Abstract
Chromobox 2 (CBX2), an epigenetic reader and component of polycomb repressor complex 1, is highly expressed in >75% of high-grade serous carcinoma. Increased CBX2 expression is associated with poorer survival, whereas CBX2 knockdown leads to improved chemotherapy sensitivity. In a high-grade serous carcinoma immune-competent murine model, knockdown of CBX2 decreased tumor progression. We sought to explore the impact of modulation of CBX2 on the tumor immune microenvironment (TIME), understanding that the TIME plays a critical role in disease progression and development of therapy resistance. Exploration of existing datasets demonstrated that elevated CBX2 expression significantly correlated with specific immune cell types in the TIME. RNA sequencing and pathway analysis of differentially expressed genes demonstrated immune signature enrichment. Confocal microscopy and co-culture experiments found that modulation of CBX2 leads to increased recruitment and infiltration of macrophages. Flow cytometry of macrophages cultured with CBX2-overexpressing cells showed increased M2-like macrophages and decreased phagocytosis activity. Cbx2 knockdown in the Trp53-null, Brca2-null ID8 syngeneic murine model (ID8 Trp53−/−Brca2−/−) led to decreased tumor progression compared with the control. NanoString immuno-oncology panel analysis suggested that knockdown in Cbx2 shifts immune cell composition, with an increase in macrophages. Multispectral immunohistochemistry (mIHC) further confirmed an increase in macrophage infiltration. Increased CBX2 expression leads to recruitment and polarization of protumor macrophages, and targeting CBX2 may serve to modulate the TIME to enhance the efficacy of immune therapies.
Significance:
CBX2 expression correlates with the TIME. CBX2 modulation shifts the macrophage population, potentially leading to an immunosuppressive microenvironment, highlighting CBX2 as a target to improve efficacy of immunotherapy.
Introduction
High-grade serous carcinoma (HGSC) is the most common histologic subtype of epithelial ovarian cancer. Understood to arise from the fallopian tube epithelium, ovarian surface, or peritoneum, HGSC is one of the most lethal gynecologic malignancies (1–3). Diagnosed at an advanced stage in most patients, HGSC is treated with a combination of surgery and chemotherapy. Although many patients will respond to primary chemotherapy, the disease has a greater than 80% risk of recurrence and subsequent development of chemoresistance (4, 5). All patients with recurrent HGSC will eventually succumb to their disease. Thus, identifying drivers of disease recurrence, progression, and platinum resistance is a critical unmet need in the care of patients with ovarian cancer.
The epigenetic reader and polycomb repressor complex 1 (PRC1) component chromobox 2 (CBX2) is highly expressed in more than 75% of HGSC tumors (6, 7). It is understood that CBX2 is involved in chromatin regulation and gene transcription and, more broadly, PRC1 specifically regulates cell-fate decisions and is essential for normal organismal development of organisms (8). Clinical studies have demonstrated that polycomb proteins are abnormally expressed many malignancies and that they play a role in cancer invasion, metastases, and progression (8). Data from previous reports suggest that increased CBX2 expression in HGSC conveys poorer disease-free and overall survival and is associated with therapy-resistant disease (6). In anchorage-independent settings, CBX2 protects cells from apoptosis via enhancing stem-like properties (6). However, beyond the concept of stemness, little is understood about how CBX2 drives HGSC disease progression.
The tumor immune microenvironment (TIME) plays a critical role in HGSC therapy response. Tumor-associated macrophages (TAM) account for up to 40% of immune cells in HGSC (9), and elevated TAMs are frequently a poor prognostic indicator and contribute to tumor progression (10). Several studies indicate that limited treatment response, and specifically platinum resistance, may be instigated by cytokines released by TAMs, particularly those traditionally defined as M2-like macrophages (11). In contrast, a subset of TAMs is predicted to have antitumor properties, classically defined as M1 (12). In prostate cancer, the PRC1 was linked to elevated CCL2-mediated tumor-promoting macrophage recruitment (13), which supports a potential role of CBX2 in TIME modulation. Although HGSC tumors are hypothesized to be enriched with tumor-promoting TAMs, there is limited understanding about how the complexity of macrophage polarization contributes to tumor progression and antitumor immunity.
Thus, in HGSC models, we sought to define the impact of modulation of CBX2 on the TIME, specifically the impact of macrophage infiltration, polarization, and activity. In human HGSC tumors, elevated CBX2 significantly correlated with macrophage infiltration. In vitro and in vivo HGSC models highlight that CBX2 expression leads to differential macrophage infiltration and phagocytosis. Given efforts underway to target CBX2 (14, 15), understanding the impact of CBX2 on the TIME and macrophage activity may allow for therapeutic optimization or novel therapeutic combinations.
Materials and Methods
Cell culture
As described in our previous work (15), human HGSC cell lines (PEO1 RRID: CVCL_2686, OVCAR4 RRID: CVCL_1627, ID8 Trp53−/− Brca2−/− RRID: CVCL_IU14) were obtained from the Gynecologic Tissue and Fluid Bank, and a syngeneic (murine) cell line (ID8) was generously provided by Dr. Ian McNeish (Imperial College London, London, United Kingdom). All cell lines were authenticated at The University of Arizona Genomic Shared Resource via short tandem repeat profiling. OVCAR4 and PEO1 cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated FBS and 100 U/mL penicillin/streptomycin. ID8 cells were cultured in DMEM supplemented with 4% heat-inactivated FBS, 100 U/mL penicillin/streptomycin, 5 μg/mL insulin, 5 μg/mL transferrin, and 5 ng/mL sodium selenite. All cells were cultured at 37°C in 5% CO2. Cells were routinely tested for Mycoplasma using LookOut (Millipore-Sigma, Darmstadt Germany), most recently on March 13, 2024.
Small hairpin RNA knockdown
As described previously (6), CBX2-specific small hairpin RNAs (shRNA) were obtained from the University of Colorado Functional Genomics Facility (human CBX2 #1: TRCN0000020327 and human CBX2 #2: TRCN0000232722, Mouse Cbx2 #1: TRCN0000334429, #2 TRCN0000096264). An empty pLKO.1-puro was utilized as shControl (shCTRL). Plasmid isolation was performed using Plasmid Midi-Prep Kit (Qiagen Hilden, Germany). HEK293T cells were transfected with lentivirus construct with packaging plasmids with Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instruction. Cells were incubated overnight and transitioned to DMEM the following morning. The viral supernatant was collected 72 hours posttransfection, applied to OVCAR4, PEO1, or ID8 Trp53−/− Brca1−/− for 24 hours with polybrene. Infected cells were selected using 1.0 μg/mL puromycin.
Small interfering RNA knockdown
As described in ref. 16, cells were plated the day before transfection in RPMI supplemented with 10% FBS but without antibiotics, to be 60% to 80% confluent at the time of transfection. Cells were transfected using Lipofectamine RNAiMAX Reagent (Thermo Fisher Scientific, Cat#13778-075) according to the manufacturer’s protocol. Briefly, for a six-well plate, each well contained 200 µL small interfering RNA (siRNA)–lipid complex in Opti-MEM, including 50 pmol siRNA and 6 µL RNAiMAX. Six hours after transfection, medium was changed to nonantibiotic RPMI. The siRNAs used were siCXCL1 (Dharmacon on target plus SMART pool, CXCL1 Cat. # L-003898-00-0005), siCXCL8 (Dharmacon on target plus SMART pool, CXCL8 Cat. # L-004756-00-0005), or negative control siRNA (Dharmacon on target plus nontargeting pool, Cat. # D-001810-10-05).
CRISPR knockout of CBX2
CBX2 CRISPR knocked out OVCAR4 cells were created by the University of Colorado Functional Genomics Facility using the IDT Alt-R RNP system. ALT-R crRNA and ATL-R tracrRNA were suspended at 100 μmol/L in nuclease-free IDTE pH7.5. The same volumes of ALT-R crRNA and ATL-R tracrRNA were mixed to prepare gRNA complex at 50 μmol/L, heated at 95°C for 5 minutes, and then were cooled to room temperature. The RNP complex was prepared with 150 pmol of gRNA and 125 pmol of Alt-R Cas9-NLS in final volume of 5 μL in PBS. Cells were harvested in Necleofector solution SF with supplement (Lonza, Basel, Switzerland) at a concentration of 1 × 106 cells per 100 μL. The transfection mix was made with 100 μL of cell suspension, 2.5 μL each of RNP complex, and 0.6 μL of 100 μL of Alt-R Cas9 Electroporation enhancer. For OVCAR4, program FE-132 was used. After pulsing, prewarmed medium was added into the Nucleocuvette vessel, and cells were plated for further culturing and isolating clonal populations. The following guide RNAs were used: Hs.Cas9.CBX2.1.AC:CCGAGTGCATCCTGAGCAAG and Hs.Cas9.CBX2.1.AA:GAGTACCTGGTCAAGTGGCG. PCR primers to screen for deletion included CBX2-F1: AGCATGGAGGAGCTGAGCA, CBX2-R2: GGTTACAGCGGGGAGAATCTG, and CBX2-R3: GGAGAATCTGGCCAAGAGGAG.
CBX2 overexpression
Full-length CBX2 (NM_005189) cDNA was cloned from OVCAR4 cDNA library, made by oligo dT primer and SuperScriptIII (Thermo Fisher). Full-length CBX2 was amplified using primers Fwd GGT GCT TTG TGT GCT GC and Rev TCA GTA ATG CCT CAG GTT GAA G. Using the PCR product above as a template, a restriction enzyme compatible clone was amplified with nested PCR using primers. For full-length, XbaI Fwd ATTTCTAGAATGGAGGAGCTGAGCAGCG.
EcoRI Rev ATTGAATTCTCAGTAATGCCTCAGGTTGAAG. PCR products were gel-purified, cut with restriction enzyme XbaI/EcoRI or EcoRI/NotI, and ligated into pCDH-CMV-MCS-EF1-Puro vector that had been cut with XbaI/EcoRI or EcoRI/NotI. Positive clones were confirmed by Sanger sequencing, transformed into Stbl3-competent cells (Thermo Fisher), and amplified with Qiagen MIDI plasmid preparation. Lentiviral particles were generated by transfecting pCDH-CMV-MCS-EF1-Puro-CBX2 into 293FT cells, and viral particles were collected. OVCAR4 cells were transduced with pCDH-CMV-MCS-EF1-Puro or pCDH-CMV-MCS-EF1-Puro-CBX2. Cells were selected with 1 μg/mL puromycin for 48 hours and CBX2 expression was confirmed via qPCR and immunoblot.
Immunoblotting
As described in ref. 17., protein was extracted with radioimmunoprecipitation assay buffer (150 mmol/L NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mmol/L Tris pH 8.0) supplemented with complete EDTA‐free protease inhibitors (Roche, Cat. #4693132001), 5 mmol/L NaF, and 1 mmol/L Na3VO4. Nuclear extraction was performed by suspending cells in a hypotonic buffer (10 mmol/L HEPES-KOH pH 7.9, 1.5 mmol/L MgCl2, 10 mmol/L KCl, 1 mmol/L DTT, and 1× halt protease inhibitor). After dounce homogenization and centrifugation, the resulting nuclear pellets were suspended in a hypertonic buffer (20 mmol/L HEPES-KOH pH 7.9, 25% glycerol, 1.5 mmol/L MgCl2, 0.6 mol/L KCl, 0.2 mmol/L EDTA, 1 mmol/L DTT, and 1× halt protease inhibitor). Protein was separated on an SDS-PAGE and transferred to the polyvinylidene fluoride membrane. Incubation with primary antibodies anti-CBX2 (Diagenode, Cat #C15410339, RRID: AB_3099491) and anti–β-actin (Abcam Cat# ab6276, RRID: AB_2223210) was performed overnight at 4°C. Secondary goat anti‐rabbit (IRDye 680RD or IRDye 800CW, LI‐COR, Cat. #92568071; RRID: AB_2721181 or Cat. #926‐32211; RRID: AB_621842; 1:20,000) and goat anti‐mouse (IRDye 680RD or IRDye 800CW, LI‐COR, Cat. # 926‐68070; RRID: AB_10956588 or Cat# 925‐32210; RRID: AB_2687825; 1:20,000) antibodies were applied for 1 hour at room temperature. Blots were visualized using the LI‐COR Odyssey Imaging System and Image Studio software (V4).
RNA sequencing and analysis
Total RNA was extracted using the Quick-RNA Miniprep Plus Kit (Zymo Research, Cat. #D4209) from cells cultured in attachment settings. Ribosome depletion and library preparation were performed using the Qiagen FastSelect (Cat. #334375) and KAPA BioSystems mRNA HyperPrep Kit (Cat. #KK8581) according to the manufacturer’s protocols. Briefly, 500 ng of RNA was used as the input, and KAPA BioSystems single-index adapters were added at a final concentration of 1.5 mmol/L. A purified, adapter-ligated library was amplified following the manufacturer’s protocol. The final libraries were pooled and sequenced on an Illumina NovaSeq 6000 (University of Colorado Genomics Core; RRID: SCR_021984) as 150-bp paired-end reads.
RNA sequencing (RNA-seq) data analysis was performed by the Biostatistics and Bioinformatics Shared Resource of the University of Colorado Cancer Center (RRID: SCR_021983). FASTQ files generated using bcl2fastq (version 2.20.0.422) were processed using the nf-core/rnaseq pipeline (version 3.9, https://nf-co.re/rnaseq/3.9; ref. 18). Sequencing adapters were removed using Cutadapt (version 3.4; ref. 19) as part of the Trim Galore package (version 0.6.7). Reads were aligned using STAR (version 2.7.10a; ref. 20) to the human transcriptome (GRCh38, gene annotation from Ensembl release 104). Gene quantification was performed using Salmon (version 1.5.2; ref. 21). Tables with transcript and gene counts and TPM were generated using the tximport R package (version 1.18.0; ref. 22) under R version 4.0.3. Normalized data were processed to counts per million (23). Differential expression was calculated using the limma R package (24). Heatmaps were generated by Microsoft Excel based on the 10th and 90th percentile of expression.
Pathway analysis of the differentially expressed genes that overlapped between CBX2 knockout and overexpression was conducted via gene set enrichment analysis and overlap with hallmark pathways.
In vivo model
All mouse experiments were approved by the Institutional Animal Care and Use Committee (IACUC protocol #569). Six- to eight-week-old C57BL/6J mice were purchased from The Jackson Laboratory (strain #000664), and each mouse was intraperitoneally injected with 1 × 106 ID8 Trp53−/−Brca2−/− cells, which either had Cbx2 knocked down using lentivirus (shCbx2 #1 and shCbx2 #2) or were treated with an empty lentivirus (shCTRL) leaving Cbx2 intact, as described above. Thirty-six days after cell injection, mice were euthanized via CO2 inhalation and cervical dislocation and necropsied. Omental tissue was resected for further processing including multispectral IHC (mIHC) and NanoString.
Multispectral IHC (mIHC) and image analysis
mIHC analyses were performed using Vectra Automated Quantitative Pathology Systems (Akoya Biosciences, Menlo Park, CA) as described previously (25). Tissues were formalin-fixed, paraffin-embedded, and sectioned at four microns onto slides. Slides were sequentially stained with antibodies specific for F480 (Cell Signaling Technology, Cat. #30325S) and DAPI. All antibody details are provided in Supplementary Table S1. All slides were de-identified and imaged by the Human Immune Monitoring Shared Resource core (RRID: SCR_021985) on Akoya Biosciences Vectra Polaris scanner. Regions of interest were selected, and multispectral images were collected with a 20x objective. A training set of nine representative images was used to train analysis algorithms for tissue and cell segmentation and phenotyping using inForm software (Akoya Biosciences, Menlo Park, CA). Representative autofluorescence was measured on an unstained control slide and subtracted from study slides. Total tumor area, total cell count, and cell densities of positive and negative cells for each phenotype were graphed and compared in GraphPad Prism 9. Statistical analyses were performed using a multiple comparison one-way ANOVA test.
Quantitative PCR (qPCR)
RNA was isolated using RNeasy Plus Mini Kit (Qiagen, Hilden Germany) according to the manufacturer’s protocol. NanoDrop spectrophotometry was used to measure concentration of extracted RNA. Luna Universal One-step RT qPCR kit (New England BioLabs, Ipswich, MA) was used on Bio-Rad CFX96 (Bio-Rad, Hercules, CA). HPRT or GAPDH were used as internal control as stated in the figure legend. All primer sequences are in Supplementary Table S2.
NanoString and analysis
As described in ref. 16, formalin-fixed, paraffin-embedded omental tumors from the shCTRL and shCbx2 ID8 study were sectioned, and 10-micron tissue–containing paraffin scrolls were collected. RNA was extracted using the High Pure FFPET RNA Isolation kit (Roche) according to the manufacturer’s instructions and eluted in 40 μL elution buffer. RNA quantity and quality were assessed using an RNA ScreenTape on TapeStation 4150 (Agilent). The concentration of RNA was determined by comparison with the RNA ladder.
Extracted RNA was diluted to 30 ng/μL and 5 μL (150 ng) was combined with hybridization buffer and the Reporter CodeSet for the Murine PanCancer IO 360 Panel (NanoString) and incubated for 20 hours at 65°C. The hybridized reaction was analyzed on an nCounter SPRINT Profiler (NanoString). nSolver calculated normalization factors for each sample using raw gene counts and 14 housekeeping genes. Differential gene expression was calculated from normalized gene counts data and an FDR with a Benjamini–Hochberg multicomparison test.
An nSolver Advanced Analysis tool was used for pathway analysis of the genes expressed in shCTRL (n = 3) and shCbx2 (n = 7) samples. The expression profile of the genes expressed was used to generate a pathway score for 25 different pathways (e.g., hypoxia). Genetic signature analysis was performed by NanoString and previously described in ref. 26–30. NanoString counts are available in Supplementary Table S3.
Human primary monocyte isolation
Healthy female donor blood samples (age 21–50 years, in total 25 unique donors) were obtained from the Children’s Hospital Colorado Blood Donor Center at the University of Colorado Anschutz Medical Campus. Collections were conducted according to ethical guidelines (Declaration of Helsinki). Participants were consented with verbal and written consent at the Children’s Hospital Colorado Blood Donor Center. Blood was isolated from leukocyte reduction system (LRS) chambers, which is considered a waste product from blood donor collections. The use of LRS chambers as a discarded material is covered in the consent signed by donors when they give blood. Serum was collected from LRS chambers, and peripheral blood mononuclear cells (PBMC) were isolated using a Leucosep tube (STEMCELL Technologies, Vancouver, Canada) with Ficoll-Paque (Cytiva, Marlborough, MA). The cells were treated with red blood cell lysis buffer (0.832% NH4Cl, 0.1% NaHCO3, and 0.02% EDTA) for 30 minutes at room temperature. Monocytes were isolated using EasySep Human Monocyte Isolation Kit (STEMCELL Technologies, Vancouver, Canada) following the manufacturer’s protocol.
Primary monocytes direct co-culture/invasion with cancer cells
OVCAR4 cells were stained with CellTracker Deep Red (#C34565 Invitrogen, Waltham MA) for 30 minutes. A total of 2 × 105 stained cells per well were seeded in six-well ultralow attachment plates (#3471 Corning Inc., Corning NY) in 1 mL CSC media (DMEM/F12, 1x B27, 4% FBS, 100 U/mL penicillin/streptomycin, 20 ng/mL human EGF, and 20 ng/mL human FGF; ref. 31). On day 3, human primary monocytes were stained with CellTracker Green CMFDA (#C7025, Invitrogen, Waltham MA) for 30 minutes, and 50,000 cells per plate were added to the spheres in 1 mL CSC media and cultured for an additional 7 days (CSC media were added every 3 days). For flow cytometry analysis, after 7 days of co-culture, the cells were collected using 40-micron filter, washed with PBS, and then digested with TrypLE (Gibco, Billings, MT) to make single cells. The cells were analyzed using Penteon (NovoCyte Agilent, Santa Clara, CA), and cytometry data were analyzed using FlowJo software (Tree Star, Ashland, OR). The picture images were taken using a Zeiss Axio Observer Z1 inverted microscope (Zeiss software Rel. 4.8) and quantified using CellProfiler 2.2.0 software.
Indirect polarization assay
Primary monocytes were cultured for 1 day with monocyte medium (RPMI 1640, 10% heat-inactivated FBS, 100 U/mL penicillin/streptomycin, 2.5 mmol/L HEPES buffer, 2 mmol/L GlutaMAX) at 37°C in 5% CO2. The assay was performed using a transwell chamber (0.4 μm pore size PET membrane) in 12-well plates (Corning, Corning NY). A total of 4 × 105 primary monocytes were plated in the bottom of the plate in 1 mL of 1% FBS monocyte medium with human M-CSF 50 ng/mL (PeproTech, Cranbury, NJ). A total of 1 × 105 monolayer OVCAR4 cells were plated in the transwell chamber in 500 μL of 1% FBS monocyte medium. The monocytes were cultured for 8 to 10 days; OVCAR4 cells were replaced every 3 to 4 days. As controls, INFγ (20 ng/mL) and LPS (100 ng/mL) were used for polarizing M1 macrophages; 50 ng/mL IL4, IL10, and IL13 (PeproTech, Cranbury, NJ) were used for polarizing M2 macrophages.
Phagocytosis assay
As similar to that described above, 2 × 105 primary monocytes were cultured in 200 μL 1% heat-inactivated FBS monocyte medium (RPMI 1640, 100 U/mL penicillin/streptomycin, 2.5 mmol/L HEPES buffer, and 2 mmol/L GlutaMAX) with 50 ng/ml M-CSF with cancer cells in the transwell chamber (0.4 μm pore size PET membrane) in 24 wells for 10 days. A total of 4 × 104 monolayer OVCAR4 cells were plated in the transwell chamber in 500 μL of 1% FBS monocyte medium. The cancer cells in transwell chamber were replaced every 3 to 4 days. A total of 30 μL FITC-conjugated microbeads were added directly in the TC plate and incubated at 37°C in 5% CO2 for 90 minutes. After the incubation, the supernatant was removed and 200 μL fixation buffer (BD Cytofix Fixation buffer) added and then incubated on ice for 20 minutes. The cells were gently scraped and filtered with 40-μm filter, analyzed by Penteon (NovoCyte Agilent, Santa Clara, CA).
Flow cytometry
As previously described (32, 33), prior to the staining, cells were treated with Fc blocker (#163404 BioLegend, San Diego, CA) for 20 minutes on ice to block nonspecific binding of antibodies. The cells were incubated with the antibodies for 30 minutes (Supplementary Table S1). Subsequently, cells were washed twice with buffer (2% FBS HBSS). Stained cells were analyzed using Penteon (Agilent, Santa Clara, CA); cytometry data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Publicly available datasets
The Cancer Genome Atlas (TCGA) Ovarian Serous Cystadenocarcinoma (PanCancer Atlas) was examined for CBX2 mRNA expression. Various imputation analyses were performed, including XCELL, CIBERSORT, CIBERSORT-ABS, EPIC, QUANTISEQ, and TIDE, using the TIMERv2 platform (34). Carcinoma EcoTyper analysis was performed to describe cell types (35). Raw data for TIMERv2 analysis using different imputation models are available in Supplementary Table S4.
Statistical analysis
All statistical analyses were conducted in Prism GraphPad v9. Survival comparison used Kaplan–Meier with log-rank, pairwise comparison used t test, multicomparison used ANOVA with multiple test correction, and pairwise comparison over time was analyzed using a mixed model effect. A P < 0.05 was considered significant and when required FDR multicomparison correction (q < 0.05) was made. Error bars are shown as standard error of the mean (SEM). All in vitro experiments were performed in triplicate, and the in vivo experiment was run in duplicate.
Data availability
All data are available within the article and its supplementary materials. RNA-seq data sets are available from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE251947.
Results
CBX2 regulates immune transcriptional profiles
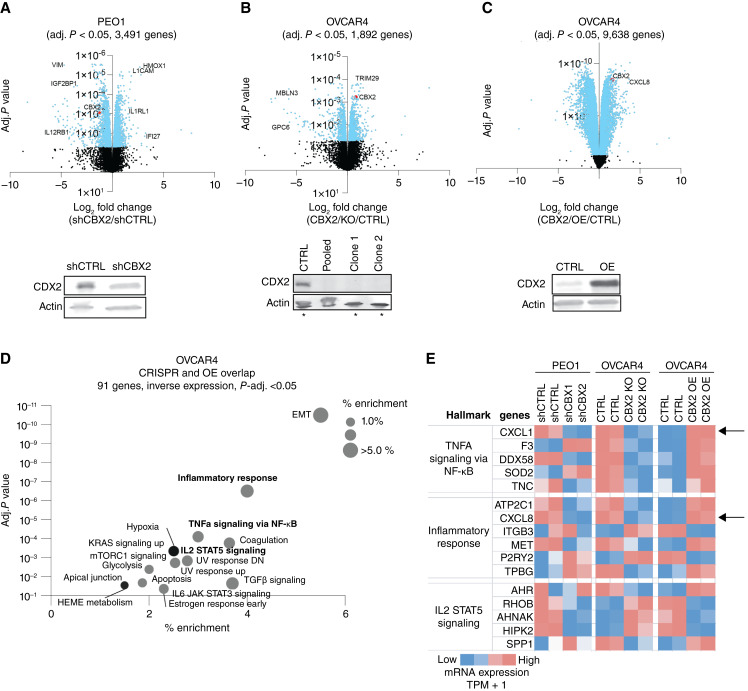
To understand the regulatory role of CBX2, RNA-seq was performed on multiple CBX2-modulated HGSC cell lines. RNA-seq was first performed in CBX2 knockdown (shCBX2) versus CBX2 intact (shCTRL) in the PEO1 cell line. CBX2 knockdown was confirmed via immunoblotting (Fig. 1A). Differential expression analysis identifies decreased expression of immune regulatory genes VIM, IGF2BP1, and IL12RB1 and increased expression of L1CAM, IL1RL1, and IFI27 in shCBX2 cells compared with shCTRL cells (Fig. 1A). RNA-seq was similarly performed on OVCAR4 cell lines, comparing CBX2 CRISPR knockout (CBX2 KO) with CBX2 intact. CBX2 KO was confirmed via immunoblotting (Fig. 1B). In CBX2 KO OVCAR4 cells compared with CBX2 intact (CTRL), there was differential expression of MBLN3, GPC6, and TRIM29 (Fig. 1B). When the OVCAR4 cell line with CBX2 overexpression (CBX2 OE) was compared with CTRL, there was differential expression of PRMT6, IGFL1, and CXCL8 (Fig. 1C). CBX2 overexpression was confirmed via immunoblotting (Fig. 1C). A hypergeometric overlap analysis was performed on OVCAR4 CBX2 KO and CBX2 overexpression (CBX2 OE) overlap (adj. P < 0.05, opposite direction), identifying 91 genes with statistically significant inverse expression. Pathway analysis demonstrated that epithelial–mesenchymal transition (EMT), inflammatory response, TNFα signaling, and IL2/STAT5 signaling are among the top hits enriched in CBX2-overexpressed cells compared with knockout (Fig. 1D; Supplementary Fig. S1A). Additionally, this analysis demonstrated that EMT is one of the most enriched pathways in CBX2 overexpressed cells, consistent with existing literature, thus serving as confirmation of our findings (36). Specific genes from three highlighted hallmark pathways highlight consistent CBX2-mediated regulation across three independent models, including PEO1 CBX2 knockdown versus control, OVCAR4 CBX2 CRISPR knockout versus control, and OVCAR4 CBX2 overexpression versus control. Specifically, we found that CXCL1, CXCL5, and CXCL8 were directly associated with CBX2 expression (Fig. 1E; Supplementary Fig S1B).
Figure 1.
CBX2 regulates immune transcriptional profiles. A, Volcano plot of differential gene expression based on shCBX2 vs. shCTRL in the PEO1 cell line. Immunoblot against CBX2 in PEO1 shCTRL and shCBX2 cells. Loading control, β-actin. B, Volcano plot of differential gene expression based on CBX2 KO vs. CTRL in the OVCAR4 cell line. Immunoblot against CBX2 in OVCAR4 CTRL and CBX2 KO. Loading control, β-actin. Asterisks indicate cell lines used for RNA-seq. C, Volcano plot of differential gene expression based on CBX2 OE vs. CTRL in the OVCAR cell line. Immunoblot against CBX2 in OVCAR4 control (CTRL) and CBX2 OE. Loading control, β-actin. D, Overlap of OVCAR4 CBX2 CRISPR knockout vs. OVCAR4 CBX2 OE. Inverse expression top hits: EMT and inflammatory response. E, Evaluation of hallmark pathways identified TNFα signaling via NF-κB inflammatory responses, and IL2 STAT5 signaling pathways. Direct relationship between CBX2 status and CXCL1 and CXCL8 (IL8) noted (black arrows).
We subsequently analyzed an existing murine-derived CBX2-directed CUT&RUN (cleavage under targets and release using nuclease) dataset (GSE210367). Compared with IgG and HA protein-tagged controls, we observed distinct CBX2-binding sites proximal to numerous genes, including CXCL1 and CXCL5 (Supplementary Fig. S1C). Of note, there is no murine CXCL8 for comparison, as no homologous murine CXCL8 equivalent exists in mice. Overall, interrogating the 91 overlapping genes identified from the transcriptomic analysis against the CBX2 chromatin profiling, we identified 62.4% of genes (53 of 85, six genes were not found in the mouse) with distinct CBX2-binding sites within 10 kilobases of the promoter region. Taken together, the transcriptional evaluation of CBX2-modulated cell lines identified multiple immune-related signatures, and both the transcriptional evaluation and CUT&RUN analysis specifically found a direct relationship between CBX2 expression and CXCL1, CXCL5, and CXCL8 expression, highlighting the potential regulatory role of CBX2.
CBX2 regulates immune-related pathways in a tumor cell–intrinsic fashion
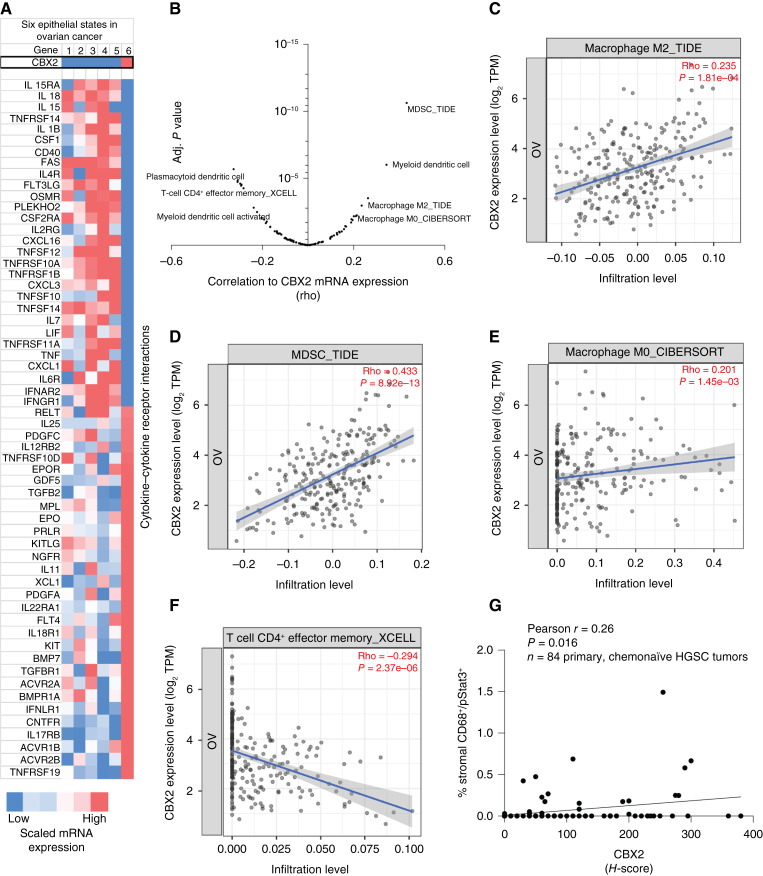
To understand the impact of CBX2 on the TIME of human HGSC tumors, we next examined the data derived from TCGA and the Carcinoma EcoTyper. Upon initial analysis, we discovered that CBX2 expression identifies a unique tumor cell type and its expression is closely associated with “Epithelial State 6” (Fig. 2A). State 6 is defined by increased expression of CBX2 and accounts for 30% of all tumor cells in HGSC tumors, thus contributing to one of the largest proportions of cells (Fig. 2A; Supplementary Fig. S2A). Like the RNA-seq data presented in Fig. 1, Epithelial State 6 also strongly correlates with hallmark pathway cytokine–cytokine interaction. Note, unlike the in vitro data, State 6 is associated with lower CXCL1 expression, suggesting a context-dependent regulation of CXCL1. Taken together, tumors with elevated CBX2 expression are significantly correlated to a differential TIME.
Figure 2.
CBX2 regulates immune-related pathways in a tumor cell–intrinsic fashion. A, Carcinoma EcoTyper epithelial states (S1–6) compared with CBX2 expression (top panel) and cytokine–cytokine receptor interaction pathway. B, Correlation (x-axis, rho) of CBX2 mRNA expression in 303 HGSC tumors with imputed cell type. Adjusted P value shown on y-axis. Imputation pipeline (e.g., CIBERSORT and TIDE) utilized follows underscore in title of plot (details included in Supplementary Table S4). C, Scatter plot of CBX2 mRNA expression and macrophage M2_TIDE. D, Scatter plot of CBX2 mRNA expression and MDSC_TIDE. E, Scatter plot of CBX2 mRNA expression and Macrophage M0_CIBERSORT. F, Scatter plot of CBX2 mRNA expression and T cell CD4+ effector memory_XCELL. G, An annotated tissue microarray was used to perform mIHC evaluating CBX2 expression and immune cell types. Plot demonstrating the relationship between % stromal CD68+/pStat3+ and CBX2 H-score. Statistical tests, unpaired t test (C) and Pearson correlation (D–G).
Considering our tumor cell–intrinsic data, TCGA data, and that modulation of CBX2 is directly involved in immune transcriptional signatures, we evaluated CBX2 expression using immune cell imputation analyses (e.g., CIBERSORT, XCELL, and TIDE) to define the correlation between CBX2 expression and multiple immune cell types in 303 ovarian cancer tumors within TCGA database (37). We identified several immune cell types that correlated with high CBX2 expression, including M0 macrophages, M2 macrophages, myeloid-derived stem cells (MDSC), as well as negative correlation to T cells (Fig. 2B–F; Supplementary Table S4). In addition, CIBERSORT analysis of M0 macrophages identified a positive correlation with CXCL5 and CXCL8 (Supplementary Fig. S2B).
Given that the imputation data depend on mRNA expression from TCGA, we used a well-annotated tissue microarray of HGSC tumors to further explore the density and distribution of immune cells (38). Vectra Polaris mIHC allowed for identification and quantification of CD8 T cells (CD3+/CD8+), T regulatory (Treg) cells (CD3+/FOXP3+), and total macrophages (CD68+) within the HGSC microenvironment, and there was no correlation with CBX2 expression (Supplementary Fig. S2C). Importantly, however, high CBX2 expression in chemonaïve HGSC tumors was positively correlated with active macrophages (CD68+/pSTAT3+; r = 0.26; P = 0.016; Fig. 2G). Altogether, we found direct links between CBX2 and the TIME using both transcriptional and protein analyses, and we specifically observed correlations between CBX2 and M0, M2, and active macrophages, as well as MDSCs. We next sought to understand CBX2-dependent macrophage recruitment and polarization.
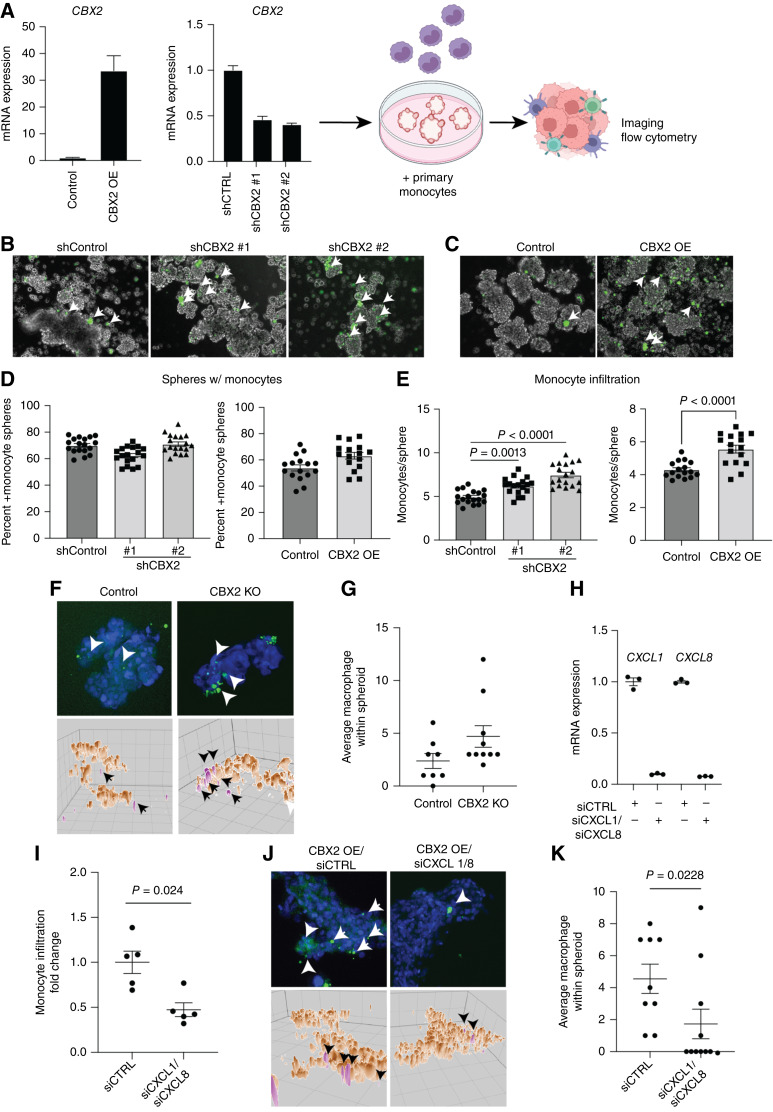
Modulation of CBX2 leads to a shift in macrophage recruitment
In an in vitro system, we wanted to determine if there is differential macrophage recruitment with changes in CBX2 expression. OVCAR4 cells with CBX2 overexpression (OE) construct or with CBX2 knockdown (shCBX2#1 and shCBX2#2; Fig. 3A) were cultured in low attachment setting to form spheroids over 2 days. In parallel, monocytes (CD14+CD16−) were isolated using negative selection from peripheral blood from healthy donors and were stained with CellTracker. Stained monocytes were then added to the spheroid cultures to monitor infiltration (Fig. 3A). After 5 days in co-culture, monocyte infiltration into the spheroids was observed with confocal microscopy (Fig. 3B and C). Although we noted that there was no difference in the number of spheroids with CellTracker-positive monocytes (Fig. 3D), we did observe that modulating CBX2 expression led to a significant increase in the number of CellTracker-positive monocytes in spheroids (Fig. 3E; Supplementary Fig. S3A–S3C). Using the CBX2 KO OVCAR4 cells, we examined spheroid infiltration via confocal microscopy and observed that compared with control, CBX2 knockout led to a modest increase in macrophage infiltration (Fig. 3F–G). These data suggest that CBX2 regulates macrophage infiltration.
Figure 3.
Modulation of CBX2 leads to shift in macrophage recruitment. A, OVCAR4 CBX2 OE and knockdown confirmation by qPCR. Diagram of protocol with direct co-culture of spheroids with primary monocytes. B, Confocal microscopy with infiltration of monocytes (green) increased with knockdown of CBX2. C, Confocal microscopy with infiltration of monocytes increased with overexpression of CBX2. D, Plots demonstrating the percentage of spheres with monocytes in shCBX2 or overexpression constructs. E, Plots demonstrating the number of spheres with CellTracker-positive monocytes in shCBX2 and overexpression. F, OVCAR4 (control) and CBX2 KO cells co-cultured with fluorescence-tagged primary monocytes (green, white arrowheads). Blue = nuclei. Images (top) are maximum projections of confocal z-stack and form-filling rendering [brown, cancer cells; pink, monocytes (black arrowheads)]. 50-micron grid. G, Quantification of F. H, qPCR of CXCL1 and CXCL8 in siControl (siCTRL) and siCXCL1/siCXCL8. Internal control, HPRT. I, Flow cytometric analysis of digested co-culture spheroids to measure monocyte infiltration. J, Same as F, but with siControl and siCXCL1/8 in cells overexpressing CBX2. K, Quantification of J. Error bars, SEM. Statistical test, unpaired t test and multicomparison ANOVA.
Based on transcriptomic findings that CBX2 potentially regulates CXCL1 and CXCL8 expression, we performed a combination siRNA-mediated knockdown of CXCL1 and CXCL8 in CBX2 OE cells (Fig. 3H). Using co-culture spheroid as noted above, we examined macrophage infiltration via flow cytometry and confocal imaging. Compared with siControl, siCXCL1/8 spheroids showed a significant reduction in macrophage infiltration (Fig. 3I–K), suggesting cancer cell CBX2-mediated regulation of CXCL1/8 expression contributes to differential macrophage infiltration. Overall, we observed that CBX2 modulation led to differential macrophage infiltration, which led us to question the CBX2-dependent polarization status of the macrophages.
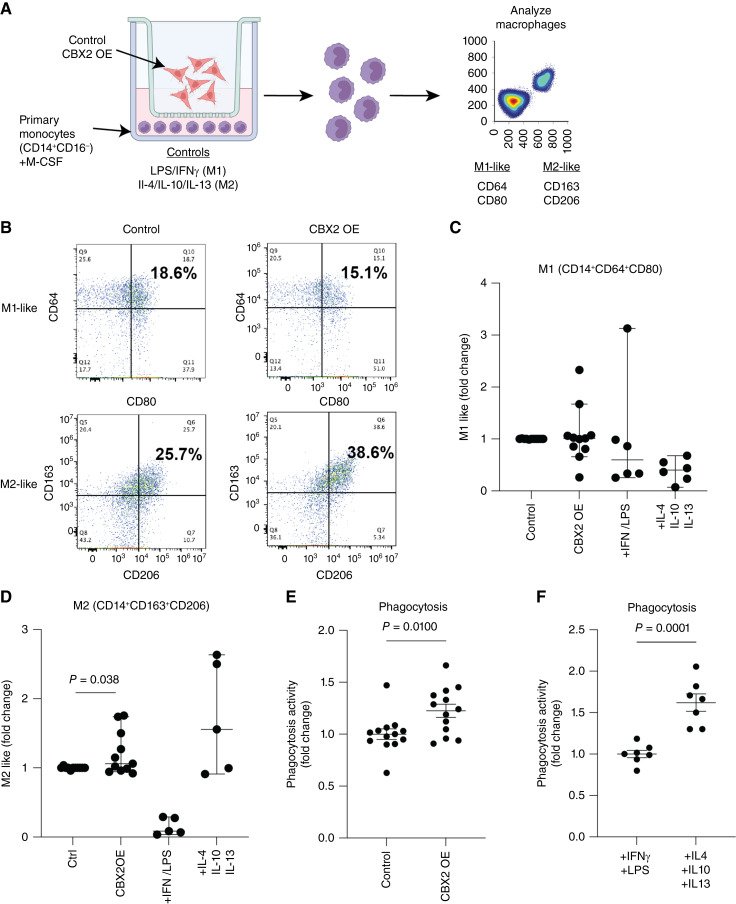
CBX2 overexpression increases tumor-promoting macrophages and phagocytic activity
Macrophages exist classically on a spectrum of differentiation states from M1 to M2. Within the tumor microenvironment (TME), M1 macrophages are proposed to be tumor-suppressive and M2 macrophages tumor-promoting; however, the expression of M1 and M2 protein markers is not mutually exclusive for each type and the complexity of this spectrum remains a significant focus in the field (39–41). In HGSC tumors, the differentiation status of macrophages is important, as elevated M1-like macrophages and depleted M2-like macrophages in the TIME correlate to improved overall survival (Supplementary Fig. S4A). These findings highlight the complex relationship between macrophages and tumor progression. Thus, we sought to assess CBX2-dependent M1/M2-mediated polarization and phagocytosis activity. Using an indirect co-culture system, OVCAR4 control and CBX2 OE cells were cultured with primary human M-CSF-stimulated monocytes. The indirect system only allows for the passage of metabolites and cytokines between the cancer cells and monocytes (Fig. 4A). LPS/IFNγ and IL4/IL10/IL13 were used as controls to differentiate the monocytes into M1- or M2-like macrophages, respectively. After 8 to 10 days of co-culture, macrophage polarization was assayed via flow cytometry. Although CBX2 OE in the cancer cells did not alter the density of M1-like macrophages (18.6% vs. 15.1%), it did promote the expression of M2-like macrophage markers (Control: 25.7% vs. CBX2 OE: 38.6%, Fig. 4B–D; gating strategy; Supplementary Fig. S4B). In co-culture conditions with monocytes and CBX2-modulated cancer cells, compared with the control cells, we observed that CBX2 overexpressing cells increased the number of CD68-positive cells, and conversely, CBX2 knockdown and knockout attenuated the number of CD68 positive cells (Supplementary Fig. S4C), suggesting that CBX2 expression in the cancer cells is promoting monocyte differentiation into macrophages. Moreover, these data highlight the impact of increased CBX2 expression in HGSC cells in driving a more M2-like macrophage polarization.
Figure 4.
Overexpression of CBX2 leads to an increase in M2-like macrophage activity. A, Diagram of co-culture experimental setup with OVCAR4 CBX2 OE exposed to primary monocytes (from eight unique donors), then analyzed by flow utilizing M1- and M2-like markers. B, Flow cytometry analysis of CBX2 OE compared with control, increase in M2-like macrophages (CD163 and CD206). C and D, Plots of the fold change of M1- and M2-like receptors in control, CBX2 OE, +LPS/INFγ, and IL4/IL10/IL13, in which IL4/IL10/IL13 serves as the control for M2-like stimulation. E, Functional phagocytosis assay of monocytes co-cultured with control OVCAR4 compared with CBX2 OE. F, Functional phagocytosis assay, +LPS (M1 stimulatory) compared with +IL4, +IL10, and +IL13 (M2 stimulatory). Error bars, SEM. Statistical test, unpaired t test. Note: Each dot represents a unique PBMC donor.
Phagocytosis, or the engulfment cells and debris within the microenvironment, is a critical activity of macrophages. Phagocytosis can enhance the presentation of antigens to drive further engagement of the immune system; however, in malignancy increased phagocytosis of tumor cells has also been linked to driving immune suppression and tolerance (42). Using macrophages generated via the indirect co-culture system described above, we observed that compared with macrophages co-cultured with OVCAR4 cells in a Boyden chamber, macrophages co-cultured with OVCAR4 CBX2 OE cells demonstrated elevated phagocytosic activity (Fig. 4E and F; Supplementary Fig. S4D). Taken together, elevated CBX2 expression leads to the recruitment of macrophages and increased macrophage phagocytic activity.
Knockdown of Cbx2 in a syngeneic mouse model of ovarian cancer shifts the composition of the TIME
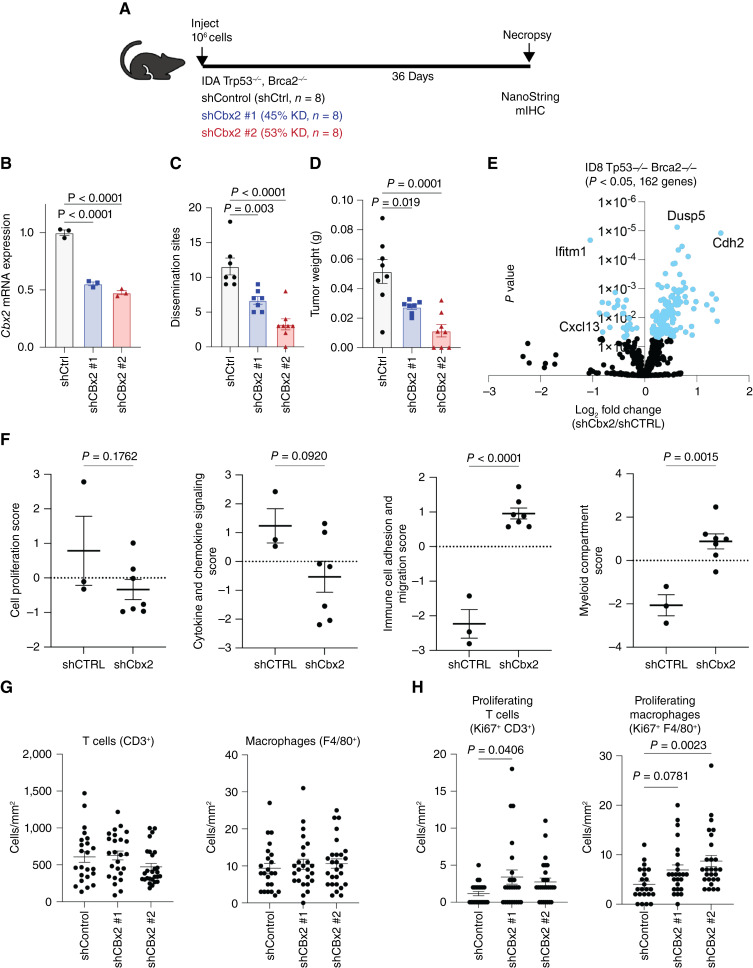
Utilizing a Trp53- and Brca2-null ID8 syngeneic mouse model (ID8 Trp53−/−Brca2−/−; ref. 43), we next examined the effect of Cbx2 knockdown on tumor progression (Fig. 5A) with three experimental arms: Cbx2 intact (shCTRL), Cbx2 knockdown #1 (shCbx2#1), and Cbx2 knockdown #2 (shCbx2#2). Cbx2 mRNA expression was evaluated with a statistically significant decrease in Cbx2 expression in shCbx2 #1 (45% knockdown) and shCbx2 #2 (53% knockdown) confirming appropriate knockdown (Fig. 5B). All mice were injected with 1 × 106 tumor cells, which were allowed to grow for 36 days at which time euthanasia and necropsy were performed. Relative to shCTRL, the number of dissemination sites and total tumor weight were significantly reduced in the Cbx2 knockdown tumor-bearing mice, and tumor weight was measured with a similar significant decrease in both knockdown lines (Fig. 5C and D). These effects were reproduced in a biological replicate (Supplementary Fig. S5A–S5F). These data confirm that loss of Cbx2 is sufficient to significantly decrease tumor growth in an immune-competent syngeneic murine model of HGSC.
Figure 5.
Knockdown of Cbx2 in a syngeneic mouse model of HGSC leads to shift in composition of the TIME. A, Diagram of ID8 syngeneic mouse model. B, RT-PCR analysis confirming Cbx2 knockdown in ID8 Trp53−/−Brca2−/− cells (internal control, 18S), C, Tumor dissemination sites and (D) total tumor weight of knockdown vs. control tumors. E, NanoString transcriptomic differential expression of shCbx2 compared with control, derived from solid tumors. F, NanoString pathway scores for cell proliferation, cytokine signaling score, immune cell and migration score, and myeloid comparison score between control and shCbx2. G, Comparison of CD3+ (T cells) and F480 (macrophages) without Ki67 positivity and (H) with Ki67 positivity in control vs. shCbx2. Error bars, SEM. Statistical test, unpaired t test and multicomparison ANOVA.
NanoString transcriptomic analysis was performed on the tumor specimens, comparing shCbx2 with intact Cbx2 and 162 differentially expressed genes were found (Fig. 5E). All NanoString raw count data can be found in Supplementary Table S3. Notably, Ifitm1 and Cxcl13 were downregulated in the setting of Cbx2 knockdown, whereas Dusp5 and Cdh2 were associated with intact Cbx2 (Fig. 5E). NanoString transcriptional profiles were scored with a significant increase in immune cell adhesion and migration score, as well as myeloid compartment score in the setting of knockdown of Cbx2 (Fig. 5F; Supplementary Fig. S5G). Of note, there were trends toward a decrease in cell proliferation score and cytokine and chemokine signaling score in the setting of shCbx2. These data demonstrate that modulation of Cbx2 impacts transcriptional profiling in various models and that the resulting transcriptional changes aid in remodeling the TIME.
To further confirm CBX2-dependent TIME remodeling, tumors derived from the mice were evaluated using Vectra Polaris mIHC. All immune cells were quantified, and there was no difference in total T cells (CD3+) or macrophages (F480+) between Cbx2 knockdown and control (Fig. 5G). However, there was an increase in macrophages when proliferation (Ki67+) was considered, highlighting a proliferative or active state (Fig. 5H; ref. 44). In shCbx2 #1, there was an increase in proliferating T cells (CD3+/Ki67+), a marker of T-cell activation (Fig. 5H). In shCbx2 tumors compared with shCtrl, there was a trend (shCbx2 #1, P = 0.0781) and significant (shCbx2#2 P = 0.0023) increase in proliferating macrophages F480+/Ki67+ (Fig. 5H). These data confirm the impact of Cbx2 modulation on the TIME in an immune-competent HGSC murine model and suggest that loss of Cbx2 expression leads to an increased T-cell activation and infiltration of macrophages.
Discussion
The HGSC TME is heterogeneous, including multiple nonmalignant cell types that support tumor progression, such as fibroblasts and immune cells. For instance, some TAMs support tumor progression by promoting chemoresistance and reinforcing immune suppression. In this series of investigations, we observed that an understudied epigenetic reader protein, CBX2, promotes the transcriptional reprogramming of tumor cells to support the differentiation of TAMs. We observed that CBX2 expression delineates a unique HGSC epithelial cell type and modulating CBX2 in tumor cells leads to a differential immune microenvironment. Furthermore, modulation of CBX2 leads to differential macrophage infiltration and polarization in both spheroid cultures and in vivo syngeneic murine tumors. Using primary human-derived macrophages co-cultured with CBX2 overexpressing HGSC cells, we observed macrophages exhibiting increased M2-like differentiation and elevated phagocytosis.
Targeting the macrophage compartment to inhibit tumor progression and support immune surveillance of the tumor is an attractive approach to promote durable antitumor responses. Rodriguez-Garcia and colleagues (45) demonstrated the utility of a chimeric antigen receptor T-cell targeting TAMs enhances the antitumor response of traditional chimeric antigen receptor T-cell, highlighting that TAM-mediated immune suppression is a critical target in overcoming immune evasion. Another example is macrophage activity and differentiation are dependent on colony-stimulating factor (CSF) stimulation. Thus, several CSF receptor (CSF1Ri) antagonists have been developed to date (46). In HGSC models, the use of CSF1Ri promoted the maintenance of vascular integrity, thereby reducing malignant ascites accumulation and alleviating chemotherapy resistance (47, 48). Taken in the context of our current and prior findings showing CBX2 is critical for HGSC progression (6), targeting CBX2 in tumor cells provides an opportunity to limit tumor progression and potentially alleviate an immune-suppressed microenvironment.
The biology and function of TAMs remain incompletely understood. When initially described, TAMs were defined as a subset of macrophages that contributed to immunosuppression and promoted tumor growth and progression (49). However, further research has demonstrated that TAMs have different states, ranging on the spectrum from M2-like to M1-like, both of which can promote or inhibit cancer progression, respectively (50). Therefore, finding treatments that tip the balance of TAM polarization to a more M1-like and less M2-like phenotype is important moving forward. In the work described, we observed that indirect co-culture of CBX2 expression in cancer cells led to the increase of CD163- and CD206-expressing TAM, highlighting the contribution of secreted factors from the cancer cells regulating the macrophage phenotype toward tumor promotion. Through RNA-seq and qPCR analysis, modulation of CBX2 led to the differential expression of immune regulatory cytokines, including CXCL1. Although multiple cell types express CXCL1 in the TME, several studies demonstrate that elevated CXCL1-mediated signaling supports tumor-promoting macrophage differentiation and cancer progression in a macrophage-dependent manner (51–53). Although we predict that CBX2-mediated regulation of TAM differentiation and activity is highly complex, our findings suggest a unique CBX2/CXCL1 or CBX2/CXCL8 axis in tumor cells that aid in remodeling the HGSC TIME.
Interestingly, in our in vivo murine model of HGSC, we observed no difference in the total number of macrophages, contrary to our prediction that CXCL1 signaling in shCTRL (CBX2 intact) tumor cells would increase the number of TAMs present. However, in the CBX2 knockdown tumors, we observed an increase in proliferating macrophages, marked by Ki67 expression. Recent reports have indicated that both tissue resident as well as recruited macrophages can undergo proliferation and self-renewal. Taken together, our data would suggest that inhibition of CBX2 increases the proliferation of macrophages and polarizes them away from an M2-like phenotype. Given the limited understanding of proliferating macrophages in the field, understanding the phenotype of these cells and their plasticity in the context of HGSC is an exciting new direction for our research, particularly given their potential role in therapy resistance (54–56).
In this report, various HGSC models and primary macrophages were used to dissect CBX2-dependent TIME remodeling, however there are limitations. We opted to use human-derived monocytes, rather than THP1 cells; however, this leads to the additional limitation of heterogeneity between samples. Additionally, the use of macrophage receptor status is an imperfect approach to determining macrophage activity as tumor promoting or tumor inhibiting. Thus, we attempted to utilize functional assays to further support our findings. We acknowledge that an additional limitation of this work is that the mechanism is not fully elucidated, and further work should assess chromatin profiling of CBX2 genomic occupancy and exploring the necessity of the adaptive immune response when targeting the CBX2–macrophage interplay.
Overall, the work reported is one of only a few studies to interrogate tumor cell–dependent epigenetic mechanisms that directly contribute to the remodeling of the TIME. CBX2 is a new potential target in HGSC and other malignancies, with the hope of limiting tumor progression and resensitizing to therapy. Thus, understanding the impact of CBX2 on the TIME will allow for optimization of CBX2 inhibition and therapeutic approaches.
Supplementary Material
Overview of supplementary data
CBX2 binds to promoter regions of cytokine genes and CBX2 is significantly associated with immune signatures.
CBX2 expression associates with epithelial state 6. CBX2 protein expression does not correlate with T cell infiltration. CXCL1, 5, and CXCL8 expression correlation with Macrophage M0_CIBERSORT infiltration.
Modulation of CBX2 enhances monocyte infiltration
M1/M2 macrophages convey differential survival outcomes. M1/M2 gating strategy. CD68 gating of monocytes in culture system.
In vivo modeling with loss of CBX2 expression and Nanostring pathway analysis
Antibodies
Primers
Nanostring
TIMERv2
Acknowledgments
We acknowledge philanthropic contributions from D. Thomas and Kay L. Dunton Endowed Memorial Chair in Ovarian Cancer Research, the McClintock-Addlesperger Family, Karen M. Jennison, Don and Arlene Mohler Johnson Family, Michael Intagliata, Duane and Denise Suess, Mary Normandin, and Donald Engelstad. This work was made possible by the Children’s Hospital Blood Donor Center and their provision of donor blood, as well as the many shared resources available through the University of Colorado Cancer Center, including the Flow Shared Resource and Functional Genomics Shared Resource. This work was supported by the Ovarian Cancer Research Alliance (L.W. Brubaker and B.G. Bitler Collaborative, 889342), The American Cancer Society (B.G. Bitler, RSG-19-129-01-DDC), the Department of Defense (B.G. Bitler, OC170228, OC200302, and OC200225; L.W. Brubaker OC230224), NIH/NCI (B.G. Bitler, R37CA261987), and the University of Colorado Cancer Center Support Grant (P30CA046934 - Bioinformatics and Biostatistics, Functional Genomics, Genomics, and Flow Cytometry). We also thank the Human Immune Monitoring Shared Resource within the University of Colorado Human Immunology and Immunotherapy Initiative for their expert assistance with the design and execution of the mIHC. This work was supported by the Alpine HPC system, which is jointly funded by the University of Colorado Boulder, the University of Colorado Anschutz, Colorado State University, and the National Science Foundation (award 2201538).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Communications Online (https://aacrjournals.org/cancerrescommun/).
Authors’ Disclosures
T. Danhorn reports grants from NIH during the conduct of the study. B.G. Bitler reports grants from NCI/NIH, ACS, DOD, and OCRA during the conduct of the study. L.W. Brubaker reports grant from OCRA during the conduct of the study and consulting work with AstraZeneca outside the submitted work, as well as a patent to CBX2 inhibitor pending. No disclosures were reported by the other authors.
Authors’ Contributions
R. Iwanaga: Data curation, formal analysis, investigation, methodology, writing-original draft, writing-review and editing. T.M. Yamamoto: Data curation, formal analysis, investigation, methodology, writing-original draft, writing-review and editing. K. Gomez: Data curation, formal analysis, investigation, writing-review and editing. L.L. Nguyen: Data curation, formal analysis, investigation, methodology, writing-review and editing. E.R. Woodruff: Data curation, formal analysis, investigation, methodology, writing-review and editing. M.D. Post: Data curation, methodology, writing-review and editing. R.G. Mikeska: Data curation, formal analysis, writing-review and editing. E. Danis: Data curation, formal analysis, writing-review and editing. T. Danhorn: Data curation, formal analysis, investigation, methodology, writing-review and editing. M.P. Boorgula: Data curation, formal analysis, methodology, writing-review and editing. S.S. Mitra: Supervision, methodology. N.A. Marjon: Supervision, methodology, writing-review and editing. B.G. Bitler: Conceptualization, data curation, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing-original draft, writing-review and editing. L.W. Brubaker: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, writing-original draft, project administration, writing-review and editing.
References
- 1. Bowtell DDL. The genesis and evolution of high-grade serous ovarian cancer. Nat Rev Cancer 2010;10:803–8. [DOI] [PubMed] [Google Scholar]
- 2. McPherson A, Roth A, Laks E, Masud T, Bashashati A, Zhang AW, et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat Genet 2016;48:758–67. [DOI] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17–48. [DOI] [PubMed] [Google Scholar]
- 4. Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet 2014;384:1376–88. [DOI] [PubMed] [Google Scholar]
- 5. Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer 2003;3:502–16. [DOI] [PubMed] [Google Scholar]
- 6. Wheeler LJ, Watson ZL, Qamar L, Yamamoto TM, Post MD, Berning AA, et al. CBX2 identified as driver of anoikis escape and dissemination in high grade serous ovarian cancer. Oncogenesis 2018;7:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo Y, Zhao S, Wang GG. Polycomb gene silencing mechanisms: PRC2 chromatin targeting, H3K27me3 ‘Readout’, and phase separation-based compaction. Trends Genet 2021;37:547–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gui T, Bai H, Zeng J, Zhong Z, Cao D, Cui Q, et al. Tumor heterogeneity in the recurrence of epithelial ovarian cancer demonstrated by polycomb group proteins. Onco Targets Ther 2014;7:1705–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Launonen IM, Lyytikainen N, Casado J, Anttila EA, Szabo A, Haltia UM, et al. Single-cell tumor-immune microenvironment of BRCA1/2 mutated high-grade serous ovarian cancer. Nat Commun 2022;13:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med 2003;348:203–13. [DOI] [PubMed] [Google Scholar]
- 11. Yang Y, Yang Y, Yang J, Zhao X, Wei X. Tumor microenvironment in ovarian cancer: function and therapeutic strategy. Front Cell Dev Biol 2020;8:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiang X, Wang J, Lu D, Xu X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Signal Transduct Target Ther 2021;6:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Su W, Han HH, Wang Y, Zhang B, Zhou B, Cheng Y, et al. The polycomb repressor complex 1 drives double-negative prostate cancer metastasis by coordinating stemness and immune suppression. Cancer Cell 2019;36:139–55.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang S, Alpsoy A, Sood S, Ordonez-Rubiano SC, Dhiman A, Sun Y, et al. A potent, selective CBX2 chromodomain ligand and its cellular activity during prostate cancer neuroendocrine differentiation. Chembiochem 2021;22:2335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brubaker LW, Backos DS, Nguyen VT, Reigan P, Yamamoto TM, Woodruff ER, et al. Novel chromobox 2 inhibitory peptide decreases tumor progression. Expert Opin Ther Targets 2023;27:361–71. [DOI] [PubMed] [Google Scholar]
- 16. Jordan KR, Sikora MJ, Slansky JE, Minic A, Richer JK, Moroney MR, et al. The capacity of the ovarian cancer tumor microenvironment to integrate inflammation signaling conveys a shorter disease-free interval. Clin Cancer Res 2020;26:6362–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McMellen A, Yamamoto TM, Qamar L, Sanders BE, Nguyen LL, Ortiz Chavez D, et al. ATF6-Mediated signaling contributes to PARP inhibitor resistance in ovarian cancer. Mol Cancer Res 2023;21:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 2020;38:276–8. [DOI] [PubMed] [Google Scholar]
- 19. Kechin A, Boyarskikh U, Kel A, Filipenko M. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol 2017;24:1138–43. [DOI] [PubMed] [Google Scholar]
- 20. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 2017;14:417–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 2015;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lang J, Capasso A, Jordan KR, French JD, Kar A, Bagby SM, et al. Development of an adrenocortical cancer humanized mouse model to characterize anti-PD1 effects on tumor microenvironment. J Clin Endocrinol Metab 2020;105:26–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Erratum for the research article “Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy” by R. Cristescu, R. Mogg, M. Ayers, A. Albright, E. Murphy, J. Yearley, X. Sher, X. Q. Liu, H. Lu, M. Nebozhyn, C. Zhang, J. K. Lunceford, A. Science 2019;363:eaax1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Danaher P, Warren S, Lu R, Samayoa J, Sullivan A, Pekker I, et al. Pan-cancer adaptive immune resistance as defined by the Tumor Inflammation Signature (TIS): results from The Cancer Genome Atlas (TCGA). J Immunother Cancer 2018;6:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Voorwerk L, Slagter M, Horlings HM, Sikorska K, van de Vijver KK, de Maaker M, et al. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: the TONIC trial. Nat Med 2019;25:920–8. [DOI] [PubMed] [Google Scholar]
- 31. Keysar SB, Le PN, Miller B, Jackson BC, Eagles JR, Nieto C, et al. Regulation of head and neck squamous cancer stem cells by PI3K and SOX2. J Natl Cancer Inst 2017;109:djw189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mohanty S, Yerneni K, Theruvath JL, Graef CM, Nejadnik H, Lenkov O, et al. Nanoparticle enhanced MRI can monitor macrophage response to CD47 mAb immunotherapy in osteosarcoma. Cell Death Dis 2019;10:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang M, Hutter G, Kahn SA, Azad TD, Gholamin S, Xu CY, et al. Anti-CD47 treatment stimulates phagocytosis of glioblastoma by M1 and M2 polarized macrophages and promotes M1 polarized macrophages in vivo. PLoS One 2016;11:e0153550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luca BA, Steen CB, Matusiak M, Azizi A, Varma S, Zhu C, et al. Atlas of clinically distinct cell states and ecosystems across human solid tumors. Cell 2021;184:5482–96.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu W, Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell 2019;49:361–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cancer Genome Atlas Research Network . Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Watson ZL, Yamamoto TM, McMellen A, Kim H, Hughes CJ, Wheeler LJ, et al. Histone methyltransferases EHMT1 and EHMT2 (GLP/G9A) maintain PARP inhibitor resistance in high-grade serous ovarian carcinoma. Clin Epigenetics 2019;11:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci 2021;22:6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheng H, Wang Z, Fu L, Xu T. Macrophage polarization in the development and progression of ovarian cancers: an overview. Front Oncol 2019;9:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity 2005;23:344–6. [DOI] [PubMed] [Google Scholar]
- 42. Germic N, Frangez Z, Yousefi S, Simon HU. Regulation of the innate immune system by autophagy: monocytes, macrophages, dendritic cells and antigen presentation. Cell Death Differ 2019;26:715–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walton J, Blagih J, Ennis D, Leung E, Dowson S, Farquharson M, et al. CRISPR/Cas9-Mediated Trp53 and Brca2 knockout to generate improved murine models of ovarian high-grade serous carcinoma. Cancer Res 2016;76:6118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Herwig MC, Holz FG, Loeffler KU. Distribution and presumed proliferation of macrophages in inflammatory diseases of the ocular adnexae. Curr Eye Res 2015;40:604–10. [DOI] [PubMed] [Google Scholar]
- 45. Rodriguez-Garcia A, Palazon A, Noguera-Ortega E, Powell DJ Jr, Guedan S. CAR-T cells hit the tumor microenvironment: strategies to overcome tumor escape. Front Immunol 2020;11:1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cannarile MA, Weisser M, Jacob W, Jegg AM, Ries CH, Rüttinger D. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Cancer 2017;5:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moughon DL, He H, Schokrpur S, Jiang ZK, Yaqoob M, David J, et al. Macrophage blockade using CSF1R inhibitors reverses the vascular leakage underlying malignant ascites in late-stage epithelial ovarian cancer. Cancer Res 2015;75:4742–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu R, Jin H, Jin C, Huang X, Lin J, Teng Y. Inhibition of the CSF-1 receptor sensitizes ovarian cancer cells to cisplatin. Cell Biochem Funct 2018;36:80–7. [DOI] [PubMed] [Google Scholar]
- 49. Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity 2014;41:49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pittet MJ, Michielin O, Migliorini D. Clinical relevance of tumour-associated macrophages. Nat Rev Clin Oncol 2022;19:402–21. [DOI] [PubMed] [Google Scholar]
- 51. Wang N, Liu W, Zheng Y, Wang S, Yang B, Li M, et al. CXCL1 derived from tumor-associated macrophages promotes breast cancer metastasis via activating NF-κB/SOX4 signaling. Cell Death Dis 2018;9:880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miyake M, Hori S, Morizawa Y, Tatsumi Y, Nakai Y, Anai S, et al. CXCL1-Mediated interaction of cancer cells with tumor-associated macrophages and cancer-associated fibroblasts promotes tumor progression in human bladder cancer. Neoplasia 2016;18:636–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu H, Jiang N, Li J, Jin Q, Jin J, Guo J, et al. Tumor cell SPTBN1 inhibits M2 polarization of macrophages by suppressing CXCL1 expression. J Cell Physiol 2024;239:97–111. [DOI] [PubMed] [Google Scholar]
- 54. Wang S, Wang J, Chen Z, Luo J, Guo W, Sun L, et al. Targeting M2-like tumor-associated macrophages is a potential therapeutic approach to overcome antitumor drug resistance. NPJ Precis Oncol 2024;8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell 2015;27:462–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. El-Arabey AA, Alkhalil SS, Al-Shouli ST, Awadalla ME, Alhamdi HW, Almanaa TN, et al. Revisiting macrophages in ovarian cancer microenvironment: development, function and interaction. Med Oncol 2023;40:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of supplementary data
CBX2 binds to promoter regions of cytokine genes and CBX2 is significantly associated with immune signatures.
CBX2 expression associates with epithelial state 6. CBX2 protein expression does not correlate with T cell infiltration. CXCL1, 5, and CXCL8 expression correlation with Macrophage M0_CIBERSORT infiltration.
Modulation of CBX2 enhances monocyte infiltration
M1/M2 macrophages convey differential survival outcomes. M1/M2 gating strategy. CD68 gating of monocytes in culture system.
In vivo modeling with loss of CBX2 expression and Nanostring pathway analysis
Antibodies
Primers
Nanostring
TIMERv2
Data Availability Statement
All data are available within the article and its supplementary materials. RNA-seq data sets are available from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE251947.