Abstract
Persistent reovirus infections of murine L929 cells select cellular mutations that inhibit viral disassembly within the endocytic pathway. Mutant cells support reovirus growth when infection is initiated with infectious subvirion particles (ISVPs), which are intermediates in reovirus disassembly formed following proteolysis of viral outer-capsid proteins. However, mutant cells do not support growth of virions, indicating that these cells have a defect in virion-to-ISVP processing. To better understand mechanisms by which viruses use the endocytic pathway to enter cells, we defined steps in reovirus replication blocked in mutant cells selected during persistent infection. Subcellular localization of reovirus after adsorption to parental and mutant cells was assessed using confocal microscopy and virions conjugated to a fluorescent probe. Parental and mutant cells did not differ in the capacity to internalize virions or distribute them to perinuclear compartments. Using pH-sensitive probes, the intravesicular pH was determined and found to be equivalent in parental and mutant cells. In both cell types, virions localized to acidified intracellular organelles. The capacity of parental and mutant cells to support proteolysis of reovirus virions was assessed by monitoring the appearance of disassembly intermediates following adsorption of radiolabeled viral particles. Within 2 h after adsorption to parental cells, proteolysis of viral outer-capsid proteins was observed, consistent with formation of ISVPs. However, in mutant cells, no proteolysis of viral proteins was detected up to 8 h postadsorption. Since treatment of cells with E64, an inhibitor of cysteine-containing proteases, blocks reovirus disassembly, we used immunoblot analysis to assess the expression of cathepsin L, a lysosomal cysteine protease. In contrast to parental cells, mutant cells did not express the mature, proteolytically active form of the enzyme. The defect in cathepsin L maturation was not associated with mutations in procathepsin L mRNA, was not complemented by procathepsin L overexpression, and did not affect the maturation of cathepsin B, another lysosomal cysteine protease. These findings indicate that persistent reovirus infections select cellular mutations that affect the maturation of cathepsin L and suggest that alterations in the expression of lysosomal proteases can modulate viral cytopathicity.
Mammalian reoviruses are nonenveloped, icosahedral viruses that contain a genome of 10 double-stranded RNA gene segments. Many viruses, including reovirus, require endocytic uptake and exposure to acidic pH or acid-dependent proteases to productively infect host cells. Following reovirus attachment to cells, virions are observed by electron microscopy in clathrin-coated pits, which suggests that viral entry occurs by receptor-mediated endocytosis (12, 13, 47, 55). Within late endosomes or lysosomes, viral outer-capsid proteins ς3 and μ1 are subject to proteolysis by vacuolar proteases, yielding infectious subvirion particles (ISVPs) (13, 17, 52, 55). ISVPs generated in the endocytic compartment are indistinguishable from those generated either in the intestinal lumen of perorally infected mice (9, 10, 19) or in vitro by treatment of virions with chymotrypsin or trypsin (13, 17, 44, 52, 55). ISVPs are obligate intermediates in reovirus disassembly that mediate penetration of the virus into the cytoplasm (12, 30, 31, 36, 58).
Treatment of cells with ammonium chloride (10, 22, 55) or inhibitors of the vacuolar proton ATPase, such as bafilomycin or concanamycin A (38), blocks infection by virions but not by ISVPs, which indicates that intracellular proteolysis of the ς3 and μ1 proteins is acid dependent. The vacuolar proteases that mediate cleavage of reovirus outer-capsid proteins have not been identified. However, treatment of cells with E64, a specific inhibitor of proteases containing active-site cysteine residues (7), blocks proteolysis of ς3 and μ1 during viral entry (6, 16). In contrast, pepstatin, a specific inhibitor of aspartyl proteases (20), does not inhibit proteolysis of ς3 and μ1 and does not inhibit reovirus growth (35). These findings indicate that a cysteine protease is required for endocytic proteolysis of the reovirus outer capsid.
Studies of persistent reovirus infections of murine L929 (L) cells have contributed significantly to an understanding of reovirus entry and disassembly (reviewed in reference 21). In contrast to wild-type (wt) viruses, viruses isolated from persistently infected cultures can grow in cells treated with ammonium chloride (22, 62) and E64 (6). These findings suggest that mutant viruses have altered requirements for decreased pH and proteolysis to complete the steps in entry required for generation of ISVPs. When persistently infected cultures of L cells are cured of reovirus infection by treatment with an antireovirus antiserum, the resulting cells do not fully support growth of wt viruses (22). However, when virions of wt viruses are first converted to ISVPs by protease treatment in vitro, they grow efficiently in cured cells (22, 61). These findings indicate that cellular mutations selected during persistent reovirus infection affect steps in reovirus replication required for formation of ISVPs. The nature of the cellular mutation that leads to a block in virion-to-ISVP conversion is not known.
In this study, we used reovirus virions and ISVPs as probes to define steps in reovirus replication blocked in mutant cells selected during persistent reovirus infection. We compared the ability of parental and mutant cells to support each step in the reovirus entry pathway and found that mutant cells are defective in maturation of the lysosomal cysteine-containing protease cathepsin L. These findings indicate that persistent reovirus infection selects a novel class of mutant cells defective in cathepsin L processing or transport and suggest that cathepsin L is required for disassembly of reovirus virions.
MATERIALS AND METHODS
Cells and viruses.
Murine L cells were grown in either suspension or monolayer cultures in Joklik’s modified Eagle’s minimal essential medium (Irvine Scientific, Santa Ana, Calif.) supplemented to contain 5% fetal bovine serum (Intergen, Purchase, N.Y.), 2 mM l-glutamine, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 0.25 μg of amphotericin (Irvine Scientific) per ml. Mutant LX cells were grown in monolayer cultures in Joklik’s modified Eagle’s minimal essential medium supplemented to contain 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 0.25 μg of amphotericin per ml. Reovirus strains type 1 Lang (T1L) and type 3 Dearing (T3D) are laboratory stocks. Purified virion preparations were made with second-passage L-cell lysate stocks of twice-plaque-purified reovirus, as previously described (23). Purified virions containing 35S-labeled proteins were obtained by adding Easy Tag Express-[35S] protein labeling mix (NEN, Boston, Mass.) to cell suspensions (∼12.5 μCi per ml) at the initiation of infection. ISVPs were prepared by treating purified reovirus virions with Nα-p-tosyl-l-lysine chloromethyl ketone-treated bovine α-chymotrypsin (Sigma Chemical Co., St. Louis, Mo.) as previously described (6). T1L was used in experiments represented by Fig. 1, 3, 5, 6, and 10 since the ratio of particles to PFU is the same for virions and ISVPs of this strain (44). Particle-to-PFU ratios of the T1L virion and ISVP preparations used in this study were 100 to 1.
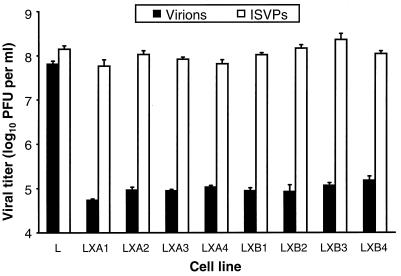
FIG. 1.
Viral titers in parental L cells and independent mutant LX cell clones after infection by virions and ISVPs. Monolayers of parental L cells and eight independent mutant LX-cell clones (5 × 105 cells) were infected with either virions or ISVPs of reovirus T1L at an MOI of 2 PFU per cell. After a 1-h adsorption period, the inoculum was removed, fresh medium was added, and the cells were incubated at 37°C for 24 h. Cells were frozen and thawed twice, and viral titers in cell lysates were determined by plaque assay using L-cell monolayers. The results are presented as mean viral titers for four independent experiments. Error bars indicate standard deviations of the means.
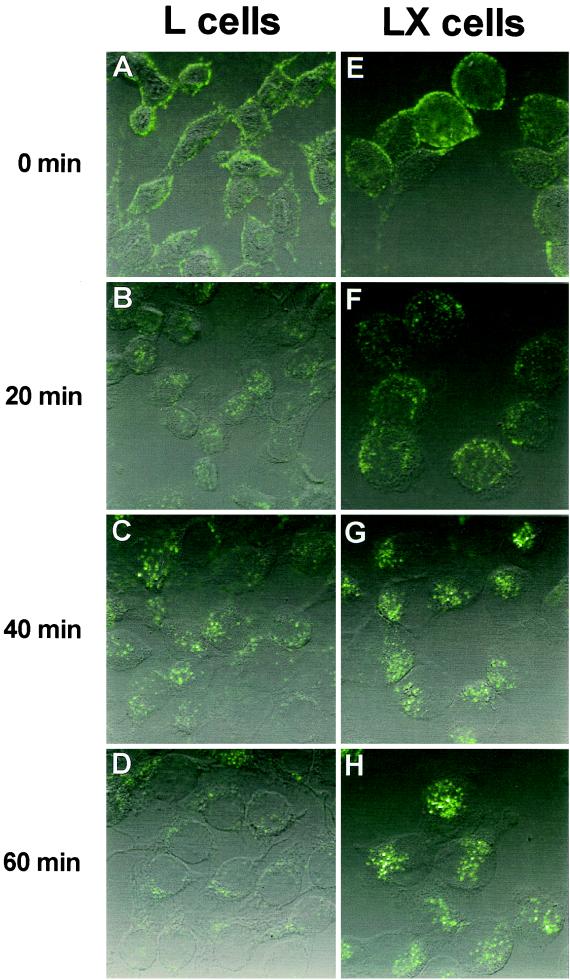
FIG. 3.
Uptake of reovirus virions following adsorption to parental L cells and mutant LXA1 cells. Monolayers of L cells (A to D) and LXA1 cells (E to H) were adsorbed with 10,000 particles per cell of Cy3-conjugated virions of reovirus strain T1L at 4°C for 45 min. Following removal of unbound virus, cells were incubated at 37°C for 0 (A and E), 20 (B and F), 40 (C and G), or 60 (D and H) min and then fixed. Cells were visualized by confocal fluorescence microscopy.
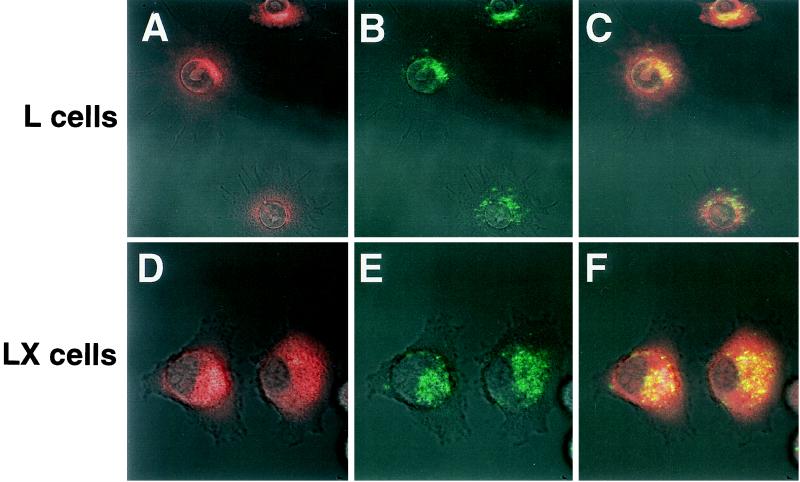
FIG. 5.
Colocalization of reovirus virions to acidic compartments in parental L cells and mutant LX cells. Virions of reovirus strain T1L conjugated to Cy5 were adsorbed to monolayers of parental L cells and mutant LXB2 cells (10,000 particles per cell) at 4°C for 45 min. Following removal of unbound virus, cells were incubated in medium containing 3 μM LysoSensor Green probe at 37°C for 1.5 h. Cells were washed and visualized by confocal fluorescence microscopy. Acidic compartments are indicated in red (A and D). Virions are indicated in green (B and E). In the merged image, yellow indicates colocalization of virions and acidic compartments (C and F).
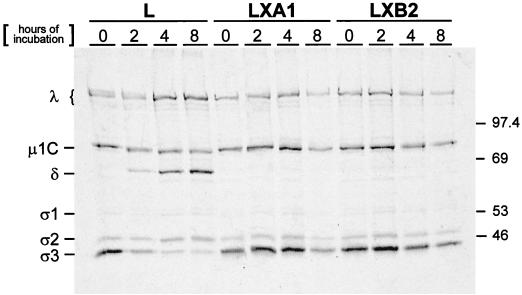
FIG. 6.
Electrophoretic analysis of reovirus structural proteins after infection of parental L cells and mutant LX cells. Monolayers of parental L cells and mutant LXA1 and LXB2 cells (107) were adsorbed with 10,000 particles per cell of purified 35S-labeled virions of reovirus strain T1L. After 1 h of adsorption at 4°C, the inoculum was removed, fresh medium was added, and the cells were incubated at 37°C for the indicated intervals. Cells then were lysed and extracted with freon. Virus particles contained in supernatants were pelleted by ultracentrifugation and solubilized in sample buffer. Equal volumes of samples were loaded into wells of a 10% polyacrylamide gel. After electrophoresis, the gel was prepared for autoradiography and exposed to film. Viral proteins are labeled, and molecular mass standards (in kilodaltons) are indicated.
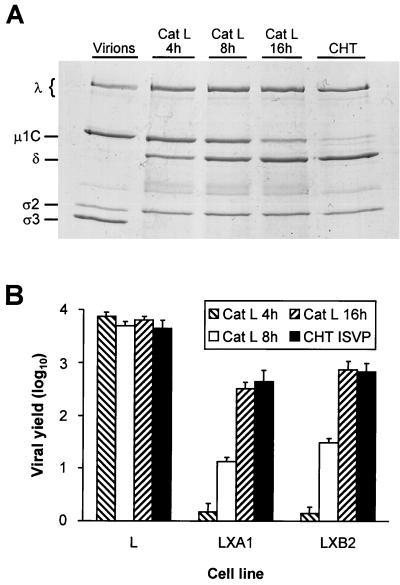
FIG. 10.
Treatment of reovirus virions with purified cathepsin L. (A) Electrophoretic analysis of viral structural proteins of reovirus virions after treatment with either cathepsin L (Cat L) or chymotrypsin (CHT). Purified virions of reovirus strain T1L were treated with either human cathepsin L (pH 5.0) at 37°C for the indicated intervals or bovine α-chymotrypsin (pH 7.4) at 37°C for 2 h. Virions also were incubated at 37°C in virion storage buffer adjusted to pH 5.0 for 16 h. Equal numbers of virus particles were loaded into wells of a 10% polyacrylamide gel. After electrophoresis, the gel was stained with Coomassie blue. Viral proteins are labeled. (B) Growth of virions treated with cathepsin L and ISVPs generated by chymotrypsin in parental L cells and mutant LX cells. Monolayers of cells (4 × 105 cells) were infected with either T1L virions treated with cathepsin L for 4 (Cat L 4 h), 8 (Cat L 8 h), or 16 (Cat L 16 h) h or ISVPs generated by treatment of T1L virions with chymotrypsin for 2 h (CHT ISVP) at an MOI of 2 PFU per cell. After a 1-h adsorption period, the inoculum was removed, fresh medium was added, and cells were incubated at 37°C for either 0 or 24 h. Cells were frozen and thawed twice, and viral titers in cell lysates were determined by plaque assay using L-cell monolayers. The results are presented as mean viral yields, calculated by dividing viral titers at 24 h by viral titers at 0 h, for three independent experiments. Error bars indicate standard deviations of the means.
Establishment of persistent reovirus infections.
Monolayer cultures of L cells (5 × 106 cells) were infected with second-passage L-cell lysate stocks of twice-plaque-purified reovirus T3D at a multiplicity of infection (MOI) of 0.1 PFU per cell. The cultures were either passaged when confluent or supplemented with fresh medium every 4th day if the cell density was not sufficient to permit passage. Cell culture lysates were collected at each passage by two cycles of freezing and thawing (−70 and 37°C). Viral titer in cell culture lysates was determined by plaque assay with L-cell monolayers (60).
Isolation of cells cured of persistent reovirus infection.
Persistently infected L-cell cultures were rendered virus free by maintenance in medium supplemented to contain 1% rabbit antireovirus antiserum (61) for 30 days. Cells cured of persistent reovirus infection were termed LX cells. Cured-cell clones were obtained by two cycles of limiting dilution into 96-well plates (Costar, Cambridge, Mass.). Individual wells were inspected visually, and only those containing a single colony were used. Cell clones presumed to be virus free were assessed by plaque assay of cell culture supernatants, infectious center assay, and immunocytochemical staining with rabbit antireovirus antiserum.
Growth of reovirus in parental L cells and mutant LX cells.
Monolayers of L cells and LX cells (5 × 105 cells) in 24-well plates (Costar) were infected with reovirus strains at an MOI of 2 PFU per cell. After 1 h of adsorption at 4°C, the inoculum was removed, cells were washed twice with phosphate-buffered saline (PBS), and 0.5 ml of fresh medium was added. After incubation at 37°C for 24 h, cells were frozen and thawed twice and viral titers in cell lysates were determined by plaque assay with L-cell monolayers (60). Independent experiments were performed with single wells of cells, which were titrated in duplicate.
Electron microscopy.
Cells were centrifuged to form a pellet (1,000 × g, 10 min) and suspended in phosphate-buffered 2% glutaraldehyde. After primary fixation, cells were again centrifuged (1,000 × g, 10 min), resuspended in 1% osmium tetroxide, dehydrated in increasing percentages of ethanol (50 to 100%) and propylene oxide, and then embedded in an epoxy resin. Ultrathin sections were prepared with an Ultratome III ultramicrotome (LKB, Piscataway, N.J.) and stained with lead citrate and uranyl acetate. Sections were examined with a Philips 300 electron microscope (Philips, Mahwah, N.J.).
Conjugation of reovirus virions to fluorescent dyes.
Purified reovirus virions of strain T1L (5 × 1013 particles per ml) were dialyzed at 4°C against 100 mM sodium bicarbonate-buffered 0.8% saline (pH 8.5 for 2 h followed by pH 9.3 for 2 h). Cy3 or Cy5 monofunctional reactive dye (Amersham, Arlington Heights, Ill.) was added to viral suspensions and incubated at 25°C for 45 min. Conjugated virus was dialyzed at 4°C for 16 h against PBS to remove free dye. Conjugation of reovirus virions with fluorescent dyes results in labeling of viral outer-capsid proteins ς1, ς3, μ1, and λ2 and a fivefold decrease in viral infectivity.
Confocal microscopy of reovirus uptake and trafficking.
L cells and LX cells (105) were grown on 12-mm glass coverslips (Fisher Scientific, Pittsburgh, Pa.) or in glass-bottomed MatTek dishes (MatTek Corp., Ashland, Mass.) for 2 days. Monolayers of cells at approximately 70% confluence were incubated at 4°C for 20 min prior to adsorption with fluorescent conjugated reovirus virions at an MOI of 10,000 particles per cell, the minimum number of reovirus particles sufficient to detect a signal. After adsorption at 4°C for 45 min, cells were incubated at 37°C for various intervals, washed three times with PBS, and fixed for 5 min in a 1:1 mixture of methanol and acetone. Cells were washed three times in PBS and mounted on glass slides with Aqua-Poly/Mount (Polysciences, Inc., Warrington, Pa.). Cells were examined with a Zeiss confocal fluorescence microscope (Carl Zeiss, New York, N.Y.).
Determination of intravesicular pH.
L cells and LX cells (105) were grown in MatTek dishes for 2 days. Cells were incubated with 0.1% (wt/vol) double-labeled fluorescein-tetramethylrhodamine dextran (Molecular Probes, Eugene, Oreg.), which contains pH-dependent (fluorescein) and pH-independent (tetramethylrhodamine) dyes, at 37°C for 16 h. To generate a standard curve correlating the fluorescein-to-tetramethylrhodamine (F/TMR) fluorescence ratio with pH, cells were fixed in 1:1 methanol and acetone and permeabilized in PBS with 1% Triton X-100. Cells were equilibrated for 1 h in buffers of various pHs—0.1 M sodium acetate (pH 4.0 and 5.0), 0.1 M sodium phosphate (pH 6.0 and 7.0), and 0.1 M Tris (pH 8.0)—and then cells were examined by confocal fluorescence microscopy. Fluorescence intensities for fluorescein and tetramethylrhodamine were determined for identical groups of 15 to 20 cells at each pH standard by using NIH Image software. A standard curve was generated by correlating the F/TMR fluorescence ratio with pH. To determine the intravesicular pH of parental L cells and mutant LX cells, cells were incubated with the double-labeled dextran at 37°C for 16 h, washed in Dulbecco’s modified Eagle’s medium (Gibco BRL, Grand Island, N.Y.), and examined with a confocal fluorescence microscope. Fluorescence intensities for fluorescein and tetramethylrhodamine were determined for identical groups of cells by using NIH Image. The intravesicular pH of parental L cells and mutant LX cells was determined by calculating the mean F/TMR fluorescence ratio for each cell type and extrapolating from the standard curve.
LysoSensor Green staining of cells.
L cells and LX cells (105) were grown in MatTek dishes for 2 days. Cells were washed once with PBS, and 2 ml of fresh Dulbecco’s modified Eagle’s medium without phenol red (Gibco BRL) was added. Cells were incubated at 37°C for 1 h, and the medium was removed and replaced with 0.5 ml of medium containing 3 μM LysoSensor Green DND-189 probe (Molecular Probes). Cells were incubated with probe for 1.5 h, washed two times with fresh medium without probe, and examined by confocal fluorescence microscopy.
Assessment of intracellular proteolysis of reovirus virions.
L cells and LX cells (107) in 75-cm2 flasks (Costar) were adsorbed with purified, 35S-labeled reovirus virions at 10,000 particles per cell. After incubation at 4°C for 1 h, the inoculum was removed, cells were washed twice with PBS, and 10 ml of fresh medium was added. After incubation at 37°C for various intervals, cells were harvested, resuspended in 0.5 ml of lysis buffer (150 mM NaCl, 10 mM Tris [pH 7.4], 0.5% Nonidet P-40 [NP-40], 1 mM EDTA, 1 mM benzamidine, 100 mM leupeptin, and 2.5 mM phenylmethylsulfonyl fluoride), and placed on ice for 30 min. After the addition of 4.5 ml of homogenization buffer (250 mM NaCl, 10 mM Tris [pH 7.4], 0.067% 2-mercaptoethanol), samples were sonicated for 1 min, 2.5 ml of freon (EM Science, Gibbstown, N.J.) was added, and samples were again sonicated for 1 min. Samples were centrifuged at 9,700 × g for 10 min, and viral particles in the aqueous fraction were pelleted by centrifugation at 210,000 × g for 1 h.
Virus particles were solubilized by incubation in sample buffer (125 mM Tris, 2% 2-mercaptoethanol, 1% sodium dodecyl sulfate [SDS], 0.01% bromphenol blue) at 100°C for 5 min. Samples were loaded into wells of 10% polyacrylamide gels and electrophoresed at 200 V of constant voltage for 1 h. Following electrophoresis, gels were fixed, dried onto filter paper (Bio-Rad Laboratories, Richmond, Calif.) under vacuum, and exposed to BioMax-MR film (Eastman Kodak Co., Rochester, N.Y.).
Immunoblot analysis for cathepsin L and cathepsin B.
L cells and LX cells (107) in 75-cm2 flasks were pretreated overnight with 0 or 200 μM E64 (Sigma). Following 6 h of incubation in 3 ml of serum-free medium supplemented to contain 0 or 200 μM E64, culture supernatants were removed and cells were harvested in 5 ml of PBS. Cells were pelleted and washed in PBS, followed by incubation in RIPA buffer (1% NP-40, 0.5% deoxycholate, 0.1% SDS) with 2.5 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, and protease inhibitor cocktail (Boehringer Mannheim, Indianapolis, Ind.) at 4°C for 5 min. Samples were vortexed vigorously for 10 s, and membranes and nuclei were pelleted by centrifugation at 13,000 × g. Supernatants were assayed for protein content by using the Bio-Rad DC protein assay and diluted in immunoblot-polyacrylamide gel electrophoresis (PAGE) sample buffer (3.3% SDS, 80 mM Tris [pH 7], 20% sucrose, 0.008% bromphenol blue, 17 mM EDTA, 17 mM dithiothreitol [DTT]) (18). Secreted proteins in culture supernatants were precipitated with 20% trichloroacetic acid containing salmon sperm DNA (25 μg per ml), centrifuged at 12,000 × g for 10 min, and resuspended in immunoblot-PAGE sample buffer.
Protein samples, normalized for either protein content (cellular proteins) or cell number (secreted proteins), were loaded into lanes of 12% polyacrylamide gels and electrophoresed at 200 V of constant voltage for 50 min. Following equilibration of the gel in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol) for 20 min, proteins were transferred to nitrocellulose membranes either overnight at 30 V or for 1 h at 100 V. After removal from the transfer apparatus, membranes were air dried for 5 min and treated for 1 h with agitation in Tris-buffered saline (TBS) (50 mM Tris [pH 7.5], 150 mM NaCl) containing 0.05% Tween 20 and 5% low-fat dry milk. Membranes were incubated at 37°C for 2 h with agitation in antiserum against murine cathepsin L (45) or human cathepsin B (Athens Research and Technology, Athens, Ga.) diluted 1:15,000 (anti-cathepsin L) or 1:2,000 (anti-cathepsin B) in TBS plus Tween 20 and milk. After three washes in TBS plus Tween 20, membranes were incubated at 25°C for 1 h with agitation in horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Amersham) diluted 1:2,500 in TBS plus Tween 20 and milk. Membranes were washed three times in TBS plus Tween 20, incubated with Enhanced chemiluminescent reagent (Amersham) for 1 min, and exposed to Biomax-MR film.
Cloning and sequencing of procathepsin L-encoding cDNA.
Cellular mRNA was purified from cell lysates with oligo(dT) magnetic beads (Dynal, Oslo, Norway). Oligodeoxynucleotide primers 5′-AGACTTCTTGTGCGCACGTA and 5′-CGACACACACACTGAGCTAA, which correspond to the 5′ and 3′ nontranslated regions of the procathepsin L cDNA, were used to generate PCR products from isolated mRNA. Purified mRNA was melted in 90% dimethyl sulfoxide at 95°C for 5 min. Ice-cold primers were annealed to the melted template, and cDNA was generated with avian myeloblastosis virus reverse transcriptase (RT) (Boehringer Mannheim). PCR was performed with Taq polymerase (Perkin-Elmer, Branchburg, N.J.) for 39 cycles with a program of template denaturation at 95°C for 2 min, primer annealing at 55°C for 2 min, and polynucleotide synthesis at 72°C for 5 min. PCR was completed by a synthesis step at 72°C for 1 h. Resultant cDNAs were cloned into the pCRII vector (Invitrogen, San Diego, Calif.). Unambiguous sequences of 1,354 nucleotides of the procathepsin L cDNA, including the entire open reading frame of the preprocathepsin L protein, were determined by dideoxy chain termination with T7 DNA polymerase (United States Biochemical, Cleveland, Ohio). Two cDNA clones generated from independent RT-PCRs were used as template in these experiments.
Transfection of cells with a procathepsin L-encoding cDNA.
A cDNA encoding murine procathepsin L was cloned into eukaryotic expression vector pSG5 (Stratagene, La Jolla, Calif.) and introduced into L cells and LX cells in 85-mm plates (Costar) by using LipofectAMINE (Life Technologies, Inc., Rockville, Md.) according to previously described techniques (18). After incubation at 37°C for 48 h, the medium was replaced with serum-free medium supplemented with insulin, ferritin, and sodium selenite. After an additional 24 h of incubation, cell lysates and culture supernatants were prepared and immunoblot analysis was performed with cathepsin L-specific antiserum (45) as described.
Treatment of reovirus virions with cathepsin L.
Purified virions of reovirus strain T1L at a concentration of 3 × 1012 particles per ml in virion storage buffer (100 mM NaCl, 15 mM MgCl2, 50 mM sodium acetate) adjusted to pH 5.0 were treated with 150 μg of purified recombinant human cathepsin L (15) per ml in the presence of 5 mM DTT at 37°C for various intervals. Cathepsin L treatment was stopped by adding 500 μM E64 to the treatment mixtures and freezing at −20°C. Aliquots of the treatment mixtures were mixed 5:1 with 6× sample buffer (350 mM Tris [pH 6.8], 9.3% DTT, 10% SDS, 0.012% bromphenol blue) and incubated at 100°C for 5 min. Samples were loaded into wells of 10% polyacrylamide gels and electrophoresed at 200 V of constant voltage for 1 h. Gels were stained with Coomassie blue R-250 (Sigma) and dried between cellophane. Aliquots of the treatment mixtures also were used to inoculate L cells and LX cells at an MOI of 2 PFU per cell. After incubation at 37°C for 24 h, cells were frozen and thawed twice and viral titers in cell lysates were determined by plaque assay with L-cell monolayers (60).
Nucleotide sequence accession numbers.
The procathepsin L-encoding cDNA sequences determined in this study have been deposited in GenBank and assigned accession no. AF121837 (L cells), AF121838 (LXA1 cells), and AF121839 (LXB2 cells).
RESULTS
Establishment of L-cell cultures persistently infected with reovirus.
Persistently infected cultures of L cells have been established previously with virus stocks passaged serially at high MOI (3, 4, 14, 22). Such stocks contain a variety of viral mutants (2, 5), and these mutants are postulated to facilitate establishment of persistent infection (14). To determine whether persistent infection also can be established with reovirus stocks passaged at low MOI, and to select cells containing blocks to reovirus infection, murine L cells were infected with independent, second-passage L-cell lysate stocks of reovirus strain T3D at an MOI of 0.1 PFU per cell. Two independent, persistently infected cultures, termed LDG and LDV, were established. Following approximately 4 days in culture, both cell lines underwent an intense period of crisis in which most of the cells in the cultures were lysed. Over the next 2 to 3 weeks, small colonies of cells became apparent and these colonies eventually reached sufficient density to permit passage. The LDG and LDV cultures were maintained for approximately 1 year and produced titers of infectious virus of between 106 and 108 PFU per ml throughout their maintenance period (data not shown). These observations confirm that L-cell cultures persistently infected with reovirus produce high titers of infectious virus for prolonged periods (1, 4, 22) and demonstrate that wt reovirus can initiate a persistent infection when cultures are inoculated at low MOI.
Growth of reovirus virions and ISVPs in parental L cells and mutant LX cells cured of persistent infection.
To isolate virus-free mutant cells for studies of reovirus entry, a neutralizing antireovirus antiserum (61) was added to the medium of subcultures of both the LDG and LDV cell lines at 230 days of culture maintenance. Antiserum treatment was continued for a 30-day period, during which time the viral titer decreased to undetectable levels (<10 PFU per ml of culture supernatant) for each culture. Following antiserum treatment of LDG and LDV, the resulting cultures, termed LXDG and LXDV, respectively, were passaged in medium without antiserum. The cured cell lines were confirmed to be virus free by the absence of virus in culture lysates, negative infectious center assays, and absence of detectable viral antigen by immunocytochemistry (data not shown).
To obtain a homogeneous population of cells for characterization of blocks to reovirus entry, cells were cloned from the cured LXDG culture by two cycles of limiting dilution. Eight LXDG subclones were established, and these subclones were tested for the capacity to support reovirus entry. Parental L cells and the eight LXDG subclones were infected with either virions or ISVPs of reovirus strain T1L at an MOI of 2 PFU per cell, and viral titers in cell lysates were determined after 24 h of viral growth (Fig. 1). Viral titers after infection of parental L cells with virions were equivalent to those after infection with ISVPs. However, viral titers after infection of the mutant cell lines with ISVPs were approximately 1,000-fold greater than those produced after infection with virions. For all mutant LXDG subclones examined, yields of viral progeny after infection with virions were <10 PFU per input PFU (Fig. 1). Similar results were obtained with strain T3D (data not shown), and the block to growth of both T1L and T3D was apparent even after adsorption with viral inocula of 100 PFU per cell (data not shown). These results indicate that cellular mutations selected during persistent infection block steps in the viral growth cycle prior to generation of ISVPs. The LXA1 and LXB2 cloned cell lines were used for subsequent studies.
Ultrastructure of parental, persistently infected, and mutant L cells.
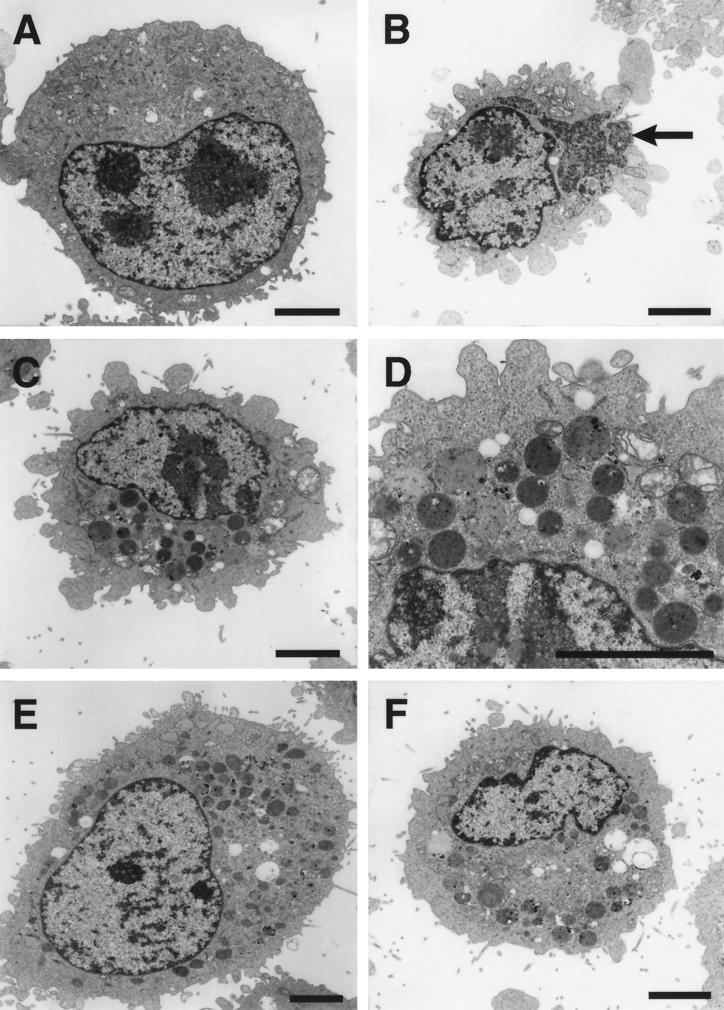
The finding that mutant cells selected during persistent infection do not support steps in viral replication required to generate ISVPs suggested that mutant cells are altered in endocytic function. To characterize changes in cellular ultrastructure associated with blocks to reovirus entry, we used electron microscopy to examine the morphology of uninfected L cells, persistently infected LDG cells, and cured LXA1 cells (Fig. 2). Parental L cells had an unremarkable appearance consisting of a large nucleus and uniform cytoplasm (Fig. 2A). No reovirus particles were found in thin sections of either uninfected L cells or cured LXA1 cells. The most striking aspect of viral infection observed in the persistently infected cells was the presence of large, perinuclear inclusions of virions (Fig. 2B, arrow). In a given plane of section, viral inclusions were found in approximately 15% of the persistently infected cells. All persistently infected LDG cells examined contained large numbers of electron-dense, membrane-bound vesicles (Fig. 2C and D). Cured cells also contained numerous vesicular structures similar to those observed in the persistently infected cells (Fig. 2E and F). Such structures were not observed in uninfected L cells. Therefore, mutant cells selected during persistent reovirus infection accumulate electron-dense, membrane-bound vesicles.
FIG. 2.
Ultrastructural morphologies of uninfected, persistently infected, and cured L cells. (A) Uninfected parental L cells. (B, C, and D) Persistently infected LDG cells. Note in panel B the large inclusion of virions (arrow). (D) Increased magnification of electron-dense vesicles from panel C. (E and F) Cured LXA1 cells. Note the presence of electron-dense vesicles in cured cells. Bars, 5 μm.
Internalization and intracellular transport of reovirus virions in parental L cells and mutant LX cells.
To determine whether L cells and LX cells differ qualitatively in reovirus uptake and intracellular transport, purified reovirus virions were conjugated to the fluorescent dye Cy3 and adsorbed to parental L cells and mutant LX cells at 4°C. Cells were washed to remove unbound virus, warmed to 37°C, and examined by confocal microscopy (Fig. 3). At 0 min postadsorption, reovirus virions were observed at the periphery of both parental L cells (Fig. 3A) and mutant LX cells (Fig. 3E). By 20 min postadsorption, virions were internalized and noted to be distributed throughout the cytoplasm (Fig. 3B and F). By 40 and 60 min postadsorption, virions were observed to coalesce in perinuclear regions of both cell types (Fig. 3C, D, G, and H), consistent with the location of late endosomes and lysosomes (34). Therefore, the internalization and subcellular localization of reovirus virions are similar in both parental L cells and mutant LX cells.
Intravesicular pHs of parental L cells and mutant LX cells.
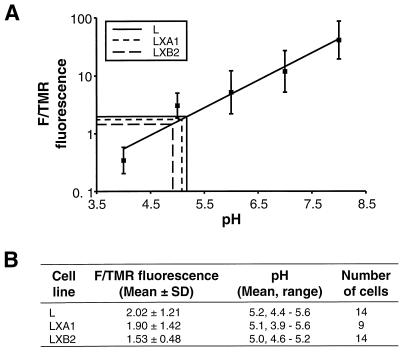
To determine whether mutant LX cells contain a defect in vesicular acidification, we measured the intravesicular pHs of both parental L cells and mutant LX cells by using a double-labeled fluorescein-tetramethylrhodamine dextran that is taken into cells by receptor-mediated endocytosis. Upon entry of the double-labeled dextran into acidified compartments, the fluorescence of fluorescein is quenched while the fluorescence of tetramethylrhodamine is unchanged (29, 57, 59). We first generated a standard curve to correlate the F/TMR fluorescence ratio with pH by equilibrating permeabilized dextran-labeled cells with buffers of known pH (Fig. 4A). We then incubated both parental and mutant cells with the dual-labeled dextran and examined cells by confocal microscopy. By extrapolation from the standard curve, the intravesicular pH of parental L cells was determined to be 5.2 while those of mutant LXA1 and LXB2 cells were 5.1 and 5.0, respectively (Fig. 4B). Thus, parental L cells and mutant LX cells have similar intravesicular pHs, which suggests that the block to reovirus entry exhibited by mutant LX cells is not due to a general defect in acidification.
FIG. 4.
Determination of intravesicular pHs of parental L cells and mutant LX cells. (A) Standard curve correlating the F/TMR fluorescence ratio with pH. Cells were incubated with double-labeled fluorescein-tetramethylrhodamine dextran (0.1% [wt/vol]) at 37°C for 16 h. Cells were fixed, equilibrated for 1 h with buffers of known pH (0.1 M sodium acetate [pH 4.0 and pH 5.0], 0.1 M sodium phosphate [pH 6.0 and pH 7.0], and 0.1 M Tris [pH 8.0]), and visualized by confocal fluorescence microscopy. Fluorescence intensities for fluorescein and tetramethylrhodamine were determined for identical groups of 15 to 20 cells at each pH standard. (B) Intravesicular pHs of parental L cells and mutant LX cells determined by calculating the mean F/TMR fluorescence ratio for each cell type and extrapolating from the standard curve, as indicated in panel A.
Localization of reovirus virions to acidic intravesicular compartments in parental L cells and mutant LX cells.
To determine whether reovirus virions are transported to acidified organelles, we used the pH-sensitive LysoSensor Green DND-189 probe, which is an acid-sensitive probe that accumulates and fluoresces in acidified organelles of living cells (28). Both parental L cells and mutant LX cells were stained with this probe (data not shown), which confirms that both cell types have an acidic intravesicular pH. In addition, treatment of both parental L cells and mutant LX cells with ammonium chloride quenched LysoSensor probe fluorescence (data not shown), indicating that the fluorescence observed in parental and mutant cells incubated with this probe is due to its accumulation in acidified intracellular organelles.
To determine whether parental L cells and mutant LX cells differ in the capacity to transport reovirus virions to acidified organelles, both cell types were examined by confocal microscopy after incubation with the LysoSensor probe and Cy5-conjugated reovirus virions (Fig. 5). Both the LysoSensor probe and reovirus virions were observed to rapidly accumulate in perinuclear vesicular structures in both cell types (Fig. 5A, B, D, and E). When images of the fluorescent probe and reovirus virions were merged, virions were observed to colocalize with the acid-sensitive probe in both parental and mutant cells (Fig. 5C and F). These findings indicate that reovirus virions localize to acidified compartments in both parental L cells and mutant LX cells and suggest that the block to reovirus growth in mutant cells is not due to a defect in transport of virions to acidified intracellular compartments where viral disassembly occurs (22, 55).
Disassembly of reovirus virions in parental L cells and mutant LX cells.
To determine whether mutant LX cells are altered in the capacity to support proteolysis of the reovirus outer capsid, radiolabeled virions of reovirus strain T1L were adsorbed to parental L cells and mutant LX cells, and viral structural proteins were analyzed by SDS-PAGE and autoradiography at various times postadsorption (Fig. 6). In parental L cells infected with T1L, degradation of the ς3 protein and generation of the 59-kDa δ cleavage fragment of the μ1C protein were observed within 2 h postadsorption, consistent with the formation of ISVPs (13, 17, 19, 44, 52, 55). However, in both mutant LX cell lines, we did not detect generation of the δ cleavage fragment of μ1C even after incubation for up to 8 h postadsorption. A gradual decrease in the intensity of all protein bands was noted at late time points after viral adsorption to LX cells (Fig. 6). This was a consistent finding (data not shown) and suggests that reovirus virions do not undergo orderly degradation after uptake into mutant cells. Therefore, these results demonstrate that persistent reovirus infection selects a mutation in cells that results in failure to support proteolytic disassembly of virions to ISVPs.
Detection of lysosomal protease cathepsin L in parental L cells and mutant LX cells.
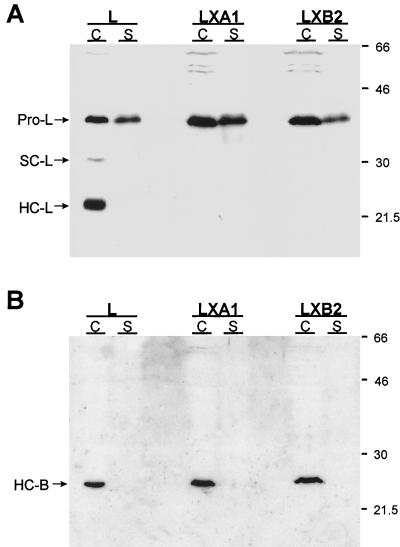
Reovirus disassembly is inhibited by protease inhibitor E64 (6, 16), a noncompetitive inhibitor of cysteine-containing proteases (7). This observation suggests that the protease responsible for reovirus disassembly is a lysosomal cysteine protease. The major cysteine proteases in lysosomes are cathepsins B, H, and L, with cathepsin L being the most abundant cysteine protease in lysosomes of several cell types (8, 11, 24, 27, 32). To assess whether parental and mutant cells differ in cathepsin L expression, we used immunoblot assays to probe for cathepsin L in cytoplasmic extracts and culture supernatants from both cell types (Fig. 7A). Cathepsin L is synthesized as a 38-kDa inactive proenzyme precursor that is either secreted from cells or processed to a 30-kDa single-chain intermediate form, which is subsequently cleaved in lysosomes to a two-chain mature form consisting of a 23-kDa heavy chain and a 5-kDa light chain (25, 39, 49). The proenzyme, single-chain, and heavy-chain forms of cathepsin L can be detected by using an antiserum raised against murine cathepsin L (18, 40, 41, 45). In parental L cells, the procathepsin L precursor, the single-chain intermediate, and the heavy-chain mature form were detected in cytoplasmic extracts, and the procathepsin L precursor was detected in the culture supernatant (Fig. 7A). In sharp contrast, only the procathepsin L precursor was detected in cytoplasmic extracts and culture supernatants from the LXA1 and LXB2 cell lines. Bands corresponding to single chain and heavy chain were not detected in either cytoplasmic extracts or culture supernatants from either mutant cell line (Fig. 7A). These findings indicate that mutant LX cells are altered in the generation of the mature, proteolytically active form of cathepsin L.
FIG. 7.
Immunoblot analysis of cathepsin L and cathepsin B expression in cytoplasmic extracts and culture supernatants from parental L cells and mutant LX cells. Monolayers of parental L cells and mutant LXA1 and LXB2 cells (107) were incubated in serum-free medium for 6 h. Cellular proteins (C), normalized for protein content, and secreted proteins (S), normalized for cell number, were resolved in a 12% polyacrylamide gel, electroblotted onto a nitrocellulose membrane, and immunoblotted with rabbit antisera raised against either murine cathepsin L (A) or human cathepsin B (B). Horseradish peroxidase-conjugated anti-rabbit immunoglobulin G antiserum was used as secondary antibody, and proteins were visualized by chemiluminescence. Bands corresponding to procathepsin L (Pro-L), the single-chain form of cathepsin L (SC-L), the heavy-chain form of cathepsin L (HC-L), and the heavy-chain form of cathepsin B (HC-B) are indicated. Molecular mass standards (in kilodaltons) are shown.
Detection of lysosomal protease cathepsin B in parental L cells and mutant LX cells.
To determine whether the absence of the mature form of cathepsin L is due to a general defect in the processing of lysosomal enzymes, we determined the status of cathepsin B in parental and mutant cells by using immunoblot assays. Cathepsin B is synthesized as a 37-kDa inactive proenzyme precursor that is either secreted from cells or processed to a 31- to 34-kDa single-chain intermediate form, which is subsequently cleaved in lysosomes to a two-chain mature form consisting of a 24- to 25-kDa heavy chain and a 5-kDa light chain (37, 46, 48). The presence of cathepsin B heavy chain in cytoplasmic extracts and culture supernatants of parental L cells and mutant LX cells was assessed by immunoblotting with an antiserum raised against human cathepsin B (Fig. 7B). To ensure consistent conditions for the cathepsin L and cathepsin B immunoblots, the cathepsin L blot was stripped of cathepsin L immunoreactivity and reprobed with the anti-cathepsin B antiserum. In both parental L cells and mutant LX cells, the mature heavy-chain form of cathepsin B was detected in cytoplasmic extracts (Fig. 7B). However, in contrast to immunoblot analysis of cathepsin L expression, we did not detect procathepsin B or the single-chain cathepsin B intermediate in either cytoplasmic extracts or culture supernatants of either cell type. It is possible that differences in the expression levels of the two enzymes or differences in the half-lives of their respective precursors account for the inability to detect cathepsin B intermediates. Nonetheless, these findings indicate that the mutant cell lines are able to process cathepsin B precursors to the mature form of the enzyme. Thus, the defect in cathepsin L maturation in mutant cells selected during persistent reovirus infection is not due to a general defect in lysosomal enzyme processing.
Inhibition of cathepsin L processing by E64.
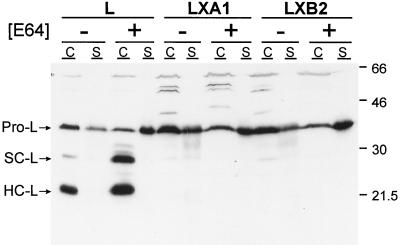
To determine whether the absence of mature cathepsin L in mutant LX cells is due to alterations in its turnover, we treated parental L cells and mutant LX cells with protease inhibitor E64 and used immunoblot assays to probe for cathepsin L in cytoplasmic extracts and culture supernatants from both cell lines (Fig. 8). Treatment of cells with E64 leads to an accumulation of the single-chain and heavy-chain forms of cathepsin L (49). In parental L cells treated with E64, the single-chain and heavy-chain forms accumulated in cytoplasmic extracts, and the procathepsin L precursor accumulated in the culture supernatant (Fig. 8). In mutant LXA1 and LXB2 cells treated with E64, no mature forms of cathepsin L were detected in cytoplasmic extracts. Similar to findings with parental L cells, there was an accumulation of the procathepsin L precursor in the culture supernatants of both mutant cell lines after treatment with E64 (Fig. 8). Thus, the defect in expression of mature cathepsin L in mutant LX cells does not appear to be caused by accelerated degradation of the enzyme.
FIG. 8.
Effect of protease inhibitor E64 on steady-state levels and secretion of cathepsin L from parental L cells and mutant LX cells. Monolayers of parental L cells and mutant LXA1 and LXB2 cells (107) were incubated at 37°C for 18 h in the presence or absence of 200 μM E64. Cellular proteins (C), normalized for protein content, and secreted proteins (S), normalized for cell number, were resolved in a 12% polyacrylamide gel, electroblotted onto a nitrocellulose membrane, and immunoblotted with rabbit anti-cathepsin L antiserum. Horseradish peroxidase-conjugated anti-rabbit immunoglobulin G antiserum was used as secondary antibody, and proteins were visualized by chemiluminescence. Bands corresponding to procathepsin L (Pro-L) and the single-chain (SC-L) and heavy-chain (HC-L) forms of cathepsin L are indicated. Molecular mass standards (in kilodaltons) are shown.
Sequence analysis of cDNA clones of procathepsin L-encoding mRNA isolated from parental L cells and mutant LX cells.
Alterations within the proregion of cathepsin L are known to inhibit processing steps required to generate the mature enzyme (18, 56). To determine whether the defect in cathepsin L maturation in mutant LX cells is due to a mutation within cathepsin L, we analyzed full-length sequences of cDNAs encoding preprocathepsin L obtained from parental L cells and two mutant LX cell lines. The cDNAs encoding cathepsin L derived from parental L cells and mutant LX cells were found to be identical. Therefore, the defect in cathepsin L processing cannot be ascribed to an intrinsic defect in cathepsin L but rather to a defect in either its transport to the organelle in which processing occurs or in the proteolytic machinery required to generate the mature enzyme.
Overexpression of murine procathepsin L in parental L cells and mutant LX cells.
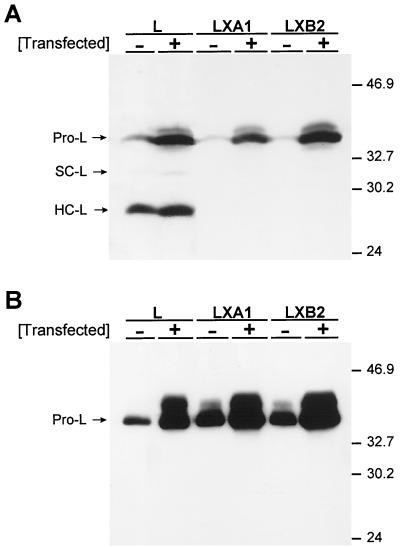
To determine whether the defect in cathepsin L maturation can be complemented by overexpression of procathepsin L, parental L cells and mutant LX cells were transfected with a plasmid encoding murine procathepsin L. Following transfection, cytoplasmic extracts and culture supernatants were analyzed for mature forms of cathepsin L by immunoblot assay (Fig. 9). Overexpression of procathepsin L in parental L cells resulted in a modest accumulation of the heavy-chain form of cathepsin L in cytoplasmic extracts (Fig. 9A) and a substantial increase in procathepsin L in the culture supernatant (Fig. 9B). Overexpression of procathepsin L in mutant LXA1 and LXB2 cells, however, did not result in detection of mature forms of cathepsin L in either cytoplasmic extracts or culture supernatants. Similar to transfected parental L cells, a substantial increase in procathepsin L was detected in the culture supernatants of transfected mutant LX cells (Fig. 9B). In both parental and mutant cells, overexpression of procathepsin L resulted in detection of a protein band migrating slightly slower than procathepsin L. This band likely corresponds to a form of procathepsin L that has altered carbohydrate modification (18). Thus, the defect in processing or transport of cathepsin L cannot be overcome by procathepsin L overexpression.
FIG. 9.
Effect of procathepsin L overexpression on steady-state levels and secretion of cathepsin L from parental L cells and mutant LX cells. Monolayers of parental L cells and mutant LXA1 and LXB2 cells at approximately 80% confluence were transfected with a plasmid encoding murine procathepsin L and incubated at 37°C for 72 h. Equal amounts of cellular proteins (A) and equal amounts of secreted proteins (B) were resolved in 12% polyacrylamide gels, electroblotted onto nitrocellulose membranes, immunoblotted with rabbit anti-cathepsin L antiserum, and visualized by chemiluminescence. Bands corresponding to procathepsin L (Pro-L) and the single-chain (SC-L) and heavy-chain (HC-L) forms of cathepsin L are indicated. Molecular mass standards (in kilodaltons) are shown.
Treatment of reovirus virions with purified cathepsin L.
The finding that mutant cells selected during persistent reovirus infection do not express mature cathepsin L suggests that cathepsin L is required for reovirus disassembly. To test directly whether cathepsin L treatment results in conversion of reovirus virions to ISVPs, purified virions of reovirus strain T1L were incubated for various times with purified human cathepsin L (15). Treated virions were analyzed by SDS-PAGE for changes in viral outer-capsid proteins indicative of ISVP formation (Fig. 10A). Treatment with cathepsin L resulted in loss of ς3 protein and generation of the δ fragment of μ1C protein. These changes in viral outer-capsid proteins are observed in ISVPs generated by treatment of reovirus virions with intestinal proteases in vitro (13, 17, 19, 44, 52, 55) (Fig. 10A) or after infection of cells (6, 53–55) (Fig. 6). Thus, cathepsin L is capable of mediating disassembly of reovirus virions to ISVPs.
We next performed experiments to determine whether ISVPs generated by treatment of virions with cathepsin L display growth characteristics like those of ISVPs generated by treatment with intestinal proteases. In contrast to virions, ISVPs generated by chymotrypsin treatment in vitro are capable of bypassing blocks to viral disassembly exhibited by mutant LX cells (22) (Fig. 1). Parental L cells and mutant LXA1 and LXB2 cells were infected with either virions treated for various times with cathepsin L or ISVPs generated by treatment of virions with chymotrypsin, and viral yields were determined after 24 h of virus growth (Fig. 10B). Viral yields in parental L cells after infection with virions treated with cathepsin L for 0, 4, 8, and 16 h were equivalent to those after infection with chymotrypsin-generated ISVPs. Viral yields in mutant LXA1 and LXB2 cells after infection with virions treated with cathepsin L for 16 h were equivalent to those after infection with chymotrypsin-generated ISVPs. However, virions treated with cathepsin L for 4 and 8 h produced viral yields in both mutant LX cell lines that correlated with the extent of μ1C-to-δ cleavage but not with the loss of ς3. Treatment of virions with cathepsin L for 4 h resulted in complete degradation of ς3 but minimal μ1C-to-δ cleavage, as judged by Coomassie blue staining (Fig. 10A), yet these particles produced <2 PFU per input PFU after 24 h of growth in LX cells. Therefore, these results indicate that treatment of reovirus virions with purified cathepsin L leads to generation of particles that have the biochemical and growth properties of authentic ISVPs and suggest that μ1C-to-δ cleavage is required for efficient growth of reovirus in mutant LX cells.
DISCUSSION
Mutant cells selected during persistent reovirus infections of murine L cells do not support growth of reovirus after infection by virions but do so after infection by ISVPs generated in vitro (reference 22 and this report). This observation suggests that mutant cells do not support entry steps leading to formation of ISVPs. Virions and ISVPs have identical requirements for binding to reovirus receptors (44); however, ISVPs do not require acid-dependent proteolysis to facilitate penetration into the cytoplasm (6, 22, 55). In contrast to virions, ISVPs likely penetrate membranes at the cell surface. ISVPs generated in vitro mediate release of 51Cr from preloaded L cells (12, 30, 31, 36), and these particles induce conductance through artificial planar lipid bilayers (58). Therefore, our finding that mutant cells support growth of reovirus when infection is initiated with ISVPs but not virions indicates that cellular mutations selected during persistent reovirus infection affect steps in reovirus entry required for the proteolytic disassembly of the viral outer capsid.
We compared parental L cells and mutant LX cells for the capacity to support internalization of reovirus virions, intracellular virion transport, acidification of endosomal organelles, and proteolytic activity required for outer capsid disassembly. Parental and mutant cells did not differ in the capacity to internalize virions or distribute them to a perinuclear compartment. Intravesicular pHs were found to be equivalent in both cell types, and virions were observed in both parental and mutant cells to colocalize with an acid-sensitive probe. However, we found a striking alteration in the capacity of mutant cells to support generation of ISVPs. After adsorption of reovirus virions to LX cells, we did not detect changes in viral outer-capsid proteins indicative of ISVP formation, even after prolonged periods of incubation. From these results, we concluded that mutant cells are altered in the expression of a protease that mediates cleavage of viral outer-capsid proteins leading to formation of ISVPs.
We used immunoblot analysis to evaluate the expression of lysosomal proteases in parental and mutant cells. We found that both cell types synthesize and secrete the proenzyme form of cathepsin L; however, only parental cells express the mature two-chain form of this enzyme. This finding demonstrates that mutant cells do not support maturation steps required for formation of the active protease. Sequence analysis of procathepsin L-encoding cDNAs corresponding to mRNA isolated from two mutant cell lines revealed no mutations. Moreover, the defect in cathepsin L maturation in mutant cells was not complemented by transfection of a cDNA encoding procathepsin L. These findings indicate that the defect in cathepsin L maturation is extrinsic, most likely due to a mutation affecting a protein involved in cathepsin L processing or transport.
Cathepsin L is translated as a proenzyme precursor, glycosylated on a single asparagine residue, and sorted for targeting to lysosomes or secretory vesicles (reviewed in references 11, 33, and 34). In prelysosomes or lysosomes, procathepsin L is processed to an enzymatically active single-chain intermediate and finally to a two-chain mature form (25). The proteases that mediate these processing steps have not been identified. However, it has been suggested that initial cleavage of the propeptide is not autocatalytic (49), which is consistent with our observation that E64 treatment does not induce cellular accumulation of the proenzyme. Thus, in mutant cells, either the enzyme that initially activates cathepsin L by cleavage of the propeptide is altered or procathepsin L is not transported to the compartment in which activation normally occurs.
The defect in cathepsin L maturation exhibited by mutant cells selected during persistent reovirus infection likely results from an alteration of a protein specifically required for cathepsin L activation or transport. This conclusion is supported by analysis of steady-state levels of cathepsin B, which demonstrates that mutant cells are not defective in generation of the mature two-chain form of this related lysosomal protease. Alterations in phosphotransferase activity, mannose 6-phosphate receptor expression, or mannose 6-phosphate receptor intracellular transport would be expected to affect processing of all lysosomal enzymes, as would a defect in acidification of the prelysosomal compartment. Similarly, alterations in the activity of chaperonins responsible for the proper folding of cathepsin L would be anticipated to affect the folding and therefore activities of many proteins. The fact that mutant cells lack the lysosomal membrane whorls characteristic of lysosomal-storage-disease lysosomes (43) suggests that most lysosomal enzymes reach lysosomes in the mutant cells. Therefore, the defect in generation of mature cathepsin L in mutant cells is probably not due to a global alteration in lysosomal enzyme processing or transport. The relationship between the alteration in cathepsin L maturation and the accumulation of electron-dense, membrane-bound vesicles in the cytoplasm of mutant cells is not apparent from our studies. It is possible that these organelles result from abnormalities in cathepsin L function or vesicular transport.
Our previous studies of persistent reovirus infections indicate that viruses and cells coevolve by selection of mutations that affect acid-dependent proteolysis of the viral outer capsid during virus entry (6, 22, 61, 62). In this study, we found that mutant cells selected during persistent infection do not generate the mature, proteolytically active form of cathepsin L. This finding suggests that cathepsin L is required for conversion of reovirus virions to ISVPs. We tested whether cathepsin L is sufficient to mediate virion-to-ISVP conversion by treating virions with purified, recombinant cathepsin L. Both SDS-PAGE analysis of viral proteins and assays of viral growth in mutant LX cells indicate that cathepsin L treatment of virions leads to generation of ISVPs. The first step in virion-to-ISVP conversion appears to be the proteolysis of ς3 protein (13, 17, 44, 52, 55). Sequences in ς3 protein adjacent to amino acid 220 confer sensitivity to a variety of proteases (42, 50), and this region of ς3 is postulated to be cleaved by lysosomal proteases during viral disassembly (51). Cathepsin L requires hydrophobic residues at the P2 and P3 positions for efficient proteolysis (8), and sequences adjacent to position 220 in the deduced amino acid sequence of ς3 (valine220, methionine221, and valine222) (26) conform to these requirements. Mutant cells might manifest alterations in the expression of other lysosomal proteases capable of converting virions to ISVPs. However, our results are consistent with the hypothesis that cathepsin L is sufficient to mediate reovirus disassembly in murine L cells.
Results reported here can be used to clarify mechanisms by which mutant viruses and cells altered in virus entry are selected during persistent reovirus infection. We propose that during establishment of persistent infection, cytolytic reoviruses select mutant cells defective in maturation of cathepsin L. These cells, which do not support conversion of reovirus virions to ISVPs, in turn select mutant viruses having altered requirements for lysosomal proteases to facilitate viral disassembly. In support of this model, mutant viruses selected during persistent reovirus infection can grow in the presence of protease inhibitor E64 (6). Thus, steps in reovirus disassembly mediated by cathepsin L appear to be a focal point for virus-cell coevolution during persistent reovirus infections of murine L cells.
In this report, we describe the characterization of cells defective in maturation of the lysosomal protease cathepsin L. To our knowledge, this is the first description of a defect in a cellular protease selected by persistent viral infection. This observation suggests that modulation of proteolytic activity in cellular endosomes plays a critical role in determining host susceptibility to intracellular pathogens. Mutant cells defective in cathepsin L maturation will be useful in our ongoing efforts to dissect viral entry mechanisms and lysosomal enzyme processing and transport pathways.
ACKNOWLEDGMENTS
We express our appreciation to Chris Aiken, Neil Green, Patrick Green, Joachim Osterman, and Earl Ruley for essential discussions and to Erik Barton, Jim Chappell, Denise Wetzel, and Greg Wilson for reviews of the manuscript. We thank Cheryl Marcum for assistance with electron microscopy and John Mort for the kind gift of purified cathepsin L.
This work was supported by Public Health Service award T32 GM07347 from the National Institute of General Medical Studies for the Vanderbilt Medical-Scientist Training Program (G.S.B. and D.H.E.), by award MCB9604139 from the National Science Foundation (A.H.E.), by Public Health Service award AI32539 from the National Institute of Allergy and Infectious Diseases (T.S.D.), and by the Elizabeth B. Lamb Center for Pediatric Research. Additional support was provided by Public Health Service awards DK20593, for the Vanderbilt Diabetes Research and Training Center, and CA68485 and DK20593, for the Vanderbilt Cell Imaging Resource.
REFERENCES
- 1.Ahmed R, Canning W M, Kauffman R S, Sharpe A H, Hallum J V, Fields B N. Role of the host cell in persistent viral infection: coevolution of L cells and reovirus during persistent infection. Cell. 1981;25:325–332. doi: 10.1016/0092-8674(81)90050-7. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed R, Chakraborty P R, Graham A F, Ramig R F, Fields B N. Genetic variation during persistent reovirus infection: presence of extragenically suppressed temperature-sensitive lesions in wild-type virus isolated from persistently infected L cells. J Virol. 1980;34:383–389. doi: 10.1128/jvi.34.2.383-389.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed R, Fields B N. Role of the S4 gene in the establishment of persistent reovirus infection in L cells. Cell. 1982;28:605–612. doi: 10.1016/0092-8674(82)90215-x. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed R, Graham A F. Persistent infections in L cells with temperature-sensitive mutants of reovirus. J Virol. 1977;23:250–262. doi: 10.1128/jvi.23.2.250-262.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed R, Kauffman R S, Fields B N. Genetic variation during persistent reovirus infection: isolation of cold-sensitive and temperature-sensitive mutants from persistently infected L cells. Virology. 1983;131:71–78. doi: 10.1016/0042-6822(83)90534-2. [DOI] [PubMed] [Google Scholar]
- 6.Baer G S, Dermody T S. Mutations in reovirus outer-capsid protein ς3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. J Virol. 1997;71:4921–4928. doi: 10.1128/jvi.71.7.4921-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett A J, Kembhavi A A, Brown M A, Kirschke H, Knight C G, Tamai M, Hanada K. l-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem J. 1982;201:189–198. doi: 10.1042/bj2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrett A J, Kirschke H. Cathepsin B, cathepsin H, and cathepsin L. Methods Enzymol. 1981;800:535–561. doi: 10.1016/s0076-6879(81)80043-2. [DOI] [PubMed] [Google Scholar]
- 9.Bass D M, Bodkin D, Dambrauskas R, Trier J S, Fields B N, Wolf J L. Intraluminal proteolytic activation plays an important role in replication of type 1 reovirus in the intestines of neonatal mice. J Virol. 1990;64:1830–1833. doi: 10.1128/jvi.64.4.1830-1833.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodkin D K, Nibert M L, Fields B N. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J Virol. 1989;63:4676–4681. doi: 10.1128/jvi.63.11.4676-4681.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bond J S, Butler P E. Intracellular proteases. Annu Rev Biochem. 1987;56:333–364. doi: 10.1146/annurev.bi.56.070187.002001. [DOI] [PubMed] [Google Scholar]
- 12.Borsa J, Morash B D, Sargent M D, Copps T P, Lievaart P A, Szekely J G. Two modes of entry of reovirus particles into L cells. J Gen Virol. 1979;45:161–170. doi: 10.1099/0022-1317-45-1-161. [DOI] [PubMed] [Google Scholar]
- 13.Borsa J, Sargent M D, Lievaart P A, Copps T P. Reovirus: evidence for a second step in the intracellular uncoating and transcriptase activation process. Virology. 1981;111:191–200. doi: 10.1016/0042-6822(81)90664-4. [DOI] [PubMed] [Google Scholar]
- 14.Brown E G, Nibert M L, Fields B N. The L2 gene of reovirus serotype 3 controls the capacity to interfere, accumulate deletions and establish persistent infection. In: Compans R W, Bishop D H L, editors. Double-stranded RNA viruses. New York, N.Y: Elsevier Biomedical; 1983. pp. 275–287. [Google Scholar]
- 15.Carmona E, Dufour É, Plouffe C, Takebe S, Mason P, Mort J S, Ménard R. Potency and selectivity of the cathepsin L propeptide as an inhibitor of cysteine proteases. Biochemistry. 1996;35:8149–8157. doi: 10.1021/bi952736s. [DOI] [PubMed] [Google Scholar]
- 16.Chandran K, Nibert M L. Protease cleavage of reovirus capsid protein μ1/μ1C is blocked by alkyl sulfate detergents, yielding a new type of infectious subvirion particle. J Virol. 1998;72:467–475. doi: 10.1128/jvi.72.1.467-475.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang C T, Zweerink H J. Fate of parental reovirus in infected cell. Virology. 1971;46:544–555. doi: 10.1016/0042-6822(71)90058-4. [DOI] [PubMed] [Google Scholar]
- 18.Chapman R L, Kane S E, Erickson A H. Abnormal glycosylation of procathepsin L due to N-terminal point mutations correlates with failure to sort to lysosomes. J Biol Chem. 1997;272:8808–8816. doi: 10.1074/jbc.272.13.8808. [DOI] [PubMed] [Google Scholar]
- 19.Chappell J D, Barton E S, Smith T H, Baer G S, Duong D T, Nibert M L, Dermody T S. Cleavage susceptibility of reovirus attachment protein ς1 during proteolytic disassembly of virions is determined by a sequence polymorphism in the ς1 neck. J Virol. 1998;72:8205–8213. doi: 10.1128/jvi.72.10.8205-8213.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean R T, Barrett A J. Lysosomes. Essays Biochem. 1976;12:1–40. [PubMed] [Google Scholar]
- 21.Dermody T S. Molecular mechanisms of persistent infection by reovirus. Curr Top Microbiol Immunol. 1998;233:1–22. doi: 10.1007/978-3-642-72095-6_1. [DOI] [PubMed] [Google Scholar]
- 22.Dermody T S, Nibert M L, Wetzel J D, Tong X, Fields B N. Cells and viruses with mutations affecting viral entry are selected during persistent infections of L cells with mammalian reoviruses. J Virol. 1993;67:2055–2063. doi: 10.1128/jvi.67.4.2055-2063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furlong D B, Nibert M L, Fields B N. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gal S, Gottesman M M. The major excreted protein (MEP) of transformed mouse cells and cathepsin L have similar protease specificity. Biochem Biophys Res Commun. 1986;139:156–162. doi: 10.1016/s0006-291x(86)80093-6. [DOI] [PubMed] [Google Scholar]
- 25.Gal S, Willingham M C, Gottesman M M. Processing and lysosomal localization of a glycoprotein whose secretion is transformation stimulated. J Cell Biol. 1985;100:535–544. doi: 10.1083/jcb.100.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giantini M, Seliger L S, Furuichi Y, Shatkin A J. Reovirus type 3 genome segment S4: nucleotide sequence of the gene encoding a major virion surface protein. J Virol. 1984;52:984–987. doi: 10.1128/jvi.52.3.984-987.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottesman M M, Sobel M E. Tumor promoters and Kirsten sarcoma virus increase synthesis of a secreted glycoprotein by regulating levels of translatable mRNA. Cell. 1980;19:449–455. doi: 10.1016/0092-8674(80)90519-x. [DOI] [PubMed] [Google Scholar]
- 28.Haughland R P. Probes for organelles. In: Spence M T Z, editor. Molecular Probes catalog: handbook of fluorescent probes and research chemicals. 6th ed. Eugene, Oreg: Molecular Probes; 1996. pp. 265–286. [Google Scholar]
- 29.Haughland R P. pH indicators. In: Spence M T Z, editor. Molecular Probes catalog: handbook of fluorescent probes and research chemicals. 6th ed. Eugene, Oreg: Molecular Probes; 1996. pp. 551–570. [Google Scholar]
- 30.Hooper J W, Fields B N. Monoclonal antibodies to reovirus ς1 and μ1 proteins inhibit chromium release from mouse L cells. J Virol. 1996;70:672–677. doi: 10.1128/jvi.70.1.672-677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hooper J W, Fields B N. Role of the μ1 protein in reovirus stability and capacity to cause chromium release from host cells. J Virol. 1996;70:459–467. doi: 10.1128/jvi.70.1.459-467.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirschke H, Langner J, Wiederanders B, Ansorge S, Bohley P. Cathepsin L. A new proteinase from rat-liver lysosomes. Eur J Biochem. 1977;74:293–301. doi: 10.1111/j.1432-1033.1977.tb11393.x. [DOI] [PubMed] [Google Scholar]
- 33.Kornfeld S. Trafficking of lysosomal enzymes. FASEB J. 1987;1:462–468. doi: 10.1096/fasebj.1.6.3315809. [DOI] [PubMed] [Google Scholar]
- 34.Kornfeld S, Mellman I. The biogenesis of lysosomes. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- 35.Kothandaraman S, Hebert M C, Raines R T, Nibert M L. No role for pepstatin-A-sensitive acidic proteinases in reovirus infections of L or MDCK cells. Virology. 1998;251:264–272. doi: 10.1006/viro.1998.9434. [DOI] [PubMed] [Google Scholar]
- 36.Lucia-Jandris P, Hooper J W, Fields B N. Reovirus M2 gene is associated with chromium release from mouse L cells. J Virol. 1993;67:5339–5345. doi: 10.1128/jvi.67.9.5339-5345.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mach L, Stuwe K, Hagen A, Ballaun C, Glossl J. Proteolytic processing and glycosylation of cathepsin B. The role of the primary structure of the latent precursor and of the carbohydrate moiety for cell-type-specific molecular forms of the enzyme. Biochem J. 1992;282:577–582. doi: 10.1042/bj2820577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez C G, Guinea R, Benavente J, Carrasco L. The entry of reovirus into L cells is dependent on vacuolar proton-ATPase activity. J Virol. 1996;70:576–579. doi: 10.1128/jvi.70.1.576-579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mason R W. Interaction of lysosomal cysteine proteinases with alpha-2-macroglobulin: conclusive evidence for the endopeptidase activities of cathepsins B and H. Arch Biochem Biophys. 1989;273:367–374. doi: 10.1016/0003-9861(89)90495-5. [DOI] [PubMed] [Google Scholar]
- 40.McIntyre G F, Erickson A H. Procathepsins L and D are membrane-bound in acidic microsomal vesicles. J Biol Chem. 1991;266:15438–15445. [PubMed] [Google Scholar]
- 41.McIntyre G F, Erickson A H. The lysosomal proenzyme receptor that binds procathepsin L to microsomal membranes at pH 5 is a 43-kDa integral membrane protein. Proc Natl Acad Sci USA. 1993;90:10588–10592. doi: 10.1073/pnas.90.22.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller J E, Samuel C E. Proteolytic cleavage of the reovirus sigma 3 protein results in enhanced double-stranded RNA-binding activity: identification of a repeated basic amino acid motif within the C-terminal binding region. J Virol. 1992;66:5347–5356. doi: 10.1128/jvi.66.9.5347-5356.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neufeld E F, McKusik V A. Disorders of lysosomal enzyme synthesis and localization: I-cell disease and pseudo-Hurler polydystrophy. In: Stanbury J B, Wyngaarden J B, Frederickson D S, Goldstein J L, Brown M S, editors. The metabolic basis of inherited disease. New York, N.Y: McGraw-Hill; 1983. p. 778. [Google Scholar]
- 44.Nibert M L, Chappell J D, Dermody T S. Infectious subvirion particles of reovirus type 3 Dearing exhibit a loss in infectivity and contain a cleaved ς1 protein. J Virol. 1995;69:5057–5067. doi: 10.1128/jvi.69.8.5057-5067.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Portnoy D A, Erickson A H, Kochan J, Ravetch J V, Unkeless J C. Cloning and characterization of a mouse cysteine proteinase. J Biol Chem. 1986;26:14697–14703. [PubMed] [Google Scholar]
- 46.Rowan A D, Mason P, Mach L, Mort J S. Rat procathepsin B. Proteolytic processing to the mature form in vitro. J Biol Chem. 1992;267:15993–15999. [PubMed] [Google Scholar]
- 47.Rubin D H, Weiner D B, Dworkin C, Greene M I, Maul G G, Williams W V. Receptor utilization by reovirus type 3: distinct binding sites on thymoma and fibroblast cell lines result in differential compartmentalization of virions. Microb Pathog. 1992;12:351–365. doi: 10.1016/0882-4010(92)90098-9. [DOI] [PubMed] [Google Scholar]
- 48.Ryan R E, Sloane B F, Sameni M, Wood P L. Microglial cathepsin B: an immunological examination of cellular and secreted species. J Neurochem. 1995;65:1035–1045. doi: 10.1046/j.1471-4159.1995.65031035.x. [DOI] [PubMed] [Google Scholar]
- 49.Salminen A, Gottesman M M. Inhibitor studies indicate that active cathepsin L is probably essential to its own processing in cultured fibroblasts. Biochem J. 1990;272:39–44. doi: 10.1042/bj2720039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schiff L A, Nibert M L, Co M S, Brown E G, Fields B N. Distinct binding sites for zinc and double-stranded RNA in the reovirus outer capsid protein ς3. Mol Cell Biol. 1988;8:273–283. doi: 10.1128/mcb.8.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shepard D A, Ehnstrom J G, Schiff L A. Association of reovirus outer capsid proteins ς3 and μ1 causes a conformational change that renders ς3 protease sensitive. J Virol. 1995;69:8180–8184. doi: 10.1128/jvi.69.12.8180-8184.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silverstein S C, Astell C, Levin D H, Schonberg M, Acs G. The mechanism of reovirus uncoating and gene activation in vivo. Virology. 1972;47:797–806. doi: 10.1016/0042-6822(72)90571-5. [DOI] [PubMed] [Google Scholar]
- 53.Silverstein S C, Dales S. The penetration of reovirus RNA and initiation of its genetic function in L-strain fibroblasts. J Cell Biol. 1968;36:197–230. [PubMed] [Google Scholar]
- 54.Silverstein S C, Levin D H, Schonberg M, Acs G. The reovirus replicative cycle: conservation of parental RNA and protein. Proc Natl Acad Sci USA. 1970;67:275–281. doi: 10.1073/pnas.67.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sturzenbecker L J, Nibert M, Furlong D, Fields B N. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J Virol. 1987;61:2351–2361. doi: 10.1128/jvi.61.8.2351-2361.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tao K, Stearns N A, Dong J, Wu Q L, Sahagian G G. The proregion of cathepsin L is required for proper folding, stability, and ER exit. Arch Biochem Biophys. 1994;31:19–27. doi: 10.1006/abbi.1994.1203. [DOI] [PubMed] [Google Scholar]
- 57.Thiebaut F, Currier S J, Whitaker J, Haugland R P, Gottesman M M, Pastan I, Willingham M C. Activity of the multidrug transporter results in alkalinization of the cytosol: measurement of cytosolic pH by microinjection of a pH-sensitive dye. J Histochem Cytochem. 1990;38:685–690. doi: 10.1177/38.5.1692055. [DOI] [PubMed] [Google Scholar]
- 58.Tosteson M T, Nibert M L, Fields B N. Ion channels induced in lipid bilayers by subvirion particles of the nonenveloped mammalian reoviruses. Proc Natl Acad Sci USA. 1993;90:10549–10552. doi: 10.1073/pnas.90.22.10549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tycko B, Maxfield F R. Rapid acidification of endocytic vesicles containing alpha-2 macroglobulin. Cell. 1982;28:643–651. doi: 10.1016/0092-8674(82)90219-7. [DOI] [PubMed] [Google Scholar]
- 60.Virgin H W, IV, Bassel-Duby R, Fields B N, Tyler K L. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing) J Virol. 1988;62:4594–4604. doi: 10.1128/jvi.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wetzel J D, Chappell J D, Fogo A B, Dermody T S. Efficiency of viral entry determines the capacity of murine erythroleukemia cells to support persistent infections by mammalian reoviruses. J Virol. 1997;71:299–306. doi: 10.1128/jvi.71.1.299-306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wetzel J D, Wilson G J, Baer G S, Dunnigan L R, Wright J P, Tang D S H, Dermody T S. Reovirus variants selected during persistent infections of L cells contain mutations in the viral S1 and S4 genes and are altered in viral disassembly. J Virol. 1997;71:1362–1369. doi: 10.1128/jvi.71.2.1362-1369.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]