We decided to use this concentration since we successfully observed nanoalgosomes internalization at 20 µg/mL25 (Figs. 2b and 3c). We did not observe any increase in the number of apoptotic corpses in the germline compared to mock (Fig. 4b). Differently, animals treated with doxorubicin showed a higher number of apoptotic corpses compared to animals treated with mock, thus confirming that nanoalgosomes have no genotoxic effect at the concentration tested neither in vitro nor in vivo.
In vivo biocompatibility of nanoalgosomes in C. elegans
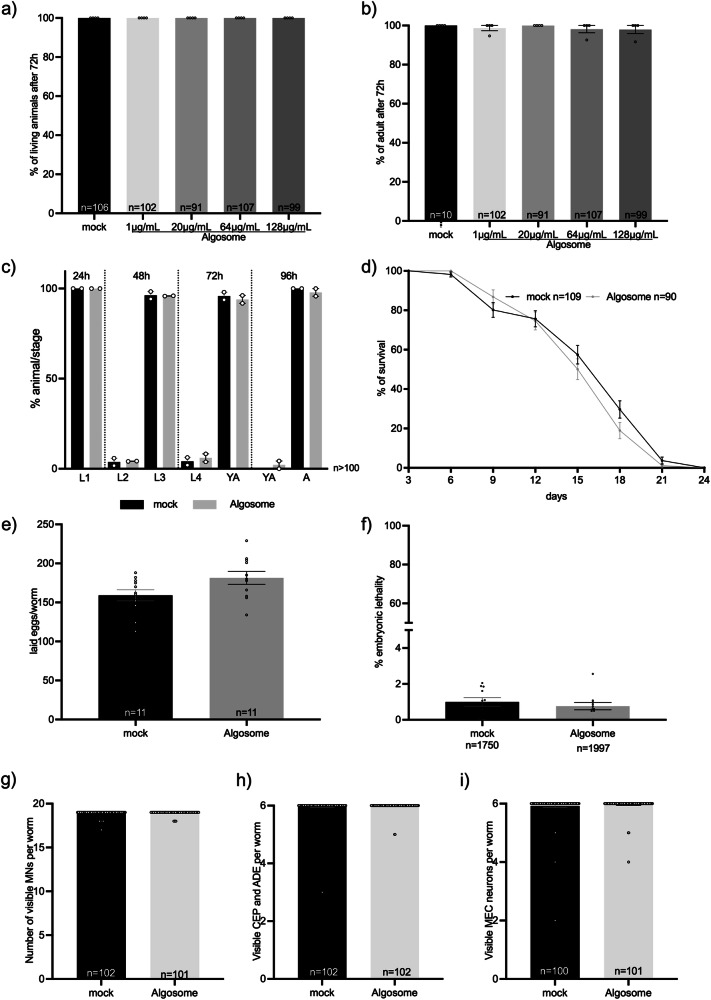
C. elegans offers a comprehensive set of experiments that allow to address the nanoparticle biocompatibility at different levels checking their effect on animal survival, growth, lifespan, fertility, lethality, and neuron viability, among others. Moreover, the short life cycle (three days) and lifespan (three weeks) of this model permits to exploit all these phenotypes in a very short period of time on large number of animals. Thus, we treated wild-type animals through in liquido culturing for 72 h with increasing concentrations of nanoalgosomes (1, 20, 64, 128 µg/mL), first evaluating animal viability and capability of reaching adulthood. After treatment for three days in liquido, we did not observe any toxic effect of nanoalgosomes, as all the animals were alive and reached adulthood, similarly to the animals treated with PBS as mock (Fig. 5a, b). Since we successfully observed nanoalgosome internalization at 20 µg/mL, we decided to use this concentration to further evaluate nanoalgosome effects on animal fitness45.
Fig. 5. Nanoalgosome in vivo biocompatibility in C. elegans.
a Quantification of the number of living animals after treatment with mock (PBS) or different nanoalgosome concentrations. b Quantification of the number of animals reaching adult stage after treatment with mock or different nanoalgosome concentrations. c Quantification of the percentages of animals at each developmental stage after treatment with mock and nanoalgosomes (20 µg/mL). d Lifespan of animals treated with mock and nanoalgosomes (20 µg/mL). e Brood size of animals after treatment with mock and nanoalgosomes (20 µg/mL). f Embryonic lethality of animals after treatment with mock and nanoalgosomes (20 µg/mL). g–i Quantification of the number of visible motoneurons (MNs), dopaminergic neurons in the head (CEP and ADE) and mechanosensory neurons (MEC) after treatment with mock and nanoalgosomes (20 µg/mL). Bars represent the means and dots the replicates (in a–d), P0 (in e, f), or single animals (in g–i); error bars are SEM. n is the number of animals analyzed (in a–d, g–i), the number of P0 animals (in e), or the number of eggs analyzed (f). In all graphs the statistical significance of the differences between treatments with mock and nanoalgosomes were assessed with Kruskal-Wallis One-way ANOVA (a, b), non-parametric Compare two proportions test (c), Mann Whitney t-test (e–i) or Log-rank (Mantel-Cox) test (d) and never found significant (p > 0.05).
C. elegans’ development and growth are finely regulated in four larval stages followed by a fertile adult stage, which can be altered when the animals are exposed to toxic agents46. To evaluate animal growth, we treated animals chronically with nanoalgosomes and tested the percentage of animals at each developmental stage every 24 h, without observing any alterations or delays compared to animals treated with mock (Fig. 5c). We treated animals over their entire lifespan and, also in this case, nanoalgosomes had no effect on their survival curve (Fig. 5d).
Another aspect used to assess the biocompatibility of nanoparticles is to evaluate their effects on animal fertility and offspring survival. Therefore, after chronic treatment of animals from eggs to L4 larval stage, we assessed the animals’ egg-laying ability and egg hatching (Fig. 5e, f), without observing any effects of nanoalgosome treatment compared to mock. This result confirms that nanoalgosomes do not affect animal fertility, offspring release, or embryonic survival. Moreover, we decided to evaluate the putative neurotoxicity of nanoalgosomes. C. elegans’ transparency makes it possible to observe neurons in living animals by using fluorescent proteins that are only expressed in the cells of interest. We thus evaluated the effects of nanoalgosomes on three different neuronal classes: GABAergic motoneurons, dopaminergic sensory neurons, and glutamatergic mechanosensory neurons. Interestingly, the invariant cell lineage in C. elegans determines a fixed number of neurons in adults, with 19 GABAergic motoneurons in the ventral cord, 6 dopaminergic neurons in the head, and 6 mechanosensory glutamatergic neurons along the body. Chronic treatment with nanoalgosomes did not cause any death of the neurons analyzed (Fig. 5g–i; Supplementary Fig. 3a–c) as the number of visible neurons was not affected by the treatment. Moreover, all these neurons exhibited normal morphology.
Taken together, our results demonstrate that nanoalgosomes are very well tolerated in vivo by C. elegans, and no toxicity was observed after chronic exposure from eggs to adult stage and throughout animal life at these concentrations.
In vivo biocompatibility of nanoalgosomes in wild-type mice
Biochemical and hematological studies were conducted on wild-type mice to thoroughly evaluate the in vivo biocompatibility in immune-competent mice, after a single intravenous (I.V.) administration of nanoalgosomes. Wild-type BALB/c mice (n = 4, 2 male and 2 female) were injected with nanoalgosomes and a negative control (i.e., PBS), via intravenous injection into the retroorbital sinus. The starting nanoalgosome doses were based on literature; in different pre-clinical studies, the dose of EVs per kg of body weight ranged from 0.10 to 100 mg of total EV proteins, with an average of 2.75 mg/kg47,48. Based on the estimation that the body weight of 6-week-old BALB/c mouse is approximately 20 grams, the average I.V. dose of EVs in mice corresponds to 55 µg/mouse49,50. Thus, in our pre-clinical studies we used the following doses: Dose 1 (low dose) = 10 µg/mouse, corresponding to 4 × 1010 EVs/mouse; Dose 2 (high dose) = 50 µg/mouse, corresponding to 2 × 1011 EVs/mouse. Blood samples were collected from mice to analyze hematocrit, creatinine, blood urea nitrogen (BUN), liver transaminase enzymes (Serum Aspartate Transaminase, AST, and Serum Alanine Transaminase, ALT), and lymphocyte numbers. Table 1 shows slight changes in white and red blood cell counts, hemoglobin, or hematocrit in nanoalgosome-treated mice compared to control mice (i.e., injected with PBS) at 48 h post-IV injection; these differences are still within the normal range of values reported for BALB/c mice (Janvier Labs, France), thus suggesting the absence of potential immune-reaction to nanoalgosomes after a single I.V. administration of nanoalgosomes.
Table 1.
Hematological and biochemical analyses of peripheral blood from BALB/c mice (n = 4 biologically independent mice/group)
| Parameters | Dose 1 | Dose 2 | Control |
|---|---|---|---|
| Hematological | |||
| Red blood cells (x106/μL) | 10.1 ± 0.0 | 9.9 ± 1.0 | 10.1 ± 0.1 |
| Hemoglobin (g/dL) | 15.2 ± 0.0 | 15.0 ± 1.4 | 15.5 ± 0.5 |
| MCV (fL) | 47.0 ± 0.2 | 45.5 ± 0.5 | 47.5 ± 0.5 |
| Hematocrit (%) | 47.0 ± 0.2 | 46.0 ± 5.0 | 48.0 ± 1.0 |
| Reticulocytes (%) | 3.4 ± 0.1 | 2.1 ± 0.0 | 2.2 ± 0.3 |
| MCH (pg) | 15.0 ± 0.0 | 15.0 ± 0.0 | 15.5 ± 0.5 |
| Leukocytes (x103/μL) | 3.1 ± 0.0 | 3.1 ± 2.0 | 4.8 ± 0.9 |
| Lymphocytes (%) | 72.0 ± 0.3 | 67.0 ± 2.0 | 78.0 ± 2.0 |
| Neutrophils (%) | 20.0 ± 0.1 | 25.0 ± 1.0 | 16.0 ± 2.0 |
| Monocytes (%) | 2.0 ± 0.0 | 3.0 ± 1.0 | 1.5 ± 0.5 |
| Eosinophils (%) | 3.0 ± 0.0 | 3.4 ± 0.5 | 2.5 ± 0.5 |
| Basophils (%) | 0.0 ± 0.0 | 0.5 ± 0.0 | 0.5 ± 0.0 |
| Biochemical | |||
| Creatinine (mg/L) | 4.0 ± 0.0 | 4.0 ± 0.8 | 4.2 ± 0.4 |
| BUN (mg/dL) | 23.0 ± 5.7 | 20.6 ± 6.6 | 27.0 ± 2.2 |
| Urea (g/L) | 0.5 ± 0.1 | 0.4 ± 0.1 | 0.5 ± 0.0 |
| ALT (x103 UI/L) | 2.8 ± 1.5 | 2.6 ± 0.4 | 2.1 ± 0.4 |
| AST (x103UI/L) | 2.5 ± 0.6 | 4.0 ± 0.6 | 3.4 ± 0.3 |
This finding was corroborated by the ex vivo basophil activation test using whole blood from human healthy subjects (n = 3), which assesses the activation state of these cells with or without stimulation51. In this test, a specific monoclonal antibody (anti-FcɛRI), binding to the high affinity IgE receptor, was used as positive control. Flow cytometry characterization showed no basophil activation following nanoalgosome treatments at different doses (0.25–2 µg/mL), with a percentage of basophil activation similar to the negative control (Supplementary Fig. 4; Supplementary Table 1), thus demonstrating the absence of acute immediate allergic reactions induced by nanoalgosomes.
Further, creatinine, urea and BUN, as well as AST and ALT values were similar to the control group, suggesting that the nanoalgosome treatment does not impair normal kidney and liver functions. These results were paralleled with the lack of any obvious histological changes in the liver and spleen architecture after 48 h of nanoalgosome administration (Supplementary Fig. 5). Thus, both two doses of nanoalgosomes did not exhibit toxic effects, blood parameter alterations after 48 h following a single acute I.V. administration.
The effects of nanoalgosomes in mice were also evaluated through clinical signs, body weight, and visual observations. Daily clinical examinations were performed to check behavior and signs of suffering, such as cachexia, weakness, difficulty in moving or feeding, hunching and convulsions. Supplementary Table 2 shows that there was not significant body weight loss, indicating that nanoalgosome administration was well-tolerated by the animals. Our results demonstrate that a single dose of up to 2 × 1011 nanoalgosomes per mouse did not elicit any noticeable local and systemic toxicity in immune-competent BALB/c mice, allowing for dose escalation or repeated administrations. This is in line with our C. elegans results and with previously published data on I.V. administration in mice of xenogeneic milk-, human- and plant-derived EVs8,10,11. However, additional safety evaluations are necessary, since it is important to further expand in vivo studies on nanoalgosome tolerability with repeated administration in mice and larger animals, along with biodistribution information to fully understand the potential and future applications48.
Biodistribution of nanoalgosomes in nude mice
Due to the presence of a lipid bilayer membrane, EVs can be labeled with fluorescent lipid dyes and their biodistribution has been evaluated in pre-clinical studies mainly using mouse models and recently using larger animals, including the pig-tailed macaque (Macaca nemestrina)48,52,53. To assess the biodistribution of nanoalgosomes systemically delivered in mice, the near-infrared dye DiR was used for nanoalgosome labeling. DiR is a lipophilic dye that fluoresces intensely only when inserted into a lipid membrane. The fluorescence spectrum of this dye (emission peak of 790 nm) allows for efficient penetration through bones and tissues with low autofluorescence, making it ideal for imaging in living animals.
Nanoalgosomes were labeled with the DiR probe, while the same amount of DiR diluted in PBS solution was used as the unbound dye negative control (Supplementary Fig. 6a). After extensive washing steps for the removal of free/self-aggregates unbound dye by ultracentrifugation, both the DiR-labeled nanoalgosome and unbound dye control samples underwent NTA analysis. The size distribution of the non-labeled and DiR-labeled nanoalgosome samples, reported in Supplementary Fig. 6b, showed that DiR labeling does not affect the nanoalgosome size distribution. Indeed, a typical nanoalgosome size distribution with a mode size of 100 ± 10 nm in diameter was observed after DiR labeling (Supplementary Fig. 6b). In free dye control samples, NTA did not detect any not-EV-associated fluorescent nano-structures, rendering the measurement unreliable. The presence of DiR fluorescence in the nanoalgosome-labeled samples was also verified by IR measurements using an Odyssey scanner, while no fluorescence signal was detected for the free dye control (Supplementary Fig. 6c).
Furthermore, to corroborate the specificity and purity of DiR labeled-nanoalgosome samples, sucrose density gradient was performed to exclude the presence of false-positive artifacts arising from EV-unbound label. As reported in Supplementary Fig. 6d, the presence of fluorescence signals, which is relative to the dye when bound to a lipophilic environment, correlates with the fractions that show positivity to the nanoalgosome marker H+-ATPase, as observed by immunoblot analysis (Supplementary Fig. 6f). In contrast, the free DiR control exhibits almost no fluorescence (Supplementary Fig. 6e), indicating minimal EV-unbound DiR background and confirming the efficiency of the free dye removal step. This suggests that DiR-nanoalgosome samples, post ultracentrifugation purification, contain DiR bound to nanoalgosomes, with negligible unbound DiR. These results are in agreement with previously shown results for EV labeling with DiR53,54.
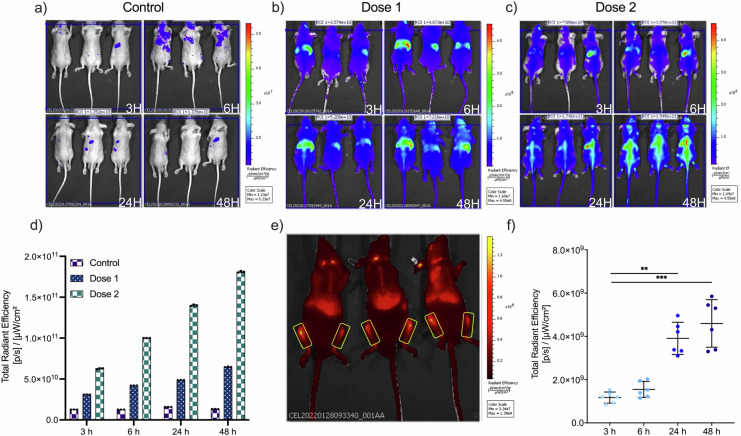
Following validation of the DiR labeling protocol, athymic nude mice (n = 6; 3 male, 3 female) were used for the IVIS Spectrum Imaging studies, to minimize the possible fluorescence background and as they are largely used for xenogeneic EV administration; these mice were intravenous injected with aforementioned DiR-labeled nanoalgosomes at two doses (or the same volume of the free dye control), dose 1 is 10 µg nanoalgosomes/mouse; dose 2 is 50 µg nanoalgosomes/mouse52,55. The biodistribution of the DiR-labeled nanoalgosomes was examined in live animals using IVIS. Prone and supine mice (prone male in Fig. 6a–c; prone female and supine male in Supplementary Fig. 7a, b) were imaged at 3, 6, 24 and 48 h post-IV injection. At 3 h post-injection, the DiR-nanoalgosome fluorescent signals were accumulating mainly in the liver (as commonly observed for other EVs). Fluorescent signals increased in a time- and dose-dependent manner, as shown in the total radiant efficiency plot (Fig. 6d). In contrast, lower and more constant fluorescent background signals were detected for the unbound dye control-treated mice after each imaging, highlighting the specificity of the DiR-nanoalgosome signal (Fig. 6d).
Fig. 6. IVIS imaging of DiR-labeled nanoalgosome biodistribution in nude mice after IV injection.
Nude mice (n = 3; male, prone) were injected with (a) v/v of unbound dye control/200 µL/mouse; (b) dose 1: 10 µg nanoalgosomes/200 µL/mouse; (c) dose 2: 50 µg nanoalgosomes/200 µL/mouse and control. Nanoalgosome biodistribution was measured at 3, 6, 24 and 48 h post-injection using an IVIS in vivo imaging system at ex/em 644/665. d Total radiant efficiency plot of DiR-labeled nanoalgosomes (doses 1 and 2) and the free dye control 3, 6, 24 and 48 h post-injection. e, f Representative images of DiR-nanoalgososome fluorescence in the femur (48 h post I.V. of 50 µg nanoalgosomes/mouse) and respective radiance efficiency quantification graph at 3, 6, 24 and 48 h. Kruskal-Wallis one-way ANOVA was used to assess the statistical significance of the differences: 3 h vs 24 h**, p < 0.01; 3 h vs 48 h***, p < 0.001.
In line with our findings, a side-by-side comparison study of three different bioimaging modalities using the fluorescent tracers DiR dye and mCherry fluorescent protein, the bioluminescent tracers Firefly (Fluc) and NanoLuc (Nluc) luciferases, as well as nuclear imaging using the [111In] radioisotope, demonstrated that DiR (unlike mCherry protein or other non-near-infrared probes) allowed detection of EVs in vivo with better sensitivity, signal-to-noise ratio, and no background fluorescence at expected tissue locations, which are mostly in the liver, followed by the spleen, and to a lesser extent the lungs and kidney53.
Unexpectedly, nanoalgosomes were localized in the bones 24 h post-IV injection, when injected at 50 µg nanoalgosomes/mouse, and, as shown in Fig. 6e, f, their concentration significantly increased and significantly accumulated in femur by 48 h (~2.5% of total radiance signal). This organotropism is peculiar for nanoalgosomes, as most mouse studies with MSC-derived EVs have reported that EVs accumulate to the liver, spleen, and sometimes kidney and lung, with rapid clearance from blood circulation after 24 h systemic injection47,52,53. Our data suggest a specific organotropism for bone compartments. Furthermore, the increasing fluorescent signals for up to 48 h in mice and in individual areas (i.e., backbone and femur) suggests that nanoalgosomes are stable in body fluids and that have sustained retention and potential accumulation from the bloodstream in mouse tissue (e.g., bone), as evidenced by detectable levels of the DiR-nanoalgosomes up to 48 h in mouse tissues. The supposed stability of nanoalgosomes was previously observed in biologic fluid23, and is also supported by evidence from in vitro and C. elegans up-take analyses (Supplementary Fig. 6g–l). Indeed, labeled-nanoalgosomes (specifically a pull of fractions 1-4 of Di-8-ANEPPS-labeled nanoalgosomes after sucrose density gradient separation) are taken up by 1-7 HB2 cells in a time-dependent manner and their presence inside cells persist up to 72 h (Supplementary Fig. 6g, h). These results are also confirmed in vivo in C. elegans intestinal cells in which the red fluorescent signal relative to labeled nanoalgosomes persists up to 72 h (Supplementary Fig. 6i). Conversely, when 1–7 HB2 cells are incubated with a pull of fractions 5–7 and fractions 8–10, that hypothetically could contain trace of dye aggregates even after washing steps, show no fluorescence when incubated with cells (Supplementary Fig. 6l).
Nanoalgosome bioactivity in vitro and in vivo
Anti-inflammatory activity of nanoalgosomes
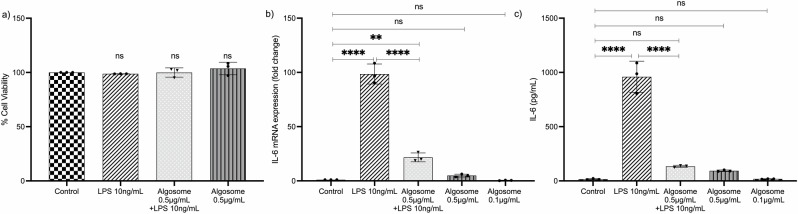
The bioactivity of nanoalgosomes was investigated in immune-responsive macrophage cells. Initially, we checked the viability of THP-1 cells treated with different concentrations of nanoalgosomes for up to 72 h, confirming that nanoalgosomes (up to 2 µg/mL) did not induce any cell toxicity (Supplementary Fig. 8). Subsequently, THP-1 cells were pre-treated with 0.5 µg/mL of nanoalgosomes for 4 h and afterwards exposed to lipopolysaccharide (LPS)-induced inflammation for 20 h. We first excluded a cytotoxic effect induced by these experimental conditions (Fig. 7a), then we monitored interleukin-6 (IL-6), which is a marker of inflammation, expression using qRT-PCR and ELISA tests. The results in Fig. 7b, c show a slight difference in IL-6 induction following 24 h of nanoalgosome treatment compared to untreated cells; this result is in line with the in vivo data, previously shown, and is indicative that nanoalgosomes have a limited, if any, inflammatory effect following single I.V. administration. Further, Fig. 7b, c shows that nanoalgosomes significantly reduced IL-6 induction in LPS-treated THP-1 cells, leading to a 4.5-fold reduction in mRNA level and a 7-fold reduction in IL-6 production, and indicating their anti-inflammatory activity in vitro.
Fig. 7. Anti-inflammatory effect of nanoalgosomes in immune-responsive cells.
a Cell viability after exposure with LPS (10 ng/mL) and nanoalgosomes (0.5 µg/mL) for 24 h, in THP-1 cells. By one-way ANOVA statistical test, differences of treated cells were not statistically significant (ns) when compared with the control (p > 0.9). b Real-time PCR quantification of IL-6 mRNA relative levels after exposure with LPS (10 ng/mL) and nanoalgosomes (0.5 µg/mL) for 24 h, in THP-1 cells. c ELISA results of IL-6 induction after exposure with LPS (10 ng/mL) and nanoalgosomes (0.5 µg/mL) for 24 h, in THP-1 cells. One-way ANOVA statistical test was used to assess the statistical significance of the differences: Control vs LPS (10 ng/mL), LPS (10 ng/mL) vs Algosomes (0.5 µg/mL) + LPS (10 ng/mL), ****p < 0.0001; Control vs Algosomes (0.5 µg/mL) + LPS (10 ng/mL), **p < 0.01; Control vs Algosomes (0.5 µg/mL) are not statistically significant (ns, p > 0.9); Control vs Algosomes (0.1 µg/mL) are ns (p > 0.9). Representative results of three independent biological replicates (n = 3 biologically independent samples).
Antioxidant activity of nanoalgosomes
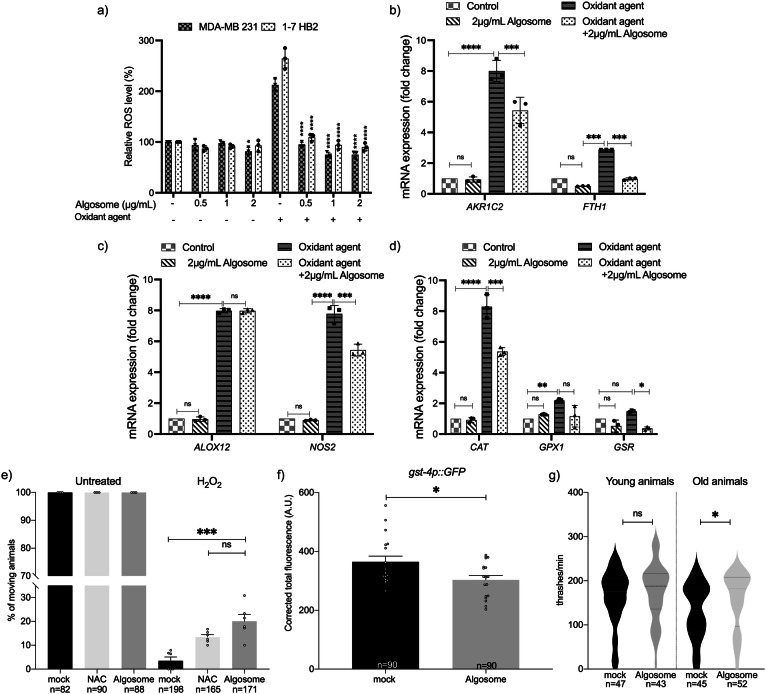
Microalgae are sources of antioxidant compounds so we sought to determine the potential antioxidant role of microalgae-derived EVs56,57. Nanoalgosome antioxidant activity was evaluated in two different cell lines: tumoral (MDA-MB 231) and normal (1-7 HB2), using the cell-permeable 2′, 7′- dichlorofluorescein diacetate (DCF-DA) fluorescent probe that emits fluorescence proportionally to intracellular ROS content. Cell viability analyses showed that none of the concentrations of the oxidant agents used (i.e., H2O2 and tert-butylhydroperoxide, TBH) were toxic to the cells (Supplementary Fig. 9b, c). Figure 8a shows the percentage of ROS levels in cells treated with different nanoalgosome concentrations (0.5, 1, and 2 μg/mL) for 24 h, with and without oxidative stress induction. As shown, treatments with nanoalgosomes per se did not induce oxidative stress, and the percentage of ROS levels was comparable to the negative control (untreated cells). After treatment with oxidant agents, ROS levels significantly increased in both cell lines. However, this increase in ROS levels was significantly lower in stressed cells which had been pre-treated with nanoalgosomes for 24 h, suggesting that nanoalgosomes significantly reduced ROS levels and rebalanced the physiological ROS levels in both cell lines. To further investigate the antioxidant abilities of nanoalgosomes, we analyzed whether they could counteract ROS, directly or indirectly modulating the expression of oxidative stress responsive genes. We selected a panel of genes that play important roles in regulating oxidative stress and maintaining cellular homeostasis. For instance, AKR1C2 (aldo-keto reductase family 1 member C2) plays a role in detoxifying lipid peroxidation products, which can contribute to the production of ROS. FTH1 (ferritin heavy chain1) helps to regulate iron levels, which is important for ROS regulation since iron can catalyze the production of free radicals. Alox12 (arachidonate 12-Lipoxygenase) can contribute to oxidative stress by producing leukotrienes, which are inflammatory mediators that can increase ROS production. NOS2 (nitric oxide synthase) produces nitric oxide, which can have harmful effects on oxidative stress. Further, to counteract the effects of ROS, CAT, GPX1 and GSR (catalase, glutathione peroxidase and reductase, respectively) work together to neutralize ROS by converting them into less harmful products and to maintain the redox balance by reducing glutathione disulfide, which can be formed as a result of ROS exposure58. Thus, these proteins are interconnected and work together to regulate ROS levels in cells and maintain cellular homeostasis. After incubating 1-7 HB2 cells with 2 µg/mL of nanoalgosomes for 24 h, with and without oxidant agent treatment, real-time PCR analyses were carried out to evaluate the mRNA expression levels of the selected genes involved in the oxidative stress cellular signaling (Fig. 8b–d). The results showed that the expression of oxidative stress-related genes in cells treated with nanoalgosomes for 24 h was similar to that of untreated cells, confirming that nanoalgosomes did not induce expression alterations of genes related to oxidative stress. As expected, after oxidative stress induction, these genes were upregulated, while the expression levels of most of the genes analyzed were significantly re-established or lowered in stressed cells pre-treated for 24 h with nanoalgosomes. These results suggested that nanoalgosomes have potent antioxidant abilities, likely due to their antioxidant cargo and ability to neutralize free radicals, promoting protective mechanisms inside cells.
Fig. 8. Antioxidant bioactivity of nanoalgosomes in cellular systems and in C. elegans.
a Percentage of ROS production in 1-7 HB2 and MDA-MB 231 cells treated with nanoalgosomes (0.5–1-2 µg/mL) for 24 h, with/without oxidant agent (250 µM TBH), normalized to untreated cells. Values are means ± SEM of three independent experiments. One-way ANOVA was used to assess the statistical significance of the differences: MDA-MB 231 vs MDA-MB 231 2 µg/mL nanoalgosome, *p < 0.05; 1-7HB2 vs 1-7 HB2 2 µg/mL nanoalgosome, *p < 0.05; MDA-MB 231 + oxidant agent vs MDA-MB 231 0.5-1-2 µg/mL nanoalgosome + oxidant agent, ****p < 0.0001; 1-7 HB2 + oxidant agent vs 1-7 HB2 0.5-1-2 µg/mL nanoalgosome + oxidant agent, ****p < 0.0001. b–d Real-time PCR quantification of antioxidant enzyme expression levels after exposure to nanoalgosomes (2 µg/mL) for 24 h, with/without oxidant agent (250 µM H2O2) in 1-7 HB2 cells. Each experiment was performed in triplicate (n = 3 biologically independent experiments). Three-way ANOVA statistical test was used to assess the statistical significance of the differences: Control vs 2 µg/mL nanoalgosome; Control vs Oxidant agent; Oxidant agent vs Oxidant agent + 2 µg/mL nanoalgosome, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns=not significant (p > 0.9). e In vivo response to exogenous oxidative stress. Animals have been pre-treated with mock, NAC (2 mM) and nanoalgosome (20 µg/mL). Animal movement was assessed right after acute exposure with water (untreated) or H2O2 5 mM. One-way ANOVA statistical test was used to assess the statistical significance of the differences: H2O2 mock vs H2O2 nanoalgosome ***p = 0.0009; H2O2 nanoalgosome vs H2O2 NAC ns=not significant (p > 0.5). f Quantification of the fluorescence in gst-4p::GFP expressing animals after chronic treatment with mock or nanoalgosomes (20 µg/mL). Each dot represents the total fluorescence in a picture corrected for the background. Bars represent the mean; error bar is the SEM. n is the total number of animals analyzed. Mann Whitney t-test was used for comparing: mock vs nanoalgosome *p = 0.0435. g Thrashing assay on young and old animals after acute treatments with mock and nanoalgosomes (10 µg/mL). Violin plots show the distribution of thrashes performed by the animals in a minute. Bold dashed lines in the center correspond to the medians, while upper and lower dashed lines correspond to the quartiles. n is the number of animals analyzed. Mann Whitney t-test was used for comparing: mock vs nanoalgosome treatment of young animals, ns=not significant (p > 0.3); mock vs nanoalgosome treatment of old animals *p = 0.0362.
To validate the nanoalgosome antioxidant effect in vivo, we evaluated the response of treated C. elegans animals to exogenous oxidative stress. Acute treatment of C. elegans with H2O2 can induce a nearly complete loss of mobility. In fact, 2 h of treatment with H2O2 caused a strong decrease in movement (Fig. 8e, mock), in contrast to the untreated animals. Interestingly, nanoalgosome treatment counteracted oxidative stress by increasing movement (Fig. 8e). This rescue was similar to one obtained with a positive control, N-AcetylCysteine (NAC) (Fig. 8e), a widely used antioxidant agent59. C. elegans copes with changes in ROS levels by expressing detoxifying genes, such as glutathione S-transferase gst-460.
To determine whether nanoalgosomes could also directly affect detoxification gene expression, we used a transgenic strain expressing GFP under the control of the gst-4 promoter61. After chronic treatment with nanoalgosomes, we observed a significant decrease in gst-4 expression levels, suggesting that nanoalgosomes can modulate detoxification gene expression (Fig. 8f and Supplementary Fig. 10). Moreover, antioxidant molecules are capable of counteracting aging and motility decline62. In fact, the animal movement is influenced by aging and starting from three days from hatching (young adult stage) it slowly declines63. Therefore, we decided to assess nanoalgosome antioxidant effects by performing an acute treatment at low concentration (10 µg/mL for 16 h), and we analyzed the locomotion of young adult animals (right after the treatment) and older animals (10 days from hatching, 7 days after treatment). As expected, we observed a physiological decline in locomotion between young and old animals (Fig. 8g). Interestingly, nanoalgosomes prevented aging effects by preserving the movement capability in older animals and keeping it similar to the one observed in younger animals (Fig. 8g). Taken together, our results indicate that nanoalgosomes are bioactive with a clear antioxidant effect both in vitro and in vivo, supporting their potential role in counteracting aging at molecular and functional level, having the potential to be used as natural and innovative antioxidant effectors.
Conclusions
In conclusion, this study demonstrates that nanoalgosomes are a new type of EVs efficiently uptaken by cells through a clathrin-dependent endocytosis and endowed with anti-inflammatory bioactivity. In addition, nanoalgosomes reduced the levels of reactive oxygen species and prevented oxidative stress in tumoral and normal cell lines and in C. elegans, where they counteract aging, thus suggesting that they are enriched with antioxidant compounds. Nanoalgosomes show also biocompatibility and unique bone tropism in living organisms, after a single I.V. administration. Furthermore, they come with the capacity to be produced at mass scale, with a renewable and sustainable bioprocess from an edible source. These features position nanoalgosomes well to be explored as a potential novel therapeutic tool.
Methods
Microalgae cultivation and nanoalgosome separation from microalgae-conditioned media
A stock culture of the marine chlorophyte Tetraselmis chuii CCAP 66/21b was cultivated in F/2 medium (using 20 nm-ultrafiltered clear sea-water) and the purification of nanoalgosomes was carried out following previously described methods24.
After a growth period of 30 days, 6 liters of microalgae cultures were clarified using a 450 nm hollow fiber cartridge in the KrosFlo® KR2i TFF System to remove larger particles. The feed flow and transmembrane pressure (TMP) were maintained at constant values of 750 mL/min and 0.05 bar, respectively. The permeate, containing particles smaller than 450 nm, was then subjected to a second filtration step using a 200 nm hollow fiber cartridge, again with a feed flow of 750 mL/min and 0.05 bar TMP. The resulting permeate, containing particles smaller than 200 nm, was subjected to ultrafiltration using a 500 kDa MWCO hollow fiber cartridge, with a feed flow of 750 mL/min and 0.05 bar TMP, and concentrated to a final volume of 150 mL. Subsequently, the samples were concentrated and diafiltered seven times using a smaller 500 kDa cutoff TFF filter module, with a feed flow of 75 mL/min and 0.25 bar TMP, with PBS without calcium and magnesium (Sigma-Aldrich) as the diafiltration solution. This process resulted in a final volume of approximately 5 mL.
Microalgae Lysates
5 mL of Tetraselmis chuii culture with an OD600 = 1.280 underwent centrifugation at 2000 g for 5 min. The resulting supernatant was discarded, and 2 mL of complete Lysis RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1.0% (v/v) NP-40, 0.5% (w/v) Sodium Deoxycholate, 0.1% (w/v) and protease (25X) and phosphatase (100X) inhibitor cocktails) was added to the microalgae pellet. Subsequently, the sample underwent 9 cycles of sonication on ice (8 s each at 70% power). Following this, the sample was centrifuged at 10,000 rpm for 15 min at 4 °C. The supernatant was collected, and protein quantification was carried out using BCA Protein Assay Kit.
Nanoalgosome characterization
BCA assays
Nanoalgosome and microalgae lysate protein content were quantified using the micro-bicinchoninic BCA Protein Assay Kit from Thermo Fisher Scientific. This colorimetric method involves comparing the relative concentration of the sample to a protein standard (bovine serum albumin, BSA) and preparing a calibration curve. The relative absorbance of the BCA soluble compound was measured at 562 nm using a GloMax Discover Microplate Reader. The protein content of the nanoalgosomes was determined using the Micro BCA Protein Assay Kit (Thermo Fisher Scientific).
Nanoparticle tracking analysis (NTA)
The size distribution and concentration of nanoparticles were determined using a NanoSight NS300 (Malvern Panalytical, UK). Nanoalgosomes samples were diluted in particle-free water (HPLC-grade Sigma-Aldrich Water filtered by 20 nm using Whatman Anotop filters) to generate a dilution with 20-120 particles per frame. This ensured a concentration within the recommended measurement range of 1–10 × 108 particles/ml. NTA 3.4 Build 3.4.003 (camera level 15–16) was used to analyze five 60-second videos per sample at a syringe speed of 60 in light scattering. Data were further processed using the NanoSight Software NTA 3.1 Build 3.1.46 with a detection threshold of 5.
Fluorescence nanoparticle tracking analysis (F-NTA)
The Fluorescence nanoparticle tracking analysis (F-NTA) was carried out as describe in Adamo et al.23. Briefly, the Di-8-ANEPPS dye was used to label nanoalgosomes, which fluoresces when activated in apolar environments and is specifically enhanced when attached to the lipid membrane of EVs. The fluorescent nanoalgosome (f-algosome) were produced as follows: 5 × 1010 EV particles/ml were stained with 500 nM of 4-(2-[6-(dioctylamino)-2-naphthalenyl]ethenyl)-1-(3-sulfopropyl)pyridinium, DI-8-ANEPPS (Ex/Em: 467/631 nm, ThermoFisher Scientific), previously filtered by 20 nm filters (Whatmann Anotop filters). Following a 1-h incubation at room temperature, NTA analyses were performed using a NanoSight NS300 instrument (Malvern Panalytical, UK) equipped with a 500LP filter (laser wavelength 488 nm). The camera level was manually adjusted to optimal settings, and the flow rate for the syringe pump was set at 150 μl/s to ensure that f-algosome crossed the main NTA screen field of view in 5 to 10 s. Additional settings were adjusted as described in the previous NTA section. To confirm specificity, a negative control was conducted to verify that the probe alone did not emit a fluorescence signal with F-NTA.
Atomic force microscopy (AFM)
Atomic Force Microscopy images were performed by using a Nanowizard III scanning probe microscope (JPK instruments) equipped with a 15 μm z-range scanner. Nanoalgosomes were initially concentrated by ultracentrifugation and resuspended in MilliQ water to a final concentration of 5 × 1011 particles/ml, as estimated by NTA. A 30 μl drop of the sample was incubated on freshly cleaved mica for 10 min, and then gently dried under nitrogen flow. AFM images were acquired in tapping mode by using a NSC-15 (Mikromasch) cantilever (typical spring constant 40 N/m, typical tip radius 8 nm).
Cryo-transmission electron microscopy (cryo-TEM)
For imaging, 3 μl of the nanoalgosome preparation (4 × 1011 particle/ml) were applied onto a 400 mesh copper grid covered with quantifoil R2/1 carbon film. Prior to application, the grid was treated with oxygen/hydrogen plasma to enhance hydrophilicity and plunged into liquid ethane using a Vitrobot Mark V (Thermo Fisher). Inspection of the specimen was done using a Titan Krios G4 (Thermo Fisher) transmission electron microscope, operated at 300 kV acceleration voltage. Acquisition was done using a Gatan K3 (Gatan, Pleasonton) camera.
Multi angle dynamic light scattering (DLS)
An aliquot of vesicle solution was pipetted and centrifuged at 1000 g for 10 min at 4 °C in order to remove any dust particle. The supernatant was withdrawn by pipet tips (previously washed by MilliQ water), put directly into a quartz cuvette and incubated at 20 °C in a thermostated cell compartment of a BI200-SM goniometer (Brookhaven Instruments) equipped with a He-Ne laser (JDS Uniphase 1136 P) with wavelength λ = 633 nm and a single pixel photon counting module (Hamamatsu C11202-050). The autocorrelation function g2(t) of scattered light intensity was measured at a scattering angle ϑ = 90° by using a BI-9000 correlator (Brookhaven Instruments).
The intensity autocorrelation function g2(t) is related to the size σ of diffusing particles and to their size distribution Pq(σ), by the relation g2(t) = 1 + |β ∫ Pq(σ) exp[−D(σ)q2t]|2, where β is an instrumental parameter, q = 4πñλ − 1 sin[ϑ/2] is the scattering vector, with ñ being the refractive index of the medium (ñ = 1.3336), and D(σ) is the diffusion coefficient of a particle of hydrodynamic diameter Dh = σ, determined by the Stokes-Einstein relation D(σ) = kBT [3πησ] − 1, with T being the temperature, η the medium viscosity, and kB the Boltzmann constant.. The size distribution Pq(σ) is calculated by assuming that the diffusion coefficient distribution is shaped as a Schultz distribution, which is a two-parameter asymmetric distribution, determined by the average diffusion coefficient <D> and its variance V. Two robust parameters may be derived from this analysis: Dz, the z-averaged hydrodynamic diameter (corresponding to the average diffusion coefficient <D > ), and PDI, the polydispersity index (PDI = V < D > − 2), which is an estimate of the distribution width.
Immunoblotting analysis
Proteins of nanoalgosome sample and microalgal lysates were separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (10%). A total of 5 μg of nanoalgosome sample (in PBS) and microalgae lysates (in Lysis RIPA buffer) were mixed with proper volumes of 5X loading buffer (0.25 M Tris-Cl pH 6.8, 10% SDS, 50% glycerol, 0.25 M dithiothreitol (DTT), 0.25% bromophenol blue). Then, the samples were heated at 100 °C for 5 min and loaded in a 10% sodium dodecyl sulfate-polyacrylamide gel for electrophoretic analyses. Proteins were blotted onto polivinilidenfluoro-membranes (PVDF), which were blocked with 3% bovine serum albumin (BSA) in TBS-T solution (50 mM Tris HCl pH 8.0, 150 mM NaCl with 0.05% Tween 20) for 1 h at room temperature. The antibodies anti H+-ATPase (dil. 1:1000 in 3% BSA/TBS-T, Agrisera), anti-Alix (clone 3A9, dil. 1:150 in 3% BSA/TBS-T, Santa Cruz) and anti-β-actin (clone 10-B3 dil. 1:500 in 3% BSA/TBS-T, Sigma Aldrich), were incubated over night at 4 °C and 2 h at room temperature. After washing, the membrane was incubated for 1 h with secondary antibodies according to the manufacturer’s instructions (horseradish peroxidase-conjugated secondary anti-mouse or anti-rabbit antibodies, Cell Signaling). The membrane was washed four times in TBS-T for 20 min. The immunoblots were revealed using SuperSignal Pierce ECL (Thermo Fisher Scientific).
Cell lines
The following cell lines were used for the gene expression, bioactivity, and intracellular trafficking analyses: (i) 1–7 HB2, a normal mammary epithelial and (ii) MDA-MB 231, an epithelial human breast cancer and (iii) THP-1 human monocytic leukemia cell lines (ECACC 88081201). All cell lines were maintained at 37 °C in a humidified atmosphere (5% CO2) in Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma-Aldrich) containing 10% (v/v) Fetal Bovine Serum (FBS) (Gibco, Life Technologies) plus 2 mM L-glutamine, 100 U/ml Penicillin and 100 mg/ml Streptomycin (Sigma-Aldrich) for the MDA-MB-231; DMEM low glucose, containing 10% (v/v) FBS, plus 2 mM L-glutamine, 100 U/ml Penicillin and 100 mg/ml Streptomycin plus 5 μg/ml Hydrocortisone and 10 μg/ml, Bovine Insulin (Sigma-Aldrich) for 1–7 HB2 cell line; RPMI 1640 Medium containing 10% (v/v) inactivated FBS plus mM L-glutamine, 100 U/ml Penicillin and 100 mg/ml Streptomycin was used for THP-1 cell line23,25.
C. elegans maintenance and strains
Nematodes have been grown and handled following standard procedures under uncrowded conditions on nematode growth medium (NGM) agar plates seeded with Escherichia coli strain OP5064. Strains used in this work have been provided by the Caenorhabditis Genetics Center (CGC): wild-type strain N2 (Bristol variety); EG1285 oxIs12 X [unc-47p::GFP; lin-15(+)](expression of the Green Fluorescent Protein, GFP, in GABA neurons); VC2405 chc-1(ok2369) III/hT2 [bli-4(e937) let-?(q782) qIs48] (I;III) (knockout mutant of chc-1); RT1315 unc-119(ed3) III, pwIs503 [vha-6p::mans::GFP + Cbr-unc-119(+)] (expression of an intestine-specific GFP marker for the Golgi apparatus); RT476 unc-119(ed3) III; pwIs170 [vha6p::GFP::rab-7 + Cbr-unc-119(+)] (expression of an intestine-specific GFP marker for late endosomes); CL2166 dvIs19 III [(pAF15) gst-4p::GFP::NLS] (expression of an oxidative stress-responsive GFP). Strain QH3803 vdEx263 [mec-4p::mCherry; odr-1p::dsRed] expressing mCherry in mechanosensory neurons was kindly provided by M.A. Hilliard (QBI, University of Queensland, Australia)65. BY250 vtIs7 [dat-1p::GFP] expressing GFP in dopaminergic neurons was kindly provided by M. Aschner (Albert Einstein College of Medicine, NY, USA)66.
Nanoalgosome staining
Nanoalgosomes were labeled with lipophilic dyes: Di-8-ANEPPS (Sigma-Aldrich) for endocytosis inhibition study and in vivo studies in C. elegans; PKH26 (Sigma-Aldrich) for intracellular trafficking studies; DiR (Sigma-Aldrich) for in vivo studies in mice.
Di-8-ANEPPS-, PKH-26- and DiR-nanoalgosome staining were performed as described23,25.
Briefly, a concentration of 500 nM Di-8-ANEPPS (Invitrogen, filtered with 20 nm Anotop filter) was incubated with 5 × 1010 particles/mL for 30 min at room temperature. For the PKH26 fluorescent staining of nanoalgosomes, PKH26 was diluted in Diluent C (provided by Sigma-Aldrich with PKH26) at a final concentration of 3 μM (dye solution). This staining solution was incubated with 5 × 1010 particles/mL at 37 °C for 1 h.
For in vivo studies in mice, DiR labeling was carried out by incubating 30 min at room temperature nanoalgosomes (5 × 1010 particles/mL) with 3.5 µM of DiR.
Concurrently, nanoalgosome-free PBS with the same amount of fluorescent dyes (e.g., DI-8-ANEPPS, PKH26, and DiR) were incubated and served as negative control for the three dyes (e.g., control for free/self-aggregate dye not incorporated into EVs).
For free/aggregate label removal, stained nanoalgosomes and the nanoalgosome-free PBS solution with the dyes underwent: (1) for Di-8-ANEPPS dialysis against PBS using a Slide-A-Lyzer MINI Dialysis Device with a 3.5 KDa MWCO (Thermo Fisher Scientific) and (2) for PKH26 and DiR - the samples were diluted to 30 mL with PBS and pelleted by ultracentrifugation at 118,000 g for 70 min at 4 °C using a Beckman SW55Ti rotor, the same procedure was repeated 2-times. The resulting pellet was gently resuspended in PBS overnight at 4 °C.
Next, all samples were analyzed by nanoparticle tracking analysis, to check nanoparticle size distribution and concentration, following the removal of free dye23. The presence of DiR-fluorescence in nanoalgosome-labeled samples was verified by Odyssey scanner IR-measurements (Supplementary Fig. 6a–c).
Sucrose density gradient
DiR/Di-8-ANEPPS labeled nanoalgosomes or unbound dye negative control were layered on top of a discontinuous gradient. Different sucrose solutions were slowly layered at the bottom of an ultracentrifuge tube (Open-Top Thickwall Polycarbonate Tube, Beckman Coulter) to prepare a discontinuous gradient containing 0.5 ml of 0.0, 0.5, 1.5, 2, and 2.5 M sucrose. After ultracentrifugation at 118,000 g for 16 h at 4 °C (rotor SW55Ti, Beckman Coulter), ten fractions of 300 μl each were collected from the top of the tubes, from fraction 10 to fraction 1. Each fraction was washed once with 3 ml of PBS (118,000 g for 70 min at 4 °C, using SW55Ti rotor, Beckman Coulter). For fractions 1-10 of DiR labeled nanoalgosomes and unbound dye negative control, the fluorescence intensity was measured using infra-red scanner (Odissey, λ 800 nm). Additionally, each fraction was subjected to subsequent western blotting analysis by loading the same volume of each fraction to assess H+-ATPase positivity (see relative Materials and Methods described for Immunoblotting analysis). Pools of fractions 1–4, 5–7, and 8–10 of Di-8-ANEPPS-labeled nanoalgosomes were used for uptake studies; a pool of fractions 1–4 of unbound Di-8-ANEPPS was included as negative control. Specifically, 1-7 HB2 cells were seeded at a density of 5 × 103 cells/well in 12-well plates containing sterile coverslips in complete medium for 24 h. Next, the cell lines were incubated with the same volume of each pool for 48 and 72 h. Afterward, cells were washed twice with PBS, fixed with 3.7% paraformaldehyde for 15 min, and washed again twice with PBS. Subsequently, the nuclei were stained with VECTASHIELD Mounting Medium with DAPI. The cells were monitored by fluorescence microscopy analysis (IX3 microscope, Olympus) and analysed using ImageJ 1.52t.
Endocytosis inhibition study in cellular models
To study the specific endocytic mechanism involved during nanoalgosome uptake, 1-7 HB2 cells (5 × 103/wells) were seeded in a 96-well plate in DMEM low glucose, containing 10% (v/v) FBS, plus 2 mM L-glutamine, 100 U/ml Penicillin and 100 mg/ml Streptomycin plus 5 μg/ml Hydrocortisone, and 10 μg/ml Bovine Insulin. After 24 h, cells were pre-incubated with pharmacological/chemical inhibitors before nanoalgosome addition. Next, the dose- and time-dependent effect of each inhibitor on nanoalgosome cellular uptake was determined. In particular, cells were pre-incubated with the following inhibitors: dynasore (a clathrin-mediated endocytosis inhibitor) at 10, 30, 60, 80 µM; EIPA (5-[N-ethyl-N-isopropyl] amiloride, a pinocytosis/macropinocytosis inhibitor) at 1, 5, 10, 25 µM; nystatin (a caveola/lipid raft-mediated endocytosis inhibitor) at 5, 10, 25, 50 µM. Cells were pre-incubated with dynasore and EIPA for 60 min at 37 °C and washed prior to nanoalgosome addition. Cells were pre-incubated with nystatin for 30 min at 37 °C. Next, 10 µg/mL Di-8-ANEPPS-labeled nanoalgosomes were added to cells in the presence of these blocking agents and were subsequently kept at 37 °C up to 3 h. The cells treated with nanoalgosomes without inhibitor treatment were used as negative control, while positive control cells were incubated at 4 °C to inhibit all energy-dependent mechanisms. Intracellular fluorescence was monitored after 2 and 3 h of nanoalgosome incubation by spectrofluorimetric analysis using a microplate reader (Glomax, Promega). As vehicle-control, cells were cultured in the presence of methanol. A cell viability assay was performed after each treatment (Supplementary Fig. 1), and treatments were performed in triplicate.
Nanoalgosome intracellular localization in vitro
The MDA-MB 231 and 1-7 HB2 cell lines were grown at a density of 5 × 103 cells/well in 12-well plates containing sterile coverslips in complete medium for 24 h. Cells were incubated with 2 µg of PKH-26-labeled nanoalgosomes at 37 °C for 24 h. Cells were then washed twice with PBS, fixed with 3.7% paraformaldehyde for 15 min, and washed twice with PBS. Afterwards, cells were permeabilized with 0.1% Triton 100-X in PBS with Ca++ and Mg++ for 10 min at room temperature. Next, cells were incubated with 1% bovine serum albumin (Sigma-Aldrich) in PBS (blocking solution) for 30 min at room temperature to block unspecific binding of the antibodies. Cells were incubated in a primary antibody (CD63 for the endosomal system, clone MX-49.129.5, VWR, anti-LAMP1 for lysosomes, clone 1D4B, Sigma-Aldrich, and anti-calnexin for ER, clone NB100-1965) diluted in blocking solution for 1 h at 37 °C. Cells were then incubated with a secondary antibody (AlexaFluor-488; Thermo Fischer Scientific) and diluted in blocking solution (1:50) for 2 h at room temperature. Coverslips were mounted with a drop of Vectashield Mounting Medium with DAPI (Sigma-Aldrich). Nanoalgosome intracellular localization was monitored by fluorescent microscopy (IX3 microscope, Olympus) and confocal laser scanning microscopy (Olympus FV10i, 1 μm thickness optical section was taken on a total of about 15 sections for each sample).
In vivo subcellular localization and internalization mechanism in C. elegans
For subcellular localization and to identify the internalization mechanism, transgenic or mutant animals were treated in solido. ~20–30 synchronized L4 larvae were transferred on freshly prepared NGM plates seeded with heat-killed OP50 bacteria or in 96-well plates with LGM (OP50 bacteria, M9 buffer, antibiotic antimycotic solution 2x cat. n. A5955 Sigma-Aldrich and cholesterol 5 ng/ml)(see below), and Di-8-ANEPPS labeled nanoalgosome (20 μg/mL final concentration) as described in Picciotto et al.25. After 24 h of treatment in the dark, young adult animals were transferred to clean NGM plates with OP50 bacteria to let the animals crawl for 1 h and remove the excess of dye. Animals were immobilized with 0.01% tetramisole hydrochloride (Sigma-Aldrich, T1512) on 4% agar pads for microscopy analysis and confocal images were collected with a Leica TCS SP8 AOBS microscope using a 40x objective. Leica settings: laser intensity 10, excitation 470 nm; emission 657–705 nm. 2–3 Z-stack 1 µm pictures have been projected with LAS X Leica software, to include multifocal planes of the intestinal cells. Pictures of mock and treated animals have been modified post-acquisition using the same parameters with macromedia Firework software increasing intensity and contrast to improve fluorescence visualization also with low-resolution monitors.
DNA damage in vitro and in vivo
Gene expression analyses in vitro
Quantitative analysis of mRNA expression was performed in 1-7 HB2 cells. Specifically, 5 × 103 cells were treated with 2 µg/mL of nanoalgosomes for 24 h for genotoxicity study, in which ATR, CHECK1, RAD51 mRNAs were chosen to assess the expression of DNA damage-related genes. RNA extraction, first-strand cDNA synthesis and gene expression by real-time qPCR are reported in detail in Supplementary methods. Treatments were performed in triplicate.
Genotoxicity assay in C. elegans
For germline apoptosis quantification, animals were treated in liquido as described afterwards, with nanoalgosome at a final concentration of 20 μg/mL, PBS as mock and doxorubicin 2.4 μM as positive control. After 72 h of treatment animals were stained by incubation with 33 µM SYTOTM12 (Invitrogen, S7574) for 1 h and 30 min at room temperature in the dark. Animals were cleaned letting them crawl in fresh plates for 30’. For microscopy analysis animals were immobilized as above and the quantification of apoptosis after treatment was calculated as the average number of apoptotic corpses per gonadal arm.
Biocompatibility in vivo assays in C. elegans
For the assessment of nanoalgosome toxicity in vivo, C. elegans synchronized animals have been treated with PBS as mock or nanoalgosomes at different concentrations (1, 20, 64 and 128 µg/mL), through in liquido culturing in 96-multiwell plates for 72 h25. Synchronized eggs were obtained by bleaching and resuspended in LGM: M9 buffer (3 g KH2PO4; 6 g Na2HPO4; 5 g NaCl; 1 M MgSO4; H2O to 1 L) with 2x antibiotic/antimycotic solution (Sigma-Aldrich, A5955), 5 ng/mL cholesterol and OP50 bacteria. 60 µL of this mix containing approximately a total of 30 eggs were aliquoted in each well. After 72 h the number of living animals and of animals reaching the adult stage was quantified per each replicate, with a minimum number of replicates corresponding to three. For the growth assay, synchronized animals have been treated in solido on NGM plates seeded with heat-killed (45 min at 65 °C) OP50 bacteria and nanoalgosomes (20 μg/mL final concentration), or PBS buffer as mock. The dilutions were performed considering a final volume of NGM in the plate of 4 mL25. ~20 adults per plate in triplicate were let lay eggs for 2 h to obtain synchronized eggs. Animal stage was quantified in each plate every 24 h for 4 days, and the percentage of animals at each stage was calculated on the total number of animals present on the plate. For the lifespan assay, synchronized animals have been treated in solido, as described above, with nanoalgosome at a final concentration of 20 μg/mL and PBS as mock. Viability was scored each 3 days and animals were transferred every 3 days in fresh plates with nanoalgosome or mock, respectively. To test the nanoalgosome effect on brood size and embryonic viability, 20 animals were treated in solido in triplicate, as described above, until L4 larval stage and then transferred to new plates without any treatment. Then, every 24 h animals were moved to new plates without any treatment for all the fertile period of the animals (4 days) and the number of laid and hatched eggs counted every day67. To assess a putative nanoalgosome neurotoxicity, animals expressing fluorescent protein in GABAergic, mechanosensory and dopaminergic neurons were treated in liquido in triplicate, as described above.
After 72 h of treatment animals were immobilized as above and the morphology and the total number of visible fluorescent neurons was quantified in each animal65,66,68,69. Confocal images were collected with a Leica TCS SP8 AOBS microscope using a 20x objective with laser setting: laser intensity 10; GFP excitation 488 nm emission 509-555 nm; mCherry excitation 605 nm emission 611–670 nm.
In vivo experiments in mice
All animal experiments were performed on wild type BALB/c and athymic nude mice, male and female, which were purchased from Janvier Labs (France) at 5 weeks of age. Animals were used in accordance with Cellvax approved standard operation procedures and with all national or local guidelines and regulations. Mice were housed in specific pathogen-free conditions with food and water were provided ad libitum, in compliance with the Federation of European Laboratory Animal Science Association (FELASA) guidelines. Housing conditions entailed a 12:12 light:dark cycle, room temperature at 20 ± 2 °C, and 70% relative humidity. Mice were acclimatized for one week and were regularly handled by personnel for gentling and habituating to the procedures. On the day of the experiment, animals were randomly allocated into 3 different treatment groups. For the biocompatibility study, groups contained 2 male and 2 female BALB/c mice (n1 = 4); for the biodistribution study, groups consisted of 3 male and 3 female athymic nude mice (n2 = 6). Each group received a different treatment. The results were presented as mean ± standard deviation (SD).
USE OF RESEARCH ANIMALS: The methods were performed in accordance with relevant guidelines and regulations and approved by Animal Facility of Hospital Paul Brousse agreed by French authorities (Animalerie INSERM -Avenue Paul Vaillant Couturier 94800, Villejuif France), with the protocol approval study number cellvax 20300.
Nanoalgosome biocompatibility study in mice
BALB/c mice (n = 4) were injected via intravenous injection into the retroorbital sinus (using 1 mL insulin syringes with a 25 G needle) with nanoalgosome formulations (dose 1: 10 µg corresponding to 4 × 1010 nanoalgosomes/200 µL/mouse; dose 2: 50 µg corresponding to 2 × 1011 nanoalgosomes/200 µL/mouse and control: v/v of PBS/200 µL/mouse).
Animals were examined clinically daily, including clinical signs, behavior, signs of suffering (cachexia, weakening, and difficulty of moving or feeding, etc.) and nanoalgosome toxicity (hunching, convulsions). Body weight was monitored at day 3 and 6 post I.V. injection and a body weight curve were designed (Mean ± SD).
Biochemical and hematological analysis
For each animal, 100 μL of blood (in compliance with the National Centre for the Replacement, Refinement and Reduction of Animals in Research guidelines) was collected from the lateral tail vein. Animals were euthanized after the last sampling point with CO2 (slow fill rate and organ harvesting performed after confirmation of death). Blood was collected and stored in K2-EDTA BD-Microtainers (Thermo Fisher Scientific), centrifuged at 4 °C, at 3000 g for 5 min and the obtained plasma (25 μL) stored at −20 °C. The blood parameters analyzed were hematocrit, lymphocyte count, creatinine, blood urea nitrogen (BUN), liver transaminase enzymes.
Nanoalgosome biodistribution in the mouse model
IVIS spectrum studies: The biodistribution of the fluorescently labeled nanoalgosomes was determined by in vivo near-infrared fluorescence imaging using an IVIS Spectrum scanner (IVIS Spectrum CT; PerkinElmer). Athymic nude mice (n = 6) were anesthetized with 5% isoflurane in 100% O2 at 1 L/min for induction, and anesthesia was maintained with 1.2 minimum alveolar concentration via face mask while mice were injected into the retroorbital sinus (as above) with the DiR labeled formulations (listed above) and images were acquired at different post-injection time points (3, 6, 24, 48 h). The IVIS Spectrum scanner was set to fluorescence imaging mode, time and an emission filter positioned at 650 nm. The focus was kept stable using subject high of 1.5 cm whereas the temperature of the chamber was set at 37 °C. Image analyses (set with a fixed count scales from 1000 to 40,000 and acquired with a 0.2 s exposure time) were computed by first defining the regions of interest (ROI; a representative image with the ROIs are shown in Fig. 5), which were kept consistent across images, and then the sum of all counts for all pixels inside the ROI (Total Fluorescence Counts-TFqC; photons/second) was recorded.
Flow cytometry-based basophil activation test
Heparinized peripheral blood was obtained from 3 volunteers. The potential allergenic activity of nanoalgosomes was studied by basophil activation in whole blood samples using flow cytometry to detect the combination of CCR3 and CD63 markers by means of the Flow CAST Basophil Activation Test (Buhlmann Laboratories AG, Switzerland). Blood aliquots (100 µl) were incubated with nanoalgosome dilutions (0.025–0.5–1–2 µg/mL, according to the BCA analysis) in RPMI for 15 min at 37 °C. PBS and anti-Fc Epsilon RI were used as negative and positive controls, respectively. Results were analyzed using WinMDI 2.8 software (Scripps Research Institute, Jupiter). Basophil activation was reported when more than 5% of activated cells were detected (according to the manufacturer’s instructions). The study was approved by the local Ethics Committee (Comitato Etico Palermo 1, 24 February 2021, resolution n. 02/2021). Verbal informed consent was obtained from all research participants.
Anti-inflammatory activity assay
The THP-1 cell line was maintained in culture with RPMI 1640 medium (Gibco Life Technologies, Italy) supplemented with heat inactivated 10% Fetal Bovine Serum (FBS, Gibco Life Technologies, Italy) and 1% antibiotic (penicillin 5000 U/mL, Streptomycin sulfate 5000 µg/mL, Gibco Life Technologies, Italy). First, cells were differentiated into macrophages by 72 h incubation with 200 nM phorbol 12-myristate 13-acetate (PMA). Next, 1 × 106 cells/mL THP-1 cells were seeded in 24-well in complete medium. After 24 h, cells were pre-incubated with nanoalgosomes (0.5 µg/mL) for 4 h. After 4 h of pre-incubation, cells were subjected to an inflammatory stimulus, by adding lipopolysaccharide (10 ng/mL LPS from E. coli O55: B5, Sigma-Aldrich) for 20 h. Cells stimulated with LPS for 20 h (without pre-incubation with nanoalgosomes) represented the positive controls. In addition, cells were incubated with different amounts of nanoalgosomes (0.5 and 0.1 µg/mL) for 24 h. Untreated cells are used as negative controls.
Cell viability in vitro
Cell viability was evaluated using the CellTiter 96® AQueous one solution reagent (Promega) according to the manufacturer’s instructions. The mean optical density at 490 nm (OD, absorbance) of each well was used to calculate the percentage of cell viability relative to negative control. Values were expressed as means±standard deviation.
Gene expression analyses in vitro
Quantitative analysis of mRNA expression was performed in THP-1 cells, in which Interleukin-6 mRNA was chosen to assess the expression of inflammatory cytokine related gene. RNA extraction, first-strand cDNA synthesis and gene expression by real-time qPCR are reported in detail in Supplementary methods. Treatments were performed in triplicate.
Inflammatory cytokine production in vitro
Interleukin-6 levels in supernatants were determined using commercially available human IL-6 ELISA kits according to the manufacturer’s protocol (Invitrogen). A detailed description of the procedure is reported in Supplementary methods. Experiments were performed in triplicate.
Antioxidant bioactivity study
Antioxidant bioactivity assay in cellular models
Intracellular ROS levels of living cells were determined using 2′, 7′- dichlorofluorescein diacetate (DCF-DA; Sigma-Aldrich). DCF-DA is oxidized to fluorescent DCF (2′, 7′- dichlorofluorescein) in the presence of ROS, which can be readily detected by a spectrofluorometer. We performed an antioxidant assay on tumoral (MDA-MB 231) and normal (HB2 1-7) cell lines. Briefly, 4 ×103 cells were cultured in 96-well microplates for 24 h. Cells were then incubated with different concentrations of nanoalgosomes (0.5, 1 and 2 μg/mL) for 24 h. The medium was removed, and cells were exposed to PBS containing 40 μM of DCF-DA and kept in a humidified atmosphere (with 5% CO2 at 37 °C) for 1 h. Next, cells were treated with/without the oxidative agent H2O2 (250 μM for 3 h) or TBH (250 μM for 1 h) (tert-butyl hydroperoxide solution, Sigma-Aldrich) in the absence/presence of nanoalgosomes. Untreated cells were used as a control to set the percentage of basal intracellular ROS. After extensive washing steps, fluorescence intensity was quantified using a fluorescence plate reader at an excitation of 485 nm and an emission of 538 nm (GloMax® Discover Microplate Reader, Promega).
The relative percentage of intracellular ROS was normalized with respect to untreated cells (control). A cell viability assay was performed after each treatment (see Supplementary methods). For gene expression analysis related to antioxidant activity of nanoalgosomes, quantitative analysis of mRNA expression was performed on 1-7 HB2 cells (as described above) and included CAT, GPX, GSR, AKR, FTH, ALOX, and NOS mRNAs. 5 × 103 cells were incubated with nanoalgosomes (2 μg/mL) for 24 h, then, the medium was removed, and cells were exposed to oxidative stress (250 μM of H2O2, Sigma-Aldrich, Germany) for 3 h. Untreated cells were used as negative control.
In vivo antioxidant bioactivity assays in C. elegans
For the assessment of exogenous H2O2 stress response, animals have been treated in liquido as described above for 72 h with nanoalgosome at 20 μg/mL final concentration, PBS as mock and N-AcetylCysteine (NAC) (Sigma-Aldrich, A7250) 2 mM as positive control. After treatment, animals were exposed to H2O2 5 mM and water as control for 2 h, then transferred to fresh NGM plates with OP50 bacteria and the number of moving animals on the total number of animals on the plate was scored.
gst-4p::GFP expression was quantified after 72 h of treatment in liquido, as described above. 5 animals on each slide were immobilized as above, this time using 100 mM NaN3 (Sigma-Aldrich, S8032) to allow the lineup of animals. Epi-fluorescence images were collected with a Leica TCS SP8 AOBS inverted microscope, using a 10x objective and FITC filter. Fluorescence quantification was performed using ImageJ, and the Corrected Total Fluorescence (CTF) was calculated for each image using this formula: (integrated density of the area containing the animals) – [(area containing the animals) x (mean fluorescence of background)]67. Representative pictures in Supplementary Fig. 10 were pseudo-colored using Image J.
The anti-aging effect of nanoalgosome on animal movement was assessed through the thrashing assay on young (3 days from hatching) and old animals (10 days from hatching). Animals at young adult stage have been treated for 16 h in liquido with nanoalgosome at a final concentration of 10 μg/mL and PBS used as mock. After treatment animals were transferred to fresh NGM plates and young animals were immediately analyzed, while old animals were transferred every 2 days in fresh plates until day 10 from hatching. For video recording animals were transferred in 7 µL of M9 buffer, left 5 min to acclimate and then recorded for 30 s. The measurement of thrashing was done counting every change of direction respect to the longitudinal axis of the body70. Treatments were performed in triplicate (biologically independent experiments).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Statistics and reproducibility
Statistical analyses were performed with GraphPad Prism software. The experimental replicates, the sample size used to derive statistics (n= number of biologically independent samples, animals or independent experiments) and statistical analyses used for each experiment are described in the figure legends. Exact p-values were added when possible, the name of the statistical test included and differences were accepted as significant when p > 0.05. Statistics and error bars have been derived only with n ≥ 3 biological replicates, without conflating technical and biological variability. We considered as independent biological replicates: the different plates containing cells and treated in parallel in the in vitro experiments; the different plates/wells containing hundreds of animals and treated in parallel in the in vivo experiments with C. elegans; the different animals in the in vivo experiments with mice.
One-way ANOVA statistical test was used to: establish the effect of metabolic inhibitors of endocytosis on nanoalgosome uptake (Fig. 2a); quantify by Real-time PCR the DNA-damage sensors expression levels following exposure to nanoalgosomes (Fig. 4a); quantify SYTO12 positive apoptotic corpses in the C. elegans germline after treatment (Fig. 4b); verify nanoalgosome in vivo biocompatibility in C. elegans (Fig. 5a–b); compare the quantifications of the radiance efficiency (Fig. 6f); test the anti-inflammatory effect of nanoalgosomes in immune-responsive cells (Fig. 7a–c); compare the percentage of ROS production in 1-7 HB2 and MDA-MB 231 cells (Fig. 8a); analyse in vivo response to exogenous oxidative stress in C. elegans (Fig. 8e). Non-parametric compare two proportions test was used for the quantification of the percentage of C. elegans animals at each developmental stage after treatment (Fig. 5c). Mann Whitney t-test was used for comparing the brood size, embryonic lethality and the number of visible neurons in C. elegans (Fig. 5e–i), the fluorescence in gst-4p::GFP expressing animals (Fig. 8f) and the thrashing assays (Fig. 8g). Log-rank (Mantel-Cox) test was used for comparing the lifespan of C. elegans animals after treatment (Fig. 5d). Three-way ANOVA statistical test was used to compare the Real-time PCR quantification of antioxidant enzyme expression levels after exposure to nanoalgosomes (Fig. 8b–d). In case of experiments lacking statistical analysis, we repeated them >3 times and verified if we always got similar results.
Associated content
Supplementary information contains additional figures, tables and detailed supplementary methods, this file is available free of charge.
Data and resource availability
All data supporting the findings of this study are available within the paper and its Supplementary Information. All raw data evaluated and used in Figs. 1–8 and Supplementary Figs. 1–10 are available in files Supplementary Data 1_and Supplementary Data 2, respectively._RawData are also available in figshare repository (https://figshare.com/s/3359ba02cdeea1d9d881). The uncropped and unedited gel images supporting Fig. 1f and Supplementary Fig. 6 are also available in Supplementary Fig. 11. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Antonella Bongiovanni (antonella.bongiovanni@cnr.it).
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
The authors thank for C. elegans strains M.A. Hilliard (QBI, University of Queensland, Australia), M. Aschner (Albert Einstein College of Medicine, NY, USA), S. Martinelli (ISS, Rome, Italy) and CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440); for cryo-TEM images Ingo Lieberwirth (Max Planck Institute for Polymer Research, Mainz, Germany); for histological evaluation Yvan Campos (St. Jude CRH, Memphis, USA). This work was supported by the VES4US and the BOW projects funded by the European Union’s Horizon 2020 research and innovation program, under grant agreements nos. 801338 and 952183; MUR PNRR “National Center for Gene Therapy and Drugs based on RNA Technology” (Project no. CN00000041 CN3 RNA); AFM-Telethon project number #24401; MUR National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.3 - Call for proposals No. 341 of 15 March 2022 funded by the European Union – NextGenerationEU (Project code PE00000003, Concession Decree No. 1550 of 11 October 2022, CUP B83C22004790001, “ON Foods - Research and innovation network on food and nutrition Sustainability, Safety and Security – Working ON Foods); CNR project FOE-2021 DBA.AD005.225.
Author contributions
Study concepts and design: G.A., P.S., S.P., E.D., and A.B.; microalgae cultures and nanoalgosome isolation and characterization: G.A., S.P., P.G., D.P.R., A.P., E.R., S.R., V.L., P.C., N.T., R.N., K.L., S.M., P.B., and M.M.; nanoalgosome in vitro studies and data evaluation: G.A., S.P., P.G., C.D.S., M.S., A.B.; C. elegans experiments and data evaluation: P.S., G.Z., and E.D.; gene expression analyses: A.N., S.C., P.C., V.L., N.A.; mice experiments and data evaluation: M.W., G.A., I.D., A.B.; manuscript preparation: G.A., P.S., S.P., A.B., and E.D.; approval of the final version of the manuscript submitted: all authors.
Peer review
Peer review information
Communications Biology thanks Max Piffoux and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Toshiro Moroishi and David Favero. A peer review file is available.
Competing interests
The authors AB, MM, and NT declare the following financial competing interests: AB, MM, and NT have filed the patent (PCT/EP2020/086622) related to microalgal-derived extracellular vesicles described in the paper. AB, MM, and NT are co-founders and AB CEO of EVEBiofactory s.r.l. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Giorgia Adamo, Pamela Santonicola, Sabrina Picciotto.
These authors jointly supervised this work: Elia Di Schiavi, Antonella Bongiovanni.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06612-9.
References
- 1.Wiklander, O. P. B., Brennan, M. Á., Lötvall, J., Breakefield, X. O. & Andaloussi, S. E. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med.11, eaav8521 (2019). 10.1126/scitranslmed.aav8521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vizoso, F. J., Eiro, N., Cid, S., Schneider, J. & Perez-Fernandez, R. Mesenchymal stem cell secretome: toward cell-free therapeutic strategies in regenerative medicine. Int. J. Mol. Sci.18, 1852 (2017). 10.3390/ijms18091852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yáñez-Mó, M. et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles, 4, 10.3402/jev.v4.27066 (2015). [DOI] [PMC free article] [PubMed]
- 4.Li, M. et al. Extracellular vesicles as bioactive nanotherapeutics: an emerging paradigm for regenerative medicine. Theranostics12, 4879–4903 (2022). 10.7150/thno.72812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan, K. et al. Extracellular vesicles as a cell-free therapy for cardiac repair: a systematic review and meta-analysis of randomized controlled preclinical trials in animal myocardial infarction models. Stem Cell Rev. Rep.18, 1143–1167 (2022). 10.1007/s12015-021-10289-6 [DOI] [PubMed] [Google Scholar]
- 6.Berumen Sánchez, G., Bunn, K. E., Pua, H. H. & Rafat, M. Extracellular vesicles: mediators of intercellular communication in tissue injury and disease. Cell Commun. Signal.19, 104 (2021). 10.1186/s12964-021-00787-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phelps, J., Sanati-Nezhad, A., Ungrin, M., Duncan, N. A. & Sen, A. Bioprocessing of mesenchymal stem cells and their derivatives: toward cell-free therapeutics. Stem Cells Int.2018, e9415367 (2018). 10.1155/2018/9415367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saleh, A. F. et al. Extracellular vesicles induce minimal hepatotoxicity and immunogenicity. Nanoscale11, 6990–7001 (2019). 10.1039/C8NR08720B [DOI] [PubMed] [Google Scholar]
- 9.Sanwlani, R., Fonseka, P., Chitti, S. V. & Mathivanan, S. Milk-derived extracellular vesicles in inter-organism, cross-species communication and drug delivery. Proteomes8, 11 (2020). 10.3390/proteomes8020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Somiya, M., Yoshioka, Y. & Ochiya, T. Biocompatibility of highly purified bovine milk-derived extracellular vesicles. J. Extracell. Vesicles7, 1440132 (2018). 10.1080/20013078.2018.1440132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urzì, O., Raimondo, S. & Alessandro, R. Extracellular vesicles from plants: current knowledge and open questions. Int. J. Mol. Sci.22, 5366 (2021). 10.3390/ijms22105366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kameli, N., Dragojlovic-Kerkache, A., Savelkoul, P. & Stassen, F. R. Plant-derived extracellular vesicles: current findings, challenges, and future applications. Membranes11, 411 (2021). 10.3390/membranes11060411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, H. et al. Bacterial extracellular vesicles as bioactive nanocarriers for drug delivery: advances and perspectives. Bioact. Mater.14, 169–181 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann, I. K., Wood, M. J. A. & Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol.16, 748–759 (2021). 10.1038/s41565-021-00931-2 [DOI] [PubMed] [Google Scholar]
- 15.Elsharkasy, O. M. et al. Extracellular vesicles as drug delivery systems: why and how? Adv. Drug Deliv. Rev.159, 332–343 (2020). 10.1016/j.addr.2020.04.004 [DOI] [PubMed] [Google Scholar]
- 16.Witwer, K. W. & Wolfram, J. Extracellular vesicles versus synthetic nanoparticles for drug delivery. Nat. Rev. Mater.6, 103–106 (2021). 10.1038/s41578-020-00277-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsiapalis, D. & O’Driscoll, L. Mesenchymal stem cell derived extracellular vesicles for tissue engineering and regenerative medicine applications. Cells9, 991 (2020). 10.3390/cells9040991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun, Y. et al. Mesenchymal stem cells-derived exosomes for drug delivery. Stem Cell Res. Ther.12, 561 (2021). 10.1186/s13287-021-02629-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warnecke, A. et al. First-in-human intracochlear application of human stromal cell-derived extracellular vesicles. J. Extracell. Vesicles10, e12094 (2021). 10.1002/jev2.12094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paganini, C. et al. Scalable production and isolation of extracellular vesicles: available sources and lessons from current industrial bioprocesses. Biotechnol. J.14, e1800528 (2019). 10.1002/biot.201800528 [DOI] [PubMed] [Google Scholar]
- 21.Zarovni, N. et al. Standardization and commercialization of extracellular vesicles. 10.1039/9781839164552-00303 (2021).
- 22.Gerritzen, M. J. H., Salverda, M. L. M., Martens, D. E., Wijffels, R. H. & Stork, M. Spontaneously released neisseria meningitidis outer membrane vesicles as vaccine platform: production and purification. Vaccine37, 6978–6986 (2019). 10.1016/j.vaccine.2019.01.076 [DOI] [PubMed] [Google Scholar]
- 23.Adamo, G. et al. Nanoalgosomes: introducing extracellular vesicles produced by microalgae. J. Extracell. Vesicles10, e12081 (2021). 10.1002/jev2.12081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Picciotto, S. et al. Isolation of extracellular vesicles from microalgae: towards the production of sustainable and natural nanocarriers of bioactive compounds. Biomater. Sci.9, 2917–2930 (2021). 10.1039/D0BM01696A [DOI] [PubMed] [Google Scholar]
- 25.Picciotto, S. et al. Extracellular vesicles from microalgae: uptake studies in human cells and caenorhabditis elegans. Front. Bioeng. Biotechnol.10, 830189 (2022). 10.3389/fbioe.2022.830189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paterna, A. et al. Isolation of extracellular vesicles from microalgae: a renewable and scalable bioprocess. Front. Bioeng. Biotechnol.10, 836747 (2022). 10.3389/fbioe.2022.836747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Théry, C. et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): a position statement of the international society for extracellular vesicles and update of the MISEV2014 Guidelines. J. Extracell. Vesicles7, 1535750 (2018). 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welsh, J. A. et al. Minimal Information for Studies of Extracellular Vesicles (MISEV2023): from basic to advanced approaches. J. Extracell. Vesicles13, e12404 (2024). 10.1002/jev2.12404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sverdlov, E. D. Amedeo Avogadro’s Cry: What Is 1 Μg of Exosomes? BioEssays34, 873–875 (2012). 10.1002/bies.201200045 [DOI] [PubMed] [Google Scholar]
- 30.Mulcahy, L. A.; Pink, R. C.; Carter, D. R. F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell Vesicles, 3. 10.3402/jev.v3.24641 (2014). [DOI] [PMC free article] [PubMed]
- 31.Macia, E. et al. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell10, 839–850 (2006). 10.1016/j.devcel.2006.04.002 [DOI] [PubMed] [Google Scholar]
- 32.Banizs, A. B., Huang, T., Nakamoto, R. K., Shi, W. & He, J. Endocytosis pathways of endothelial cell derived exosomes. Mol. Pharm.15, 5585–5590 (2018). 10.1021/acs.molpharmaceut.8b00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian, T. et al. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem.289, 22258–22267 (2014). 10.1074/jbc.M114.588046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delenclos, M. et al. Investigation of endocytic pathways for the internalization of exosome-associated oligomeric alpha-synuclein. Front. Neurosci.11, 172 (2017). 10.3389/fnins.2017.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eom, H.-J. & Choi, J. Clathrin-mediated endocytosis is involved in uptake and toxicity of silica nanoparticles in caenohabditis elegans. Chem. Biol. Interact.311, 108774 (2019). 10.1016/j.cbi.2019.108774 [DOI] [PubMed] [Google Scholar]
- 36.Behzadi, S. et al. Cellular uptake of nanoparticles: journey inside the cell. Chem. Soc. Rev.46, 4218–4244 (2017). 10.1039/C6CS00636A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao, Z. et al. Exosomes exploit the virus entry machinery and pathway to transmit alpha interferon-induced antiviral activity. J. Virol.92, e01578–18 (2018). 10.1128/JVI.01578-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maréchal, A. & Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol.5, a012716 (2013). 10.1101/cshperspect.a012716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rx, H.; Pk, Z. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal transduction and targeted therapy, 5. 10.1038/s41392-020-0150-x (2020). [DOI] [PMC free article] [PubMed]
- 40.Zhou, B.-B. S. & Elledge, S. J. The DNA damage response: putting checkpoints in perspective. Nature408, 433–439 (2000). 10.1038/35044005 [DOI] [PubMed] [Google Scholar]
- 41.Gumienny, T. L., Lambie, E., Hartwieg, E., Horvitz, H. R. & Hengartner, M. O. Genetic control of programmed cell death in the caenorhabditis elegans hermaphrodite germline. Development126, 1011–1022 (1999). 10.1242/dev.126.5.1011 [DOI] [PubMed] [Google Scholar]
- 42.Silva, N., Adamo, A., Santonicola, P., Martinez-Perez, E. & La Volpe, A. Pro-crossover factors regulate damage-dependent apoptosis in the caenorhabditis elegans germ line. Cell Death Differ.20, 1209–1218 (2013). 10.1038/cdd.2013.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santonicola, P., Germoglio, M., d’Abbusco, D. S. & Adamo, A. Functional characterization of caenorhabditis elegans Cbs-2 gene during Meiosis. Sci. Rep.10, 20913 (2020). 10.1038/s41598-020-78006-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papaluca, A. & Ramotar, D. A novel approach using C. Elegans DNA damage-induced apoptosis to characterize the dynamics of uptake transporters for therapeutic drug discoveries. Sci. Rep.6, 36026 (2016). 10.1038/srep36026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yao, Y., Zhang, T. & Tang, M. A critical review of advances in reproductive toxicity of common nanomaterials to caenorhabditis elegans and influencing factors. Environ. Pollut.306, 119270 (2022). 10.1016/j.envpol.2022.119270 [DOI] [PubMed] [Google Scholar]
- 46.Hunt, P. R. The C. Elegans model in toxicity testing. J. Appl. Toxicol.37, 50–59 (2017). 10.1002/jat.3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi, H. et al. Biodistribution of exosomes and engineering strategies for targeted delivery of therapeutic exosomes. Tissue Eng. Regen. Med.18, 499–511 (2021). 10.1007/s13770-021-00361-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Driedonks, T. et al. Pharmacokinetics and biodistribution of extracellular vesicles administered intravenously and intranasally to macaca nemestrina. J. Extracell. Bio.1. 10.1002/jex2.59 (2022). [DOI] [PMC free article] [PubMed]
- 49.Gupta, D., Zickler, A. M. & El Andaloussi, S. Dosing extracellular vesicles. Adv. Drug Deliv. Rev.178, 113961 (2021). 10.1016/j.addr.2021.113961 [DOI] [PubMed] [Google Scholar]
- 50.Tieu, A. et al. An analysis of mesenchymal stem cell-derived extracellular vesicles for preclinical use. ACS Nano14, 9728–9743 (2020). 10.1021/acsnano.0c01363 [DOI] [PubMed] [Google Scholar]
- 51.Santos, A. F., Alpan, O. & Hoffmann, H.-J. Basophil activation test: mechanisms and considerations for use in clinical trials and clinical practice. Allergy76, 2420–2432 (2021). 10.1111/all.14747 [DOI] [PubMed] [Google Scholar]
- 52.Lai, C. P. et al. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano8, 483–494 (2014). 10.1021/nn404945r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lázaro-Ibáñez, E. et al. Selection of fluorescent, bioluminescent, and radioactive tracers to accurately reflect extracellular vesicle biodistribution in vivo. ACS Nano15, 3212–3227 (2021). 10.1021/acsnano.0c09873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiklander, O. P. B. et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles, 4, 10.3402/jev.v4.26316 (2015). [DOI] [PMC free article] [PubMed]
- 55.Gupta, D. et al. Quantification of extracellular vesicles in vitro and in vivo using sensitive bioluminescence imaging. J. Extracell. Vesicles9, 1800222 (2020). 10.1080/20013078.2020.1800222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borowitzka, M. A. High-value products from microalgae—their development and commercialisation. J. Appl. Phycol.25, 743–756 (2013). 10.1007/s10811-013-9983-9 [DOI] [Google Scholar]
- 57.Plaza, M., Herrero, M., Cifuentes, A. & Ibáñez, E. Innovative natural functional ingredients from microalgae. J. Agric Food Chem.57, 7159–7170 (2009). 10.1021/jf901070g [DOI] [PubMed] [Google Scholar]
- 58.Das, K., Roychoudhury, A. Reactive Oxygen Species (ROS) and response of antioxidants as ROS-Scavengers during environmental stress in plants. Front. Environ. Sci.2, 53 (2014).
- 59.Ezeriņa, D., Takano, Y., Hanaoka, K., Urano, Y. & Dick, T. P. N-Acetyl cysteine functions as a fast-acting antioxidant by triggering intracellular H2S and sulfane sulfur production. Cell Chem. Biol.25, 447–459.e4 (2018). 10.1016/j.chembiol.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu, Q., D’Amora, D. R., MacNeil, L. T., Walhout, A. J. M. & Kubiseski, T. J. The caenorhabditis elegans oxidative stress response requires the NHR-49 transcription factor. G3 Genes|Genomes|Genet.8, 3857–3863 (2018). 10.1534/g3.118.200727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cd, L., Cj, J. Reporter transgenes for study of oxidant stress in caenorhabditis elegans. Methods Enzymol. 353. 10.1016/s0076-6879(02)53072-x (2002). [DOI] [PubMed]
- 62.Sugawara, T. & Sakamoto, K. Quercetin enhances motility in aged and heat-stressed caenorhabditis elegans nematodes by modulating both HSF-1 Activity, and insulin-like and P38-MAPK signalling. PLoS ONE15, e0238528 (2020). 10.1371/journal.pone.0238528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herndon, L. A., Wolkow, C. A., Driscoll, M., Hall, D. H. Effects of ageing on the basic biology and anatomy of C. Elegans. In Ageing: Lessons from C. elegans; Olsen, A., Gill, M. S., (Eds) (Healthy Ageing and Longevity; Springer International Publishing: Cham, 2017).
- 64.Brenner, S. The genetics of caenorhabditis elegans. Genetics77, 71–94 (1974). 10.1093/genetics/77.1.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirszenblat, L., Neumann, B., Coakley, S. & Hilliard, M. A. A dominant mutation in Mec-7/β-Tubulin Affects axon development and regeneration in Caenorhabditis Elegans Neurons. MBoC24, 285–296 (2013). 10.1091/mbc.e12-06-0441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.González-Hunt, C. P. et al. Exposure to mitochondrial genotoxins and dopaminergic neurodegeneration in caenorhabditis elegans. PLoS ONE9, e114459 (2014). 10.1371/journal.pone.0114459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Romano, M. et al. Synthesis and characterization of a biocompatible nanoplatform based on silica-embedded SPIONs functionalized with polydopamine. ACS Biomater. Sci. Eng. 9. 10.1021/acsbiomaterials.2c00946 (2022). [DOI] [PubMed]
- 68.Hensel, N. et al. Impairment of the neurotrophic signaling hub B-Raf contributes to motoneuron degeneration in spinal muscular atrophy. Proc. Natl Acad. Sci. USA.118, e2007785118 (2021). 10.1073/pnas.2007785118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Illiano, P. et al. A caenorhabditis elegans model to study dopamine transporter deficiency syndrome. Eur. J. Neurosci.45, 207–214 (2017). 10.1111/ejn.13366 [DOI] [PubMed] [Google Scholar]
- 70.Di Giorgio, M. L. et al. WDR79/TCAB1 plays a conserved role in the control of locomotion and ameliorates phenotypic defects in SMA Models. Neurobiol. Dis.105, 42–50 (2017). 10.1016/j.nbd.2017.05.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files