Abstract
The amino acid residues in adenovirus type 5 (Ad5) fiber that interact with its cellular receptor, the coxsackie B virus and Ad receptor (CAR), have not been defined. To investigate this, multiple mutations were constructed in the region between residues 479 and 497 in Ad5 fiber (β-strands E and F and the adjacent region of the DG loop). The effects of these mutations on binding to CAR were determined by use of cell-binding competition experiments, surface plasmon resonance, and direct binding studies. The mutation effects on the overall folding and secondary structure of the protein were assessed by circular dichroism (CD) spectroscopy. Deletions of two consecutive amino acids between residues 485 and 493 abolished high-affinity binding to CAR; the CD spectra indicated that although there was no disruption of the overall folding and secondary structure of the protein, local conformational changes did occur. Moreover, single site mutations in this region of residues with exposed, surface-accessible side chains, such as Thr492, Asn493, and Val495, had no effect on receptor binding, which demonstrates that these residues are not in contact with CAR themselves. This implies the involvement of residues in neighboring loop regions. Replacement of the segment containing the two very short β-strands E and F and the turn between them (residues 479 to 486) with the corresponding sequence from Ad3 (βEFAd3→5 mutation) resulted in the loss of receptor binding. The identical CD spectra for βEFAd3→5 and wild-type proteins suggest that these substitutions caused no conformational rearrangement and that the loss of binding may thus be due to the substitution of one or more critical contact residues. These findings have implications for our understanding of the interaction of Ad5 fiber with CAR and for the construction of targeted recombinant Ad5 vectors for gene therapy purposes.
The first step in human adenovirus (Ad) infection consists of virus-cell recognition and attachment, involving the fiber protein and cell surface receptor(s) (7, 15, 30). A second step of virus internalization through receptor-mediated endocytosis depends on an interaction between the conserved Arg-Gly-Asp (RGD) motif in Ad penton base protein and cell surface integrins (1, 2, 6, 36, 37). These two distinct but cooperative events facilitate Ad uptake into cells and may represent an important determinant of Ad tissue tropism.
The existence of three groups of Ad fiber cell receptors has been suggested (12, 19, 21, 28, 30). The coxsackie B virus and Ad receptor (CAR), a 46-kDa protein, was initially identified as a cellular receptor involved in Ad type 2 (Ad2) and Ad5 attachment (3, 4, 31). In addition to subgroup C Ad fibers, CAR was also shown to bind to subgroup A, D, E, and F Ad fibers but not to subgroup B Ad fibers such as serotype 3 (25, 26) or to the short fiber of subgroup F Ad (28). The CAR extracellular domain is sufficient to allow virus attachment and infection (11, 28), while the transmembrane and intracellular regions appear to be dispensable for these functions (32). The major histocompatibility complex (MHC) class I α2 domain was also proposed as a high-affinity cell receptor for Ad2 and Ad5 fibers (18). However, when expressed in hamster cells, MHC class I allele HLA-A*0201 bound to Ad5 fiber with low affinity (7). When CAR and HLA-A*0201 were coexpressed on the same cell surface, Ad5 fiber bound to a single high-affinity receptor which was CAR (7). Based on these findings, we hypothesized that HLA class I-dependent Ad5 attachment and permissivity may only be observed when there is low or no surface expression of CAR (7). The broad tissue tropism of human serotype C Ads was initially explained on the basis of ubiquitous distribution of fiber cell surface receptor(s) (8). However, the ability of multiple Ad, as well as coxsackie B viruses, to bind to CAR led to the hypothesis that the Ad fiber-receptor interaction is not the sole determinant of viral tissue tropism (28). It has been proposed that the length of the Ad fiber shaft is a major determinant of Ad attachment strategy; eight or fewer β-repeats in the shaft result in attachment being enhanced by an interaction between penton base protein with cell surface integrins (28).
The Ad5 fiber is a trimeric protein which protrudes outward from the adenoviral particle. It is divided into three domains: the tail, which binds to the penton-base; the shaft, whose length varies among various serotypes and which is characterized by a repeating motif of approximately 15 residues (14); and the knob, which is essential and sufficient for the binding of the Ad virion to host cells (17, 40). Ad fiber is also involved in the assembly and/or stabilization of the virion; it is immunogenic and can shut off host cell division (23, 24, 26). Analysis of fiberless Ad species showed that fiber is dispensable for particle formation but necessary for the production of fully infectious and correctly assembled virions (22).
The crystal structure of the knob domain of Ad5 fiber has been determined from recombinant protein expressed in Escherichia coli (17). It is a trimer with a three-bladed “propeller” structure with a central surface depression. The structure of Ad5 fiber knob monomer is an eight-stranded antiparallel β-sandwich composed of two β-sheets, with loops and turns connecting the β-strands (17, 40). The two β-sheets appear to be functionally distinct: the “R-sheet” (strands G, H, I, and D) has been proposed to be involved in receptor recognition, and the “V-sheet” (strands A, B, C, and J) appears to be involved in trimerization (17, 40). Based on the structure and the alignment of sequences of the knob domains from various Ad proteins, two possible receptor binding modes have been proposed for Ad5 fiber knob (40). In the first, receptor binding occurs through an interaction between the cellular receptor and the central surface depression of the Ad5 fiber trimer. Alternatively, binding to the cellular receptor employs the surface formed by the R-sheet and the HI loop on each blade of the trimer. In this case, three receptor-binding sites could be utilized by each fiber trimer.
We recently showed that the R-sheet of Ad5 fiber and the β-strands E and F, or regions close to them, may be involved in Ad5 fiber knob binding to CAR (29). In the present study, we assessed the contribution of the region between amino acid residues 479 and 497 (β-strands E and F and the adjacent region of the DG loop) of the Ad5 fiber knob in receptor recognition.
MATERIALS AND METHODS
Site-directed mutagenesis of Ad5 fiber knob.
Ad5 sequences from nucleotide 32197 to nucleotide 32783, corresponding to the Ad5 fiber knob and 15 residues of the terminal repeating unit of the shaft (40), was synthesized by PCR on Ad5 DNA with the primers 5′-CCC GAA TTC TAT GGG TGC CAT TAC AGT AGG AAA-3′ (5′ oligonucleotide) and 5′-CCC AAG CTT ATT CTT GGG CAA TGT ATG A-3′ (3′ oligonucleotide), which contain an EcoRI and a HindIII site, respectively. The amplified product was ligated in a EcoRI/HindIII-restricted pKK233-3, an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible procaryotic expression vector (Pharmacia), to yield pF5knob. Plasmid pF5knob was used as the basis for the mutagenesis of the Ad5 fiber knob. Single amino acid substitutions, small deletions, and a single domain switch between Ad5 and Ad3 fibers were introduced in pF5knob by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). All mutations and the integrity of the remaining sequence were confirmed by DNA sequencing. The following pairs of complementary oligonucleotide primers were used to define each mutation in Ad5 fiber: (i) dl485-86 (deletion of residues Leu485 and Thr486 corresponding to β-strand F; 5′-AGAAATGGAGATGAAGGCACAGCCTAT and 5′-ATAGGCTGTGCCTTCATCTCCAT TTCT); (ii) dl487-88 (deletion of residues Glu487 and Gly488 in the DG loop; 5′-AATGGAGATCTTACTACAGCCTATACAAAC and 5′ GTTTGTATAGGCTGTAGTAAGATCTCCATT); (iii) dl489-90 (deletion of residues Thr489 and Ala490 in the DG loop; 5′-CTTACTGAAGGCTATACAAACGCTGTT and 5′-AACAGCGTTTGTATAGCCTTCAGTAAG); (iv) dl491-92 (deletion of residues Tyr491 and Thr492 in the DG loop; 5′-AAATCCAACAGCGTTGGCTGTGCCTTCAGTAAG and 5′-CTTACTGAAGGCACAGCCAACGC TGTTGGATTT); (v) dl492-93 (deletion of residues Thr492 and Asn493 in the DG loop; 5′-GGCACAGCCTATGCTGTTGGATTTATG and 5′-CATAAATCCAACAGCATAGGCTGTGCC); (vi) dl494-95 (deletion of residues Ala494 and Val495 in the DG loop; 5′-GCCTATACAAACGGATTTATGCCTAAC and 5′-GTTAGGCATAAATCCGTTTGTATAGGC); (vii) dl496-97 (deletion of residues Gly496 and Phe497 in the DG loop; 5′-ACAAACGCTGTTATGCCTAACCTATCA and 5′-TGATAGGTTAGGCATAACAGCGTTTGT); (viii) Tyr491Gly (substitution of Tyr491 with a glycine residue at position 491; 5′-AACAGCGTTTGTACCGGCTGTGCCTTCAGT and 5′-ACTGAAGGCACAGCCGGTACAAACGCTGTT); (ix) Tyr491Cys (substitution of Tyr491 with a cysteine residue at position 491; 5′-AACAGCGTTTGTGCAGGCTGTGCCTCC and 5′-GAAGGCACAGCCTGCACAAACGCTGTT); (x) Thr492Val (substitution of Thr492 with a valine residue at position 492; 5′-AACAGCGTTGACATAGGCTGTGCCTTC and 5′-GAAGGCACAGCCTATGTCAACGCTGTT); (xi) Thr492Phe (substitution of Thr492 with a phenylalanine residue at position 492; 5′-GAAGGCACAGCCTATTTCAACGCTGTTGGATTT and 5′-AAATCCAACAGCGTTGAAATAGGCTGTGCCTTC); (xii) Asn493Ser (substitution of Asn493 with a serine residue at position 493; 5′-GGCACAGCCTATACAAGCGCTGTTGGATTTATG and 5′-CATAAATCCAACAGCGCTTGTATAGGCTGTGCC); (xiii) Val495Arg (substitution of Val495 with an arginine residue at position 495; 5′-GCCTATACAAACGCTAGAGGATTTATGCCTAAC and 5′-GTTAGGCATAAATCTCCTAGCGTTTGTATAGGC); and (xiv) βEFAd3→5 (substitution of residues 479 to 486 that incorporate β-strands E and F of Ad5 fiber with those of Ad3 fiber; 5′-GACCCAGAATATTGGAAAACTGATCTCGAGCTTAAGTATGAAGGCACAGCCTATACAAAC and 5′-GTTTGTATAGGCTGTGCCTTCATACTTAAGCTCGAGATCAGTTTTCCAATATTCTGGGTC).
Cell-binding competition experiments.
The capacity for cell attachment of recombinant mutant fibers to functional receptors on CHO-CAR cells (3) was estimated from cell-binding competition assays between Ad5Luc3 and recombinant fibers by using the level of luciferase gene expression as the endpoint assay. This assay measures fiber attachment to functional cell surface receptors and dissociates virus attachment from internalization because at low temperatures only virus-cell attachment occurs, whereas endocytosis requires physiological temperatures. Ad5Luc3, obtained from P. Boulanger, is a replication-competent virus which contains the luciferase gene under the control of the simian virus 40 early promoter inserted in the E3 region of the Ad5 genome. Confluent monolayers of cells were preincubated with recombinant proteins (0 to 100 μg/ml, final concentration, in serum-free medium) at 4°C, and the mixture was added to CHO-CAR cells precooled on ice for 10 min prior to the addition of Ad5Luc3 at a multiplicity of infection (MOI) of 10. After incubation for 1 h at 0°C, unabsorbed virus was rinsed off, and the cell monolayers were covered with prewarmed medium, transferred to 37°C, and further incubated at that temperature for 18 h; they were then processed for the luciferase assay. Luciferase activity, expressed in relative light units (RLUs), was assayed in cell lysates by using the luciferase assay system (Promega).
The efficiency and affinity of recombinant wild-type and mutant Ad5 fiber knob proteins for CHO-CAR cell receptors was estimated on lysates of cells (105 cells per sample) infected with Ad5Luc3 in the presence of increasing amounts of recombinant proteins. The efficiency with which mutant Ad5 fiber bound to CHO-CAR cell receptors was assessed by measuring the reduction in luciferase activity in the presence of a large excess of mutant fiber protein. The relative affinities of wild-type and mutant fiber binding to CAR was assessed by measuring the amount of each protein required to achieve 50% of maximal inhibition of luciferase activity (IC50).
Biomolecular interactions between wild-type and mutant Ad5 fibers with soluble CAR as measured using surface plasmon resonance (SPR). (i) Preparation of SPR sensor surface.
Purified soluble recombinant CAR (sCAR) (28) was coupled to the CM5 sensor chip by using the amine coupling reaction according to the manufacturer’s instructions. Briefly, the CM sensor chip was activated with a solution of 50 mM N-hydroxysuccinimide and 200 mM N-ethyl-N′-(dimethylaminopropyl)carbonide to activate the esters on the chip surface. sCAR was then injected over the activated surface until a sufficient quantity had bound to the activated esters on the chip surface. A range of 100 to 6,000 resonance units (RUs) were tested, with immobilization densities of 600 to 1,000 RU used in all of the final experiments. Residual esters were inactivated by injection of 1 M ethanolamine hydrochloride (pH 8.5). The BIAcore System, CM5 sensor chip, and the amine coupling kit were supplied by Biacore AB.
SPR analysis of wild-type and mutant Ad5 fibers.
SPR analysis of immobilized sCAR with recombinant wild-type and mutant Ad5 fiber knobs was performed as follows. All interactions were carried out at 25°C with HBS (10 mM HEPES, pH 7.4; 150 mM NaCl; 3 mM EDTA; 0.005% P-20 surfactant) as the continuous flow buffer at a flow rate of 10 μl/min. A wide range of concentrations (0.1 to 500 μg/ml) of each protein was used as the analyte, with the analyte diluted in HBS and injected for 300 s followed by the injection of HBS for approximately 600 s to monitor the dissociation of the bound analyte. The chip was then regenerated with four 2-s pulses of 1 M MgCl2 to remove the analyte bound to the chip surface. Nonspecific binding of the analyte to the sensor chip surface was assessed by performing sample injections onto a sensor chip with immobilized bovine serum albumin as an irrelevant protein. Nonspecific binding to the sensor chip was assessed by use of an injection onto an uncoated sensor chip. Nonspecific binding was subtracted from the total binding for each case.
Cell-binding assays.
Wild-type and mutant Ad5 fiber attachment to CHO-CAR cells (3) was also assessed by direct binding experiments. 125I labeling of wild-type and mutant Ad5 fibers was performed by using iodination beads (Pierce) via standard methods. Confluent monolayers of 105 CHO-CAR cells in 24-well plates were washed in serum-free medium and prechilled on ice for 1 h. Monolayers were incubated with increasing concentrations of 125I-labeled wild-type and mutant Ad5 fiber knob in the presence (nonspecific binding) and absence (total binding) of a 100-fold excess of unlabelled fiber knob. The cell monolayers were then washed three times with phosphate-buffered saline and, after being removed from tissue culture dishes, the samples were placed into 5 ml of Ecoscint O (National Diagnostics) read in a 1900CA Tri-Carb liquid scintillation analyzer (Packard) to determine the β-counts emitted per minute. Scatchard analysis was performed by using the Prism program as the standard method for determining the dissociation constant (Kd) for each recombinant protein.
Phenotypic analysis of recombinant mutated fibers.
Circular dichroism (CD) measurements were performed on a Jobin-Yvon CD6 spectrophotometer (Longjumeau, France) with cylindrical quartz cells of 0.2-cm path lengths. The spectrophotometer was calibrated for wavelength and ellipticity by using d-10-camphorsulfonic acid. Samples were measured in the concentration range of 0.9 of 1.4 mg/ml in 50 mM sodium perchlorate (pH 6.5) at a constant temperature in a thermostated cell holder. The CD data were analyzed for percentages of α-helix and β-sheet as previously described (27).
Cells and recombinant proteins.
CHO cells and CHO-CAR cells (3) were maintained in alpha-medium (Gibco) supplemented with 10% fetal calf serum. Ad5Luc3 was plaque purified twice and propagated in 293 cells in accordance with established methods (13). Expression and purification of wild-type and mutated Ad5 fiber knobs in E. coli and of sCAR in insect cells was performed as previously described (28, 29, 40).
RESULTS
Selection of mutations.
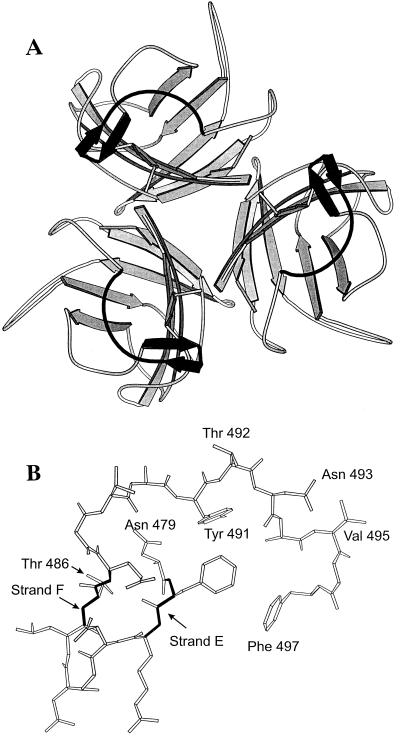
We have previously shown that the β-strands E and F, or regions close to them in Ad5 fiber, may be involved in receptor recognition (29). In addition, monoclonal antibody 7A2.7, an Ad5 neutralizing antibody, blocked Ad5 cell attachment by recognizing an epitope formed in part by β-strands E and F (18). The region between residues Asn479 and Phe497 was therefore extensively mutated to examine its contribution to the interaction between Ad5 fiber knob and CAR. This region of the Ad5 fiber is an extensive surface loop containing many residues that are exposed on the surface of the molecule (e.g., Thr492, Asn493, and Val495) (Fig. 1), as well as residues that contribute to the hydrophobic core (e.g., Tyr491) (Fig. 1) (40).
FIG. 1.
(A) Schematic representation of the trimeric structure of the fiber knob protein, viewed along the threefold axis from the side connected to the fiber shaft. Sheets are indicated by arrows, and the segment in solid black in each protein subunit corresponds to the part of the DG loop (residues 479 to 497) that was mutagenized in these experiments. This region is shown in detail in Fig. 1B. The coordinates were obtained from the crystal structure of Ad5 fiber knob (40). (B) Representation of the conformation adopted in the native structure by the peptide sequence Asn479 to Phe497, showing the short β-strands E and F, and the side chains of residues discussed in the text. Thr492, Asn493, and Val495 residues are all exposed, in contrast to Tyr491, whose bulky aromatic side chain is almost totally buried. For example, the surface accessibility of the Tyr491 side chain was calculated to be 10.2% of the maximum possible compared to the surface accessibility of the adjacent Thr492, which was calculated to be 99.7% of the maximum possible (18a).
We introduced 14 mutations between residues Asn479 and Phe497. These include (i) the substitution of β-strands E and F in the R-sheet of Ad5 fiber with the corresponding β-strands of Ad3 to generate the chimaeric Ad5 fiber knob βEFAd3→5; (ii) six single amino acid substitutions in which residues Tyr491, Thr492, Asn493, and Val495 were mutated; and (iii) seven double deletions, each bearing the deletion of two amino acid residues between Leu485 and Phe497 (dl485-486, dl487-488, dl489-490, dl491-492, dl492-493, dl494-495, and dl496-497). The nucleotide sequence of all mutant fibers was determined; each contained the appropriate mutation without any additional sequence alterations.
All mutant fiber knobs were expressed at high levels in bacteria (∼10 mg/liter). Mutants dl494-495 and dl496-497 were found to be insoluble, presumably because of disruption of the highly conserved region between residues 494 and 505, which includes amino acid side chains that pack between the two β-sheets within the hydrophobic core of the molecule (40). These mutants were therefore excluded from all further studies. The remainder of the mutants all accumulated as soluble trimers (data not shown).
Analysis of wild-type and mutant Ad5 fiber knob binding to CAR.
Cell-binding competition assays SPR, and conventional cell-binding assays were used to determine the consequences of these mutations on Ad5 fiber knob binding to CAR.
(i) Cell-binding competition experiments.
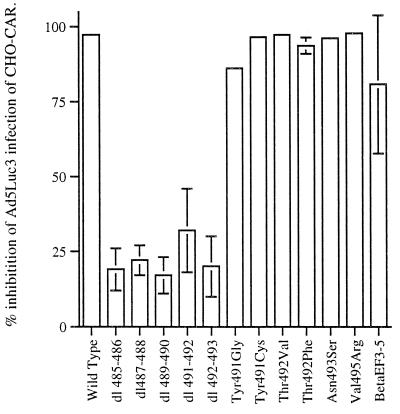
First, the effect of the various mutations on receptor recognition was assessed in cell-binding competition experiments with Ads encoding luciferase (Ad5Luc3) by using CHO-CAR cells. CHO-CAR cells were infected with Ad5Luc3 at a constant MOI (10 PFU/cell), and the efficiency with which mutant Ad5 fiber bound to CHO-CAR cell receptors was evaluated by measuring the reduction in luciferase activity in the presence of maximal concentrations of recombinant proteins (100 μg/ml). In the absence of competing Ad fibers, luciferase gene expression in CHO-CAR cells infected with 10 PFU/cell of Ad5Luc3 was 38,923 RLU compared to 428 RLU obtained with CHO cells infected with Ad5Luc3 at the same MOI. At the highest concentration of wild-type Ad5 fiber, the luciferase activity in CHO-CAR cells was inhibited by 98% (Fig. 2).
FIG. 2.
Reduction in luciferase activity in CHO-CAR cells in the presence of maximal amounts of wild-type and mutant Ad5 fiber knobs. Cells were infected at constant MOIs of Ad5Luc3 (MOI = 10) in the presence of large excess of recombinant full-length Ad5 fiber proteins (100 μg/ml). Ad5Luc3 was preincubated with recombinant proteins at room temperature, and the mixture was added to CHO-CAR cells precooled on ice. After incubation for 1 h at 0°C, unabsorbed virus was rinsed off, and the cell monolayers were covered with prewarmed medium, transferred to 37°C, and further incubated at that temperature for 18 h; they were then processed for the luciferase assay. The luciferase activity, expressed in RLUs, was assayed in cell lysates by using luciferase substrate solution. Results were expressed as percentages of the control cells (i.e., no recombinant fiber = 100%). The data presented are means and standard errors of the means (n = 3) of three representative experiments.
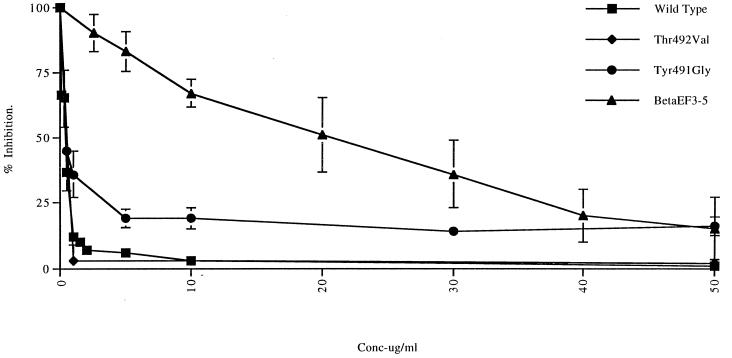
The most marked effect on the efficiency of the interaction between Ad5 fiber knobs and CHO-CAR cell receptors was observed with the five deletion mutants. These failed to compete efficiently with adenovirions at concentrations of up to 100 μg/ml, which would indicate that each of these deletions significantly reduced Ad5 fiber knob binding to CAR (Fig. 2). Two other fiber mutants, the chimeric fiber knob βEFAd3→5 and Tyr491Gly had a less-profound effect on Ad5 fiber knob binding to CHO-CAR cell receptors. When used in large excess (100 μg/ml), both mutants reduced luciferase activity by approximately 80%, which indicates that they bound to CAR efficiently (Fig. 2). However, the IC50 of βEFAd3→5 (21 μg/105 cells) was 180-fold higher than the IC50 of wild-type protein (0.115 μg/105 cells), while the IC50 of Tyr491Gly (0.25 μg/105 cells) was 2-fold higher (Fig. 3). This would suggest that both mutant fiber knobs bound to CHO-CAR cell receptors with reduced affinity. In contrast, mutant fiber knob proteins Tyr491Cys, Thr492Val, Thr492Phe, Asn493Ser, and Val495Arg appeared to bind to CAR with the same efficiency and affinity as the wild-type protein since, in the presence of large excess of each of these four mutants, luciferase activity was reduced by 95%, a value comparable to the reduction observed in the presence of the same concentration of Ad5 fiber knob (Fig. 2). The IC50s of Tyr491Cys (0.13 μg/105 cells), Thr492Val (0.09 μg/105 cells) (Fig. 3), Thr492Phe (0.124 μg/105 cells), Asn493Ser (0.11 μg/105 cells), and Val495Arg (0.135 μg/105 cells) were similar to the IC50 of wild-type Ad5 fiber for CHO-CAR cell receptors.
FIG. 3.
Inhibition of luciferase activity in CHO-CAR cells infected with Ad5Luc3 (MOI = 10) in the presence of increasing concentrations (0 to 50 μg/ml) of wild-type Ad5 fiber knob and of mutant fibers Tyr491Gly, Thr492Val, and βEFAd3→5. Luciferase activity was assayed as described in Fig. 2. The IC50 for wild-type and mutant fibers was obtained from individual curves by calculating the amount of each protein required to achieve 50% of maximal inhibition of luciferase activity.
(ii) SPR analysis of wild-type and mutant Ad5 fibers.
SPR was successfully utilized in previous studies to define contact residues in protein-protein interactions that involve a large interface (5, 16). Therefore, we used this approach to study the interaction between Ad5 fiber knob and CAR. Assays were performed with the soluble extracellular domains of CAR (sCAR), which were immobilized on the sensor surface. Binding of the Ad5 fiber knob, used as the analyte, to sCAR was shown to be specific, saturable, and concentration dependent (data not shown). This demonstrates that the extracellular domains of CAR bind to Ad5 fiber knob in the absence of other membrane cofactors.
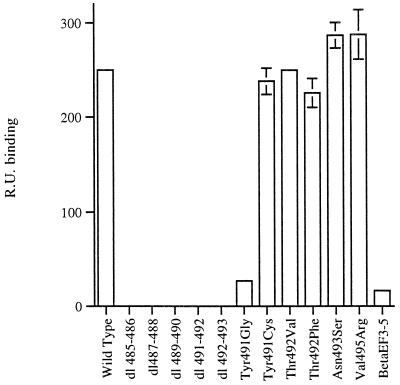
The effect of the different mutations on the affinity of Ad5 fiber knob binding to sCAR was assessed by analysis of the sensograms generated for the interaction between each of the mutants and the immobilized sCAR. The maximal response at equilibrium (which was attained in all cases), as measured in RUs for each mutant, was compared to the response obtained for wild-type Ad5 fiber knob bound to sCAR (Fig. 4). We found that all deletion mutants that failed to compete efficiently with adenovirions for CHO-CAR cell receptors failed to bind to sCAR, as assessed by SPR (Fig. 4). However, the fact that these mutants reduced Ad5Luc3 infection of CHO-CAR cells by up to 25% (Fig. 2) would indicate that they may still bind to CAR with very low affinity. Fiber mutants βEFAd3→5 and Tyr491Gly showed a reduced level of binding to sCAR (15- and 9-fold lower levels, respectively), whereas all other mutant fiber knobs bound at a level comparable to the wild-type protein (Fig. 4).
FIG. 4.
SPR analysis of the biomolecular interactions between the soluble extracellular domains of CAR (sCAR) and wild-type and mutant fiber knobs. sCAR was immobilized on the CM5 chip. The signal correlates with the degree of surface binding of the fiber knob protein to the immobilized sCAR and was expressed in RU. Sensograms were used to obtain a quantitative indication of biomolecular interactions in RUs.
(iii) Conventional cell-binding assays.
The kinetics of the binding of wild-type fiber knob and of mutant fiber knobs was assessed further in conventional binding experiments. Four representative fiber knob mutants (dl491-492, Tyr491Gly, βEFAd3→5, and Thr492Val) were selected for kinetic analysis. We found that mutant dl491-492 failed to bind specifically to CHO-CAR receptors, a result which is in accordance with our observation that this mutant also failed to bind to sCAR and did not compete efficiently with adenovirions for CHO-CAR cell receptors. Mutant Thr492Val bound to CHO-CAR receptors with similar affinity (Kd = 1.2 × 10−9 M) to that of wild-type fiber knob (Kd = 4.75 × 10−9 M). In contrast, Tyr491Gly bound to CHO-CAR cell receptors with 10-fold lower affinity (Kd = 3.6 × 10−8 M), and βEFAd3→5 bound with markedly lower affinity than the wild-type protein (Kd = 10−7 M). These findings are in agreement with the results from cell-binding competition experiments and SPR analysis.
Structural analysis of mutant Ad5 fiber knobs.
The effect of the mutations described above on the interaction between Ad5 fiber knob and CAR may be due to deletion of residues in contact with CAR or to an indirect effect upon the conformation of distant residues that makes contact with the receptor. The consequence of these mutations on the folding and secondary structure of the Ad5 fiber knob were therefore analyzed by monitoring the ability of mutant fibers to form stable trimers and also by CD spectroscopy.
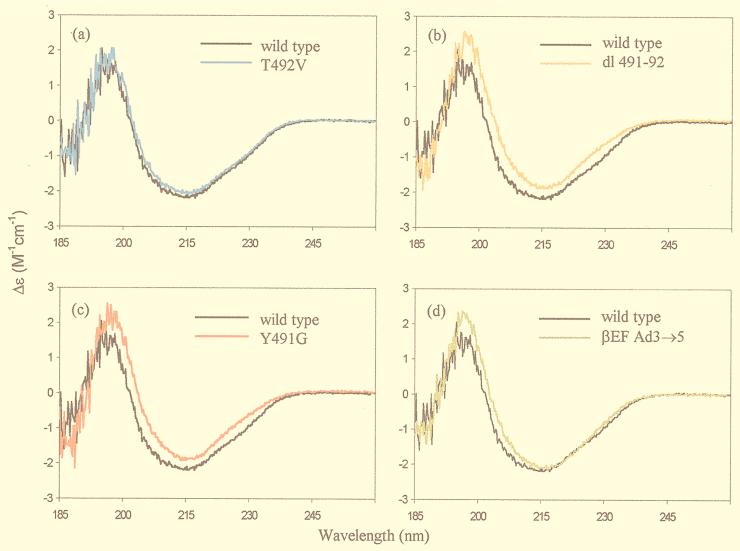
All mutants formed stable trimers as determined by gel filtration chromatography and native gel electrophoresis. The wild-type and mutant fiber knobs were distributed by gel filtration in a single sharp peak at concentrations of from 50 to 500 μg/ml (data not shown). This result shows that these mutations had no significant effect on the affinity of self-assembly of wild-type and mutant fiber knob trimers. We used CD spectroscopy to assess the structure of three types of Ad5 fiber knob mutants. Mutants analyzed by CD spectroscopy included those that bound to CAR normally (Thr492Val, Asn493Ser, and Val495Arg), mutants that bound with reduced affinity (Tyr491Gly and βEFAd3→5), and mutants that showed no specific binding to CAR (dl485-486, dl487-488, dl489-490, dl491-492, and dl492-493). The secondary structure of Ad5 fiber knob monomer is primarily a β-sheet, with connecting loops and turns and no α-helix (15, 37). The positive signal at 203 nm and the negative signal at 215 nm, observed in the spectrum of the wild-type Ad5 fiber knob (Fig. 5), are in accord with the known structure. The percent α-helix and β-sheet for each protein showed small differences (up to 5%, the error of the measurements being up to 10%) between wild-type and some mutant fiber knobs, indicating that the β-sheet structure of all mutants had not been significantly disrupted (27). In support of this, the CD spectra of all the Ad5 fiber mutants examined was almost identical to that of the wild-type protein over the entire recorded spectrum (Fig. 5). The spectra for Thr492Val, Asn493Ser, and Val495Arg were identical to wild-type protein (e.g., Thr492Val [Fig. 5a]). However, slight but significant differences were observed for the double deletions (e.g., dl491-492 [Fig. 5b]) and the Tyr491Gly mutation (Fig. 5C). There was a shift in the position of the peaks at ca. 200 nm, which indicates changes in the secondary structure of the protein. In addition, characteristic changes at ∼230 nm may reflect conformational changes involving buried aromatic residues. The mutation βEFAd3→5 appeared to cause no disruption (Fig. 5d), suggesting that despite the large number of nonconservative substitutions, the overall fold in this region may be unaffected. This would suggest that the region involved in this domain switch may contain residues in contact with CAR.
FIG. 5.
Comparison of the CD spectrum of wild-type Ad5 fiber knob with those of mutant fibers (a) Thr492Val (a), Tyr491Gly (b), βEFAd3→5 (c), and dl491-492 (d). All samples were measured in the concentration range of 0.9 to 1.4 mg/ml in 50 mM sodium perchlorate (pH 6.5) in a 0.2-cm-path-length cell at a constant temperature.
DISCUSSION
The receptor binding sites in Ad5 fiber knob have not yet been identified. In this study, multiple mutations were constructed in the region between residues Asn479 and Phe497 in order to assess the contribution of this region to receptor recognition. This region, which corresponds to β-strands E and F and part of the DG loop, is located at the base of the knob structure (Fig. 1A). It contains residues that are predicted to lie on the outer surface of the knob trimer (40) and is highly antigenic (10). This region of the protein was selected for detailed mutagenesis after deletion analysis of putative receptor binding regions of the protein indicated that β-strands E and F, or a region close to them, may be involved in binding to CAR (29). In addition, indirect evidence from epitope mapping of two monoclonal antibodies that neutralized Ad5 infection by blocking the virus-cell attachment interaction indicated that a region spanning residues 438 to 486 contained sequences required for receptor binding (18).
We found that all of the deletions of two consecutive amino acids between residues 485 and 493 abolished high-affinity binding to CAR, suggesting that the reorganization of this loop region that must result in each case has an effect, direct or indirect, upon residues involved in receptor binding. However, the fact that the resulting proteins form stable, soluble trimers, as assessed by gel filtration chromatography and native gel electrophoresis, and display CD spectra that are almost identical to wild-type protein shows that they are correctly folded and that loss of receptor binding is not due to global disruption of the structure. However, single site mutations in this region of residues with exposed, surface-accessible side chains, such as Thr492, Asn493, and Val495 (Fig. 1B), had no effect on receptor binding. Also, none of these mutations had any effect upon the CD spectra and therefore no effect on the conformation of the protein (e.g., Thr492Val [Fig. 5a]). It is therefore likely that these are not contact residues themselves.
Mutation of the adjacent Tyr491 to glycine did reduce binding, but the tyrosine side chain is almost entirely buried in the native structure and deletion of the bulky, hydrophobic side chain would be expected to cause a local destabilization and conformational rearrangement. In fact, the CD spectrum for Tyr491Gly shows small but significant difference in curve shape at 230 nm from the CD spectrum of the wild-type protein that is characteristic of changes to the environment of buried aromatic residues (Fig. 5c). Interestingly, the double deletions had the same effect on the CD spectra (Fig. 5b) as did the Tyr491Gly mutation, perhaps reflecting some local structural rearrangement. When replaced by cysteine, another hydrophobic residue of similar size to tyrosine, receptor binding was unaffected, a finding consistent with this interpretation.
Taken together, these results suggest that contact residues for receptor binding must lie in a region of the structure immediately adjacent to the part of the DG loop between residues 485 to 493. The E and F β-strand region is clearly one possibility, and replacement of the segment containing these two very short strands and the turn between them (residues 479 to 486) with the corresponding sequence from Ad3 did result in a loss of receptor binding. However, all of these residues differed between the two sequences, and many were nonconservative substitutions. Yet the almost identical CD spectra for wild-type and βEFAd3→5 protein (Fig. 5d) show that these substitutions may have caused no conformational rearrangement and that the loss of binding may therefore be due to the substitution of one or more critical contact residues. Single-site mutagenesis is being carried out in this region to explore this possibility, as well as in other adjacent loop regions that may have been affected by local perturbations caused by the deletion mutants.
Our results have potential implications for the construction of novel Ad vectors for targeted gene delivery. Attempts in this direction have so far involved the coupling of an asialoglycoprotein-polylysine conjugate to wild-type Ad5 (39); the insertion of a 10-amino-acid peptide linker sequence and the N-terminal decapeptide of the gastrin releasing peptide (25), polylysine (34), or a high-affinity RGD motif (38) at the carboxyl terminus of Ad5 fiber protein; and the insertion of heterologous peptides in the HI loop of the fiber knob (20). In addition, a bispecific antibody with primary specificity to αv integrin and a second specificity to an epitope incorporated into the penton base coat protein (34, 35) and bispecific antibodies that simultaneously block binding of Ad5 fiber to its cellular receptor and target-specific cell surface molecules (9, 33) have also been used. The interaction between these chimeric fiber proteins or recombinant Ad5 vectors and CAR was not addressed. Genetic modification of the fiber knob of Ad5 to abolish binding to CAR will constitute an important alternative to these strategies. Our finding that mutations within the Ad5 fiber knob can abolish binding to CAR without adversely affecting the secondary structure and overall conformation of the protein represents an important step in this direction. In addition, by identifying such mutations, it should be possible to dissect receptor binding from other biological functions of the fiber protein. Moreover, the contribution of primary attachment to viral tropism could be assessed by the construction of recombinant Ad bearing mutations in the fiber gene that abolish binding to CAR.
ACKNOWLEDGMENTS
We thank J. M. Bergelson and R. W. Finberg for CHO-CAR cells and S. S. Hong and P. Boulanger for Ad5Luc3.
This work was funded by grants from the Wellcome Trust and the Special Trustees for Guy’s and St. Thomas’ Hospitals to G. Santis. B. J. Sutton and A. J. Beavil also thank the Wellcome Trust for its support.
REFERENCES
- 1.Bai M, Campisi L, Freimuth P. Vitronectin receptor antibodies inhibit infection of HeLa and A549 cells by adenovirus type 12 but not by adenovirus type 2. J Virol. 1994;68:5925–5932. doi: 10.1128/jvi.68.9.5925-5932.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belin M, Boulanger P A. Involvement of cellular adhesion sequences in the attachment of adenovirus to the HeLa cell surface. J Gen Virol. 1994;74:1485–1497. doi: 10.1099/0022-1317-74-8-1485. [DOI] [PubMed] [Google Scholar]
- 3.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson J M, Krithivas A, Celi L, Droguett G, Horwitz M S, Wickham T J, Crowell R L, Finberg R W. The murine CAR homolog is a receptor for coxsackie B viruses and adenoviruses. J Virol. 1998;72:415–419. doi: 10.1128/jvi.72.1.415-419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook J P D, Henry A J, McDonnell J M, Owens R J, Sutton B J, Gould H J. Identification of contact residues in the IgE binding sites of human FceRIa. Biochemistry. 1997;36:15579–15588. doi: 10.1021/bi9713005. [DOI] [PubMed] [Google Scholar]
- 6.Davison E, Diaz R M, Hart I R, Santis G, Marshall J F. Integrin α5β1-mediated adenovirus infection is enhanced by the integrin activating antibody TS2/16. J Virol. 1997;71:6204–6207. doi: 10.1128/jvi.71.8.6204-6207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davison E, Kirby I, Elliot T, Santis G. The human HLA-AO201 allele, when expressed in hamster cells, is not a high-affinity receptor for adenovirus type 5 fiber. J Virol. 1999;73:4513–4517. doi: 10.1128/jvi.73.5.4513-4517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Defer C, Belin M-T, Caillet-Boudin M-L, Boulanger P A. Human adenovirus-host cell interactions: comparative study with members of subgroups B and C. J Virol. 1990;64:3661–3673. doi: 10.1128/jvi.64.8.3661-3673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas J, Rogers B, Rosenfeld M, Michael S, Feng M, Curiel D. Targeted gene delivery by tropism-modified adenoviral vectors. Nat Biotechnol. 1996;14:1574–1578. doi: 10.1038/nbt1196-1574. [DOI] [PubMed] [Google Scholar]
- 10.Fender P, Kidd A, Brebant R, Oberg M, Drouet E, Chroboczek J. Antigenic sites on the receptor-binding domain of human adenovirus type 2 fiber. Virology. 1995;214:110–117. doi: 10.1006/viro.1995.9949. [DOI] [PubMed] [Google Scholar]
- 11.Freimuth P, Springer K, Berard C, Hainfeld J, Bewley M, Flanagan J. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J Virol. 1999;73:1392–1398. doi: 10.1128/jvi.73.2.1392-1398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gall J, Kass-Eisler A, Leinwand L, Falck-Pedersen E. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J Virol. 1996;70:2116–2123. doi: 10.1128/jvi.70.4.2116-2123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham F, Prevec L. Adenovirus based expression vectors and recombinant veccines. In: Ellis R, editor. Vaccine: new approaches to immunological problems. London, United Kingdom: Butterworth-Heinemann; 1992. pp. 363–390. [DOI] [PubMed] [Google Scholar]
- 14.Green N M, Wringley N G, Russell W C, Martin S R, MacLachlan A D. Evidence of a repeating cross-beta sheet structure in the adenovirus fibre. EMBO J. 1983;2:1357–1365. doi: 10.1002/j.1460-2075.1983.tb01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennache B, Boulanger P. Biochemical study of KB-cell receptor for adenovirus. Biochem J. 1977;166:237–247. doi: 10.1042/bj1660237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry A J, Cook J P D, McDonnell J M, McCay G A, Shi J, Sutton B J, Gould H J. Identification of contact residues in the IgE binding site of human Fcepsilon RIalpha. Biochemistry. 1997;36:15568–15578. doi: 10.1021/bi9713005. [DOI] [PubMed] [Google Scholar]
- 17.Henry L J, Xia D, Wilke M E, Deisenhofer J, Gerard R D. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J Virol. 1994;68:5239–5246. doi: 10.1128/jvi.68.8.5239-5246.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong S S, Karayan L, Tournier J, Curiel D T, Boulanger P A. Adenovirus type 5 fiber knob binds to MHC class I alpha 2 domain at the surface of human epithelial and B lymphoblatoid cells. EMBO J. 1997;16:2294–2306. doi: 10.1093/emboj/16.9.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Hubbard S J, Thornton J M. NACCESS computer program. London, United Kingdom: University College London; 1999. [Google Scholar]
- 19.Karayan L, Gay B, Gerfaux J, Boulanger P A. Oligomerization of recombinant penton base of adenovirus type 2 and its assembly with fiber in baculovirus-infected cells. Virology. 1994;202:782–795. doi: 10.1006/viro.1994.1400. [DOI] [PubMed] [Google Scholar]
- 20.Krashnykh V, Dmitriev I, Mikheeva G, Millers C R, Belousova N, Curiel T D. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J Virol. 1998;72:1844–1852. doi: 10.1128/jvi.72.3.1844-1852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krasnykh V, Mikheeva G, Douglas J, Curiel D. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Legrand V, Spehner D, Schlesinger Y, Settelen N, Pavirani A, Mehtali M. Fiberless recombinant adenoviruses: virus maturation and infectivity in the absence of fiber. J Virol. 1999;73:907–913. doi: 10.1128/jvi.73.2.907-919.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levine A, Ginsberg H. Mechanism by which fiber antigen inhibit multiplication of type 5 adenovirus. J Virol. 1967;1:747–757. doi: 10.1128/jvi.1.4.747-757.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mautner V, Willcox H. Adenovirus antigens: a model system in mice for subunit vaccination. J Gen Virol. 1974;25:325–336. doi: 10.1099/0022-1317-25-3-325. [DOI] [PubMed] [Google Scholar]
- 25.Michael S I, Hong J S, Curiel D T, Engler J A. Addition of a short peptide ligand to the adenovirus fiber protein. Gene Ther. 1995;2:660–668. [PubMed] [Google Scholar]
- 26.Novelli A, Boulanger P A. Deletion analysis of functional domains in baculovirus-expressed adenovirus type 2 fiber. Virology. 1991;185:365–376. doi: 10.1016/0042-6822(91)90784-9. [DOI] [PubMed] [Google Scholar]
- 27.Provencher S W, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 28.Roelvink P W, Lizonova A, Lee J G M, Li Y, Bergelson J M, Finberg R W, Brough D E, Kovesdi I, Wickham T J. The coxsackie-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santis G, Legrand V, Hong S, Davison E, Kirby I, Imler J-L, Bergelson J M, Finberg R W, Mehtali M, Boulanger P A. Molecular determinants and serotype specificity of Ad5 fiber binding to its high affinity receptor CAR. J Gen Virol. 1999;80:1519–1527. doi: 10.1099/0022-1317-80-6-1519. [DOI] [PubMed] [Google Scholar]
- 30.Svensson U, Persson R, Everitt E. Virus-receptor interaction in the adenovirus system: identification of virion attachment proteins of the Hela cell plasma membrane. J Virol. 1981;38:70–81. doi: 10.1128/jvi.38.1.70-81.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomko R, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Bergelson J M. Coxsackievirus and adenovirus receptor cytoplasmic and transmembrane domains are not essential for coxsackievirus and adenovirus infection. J Virol. 1999;73:2559–2562. doi: 10.1128/jvi.73.3.2559-2562.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watkins S J, Mesyanzhinov V V, Kurochkina L P, Hawkins R E. The ‘adenobody’ approach to viral targeting: specific and enhanced adenoviral gene delivery. Gene Ther. 1997;4:1004–1012. doi: 10.1038/sj.gt.3300511. [DOI] [PubMed] [Google Scholar]
- 34.Wickham T J, Roelvink P, Brough D, Kovesdi I. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14:1570–1578. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 35.Wickham T, Segal D, Roelvink P, Carrion M, Lizonova A, Lee G, Kovesdi I. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J Virol. 1996;70:6831–6838. doi: 10.1128/jvi.70.10.6831-6838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wickham T J, Filardo E J, Cheresh D A, Nemerow G R. Integrin αvβ5 selectively promotes adenovirus cell membrane permeability. J Cell Biol. 1994;127:257–264. doi: 10.1083/jcb.127.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins αv β3 and αvβ5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 38.Wickham T J, Tzeng D M, Roelvink P W, Li Y, Lee G M, Brough D E, Lizonova A, Kovesdi I. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu G Y, Zhan P, Sze L L, Rosenberg A R, Wu C H. Incorporation of adenovirus into a ligand-based DNA carrier system results in retenion of original receptor specificity and enhances targeted gene expression. J Biol Chem. 1994;269:11542–11546. [PubMed] [Google Scholar]
- 40.Xia D, Henry L, Gerard R, Desenhofer J. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure. 1994;2:1259–1270. doi: 10.1016/s0969-2126(94)00126-x. [DOI] [PubMed] [Google Scholar]