Abstract
Glycoprotein D (gD) of herpes simplex virus type 1 (HSV-1) was modified to encode targeting signals known to localize proteins to either the endoplasmic reticulum (ER) or the trans-Golgi network. These motifs conferred the predicted targeting properties on gD in transfected cells as judged by immunofluorescence staining, and the exclusion of targeted gD from the cell surface was confirmed by the fact that these molecules exhibited substantially reduced activity in cell-cell fusion assays. Recombinant viruses expressing Golgi-targeted forms of gD grew to wild-type levels in noncomplementing cells, exhibited unaltered particle/infectivity ratios, and were found to contain wild-type levels of gD, whereas a recombinant expressing ER-retained gD was helper cell dependent and, when grown on noncomplementing cells, produced virions of low specific infectivity with greatly reduced levels of gD. These data imply that HSV-1 acquires its final membrane from a post-ER compartment and lend support to the view that the virus undergoes de-envelopment and reenvelopment steps during virus egress.
Herpesvirus nucleocapsids assemble in the nuclei of infected cells and acquire an envelope by budding through the inner nuclear membrane, but the subsequent route of virus maturation and egress is uncertain. Johnson and Spear (23) reported that monensin treatment of herpes simplex virus type 1 (HSV-1)-infected cells prevented the maturation of viral glycoproteins and the secretion of virus particles but did not abrogate the production of intracellular infectivity. This suggested that, after budding through the inner nuclear membrane, the enveloped virions are infectious, and the authors therefore argued that these virions are transported via the secretory pathway to the cell surface and that the envelope glycoproteins are processed in situ. This model of HSV-1 maturation following a single envelopment event has the virtue of economy and receives support from some electron-microscopic studies (36). Proponents of this model argue that the naked nucleocapsids that are frequently observed in the cytoplasm of infected cells must represent aberrant fusion events and are dead ends, a view which is supported by the properties of an HSV-1 mutant which accumulates large numbers of cytoplasmic nucleocapsids (8).
An alternative pathway, originally proposed by Stackpole (32), involves the release of naked nucleocapsids into the cytoplasm by fusion of “primary enveloped virions” with the outer nuclear membrane. Final envelopment then occurs by budding into a cytoplasmic compartment. This route of alphaherpesvirus maturation is supported by a number of electron-microscopic studies which have been interpreted as showing final envelopment by budding into a late Golgi compartment or into cytoplasmic vesicles (18, 19, 24, 40) and by the claim that increased numbers of naked nucleocapsids are observed in infected cells treated with brefeldin A (11). Furthermore, the phospholipid composition of secreted virions most closely resembles that of Golgi membranes (38), a finding difficult to reconcile with the “single-envelopment” route of virion egress. A prediction of this two-stage envelopment model is that the envelope proteins of alphaherpesviruses accumulate in the Golgi compartment or in Golgi-derived vesicles where final envelopment occurs. Glycoprotein E (gE) of HSV-1 and its homologues in varicella-zoster virus (VZV) and pseudorabies virus (PRV) contain endocytosis motifs and after internalization are sorted to the trans-Golgi network (TGN) (1, 34, 35, 42, 43), but localization of other virion membrane proteins to this compartment has not been observed.
The resolution of this controversy is of some consequence in defining future cell-biological questions relating to alphaherpesvirus replication. If the single-envelopment route is correct, then all the tegument proteins must accumulate and assemble in the nucleus and all the virion membrane proteins must presumably traverse the nuclear pore to the inner nuclear membrane. If the two-stage process is correct, then the same proteins must accumulate and assemble in a late cytoplasmic compartment. If the single-stage envelopment route is correct, then we know the composition of enveloped viruses in the periplasmic space: it is the same as that of mature virions. If the two-stage process is correct, then our knowledge of the enveloped virion in the periplasmic space is superficial. Indeed Granzow et al. (19) consider that the structure of the tegument in PRV virions in the periplasmic space is quite different from its structure in virions found in cytoplasmic vesicles. Elliott and O’Hare (14) studied the localization of the abundant tegument protein VP22 in live infected cells and reported that the protein was present exclusively in the cytoplasm, a finding which strongly supports final envelopment in a cytoplasmic compartment. Others, however, report the presence of VP22 in the nucleus during the productive phase of infection (31). The evidence for and against the two alternative routes of alphaherpesvirus egress and maturation has recently been reviewed comprehensively by Enquist et al. (15).
In a previous report (6) we showed that the retention of HSV-1 gH in the endoplasmic reticulum (ER) prevented the incorporation of the molecule into the envelopes of mature virions and we argued that these observations favored a two-stage envelopment process in which the final envelope was acquired from a “post-ER” compartment. We have repeated and extended these observations by introducing an ER retrieval signal, a Golgi retrieval signal, or a Golgi retention signal into HSV-1 gD. We report that Golgi retrieval or retention of gD allows incorporation of wild-type levels of gD into mature virions, whereas ER retrieval excludes gD from the virion.
MATERIALS AND METHODS
Cells and viruses.
BHK21, Vero, and VD60 cells were grown in Glasgow’s modified Eagle’s medium supplemented with 10% newborn calf serum and tryptose phosphate broth. Cos7 cells were maintained in Glasgow’s modified Eagle’s medium containing 10% fetal calf serum. VD60 is a Vero-derived cell line (26) which expresses gD upon infection with HSV-1. The gD-negative parental virus used to generate recombinants expressing targeted gD molecules was based on HSV-1 strain SC16 and was named SC16gDdel-Z. The construction of SC16gDdel-Z involved subcloning a 4.1-kb HincII fragment (nucleotides 136449 to 140555) of HSV-1 strain Patton into EcoRI-HincII-digested pING to generate pING-HincII-gD. To facilitate the removal of the entire gD gene and its promoter, EcoRV restriction sites were introduced at positions 138019 and 139606 by site-directed mutagenesis (25), and the gD gene was replaced with an end-repaired HindIII fragment of pMV10, which contains the lacZ gene under the control of the human cytomegalovirus (HCMV) immediate-early promoter (17). This construct is called pING-HincII β-gal and was cotransfected into VD60 cells with wild-type strain SC16-infected cell DNA according to the method of Chen and Okayama (10). Recombinant gD-negative progeny were identified by their blue phenotype after staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and one isolate, SC16gDdel-Z was plaque-purified and cloned by limiting dilution on the VD60 cell line. This isolate was found to be helper cell dependent, and the correct insertion of the HCMV immediate-early promoter-lacZ cassette at the gD locus was confirmed by Southern hybridization. All virus titers were determined by plaque assay on VD60 or Vero monolayers.
Plasmids and mutagenesis.
Site-directed mutagenesis was used to introduce targeting motifs into gD, and the mutagenesis reactions were carried out on single-stranded templates prepared from pING-HincII-gD. The ER retention motif, KKXX (where X is any amino acid), the KKXXXX (KKX4) control motif which overrides KKXX, and the TGN retrieval signal, YQRL, were each appended to the carboxy terminus of gD. All mutagenic oligonucleotides were designed to introduce a diagnostic XbaI restriction site in addition to the targeting signal and a translation stop codon. The oligonucleotides used for this were KKXX (5′TCGCACCAGCCCTTGTTTTACTCTAAGAAGTCTCTATAGAGAATACCCCCCCTT3′), which appends the amino acids SKKSL to the carboxy terminus of gD, KKX4 (5′TCGCACCAGCCCTTGTTTTACTCTAAGAAGTCTCTAGCTCTATAGTCTAGAATACCCCCCCTT3′), which appends SKKSLAL to the carboxy terminus of gD, and YQRL (5′CCCTTGTTTTACTCGGACTACCAGCGTCTTAACTAGTAGAATACCCCCCCTT3′), which appends the amino acids SDYQRLN to the carboxy terminus of gD.
The resulting mutagenized plasmids were named pING-HincII-gDKKXX, pING-HincII-gDKKX4, and pINGHincII-gDYQRL, respectively. For transient-expression studies, the coding sequences for these targeted gD molecules were each transferred as HindIII-XbaI fragments into HindIII-XbaI-digested pCDNA3 (Invitrogen) to give plasmids pCDNA3gD-KKXX, pCDNA3gD-KKX4, and pCDNA3gD-YQRL, respectively. To replace the transmembrane domain of gD with that of the Golgi resident enzyme, α-2-6-sialyltransferase (2ST), an HpaI restriction site was introduced into the gD gene in pING-HincII-gD between the sequence encoding the gD ectodomain and that predicted to encode the transmembrane domain (i.e., between the codons for amino acids 340 and 341 of gD). The oligonucleotide used was 5′ACCCCGAACAACATGGTTAACGGCCTGATCGCCGGC3′. The gD external domain was subsequently removed as a HindIII-HpaI fragment and transferred to HindIII-EcoRV-digested pS85. pS85 was a gift from Sean Munro, Cambridge, and is derived from a plasmid called CD8-D, which is described by Chapman and Munro (9). pS85 contains sequences encoding the transmembrane domain of 2ST and the cytoplasmic tail of CD8 under the control of the adenovirus major late promoter. The resulting construct was called pS85gD-2ST and was used in transient transfection experiments. In this construct, the transmembrane region of gD is replaced with that of 2ST and the cytoplasmic tail of gD is replaced with that of CD8. Previous studies have shown, however, that the CD8 cytoplasmic tail does not influence the Golgi-targeting activity of the 2ST transmembrane region (29) and that the cytoplasmic tail of gD is not required for the incorporation of functional gD into virus particles (16). We therefore considered that the presence of the CD8 cytoplasmic tail should not affect gD targeting or assembly of the molecule into virus particles. To generate a plasmid for making a recombinant virus expressing gD-2ST, a HindIII-XbaI fragment of pS85gD-2ST (which encodes the gD ectodomain fused to the 2ST transmembrane domain and the CD8 cytoplasmic tail) was ligated into pING-HincII-gD in place of wild-type gD sequences. The resulting construct was called pING-HincII-gD2ST.
Plasmids expressing HSV-1 gH, gB, and gL have been described previously (37).
Antibodies.
The expression of targeted gD molecules in transfected Cos cells was detected by immunofluorescence staining and confocal microscopy with antibody LP2 (12). Antibodies used to detect virion proteins in Western blots were LP1 (27), which detects VP16, LP14 (28), which detects gD, and R69 (13), which detects gB.
Transfection and immunofluorescence of Cos cells.
Cos cells seeded on glass coverslips were transfected by a DEAE-dextran method as described previously (41). For immunofluorescence staining, the cells were fixed 48 h after transfection in 2% formaldehyde in phosphate-buffered saline (PBS) for 5 min at room temperature and washed three times in PBS containing 1% fetal calf serum (FCS). The cells were permeabilized with 1% Triton X-100–10% sucrose–1% FCS for 5 min and then washed three times in PBS–1% FCS. Coverslips were incubated for 1 h in LP2 (hybridoma supernatant diluted 1:3), washed three times, incubated with fluorescein-conjugated rabbit anti-mouse immunoglobulin G at a dilution of 1:50 for 45 min, and washed three times before visualization with a Leica confocal microscope.
Virus-free fusion assay.
This assay was carried out as described by Turner et al. (37). Briefly, 5 × 104 Cos cells seeded into six-well tissue culture plates were transfected by using DEAE-dextran with plasmids expressing wild-type forms of HSV-1 gB, gH, and gL together with a plasmid expressing either wild-type or targeted forms of gD. Two days after transfection the cells were overlaid with 2 × 105 Vero cells/well. After a further 24 h cell monolayers were fixed in 0.5% glutaraldehyde and examined by phase-contrast microscopy for multinucleate cells. The numbers of nuclei in syncytia containing 11 or more nuclei were determined for each cotransfection.
Analysis of progeny virions.
BHK cells (108) were infected at 5 PFU/cell with recombinant viruses. After virus adsorption, unpenetrated virions were inactivated with a citrate wash (135 mM NaCl, 10 mM KCl, 40 mM citric acid, pH 3), and after two washes with PBS the cells were incubated for 24 h. Supernatants were harvested and centrifuged at 2,500 rpm (Mistral 6000 rotor) for 10 min to remove cellular debris. Virions were then pelleted in an SW28 rotor at 25,000 rpm for 90 min. Each pellet was resuspended in 200 μl of PBS and stored at −70°C before the plaque assay, particle counting, and Western blot analysis. Particle counts were done by the loop-drop method (39). Immunoblotting was performed on samples of virions, usually containing approximately 108 particles, which were boiled in Laemmli buffer and electrophoresed in sodium dodecyl sulfate (SDS)–10% polyacrylamide gels. Proteins were transferred to nitrocellulose, and, after being blocked in 5% milk powder, the filters were incubated with primary antibodies for 1 h. The membranes were washed in 0.1% Tween 20 in PBS, and antibody binding was detected by incubation with a mixture of biotinylated protein A, streptavidin peroxidase, and enhanced chemiluminescence reagents supplied by Amersham plc.
Endoglycosidase H treatment of virions.
Virion samples were diluted as appropriate in PBS to give a volume of 10 μl, and this mixture was boiled for 5 min after the addition of 1 μl of 5% SDS–10% 2-mercaptoethanol. Subsequently, 1 μl of 0.5 M sodium citrate, pH 5.5, and 500 U of endoglycosidase H were added, and the samples were incubated for 1 h at 37°C before the addition of Laemmli sample buffer and analysis by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
RESULTS
Characterization of targeted gD molecules in transfected cells.
The signals used to target gD to specific cellular compartments are well characterized. The KKXX motif acts as a signal for retrieval of proteins from post-ER sorting compartments to the ER, whereas the addition of two further residues overrides this signal (22). The YQRL motif results in retrieval of protein from the plasma membrane or the endosomal sorting compartments to the TGN (4), and the transmembrane sequence of the Golgi resident enzyme 2ST is responsible for Golgi retention of the enzyme (29).
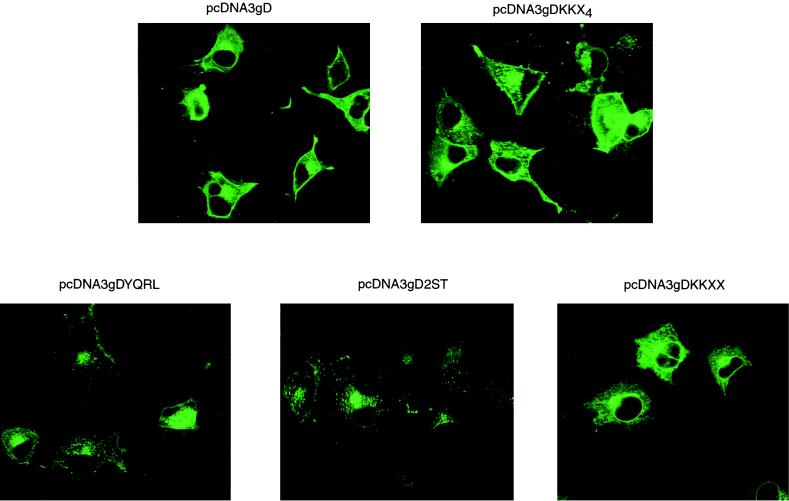
The distribution of gD or targeted forms of gD was examined by standard fluorescence microscopy or confocal microscopy following transfection of Cos7 cells with plasmids encoding wild-type or mutated forms of the molecule. In nonpermeabilized cells expressing wild-type gD, the molecule was readily detected on the cell surface, whereas expression of gD-KKXX, gD-YQRL, or gD-2ST gave no detectable cell surface expression. Cells expressing gD-KKX4, in which the dilysine ER-targeting motif is abrogated by the addition of two further residues, exhibited cell surface fluorescence similar to that seen in cells expressing wild-type gD (data not shown). Confocal microscopy of transfected, permeabilized cells (Fig. 1) showed that wild-type gD was widely distributed, most cells showing fluorescence throughout the cytoplasm and on the cell surface and some showing pronounced Golgi staining. The addition of the dilysine motif KKXX to the C terminus of gD resulted in a perinuclear staining pattern characteristic of resident ER proteins, whereas the addition of two further residues, in gD-KKX4, resulted in a pattern similar to that for wild-type gD. In cells expressing gD-YQRL or gD-2ST, staining was limited to a region adjacent to the nucleus characteristic of Golgi staining. The distribution of these molecules is therefore consistent with the well-established targeting characteristics of the retrieval and retention signals used in their construction.
FIG. 1.
Cos7 cells were transfected with plasmids expressing HSV-1 gD or targeted forms of gD. After 48 h the cells were fixed, permeabilized, and stained with monoclonal antibody LP2, specific for gD. Bound primary antibody was detected with fluorescein-conjugated rabbit anti-mouse IgG, and images were recorded by confocal microscopy.
The expression of HSV-1 gD, in combination with gB and the gH-gL complex is necessary and sufficient to induce cell-cell fusion (37). Since gD-KKXX, gD-YQRL, and gD-2ST were not detected at the cell surface we predicted that these molecules would not function in a cell fusion assay. We therefore transfected Cos7 cells with plasmids expressing wild-type gD or each targeted form of gD together with plasmids expressing gB, gH, and gL. After Vero cells were overlaid, the extent of cell fusion was assessed by counting the number of nuclei recruited into syncytia. As shown in Table 1, the ER-targeted gD molecule was unable to mediate membrane fusion in this assay, giving scores close to background, whereas the control construct expressing gD-KKX4 induced fusion as efficiently as the wild-type gD construct. The Golgi-targeted forms of gD, gD-YQRL, and gD-2ST were severely compromised in their ability to cooperate with gB and gH-gL in inducing fusion but gave scores above background. It would be difficult to demonstrate that these values are significant, but the results suggest that these molecules are present, perhaps transiently, at low levels on the cell surface, despite our inability to detect them by surface immunofluorescence. Of course, it could be argued that modifications to the cytoplasmic tail of gD resulted in a failure in fusion function rather than a failure in cell surface presentation of otherwise functional molecules, but this seems unlikely in view of the observation that gD molecules containing no cytoplasmic tail are functional (16). The failure of the molecules to function efficiently in the cell fusion assay is consistent with their observed cellular location.
TABLE 1.
The ability of targeted gD molecules to function in a virus-free fusion assaya
| Expt no. | Combination of glycoproteins expressed | No. of nuclei in syncytia containing 11 or more nuclei in transfection:
|
|
|---|---|---|---|
| 1 | 2 | ||
| 1 | gB, gH, gL, gD | 1,140 | 987 |
| gB, gH, gL, gDKKX4 | 1,200 | 1,131 | |
| No DNA | 37 | 45 | |
| 2 | gB, gH, gL, gD | 1,913 | 1,731 |
| gB, gH, gL, gDKKXX | 157 | 106 | |
| No DNA | 79 | 61 | |
| 3 | gB, gH, gL, gD | 1,146 | 1,007 |
| gB, gH, gL, gDYQRL | 259 | 198 | |
| gB, gH, gL, gD2ST | 124 | 152 | |
| No DNA | 23 | 22 | |
Cos7 cells were transfected with plasmids expressing HSV-1 gB, gH, and gL together with plasmids expressing wild-type or targeted forms of gD. After 48 h the cells were overlaid with Vero cells, and after a further 24 h the extent of cell fusion was assessed by counting nuclei recruited into syncytia containing more than 10 nuclei. Each transfection was performed in duplicate, and each number is the score recorded in a 4-cm2 area.
Construction and characterization of recombinant viruses expressing targeted gD molecules. (i) Construction of recombinant viruses.
Viruses expressing targeted forms of gD were constructed by cotransfecting VD60 cells with 10 μg of SC16gDdel-Z DNA and 2.5 μg of pING-HincII-derived plasmids containing modified gD sequences. Since SC16gDdel-Z lacks the entire gD coding sequence and forms blue-staining plaques on complementing cell lines when stained with X-Gal, recombinant progeny could be identified by their β-galactosidase-negative phenotype on VD60 monolayers and were subsequently cloned by limiting dilution. The recombinant viruses were designated SCgD-KKXX, SCgD-KKX4, SCgD-YQRL, and SCgD-2ST, and the genome of each virus was analyzed by Southern hybridization to confirm the correct insertion of the modified forms of the gD gene.
(ii) Characteristics of recombinant viruses.
With the exception of SCgD-KKXX all the recombinant viruses grew in BHK or Vero cells to titers equivalent to those for the wild-type parent, HSV-1-SC16, and produced plaques of similar size and morphology. SCgD-KKXX, in contrast, was dependent for propagation on the VD60 helper cell line, formed very small foci of cytopathic effect on noncomplementing cells, and gave low yields of infectivity following high multiplicity of infection on these cells. In a series of experiments in which wild-type virus and SCgD-KKXX were used to infect parallel cultures of BHK or Vero cells, the yields of SCgD-KKXX ranged from as low as 1% to as high as 10% by comparison with yields of wild-type virus. We think that the variability reflects the state of the host cells, but despite repeating the experiments using cells at different densities and passage numbers, we were unable to identify a factor which correlated with the variable relative yield. Particle/infectivity ratios of preparations of each mutant virus grown on noncomplementing cells were determined. Values for SCgD-YQRL, SCgD-2ST, and SCgD-KKX4 were 20, 33, and 45, respectively, and these lie within the range of values reported for preparations of HSV-1-SC16 (3, 20). However, when gD expression was restricted to the ER, in cells infected with SCgD-KKXX, the progeny virions had a particle/infectivity ratio of 870, establishing that the low infectivity yield was due to reduced specific infectivity of virus particles rather than a reduced particle yield and suggesting that the consequence of ER retrieval is to exclude gD from progeny virions.
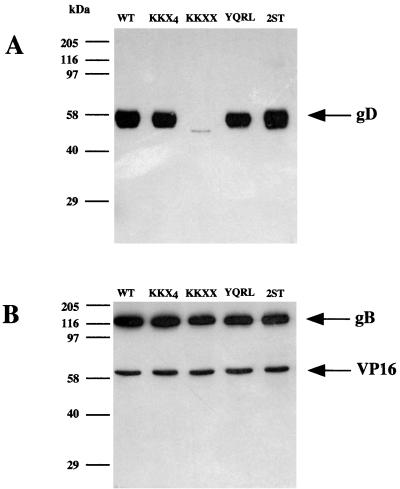
In order to examine the gD content of progeny virions, parallel cultures of approximately 108 BHK cells were infected at a multiplicity of infection of 5 with HSV-1-SC16, SCgD-KKXX, SCgD-KKX4, SCgD-YQRL, or SCgD-2ST. After 24 h progeny virions were pelleted from the supernatant and an equal proportion of each preparation, corresponding to approximately 108 virions, was denatured in Laemmli buffer and subjected to PAGE. After transfer to nitrocellulose VP16, gB and gD were detected with monoclonal antibodies to each protein. As shown in Fig. 2 each preparation contained approximately equal amounts of VP16 and gB, and the SCgD-KKX4, SCgD-YQRL, and SCgD-2ST progeny contained amounts of gD more or less similar to that for the wild-type preparation. SCgD-KKXX progeny, however, contained barely detectable levels of gD. The reduced infectivity of SCgD-KKXX virions thus correlates with their greatly reduced gD content. The residual infectivity in SCgD-KKXX preparations must be due to a subpopulation of gD-containing virions (or to a uniformly lower level of gD in all virions) because SC16gDdel-Z virions, which lack gD, are entirely noninfectious (particle/infectivity ratio > 105) and the residual SCgD-KKXX infectivity was neutralizable by monoclonal antibody LP2, specific for gD.
FIG. 2.
gD content of recombinant virions. BHK cells were infected with wild-type (WT) HSV-1 or with recombinant viruses expressing different targeted forms of gD. After 24 h progeny particles were pelleted from the medium and approximately 108 virions from each preparation were subjected to SDS-PAGE. Separated proteins were transferred to nitrocellulose filters and detected with monoclonal antibodies. (A) gD was detected with antibody LP14. (B) gB and VP16 were detected with antibodies R69 and LP1, respectively.
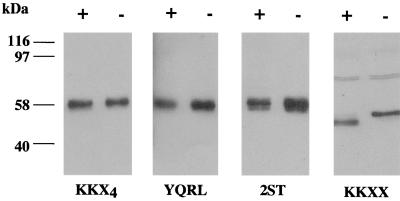
These observations are consistent with a model of egress in which the virus acquires its final membrane from a compartment which contains Golgi-targeted gD but from which ER-retrieved gD is excluded. A longer exposure of the autoradiograph shown in Fig. 2 confirmed the presence of a gD-specific species in SCgD-KKXX virions, and this molecule migrated with slightly higher mobility than gD from wild-type virions. This suggested that the residual SCgD-KKXX infectivity represents virions derived from the primary envelopment compartment containing immature, ER-retrieved gD. We tested this possibility by examining the endoglycosidase H sensitivities of the gD present in different virion preparations. Equal proportions of the SCgD-KKXX, SCgD-KKX4, SCgD-YQRL, and SCgD-2ST virion preparations shown in Fig. 2 were incubated with endoglycosidase H prior to SDS-PAGE and immunoblotting with a gD-specific antibody. With the exception of the SCgD-KKXX sample, each track was loaded with 1/20 the sample shown in Fig. 2 in order to achieve more-similar gD loads. The results (Fig. 3) show that, as expected, SCgD-KKX4, SCgD-YQRL, and SCgD-2ST virions contain mature endoglycosidase H-resistant gD. The SCgD-KKXX virion preparation, however, contains immature endoglycosidase H-sensitive gD, implying that the gD-containing virions in this preparation are derived from the ER compartment.
FIG. 3.
Endoglycosidase H treatment of virion gD. Approximately 2 × 108 virions from the same preparations used for Fig. 2 were treated with endoglycosidase H (+) or incubated in the absence of the enzyme (−) and were then subjected to SDS-PAGE. The SCgD-KKXX tracks were loaded with approximately 108 virions, whereas all other tracks were loaded with approximately 5 × 106 virions. The separated proteins were transferred to nitrocellulose, and gD was detected with antibody LP14.
DISCUSSION
We have previously reported that an HSV-1 mutant expressing an ER-restricted gH released wild-type numbers of virions but that these particles contained no detectable gH, and we interpreted these observations as evidence that the virus acquires its final membrane from a post-ER compartment (6). We report here that restricting another essential glycoprotein, gD, to this compartment gives rise to a recombinant virus with a similar phenotype. The recombinant SCgD-KKXX is dependent on a gD helper cell line for its propagation, and virion preparations grown in noncomplementing cells contain greatly reduced levels of gD. A potential flaw in our interpretation of these results arises from the possibility that the addition of the KKSL motif to the cytoplasmic tail of gD or gH might exclude these molecules from the envelopment process, either directly or by excluding the molecule from the inner nuclear membrane. For gH this seemed a possibility because modifications to the cytoplasmic tail have been shown to affect the function of the molecule (7, 41). The cytoplasmic tail of gD, however, is apparently irrelevant to the function of the molecule (16). Furthermore, as shown in this report, a variety of modifications to the cytoplasmic domain of gD—addition of the sequence KKSLAL or YQRL or the substitution of the CD8 cytoplasmic domain—have no discernible effect on the incorporation of gD into virions or on the infectivity of virions. It therefore seems most unlikely that the KKSL sequence could exclude gD from the envelopment process or from the inner nuclear membrane and much more probable that the retrieval of gD-KKSL to the ER excludes the molecule from the cytoplasmic compartment where final envelopment takes place.
The residual infectivity in preparations of SCgD-KKXX virions is due to the presence of residual gD since this infectivity is neutralizable with antibodies to gD. The small amount of gD detectable in SCgD-KKXX virions is endoglycosidase H sensitive, confirming that its source is a pre-Golgi compartment and implying that the infectivity corresponds to virions at the primary envelopment stage. We recognize that this interpretation, also, is open to question. The very small amount of gD present in SCgD-KKXX virions (shown in Fig. 2 and 3) could be due to contaminating ER membranes, and, since it is impossible to demonstrate purity of the virions at the 99% level, it is impossible formally to refute this argument. Nevertheless, we know that these virions must contain some gD because they are neutralized with an anti-gD antibody, the amount of gD detected is consistent with the infectivity of the preparation (about 1% of that for wild-type virus), and all the detectable gD is endoglycosidase H sensitive. Our data and these arguments do not represent formal proof, but we consider that, taken together, they provide good evidence for a two-stage envelopment process in which an ER-retrieved gD is restricted to primary envelopment and is absent from final envelopment.
The addition of the YQRL TGN retrieval motif to the carboxy terminus of gD or the replacement of the transmembrane domain with that of the Golgi-resident enzyme 2ST was effective in targeting the glycoproteins to the Golgi network and severely reduced the ability of these molecules to participate in induction of cell-cell fusion. We were, however, unable to discern a phenotype when these targeted gD molecules were introduced into recombinant viruses. Viruses expressing TGN-targeted gD were unaltered in plaque size or morphology, grew to wild-type titers, exhibited wild-type particle/infectivity ratios, and released virions which contained levels of gD similar to those of wild-type virions. These observations show that these targeted forms of gD are functional and suggest that the final envelope of HSV-1 is obtained from the Golgi compartment or a Golgi-derived vesicle, a view consistent with the phospholipid composition of the envelope (38).
If the proposal that HSV has both primary and final envelopment stages is correct, it raises many questions. Our data and the observations that infectivity accumulates in monensin-treated cells (23) suggest that primary envelopment yields infectious virions in the perinuclear space, but we do not know the composition of these virions, and, at present, there are no satisfactory methods available for purifying enveloped virions from this compartment. Virions in the perinuclear space must fuse with the outer nuclear membrane, but this cannot require those glycoproteins essential for plasma membrane fusion (37) because virions lacking these proteins are processed and secreted. The UL20 gene product is a possible candidate for this function because HSV-1 mutants lacking this gene accumulate in the perinuclear space (2). If a late cytoplasmic compartment is the site of final envelopment, then the tegument proteins and all the virion membrane proteins must accumulate on the membranes of this compartment. Electron-microscopic evidence suggests that cytoplasmic nucleocapsids are associated with a dense “protein coat”, probably corresponding to the tegument protein (32), and, in circumstances where envelopment fails to occur, cytoplasmic nucleocapsids associate with accumulations of dense material thought to comprise the tegument protein (5, 40). The existence of tegument-containing “light particles” (33), however, implies that tegument proteins can accumulate at membranes containing viral glycoproteins and induce budding in the absence of nucleocapsids. It appears, therefore, that tegument proteins can associate with nucleocapsids in the absence of membranes, and with membranes in the absence of nucleocapsids, but the details of these processes are obscure and the recognition signals involved are entirely unknown.
The mechanism whereby some 10 or more virion membrane proteins accumulate at the final envelopment compartment presents a similar problem. Of course, it could be argued that HSV-1 glycoproteins achieve a sufficient density on all membranes and that budding in any compartment will result in glycoprotein acquisition, but this seems an intuitively unsatisfactory solution. Where viruses are known to bud at internal membranes, targeting signals have been identified in the relevant virion membrane proteins (21, 30), but of the HSV-1 glycoproteins only gE has been shown to be Golgi targeted (1). This protein alone cannot, however, play a key role in directing virus assembly and envelopment because gE-negative mutants produce normal yields of infectious virions (3). Recently Brack et al. (5) have reported that PRV mutants lacking gE, gI, and gM fail to envelope cytoplasmic nucleocapsids or secrete enveloped virions but that this phenotype can be reversed by supplying either gE-gI or gM in trans. These findings suggest that there is functional redundancy in the alphaherpesvirus genome and that our concept of “dispensable genes” needs reassessment. Perhaps the gE-gI complex plays a role in the final envelopment process, but gM can also play this role. While it is tempting to speculate that the targeting of alphaherpesvirus gE to an internal compartment plays some role in virus assembly, it is notable that, in PRV mutants containing a gE lacking the endocytosis motif, the molecule is nevertheless incorporated into virus particles (34, 35).
Finally, it may be that interactions between virion glycoproteins or between these molecules and tegument proteins result in the formation of compartment-restricted complexes, and studies involving coexpression of these molecules may resolve this issue.
ACKNOWLEDGMENTS
We thank S. Munro for the gift of plasmid pS85, G. Cohen for antibody R69, and P. Luzio and D. Wilson for advice. We thank Susanne Bell for excellent technical assistance.
We thank the Medical Research Council, United Kingdom, for a Co-operative Group Grant and for a studentship to A.W. This work was supported by the Wellcome Trust, United Kingdom.
REFERENCES
- 1.Alconada A, Bauer U, Sodeik B, Hoflack B. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J Virol. 1999;73:377–387. doi: 10.1128/jvi.73.1.377-387.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Ward P L, Campadelli-Fiume G, Roizman B. The UL20 gene of herpes simplex virus type 1 encodes a function necessary for viral egress. J Virol. 1991;65:6414–6424. doi: 10.1128/jvi.65.12.6414-6424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balan P, Davis-Poynter N, Bell S, Atkinson H, Browne H, Minson T. An analysis of the in vitro and in vivo phenotypes of mutants of herpes simplex virus type 1 lacking glycoprotein gG, gE, gI or the putative gJ. J Gen Virol. 1994;75:1245–1258. doi: 10.1099/0022-1317-75-6-1245. [DOI] [PubMed] [Google Scholar]
- 4.Bos K, Wraight C, Stanley K K. TGN 38 is maintained in the trans-Golgi network by a tyrosine-containing motif in the cytoplasmic domain. EMBO J. 1993;12:2219–2228. doi: 10.1002/j.1460-2075.1993.tb05870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brack A R, Dijkstra J M, Granzow H, Klupp B G, Mettenleiter T C. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J Virol. 1999;73:5364–5372. doi: 10.1128/jvi.73.7.5364-5372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne H, Bell S, Minson T, Wilson D. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J Virol. 1996;70:4311–4316. doi: 10.1128/jvi.70.7.4311-4316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browne H M, Bruun B C, Minson A C. Characterisation of herpes simplex virus type 1 recombinants with mutations in the cytoplasmic tail of glycoprotein H. J Gen Virol. 1996;77:2569–2573. doi: 10.1099/0022-1317-77-10-2569. [DOI] [PubMed] [Google Scholar]
- 8.Campadelli-Fiume G, Farabegoli F, Di Gaeta S, Roizman B. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J Virol. 1991;65:1589–1595. doi: 10.1128/jvi.65.3.1589-1595.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman R E, Munro S. Retrieval of TGN proteins from the cell surface requires endosomal acidification. EMBO J. 1994;13:2305–2312. doi: 10.1002/j.1460-2075.1994.tb06514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung P, Banfield B, Tufaro F. Brefeldin A arrests the maturation and egress of herpes simplex virus particles during infection. J Virol. 1991;65:1893–1904. doi: 10.1128/jvi.65.4.1893-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cranage M P, McLean C S, Buckmaster E A, Minson A C, Wildy P, Coombs R R. The use of monoclonal antibodies in (reverse) passive haemagglutination tests for herpes simplex virus antigens and antibodies. J Med Virol. 1983;11:295–306. doi: 10.1002/jmv.1890110405. [DOI] [PubMed] [Google Scholar]
- 13.Eisenberg R J, Ponce de Leon M, Friedman H M, Fries L F, Frank M M, Hastings J C, Cohen G H. Complement component C3b binds directly to purified glycoprotein C of herpes simplex virus types 1 and 2. Microb Pathog. 1987;3:423–435. doi: 10.1016/0882-4010(87)90012-x. [DOI] [PubMed] [Google Scholar]
- 14.Elliott G, O’Hare P. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J Virol. 1999;73:4110–4119. doi: 10.1128/jvi.73.5.4110-4119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enquist L W, Husak P J, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1999;51:237–247. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 16.Feenstra V, Hodaie M, Johnson D C. Deletions in herpes simplex virus glycoprotein D define nonessential and essential domains. J Virol. 1990;64:2096–2102. doi: 10.1128/jvi.64.5.2096-2102.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–348. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gershon A A, Sherman D L, Zhu Z, Gabel C A, Ambron R T, Gershon M D. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J Virol. 1994;68:6372–6390. doi: 10.1128/jvi.68.10.6372-6390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Granzow H, Weiland F, Jons A, Klupp B G, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths A, Renfrey S, Minson A C. Glycoprotein-C deficient mutants of two strains of herpes simplex virus type 1 exhibit unaltered adsorption characteristics on polarised or non-polarised cells. J Gen Virol. 1998;79:807–812. doi: 10.1099/0022-1317-79-4-807. [DOI] [PubMed] [Google Scholar]
- 21.Griffiths G, Rottier P. Cell biology of viruses that assemble along the biosynthetic pathway. Semin Virol. 1992;3:367–381. doi: 10.1016/1043-4682(92)90022-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson M R, Nilsson T, Peterson P A. Retrieval of transmembrane proteins to the endoplasmic reticulum. J Cell Biol. 1993;121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson D C, Spear P G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komuro M, Tajima M, Kato K. Transformation of Golgi membrane into the envelope of herpes simplex virus in rat anterior pituitary cells. Eur J Cell Biol. 1989;50:398–406. [PubMed] [Google Scholar]
- 25.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic detection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ligas M W, Johnson D C. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J Virol. 1988;62:1486–1494. doi: 10.1128/jvi.62.5.1486-1494.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLean C, Buckmaster A, Hancock D, Buchan A, Fuller A, Minson A C. Monoclonal antibodies to three non-glycosylated antigens of herpes simplex virus type 2. J Gen Virol. 1982;63:297–305. doi: 10.1099/0022-1317-63-2-297. [DOI] [PubMed] [Google Scholar]
- 28.Minson A C, Hodgman T C, Digard P, Hancock D C, Bell S E, Buckmaster E A. An analysis of the biological properties of monoclonal antibodies against glycoprotein D of herpes simplex virus and identification of amino acid substitutions that confer resistance to neutralisation. J Gen Virol. 1986;67:1001–1013. doi: 10.1099/0022-1317-67-6-1001. [DOI] [PubMed] [Google Scholar]
- 29.Munro S. Sequences within and adjacent to the transmembrane segment of alpha-2,6-sialyltransferase specify Golgi retention. EMBO J. 1991;10:3577–3588. doi: 10.1002/j.1460-2075.1991.tb04924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersson R F. Protein localisation and virus assembly at intracellular membranes. Curr Top Microbiol. 1991;170:69–106. doi: 10.1007/978-3-642-76389-2_3. [DOI] [PubMed] [Google Scholar]
- 31.Pomeranz L E, Blaho J A. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J Virol. 1999;73:6769–6781. doi: 10.1128/jvi.73.8.6769-6781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stackpole C W. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J Virol. 1969;4:75–93. doi: 10.1128/jvi.4.1.75-93.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szilagyi J F, Cunningham C. Identification and characterisation of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 34.Tirabassi R S, Enquist L W. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J Virol. 1999;73:2717–2728. doi: 10.1128/jvi.73.4.2717-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tirabassi R S, Enquist L W. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J Virol. 1998;72:4571–4579. doi: 10.1128/jvi.72.6.4571-4579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torrisi M R, Di Lazzara C, Paues A, Pereira L, Campadelli-Fiume G. Herpes simplex virus envelopment and maturation studies by fracture label. J Virol. 1992;66:554–561. doi: 10.1128/jvi.66.1.554-561.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner A, Bruun B, Minson T, Browne H. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J Virol. 1998;72:873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Genderen I, Brandimarti R, Torrisi M, Campadelli-Fiume G, van Meer G. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology. 1994;200:831–836. doi: 10.1006/viro.1994.1252. [DOI] [PubMed] [Google Scholar]
- 39.Watson D H, Russell W C, Wildy P W. Electron microscope particle counts on herpes virus using the phosphotungstate negative staining technique. Virology. 1963;19:250–260. doi: 10.1016/0042-6822(63)90062-x. [DOI] [PubMed] [Google Scholar]
- 40.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson D W, Davis-Poynter N, Minson A C. Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J Virol. 1994;68:6985–6993. doi: 10.1128/jvi.68.11.6985-6993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Z, Gershon M D, Hao Y, Ambron R T, Gabel C A, Gershon A A. Envelopment of varicella-zoster virus: targeting of viral glycoproteins to the trans-Golgi network. J Virol. 1995;69:7951–7959. doi: 10.1128/jvi.69.12.7951-7959.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Z, Hao Y, Gershon M D, Ambron R T, Gershon A A. Targeting of glycoprotein I (gE) of varicella-zoster virus to the trans-Golgi network by an AYRV sequence and an acidic amino acid-rich patch in the cytosolic domain of the molecule. J Virol. 1996;70:6563–6575. doi: 10.1128/jvi.70.10.6563-6575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]