Abstract
Aging poses a global public health challenge, which is linked to the rise of age-related lung diseases. The precise understanding of the molecular and genetic changes in the aging lung that elevate the risk of acute and chronic lung diseases remains incomplete. Alveolar type II (AT2) cells are stem cells that maintain epithelial homeostasis and repair the lung after injury. AT2 progenitor function decreases with aging. The maintenance of AT2 function requires niche support from other cell types, but little has been done to characterize alveolar alterations with aging in the AT2 niche. To systematically profile the genetic changes associated with age, we present a single-cell transcriptional atlas comprising nearly half a million cells from the healthy lungs of human subjects spanning various ages, sexes, and smoking statuses. Most annotated cell lineages in aged lungs exhibit dysregulated genetic programs. Specifically, the aged AT2 cells demonstrate loss of epithelial identities, heightened inflammaging characterized by increased expression of AP-1 (Activator Protein-1) transcription factor and chemokine genes, and significantly increased cellular senescence. Furthermore, the aged mesenchymal cells display a remarkable decrease in collagen and elastin transcription and a loss of support to epithelial cell stemness. The decline of the AT2 niche is further exacerbated by a dysregulated genetic program in macrophages and dysregulated communications between AT2 and macrophages in aged human lungs. These findings highlight the dysregulations observed in both AT2 stem cells and their supportive niche cells, potentially contributing to the increased susceptibility of aged populations to lung diseases.
Keywords: aging, human lung, alveolar type II cell, niche, macrophage
Clinical Relevance
This study has drawn a comprehensive transcriptomics map of healthy human lungs. A deep excavation of this map will hopefully improve our understanding of lung degeneration and exhaustion in elderly people, as well as age-associated deficiency and pathogenesis in respiratory diseases.
In the past century, human life expectancy has nearly doubled globally, with an increase more significant than that in all previous millennia combined. By 2019, more than 600 million people were 65 years old or older, and this number will likely triple by 2050 (1). Increases in human age necessitate new scientific understanding of how to both extend and enhance health over the course of longer human lives (2). The respiratory system, representing a unique interface with the outside environment, is one of the major organs that is reported to be most affected by human aging (3). Several lung diseases have aging as a major risk factor, including chronic obstructive pulmonary disease (COPD), interstitial fibrotic lung disease, lung cancer, and inflammatory lung diseases such as pneumonia and coronavirus disease (COVID-19) (3, 4). However, it is not fully appreciated what the mechanistic, molecular, and genetic drivers of lung aging are and how aging increases susceptibility to these acute and chronic lung diseases.
Alveoli, the essential units for lung gas exchange, consist of epithelial and mesenchymal populations. Alveolar type II epithelial cells (AT2s) have been well accepted as progenitor cells that maintain alveolar homeostasis and repair damaged epithelium after injury (5). However, AT2s exhibit reduced self-renewal and differentiation capacity with aging (6). Endothelial and mesenchymal cells, as well as the immune components, especially the macrophages (7), participate in the alveolar niches (8–10) and are also affected by aging. As an example, the aging of lung mesenchymal cells, vital for supporting AT2 renewal (11), further impairs AT2 cell function (9). However, a comprehensive dissection of the AT2 niches during lung aging using a large dataset is lacking.
Recent studies have greatly benefited from the advancements in next-generation sequencing technologies, especially single-cell RNA sequencing (scRNA-seq), which has profiled healthy and diseased human lungs (12–16). Specifically, progressive lung diseases with increased prevalence with aging, including idiopathic pulmonary fibrosis (IPF), COPD, COVID-19, and lung cancers, have been better described, with known cell population delineated, novel cell types identified, and changes in cellular phenotypes and gene expression patterns better characterized (17). The hallmarks of these diseases, such as metabolic defects, genomic instability, telomere attrition, cellular senescence, and genomic and epigenetic alterations, overlap with those that occur during aging and may act as sensitizers to these diseases (18–28). Although the single-cell transcriptomics atlas on these diseases has generated a huge amount of information on molecular and cellular profiles and disease-specific alterations in the lungs, limited studies have focused on aging of human lungs (29, 30), and longitudinal transcriptomics profiling of human lungs at the single-cell level across all adult age ranges is not available.
To fill the gap, here we generated a single-cell transcriptomics atlas of 491,187 human lung cells from 14 datasets with 92 adult healthy subjects of different ages, sexes, and smoking statuses. The comprehensive analyses elucidated the changes of transcriptional programs in all cell types in the lung during aging. Strikingly, we found a significant decline in the AT2 niches, characterized by the aberrantly inflammaging profile of AT2s themselves, and a decay of aged stromal cells in supporting alveolar epithelium. Dysregulated alveolar niches were further presented by dysregulated transcriptional programs in the immune systems in aged lungs. Moreover, cell–cell communication analyses suggested a strong proinflammatory niche between AT2 cells and macrophages in aged subjects. Thus, these detailed observations highlight significant dysregulations in the aging of human lungs, shedding light on the mechanisms behind the decline in lung capacity and resilience and the increased susceptibility to diseases in older populations. The human lung atlas presented here serves as a valuable resource, resembling a pathogenetic-like library of aging lungs, and targeting specific molecular or cellular factors could potentially offer novel insights for intervening in age-related lung diseases.
Some of the results of these studies have been previously reported in the form of a preprint (31).
Methods
Subject Details
All human lung experiments were approved by the Cedars-Sinai Medical Center Institutional Review Board (IRB) and were in accordance with the guidelines outlined by the IRB. Informed consent was obtained from each subject (IRB: Pro00032727). For histological verifications on human lungs, slides from three young and three aged donors were used: CC02-19, 37-year-old male with no smoking history; CC07-19, 18-year-old male with no smoking history; CC07-21, 32-year-old female with no smoking history; CC06-19 (male) and CC08-19 (female), both donors 78 years old with no smoking history; and CC01-20, 72-year-old female with no smoking history.
Data Collection
The original processed data on human lungs of healthy donors were reaccessed either from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) or LungMAP (https://www.lungmap.net/). These data included scRNA-seq (LungMAP, GSE135893, GSE128033, GSE122960, GSE136831, GSE132771, GSE146981, GSE190889, GSE157996, GSE135851, GSE158127, and GSE168191) and single-nucleus RNA sequencing (GSE161382 and GSE171524). All or specific cell or nucleus types were enriched in each dataset and the detailed cell or nucleus types of the original studies were described (see Table E1 in the data supplement).
Data Integration, Visualization, and Comparative Analysis
Quality control, cell clustering, and cell type annotation were conducted and the bioinformatic quantification and data visualization were performed following the computational, quantificational, and statistical methods as described previously (32). Detailed methods can be found in the data supplement.
Ingenuity Pathway Analysis and Fast Gene-Set Enrichment Analysis
Ingenuity Pathway Analysis (IPA) was performed as described previously (33) and in the data supplement. Gene-set enrichment analysis (GSEA) was conducted with fgsea, an R-package for fast preranked GSEA, following authors’ detailed vignettes (https://github.com/ctlab/fgsea). In brief, the differentially expressed genes in aged versus young AT2 cells were calculated, and hallmark gene sets were accessed from GSEA web (https://www.gsea-msigdb.org/gsea/index.jsp). The top signaling pathways were determined based on the expression levels (avg_log2FC) of the genes they accessed, and then normalized enrichment scores were calculated. The significantly activated signaling pathways on aged AT2 cells were ranked based on the normalized enrichment scores, and the top pathways were presented by bar plots. Individual pathways were visualized by Enrichment Plots.
Inference and Analysis of Cell–Cell Communication by CellChat
The cell–cell communications or interactions among different annotated cell types were performed and visualized by newly updated CellChat following the generator’s vignette (https://github.com/sqjin/CellChat) (34). Detailed methods can be found in the data supplement.
Histology and Immunofluorescence Staining
Histology and immunofluorescence analysis were done mainly as verifications of the bioinformatic analysis. Immunofluorescence was performed on both paraffin-embedded and optimal cutting temperature compound–embedded sections as described previously (32). Detailed methods can be found in the data supplement.
Quantification and Statistical Analysis
For the subject and cell numbers of the integrated data, the percentages were calculated and visualized in GraphPad, and the exact values were shown. The gene-expression–positive cells were counted by their actual gene transcription >0, and the percentage was determined by normalization of the positive cell number to total cell number.
Results
Generation of the Single-Cell Transcriptional Atlas of Healthy Human Lungs
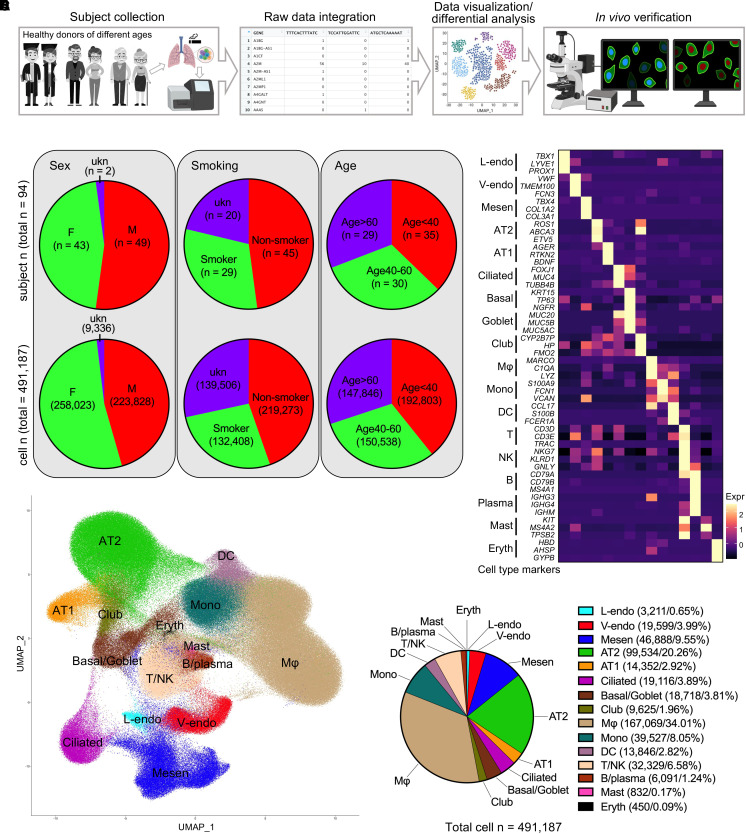
To generate a single-cell transcriptomics map of healthy human lungs and to study the mechanisms underlying lung aging, we used a substantial number of single-cell multiomics databases and isolated data on healthy human lungs from 14 published databases (13, 14, 16, 18, 28, 35–42) containing 94 healthy subjects and 491,187 cells (Figure 1A and Table E1). The donors spanned wide age ranges (Figure E1A), included both sexes, had varying smoking histories, and represented different ethnic races (Figure E1B).
Figure 1.
Integration of single-cell RNA sequencing data on human lungs from healthy subjects. (A) Schematic overview of the data collection, integration, differential analysis, and histological verifications of the healthy human lungs. The donors were all adults from different sex, age, and smoking history statuses. (B) The subjects and the cells were divided regarding donor sex, smoking history, and age stages. For grouping by ages, the subjects were evenly divided into three groups: age < 40 years, age 40–60 years, and age > 60 years. Pie charts are used to illustrate the corresponding subject and cell numbers and proportions. (C) The average transcriptions of the canonical cell type marker genes (rows) in cell types (columns) are determined and visualized by heatmap in the integrated data. (D) The distribution of the cells in the integrated data is visualized by Uniform Manifold Approximation and Projections (UMAP), and major cell types are identified based on the expression of the cell type marker genes in C and Figure E1J. (E) Cell number of each major cell type in the integrated data was determined, and the proportion of each cell type is quantified and visualized by pie chart. Color codes are followed by cell type annotations and cell numbers and percentages. AT1/2 = alveolar type I/II epithelial cells; DC = dendritic cells; Eryth = erythrocytes; F = female; L/V-endo = lymphatic/vascular endothelial cells; M = male; Mesen = mesenchymal cells; Mono = monocytes; Mφ = macrophages; ukn = unknown.
To study the lung transcriptional profiles under different conditions, the subjects and cells were divided based on donor identities, including sex (female, male, and unknown sex), smoking history (smoker, nonsmoker, and smoking history unknown), and age range (<40 yr, 40–60 yr, and >60 yr) (Figure 1B). The cells were then integrated, and cell distributions were visualized using Uniform Manifold Approximation and Projections (UMAPs). The comparable sequencing depth of different datasets was confirmed, although the transcription of some genes in GSE135893 was far beyond the scale of other datasets (Figure E1C). Therefore, the following comparative analyses were conducted with this dataset separated, unless mentioned otherwise. Well-performed batch corrections (Figures E1D–E1H) were confirmed, and the cells were then clustered (Figure E1I) in the integrated data. We then cataloged the cells into 15 major cell types annotated with the transcriptions of canonical marker genes (Figures 1C and E1J), composed of five distinct lineages, including endothelial (lymphatic and vascular endothelial cells), mesenchymal, epithelial (AT2, AT1, ciliated, basal/goblet, and club cells), myeloid (macrophages, monocytes, dendritic cells, mast cells, and erythrocytes), and lymphoid (T/NK and B/plasma cells) lineages (Figures 1D and 1E).
Accelerated Cellular Senescence and Loss of Epithelial Identities in Aged AT2s
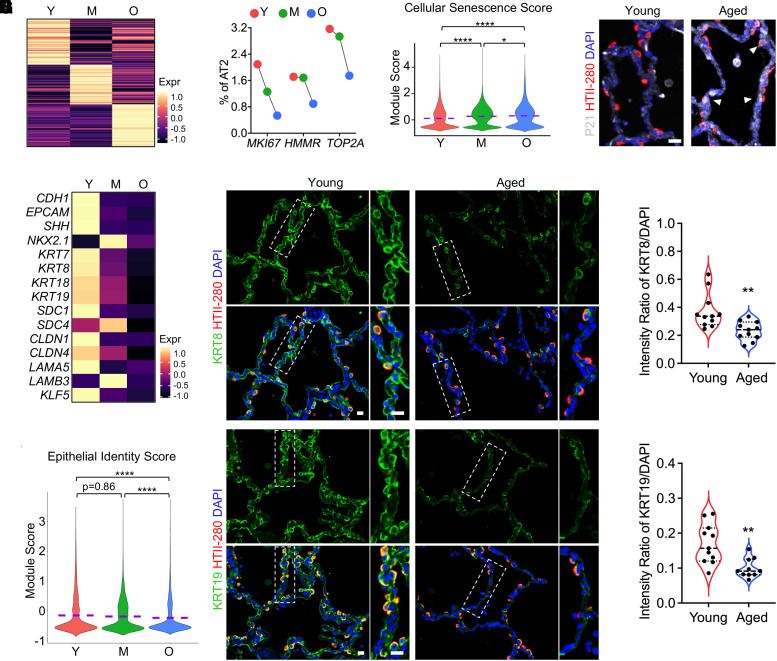
The respiratory epithelium is one of the major cellular components in human lungs, with alveolar epithelial cells being particularly impacted by the aging (3). AT2 cells, surfactant-secreting cells, serve as the epithelial lineage progenitors in the alveoli (5). To examine the aging process of human alveolar epithelial cells, AT2 cells were isolated, and differential expression analysis revealed a unique gene expression profile of AT2 cells from each age group (Figure 2A). Notably, aged AT2 cells underwent a significant decrease in cell proliferation rate, as indicated by a reduction in the proportion of cells that were positive for MKI67, HMMR, or TOP2A (Figure 2B). Furthermore, elevated cellular senescence, a hallmark of aging (43), was found in the aged AT2 cells. This was confirmed by both bioinformatically increased senescence marker genes (Figures E2A and E2B) and the cellular senescence score returned by the average expression of core senescence genes, as described previously (38) (Figure 2C), and histologically upregulated P21 (CDKN1A) protein expression, a commonly recognized cellular senescence marker, in HTII-280+ AT2 cells in human lung sections from aged donor (Figure 2D).
Figure 2.
Elevated cellular senescence and loss of epithelial identities in AT2 cells of aged human lungs. (A) Heatmaps show the expression of top 100 genes (rows) with distinct gene profiles of AT2 cells from subjects of different age stages (columns). (B) Percentages of AT2 cells positive for cell-proliferating marker genes MKI67, HMMR, and TOP2A are quantified from subjects of different age stages. (C) Violin plots show the cellular senescence scores of AT2 cells from different age stages. The cellular senescence score is defined as the average transcription of 130 core senescence genes from CSGene that were detectable in the AT2 cells. (D) Representative immunofluorescence staining of cell senescence marker (CDKN1A/P21) and human AT2 cell marker (HTII-280) on human lung sections from young and aged donors. White arrowheads indicate the senescent AT2 cells. (E and F) Heatmaps compare the relative transcriptions of the epithelial identity genes (E), and violin plots show the average expression of the epithelial identity genes (F) in AT2 from subjects of different age stages. (G) Immunofluorescence costaining of epithelial identity genes KRT8 and KRT19 with human AT2 cell marker (HTII-280) in alveolar regions of human lung sections from young and aged donors. Boxed regions are magnified. (H) Quantified relative intensity ratios for KRT8 and KRT19 staining compared with nuclear DAPI on human lung sections from young and aged donors (n = 11 sections from three subjects per group). Purple dashed lines in the violin plots indicate the mean levels of module score and gene transcriptions. Scale bars, 20 μm (D and G). *P < 0.05, **P < 0.01, and ****P < 0.0001. M = middle-aged donors (age 40–60 yr); O = aged (old) donors (age > 60 yr); Y = young donors (age < 40 yr).
The transcription of surfactant protein genes, together with the lamellar body marker gene ABCA3, signifies the typical functions of AT2 cells (44). Most of these genes showed decreased transcription in the aged AT2 cells in most individual datasets, although there was some inconsistency in other datasets (Figure E2C). ETV5, a transcription factor essential for the maintenance of AT2 cells (45), showed slightly impaired transcription in aged AT2 cells (Figure E2C).
The other downregulated genes in aged AT2s were represented by Small Nucleolar RNA Host Genes (SNHG7 and SNHG8), Keratin genes (KRT7, KRT8, KRT18, and KRT19), S100 Protein genes (S100A10, S100A14, and S100A16), Serpin family genes (SERPINA1, SERPINB1, and SERPINH1), and Solute carrier family genes (SLC1A5, SLC25A3, SLC25A5, SLC25A6, SLC34A2, and SLC39A8) (Figures E2D and E2E). Among these downregulated genes, those representing the epithelial identities were dominant (Figure E2D). Defective transcription of these genes, together with other downregulated epithelial identity genes (Figure 2E) and a decreased epithelial identity score indicated by the average expression of these genes (Figure 2F), suggested a loss of epithelial identity in aged AT2 cells in human lungs. This was further confirmed by histological loss of KRT8 and KRT19, representative epithelial feature proteins, in alveolar regions of aged human lungs (Figures 2G and 2H). Among these AT2-specific and dysregulated genes, we reported recently that SLC39A8 (ZIP8) is required for AT2 cell renewal in murine and human lungs (18).
Aberrantly Activated AP-1 and Elevated Inflammation in Human AT2s of Aged Lungs
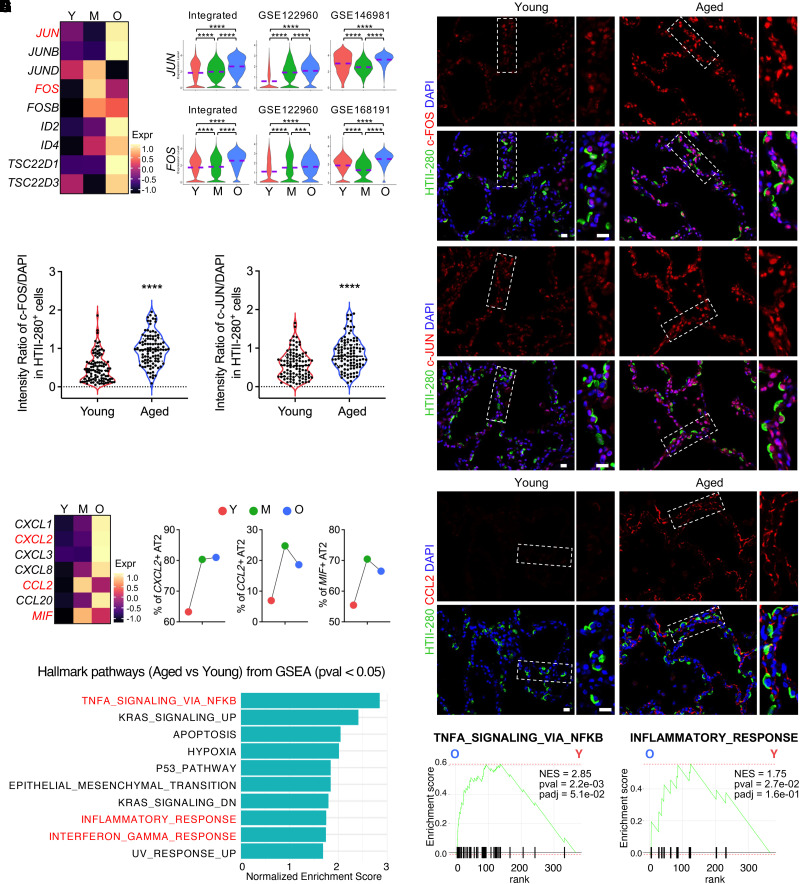
Surprisingly, a set of AP-1 (Activator Protein-1) transcription factor subunit genes, including JUN, JUNB, JUND, FOS, and FOSB, were found to be elevated both bioinformatically (Figures 3A and 3B) and histologically (Figures 3C and 3D) in the aged AT2 cells. AP-1 has been demonstrated to lead a transcription factor network that drives the transcriptional program of cellular senescence in cell culture (46) and immune aging in murine tissues (47, 48), implying a relevance of the activation of these genes with AT2 cell senescence and inflammation in human lungs. The other significantly activated transcription factor genes included ID genes (ID2 and ID4) and TSC22 domain family genes (TSC22D1 and TSC22D3) (Figure 3A), the roles of which in AT2 cells are to be determined.
Figure 3.
Activated AP-1 (Activator Protein-1) and elevated inflammation in AT2 cells of aged human lungs. (A and B) Heatmap (A) and violin plot (B) visualization of the upregulated AP-1 transcriptional factor genes in aged AT2 cells in the integrated data and representative individual datasets. (C) Immunofluorescence staining presented the protein levels of representative AP-1 transcription factor gene, c-FOS (FOS) and c-JUN (JUN), in AT2 cells (HTII-280+) on human lung sections from young and aged donors. Boxed regions are magnified. (D) Quantified relative intensity ratios for c-FOS and c-JUN staining in AT2 cells (HTII-280+) compared with nuclear DAPI on human lung sections from young and aged donors (n = 100 cells from three subjects per group). (E) Heatmaps show the transcriptional levels of chemokine and cytokine genes in aged AT2 cells in the integrated data. (F) Percentage quantifications of AT2 cells positive for the representative chemokine and cytokine genes, CXCL2, CCL2, and MIF, in lungs from subjects of different age stages. (G) Immunofluorescence staining for the representative chemokine protein, CCL2, in alveolar regions on human lung sections from young and aged donors. Boxed regions are magnified. (H and I) Top canonical Hallmark pathways from GSEA (pval < 0.05) on genes upregulated in aged AT2 cells identified by Fast GSEA (FGSEA) are illustrated (H) and representative pathways are visualized by enrichment plots (I). Purple dashed lines in the violin plots indicate the mean levels of module score and gene transcriptions. Scale bars, 20 μm (C and G). ***P < 0.001 and ****P < 0.0001. GSEA = gene-set enrichment analysis; NES = normalized enrichment score.
Alveolar epithelial cells have been reported to secrete chemokines in response to some pathogenetic stimulations (49, 50). In the aged AT2 cells, several chemokine and cytokine genes, such as CXCL1, CXCL2, CXCL3, CXCL8, CCL2, CCL20, and MIF (macrophage migration inhibitory factor), were found to be remarkably upregulated (Figure 3E). Among these chemokine and cytokine genes, CXCL2, CCL2, and MIF-positive AT2 cells were representatively quantified and were found to be increased with aging (Figure 3F). CCL2 was chosen as the representative gene to be histologically verified in the aged AT2 cells on human lung sections (Figure 3G). These secreted chemokines and cytokines suggested the activated cell–cell communications of aged AT2 within the alveolar microenvironments, although most of their receptor genes were of low transcriptional levels except CD74 (Figure E3A), the canonical receptor for MIF (51). To study the potential pathways regulating the DE genes of the aged AT2 cells, we used an R-based FGSEA for the pathway analysis on the aged versus young AT2 cells. Remarkably, among the top hallmark pathways of aged AT2 cells, many are related to activated inflammatory response (Figures 3H and 3I). To further confirm this, we input the significantly upregulated genes of aged AT2 cells into another leading pathway analysis application, IPA. Consistently, most of the top activated signaling pathways were myeloid- or lymphoid-related inflammation pathways (Figure E3B). These data suggest an activated inflammatory reaction in AT2 cells from aged human lungs.
Diminished Collagen and Elastin Gene Transcription in Aged Lung Mesenchyme
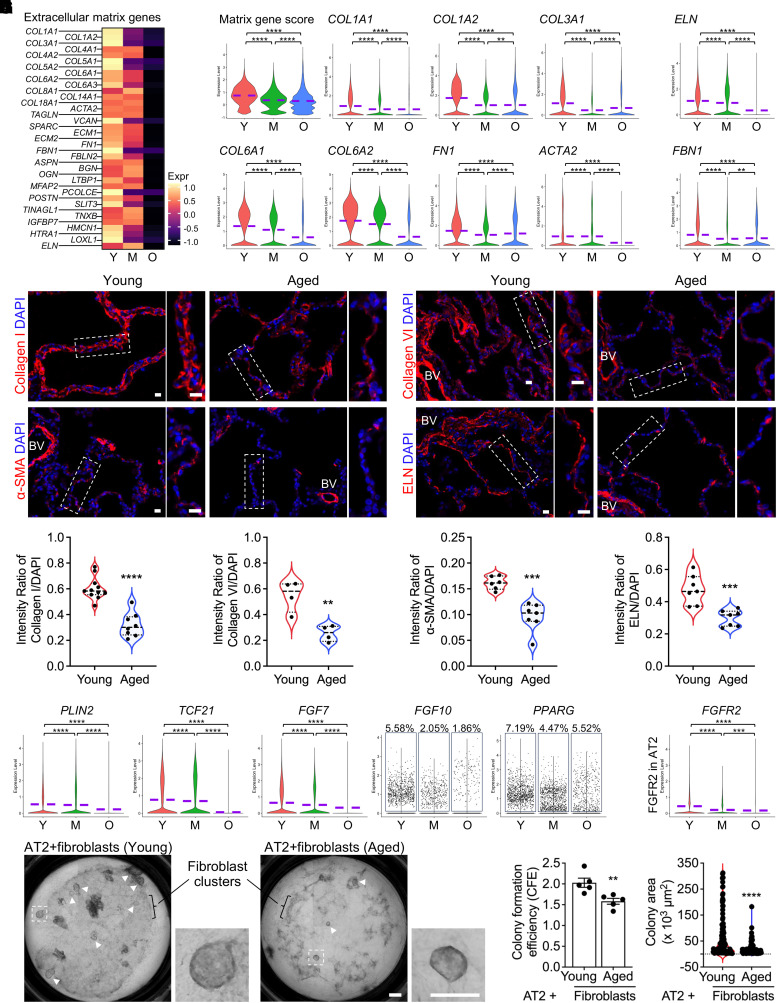
Aging has been considered as the driving force of progressive interstitial lung diseases such as IPF, a disease with accumulated fibroblasts and excessive extracellular matrix (ECM) production as the hallmarks (52). To comparatively profile the fibroblasts from lungs of donors at different ages, the mesenchymal cells were isolated from the integrated data and subjected to differential analysis. Intriguingly, a declined ECM transcriptional program was observed in the aged fibroblasts (Figure 4A). More specifically, decreased transcription of matrix genes, including COL1A1, COL1A2, COL3A1, COL6A1, COL6A2, FN1, and ACTA2 (Figure 4B), as well as a reduced level of the Matrix gene score, defined by the average expression of these genes, were identified in the aged fibroblasts (Figure 4B). To eliminate the potential bias caused by the smoking history of the subjects, the fibroblasts from lungs of nonsmokers and smokers were examined separately. Downregulated collagen gene expression was observed in aged fibroblasts from both nonsmokers (Figure E4A) and smokers (Figure E4B) compared with fibroblasts from young donors. These observations were verified by the immunostainings for Collagen I, Collagen VI, and α-SMA (Figure 4D) on lung sections from young and aged donors and their protein intensity quantifications (Figure 4E).
Figure 4.
Decreased extracellular matrix gene transcription in aged lung mesenchymal cells. (A) Heatmaps show the expression of the extracellular matrix genes in the mesenchymal cells from subjects of different age stages. (B) The Matrix gene score (the average transcription of COL1A1, COL1A2, COL3A1, COL6A1, COL6A2, FN1, and ACTA2) and transcriptions of representative collagen genes in mesenchymal cells from subjects of different age stages are visualized by violin plots. (C) Violin plots show the transcriptions of genes specific to elastic fiber components and assembly in mesenchymal cells from subjects of different age stages. (D) Immunofluorescence staining for Collagen I, Collagen VI, α-SMA (ACTA2), and ELN (Elastin) on human lung sections from young and aged donors. Boxed regions are magnified. BV = blood vessel. Scale bars, 20 μm. (E) Quantified intensity ratios for Collagen I, Collagen VI, α-SMA, and ELN staining compared with nuclear DAPI on human lung sections from young and aged donors. (n = 4–11 sections from three subjects per group). (F) The quantification of PLIN2, TCF21, and FGF7 transcriptions and FGF10- and PPARG-positive cell percentages in total lung mesenchymal cells from donors of different age stages. (G) Visualization of FGF receptor gene, FGFR2, in AT2 cells from donors of different age stages. (H–J) Representative images (H), colony formation efficiency (CFE) (I; n = 5), and colony size (J; Young, n = 130; Aged, n = 99) of human AT2s cultured with primary fibroblasts from young and aged donors. Boxed colonies are magnified, and visible colonies are indicated by arrowheads (H). Scale bars, 500 μm (H). Purple dashed lines in the violin plots indicate the mean levels of gene transcriptions. **P < 0.01, ***P < 0.001, and ****P < 0.0001.
The synthesis and function of ELN (Elastin), as a unique ECM component with a specific function to provide elasticity (both compliance and recoil) to the lungs, rapidly decline with age (53). As one of the decreased ECM genes in aged fibroblasts (Figure 4A), the transcription of Elastin, as well as other elastic fiber component genes, was found consistently downregulated (Figure 4C). Consistent with the RNA levels in the scRNA-seq datasets, the protein level of ELN in aged lung sections was significantly declined (Figures 4D and 4E).
Lung mesenchymal cells exhibit significant heterogeneity. To assess changes in the ECM within fibroblast subtypes, the total mesenchymal population was reclustered, and distinct fibroblast subpopulations were delineated (Figures E4C and E4D) using similar approaches as described recently (32). Interestingly, reduced transcription of ECM and ELN genes was observed across most fibroblast subtypes in aged human lungs (Figures E4E and E4F).
In addition to the loss of ECM and elasticity, decayed functions of aged mesenchymal cells were further reflected by decreased support to AT2 niches. This was evidenced by downregulated genes required for the support of AT2 renewal and stemness, such as PLIN2, TCF21, FGF10, FGF7, and PPARG (54), in aged mesenchymal cells (Figure 4F), as well as a loss of FGF receptor (mainly FGFR2) in aged AT2s (Figure 4G). Using a three-dimensional (3D)-organoid system presents a significant advancement in studying how alveolar epithelial cells maintain stemness and regenerate as well as how lung mesenchymal cells support alveolar epithelial cell stemness (55). By using the 3D-organoid assay, we observed that AT2s cocultured with primary fibroblasts from aged donors exhibited lower colony formation efficiency (CFE) (Figures 4H and 4I) and smaller organoid sizes (Figures 4H and 4J). This suggests that fibroblasts from aged donors possess a reduced capacity to support human AT2 renewal compared with those from young donors.
Aberrantly Activated Inflammation in Aged Lung Macrophages
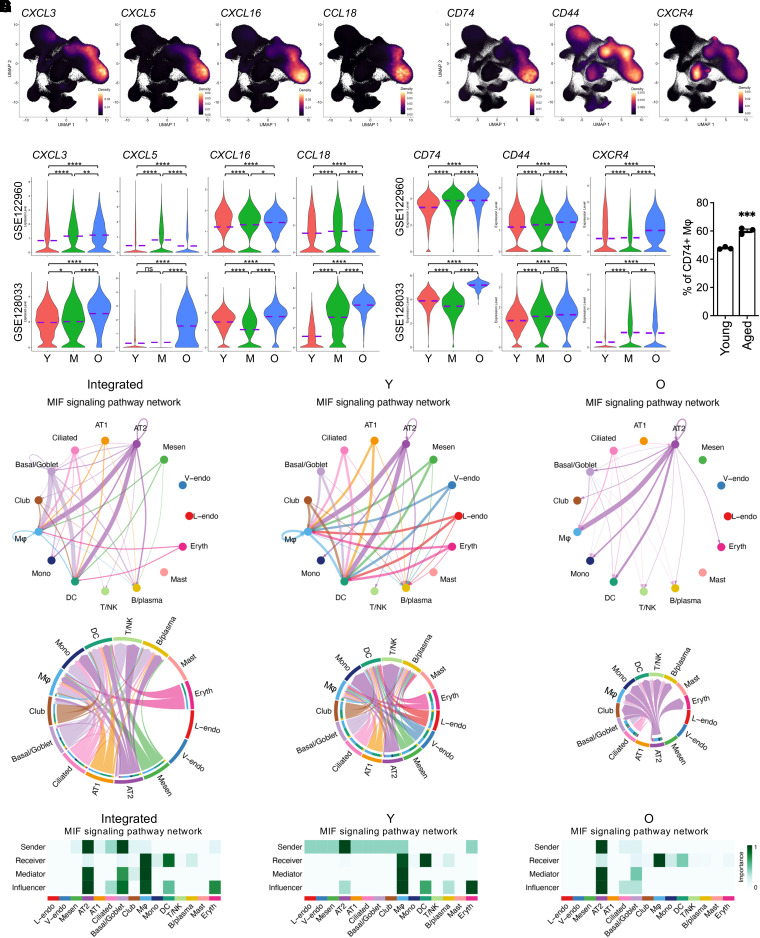
Pulmonary immune homeostasis in human lungs is maintained by a complex network of immunocytes derived from myeloid and lymphoid lineages (56). To profile the immunocytes from human lungs of different age stages, we isolated the myeloid and lymphoid lineages identified above (Figure 1D). Macrophages, the largest myeloid population in the integrated data, have been recently suggested to present the transcriptomic age-related changes, including a reduced number and compromised proliferation induced by the aging microenvironment in human and mouse lungs (7). Similarly, here macrophages were isolated, and comparative analysis revealed dramatically differentially expressed gene profiles between the young and aged lung macrophages (Figure E5A). Many of the DE genes were functional genes specific to macrophages (Figures E5B and E5C). The downregulated genes included several gene families, such as LAIRs (Leukocyte associated immunoglobulin like receptor genes), CTSs (Cathepsin genes), CSTs (Cystatin genes), SH3BGRL (SH3 domain binding glutamate rich protein like genes), MS4As (Membrane spanning 4-domains genes), and CD68 (Figure E5B). All these genes have shown some relevance with either inflammation inhibition or M2 macrophage polarization, suggesting an age-associated decay in antiinflammation of the lung macrophages from aged subjects. Together with these data, ectopic activation of macrophage-mediated inflammatory genes (MSR1, ALOX5, and SCD) and macrophage-specific antigen presentation genes (HLA-DQA1, HLA-DRB5, and HLA-DRB6) (Figure E5C) further revealed an activated inflammatory genetic program modulated by aged macrophages in human lungs. CD81, one of the representative DE genes, was verified via flow cytometry and was found to be decreased in aged lung macrophages (Figure E5D). Remarkably, the elevated genes in aged lung macrophages included a set of macrophage-specific chemokine genes, such as CXCL3, CXCL5, CXCL16, and CCL18 (Figure 5A), and the transcription of these chemokine genes was consistently increased in the age macrophages across most individual datasets (Figure 5B). CD74, CD44, and CXCR4, canonical receptor genes of MIF predominantly expressed in macrophages, as mentioned above, were also significantly elevated in the aged macrophages (Figures 5C and 5D). Flow cytometry analysis validated the upregulation of CD74 expression in lung macrophages from aged donors (Figure 5E). These data suggest a growing MIF-receptor interaction between epithelial cells and macrophages with the aging of lungs.
Figure 5.
Hyperactivated chemokine expression in macrophages from aged and smoking lungs. (A and C) Density plots show the specification of macrophage chemokine and MIF (macrophage migration inhibitory factor) receptor genes in lungs from aged subjects. (B and D) Violin plots present the elevated transcriptions of macrophage chemokine and MIF receptor genes in lungs from aged subjects. (E) Flow cytometry analysis for CD74+ macrophages from lungs of young and aged human donors (n = 3). (F) Circle plot (top panels) and Chord diagram (bottom panels) visualization of cell–cell communications by MIF signaling pathway network among cell types in the integrated, young and aged datasets. The communications between AT2 and macrophages are strong and are stronger in aged lungs. (G) Identification of dominant senders, receivers, mediators, and influencers in the intercellular communication network by computing MIF signaling pathway network centrality measures for each cell type in the integrated, young, and aged datasets. The communications by MIF signaling pathway network are wide, with several cell types as senders in young lungs, but are more specific to AT2–macrophage, with AT2 as the only sender in the age lungs. Purple dashed lines in the violin plots indicate the mean levels of gene transcriptions. *P < 0.05, **P < 0.01, and ****P < 0.0001.
Constitutively Activated Cell–Cell Communications between AT2s and Macrophages in Aged Lungs
Qualitative deficits during aging are believed to lead to abnormal cell–cell communication mechanisms in lungs (3). To determine the alterations in cell–cell communications during aging, we used CellChat, an R toolkit for inference, visualization, and analysis of cell–cell communication (34), and elucidated a dynamic change in cell–cell communications with the aging of human lungs. We noticed an increased cell–cell communication profile between AT2 cells and macrophages in aged lungs when the interaction weights among other cell types were mostly compromised (Figure E6A). The increased cell–cell communication was even more obvious when AT2s were set as the sender cells (Figure E6B). Visualizations of all the individual L–R (Ligand-Receptor) pairs emanated from AT2s suggested that the dominant significant interaction in the aged lungs was MIF and its receptor, CD74 (and CD44, CXCR4). The interaction strength of MIF–CD74 among cell types was relatively even in lungs of young subjects but became more condensed between AT2s and macrophages in the lungs from aged subjects (Figure 5F). Heatmap quantification further demonstrated that in the aged lungs the AT2s were the only dominant originator cells of the MIF signaling pathway network (Figure 5G). This may be explained by the more dominant and elevated expression of MIF in AT2s and CD74 in macrophages in aged lungs (Figures 3E, 3F, and E6C). This observation was in alignment with a previous review discussing the complex interaction between MIF-mediated inflammation and host defense and age-related lung biology via binding to its receptors (57). Together, with the aberrant MIF-CD74 signaling pathway in aged AT2s and macrophages as an example, we established a dysregulated cell–cell interactome in aged human lungs that may interlink the exhausted cellular niches with compromised supportiveness among alveolar cell types and subsequent accumulated cellular deficits together with aging.
Discussion
With the rapid increase of the aging population worldwide, degenerative changes in lungs have been found to be strongly correlated with the development and incidence of chronic respiratory diseases, such as COPD, IPF, and lung cancers. The cellular and molecular hallmarks of aging, including genomic instability, telomere attrition, epigenetic alterations, loss of proteostasis, deregulated nutrient sensing, mitochondrial dysfunction, cellular senescence, stem cell exhaustion, and altered intercellular communication, have been summarized (43). A number of transcriptomics studies of murine lung aging have been recently performed (29, 30); however, a longitudinal transcriptomics profiling of human lungs at the single-cell level across all adult age ranges is not available. To systemically characterize the effect of aging on the transcriptomics changes of human lungs, we collected and integrated the open-access single-cell transcriptomics studies on adult human lungs of all age ranges and applied a comprehensive analysis covering all the lung cell lineages. An overall dysregulated genetic program has been demonstrated in the aged lungs, with the abnormal transcriptomics changes within all lineages, including mesenchymal cells, epithelial cells, and myeloid cells.
The lung epithelium in aging shows deficits in progenitor cells in the airway and alveolar parenchyma. One aging hallmark of the AT2 cells, which are the major progenitor cells in the alveoli, is the depletion of adult stem cell reservoirs and failure in self-renewal and differentiation capacity (3). This depletion and dysfunction of AT2 cells may contribute to pulmonary diseases like emphysema and pulmonary fibrosis (18, 38). However, most of the existing data are derived from rodent studies. Thus, questions remain about how aging reshapes the lung stem niches and how advanced age alters the contributions of epithelial populations to lung regeneration (3). In this study, we find that the genes driving cellular senescence are elevated, and the genetic programs deriving immune aging, such as AP-1 transcription factor subunits, a complex recently found to be a conserved signature of immune aging that contributes to inflammaging in mouse tissues (47, 48), are aberrantly activated in AT2 cells in aged lungs. Another consistent feature of both AT2 and AT1 cells from aged lungs is the loss of epithelial identities. These dysregulated genetic programs of aged AT2 cells would trigger some pathogenetic signaling cascades, ultimately leading to defects in respiratory epithelium and increased susceptibility to the development of chronic diseases. AT2 cells from aged lungs produce higher levels of chemokines. This has been manifested by a previous study on an influenza murine model, in which neutrophils induced by alveolar epithelial cell–secreted chemokines increased the mortality of aged animals after infection (58). Although we do not have any direct evidence, we assumed that the loss of epithelial features and structures induced by long-term exposure to the environment might cause the heightened inflammatory program and chemokine secretion as well as increased cellular senescence in aged human lungs. Increasing statistical and metastudies have claimed that cumulative inhalational exposures over the lifespan introduce accumulating inflammatory oxidative stress and structure damage to the epithelium and induce widespread pulmonary cellular senescence, which in turn accelerates lung aging (59–61). Thus, aging not only enhances the expression of the genes related to inflammation, stress responses, and senescence in a normal state but also blunts the recovery of impaired AT2s in mouse lungs after injury (62). These data collectively propose the effects of aging on pulmonary innate immune dysfunction and the compromised response to invading microorganisms.
The AT2 niche is supported by lung mesenchymal cells, mainly fibroblasts, which have critical roles in lung development, homeostasis, and lung repair to chronic injuries. Aging increases the capacities of lung fibroblasts in differentiation into myofibroblasts and restricts their effects in supporting alveolar epithelial renewal (52, 54). Increased collagen production in fibroblasts is linked to interstitial lung diseases, which tend to have higher morbidity rates in older individuals. Here we reported a decrease in the transcription of collagen genes by the total mesenchymal cells and by multiple mesenchymal populations from aged human lungs. However, a recent comparative analysis by transcriptomics and proteomic data identified increased expression of several ECM genes in specific regions of aged healthy human lungs (63). This inconsistency may arise from either unpurified mesenchymal cell isolation or unbiased selection of lung regions during transcriptomics and proteomic analysis at bulk levels by the previous study. The mechanisms or consequences of collagen loss in aged lung fibroblasts have rarely been reported, but decreased collagen production in aged skin fibroblasts has been frequently demonstrated to result in fragmentation of the dermal collagen matrix and impairment in the structural integrity of the dermis (64). Whether the fibroblasts from these two organs share similar roles or underlying mechanisms remains to be further investigated. Another observation in the aged lungs was the loss of ELN expression by fibroblasts. Elastin is required for normal lung development, and destruction of elastin or abnormality in elastic fiber assembly are major factors in emphysema and destructive lung diseases. In adults, Elastin conveys elasticity to the lungs, and loss of elasticity is associated with reduced lung compliance and recoil, leading to an increasing risk of emphysema (53). Impaired lung elasticity due to defected Elastin production and elastic fiber assembly might explain the compromised forced vital capacity and expiratory volume in aged human populations. Some specific mesenchymal subpopulations, such as lipofibroblasts, are lipid droplet and growth factor contributor cells to AT2 homeostasis and stemness in adult human lungs (54). Dysregulated transcriptional programs in aged mesenchymal cells also revealed a decayed support to AT2 cells, as observed in the in vitro organoid assay by us and another recent publication (9).
The immune response is required to protect against invading pathogens and external material, and well-coordinated control of proinflammatory and antiinflammatory mediators is critical to prevent uncontrolled injury and fibrotic deposition (56). The lung myeloid cells, in conjunction with lymphoid lineages, play the central role in orchestrating the pulmonary immune response. However, we found a dysregulated genetic program in the immune system of the aged human lungs. Most significantly, the aged lung macrophages exhibited a sustained proinflammatory effect and aberrant cytokine gene expression. These observations partially align with a recent study indicating a decrease in “cell cycle” and “translation” related genes and an increase in “inflammatory response” genes in macrophages from aged human and mouse lungs (7). Together, the dysregulated gene expression profiles in the immunocytes, consistent with a recent review (3), might indicate an age-associated decline in immune function efficiency in the lungs of older individuals.
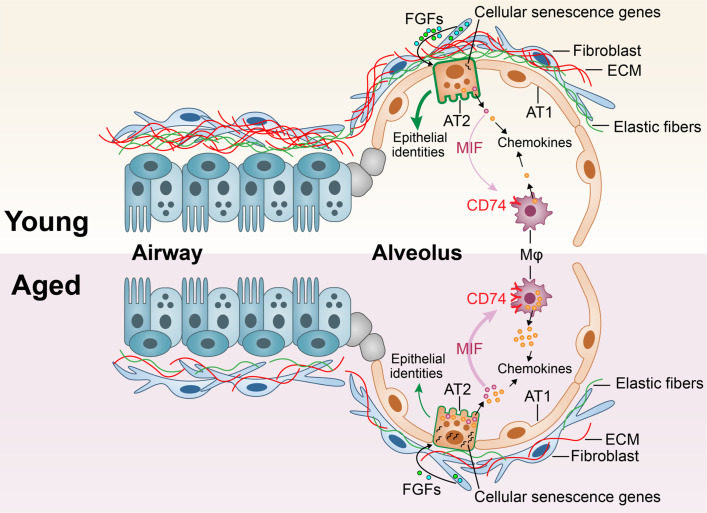
Overall, we have presented a comprehensive single-cell multiomics atlas of healthy human lungs, encompassing distinct cellular lineages across all age groups. The progressive dysregulation of genetic profiles in major cellular lineages suggests a general decline in lung function with age, particularly highlighting a significant deterioration of the AT2 niches. The dysregulated alveolar niche is further compromised by impaired macrophages and mesenchymal cells in aged lungs (Figure 6). This proposition raises the hypothesis that the increased susceptibility to lung diseases in aging populations may stem from the accumulation of deficits across multiple cell lineages and the resultant miscommunication among these affected cell populations. This study offers a comprehensive depiction of healthy human lungs, providing a valuable resource for investigating age-related cellular deficiencies in lung diseases. Although many observations require further functional and mechanistic verification, the current study has drawn a comprehensive transcriptomics map of healthy human lungs. Further exploration of this map holds the potential to enhance our understanding of lung degeneration and functional decline in elderly individuals, as well as age-associated deficiencies and pathogenesis in respiratory diseases.
Figure 6.
Constitutively activated cell–cell communications between AT2 and macrophages in aged lungs. Schematic summarization of the histopathological, cellular, and molecular alterations in aged human lungs. The aged lungs have a general aged-associated decay in multiple cell types, suggesting a general compromised lung function. The basement membranes are represented by elastic fibers.
Acknowledgments
Acknowledgment
The authors thank Drs. Tatsuya Tsukui and Dean Sheppard for sharing the metadata of the donors in GSE132771. The authors thank the lung research community for the open sharing of datasets available.
Footnotes
Supported by National Institutes of Health grants R35-HL150829 (P.W.N.), P01-HL108793 (P.W.N. and D.J.), and R01-AG078655 (J.L.).
Author Contributions: D.J. and P.W.N. conceived and supervised the study. X.L. and C.Y. performed the bioinformatic analysis. X.L., X.Z., and J.L. designed and performed the experiments. X.L., X.Z., C.Y., J.L., P.W.N., and D.J. prepared the figures and wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
This article has a data supplement, which is accessible at the Supplements tab.
Originally Published in Press as DOI: 10.1165/rcmb.2023-0363OC on April 18, 2024
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Dzau VJ, Inouye SK, Rowe JW, Finkelman E, Yamada T. Enabling healthful aging for all: the National Academy of Medicine grand challenge in healthy longevity. N Engl J Med . 2019;381:1699–1701. doi: 10.1056/NEJMp1912298. [DOI] [PubMed] [Google Scholar]
- 2. Fried LP, Rowe JW. Health in aging: past, present, and future. N Engl J Med . 2020;383:1293–1296. doi: 10.1056/NEJMp2016814. [DOI] [PubMed] [Google Scholar]
- 3. Schneider JL, Rowe JH, Garcia-de-Alba C, Kim CF, Sharpe AH, Haigis MC. The aging lung: physiology, disease, and immunity. Cell . 2021;184:1990–2019. doi: 10.1016/j.cell.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rojas M, Mora AL, Kapetanaki M, Weathington N, Gladwin M, Eickelberg O. Aging and lung disease: clinical impact and cellular and molecular pathways. Ann Am Thorac Soc . 2015;12:S222–S227. doi: 10.1513/AnnalsATS.201508-484PL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest . 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watson JK, Sanders P, Dunmore R, Rosignoli G, Julé Y, Rawlins EL, et al. Distal lung epithelial progenitor cell function declines with age. Sci Rep . 2020;10:10490. doi: 10.1038/s41598-020-66966-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McQuattie-Pimentel AC, Ren Z, Joshi N, Watanabe S, Stoeger T, Chi M, et al. The lung microenvironment shapes a dysfunctional response of alveolar macrophages in aging. J Clin Invest . 2021;131:e140299. doi: 10.1172/JCI140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi J, Park JE, Tsagkogeorga G, Yanagita M, Koo BK, Han N, et al. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell . 2020;27:366–382.e7. doi: 10.1016/j.stem.2020.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chanda D, Rehan M, Smith SR, Dsouza KG, Wang Y, Bernard K, et al. Mesenchymal stromal cell aging impairs the self-organizing capacity of lung alveolar epithelial stem cells. eLife . 2021;10:e68049. doi: 10.7554/eLife.68049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee JH, Bhang DH, Beede A, Huang TL, Stripp BR, Bloch KD, et al. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell . 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, et al. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell . 2017;170:1134–1148.e10. doi: 10.1016/j.cell.2017.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature . 2020;587:619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med . 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv . 2020;6:eaba1972. doi: 10.1126/sciadv.aba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu X, Rowan SC, Liang J, Yao C, Huang G, Deng N, et al. Categorization of lung mesenchymal cells in development and fibrosis. iScience . 2021;24:102551. doi: 10.1016/j.isci.2021.102551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv . 2020;6:eaba1983. doi: 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Justet A, Zhao AY, Kaminski N. From COVID to fibrosis: lessons from single-cell analyses of the human lung. Hum Genomics . 2022;16:20. doi: 10.1186/s40246-022-00393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang J, Huang G, Liu X, Taghavifar F, Liu N, Wang Y, et al. The ZIP8/SIRT1 axis regulates alveolar progenitor cell renewal in aging and idiopathic pulmonary fibrosis. J Clin Invest . 2022;132:e157338. doi: 10.1172/JCI157338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raghu G, Weycker D, Edelsberg J, Bradford WZ, Oster G. Incidence and prevalence of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med . 2006;174:810–816. doi: 10.1164/rccm.200602-163OC. [DOI] [PubMed] [Google Scholar]
- 20. Singh D, Agusti A, Anzueto A, Barnes PJ, Bourbeau J, Celli BR, et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease: the GOLD science committee report 2019. Eur Respir J . 2019;53:1900164. doi: 10.1183/13993003.00164-2019. [DOI] [PubMed] [Google Scholar]
- 21. Brandsma CA, de Vries M, Costa R, Woldhuis RR, Königshoff M, Timens W. Lung ageing and COPD: is there a role for ageing in abnormal tissue repair? Eur Respir Rev . 2017;26:170073. doi: 10.1183/16000617.0073-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. MacNee W. Is chronic obstructive pulmonary disease an accelerated aging disease? Ann Am Thorac Soc . 2016;13:S429–S437. doi: 10.1513/AnnalsATS.201602-124AW. [DOI] [PubMed] [Google Scholar]
- 23. Sacher AG, Dahlberg SE, Heng J, Mach S, Jänne PA, Oxnard GR. Association between younger age and targetable genomic alterations and prognosis in non-small-cell lung cancer. JAMA Oncol . 2016;2:313–320. doi: 10.1001/jamaoncol.2015.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature . 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guan WJ, Liang WH, Zhao Y, Liang HR, Chen ZS, Li YM, et al. China Medical Treatment Expert Group for COVID-19 Comorbidity and its impact on 1590 patients with COVID-19 in China: a nationwide analysis. Eur Respir J . 2020;55:2000547. doi: 10.1183/13993003.00547-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sauler M, McDonough JE, Adams TS, Kothapalli N, Barnthaler T, Werder RB, et al. Characterization of the COPD alveolar niche using single-cell RNA sequencing. Nat Commun . 2022;13:494. doi: 10.1038/s41467-022-28062-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun . 2020;11:2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Melms JC, Biermann J, Huang H, Wang Y, Nair A, Tagore S, et al. A molecular single-cell lung atlas of lethal COVID-19. Nature . 2021;595:114–119. doi: 10.1038/s41586-021-03569-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Angelidis I, Simon LM, Fernandez IE, Strunz M, Mayr CH, Greiffo FR, et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat Commun . 2019;10:963. doi: 10.1038/s41467-019-08831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tabula Muris Consortium. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature . 2020;583:590–595. doi: 10.1038/s41586-020-2496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu X, Zhang X, Yao C, Liang J, Noble PW, Jiang D.2023. www.biorxiv.org/content/10.1101/2023.06.16.545378v1
- 32. Liu X, Dai K, Zhang X, Huang G, Lynn H, Rabata A, et al. Multiple fibroblast subtypes contribute to matrix deposition in pulmonary fibrosis. Am J Respir Cell Mol Biol . 2023;69:45–56. doi: 10.1165/rcmb.2022-0292OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu X, Geng Y, Liang J, Coelho AL, Yao C, Deng N, et al. HER2 drives lung fibrosis by activating a metastatic cancer signature in invasive lung fibroblasts. J Exp Med . 2022;219:e20220126. doi: 10.1084/jem.20220126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun . 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang A, Chiou J, Poirion OB, Buchanan J, Valdez MJ, Verheyden JM, et al. NHLBI LungMap Consortium Single-cell multiomic profiling of human lungs reveals cell-type-specific and age-dynamic control of SARS-CoV2 host genes. eLife . 2020;9:e62522. doi: 10.7554/eLife.62522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morse C, Tabib T, Sembrat J, Buschur KL, Bittar HT, Valenzi E, et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J . 2019;54:1802441. doi: 10.1183/13993003.02441-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsukui T, Sun KH, Wetter JB, Wilson-Kanamori JR, Hazelwood LA, Henderson NC, et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat Commun . 2020;11:1920. doi: 10.1038/s41467-020-15647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yao C, Guan X, Carraro G, Parimon T, Liu X, Huang G, et al. Senescence of alveolar type 2 cells drives progressive pulmonary fibrosis. Am J Respir Crit Care Med . 2021;203:707–717. doi: 10.1164/rccm.202004-1274OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heinzelmann K, Hu Q, Hu Y, Dobrinskikh E, Ansari M, Melo-Narváez MC, et al. Single-cell RNA sequencing identifies G-protein coupled receptor 87 as a basal cell marker expressed in distal honeycomb cysts in idiopathic pulmonary fibrosis. Eur Respir J . 2022;59:2102373. doi: 10.1183/13993003.02373-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guo M, Yu JJ, Perl AK, Wikenheiser-Brokamp KA, Riccetti M, Zhang EY, et al. Single-cell transcriptomic analysis identifies a unique pulmonary lymphangioleiomyomatosis cell. Am J Respir Crit Care Med . 2020;202:1373–1387. doi: 10.1164/rccm.201912-2445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bharat A, Querrey M, Markov NS, Kim S, Kurihara C, Garza-Castillon R, et al. Lung transplantation for patients with severe COVID-19. Sci Transl Med . 2020;12:eabe4282. doi: 10.1126/scitranslmed.abe4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Basil MC, Cardenas-Diaz FL, Kathiriya JJ, Morley MP, Carl J, Brumwell AN, et al. Human distal airways contain a multipotent secretory cell that can regenerate alveoli. Nature . 2022;604:120–126. doi: 10.1038/s41586-022-04552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell . 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shulenin S, Nogee LM, Annilo T, Wert SE, Whitsett JA, Dean M. ABCA3 gene mutations in newborns with fatal surfactant deficiency. N Engl J Med . 2004;350:1296–1303. doi: 10.1056/NEJMoa032178. [DOI] [PubMed] [Google Scholar]
- 45. Zhang Z, Newton K, Kummerfeld SK, Webster J, Kirkpatrick DS, Phu L, et al. Transcription factor Etv5 is essential for the maintenance of alveolar type II cells. Proc Natl Acad Sci USA . 2017;114:3903–3908. doi: 10.1073/pnas.1621177114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martínez-Zamudio RI, Roux PF, de Freitas JANLF, Robinson L, Doré G, Sun B, et al. AP-1 imprints a reversible transcriptional programme of senescent cells. Nat Cell Biol . 2020;22:842–855. doi: 10.1038/s41556-020-0529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu X, Wang Y, Song Y, Gao X, Deng H. AP-1 is a regulatory transcription factor of inflammaging in the murine kidney and liver. Aging Cell . 2023;22:e13858. doi: 10.1111/acel.13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Karakaslar EO, Katiyar N, Hasham M, Youn A, Sharma S, Chung CH, et al. Transcriptional activation of Jun and Fos members of the AP-1 complex is a conserved signature of immune aging that contributes to inflammaging. Aging Cell . 2023;22:e13792. doi: 10.1111/acel.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Manzer R, Wang J, Nishina K, McConville G, Mason RJ. Alveolar epithelial cells secrete chemokines in response to IL-1beta and lipopolysaccharide but not to ozone. Am J Respir Cell Mol Biol . 2006;34:158–166. doi: 10.1165/rcmb.2005-0205OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao MQ, Stoler MH, Liu AN, Wei B, Soguero C, Hahn YS, et al. Alveolar epithelial cell chemokine expression triggered by antigen-specific cytolytic CD8(+) T cell recognition. J Clin Invest . 2000;106:R49–R58. doi: 10.1172/JCI9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, et al. MIF signal transduction initiated by binding to CD74. J Exp Med . 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pardo A, Selman M. Lung fibroblasts, aging, and idiopathic pulmonary fibrosis. Ann Am Thorac Soc . 2016;13:S417–S421. doi: 10.1513/AnnalsATS.201605-341AW. [DOI] [PubMed] [Google Scholar]
- 53. Heinz A. Elastic fibers during aging and disease. Ageing Res Rev . 2021;66:101255. doi: 10.1016/j.arr.2021.101255. [DOI] [PubMed] [Google Scholar]
- 54. El Agha E, Thannickal VJ. The lung mesenchyme in development, regeneration, and fibrosis. J Clin Invest . 2023;133:e170498. doi: 10.1172/JCI170498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liang J, Zhang Y, Xie T, Liu N, Chen H, Geng Y, et al. Hyaluronan and TLR4 promote surfactant-protein-C-positive alveolar progenitor cell renewal and prevent severe pulmonary fibrosis in mice. Nat Med . 2016;22:1285–1293. doi: 10.1038/nm.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Iwasaki A, Foxman EF, Molony RD. Early local immune defences in the respiratory tract. Nat Rev Immunol . 2017;17:7–20. doi: 10.1038/nri.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sauler M, Bucala R, Lee PJ. Role of macrophage migration inhibitory factor in age-related lung disease. Am J Physiol Lung Cell Mol Physiol . 2015;309:L1–L10. doi: 10.1152/ajplung.00339.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kulkarni U, Zemans RL, Smith CA, Wood SC, Deng JC, Goldstein DR. Excessive neutrophil levels in the lung underlie the age-associated increase in influenza mortality. Mucosal Immunol . 2019;12:545–554. doi: 10.1038/s41385-018-0115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Eckhardt CM, Wu H. Environmental exposures and lung aging: molecular mechanisms and implications for improving respiratory health. Curr Environ Health Rep . 2021;8:281–293. doi: 10.1007/s40572-021-00328-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vrijheid M. The exposome: a new paradigm to study the impact of environment on health. Thorax . 2014;69:876–878. doi: 10.1136/thoraxjnl-2013-204949. [DOI] [PubMed] [Google Scholar]
- 61. Zhou S, Zhu J, Zhou PK, Gu Y. Alveolar type 2 epithelial cell senescence and radiation-induced pulmonary fibrosis. Front Cell Dev Biol . 2022;10:999600. doi: 10.3389/fcell.2022.999600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liang J, Huang G, Liu X, Liu N, Taghavifar F, Dai K, et al. Reciprocal interactions between alveolar progenitor dysfunction and aging promote lung fibrosis. eLife . 2023;12:e85415. doi: 10.7554/eLife.85415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Koloko Ngassie ML, De Vries M, Borghuis T, Timens W, Sin DD, Nickle D, et al. Age-associated differences in the human lung extracellular matrix. Am J Physiol Lung Cell Mol Physiol . 2023;324:L799–L814. doi: 10.1152/ajplung.00334.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Varani J, Dame MK, Rittie L, Fligiel SE, Kang S, Fisher GJ, et al. Decreased collagen production in chronologically aged skin: roles of age-dependent alteration in fibroblast function and defective mechanical stimulation. Am J Pathol . 2006;168:1861–1868. doi: 10.2353/ajpath.2006.051302. [DOI] [PMC free article] [PubMed] [Google Scholar]