Abstract
Porcine reproductive and respiratory syndrome (PRRS) is a globally prevalent contagious disease caused by the positive-strand RNA PRRS virus (PRRSV), resulting in substantial economic losses in the swine industry. Modifying the CD163 SRCR5 domain, either through deletion or substitution, can eff1ectively confer resistance to PRRSV infection in pigs. However, large fragment modifications in pigs inevitably raise concerns about potential adverse effects on growth performance. Reducing the impact of genetic modifications on normal physiological functions is a promising direction for developing PRRSV-resistant pigs. In the current study, we identified a specific functional amino acid in CD163 that influences PRRSV proliferation. Viral infection experiments conducted on Marc145 and PK-15CD163 cells illustrated that the mE535G or corresponding pE529G mutations markedly inhibited highly pathogenic PRRSV (HP-PRRSV) proliferation by preventing viral binding and entry. Furthermore, individual viral challenge tests revealed that pigs with the E529G mutation had viral loads two orders of magnitude lower than wild-type (WT) pigs, confirming effective resistance to HP-PRRSV. Examination of the physiological indicators and scavenger function of CD163 verified no significant differences between the WT and E529G pigs. These findings suggest that E529G pigs can be used for breeding PRRSV-resistant pigs, providing novel insights into controlling future PRRSV outbreaks.
Keywords: PRRSV, CD163, Point mutation, E529G, Pigs
INTRODUCTION
Porcine reproductive and respiratory syndrome (PRRS), commonly known as pig blue-ear disease, is caused by the PRRS virus (PRRSV) (Lunney et al., 2016). This contagious disease is characterized by reproductive disorders in pregnant sows and respiratory disease in pigs of all ages, particularly piglets (Ma et al., 2021b). PRRS has become one of the most economically devastating diseases affecting the swine industry worldwide (Holtkamp et al., 2013; Renken et al., 2021). While current prevention and control methods, such as vaccine immunization, are somewhat effective (Kick et al., 2023), challenges remain in overcoming the virus’s immune escape mechanisms, recombination diversity, and antibody-dependent enhancement.
Research has identified CD163 as a crucial host factor for PRRSV, serving as a specific and indispensable receptor for viral entry and uncoating (Calvert et al., 2007; Van Gorp et al., 2008; Welch & Calvert, 2010; Yu et al., 2020). Advances in CRISPR/Cas9 technology have made gene editing more feasible, offering potential solutions for eradicating various diseases through genetic modification (Jiang & Doudna, 2017). Specifically, CD163 gene knockout using the CRISPR/Cas9 system has been shown to confer complete resistance to PRRSV infection (Yang et al., 2018).
The fifth scavenger receptor cysteine-rich domain (SRCR5) of CD163 is vital for mediating PRRSV invasion into host cells by binding to viral GP2a and GP4 protein dimers (Stoian et al., 2022a). Recent studies have demonstrated that deletion or substitution of the CD163 SRCR5 domain confers resistance to PRRSV infection (Burkard et al., 2017; Prather et al., 2017; Wells et al., 2017; Whitworth et al., 2014, 2016; Xu et al., 2020). The SRCR5 domain contains four disulfide bonds necessary for PRRSV infection (Stoian et al., 2022b). Liang Guang Small Spotted and Large White pigs lacking 41 amino acids in the SRCR5 domain, including the ligand binding pocket (LBP), are not susceptible to PRRSV infection (Guo et al., 2019). Additionally, knockout of the PSTII domain of CD163 has been found to confer complete resistance to PRRSV infection (Salgado et al., 2024).
Beyond its role as a host factor for PRRSV, CD163 functions as a scavenger receptor, participating in various physiological functions (Etzerodt & Moestrup, 2013; Moestrup & Møller, 2004). For instance, the CD163 SRCR2 domain contributes to erythroblast adhesion (Fabriek et al., 2007), while the SRCR3 domain is vital for the calcium-sensitive coupling of hemoglobin (Hb)-haptoglobin (Hp) complexes (Madsen et al., 2004). Xu et al. (2020) found that pigs with SRCR5 knockout exhibited significantly higher meat color scores, increased Fe concentrations, and higher Hp content, likely due to decreased Hb metabolism and subsequent mild Fe accumulation in the meat. Consequently, large fragment modification of CD163 raises concerns regarding their potential effects on growth performance. Thus, minimizing the impact of CD163 gene modifications on its normal physiological function, while still contributing to anti-PRRSV breeding, represents a promising direction for future research.
Previous studies have shown that PRRSV exhibits strict host specificity for swine, with an exclusive affinity for porcine primary alveolar macrophages (PAMs) (Xie et al., 2021). Additionally, artificially induced African green monkey (Chlorocebus sabaeus) kidney cells, such as MA-104, Marc145, and CL2621, are also susceptible to PRRSV (Shi et al., 2015), showing persistent infection in epidemiological studies. Given that CD163 performs similar biological functions in vivo, we hypothesized that specific amino acids in CD163 affect PRRSV proliferation. Ma et al. (2021a) discovered significant discrepancies in surface electrostatic potential among the crystal structures of the monkey CD163 SRCR5 domain, pig CD163 SRCR5 domain, and human CD163L1 SRCR8 domain, and identified several potential site mutations, including E534 (referred to as E529 in this study), that affect PRRSV proliferation in PK-15 cells.
Building upon this, we investigated the role of the pig CD163 E529 and corresponding E535 in monkeys in PRRSV proliferation. We initially verified that E535G mutation in Marc145 cells and E529G mutation in PK-15CD163 cells inhibited PRRSV infection by preventing viral binding and entry. Subsequently, we generated genetically modified pigs with the CD163 E529G mutation, with physiological examination confirming normally maintained physiological functions. Individual PRRSV challenge experiments verified that E529G pigs were resistant to PRRSV infection, emphasizing the feasibility of the E529G mutation strategy for commercial anti-PRRSV breeding. In future research, identifying additional key amino acids in CD163 that affect PRRSV proliferation without disrupting its physiological function and developing pigs with these specific CD163 amino acid mutations may provide an effective solution to prevent PRRSV infection.
MATERIALS AND METHODS
Ethics statement
All animal work was approved by the Animal Welfare and Research Ethics Committee at Jilin University (Approval No. SY202403012). All procedures were conducted strictly in accordance with the Guide for the Care and Use of Laboratory Animals.
Cells and viruses
African green monkey embryonic kidney (Marc145) cells and the porcine kidney cell line 15 (PK-15) were cultured in RPMI Medium 1640 basic (RPMI, Gibco, USA) or Dulbecco’s Modified Eagle Medium (DMEM, Gibco, USA), both supplemented with 5% fetal bovine serum (FBS, Gibco, USA). The cells were incubated at 37°C in a 5% CO2 atmosphere. Porcine fetal fibroblasts (PFFs) were cultured in DMEM supplemented with 10% FBS.
Highly pathogenic PRRSV (HP-PRRSV) strains JXA1, GD, and JXA1-GFP were kindly provided by Prof. Shuqi Xiao (Lanzhou Veterinary Research Institute, Lanzhou, China).
Plasmid construction
Single-guide RNAs (sgRNAs) were designed using Zhang Lab Guide Design Resources and synthesized by Comate Bioscience (China). The plasmid pBluescriptSKII+U6-sgRNA(F+E) empty (#74707, Addgene, USA) was digested with the BbsI restriction enzyme (NEB, USA) at 37°C for 1 h. The sgRNA oligonucleotides were annealed into double-stranded DNA, then inserted into the BbsI site of the 74707-vector backbone. The target plasmid was named 74707-sgRNA-mE535G or 74707-sgRNA-pE529G.
Total RNA was extracted from lung tissue and reverse transcribed into cDNA. The CD163 encoding sequence was amplified from the cDNA using long-sequence polymerase chain reaction (PCR) with specific primers (CD163-CDS-F/R) and cloned into the pcDNA3.1 vector. The recombinant plasmid pcDNA3.1-CD163 and PB513B-Blastin vector were each digested with XbaI and AgeI (NEB, USA), then ligated together. The constructed plasmid with a FLAG tag was named PB513B-Blastin-CD163.
The NG-ABE8e plasmid was purchased from Addgene (#138491, USA) (Jing et al., 2023).
Construction and verification of PK-15 cells with stable CD163 expression
The PB513B-Blastin-CD163 plasmid (200 ng), used as a transposon, was mixed with transposase (20 μg) in electroporation cuvettes plus (BTX, USA) and electrotransferred into 2×106 PK-15 cells at 280 V (1 ms, three pulses) using an ECM 2001 Electro Cell Manipulator (BTX, USA). Three days after electroporation, blasticidin S was used to screen resistant cells by gradually increasing the concentration daily until it reached 15 ng/μL. Four monoclonal cells were then selected by limiting dilution. The expression of CD163 was verified by quantitative real-time polymerase chain reaction (qPCR) and western blotting (WB). Both qPCR and indirect immunofluorescence assays (IFA) were used to determine PRRSV susceptibility of the four clones.
Mutation efficiency assessment of guide RNAs and selection of cell clones
Mutation efficiency of the guides was assessed by transfecting the 74707-sgRNA-mE535G target plasmid into 2×106 Marc145 cells or 74707-sgRNA-pE529G into PK-15CD163 cells and PFFs, along with the NG-ABE8e base editor, using a Neon transfection system (Invitrogen, USA) (1 400 mV, one pulse, 30 ms), with a final concentration of 30 μg per component. After 72 h, one-third of the cells were collected, and genomic DNA was extracted using a TIANamp Genomic DNA Kit (Tiangen, China). The remaining cells were cultured continuously. PCR across the target sites of genomic DNA was performed using primers (Supplementary Table S1) with 2×Taq Plus PCR Mix (Tiangen, China) to produce 1 261 bp and 1 030 bp products, respectively. Following this, 1% agarose gel electrophoresis and Sanger sequencing were conducted to analyze editing efficiency.
The reserved cells were inoculated into several 100 mm dishes after efficiency detection, with an inoculation density of 600 cells/dish for Marc145/PK-15CD163 cells or 2 000 cells/dish for PFFs, using the limiting dilution method. Single-cell clones were selected and cultured in 24-well plates 10–12 days later. When confluence reached 80% or more, 20% of each single clone was digested and lysed with 10 μL of NP40 lysis buffer (NP40 stock solution: proteinase K: 10×standard buffer: ddH2O, 1:1:10:90) for 1 h at 56°C and 10 min at 95°C. The lysate was then used as the PCR template for electrophoresis. Additionally, single point mutation events were confirmed by Sanger sequencing.
Cell proliferation detection
Wild-type (WT) cells and clones were inoculated into 96-well plates (n=3) after cell counting to ensure consistent cell numbers. Cell proliferation was detected using a CCK-8 Kit (Boster, China) and absorbance was measured at an optical density (OD) of 450 nm using an Infinite® 200 PRO Multifunctional microplate reader (TECAN, Switzerland).
Quantification of CD163 or PRRSV mRNA
RNA was extracted from 1×105 cells using TRNzol Universal Reagent (Tiangen, China) and RNA concentration was measured using a NanoDrop™ 1000 (Thermo Scientific, USA). Subsequently, RNA of equal quality was reverse transcribed into cDNA using a FastKing One Step RT-PCR Kit (Tiangen, China) according to the manufacturer’s instructions. The mRNA levels of CD163 and PRRSV and the reference mRNA levels of GAPDH were quantified using Talent qPCR PreMix (Tiangen, China) and a fluorescence quantitative PCR detection instrument q225 (Nevogene, China) with primers (Supplementary Table S1).
Virus titration
Marc145 cells were inoculated into 96-well plates and infected with 10-fold serial dilutions of sample supernatants before incubation for 1 h at 37°C. The supernatants were then replaced with DMEM containing 5% FBS. The cytopathic effect (CPE) at 96 hours post-infection (hpi), characterized by clumping and shrinkage of cells, was evaluated. Viral titers were determined using the 50% tissue culture infective dose (TCID50) and calculated following the Reed-Muench formula.
Expression levels of CD163 and PRRSV protein by western blotting
Approximately 1×107 cells were suspended in RIPA buffer, lysed on ice for 30 min, and centrifuged at 3 000 r/min for 20 min at 4°C. The supernatant was collected, and the protein concentration was measured using an Enhanced BCA Protein Assay Kit (Beyotime, China). Equal amounts of protein were mixed with loading buffer prior to boiling for 10 min, then subjected to electrophoresis on 10% acrylamide gels. The gels were transferred to 0.22 μm polyvinylidene fluoride (PVDF) membranes (Boster, China). Proteins were divided into several parts and separately probed with antibodies against CD163 (mouse mAb, non-reducing, Bio-Rad, China, 1 μg/mL), PRRSV-N (mouse mAb, Shandong Lvdu, China, 1:100), β-actin (mouse mAb, Proteintech, China, 1:2 000), or FLAG (Proteintech, China, 1:4 000) at 4°C for 12 h. The membranes were subsequently incubated with horseradish peroxidase (HRP)-conjugated Affinipure goat anti-mouse IgG (H+L) (Proteintech, China, 1:5 000), followed by HRP-conjugated Affinipure goat anti-rabbit IgG (H+L) (Proteintech, China, 1:5 000) for 1h at room temperature. Protein bands were visualized using a Western Blotting Visualizer C600 (Azure Biosystems, USA), according to the manufacturer’s instructions.
IFA and flow cytometry analysis
Approximately 5×104 cells and 2×106 cells were seeded into 12- and 6-well plates, respectively. After 12 h, the cells were inoculated at a multiplicity of infection (MOI) of 0.1 with the viruses (PRRSV JXA1/GD/JXA1-GFP) or not and cultured in RPMI for 48 h at 37°C. Cells for flow cytometry (FACS) analysis were digested into individual cells and fixed in phosphate-buffered saline (PBS) with 4% formaldehyde (Boster, China) for 15 min at room temperature. Cells for IFA were directly added to 4% formaldehyde. Subsequently, all cells were permeabilized in PBS containing 0.1% Triton-X-100 (Solarbio, China) for 10 min. Cells infected with viruses were incubated overnight with antibodies against PRRSV-N or β-actin at a 1:50 dilution in PBS with 10% FBS, then incubated with fluorescein (FITC)-conjugated Affinipure goat anti-mouse IgG (H+L) (Proteintech, China, 1:500). Cells infected with viruses or not were incubated overnight with antibodies against CD163 or β-actin at a 1:50 dilution in PBS with 10% FBS, then incubated with goat anti-mouse IgG/PE antibody (Absin, China, 1:500). Three washes with PBS were performed after each step. Expression levels of viruses and CD163 were determined by antibody labeling and assessed by IFA and FACS analysis using a fluorescence microscope EVOS (AMG, USA) or a FCM SH800S (Sony, Japan) with FlowJo software.
Binding and entry detection of PRRSV
Approximately 5×105 cells were seeded into two 60 mm dishes for binding and entry. After 12 h, the cells were inoculated with the PRRSV GD strain at MOI=1 for 1 h at 4°C for binding. Cells were subsequently washed three times with PBS, with the binding dish stored at −80°C. Another dish was continuously cultured in RPMI 1640 for 3 h at 37°C for entry. Cells were washed three times with PBS, then dipped in an alkaline high salt solution (1 mol/L NaCl and 50 mmol/L NaHCO3, pH 9.5) (Zhu et al., 2023) for 3 min, followed by washing as above. The cells were collected for RNA extraction and viral RNA was quantified by qPCR to characterize binding and entry.
Detection of blastocyst development rate
PFFs from Large White pigs with the E529G mutation were transplanted into enucleated oocytes by microinjection. The rate of recombinant embryo development to the blastocyst stage was examined under a micromanipulator (Nikon, Japan).
Generation of E529G pigs
E529G mutation PFFs were used for somatic cell nuclear transfer (SCNT), followed by embryo transplantation, as described previously (Xie et al., 2018). The genotype was identified by PCR amplification and Sanger sequencing.
Assessment of free Hp in serum
Blood samples from WT and E529G pigs were collected at 2 weeks of age, and serum was separated for free Hp examination using the Porcine (Pig) ELISA Kit for Haptoglobin (Hpt) (Cloud-Clone, USA) following the manufacturer’s instructions.
Visualization and quantification of Hb-Hp uptake
PAMs were flushed out of the lung and seeded on a 24-well plate. FITC-Hb (Ruixibio, China) and Hp (BioVISION, USA) were mixed at a 1:1 wt/wt ratio in PBS for 15 min in a centrifuge tube before experimentation. PAMs were incubated with 100 μg/mL FITC-Hb-Hp in RPMI for 4 h at 37°C, washed three times with PBS, then treated with Hoechst 33342 for 5 min followed by additional washing. Fluorescence was detected and analyzed using a fluorescence microscope EVOS (AMG, USA). The cells were also collected for quantification by FACS assay.
Viral challenge assessment of pigs
Both WT (n=3) and E529G pigs (n=3) (5 weeks of age) were challenged in one room with approximately 4×107 TCID50 of JXA1. One-half of the inoculum was delivered intramuscularly and the remainder intranasally. The in vivo viral challenge assay was strictly performed at a designated safe place. Clinical signs, rectal temperature, and body weight gain were monitored daily. Blood samples from pigs were collected on days 0, 3, 7, 10, and 14 post injection. Serum viral load was measured by qPCR. A commercial Porcine Reproductive and Respiratory Syndrome Virus Antibody ELISA Test Kit (Jndiag, China) was used to evaluate serum virus antibody levels according to the manufacturer’s instructions. The animals were then euthanized by sodium pentobarbital (100 mg/kg) at 14 days post-infection. Tissue samples from the pigs were photographed and then fixed in formalin, followed by routine paraffin sectioning, hematoxylin & eosin (H&E) staining, and Immunohistochemical (IHC) analysis. Histopathological lesions were observed under a High-Resolution Microscope Digital Camera (Olympus, Japan).
Statistical analyses
All data are presented as mean±standard error of the mean (SEM) from three independent experiments. Statistical analyses were performed using untailed t-test with GraphPad Prism software v.8.0. P<0.05 was considered statistically significant. *: P<0.05; **: P<0.01; ***: P<0.001; ****: P<0.0001; ns: no significance.
RESULTS
Preparation of Marc145 cells with E535G mutation
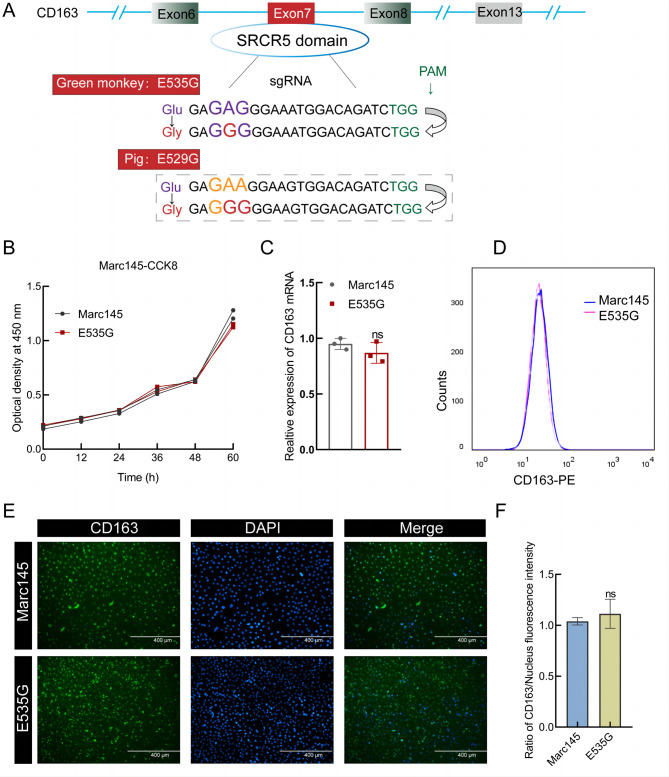
To affirm the feasibility of the CD163 functional amino acid site mutation strategy, sgRNAs introducing monkey E535G and pig E529G mutations were designed and electroporated into Marc145 cells along with NG-ABE8e (Figure 1A). Ten single-cell clones were identified, and four homozygous clones (Clone-#7 referred to as E535G cells) were obtained (Supplementary Figure S1). The E535G cells grew similarly to the WT cells, with no obvious phenotypic changes or effects on cell viability, suggesting that the E535G mutation is not essential for cell growth (Figure 1B). To further ascertain whether the E535G mutation resulted in the loss of CD163 expression at the RNA and protein levels, extracts from WT and E535G mutation cells were collected and subjected to qPCR, FACS, and IFA (Figure 1C–F). Results indicated that the E535G mutation did not affect Marc145 cell viability or CD163 expression.
Figure 1.
Preparation of Marc145 cells with E535G mutation
A: Schematic of sgRNA positions and sequences in green monkey and pig CD163. PAM and sgRNA sequences are shown in green and purple, respectively. B: CCK-8 assay of WT and E535G Marc145 cells. C: Relative expression levels of CD163 RNA were detected using qPCR with primers mCD163-F/R (Supplementary Table S1) and normalized by GAPDH. Data were analyzed using an unpaired t-test. Error bars represent SEM, n=3. Significant differences in results compared to WT are indicated as follows: ns means no significance. D, E: Expression levels of CD163 protein in WT or E535G Marc145 cells were determined by FACS (D) and IFA (E). F: Fluorescence intensity in (E) was quantified by ImageJ analysis.
E535G mutation of monkey CD163 inhibited PRRSV proliferation in Marc145 cells
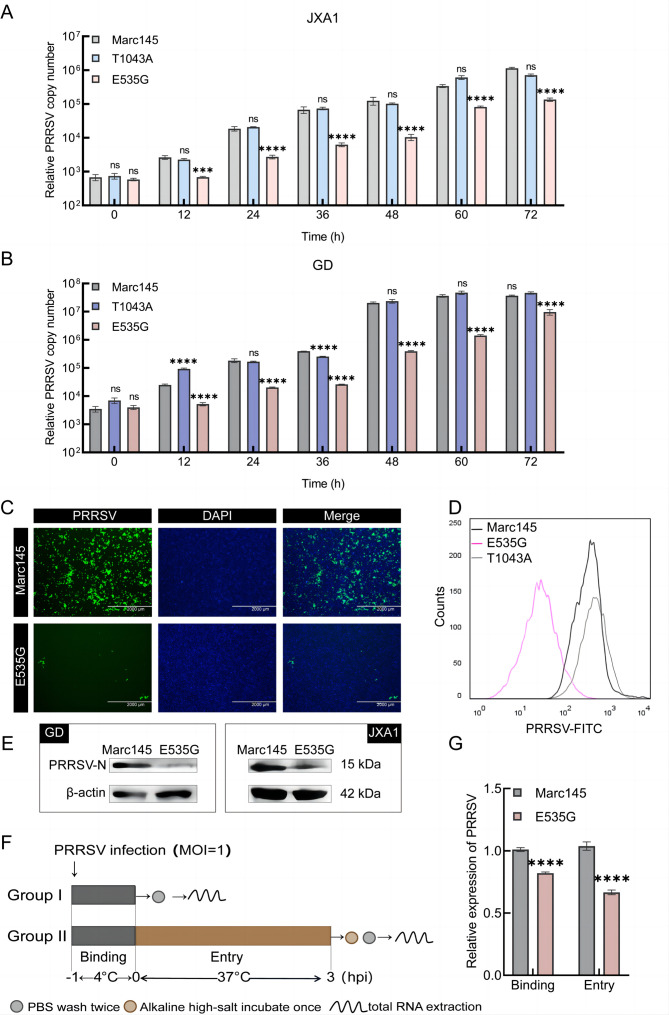
To assess whether E535 is a key amino acid site affecting PRRSV proliferation, we inoculated cells with HP-PRRSV, including Marc145, E535G cells, and control cells (T1043A, located on CD163 exon 13, which is not associated with the SRCR5 domain). Viral RNA levels in E535G cells post JXA1 (Figure 2A) and GD strain (Figure 2B) infection demonstrated marked inhibition, while no significant differences in T1043A cells compared with WT cells were observed. Subsequently, IFA, WB, FACS, and virus titer detection revealed a significant decrease in viral load in E535G cells (Figure 2C–E, Supplementary Figure S2A). Overall, these results suggest that the E535G mutation within CD163 can resist HP-PRRSV infection in Marc145 cells.
Figure 2.
E535G mutation of monkey CD163 inhibited PRRSV proliferation in Marc145 cells
A, B: WT or E535G Marc145 cells were seeded into 12-well plates and inoculated with JXA1 and GD strain at MOI=0.1, respectively. Cells were collected at 0, 12, 24, 36, 48, 60, and 72 hours post infection (hpi) to quantify viral RNA using qPCR with specific primers PRRSV-ORF7-F and PRRSV-ORF7-R. PRRSV copy numbers were statistically analyzed using unpaired t-test. C: Cells were inoculated with JXA1-GFP at MOI=0.1. PRRSV infection was characterized by IFA at 48 hpi. D: WT or E535G Marc145 cells were seeded into 60 mm dishes and inoculated with GD at MOI=0.1. PRRSV infection was assessed by FACS at 48 hpi. E: WT or E535G Marc145 cells were inoculated with GD and JXA1 strain at MOI=0.1, respectively. Viral proteins were determined using WB at 48 hpi. F: Binding and entry determination schematic of PRRSV. G: Relative expression of GD RNA was measured by qPCR for binding and entry assay. **: P<0.01; ****: P<0.0001; ns: No significance.
As CD163 is an imperative host factor for PRRSV infection, we further analyzed whether the inhibitory effect on PRRSV in E535G cells was achieved by reducing CD163 expression. As demonstrated in Supplementary Figure S2B–D, no significant differences in the expression levels of CD163 mRNA or protein were detected. Binding and entry assays were performed (Figure 2F), with relative viral RNA expression indicating that binding and entry were significantly prevented, as expected (Figure 2G). Taken together, these results suggest that E535 is a key amino acid site that suppresses HP-PRRSV infection by inhibiting PRRSV binding and entry.
E529G mutation inhibited PRRSV proliferation in PK-15CD163 cells
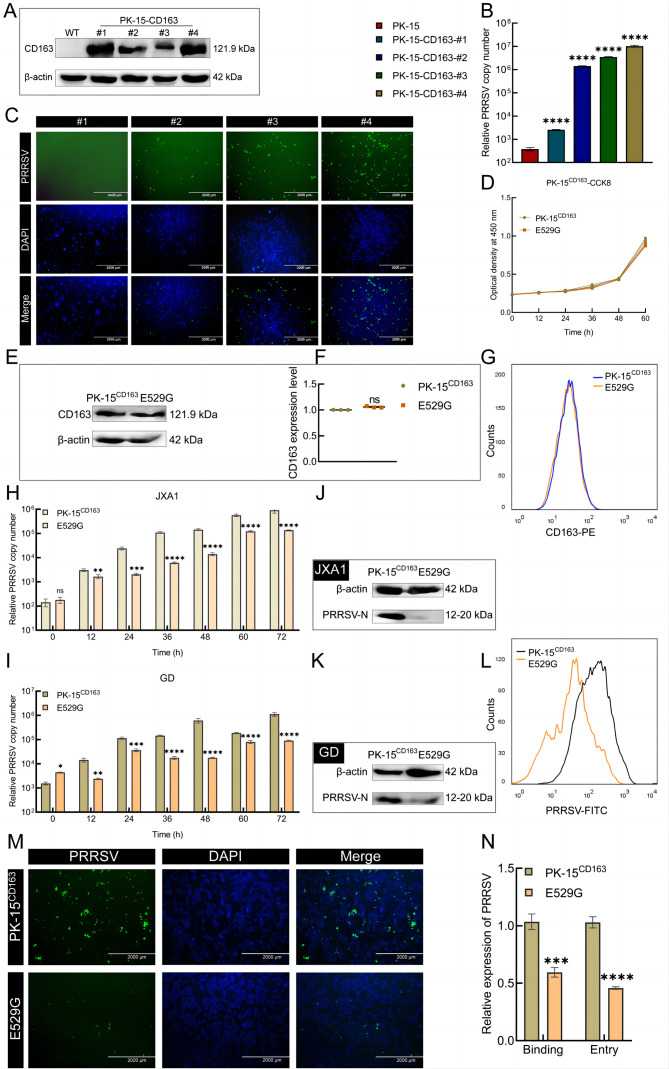
We confirmed that the mE535G mutation significantly inhibited PRRSV infection in Marc145 cells. Thus, we further hypothesized that the corresponding E529 site on porcine CD163 may play the same role in PRRSV infection. To better characterize the antiviral ability of the E529G mutation, we established PRRSV-permissive PK-15 cells that stably integrated pCD163. The expression of CD163 in monoclonal cells (#1-4) showed that CD163 was successfully integrated into PK-15 cells (Figure 3A). Subsequent examination of viral RNA (Figure 3B) and protein (Figure 3C) levels in these monoclonal cells indicated that clone #4 (referred to as PK-15CD163) was most susceptible to PRRSV. To ensure consistent CD163 copy numbers, PK-15CD163 E529G cells (named E529G cells) were selected using NG-ABE8e (Supplementary Figure S3A). The CCK-8 assay results indicated that the E529G mutation had no impact on cell proliferation (Figure 3D). Moreover, to further investigate CD163 expression after E529G mutation, cell extracts were subjected to qPCR, WB, and FACS (Supplementary Figure S3B; Figure 3E–G), revealing no marked differences in CD163 expression between the PK-15CD163 and E529G cells.
Figure 3.
E529G mutation restrained PRRSV proliferation in PK-15CD163 cells
A: Expression of CD163 protein in monoclonal cells (#1-4) was measured by WB with FLAG antibody. B, C: PRRSV susceptibility judging of four PK-15-CD163 cell clones was determined by JXA1-GFP proliferation using qPCR (B) and IFA (C). D: CCK-8 assay of PK-15CD163 and E529G cells. E, F: The expression of CD163 protein was further characterized by WB with FLAG antibody (E) and quantified by ImageJ analysis (F). G: The expression levels of CD163 were texted by FACS, orange and blue lines are on behalf of PK-15CD163 and E529G cells severally. H, I: Quantification of PRRSV mRNA were measured using qPCR at 0, 12, 36, 48, 60, and 72 hours post infection of JXA1 (H) and GD (I) strain at MOI=0.1. J, K: The cells were collected for WB assay at 48 hours post infection of JXA1 (J) and GD (K) strain (MOI=0.1) for PRRSV-N protein detecting, respectively. L: Viral protein was further determined at 48 hours post infection of GD strain at MOI=0.1 by FACS. Black line acts PK-15CD163 cells and orange line is E529G cells. M: IFA judged viral protein expression at 72 h post JXA1-GFP infection at MOI=0.1. N: Relative expression quantity of viral RNA was examined for the process of binding and entry post infection of GD strain at MOI=1. ***: P<0.001; ****: P<0.0001; ns: No significance.
To evaluate whether E529G cells can resist PRRSV infection, we detected viral RNA based on qPCR. The PRRSV copy numbers at 0, 12, 36, 48, 60, and 72 h as well as viral protein levels post JXA1 and GD strain infection showed a prominent PRRSV-inhibiting effect on the E529G cells (Figure 3H–K). Simultaneously, viral protein assessment using FACS and IFA showed clear inhibition of PRRSV in E529G cells (Figure 3L, M). These findings suggest that E529G cells can effectively resist PRRSV infection. As determined above in Marc145 cells, the mE535G mutation blocked PRRSV replication by inhibiting the binding and entry of PRRSV, rather than by down-regulating CD163 expression. Consequently, we posited that resistance to PRRSV infection in E529G cells may be achieved in the same way. As expected, qPCR analysis indicated that the E529G cells resisted PRRSV infection by suppressing the binding and entry of PRRSV (Figure 3N). We also detected CD163 expression based on qPCR, FACS, and WB after exposure of cells to HP-PRRSV. Results showed no significant differences in CD163 expression between the PK-15CD163 and E529G cells (Supplementary Figure S3D–G). In summary, these findings substantiate that the mE535G and pE529G mutations significantly prevent PRRSV infection, highlighting the importance of these specific amino acid positions within CD163 as vital sites associated with PRRSV infection.
Generation of CD163-E529G mutation pigs
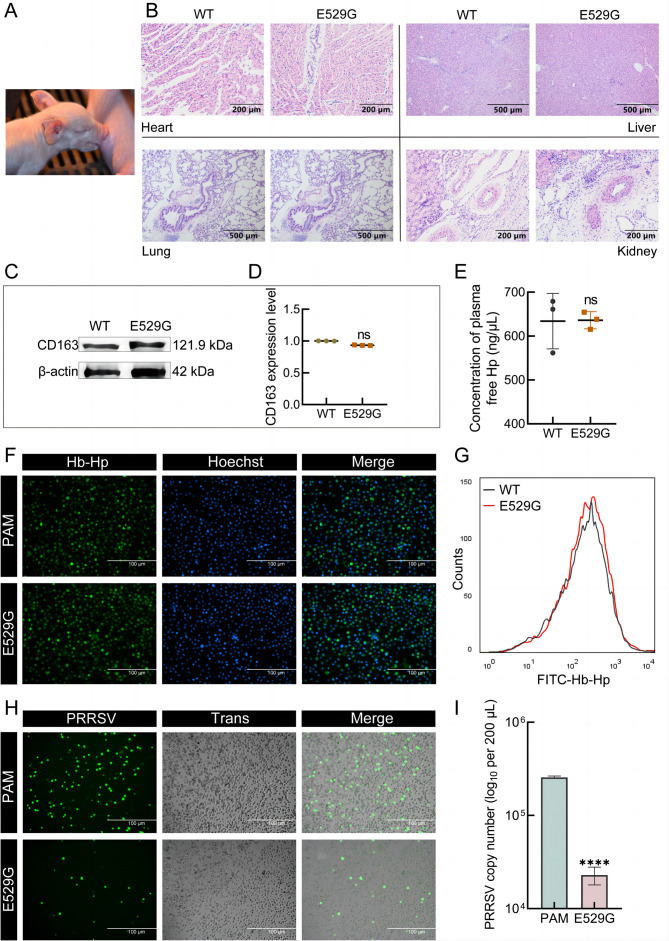
To further investigate whether the E529G mutation resists PRRSV infection in vivo, we generated E529G mutation pigs. Initially, PFFs from four Large White pigs with the E529G mutation (Supplementary Figure S4A) were identified and transplanted into enucleated oocytes by microinjection. Blastocyst development analysis indicated that the recombinant embryos developed normally to the blastocyst stage in vitro (Supplementary Table S2), substantiating that E529G PFFs may serve as SCNT donor cells. Four healthy F0 generation E529G pigs were obtained (Figure 4A). Genotype identification (Supplementary Figure S4B) revealed that all four genetically modified pigs produced the desired E529G mutation with no off-target effects (Supplementary Figure S4C).
Figure 4.
Generation of CD163-E529G mutation pigs
A: Picture of E529G piglets. B: H&E staining indicated normal physiological morphology of tissues from WT and E529G pigs. C: Identification of CD163 protein expression in lungs from WT and E529G pigs by WB with pCD163 antibody. D: Intensity of bands in (C) was quantified by ImageJ analysis. E: Free Hp in serum from WT and E529G pigs was detected by ELISA. SEM: WT, 634.10±63.18; E529G, 636.10±19.56, n=3. F, G: PAMs were separated from WT or E529G pigs and FITC-Hb-Hp complex uptake in PAMs was characterized by IFA (F) and FACS (G). H: PAMs from WT and E529G pigs were then challenged by JXA1-GFP at MOI=0.1 and viral protein expression was determined by IFA at 12 hpi. I: Viral RNA in 200 μL of supernatant in (H) was extracted for qPCR. ****: P<0.0001; ns: No significance. WT PAMs in F–I were from the same WT pig and E529G PAMs were from the same E529G pig.
Blood samples were collected to determine whether physiological indicators are normal in E529G pigs. Full blood counts showed that the E529G mutation had negligible impact on porcine growth (Supplementary Table S3). Concurrently, organs from both WT and E529G pigs were subjected to H&E staining, confirming the normal tissue morphology of cloned pigs (Figure 4B). Additionally, CD163 expression levels in the lung displayed no significant differences between WT and E529G pigs (Figure 4C, D). As CD163 is involved in anti-inflammatory and anti-oxidative processes via Hb-Hp complex uptake (Subramanian et al., 2013), free Hp levels in serum were measured. Results indicated no effect after E529G mutation (Figure 4E), indirectly confirming that CD163 from E529G pigs can remove free Hp normally. Furthermore, PAMs were isolated from WT and E529G pigs, and their uptake of FITC-Hb-Hp was determined using fluorescence microscopy (Figure 4F) and FACS (Figure 4G). Results indicated that PAMs from E529G pigs retained normal scavenger function. Additionally, inoculation of PAMs with HP-PRRSV-JXA1-GFP followed by fluorescence observation confirmed efficient PRRSV defense in E529G pigs (Figure 4H). Viral RNA and titer assays (Figure 4I; Supplementary Figure S4D) demonstrated that the antiviral intensity reached an order of magnitude. Overall, these in vitro results suggest that E529G pigs maintain normal CD163 scavenger function, exhibit healthy growth, and acquire antiviral ability against PRRSV.
CD163-E529G mutation pigs exhibited resistance to HP-PRRSV infection
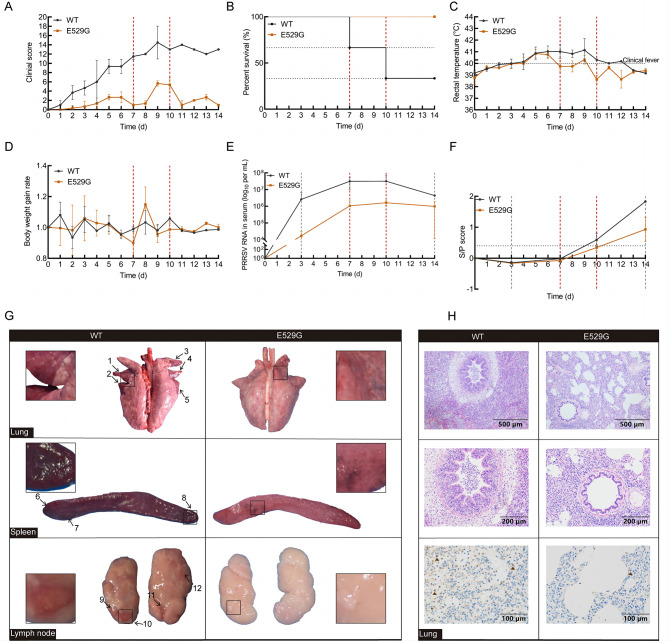
PRRS is a highly contagious and devastating disease affecting the pig industry. To investigate the antiviral activity of E529G pigs, three WT and three E529G pigs (5 weeks of age) were cohoused throughout the experiment and allowed to acclimate for three days prior to the initiation of the challenge. All pigs were confirmed to be PRRSV-specific antibody negative. The experiment lasted 14 days following inoculation of each pig with 4×107 TCID50 of JXA1, with half of the inoculum delivered intramuscularly and the other half intranasally. Clinical parameters, including clinical score, percent survival, rectal temperature, and body weight gain rate, were recorded daily during the infection monitoring period and scored (Supplementary Table S4). Serum samples were collected on days 0, 3, 7, 10, and 14 to detect viral load and viral antibody levels.
During the viral challenge, WT pigs developed characteristic clinical symptoms of PRRSV infection, including respiratory disorders, high fever, slow growth, high morbidity, and mortality (Figure 5A). The average occurrence time for typical PRRSV symptoms in WT and E529G pigs was 2 dpi and 5 dpi, respectively (Figure 5A). Notably, while E529G pigs exhibited most PRRSV-related clinical signs, the symptoms were less severe compared to those in WT pigs. In addition, two of the three WT pigs died at 7 dpi and 10 dpi, respectively, whereas all E529G pigs survived until euthanasia (Figure 5B). Rectal temperatures were significantly elevated on day 4 in all challenged pigs; however, E529G pigs began to recover by 7 dpi, while WT pigs maintained a high fever throughout the monitoring period (Figure 5C). Daily body weights indicated that the E529G pigs exhibited faster weight gain than the WT pigs (Figure 5D). These findings suggest that PRRSV infection severity is mitigated in E529G pigs compared to WT pigs. Furthermore, as illustrated in Figure 5E, serum viral load in E529G pigs was two orders of magnitude lower than that in WT pigs. Concurrently, the antibody response was significantly higher in the WT pigs than in the E529G pigs (Figure 5F). Collectively, these results indicate that E529G pigs can resist PRRSV infection.
Figure 5.
CD163-E529G mutation pigs were resistant to HP-PRRSV infection
A–D: Relative clinical symptoms (A), survival rate (B), rectal temperature (C), and body weight gain rate (D) in challenged WT (black line) and E529G (brown line) pigs were monitored daily. Survival rate showed that two WT pigs died at 7 and 10 days, respectively (red dashed line). E, F: Blood samples from challenged pigs were collected on days 0, 3, 7, 10, and 14. Viral load (E) and viral antibody levels (F) in serum were separately determined by qPCR and ELISA. G: Each photo is from a single representative pig. Macroscopic observation of lung (above), spleen (medium), and lymph node (below) from WT (left) and E529G (right) pigs, and locations of tissue lesions are marked with black arrows, partial lesion locations are enlarged. H: Histopathological diagnosis in lungs between WT (left) and E529G (right) pigs was indicated by H&E staining, with 10× magnification shown above and 20× magnification shown in the middle. Viral infection in lungs from WT and E535G pigs was determined by IHC (below).
Organs were collected for histopathological analysis at the time of death for WT pigs or following euthanasia of all challenge pigs at 14 dpi. Macroscopic observation revealed significant lesions in the lung, spleen, lymph node, liver, and kidney (Figure 5G; Supplementary Figure S5A) of WT pigs (left) but not in E529G pigs (right). To further evaluate viral resistance of E529G pigs, H&E staining was conducted, which identified interstitial edema with infiltration of mononuclear cells and marked hemorrhage in lung sections of WT pigs but not in E529G pigs (Figure 5H). Pathological examination revealed significant lesions in the liver and obvious thrombi in the kidneys of WT pigs, which were absent in E529G pigs (Supplementary Figure S5B). IHC analysis of the lung indicated more severe viral infection in WT pigs than in E529G pigs (Figure 5H). Overall, these findings demonstrate significant differences in typical symptoms, mortality, viral load, viral antibody levels, and tissue lesions between WT and E529G pigs during the viral challenge.
DISCUSSION
Genetically modified pigs with CD163 SRCR5 deletions and functional region substitutions have been shown to be resistant to PRRSV infection. However, the CD163 SRCR2 domain is implicated in erythroblast adhesion (Fabriek et al., 2007), while the SRCR3 domain is considered to be critical for the calcium-sensitive coupling of Hb-Hp complexes (Madsen et al., 2004). Xu et al. (2020) reported that SRCR5 knockout pigs exhibit significantly higher meat color scores, Fe concentrations, and Hp content than WT pigs, attributing the darker red color in SRCR5 knockout meat to decreased Hb metabolism and mild Fe-containing Hb accumulation. However, large fragment editing of CD163 raises concerns about its impact on growth performance. PRRSV is highly specific to pigs and preferentially infects PAMs (Xie et al., 2021). Additionally, MA-104 cells and their derivatives, Marc145 and CL2621, are also susceptible to PRRSV (Shi et al., 2015). As CD163 performs similar physiological functions across species, we hypothesized that certain key amino acids in CD163 may affect PRRSV proliferation. Hence, identifying these key amino acids and establishing pigs with specific CD163 amino acid mutation may provide an effective solution to prevent PRRSV infection.
Ma et al. (2021a) suggested that E529 could be a potential site for suppressing PRRSV proliferation, although their verification was conducted only in cells, not in whole organisms. Further research is needed to determine whether the E529 point mutation affects pig growth, development, and resistance to PRRSV. Virus infection experiments on Marc145 cells demonstrated that the mE535G mutation significantly inhibited HP-PRRSV proliferation by preventing PRRSV binding and entry (Figure 2).
To exclude the influence of varying CD163 copy numbers on the assessment of cell antiviral capacity, we performed single-base editing in PK-15 cells stably integrated with CD163 using NG-ABE8e, resulting in PK-15CD163 cells with the pE529G mutation and consistent CD163 copy numbers (Figure 3). Similarly, virus infection experiments on PK-15CD163 cells showed that the E529G mutation markedly inhibited PRRSV binding and entry, thereby reducing HP-PRRSV proliferation (Figure 3). Interestingly, although the inhibition trends in E535G and E529G cells were consistent, the degree of inhibition varied considerably. We hypothesize that pigs, being the only natural host of PRRSV, may possess auxiliary pathways that enhance CD163-induced virus infection in PK-15 cells stably expressing CD163. In contrast, Marc145 cells derived from monkeys, which are not natural hosts of PRRSV, can only be infected with PRRSV following artificial induction, but lack additional mechanisms to promote virus infection. This distinction may explain the differences observed in the antiviral effects between the two types of mutations.
To further explore the effects of the CD163 E529G mutation on individual growth and development, E529G PFF clones were screened, and blastocyst development was assessed. Results indicated that the E529G mutation did not affect the development of reconstructed embryos (Supplementary Table S2). Subsequently, four healthy F0 generation individuals were produced (Supplementary Figure S4B). Compared to the WT controls, the physiological parameters of the E529G pigs remained normal (Figure 4A, B; Supplementary Table S3). Notably, the E529G mutation did not affect the expression of the CD163 protein (Figure 4C, D), nor did it influence the ability to remove free Hp or clear Hb-Hp complexes in PAMs (Figure 4E–G). Additionally, PAMs derived from E529G pigs effectively inhibited the proliferation of HP-PRRSV in vitro (Figure 4H, I).
In individual challenge experiments, E529G pigs demonstrated effective defense against PRRSV infection (Figure 5). Notably, E529G pigs exhibited better anti-PRRSV efficacy than cells, likely due to the presence of both innate and acquired immunity in pigs, whereas cells only display innate immunity. None of the E529G pigs died, although the PRRSV genome was consistently detected in serum (Figure 5), which may be attributed to the high viral infective dose (4×107 TCID50) compared to previously reported ranges (1×105 to 2×106 TCID50) (Burkard et al., 2017; Prather et al., 2017; Xu et al., 2020; Yang et al., 2018).
The recent application of ABE/CBE base-editing libraries (Base Editor, ABE (A·T→G·C), CBE (C·G→T·A)) has shown promise in identifying causal mutation sites associated with disease resistance traits. Xu et al. (2021) identified several OsACC resistance loci in rice using CBE and ABE base-editing libraries. Our lab also utilized CBE base-editing libraries to identify multiple pathogenic loci in the human LDLR associated with familial hypercholesterolemia (Li et al., 2023). In the future, exploring additional key amino acids within CD163 that impact PRRSV proliferation without compromising the physiological function of CD163 via CBE and ABE base-editing libraries, with subsequent development of CD163 multi-amino acid mutant pigs, may offer an effective solution for preventing PRRSV infection.
CONCLUSIONS
PRRS has become one of the most economically devastating diseases due to its long incubation period and multiple transmission routes, with the recombination and pathogenic diversity of PRRSV presenting additional challenges for the pig industry. Reducing the impact of CD163 gene modification on normal physiological functions is essential for commercial anti-PRRSV breeding. In this study, we produced the first pig with a single amino acid mutation in CD163, thereby minimizing the adverse effects of CD163 modification. Our identification of key amino acids within CD163 has established another feasible route for CD163 gene modification, providing new insights into accelerating the development of PRRSV prevention and control.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
Acknowledgments
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
Conceived and designed the experiments: D.X.P., H.S.O.Y., and H.M.Y.; performed the experiments: Y.L., L.Y., H.Y.X., M.N., J.C.D., X.Y.L., W.J.H., T.Y.L., and X.C.T.; wrote the manuscript: Y.L., H.M.Y., and L.Y. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the Major Scientific and Technological Projects in Agricultural Biological Breeding of China (2023ZD0404302) and Youth Program of National Natural Science Foundation of China (32202754)
Contributor Information
Da-Xin Pang, Email: pdx@jlu.edu.cn.
Hong-Ming Yuan, Email: yuanhongming@jlu.edu.cn.
References
- Burkard C, Lillico SG, Reid E, et al Precision engineering for PRRSV resistance in pigs: macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathogens. 2017;13(2):e1006206. doi: 10.1371/journal.ppat.1006206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert JG, Slade DE, Shields SL, et al CD163 expression confers susceptibility to porcine reproductive and respiratory syndrome viruses. Journal of Virology. 2007;81(14):7371–7379. doi: 10.1128/JVI.00513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etzerodt A, Moestrup SK CD163 and inflammation: biological, diagnostic, and therapeutic aspects. Antioxidants & Redox Signaling. 2013;18(17):2352–2363. doi: 10.1089/ars.2012.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabriek BO, Polfliet MMJ, Vloet RPM, et al The macrophage CD163 surface glycoprotein is an erythroblast adhesion receptor. Blood. 2007;109(12):5223–5229. doi: 10.1182/blood-2006-08-036467. [DOI] [PubMed] [Google Scholar]
- Guo CH, Wang M, Zhu ZB, et al Highly efficient generation of pigs harboring a partial deletion of the CD163 SRCR5 domain, which are fully resistant to porcine reproductive and respiratory syndrome virus 2 infection. Frontiers in Immunology. 2019;10:1846. doi: 10.3389/fimmu.2019.01846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp DJ, Kliebenstein JB, Neumann EJ, et al Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. Journal of Swine Health and Production. 2013;21(2):72–84. [Google Scholar]
- Jiang FG, Doudna JA CRISPR–Cas9 structures and mechanisms. Annual Review of Biophysics. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- Jing QQ, Liu WW, Jiang HY, et al Highly efficient A-To-G editing in pffs via multiple abes. Genes. 2023;14(4):908. doi: 10.3390/genes14040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kick AR, Grete AF, Crisci E, et al Testable candidate immune correlates of protection for porcine reproductive and respiratory syndrome virus vaccination. Vaccines. 2023;11(3):594. doi: 10.3390/vaccines11030594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Ma LR, Chen YW, et al Large-scale CRISPR screen of LDLR pathogenic variants. Research. 2023;6:0203. doi: 10.34133/research.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunney JK, Fang Y, Ladinig A, et al Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annual Review of Animal Biosciences. 2016;4:129–154. doi: 10.1146/annurev-animal-022114-111025. [DOI] [PubMed] [Google Scholar]
- Ma HF, Li R, Jiang LG, et al Structural comparison of CD163 SRCR5 from different species sheds some light on its involvement in porcine reproductive and respiratory syndrome virus-2 infection in vitro. Veterinary Research. 2021a;52(1):97. doi: 10.1186/s13567-021-00969-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Ma LL, Yang MT, et al The function of the PRRSV–host interactions and their effects on viral replication and propagation in antiviral strategies. Vaccines. 2021b;9(4):364. doi: 10.3390/vaccines9040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen M, Møller HJ, Nielsen MJ, et al Molecular characterization of the haptoglobin•hemoglobin receptor CD163: ligand binding properties of the scavenger receptor cysteine-rich domain region. Journal of Biological Chemistry. 2004;279(49):51561–51567. doi: 10.1074/jbc.M409629200. [DOI] [PubMed] [Google Scholar]
- Moestrup S, Møller H CD163: a regulated hemoglobin scavenger receptor with a role in the anti‐inflammatory response. Annals of Medicine. 2004;36(5):347–354. doi: 10.1080/07853890410033171. [DOI] [PubMed] [Google Scholar]
- Prather RS, Wells KD, Whitworth KM, et al Knockout of maternal CD163 protects fetuses from infection with porcine reproductive and respiratory syndrome virus (PRRSV) Scientific Reports. 2017;7(1):13371. doi: 10.1038/s41598-017-13794-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renken C, Nathues C, Swam H, et al Application of an economic calculator to determine the cost of porcine reproductive and respiratory syndrome at farm-level in 21 pig herds in Germany. Porcine Health Management. 2021;7(1):3. doi: 10.1186/s40813-020-00183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado B, Rivas RB, Pinto D, et al Genetically modified pigs lacking CD163 PSTII-domain-coding exon 13 are completely resistant to PRRSV infection. Antiviral Research. 2024;221:105793. doi: 10.1016/j.antiviral.2024.105793. [DOI] [PubMed] [Google Scholar]
- Shi CX, Liu YL, Ding YZ, et al PRRSV receptors and their roles in virus infection. Archives of Microbiology. 2015;197(4):503–512. doi: 10.1007/s00203-015-1088-1. [DOI] [PubMed] [Google Scholar]
- Stoian AMM, Rowland RRR, Brandariz-Nuñez A Identification of CD163 regions that are required for porcine reproductive and respiratory syndrome virus (PRRSV) infection but not for binding to viral envelope glycoproteins. Virology. 2022a;574:71–83. doi: 10.1016/j.virol.2022.07.012. [DOI] [PubMed] [Google Scholar]
- Stoian AMM, Rowland RRR, Brandariz-Nuñez A Mutations within scavenger receptor cysteine-rich (SRCR) protein domain 5 of porcine CD163 involved in infection with porcine reproductive and respiratory syndrome virus (PRRS) Journal of General Virology. 2022b;103(5):001740. doi: 10.1099/jgv.0.001740. [DOI] [PubMed] [Google Scholar]
- Subramanian K, Du RJ, Tan NS, et al CD163 and igg codefend against cytotoxic hemoglobin via autocrine and paracrine mechanisms. The Journal of Immunology. 2013;190(10):5267–5278. doi: 10.4049/jimmunol.1202648. [DOI] [PubMed] [Google Scholar]
- Van Gorp H, Van Breedam W, Delputte PL, et al Sialoadhesin and CD163 join forces during entry of the porcine reproductive and respiratory syndrome virus. Journal of General Virology. 2008;89(12):2943–2953. doi: 10.1099/vir.0.2008/005009-0. [DOI] [PubMed] [Google Scholar]
- Welch SKW, Calvert JG A brief review of CD163 and its role in PRRSV infection. Virus Research. 2010;154(1-2):98–103. doi: 10.1016/j.virusres.2010.07.018. [DOI] [PubMed] [Google Scholar]
- Wells KD, Bardot R, Whitworth KM, et al Replacement of porcine CD163 scavenger receptor cysteine-rich domain 5 with a CD163-like homolog confers resistance of pigs to genotype 1 but not genotype 2 porcine reproductive and respiratory syndrome virus. Journal of Virology. 2017;91(2):e01521–16. doi: 10.1128/JVI.01521-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth KM, Lee K, Benne JA, et al Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biology of Reproduction. 2014;91(3):78. doi: 10.1095/biolreprod.114.121723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth KM, Rowland RRR, Ewen CL, et al Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nature Biotechnology. 2016;34(1):20–22. doi: 10.1038/nbt.3434. [DOI] [PubMed] [Google Scholar]
- Xie JX, Vereecke N, Theuns S, et al Comparison of primary virus isolation in pulmonary alveolar macrophages and four different continuous cell lines for type 1 and type 2 porcine reproductive and respiratory syndrome virus. Vaccines. 2021;9(6):594. doi: 10.3390/vaccines9060594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie ZC, Pang DX, Yuan HM, et al Genetically modified pigs are protected from classical swine fever virus. PLoS Pathogens. 2018;14(12):e1007193. doi: 10.1371/journal.ppat.1007193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhou YR, Mu YL, et al CD163 and pAPN double-knockout pigs are resistant to PRRSV and TGEV and exhibit decreased susceptibility to pdcov while maintaining normal production performance. eLife. 2020;9:e57132. doi: 10.7554/eLife.57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RF, Liu XS, Li J, et al Identification of herbicide resistance OsACC1 mutations via in planta prime-editing-library screening in rice. Nature Plants. 2021;7(7):888–892. doi: 10.1038/s41477-021-00942-w. [DOI] [PubMed] [Google Scholar]
- Yang HQ, Zhang J, Zhang XW, et al CD163 knockout pigs are fully resistant to highly pathogenic porcine reproductive and respiratory syndrome virus. Antiviral Research. 2018;151:63–70. doi: 10.1016/j.antiviral.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Yu P, Wei RP, Dong WJ, et al CD163ΔSRCR5 MARC-145 cells resist PRRSV-2 infection via inhibiting virus uncoating, which requires the interaction of CD163 with calpain 1. Frontiers in Microbiology. 2020;10:3115. doi: 10.3389/fmicb.2019.03115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ZB, Zhang M, Yuan LL, et al LGP2 promotes type I interferon production to inhibit PRRSV infection via enhancing MDA5-mediated signaling. Journal of Virology. 2023;97(1):e0184322. doi: 10.1128/jvi.01843-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.