Abstract
Sin Nombre virus (SNV) is thought to establish a persistent infection in its natural reservoir, the deer mouse (Peromyscus maniculatus), despite a strong host immune response. SNV-specific neutralizing antibodies were routinely detected in deer mice which maintained virus RNA in the blood and lungs. To determine whether viral diversity played a role in SNV persistence and immune escape in deer mice, we measured the prevalence of virus quasispecies in infected rodents over time in a natural setting. Mark-recapture studies provided serial blood samples from naturally infected deer mice, which were sequentially analyzed for SNV diversity. Viral RNA was detected over a period of months in these rodents in the presence of circulating antibodies specific for SNV. Nucleotide and amino acid substitutions were observed in viral clones from all time points analyzed, including changes in the immunodominant domain of glycoprotein 1 and the 3′ small segment noncoding region of the genome. Viral RNA was also detected in seven different organs of sacrificed deer mice. Analysis of organ-specific viral clones revealed major disparities in the level of viral diversity between organs, specifically between the spleen (high diversity) and the lung and liver (low diversity). These results demonstrate the ability of SNV to mutate and generate quasispecies in vivo, which may have implications for viral persistence and possible escape from the host immune system.
Many RNA viruses are recognized as having highly mutable and genetically diverse genomes, largely due to their high replication rates and error-prone polymerases (18). Populations of closely related viral genomes found in individual hosts are termed quasispecies, and the consensus sequence of all viral variants represents the wild-type or master sequence. The consequences of this phenomenon include the ability of RNA viruses to escape host immunosurveillance and generate drug-resistant variants, most notably in patients infected with human immunodeficiency virus (HIV) (9, 10, 49). High mutation rates have hindered the development of vaccines against some RNA viruses due to the continued appearance of variants with new epitopes found on exposed antigenic viral proteins (18). The tremendous diversity seen in RNA viruses may also explain the emergence of new strains from old viruses, thereby generating new disease patterns and promoting expansion into unique host ranges (16).
Hantaviruses are enzootic viruses of wild rodents that cause persistent infections in their natural hosts in the presence of an apparent immune response (21). Many hantaviruses are pathogenic in humans, and infection is thought to occur via contaminated rodent excreta (17). Old World hantaviruses, including Hantaan, Seoul, Dobrava, and Puumala viruses, are associated with hemorrhagic fever with renal syndrome, while many New World hantaviruses, such as Sin Nombre virus (SNV), are responsible for a severe pulmonary disease now termed hantavirus pulmonary syndrome (HPS) (64). These viruses consist of a tripartite negative-sense single-stranded RNA genome encoding at least four structural proteins. The large segment (L) encodes the viral RNA polymerase, the medium (M) segment encodes the precursor of two glycoproteins (G1 and G2), and the small (S) segment encodes the nucleocapsid protein (65). Some hantaviruses also encode a putative open reading frame of unknown function, designated the nonstructural protein of the S segment (NSS) (66).
SNV is a recently identified member of the genus Hantavirus (8) that exhibits many of the properties associated with emerging RNA viruses (16). SNV was first identified as the agent responsible for an HPS outbreak initially observed in the Four Corners region of the United States in 1993 (47). The disease is characterized by a rapid onset of interstitial pulmonary edema and respiratory failure that contributes to a mortality rate of up to 50% in infected humans (31). β3 integrins, found on a wide variety of cell types, have recently been identified as receptors for SNV (20). The deer mouse (Peromyscus maniculatus) is recognized as the primary natural reservoir for SNV, although spillover infection into other animals has been well documented. Additional hantaviruses found in both North and South America are each associated with distinct rodent members of the sigmodontine genera, and many have also been associated with HPS (4, 28, 30, 36).
SNV and SNV-like hantaviruses are known to exhibit tremendous diversity with respect to geographic distribution and are thought to coevolve with their rodent hosts (41, 43, 61). In addition, SNV has been shown to generate diversity by reassortment, both naturally and in tissue culture (24, 37, 60). Viral persistence in SNV-infected deer mice has not been explained but may occur through several mechanisms. Viral load in both tissues and blood may drop drastically following humoral and cellular immune responses, although low levels of virus shedding occur continuously (26). The generation of defective interfering viral particles has been postulated and may play a role in persistence; however, none have ever been detected either in tissue culture or infected rodents (65). Recent reports indicate the presence of viral quasispecies in rodents infected with either Tula or Puumala hantavirus (56, 57). Also, the ability of Puumala virus to accumulate changes in the noncoding region of the S segment during passage in cell culture (39) and the capacity of Hantaan virus to escape antibody neutralization in vitro (67) demonstrate the adaptability and plasticity of hantaviruses.
We hypothesized that SNV may be present in persistently infected rodents as a population of heterogeneous viral RNA genomes, which may help the virus evade the host immune system and establish persistence (48). Deer mice were found to produce neutralizing antibodies specific for SNV following infection; however, no correlation was seen between the presence of neutralizing antibodies and the disappearance of viral RNA. To evaluate the presence of SNV quasispecies in infected deer mice temporally, animals were trapped multiple times, bled, and released back into their natural environment. Analysis of infected blood demonstrated a complex mixture of SNV variants within one time point and over subsequent time points in both coding and noncoding regions of the genome. Many of the amino acid changes occurred in a region previously identified as the immunodominant domain of G1 (27). Differences in the level of SNV diversity were observed when virus sequences amplified from various organs of infected deer mice were compared. The high percentage of A→G or U→C mutations in viral clones isolated from certain tissues suggests a role for the cellular RNA-editing enzyme, double-stranded RNA adenosine deaminase (dsRAD), in generating SNV genetic variation (6). These results may provide an indication of how SNV persists in deer mice over time and whether genetic diversity may allow the virus to exploit and jump between different host species.
MATERIALS AND METHODS
Sample collection.
Details of our field sampling methods have been published elsewhere (5). Briefly, rodent blood and tissue samples were collected in western Nevada and eastern California on plots that were sampled at monthly intervals. On these plots, deer mice were marked with individually numbered ear tags to allow identification of animals upon subsequent recapture. Blood samples were collected from these animals by retro-orbital puncture with a heparinized capillary tube, after which they were released at the point of capture. On some plots, deer mice were sacrificed for tissue analysis. All samples were immediately placed on dry ice for transport to a biosafety level 3 laboratory for RNA extraction.
Serological screening and enzyme-linked immunosorbent assay (ELISA) analysis.
Procedures for serological screening have been published elsewhere (52). Briefly, microtiter plates (Dynatech Laboratories, Chantilly, Va.) were coated overnight at 4°C with recombinant nucleocapsid antigen diluted 1:2,000 in phosphate-buffered saline (PBS; pH 7.4). Following incubation, the plates were washed three times with wash buffer (PBS [pH 7.4], 0.5% Tween 20, 0.01% thimerosal). Heat-inactivated mouse sera were diluted 1:50 with serum dilution buffer (PBS [pH 7.4], 0.5% Tween 20, 0.01% thimerosal, and 5% skim milk) and added to each well for 60 min at 37°C. The wells were then washed three times with wash buffer and incubated with the specific secondary antibody, horseradish peroxidase-labeled goat anti-Peromyscus leucopus antibody (Kirkegaard and Perry Laboratories, Gaithersburg, Md.), at 37°C for 60 min. The plates were washed again as before and incubated with 100 μl of 2,2′-azino-di-(3-ethyl-benzthiozaline-sulfonate) microwell peroxidase substrate solution (Kirkegaard and Perry Laboratories) for 30 min at 37°C. The absorbance at 405 nm was determined and compared with those of negative and positive controls. Samples with titers less than 1:400 were considered negative.
Cell culture and virus strains.
Vero E6 cells were grown in Iscove’s modified Dulbecco’s medium (IMDM) containing 10% fetal bovine serum. The Convict Creek 107 (CC107) viral strain was initially provided by Connie Schmaljohn (U.S. Army Medical Research Institute of Infectious Disease, Fort Detrick, Frederick, Md.) and propagated on Vero E6 cells. The titer of CC107 viral stock used for focus reduction neutralization assays was determined by counting plaques on Vero E6 cells following immunostaining.
Focus reduction neutralization assay.
Deer mouse serum was serially diluted with IMDM plus 1% fetal bovine serum. Neutralizing antibodies were detected in sera by incubating 100 PFU of CC107 virus (150 μl) in serially diluted (1:20, 1:80, 1:320, 1:1,280, 1:5,120, and 1:20,480) deer mouse sera (150 μl) for 1 h at 37°C. Following incubation, the 300-μl virus-antibody mixture was added to 24-well dishes containing confluent monolayers of Vero E6 cells for 1 h at 37°C. Subsequently, the dishes were overlaid with 0.6% agarose and incubated for 10 days at 37°C. After the agarose plug was removed, the cells were washed two times with PBS and fixed in 75:25 methanol-acetone for 10 min. The dishes were air dried and stored at −20°C until immunostaining was carried out, and the number of virus plaques was determined.
Immunostaining.
Air-dried dishes were washed three times for 5 min each time with PBS containing 0.1% Tween 20 (PBS-Tween). Pooled convalescent human sera (from SNV-infected patients) at a 1:300 dilution in PBS-Tween was used as the primary antibody. The primary antibody was incubated with cells for 1 h at 37°C and then washed three times with PBS-Tween for 5 min each time. After being washed, the dishes were incubated with anti-human alkaline phosphatase-conjugated antibody at a 1:100 dilution in PBS-Tween. The secondary antibody was incubated and washed as described above. Virus plaques were visualized with the Vector Red alkaline phosphatase substrate kit (Vector Laboratories Inc., Burlingame, Calif.) as specified by the manufacturer. The plaques were counted under a light microscope.
Oligonucleotide primer design.
Primers specific for eastern California SNV lineages were synthesized for the N-terminal domain of G1 and for the S segment noncoding variable region (SVAR) found 3′ proximal to the nucleocapsid gene. For first-round amplification, the G1 primers used were 5′ACTCCGCA(A/C)GAAGAAGCAA3′ (corresponding to M segment positions 10 to 28 of SNV isolate CC107 plus strand [63]) and 5′T(A/T)GATAGCAGACCTATCATACAGCT3′ (corresponding to M segment positions 529 to 553 of SNV isolate CC107 minus strand [63], except that residue 547 is A in CC107 instead of G) while the SVAR primers used were 5′CAGGGTAATGGGCAC(C/T)A3′ (corresponding to S segment positions 1373 to 1389 of SNV isolate CC107 plus strand [63]) and 5′GTCATGTACTATTAACGGAACGAA3′ (corresponding to S segment positions 1921 to 1944 of SNV isolate CC107 minus strand [63]). For second-round amplification, the G1 primers used were 5′TGAATAAAGGA(G/T)ATACAGAATGGT3′ (corresponding to M segment positions 33 to 56 of SNV isolate CC107 plus strand [63]) and 5′GTTTGATTACAGGC(C/T)AAATCATAAC3′ (corresponding to M segment positions 446 to 470 of SNV isolate CC107 minus strand [63]) while the SVAR primers were 5′AAGGGCCAATTATAT(C/T)ACAGG3′ (corresponding to S segment positions 1419 to 1439 of SNV isolate CC107 plus strand [63]) and 5′AA(C/T)GGTTAATAG(A/G)ACAATC(C/T)TC3′ (corresponding to S segment positions 1829 to 1850 of SNV isolate CC107 minus strand [63]).
RT reaction.
RNA was extracted from samples in a designated PCR “clean” room with TRIzol reagent (Gibco-BRL, Gaithersburg, Md.). Tissues were first washed with PBS (pH 7.4) to limit blood contamination. Approximately 100 mg of tissue or 100 μl of blood was homogenized with 1 ml of TRIzol reagent. RNA was then isolated as specified by the manufacturer. The RNA was resuspended in diethyl pyrocarbonate-treated water, quantified, and stored at −80°C. Reverse transcription (RT) of SNV RNA was performed with SuperScript II reverse transcriptase (Gibco-BRL) as specified by the manufacturer. Briefly, approximately 100 ng of total RNA was added to 10 μl of diethyl pyrocarbonate-treated water plus 30 pmol of G1- or SVAR-specific primers. The mixture was heated to 70°C for 10 min and chilled on ice. Next, 5× First Strand buffer, 0.01 M dithiothreitol, and 5 nmol of each deoxynucleotide were added to bring the reaction mixture to 20 μl total, and the contents were incubated at 42°C for 2 min. Subsequently, 200 U of Superscript II reverse transcriptase enzyme were added and the mixture was incubated at 42°C for an additional 50 min. Prior to amplification, the reaction mixture was incubated at 70°C for 15 min to inactivate the Superscript II RT enzyme.
PCR amplification.
Nested PCR was employed to amplify hantavirus sequences with a Perkin-Elmer 9600 GeneAmp PCR system. A 50 μl-reaction mixture contained 2.5 U of Taq polymerase (Promega, Madison, Wis.), first-round primers (30 pmol each), 10 nmol of each deoxynucleotide, 1.85 mM MgCl2, and 1/10 of the RT mixture (2 μl). For second-round amplification, 2 μl of the first-round mixture was added to another 50-μl reaction mixtures containing identical components except for the addition of second-round primers. Each amplification was carried out for 30 cycles under the following conditions: an initial denaturation of 95°C for 2 min; 30 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 1 min; and a final extension at 72°C for 10 min. RNA isolated from uninfected Vero E6 cells was run in parallel as a negative control with each RT-PCR. The PCR amplification products (389 bp for G1 products and approximately 389 bp for SVAR products, depending on the viral strain) were detected by electrophoresis on a 1.5% agarose gel.
Isolation, cloning, and sequencing of PCR products.
The amplified products were excised from the gel and isolated with a QIAEX II gel extraction kit (Qiagen, Valencia, Calif.). The products were ligated into the pGEM-T Easy vector (Promega) and transformed into DHFα competent Escherichia coli cells. Colonies containing PCR product inserts were identified by blue-white selection in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside). Only one positive colony was picked from each PCR and cloning procedure to limit the possibility of resampling in low-viral-load samples (38). Positive colonies were grown, and plasmids were isolated with the QIAprep Spin Miniprep kit (Qiagen). All isolated plasmids were analyzed for the presence or absence of viral inserts on a 1.5% agarose gel after digestion with EcoRI. Plasmids (400 ng each) were sequenced with an ABI Prism 310 genetic analyzer with both M13 forward and reverse primers (Perkin Elmer, Norwalk, Conn.). Amplification primer sequences were removed prior to analysis. A reamplification control was carried out to determine the mutation frequency contributed by both Superscript II RT enzyme and Taq polymerase for our system. A G1 SNV plasmid of known sequence was linearized with EcoRI and transcribed with an RNA transcription kit (Stratagene, La Jolla, Calif.). RNA was isolated, reverse transcribed, and amplified as before. In order to exclude any increase in mutation frequency due to low viral copy numbers found in weakly positive samples, the highest dilution of in vitro-synthesized RNA which generated a visible band after amplification was used for subsequent error analysis. Reamplification clones were analyzed for reverse transcriptase- and Taq-specific nucleotide changes.
Sequence alignments, phylogenetic analysis, and variability analysis.
Sequences were aligned by using both Clustal and Jotun-Hein algorithms found on the MegAlign module of the Lasergene 99 program (DNASTAR Inc., Madison, Wis.). Phylogenetic analysis was performed with the distance-based neighbor-joining (NJ) and unweighted pair group method with arithmetic averages (UPGMA) methods (MEGA software [35]). Both Jukes-Cantor distances and gamma distances for the Tamura-Nei estimation methods were used for phylogenetic inference. Bootstrap confidence limits were calculated by 500 search repetitions for the phylogenetic tree shown in Fig. 1. The tree was imported into the TreeView (version 1.52) tree-editing program (54) for text editing and printing purposes. VarPlot for Windows software was used (kindly provided by Stuart Ray, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Md.) to calculate ratios of nonsynonymous to synonymous substitutions (dN/dS) or dN − dS values in a “sliding window” of nucleotide sequence (59). A 75-bp segment was analyzed to determine the number of mutations per site, and this process was repeated for overlapping segments of the same size shifted by 3 bp (step size) and continued through the length of the region sequenced. The results were plotted to determine areas of high and low dN/dS ratios or dN − dS values within the G1 region analyzed.
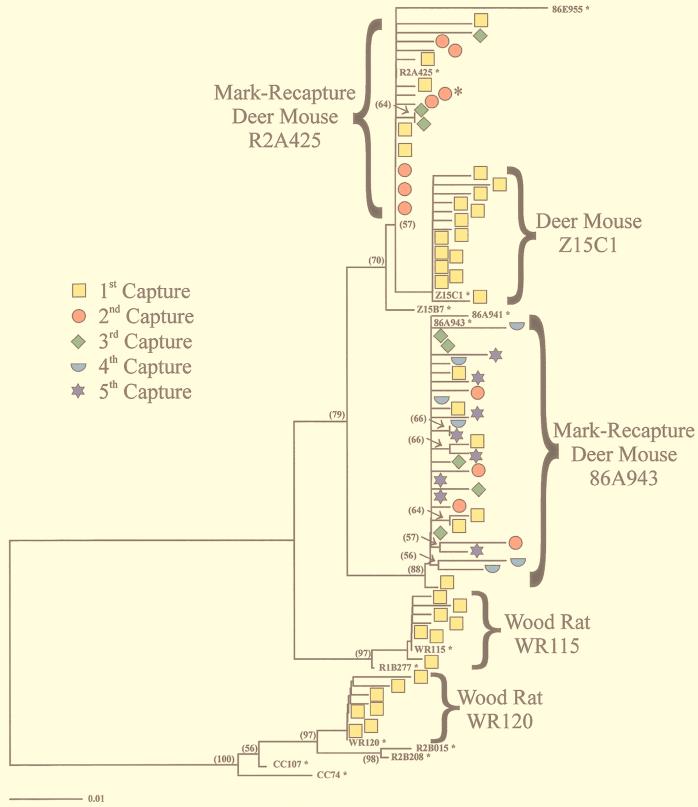
FIG. 1.
Phylogenetic analysis of SNV G1 consensus sequences from deer mice analyzed in Table 2. Also shown are all G1 clones isolated from deer mouse Z15 C1, mark-recapture deer mice (86A943 and R2A425), and two wood rats (WR115 and WR120). The consensus sequence for each deer mouse is indicated with the animal ID number and an asterisk. SNV clones isolated from particular rodents are grouped, and sequential time points are indicated with distinct symbols and colors. NJ analysis was performed with MEGA software and the Jukes-Cantor distance estimation model. Qualitatively similar trees were generated with gamma distances (a = 0.5) for the Tamura-Nei or Kimura two-parameter distance method with either NJ or UPGMA analysis. Bootstrap analysis was carried out with 500 replicates. The percentage of bootstrap support exceeding 50% is indicated by the numbers in parenthesis next to the corresponding branches. SNV heterogeneity was demonstrated in both deer mouse and wood rat species, with similar mutation frequencies seen in all animals. Deer mouse R2A425 was seropositive for SNV for all time points analyzed. Deer mouse 86A943 was captured before (first time point) and after (all other time points) seroconversion. The deer mouse seroconverted after its first capture; however, viral sequences were amplified from all time points analyzed. The asterisk next to a viral clone from R2A425 indicates the presence of a mutation (underlined) (codon TGG→TGA) which gave rise to a stop codon at amino acid residue 81 (tryptophan), suggesting either that this clone corresponds to a defective SNV genome or that the mutation represents an error incorporated during the isolation procedure. SNV isolates CC107 and CC74 (GenBank accession no. L33474 and L33684, respectively) were also included to rule out possible contamination with viral isolates grown in the laboratory (63).
Statistical analysis.
The effects of organ type, mutation type (A→G and U→C versus all others), and region (G1 versus SVAR) on the mutation frequency of SNV in deer mouse Z15 B7 were tested by a three-way analysis of variance with the different mutation frequencies within each clone serving as replicates. The number of replicates for each region-organ-mutation type combination varied from three to eight clones. Non-A→G and -U→C mutation frequencies were weighted by a factor of 0.2 to account for the fact that they would be expected to occur five times more frequently than A→G and U→C mutations in the absence of a dsRAD-like activity. Thus, the comparison between the two mutation types was a test of whether A→G and U→C mutations were more or less frequent than would be predicted under a hypothesis of purely random mutations. A paired t test (SAS ver 6.12; PROC MEANS) was performed on clones derived from all mice to test for overall differences in A→G and U→C versus G→A and C→U mutation rates, irrespective of organ or region (G1 or SVAR). For a fair comparison, only transitional mutations were analyzed, rather than transversions, due to their scarcity during mutagenesis and evolution.
RESULTS
Rodents analyzed for neutralizing antibodies.
Sera from SNV-infected deer mice were chosen randomly from previous captures and tested for their ability to neutralize an SNV laboratory isolate (CC107). The deer mice were initially tested by both ELISA and RT-PCR for SNV infection. All mice analyzed were positive for SNV as determined by ELISA (Table 1). Four of nine rodents had viral RNA in the blood, while eight of nine rodents were positive for viral RNA in the lungs. We detected high levels of neutralizing antibodies in three rodents, all of which were positive for viral RNA. There was no correlation between the lack of viral RNA in either the blood or lungs and high levels of neutralizing antibodies.
TABLE 1.
Neutralization titers of sera from SNV-infected deer micea
| Rodent ID no. | ELISA | RT-PCR
|
Neutralization titerb | |
|---|---|---|---|---|
| Blood | Lung | |||
| PS 3-20 | + | + | + | 1,280 |
| TC 4-52 | + | + | + | 80 |
| TC 4-26 | + | + | + | 5,120 |
| TC 3-44 | + | + | + | 80 |
| LS 4-7 | + | − | + | 1,280 |
| TC 3-29 | + | − | + | 80 |
| TC 4-53 | + | − | +/− | 80 |
| TC 3-33 | + | − | +/− | 80 |
| PS 3-22 | + | − | − | <20 |
| Pooled normal deer mouse | − | − | − | <20 |
Rodents were tested for both the S and M segments of SNV by RT-PCR as described previously (61); +, positive for both S and M segments; +/−, positive for S segment only; −, below detection limits.
Neutralization titers represent the reciprocal of the highest dilution of serum which neutralized 50% of 100 PFU of virus.
Rodents analyzed by ELISA and RT-PCR.
Deer mice captured multiple times were bled and tested by ELISA for antibodies against SNV nucleocapsid antigen. Blood samples were taken over a period of months, and many animals were captured on more than two occasions. Those animals with high ELISA titers were examined for SNV nucleic acid by RT-PCR with primers specific for G1. Detection of viral RNA in sequential blood samples was variable with some animals, while other animals were consistently positive over a time course of months, reflecting the persistence of SNV in deer mice (Table 2). Two deer mice were found to be positive for viral RNA even before seroconversion, indicating that these mice were captured at an early stage of infection. Although many ELISA-positive rodents were tested by RT-PCR, only those animals tested that resulted in amplification of SNV sequences from all time points could be analyzed sequentially for viral diversity. In addition to the live trapped rodents, 35 deer mice captured in an area previously known to have high numbers of SNV-infected rodents were first bled and then sacrificed. Analysis of blood samples revealed that 7 of 35 animals were positive for SNV by ELISA and RT-PCR. Organs from two of these deer mice were dissected and analyzed for the presence of SNV nucleic acid. Two wood rats (Neotoma lepida) previously found to be positive for SNV by ELISA and RT-PCR were also analyzed for SNV diversity in the blood (11).
TABLE 2.
Rodents analyzed by ELISA and RT-PCRa
| Rodent | ID no. and source | Date of capture (mo-day-yr) | ELISAd | RT-PCRe |
|---|---|---|---|---|
| Deer mouse | R2A 425 Ac | 6-2-96 | 5 | + |
| Bc | 8-25-96 | 5 | + | |
| Cc | 9-18-96 | 4 | + | |
| Deer mouse | 86A 943 Ac | 7-6-97 | 0 | + |
| Bc | 8-3-97 | 5 | + | |
| Cc | 8-27-97 | 5 | +/− | |
| Dc | 9-21-97 | 4 | + | |
| Ec | 10-13-97 | 5 | + | |
| Deer mouse | R1B 277 A | 6-22-95 | 0 | ND |
| B | 8-4-95 | 0 | ND | |
| C | 9-13-95 | 5 | + | |
| D | 10-12-95 | 5 | − | |
| Deer mouse | R2B 015 A | 6-22-95 | 1 | + |
| B | 8-4-95 | 5 | − | |
| Deer mouse | R2B 208 A | 6-22-95 | 0 | ND |
| B | 8-4-95 | 5 | + | |
| C | 9-13-95 | 5 | − | |
| Deer mouse | 86A 941 A | 7-6-97 | 0 | +/− |
| B | 8-3-97 | 5 | + | |
| C | 8-27-97 | 4 | − | |
| D | 9-21-97 | 4 | +/− | |
| E | 10-13-97 | 4 | + | |
| Deer mouse | 86E 955 A | 7-9-97 | 0 | − |
| B | 9-21-97 | 5 | + | |
| C | 11-2-97 | 4 | + | |
| Deer mouseb | Z15 B7 Bloodc | 7-19-96 | 5 | + |
| Lungc | + | |||
| Liverc | + | |||
| Kidneyc | + | |||
| Spleenc | + | |||
| Salivaryc | + | |||
| Bladderc | +/− | |||
| Deer mouseb | Z15 C1 Bloodc | 7-19-96 | 3 | + |
| Lung | + | |||
| Liver | + | |||
| Kidney | +/− | |||
| Spleen | − | |||
| Salivary | − | |||
| Bladder | − | |||
| Wood ratb | WR 115c | 4-3-96 | 4 | + |
| Wood ratb | WR 120c | 4-3-96 | 5 | + |
Unless otherwise indicated, all sample are derived from blood.
Sacrificed.
Analyzed for viral diversity.
An ELISA value of 0 indicates seronegativity, while a value of 5 represents high amounts of SNV antibodies in deer mouse sera.
Rodents were tested for the G1 region of the M segment of SNV by RT-PCR as described in Materials and Methods; +, strong signal; +/−, weak signal; −, below detection limits; ND, not determined.
Regions of SNV cloned for sequence analysis.
Analysis of virus sequence variation was conducted on SNV amplified from six rodents. Two regions of the SNV genome were chosen based on their possible antigenic potential (25) and high degree of sequence variation (34). A portion of the G1 protein near the N-terminal domain corresponding to amino acid positions 3 to 131 of SNV isolate CC107 was chosen for analysis (63). This region included the immunodominant domain of G1 (27) and regions of high surface probability (determined by the Emini method [Protean Module; DNASTAR Inc.]) and high antigen index values (determined by the Jameson-Wolf method [Protean Module]). Also, a noncoding region of the S segment corresponding roughly to nucleotide positions 1440 to 1828 of SNV isolate CC107 was analyzed (63). This area of the SNV genome was found to have sequence heterogeneity and insertions or deletions between different published viral isolates.
Mutation frequencies for all clones isolated.
SNV diversity was seen in the G1 region of the genome at a level 8- to 10-fold higher than that seen for errors incorporated by both reverse transcriptase and Taq polymerase detected in control reamplifications (Table 3). Mutation frequencies for SVAR clones were found to be quite variable, depending on the deer mouse and the organ analyzed. Overall, similar mutation frequencies were observed for all rodents when G1 clones derived from the blood were analyzed. A total of 68,685 nucleotides were sequenced which contained 222 nucleotide changes, giving a mutation frequency of 3.2 × 10−3 for all clones analyzed. The majority of these changes (71%) were specifically A→G or U→C mutations, both in G1 and SVAR clones. A→G or U→C transitions were significantly more frequent than G→A or C→U transitions (mean rate, 2.33 × 10−3 versus 0.38 × 10−3; P = 0.0001) for clones derived from all rodents (see Materials and Methods). A large percentage (62%) of G1 nucleotide mutations were nonsynonomous, with many identical amino acid changes observed in multiple clones. Mutations which occurred repeatedly in viral clones either from the same animal or other animals, or when compared to published SNV isolates (NMR11, CC107, and CC74) (8, 63), are presented as quasineutral mutations (56). Alignment of SVAR viral clones demonstrated possible fixed mutations in bladder-, spleen-, and lung-specific clones, although the master sequence was found in these tissues as well (data not shown). Many SVAR nucleotide changes (58%) were found to be well tolerated or quasineutral, which may reflect structural or promoter constraints for this region. The mutation frequency for SVAR viral clones from the bladder of deer mouse Z15 B7 was found to be quite high (5.1 × 10−3), while one SVAR viral clone isolated from the salivary gland contained a single-base deletion.
TABLE 3.
Rodents analyzed for SNV diversity
| Rodent ID no. | Region | Mutation frequencya (10−3) | Nucleotide changes/total | A→G or U→Cb (%) | Amino acid changes (%) | Quasineutral mutationsc (%) |
|---|---|---|---|---|---|---|
| R2A 425 A | G1 | 3.1 | 6/1,945 | 50 | 67 | 33/25 |
| R2A 425 B | G1 | 1.8 | 5/2,723 | 60 | 80 | 20/25 |
| R2A 425 C | G1 | 5.1 | 6/1,167 | 67 | 67 | 17/25 |
| R2A 425 total | G1 | 2.9 | 17/5,835 | 59 | 71 | 24/25 |
| R2A 425 A | SVAR | 4.7 | 16/3,429 | 69 | NAf | 38/NA |
| R2A 425 B | SVAR | 6.6 | 15/2,286 | 53 | NA | 53/NA |
| R2A 425 C | SVAR | 4.4 | 10/2,286 | 80 | NA | 40/NA |
| R2A 425 total | SVAR | 5.1 | 41/8,001 | 66 | NA | 44/NA |
| 86A 943 A | G1 | 3.0 | 7/2,334 | 86 | 86 | 57/50 |
| 86A 943 B | G1 | 5.8 | 9/1,556 | 56 | 56 | 22/20 |
| 86A 943 C | G1 | 2.1 | 4/1,945 | 50 | 100 | 0/0 |
| 86A 943 D | G1 | 5.1 | 12/2,334 | 67 | 89 | 58/50 |
| 86A 943 E | G1 | 3.9 | 12/3,112 | 67 | 58 | 42/43 |
| 86A 943 total | G1 | 3.9 | 44/11,281 | 66 | 68 | 41/37 |
| Z15 B7 | G1e | 3.5 | 42/12,059 | 83 | 70 | 60/68 |
| Z15 B7 | SVARe | 2.5 | 52/20,617 | 75 | NA | 58/NA |
| Z15 C1 | G1 | 2.4 | 13/5,446 | 76 | 54 | 30/0 |
| WR115 | G1 | 2.2 | 6/2,723 | 33 | 50 | 33/67 |
| WR120 | G1 | 2.6 | 7/2,723 | 71 | 86 | 14/40 |
| TOTAL | Both | 3.2 | 222/68,685 | 71 | 62 | 46/41 |
| Reamplification controld | G1 | 0.51 | 3/5,835 | 33 | 67 | 33/0 |
Mutation frequency is defined as the proportion of mutations relative to the master or consensus nucleotide sequence for each rodent and calculated by dividing the number of mutations relative to the consensus by the total number of nucleotides sequenced in each animal. The mean mutation frequencies ± standard deviations for the G1 and the 3′ noncoding region (SVAR) were 2.9 ± 0.6 and 3.8 ± 1.3, respectively.
Percentage of A→G or U→C transitional mutations.
Quasineutral mutations are defined as redundant mutations found within the mutant spectra as a minority or in master sequences of the represented rodents or published SNV isolates NMR11, CC107, or CC74 (8, 63). The first value refers to nucleotide percentages, while the second refers to amino acid percentages.
A G1 reamplification control determined the level of Superscript II reverse transcriptase and Taq polymerase errors incorporated during the amplification and isolation of SNV clones.
All organ clones included for analysis.
NA, not applicable.
Roughly equal numbers of nucleotide mutations were observed between the first, second, and third base codons for all variants (45, 34, and 50, respectively). All rodents exhibited SNV diversity in both coding and noncoding regions; however, no substitutions predominated at later time points for mark-recapture deer mouse samples. Three mutations distributed randomly were observed in RT-PCR reamplification control samples at nucleotide positions 11, 168, and 271 of our G1 clone. The introduction of errors by reverse transcriptase and Taq polymerase (0.5 × 10−3) determined for our system is comparable to other published values (56). This value should represent the upper limit of errors introduced by enzymatic manipulation of viral sequences but may also include T7 polymerase errors generated during control G1 RNA production.
SNV diversity comparison between the reservoir host and presumed “dead-end” rodent hosts.
Many rodents other than deer mice are thought to be dead-end hosts for SNV replication (7). To determine whether differences in viral diversity could be seen between deer mice and nonreservoir host rodents, blood from an SNV-infected deer mouse (Z15 C1) and two wood rats (WR115 and WR120) was obtained and analyzed for virus sequence heterogeneity in the G1 region. All of the rodents carried circulating antibodies specific for SNV, as indicated by high ELISA titers. Viral diversity at the nucleotide and amino acid level was seen in both rodent species for the G1 region of the genome, with all of the animals exhibiting similar mutation frequencies. A predominant viral strain or master sequence was observed in the presence of variant SNV genomes for each rodent. Figure 1 depicts the phylogenetic relationships of viral consensus sequences isolated from all deer mice and wood rats represented in Table 2. In addition, all viral clones isolated from deer mouse Z15 C1, mark-recapture deer mice R2A425 and 86A943, and wood rats WR115 and WR120 are represented.
G1 and SVAR sequence analysis from a deer mouse captured on multiple occasions.
Blood from a deer mouse captured on three different occasions was analyzed to determine if levels of quasispecies remained constant over time and whether progressive changes in SNV sequence could be observed. ELISA titers from deer mouse R2A425 were positive for all time points taken, indicating an active antibody response to SNV (Table 2). Varying levels of virus sequence heterogeneity in the G1 region were found for each infected blood sample, suggesting that SNV exists as a mixture of variants over sequential time points in infected deer mice (Fig. 1). A slightly higher total mutation frequency (5.1 × 10−3) was seen for SVAR clones from the same mouse; however, no insertions or deletions were observed in this region. Despite the viral diversity seen in deer mouse R2A425, no mutations at the nucleotide or amino acid level predominated at later time points. However, certain nucleotide and amino acid positions were preferentially mutated in many clones (quasineutral mutations). As with viral clones analyzed from other deer mice, the majority of the mutations involved either A→G or U→C specific changes.
G1 sequence analysis of a deer mouse before and after seroconversion.
To determine if host immune responses were influencing the level of SNV quasispecies, deer mouse blood from an RT-PCR-positive rodent (86A943) captured before and after seroconversion was analyzed for viral sequence variation. An average mutation frequency of 3.9 × 10−3 was observed for G1 viral clones calculated from all time points in this deer mouse, similar to the mutation frequency in the G1 region for other deer mice. Viral clones isolated before seroconversion indicate SNV diversity prior to the generation of an active antibody response, at both the nucleotide and amino acid levels (Fig. 1). Subsequent time points with high ELISA titers also revealed virus sequence heterogeneity. As with deer mouse R2A425, no particular mutations became fixed in the SNV population over sequential time points, although certain nucleotide and amino acid positions were preferentially mutated.
Identification of organ-specific levels of SNV quasispecies.
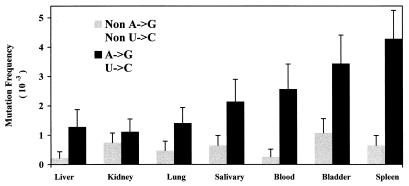
To determine if viral diversity and SNV tropisms exist for different tissues of infected deer mice, animals were sacrificed and examined for the presence of SNV nucleic acid. For one deer mouse (Z15 B7), viral RNA was detected in all organs examined by RT-PCR. G1 viral clones isolated from various organs of deer mouse Z15 B7 revealed no organ-specific nucleotide or amino acid substitutions. However, the mutation frequency was very different for each organ, especially when the spleen (high viral diversity) was compared with the lungs and liver (low viral diversity) (Fig. 2). G1 sequence heterogeneity in these organs ranged from a mutation frequency of 4.7 × 10−3 for spleen-specific G1 viral clones to 1.5 × 10−3 for liver-specific G1 viral clones. As with G1 viral clones, SVAR viral clones exhibited higher diversity in the spleen than in the lung and liver. It has been suggested that the double-stranded-RNA (dsRNA)-editing enzyme, dsRAD, may be responsible for A→G or U→C viral hypermutations (6) and may have a higher activity in one tissue than another (55). Including only these mutations for analysis revealed major disparities in the mutation frequencies between different organs (Fig. 2). In contrast, non-A→G, non-U→C mutation frequencies were fairly uniform between organs (Fig. 2). These results suggest that a mutational activity (perhaps due to dsRAD or a similar enzyme) which specifically changes A to G or U to C is greater in some organs than others (55).
FIG. 2.
Comparison of the levels of SNV diversity in different organs of a deer mouse (Z15 B7). The mutation frequencies represented in the graph are the mean mutation frequencies observed in both G1 and SVAR clones (with respect to the consensus sequence) ± standard errors, with each viral clone serving as a replicate. Statistical analysis was done on mutation frequencies with respect to organ, region, and mutation type (see Materials and Methods). A→G and U→C mutations were shown to be statistically more common than expected by chance (P < 0.0001). Non-A→G and non-U→C mutation frequencies for all organs were very similar (0.59 ± 0.22 [mean mutation frequency ± standard deviation]).
Three-way analysis of variance indicated that mutation frequencies for the SVAR versus G1 region within each organ were statistically equivalent. However, there were significantly different mutation frequencies among the organs (P = 0.02), particularly between the spleen and the lungs and liver (P = 0.0021), and A→G or U→C mutations were much more prevalent than would be expected by chance alone (P = 0.0001). Additionally, examination of the interaction term for organ and mutation type showed that the distribution of mutations among organs differed for A→G or U→C mutations versus non-A→G or non-U→C mutations (P = 0.017). Specifically, A→G or U→C mutation frequencies varied noticeably among organs while other kinds of mutations appeared to occur at a more constant (and overall lower) rate among the different organs.
Amino acid alignment of organ-specific G1 clones.
SNV sequences isolated from various organs from a deer mouse were analyzed at the amino acid level to determine regions of high and low antigenic diversity for the G1 domain (Fig. 3). Many of the amino acid mutations were found within certain areas of the protein, including the known immunodominant domain. These areas correlated well with regions of high surface probabilities and antigen index values. A large number of amino acid changes in G1 viral clones from the spleen and salivary gland were observed, whereas lung and liver viral clones have relatively few amino acid changes. As with mark-recapture deer mice, preferential sites of nucleotide and amino acid changes between viral clones were observed. Values were determined for dN/dS and dN − dS with VarPlot (see Materials and Methods) for deer mouse Z15 B7 (Fig. 3), as well as for other rodents analyzed for viral diversity. High dN/dS ratios or dN − dS values have been used as surrogate indicators for selection pressure during virus evolution (59). dN/dS ratios were highest in only certain areas of G1 (including a portion of the immunodominant domain) of deer mouse Z15 B7; however, these values were relatively low compared to values determined by those studying hypervariable regions of hepatitis C or HIV (51, 59). dN − dS values were highest in the flanking portions of the G1 region analyzed for deer mouse Z15 B7. Among different rodents, including deer mouse 86A943 (captured before and after seroconversion), dN/dS ratios and dN − dS values varied within regions of G1, with no apparent common hypervariable domain in viral clones isolated from each animal and no apparent increase in values after seroconversion (data not shown).
FIG. 3.
Inferred amino acid alignment of G1 organ-specific SNV clones from deer mouse Z15 B7. All organ-specific viral clones are shown aligned, with the consensus sequence given at the top. Each G1 clone is listed, with the organ description on the right side of the alignment. The relative abundance of each particular sequence is given as the percentage of the total number of clones analyzed (spleen, six; liver and salivary gland, five; kidney, lung, and bladder, four; and blood, three). The immunodominant domain of G1 is shown above the consensus sequence. Amino acid mutations found in viral clones from all animals are indicated as bars above the consensus sequence for deer mouse Z15 B7 (light gray, one mutation; dark gray, two mutations; black, three mutations). The antigenic index (Jameson-Wolf; DNASTAR Inc.) for the SNV G1 region is given directly below the alignment. Possible hypervariable regions of G1 for deer mouse Z15 B7 viral clones are boxed with heavy lines, and quasineutral changes are boxed with light lines. The variability plot (VarPlot for Windows) for deer mouse Z15 B7 is shown below the antigenic index graph. Both dN/dS ratios and dN − dS values were included in the analysis. aa, amino acids.
DISCUSSION
Viruses employ many different strategies to evade the immune system long enough to find a new host (40). Size constraints for RNA viruses limit their ability to establish persistence through the expression of immune evasion genes common in large DNA viruses (13, 22), and they must therefore rely on other mechanisms, such as antigenic variation (49). Hantavirus infection in rodents is characterized by an acute phase followed by a chronic infection in which virus is continuously being shed in the presence of an active immune response (17, 26).
Neutralizing antibodies were observed in deer mice despite the presence of SNV RNA in the blood and lungs. These results suggest that viral replication is occurring in deer mice even in the presence of a strong immune response. The neutralizing values determined for the deer mice tested may be an underestimation, due to possible antigenic differences between the laboratory isolate CC107 and the particular viral strain circulating within each deer mouse. However, the presence of neutralizing antibodies in infected deer mice raised the question of the mechanism of viral persistence in these animals.
The G1 region analyzed in our studies was chosen specifically for its antigenic potential and its possible importance in cell receptor binding and antibody neutralization (25, 27, 67). SNV diversity was seen in the immunodominant domain of G1, as well as in flanking regions of the domain. Over 60% of all nucleotide substitutions in the G1 region of SNV were found to be nonsynonomous, correlating with what has been found with Tula virus quasispecies (56). Many of the amino acid changes (41%) were encountered in the mutant spectra from different animals or in published isolates of SNV, suggesting that these particular mutations are well tolerated or quasineutral and may not adversely affect protein function (56). A slightly higher rate of viral diversity was observed in SVAR clones derived from the blood of one deer mouse and in certain organs of another deer mouse, suggesting that greater SNV diversity may be present in viral regions not subject to protein functional constraints.
Our phylogenetic analysis indicates that SNV variants cannot easily be divided into related clusters over sequential time points for mark-recapture deer mice. These results suggest that a single viral strain predominates in deer mice because of its greater replicative potential or fitness (16). Our results parallel studies of HIV and hepatitis C quasispecies, which exhibit stability and evolutionary stasis with optimally adapted strains, even in the presence of diverse viral forms (3, 68). Despite the lack of progressive nucleotide or amino acid shifts in the SNV population over time, these variants may be contributing to persistence by yielding analogous peptide epitopes which can still interact with SNV-specific T-cell receptors without delivering a full stimulatory signal (32, 33).
Consequently, SNV evolution may be proceeding over a time course longer than was studied for our catch-and-release rodents. We are currently determining whether SNV evolution within an individual deer mouse can be observed in sequential blood samples separated by a period of a year or more. Perhaps, the evolution of SNV variants with respect to progressive and accumulated amino acid changes in the viral genome over time may occur only after a genetic bottleneck, such as transmission of a minor variant to another deer mouse. Alternatively, fixation of nucleotide or amino acid changes may be occurring temporally and spatially for SNV-infected rodents in regions of the viral genome which were not chosen for this study.
It is important to note that the ability of RNA viruses to form quasispecies due to their error-prone polymerases or other environmental factors is independent of any immune response generated by the host (9, 14, 44). Rather, selection of a preexisting pool of variant genomes during a prolonged immune response or antiviral therapy may be responsible for an increase of certain mutants in the quasispecies population, which would normally have a lower replicative fitness (16). Although positive Darwinian selection forces acting on viral diversity during persistent infections have been well documented (68), the dynamics of quasispecies populations predicts that genetic variation will occur even in the absence of immune selection (13, 44). This may explain the SNV sequence heterogeneity seen in one particular deer mouse even before seroconversion. High viral replication rates during early infection may also be contributing to the viral diversity seen for this deer mouse.
The locations of certain amino acid substitutions may be more critical for the characterization of SNV variants evading the host immune system. Although SNV-specific CD4+- and CD8+-T-cell epitopes and an antibody immunodominant region have been determined from human studies (19, 27), rodent immune responses may target completely different regions, making such analysis difficult. We attempted to scan for regions of G1 undergoing selective pressure by using VarPlot, which calculates dN/dS ratios and dN − dS values in a sliding window. Analyses based on these surrogate indicators of immune pressure make the assumption that synonomous changes occur at a constant rate and are not selected against, whereas nonsynonomous mutations are selected by immune pressure. Although regions of high and low values were seen in G1 after plotting dN/dS and dN − dS, these values were lower than those previously seen for hypervariable domains of hepatitis C and HIV (51, 59). It is unclear whether synonomous mutations are completely neutral with respect to viral fitness; however, hantavirus genomes in general have extremely biased AT nucleotide compositions. This large nucleotide composition discrepancy suggests constraints against G or C mutations which may lead to variants with lower replicative fitness than wild-type sequences. Such nucleotide selection pressures would affect dN/dS and dN − dS values.
Significant differences in the level of SNV quasispecies were evident when organs from an infected deer mouse were compared. Higher SNV diversity was seen in the spleen, at both the nucleotide and amino acid levels. Low viral diversity was seen in lung and liver viral clones. The reasons for these differences may lie in the fact that the spleen is a major site of immune responses to bloodborne antigens (1). Viral immunological escape through the generation of viral diversity may be more critical in anatomical sites known to “sample” and respond to foreign antigens, such as the spleen and lymph nodes, where lymphocytes are found in high numbers. Alternatively, tissue-specific levels of a mutation-generating enzyme, such as dsRAD, may contribute to the observed spatial differences in SNV diversity (55).
No fixation of amino acid changes was apparent over time or with respect to different organs, although preferential sites of change were found in different viral clones. Only one SNV clone from the salivary gland of deer mouse Z15 B7 was found with a single-base deletion present in the SVAR region despite a large number of insertions and deletions between published sequences of SNV isolates (34). Our observation that SNV heterogeneity exists in the salivary gland and bladder is significant, since virus shedding and transmission may occur through contaminated saliva or urine (64). Viral transmission into new hosts is thought to be dependent on the transfer of a minor variant of the virus population (2, 42, 70). Also, the emergence of new viral pathogens is favored by viral diversity and the genetic plasticity of RNA viruses (16). With this in mind, it is notable that SNV spillover to other rodent species is a common occurrence (64). SNV variants found in abundance in the bladder may be contributing to mouse-to-human transmission and may increase the disease potential of the virus (2).
The large number of A→G or U→C transitional mutations found in SNV clones mirrors what has been found with other RNA viruses (46, 56). These specific mutations suggest a role for dsRAD, or a similar enzyme, in generating many of the changes identified in the SNV genome (6, 12). dsRAD has been demonstrated to be the enzyme responsible for editing the hepatitis delta virus antigenome RNA and has been implicated in the hypermutation of viral RNA genomes (6, 58). Interestingly, this enzyme has been found to be interferon inducible. It has been suggested that dsRAD, along with inosine-RNase, may form a cellular antiviral defense mechanism which acts to degrade dsRNA (62). dsRNA is routinely found in abundance during RNA viral infections as a consequence of either viral replication or transcription, and SNV RNA may be a target for a dsRAD-like enzyme in deer mice. By including only A→G or U→C mutations for analysis, viral diversity was found to be very different among organs, indicating potential tissue-specific levels of a dsRAD-like activity. Others have recently identified tissue-specific levels of dsRAD in various mammalian tissues (55). Alternatively, viral polymerases may be biased toward specific mutations (29).
The average mutation frequency seen for all viral clones isolated is similar to what has been found for other RNA viruses (15, 57), although slightly higher than that previously reported for Tula virus (56). This discrepancy may reflect the fact that each viral clone in our study was isolated from a different amplification reaction. Viral clones obtained from rodents with low viral burden and isolated from a single amplification may artificially limit true viral diversity due to resampling of the same input templates (38).
There exists the possibility that viral clones isolated from infected deer mouse organs may be contaminated with blood-specific SNV variants circulating through the organs. However, previous studies have shown both SNV genomic RNA and antigens present in resident cells of various tissues in both infected humans and deer mice, making it likely that these viral clones represent true organ-specific SNV variants (23, 45, 50, 69). Our analysis does not differentiate between SNV clones isolated from infected resident macrophages and those isolated from endothelial cells, although both cell types may be important in describing organ-specific SNV variants. Others have shown that HIV-infected resident macrophages isolated from various tissues may be contributing to the microevolution of HIV, both temporally and spatially (70).
These studies may help in understanding SNV persistence in deer mice and explain the SNV diversity found across different geographical areas (41). Viral diversity may also have implications for SNV pathogenesis and adaptability, which is particularly relevant with new findings that certain South American hantaviruses are likely to be transmitted from person to person (53). Variants found within the SNV population may facilitate the spread of the virus, both within the host and between species (42). In addition, these studies may give predictions of conserved epitopes in viral antigens for vaccine development. Future experiments will focus on the generation and characterization of antiviral and antibody SNV virus escape mutants generated in vitro.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI36418 and AI39808 and by National Cancer Institute grant CA09563.
We thank Elmer Otteson, Pascal Villard, Joan Rowe, and Kathryn Saxton for technical assistance and Gerold Feuer and Albert van Geelen for helpful comments and critical reading of the manuscript. We are also grateful to Stuart Ray for providing his VarPlot software program and for advice on how to use it.
REFERENCES
- 1.Abbas A K, Lichtman A H, Pober J S. Cellular and molecular immunology. Philadelphia, Pa: W. B. Saunders Company; 1998. pp. 14–31. [Google Scholar]
- 2.Baric R S, Sullivan E, Hensley L, Yount B, Chen W. Persistent infection promotes cross-species transmissibility of mouse hepatitis virus. J Virol. 1999;73:638–649. doi: 10.1128/jvi.73.1.638-649.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassett S E, Thomas D L, Brasky K M, Lanford R E. Viral persistence, antibody to E1 and E2, and hypervariable region 1 sequence stability in hepatitis C virus-inoculated chimpanzees. J Virol. 1999;73:1118–1126. doi: 10.1128/jvi.73.2.1118-1126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bharadwaj M, Botten J, Torrez-Martinez N, Hjelle B. Rio Mamore virus: genetic characterization of a newly recognized hantavirus of the pygmy rice rat, Oligoryzomys microtis, from Bolivia. Am J Trop Med Hyg. 1997;57:368–374. doi: 10.4269/ajtmh.1997.57.368. [DOI] [PubMed] [Google Scholar]
- 5.Boone J D, Otteson E W, McGwire K C, Villard P, Rowe J E, St. Jeor S C. Ecology and demographics of hantavirus infections in rodent populations in the Walker River Basin of Nevada and California. Am J Trop Med Hyg. 1998;59:445–451. doi: 10.4269/ajtmh.1998.59.445. [DOI] [PubMed] [Google Scholar]
- 6.Cattaneo R. Biased (A→I) hypermutation of animal RNA virus genomes. Curr Opin Genet Dev. 1994;4:895–900. doi: 10.1016/0959-437x(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 7.Childs J E, Ksiazek T G, Spiropoulou C F, Krebs J W, Morzunov S, Maupin G O, Gage K L, Rollin P E, Sarisky J, Enscore R E. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J Infect Dis. 1994;169:1271–1280. doi: 10.1093/infdis/169.6.1271. [DOI] [PubMed] [Google Scholar]
- 8.Chizhikov V E, Spiropoulou C F, Morzunov S P, Monroe M C, Peters C J, Nichol S T. Complete genetic characterization and analysis of isolation of Sin Nombre virus. J Virol. 1995;69:8132–8136. doi: 10.1128/jvi.69.12.8132-8136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 10.Condra J H, Schleif W A, Blahy O M, Gabryelski L J, Graham D J, Quintero J C, Rhodes A, Robbins H L, Roth E, Shivaprakash M. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 11.Dearing M D, Mangione A M, Karasov W H, Morzunov S, Otteson E, St. Jeor S. Prevalence of hantavirus in four species of Neotoma from Arizona and Utah. J Mammal. 1998;79:1254–1259. [Google Scholar]
- 12.de la Torre J C, Giachetti C, Semler B L, Holland J J. High frequency of single-base transitions and extreme frequency of precise multiple-base reversion mutations in poliovirus. Proc Natl Acad Sci USA. 1992;89:2531–2535. doi: 10.1073/pnas.89.7.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Domingo E, Baranowski E, Ruiz-Jarabo C M, Martin-Hernandez A M, Saiz J C, Escarmis C. Quasispecies structure and persistence of RNA viruses. Emerg Infect Dis. 1998;4:521–527. doi: 10.3201/eid0404.980402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domingo E, Diez J, Martinez M A, Hernandez J, Holguin A, Borrego B, Mateu M G. New observations on antigenic diversification of RNA viruses. Antigenic variation is not dependent on immune selection. J Gen Virol. 1993;74:2039–2045. doi: 10.1099/0022-1317-74-10-2039. [DOI] [PubMed] [Google Scholar]
- 15.Domingo E, Holland J J. Mutation rates and rapid evolution of RNA viruses. In: Morse S S, editor. The evolutionary biology of viruses. New York, N.Y: Raven Press; 1994. pp. 161–184. [Google Scholar]
- 16.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 17.Doyle T J, Bryan R T, Peters C J. Viral hemorrhagic fevers and hantavirus infections in the Americas. Infect Dis Clin N Am. 1998;12:95–110. doi: 10.1016/s0891-5520(05)70411-6. [DOI] [PubMed] [Google Scholar]
- 18.Eigen M. Viral quasispecies. Sci Am. 1993;269:42–49. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- 19.Ennis F A, Cruz J, Spiropoulou C F, Waite D, Peters C J, Nichol S T, Kariwa H, Koster F T. Hantavirus pulmonary syndrome: CD8+ and CD4+ cytotoxic T lymphocytes to epitopes on Sin Nombre virus nucleocapsid protein isolated during acute illness. Virology. 1997;238:380–390. doi: 10.1006/viro.1997.8827. [DOI] [PubMed] [Google Scholar]
- 20.Gavrilovskaya I N, Shepley M, Shaw R, Ginsberg M H, Mackow E R. β3 integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc Natl Acad Sci USA. 1998;95:7074–7079. doi: 10.1073/pnas.95.12.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Scarano F, Nathanson N. Bunyaviridae. 1996. pp. 1473–1504. . In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 22.Gooding L R. Virus proteins that counteract host immune defenses. Cell. 1992;71:5–7. doi: 10.1016/0092-8674(92)90259-f. [DOI] [PubMed] [Google Scholar]
- 23.Green W, Feddersen R, Yousef O, Behr M, Smith K, Nestler J, Jenison S, Yamada T, Hjelle B. Tissue distribution of hantavirus antigen in naturally infected humans and deer mice. J Infect Dis. 1998;177:1696–1700. doi: 10.1086/515325. [DOI] [PubMed] [Google Scholar]
- 24.Henderson W W, Monroe M C, St. Jeor S C, Thayer W P, Rowe J E, Peters C J, Nichol S T. Naturally occurring Sin Nombre virus genetic reassortants. Virology. 1995;214:602–610. doi: 10.1006/viro.1995.0071. [DOI] [PubMed] [Google Scholar]
- 25.Hjelle B, Chavez-Giles F, Torrez-Martinez N, Yamada T, Sarisky J, Ascher M, Jenison S. Dominant glycoprotein epitope of four corners hantavirus is conserved across a wide geographical area. J Gen Virol. 1994;75:2881–2888. doi: 10.1099/0022-1317-75-11-2881. [DOI] [PubMed] [Google Scholar]
- 26.Hutchinson K L, Rollin P E, Peters C J. Pathogenesis of a North American hantavirus, Black Creek Canal virus, in experimentally infected Sigmodon hispidus. Am J Trop Med Hyg. 1998;59:58–65. doi: 10.4269/ajtmh.1998.59.58. [DOI] [PubMed] [Google Scholar]
- 27.Jenison S, Yamada T, Morris C, Anderson B, Torrez-Martinez N, Keller N, Hjelle B. Characterization of human antibody responses to four corners hantavirus infections among patients with hantavirus pulmonary syndrome. J Virol. 1994;68:3000–3006. doi: 10.1128/jvi.68.5.3000-3006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson A M, Bowen M D, Ksiazek T G, Williams R J, Bryan R T, Mills J N, Peters C J, Nichol S T. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology. 1997;238:115–127. doi: 10.1006/viro.1997.8840. [DOI] [PubMed] [Google Scholar]
- 29.Keulen W, Back N K, van Wijk A, Boucher C A, Berkhout B. Initial appearance of the 184Ile variant in lamivudine-treated patients is caused by the mutational bias of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 1997;71:3346–3350. doi: 10.1128/jvi.71.4.3346-3350.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan A S, Gaviria M, Rollin P E, Hlady W G, Ksiazek T G, Armstrong L R, Greenman R, Ravkov E, Kolber M, Anapol H, Sfakianaki E D, Nichol S T, Peters C J, Khabbaz R F. Hantavirus pulmonary syndrome in Florida: association with the newly identified Black Creek Canal virus. Am J Med. 1996;100:46–48. doi: 10.1016/s0002-9343(96)90010-8. [DOI] [PubMed] [Google Scholar]
- 31.Khan A S, Khabbaz R F, Armstrong L R, Holman R C, Bauer S P, Graber J, Strine T, Miller G, Reef S, Tappero J, Rollin P E, Nichol S T, Zaki S R, Bryan R T, Chapman L E, Peters C J, Ksiazek T G. Hantavirus pulmonary syndrome: the first 100 US cases. J Infect Dis. 1996;173:1297–1303. doi: 10.1093/infdis/173.6.1297. [DOI] [PubMed] [Google Scholar]
- 32.Klenerman P, Rowland-Jones S, McAdam S, Edwards J, Daenke S, Lalloo D, Koppe B, Rosenberg W, Boyd D, Edwards A. Cytotoxic T-cell activity antagonized by naturally occurring HIV-1 Gag variants. Nature. 1994;369:403–407. doi: 10.1038/369403a0. [DOI] [PubMed] [Google Scholar]
- 33.Klenerman P, Zinkernagel R M. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature. 1998;394:482–485. doi: 10.1038/28860. [DOI] [PubMed] [Google Scholar]
- 34.Kukkonen S K, Vaheri A, Plyusnin A. Completion of the Tula hantavirus genome sequence: properties of the L segment and heterogeneity found in the 3′ termini of S and L genome RNAs. J Gen Virol. 1998;79:2615–2622. doi: 10.1099/0022-1317-79-11-2615. [DOI] [PubMed] [Google Scholar]
- 35.Kumar, S., K. Tamura, and M. Nei. MEGA: molecular evolutionary genetics analysis, version 1.0. 1993. The Pennsylvania State University, University Park, Pa.
- 36.Levis S, Morzunov S P, Rowe J E, Enria D, Pini N, Calderon G, Sabattini M, St. Jeor S C. Genetic diversity and epidemiology of hantaviruses in Argentina. J Infect Dis. 1998;177:529–538. doi: 10.1086/514221. [DOI] [PubMed] [Google Scholar]
- 37.Li D, Schmaljohn A L, Anderson K, Schmaljohn C S. Complete nucleotide sequences of the M and S segments of two hantavirus isolates from California: evidence for reassortment in nature among viruses related to hantavirus pulmonary syndrome. Virology. 1995;206:973–983. doi: 10.1006/viro.1995.1020. [DOI] [PubMed] [Google Scholar]
- 38.Liu S L, Rodrigo A G, Shankarappa R, Learn G H, Hsu L, Davidov O, Zhao L P, Mullins J I. HIV quasispecies and resampling. Science. 1996;273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- 39.Lundkvist A, Cheng Y, Sjolander K B, Niklasson B, Vaheri A, Plyusnin A. Cell culture adaptation of Puumala hantavirus changes the infectivity for its natural reservoir, Clethrionomys glareolus, and leads to accumulation of mutants with altered genomic RNA S segment. J Virol. 1997;71:9515–9523. doi: 10.1128/jvi.71.12.9515-9523.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marrack P, Kappler J. Subversion of the immune system by pathogens. Cell. 1994;76:323–332. doi: 10.1016/0092-8674(94)90339-5. [DOI] [PubMed] [Google Scholar]
- 41.Monroe M C, Morzunov S P, Johnson A M, Bowen M D, Artsob H, Yates T, Peters C J, Rollin P E, Ksiazek T G, Nichol S T. Genetic diversity and distribution of Peromyscus-borne hantaviruses in North America. Emerg Infect Dis. 1999;5:75–86. doi: 10.3201/eid0501.990109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morimoto K, Hooper D C, Carbaugh H, Fu Z F, Koprowski H, Dietzschold B. Rabies virus quasispecies: implications for pathogenesis. Proc Natl Acad Sci USA. 1998;95:3152–3156. doi: 10.1073/pnas.95.6.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morzunov S P, Rowe J E, Ksiazek T G, Peters C J, St. Jeor S, Nichol S T. Genetic analysis of the diversity and origin of hantaviruses in Peromyscus leucopus mice in North America. J Virol. 1998;72:57–64. doi: 10.1128/jvi.72.1.57-64.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Najera I, Holguin A, Quinones-Mateu M E, Munoz-Fernandez M A, Najera R, Lopez-Galindez C, Domingo E. Pol gene quasispecies of human immunodeficiency virus: mutations associated with drug resistance in virus from patients undergoing no drug therapy. J Virol. 1995;69:23–31. doi: 10.1128/jvi.69.1.23-31.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Netski D, Thran B H, St. Jeor S C. Sin nombre virus pathogenesis in Peromyscus maniculatus. J Virol. 1999;73:585–591. doi: 10.1128/jvi.73.1.585-591.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nichol S. RNA viruses. Life on the edge of catastrophe. Nature. 1996;384:218–219. doi: 10.1038/384218a0. [DOI] [PubMed] [Google Scholar]
- 47.Nichol S T, Spiropoulou C F, Morzunov S, Rollin P E, Ksiazek T G, Feldmann H, Sanchez A, Childs J, Zaki S, Peters C J. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–917. doi: 10.1126/science.8235615. [DOI] [PubMed] [Google Scholar]
- 48.Nowak M A, Bangham C R. Population dynamics of immune responses to persistent viruses. Science. 1996;272:74–79. doi: 10.1126/science.272.5258.74. [DOI] [PubMed] [Google Scholar]
- 49.Nowak M A, May R M, Phillips R E, Rowland-Jones S, Lalloo D G, McAdam S, Klenerman P, Koppe B, Sigmund K, Bangham C R. Antigenic oscillations and shifting immunodominance in HIV-1 infections. Nature. 1995;375:606–611. doi: 10.1038/375606a0. [DOI] [PubMed] [Google Scholar]
- 50.Nuovo G J, Simsir A, Steigbigel R T, Kuschner M. Analysis of fatal pulmonary hantaviral infection in New York by reverse transcriptase in situ polymerase chain reaction. Am J Pathol. 1996;148:685–692. [PMC free article] [PubMed] [Google Scholar]
- 51.Ostrowski M A, Krakauer D C, Li Y, Justement S J, Learn G, Ehler L A, Stanley S K, Nowak M, Fauci A S. Effect of immune activation on the dynamics of human immunodeficiency virus replication and on the distribution of viral quasispecies. J Virol. 1998;72:7772–7784. doi: 10.1128/jvi.72.10.7772-7784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otteson E W, Riolo J, Rowe J E, Nichol S T, Ksiazek T G, Rollin P E, St. Jeor S C. Occurrence of hantavirus within the rodent population of northeastern California and Nevada. Am J Trop Med Hyg. 1996;54:127–133. doi: 10.4269/ajtmh.1996.54.127. [DOI] [PubMed] [Google Scholar]
- 53.Padula P J, Edelstein A, Miguel S D, Lopez N M, Rossi C M, Rabinovich R D. Hantavirus pulmonary syndrome outbreak in Argentina: molecular evidence for person-to-person transmission of Andes virus. Virology. 1998;241:323–330. doi: 10.1006/viro.1997.8976. [DOI] [PubMed] [Google Scholar]
- 54.Page R D M. TREEVIEW: An application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 55.Paul M S, Bass B L. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plyusnin A, Cheng Y, Lehvaslaiho H, Vaheri A. Quasispecies in wild-type tula hantavirus populations. J Virol. 1996;70:9060–9063. doi: 10.1128/jvi.70.12.9060-9063.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plyusnin A, Vapalahti O, Lehvaslaiho H, Apekina N, Mikhailova T, Gavrilovskaya I, Laakkonen J, Niemimaa J, Henttonen H, Brummer-Korvenkontio M. Genetic variation of wild Puumala viruses within the serotype, local rodent populations and individual animal. Virus Res. 1995;38:25–41. doi: 10.1016/0168-1702(95)00038-r. [DOI] [PubMed] [Google Scholar]
- 58.Polson A G, Bass B L, Casey J L. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 59.Ray S C, Wang Y M, Laeyendecker O, Ticehurst J R, Villano S A, Thomas D L. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J Virol. 1999;73:2938–2946. doi: 10.1128/jvi.73.4.2938-2946.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez L L, Owens J H, Peters C J, Nichol S T. Genetic reassortment among viruses causing hantavirus pulmonary syndrome. Virology. 1998;242:99–106. doi: 10.1006/viro.1997.8990. [DOI] [PubMed] [Google Scholar]
- 61.Rowe J E, St. Jeor S C, Riolo J, Otteson E W, Monroe M C, Henderson W W, Ksiazek T G, Rollin P E, Nichol S T. Coexistence of several novel hantaviruses in rodents indigenous to North America. Virology. 1995;213:122–130. doi: 10.1006/viro.1995.1552. [DOI] [PubMed] [Google Scholar]
- 62.Scadden A D, Smith C W. A ribonuclease specific for inosine-containing RNA: a potential role in antiviral defence? EMBO J. 1997;16:2140–2149. doi: 10.1093/emboj/16.8.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmaljohn A L, Li D, Negley D L, Bressler D S, Turell M J, Korch G W, Ascher M S, Schmaljohn C S. Isolation and initial characterization of a newfound hantavirus from California. Virology. 1995;206:963–972. doi: 10.1006/viro.1995.1019. [DOI] [PubMed] [Google Scholar]
- 64.Schmaljohn C, Hjelle B. Hantaviruses: a global disease problem. Emerg Infect Dis. 1997;3:95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmaljohn C S. Bunyaviridae: The viruses and their replication. 1996. pp. 1447–1471. . In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 66.Spiropoulou C F, Morzunov S, Feldmann H, Sanchez A, Peters C J, Nichol S T. Genome structure and variability of a virus causing hantavirus pulmonary syndrome. Virology. 1994;200:715–723. doi: 10.1006/viro.1994.1235. [DOI] [PubMed] [Google Scholar]
- 67.Wang M, Pennock D G, Spik K W, Schmaljohn C S. Epitope mapping studies with neutralizing and non-neutralizing monoclonal antibodies to the G1 and G2 envelope glycoproteins of Hantaan virus. Virology. 1993;197:757–766. doi: 10.1006/viro.1993.1652. [DOI] [PubMed] [Google Scholar]
- 68.Wolinsky S M, Korber B T, Neumann A U, Daniels M, Kunstman K J, Whetsell A J, Furtado M R, Cao Y, Ho D D, Safrit J T. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 69.Zaki S R, Greer P W, Coffield L M, Goldsmith C S, Nolte K B, Foucar K, Feddersen R M, Zumwalt R E, Miller G L, Khan A S. Hantavirus pulmonary syndrome. Pathogenesis of an emerging infectious disease. Am J Pathol. 1995;146:552–579. [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu T, Wang N, Carr A, Nam D S, Moor-Jankowski R, Cooper D A, Ho D D. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J Virol. 1996;70:3098–3107. doi: 10.1128/jvi.70.5.3098-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]