Abstract
Circulating plasma miRNAs have emerged as potential early predictors of glucometabolic disorders. However, their biomarker potential remains unvalidated in populations with diverse genetic backgrounds, races, and ethnicities. This study aims to validate the biomarker potential of plasma miR-9, miR-29a, miR-192, and miR-375 for early detection of prediabetes and type 2 diabetes mellitus (T2DM) in Nepali populations that represent distinct genetic backgrounds, races, and ethnicities. A total of 46 adults, categorized into healthy controls (n = 25), prediabetes (n = 9), and T2DM (n = 12) groups, were enrolled. Baseline sociodemographic, anthropometric, and clinical characteristics were collected. Fold change in plasma expression of all four miRNAs was quantified using RT-qPCR against the RNU6B reference gene. Their biomarker potential was determined by receiver operating characteristic (ROC) curve analysis. Multivariate discriminant function and hierarchical cluster analyses were used to evaluate the effectiveness of the miRNA panel in reclassifying study participants who were initially categorized according to their glucose tolerance status. Plasma expression of all four miRNAs was significantly upregulated in T2DM patients compared to normoglycemic controls. Furthermore, the expression of only miR-29a and miR-375 was upregulated in T2DM patients than in prediabetic individuals. Notably, only miR-192 expression was significantly upregulated in prediabetic individuals than in the normoglycemic controls. The miRNA expression profiles had the potential of reclassifying the participants into three original groups with an accuracy of 69.6 %. ROC curve analysis identified miR-192 as the predictor for both prediabetes and T2DM, while miR-9, miR-29a, miR-192, and miR-375 were predictive only for T2DM. The specific set of miRNA combinations significantly improved their predictive accuracy. This study validates the early predictive biomarker potential of plasma miR-9, miR-29a, miR-192, and miR-375 also in the Nepali population and paves the way for future translational studies to validate their utility in clinical laboratories.
Keywords: Biomarkers, miRNAs, Type 2 diabetes, RT‒qPCR, Manipal teaching hospital, Nepal
Highlights
-
•
Identification of Early Predictive Biomarkers: This study identifies circulating miR-192 as an early predictive biomarker for both prediabetes and T2DM, while miR-9, miR-29a, and miR-375 as biomarkers only for T2DM , with miR-29a having the strongest prediction potential.
-
•
Distinctive Population Focus: It targets the Nepali population, a South Asian group with unique genetic and phenotypic traits, to underscore the importance of population-specific variations in diabetes biomarkers.
-
•
Comprehensive Methodology: The study employs a thorough methodology, including clinical characterization of the participants, quantification of miRNA expression, and detailed statistical analyses such as multivariate discriminant, cluster, and ROC curve analyses to evaluate the biomarker potential.
-
•
Differentiationbetween Prediabetes and T2DM: The study distinguishes between prediabetes and T2DM by idenitfying key miRNA expression changes as glucose tolerance deteriorates, offering insights for targeted interventions.
List of abbreviations
- AUC
Area under the curve
- BMI
Body mass index
- cDNA
Complimentary Deoxyribonucleic acid
- eGFR
Estimated glomerular filtration rate
- FIN
Fasting serum insulin
- FSG
Fasting serum glucose
- HOMA1-IR
Homeostatic model assessment for insulin resistance
- HPLC
High-performance liquid chromatography
- K3EDTA
Tripotassium Ethylene Diamine Tetraacetic Acid
- MAP
Mean arterial pressure
- MDRD
Modificationof diet in renal disease
- miR/miRNA
MicroRNA
- PD
Prediabetes
- PMG
Post-meal glucose
- ROC curve
Reciever Operating Characteristics curve
- RPM
Rotation per minute
- SPSS
Statistical Package for the Social Sciences
- T2DM
Type 2 diabetes mellitus
- WC
Waist circumference
- WHR
Waist-hip ratio
- YI
Youden index
1. Introduction
Prediabetes and type 2 diabetes mellitus (T2DM) represent a spectrum of glucometabolic disorders associated with various abnormalities in glucose metabolism. These conditions are associated with insulin resistance and/or impaired insulin secretion, which results in persistent hyperglycemia and associated acute and chronic complications [1]. T2DM is a major global health problem, as it affects more than 400 million people worldwide, with a higher prevalence and earlier onset in Nepali and other South Asian populations [2,3]. It is often preceded by an intermediate state of hyperglycemia called prediabetes, which increases the risk of developing T2DM and cardiovascular diseases [4]. Early detection and intervention of prediabetes can prevent or delay the progression to T2DM and reduce the burden of morbidity and mortality. However, current diagnostic markers, such as fasting glucose and glycated hemoglobin (HbA1c), have limited sensitivity and specificity for identifying individuals at high risk of developing T2DM, especially in its early stages [5]. Therefore, there is an urgent need to discover and validate novel biomarkers that can accurately predict or detect the early onset and progression of prediabetes and T2DM.
MiRNAs are a class of small noncoding RNAs that regulate gene expression at the posttranscriptional level by binding to the 3’ untranslated regions of target messenger RNAs. They are involved in various biological processes, such as cell differentiation, proliferation, apoptosis, metabolism, and inflammation [6]. Recently, several miRNAs (e.g., let-7, miR-9, miR-29a, miR-124, miR-126, miR-143, miR-155, miR-192, miR-375, and miR-378) have been shown to regulate the basic biology and metabolic functions of the pancreas, liver, adipose tissue, and skeletal muscle and the pathogenesis of T2DM [7,8]. In addition to their usual site of expression, miRNAs are also released into extracellular fluids, such as plasma, where they are protected from degradation by binding to proteins or being encapsulated in vesicles [9]. Circulating miRNAs have emerged as promising biomarkers for various diseases, including prediabetes and T2DM, as they reflect the physiological and pathological conditions of the tissues from which they originate [10].
Several studies have shown that circulating plasma miRNAs are dysregulated in patients with T2DM compared to healthy controls, suggesting their potential role in the pathogenesis and diagnosis of T2DM [11]. For example, the expression profiles of miR-9, miR-29a, miR-192, and miR-375, which are involved in the regulation of insulin secretion, glucose metabolism, and inflammation, are shown to be altered in plasma samples from prediabetic and T2DM patients [12]. However, their expression levels and potential for early detection of prediabetes and T2DM patients have been shown to vary with population characteristics, such as ethnicity, genetic background, environmental factors, and comorbidities [13,14]. Therefore, it is important to clinically validate these findings from different external populations and to establish our population-specific diagnostic measures for these circulating miRNAs.
The expression profiles and biomarker potential of circulating miRNAs for glucometabolic disorders have not yet been explored in the Nepali population. The population of Nepal is a heterogeneous group of people with diverse ethnicities, cultures, and lifestyles. Due to its distinct South Asian phenotype, which includes early-stage hyperinsulinemia, a greater degree of insulin resistance, and possibly an early decline in β-cell functions, the Nepali population also has a high susceptibility to and prevalence of obesity, prediabetes, and T2DM [2]. Therefore, using a representative sample of adult Nepali individuals with varying levels of glucose tolerance, this study aimed to evaluate the biomarker potential of plasma miR-9, miR-29a, miR-192, and miR-375 for the early detection of prediabetes and T2DM.
2. Materials and methods
2.1. Study participants and settings
A total of 46 Nepali adults who visited Manipal Teaching Hospital, Pokhara, Nepal, either as caretakers or patients were enrolled and divided into three study groups: (a) healthy controls with normal glucose tolerance and a BMI <27.5 kg/m2 (n = 25), (b) patients with prediabetes and impaired fasting glycemia and/or glucose tolerance (n = 9), and (c) patients with T2DM who were diagnosed within the last three months with no known complications (n = 12). Prediabetes and T2DM were diagnosed according to the American Diabetes Association criteria [15]. Prediabetes was defined as a fasting glucose level between 100 and 126 mg/dL (impaired fasting glucose, IFG) and/or a 2-h postprandial glucose level between 140 and 200 mg/dL (impaired glucose tolerance, IGT) and a glycated hemoglobin (HbA1c) level between 5.7 and 6.4 %. T2DM was defined as a fasting serum glucose level ≥ 126 mg/dL or a 2-h post-meal glucose level ≥ 200 mg/dL and an HbA1c level >6.5 %.
2.2. Collection of baseline sociodemographic, anthropometric, and clinical data
Baseline sociodemographic, anthropometric, and clinical data were collected using a set of pre-validated structured questionnaires, measurement tools such as measuring tape, stadiometer, digital scale, sphygmomanometer, and personal medical records by trained laboratory personnel and postgraduate medical students. Participants with a known history or clinical evidence of active inflammatory diseases, vascular events (e.g., stroke, unstable angina pectoris, acute myocardial infarction, or coronary artery disease), cancer, Cushing's syndrome, hyperpituitarism, hepatic/renal dysfunctions, and active treatment with plasma glucose-lowering drugs (e.g., sulfonyl and non-sulfonyl urea, thiazolidinediones, sodium-glucose transporter-2 inhibitors, incretin mimetics, dipeptidyl peptidase-4 inhibitors, and α-glucosidase inhibitors) and lipid-lowering drugs (e.g., statins, fibrates, bile acid sequestrants, vitamin B3, microsomal transfer protein inhibitors, omega-3 acids and 2-azetidione) were excluded from the study due to their potential influence on β-cell functions and glucose metabolism. Participants who were diagnosed with T2DM three months ago or who had evidence of any acute or chronic complications of diabetes were also excluded from the study.
2.3. Sample collection, processing, and storage
Venous blood samples (∼10 ml) were collected from all participants early in the morning after an 8–12 h overnight fasting and divided into three aliquots: (a) one in a plain gel tube for serum separation, (b) one in a K3EDTA tube for plasma separation and (c) one in another K3EDTA tube for whole blood separation. The first two aliquots were centrifuged at 4000 rpm for 10 min, and the separated serum or plasma was stored in DNase/RNase-free tubes at −20 °C until analysis. A whole blood aliquot was used to determine HbA1c.
2.4. Biochemical measurements
All the baseline biochemical analyses were carried out at the Department of Clinical Biochemistry, Manipal Teaching Hospital, Pokhara, Nepal. HbA1c level was estimated using a Bio-Rad HPLC D-10 hemoglobin assay system (Bio-Rad, USA). Serum levels of glucose, creatinine and lipid profile parameters were estimated using a VITROS 350 chemistry analyzer (Ortho Clinical Diagnostics, UK). The Friedewald equation, LDL-C = TC-HDL-C-TG/5, was used to estimate low-density lipoprotein cholesterol (LDL-C) level [16]. Fasting serum insulin level was measured by electrochemiluminescence immunoassay (eCLIA) using a Vitros ECiQ immunoassay analyzer (Ortho Clinical Diagnostics, UK). The homeostasis model assessment estimated insulin resistance (HOMA1-IR) index was calculated using the following formula: HOMA1-IR = fasting plasma insulin (μU/ml) × fasting plasma glucose (mmol/L)/22.5 [17]. The estimated glomerular filtration rate (eGFR) was estimated using the serum creatinine level and modified diet in renal disease (MDRD) equation [18]. Analytical variability was minimized by using the same lot of reagents, instrument calibration, and quality control measures.
2.5. Isolation and reverse transcription of plasma miRNAs
Isolation and reverse transcription of plasma miRNAs were carried out at the Central Department of Biotechnology, Tribhuvan Univesity, Kathmandu, Nepal. Total RNA, including miRNAs, was isolated from the thawed plasma samples using Direct-zol RNA Microprep Kits according to the protocol provided by the reagent kit manufacturer (Zymo Research, catalog number: R2060, USA). The concentrations and purities of the isolated RNAs were assessed using a NanoDrop one/one C Microvolume UV–Vis spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). All isolated RNA samples were stored at −80 °C until use.
The isolated miRNAs were reverse transcribed using an iScript cDNA Synthesis Kit (Bio-Rad, USA). Briefly, the reaction mixture contained 4 μL of 5x iScript reaction mix, 1 μL of iScript reverse transcriptase, 10 μL of nuclease-free water, and 5 μL of RNA template. These reaction mixtures were then incubated in a thermal cycler (Bio-Rad T100 Thermal Cycler), and cDNA was synthesized by the following steps: (i) priming at 25 °C for 5 min, (ii) reverse transcription at 45 °C for 20 min, (iii) RNA inactivation for 2 min at 95 °C, and (iv) an optional step of holding the mixture at 40 °C. The concentrations and purity of the cDNA were assessed using a NanoDrop One/One C Microvolume UV–Vis spectrophotometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA), and the cDNA was stored at −80 °C until use.
2.6. Real-time RT‒qPCR
The forward and reverse primer sequences (Table 1) targeting the four miRNAs and RNU6B genes (used for normalization) were obtained from published literature [[19], [20], [21]] and purchased from Macrogen, Inc. (Seoul, South Korea). The lyophilized primers were serially diluted with nuclease-free water to prepare stock (100 μM) and working (10 μM) solutions, which were stored at −20 °C until use.
Table 1.
Forward and reverse primer sequences used for qPCR amplification of miRNAs.
| RNAs | Forward Sequence, 5′-3′ | Reverse Sequence, 5′-3′ | Ref. |
|---|---|---|---|
| miR-9 | GCCCGCTCTTTGGTTATCTAG | CCAGTGCAGGGTCCGAGGT | [19] |
| miR-29a | CGCGGATCCTGGATTTAGTAAGATTTGGGC | CCGGAATTCACATGCAATTCAGGTCAGTG | [20] |
| miR-192 | CTGACCTATGAATTGACAGCCA | GCTGTCAACGATACGCTACGT | [21] |
| miR-375 | GAGCATTTTGTTCGTTCGGC | AGTGCAGGGTCCGAGG | [19] |
| RNU6B | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT | [19] |
RT-qPCRs of the reverse transcribed miRNAs were performed in duplicate using the Azure cielo DX real-time PCR system (Azure Biosystems, USA). qPCR was performed at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min with a melting curve from 50 °C to 95 °C. The specificity of the PCR for each miRNA was verified using its melting curve and amplification plot. Only the Ct values with bell-shaped melting peaks at approximately 80 °C were accepted. Ct values from qPCR assays with >35 cycles were treated as not expressed. qPCR amplification of the genes for RNU6B and the four miRNAs was also performed, and the results were visualized by 1.5 % agarose gel electrophoresis using a 100-bp ladder (Solis BioDyne) for 30 min.
2.7. Calculation of the fold change in plasma miRNA expression
The expression levels of plasma miRNAs were normalized to those of the RNU6B gene, which encodes a small noncoding nuclear RNA that is abundantly expressed in various human tissues and organs [21]. The fold change in expression values was calculated using the 2−ΔΔCt (cycle threshold) method as previously described [22]. ΔΔCt denotes the difference in ΔCt values between test groups (T2DM or prediabetes) and the control group (normoglycemic healthy controls) (ΔΔCt = ΔCtTest-ΔCtControl), and ΔCt is the difference in Ct values between the target groups and RNU6B (ΔCt = Ct Target-Ct RNU6B). A fold change <1 was defined as underexpressed, and >1 was defined as overexpressed.
2.8. Classification potential and cluster analysis of differentially expressed miRNAs
Agglomerative hierarchical cluster analysis of the study participants was carried out using a heatmap created with OriginLab Pro 2023b. The cluster rows and columns were adjusted using the group average method. The hierarchical clustering of genes and the construction of sample trees were based on the Euclidean distance metric. The probability of how accurately participants were assigned to their predefined study groups based on plasma miRNA levels was determined using multivariate discriminant function analysis.
2.9. Statistical analysis
Statistical analysis was performed using SPSS version 26.0 (IBM Corp, Armonk, NY, USA), R version 4.2.3, and OriginLab Pro 2023b. Data normality was tested using the Kolmogorov‒Smirnov test. As the majority of the data were non-normally distributed, nonparametric tests were used for all analyses. Descriptive statistics are presented as means, medians, and standard deviations. The nonparametric Mann‒Whitney U test was used to compare values between any two study groups. Spearman bivariate correlation analysis was performed to evaluate the strength of linear monotonic associations between plasma expression levels of miRNA and T2DM risk factors. Hierarchical cluster analysis was performed using group average and Euclidean distance metric methods. To assess the accuracy of participant classification based on plasma miRNA expression profiles, canonical discriminant function analysis was performed. Receiver operating characteristic (ROC) curve analysis was used to evaluate the predictive biomarker potential of individual and combined miRNAs. The 95 % confidence intervals (CIs) for the area under the curves (AUCs) were determined using the binomial exact confidence interval method. All hypothesis tests were two-tailed, with p-values <0.05 considered statistically significant.
3. Results
3.1. Baseline characteristics of the study participants
Of the 46 adult participants enrolled, 54.4 % (n = 25) had normal glucose tolerance, 19.6 % (n = 9) had prediabetes, and 26.1 % (n = 12) had newly diagnosed T2DM. Importantly, the gender distribution was almost equal, with males comprising 52.2 % (n = 24) and females making up 47.8 % (n = 22) of the cohort. Pairwise nonparametric hypothesis testing revealed that participants with prediabetes and T2DM had significantly greater (p < 0.05) distributions of several baseline characteristics, such as BMI, waist circumference, waist-to-hip ratio, mean arterial blood pressure, HbA1C, fasting serum insulin, glucose, and triglyceride levels, as well as HOMA1-IR, than healthy controls. A similar pairwise hypothesis test between the prediabetes and T2DM groups revealed that the former had significantly (p < 0.05) different distribution values, specifically for serum total cholesterol and estimated glomerular filtration rate (eGFR). However, the three study groups did not differ significantly in terms of their mean age or serum LDL-C and creatinine levels (p > 0.05) (Table 2).
Table 2.
Summary statistics on baseline demographic, clinical and metabolic characteristics by the glycemic status.
| Test variables | Control (I) (n = 25) |
Prediabetes (II) (n = 9) |
T2DM (III) (n = 12) |
p-values |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Median | Mean ± SD | Median | Mean ± SD | Median | Mean ± SD | I vs II | I vs III | II vs III | |
| Age (years) | 42.0 | 42.2 ± 7.7 | 50.0 | 49.1 ± 8.5 | 43.0 | 43.1 ± 8.6 | 0.086 | 0.953 | 0.221 |
| Sex Male, n (%) | 12 (26.1) | 5 (10.9) | 7 (15.2) | ||||||
| Female, n (%) |
13 (28.3) |
4 (8.7) |
5 (10.9) |
||||||
|
Anthropometry | |||||||||
| BMI (kg/m2) | 22.5 | 24.1 ± 3.9 | 27.6 | 28.8 ± 5.7 | 27.1 | 28.7 ± 3.4 | 0.012 | 0.004 | 0.803 |
| WC (cm) | 85.0 | 87.3 ± 10.2 | 99.0 | 97.4 ± 11.0 | 97.5 | 98.8 ± 5.4 | 0.018 | 0.001 | 0.972 |
| WHR |
0.91 |
0.91 ± 0.07 |
0.94 |
0.97 ± 0.07 |
0.96 |
0.98 ± 0.06 |
0.105 |
0.011 |
0.498 |
|
Blood pressure | |||||||||
| SBP (mmHg) | 116.0 | 114.0 ± 10.2 | 120.0 | 123.8 ± 10.9 | 122.0 | 121.5 ± 11.5 | 0.031 | 0.068 | 0.748 |
| DBP (mmHg) | 76.0 | 76.4 ± 7.5 | 86.0 | 85.8 ± 8.9 | 79.5 | 80.1 ± 7.9 | 0.011 | 0.187 | 0.126 |
| MAP (mmHg) |
89.7 |
88.9 ± 8.0 |
100.7 |
98.5 ± 8.0 |
92.0 |
93.9 ± 8.6 |
0.012 |
0.205 |
0.419 |
|
Metabolic profile | |||||||||
| FSG (mm/dl) | 89.0 | 88.0 ± 7.3 | 106.0 | 106.7 ± 8.6 | 112.0 | 110.7 ± 18.3 | 0.000 | 0.001 | 0.477 |
| 2h PMG (mg/dl) | 99.0 | 7.6 ± 16.0 | 136.0 | 134.8 ± 36.5 | 140.0 | 146.6 ± 38.7 | 0.008 | 0.000 | 0.702 |
| HbA1C (%) | 5.2 | 5.1 ± 0.4 | 5.8 | 5.8 ± 0.6 | 6.4 | 6.6 ± 1.3 | 0.001 | 0.000 | 0.087 |
| FIN (mU/L) | 7.0 | 9.5 ± 9.6 | 9.2 | 10.8 ± 3.9 | 9.7 | 11.7 ± 6.6 | 0.072 | 0.048 | 0.862 |
| HOMA1-IR |
1.5 |
2.1 ± 2.31 |
2.3 |
2.84 ± 1.04 |
2.6 |
3.15 ± 1.68 |
0.009 |
0.007 |
0.776 |
|
Lipid profile | |||||||||
| TC (mg/dl) | 177.0 | 176.6 ± 29.9 | 206.0 | 193.3 ± 28.7 | 181.0 | 167.4 ± 32.4 | 0.056 | 0.77 | 0.043 |
| HDL-C (mg/dl) | 51.0 | 49.4 ± 13.3 | 42.0 | 43.7 ± 8.4 | 36.5 | 38.6 ± 10.6 | 0.348 | 0.02 | 0.225 |
| LDL-C (mg/dl) | 103.0 | 101.8 ± 25.6 | 113.0 | 104.6 ± 26.7 | 92.0 | 93.8 ± 27.6 | 0.494 | 0.581 | 0.374 |
| TG (mg/dl) |
105.0 |
127.0 ± 65.4 |
223.0 |
224.7 ± 80.9 |
183.5 |
207.4 ± 98.8 |
0.002 |
0.005 |
0.382 |
|
Renal function tests | |||||||||
| Creatinine (mg/dl) | 0.7 | 0.76 ± 0.18 | 0.8 | 0.87 ± 0.17 | 0.7 | 0.74 ± 0.16 | 0.068 | 0.853 | 0.078 |
| eGFRCr (ml/min/1.73m2) | 97.0 | 99.9 ± 18.7 | 75.9 | 79.3 ± 12.1 | 100.1 | 106.3 ± 21.9 | 0.002 | 0.471 | 0.002 |
The values are presented either as numbers and percentages (%) for categorical data, or median and mean ± standard deviation for continuous data. Mean arterial pressure (MAP) was calculated using the formula: MAP = DBP + 1/3(SBP – DBP). PD: Prediabetes, T2DM: Type 2 diabetes mellitus, BMI: Body mass index, WC: Waist circumference, WHR: Waist-to-hip ratio, SBP: Systolic blood pressure, DBP: Diastolic blood pressure, FSG: Fasting serum glucose, 2hPMG: Two hours post-meal glucose, HbA1C: Glycated hemoglobin, FIN: Fasting serum insulin, HOMA1-IR: Homeostasis model assessment of insulin resistance, TC: Total cholesterol, HDL-C: High-density lipoprotein cholesterol, LDL-C: Low-density lipoprotein cholesterol, TG: Triglycerides, eGFRcr: Serum creatinine-based glomerular filtration rate estimated using a modification of diet in renal disease (MDRD) equation. The group variables were compared pairwise using Mann-Whitney U tests.
3.2. Detection of circulating plasma miRNAs
The mean concentrations of plasma total RNA in healthy controls, prediabetic and T3DM patients were 6.04 ± 3.83 ng/μl, 7.04 ± 3.37 ng/μl, and 7.13 ± 3.41 ng/μl, respectively, and did not differ significantly (p > 0.05) from each other (Supplementary Figure 1). The average Ct values determined for miR-9, miR-29a, miR-192, and miR-375 and a control gene (RNU6B) used for normalization were 35.99, 38.21, 36.94, 40.77, and 31.39, respectively. The amplification curves were generated for all 46 samples for RNUB6, 44 for miR-9 and miR-192, 43 for miR-375, and 18 for miR-29a (Supplementary Table 1). The presence of amplified RNUB6 and four miRNA genes was further confirmed by 1.5 % agarose gel electrophoresis. A band corresponding to RNU6B was detected near the 100-bp ladder, and miRNA bands were detected even below RNU6B, confirming the expression of all four miRNAs (Supplementary Figure 2).
3.3. Fold change in plasma miRNA expression according to glucose tolerance status
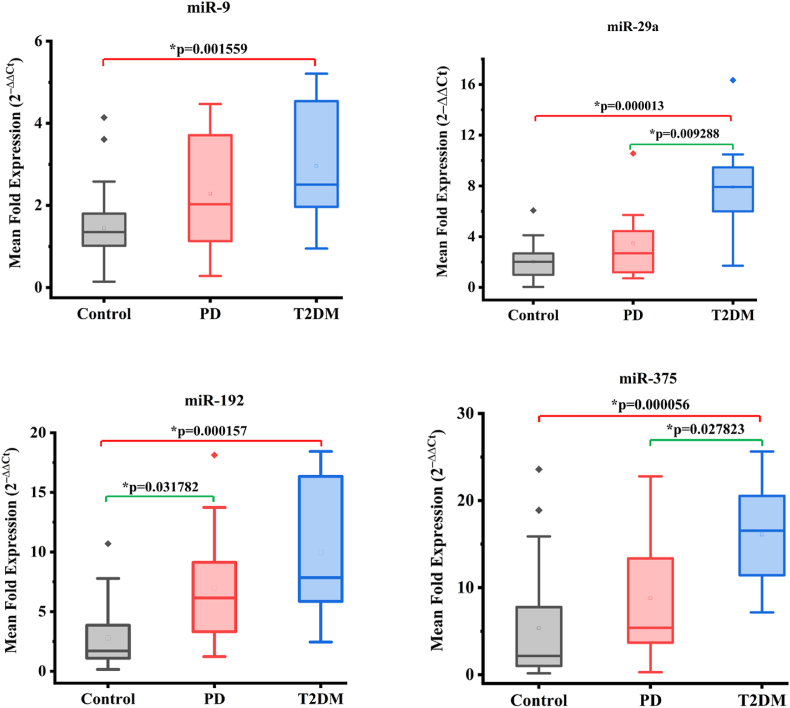
The relative expression levels of each normalized miRNA for all three study groups are presented in the boxplots (Fig. 1). Compared to healthy controls, T2DM patients showed significantly upregulated expression of plasma miR-9 (1.45 ± 0.95 vs 2.96 ± 1.46, p = 0.001559), miR-29a (2.02 ± 1.42 vs 7.93 ± 3.60, p = 0.000013), miR-375 (5.37 ± 6.39 vs 16.11 ± 5.80, p = 0.000056), and miR-192 (2.80 ± 2.67 vs 9.97 ± 6.03, p = 0.000157). Furthermore, T2DM patients also exhibited significantly greater expression of miR29a (3.46 ± 3.16 vs 7.93 ± 3.60, p = 0.009288) and miR-375 (8.82 ± 7.44 vs. 16.11 ± 5.80, p = 0.027823) than did prediabetes patients (Fig. 1). There was no significant difference in the plasma mean fold change in the expression of miR-9 (1.45 ± 0.95 vs. 2.29 ± 1.54, p = 0.128), miR-29a (2.02 ± 1.42 vs. 3.46 ± 3.16, p = 0.274), or miR-375 (5.37 ± 6.39 vs. 8.82 ± 7.44, p = 0.130) between healthy controls and prediabetic participants. Only miR-192 expression was significantly greater in prediabetic participants (2.80 ± 2.67 vs. 6.98 ± 5.77, p = 0.0318) than in healthy controls (Fig. 1).
Fig. 1.
Box plots for the mean fold change in the expression levels of normalized miRNAs by the glycemic status. Control refers to healthy individuals with normal glucose tolerance, PD refers to prediabetes with impaired fasting glucose and/or impaired glucose tolerance, and T2DM refers to the patients with type 2 diabetes mellitus. The group means were compared pairwise using Mann-White U tests.
3.4. Linear association of T2DM risk factors with plasma expression levels of miRNAs
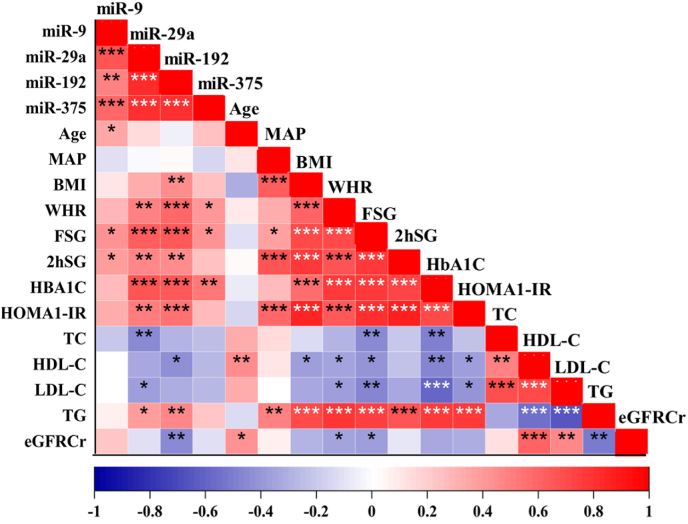
All four miRNAs showed a strong positive correlation with each other (p < 0.001). miR-29a and miR-192 showed significant positive correlations (p < 0.05) with several risk factors for insulin resistance and T2DM, including the waist-hip ratio, serum fasting and postprandial glucose levels, HbA1C%, HOMA1-IR, and plasma triglyceride level. However, miR-29a showed a negative correlation (p < 0.05) with serum total cholesterol and LDL-cholesterol levels. Similarly, miR-192 showed a negative correlation with HDL-cholesterol levels and estimated glomerular filtration rate (eGFRcr) values. In comparison, plasma miR-9 and miR-375 correlated with fewer T2DM risk factors than did miR-29a and miR-192. miR-9 showed significant positive correlations with age and serum glucose levels, while miR-375 showed significant positive correlations with WHR, fasting serum glucose, and HbA1c levels (Fig. 2).
Fig. 2.
The correlation matrix showing the monotonic relationship between the mean fold expression of plasma miRNAs and baseline test variables. The bright red color indicates the highest positive correlation while the dark blue color indicates the lowest negative correlation. The significant correlation coefficients are flagged with × p < 0.05, **p < 0.01, ***p < 0.001.
3.5. Plasma miRNA-based clustering and classification of study participants
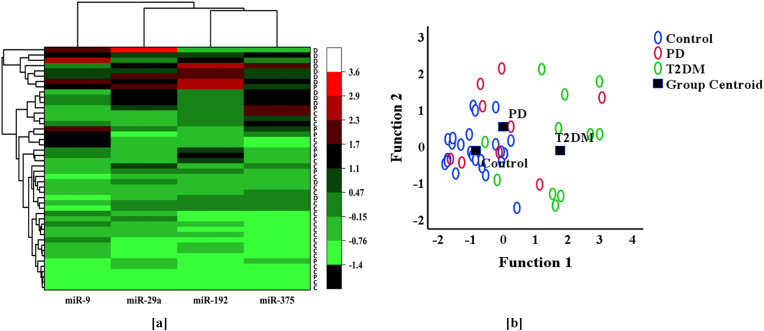
Based on the expression pattern of each miRNA, the study participants could be divided into three clusters (Fig. 3a), which were largely consistent with their predefined groups. Discriminant analysis revealed that the plasma expression levels of each miRNA could serve as a significant discriminant of the original grouping based on glycemic status. Among the four miRNAs, miR-29a had the highest correlation coefficient, and miR-9 had the lowest standardized canonical discriminant function coefficient (Table 3).
Fig. 3.
[a] Hierarchical clustering of study groups based on their specific miRNA expression. Heatmap analyses showing the differential expression pattern of plasma circulating miRNAs in the study subjects. The color scale on the right-hand side shows saturation and brightness based on the mean values of the fold change in miRNA expression. The red color with maximum intensity indicates the highest fold change in the plasma expression while the green color with maximum intensity indicates the reverse. C: Normoglycemic healthy controls, P: Prediabetic individuals, and D: Patients with type 2 diabetes mellitus. [b] Scatter plot showing canonical discriminant functions, Control: Normoglycemic healthy control, PD: Prediabetes, T2D: Type 2 diabetes.
Table 3.
Data of canonical discriminant analysis.
| miRNAs |
Wilkis lambda |
p-value |
STD CDF1 Coefficients* |
Classification function coefficients |
||
|---|---|---|---|---|---|---|
| Control | PD | T2DM | ||||
| miR-9 | 0.766 | 0.003 | 0.132 | 0.825 | 1.189 | 1.106 |
| miR-29a | 0.488 | 0.000 | 0.684 | 0.093 | 0.145 | 0.803 |
| miR-192 | 0.654 | 0.000 | 0.223 | 0.004 | 0.177 | 0.136 |
| miR-375 | 0.657 | 0.000 | 0.284 | 0.093 | 0.102 | 0.207 |
| Constant | −2.043 | −3.778 | −8.264 | |||
STD CDF1=Standardized canonical discriminant function 1, PD=Prediabetes, T2DM = Type 2 diabetes mellitus.
Overall, 69.6 % of the originally grouped cases were correctly classified. Specifically, 72.0 % of the participants were accurately assigned to the control group, 55.6 % to the prediabetes group, and 75.0 % to the type 2 diabetes group. The scatter plot between function 1 and function 2 is shown in Fig. 3b. It was found that in Function 1 (p = 0.000, Wilks Lambda), the centroid of normoglycemic controls (−0.845) was relatively closer to the centroid of prediabetes (0.000) but had greater controls (−0.845) and was relatively closer to the centroid of prediabetes (0.000) but had a greater difference from the centroid (1.761) of type 2 diabetes.
3.6. Biomarker potential of plasma miRNAs for the detection of prediabetes and T2DM
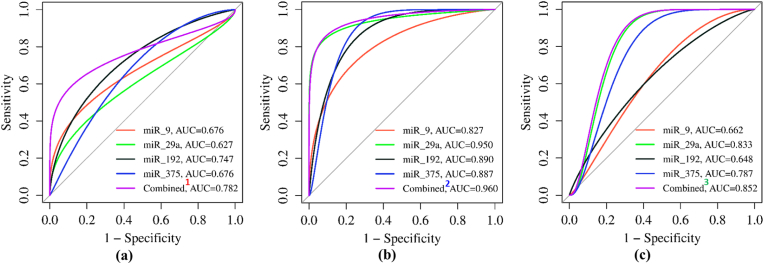
We evaluated the biomarker potential of individual plasma miRNAs and their combinations using receiver operating characteristic (ROC) curve analysis (Fig. 4a–c). Of the four miRNAs examined, only miR-192 showed significant potential in distinguishing prediabetic patients from normoglycemic healthy controls (AUC value: 0.747, p = 0.032). This distinction was supported by its YI (0.498), sensitivity (77.8 %), and specificity (72.0 %). Combining the information from all four plasma miRNAs further improved the AUC (0.782, p = 0.013), YI (0.681), sensitivity (77.8 %), and specificity (84.0 %) (Fig. 4, Table 4).
Fig. 4.
The Receiver Operating Characteristic (ROC) curve analyses of individual and combined plasma miRNA for their potential to discriminate between (a) normoglycemia vs prediabetes, (b) normoglycemia vs type 2 diabetes, and (c) prediabetes vs type 2 diabetes states. Combined1: Combination of miRs-9+29a+192 + 375; Combined2: Combination of miRs-29a+375; Combined3: Combination of miRs-9+29a+375, AUC=Area under the curve.
Table 4.
The AUCs, sensitivity, and specificity of plasma miRNAs for detecting prediabetes and type 2 diabetes.
| Target gene | AUC | 95 % CI | p-value | YI | Criterion | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|
|
Normal vs Prediabetes | |||||||
| miR-9 | 0.676 | 0.438–0.913 | 0.123 | 0.436 | ≥1.87 | 55.6 | 88.0 |
| miR-29a | 0.627 | 0.391–0.862 | 0.266 | 0.324 | ≥3.36 | 44.4 | 88.0 |
| miR-192 | 0.747 | 0.556–0.937 | 0.032 | 0.498 | ≥3.01 | 77.8 | 72.0 |
| miR-375 | 0.676 | 0.471–0.881 | 0.123 | 0.489 | ≥2.57 | 88.9 | 60.0 |
| miR-9+29a+192+375 |
0.782 |
0.555–1.00 |
0.013 |
0.618 |
≥0.33 |
77.8 |
84.0 |
|
Normal vs Diabetes | |||||||
| miR-9 | 0.827 | 0.677–0.977 | 0.000 | 0.630 | ≥2.02 | 75.0 | 88.0 |
| miR-29a | 0.950 | 0.864–1.000 | 0.000 | 0.877 | ≥4.19 | 91.7 | 96.0 |
| miR-192 | 0.890 | 0.787–0.993 | 0.000 | 0.673 | ≥5.61 | 83.3 | 84.0 |
| miR-375 | 0.887 | 0.781–0.992 | 0.000 | 0.673 | ≥11.01 | 83.3 | 84.0 |
| miR-29a+375 |
0.960 |
0.892–1.000 |
0.000 |
0.877 |
≥0.25 |
91.7 |
96.0 |
|
Prediabetes vs Diabetes | |||||||
| miR-9 | 0.662 | 0.421–0.903 | 0.214 | 0.334 | ≥2.18 | 66.7 | 66.7 |
| miR-29a | 0.833 | 0.632–1.000 | 0.009 | 0.639 | ≥6.16 | 75.0 | 88.9 |
| miR-192 | 0.648 | 0.406–0.890 | 0.256 | 0.277 | ≥4.77 | 83.3 | 44.4 |
| miR-375 | 0.787 | 0.570–1.000 | 0.028 | 0.556 | ≥6.28 | 100.0 | 55.6 |
| miR-9+29a+375 | 0.852 | 0.671–1.000 | 0.007 | 0.695 | ≥0.37 | 91.7 | 77.8 |
The AUCs for each plasma miRNA were determined using non-parametric methods. The 95 % confidence intervals for AUCs were determined using the binomial exact confidence interval method. The Youden index (YI) values were calculated using the formula: YI= Sensitivity-(1-Specificity). The sets of combined microRNAs included under each study group represent the combinations that yielded the highest AUCs out of all possible combinations. All tests were two-tailed and a p-value <0.05 was considered statistically significant. ROC: Repeater Operating Characteristic, AUC: Area under the curve, CI: Confidence interval, YI: Youden index.
In the context of distinguishing T2DM patients from healthy controls, all four miRNAs demonstrated significant potential (AUC: >0.82) (Fig. 4, Table 4). Notably, miR-29a was found to be the most promising candidate with the highest AUC (0.950, p = 0.000), YI (0.877), sensitivity (91.7 %), and specificity (96.0 %). Although the combination of miR-29a with miR-375 increased the discriminatory potential, with an AUC of 0.960 (p = 0.000), this combination did not improve the YI (0.877), sensitivity (91.7 %), or specificity (96.0 %). Furthermore, both miR-29a and miR-375 showed significant potential in distinguishing T2DM patients from prediabetes patients. miR-29a showed the highest discriminatory potential based on its AUC (0.833, p = 0.009), YI (0.639), sensitivity (75.0 %), and specificity (88.9 %). Combining miR-29a with miR-9 and miR-375 further improved the AUC (0.852, p = 0.007), YI (0.695), sensitivity (91.7 %), and specificity (77.8 %)
4. Discussion
This study is the first to validate the biomarker potential of plasma-circulating miR-9, miR-29a, miR-192, and miR-375 for the early detection of glucometabolic disorders in the Nepalese population. Previous research has shown that ethnicity, genetic background, and dietary habits can significantly influence plasma miRNA expression profiles, necessitating validation across diverse populations before implementation in clinical diagnostics [13,14]. The Nepali population, with its distinct ethnicities, genetic backgrounds, dietary habits, and unique geographic locations, presents an important cohort for such validation. Our findings demonstrate that expression of all four plasma miRNAs is significantly upregulated in T2DM patients, while only miR-192 in prediabetic individuals when compared to normoglycemic controls.
miR-9, a pancreas-specific miRNA, plays an important role in glucose-stimulated insulin secretion by directly down-regulating the expression of target genes for Onecut-2 (OC-2) and Sirtuin 1(Sirt1) which are the negative regulator of insulin secretion [23,24]. Its overexpression in our T2DM patients, but not in prediabetics, suggests that impairment of glucose-stimulated insulin secretion and associated metabolic derangement occur lately when the prediabetes state progresses to overt T2DM. This hypothesis is further supported by the significant correlation of plasma miR-9 levels with a limited number of T2DM risk factors, specifically age and plasma glucose level, but not with obesity, long-term glycemic status, lipid profile parameters, or insulin resistance indices. A similar study on the Chinese population corroborated our findings, suggesting that plasma miR-9 levels in prediabetic individuals remain relatively stable compared to those with normal glycemia, indicating a potential plateau in expression during the early stages of glucose intolerance [25].
Our study showed a significant correlation of miR-29a with markers of obesity, insulin resistance, glycemic status, dyslipidemia, and overexpression in T2DM patients relative to both healthy and prediabetic individuals. Studies have demonstrated that miR-29a is abundantly expressed in metabolically active organs, and regulates glucose-stimulated insulin secretion, beta cell function, and glucose uptake in peripheral tissues. Its upregulated expression in T2DM patients thus potentially contributes to insulin resistance and T2DM development [[26], [27], [28]]. A similar study in the Chinese population has also shown almost five-fold higher expression of serum miR-29a in type 2 diabetic patients when compared to individuals with normal glucose tolerance [25].
miR-192 is expressed by many organs but is expressed mostly in the liver and pancreas. It was found significantly overexpressed in both prediabetes and T2DM patients relative to healthy controls. It showed a significant correlation with several markers of insulin resistance such as BMI, WHR, blood glucose and HDL-C levels, HbA1c%, HOMA1-IR, and renal function marker such as eGFRcr value. Its significant overexpression in both prediabetic and T2DM patients suggests its early involvement in the development of glucose intolerance and progression to T2DM and associated complications [29,30]. While some studies show increased miR-192 expression in prediabetes but not T2DM, others report higher folds of expression in T2DM compared to normal or prediabetic individuals [30,31].
The miR-375, expressed abundantly in the pancreas, regulates α and β cell mass and insulin [[32], [33], [34]]. Its overexpression both in animal models and humans leads to hyperglycemia and a decrease in β-cell mass suggesting it to be another potential biomarker of type 2 diabetes [32,35]. We also found miR-375 significantly correlated with the markers of insulin resistance such as WHR, plasma glucose level, HbA1C%, and number of components of MetS and overexpression in T2DM patients compared to both healthy and prediabetic individuals. Studies in Chinese and Bahraini populations have linked plasma miR-375 levels to insulin resistance and diabetes onset [19,36]. A five-year follow-up study in Spanish adults confirmed that miR-375 dysregulation precedes prediabetes and T2DM, suggesting its potential as a predictive biomarker for high-risk individuals [37].
The collective dysregulated expression of these miRNAs could disrupt their harmonious interaction and synergistic roles in regulating insulin release and glucose metabolism, potentially contributing to the onset of prediabetes and eventual progression to T2DM [[38], [39], [40]]. This underscores the complex nature of T2DM development and emphasizes the potential use of these miRNAs as early biomarkers for glucometabolic disorders.
Despite alignment in the direction of dysregulation between our study and others, we observed slight discrepancies in the expression patterns of these miRNAs among the three study groups. These discrepancies may be attributed to variations in race, ethnicity, and age demographics of study participants, the complexity and duration of disease progression, criteria employed for prediabetes and T2DM classification, and the types of samples used, along with the possible existence of undiagnosed comorbidities. Previous studies have indeed documented that race, ethnicity, genetic background or geographical location can confound plasma miRNA expression profiles [13,14].
To assess the diagnostic potential of these four miRNAs and their selected combinations, we employed ROC curve analysis. MiR-9 did not emerge as a strong early predictive marker for either prediabetes or T2DM due to its comparatively lower AUC, Youden index, and sensitivity. Conversely, miR-29a showed strong potential to distinguish T2DM patients from both prediabetic and healthy individuals, exhibiting the highest AUC, Youden index, sensitivity, and specificity. This indicates that miR-29a could act as an early predictive biomarker for T2DM diagnosis. The AUC value of 0.950 for miR-29a in T2DM patients versus healthy controls surpasses reported values for other miRNAs or traditional markers for T2DM diagnosis [41]. Although combining miR-29a with miR-375 marginally improved the AUC value to 0.960, it did not alter the sensitivity or specificity, suggesting that miR-29a alone might be adequate for accurately identifying patients with T2DM.
The significant ability of plasma miR-192 to distinguish prediabetes patients from healthy controls suggests its potential as a biomarker for detecting prediabetes. Its AUC value of 0.747 is comparable to reported values for other miRNAs or traditional markers for prediabetes detection [31]. The combination of all four miRNAs increased the AUC to 0.782 but did not enhance sensitivity or specificity, indicating that miR-192 alone might be effective in moderately accurate identification of prediabetes patients. Plasma miR-375 levels demonstrated excellent potential to differentiate T2DM patients from healthy controls with relatively high sensitivity and specificity, and from prediabetic individuals with 100 % sensitivity. This finding implies that miR-375 could be another crucial biomarker for predicting the early onset of T2DM in the Nepali population.
While the plasma levels of all four miRNAs were elevated in T2DM patients compared to prediabetes patients and healthy controls, there were similar expression patterns among these groups. This suggests that the potential of individual miRNA expression profiles for the differential diagnosis or staging of T2DM pathogenesis might not be strong. However, when the four miRNAs were grouped as a panel, a significant majority of the study participants across the three study groups could be classified and differentiated with an accuracy of 69.6 %, suggesting enhanced identification using the panel. Furthermore, integrating the information of all four miRNAs in the ROC curve analysis led to superior differentiation of prediabetes participants compared to individual miRNAs. However, distinguishing between healthy controls and individuals with prediabetes remained challenging due to increased overlap in miRNA expression profiles, a finding similar to a previous study in Chinese adults analyzing the expression profiles of miR-9, miR-29a, and miR-375 [25].
Our study has several strengths, including the use of a representative sample of Nepali adults with diverse glucose tolerance, robust miRNA isolation and quantification techniques, and comprehensive statistical analyses. However, it is important to acknowledge certain limitations. The cross-sectional design prevents causal inference and longitudinal evaluation of changes in miRNA expression. Additionally, we could not adjust the impact of potential confounding factors such as diet, physical activity, smoking, alcohol intake, medication usage, or genetic variations in the reported miRNA expression levels. Furthermore, our study did not investigate the functional mechanisms or target genes of these miRNAs concerning T2DM pathophysiology.
5. Conclusion
In conclusion, this study validates the biomarker potential of plasma miR-9, miR-29a, miR-192, and miR-375 for early detection of glucometabolic disorders in Nepali adults. MiR-29a emerged as the most robust biomarker for differentiating T2DM patients from healthy and prediabetic individuals, while miR-192 effectively distinguished both prediabetic and T2DM individuals from healthy controls. MiR-375 demonstrated promise in identifying prediabetic and T2DM individuals with high accuracy. These findings provide novel insights into the utility of these plasma-circulating miRNAs for early detection of prediabetes and T2DM. Future large-scale, longitudinal studies with functional validation are warranted to confirm these results and elucidate the molecular mechanisms underlying these miRNAs' associations with glucometabolic disorders across diverse populations.
Funding
This work was supported by the UGC, Nepal, through its Faculty Research Grants Scheme (FRG-75/76-HS-8). The UGC, Nepal, did not influence the design or performance of this study or the interpretation of the research data.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical statement
The study received approval (MEMG/IRC/241/GA) from the Research and Ethics Committee of the Manipal College of Medical Sciences, Pokhara, Nepal, and conformed to the principles outlined in the Declaration of Helsinki for human participants. The study was conducted without disturbing the routine clinical care and treatment of the enrolled participants.
Informed consent
All participants provided written informed consent before the collection of test specimens and personal sociodemographic, physiological, and clinical data.
CRediT authorship contribution statement
Daya Ram Pokharel: Research conceptualization, Funding acquisition, Project supervision, Data management and curation, Manuscript writing and editing, journal correspondence, manuscript editing Abhishek Maskey and Ram Chandra Kafle: Fund Acquisition, Medical screening and grouping of study participants, Clinical interpretation of the study results Ashim Bataju and Prajwal Dahal: Enrollment of screened study participants, Collection of sociodemographic and clinical data, Data compilation Shailesh Adhikari and Roji Raut: Primer design, Method validation, RT-qPCR assay, Calculation of fold change in expression, Data compilation, and interpretation Binod Manandhar: Detailed statistical analysis and interpretation of study data, Writing of the results section, Critical reading and review of manuscript Krishna Das Manandhar: Supervision for RT-qPCR analysis, Quality check of RT-qPCR data, Critical reading, review and improvement of the manuscript.
Acknowledgments
The authors would like to thank all the investigators, staff, study participantsand the University Grants Commission (UGC), Nepal, for their unwavering support in carrying out this research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2024.07.001.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Galicia-Garcia U., Benito-Vicente A., Jebari S., Larrea-Sebal A., Siddiqi H., Uribe K.B., Ostolaza H., Martín C. Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci. 2020;21(17):6275. doi: 10.3390/ijms21176275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shrestha D.B., Budhathoki P., Sedhai Y.R., Marahatta A., Lamichhane S., Nepal S., Adhikari A., Poudel A., Nepal S., Atreya A. Type 2 Diabetes Mellitus in Nepal from 2000 to 2020: a systematic review and meta-analysis. F1000Res. 2021;10:543. doi: 10.12688/f1000research.53970.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Narayan K.M.V., Kanaya A.M. Why are South Asians prone to type 2 diabetes? A hypothesis based on underexplored pathways. Diabetologia. 2020;63:1103–1109. doi: 10.1007/s00125-020-05132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tabák A.G., Herder C., Rathmann W., Brunner E.J., Kivimäki M. Prediabetes: a high-risk state for diabetes development. Lancet. 2012;16(9833):2279–2290. doi: 10.1016/S0140-6736(12)60283-9. 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortiz-Martínez M., González-González M., Martagón A.J., Hlavinka V., Willson R.C., Rito-Palomares M. Recent developments in biomarkers for diagnosis and screening of type 2 diabetes mellitus. Curr. Diabetes Rep. 2022;22(3):95–115. doi: 10.1007/s11892-022-01453-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landrier J.F., Derghal A., Mounien L. MicroRNAs in obesity and related metabolic disorders. Cells. 2019;8(8):859. doi: 10.3390/cells8080859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aghaei-Zarch S.M. Crosstalk between miRNAs/lncRNAs and PI3K/AKT signaling pathway in diabetes mellitus: mechanistic and therapeutic perspectives. Noncoding RNA Res. 2024 Jan 14;9(2):486–507. doi: 10.1016/j.ncrna.2024.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu D., Di K., Fan B., Wu J., Gu X., Sun Y., Khan A., Li P., Li Z. MicroRNAs in extracellular vesicles: sorting mechanisms, diagnostic value, isolation, and detection technology. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.948959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J., Chen J., Sen S. MicroRNA as biomarkers and diagnostics. J. Cell. Physiol. 2016;231(1):25–30. doi: 10.1900/RDS.2012.9.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afsharmanesh M.R., Mohammadi Z., Mansourian A.R., Jafari S.M. A Review of microRNAs changes in T2DM in animals and humans. J. Diabetes. 2023;15:649–664. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González-Sánchez L.E., Ortega-Camarillo C., Contreras-Ramos A., Barajas-Nava L.A. miRNAs as biomarkers for diagnosis of type 2 diabetes: a systematic review. J. Diabetes. 2021;13(10):792–816. doi: 10.1111/1753-0407.13166. [DOI] [PubMed] [Google Scholar]
- 13.Zhu Z., Zhang Y., Bai R., Yang R., Shan Z., Ma C., Yang J., Sun D. Association of genetic polymorphisms in microRNAs with type 2 diabetes mellitus in a Chinese population. Front. Endocrinol. 2021;11 doi: 10.3389/fendo.2020.587561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flowers E., Kanaya A.M., Zhang L., Aouizerat B.E. The role of racial and ethnic factors in microRNA expression and risk for type 2 diabetes. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.853633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ElSayed N.A., Aleppo G., Aroda V.R., Bannuru R.R., Brown F.M., Bruemmer D., Collins B.S., Hilliard M.E., Isaacs D., Johnson E.L., Kahan S., Khunti K., Leon J., Lyons S.K., Perry M.L., Prahalad P., Pratley R.E., Seley J.J., Stanton R.C., Gabbay R.A., on behalf of the American Diabetes Association 2 Classification and diagnosis of diabetes: standards of care in diabetes-2023. Diabetes Care. 2023;46(1):S19–S40. doi: 10.2337/dc23-S002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedewald W.T., Levy R.I., Fredrickson D.S. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without the use of the preparative ultracentrifuge. Clin. Chem. 1972;18(6):499–502. [PubMed] [Google Scholar]
- 17.Matthews D.R., Hosker J.P., Rudenski A.S., Naylor B.A., Treacher D.F., Turner R.C. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations. man Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 18.Levey A.S., Stevens L.A., Schmid C.H., Zhang Y.L., Castro A.F., Feldman H.I., Kusek J.W., Eggers P., Van Lente F., Greene T., Coresh J. CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration). A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009;150(9):604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Muhtaresh H.A., Al-Kafaji G. Evaluation of two-diabetes related microRNAs suitability as earlier blood biomarkers for detecting prediabetes and type 2 diabetes mellitus. J. Clin. Med. 2018;7:12. doi: 10.3390/jcm7020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang Y., Song Y., Wang Z., Liu Z., Gao P., Liang J., Zhu J., Xing C., Xu H. microRNA-192, -194 and -215 are frequently downregulated in colorectal cancer. Exp. Ther. Med. 2012;3:560–566. doi: 10.1159/000455869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarzenbach H., da Silva A.M., Calin G., Pantel K. Data normalization strategies for MicroRNA quantification. Clin. Chem. 2015;61(11):1333–1342. doi: 10.1373/clinchem.2015.239459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Plaisance V., Abderrahmani A., Perret-Menoud V., Jacquemin P., Lemaigre F., Regazzi R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J. Biol. Chem. 2006;281(37):26932–26942. doi: 10.1074/jbc.M601225200. [DOI] [PubMed] [Google Scholar]
- 24.Ramachandran D., Roy U., Garg S., Ghosh S., Pathak S., Kolthur-Seetharam U. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic β-islets. FEBS J. 2011;278(7):1167–1174. doi: 10.1111/j.1742-4658.2011.08042.x. [DOI] [PubMed] [Google Scholar]
- 25.Kong L., Zhu J., Han W., Jiang X., Xu M., Zhao Y., Dong Q., Pang Z., Guan Q., Gao L., Zhao J., Zhao L. Significance of serum microRNAs in prediabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011;48(1):61–69. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]
- 26.Bagge A., Clausen T.R., Larsen S., Ladefoged M., Rosenstierne M.W., Larsen L., Vang O., Nielsen J.H., Dalgaard L.T. MicroRNA-29a is up-regulated in beta-cells by glucose and decreases glucose-stimulated insulin secretion. Biochem. Biophys. Res. Commun. 2012;426(2):266–272. doi: 10.1016/j.bbrc.2012.08.082. [DOI] [PubMed] [Google Scholar]
- 27.Dalgaard L.T., Sørensen A.E., Hardikar A.A., Joglekar M.V. The microRNA-29 family: role in metabolism and metabolic disease. Am. J. Physiol. Cell Physiol. 2022;323(2):C367–C377. doi: 10.1152/ajpcell.00051.2022. [DOI] [PubMed] [Google Scholar]
- 28.Yesuf H.A., Molla M.D., Malik T., Seyoum Wendimagegn Z., Yimer Y. MicroRNA-29-mediated cross-talk between metabolic organs in the pathogenesis of diabetes mellitus and its complications: a narrative review. Cell Biochem. Funct. 2024;42(4) doi: 10.1002/cbf.4053. [DOI] [PubMed] [Google Scholar]
- 29.Shah R., Murthy V., Pacold M., Danielson K., Tanriverdi K., Larson M.G., Hanspers K., Pico A., Mick E., Reis J., de Ferranti S., Freinkman E., Levy D., Hoffmann U., Osganian S., Das S., Freedman J.E. Extracellular RNAs are associated with insulin resistance and metabolic phenotypes. Diabetes Care. 2017;40(4):546–553. doi: 10.2337/dc16-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Párrizas M., Brugnara L., Esteban Y., González-Franquesa A., Canivell S., Murillo S., Gordillo-Bastidas E., Cussó R., Cadefau J.A., García-Roves P.M., Servitja J.M., Novials A. Circulating miR-192 and miR-193b are markers of prediabetes and are modulated by an exercise intervention. J. Clin. Endocrinol. Metab. 2015;100(3):E407–E415. doi: 10.1210/jc.2014-2574. [DOI] [PubMed] [Google Scholar]
- 31.Jaeger A., Zollinger L., Saely C.H., Muendlein A., Evangelakos I., Nasias D., Charizopoulou N., Schofield J.D., Othman A., Soran H., Kardassis D., Drexel H., Eckardstein A.V. Circulating microRNAs -192 and -194 are associated with the presence and incidence of diabetes mellitus. Sci. Rep. 2018 Sep 24;8(1) doi: 10.1038/s41598-018-32274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poy M.N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P., Zavolan M., Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. U. S. A. 2009;106(14):5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolmeson C., Esguerra J.L., Salehi A., Speidel D., Eliasson L., Cilio C.M. Differences in islet-enriched miRNAs in healthy and glucose intolerant human subjects. Biochem. Biophys. Res. Commun. 2011;404(1):16–22. doi: 10.1016/j.bbrc.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 34.Li Y., Xu X., Liang Y., Liu S., Xiao H., Li F., Cheng H., Fu Z. miR-375 enhances palmitate-induced lipoapoptosis in insulin-secreting NIT-1 cells by repressing myotrophin (V1) protein expression. Int. J. Clin. Exp. Pathol. 2010;3(3):254–264. [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao H., Guan J., Lee H.M., Sui Y., He L., Siu J.J., et al. Up-regulated pancreatic tissue microRNA-375 associated with human type 2 diabetes through beta-cell deficit and islet amyloid deposition. Pancreas. 2010;39(6):843–846. doi: 10.1097/MPA.0b013e3181d12613. [DOI] [PubMed] [Google Scholar]
- 36.Wu X., Li Y., Man B., Li D. Assessing MicroRNA-375 levels in type 2 diabetes mellitus (T2DM) patients and their first-degree relatives with T2DM. Diabetes Metab Syndr Obes. 2021;14:1445–1451. doi: 10.2147/DMSO.S298735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiménez-Lucena R., Camargo A., Alcalá-Diaz J.F., Romero-Baldonado C., Luque R.M., van Ommen B., Delgado-Lista J., Ordovás J.M., Pérez-Martínez P., Rangel-Zúñiga O.A., López-Miranda J. A plasma circulating miRNAs profile predicts type 2 diabetes mellitus and prediabetes: from the CORDIOPREV study. Exp. Mol. Med. 2018;50(12):1–12. doi: 10.1038/s12276-018-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parrizas M., Novials A. Circulating microRNAs as biomarkers for metabolic disease. Best Pract. Res. Clin. Endocrinol. Metabol. 2016;30(5):591–601. doi: 10.1016/j.beem.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Aghaei-Zarch S.M. Crosstalk between MiRNAs/lncRNAs and PI3K/AKT signaling pathway in diabetes mellitus: mechanistic and therapeutic perspectives. Noncoding RNA Res. 2024;9(2):486–507. doi: 10.1016/j.ncrna.2024.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu T., Zheng Y., Chen X., Lin Z., Liu C., Yuan C. The role of exosome derived miRNAs in inter-cell crosstalk among insulin-related organs in type 2 diabetes mellitus. J. Physiol. Biochem. 2024 doi: 10.1007/s13105-024-01026-x. [DOI] [PubMed] [Google Scholar]
- 41.Dzung P.T., Trung N.T., Van Khanh L., Chinh D.D., VanDe D., Van Tong H., Toan N.L. Clinical association and diagnostic significance of miRNA-29a and miRNA-147b in type 2 diabetes mellitus. Int. J. Med. Sci. 2023;20(10):1316–1325. doi: 10.7150/ijms.84899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.