Abstract
Litopenaeus vannamei is the most extensively cultured shrimp species globally, recognized for its scale, production, and economic value. However, its aquaculture is plagued by frequent disease outbreaks, resulting in rapid and massive mortality. etiological research often lags behind the emergence of new diseases, leaving the causal agents of some shrimp diseases unidentified and leading to nomenclature based on symptomatic presentations, especially in cases involving co- and polymicrobial pathogens. Comprehensive data on shrimp disease statuses remain limited. In this review, we summarize current knowledge on shrimp diseases and their effects on the gut microbiome. Furthermore, we also propose a workflow integrating primary colonizers, “driver” taxa in gut networks from healthy to diseased states, disease-discriminatory taxa, and virulence genes to identify potential polymicrobial pathogens. We examine both abiotic and biotic factors (e.g., external and internal sources and specific-disease effects) that influence shrimp gut microbiota, with an emphasis on the “holobiome” concept and common features of gut microbiota response to diverse diseases. After excluding the effects of confounding factors, we provide a diagnosis model for quantitatively predicting shrimp disease incidence using disease common-discriminatory taxa, irrespective of the causal agents. Due to the conservation of functional genes used in designing specific primers, we propose a practical strategy applying qPCR-assayed abundances of disease common-discriminatory functional genes. This review updates the roles of the gut microbiota in exploring shrimp etiology, polymicrobial pathogens, and disease incidence, offering a refined perspective for advancing shrimp aquaculture health management.
Keywords: Shrimp disease, Gut microbiota, Polymicrobial pathogens, Diagnosis model, Disease common-discriminatory taxa, Disease prediction
INTRODUCTION
Litopenaeus vannamei has emerged as one of the most successful aquaculture species due to its low food conversion ratio, contributing significantly to the “Blue Revolution”. Over the past 40 years, its production has increased from less than 0.075 million tons in 1980 to over 7.3 million tons in 2021 (FAO, 2023). This remarkable achievement is partially attributed to the development of large-scale production facilities, high-density farming practices, and intensive feeding regimens, which have also created stressful conditions that facilitate the frequent occurrence and rapid spread of diverse infectious diseases (Table 1). Although antibiotics are commonly applied as prophylactic and therapeutic agents, their overuse and abuse has led to bacterial resistance, increased disease susceptibility, and the emergence of new diseases (Lu et al., 2021; Rodgers & Furones, 2009; Sha et al., 2024a; Zhou et al., 2019a). Vaccination represents an alternative strategy for controlling infectious diseases; however, its efficacy is often limited in juvenile shrimp due to their inability to develop long-term acquired immunity (Falaise et al., 2016). Importantly, shrimp diseases cause rapid and massive mortality within days, often without noticeable symptoms at onset stage of disease (Kumar et al., 2020; Thitamadee et al., 2016; Xiong et al., 2017a). Several methods are currently used to evaluate disease risk in shrimp aquaculture, including water quality indices, Vibrio species density, pathogen diagnostics, as well as artificial intelligence and machine learning techniques (Bohara et al., 2024; Ferreira et al., 2011; MacAulay et al., 2022). However, these approaches are predominantly qualitative, rather than quantitative, and lag behind the detection of early stages of disease onset stage. Therefore, there is an urgent need to establish quantitative strategies for predicting the incidence of shrimp disease.
Table 1. Summary of shrimp diseases, potential causative agents, and typical symptoms.
| Disease name | Causative agent | Typical symptoms | References |
| White spot disease | White spot syndrome virus | Sharp reduction in food consumption, listlessness, loose cuticle, white spots 0.5–2.0 mm in diameter | Liu et al., 2009 |
| Taura syndrome | Taura syndrome virus | Soft cuticle, empty guts, extreme lethargy, and expansion of reddish chromatophores | Brock, 1997 |
| Yellow head disease | Yellow head virus | Nuclear pyknosis and heterokaryosis, bleached or pale-yellow cephalothorax, discoloration of underlying hepatopancreas | Boonyaratpalin et al., 1993 |
| Tetrahedral baculovirosis | Baculovirus penaei | Whitish midguts in severe infections. Polyhedral occlusion bodies | Couch, 1974 |
| Spherical baculovirosis | Monodon baculovirus | Hypertrophied nuclei with single or multiple eosinophilic occlusion bodies, chromatin diminution and margination | Chen et al., 1989 |
| Infectious hypodermal and hematopoietic necrosis (IHHN) | IHHN virus | Buff-colored spots at carapace and abdominal plates, bluish, necrotic, melanized cuticle in serious disease | Lightner et al., 1983 |
| Infectious myonecrosis (IMN) | IMN virus | Opaque skeletal muscle | Lightner et al., 1983 |
| White tail disease | Macrobrachium nodavirus | Tails exhibiting white lesions | Arcier et al., 1999 |
| Hepatopancreatic parvovirus disease | Hepatopancreatic parvovirus | Slowed growth rate | Chong & Loh, 1984 |
| Hemocyte iridescent virus disease | Hemocyte iridescent virus | Hepatopancreatic atrophy with color fading, empty stomach and guts, and soft shell | Qiu et al., 2017 |
| Running mortality syndrome | Nodavirus | Enlarged nuclei in hepatopancreas, coagulative muscle necrosis, whitening muscle, slow growth and softening shell | Zhang et al., 2014 |
| Abdominal segment deformity disease | Unknown retrovirus-like agent | Enlarged or twisted abdominal segments, either laterally and/or dorsoventrally, sometimes opaque muscles | Sakaew et al., 2008 |
| Slow growth syndrome | Laem Singh virus and a cryptic integrase containing element | Slow growth and coefficients of size variation > 35% | Chayaburakul et al., 2004 |
| Acute hepatopancreatic necrosis disease | Vibrio anguillarum, Vibrio parahaemolyticus | Sloughing tubule epithelial cells, pale hepatopancreas, massive and rapid mortality | Tran et al., 2013 |
| Necrotizing hepatopancreatitis | Rickettsia-like bacterium | Reduced feeding activity, empty guts, pale hepatopancreas | Chayaburakul et al., 2004 |
| “Bright-red” syndrome | Vibrio harveyi | Lethargy, anorexia, flaccid body, multifocal reddish discoloration spots on abdominal cuticle, melanized erosions around spots | Soto-Rodriguez et al., 2010 |
| Luminous vibriosis | Vibrio campbellii | Symptoms include fluorescent, less food consumption and high mortality | Wang et al., 2015 |
| White feces syndrome | Uncertain Vibrio | White-golden gut contents and white fecal strings | Durai et al., 2015 |
| Black gill disease | Fusarium solani, Fusarium moniliforme, Aspergillus awamori | Changes from “gill discoloration” to “blackened gill” with disease progression, with subsequent death | Hatai & Egusa, 1978 |
| Balantidiasis | Zoothamnium | Reduction in feeding and growth. Increased risk of secondary bacterial infections and massive mortality | Nurlatiffah et al., 2018 |
| Hepatopancreatic microsporidiosis | Enterocytozoon hepatopenaei | Severely stunted growth | Chayaburakul et al., 2004 |
| Amoebic gill disease | Amphizoic amoeba | Reduced appetite, lethargy, respiratory distress, eroded carapace, and blackened gill | Han, 2019 |
| “Cotton shrimp-like” disease | Uncertain fungi | Reduced growth, atrophic hepatopancreas, empty digestive tract, and inactivity. Soft shell with slightly white, opaque muscle | Zhou et al., 2019b |
| Blue body syndrome | Unknown | Slow growth, reduced or no feed intake, thin, partially or fully blue bodies | Liang et al., 2020 |
The gut microbiota plays an indispensable role in maintaining shrimp fitness and health by aiding in digestion, vitamin synthesis, immunity regulation, toxin degradation, and pathogen suppression (Cornejo-Granados et al., 2018; Holt et al., 2021; Sha et al., 2024b; Xiong, 2018). As such, the gut microbiota acts as a central hub, integrating environmental exposure with genetic and immune signals to influence shrimp fitness. In healthy shrimp, optimal balance of gut symbionts provides numerous benefits, while an imbalanced gut microbiota increases disease risk (Holt et al., 2021; Xiong, 2018). Researches on human and terrestrial animal microbiomes have established cause-and-effect relationships, demonstrating that a healthy gut microbiome influences the colonization, growth, and virulence of pathogens, thereby affecting disease outcomes (Bäumler & Sperandio, 2016; Kamada et al., 2013). However, comparable studies on the causal roles of the gut microbiota in aquatic invertebrates such as shrimp are scarce (Sha et al., 2024b). High-throughput sequencing (HTS) and metagenomics have provided increasing evidence that the gut microbiomes of diseased shrimp differ significantly from those of healthy individuals (Holt et al., 2021; Mao et al., 2023; Sha et al., 2022b), a condition known as “dysbiosis”. This suggests that sensitive gut commensals could serve as reliable indicators for diagnosing shrimp health. Growing evidence suggests that the extent of deviation from the healthy gut microbiota may be linearly correlated with the severity of shrimp disease (Chen et al., 2017; Lu et al., 2023; Xiong et al., 2017a). Consequently, gut bacterial indicators could be used to diagnose the onset and severity of shrimp diseases.
The causal links between gut microbiota and shrimp health remain relatively unexplored compared to the extensive research conducted on humans. Advances in sequencing technologies have significantly enhanced our understanding of the interactions between shrimp health and their gut microbiota. Nevertheless, much remains to be discovered regarding the shared and unique responses of the gut microbiome to diverse shrimp diseases. Recent reviews have highlighted external factors influencing shrimp microbiota (Cornejo-Granados et al., 2018; Holt et al., 2021) and diagnostic approaches ranging from traditional visual inspection and histopathology to portable sequencing and artificial intelligence for early disease detection (Bohara et al., 2024; MacAulay et al., 2022). In contrast, this review focuses on the current state of shrimp diseases and their effects on the gut microbiota. Going a step further, we conclude the common features of gut microbiota responses to diverse shrimp diseases and their potential applications in predicting disease incidence. While the concept of co- and polymicrobial pathogens is accepted in human and veterinary medicine (Frisan, 2021), it has received little attention in aquaculture. Given the challenges of identifying polymicrobial pathogens using Koch’s postulates, we propose an integrated workflow that incorporates diverse ecological features to identify potential polymicrobial pathogens.
CURRENT STATE OF SHRIMP DISEASES
One of the primary challenges in shrimp aquaculture is the frequent occurrence of disease outbreaks, leading to rapid and massive mortality (Seibert & Pinto, 2012; Thitamadee et al., 2016). Over the past two decades, scientific research on shrimp diseases has intensified in response to the global expansion of shrimp production. Shrimp are susceptible to infections from a wide range of pathogens across microbial kingdoms, including viruses, bacteria, fungi, and parasites, throughout the entire aquaculture cycle (Table 1). Despite periodic updates from the World Organization for Animal Health (WOAH) on the status on shrimp diseases, a comprehensive understanding is still lacking. The following section provides a brief summary of current knowledge on shrimp diseases and their impact on the gut microbiome.
Viral pathogens
Approximately 20 viruses have been identified as causative agents of shrimp diseases, including members of the Parvoviruses, Baculoviruses, Picornaviruses, Toga-like viruses, as well as several newly identified viral families (Arulmoorthy et al., 2020). Among these, white spot syndrome virus (WSSV) poses the most significant threat to shrimp health globally, leading to substantial production losses (Flegel, 2006; Liu et al., 2009). WSSV, an ovaloid, double-stranded DNA virus with a lipid envelope (120–150 nm×270–290 nm), infects the nuclei of mesodermal- and ectodermal-derived tissues, resulting in reduced food intake and lethargy in infected shrimp (Pradeep et al., 2012). Intriguingly, WSSV can attach to abiotic microplastics, thereby prolonging their survival and virulence under varying water pH and temperatures (Shan et al., 2023). This persistence in abiotic hosts may contribute to the recurrence of WSSV infections. Since its first report in the early 1990s, global economic losses attributed to WSSV have been established at approximately $15 billion USD, increasing at an alarming rate of $1 billion USD per year and accounting for a 10% loss in global shrimp production (Panchal et al., 2021). Currently, there is no effective strategy to prevent and control this disease. WSSV infection significantly alters the composition of the shrimp gut microbiota, leading to enrichment in Proteobacteria and Fusobacteria phyla, as well as pathogenic bacteria belonging to the Arcobacter and Flavobacterium genera (Wang et al., 2019). While gut bacterial diversity and richness remain unaffected, WSSV infection increases the heterogeneity of gut bacterial communities (de Souza Valente et al., 2020). Additionally, prior WSSV infection predisposes shrimp to subsequent Vibrio campbellii infections, resulting in more rapid and severe mortality compared to V. campbellii alone (Phuoc et al., 2008).
In addition to WSSV, yellow head virus (YHV), specifically genotype one among six genetically related genotypes, is a rod-shaped, enveloped positive-sense single-stranded RNA virus (40–60 nm×150–200 nm). YHV induces persistent and severe disease in shrimp (Boonyaratpalin et al., 1993), although major outbreaks have not been reported in recent years. YHV has been detected exclusively in outdoor ponds, but not in covered ponds, suggesting an airborne mode of transmission (Thitamadee et al., 2016). However, potential carriers and non-aquatic insects have tested negative for YHV, casting doubt on the airborne vector hypothesis. The rapid dissemination of YHV may be attributed to the cannibalistic behavior of YHV-infected individuals (Tuyen et al., 2014). The initial source of YHV infection remains unclear, but the close genetic relationship among YHV genotypes suggests that it may naturally infect wild penaeid or palemonid shrimp (Wijegoonawardane et al., 2008). Surprisingly, limited research has been conducted on the response of the shrimp gut microbiota to YHV infection.
Decapod iridescent virus 1 (DIV1), a recently identified icosahedral linear double-stranded DNA virus, induces stunted growth and 100% cumulative mortality in shrimp (Qiu et al., 2017). DIV1 infection reduces the abundance of potentially beneficial populations of Bacteroidetes, Firmicutes, and Actinobacteria, while significantly enriching pathogenic bacteria such as Vibrio and Photobacterium. These microbial shifts elevate the risk of secondary bacterial infections. Furthermore, the proliferation of pathogenic bacteria triggers the expression of NF-κB inhibitors, which suppress the TLR-mediated immune response, exacerbating DIV1 infection (Liao et al., 2023).
In summary, viral infections frequently disrupt the composition of shrimp gut microbiota, increasing susceptibility to secondary bacterial infections (Liao et al., 2023; Shan et al., 2023). However, the specific alterations in bacterial populations appear to be dependent on the viral pathogen involved.
Bacterial pathogens
Vibriosis is the most prevalent and severe bacterial disease endangering shrimp aquaculture, caused by gram-negative Vibrio bacteria such as V. harveyi, V. parahaemolyticus, V. vulnificus, and V. anguillarum (Kumar et al., 2020; Li et al., 2022; Shen et al., 2021a). These diseases are typically elicited by external stressors and, if left untreated, can result in rapid and extensive mortality. Although many Vibrio species are opportunistic pathogens, certain strains, such as V. harveyi and V. parahaemolyticus, can act as acute pathogens (Dhar et al., 2019; Zhou et al., 2012). The V. harveyi clade comprises luminous bacteria commonly detected in coastal and marine waters, as well as in the skin and gut of aquatic organisms (Ina‐Salwany et al., 2019; Vandenberghe et al., 2003). Vibrio harveyi infections can cause multiple shrimp diseases, including “bright-red syndrome” (Soto-Rodriguez et al., 2010), “bacterial white tail disease” (Zhou et al., 2012), and luminescent vibriosis (Mandal & Das, 2018; Wang et al., 2015). In addition, V. parahaemolyticus carrying the pirAB gene can cause acute hepatopancreatic necrosis disease (AHPND) (Han et al., 2015; Tran et al., 2013) and red disease in shrimp (Jayasree et al., 2006). In addition to Vibrio, other pathogenic genera have been reported, such as Shewanella algae, which causes black spot disease (Cao et al., 2018), Acinetobacter venetianus, which causes red leg disease (Huang et al., 2020a), and Streptocephalus sirindhornae and Branchinella thailandensis, which cause black disease (Saejung et al., 2011). Notably, some Vibrio species, such as V. hepatarius and V. diabolicus, are non-pathogenic and serve as probiotics that protect shrimp from infection (Ramirez et al., 2022; Restrepo et al., 2021). Consequently, traditional monitoring of Vibrio species density may erroneously suggest an elevation of disease risk.
Gut bacterial communities in shrimp are disrupted upon exposure to V. harveyi, with amplicon sequence variants (ASVs) from the Harveyi and Vulnificus clades becoming enriched (Angthong et al., 2023). Interestingly, the resilience of the gut microbiota can mitigate shrimp mortality during V. harveyi infection (Rungrassamee et al., 2016). Vibrio strains carrying the pVA1 plasmid, which encodes the binary pirAB toxin genes, are the causal agents of shrimp AHPND (Han et al., 2015). The virulent plasmid pVA1 harbors a post-segregational killing system, enabling its stable inheritance and dissemination among Vibrio species (Kumar et al., 2020) and non-Vibrio and non-pathogenic strains, such as Algoriphagus species, via conjugative transfer (Muthukrishnan et al., 2019). These molecular characteristics partially explain the difficulty in controlling shrimp AHPND. Indeed, AHPND poses a serious threat to the shrimp industry worldwide, often causing extensive or complete mortality within days (Kumar et al., 2020). The occurrence of AHPND substantially reduces α-diversity and the stability of the gut bacterial community, while increasing the variation coefficient and ecological dysbiosis indices compared to healthy shrimp (Dong et al., 2021). Of note, the gut microbiota exhibits different responses to infection by AHPND-causing V. parahaemolyticus (Vp) compared to common pathogenic Vp. Specifically, AHPND-causing Vp infection results in rapid shifts in the gut bacterial community, with enrichment of Photobacterium and Vibrio genera. In contrast, common Vp infection increases the abundance of gut Motilimonas and Ruegeria genera (Chang et al., 2023). Furthermore, variations in the bacterioplankton community in rearing water correlate with the virulence of Vp (Aguilar-Rendón et al., 2022). Similarly, elevated nutrient levels enhance the virulence of AHPND-causing Vp (Li et al., 2022), leading to quicker dysbiosis in the gut microbiota and earlier and higher mortality in shrimp compared to AHPND-causing Vp precultured at oligotrophic condition (Sha et al., 2022a). Intriguingly, the gut microbiota responds linearly to increasing shrimp AHPND severity, enabling accurate diagnosis of progressed stages of AHPND using gut indicators (Lu et al., 2023; Shen et al., 2021a).
Fungal pathogens
Fungi are prevalent in aquatic environments, with approximately 500 species isolated from marine and estuarine habitats, including several that act as opportunistic pathogens of shrimp (Karunasagar et al., 2004). Fungal infections typically result in chronic and steady losses, causing lower mortality rates compared to viral and bacterial infections (Paulraj et al., 2016). Common pathogenic fungi include Lagenidium callinectes and Sirolpidium species (Karunasagar et al., 2004), which preferentially infect larval shrimp. These pathogens enter larvae through antennae, eyestalks, and body tissues, ultimately causing death. Fungal spores and mycelia are detectable in targeted tissues, particularly the gills and appendages, rather than in the shrimp gut and hepatopancreas (Karunasagar et al., 2004). Endophytic Fusarium species, known plant pathogens producing fumonisins and trichothecenes that affect human and animal health (Munkvold, 2017), are also causal agents of shrimp black gill disease (BG), leading to chronic mortality over prolonged infections (Khoa et al., 2004). Research on the effects of fungal pathogens on the shrimp gut microbiota is limited compared to studies on viral and bacterial infections. Some studies indicate that fungal infections alter gut microbiota composition. For example, shrimp with BG harbor higher levels of the pathogenic fungus Candida, and reduced abundances of Didymella and Filobasidium in the gut (Li et al., 2019). However, gut microbial diversity remains comparable between healthy and BG-affected cohorts (Song et al., 2022). Although many fungi are saprophytic or parasitic, certain strains, such as Aspergillus niger and A. fumigatus are beneficial for shrimp growth, health, and antifungal immunity (Brzezinska & Jankiewicz, 2012; Li et al., 2012; Zhang et al., 2023).
Parasitic pathogens
Parasites are detrimental to their hosts as they exploit host nutrients for their own utilization, adversely affecting host immunity and growth. Various parasites, particularly protozoa, impact shrimp at different developmental stages (Table 1). Among these, Enterocytozoon hepatopenaei (EHP) has emerged as an important pathogen in shrimp aquaculture (Aranguren et al., 2017). EHP is an intracellular and microsporidian parasite that causes shrimp hepatopancreatic microsporidiosis, leading to stunted growth (Chayaburakul et al., 2004). Shrimp size is inversely associated with EHP load (Shen et al., 2021b). Environmental reservoirs can harbor EHP, resulting in recurrent infections in shrimp populations (Raj et al., 2023). EHP infection follows a cyclic pattern, where shrimp with severe infections can regress to milder infection, but never fully recover. This parasite-host interaction impairs shrimp health, leading to stunted growth and low mortality (López-Carvallo et al., 2022). Although EHP infection does not cause acute mortality, it increases shrimp susceptibility to subsequent microbial infections, including Vibrio species (Shen et al., 2021a), and WSSV (Suryakodi et al., 2022). The impact of EHP on the shrimp gut microbiota is less understood compared to bacterial and viral pathogens. The gut microbiota is affected by EHP infection severity, with enrichment of Vibrio, Pseudomonas, and Bradyrhizobium species (Shen et al., 2021a). EHP infection progression displaces beneficial microbes with opportunistic/pathogenic bacteria and fungi, depleting energy reserves. Specifically, the late stages of EHP infection are marked by an increase in gut bacterial genera such as Leadbetterella, Aquimarina, and Vibrio, and the fungal genus Malassezia (López-Carvallo et al., 2022). EHP infection causes greater dispersion, lower modularity, and a decrease in the proportion of negative associations and keystone taxa within the gut network, resulting in an unstable gut microbiota (Shen et al., 2022). These alterations, in turn, compromise the capacity of shrimp to combat invading pathogens (Xiong, 2018). Chronic EHP infection progressively disrupts the bacterial communities in the gut and hepatopancreas (López-Carvallo et al., 2022; Shen et al., 2022). Consequently, gut EHP-discriminatory taxa can accurately distinguish healthy shrimp from EHP-infected individuals with infection progression (Shen et al., 2022). It is important to note that not all parasitic protozoa are harmful; some, such as certain phagotrophs, can be commensal or beneficial to shrimp health. For example, gut phagotrophs can directly prey on bacterial pathogens, thereby supporting shrimp health through interkingdom predator-prey interactions (Lu et al., 2022).
Emerging and syndromic diseases
Given the lag in etiological studies, the causal agents of various shrimp diseases are unknown. In such cases, the emerging and syndromic diseases are named based on typical symptoms, such as white feces syndrome (WFS) (Durai et al., 2015), “blue body syndrome” (Liang et al., 2020), and “cotton shrimp-like” disease (Zhou et al., 2019b) (Table 1).
Shrimp WFS typically manifests 50 to 60 days post-inoculation, causing persistent mortality, reducing survival rates by 20% to 30% and decreasing yield by up to 60% (Durai et al., 2015). WFS is characterized by white-golden gut contents and white fecal strings, but its complex etiology remains inconclusive. In this context, efforts devoted to the exploration of the etiological factors of WFS could significantly contribute to the current paradigm. The etiology of WFS has been proposed as a co-infection involving EHP and opportunistic marine pathogens, including Vibrio and Propionigenium species (Sriurairatana et al., 2014; Subash et al., 2023). Nevertheless, experimental challenges using these pathogens have failed to satisfy Koch’s postulates (Aranguren Caro et al., 2021; Munkongwongsiri et al., 2022). Transplantation of gut microbiota from WFS-affected shrimp to healthy subjects induces WFS in recipients, suggesting a causal role for gut dysbiosis in WFS (Huang et al., 2020b). Shrimp infected with WFS show enrichment of various gut bacteria, such as Candidatus Bacilloplasma, Phascolarctobacterium, and Vibrio, but decreased levels of Paracoccus and Lactococcus based on field and microcosm studies (Boopathi et al., 2023; Hou et al., 2018; Huang et al., 2020b). Both gut bacterial and microeukaryotic disease-discriminatory taxa indicate the progression of WFS in shrimp (Dai et al., 2019a; Xiong et al., 2017a). While these findings collectively suggest that gut microbiota dysbiosis is implicated in WFS etiology, the causal pathogens involved in WFS remain uncertain. Recent studies integrating primary colonizers, “driver” taxa in gut networks from healthy and WFS-affected shrimp, WFS-discriminatory taxa, and encoding virulence genes have identified V. fluvialis, V. coralliilyticus, and V. tubiashii as potential WFS pathogens (Dai et al., 2018; Lu et al., 2020). The causal roles of the three pathogens in shrimp WFS have been further validated through pure culture experiments (Sha et al., 2024b). Given ongoing debates, other pathogens implicated in shrimp WFS cannot be ruled out, meriting further corroborative research.
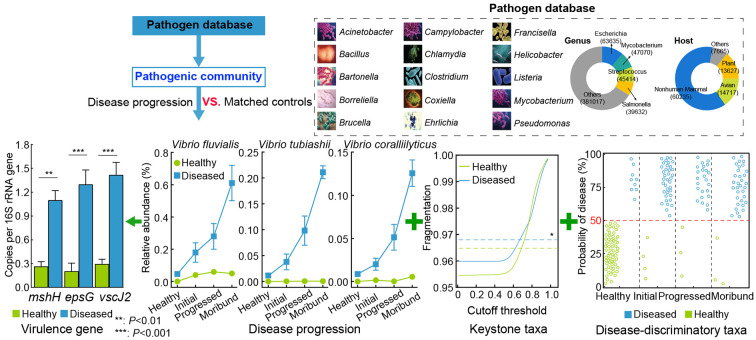
Based on previous studies, we propose a workflow for identifying potential polymicrobial pathogens (Figure 1). Initially, the taxonomies of the microbial community are assigned by blasting against an up-to-date pathogen database with strict criteria, including 100% similarity (Lo et al., 2023). Due to the high temporal dynamics of the gut microbiota as shrimp age (Figure 2A) (Xiong et al., 2020), age-discriminatory taxa are excluded to account for host age effects (Figure 2B). After this optimization, primary responders, which are absent from healthy individuals and show linear enrichment during disease progression, are extracted from the community. Next, keystone taxa are identified using a causal inference algorithm integrated with dynamic intervention simulation (Wu et al., 2022). The removal of these keystone taxa results in network fragmentation (Dai et al., 2019b). These potential pathogens can serve as biomarkers for accurately diagnosing shrimp health status. Finally, candidate polymicrobial pathogens are confirmed using molecular approaches, such as pathogen-specific virulence genes. The infection order and ratio of the polymicrobial pathogens can be inferred by tracing back to the gut microbiota in disease-progressed individuals (Figure 1). This workflow provides a robust solution for identifying polymicrobial pathogens (Dai et al. 2018; Lu et al. 2020). The efficacy of this workflow is exemplified in our recent work, which identified V. fluvialis, V. coralliilyticus, and V. tubiashii as the causative pathogens of shrimp WFS (Sha et al., 2024b).
Figure 1.
Proposed workflow for identifying potential polymicrobial pathogens
Polymicrobial pathogens are identified based on high similarity with pathogens, linear enrichment over disease progression, keystone taxa, disease-discriminatory taxa, and pathogen-specific virulence genes.
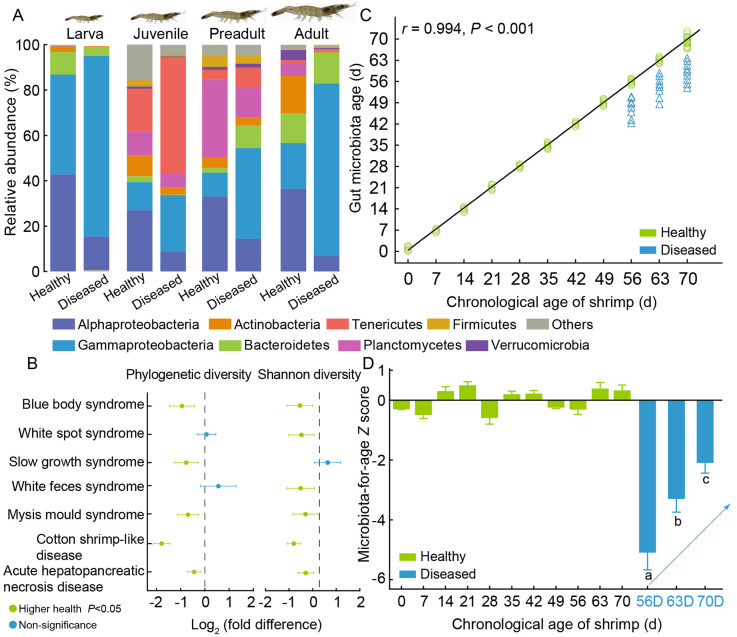
Figure 2.
Gut microbiota between shrimp health status and over life stages
A: Comparison of dominant gut bacterial phyla or classes between healthy and diseased shrimp at each life stage. B: Gut microbial diversity showing no consistent change between shrimp health status. C: Regression between actual age of healthy shrimp against predicted age using age-discriminatory taxa profiles. Predicted age of diseased shrimp significantly deviated from linear fit. D: Microbiota maturity expressed as a microbiota-for-age Z score, with mean ± standard deviation. Differences in microbiota maturity at different ages between healthy (green bar) and diseased (blue bar) shrimp based on post-hoc Tukey’s multiple comparison.
Running mortality syndrome (RMS) leads to persistent low-level mortality throughout the shrimp culture period (Zhang et al., 2014). RMS-affected shrimp are negative for all major WOAH-listed pathogens as well as common pathogens, such as WSSV, YHV, and Vibrio species. Moreover, cohabitation of healthy and RMS-affected shrimp does not result in RMS transmission (Alavandi et al., 2019). The incidence of RMS is positively associated with high stocking density, elevated nitrite levels, and increased turbidity, indicating that RMS may be related to pond ecosystem conditions rather than being infectious in nature. However, recent research has identified covert mortality nodavirus (CMNV) as the causal agent of RMS. CMNV infection suppresses the expression of genes involved in digestion, absorption, and growth hormones, which is consistent with the stunted growth observed in shrimp infected with CMNV (Liu et al., 2022).
The two aforementioned diseases reinforce that our understanding of the etiology of newly emerging shrimp diseases is still lacking. Given deteriorating water quality, increasing stocking density, and extreme weather events, this knowledge gap is likely to persist. However, the identification of causal agents for emerging and syndromic diseases can be achieved through the dedicated efforts of aquatic scientists and the advancement of new technologies.
OVERVIEW OF FACTORS AFFECTING GUT MICROBIOTA OF SHRIMP
Bacterioplankton and shrimp coexist in their rearing environments, forming a mutually beneficial equilibrium characterized by synergistic interactions. The most significant association occurs in the gut, where the microbiota begins to establish from the egg stage, acquiring microbes from the surrounding water, initial feed, and younger individuals (Xiong, 2018; Zhang et al., 2021a). It is increasingly clear that the gut microbiota plays indispensable roles in shrimp fitness, immunity, and health (Holt et al., 2021; Xiong et al., 2019a). Regulating the gut microbiome offers a promising strategy for disease prevention in shrimp aquaculture. However, developing effective approaches requires knowledge of the factors influencing the shrimp gut microbiota. Extensive research has identified various factors affecting the shrimp gut microbiota, including diet (Arce et al., 2021), rearing water salinity (Deris et al., 2022; Hou et al., 2020), temperature (Xiong et al., 2015), nitrite exposure (Huang et al., 2020c), chronic hypoxia (Sun et al., 2020), and stock density (Deng et al., 2019). In addition to these abiotic variables, biotic factors also strongly influence the gut microbiota, including life stage (Xiong et al., 2020), pathogen exposure (Angthong et al., 2023), disease severity (Xiong et al., 2017a), causal agents (Song et al., 2022), and interkingdom predator-prey interactions (Lu et al., 2022). Meta-analysis has shown that water type (seawater or freshwater) is the primary factor shaping the shrimp gut microbiota, followed by lifestyle (wild-type, farm, or laboratory), life stage, diet, and health status (Cornejo-Granados et al., 2018). Correspondingly, our recent research on marine-cultured shrimp highlighted the impact of life stage and different diseases on shaping the gut microbiota (Mao et al., 2023). While it has been suggested that microbial communities exhibit functional redundancy due to their high diversity (Miki et al., 2014), this redundancy diminishes when disturbances, such as abrupt changes in geochemical variables or host diseases, exceed the buffer capacity of the gut microbiome (Lu et al., 2022; Zhu et al., 2016). Thus, the gut microbiome is frequently altered by disturbances, leading to physiological disorders in shrimp and increased susceptibility to infections (Chen et al., 2017; Holt et al., 2021; Huang et al., 2021; Xiong et al., 2015). These lines of evidence, among others, demonstrate that the shrimp gut microbiota is highly dynamic, rather than static, and is consistently governed by both biotic and abiotic variables.
Koch’s postulates have been updated and reformulated to encompass pathogens, local milieu, and susceptible individuals as collective contributors to disease (Sultana et al., 2017; Xiong & Shi, 2024). Under this scenario, it is necessary to consider the shrimp farming system holistically, using the “holobiome” concept, which includes microbiomes in rearing water, sediment, feed, and host, along with their interactions (Gutiérrez-Pérez et al., 2022). Following this logic, disease occurrence can be attributed to an imbalance among pathogens, hosts, and the environment, with the gut microbiota serving as a critical link between the host and the whole system (Engering et al., 2013). Recent research on the shrimp microbiome has thus focused on optimizing the gut microbiome through dietary supplements, probiotics, and prebiotics (Goh et al., 2022). To date, however, available approaches have proven inefficient and unsustainable in the field, despite promising results in microcosm and mesocosm studies. A plausible explanation is that healthy shrimp also exert strong selection on external taxa (Kamada et al., 2013; Xiong, 2018; Xiong et al., 2019a), hindering the successful establishment of applied probiotics. Additionally, as diseases cause a gradual deviation from a healthy state, the pathobiome is expected to evolve, leading to specific symbiont profiles (pathobiomes) at the onset, progression, and moribund stages of shrimp diseases (Lu et al., 2023; Shen et al., 2021a; Xiong et al., 2017a). Addressing this challenge requires the design of therapeutic strategies tailored to shrimp age and disease stage (Xiong et al., 2016). From this perspective, integrative experimental designs are essential to identify the “holobiome” components that significantly influence the shrimp gut microbiome and drive disease progression. Understanding these factors will enable the development of targeted strategies to optimize shrimp cultivation through effective gut microbiome management.
SOURCES OF SHRIMP GUT MICROBIOTA
Aquatic animals are continuously exposed to their rearing water, and shrimp, being filter feeders, are assumed to acquire their gut commensals primarily from bacterioplankton communities in rearing water (De Schryver & Vadstein, 2014). However, research indicates that the microbial communities in rearing water and shrimp gut are distinct (Chen et al., 2017; Cornejo-Granados et al., 2017; Xiong et al., 2019b). As benthic animals, shrimp also interact with sediment microbial communities, which serve as an important external species pool for the gut microbiota. Compared to rearing water, sediment bacterial communities are more similar to those in the shrimp gut (Cornejo-Granados et al., 2017; Fan et al., 2019; Huang et al., 2021), contributing 36% and 75% of gut symbionts in juveniles and adults, respectively (Zhang et al., 2021a). Interestingly, the gut microbiota in adult shrimp is primarily inherited from the gut commensals of juveniles, forming an internal species pool (Zhang et al., 2021a, 2021b). Indeed, the shrimp gut microbiota is only weakly influenced by the geochemical variables and microbial communities in the rearing water (Xiong et al., 2019b; Zhang et al., 2021b). One possible explanation is that shifts in the gut microbiota lag behind shifts in the bacterioplankton community and geochemical variables. Alternatively, healthy shrimp may be resilient to mild stressors, with their gut microbiota primarily affected by host physiological parameters (Huang et al., 2018). This assertion is supported by observations that the proportion of gut commensals sourced from the ambient bacterioplankton community increases gradually during disease progression, due to compromised selection on surrounding taxa over the same timeframe (Lu et al., 2023; Xiong et al., 2018). Given the open circulatory system of invertebrates, shrimp gut symbionts may also disperse from other organs, such as the hepatopancreas. A high proportion of bacterial taxa are shared between the shrimp gut and hepatopancreas, serving as species pools for each other (Bao et al., 2024). Notably, dramatic changes in rearing conditions, such as elevated temperature and eutrophication, can markedly enhance pathogen virulence, resulting in earlier and higher shrimp mortality (Li et al., 2022; Sha et al., 2022a). Considering that disease results from an imbalance among the host, environmental variables, and ambient microbes (Mallon et al., 2015; Xiong, 2018), it is crucial to systematically explore the microbiomes across rearing water, sediment, and gut in relation to shrimp health status. Such exploration is essential for understanding pathogen transmission and etiological mechanisms.
DIVERSITY AS A BIOMARKER OF DISEASE
Ecologically, a diversity-poor community is more susceptible to invasion by external species compared to a diversity-rich community, known as the diversity-stability relationship (Wang & Loreau, 2014). The rationale behind a positive diversity-stability relationship is that a diverse community confers an insurance effect, where high diversity increases the presence of functionally versatile and redundant species against disturbances, and a negative covariance effect, where losses in some taxa due to disturbance are compensated by gains in others (Cadotte et al., 2012). Given the crucial role of the gut microbiota in antagonizing invading pathogens, numerous studies have compared the gut microbial diversity between healthy and diseased shrimp, revealing a decrease in alpha-diversity in infected shrimp (Shen et al., 2021a; Song et al., 2022; Yao et al., 2018). However, a reduction in diversity is not universally observed in diseased individuals, with some studies reporting comparable (Wang et al., 2019; Xiong et al., 2017b) or significantly increased diversity in infected shrimp (Zhou et al., 2019b) (Figure 2C). These discrepancies can be attributed to the type (e.g., causal agents) and strength (e.g., disease severity) of disturbance and the metrics used to measure diversity (e.g., weighted Shannon diversity, unweighted observed species). For example, we recently found that gut bacterial diversity is comparable between healthy and diseased shrimp at the disease onset stage (Chen et al., 2017; Lu et al., 2020; Xiong et al., 2017a). As the disease progresses, pathogen colonization outcompetes resident gut microbes, resulting in reduced microbial diversity. However, some resident species may benefit from the ecological niches created by the infection. In addition, diseased shrimp may attenuate their selective effect on external taxa, allowing for the random establishment of surrounding species (Mao et al., 2023; Sha et al., 2022b). Consequently, there is no consistent pattern of decreased gut microbial diversity in diseased shrimp (Figure 2C). Moreover, diversity is strongly affected by sequencing depth, making it challenging to compare diversity across studies. Therefore, declining gut microbial diversity is not a reliable indicator of shrimp health.
COMMON FEATURES OF GUT MICROBIOTA IN RESPONSE TO SHRIMP DISEASES
A growing body of evidence indicates that shrimp diseases significantly alter the gut microbiota, supporting the notion that gut dysbiosis may be a causal agent of shrimp diseases (Angthong et al., 2023; Chen et al., 2017; Cornejo-Granados et al., 2017; Huang et al., 2020b). However, this “chicken-and-egg” dilemma requires sophisticated experiments to disentangle the causes and mechanisms. According to the Anna Karenina principle, all healthy microbiomes are alike, whereas each disease-associated microbiome is unique to the diseased individual (Zaneveld et al., 2017). This principle suggests that host diseases induce heterogeneity and stochasticity in the gut microbiota. Following this logic, extensive studies have compared the heterogeneity of gut microbiota between healthy and diseased shrimp. Generally, shrimp diseases increase beta-dispersion, average variation degree, temporal turnover rate, and dysbiosis indices of the gut microbiota (Lu et al., 2023; Mao et al., 2023; Yao et al., 2018). However, the consistency of these signatures remains controversial. Shrimp with severe disease appear to exhibit a more stable gut microbiota over time compared to healthy controls (Lu et al., 2020; Xiong et al., 2017a). A plausible explanation is that the gut microbiota is not resilient to intense disturbance, such as severe disease.
According to the holobiont theory, hosts selectively acquire gut commensals from their surrounding environments to improve their fitness (Bordenstein & Theis, 2015). This suggests that the gut microbiota of healthy shrimp is predominantly shaped by deterministic processes, including selection pressures from abiotic environmental variables, biotic host filtering, and interspecies interactions (Cavender-Bares et al., 2009; Nemergut et al., 2013), leading to predictable community structures over space and time. Accordingly, the gut microbiota exhibits a time-similarity decay relationship throughout shrimp development (Xiong et al., 2020). This pattern is fundamental, as deviations from the temporally defined trend are positively associated with disease severity. Conversely, the occurrence of shrimp diseases weakens the relative importance of deterministic processes governing the gut microbiota, regardless of the causal pathogens (Mao et al., 2023; Sha et al., 2022b). Diseased shrimp typically exhibit a sharp reduction in food consumption (Table 1) and may redirect energy towards immune responses. Consequently, the ability of diseased shrimp to selectively filter external taxa becomes compromised, resulting in increased inter-individual variation and a more stochastic assembly of the gut microbiota.
Although extensive studies have illustrated the effect of environmental and host selection on the gut microbiota, few have explored the role of interspecies interactions during shrimp disease progression (Bäumler & Sperandio, 2016; Lu et al., 2023; Xiong et al., 2018). Characterizing these interactions is critical, as resident gut microbes can directly compete for nutrients or facilitate resistance against invading pathogens (Coyte et al., 2015). The strength of these interspecies interactions is a good predictor of susceptibility to pathogenic disease (Banerjee et al., 2018). In general, gut network stability and complexity are disrupted by shrimp diseases, as evidenced by diverse topological properties, including reduced robustness, modularity and proportion of negative associations, as well as increased vulnerability (Sha et al., 2024b; Xiong et al., 2018; Yao et al., 2018). Among network nodes, gut keystone species exert disproportional roles in sustaining the stability, dynamics, and function of the gut microbiota, making them excellent candidate targets for gut microbiota-based interventions (Dai et al., 2019b; Shetty et al., 2017; Wu et al., 2022). Case studies have shown that one keystone strain can effectively inhibit or eliminate one pathogen, such as Clostridium scindens against C. difficile (Buffie et al., 2015) and Bacillus subtilis against Staphylococcus aureus (Piewngam et al., 2018) in humans. Inspired by these findings, we recently designs a probiotic consortium that efficiently antagonizes the casual pathogens of shrimp WFS (Sha et al., 2024b). Nevertheless, comparable work in aquaculture species is still limited, warranting further study.
In conclusion, shrimp diseases typically induce dysbiosis, elevated heterogeneity, and stochasticity in the gut microbiota, although the causal relationships require rigorous validation. In addition, shrimp diseases disrupt the stability of the gut microbial network, potentially increasing susceptibility to invading pathogens. These common features across diverse diseases highlight the potential of sensitive gut microbial markers as indicators of shrimp health, irrespective of the causal agents. Early detection of these adverse changes in the gut microbiota is important for predicting and managing the risk of shrimp diseases.
COMMON DISEASE-DISCRIMINATORY TAXA PREDICTING SHRIMP HEALTH STATUS
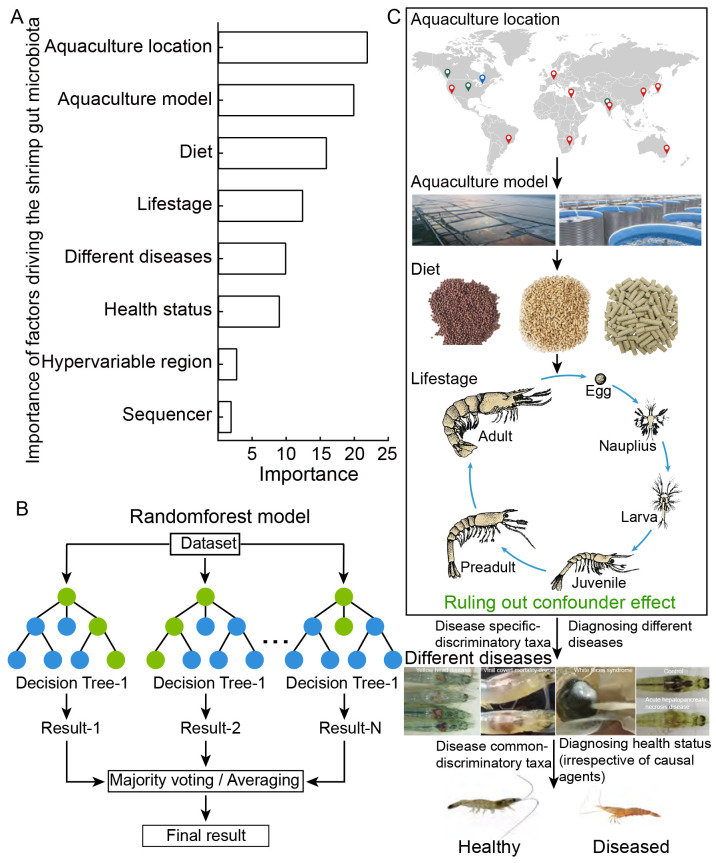
As discussed above, the shrimp gut microbiota displays common features in response to diverse diseases. The underlying mechanism involves pathogenic infections triggering the production of reactive oxygen species, which facilitate aerobic respiration. Facultative anaerobic bacteria can utilize nonfermentable substrates as carbon sources, enabling them to outcompete obligate anaerobic Bacteroidia for fermentable nutrients (Winter & Bäumler, 2014). Consistent with this mechanism, shrimp diseases frequently increase the abundance of facultative anaerobic Proteobacteria in the gut, while reducing obligate anaerobic Bacteroidia (Mao et al., 2023; Sha et al., 2022b). It is important to note that the shrimp gut microbiota is affected by diverse biotic and abiotic variables, including shrimp life stage (Xiong et al., 2020), different diseases (Yu et al., 2018a), hypervariable regions (Mao et al., 2023), health status (Angthong et al., 2023; Chen et al., 2017), aquaculture location, and sequencing platform (Cornejo-Granados et al., 2018; Mao et al., 2023) (Figure 3A).
Figure 3.
Flowchart screening disease common-discriminatory taxa for diagnosing shrimp health status
A: Quantifying relative importance of factors governing shrimp gut microbiota in descending order. B: Identifying discriminatory taxa for each factor using a random forest model. C: Identifying disease common-discriminatory taxa after progressive removal of effects of factors more important than health status in governing gut microbiota.
Shrimp age is a robust predictor of variance in the gut microbiota, leading to a predictable microbial assembly as shrimp age (Xiong et al., 2020). The profiles of age-discriminatory taxa in the gut accurately diagnose the chronological age of healthy shrimp. However, the onset of disease can disrupt this defined trend (Figure 2B). Specifically, the microbiota-for-age Z (MAZ) score quantifies the variance in predicted microbiota age across different host age ranges and remains relatively stable in healthy shrimp. In contrast, the MAZ value sharply decreases with the onset of shrimp disease (Figure 2D) (Xiong et al., 2017a). A key challenge is distinguishing gut microbiota features between healthy and diseased shrimp, considering life stage and other confounding variables (Figure 3). To address this, we propose a workflow for identifying disease common-discriminatory taxa. Initially, a parametric permutational multivariate analysis of variance quantitatively evaluates the effect of various factors on gut microbiota variance, ranking them based on their relative contributions. Subsequently, a random forest model or comparable method, such as statistical logistic regression, identifies indicators characterizing the most influential driving factors, with aquaculture location often being paramount (Figure 3A). To minimize the location effect, the identified location-discriminatory taxa are excluded from the gut microbiota. This process is repeated for subsequent factors until shrimp health status is the primary focus. Finally, disease common-discriminatory taxa serve as biomarkers for diagnosing shrimp health status, independent of the causal agents (Figure 3C). It is also possible to infer potential disease or causal pathogens through disease specific-discriminatory taxa, dependent on the causal agents. These optimized procedures can significantly improve diagnostic accuracy, as demonstrated in our recent studies (Mao et al., 2023; Sha et al., 2022b). Current research often compares gut microbiotas between healthy and severely diseased cohorts, leaving uncertainty about whether disease common-discriminatory taxa can predict disease onset. However, case studies provide supportive evidence for this potential (Lu et al., 2023; Shen et al., 2021a; Xiong et al., 2017a), although further validation is needed before practical application.
Disease common-discriminatory taxa are predominantly identified using high-throughput sequencing (HTS), which is time-consuming, expensive, and technically demanding, limiting its application in routine rapid-turnaround disease monitoring and early warning. An attractive solution to these limitations is proposed using quantitative real-time polymerase chain reaction (qPCR) to detect the relative abundances of gut microbial indicators (Yu et al. 2018b). In this case study, using disease-discriminatory phyla, diagnostic accuracy (93.2%) slightly lower than disease-discriminatory taxa (95.1%) and designed specific primers for these phyla for qPCR analysis, showing high consistency between qPCR and HTS assays in diagnosing shrimp WFS (Yu et al., 2018b). Nevertheless, subsequent meta-analyses have reported bacterial taxonomic effects on the diagnostic model, demonstrating that disease biomarkers at the bacterial species level provide the highest diagnostic accuracy (Mao et al., 2023; Sha et al., 2022b). However, due to confounding variables among studies, the identified gut disease-discriminatory taxa rarely overlap. Additionally, the bacterial 16S rDNA gene is insufficient to distinguish bacteria at the species level, except for a few well-studied pathogens. In contrast, functional genes are more conserved than 16S rDNA genes among phylogenic species that encode specific traits (Yang, 2021). Genes encoding a functional trait share conserved regions for maintaining topological structures and active sites, enabling the design of specific primers, such as those used in qPCR-based SmartChip assays. Following this logic, detecting disease-discriminatory functional genes using qPCR is a promising approach. Considering the rapid decrease in the cost of metagenomic sequencing, researchers can explore the gut functional structures across different shrimp health statuses and during disease progression. Importantly, gut microbiome data should be deposited into publicly accessible databases, allowing researchers to screen for common disease-discriminatory functional genes using meta-analysis in an unbiased manner.
CONCLUSIONS AND PERSPECTIVES
Due to increasing stocking densities and deteriorating water quality, the emergence of new and frequent shrimp diseases is expected. The etiological and molecular mechanisms of these newly emerged diseases remain poorly understood (Table 1), posing significant challenges to shrimp production. It is now recognized that certain infectious diseases are caused by co-infections with polymicrobial pathogens that synergistically disrupt host homeostasis, rather than by a single pathogen. This complexity challenges the traditional “one pathogen, one disease” concept, especially as uncultivable bacteria may significantly contribute to pathogenicity. In addition, Koch’s postulates are difficult to satisfy when the composition and ratios of polymicrobial pathogens are unknown. In this context, ecological approaches that integrate primary colonizers, keystone taxa, and disease-discriminatory taxa offer a promising alternative for inferring polymicrobial pathogens (Figure 1). Candidate pathogens can be selectively isolated using inferred conditions for subsequent validation (Oberhardt et al., 2015). Shrimp diseases, such as AHPND, generally cause rapid and massive mortality, making early disease prediction crucial for preventing disease progression. Disease common-discriminatory taxa and disease specific-discriminatory taxa hold potentials for predicting disease incidence and inferring causal agents (Figure 2), guiding timely and allopathic treatments. However, current studies typically use static comparisons between diseased and healthy shrimp, resulting in limited data on gut microbiota over the course of the disease and limited confidence in diagnosing disease onset (Yu et al., 2018b). Additionally, abiotic and biotic information is often insufficient when depositing shrimp disease research data into public databases. To address these issues, we propose that detailed relevant data should be collected throughout the course of the disease to construct reliable models for early disease prediction. As our understanding of the gut microbiome and shrimp diseases advances, it becomes increasingly clear that innovative solutions, such as antagonistic probiotic consortia and biomarkers, can be systematically developed to enhance the sustainability of shrimp aquaculture.
Acknowledgments
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
J.C. and J.B.X. conceptualized the manuscript. J.B.X. and H.N.S. wrote the manuscript. H.N.S. and J.B.X. collected the references and prepared the figures. All authors read and approved the final version of the manuscript.
Funding Statement
This work was supported by the National Natural Science Foundation of China (32371596, 32071549), Key Research and Development Project of Zhejiang Province (2021C02062), Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural Varieties (2021C02069-5-2), Key Project of Ningbo Science and Technology Bureau (2023S003), and One Health Interdisciplinary Research Project of Ningbo University (HZ202404)
Contributor Information
Jin-Bo Xiong, Email: xiongjinbo@nbu.edu.cn.
Jiong Chen, Email: jchen1975@163.com.
References
- Aguilar-Rendón KG, Soto-Rodriguez SA, Gomez-Gil B, et al Water microbiome dynamics of pacific white shrimp Penaeus vannamei infected with Vibrio parahaemolyticus strains responsible for acute hepatopancreatic necrosis disease. Aquaculture. 2022;551:737871. doi: 10.1016/j.aquaculture.2021.737871. [DOI] [Google Scholar]
- Alavandi SV, Muralidhar M, Syama Dayal J, et al Investigation on the infectious nature of running mortality syndrome (RMS) of farmed Pacific white leg shrimp, Penaeus vannamei in shrimp farms of India. Aquaculture. 2019;500:278–289. doi: 10.1016/j.aquaculture.2018.10.027. [DOI] [Google Scholar]
- Angthong P, Uengwetwanit T, Uawisetwathana U, et al. 2023. Investigating host-gut microbial relationship in Penaeus monodon upon exposure to Vibrio harveyi. Aquaculture, 567 : 739252.
- Aranguren Caro LF, Mai HN, Cruz-Florez R, et al Experimental reproduction of white feces syndrome in whiteleg shrimp. Penaeus vannamei. PLoS One. 2021;16(12):e0261289. doi: 10.1371/journal.pone.0261289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranguren LF, Han JE, Tang KFJ. 2017. Enterocytozoon hepatopenaei (EHP) is a risk factor for acute hepatopancreatic necrosis disease (AHPND) and septic hepatopancreatic necrosis (SHPN) in the Pacific white shrimp Penaeus vannamei. Aquaculture, 471 : 37–42.
- Arce KS, de Souza Valente C, do Vale Pereira G, et al Modulation of the gut microbiota of Pacific white shrimp (Penaeus vannamei boone, 1931) by dietary inclusion of a functional yeast cell wall‐based additive. Aquaculture Nutrition. 2021;27(4):1114–1127. doi: 10.1111/anu.13252. [DOI] [Google Scholar]
- Arcier JM, Herman F, Lightner DV, et al. 1999. A viral disease associated with mortalities in hatchery-reared postlarvae of the giant freshwater prawn Macrobrachium rosenbergii. Diseases of Aquatic Organisms, 38 (3): 177–181.
- Arulmoorthy MP, Anandajothi E, Vasudevan S, et al Major viral diseases in culturable penaeid shrimps: a review. Aquaculture International. 2020;28(5):1939–1967. doi: 10.1007/s10499-020-00568-3. [DOI] [Google Scholar]
- Banerjee S, Schlaeppi K, van der Heijden MG Keystone taxa as drivers of microbiome structure and functioning. Nature Reviews Microbiology. 2018;16(9):567–576. doi: 10.1038/s41579-018-0024-1. [DOI] [PubMed] [Google Scholar]
- Bao SC, Wang WJ, Deng ZX, et al Changes of bacterial communities and bile acid metabolism reveal the potential "intestine-hepatopancreas axis" in shrimp. Science of the Total Environment. 2024;938:173384. doi: 10.1016/j.scitotenv.2024.173384. [DOI] [PubMed] [Google Scholar]
- Bäumler AJ, Sperandio V Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535(7610):85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohara K, Joshi P, Acharya KP, et al Emerging technologies revolutionising disease diagnosis and monitoring in aquatic animal health. Reviews in Aquaculture. 2024;16(2):836–854. doi: 10.1111/raq.12870. [DOI] [Google Scholar]
- Boonyaratpalin S, Supamattaya K, Kasornchandra J, et al Non-occluded baculo-like virus, the causative agent of yellow head disease in the black tiger shrimp (Penaeus monodon) Fish Pathology. 1993;28(3):103–109. doi: 10.3147/jsfp.28.103. [DOI] [Google Scholar]
- Boopathi S, Meenatchi R, Brindangnanam P, et al Microbiome analysis of Litopenaeus vannamei reveals Vibrio as main risk factor of white faeces syndrome. Aquaculture. 2023;576:739829. doi: 10.1016/j.aquaculture.2023.739829. [DOI] [Google Scholar]
- Bordenstein SR, Theis KR Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biology. 2015;13(8):e1002226. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock JA Taura syndrome, a disease important to shrimp farms in the americas. World Journal of Microbiology & Biotechnology. 1997;13(4):415–418. [Google Scholar]
- Brzezinska MS, Jankiewicz U Production of antifungal chitinase by Aspergillus niger LOCK 62 and its potential role in the biological control. Current Microbiology. 2012;65(6):666–672. doi: 10.1007/s00284-012-0208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, et al. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature, 517 (7533): 205–208.
- Cadotte MW, Dinnage R, Tilman D Phylogenetic diversity promotes ecosystem stability. Ecology. 2012;93(sp8):S223–S233. [Google Scholar]
- Cao HP, Chen SJ, Lu LQ, et al Shewanella algae: an emerging pathogen of black spot disease in freshwater-cultured whiteleg shrimp (Penaeus vannamei) The Israeli Journal of Aquaculture-Bamidgeh. 2018;70:1472. [Google Scholar]
- Cavender-Bares J, Kozak KH, Fine PVA, et al The merging of community ecology and phylogenetic biology. Ecology Letters. 2009;12(7):693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- Chang YT, Ko HT, Wu PL, et al. 2023. Gut microbiota of Pacific white shrimp (Litopenaeus vannamei) exhibits distinct responses to pathogenic and non-pathogenic Vibrio parahaemolyticus. Microbiology Spectrum, 11 (5): e01180-23.
- Chayaburakul K, Nash G, Pratanpipat P, et al Multiple pathogens found in growth-retarded black tiger shrimp Penaeus monodon cultivated in thailand. Diseases of Aquatic Organisms. 2004;60(2):89–96. doi: 10.3354/dao060089. [DOI] [PubMed] [Google Scholar]
- Chen SN, Chang PS, Kou GH, et al Studies on virogenesis and cytopathology of penaeus monodon baculovirus (MBV) in the giant tiger prawn (Penaeus monodon) and the red tail prawn (Penaeus penicillatus) Fish Pathology. 1989;24(2):89–100. doi: 10.3147/jsfp.24.89. [DOI] [Google Scholar]
- Chen WY, Ng TH, Wu JH, et al Microbiome dynamics in a shrimp grow-out pond with possible outbreak of acute hepatopancreatic necrosis disease. Scientific Reports. 2017;7(1):9395. doi: 10.1038/s41598-017-09923-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong Y, Loh H Hepatopancreas clamydial and parvoviral infections of farmed marine prawns in Singapore. Singapore Veterinary Journal. 1984;9:51–56. [Google Scholar]
- Cornejo-Granados F, Gallardo-Becerra L, Leonardo-Reza M, et al A meta-analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota. PeerJ. 2018;6:e5382. doi: 10.7717/peerj.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo-Granados F, Lopez-Zavala AA, Gallardo-Becerra L, et al Microbiome of Pacific whiteleg shrimp reveals differential bacterial community composition between wild, aquacultured and AHPND/EMS outbreak conditions. Scientific Reports. 2017;7(1):11783. doi: 10.1038/s41598-017-11805-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couch JA An enzootic nuclear polyhedrosis virus of pink shrimp: ultrastructure, prevalence, and enhancement. Journal of Invertebrate Pathology. 1974;24(3):311–331. doi: 10.1016/0022-2011(74)90139-6. [DOI] [PubMed] [Google Scholar]
- Coyte KZ, Schluter J, Foster KR The ecology of the microbiome: networks, competition, and stability. Science. 2015;350(6261):663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- Dai WF, Qiu QF, Chen J, et al Gut eukaryotic disease-discriminatory taxa are indicative of Pacific white shrimp (Litopenaeus vannamei) white feces syndrome. Aquaculture. 2019a;506:154–160. doi: 10.1016/j.aquaculture.2019.03.034. [DOI] [Google Scholar]
- Dai WF, Chen J, Xiong JB Concept of microbial gatekeepers: positive guys? Applied Microbiology & Biotechnology. 2019b;103(2):633–641. doi: 10.1007/s00253-018-9522-3. [DOI] [PubMed] [Google Scholar]
- Dai WF, Yu WN, Xuan LX, et al Integrating molecular and ecological approaches to identify potential polymicrobial pathogens over a shrimp disease progression. Applied Microbiology & Biotechnology. 2018;102(8):3755–3764. doi: 10.1007/s00253-018-8891-y. [DOI] [PubMed] [Google Scholar]
- De Schryver P, Vadstein O Ecological theory as a foundation to control pathogenic invasion in aquaculture. The ISME Journal. 2014;8(12):2360–2368. doi: 10.1038/ismej.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Valente C, Rodiles A, Marques MRF, et al White spot syndrome virus (WSSV) disturbs the intestinal microbiota of shrimp (Penaeus vannamei) reared in biofloc and clear seawater. Applied Microbiology & Biotechnology. 2020;104(18):8007–8023. doi: 10.1007/s00253-020-10816-4. [DOI] [PubMed] [Google Scholar]
- Deng YL, Xu XY, Yin XW, et al Effect of stock density on the microbial community in biofloc water and Pacific white shrimp (Litopenaeus vannamei) gut microbiota. Applied Microbiology & Biotechnology. 2019;103(10):4241–4252. doi: 10.1007/s00253-019-09773-4. [DOI] [PubMed] [Google Scholar]
- Deris ZM, Iehata S, Gan HM, et al. 2022. Understanding the effects of salinity and Vibrio harveyi on the gut microbiota profiles of Litopenaeus vannamei. Frontiers in Marine Science, 9 : 974217.
- Dhar AK, Piamsomboon P, Caro LFA, et al First report of acute hepatopancreatic necrosis disease (AHPND) occurring in the USA. Diseases of Aquatic Organisms. 2019;132(3):241–247. doi: 10.3354/dao03330. [DOI] [PubMed] [Google Scholar]
- Dong PS, Guo HP, Wang YT, et al Gastrointestinal microbiota imbalance is triggered by the enrichment of Vibrio in subadult Litopenaeus vannamei with acute hepatopancreatic necrosis disease. Aquaculture. 2021;533:736199. doi: 10.1016/j.aquaculture.2020.736199. [DOI] [Google Scholar]
- Durai V, Gunalan B, Johnson PM, et al Effect on white gut and white feces disease in semi intensive Litopenaeus vannamei shrimp culture system in south Indian state of Tamilnadu. International Journal of Marine Science. 2015;5(14):1–5. [Google Scholar]
- Engering A, Hogerwerf L, Slingenbergh J Pathogen-host-environment interplay and disease emergence. Emerging Microbes & Infections. 2013;2(1):e5. doi: 10.1038/emi.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaise C, François C, Travers MA, et al Antimicrobial compounds from eukaryotic microalgae against human pathogens and diseases in aquaculture. Marine Drugs. 2016;14(9):159. doi: 10.3390/md14090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan LF, Wang ZL, Chen MS, et al Microbiota comparison of Pacific white shrimp intestine and sediment at freshwater and marine cultured environment. Science of the Total Environment. 2019;657:1194–1204. doi: 10.1016/j.scitotenv.2018.12.069. [DOI] [PubMed] [Google Scholar]
- FAO. 2023. Global aquaculture production data. Rome: FAO.
- Ferreira NC, Bonetti C, Seiffert WQ Hydrological and water quality indices as management tools in marine shrimp culture. Aquaculture. 2011;318(3-4):425–433. doi: 10.1016/j.aquaculture.2011.05.045. [DOI] [Google Scholar]
- Flegel TW Detection of major penaeid shrimp viruses in asia, a historical perspective with emphasis on thailand. Aquaculture. 2006;258(1-4):1–33. doi: 10.1016/j.aquaculture.2006.05.013. [DOI] [Google Scholar]
- Frisan T Co‐ and polymicrobial infections in the gut mucosa: the host–microbiota–pathogen perspective. Cellular Microbiology. 2021;23(2):e13279. doi: 10.1111/cmi.13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh JXH, Tan LTH, Law JWF, et al Harnessing the potentialities of probiotics, prebiotics, synbiotics, paraprobiotics, and postbiotics for shrimp farming. Reviews in Aquaculture. 2022;14(3):1478–1557. doi: 10.1111/raq.12659. [DOI] [Google Scholar]
- Gutiérrez-Pérez ED, Vázquez-Juárez R, Magallón-Barajas F, et al How a holobiome perspective could promote intensification, biosecurity and eco-efficiency in the shrimp aquaculture industry. Frontiers in Marine Science. 2022;9:975042. doi: 10.3389/fmars.2022.975042. [DOI] [Google Scholar]
- Han JE Detection of the amoebic parasite (order Dactylopodida) in cultured Pacific white shrimp (Litopenaeus vannamei) Aquaculture. 2019;507:246–250. doi: 10.1016/j.aquaculture.2019.04.036. [DOI] [Google Scholar]
- Han JE, Tang KFJ, Tran LH, et al Photorhabdus insect-related (Pir) toxin-like genes in a plasmid of Vibrio parahaemolyticus, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp. Diseases of Aquatic Organisms. 2015;113(1):33–40. doi: 10.3354/dao02830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatai K, Egusa S Studies on the pathogenic fungus associated with black gill disease of kuruma prawn, Penaeus japonicus-II: some of the note on the BG-Fusarium. Fish Pathology. 1978;12(4):225–231. doi: 10.3147/jsfp.12.225. [DOI] [Google Scholar]
- Holt CC, Bass D, Stentiford GD, et al Understanding the role of the shrimp gut microbiome in health and disease. Journal of Invertebrate Pathology. 2021;186:107387. doi: 10.1016/j.jip.2020.107387. [DOI] [PubMed] [Google Scholar]
- Hou DW, Huang ZJ, Zeng SZ, et al Intestinal bacterial signatures of white feces syndrome in shrimp. Applied Microbiology & Biotechnology. 2018;102(8):3701–3709. doi: 10.1007/s00253-018-8855-2. [DOI] [PubMed] [Google Scholar]
- Hou DW, Zhou RJ, Zeng SZ, et al Intestine bacterial community composition of shrimp varies under low-and high-salinity culture conditions. Frontiers in Microbiology. 2020;11:589164. doi: 10.3389/fmicb.2020.589164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Pan LQ, Song MS, et al Microbiota assemblages of water, sediment, and intestine and their associations with environmental factors and shrimp physiological health. Applied Microbiology & Biotechnology. 2018;102(19):8585–8598. doi: 10.1007/s00253-018-9229-5. [DOI] [PubMed] [Google Scholar]
- Huang MX, Xie J, Yu QR, et al. 2020c. Toxic effect of chronic nitrite exposure on growth and health in Pacific white shrimp Litopenaeus vannamei. Aquaculture, 529 : 735664.
- Huang XD, Gu Y, Zhou HH, et al. 2020a. Acinetobacter venetianus, a potential pathogen of red leg disease in freshwater-cultured whiteleg shrimp Penaeus vannamei. Aquaculture Reports, 18 : 100543.
- Huang ZJ, Hou DW, Zhou RJ, et al Environmental water and sediment microbial communities shape intestine microbiota for host health: the central dogma in an anthropogenic aquaculture ecosystem. Frontiers in Microbiology. 2021;12:772149. doi: 10.3389/fmicb.2021.772149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Zeng SZ, Xiong JB, et al Microecological Koch's postulates reveal that intestinal microbiota dysbiosis contributes to shrimp white feces syndrome. Microbiome. 2020b;8(1):32. doi: 10.1186/s40168-020-00802-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ina‐Salwany MY, Al‐Saari N, Mohamad A, et al Vibriosis in fish: a review on disease development and prevention. Journal of Aquatic Animal Health. 2019;31(1):3–22. doi: 10.1002/aah.10045. [DOI] [PubMed] [Google Scholar]
- Jayasree L, Janakiram P, Madhavi R Characterization of Vibrio spp. Associated with diseased shrimp from culture ponds of Andhra Pradesh (India) Journal of the World Aquaculture Society. 2006;37(4):523–532. doi: 10.1111/j.1749-7345.2006.00066.x. [DOI] [Google Scholar]
- Kamada N, Chen GY, Inohara N, et al Control of pathogens and pathobionts by the gut microbiota. Nature Immunology. 2013;14(7):685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunasagar I, Karunasagar I, Umesha RK. 2004. Microbial diseases in shrimp aquaculture. Goa, India: National Institute of Oceanography.
- Khoa LV, Hatai K, Aoki T Fusarium incarnatum isolated from black tiger shrimp, Penaeus monodon fabricius, with black gill disease cultured in Vietnam. Journal of Fish Diseases. 2004;27(9):507–515. doi: 10.1111/j.1365-2761.2004.00562.x. [DOI] [PubMed] [Google Scholar]
- Kumar R, Ng TH, Wang HC Acute hepatopancreatic necrosis disease in penaeid shrimp. Reviews in Aquaculture. 2020;12(3):1867–1880. doi: 10.1111/raq.12414. [DOI] [Google Scholar]
- Li J, Jiang HY, Li LM, et al. 2019. The effect of disease and season to hepatopancreas and intestinal mycobiota of Litopenaeus vannamei. Frontiers in Microbiology, 10 : 889.
- Li LY, Lu JQ, Zhan PP, et al. 2022. RNA-seq analysis unveils temperature and nutrient adaptation mechanisms relevant for pathogenicity in Vibrio parahaemolyticus. Aquaculture, 558 : 738397.
- Li XJ, Zhang Q, Zhang AL, et al Metabolites from Aspergillus fumigatus, an endophytic fungus associated with Melia azedarach, and their antifungal, antifeedant, and toxic activities. Journal of Agricultural & Food Chemistry. 2012;60(13):3424–3431. doi: 10.1021/jf300146n. [DOI] [PubMed] [Google Scholar]
- Liang QJ, Li ZH, Ou MF, et al. 2020. Hypoimmunity and intestinal bacterial imbalance are closely associated with blue body syndrome in cultured Penaeus vannamei. Aquaculture, 522 : 735118.
- Liao MZ, Liao XZ, Long XX, et al Host-microbiota interactions and responses of Metapenaeus ensis infected with decapod iridescent virus 1. Frontiers in Microbiology. 2023;13:1097931. doi: 10.3389/fmicb.2022.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner DV, Redman RM, Bell TA Infectious hypodermal and hematopoietic necrosis, a newly recognized virus disease of penaeid shrimp. Journal of Invertebrate Pathology. 1983;42(1):62–70. doi: 10.1016/0022-2011(83)90202-1. [DOI] [PubMed] [Google Scholar]
- Liu KF, Liu WJ, Kou GH, et al Shrimp white spot syndrome-from pathology to pathogenomics. Fish Pathology. 2009;44(2):55–58. doi: 10.3147/jsfp.44.55. [DOI] [Google Scholar]
- Liu S, Xia JT, Tian Y, et al. 2022. Investigation of pathogenic mechanism of covert mortality nodavirus infection in Penaeus vannamei. Frontiers in Microbiology, 13 : 904358.
- Lo LSH, Liu X, Liu HB, et al Aquaculture bacterial pathogen database: pathogen monitoring and screening in coastal waters using environmental DNA. Water Research. 2023;20:100194. doi: 10.1016/j.wroa.2023.100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Carvallo JA, Cruz-Flores R, Dhar AK The emerging pathogen Enterocytozoon hepatopenaei drives a degenerative cyclic pattern in the hepatopancreas microbiome of the shrimp (Penaeus vannamei) Scientific Reports. 2022;12(1):14766. doi: 10.1038/s41598-022-19127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JQ, Li XH, Qiu QF, et al Gut interkingdom predator-prey interactions are key determinants of shrimp health. Aquaculture. 2022;546:737304. doi: 10.1016/j.aquaculture.2021.737304. [DOI] [Google Scholar]
- Lu JQ, Mao JN, Qi XJ, et al The assembly of gut microbiota implicates shrimp acute hepatopancreas necrosis disease progression. Applied Microbiology & Biotechnology. 2023;107(24):7489–7500. doi: 10.1007/s00253-023-12810-y. [DOI] [PubMed] [Google Scholar]
- Lu JQ, Zhang XC, Qiu QF, et al Identifying potential polymicrobial pathogens: moving beyond differential abundance to driver taxa. Microbial Ecology. 2020;80(2):447–458. doi: 10.1007/s00248-020-01511-y. [DOI] [PubMed] [Google Scholar]
- Lu JQ, Zhang XX, Wang CH, et al Responses of sediment resistome, virulence factors and potential pathogens to decades of antibiotics pollution in a shrimp aquafarm. Science of the Total Environment. 2021;794:148760. doi: 10.1016/j.scitotenv.2021.148760. [DOI] [PubMed] [Google Scholar]
- MacAulay S, Ellison AR, Kille P, et al Moving towards improved surveillance and earlier diagnosis of aquatic pathogens: from traditional methods to emerging technologies. Reviews in Aquaculture. 2022;14(4):1813–1829. doi: 10.1111/raq.12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon CA, van Elsas JD, Salles JF Microbial invasions: the process, patterns, and mechanisms. Trends in Microbiology. 2015;23(11):719–729. doi: 10.1016/j.tim.2015.07.013. [DOI] [PubMed] [Google Scholar]
- Mandal A, Das SK. 2018. Comparative efficacy of neem (Azadirachta indica) and non-neem supplemented biofloc media in controlling the harmful luminescent bacteria in natural pond culture of Litopenaeus vannaemei. Aquaculture, 492 : 157–163.
- Mao JN, Lu JQ, Chen J, et al Consistent features of the gut microbiota in response to diverse shrimp Litopenaeus vannamei diseases: a meta‐analysis. Fish & Fisheries. 2023;24(6):1103–1117. [Google Scholar]
- Miki T, Yokokawa T, Matsui K Biodiversity and multifunctionality in a microbial community: a novel theoretical approach to quantify functional redundancy. Proceedings of the Royal Society B: Biological Sciences. 2014;281(1776):20132498. doi: 10.1098/rspb.2013.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkongwongsiri N, Prachumwat A, Eamsaard W, et al. 2022. Propionigenium and Vibrio species identified as possible component causes of shrimp white feces syndrome (WFS) associated with the microsporidian Enterocytozoon hepatopenaei. Journal of Invertebrate Pathology, 192 : 107784.
- Munkvold GP Fusarium species and their associated mycotoxins. Methods in Molecular Biology. 2017;1542:51–106. doi: 10.1007/978-1-4939-6707-0_4. [DOI] [PubMed] [Google Scholar]
- Muthukrishnan S, Defoirdt T, Shariff M, et al. 2019. Horizontal gene transfer of the pirAB genes responsible for acute hepatopancreatic necrosis disease (AHPND) turns a non-Vibrio strain into an AHPND-positive pathogen. BioRxiv.
- Nemergut DR, Schmidt SK, Fukami T, et al Patterns and processes of microbial community assembly. Microbiology & Molecular Biology Reviews. 2013;77(3):342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurlatiffah N, Kismiyati I, Ulkhaq MF. 2018. Value of prevalence and intensity of ectoparasite infesting Litopenaeus vannamei. Journal of Fisheries & LIife Science, 3 (2): 17–19.
- Oberhardt MA, Zarecki R, Gronow S, et al Harnessing the landscape of microbial culture media to predict new organism–media pairings. Nature Communications. 2015;6:8493. doi: 10.1038/ncomms9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal V, Kumar S, Hossain SN, et al Structure analysis of thymidylate synthase from white spot syndrome virus reveals WSSV-specific structural elements. International Journal of Biological Macromolecules. 2021;167:1168–1175. doi: 10.1016/j.ijbiomac.2020.11.071. [DOI] [PubMed] [Google Scholar]
- Paulraj A, Musthafa MS, Altaff K, et al Chytrid Batrachochytrium dendrobatidis fungal infection in freshwater prawn, Macrobrachium rosenbergii (de Man)-A new report. Aquaculture. 2016;464:521–528. doi: 10.1016/j.aquaculture.2016.07.035. [DOI] [Google Scholar]
- Phuoc LH, Corteel M, Nauwynck HJ, et al. 2008. Increased susceptibility of white spot syndrome virus-infected Litopenaeus vannamei to Vibrio campbellii. Environmental Microbiology, 10 (10): 2718–2727.
- Piewngam P, Zheng Y, Nguyen TH, et al Pathogen elimination by probiotic Bacillus via signalling interference. Nature. 2018;562(7728):532–537. doi: 10.1038/s41586-018-0616-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradeep B, Rai P, Mohan SA, et al. 2012. Biology, host range, pathogenesis and diagnosis of White spot syndrome virus. Indian Journal of Virology, 23 (2): 161–174.
- Qiu L, Chen MM, Wan XY, et al Characterization of a new member of Iridoviridae, Shrimp hemocyte iridescent virus (SHIV), found in white leg shrimp (Litopenaeus vannamei) Scientific Reports. 2017;7(1):11834. doi: 10.1038/s41598-017-10738-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj M, Sathiyaraj G, Narayanan B, et al Characterization of Enterocytozoon hepatopenaei causing hepatopancreatic microsporidiosis in L. vannamei and a new molecular method for its detection in shrimps, and other environmental samples. Journal of Invertebrate Pathology. 2023;199:107951. doi: 10.1016/j.jip.2023.107951. [DOI] [PubMed] [Google Scholar]
- Ramirez M, Domínguez-Borbor C, Salazar L, et al. 2022. The probiotics Vibrio diabolicus (Ili), Vibrio hepatarius (P62), and Bacillus cereus sensu stricto (P64) colonize internal and external surfaces of Penaeus vannamei shrimp larvae and protect it against Vibrio parahaemolyticus. Aquaculture, 549 : 737826.
- Restrepo L, Domínguez-Borbor C, Bajaña L, et al Microbial community characterization of shrimp survivors to AHPND challenge test treated with an effective shrimp probiotic (Vibrio diabolicus) Microbiome. 2021;9(1):88. doi: 10.1186/s40168-021-01043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers CJ, Furones MD. 2009. Antimicrobial agents in aquaculture: practice, needs and issues. In: Rogers C, Basurco B. The use of veterinary drugs and vaccines in mediterranean aquaculture. Zaragoza: CIHEAM, 41–59.
- Rungrassamee W, Klanchui A, Maibunkaew S, et al Bacterial dynamics in intestines of the black tiger shrimp and the Pacific white shrimp during Vibrio harveyi exposure. Journal of Invertebrate Pathology. 2016;133:12–19. doi: 10.1016/j.jip.2015.11.004. [DOI] [PubMed] [Google Scholar]
- Saejung C, Hatai K, Wada S, et al Clinical observations of black disease in fairy shrimps, Streptocephalus sirindhornae and Branchinella thailandensis, from Thailand and pathogen verification. Journal of Fish Diseases. 2011;34(12):911–920. doi: 10.1111/j.1365-2761.2011.01314.x. [DOI] [PubMed] [Google Scholar]
- Sakaew W, Pratoomthai B, Anantasomboon G, et al Abdominal segment deformity disease (ASDD) of the whiteleg shrimp Penaeus vannamei reared in Thailand. Aquaculture. 2008;284(1-4):46–52. doi: 10.1016/j.aquaculture.2008.07.041. [DOI] [Google Scholar]
- Seibert CH, Pinto AR Challenges in shrimp aquaculture due to viral diseases: distribution and biology of the five major penaeid viruses and interventions to avoid viral incidence and dispersion. Brazilian Journal of Microbiology. 2012;43(3):857–864. doi: 10.1590/S1517-83822012000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha HN, Li LY, Lu JQ, et al High nutrient induces virulence in the AHPND-causing Vibrio parahaemolyticus, interpretation from the ecological assembly of shrimp gut microbiota. Fish & Shellfish Immunology. 2022a;127:758–765. doi: 10.1016/j.fsi.2022.07.016. [DOI] [PubMed] [Google Scholar]
- Sha HN, Lu JQ, Chen J, et al A meta‐analysis study of the robustness and universality of gut microbiota–shrimp diseases relationship. Environmental Microbiology. 2022b;24(9):3924–3938. doi: 10.1111/1462-2920.16024. [DOI] [PubMed] [Google Scholar]
- Sha HN, Song TT, Zhan PP, et al Sarafloxacin hydrochloride exposure disrupts gut microbiota and increases shrimp susceptibility to Vibrio anguillarum infection. Aquaculture. 2024a;586:740810. doi: 10.1016/j.aquaculture.2024.740810. [DOI] [Google Scholar]
- Sha HN, Lu JQ, Chen J, et al Rationally designed probiotics prevent shrimp white feces syndrome via the probiotics–gut microbiome–immunity axis. npj Biofilms & Microbiomes. 2024b;10(1):40. doi: 10.1038/s41522-024-00509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan LP, Hu Y, Hu L, et al Involvement of microplastics in the conflict between host immunity defense and viral virulence: promoting the susceptibility of shrimp to WSSV infection. Environmental Science & Technology. 2023;57(31):11634–11642. doi: 10.1021/acs.est.3c01566. [DOI] [PubMed] [Google Scholar]
- Shen HY, Song TT, Lu JQ, et al Shrimp AHPND causing Vibrio anguillarum infection: quantitative diagnosis and identifying antagonistic bacteria. Marine Biotechnology. 2021a;23(6):964–975. doi: 10.1007/s10126-021-10079-8. [DOI] [PubMed] [Google Scholar]
- Shen H, Fan XP, Qiao Y, et al. 2021b. The links among Enterocytozoon hepatopenaei infection, growth retardation and intestinal microbiota in different sized shrimp Penaeus vannamei. Aquaculture Reports, 21 : 100888.
- Shen HY, Zhang XC, Qian D, et al Pathobiology of Enterocytozoon hepatopenaei (EHP) in shrimp: diagnosis and interpretation from the gut bacterial community. Aquaculture. 2022;554:738169. doi: 10.1016/j.aquaculture.2022.738169. [DOI] [Google Scholar]
- Shetty SA, Hugenholtz F, Lahti L, et al Intestinal microbiome landscaping: insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiology Reviews. 2017;41(2):182–199. doi: 10.1093/femsre/fuw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song TT, Sha HN, Qiu QF, et al Common disease-discriminatory fungal taxa accurately diagnose shrimp white feces syndrome, black gills, and retardation diseases. Aquaculture. 2022;561:738643. doi: 10.1016/j.aquaculture.2022.738643. [DOI] [Google Scholar]
- Soto-Rodriguez SA, Gomez-Gil B, Lozano R. 2010. ‘Bright-red’syndrome in Pacific white shrimp Litopenaeus vannamei is caused by Vibrio harveyi. Diseases of Aquatic Organisms, 92 (1): 11–19.
- Sriurairatana S, Boonyawiwat V, Gangnonngiw W, et al White feces syndrome of shrimp arises from transformation, sloughing and aggregation of hepatopancreatic microvilli into vermiform bodies superficially resembling gregarines. PLoS One. 2014;9(6):e99170. doi: 10.1371/journal.pone.0099170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subash P, Chrisolite B, Sivasankar P, et al White feces syndrome in Penaeus vannamei is potentially an Enterocytozoon hepatopenaei (EHP) associated pathobiome origin of Vibrio spp. Journal of Invertebrate Pathology. 2023;198:107932. doi: 10.1016/j.jip.2023.107932. [DOI] [PubMed] [Google Scholar]
- Sultana S, Sarker SA, Brüssow H What happened to koch's postulates in diarrhoea? Environmental Microbiology. 2017;19(8):2926–2934. doi: 10.1111/1462-2920.13787. [DOI] [PubMed] [Google Scholar]
- Sun SM, Yang M, Fu HT, et al. 2020. Altered intestinal microbiota induced by chronic hypoxia drives the effects on lipid metabolism and the immune response of oriental river prawn Macrobrachium nipponense. Aquaculture, 526 : 735431.
- Suryakodi S, Ahmed AN, Badhusha A, et al First report on the occurrence of white spot syndrome virus, infectious myonecrosis virus and Enterocytozoon hepatopenaei in Penaeus vannamei reared in freshwater systems. Journal of Fish Diseases. 2022;45(5):699–706. doi: 10.1111/jfd.13595. [DOI] [PubMed] [Google Scholar]
- Thitamadee S, Prachumwat A, Srisala J, et al Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture. 2016;452:69–87. doi: 10.1016/j.aquaculture.2015.10.028. [DOI] [Google Scholar]
- Tran L, Nunan L, Redman RM, et al Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Diseases of Aquatic Organisms. 2013;105(1):45–55. doi: 10.3354/dao02621. [DOI] [PubMed] [Google Scholar]
- Tuyen NX, Verreth J, Vlak JM, et al. 2014. Horizontal transmission dynamics of white spot syndrome virus by cohabitation trials in juvenile Penaeus monodon and P. vannamei. Preventive Veterinary Medicine, 117 (1): 286–294.
- Vandenberghe J, Thompson FL, Gomez-Gil B, et al Phenotypic diversity amongst Vibrio isolates from marine aquaculture systems. Aquaculture. 2003;219(1-4):9–20. doi: 10.1016/S0044-8486(02)00312-5. [DOI] [Google Scholar]
- Wang J, Huang YJ, Xu KH, et al. 2019. White spot syndrome virus (WSSV) infection impacts intestinal microbiota composition and function in Litopenaeus vannamei. Fish & Shellfish Immunology, 84 : 130–137.
- Wang LP, Chen YW, Huang H, et al. 2015. Isolation and identification of Vibrio campbellii as a bacterial pathogen for luminous vibriosis of Litopenaeus vannamei. Aquaculture Research, 46 (2): 395–404.
- Wang SP, Loreau M Ecosystem stability in space: α, β and γ variability. Ecology Letters. 2014;17(8):891–901. doi: 10.1111/ele.12292. [DOI] [PubMed] [Google Scholar]
- Wijegoonawardane PKM, Cowley JA, Phan T, et al Genetic diversity in the yellow head nidovirus complex. Virology. 2008;380(2):213–225. doi: 10.1016/j.virol.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Bäumler AJ Why related bacterial species bloom simultaneously in the gut: principles underlying the ‘like will to like’ concept. Cellular Microbiology. 2014;16(2):179–184. doi: 10.1111/cmi.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DF, Liu L, Jiao N, et al Targeting keystone species helps restore the dysbiosis of butyrate‐producing bacteria in nonalcoholic fatty liver disease. iMeta. 2022;1(4):e61. doi: 10.1002/imt2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JB, Wang K, Wu JF, et al Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Applied Microbiology & Biotechnology. 2015;99(16):6911–6919. doi: 10.1007/s00253-015-6632-z. [DOI] [PubMed] [Google Scholar]
- Xiong JB, Dai WF, Li CH Advances, challenges, and directions in shrimp disease control: the guidelines from an ecological perspective. Applied Microbiology & Biotechnology. 2016;100(16):6947–6954. doi: 10.1007/s00253-016-7679-1. [DOI] [PubMed] [Google Scholar]
- Xiong JB, Zhu JY, Dai WF, et al Integrating gut microbiota immaturity and disease‐discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environmental Microbiology. 2017a;19(4):1490–1501. doi: 10.1111/1462-2920.13701. [DOI] [PubMed] [Google Scholar]
- Xiong JB, Dai WF, Zhu JY, et al The underlying ecological processes of gut microbiota among cohabitating retarded, overgrown and normal shrimp. Microbial Ecology. 2017b;73(4):988–999. doi: 10.1007/s00248-016-0910-x. [DOI] [PubMed] [Google Scholar]
- Xiong JB Progress in the gut microbiota in exploring shrimp disease pathogenesis and incidence. Applied Microbiology & Biotechnology. 2018;102(17):7343–7350. doi: 10.1007/s00253-018-9199-7. [DOI] [PubMed] [Google Scholar]
- Xiong JB, Dai WF, Qiu QF, et al Response of host–bacterial colonization in shrimp to developmental stage, environment and disease. Molecular Ecology. 2018;27(18):3686–3699. doi: 10.1111/mec.14822. [DOI] [PubMed] [Google Scholar]
- Xiong JB, Nie L, Chen J Current understanding on the roles of gut microbiota in fish disease and immunity. Zoological Research. 2019a;40(2):70–76. doi: 10.24272/j.issn.2095-8137.2018.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JB, Xuan LX, Yu WN, et al Spatiotemporal successions of shrimp gut microbial colonization: high consistency despite distinct species pool. Environmental Microbiology. 2019b;21(4):1383–1394. doi: 10.1111/1462-2920.14578. [DOI] [PubMed] [Google Scholar]
- Xiong JB, Li XH, Yan MC, et al. 2020. Comparable ecological processes govern the temporal succession of gut bacteria and microeukaryotes as shrimp aged. Microbial Ecology, 80 (4): 935–945.
- Xiong JB, Shi ZJ. 2024. Editorial: environments-pathogens-the gut microbiota and host diseases. Frontiers in Microbiology 14 : 1357125.
- Yang YF Emerging patterns of microbial functional traits. Trends in Microbiology. 2021;29(10):874–882. doi: 10.1016/j.tim.2021.04.004. [DOI] [PubMed] [Google Scholar]
- Yao ZY, Yang KJ, Huang L, et al Disease outbreak accompanies the dispersive structure of shrimp gut bacterial community with a simple core microbiota. AMB Express. 2018;8(1):120. doi: 10.1186/s13568-018-0644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WN, Wu JH, Zhang JJ, et al A meta-analysis reveals universal gut bacterial signatures for diagnosing the incidence of shrimp disease. FEMS Microbiology Ecology. 2018a;94(10):fiy147. doi: 10.1093/femsec/fiy147. [DOI] [PubMed] [Google Scholar]
- Yu WN, Cao JX, Dai WF, et al Quantitative PCR analysis of gut disease-discriminatory phyla for diagnosing shrimp disease incidence. Applied & Environmental Microbiology. 2018b;84(18):e01387–18. doi: 10.1128/AEM.01387-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneveld JR, McMinds R, Thurber RV Stress and stability: applying the anna karenina principle to animal microbiomes. Nature Microbiology. 2017;2:17121. doi: 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- Zhang M, Liang HF, Lei YF, et al Aspergillus niger confers health benefits and modulates the gut microbiota of juvenile Pacific white shrimp (Penaeus vannamei) under farming conditions. Frontiers in Marine Science. 2023;10:1211993. doi: 10.3389/fmars.2023.1211993. [DOI] [Google Scholar]
- Zhang QL, Liu Q, Liu S, et al A new nodavirus is associated with covert mortality disease of shrimp. Journal of General Virology. 2014;95(12):2700–2709. doi: 10.1099/vir.0.070078-0. [DOI] [PubMed] [Google Scholar]
- Zhang XC, Li XH, Lu JQ, et al Quantifying the importance of external and internal sources to the gut microbiota in juvenile and adult shrimp. Aquaculture. 2021a;531:735910. doi: 10.1016/j.aquaculture.2020.735910. [DOI] [Google Scholar]
- Zhang WQ, Zhu ZD, Chen J, et al Quantifying the importance of abiotic and biotic factors governing the succession of gut microbiota over shrimp ontogeny. Frontiers in Microbiology. 2021b;12:752750. doi: 10.3389/fmicb.2021.752750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JF, Fang WH, Yang XL, et al A nonluminescent and highly virulent Vibrio harveyi strain is associated with “bacterial white tail disease” of Litopenaeus vannamei shrimp. PLoS One. 2012;7(2):e29961. doi: 10.1371/journal.pone.0029961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Chen CZ, Xie J, et al Intestinal bacterial signatures of the “cotton shrimp-like” disease explain the change of growth performance and immune responses in Pacific white shrimp (Litopenaeus vannamei) Fish & Shellfish Immunology. 2019b;92:629–636. doi: 10.1016/j.fsi.2019.06.054. [DOI] [PubMed] [Google Scholar]
- Zhou RJ, Zeng SZ, Hou DW, et al Occurrence of human pathogenic bacteria carrying antibiotic resistance genes revealed by metagenomic approach: a case study from an aquatic environment. Journal of Environmental Sciences. 2019a;80:248–256. doi: 10.1016/j.jes.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Zhu JY, Dai WF, Qiu QF, et al Contrasting ecological processes and functional compositions between intestinal bacterial community in healthy and diseased shrimp. Microbial Ecology. 2016;72(4):975–985. doi: 10.1007/s00248-016-0831-8. [DOI] [PubMed] [Google Scholar]