Abstract
cORF, a protein encoded by the human endogenous retrovirus family HTDV/HERV-K, contains amino acid motifs which resemble the nuclear import and export signals of the viral regulatory proteins Rev (human immunodeficiency virus) and Rex (human T-cell leukemia virus [HTLV]). In this study, we demonstrated that cORF indeed has a Rev-like function and mapped the cORF-responsive RNA element to a sequence in the 3′ long terminal repeat, a localization similar to RxRE, the responsive element in HTLV type 1. Accordingly, we have given the element the designation RcRE. cORF and RcRE stabilize unspliced and incompletely spliced viral transcripts and enhance their nuclear export via the CRM1 export pathway. So far, HTDV/HERV-K is the only endogenous retrovirus family with a complex regulation at the posttranscriptional level. It may be regarded as an intermediate in the evolution from simple to complex retroviruses.
All retroviruses have three genes in common: gag, the protease/polymerase gene (prt/pol), and the envelope gene (env). Complex retroviruses possess additional accessory genes, some of which regulate the expression of viral genes at the transcriptional or the posttranscriptional level. In the human immunodeficiency virus (HIV), for example, inefficient splice sites and so-called instability sequences in the viral introns cause nuclear retention or degradation of primary transcripts (30, 34, 43) unless these transcripts are completely spliced to small mRNAs encoding inter alia the regulatory proteins Tat and Rev (6). Efficient nuclear export of full-length transcripts and subgenomic env transcripts depends on the interaction of two viral elements, the regulator of expression of viral proteins (Rev) and a Rev-responsive RNA element (RRE) (15, 42). The equivalent elements of human T-cell leukemia virus (HTLV) are designated Rex and RxRE, respectively.
Transport of Rev and Rev homologues into the nucleus is mediated by the interaction of a domain rich in basic amino acids, the nuclear localization signal (NLS), with cellular import factors (18) (for reviews, see references 6, 36, and 46). In the nucleus, the viral protein binds to its responsive element in the primary viral transcript, forming multimers. In cooperation with a plethora of cellular factors, the Rev-RRE or Rex-RxRE interaction leads to suppression of splicing, to stabilization of full-length and singly spliced transcripts in the nucleus, to increased export of such transcripts to the cytoplasm, and thus eventually to the generation of infectious progeny (reviewed, e.g., in references 19 and 41). The nuclear export signal of Rev and Rev homologues most often consists of a motif enriched in leucine residues (11). CRM1, the nuclear export factor for leucine-rich nuclear export signals (NESs), is the most important of the cellular factors which have been found to bind to this signal (12, 13, 35). CRM1 is specifically inhibited upon binding to the antibiotic leptomycin B (LMB) (4, 16, 23, 39, 48).
The responsive RNA sequences to which proteins with Rev-like function bind are located in quite different parts of the respective viral transcripts. The Rev-responsive element of HIV (RRE) is located in env (31), while the Rex-responsive element (RxRE) of HTLV type 1 (HTLV-1) is located in the 3′ U3R (1). In several test systems, Rex as well as Rev can cross-act with responsive elements of related viruses (reviewed in reference 10). In summary, the regulation of gene expression by the interaction of a Rev-like protein with a responsive RNA element is a hallmark of complex retroviruses.
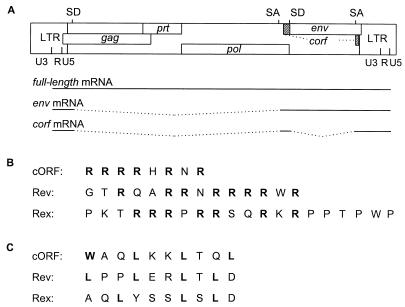
Endogenous retroviruses (ERVs) are integral parts in the genomes of many, if not of all, species. ERVs most probably resulted from infection of germ line cells with exogenous retroviruses and subsequent fixation of their genetic information in the host genome. While ERVs related to simple retroviruses have been well known for many years, it is intriguing that no endogenous counterparts of complex retroviruses have yet been detected (28). In this report, we show evidence that the human ERV (HERV) family HERV-K is an exception. This is the only HERV family known to code for retroviral particles, the human teratocarcinoma-derived virus (HTDV) particles described more than a decade ago (5, 24–26). The family comprises 30 to 50 proviruses in the genomes of humans and of Old World monkeys (38). Their closest contemporary relatives are mammalian type B and D retroviruses. A prototypic HTDV/HERV-K genome (type 2) (27) with its mRNA transcripts is depicted in Fig. 1A.
FIG. 1.
(A) Genomic organization and transcripts of HTDV/HERV-K type 2. SA, splice acceptor; SD, splice donor. (B) Comparison of arginine-rich NLSs. (C) Comparison of leucine-rich NESs. Amino acids in boldface are essential for function.
Like all hitherto identified HERVs, HTDV/HERV-K proviruses are not infectious, although complete open reading frames for all viral genes have been observed in cDNA clones and recently also in a full-length provirus on chromosome 7 (32). Their potential for encoding functional proteins has been studied in particle-producing cell lines and with recombinant expression systems (28, 45). HTDV/HERV-K mRNA expression in teratocarcinoma cell lines resembles the pattern seen with complex retroviruses: full-length transcripts and env transcripts are accompanied by alternatively spliced small mRNA species (Fig. 1A) (26). One of these transcripts, a 1.8-kb doubly spliced mRNA, codes for a protein of 14 kDa designated cORF, which contains a putative arginine-rich NLS (Fig. 1B) and a putative leucine-rich NES (Fig. 1C) and which is targeted to the nucleoli similarly to Rev and Rex (27, 29).
This similarity to regulatory proteins of complex exogenous retroviruses prompted us to investigate whether efficient protein expression in the HTDV/HERV-K family also depends on Rev-RRE-like regulatory elements. In this study, we demonstrated that cORF indeed has a Rev/Rex-like function and mapped the responsive element. We showed that cORF stabilizes incompletely spliced mRNAs in the nucleus and enhances their transport to the cytoplasm when the cORF-responsive element is present. Furthermore, we present data indicating that cORF uses the CRM1 nuclear export pathway. HTDV/HERV-K is the first example of an ERV with similarities to complex retroviruses.
MATERIALS AND METHODS
Cells.
COS7 (African green monkey kidney) and HeLa (human cervical carcinoma) cells were obtained from the American Type Culture Collection. The teratocarcinoma cell line GH had been established earlier (24). HLtat is a HeLa derived cell line stably transfected with a Tat-expressing plasmid. All cell lines were grown in Dulbecco’s minimal essential medium supplemented with 10% fetal calf serum (PAA Laboratories), 2 mM glutamine, and antibiotics. Cells were subdivided weekly as follows: COS7, 1:20; HeLa, 1:40; GH, 1:3; and HLtat, 1:30.
Plasmid constructions.
Two HIV expression plasmids were kindly provided by B. Felber, Frederick, Md.: (i) plasmid pNLcgagA2, a derivative of plasmid pCgagA2, here called pHIVgagRRE; and (ii) plasmid pBsrev, here designated pREV. Two control plasmids were constructed as follows: pHIVgag, by removing the RRE in pHIVgagRRE by the use of restriction endonucleases SacII and XhoI, treatment with the Klenow fragment of DNA polymerase to generate blunt ends, and ligation; and p(−), by removing most of the Rev-coding sequence in pBsrev by the use of HindIII and EcoRI, filling in recessive ends with Klenow enzyme, and blunt-end ligation. All enzymes were purchased from either New England Biolabs or Gibco BRL.
The plasmid pHKgagRRE was cloned by replacement of HIV gag with the sequence of HTDV/HERV-K gag (pcG3gag) (45). The gag sequence was excised with XhoI (nucleotide [nt] 1012) and SpeI (nt 3259) to preserve the first splice donor site, located at nt 1074 to 1083. Resulting fragments were blunted with Klenow enzyme and ligated into pHIVgagRRE which had been restricted with SalI and SacII and blunted to remove the HIV gag gene.
For replacement of the RRE in pHIVgagRRE, overlapping fragments of approximately 400 nt from the 3′ end of the HERV-K genome were amplified by PCR. The following templates were used: a cDNA clone that comprises the 3′ end of the integrase gene, the entire env gene, U3, and R (25), and a long terminal repeat (LTR) clone (pcK30LTR [see below]). The 3′ R regions of these two clones differ by one point mutation. Note that the experimentally determined 3′ end of R in these cDNA clones is not identical to the prediction made in reference 37.
For the fragments named U3RU5 (nt 8505 to 9380), U3RIII (nt 8720 to 9148), U3RIX (nt 8720 to 9093), and RU5 (nt 9065 to 9492), the following primers were used: 5′ataccgcggTGTGGGGAAAAGCAAG (nt 8505 to 8520), 5′atactcgagGAGGAAAGAAAGAAGGGG (nt 9363 to 9380), 5′ataccgcggACAGATGCTTGAAGGC (nt 8720 to 8735), 5′atactcgagCACATAAACATCTCAATGC (nt 9130 to 9148), 5′atactcgagCCAGCCCTAAGGCGG (nt 9079 to 9093), 5′ataccgcggGAGATAGGGAAAAACCG (nt 9065 to 9081), and 5′atactcgagCCTCCACGTTGGGCAC (nt 9474 to 9492). Nucleotides in uppercase define the HTDV/HERV-K sequence; inserted restriction sites for SacII and XhoI are underlined. Restricted PCR fragments were ligated into pHIVgagRRE after elimination of the RRE sequence by SacII and XhoI.
Construction of the following vectors was based on the eucaryotic expression vector pRC-CMV (Invitrogen) and on the HTDV/HERV-K sequence pcGPK31, kindly provided by R. R. Tönjes, Langen, Germany (45). The ERV sequences of plasmids pHK (nt 1012 [XhoI site upstream of the 5′ splice donor] to nt 8737 [SphI site in the 3′ U3 region]) and pHKRcRE (nt 1012 to the end of the proviral genome) were cloned into the HindIII and ApaI sites of pRC-CMV by several subcloning steps aimed at eliminating the pBluescript (Stratagene) multiple cloning site sequences present in pcGPK31. To insert the 3′ LTR into pHKRcRE, we used clone pcK30LTR, which harbors an LTR sequence reconstituted from the U3R region of pcK30env (EMBL accession no. X82272) and the RU5 region of pcK32cORF (EMBL accession no. X82271). The clone had been constructed by using the internal BamHI site, with subsequent PCR amplification of the sequence beginning at nt 1 of the U3 region to the primer binding site, including HindIII restriction sites at both ends for cloning into pBluescript. Further EMBL accession numbers are Y10390 for pcG3gag, Y10391 for pcP23pol, and Y10392 for pcG1pro.
The 5′ splice donor site was added by insertion of a small PCR product into the HindIII site. PCR primers used were 5′ataaagcttCTCGAGCGTGGTCATTGAG (the HTDV/HERV-K sequence starts at nt 1012) and 5′TCTTCCGAGCGCGCAAGCTTACCG (HindIII sites are underlined).
For construction of pcORF, the coding sequence (nt 6442 to 6711 and 8411 to 8470) of a cDNA clone that differs from the published sequence pcK32cORF by a point mutation (altering amino acid 52 from Thr to Ser) was amplified with primers 5′ccaagcttAGTTCTACAATGAACCCA and 5′atagggcccTCATCATGGCCCGTT. Restricted fragments were cloned into the HindIII and ApaI sites of pRC-CMV.
All plasmids were grown in Escherichia coli DH5α cells (Gibco BRL). For transfections, plasmids were prepared with an endotoxin-free plasmid preparation kit (Qiagen).
Transfections.
Transfections were performed either in six-well plates (Greiner) containing coverslips (for immunofluorescence studies), in 25-cm2 tissue culture flasks (Nunc) (for p24 quantitation), or in tissue culture dishes (diameter, 14.5 cm) (for RNA preparations). Cells were seeded at a density of 1 × 106 (six-well plates), 2 × 106 (flasks), or 2.5 × 106 (dishes) the day before transfection. On the day of transfection, the cells were 50 to 70% confluent (for RNA preparations they were less than 50% confluent). Transfections were carried out with DOTAP liposomal transfection reagent (Boehringer Mannheim) or Lipofectamine Plus (Gibco BRL) in accordance with the users’ manual. The total amount of DNA used in transfections was 4 μg in six-well plates and 8 μg in bottles with DOTAP, respectively, or 1 μg in six-well plates, 1.5 μg in bottles, and 6 μg in dishes with Lipofectamine Plus. In cotransfections, vectors were used in equimolar amounts. Cells were incubated in serum-free transfection medium for 5 to 6 h at 37°C, after which the transfection medium was changed to medium containing 10% fetal calf serum. The cells were processed for further studies 24 to 48 h after transfection.
For transfections with LMB, 2 to 6 nM LMB was added at the time of transfection and cells were treated with constant amounts of LMB until the time of fixation or lysis.
Immunocytochemistry.
Cells on coverslips were fixed 24 or 48 h posttransfection, and immunofluorescence staining was performed as described previously (5). The following antibodies and dilutions were used: anti-cORF (α-cORF; rabbit), 1:200 to 1:500 (27); α-HERV-K Gag (rabbit), 1:500 to 1:1,000 (26); α-HERV-K Gag (goat), 1:500 to 1:1,000; α-HERV-K Env TM-env (goat), 1:500 to 1:1,000; and α-HIV p24 monoclonal (mouse), 1:400 (Du Pont). The α-HERV-K Gag (goat) antibody was provided by R. R. Tönjes, Langen, Germany; the α-HERV-K Env TM-env (goat) antibody was donated by J. Denner, Langen, Germany. Secondary antibodies and their dilutions were as follows: fluorescein isothiocyanate (FITC)-conjugated goat α-rabbit immunoglobulin G (IgG), 1:400 (Cappel); FITC-conjugated donkey α-rabbit IgG, 1:25 to 1:50 (Amersham); Cy3-conjugated donkey α-goat IgG, 1:500 to 1:1,000 (Dianova); and Cy3-conjugated goat α-mouse IgG, 1:500 to 1:1,000 (Dianova). For double immunofluorescent staining, two primary as well as two secondary antibodies were mixed and incubations were carried out simultaneously. After final washes in phosphate-buffered saline (PBS), cells were mounted in Moviol (Hoechst, Frankfurt) on slides. Preparations were examined by using a Zeiss Axiophot microscope equipped with a fluorescence illuminator with appropriate filters. Kodak Ektachrome 400 X color reversal film was used for documentation.
Quantitation of p24.
Two days after transfection in the presence of DOTAP, Gag particles were prepared from cell culture supernatants of transfected cells and duplicates were combined. After centrifugation at 10,000 × g and 4°C for 15 min to remove cellular components, supernatants were centrifuged at 100,000 × g and 4°C for 80 min. The resulting pellet was dissolved in 200 μl of cell culture lysis buffer (Promega). Volumes of 200 μl of diluted cell lysate or entire particle preparations were employed in the HIV AG-1 Monoclonal p24 capture assay (Abbott). One day after transfection in the presence of Lipofectamine Plus or 2 days after transfection in the presence of DOTAP, cells were lysed with 500 μl of cell culture lysis buffer per bottle after two washes with PBS. The lysate was centrifuged for 30 s at 4°C and 10,000 rpm in a Biofuge R centrifuge (Heraeus). Total protein contents of cell lysates were determined by the use of the Bio-Rad protein assay (Bio-Rad) according to the users’ manual. Depending on the expected concentration of HIV Gag, 1 to 5 μl of the cell lysate were diluted to 240 μl with 0.5× lysis buffer. The test was carried out according to the manufacturer’s instructions.
Immunoblotting.
Cell lysates of the same transfectants as for the p24 capture assay were used for immunoblotting. Proteins were separated in 12.5 or 10% polyacrylamide gels under reducing conditions. Fifteen microliters (DOTAP) or 2 μl (Lipofectamine Plus) of lysate was applied per lane. The proteins were transferred to polyvinylidene difluoride membranes (Hybond-P; Amersham). The membranes were blocked with 5% nonfat dried milk (Marvel) (see Fig. 2) or 5% nonfat dried milk plus 2% bovine serum albumin fraction V (Boehringer Mannheim) (see Fig. 5) and 0.1% Tween 20 (Serva) in PBS. After being washed with PBS containing 0.1% detergent, membranes were incubated for 1 h at room temperature with an HIV-positive human serum specimen at a dilution of 1:500. Membranes were washed extensively and incubated with a second antibody conjugated with horseradish peroxidase: α-human IgG (Sigma) at a dilution of 1:2,000 to 1:5,000. Immunoreactive proteins were detected by the use of an enhanced chemiluminescence kit (ECL+Plus; Amersham).
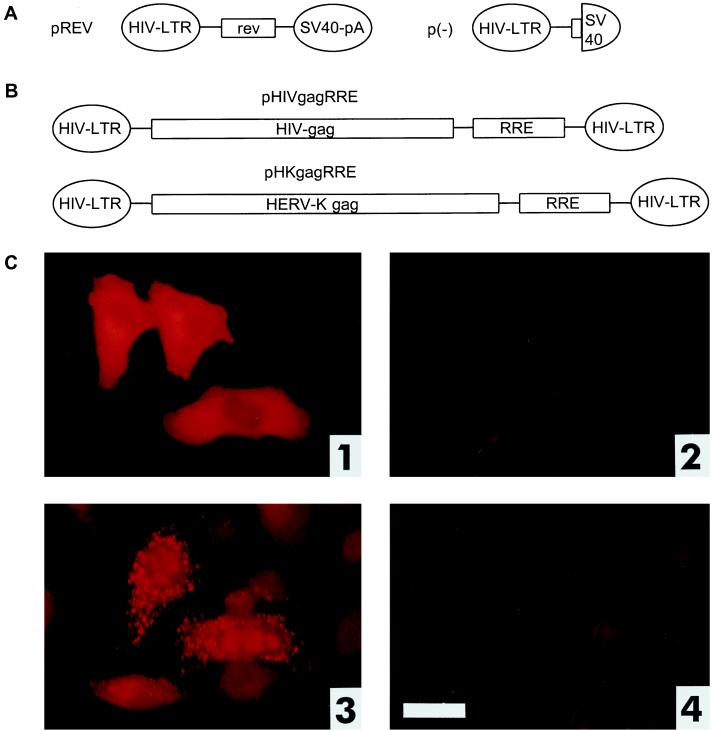
FIG. 2.
Expression of HTDV/HERV-K Gag in the presence of the viral regulatory elements Rev and RRE. (A) Mammalian expression vectors encoding HIV Rev and the respective control plasmid (effector plasmids). SV40-pA, simian virus 40 polyadenylation sequence (for plasmid construction, see Materials and Methods). (B) Mammalian expression vectors encoding HIV or HERV-K Gag proteins as well as the HIV RRE (reporter plasmids). (C) Cotransfection of reporter and effector plasmids into HLtat cells. Panel 1, pHIVgagRRE and pREV; panel 2, pHIVgagRRE and p(−); panel 3, pHKgagRRE and pREV; panel 4, pHKgagRRE and p(−). Staining was achieved with a mouse monoclonal α-HIV p24 antibody (panels 1 and 2) or with a rabbit α-HERV-K Gag antibody (panels 3 and 4). Bar, 25 μm.
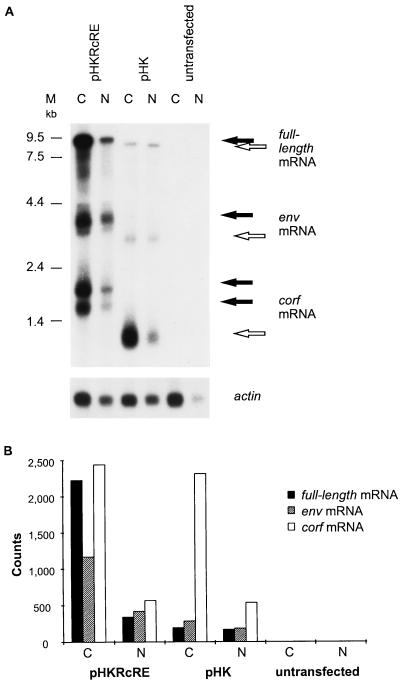
FIG. 5.
Analyses of HTDV/HERV-K nuclear export. (A) Northern blot of cytoplasmic (C) and nuclear (N) mRNAs of transfected HeLa cells. Cells were transfected with pHKRcRE (lanes 1 and 2) or pHK (lanes 3 and 4) or were not transfected (lanes 5 and 6). The probe was the first exon of cORF. The two cORF bands detected with the vector pHKRcRE are a consequence of the two polyadenylation signals present in pHKRcRE, one in the HERV-K LTR and the other in the plasmid backbone. The membrane was rehybridized with an actin mRNA probe to show that there were equal amounts of mRNA in the different cytoplasmic and nuclear fractions. The positions of molecular size markers (M) are indicated on the left. (B) Quantification of the radioactive signals by phosphorimaging. The signals were normalized to those of the actin mRNA.
Northern blotting.
Cytoplasmic and nuclear RNA was prepared with an RNeasy Mini Kit (Qiagen) in accordance with the manufacturer’s instructions. mRNA preparation and Northern blotting conditions are described in reference 27. The RNA probe was specific for the first exon of cORF.
RESULTS
HTDV/HERV-K Gag expression depends on posttranscriptional regulation.
To investigate whether the expression of HTDV/HERV-K structural proteins is controlled by viral regulatory elements, like HIV expression is regulated by Rev and its responsive RNA element, RRE, we used a well-established HIV-based reporter system with three components: (i) a Rev expression plasmid designated pREV (pBsrev in references 8 and 33) (Fig. 2A); (ii) an RRE sequence-containing HIV Gag expression vector designated pHIVgagRRE (pNLcgagA2 in references 7 and 15) (Fig. 2B); and (iii) HLtat cells (44), a subclone of HeLa cells stably transfected with a Tat expression plasmid. The Gag protein is expressed at high levels only when Rev can bind to the RRE, thereby stabilizing the gag mRNA and enhancing its nuclear export. In transient-transfection assays in HLtat cells, HIV Gag expression can be monitored either by immunofluorescence or by p24 enzyme-linked immunosorbent assay (ELISA).
Since HTDV/HERV-K proviruses are not expressed in HLtat cells, even under conditions of transfection (data not shown), we adopted this system to study the regulation of expression of HTDV/HERV-K Gag, using the vectors described below. We replaced the HIV gag sequence in pHIVgagRRE with the HTDV/HERV-K gag (pHKgagRRE) (Fig. 2B) sequence and generated a number of control plasmids: HTDV/HERV-K or HIV gag-containing plasmids without RRE (pHKgag and pHIVgag), and a plasmid lacking the coding sequence of Rev [p(−)] (Fig. 2A) (for details on plasmid construction, see Materials and Methods).
When we cotransfected pHIVgagRRE and pREV into HLtat cells, a large number of HIV Gag-positive cells were detected with a monoclonal α-HIV p24 antibody by immunofluorescence staining (Fig. 2C, panel 1). Control cotransfections with expression plasmids lacking either the Rev coding sequence (Fig. 2C, panel 2) or the RRE (data not shown) were negative for HIV Gag expression. Similarly, when pHKgagRRE and pREV were transfected into HLtat cells, HTDV/HERV-K Gag protein was expressed only when Rev was present (Fig. 2C, panel 3); cotransfections of pHKgagRRE and p(−) (Fig. 2C4) and of pHKgag and either pREV or p(−) (data not shown) were negative for Gag expression as monitored by immunofluorescence using a rabbit or a goat polyclonal antiserum raised against recombinant HTDV/HERV-K Gag. These results clearly demonstrate that HERV-K Gag expression depends on posttranscriptional regulation just as HIV Gag expression depends on a Rev-RRE-like function. The restriction of the nuclear export of HTDV/HERV-K Gag may be due to cis-acting elements as for HIV Gag.
cORF and RcRE, the Rev-RRE homologues of HTDV/HERV-K.
Having shown that regulatory elements such as Rev and RRE are essential for viral gene expression in the ERV family HTDV/HERV-K, we assumed that cORF was the most probable counterpart to Rev. Using a cORF expression vector (Fig. 3B), we confirmed that, just like Rev, cORF was efficiently expressed independently of posttranscriptional regulation. Therefore, the expression of cORF could also serve as a control for transfection efficiency.
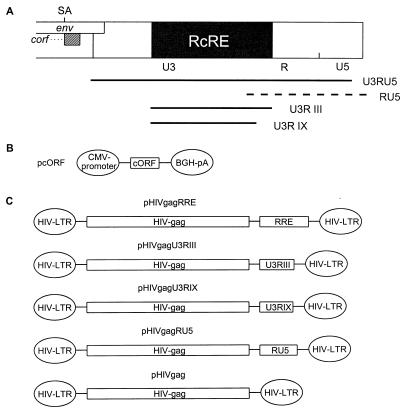
FIG. 3.
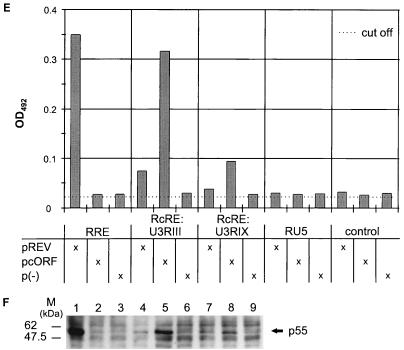
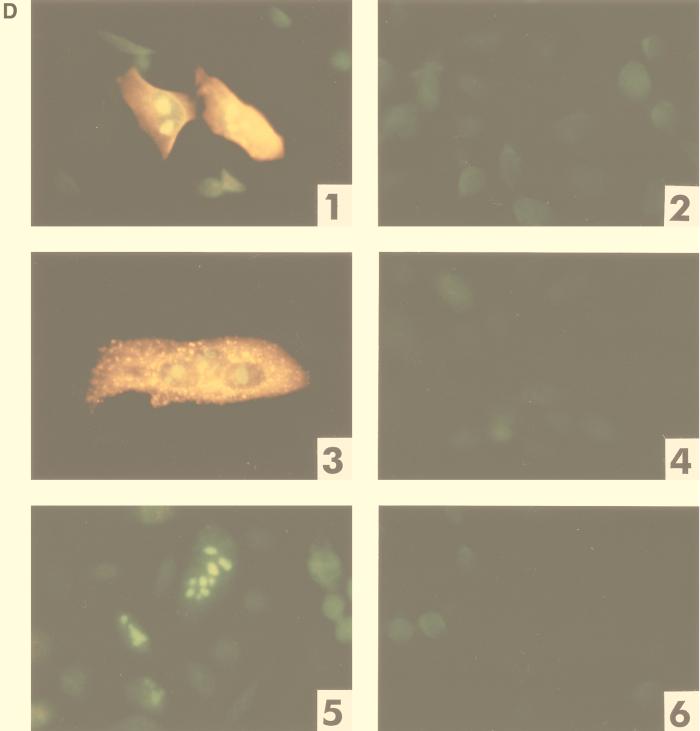
Mapping of the HTDV/HERV-K cORF-responsive element. (A) Mapping of RcRE: genomic localization of 3′-LTR subfragments tested for RRE-like activity in combination with cORF. Fragments depicted as solid lines were active; that depicted by a hatched line was inactive. (B) Mammalian expression vector encoding HTDV/HERV-K cORF (effector plasmid). BGH, bovine growth hormone; CMV, cytomegalovirus; pA, polyadenylation sequence. (C) Mammalian HIV Gag expression vectors containing the HIV RRE or different sections of the HTDV/HERV-K 3′ LTR (U3RIII, nt 8720 to 9149; U3RIX, nt 8720 to 9093; RU5, nt 9065 to 9492) and the negative-control vector lacking an RRE element (reporter plasmid). (D) Cotransfection of reporter and effector plasmids into HLtat cells. Panel 1, pHIVgagU3RIII and pcORF; panel 2, pHIVgagU3RIII and p(−); panel 3, pHIVgagU3RIX and pcORF; panel 4, pHIVgagU3RIX and p(−); panel 5, pHIVgagRU5 and pcORF; panel 6, pHIVgagRU5 and p(−). Staining was performed with a mouse monoclonal α-HIV p24 antibody (panels 1 to 6) (orange) and with a rabbit α-cORF antibody (panels 1, 3, and 5) (green). A filter suitable for both fluorescence markers was used. (E) Determination of the HIV Gag precursor levels in particle preparations by an HIV p24 capture assay (Abbott). HLtat cells were transfected with the vector combinations listed in the table below. The cutoff value was determined by the extinction of the lysis buffer. (F) Detection of the HIV Gag precursor by Western blot analysis. The p55 Gag precursor was present in combination 1 (Rev and RRE), in 5 and 8 (cORF and RcRE), and faintly in 4 and 7 (Rev and RcRE). The positions of molecular mass markers (M) are indicated on the left.
pREV was replaced by pcORF in the test system described above in a first attempt to determine whether cORF could interact with the RRE; however, this experiment failed (data not shown). This was not surprising because many Rev homologues are limited in the use of an unrelated RRE (reviewed in reference 10).
Then we searched for the authentic HTDV/HERV-K RRE equivalent by screening the entire 3′ one-third of the HTDV/HERV-K genome, starting with nt 6481. The HIV Gag expression vector continued to be used as a reporter, but the RRE sequence in pHIVgagRRE was replaced by consecutive segments of the HTDV/HERV-K genome, overlapping by roughly 200 nt. All of these vectors were cotransfected with either pcORF, pREV, or p(−) into HLtat cells.
Using immunofluorescence staining as a primary detection method, significant amounts of HIV Gag were detected only in cotransfections with a reporter plasmid containing the HTDV/HERV-K 3′ LTR (U3RU5) (Fig. 3A) as an RRE substitute and cORF as an effector (data not shown). To define the minimum size of the cORF-responsive element, smaller fragments of the 3′ LTR were tested (Fig. 3A). We mapped the most efficient fragment to the sequence U3RIII (nt 8720 to 9148) (Fig. 3A and C and Fig. 3D, panel 1). The smallest but least efficient cORF-responsive element was mapped to fragment U3RIX (nt 8720 to 9093) (Fig. 3A and C and Fig. 3D, panel 3). Cotransfections of a reporter construct carrying the fragment RU5 (nt 9065 to 9492) (Fig. 3A and C and Fig. 3D, panel 5) with pcORF were negative for HIV Gag expression, as were cotransfections with each of these reporter constructs and p(−) (Fig. 3D, panels 2, 4, and 6).
We further quantitated the degree of HIV Gag expression with a commercial p24 ELISA (HIV AG-1 Monoclonal p24 capture assay; Abbott). Cotransfections were performed with the reporter vector pHIVgagRRE, pHIVgagU3RIII, pHIVgagU3RIX, pHIVgagRU5, or pHIVgag in combination with the effector plasmid pREV, pcORF, or p(−). In 13 independent transfection experiments, Gag expression was analyzed either in cell lysates (data not shown) or in particle preparations purified by ultracentrifugation from the supernatants of transfected-cell cultures, with equivalent results. A typical experiment, shown in Fig. 3E, clearly indicates that U3RIII and the minimal fragment U3RIX contain the cORF-responsive element. The presence of the p55 HIV Gag precursor protein in the positive transfectants was confirmed by Western blot analysis with cell lysates (Fig. 3F).
The data (Fig. 3E and F) also indicate that Rev may substitute for cORF on the HTDV/HERV-K sequences, albeit with a much lower efficiency, confirming observations made in the immunofluorescence studies (data not shown). Rev function on the smaller element, U3RIX, was no longer statistically significant, indicating that Rev may bind to a slightly different sequence. Further analyses will be needed to elucidate this hypothesis and to map the exact Rev binding site.
In summary, these findings unequivocally demonstrate that cORF has a Rev-like function. In contrast to HIV, the cORF-responsive element is not localized in env but lies within the U3R region of the 3′ LTR, resembling the Rex-responsive element RxRE in HTLV-1 (1). By analogy, we designate the HTDV/HERV-K element RcRE (for regulatory cORF-responsive element).
Posttranscriptional regulation of HTDV/HERV-K in full-length constructs.
To study HTDV/HERV-K gene regulation in the context of the viral genome, we relied on a complete coding sequence (lacking the LTRs) which had recently been constructed from overlapping cDNA clones. This sequence has been shown to induce a high level of Gag expression and the formation of virus-like particles in a baculovirus expression system but does not yield any cORF or Env protein, probably due to inefficient mRNA splicing in insect cells (45).
We used this sequence to construct two expression plasmids which both contain the complete HTDV/HERV-K coding sequence but differ by the presence or absence of most of the 3′ LTR and thus in the presence (pHKRcRE) or absence (pHK) of RcRE (Fig. 4A) (for details on plasmid construction, see Materials and Methods). For transfections into primate cell lines in which HTDV/HERV-K mRNAs are not constitutively expressed, like HeLa and COS7 cells, we had to use a foreign promoter, the cytomegalovirus promoter, since HTDV/HERV-K LTRs are active only in teratocarcinoma cell lines (unpublished data). In mammalian cells, in contrast to the baculovirus expression system in insect cells, vectors containing all HTDV/HERV-K genes and splice sites give rise to the formation of cORF transcripts by two splice events (Fig. 1A) (27), alleviating the need to cotransfect pcORF. Again, cORF expression was used to monitor the transfection efficiency.
FIG. 4.
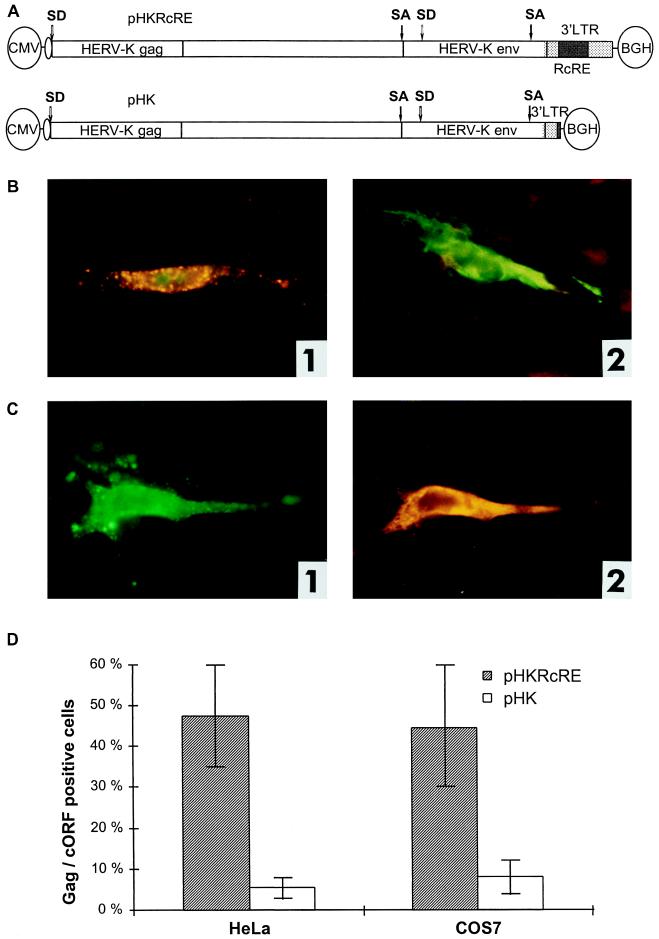
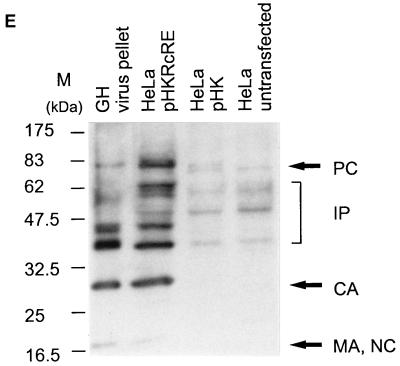
Posttranscriptional regulation of HTDV/HERV-K protein expression in full-length clones. (A) Expression vectors containing the complete HTDV/HERV-K genome with the exception of the 5′ LTR (starting with nt 1012): a cytomegalovirus (CMV) promoter was used to overcome the tissue specificity of HTDV/HERV-K LTRs. In the negative-control vector pHK, the RcRE and the HTDV/HERV-K termination signals were deleted. SA, splice acceptor; SD, splice donor; BGH, bovine growth hormone. (B) Transfections into HeLa cells. Panel 1, pHKRcRE: expression of HERV-K Gag (orange) and cORF (green); panel 2, pHK: expression of cORF (green). Staining was achieved with goat α-HERV-K Gag (panels 1 and 2) (orange) and with rabbit α-cORF (panels 1 and 2) (green). (C) Transfections of pHKRcRE into HeLa cells. Panel 1, expression of HERV-K Gag (green); panel 2, expression of Env (orange). Staining was performed with rabbit α-HERV-K Gag (panel 1) (green) and with goat α-HERV-K Env (panel 2) (orange). The same cell was photographed with an FITC filter to demonstrate Gag expression and with a Cy3 filter to show Env expression. (D) Quantification of Gag- and cORF- double-positive cells after transfection of pHKRcRE (hatched bars) or pHK (white bars) in HeLa (right) and COS7 (left) cells. cORF-positive cells were set as 100%. (E) Western blot analysis of Gag proteins in GH virus pellets (lane 1), in lysates of HeLa cells transfected with pHKRcRE (lane 2), or pHK (lane 3), and in lysates of untransfected HeLa cells (lane 4). PC, precursor; IP, intermediate proteins; CA, capsid; MA, matrix; NC, nucleocapsid. The positions of molecular mass markers (M) are indicated on the left.
Transient transfections of the full-length vectors pHKRcRE and pHK into HeLa and COS7 cells led to the following results. cORF was the protein most efficiently synthesized. In transfections with pHKRcRE, many HeLa and COS7 cells expressed Gag in addition to cORF. In these cells, cORF was localized in the nucleoli as expected. In contrast, in transfections with pHK, very few cells were Gag positive, whereas a massive cytoplasmic cORF expression was detected (Fig. 4B, panels 1 and 2, and data not shown). This unlimited cORF expression indicated that in the absence of the RcRE, splicing was not inhibited and unspliced transcripts were inefficiently exported out of the nucleus (see also below). In addition, Env protein expression was enhanced by the presence of cORF and RcRE. HeLa and COS7 cells transfected with pHKRcRE expressed high levels of Env, whereas cells transfected with pHK did not. Env was predominantly detected in the cells which were positive for Gag (Fig. 4C, panels 1 and 2).
HTDV/HERV-K Gag or Env could not be quantified by ELISA due to the lack of specific assays. Instead, we checked 300 to 400 cORF-positive cells per transfection for concomitant expression of Gag. In transfections of pHKRcRE into either HeLa or COS7 cells, approximately 50% of the cORF-positive cells were also positive for Gag, whereas in transfections with the vector lacking the RcRE, only 5 to 8% of the cORF-positive cells expressed Gag (Fig. 4D). In addition, the amounts of Gag per cell were very small when Gag was detected in pHK transfectants (data not shown). The presence of the cORF-responsive element thus leads, on average, to a 10-fold increase of Gag-expressing cells in HeLa cells and to a 6-fold increase in COS7 cells. In transfections with the vector pHKRcRE, Env protein expression was detected in about 50% of the Gag-positive HeLa cells and in 80% of the Gag-positive COS7 cells.
The presence or absence of Gag was confirmed by Western blot analyses using cell lysates of HeLa cells transfected with pHKRcRE or pHK (Fig. 4E). Gag-specific bands were detected in the cell lysate of pHKRcRE-transfected cells (lane 2), but not in that of pHK-transfected or untransfected control HeLa cells (lanes 3 and 4, respectively). As expected, the Gag precursor was the predominant product in the cell lysates, whereas in a virus pellet prepared from the supernatant of a particle-producing teratocarcinoma cell line (GH) (lane 1), the major products were intermediate, partially cleaved proteins and the capsid protein.
In summary, these findings clearly demonstrate that cORF upregulates the expression of HTDV/HERV-K structural proteins when it can interact with its responsive element, RcRE.
Stabilization and enhanced export of incompletely spliced HTDV/HERV-K mRNAs by cORF and RcRE.
To confirm the Rev-like function of cORF, we analyzed the nuclear export of HTDV/HERV-K transcripts. pHKRcRE and pHK were transfected into HeLa cells, and nuclear and cytoplasmic mRNAs of the transfectants were prepared. To demonstrate the lack of endogenous HTDV/HERV-K expression, nuclear and cytoplasmic mRNAs from untransfected HeLa cells were prepared in the same way. Equal amounts of nuclear and cytoplasmic mRNAs were analyzed by Northern blotting, using a probe specific for the first exon of cORF, which is present in all predicted mRNA species (a typical experiment is shown in Fig. 5A). HTDV/HERV-K-specific transcripts were consistently detected in the transfected HeLa cells, but not in the untransfected HeLa cells. Viral transcripts comprised the three expected species (Fig. 1A): a full-length transcript, a subgenomic env transcript, and transcripts encoding cORF (the transcripts derived from pHK are 1,050 nt shorter due to the lack of an RcRE). The two cORF bands detected in pHKRcRE transfectants are a consequence of two polyadenylation signals present in the construct, one in the HTDV/HERV-K 3′ LTR and the other in the plasmid downstream of the inserted HTDV/HERV-K sequence.
It is obvious that pHKRcRE transfectants contained much more env and full-length mRNAs in the cytoplasmic mRNA fraction than did cells transfected with pHK, while the amounts of doubly spliced cORF mRNA were very similar (Fig. 5A). The same effect was seen in the two nuclear fractions, albeit to a somewhat lesser extent. Rehybridization of the Northern blot with an actin probe indicated that equal amounts of nuclear mRNA (untransfected HeLa cell mRNA being the exception) and cytoplasmic mRNA were loaded onto the gel, with more actin mRNA existing in cytoplasmic fractions than in nuclear fractions. These data demonstrate that the presence of the RcRE significantly enhances the absolute amount of incompletely spliced transcripts.
To quantify these observations, the radioactive signals were counted by phosphorimaging (the two cORF bands in pHKRcRE transfectants were assessed as one). The results were normalized to actin mRNA levels and are depicted in Fig. 5B. In the cytoplasmic mRNA fraction of pHKRcRE-transfected cells, the amount of full-length and env mRNAs was 1.4 times the amount of cORF mRNA; the corresponding factor in pHK-transfected cells was only 0.2. Normalized to cORF mRNA, the amount of full-length and singly spliced mRNA was enhanced by a factor of 7 in the presence of the RcRE. This significant increase in unspliced or incompletely spliced transcripts containing the RcRE in the cytoplasmic fraction reflects enhanced nuclear export, as has been described for Rev and Rex (see, e.g., references 9 and 20).
For the nuclear fractions, the same type of calculation resulted in a factor of 1.4 for RcRE-containing transcripts versus 0.7 for transcripts lacking the RcRE, reflecting a twofold enhancement. This calculation may be hampered by differences in cytoplasmic contamination in nuclear preparations, although their effect should be reduced by normalization to actin mRNA. Even with this precaution in mind, the twofold enhancement may indicate that cORF stabilizes RcRE-containing mRNA in the nucleus, as do Rev and Rex (see, e.g., references 9, 14, and 21). Whether this stabilization depends on prevention of splicing, overcoming instability of sequences, or merely enhanced nuclear export remains to be determined.
Taken together, these findings clearly demonstrate the Rev/Rex-like function of cORF in enhancing stabilization and export of full-length as well as singly spliced transcripts.
cORF-mediated nuclear export is CRM1 dependent.
Next we studied whether cORF uses the same nuclear export pathway as Rev and Rex, that is, the CRM1-dependent pathway. To investigate this, we took advantage of LMB, a specific inhibitor of CRM1, which very efficiently blocks nuclear export mediated by Rev and Rex (16, 48).
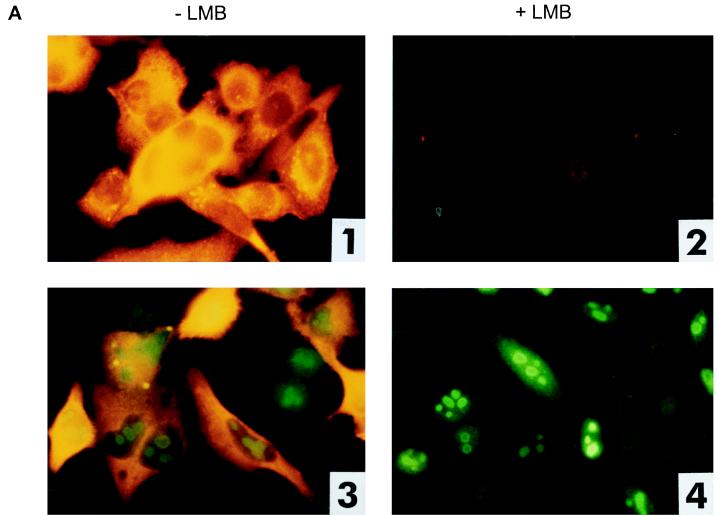
Since LMB is cytotoxic, we determined the optimal concentration of LMB that blocks nuclear export without having toxic effects, using the HIV-based reporter system as an indicator and immunofluorescence as a detection method. We cotransfected pHIVgagRRE or pHIVgagU3RIII with pREV, pcORF, or p(−) into HLtat cells. When LMB was added to the transfection and to the growth medium at a concentration of 2 nM, cORF-RcRE- as well as Rev-RRE-mediated HIV Gag expression was significantly reduced, while almost no effect on cell proliferation was seen (data not shown). At a concentration of 4 to 6 nM LMB, cORF-RcRE- and Rev-RRE-mediated HIV Gag expression was completely blocked (Fig. 6A). Cell viability was examined by staining for cORF. Using 4 to 6 nM LMB for treatment, the cells proliferated slightly more slowly, resulting in a smaller number of cells, but cORF expression in these cells was not dramatically reduced (Fig. 6A, panel 4). In LMB-treated cells, cORF was located in the nucleoli and no cORF was visible in the cytoplasm, indicating that nuclear import was not altered but nuclear export was completely inhibited. cORF was preferentially located at the rim of the nucleolus, a phenomenon which has been described for Rev after treatment with LMB as well (48).
FIG. 6.
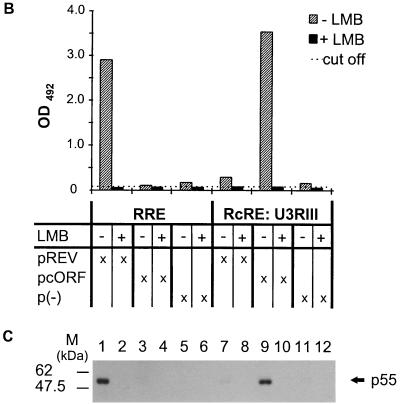
Blocking of cORF-mediated nuclear export by LMB, a CRM1 inhibitor. (A) Cotransfection of reporter and effector plasmids into HLtat cells without (−) or with (+) 4 nM LMB. Panels 1 and 2, pHIVgagRRE and pREV; panels 3 and 4, pHIVgagU3RIII and pcORF. Staining was achieved with a mouse monoclonal α-HIV p24 antibody (all panels) and with a rabbit α-cORF antibody (panels 3 and 4). (B) Determination of the levels of HIV Gag precursor in lysates of cells that were or were not treated with LMB, using the HIV p24 capture assay. HLtat cells were transfected with the vector combinations listed in the table below. Each variant was analyzed without (−) and with (+) 5 nM LMB. The cutoff value was determined by the extinction of the lysis buffer. (C) Detection of the HIV Gag precursor by Western blot analysis. The p55 Gag precursor was present only in the absence of LMB in Rev-RRE combinations (lane 1), cORF-RcRE (lane 9), and (very faintly) Rev-RcRE (lane 7). LMB completely blocked HIV Gag expression at a 5 nM concentration. The positions of molecular mass markers are indicated on the left.
To quantify the effect of 5 nM LMB, HLtat cells were transfected with the same vector combinations, and cell lysates were analyzed by the p24 antigen capture assay. As expected, we could not detect p24 in any of the LMB-treated samples (Fig. 6B). When the total protein contents of LMB-treated and untreated cells were compared, we found that LMB reduced the total amount of cellular protein to about 70%, indicating a slightly inhibitory effect on cell proliferation. At lower concentrations of LMB, the levels of p24 were reduced by 40 to 50% without lowering the total amount of protein in cell lysates (data not shown).
The specificity of the p24 capture assay was verified by Western blotting (Fig. 6C). Only in non-LMB-treated transfectants was the HIV Gag precursor protein p55 present.
We further tested whether the full-length construct pHKRcRE is sensitive to treatment with LMB. In transfections into HeLa cells, pHKRcRE showed a significant reduction in the number of Gag- and Env-positive cells (data not shown), confirming the observation that nuclear export of incompletely spliced HTDV/HERV-K transcripts is CRM1 dependent.
Taken together, these data clearly demonstrate that the cORF-mediated nuclear export of mRNA is inhibited by LMB and that this inhibition is specific. cORF uses the same export pathway as Rev and Rex and other proteins with leucine-rich NESs (13).
DISCUSSION
The HTDV/HERV-K family is a unique group of human endogenous retroviruses in so far as they contain open reading frames for all viral proteins necessary for the formation of retroviral particles. Such particles have been detected in teratocarcinoma cell lines (HTDV particles [5, 24–26]). In these cell lines, HTDV/HERV-K are expressed with an RNA pattern reminiscent of that seen for complex retroviruses (26). One of the predominant proteins (cORF), encoded by a small, doubly spliced mRNA, resembles the viral regulatory proteins Rev and Rex in having an arginine-rich NLS and a leucine-rich NES (27). The presented study demonstrates that the cORF protein, interacting with a responsive element in the HTDV/HERV-K sequence, exerts a Rev-like function.
This conclusion is supported by several lines of evidence. (i) HTDV/HERV-K Gag proteins, like HIV Gag proteins, need a Rev-RRE-like export system for proper expression. (ii) For HIV gag-containing constructs, Rev and RRE can be replaced by cORF and a responsive element in the 3′ LTR of the HTDV/HERV-K mRNA, as demonstrated by immunofluorescence, determination of p24 levels, and Western blotting. This element, designated RcRE, maps to nt 8720 to 9148 in U3R. (iii) With full-length HTDV/HERV-K constructs, the expression of structural proteins is significantly enhanced by the presence of RcRE. (iv) As shown by Northern blot analysis, the amount of cytoplasmic viral mRNA specific for the structural proteins is also significantly increased when the constructs contain RcRE and when cORF is expressed, demonstrating the stabilization and enhanced nuclear export of these mRNAs by cORF and RcRE. (v) The expression of structural proteins is inhibited by LMB, a potent inhibitor of CRM1-mediated nuclear export. This export pathway has been shown to be used by Rev and Rex (4, 16, 23, 39, 48).
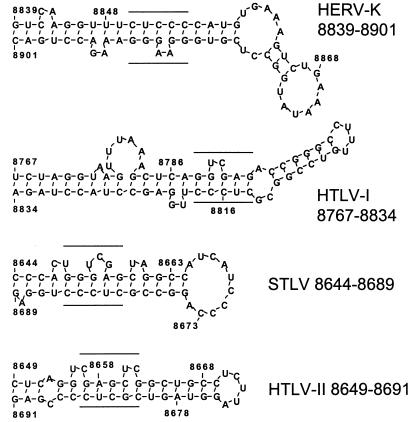
Since RRE and RxRE are complex mRNA structures, the possible existence of similar features in RcRE has been investigated by using the DNASIS RNA secondary structure prediction software. Two adjacent energetically favored stem-loop structures are consistently identified (nt 8839 to 8945). The exact binding sites for Rev and Rex on their responsive elements have been described in detail (see, e.g., references 2, 3, and 22). Rev binds to the sequence 5′UGGGCG/5′CGGUACA, forming a short stem with an internal loop located in stem-loop 2 of the RRE (17). The Rex binding core is mapped to the sequence 5′CUCAGGUCGA(G)/5′(C)UCCCUUGGAG in HTLV-1 (1) and to the sequence 5′GAGCUCG/5′CGCUC in HTLV-2. A putative binding site [5′GGGUCGA(G)/5′CUCCCC] has been identified in simian T-cell leukemia virus (STLV) PH969, a primate T-cell lymphotropic virus isolated from an African baboon (47). In the stem of the first stem-loop structure of RcRE, the sequence 5′CUCCC/5′GGAAGGG shows a striking similarity to the Rex binding domains of the HTLV/STLV group (Fig. 7). Further investigations will aim at high-resolution mapping of the biologically relevant nucleotides to define the core binding sites in RcRE of cORF and of Rev, which seems to be able to replace cORF to some extent. In addition, whether Rex can replace cORF on RcRE or cORF can substitute for Rex on the RxRE will be examined.
FIG. 7.
Comparison of primate RRE sequences. Secondary structures of the experimentally determined (HTLV-1 [HTLV-I] and HTLV-2 [HTLV-II]) or predicted (STLV and HTDV/HERV-K) Rex or cORF binding sites (relevant nucleotides are underlined) were compared by using the algorithm contained in the DNASIS software (Hitachi). Similar to the RxRE binding sites, the putative HTDV/HERV-K binding site is located on a stem-loop structure displaying a characteristic bulge.
HTDV/HERV-K genomes exist in two subgroups which differ by a deletion of 292 nt at the boundary between the pol and env reading frames. This deletion affects the splice acceptor site for the formation of the subgenomic env transcript and, most importantly, removes the first coding exon of cORF (27). The subclass of proviruses harboring the deletion has been designated type 1 in order to credit the first sequencing of a provirus of this family, the HERV-K 10 provirus (38); the subclass of the prototypic complete genomes has been designated type 2 (Fig. 1A) (27). Type 1 proviruses do not code for the cORF protein but contain RcRE. Therefore, their proper expression may still depend on a cORF-RcRE-based nuclear export system. A survey of whole-cell RNA prepared from different human tissues showed widespread expression of full-length type 1 sequences. However, spliced viral mRNAs or viral proteins could not be detected in those tissues (40). One possible explanation is that the nuclear export of viral RNA is disturbed due to the lack of cORF.
Expression of type 2 proviruses, which encode cORF, is mainly restricted to testicular cells. The precise mechanism leading to the differential expression in testicular cells is not known but may involve a cell type-specific regulation of transcription of putative type 2 specific LTRs (unpublished data). The presence of cORF, translated from type 2 specific mRNA, allows the high-level export of spliced and unspliced viral RNAs into the cytoplasm and their subsequent translation into proteins. Type 1 sequences may contribute to the pool of viral cytoplasmic mRNAs and viral proteins expressed in cases in which cORF is provided in trans. Especially in testicular tumor cell lines, both type 1 and 2 proviruses may be a source for upregulation of viral proteins to a level which allows the formation of particles (24). Surprisingly, HTDV/HERV-K proteins, predominantly cORF, can be detected in normal human testis tissue (40). This finding raises the question of whether cORF has taken on some role in the development of testes in the primate lineage. In this case, the persistence of open reading frames in this class of HERVs would be obvious. It will be interesting to study whether dysregulation of cORF expression contributes to the onset or progression of germ cell tumors.
The particles observed in teratocarcinoma cell lines may be solely Gag particles, because Env is present in only limited amounts in the cells as well as in particle preparations (unpublished data). A similar, albeit less dramatic, imbalance can be seen in transient-transfection assays with a full-length HTDV/HERV-K clone: only 50 to 80% of Gag-producing transfectants express Env. These observations do not conform with the pattern expected for a functional cORF-RcRE interaction and may be explained by as-yet-unidentified deficiencies in HTDV/HERV-K proviruses.
In sequence composition, gag and pol genes of HTDV/HERV-K proviruses resemble those of type D viruses, but their Rev-RRE-like posttranscriptional regulation resembles that of the bovine leukemia virus-HTLV and lentivirus genera. Therefore, these endogenous proviruses could be seen as intermediates in the evolution to contemporary primate retroviruses. Bearing this example in mind, ERVs in general can be regarded as remnants of the evolution buried in the genomes of living cells. Uncovering and analyzing these sequences as fossils may extend our understanding of the different steps taken during the evolution of retroviruses. Our findings, for instance, indicate that the strategy of regulation of gene expression by a viral protein which binds to a responsive element may have evolved 40 million years ago and was not first achieved during the recent evolution of contemporary retroviruses. The observation that exogenous retroviruses frequently recombine with endogenous counterparts may indicate that ERVs provide a useful reservoir of genes by which exogenous retroviruses can evolve new functions. One might even cautiously speculate that an HIV predecessor could have picked up the Rev gene by swapping sequences with an HTDV/HERV-K provirus in the course of evolution. However, other hypotheses, like derivation of HTDV/HERV-K from an even more complex ancestor or the independent evolution of strategies for nuclear export, are still possible.
Having shown the Rev-like function of the cORF protein, the abbreviation of the corresponding gene needs a new interpretation: cORF is convergent to Rev in function.
ACKNOWLEDGMENTS
We thank B. Felber and G. N. Pavlakis, Frederick, Md., for providing plasmids and cells; B. Wolff-Winiski, Novartis, for the gift of leptomycin B; and R. R. Tönjes and J. Denner for providing plasmids and antibodies. We are indebted to U. Held, H. Strobel, A. Hornung, H. Bartel, and H. Rahmouni for excellent technical assistance. We thank K. Boller, M. Chudy, J. Denner, J. Hesse, M. Knößl, R. Kurth, M. Marschall, M. Nübling, R. Seitz, and R. R. Tönjes for helpful discussions. We thank I. Plumbaum for expert help in editing the manuscript.
REFERENCES
- 1.Ahmed Y F, Hanly S M, Malim M H, Cullen B R, Greene W C. Structure-function analyses of the HTLV-I Rex and HIV-1 Rev RNA response elements: insights into the mechanism of Rex and Rev action. Genes Dev. 1990;4:1014–1022. doi: 10.1101/gad.4.6.1014. [DOI] [PubMed] [Google Scholar]
- 2.Baskerville S, Zapp M, Ellington A D. High-resolution mapping of the human T-cell leukemia virus type 1 Rex-binding element by in vitro selection. J Virol. 1995;69:7559–7569. doi: 10.1128/jvi.69.12.7559-7569.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogerd H P, Tiley L S, Cullen B R. Specific binding of the human T-cell leukemia virus type I Rex protein to a short RNA sequence located within the Rex-response element. J Virol. 1992;66:7572–7575. doi: 10.1128/jvi.66.12.7572-7575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogerd H P, Echarri A, Ross T M, Cullen B R. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boller K, König H, Sauter M, Müller-Lantzsch N, Löwer R, Löwer J, Kurth R. Evidence that HERV-K is the endogenous retrovirus sequence that codes for the human teratocarcinoma derived retrovirus HTDV. Virology. 1993;196:349–353. doi: 10.1006/viro.1993.1487. [DOI] [PubMed] [Google Scholar]
- 6.Cullen B R. HIV-1 auxillary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 7.D’Agostino D M, Felber B K, Harrison J E, Pavlakis G N. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol Cell Biol. 1992;12:1375–1386. doi: 10.1128/mcb.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Agostino D M, Ciminale V, Pavlakis G N, Chieco-Bianchi L. Intracellular trafficking of the human immunodeficiency virus type 1 Rev protein: involvement of continued rRNA synthesis in nuclear retention. AIDS Res Hum Retroviruses. 1995;11:1063–1071. doi: 10.1089/aid.1995.11.1063. [DOI] [PubMed] [Google Scholar]
- 9.Felber K B, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. rev protein of human immunodeficiency virus type 1 affects the stability and transport of viral mRNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felber K B, Pavlakis G N. Molecular biology of HIV-1: positive and negative regulatory elements important for virus expression. AIDS. 1993;7(Suppl. 1):S52–S62. [PubMed] [Google Scholar]
- 11.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 12.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM 1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 14.Gröne M, Koch C, Grassmann R. The HTLV-1 Rex protein induces nuclear accumulation of unspliced viral RNA by avoiding intron excision and degradation. Virology. 1996;218:316–325. doi: 10.1006/viro.1996.0200. [DOI] [PubMed] [Google Scholar]
- 15.Hadzopoulou-Cladaras M, Felber B K, Cladaras C, Athanassopoulos A, Tse A, Pavlakis G N. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hakata Y, Umemoto T, Matsushita S, Shida H. Involvement of human CRM1 (exportin 1) in the export and multimerization of the Rex protein of human T-cell leukemia virus type 1. J Virol. 1998;72:6602–6607. doi: 10.1128/jvi.72.8.6602-6607.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heaphy S, Dingwall C, Ernberg I, Gait M J, Green S M, Karn J, Lowe A D, Singh M, Skinner M A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990;60:685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- 18.Henderson B R, Percipalle P. Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-β. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 19.Hope T, Pomerantz R J. The human immunodeficiency virus type 1 Rev protein: a pivotal protein in the viral life cycle. Curr Top Microbiol Immunol. 1995;193:91–105. doi: 10.1007/978-3-642-78929-8_5. [DOI] [PubMed] [Google Scholar]
- 20.Inoue J-I, Itoh M, Akizawa T, Toyoshima H, Yoshida M. HTLV-1 Rex protein accumulates unspliced RNA in the nucleus as well as in the cytoplasm. Oncogene. 1991;6:1753–1757. [PubMed] [Google Scholar]
- 21.Kalland K-H, Langhoff E, Bos H J, Göttlinger H, Haseltine W A. Rex-dependent nucleolar accumulation of HTLV-I mRNAs. New Biol. 1991;3:389–397. [PubMed] [Google Scholar]
- 22.Kjems J, Brown M, Chang D D, Sharp P A. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc Natl Acad Sci USA. 1991;88:683–687. doi: 10.1073/pnas.88.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 24.Löwer R, Löwer J, Frank H, Harzmann R, Kurth R. Human teratocarcinomas cultured in vitro produce unique retrovirus-like viruses. J Gen Virol. 1984;65:887–898. doi: 10.1099/0022-1317-65-5-887. [DOI] [PubMed] [Google Scholar]
- 25.Löwer R, Löwer J, Tondera-Koch C, Kurth R. A general method for the identification of transcribed retrovirus sequences (R-U5 PCR) reveals the expression of the human endogenous retrovirus loci HERV-H and HERV-K in teratocarcinoma cells. Virology. 1993;192:501–511. doi: 10.1006/viro.1993.1066. [DOI] [PubMed] [Google Scholar]
- 26.Löwer R, Boller K, Hasenmaier B, Korbmacher C, Müller-Lantzsch N, Löwer J, Kurth R. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Löwer R, Tönjes R R, Korbmacher C, Kurth R, Löwer J. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HDTV/HERV-K. J Virol. 1995;69:141–149. doi: 10.1128/jvi.69.1.141-149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magin C. Ph.D. thesis. Frankfurt am Main, Germany: University of Frankfurt am Main; 1998. [Google Scholar]
- 30.Maldarelli F, Martin M A, Strebel K. Identification of posttranscriptionally active inhibitory sequences in human immunodeficiency virus type 1 RNA: novel level of gene regulation. J Virol. 1991;65:5732–5743. doi: 10.1128/jvi.65.11.5732-5743.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malim M H, Tiley L S, McCarn D F, Rusche J R, Hauber J, Cullen B R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 32.Mayer J, Sauter M, Rácz A, Scherer D, Mueller-Lantzsch N, Meese E. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 1999;21:257–258. doi: 10.1038/6766. [DOI] [PubMed] [Google Scholar]
- 33.Mermer B, Felber B K, Campbell M, Pavlakis G N. Identification of trans-dominant HIV-1 rev protein mutants by direct transfer of bacterially produced proteins into human cells. Nucleic Acids Res. 1990;18:2037–2044. doi: 10.1093/nar/18.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nasioulas G, Zolotukhin A S, Tabernero C, Solomin L, Cunningham C P, Pavlakis G N, Felber B K. Elements distinct from human immunodeficiency virus type 1 splice sites are responsible for the Rev dependence of env mRNA. J Virol. 1994;68:2986–2993. doi: 10.1128/jvi.68.5.2986-2993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neville M, Stutz F, Lee L, Davis I L, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 36.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 37.Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol. 1986;58:937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589–598. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otero G C, Harris M E, Donello J E, Hope T J. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phelps, R. C., J. Denner, R. Löwer, A. Hörlin, K. Boller, R. R. Tönjes, J. Löwer, and R. Kurth. Submitted for publication.
- 41.Pollard V W, Malim M H. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 42.Rosen C A, Terwilliger E, Dayton A, Sodroski J G, Haseltine W A. Intragenic cis-acting art gene responsive sequences of the human immunodeficiency virus. Proc Natl Acad Sci USA. 1988;85:2071–2075. doi: 10.1073/pnas.85.7.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider R, Campbell M, Nasioulas G, Felber B K, Pavlakis G N. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J Virol. 1997;71:4892–4903. doi: 10.1128/jvi.71.7.4892-4903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwartz S, Felber B K, Benko D M, Fenyö E-M, Pavlakis G N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tönjes R R, Boller K, Limbach C, Lugert R, Kurth R. Characterization of human endogenous retrovirus type K virus-like particles generated from recombinant baculoviruses. Virology. 1997;233:280–291. doi: 10.1006/viro.1997.8614. [DOI] [PubMed] [Google Scholar]
- 46.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 47.Van Brussel M, Goubau P, Rousseau R, Desmyter J, Vandamme A-M. Complete nucleotide sequence of the new simian T-lymphotrophic virus, STLV-PH969 from a hamadryas baboon, and unusual features of its long terminal repeat. J Virol. 1997;71:5464–5472. doi: 10.1128/jvi.71.7.5464-5472.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolff B, Sanglier J-J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]