Abstract
Herpes simplex virus type 1 (HSV-1) immediate-early protein Vmw110 stimulates the onset of virus infection in a multiplicity-dependent manner and is required for efficient reactivation from latency. Recent work has shown that Vmw110 is able to interact with or modify the stability of several cellular proteins. In this report we analyze the ability of Vmw110 to inhibit the progression of cells through the cell cycle. We show by fluorescence-activated cell sorter and/or confocal microscopy analysis that an enhanced green fluorescent protein-tagged Vmw110 possesses the abilities both to prevent transfected cells moving from G1 into S phase and to block infected cells at an unusual stage of mitosis defined as pseudo-prometaphase. The latter property correlates with the Vmw110-induced proteasome-dependent degradation of CENP-C, a centromeric protein component of the inner plate of human kinetochores. We also show that whereas Vmw110 is not the only viral product implicated in the block of infected cells at the G1/S border, the mitotic block is a specific property of Vmw110 and more particularly of its RING finger domain. These data explain the toxicity of Vmw110 when expressed alone in transfected cells and provide an explanation for the remaining toxicity of replication-defective mutants of HSV-1 expressing Vmw110. In addition to contributing to our understanding of the effects of Vmw110 on the cell, our results demonstrate that Vmw110 expression is incompatible with the proliferation of a dividing cell population. This factor is of obvious importance to the design of gene therapy vectors based on HSV-1.
Among the human pathogens, herpes simplex virus type 1 (HSV-1) is one of the most extensively studied viruses, yet biologically it remains incompletely understood. One of its most interesting features is the dual life cycle that this virus has adopted to maintain its survival. After the initial lytic infection at the periphery, the virus will evade the host immune system by infecting sensory neurons, where it can stay in a latent state lifelong (for a review, see reference 18). The lytic and latent states differ by the number of transcriptionally active genes that can be detected. All viral genes, numbering about 80, are expressed from the 152-kb double-stranded genomic DNA during lytic infection, but only one set of viral transcripts can be readily detected during latency (19). The expression of the lytic genes is temporarily regulated, with the genes classified as immediate-early (IE), early, and late, depending on the time course of their synthesis and requirement for prior viral gene expression and DNA replication (40).
Five IE proteins are encoded by HSV-1, of which four regulate gene expression during lytic infection. Vmw175 (ICP4) and Vmw63 (ICP27) have been shown to be essential for virus replication (8, 9, 32, 36, 41, 53), whereas Vmw68 (ICP22) is dispensable for virus viability in most cell types (35, 48). Vmw110 (ICP0) is a RING finger protein which activates gene expression in a strong and promiscuous manner in transfection assays and which can act synergistically with Vmw175 (12). Mutant viruses either deficient for the expression of Vmw110 or expressing an inactive form of the protein are able to grow in cell culture. However, these viruses exhibit a cell type- and multiplicity-dependent growth phenotype which affects the onset of lytic infection and strongly decreases their probability of initiating a productive infection (42, 51). A more definite role of Vmw110 in influencing the latent-lytic switch has been demonstrated in cultured cells (16, 20, 55, 57) as well as in mouse latency models (5, 6, 30). Indeed, the absence of Vmw110 causes a mutant virus to reactivate inefficiently from latency, a defect overcome in vitro by providing exogenous Vmw110 (20, 57).
The study of the multiple effects of the IE proteins on the biology of the virus as well as on the metabolism of host cells has constituted a major challenge, which became more prominent with the development of vector therapy aiming to use HSV-1 as a delivery system. The safety of such vectors is of obvious concern, and among the several criteria that have to be satisfied are lack of toxicity, genome persistence, and gene expression. The first replication-defective mutants of HSV-1 with a markedly reduced cytopathic effect independent of the multiplicity of infection (MOI) were deficient for the expression of either Vmw175 or Vmw63 (23). Infection of cells by HSV-1 mutants unable to express both Vmw175 and Vmw63 in addition to either Vmw68 (56) or Vmw110 (44) led to a prolonged cell survival and gene expression. The toxicity of other mutants unable to express the virion structural transactivator protein Vmw65 (VP16 or αTIF) (1), Vmw65 and Vmw175 (24), or Vmw65 in combination with mutations affecting both Vmw175 and Vmw110 (37, 38) were also investigated. A significant amount of cytotoxicity was still retained by all these mutant viruses, suggesting that mutation or reduction of the expression of all HSV-1 IE genes was necessary to significantly reduce adverse effects on the cell. The work of Samaniego et al. (45) showed that an HSV-1 mutant lacking all five IE proteins was nontoxic to Vero and human embryonic lung cells, but paradoxically the level of transgene expression in infected cells was dramatically decreased. One of the major pieces of information highlighted by these different studies was that although Vmw110 is dispensable for virus replication in cell culture, infection with a replication-defective mutant of HSV-1 expressing Vmw110 decreases cell survival. In addition, overproduction of Vmw110 in the absence of Vmw175, Vmw63, and Vmw68 inhibited further growth of cell cultures, suggesting that Vmw110 might inhibit cellular DNA synthesis or cell cycle progression (56).
Several aspects of cell metabolism are affected by Vmw110. Proteins as different as elongation factor 1δ (25), the cell cycle regulator cyclin D3 (26), and a ubiquitin-specific protease named HAUSP (14, 33) have been reported to interact with Vmw110. Furthermore, Vmw110 is specifically implicated in the proteasome-dependent degradation of several cellular proteins or protein isoforms. This is the case for the catalytic subunit of the DNA-dependent protein kinase, although the consequences of this activity for both virus and cell biology have not been well defined (29, 34). Specific isoforms of PML, a permanent component of nuclear domains called ND10, PML nuclear bodies, or promyelocytic oncogenic domains (PODs), are also targeted for degradation by Vmw110. This process has been shown to correlate with the disappearance of the ND10 domains in cells infected with wild-type HSV-1 or transfected with a plasmid expressing Vmw110 (2, 14, 15). Finally, Vmw110 is also directly implicated in the proteasome-dependent degradation of CENP-C (17), a 140-kDa centromeric protein component of the inner kinetochore plate which plays a critical role in cell division by establishing and/or maintaining proper kinetochore size and stabilizing microtubule attachments (10, 43, 52).
From these data, it is very likely that various aspects of the metabolism of cells infected by wild-type HSV-1 or by any replication-defective mutant expressing Vmw110 will be affected in ways that can be directly attributed to the effect of Vmw110. In this report, we investigate the effect of expression of Vmw110 on the cell cycle. We show by fluorescence-activated cell sorter (FACS) analysis that the expression of an enhanced green fluorescent protein (EGFP)-tagged Vmw110 in transfected cells blocks the G1-to-S phase progression. We also analyze the progression through mitosis of synchronized cells infected in G2 and show that Vmw110 and more precisely the RING finger domain of Vmw110 is directly implicated in blocking infected cells at a stage of mitosis defined as pseudo-prometaphase.
MATERIALS AND METHODS
Plasmids and bacteria.
Plasmids expressing wild-type Vmw110 (pEG110) or the RING finger mutant Vmw110 protein FXE (pEGFXE) linked at their N-terminal ends to EGFP were based on the pEGFP-C1 vector (Clontech). Fusion proteins synthesized from plasmids pEG110 and pEGFXE were called EG110 and EGFXE, respectively. Wild-type or RING finger mutant Vmw110 genes were cloned in frame with the EGFP open reading frame, and cloning regions were sequenced to verify the correct orientation and in-frame position of Vmw110 genes (31). Plasmids were grown in Escherichia coli DH5, and large-scale preparations were made by the boiling method and CsCl purification.
Viruses and cells.
HSV-1 strain 17 syn+ was the parental strain used in this study. Virus vEG110 expresses at both IE-1 loci the full-length Vmw110 linked at its N-terminal end to EGFP; it was constructed by cotransfection of infectious dl1403 DNA (as described previously [11]) with plasmid p111E110 containing the Vmw110 gene fused with the EGFP gene and flanked at both ends by the DNA sequences localized at each side of the Vmw110 gene in the HSV-1 genome (31). Virus vEGdl110 is a deletion mutant from which both Vmw110 gene copies have been removed and replaced at both loci by the EGFP coding sequence, and virus vEGFXE is a mutant virus expressing the EGFP version of the RING finger FXE Vmw110 mutant protein; both were constructed in a manner similar that used for vEG110 (31). Vmw110 mutant viruses dl1403 and FXE (11, 51) were also used. vEG110, vEGFXE, and vEGdl110 were genetically and biologically tested to ensure that their properties were identical to those of the non-EGFP versions. Table 1 summarizes several known biological characteristics of the wild-type, FXE, and dl1403 viruses that were tested in parallel with vEG110, vEGFXE, and vEGdl110 by immunofluorescence and/or Western blotting to ensure that the latter viruses retain the same biological properties than their non-EGFP counterparts. Furthermore, like the FXE and dl1403 viruses, vEGFXE and vEGdl110 showed a 2-log decrease in their titers in human fetal lung cells compared to baby hamster kidney cells, whereas vEG110 showed titers similar to those of wild-type HSV-1 in both cell lines (results not shown). These results indicate that EG110 was able to restore a growth phenotype similar to that of the wild-type virus and taken together indicate that EG110 retains the functions of the normal protein. All viruses were grown and titrated in baby hamster kidney cells propagated in Glasgow modified Eagle’s medium containing penicillin (10 U/ml) and streptomycin (100 μg/ml) and supplemented with 10% newborn calf serum and 10% tryptose phosphate broth. HEp-2 cells were grown at 37°C in 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and antibiotics as specified above.
TABLE 1.
Biological properties of viruses expressing EGFPa
| Virus | Localization of Vmw110 tob:
|
Degradation ofc:
|
||
|---|---|---|---|---|
| ND10 | Centromere | PML | CENP-C | |
| 17 syn+ | + | + | + | + |
| vEG110 | + | + | + | + |
| FXE | + | − | − | − |
| vEGFXE | + | − | − | − |
| dl1403 | − | − | − | − |
| vEGdl110 | − | − | − | − |
Except for the Western blot experiments, these properties have also been tested for EG110 and EGFXE proteins expressed from plasmids pEG110 and pEGFXE. Western blotting also showed that the EGFP-tagged proteins were expressed in similar amount to the corresponding non-EGFP-tagged versions in both virus infection and transfection experiments.
Tested by immunofluorescence according to the protocol described in reference 17.
Electroporation.
HEp-2 cells were trypsinized, resuspended in complete medium, pelleted, and washed once with serum-free medium before being resuspended in serum-free medium at a concentration of 7.5 × 106 cells per ml. Plasmid DNA (20 μg) and 0.8 ml of cells were added to a 4-mm electroporation cuvette, incubated on ice for 10 min, mixed again, and pulsed in a Hybaid electroporator at a setting of 400 V. Cells were incubated on ice for a further 10 min before being diluted into fresh complete medium, seeded into two 35-mm-diameter dishes, and incubated at 37°C in 5% CO2. Six to eight hours postelectroporation, the medium was taken out of the dishes to remove dead cells and was replaced with fresh medium. Cells were then left in the incubator until use.
FACS analysis.
HEp-2 cells were trypsinized at the appropriate time postinfection or postelectroporation and resuspended in complete medium. After centrifugation at 800 × g for 5 min, cells were washed with 5 ml of cold phosphate-buffered saline (PBS), centrifuged again, and resuspended in formaldehyde (1% [vol/vol] in PBS containing 2% sucrose). After fixation on ice for 10 min, cells were centrifuged, washed once with PBS, and centrifuged again. Pelleted cells were resuspended in 500 μl of a solution of 0.1% saponin (ICN), 0.5% bovine serum albumin (Sigma), propidium iodide (PI; 100 μg/ml), and RNase A (100 μg/ml) in PBS. After 30 min of incubation on ice, the total DNA content was analyzed by a FACScan analyzer using LYSYS II software (Becton Dickinson, San Jose, Calif.).
Synchronization of cells.
Sequential thymidine and aphidicolin blocking steps produced monolayers of synchronized HEp-2 cells. Cells were seeded at a density of 1.25 × 105 per 35-mm-diameter dish, usually containing four coverslips, depending on the experiment to be performed. The next day, medium containing 2 mM thymidine was substituted, and 12 h later cells were washed twice and medium containing 0.025 mM thymidine and 0.025 mM deoxycytidine was added. A further 12 h later cells were washed twice again and refed with medium containing 2.5 μM aphidicolin. After another 14 h, cells were washed three times and refed with normal medium. Infections were carried out at suitable time after release.
Immunofluorescence and confocal microscopy.
HEp-2 cell coverslips prepared during synchronization experiments were put in Linbro wells at the appropriate time after viral infection. Cells were fixed with formaldehyde (5% [vol/vol] in PBS containing 2% sucrose). If infections were carried out with viruses expressing an EGFP, cells were permeabilized with 0.5% NP-40 in PBS with 10% sucrose and 0.5 μg of PI per ml for 30 s. Cells were then washed three times with PBS, and a final wash was done with distilled water before mounting of the coverslips by using Citifluor. In the case of infections with non-EGFP-expressing viruses, cells were permeabilized for 5 min in a PBS solution containing 0.5% NP-40 and 10% sucrose. Primary antibodies were diluted in PBS containing 1% newborn calf serum. Monoclonal antibodies (MAbs) were used at dilutions of 1/1,000 (anti-Vmw110 MAb 11060 [13]) and 1/100 (anti-Vmw175 MAb 58S) [50]). After incubation at room temperature for 1 h, coverslips were washed at least three times with PBS and then treated with the fluorescein isothiocyanate-conjugated sheep anti-mouse immunoglobulin G (Sigma) secondary antibody diluted at 1/100. After a further 30-min incubation, coverslips were washed three times with PBS, incubated for 30 s in a solution of PBS containing PI (0.5 μg/ml), washed again three times with PBS, and then washed once with water before being mounted by using Citifluor. Cell samples were examined in a Zeiss LSM 510 confocal microscope with two lasers giving excitation lines at 543 and 488 nm. The data from the channels were collected simultaneously with eightfold averaging at a resolution of 1,024 by 1,024 pixels, using optical slices of 1 μm. The microscope was a Zeiss Axioplan with either a 40× or a 63× oil immersion objective lens. Data sets were processed with the LSM 510 software and then exported for preparation for printing by using Adobe Photoshop.
Western blotting.
Sodium dodecyl sulfate-polyacrylamide gels (10%) were prepared and run in the Bio-Rad MiniProtean II apparatus and then proteins were electrophoretically transferred to nitrocellulose membranes according to the manufacturer’s recommendations. After blocking in PBS containing 0.1% Tween 20 (PBST) and 5% dried milk overnight at 4°C, the membranes were incubated with primary antibody in PBST–5% dried milk at room temperature for 1 h and then washed in PBST at least three times before incubation with horseradish peroxidase-conjugated secondary antibody in PBST–2% dried milk at room temperature for 1 h. After extensive washing, filters were soaked in Amersham ECL reagent and exposed to film.
RESULTS
Vmw110 blocks transfected cells at the G1/S border of the cell cycle.
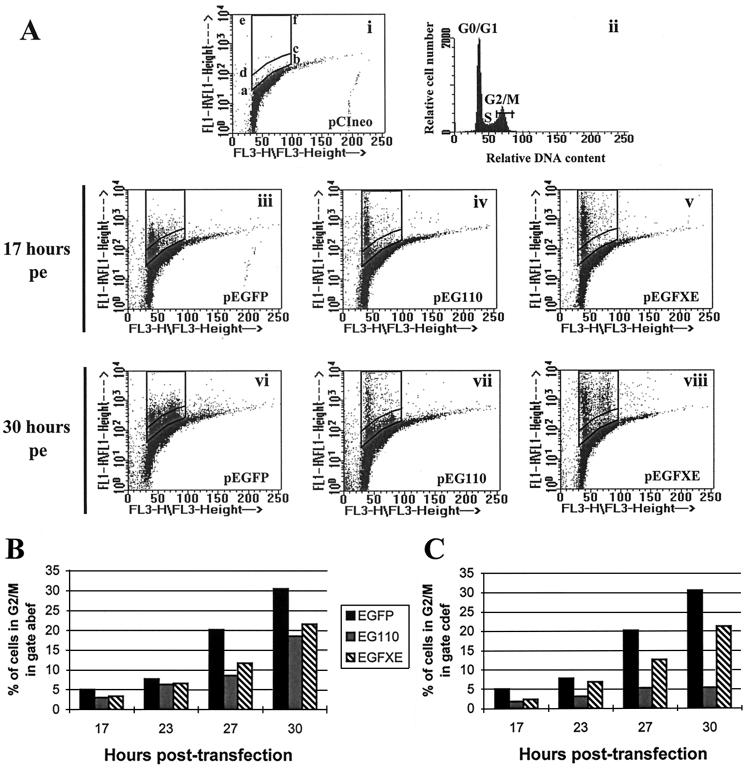
HSV-1 infection has already been reported to block G1-to-S phase progression of synchronized CV-1 cells infected in the G1 phase of the cell cycle (7). More recently, Wu et al. (56) suggested that Vmw110 might be implicated in the inhibition of cell DNA synthesis in cells infected with the replication-defective mutant of HSV-1, d95 (ICP4− ICP27− ICP22−). Based on these data, we investigated the effect of the expression of Vmw110 on the progression of cells from G1 into S phase of the cell cycle. To enable a simple method of studying the effect of Vmw110 on the cell cycle in the absence of all other viral components, we constructed plasmids expressing wild-type and mutant forms of Vmw110 linked to EGFP (see Materials and Methods). HEp-2 cells were electroporated with plasmid pEGFP, pEG110, or pEGFXE. Both EG110 and EGFXE had been previously tested to ensure that their biological properties were identical to those of the non-EGFP versions of the proteins (reference 31 and Table 1). Cells were harvested 17, 23, 27, and 30 h postelectroporation, and their DNA content was quantified by FACS analysis to determine their distribution in the cell cycle (Fig. 1, FL3-H). Cells positive for the expression of an EGFP were easily detectable because their position on the y axis of the dot plot diagram (Fig. 1, FL1-H) was above the bulk of negative cells. To restrict the analysis to positive cells, two selection gates were drawn above negative cells transfected with a pCIneo vector (Promega) (Fig. 1A, i). The first gate (abef) included all positive green cells, and the second gate (cdef) included only those cells with fluorescence intensities exceeding about 1/10 of the highest values above background. Cell brightness was different depending on the protein expressed, as the maximum fluorescence due to native EGFP (Fig. 1A, iii and vi) was about 10-fold less than that of EG110 (Fig. 1A, iv to vii) or EGFXE (Fig. 1A, v to viii). This correlated with immunofluorescence observations of electroporated cells expressing these proteins (results not shown). The differences in fluorescence intensity between EGFP and EG110 or EGFXE are probably due to the diffuse nuclear and cytoplasmic distribution of EGFP, compared to the concentrated localization of EG110 and EGFXE in bright nuclear dots which initially correspond to the ND10 domains. The heterogeneous distribution of the intensity of green fluorescence of positive cells expressing a particular protein correlated with the variability in the amount of expression observed from cell to cell by immunofluorescence (data not shown).
FIG. 1.
FACS analysis of cells expressing EGFP, EG110, or EGFXE. HEp-2 cells were electroporated with plasmid pEGFP, pEG110, or pEGFXE or with the control vector pCIneo. Cells were harvested (50,000 for pEGFXE and pEGdl110-transfected cells; 100,000 for pEG110-transfected cells) 17, 23, 27, and 30 h postelectroporation (pe) and checked for their distribution in the cell cycle, using the LYSYS II software (Becton Dickinson). (A) Dot plot diagrams of cells expressing EGFP (iii and vi), EG110 (iv and vii), or EGFXE (v and viii) 17 (iii to v) and 30 (vi to viii) h postelectroporation. A dot plot diagram (i) and the corresponding histogram (ii) obtained by the analysis of control cells are also shown representing the distribution of control cells in the cell cycle. The horizontal bar in histogram ii indicates the region selected to determine the percentage of positive cells in G2/M during the course of the experiment. Gate abef was chosen for the analysis of the total number of cells positive for the expression of an EGFP. Gate cdef restricted the analysis to cells which were highly positive. (B and C) Analysis of the percentage of G2/M cells expressing EGFP, EG110, or EGFXE in both gate abef and gate cdef at 17, 23, 27, and 30 h posttransfection.
Figure 1A illustrates the distribution of positive EGFP-expressing cells in the cell cycle at two time points corresponding to 17 h (T17) (iii to v) and 30 h (T30) (vi to viii) postelectroporation. Seventeen hours postelectroporation positive cells were almost all in G0/G1 whatever the protein expressed. This phenomenon is probably due to the transfection itself, which might result in a block of transfected cells in G0/G1 until complete recovery from the modifications undergone during the transfection process. Thirty hours postelectroporation, about 30% of cells expressing EGFP were in G2/M (Fig. 1A, vi; Fig. 1B) whereas two populations of EG110-expressing cells could be distinguished. On one hand, cells expressing low amounts of EG110 (region abcd) were not affected by the protein expression and seemed to progress partially to G2/M (Fig. 1A, vii). On the other hand, cells positive for higher expression of EG110 (region cdef) stayed almost entirely in G1/S (Fig. 1A, vii; compare Fig. 1B and C). Such a disparity was not observed in EGFXE-expressing cells, as they were able to progress to G2/M independently of the level of expression of EGFXE (Fig. 1A, viii; compare Fig. 1B and C). A putative lack of expression or a rapid degradation of EG110 in cells in S-G2/M, which would result in the absence of detection of cells expressing high amounts of EG110, was excluded by control experiments. For example, FACS analysis showed a high level of expression of EG110 in cells in G2/M when synchronized cells were transfected in S-G2/M by using a liposomal reagent (results not shown).
Since the selection gates were drawn on the basis of the results in each individual experiment, it was not possible to present averaged results from different experiments. However, data presented in Fig. 1B and C are representative of multiple independent repeat experiments. They thus give a reliable summary of the actual effect of each protein on the progression of cells to the S phase of the cell cycle. Figures 1B and C represent the percentage of cells in G2/M (Fig. 1A, ii, gate G2/M) during the course of an experiment either among the entire population of positive cells (Fig. 1A, gate abef) or among cells positive for higher expression of EG110 or EGFXE (Fig. 1A, gate cdef). The total number of EGFP-expressing cells (gate abef) was used as a control in both graphs, as the distribution of positive cells was similar in both gates. Figure 1B shows that progression of positive cells from G1/S to G2/M in gate abcd during the course of the experiment is slightly affected by the expression of EG110 or EGFXE in comparison with EGFP alone. However, whereas the percentage of G2/M cells positive for the expression of EGFXE is exactly the same in both gates (about 22% at T30), high EG110-expressing cells are dramatically affected in their progression from G1/S to G2/M (only about 5% of positive cells in G2/M at T30) (Fig. 1C). These results show that Vmw110 is able to block cell cycle progression from G1 to S-G2/M; however, its activity is clearly dependent on the amount of protein expressed in the cell, and the RING finger domain makes an important contribution to this effect.
Vmw110 is not the only viral factor implicated in the block of infected cells at the G1/S border of the cell cycle.
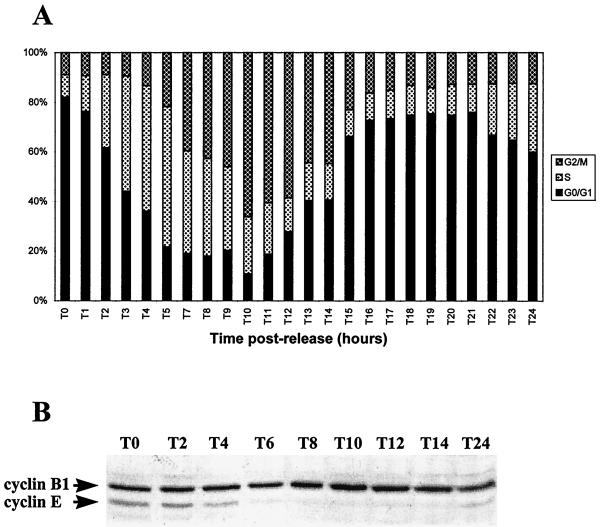
To analyze whether Vmw110 might by itself cause the previously observed block of infected cells at the G1/S border and to specifically analyze cells which had been successfully infected, FACS analyses were performed on synchronized HEp-2 cells infected with virus vEG110, vEGFXE, or vEGdl110, expressing EGFP fusion protein linked to wild-type or FXE Vmw110 or to EGFP in place of Vmw110, respectively. The construction and properties of these viruses are summarized in Materials and Methods and Table 1. Synchronized cells were infected at different stages during the G1 phase prior to entry into S phase, and their DNA content was measured by FACS at different times after infection. Before embarking on this experiment, we analyzed the progression of uninfected cells through the cell cycle after release from G0/G1-S block by aphidicolin over a period of 24 h (Fig. 2A). The distribution of the cell population in the cycle was determined by analyzing their DNA content by FACS and by measuring the percentage of cells in G0/G1, S, and G2/M. To confirm that cells were cycling correctly, the amounts of both cyclin B1 and cyclin E were monitored by Western blotting during the whole period (Fig. 2B). From the time of release onward, the level of cyclin E decreases in accordance with the cells moving from S into G2/M. After a 10-h period (T10) cells consistently enter mitosis, which correlates with the peak in the amount of cyclin B1, and the whole population completes mitosis by 14 h postrelease. Then, from T16 to T21 the majority of cells (about 75%) are in G1 and reenter S phase about 22 to 24 h postrelease (as confirmed by the increase of the level of cyclin E at T24). Cells were thus infected at T18, T20, T21, T22, and T24, and infections were stopped at T27, when a high proportion of cells would normally have progressed to S-G2.
FIG. 2.
Progression of aphidicolin-synchronized HEp-2 cells through the cell cycle. HEp-2 cells were blocked in G0/G1-S by aphidicolin and monitored for 24 h after release to follow their progression through the cell cycle. Each hour cells were harvested, and their DNA content was analyzed by FACS to determine their position in the cell cycle (A). (B) Western blotting following the variation in the amount of both cyclin B1 and cyclin E during the course of the experiment.
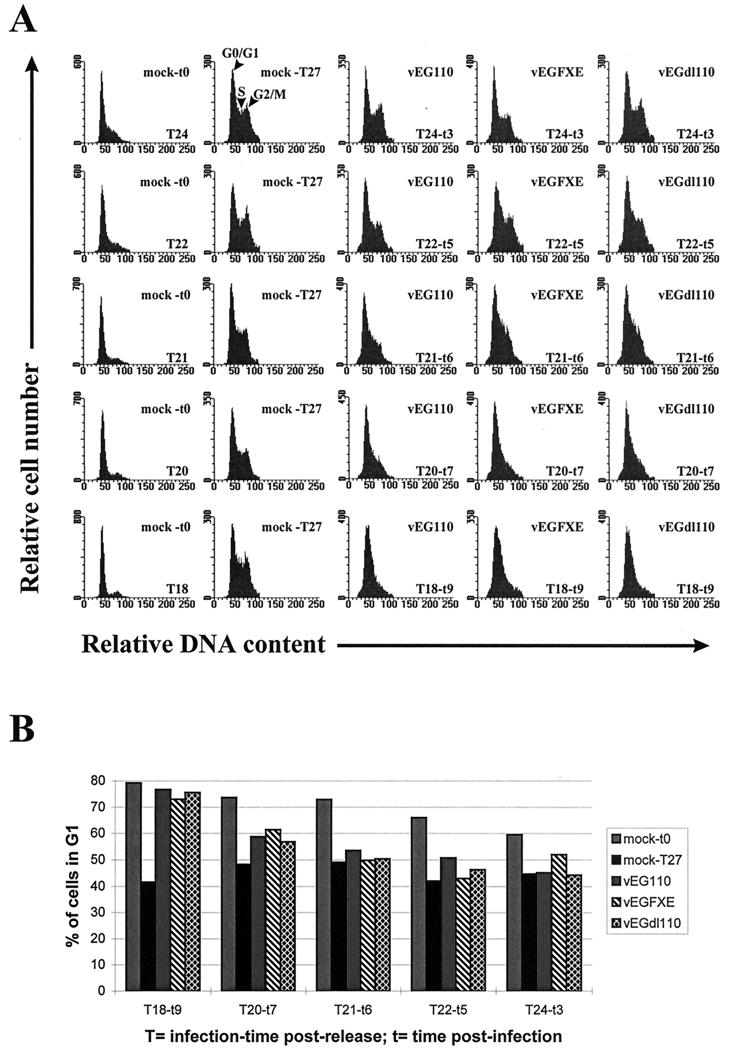
As the EFGPs are easily detectable by fluorescence 3 h postinfection and EG110 and EGFXE are already present at the ND10 domains at this time (results not shown), T27 was considered a suitable time to analyze by FACS the DNA content of infected cells in comparison with mock-infected cells (Fig. 3A). The G2/M peak of cells infected by any of the three viruses disappeared in cells infected early in G1 (T18 to T20) which suggests that the earlier cells are infected before the beginning of S phase, the less chance they have to progress from G1 into S. Figure 3B shows the percentage of infected green cells still in G1 at T27, depending on their infection time postrelease. The results show that (i) the ability of cells infected just before (T21) or at the start of (T22 and T24) S phase to move from G1 into S is not reduced by the infection, (ii) compared to mock-infected cells, a slight increase of infected cells still in G1 is noticeable when cells were infected at T20, but there were no differences between the three viruses, and (iii) the percentage of T18-infected cells still in G1 is dramatically increased compared to the mock-infected cells, but once again without any differences between the viruses. By infecting cells that early before the start of S phase, we obviously cannot rule out the possibility that viral DNA replication had already started by the time cells reached the G1/S border, which would undoubtedly affect cell cycle progression. Taken with the results of Fig. 1, these experiments suggest that although Vmw110 is able to block G1-to-S phase progression by itself, other viral proteins expressed during infection are able to do so, as shown by the results obtained with the Vmw110-deficient mutants. Furthermore, this experiment shows that the time of infection before the start of S phase is important for the block to be efficient. These data are in complete agreement with previous observations (7) and suggest that there is a critical point during G1 beyond which infection by HSV-1 will not lead to the block of infected cells at the G1/S boundary, as shown by the absence of any block of cells infected at T21 to T24. It follows that the G1/S block caused by Vmw110 in electroporated cells requires that Vmw110 be expressed in greater amounts or for longer periods than occurred in the infections initiated at T21 to T24.
FIG. 3.
FACS analysis of the progression of HSV-1-infected HEp-2 cells from G1 into S phase of the cell cycle. Aphidicolin-synchronized cells were mock infected or infected with vEG110, vEGFXE, or vEGdl110 (MOI of 5 to 10) at 18 (T18), 20 (T20), 21 (T21), 22 (T22), and 24 (T24) h postrelease for 9 (t9), 7 (t7), 6 (t6), 5 (t5), or 3 (t3) h, respectively. Cells were then harvested 27 h (T27) postrelease to measure the amount of infected cells still in G1 compared to mock-infected cells. A gate was selected on the total cell data to specifically analyze green fluorescent (infected) cells. (A) DNA distribution of mock-infected and infected synchronized HEp-2 cells 27 h after release from aphidicolin block. (B) Percentage of cells in G1 at T27 depending on the infection time postrelease. mock-t0 represents mock-infected cells at the time of infection corresponding to 24 (T24), 22 (T22), 21 (T21), 20 (T20), and 18 (T18) hours postrelease; mock-T27, vEG110, vEGFXE, and vEGdl110 represent mock-, vEG110-, vEGFXE-, and vEGdl110-infected cells at T27, respectively.
Synchronized cells infected during S-G2 phase of the cell cycle do not efficiently progress through mitosis.
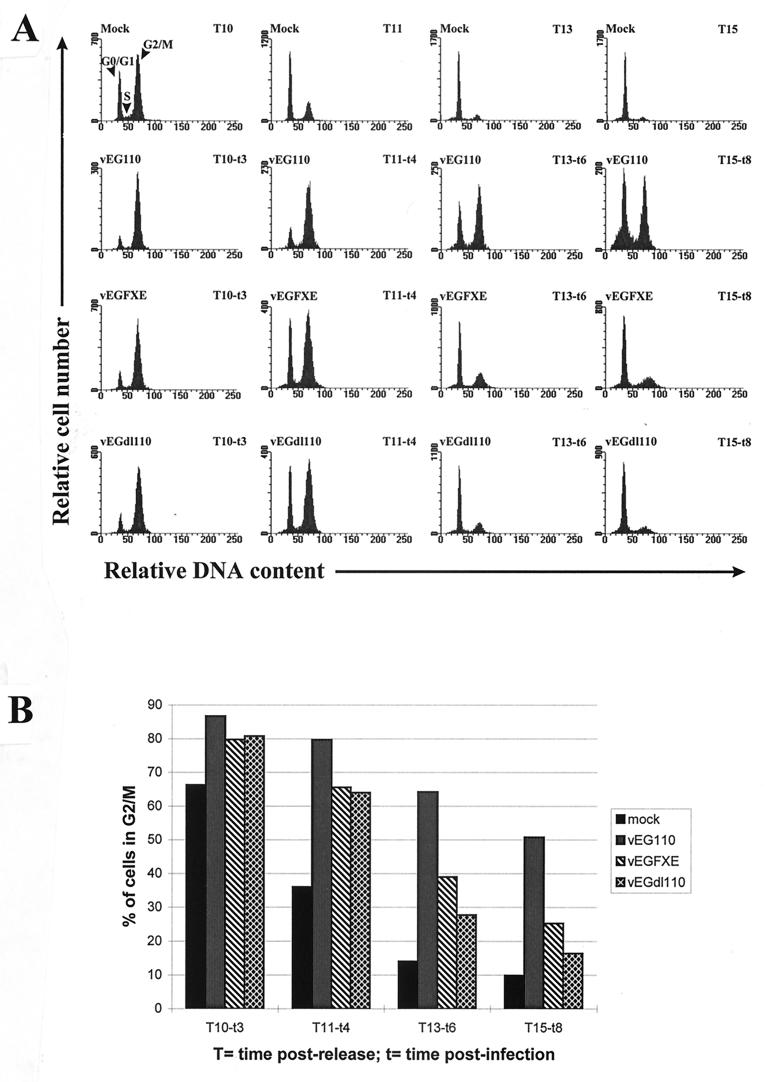
It has recently been shown that the centromeric protein CENP-C, a component of the inner plate of kinetochores which plays a key role in chromatid separation during mitosis (for a review, see reference 39), is degraded by a proteasome and Vmw110-dependent mechanism in cells infected by HSV-1 (17). These data led us to investigate in greater detail than described previously whether the degradation of CENP-C by Vmw110 could affect the progression of infected cells through mitosis. Synchronized HEp-2 cells were infected 7 h postrelease from an aphidicolin block (T7), when about 80% of the cells are in S-G2 (Fig. 2A), and then analyzed by FACS. To specifically analyze cells that were successfully infected the vEG110, vEGFXE, and vEGdl110 viruses were used. The DNA profiles of either infected or mock-infected HEp-2 cells during the course of the experiment and the percentages of cells still in G2/M at various times after infection are represented in Fig. 4A and B, respectively. Whereas mock-infected cells were going through mitosis between 10 and 11 h postrelease, a high percentage of cells infected with vEG110 remained in G2/M 15 h postrelease. Such a dramatic effect was not observed in cells infected with either vEGFXE or vEGdl110, which suggested that Vmw110 was specifically implicated in the block of infected cells in G2/M. However there was a slight delay in vEGFXE- and vEGdl110-infected cells progressing through mitosis, suggesting that infection itself might slow down (but not block) the progression of cells from G2/M to G1. This observation is in accordance with the higher amount of mock-infected than of infected cells in G1 seen at T10 (Fig. 4A). The lower portion of the peak of vEG110-infected cells in G1 at T15 seems atypically enlarged (compare vEG110 T15 with Mock T15, vEGFXE T15, and vEGdl110 T15 in Fig. 4A), which suggests that a subpopulation of these infected cells might contain an abnormal amount of DNA (see below). An important factor in the interpretation of this experiment is that the development of infection in HEp-2 cells is comparatively slow, so that few cells have developed replication centers at these times (data not shown). It would be expected that in cells in which DNA replication is established more quickly, consequential chromosomal damage would in any case halt cell cycle progression.
FIG. 4.
FACS analysis of the progression of infected cells from G2/M to G1. HEp-2 cells were blocked in G0/G1-S by aphidicolin and released for 7 h before being infected by vEG110, vEGFXE, or vEGdl110 (MOI of 10). Cells were harvested at 10 (T10), 11 (T11), 13 (T13), and 15 (T15) h postrelease, which corresponded to 3 (t3), 4 (t4), 6 (t6), and 8 (t8) h postinfection. The progression of mock-infected and infected cells from G2/M to G1 was followed by monitoring their DNA content by FACS as shown by the histograms (A). The percentage of cells in G2/M was then determined at each time point (B). mock, vEG110, vEGFXE, and vEGdl110 represent mock-, vEG110-, vEGFXE-, and vEGdl110-infected cells, respectively.
Cells infected by viruses expressing either EG110 or Vmw110 are blocked between prophase and metaphase stages of mitosis.
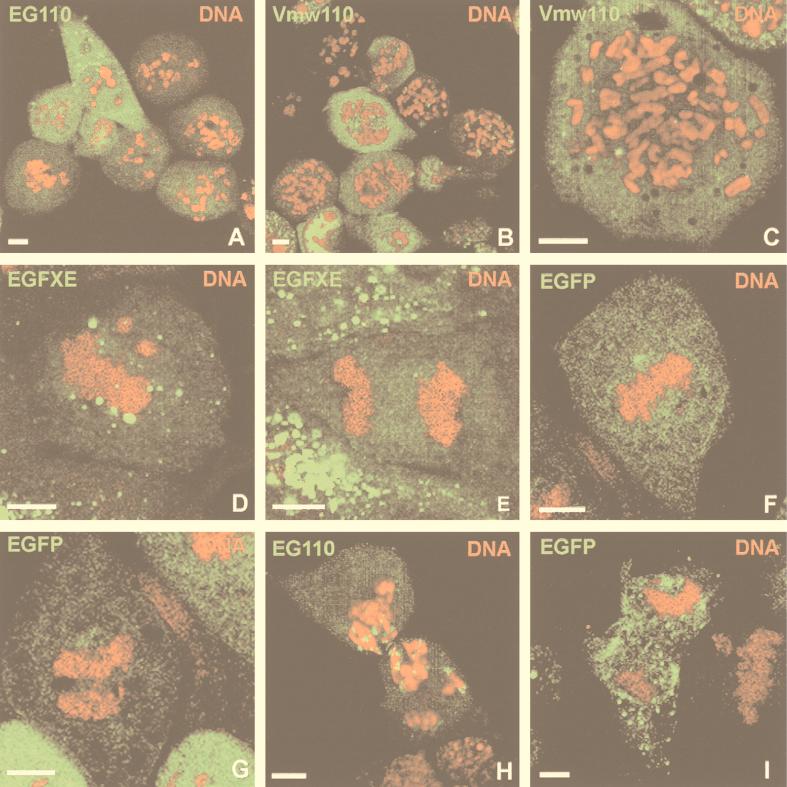
To analyze in detail the effect of infection on synchronized cells, fluorescence microscopy was performed on cells that were infected similarly to the above experiment. Cell DNA was stained by PI to visualize the distribution of the DNA in infected cells during the course of the experiment. Strikingly, 4 h postinfection, cells infected at T7 by vEG110 started to accumulate at an unusual stage of mitosis, and these cells constituted a high proportion of the total cell population 8 h postinfection (Fig. 5A). These cells showed an atypical chromosome distribution between prophase and prometaphase identical to that observed for cells infected in the same conditions by wild-type HSV-1 (Fig. 5B and C). A similar phenotype, called pseudo-prometaphase, has already been described by Bernat et al. (3) for mitotic cells which were microinjected with anticentromere antibodies at a suitable time before the start of mitosis. The atypical chromosome distribution phenotype observed in our experiments will thus be referred as to pseudo-prometaphase in accordance to the terminology used in that paper. Extremely few cells infected by either vEG110 or wild-type HSV-1 (not shown) could be seen in metaphase or anaphase, but some were proceeding to cytokinesis despite chromosomal DNA still being present at the cleavage furrow (Fig. 5H). This observation was specific for cells infected with these viruses, as mock-infected (not shown) as well as vEGFXE (or FXE [not shown])- or vEGdl110 (or dl1403 [not shown])-infected cells did not show these phenotypes and were easily found in metaphase (Fig. 5D and F), at anaphase (Fig. 5E and G), and in normal cytokinesis, as shown as an example for a vEGdl110-infected cell (Fig. 5I). The abnormal cytokinesis of vEG110-infected cells would most likely lead to aneuploid micronucleated daughter cells with an aberrant DNA content (see reference 17), which might explain the broadening of the peak corresponding to vEG110-infected cells in G1 at T15 observed by FACS analysis (Fig. 4A, vEG110 T15).
FIG. 5.
Confocal microscopy analysis of infected mitotic cells. HEp-2 cells were synchronized as described previously and infected 7 h postrelease with either vEG110 or wild-type HSV-1 or with vEGFXE or vEGdl110 (MOI of 10). Eight hours postinfection, cells were harvested, treated with PI (0.5 μg/ml) (red), and then examined by fluorescence microscopy. Vmw110 in cells infected with wild-type HSV-1 (B and C) was detected by immunofluorescence according to the protocol described in reference 17. (A and B) Cells infected with vEG110 and wild-type HSV-1, respectively, with their chromosomes stalled in pseudo-prometaphase; (C) detailed view of the chromosome distribution defined as pseudo-prometaphase; (D to G) cells infected with vEGFXE (green staining due to EGFXE) or vEGdl110 (green staining due to EGFP). Unlike cells infected with vEG110 or wild-type HSV-1, vEGFXE- and vEGdl110-infected cells can be easily found in the normal stages of metaphase (D and F), anaphase (E and G), or cytokinesis (shown as an example for a vEGdl110-infected cell [I]). Panel H shows a vEG110-infected cell going through cytokinesis although its DNA is still present at the cleavage furrow. This cell shows a close association of EG110 with the edges of the chromatin staining at this stage of mitosis. The scale bars represent 5 μm.
The block of infected cells at the pseudo-prometaphase stage is specific of Vmw110 expression.
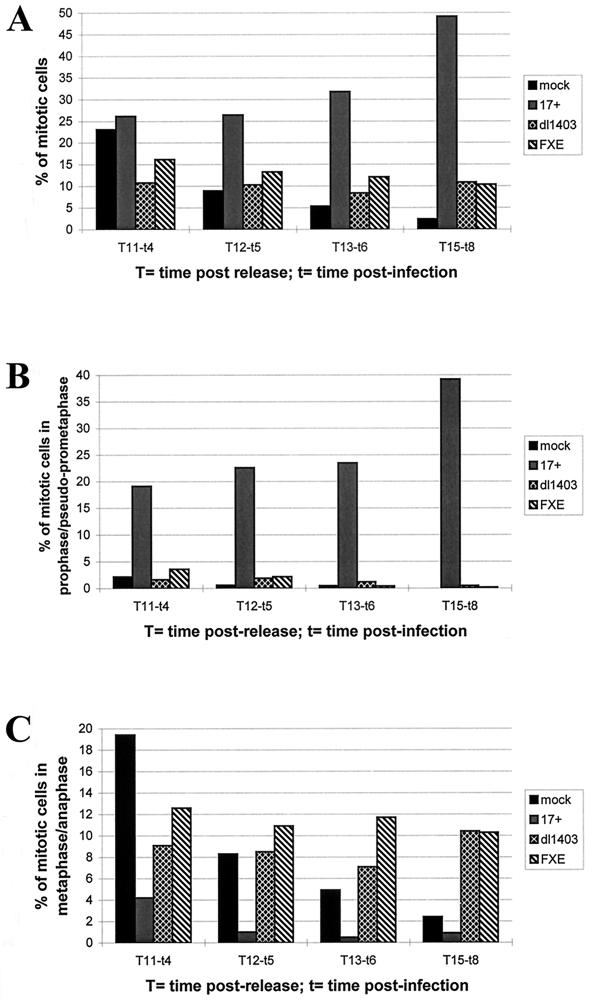
To quantify the effect described above, the percentage of cells in mitosis, in prophase/pseudo-prometaphase, and in metaphase/anaphase was calculated for mock-infected cells and for cells infected with wild-type (17 syn+ strain), FXE, or dl1403 virus. A methodology similar to that described above was used, in that HEp-2 cells were synchronized, infected at 7 h after release from the aphidicolin block, and then fixed for examination at various times thereafter. DNA was stained with PI, and infected cells were detected by using MAb 11060 (anti-Vmw110) for cells infected with the wild-type and FXE viruses and MAb 58S (anti-Vmw175) for dl1403-infected cells. Each parameter was measured by analyzing by immunofluorescence of three fields of cells taken randomly, using the 40× objective lens of the microscope (between 150 and 300 cells/field). Averages were calculated, and the results are shown in Fig. 6. The overall percentage of mitotic cells clearly accumulated in cells infected with the wild-type virus compared to mock-infected cells or cells infected with the Vmw110 mutant viruses (Fig. 6A). The analysis of their mitotic stages showed that 8 h postinfection, about 40% of cells were stalled in prophase/pseudo-prometaphase (Fig. 6B) and very few (<1%) were in metaphase/anaphase (Fig. 6C). Eight hours postinfection, FXE- and dl1403-infected cells still in mitosis were almost all in metaphase/anaphase, and in no cases did cells in prophase show a phenotype similar to the pseudo-prometaphase phenotype (Fig. 5A and B). Mock-infected cells showed a normal mitotic distribution during the course of the experiment. These results demonstrate that Vmw110 is directly implicated in the block of infected cells in an early stage of mitosis between prophase and metaphase. This effect is clearly dependent on Vmw110 and more precisely on the RING finger domain of Vmw110, as the FXE mutant virus does not block cells at the pseudo-prometaphase stage.
FIG. 6.
Analysis of the distribution of infected mitotic cells in different stages of mitosis. Synchronized HEp-2 cells 7 h after release from the aphidicolin block were mock infected (mock) or infected with wild-type HSV-1 (17 syn+ [17+]), Vmw110 deletion mutant dl1403, or a virus expressing the RING finger deletion mutant of Vmw110 (FXE) (MOI of 10). At various times after infection, immunofluorescence was performed with an anti-Vmw110 antibody to detect cells infected with either 17 syn+ or FXE or with an anti-Vmw175 antibody for cells infected with dl1403. DNA was stained by PI (0.5 μg/ml). Percentages of mitotic cells were determined at 4 (t4), 5 (t5), 6 (t6), and 8 (t8) h postinfection by counting three fields of cells taken randomly, using the 40× optical lens and by calculating the average results. Panels A, B, and C represent the percentages of cells being in any stages of mitosis, in prophase/pseudo-prometaphase, and in metaphase/anaphase, respectively.
DISCUSSION
We have shown in this study that Vmw110 affects both the G1-to-S and the G2/M-to-G1 transitions of the cell cycle. The progression of cells through the cell cycle is a complex process involving interactions between positive and negative regulators whose activities are dependent on a variety of both intra- and extracellular stimuli. These activities regulate, among other things, cellular DNA replication and division which constitute two major cell transformations responsible for the transmission of genetic information from a cell to its daughters. These two aspects of the cell cycle are strictly regulated to avoid any malfunction to be transmitted from generation to generation. In that respect key events, which have been extensively documented, are responsible for the cell to progress beyond checkpoints leading to either DNA replication or mitosis.
A large number of proteins and protein modifications are implicated in the G1-to-S transition of the cell cycle, and the product of the retinoblastoma tumor suppressor gene, pRb, plays a central role in the achievement of that process (for a review see reference 54). Activation of cyclin-dependent kinases (cdks) by G1 cyclins, among which are cyclins of the D class (D1, D2, and D3) and cyclin E, leads to the inactivation of pRb by hyperphosphorylation resulting in, among other events, the release of active transcription factor E2F. Many S-phase-specific genes are then activated by E2F to allow entry of the cell into S phase (for reviews, see references 22, 27, 28, 49, and 54). Once cells enter S phase, cyclin E is degraded and cdk2, which earlier in the cycle associates with cyclin E, forms complexes with cyclin A.
Viral oncoproteins such as simian virus 40 T antigen, adenovirus E1A, and human papillomavirus E7, facilitate the G1-S transition by binding to pRb and releasing E2F. Systematic studies on the effects of HSV-1 infection on this particular stage of the cell cycle have not yet been reported, although some relevant data are available. Previous data have shown that synchronized CV-1 cells infected by HSV-1 in G1 were unable to progress to S phase (7). However, multiple protein activities might be affected during the pre-S phase entry, which would prevent infected cells progressing beyond the G1/S phase boundary. Interestingly, Hilton et al. (21) showed that HSV-1 infection of asynchronous cultures of C33A cells induces DNA binding activities of both free E2F and the p107/E2F heterodimer. These data might seem contradictory, as an increase in free E2F would stimulate S phase entry whereas by forming a heterodimer with E2F, p107 blocks E2F activity. However, as suggested by the authors, it is reasonable to assume that efficient HSV-1 DNA replication might require cellular factors that are expressed in late G1 just prior to cellular DNA replication. If so, the virus-induced block of entry into S phase would be expected to be after the stage of E2F-activated late G1 functions. One other key component of G1-to-S phase progression that has been studied in the context of virus infection is cyclin D3. Kawaguchi et al. (26) reported the partial degradation of cyclin D3 by HSV-1 in Vero cells infected for 8 h, by a process that was accelerated by the absence of Vmw110. However, although cyclin D3 is implicated in the phosphorylation of pRb, the results described by Hilton et al. (21) suggest that it is unlikely that its downregulation would prevent infected cells reaching the G1/S border. Thus, it is also unlikely that the interaction of Vmw110 with cyclin D3 (26) directly accounts for the G1/S block of EG110-expressing transfected cells observed in our study.
In addition to these data, it has also recently been shown that the use of specific inhibitors of cdks which are required for G1-to-S phase progression results in considerable inhibition of IE and E transcription and HSV-1 replication (46, 47). It was suggested that cdk-activated cellular and/or viral transcription factors might be required for optimal transcriptional activation of the viral genome. All of these data show that HSV-1 is able to influence and is influenced by some key events occurring in G1/S, a critical phase of the cell cycle beyond which cells are irreversibly committed to mitosis. Our current results show that Vmw110 is likely to affect some factors implicated in that decision resulting in the block of the G1-to-S transition, an effect partly dependent on the RING finger domain of the protein. However, the ability of viruses either deficient for the expression of Vmw110 (vEGdl110) or expressing the RING finger mutant of the protein (vEGFXE) to block infected cells in G1/S suggests that viral proteins other than Vmw110 also cause a G1/S block. Moreover, an efficient block of infected cells at the G1/S border is observed only when cells are infected early enough before the onset of S phase (reference 7 and this study). Therefore the G1-S block observed in transfected cells expressing Vmw110 is likely to occur only after Vmw110 has been present for a number of hours, and during this time in a normal infection other viral factors could also block the cell cycle at this stage.
This study also shows that infection by HSV-1 is able to block cells at the mitotic stage of the cell cycle. Of great interest is that, unlike the G1/S block, this mitotic block is specifically due to Vmw110 and more precisely to its RING finger domain, as mutant viruses unable to express Vmw110 (dl1403 and vEG110) or expressing the RING finger Vmw110 mutant protein (FXE and vEGFXE) do not retain this activity. This function of Vmw110 can be related to recent work which showed that Vmw110 induces the proteasome-dependent degradation of the centromeric protein CENP-C (17). Indeed, it has previously been shown that microinjection of anticentromere antibodies in cells about to reach mitosis was able either to significantly delay mitosis or block chromosome distribution in an atypical stage of mitosis called pseudo-prometaphase (3, 4). The observations made in our study are fully consistent with those obtained by the microinjection of anticentromere antibodies, as HSV-1 induces similar pseudo-prometaphase distribution of chromosomes and delay of mitosis. However, microinjection of anti-CENP-C antibodies early enough before the beginning of mitosis results in a metaphase block (52), which suggests that the inhibition of CENP-C alone is not sufficient to generate a pseudo-prometaphase arrest. Therefore, in addition to CENP-C, the stability of other centromeric proteins might be affected by Vmw110 in HSV-1-infected cells. An interesting prediction from our findings is that cells infected at an appropriate stage in G2 with wild-type HSV-1 may not produce progeny virus. Indeed, the infected cells stalled in pseudo-prometaphase lack a nuclear envelope, and it seems highly likely that viral transcription, DNA replication and maturation would be severely compromised.
During the last decade, the development of therapies aiming to use virus-based vectors as delivery systems has opened new fields of interest for HSV-1. However, although its ability to persist in hosts for very long time makes it a good candidate for permanent gene expression, its high degree of toxicity in infected cells is incompatible with the use of unmodified parental genomes as vector systems. In order to reduce the toxicity of such vectors, replication-defective mutants of HSV-1 have been constructed with multiple deletions in genes encoding proteins affecting cell survival. Deletion of one or several IE genes coding for proteins implicated in virus replication dramatically decreased the vector toxicity (1, 23, 24, 37, 38, 44, 56), but survival of cells infected by these vectors was still significantly affected unless all five IE proteins were absent (45). More specifically, Wu et al. (56) suggested a putative role for Vmw110 in the inhibition of both cellular DNA synthesis and the division potential of the cells. The data presented in our study provide an explanation for these suggestions, as we showed that Vmw110 expression in infected cells would affect both pathways resulting in an incompatibility between Vmw110 expression and survival of a cell population. These data also partly explain both the failure to establish cell lines constitutively expressing Vmw110 and the remaining toxicity of any replication-defective mutants of HSV-1 still able to express Vmw110. Therefore, the aberrations in mitotic events likely due to the degradation of CENP-C exclude the expression of Vmw110 in any HSV-1-based vectors used for stable expression of foreign proteins in gene therapy of dividing cells. Over the past few years, several independent studies emphasized the possible effects of the expression of Vmw110 on cell metabolism and/or survival. Our study shows that expression of Vmw110 in cycling cells will affect both G1-to-S and G2/M-to-G1 transitions, undoubtedly creating physiological changes, which would eventually lead to cell death.
ACKNOWLEDGMENTS
We are grateful to Alan Mowat for collaboration in the use of the FACS, to the Department of Clinical Immunology for use of the equipment, to Elizabeth Allen and Susana de la Luna for helpful constructive criticisms, and to Duncan McGeoch for support.
This work was supported by the Medical Research Council. P.L. is a postdoctoral researcher funded by a European Community training project (Marie Curie research training grant) financed by the European Commission under the Biomedicine and Health Fourth Framework Programme.
REFERENCES
- 1.Ace C I, McKee T A, Ryan J M, Cameron J M, Preston C M. Construction and characterization of herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J Virol. 1989;63:2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ascoli C A, Maul G G. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernat R L, Borisy G G, Rothchild N F, Earnshaw W C. Injection of anti-centromere antibodies in interphase disrupts events required for chromosome movement at mitosis. J Cell Biol. 1990;111:1519–1533. doi: 10.1083/jcb.111.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernat R L, Delannoy M R, Rothfield N F, Earnshaw W C. Disruption of centromere assembly during interphase inhibits kinetochore morphogenesis and function in mitosis. Cell. 1991;66:1229–1238. doi: 10.1016/0092-8674(91)90045-z. [DOI] [PubMed] [Google Scholar]
- 5.Cai W, Astor T D, Liptak L M, Cho C, Coen D, Schaffer P A. The herpes simplex virus type 1 regulatory protein ICP0 enhances replication during acute infection and reactivation from latency. J Virol. 1993;67:7501–7512. doi: 10.1128/jvi.67.12.7501-7512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clements J B, Stow N D. A herpes simplex virus type 1 mutant containing a deletion within immediate-early gene 1 is latency competent in mice. J Gen Virol. 1989;70:2501–2506. doi: 10.1099/0022-1317-70-9-2501. [DOI] [PubMed] [Google Scholar]
- 7.De Bruyn Kops A, Knipe D M. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell. 1988;55:857–868. doi: 10.1016/0092-8674(88)90141-9. [DOI] [PubMed] [Google Scholar]
- 8.DeLuca N M, Courtney M A, Schaffer P A. Temperature-sensitive mutants in herpes simplex virus type 1 ICP4 permissive for early gene expression. J Virol. 1984;52:767–776. doi: 10.1128/jvi.52.3.767-776.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dixon R A F, Schaffer P A. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J Virol. 1980;36:189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earnshaw W C, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 11.Everett R D. Construction and characterisation of herpes simplex virus type 1 mutants with defined lesions in immediate-early gene 1. J Gen Virol. 1989;70:1185–1202. doi: 10.1099/0022-1317-70-5-1185. [DOI] [PubMed] [Google Scholar]
- 12.Everett R D, Preston C M, Stow N D. Functional and genetic analysis of the role of Vmw110 in herpes simplex virus replication. In: Wagner E K, editor. The control of herpes simplex virus gene expression. Boca Raton, Fla: CRC Press Inc.; 1991. pp. 50–76. [Google Scholar]
- 13.Everett R D, Cross A, Orr A. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell-type dependent manner. Virology. 1993;197:751–756. doi: 10.1006/viro.1993.1651. [DOI] [PubMed] [Google Scholar]
- 14.Everett R D, Meredith M R, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110 and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett R D, Orr A, Preston C M. A viral activator of gene expression functions via the ubiquitin-proteasome pathway. EMBO J. 1998;17:7161–7169. doi: 10.1093/emboj/17.24.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett R D, Earnshaw W C, Findlay J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fields B N, Knipe D M, Howley P M. Virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. [Google Scholar]
- 19.Fraser N W, Block T M, Spivack J G. The latency associated transcripts of herpes simplex virus: RNA in search of a function. Virology. 1992;191:1–8. doi: 10.1016/0042-6822(92)90160-q. [DOI] [PubMed] [Google Scholar]
- 20.Harris R A, Everett R D, Zhu X, Silverstein S, Preston C M. The herpes simplex virus (HSV) immediate-early protein Vmw110 reactivates latent HSV type 2 in an in vitro latency system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hilton M J, Mounghane D, McLean T, Contractor N V, O’Neil J, Carpenter K, Bachenheimer S L. Induction by herpes simplex virus of free and heteromeric forms of E2F transcription factor. Virology. 1995;213:624–638. doi: 10.1006/viro.1995.0034. [DOI] [PubMed] [Google Scholar]
- 22.Hunter T, Pines J. Cyclins and cancer II: cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 23.Johnson P A, Miyanohara A, Levine F, Cahill T, Friedmann T. Cytotoxicity of a replication-defective mutant of herpes simplex virus type 1. J Virol. 1992;66:2952–2965. doi: 10.1128/jvi.66.5.2952-2965.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson P A, Wang M J, Friedmann T. Improved cell survival by the reduction of immediate-early gene expression in replication-defective mutants of herpes simplex virus type 1 but not by mutation of the virion host shutoff function. J Virol. 1994;68:6347–6362. doi: 10.1128/jvi.68.10.6347-6362.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translation machinery. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex type 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam E W-F, La Thangue N B. DP and E2F proteins: coordinating transcription with cell cycle progression. Curr Opin Cell Biol. 1994;6:859–866. doi: 10.1016/0955-0674(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 28.La Thangue N B. DRTF1/E2F: an expanding family of heterodimeric transcription factors implicated in cell-cycle control. Trends Biochem Sci. 1994;19:108–114. doi: 10.1016/0968-0004(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 29.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leib D A, Coen D M, Bogard C L, Hicks K A, Yager D R, Knipe D M, Tyler K L, Schaffer P A. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759–768. doi: 10.1128/jvi.63.2.759-768.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lomonte, P., and R. D. Everett. Unpublished data.
- 32.McCarthy A M, McMahan L, Schaffer P A. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J Virol. 1989;63:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meredith M R, Orr A, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135kD cellular protein. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 34.Parkinson J, Lees-Miller S P, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus type 1 is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 36.Preston C M. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsk. J Virol. 1979;29:228–239. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preston C M, Nicholl M J. Repression of gene expression upon infection of cells with herpes simplex virus type 1 mutants impaired for immediate-early protein synthesis. J Virol. 1997;71:7807–7813. doi: 10.1128/jvi.71.10.7807-7813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Preston C M, Mabbs R, Nicholl M J. Construction and characterization of herpes simplex virus type 1 mutants with conditional defects in immediate early gene expression. Virology. 1997;229:228–239. doi: 10.1006/viro.1996.8424. [DOI] [PubMed] [Google Scholar]
- 39.Rieder C L, Salmon E D. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 1998;8:310–318. doi: 10.1016/s0962-8924(98)01299-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 2231–2296. [Google Scholar]
- 41.Sacks W R, Greene C C, Aschman D P, Schaffer P A. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J Virol. 1985;55:796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saitoh H, Tomkiel J, Cooke C A, Ratrie H, Maurer M, Rothfield N F, Earnshaw W C. CENP-C, and autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 44.Samaniego L A, Wu N, DeLuca N A. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J Virol. 1997;71:4614–4625. doi: 10.1128/jvi.71.6.4614-4625.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samaniego L A, Neiderhiser L, DeLuca N A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schang L M, Phillips J, Schaffer P A. Requirement for cellular cyclin-dependent kinases in herpes simplex virus replication and transcription. J Virol. 1998;72:5626–5637. doi: 10.1128/jvi.72.7.5626-5637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schang L M, Rosenberg A, Schaffer P A. Transcription of herpes simplex virus immediate-early and early genes is inhibited by roscovitine, an inhibitor specific for cellular cyclin-dependent kinases. J Virol. 1999;73:2161–2172. doi: 10.1128/jvi.73.3.2161-2172.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sears A E, Halliburton I W, Meignier B, Silver S, Roizman B. Herpes simplex virus 1 mutant deleted in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J Virol. 1985;55:338–346. doi: 10.1128/jvi.55.2.338-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherr C J. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 50.Showalter L D, Zweig M, Hampar B. Monoclonal antibodies to herpes simplex type 1 proteins including the immediate-early protein ICP4. Infect Immun. 1981;34:684–692. doi: 10.1128/iai.34.3.684-692.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stow N D, Stow E C. Isolation and characterisation of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate-early polypeptide Vmw110. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 52.Tomkiel J, Cooke C A, Saitoh H, Bernat R, Earnshaw W C. CENP-C is required for maintaining proper kinetochore size and for a timely transition to anaphase. J Cell Biol. 1994;125:531–545. doi: 10.1083/jcb.125.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson R J, Clements J B. Characterization of transcription-deficient temperature-sensitive mutants of herpes simplex virus type 1. Virology. 1978;91:364–379. doi: 10.1016/0042-6822(78)90384-7. [DOI] [PubMed] [Google Scholar]
- 54.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 55.Wilcox C L, Smith R L, Everett R D, Mysofski D. The herpes simplex virus type 1 immediate-early ICP0 is necessary for the efficient establishment of latent infection. J Virol. 1997;71:6777–6785. doi: 10.1128/jvi.71.9.6777-6785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu N, Watkins S C, Schaffer P A, DeLuca N A. Prolonged gene expression and cell survival after infection by a herpes simplex virus mutant defective in the immediate-early genes encoding ICP4, ICP27, and ICP22. J Virol. 1996;70:6358–6369. doi: 10.1128/jvi.70.9.6358-6369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu X, Chen J, Young C S H, Silverstein S. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J Virol. 1990;64:4489–4498. doi: 10.1128/jvi.64.9.4489-4498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]