Abstract
Children with cancer often endure a range of psychoneurological symptoms (PNS), including pain, fatigue, cognitive impairment, anxiety, depressive symptoms, and sleep disturbance. Despite their prevalence, the underlying pathophysiology of PNS remains unclear. Hypotheses suggest an interplay between the gut microbiome and the functional metabolome, given the immune, neurological, and inflammatory influences these processes exert. This mini-review aims to provide a synopsis of the literature that examines the relationship between microbiome–metabolome pathways and PNS in children with cancer, drawing insights from the adult population when applicable. While there is limited microbiome research in the pediatric population, promising results in adult cancer patients include an association between lower microbial diversity and compositional changes, including decreased abundance of the beneficial microbes Fusicatenibacter, Ruminococcus, and Odoribacter, and more PNS. In pediatric patients, associations between peptide, tryptophan, carnitine shuttle, and gut microbial metabolism pathways and PNS outcomes were found. Utilizing multi-omics methods that combine microbiome and metabolome analyses provide insights into the functional capacity of microbiomes and their associated microbial metabolites. In children with cancer receiving chemotherapy, increased abundances of Intestinibacter and Megasphaera correlated with six metabolic pathways, notably carnitine shuttle and tryptophan metabolism. Interventions that target the underlying microbiome–metabolome pathway may be effective in reducing PNS, including the use of pre- and probiotics, fecal microbiome transplantation, dietary modifications, and increased physical activity. Future multi-omics research is needed to corroborate the associations between the microbiome, metabolome, and PNS outcomes in the pediatric oncology population.
Keywords: Psychoneurological symptoms, Microbiome, Metabolome, Multi-omics, Pediatric oncology
Introduction
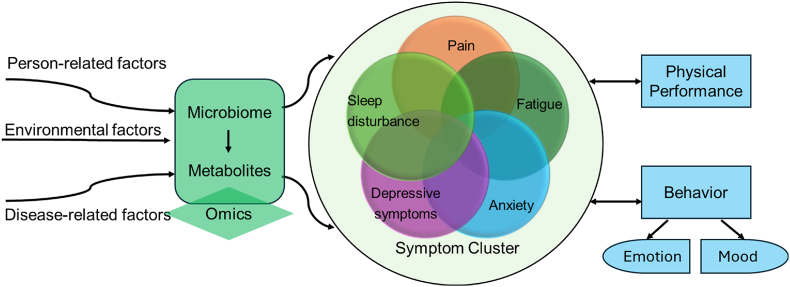
Children with cancer experience debilitating symptoms, with pain, fatigue, cognitive impairment, anxiety, depressive symptoms, and sleep disturbance being amongst the most commonly reported.1,2 These symptoms are classified as psychoneurological symptoms (PNS), or a cluster of symptoms hypothesized to stem from similar pathophysiologic pathways.3 Poor understanding of the underlying pathophysiology limits the available treatment options, resulting in reduced quality of life. A symptom cluster framework proposed by Hockenberry and Hooke outlined antecedents that influence PNS in children with cancer, including person-related, environmental, and disease-related factors.4 For example, survivors of childhood brain tumors have reported higher incidence of PNS, specifically fatigue, cognitive impairment, and anxiety.5,6 Chemotherapy and radiotherapy are also known to influence the development of PNS, with pediatric patients reporting higher levels of anxiety, depression, fatigue, and cognitive impairment after treatment.1,5 Examining the underlying physiological mechanisms of PNS, such as microbiome–metabolome pathways, within different cancer diagnoses over the course of treatment can enable the development of novel interventions that reduce symptom incidence and severity (Fig. 1).
Fig. 1.
Framework of symptom clusters in children and adolescents adapted from Hockenberry and Hooke (2007). Citation: https://doi.org/10.1016/j.soncn.2007.01.001.
The microbiome is a collection of microorganisms that assemble into communities across different body sites, including the skin, oral cavity, and gastrointestinal tract.3 The diversity and composition of microbial communities plays a crucial role in human health. For example, a healthy gut microbiome consists of symbiotic microbes (e.g., Bacteroidota [new name of Bacteroidetes], Bacillota [new name of Firmicutes], Actinomycetota [new name of Actinobacteria], and Pseudomonadota [new name of Proteobacteria]) and high diversity,3,7,8 maintaining homeostasis within the host. A dysbiotic, or unbalanced, gut microbiome is characterized by lower overall diversity and the presence of pathogenic microbes, modulating many components of the host's physiological processes, including inflammatory and immune pathways.7 These deviations from healthy biological processes cause the host to be more susceptible to diseases like autoimmune disorders, Alzheimer’s disease, and cancer.3,7,9
The metabolites that derive from the microbiome provide insight into the functional mechanisms of the microbes and how these biological mechanisms influence cancer outcomes. The use of metabolomics techniques has been increasing rapidly in the field of oncology and symptom science.10 These analyses can be conducted using samples from various origins, such as stool, urine, cerebrospinal fluid (CSF), and blood.1,10,11 Metabolites from these samples can then be mapped to specific metabolic pathways. Previous work has suggested the potential effects of metabolites and their associated pathways on cancer occurrence and prognosis, treatment efficacy, and treatment-related complications (e.g., PNS and congestive heart failure [CHF]).1,12 However, metabolomic analyses are underutilized within the pediatric oncology population.
The underlying pathophysiology of PNS remains a relevant research topic, with recent studies outlining the influence of the microbiome on the central and peripheral nervous system via bidirectional pathways, such as the microbiome–gut–brain (MGB) axis.13 Metabolites are proposed to serve as the communication channel between these two entities.1 For example, short-chain fatty acids (SCFAs) are metabolites produced by the fermentation of dietary fibers by bacteria in the gut.9 Tryptophan, an amino acid primarily obtained from dietary sources, also connects the microbiome and brain by regulating intestinal homeostasis and neuronal function. This mini-review aims to provide a synopsis of the microbiome and metabolome literature to aid in understanding the functional mechanisms of theses underlying pathways and how they influence PNS outcomes in pediatric oncology patients. Due to limited literature conducted in the pediatric oncology population, some evidence from the adult population is described to foster the development of future hypotheses and research. Table 1 provides definitions for important terms discussed in this article.
Table 1.
Microbiome and metabolome glossary.
| Term | Definition |
|---|---|
| Psychoneurological symptoms | Symptoms that often co-occur and have similar potential underlying causal pathways. Examples include pain, fatigue, sleep disturbance, anxiety, depressive symptoms, and cognitive dysfunction. |
| Microbiome | A community of microorganisms (bacteria, viruses, and fungi) that reside in different parts of the human body, such as the skin, gastrointestinal tract, oral cavity, and vagina. |
| Microbial composition | A description of the microorganisms that form the microbiome. Compositional components include microbial abundance and diversity metrics (alpha and beta diversity). |
| Microbial abundance | The quantity or proportion (relative abundance) of a specific microorganism that resides in the microbiome. |
| Alpha diversity | Measurement of microbial species diversity within a given sample or microbial site. |
| Beta diversity | Measurement of dissimilarity (or similarity) of species diversity between two or more samples or microbial sites. |
| Metabolome | The collection of small molecules (metabolites) and their interactions within a specific biological system or sample. |
| Metabolites | Small molecules that are substrates, intermediates, or products of metabolic reactions that regulate various physiological functions within a cell or organism. |
| Untargeted metabolomics | An analytical approach that aims to comprehensively detect and quantify metabolites within a sample due to limited prior knowledge; often used as an exploratory tool. |
| Targeted metabolomics | An analytical approach that aims to quantify specific types of metabolites within a sample based on known relevant metabolic pathways or physiological processes. |
| Metabolomic pathway | A sequence of chemical reactions that produce new molecules (synthesis) or breakdown complex molecules (degradation). |
The microbiome
The interplay between the microbiome and cancer treatments
The microbiome of pediatric oncology patients is a dynamic community that has the potential to influence treatment efficacy. Dysbiotic changes in the gut may exist pre-cancer,7 though the origin of this initial dysbiosis remains unclear. Hypotheses predict that this imbalance may be a cause of cancer development due to the microbiome's potential carcinogenetic effects or is a consequence of cancer-related inflammatory processes.14 Emerging evidence in adults with cancer suggests microbiome diversity and composition correlate with short- and long-term treatment response, specifically infection rates.15 Additionally, the abundance of Fusobacterium nucleatum was associated with poor treatment outcomes in patients with colorectal cancer receiving chemotherapy. Evidence also suggests this association between microbial composition and immunotherapy response, with higher abundances of beneficial microbes (e.g., Ruminococcacae, Akkermansia muciniphila, and Bifidobacteria) correlating with better responses to immune checkpoint inhibitors.16 Microbes may affect treatment efficacy through nutrient competition, toxin secretion, or modulation of host immunity.15 However, the impact of microbial effects on treatment efficacy pathways are underexplored in pediatric cancer, limiting definitive conclusions on baseline microbiome–treatment interactions.
Cancer treatments influence the microbiome's composition via inflammatory, metabolic, and dietary pathways.14,15 Chemotherapy, radiotherapy, and immunotherapy have been found to reduce microbial diversity and alter community composition by favoring pathogenic microbes. For example, children with acute myeloid leukemia and acute lymphoblastic leukemia (ALL) receiving chemotherapy showed reductions in diversity when compared to healthy controls.9,17 Compositional changes were also present, with significant reductions in Lachnospiraceae, Roseburia, Bifidobacteria, Lactobacillus, and Escherichia coli and an increased abundance of Bacteroides. These microbial changes often persist beyond treatment. Adolescents and young adult Hodgkin's lymphoma survivors were found to have sustained lower diversity within the gut microbiome, with reduced abundances of Actinobacteria Collinsella compared to twin controls.18 These treatment-induced gut microbial changes have lasting implications, particularly on symptom burden.
The microbiome and psychoneurological symptoms
Several physiological pathways through which the microbiome influences the incidence and severity of PNS in patients with cancer have been proposed. Cancer treatments may increase gut permeability,19 permitting microbes to migrate across the intestinal mucosa into systemic circulation, inducing PNS via the modulation of neuroimmune modulators.7,13 Preliminary investigations into these potential causal pathways have begun by examining how microbial diversity and composition influence PNS in adults with cancer.
Significant associations between the diversity and composition of the gut microbiome and PNS in adult cancer have been discovered. Table 2 outlines significant microbes associated with PNS in this population. In a longitudinal study by Barandouzi et al.,20 women with gynecological cancers exhibited lower reports of the PNS cluster post-treatment when they had higher alpha diversity. Similarly, beta diversity of the gut microbiome was associated with cognitive impairment in women with breast cancer undergoing chemotherapy treatment, although the directionality of this relationship was not reported.21 Moreover, reductions in bacteria possibly related to anti-inflammatory processes (e.g., Fusicatenibacter, Ruminococcus, and Odoribacter) were observed in women who reported higher PNS,20,21 supporting the hypothesis of potential microbiome-derived inflammatory pathways. When examining relationships between microbial composition and PNS in adult patients with advanced cancer, an increased abundance of Eubacterium halli and a decreased abundance of Cosenzaea were associated with more fatigue.22 (Table 2).
Table 2.
Significant microbes associated with PNS in the identified oncology literature.
| PNS | Significant microbes (abundance) |
|---|---|
| Increased fatigue | s.Eubacterium hallii (↓)22 |
| g.Cosenzaea (↑)22 | |
| g.Faecalibacterium (NR)13 | |
| g.Prevotella (NR)13 | |
| Increased anxiety | g.Coprococcus (NR)13 |
| g.Bacteroides (NR)13 | |
| f.Lachnospiraceae (↓)23 | |
| Increased cognitive impairment | g.Odoribacter (↓)21 |
| f.Erysipelotrichaceae (↑)21 | |
| g.Clostridium (↑)21 | |
| g.Intestinibacter (↓)23 | |
| Decreased cognitive impairment | f.Lachnospiraceae (↓)23 |
| Increased depressive functional interference | f.Ruminococcaceae (↓)23 |
| g.Intestinibacter (↓)23 | |
| Decreased depressive symptoms | p.Tenericutes (↑)21 |
| Increased PNS cluster | g.Atopobium (↑)20 |
| g.Parvimonas (↑)20 | |
| g.Corynebacterium (↑)20 | |
| g.Gardnerella (↑)20 | |
| g.Mageibacillus (↑)20 | |
| g.Howardella (↑)20 | |
| g.Shuttleworthia (↑)20 | |
| g.Megasphaera (↑)20 | |
| g.Gardnerella (↑)20 | |
| g.Fusicatenibacter (↓)20 | |
| g.Ruminococcus (↓)20 | |
| Decreased PNS cluster | g.Akkermansia (↑)20 |
| g.Gallicola (↑)20 | |
| g.Frisingicoccus (↑)20 | |
| g.Negativicoccus (↑)20 | |
| g.Fusicatenibacter (↑)20 | |
| g.Lachnospiraceae_NK4A136 (↑)20 | |
| g.Ruminococcus (↑)20 | |
| g.Tyzzerella (↑)20 | |
| g.Anaerostipes (↑)20 | |
| g.CAG_56 (↑)20 | |
| g.Parasutterella (↑)20 |
f, family; g, genus; NR, not reported; p, phylum; PNS, psychoneurological symptoms; s, species.
Table 3.
Significant metabolic pathways associated with PNS in the identified pediatric oncology literature.
| PNS | Significant metabolic pathways |
|---|---|
| Fatigue | Xenobiotics metabolisma10 |
| Microbial gut flora metabolisma10 | |
| Tryptophan metabolisma,c1,10 | |
| Drug metabolism (cytochrome P450)c1 | |
| Peptide metabolismb11 | |
| Amino acid metabolismb11 | |
| Nucleotide metabolismb11 | |
| Lipid metabolismb11 | |
| Carbohydrate metabolismb11 | |
| Pain | De novo fatty acid biosynthesisc1 |
| Fatty acid metabolismc1 | |
| Linoleate metabolismc1 | |
| Fatty acid activationc1 | |
| Omega-3 fatty acid metabolismc1 | |
| Bile acid biosynthesisc1 | |
| Glycerophospholipid metabolismc1 | |
| Anxiety | Glycerophospholipid metabolismc1 |
| Phosphatidylinositol phosphate metabolismc1 | |
| Sialic acid metabolismc1 | |
| Tryptophan metabolismc1 | |
| Depressive symptoms | Limonene and pinene degradationc1 |
| Phoshatidylinositol phosphate metabolismc1 | |
| PNS cluster | Tryptophan metabolismc1 |
| Carnitine shuttlec1 |
PNS, psychoneurological symptoms.
Metabolic pathway found in urine sample.
Metabolic pathway found in cerebrospinal fluid sample.
Metabolic pathway found in blood sample.
Research corroborating the associations between the microbiome and PNS clusters in the pediatric oncology population is sparce. One pilot study examined these associations within young adult (18–39 years) cancer survivors.23 In this younger population, higher alpha diversity was associated with lower depressive interference and higher cognitive function. Lower abundances of Lachnospiraceae, Ruminococcaceae, and Intestinibacter were associated with increased PNS. Notably, decreased abundance of Lachnospiraceae was also associated with higher cognitive function, highlighting the importance of understanding the bacterial taxa's underlying functional and metabolic characteristics.
The metabolome
The metabolome as a diagnostic and prognostic biomarker
Metabolic profiles have emerged as promising diagnostic and prognostic biomarkers for childhood cancer outcomes and treatment complications. For example, metabolites that may aid in the diagnosis of osteosarcoma include glutamic acid and lactic acid, while the presence of lung metastasis correlated with 5-aminopentanamide.24 In childhood brain tumors, elevated levels of myo-inositol, taurine, and glutamine tissue metabolites have been proposed as potential diagnostic biomarkers for ependymomas, medulloblastoma, and pilocytic astrocytomas, respectively.25 Metabolites also play a role in the development of treatment-related complications. Anthracyclines, a standard chemotherapy amongst childhood cancers, are known to increase the risk of late-onset CHF in cancer survivors. Metabolomic analyses revealed that lower carnitine levels were associated with poor cardiac function in this population.12 Carnitine is an essential metabolite for energy production in the myocardium, with healthy cardiac myocytes containing high levels of the metabolite. These findings provide potential metabolic pathways from which future diagnostic and prevention strategies may be developed to reduce the incidence and severity of late-onset CHF in patients who receive anthracyclines.
The metabolome and psychoneurological symptoms
Metabolomic analyses offer insights into the physiological processes underlying PNS in pediatric cancer patients. Table 3 highlights key metabolic pathways linked to PNS. Initial examinations into this relationship utilized untargeted metabolomic analyses, offering a broader, more comprehensive approach to exploring metabolites and metabolic pathways.10 In CSF analyses of children with ALL, untargeted metabolomic analyses revealed eight metabolites and five metabolic pathways related to fatigue, including amino acid (e.g., 3-methoxytyrosine, dimethylglycine), lipid (e.g., myo-inositol, dimethylmalonic acid), carbohydrate (e.g. ribitol), nucleotide (e.g., allantoin), and peptide (e.g., gamma-Glutamylglutamine) metabolism.11 A significant negative correlation between gamma-glutamylglutamine and fatigue aligns with its potential role in maintaining amino acid balance within the brain and regulating neurotransmitter passage across the blood–brain barrier. Tryptophan is another metabolite associated with neurotransmitter processes, specifically the production of serotonin and melatonin.26,27 Untargeted blood metabolomic analyses in children undergoing chemotherapy revealed associations between tryptophan (e.g., indole-3-acetaldehyde, L-kynurenine) and carnitine shuttle metabolic pathways (e.g., stearoylcarnitine, dihomo-gamma-linolenyl carnitine) with the PNS cluster.1 Examination of specific symptoms within the PNS cluster revealed that the tryptophan pathway was also associated with fatigue and anxiety, whereas fatty acid and bile acid biosynthesis pathways were associated with pain. Notably, bile acids influence microbial signaling in the gut, alluding to the potential interplay between the microbiome, metabolites, and PNS within the pediatric oncology population. Targeted metabolic analyses in childhood cancer survivors and those undergoing cancer therapy found similar associations between urinary metabolites, the microbiome, and PNS.10 Results revealed associations between indole-3-lactic acid, acetohydroxamic acid, and trimethylamine-N-oxide and self-reported fatigue. These metabolites are mapped to tryptophan, drug and environmental exposure (xenobiotic metabolism), and gut microbial metabolic pathways, respectively. The recurrence of microbial-related metabolic pathways underscores the importance of further investigating the microbiome-metabolite interplay in PNS among pediatric cancer patients.
Microbiome–metabolome pathways
Multi-omic methods, specifically investigating microbiome–metabolome pathways, provide nuanced insights into potential mechanisms underlying health outcomes in the pediatric oncology field. For example, bile acid and amino acid metabolites were linked to gut dysbiosis in pediatric patients pre-hematopoietic cell transplantation (HCT).28 Bile acid metabolites, along with dysbiotic gut microbiome, are potential biomarkers for graft-versus-host disease, a serious and life-threatening complication of HCT. Fecal microbiome–metabolome pathways are also altered in pediatric glioma patients and murine models, specifically decreased levels of 5-hydroxyindoleacetic acid (5-HIAA) and SCFAs (e.g., propionate, butyrate, and acetate) after tumor growth.29 Shifts in gut microbial abundance, such as increased Verrucomicrobia, Akkermansia, and Bacteroides, and deceased Firmicutes, were associated with 5-HIAA level changes, indicating potential communication channels between the microbiome and metabolites. Furthermore, 5-HIAA levels may indirectly influence brain functioning via its precursor serotonin, which crosses the blood–brain barrier. SCFAs serve as another metabolic communication channel between the microbiome and brain. Primarily produced from bacterial metabolic activity in the gut, SCFAs play a vital role in modulating neurotransmitters, influencing host immune response, reducing inflammation, and maintaining the integrity of the blood–brain barrier.29 These correlations between the microbiome, metabolome, and brain health support the hypothesized physiological communication channels proposed in the MGB axis.
Microbiome–metabolome pathways and psychoneurological symptoms
Limited research has investigated associations between microbiome–metabolome pathways and PNS in the pediatric oncology population. However, insights can be gathered from the adult population. In adult rectal cancer patients, associations between gut microbiome–metabolome pathways and PNS, specifically fatigue and depressive symptoms, have been demonstrated following chemotherapy and radiation therapy.30, 31, 32 High levels of fatigue correlated with more abundant Eubacterium, Streptococcus, Adlercreutzia, and Actinomyces.30 Meanwhile, increased depressive symptoms were correlated with higher levels of Gemella, Bacillales Family XI, Actinomyces, Streptococcus, Lactococcus, Weissella, and Leuconostocaceae, and decreased levels of Coprobacter, Intestinibacter, Intestimonas, Lachnospiraceae, Phascolarctobacterium, Ruminiclostridium, Ruminococcaceae (UCG-005 and uncultured), Tyzzerella, and Parasutterella.31 Rectal cancer patients exhibiting high co-occurrence of fatigue and depressive symptoms demonstrated higher abundances of Ezakiella, Clostridium sensu stricto, Porphyromonas, Barnesiella, Coriobacteriales Incertae Sedis, Synergistiaceae, Echerichia-Shigella, and Turicibacter, whereas low co-occurrence reports demonstrated increased abundances of Enterococcus and Lachnospiraceae.32 Metabolic functional pathways were predicted using the MetaCyc database based on microbial abundances, with seven pathways correlating with fatigue, 14 with depressive symptoms, and eight with the co-occurrence of fatigue and depressive symptoms.30, 31, 32 Notable relationships between functional pathways and PNS include the correlation between sucrose degradation associated with fatigue, biosynthesis pathways and depressive symptoms, and L-tryptophan and PNS co-occurrence. As previously highlighted, tryptophan plays a vital role in the production of serotonin and has been linked to disorders associated with serotonin shifts, such as depression and insomnia, suggesting potential implications for future PNS interventions.32
Despite providing insights into preliminary associations between the microbiome, metabolic activity, and PNS, predictive functional pathway methods have limitations. These methods rely on an assumption of potential metabolic activity within the microbiome and do not capture actual metabolic data that can be analyzed using targeted or untargeted metabolomic approaches.
One recent study examined these associations in children with cancer undergoing chemotherapy using an untargeted, multi-omics approach.33 Prior to cycle two of chemotherapy, decreased abundance of Lactobacillus, Bifidobacterium, and Roseburia in the gut correlated with increased PNS clusters. Associated metabolic pathways included carnitine shuttle, fatty acid metabolism and activation, and tryptophan metabolism. Following chemotherapy completion, decreased abundances of Intestinibacter and Megasphaera were associated with increased PNS clusters, which were linked to six metabolic pathways including aspartate and asparagine metabolism, carnitine shuttle, and tryptophan metabolism. The identification of these PNS pathways provides potential physiological targets for future interventions in the pediatric oncology population. Due to small sample size, future multi-omics studies are necessary to validate these findings, including in other pediatric oncology populations and contexts.
Challenges
Microbiome, metabolome, and multi-omics research in the pediatric oncology population presents study design, analysis, and generalizability challenges. The influence of cancer on microbial and metabolic processes makes obtaining baseline profiles difficult, obscuring the impact of pre-cancer profiles on future health outcomes. To address this challenge, healthy controls are often used. Cost constraints drive challenges like small sample sizes and cross-sectional designs, limiting causal inferences. Some sequencing techniques, like 16S rRNA, offer limited genus-level information of microbes, missing important species-level details.14 The vast amount of available biological data also presents a challenge, yielding unidentified microbes and metabolites with unknown functional characteristics.34 Lastly, inter- and intraindividual variations further limit the generalizability, hindering the ability to draw causal conclusions due to the difficulty in capturing an exhaustive list of confounding factors,14 such as antibiotic use, age, gender, environmental exposure, genetics, dietary habits, and neighborhood social determinants of health. Cancer-specific confounders that add to this challenge of generalizability include diagnosis (e.g., cancer type and staging) and associated treatment modalities (e.g., steroids, chemotherapy, radiotherapy, and surgery), which cause differing risks for mucosal disruption. Research that examines microbiome–metabolome pathways in pediatric populations with a singular cancer type and stage can adjust for these cancer-specific confounders.
Therapeutic interventions
Potential interventions targeting microbial and metabolic pathways for PNS treatment include nutritional supplements, fecal microbiome transplantation (FMT), and lifestyle modifications. Prebiotics and probiotics have been shown to alleviate symptoms of depression, anxiety, and fatigue in colorectal cancer patients by fostering the growth of symbiotic microbes through undigestible fiber supplementation and instillation of live, healthy microorganisms (e.g., Lactobacillus and Bifidobacterium infantis) that restore the gut microbiome.9 FMTs also aim to restore the disrupted microbiome by utilizing healthy fecal donors, but its use in immunocompromised patients causes concern for bacterial translocation and associated infections.8,9 Dietary changes and exercise promotion have dual effects on the microbiome and metabolome, serving as potential treatments for PNS. Eating kimchi, yogurts, and fiber-rich foods has been shown to increase levels of beneficial microbes.9,35 Additionally, ingesting more dietary carbohydrates that undergo bacterial fermentation in the gut can increase metabolic levels of SCFAs.30 Physical activity increases microbial diversity and reduces inflammation and oxidative stress.36 Notably, physical activity has been linked to improvements in fatigue of childhood cancer survivors and children undergoing cancer treatments.10 Future research must examine the feasibility and effects of these interventions on PNS outcomes in various pediatric oncology diagnoses and treatment contexts.
Future implications
Current research highlights numerous microbiome–metabolome pathways linked to PNS, paving the way for future hypotheses and clinical applications. While most studies have focused on the gut microbiome, there's a pressing need to explore potential interactions involving other microbial sites, specifically the oral microbiome due the hypothesized oral–microbiome–brain axis.37 Additionally, most microbiome and metabolome research has been conducted in the adult population. Research within the pediatric population is imperative due to differing microbial compositions based on age and developmental stage. Nurses play a pivotal role in translating research findings into clinical practice. Recognizing the significance of microbiome–metabolome pathways in PNS pathophysiology is essential for delivering quality nursing care. Nurses can evaluate individual risk factors for microbiome dysbiosis and associated PNS, providing early intervention strategies and suggesting treatment options through patient and family education on self-care strategies, such as pre- and probiotics, dietary adjustments, and increased physical activity.
Conclusions
The intricate nature of PNS in pediatric oncology patients underscores the significance of exploring the interplay between the microbiome, metabolome, and cancer treatments. Emerging research highlights the influence that microbial dysbiosis and metabolic shifts have on the incidence and severity of PNS. Due to the absence of literature that examines associations between the microbiome and PNS in pediatric patients, inferences must be drawn from the adult oncology population. For example, lower microbial diversity and compositional changes have been associated with an increase in PNS in young adult cancer survivors.23 In pediatric patients, peptide, tryptophan, carnitine shuttle, and gut microbial metabolism pathways have been associated with PNS outcomes.1,10,11 One multi-omics study in children with cancer connected compositional differences in the microbiome before and after chemotherapy to metabolic pathways, notably carnitine shuttle and tryptophan metabolism.33 These microbiome–metabolome pathways were associated with an increase in the PNS cluster. Findings from these studies support the hypothesized MGB axis and offer potential avenues for targeted interventions. Future research must add to the existing evidence by integrating multi-omics approaches within various microbial sites, oncologic diagnoses, and age groups, thus advancing our understanding of PNS and aiding in the development of effective interventions that enhance quality of life in pediatric oncology patients.
Ethics statement
Not required.
Funding
This study was supported by the National Institutes of Health, United States (Grant No. 1K99NR017897-01 and 4R00NR017897-03, 2018-2024). The funders had no role in considering the study design or in the collection, analysis, interpretation of data, writing of the report, or decision to submit the article for publication.
CRediT author statement
Caitlin Webster: Methodology, Visualization, Writing – Original draft preparation. Janice S. Withycombe: Methodology, Resources, Writing – Reviewing and Editing, Funding acquisition; Jessica Sheth Bhutadac: Resources, Writing – Reviewing and Editing; Jinbing Bai: Conceptualization, Methodology, Visualization, Writing – Reviewing and Editing, Funding acquisition. All authors had full access to all the data in the study, and the corresponding author had final responsibility for the decision to submit for publication. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability statement
Data availability is not applicable to this article as no new data were created or analyzed in this study.
Declaration of generative AI and AI-assisted technologies in the writing process
No AI tools or services were used during the preparation of this work.
References
- 1.Bai J., Withycombe J., Eldridge R.C. Metabolic pathways associated with psychoneurological symptoms in children with cancer receiving chemotherapy. Biol Res Nurs. 2022;24(3):281–293. doi: 10.1177/10998004211069619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin H., Dudley W.N., Bhakta N., et al. Associations of symptom clusters and health outcomes in adult survivors of childhood cancer: a report from the St Jude Lifetime Cohort Study. J Clin Orthod. 2023;41(3):497–507. doi: 10.1200/JCO.22.00361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai J., Zhang W., Barandouzi Z.A. Human microbiome: understanding the role of the gut microbiome and implications for oncology nursing care. Clin J Oncol Nurs. 2021;25(4):383–387. doi: 10.1188/21.CJON.383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hockenberry M., Hooke M.C. Symptom clusters in children with cancer. Semin Oncol Nurs. 2007;23(2):152–157. doi: 10.1016/j.soncn.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Rydén I., Fernström E., Lannering B., et al. Neuropsychological functioning in childhood cancer survivors following cranial radiotherapy – results from a long-term follow-up clinic. Neurocase. 2022;28(2):163–172. doi: 10.1080/13554794.2022.2049825. [DOI] [PubMed] [Google Scholar]
- 6.Brinkman T.M., Recklitis C.J., Michel G., Grootenhuis M.A., Klosky J.L. Psychological symptoms, social outcomes, socioeconomic attainment, and health behaviors among survivors of childhood cancer: current state of the literature. J Clin Orthod. 2018;36(21):2190–2197. doi: 10.1200/JCO.2017.76.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly D.L., Lyon D.E., Yoon S.L., Horgas A.L. The microbiome and cancer: implications for oncology nursing science. Cancer Nurs. 2016;39(3):E56–E62. doi: 10.1097/NCC.0000000000000286. [DOI] [PubMed] [Google Scholar]
- 8.Suraya R., Nagano T., Kobayashi K., Nishimura Y. Microbiome as a target for cancer therapy. Integr Cancer Ther. 2020;19 doi: 10.1177/1534735420920721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai J., Behera M., Bruner D.W. The gut microbiome, symptoms, and targeted interventions in children with cancer: a systematic review. Support Care Cancer. 2018;26(2):427–439. doi: 10.1007/s00520-017-3982-3. [DOI] [PubMed] [Google Scholar]
- 10.Withycombe J.S., Eldridge R., Jin Y., Gu H., Castellino S.M., Sears D.D. Metabolites associated with fatigue and physical activity in childhood cancer. Biol Res Nurs. 2022;24(3):350–361. doi: 10.1177/10998004221085029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown A.L., Sok P., Taylor O., et al. Cerebrospinal fluid metabolomic profiles associated with fatigue during treatment for pediatric acute lymphoblastic leukemia. J Pain Symptom Manage. 2021;61(3):464–473. doi: 10.1016/j.jpainsymman.2020.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antoniadi K., Thomaidis N., Nihoyannopoulos P., et al. Prognostic factors for cardiotoxicity among children with cancer: definition, causes, and diagnosis with omics technologies. Diagnostics. 2023;13(11):1864. doi: 10.3390/diagnostics13111864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song B., Bai J. Microbiome-gut-brain axis in cancer treatment-related psychoneurological toxicities and symptoms: a systematic review. Support Care Cancer. 2021;29(2):605–617. doi: 10.1007/s00520-020-05739-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart N.H., Wallen M.P., Farley M.J., et al. Exercise and the gut microbiome: implications for supportive care in cancer. Support Care Cancer. 2023;31(12):724. doi: 10.1007/s00520-023-08183-7. [DOI] [PubMed] [Google Scholar]
- 15.Alexander J.L., Wilson I.D., Teare J., Marchesi J.R., Nicholson J.K., Kinross J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat Rev Gastroenterol Hepatol. 2017;14(6):356–365. doi: 10.1038/nrgastro.2017.20. [DOI] [PubMed] [Google Scholar]
- 16.Zitvogel L., Ma Y., Raoult D., Kroemer G., Gajewski T.F. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science. 2018;359(6382):1366–1370. doi: 10.1126/science.aar6918. [DOI] [PubMed] [Google Scholar]
- 17.Rajagopala S.V., Yooseph S., Harkins D.M., et al. Gastrointestinal microbial populations can distinguish pediatric and adolescent Acute Lymphoblastic Leukemia (ALL) at the time of disease diagnosis. BMC Genom. 2016;17:635. doi: 10.1186/s12864-016-2965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cozen W., Yu G., Gail M.H., et al. Fecal microbiota diversity in survivors of adolescent/young adult Hodgkin lymphoma: a study of twins. Br J Cancer. 2013;108(5):1163–1167. doi: 10.1038/bjc.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahlgren D., Lennernäs H. Review on the effect of chemotherapy on the intestinal barrier: epithelial permeability, mucus and bacterial translocation. Biomed Pharmacother. 2023;162 doi: 10.1016/j.biopha.2023.114644. [DOI] [PubMed] [Google Scholar]
- 20.Barandouzi Z.A., Eng T., Shelton J., et al. Associations of the gut microbiome with psychoneurological symptom cluster in women with gynecologic cancers: a longitudinal study. Support Care Cancer. 2023;31(11):626. doi: 10.1007/s00520-023-08058-x. [DOI] [PubMed] [Google Scholar]
- 21.Bilenduke E., Sterrett J.D., Ranby K.W., et al. Impacts of breast cancer and chemotherapy on gut microbiome, cognitive functioning, and mood relative to healthy controls. Sci Rep. 2022;12 doi: 10.1038/s41598-022-23793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hajjar J., Mendoza T., Zhang L., et al. Associations between the gut microbiome and fatigue in cancer patients. Sci Rep. 2021;11:5847. doi: 10.1038/s41598-021-84783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deleemans J.M., Chleilat F., Reimer R.A., et al. The chemo-gut pilot study: associations between gut microbiota, gastrointestinal symptoms, and psychosocial health outcomes in a cross-sectional sample of young adult cancer survivors. Curr Oncol. 2022;29(5):2973–2994. doi: 10.3390/curroncol29050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv D., Zou Y., Zeng Z., et al. Comprehensive metabolomic profiling of osteosarcoma based on UHPLC-HRMS. Metabolomics. 2020;16(12):120. doi: 10.1007/s11306-020-01745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bennett C.D., Kohe S.E., Gill S.K., et al. Tissue metabolite profiles for the characterisation of paediatric cerebellar tumours. Sci Rep. 2018;8 doi: 10.1038/s41598-018-30342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Höglund E., Øverli Ø., Winberg S. Tryptophan metabolic pathways and brain serotonergic activity: a comparative review. Front Endocrinol. 2019;10 doi: 10.3389/fendo.2019.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao M., Tuo H., Wang S., Zhao L. The effects of dietary nutrition on sleep and sleep disorders. Media Inflam. 2020;2020 doi: 10.1155/2020/3142874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elgarten C.W., Tanes C., Lee J.J., et al. Early stool microbiome and metabolome signatures in pediatric patients undergoing allogeneic hematopoietic cell transplantation. Pediatr Blood Cancer. 2022;69(1) doi: 10.1002/pbc.29384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dono A., Patrizz A., McCormack R.M., et al. Glioma induced alterations in fecal short-chain fatty acids and neurotransmitters. CNS Oncol. 2020;9(2) doi: 10.2217/cns-2020-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.González-Mercado V.J., Lim J., Marrero S., Pedro E., Saligan L.N. Gut microbiota and fatigue in rectal cancer patients: a cross-sectional pilot study. Support Care Cancer. 2021;29(8):4615–4621. doi: 10.1007/s00520-021-06013-2. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Mercado V.J., Lim J., Saligan L.N., et al. Gut microbiota and depressive symptoms at the end of CRT for rectal cancer: a cross-sectional pilot study. Depress Res Treat. 2021;2021 doi: 10.1155/2021/7967552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.González-Mercado V.J., Lim J., Yu G., et al. Co-occurrence of symptoms and gut microbiota composition before neoadjuvant chemotherapy and radiation therapy for rectal cancer: a proof of concept. Biol Res Nurs. 2021;23(3):513–523. doi: 10.1177/1099800421991656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai J., Eldridge R., Houser M., et al. Multi-omics analysis of the gut microbiome and metabolites associated with the psychoneurological symptom cluster in children with cancer receiving chemotherapy. J Transl Med. 2024;22(1):256. doi: 10.1186/s12967-024-05066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smirnov K.S., Maier T.V., Walker A., et al. Challenges of metabolomics in human gut microbiota research. Inter J Med Microbiol. 2016;306(5):266–279. doi: 10.1016/j.ijmm.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Singh R.K., Chang H.W., Yan D., et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matei B., Winters-Stone K.M., Raber J. Examining the mechanisms behind exercise's multifaceted impacts on body composition, cognition, and the gut microbiome in cancer survivors: exploring the links to oxidative stress and inflammation. Antioxidants. 2023;12(7):1423. doi: 10.3390/antiox12071423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowland G.B., Weyrich L.S. The oral-microbiome-brain axis and neuropsychiatric disorders: an anthropological perspective. Front Psychiatry. 2022;13 doi: 10.3389/fpsyt.2022.810008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data availability is not applicable to this article as no new data were created or analyzed in this study.