Abstract
Thermal inactivation is a major bottleneck to the scalable production, storage, and transportation of protein-based reagents and therapies. Failures in temperature control both compromise protein bioactivity and increase the risk of microorganismal contamination. Herein, we report the rational design of fluorochemical additives that promiscuously bind to and coat the surfaces of proteins to enable their stable dispersion within fluorous solvents. By replacing traditional aqueous liquids with fluorinated media, this strategy conformationally rigidifies proteins to preserve their structure and function at extreme temperatures (≥90 °C). We show that fluorous protein formulations resist contamination by bacterial, fungal, and viral pathogens, which require aqueous environments for survival, and display equivalent serum bioavailability to standard saline samples in animal models. Importantly, by designing dispersants that decouple from the protein surface in physiologic solutions, we deliver a fluorochemical formulation that does not alter the pharmacologic function or safety profile of the functionalized protein in vivo. As a result, this nonaqueous protein storage paradigm is poised to open technological opportunities in the design of shelf-stable protein reagents and biopharmaceuticals.
Keywords: fluorine chemistry, perfluorocarbon, protein structure, protein formulation, protein stability, biotherapeutics
1. Introduction
Most protein reagents and therapies require low-temperature conditions throughout the supply chain to maintain integrity.1,2 This “cold chain” is expensive, requires significant oversight, and is difficult to operationalize in remote environments. Highlighting the significance of this problem, biopharmaceutical industries lose nearly $35 billion annually as a result of failures in temperature-controlled logistics, and breaks in the cold chain can be life-threatening.3,4 Traditional approaches to enhance the thermal stability of proteins involve combinations of freeze-drying, excipient additives, and sequence engineering to generate stable mutants.5−8 Although AI-guided sequence engineering, in tandem with advances in excipient design, have led to significant gains in protein stabilization, these approaches require customization for each biologic at considerable effort and cost.9,10 Moreover, despite these successes, cold storage is still required to maintain product sterility and minimize protein degradation.
We recently reported perfluorochemical additives that promiscuously adsorb to the surfaces of proteins to enable their dispersion within nonaqueous perfluorocarbon (PFC) liquids.11,12 Biophysical studies demonstrated that this approach restricts the conformational plasticity of the dispersed protein and alters their thermodynamic equilibrium. As a result, PFC-dispersed proteins maintain their structure and function at temperatures as high as 90 °C.13 Moreover, these nonaqueous liquid samples were impervious to contamination by bacteria and fungi, which require aqueous solvents for survival, and demonstrated an enhanced resistance to proteolytic degradation.
Despite this success, our lead dispersive agent, perfluorononanoic acid (PFNA), was not without formulation and pharmacologic liabilities. Although PFNA efficiently dispersed proteins in the short-chain PFC perfluorohexane (bp = 56 °C), we show here that its performance markedly decreases in the longer-chain and more thermally stable solvent perfluorooctane (PFOc; bp = 108 °C). The elevated boiling point of PFOc is required to access the full capabilities of this thermal stabilization approach given that melting temperatures of fluorous-dispersed proteins routinely reach >90 °C. Additional in vitro and in vivo toxicity of PFNA observed in our previous studies warranted further scaffold optimization.
Here, we report the rational design of a family of perfluorochemical dispersants with improved PFC solubilization efficiencies and biocompatibility compared with PFNA (Figure 1a). Utilizing a series of complementary biophysical, in silico, in vitro, and in vivo approaches, we reveal the mechanistic basis for the improved performance of our lead compound, PD-7, relative to the first-generation PFNA reagent. Importantly, we show that, unlike PFNA, PD-7 decouples from the protein surface in physiologic solutions to avoid bioaccumulation of the fluorinated reagents in tissues, an essential technological milestone given the growing concern over the toxicity of perfluoroalkyl substances.
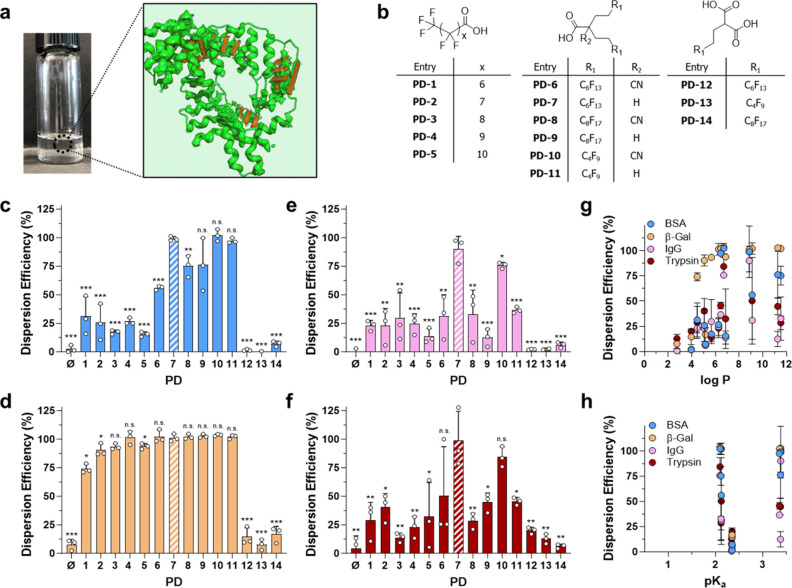
Figure 1.
Fluorous protein dispersant design and dispersion screening. (a) Optical image of a fluorous protein sample with a conceptual representation of the biologic (red) coated with a dispersant (bright green) to enable its solubilization within the fluorous solvent (light green background). Graphic adapted from an image created with the assistance of AI through DALL·E 2. (b) Structural library of protein dispersants (PDs) used to study structure–function–performance relationships of protein dispersion in PFOc. (c–f) Dispersion efficiency of (c) bovine serum albumin (BSA), (d) β-galactosidase (β-Gal), (e) rabbit serum immunoglobulin (IgG), and (f) trypsin in PFOc using the indicated dispersant ([PD]/[protein] = 1000:1). Results are represented as percent dispersed protein with respect to original loading. PD-7 is highlighted via cross-hatching to aid in indexing results to the dispersant structure. (g, h) Influence of dispersant (g) partition coefficient (log P) and (h) dissociation coefficient (pKa) on protein dispersion efficiency. Data shown in panels c–h represent the average ± s.d. of n = 3 technical replicates. Statistical significance in panels c–f was determined for each dispersant relative to the lead compound PD-7 using unpaired Student’s t test with n.s. = not significant, * p < 0.05, ** p < 0.01, and *** p < 0.001.
2. Results and Discussion
2.1. Protein Dispersant Design and Characterization
Early structure–activity studies from our lab11,13 identified three parameters important to perfluoroalkyl protein dispersants (PDs): (1) carboxylic acids are favored among other heteroatom substituents due to their ability to hydrogen bond to the protein backbone; (2) an amphiphilic character that balances interaction of the fluorochemical with polar protein surfaces and the fluorous solvent is preferred; and (3) linear perfluorocarbons outperform cyclic and aromatic species. Based on these criteria, we designed three compound families that include rational combinations of mono- or diacids as well as single or bivalent perfluorinated tails (Figure 1b). The single perfluorinated tail containing protein dispersants PD-1 to PD-5 were procured commercially, whereas bivalent perfluorinated derivatives (PD-6 to PD-11) were synthesized through modification of reported protocols (see supplementary synthetic methods in the Supporting Information).14 For example, the alkylation of ethyl cyanoacetate in the presence of perfluoroalkyl ethyl iodides (Supplementary Scheme S1, compounds 1a–c) under basic conditions gave 2,2′-dialkylated ethyl cyanoacetates (Supplementary Scheme S1, compounds 2a–c) followed by basic hydrolysis to yield dispersants PD-6, PD-8, and PD-10. Subsequent acidic hydrolysis and decarboxylation of these dispersants were performed to isolate PD-7, PD-9, and PD-11 in good yields. Similarly, monoalkylation of diethylmalonate with perfluoroalkyl ethyl iodides followed by basic hydrolysis produced diacids PD-12 to PD-14 in quantitative yields (Supplementary Scheme S2). Structures of the synthesized compounds were validated using 1H, 19F, and 13C NMR and high-resolution mass spectroscopic techniques (see supplementary synthetic methods in the Supporting Information).
Next, we screened the performance of these compounds for their ability to disperse four model proteins—bovine serum albumin (BSA), β-galactosidase (β-Gal), rabbit serum immunoglobulin (IgG), and bovine trypsin—into the PFOc solvent (Figure 1c–f). These proteins were selected for their diversity in molecular weight and metabolic function (e.g., enzymes, antibodies, and carrier proteins) to demonstrate the universal nature of this fluorous dispersion methodology. Candidate proteins were first dispersed into PFOc in the presence of each fluorochemical compound (1000:1 molar ratio with protein) followed by centrifugation to remove undissolved aggregates and quantitation of protein recovered from the supernatant using a modified Bradford assay. PD-7 (highlighted with cross-hatching in Figure 1c–f) displayed dispersion efficiencies of 90–100%, outperforming all other PDs in the test set, without apparent specificity for protein molecular weight (Supplementary Figure S1). PD-10 was a notable secondary candidate, achieving 80–100% dispersion efficiencies. Importantly, our lead compound from our previous screens, PFNA (referred to as PD-2 in this study), generally did not achieve >50% dispersion of the tested proteins in PFOc. The notable exception was β-Gal, which showed nearly quantitative dispersion across candidates PD-2 through PD-11. The most likely explanation for this is that the high molecular weight of the β-Gal tetramer complex (∼520 kDa) provides an abundant surface area for promiscuous complexation to most of the PFOc soluble dispersants. Interpreting structure–function–performance relationships from these results suggests that divalent fluorinated tails, particularly C4F9 and C6F13, outperform monovalent analogues with a similar fluorine content. Likewise, monoacid dispersants perform better than diacids, most likely because of the improved solubility of monoacid derivatives in PFOc.
To better understand the physiochemical determinants of dispersant activity, we compared the partition coefficient (log P) and dissociation constant (pKa) of each PD to its dispersion efficiency (Figure 1g,h). This analysis showed that a dispersant logP ≥ 7 generally maximizes its protein solubilizing efficiency. No clear correlation was observed for carboxy pKa and dispersion efficiency. Taken together, our data suggest that there are three properties important to PD-protein PFOc dispersion: (1) divalent fluorinated tails outperform monovalent tails; (2) monocarboxylic acid amphiphiles are preferred over diacids; and (3) dispersion efficiency increases monotonically with PD log P until a maximum at ∼7, likely as a result of the improved solubility of the dispersant in the bulk perfluorocarbon solvent.
2.2. Thermal Stabilization of Fluorous Dispersed Proteins
Before proceeding to thermal studies, we screened the dispersion compounds for toxicity against HepG2, a human liver hepatocellular cell line (Supplementary Figure S2). This cell line was chosen as liver is the primary site of metabolism and toxicity for many perfluorinated amphiphiles.15,16 Gratifyingly, PD-7, our best performing dispersant, was also the least toxic, with an IC50 ≈ 1.0 mM. Similarly, the dispersion solvent, PFOc, was biocompatible toward human liver cells (Supplementary Figure S3). Thus, PD-7 and PFOc were prioritized as the lead formulation for development of protein dispersions for follow up thermal stability testing.
Temperature-dependent protein denaturation is a thermodynamic process with enthalpic (disruption of hydrophobic interactions, van der Waals forces, and intramolecular hydrogen bonds) and entropic (increased conformational states of extended polypeptide chains) components. To evaluate the temperature-dependent structural changes of fluorous protein samples, we utilized circular dichroism (CD) spectroscopy, an optical technique that monitors protein secondary structure. Results in Figure 2a,b show minimal changes in secondary structure for the four model test proteins BSA, β-Gal, IgG, and trypsin when dispersed in PFOc using PD-7 and heated up to 85 °C. Conversely, nearly all the saline control formulations showed denaturation under the tested temperature range, as demonstrated by a significant loss of β-sheet (212 nm) and α-helical (208 and 222 nm) canonical minima. The notable exception was trypsin, which showed minimal change in structure between 40 and 85 °C.
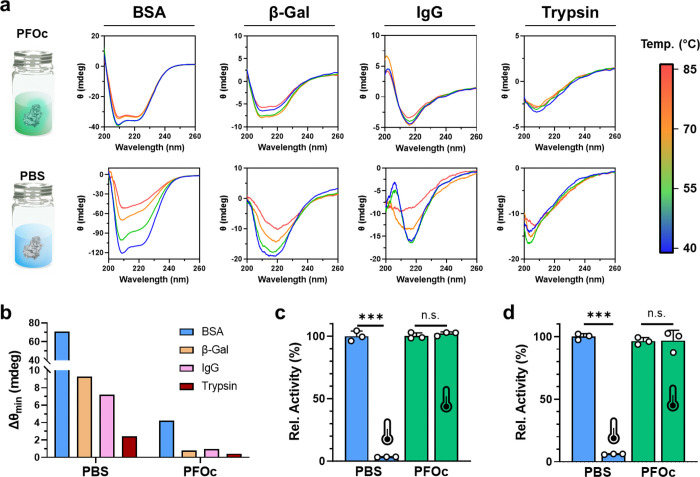
Figure 2.
Thermal stability of fluorous proteins. (a) CD spectra of the indicated protein at varying temperatures (40–85 °C) in PFOc using the PD-7 dispersant (top panel) or PBS as a control (bottom panel). (b) Variation in CD ellipticity minima (Δθmin) over the 40–85 °C temperature interval for the indicated protein dissolved in either PBS or PFOc. (c, d) Bioactivity of (c) β-gal and (d) trypsin in PBS (blue) or PFOc (green) before (25 °C) and after incubation at 90 °C (represented by the thermometer icon) for 30 min. Representative data from n = 3 technical replicates are shown in panels a and b. Panels c and d represent the average ± s.d. of n = 3 technical replicates. Horizontal lines in panels c and d represent statistical significance between treated conditions, calculated using unpaired Student’s t test, with n.s. = not significant and *** p < 0.001.
Although these results are encouraging, small changes in the protein structure can dramatically impact bioactivity. Therefore, we next examined the activity of β-Gal (Figure 2c) and trypsin (Figure 2d) after a 30 min incubation at 90 °C in either saline or PFOc. These assays required us to extract the PFOc dispersed protein samples into PBS after heating to enable reaction with the water-soluble conversion substrate. As expected, both proteins in control saline formulations lost >90% of their bioactivity after heating, whereas no statistically significant change in activity of the PFOc dispersed proteins was detected under the same conditions. Control bioactivity experiments in saline containing PD-7 demonstrated that the dispersant alone was not sufficient to confer thermal stability to proteins in an aqueous environment (Supplementary Figure S4). Together, this supports our assertion that the fluorous coating and PFC solvent, in combination, serve to constrain the conformational flexibility of the dispersed protein and, as a result, increase its melting temperature.
2.3. Biophysical Determinants of Protein Stabilization
We next evaluated the biophysical mechanisms of PD-7/protein interaction to gain a deeper understanding of its stabilizing effects using BSA as an exemplary protein. Transmission electron microscopy demonstrated that, without the PD-7 dispersant, BSA aggregates into amorphous structures when attempting to disperse it in the PFOc solvent (Figure 3a, see aqueous control in Supplementary Figure S5). Conversely, PD-7, on its own, showed an oily morphology when it was dissolved in PFOc (Figure 3b). Complexation of PD-7 and BSA in PFOc, however, led to the formation of hierarchical proteinaceous fibrils (Figure 3c), where the fiber surface was coated by the oily PD-7 dispersant (Figure 3d). We envisage that this coating provides a fluorophilic covering of the assembled protein surface that potentiates the solubilization of the nanostructures by the PFOc solvent.
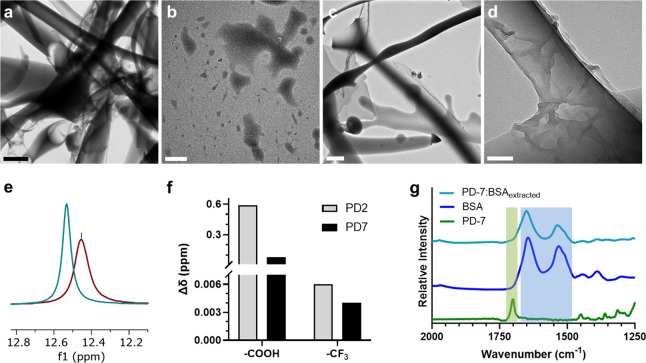
Figure 3.
Mechanistic investigation of PD-7/protein interactions. (a, b) Transmission electron microscopic images of (a) BSA or (b) PD-7 in PFOc. Scale bar in panel a = 2 μm and panel b = 500 nm. (c) Transmission electron microscopy image of BSA dispersed with PD-7 in PFOc. Scale bar = 2 μm. (d) Magnified view of PD-7 coated BSA fibrils. Scale bar: 100 nm. (e) Superimposed 1H NMR region demonstrating the downfield shift of PD-7’s −COOH proton in the absence (maroon) and presence (teal) of BSA. (f) Change in 1H (−COOH) and 19F (−CF3) NMR chemical shift (Δδ) for PD-2 (gray) and PD-7 (black) following coordination with the BSA protein. (g) Stacked FTIR spectra of PD-7 (green), BSA (blue), and the PD-7/BSA complex after elution into PBS (teal). Green and blue shading highlights PD-7 and BSA specific spectral features, respectively.
Next, we employed 1H NMR and 19F NMR spectroscopy to examine the biophysical interactions between BSA and PD-7 in a fluorous solvent. Proton NMR experiments showed a 0.08 ppm downfield shift of PD-7’s −COOH hydrogen in the presence of BSA (Figure 3e and Supplementary Figure S6), suggesting that it weakly hydrogen bonds with the solvent exposed backbone and amino acid side chains of BSA. In contrast, PD-2, our first-generation dispersant, demonstrated strong hydrogen bonding with BSA, as exemplified by a −COOH downfield shift of 0.56 ppm (Supplementary Figure S7). This is a 7-fold change in chemical shift relative to PD-7/BSA (Figure 3f). Conversely, 19F NMR showed a negligible change in the chemical environment of CF3 groups for both dispersants in the presence of BSA (PD-7 Δδ = 0.005 ppm and PD-2 Δδ = 0.006 ppm, Figure 3f, Supplementary Figures S8 and S9). Taken together, our data indicate that hydrogen bonding between the dispersant carboxylic headgroup and the protein surface is responsible for the coating phenomenon observed in TEM (Figures 3a–d), whereas the fluorinated tail(s) is/are extended into the bulk solution to promote solvation of the complex by PFOc.
Curiously, the stronger hydrogen bonding interactions of PD-2 with BSA, relative to PD-7, contradict the dispersion performance of these compounds, where our earlier studies showed that PD-7 was the superior dispersant (Figure 1). Although computational studies presented later help to explain this paradoxical finding, we hypothesized that the weak binding of PD-7 may promote rapid decoupling of the dispersant from the protein surface in a physiologic solution like blood. This is advantageous given the desire to produce protein formulations that can be directly administered to patients without additional solvent exchanges. In this clinical context, it is desirable for the dispersant to remain in the PFOc solution and the protein to partition into the serum after injection to avoid the fluorous coating interfering with protein function. Additionally, retention of the dispersant in the PFOc solvent will limit bioaccumulation of the fluorochemical in tissues and therefore reduce its potential toxic side effects because the bioinert fluorous solvent is expected to be renally excreted. To test this assertion, we performed Fourier transform infrared (FTIR) spectroscopy on BSA samples following extraction of the PD-7 dispersed protein from PFOc into a physiologic buffer. This was accomplished by adding an equal volume of PBS to the PD-7/BSA sample in PFOc and vortexing the phase separated mixture for ∼2 s. Gratifyingly, FTIR spectra of the extracted fraction showed only the presence of BSA’s amide-I (1644 cm–1) and II (1531 cm–1) bands and did not show the C=O stretching spectral feature (1701 cm–1) indicative of PD-7 (Figure 3g). Similar results were observed for extracted β-Gal and IgG formulations (Supplementary Figure S10). This suggests that PD-7 successfully dissociates from the surface of dispersed proteins during mixing in a buffer and remains in the PFOc phase. These results support our assertion that, in an in vivo setting, the PFOc liquid containing PD-7 is likely to be cleared by the kidney and excreted from patients in urine to avoid long-term accumulation of the fluorinated compounds in tissues. On the contrary, FTIR spectra of extracted PD-2 dispersed BSA samples showed the presence of C–F stretching vibrations (1500–1000 cm–1) from the dispersant (Supplementary Figure S11). This indicates that PD-2 coelutes into the aqueous layer, likely due to its stronger coupling interactions with the protein surface (as suggested by our NMR results, Supplementary Figures S6–S9 and S12). Thus, our data suggest that, unlike PD-2, PD-7 successfully balances hydrogen bonding propensity and fluorophilicity to enable its use as a transient masking agent that decouples from the protein biologic in physiologic solutions as desired.
To gain a deeper mechanistic insight into dispersant/protein complexation and further inform the observed decoupling phenomena of PD-7, we performed discrete molecular dynamics simulations (Figure 4a). Strikingly, in silico models, again using BSA as an exemplary protein, revealed a distinct morphological difference between PD-2 and PD-7 assemblies in the presence of the protein. For example, PD-2 organized into discrete clusters that predominantly absorb to solvent exposed loop regions of the protein over the 10 ns simulation time. In contrast, PD-7 assembled into fibrillar bundles that circumscribed the protein surface, ultimately forming a cage-like structure. This resembles the ropy coatings on the surface of PD-7 functionalized protein assemblies observed in our TEM experiments (Figure 3d). Molecular models further demonstrate the ability of PD-2 (Figure 4b) and PD-7 (Figure 4c) to form hydrogen bonds with amino acid side chains and amide backbone of the solvent exposed protein surface, with distances typical for protein–ligand interactions (∼1.5–2.0 Å).
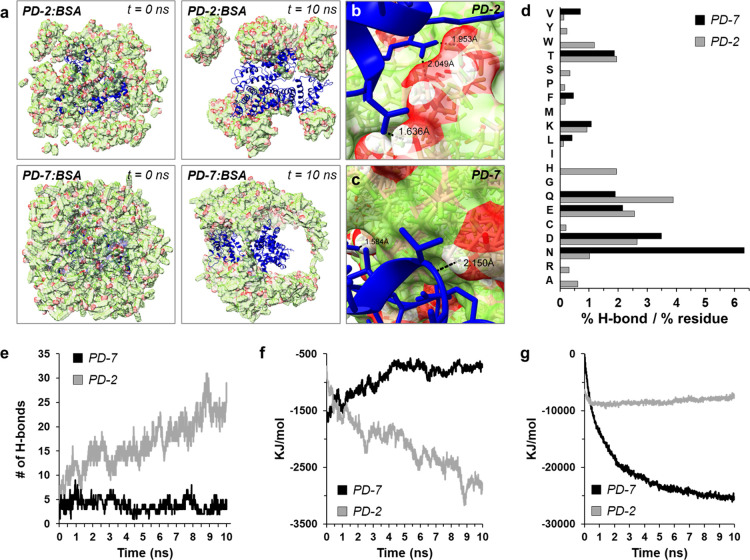
Figure 4.
In silico atomistic investigation of dispersant–protein interactions: (a) molecular organization of PD-2/BSA (top panels) and PD-7/BSA (lower panels) assemblies at the initial (t = 0 ns) and 10 ns simulation time points. The BSA protein is colored blue, and dispersant is shown in red/green. (b, c) Molecular models of (b) PD-2 and (c) PD-7 docked to the surface of BSA, with the indicated distance of representative hydrogen bonds. (d) Residue-specific hydrogen-bonding interactions of PD-7 (black) or PD-2 (gray) with BSA. Data reported as the percentage of total hydrogen bonds divided by the frequency normalized residue count in the BSA protein. H-bonding frequency plots for dispersant interaction with Hb, β-Gal, GFP, and trypsin are shown in Figure S13. (e–g) Change in (e) number of hydrogen bonds, (f) free energy after interaction with the protein surface, and (g) free energy of dispersant oligomerization for PD-7 (black) or PD-2 (gray) over the 10 ns simulation time. Free energy plots for dispersant interactions with Hb, β-Gal, GFP, and trypsin are shown in Figures S14 and S15.
Plotting the frequency normalized, amino-acid-specific, hydrogen bonding propensity of each dispersant showed a general preference for interactions of the perfluoroalkyls with residues that possess both hydrogen bond donors and acceptors in their side chains (Figure 4d), particularly asparagine (N), glutamine (Q), aspartic acid (D), and glutamic acid (E). Calculating additional thermodynamic parameters from the in silico simulations showed that, as expected, PD-2 had a greater number of hydrogen bonds formed with the protein surface (Figure 4e) and a lower predicted interaction energy (Figure 4f) compared to PD-7. This corroborates our NMR experimental data (Figure 3e,f) and further demonstrates the thermodynamically favored interactions between PD-2 and the protein surface. Conversely, analysis of dispersant oligomerization showed a rapid decrease in the interaction energy of PD-7 as it assembled with itself, whereas PD-2 self-assembly was not favored (Figure 4g). Importantly, these thermodynamic conclusions are corroborated across similar simulations using hemoglobin, β-galactosidase, GFP, and trypsin as target proteins (Supplementary Figures S13–S15). In sum, our in silico experiments indicate that homo-oligomeric assembly of PD-7 creates laminated bundles that encompass the protein surface, with more uniform coverage compared to PD-2, thereby generating a more complete fluorophilic coating that promotes protein solubilization in PFOc. PD-2, in contrast, favors hydrogen bonding with the protein surface rather than self-assembly, leading to the formation of dispersant islands that leave large areas of the protein surface exposed to the fluorous solvent. Finally, the weak van der Waals interactions calculated between PD-7 and the protein surface may explain the preferential decoupling of this dispersant in ionic solutions, as was observed during our FTIR experiments (Figures 3g and Supplementary Figure S10). It is important to note that the conclusions discussed here are drawn from simulations performed in an aqueous environment. Unfortunately, the absence of reported force fields for PFCs prevented in silico simulations in a fluorous solvent. Thus, we are careful to use these conclusions to generate hypotheses and inform the interpretation of the biophysical results.
2.4. Contamination Resistance of Fluorous Protein Formulations
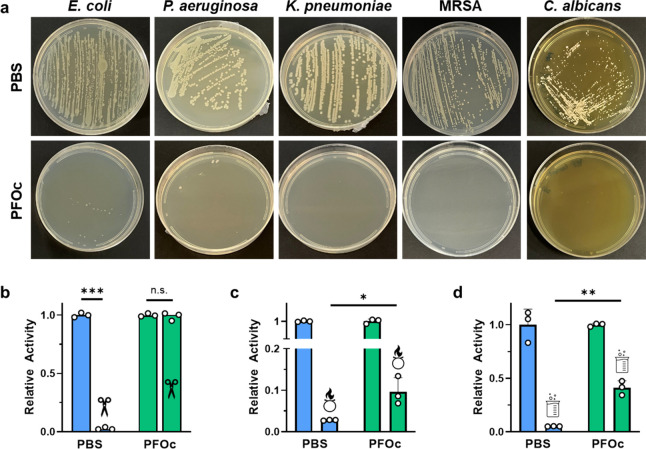
Aqueous protein formulations can be compromised by contaminating pathogens or the biologic inactivated by exposure to environmental disinfectants. In the context of microorganismal contamination, we hypothesized that the removal of the water solvent required for organism survival would make our PFOc protein formulations intrinsically sterile. To test this assertion, we contaminated a hypodermic needle with the human bacterial pathogens E. coli, P. aeruginosa, K. pneumoniae, or methicillin-resistant S. aureus (MRSA), as well as the human fungal pathogen C. albicans, and used it to inoculate BSA protein formulations prepared in either PBS or PFOc solvents. The contaminated samples were then incubated at 37 °C overnight and replated onto agar to assess the growth. Results in Figure 5a show that, as expected, BSA in PBS was readily contaminated by all five pathogens, as demonstrated by the emergence of viable colonies on the plate. In contrast, PFOc samples remained sterile despite the high pathogen inoculum. These formulations were also able to attenuate the infectivity of dispersed lentiviral vectors toward HEK293 cells, which were used as a model for viral contamination (Supplementary Figure S16). Possible explanations for this are that contaminating pathogens dehydrate when submerged in the nonaqueous PFOc formulation and/or have their lipid and protein membranes solubilized by the lipophilic fluorous media, collectively leading to their destruction. Yet, the independent addition of each component, that is, mixing the PFOc solvent with bacterial media or addition of PD-7 to PBS, did not compromise bacterial viability (no MIC could be calculated in either control experiment). This suggests that the combination of PD-7 and PFOc, as well as the exclusion of an aqueous phase, is necessary to maintain the sterility of fluorescence dispersed proteins.
Figure 5.
Fluorous protein sterility and stability. (a) Representative photographs of agar plates streaked with the indicated pathogen from contaminated BSA formulations in PBS (top panel) or PFOc (lower panel). MRSA = methicillin-resistant Staphylococcus aureus. (b–d) Relative activity of β-Gal in PBS (blue) or PFOc (green) solvents without (no icon) and with (icon) contamination by (b) proteinase K, (c) bleach, or (d) hydrochloric acid. Data shown as average ± s.d. of n = 3 technical replicates. Statistical significance between conditions is indicated by a line using unpaired Student’s t test with n.s. = not significant, * p < 0.05, ** p < 0.01, and *** p < 0.001.
In addition to microorganismal contamination, protein formulations can be compromised by incident contact with environmental proteases, oxidizing cleaners, and acidic disinfectants. To empirically model these conditions, we separately added aliquots of proteinase K, bleach (chlorine and sodium hydroxide mixture), or hydrochloric acid to PFOc and PBS protein samples (Figures 5b–d). Here, we use β-Gal as an exemplary biologic and substrate conversion assay to evaluate protein activity after exposure to each denaturant. Whereas β-Gal in PBS was completely inactivated by proteinase K, there was no statistically significant change in activity for PFOc formulations under similar conditions (Figure 5b). This is most likely due to the coating of the proteinase K enzyme by the PD-7 additive after addition to PFOc, leading to its segregation from the codispersed β-Gal protein. Moreover, enzymatic hydrolysis requires water solvent molecules, which, if still present in the PFOc mixture, are expected to be tightly bound to the protein structure. Although these results are encouraging, the fluorous formulations were less resistant to chemical denaturants (Figures 5c,d). Although the activity of oxidized (Figure 5c) or acidified (Figure 5d) β-Gal PFOc samples was statistically higher than that of PBS controls, both conditions lead to a ≥50% loss in functionality after only a few minutes of incubation. It should be noted that, in a typical healthcare exposure, protein solutions would be contaminated by noxious vapors and not directly spiked with a solution of the denaturant as done in our studies. Despite these stringent conditions, PFOc samples still improved protein stability relative to standard saline control solutions.
2.5. In Vivo Serum Half-Life and Toxicity
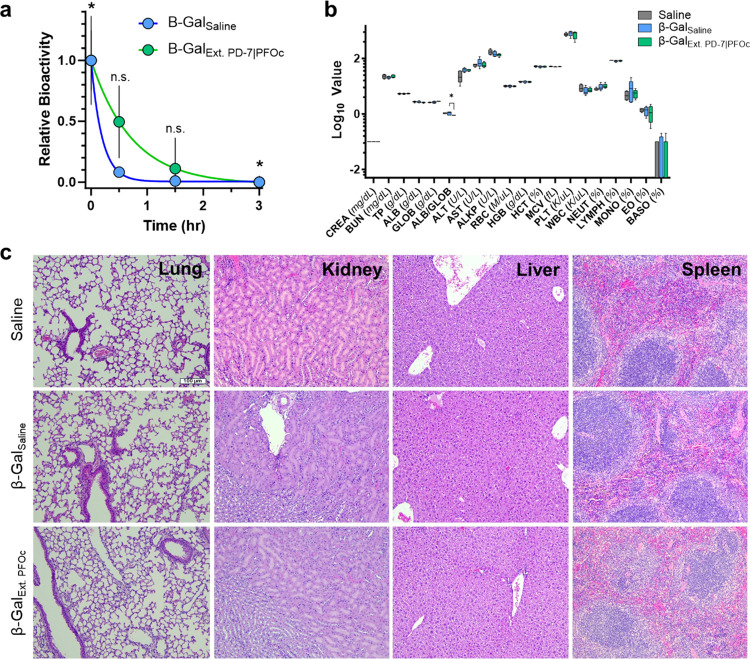
To test whether PD-7 coating and PFOc dispersion influence the circulatory half-life of formulated proteins, we intravenously administered β-Gal formulations to C57BL/6 mice and monitored time-dependent serum bioavailability (Figure 6a). This experiment utilized a fluorescent conversion substrate to monitor the amount of functional protein in serum, thereby investigating changes to both bioavailability and bioactivity. Because of the large injection volumes (100 μL, ∼10% blood volume), we decided to extract the PFOc protein samples into saline before injection to avoid embolism and/or hyponatremia. We anticipate that future studies in large animals would allow direct injection of the fluorous solution, without pre-extraction, due to the increased blood volumes and bioinert nature of most perfluorocarbon solvents.17 Results in Figure 6a show that the serum half-life of β-Gal from PFOc formulations was slightly extended (t1/2 = 0.50 h) relative to the native β-Gal administered in saline (t1/2 = 0.12 h). However, variance in the data yielded statistical significance only at the t = 0 and 3 h time points.
Figure 6.
In vivo half-life and safety of fluorous proteins. (a) Time-dependent activity of β-Gal in plasma delivered intravenously in either saline (β-GalSaline, blue) or extracted PFOc (β-GalExt. PFOc, green). Data shown as average ± s.d. of n = 5 technical replicates. Statistical significance calculated with Student’s t test, with n.s. = not significant and *p < 0.05. (b) Serum toxicology results from C57BL/6J mice 24 h after injection of saline (control), β-GalSaline, or β-GalExt. PFOc. Data shown as box and whisker plot ± s.d. of n = 5 technical replicates. Statistical significance calculated using Student’s t test and represented as * p < 0.05; data subsets with p > 0.05 represented nonsignificant changes. (c) Representative histology images of lung, kidney, liver, and spleen tissue section from C57BL/6J mice 24 h after administration of saline (control), β-GalSaline, or β-GalExt. PFOc. Each imaging group consisted of n = 4 mice, with detailed tissue section evaluation conducted by an unblinded board-certified veterinary anatomic pathologist. Scale bar: 100 μm.
Finally, to assess the potential for acute toxicity of the fluorous dispersant, serologic chemistry analysis and histology were performed 24 h after treating mice with β-Gal delivered from saline or PFOc extractions. Hematologic, renal, and hepatic blood toxicity markers showed a statistically significant reduction in the albumin/globulin (ALB/GLOB) ratio between β-Gal delivered from PFOc and saline (Figure 6b). Reduction of this marker can indicate a transient impairment of kidney or liver function. However, the absence of statistically significant changes in all other renal (e.g., CREA, BUN, and TP) and hepatic (e.g., ALT, AST, and ALKP) markers suggests that the ALB/GLOB finding is not likely to be clinically meaningful. This is further corroborated by unblinded histologic analysis of vital organs by a board-certified veterinary anatomic pathologist, which did not identify pathologic evidence of necrosis, noninflammatory cell infiltration, or antemortem hemorrhage (Figure 6c). Minimal background histologic findings were similar in all three groups and consisted solely of nominal focal inflammation within the liver and minimal peracute collection-associated intraalveolar hemorrhage. Slight focal inflammation in the liver of all three groups was lymphohistiocytic with occasional few neutrophils. Because of the similar frequency in all three groups, these were interpreted as background lesions and not test article-related findings. Collectively, these results suggest that the perfluorinated reagents used in our protein formulations are unlikely to induce acute toxic side effects.
3. Conclusions
Production of shelf-stable and intrinsically sterile protein formulations holds promise to reduce or altogether eliminate the need for cold-chain logistics. Such a technological advance may also expand the deployment of important protein reagents and therapeutics in resource-limited settings. Toward this goal, prior work from our group demonstrated that proteins coated by the amphiphilic perfluorinated dispersant PD-213 and dissolved in nonaqueous fluorous solvents restricted the conformational plasticity of proteins and yielded biologics that were structured and functional at temperatures as high as 90 °C. Yet, our first-generation dispersant, PD-2 (PFNA), showed low dispersion efficiencies in long-chain perfluorocarbon solvents (important for creating formulations with high boiling points) and coeluted with proteins into physiologic solutions. The latter was particularly concerning given the emerging evidence that perfluorinated compounds bioaccumulate in tissues and cause long-term toxicity in mammals.
To address these formulaic and pharmacologic liabilities, we synthesized, screened, and validated a library of novel dispersants designed to improve dispersion efficiencies and protein–dispersant decoupling rates. We identified the bivalent perfluorinated carboxylic acid PD-7 as a lead second-generation candidate due to its high dispersion efficiencies (>95%) and low cytotoxicity (IC50 ≈ 1.0 mM). Subsequent thermostability experiments showed that proteins dispersed into PFOc using PD-7 had near-complete retention of bioactivity at temperatures up to 90 °C. A series of biophysical and in silico assays elucidated the mechanistic basis for thermal stabilization. Finally, in vitro and in vivo experiments show that fluorous-dispersed proteins remain sterile after intentional contamination by bacterial, fungal, and viral pathogens, which require aqueous solvents for survival, and display equivalent pharmacologic properties, bioactivity, and safety profiles to standard saline protein formulations in mice. These findings highlight the promise of PD-7 as a clinically relevant protein coating technology that, when paired with intrinsically sterile fluorous solvents, can produce a novel storage paradigm to generate extremophilic protein formulations. Future work will seek to validate these claims, advance new formulations of clinically relevant protein therapies, and study the potential toxicity and environmental impact of the employed fluorous media.
4. Experimental Section
4.1. Materials
Commercially available chemicals and reagents utilized for synthetic procedures were procured from different vendors and were used as received unless otherwise stated. Perfluoroalkyl carboxylic acids (PD-1 to PD-5) and perfluoroalkyl ethyl iodides (1a–1c) were purchased from Oakwood Chemicals. CaCl2, Hepes buffer solution (HBS 2×), polyethylene glycol 6000 (PEG6000), anhydrous DMF, and anhydrous THF were purchased from ThermoFisher Scientific. Silica gel 60 F254 coated aluminum TLC sheets were obtained from Merck, Darmstadt. Chromatographic separations of crude products were accomplished on a Buchi FlashPure C-815 flash chromatography system having HPLC standard ethyl acetate and hexanes as the mobile phase. The Pierce Coomassie Plus (Bradford) assay reagent was purchased from ThermoFisher Scientific. NMR spectra of synthesized materials were recorded on a Bruker NEO-400 spectrometer operating at 400 MHz for 1H and 100 MHz for 13C, respectively, using deuterated solvents (CDCl3 and methanol-d4). TMS and deuterated solvents were used as internal references for 1H and 13C NMR experiments. Raw data were processed on the Mnova software, and resultant chemical shifts were reported as ppm (δ). The splitting patterns for respective peaks were abbreviated as s = singlet, bs = broad singlet, d = doublet, t = triplet, q = quartet, m = multiplet, tt = triplet of triplets, dt = doublet of triplets, and qd = quartet of doublets. Thiozolyl blue tetrazolium bromide (MTT) was acquired from Chem-Impex. DMEM culture medium, FBS, PBS, and trypsin were purchased from Corning. The HepG2 cell line used for the cytotoxicity assay was a generous gift from the National Cancer Institute (NCI). HEK293 cells, lentiviral packaging plasmids (REV, TAT, Gag Pol, VSV-G), and a recombinant GFP vector were provided as generous gifts from the laboratory of Dr. Justin Pritchard, Pennsylvania State University.
4.2. Synthetic Procedures
4.2.1. General Synthetic Procedure for PD-6, PD-8, and PD-10
Cyanoacetate derivative, 2a/2b/2c (1.0 equiv, 0.31 mmol), and aq. KOH (3.1 equiv, 0.962 mmol) were taken in a round-bottomed flask containing 20 mL of an ethanol/H2O (1:1) mixture (Supplementary Scheme S1). The resultant reaction mixture was stirred with heating at 80 °C for 3 h. During this time, the completion of the reaction was evidenced by TLC. The reaction mixture was poured into 1 M HCl solution to obtain a white precipitate followed by extraction using diethyl ether as the organic phase. The obtained ether layer was washed with brine solution and concentrated to give cyano-acid derivative PD-6/PD-8/PD-10 as a white solid in quantitative yield. See corresponding 1H, 19F, and 13C NMR and mass spectra in Supplementary Figures S17–S25, S35–S38, S43–S45, and S49–S52.
4.2.1.1. PD-6
Yield = 220 mg (91%); 1H NMR (400 MHz, MeOD) δ 2.58–2.43 (m, 2H), 2.43–2.23 (m, 6H) ppm; 19F NMR (376 MHz, MeOD) δ −82.43 (t, J = 10.2 Hz, 6F), −115.36 (dt, J = 14.9, 4.8 Hz, 4F), −122.78 to −123.06 (m, 4F), −123.82 to −124.02 (m, 4F), −124.28 to −124.46 (m, 4F), −127.28 to −127.42 (m, 4F) ppm; 13C NMR (100 MHz, CDCl3) δ 168.31, 117.47, 27.22, 27.06, 26.84 ppm. HRMS-ESI calculated: C19H9F26NO2, m/z = 777.0218; experimental = 776.0147 [M – H]−.
4.2.1.2. PD-8
Yield = 260 mg (85%); 1H NMR (400 MHz, MeOD) δ 2.45–2.30 (m, 2H), 2.30–2.06 (m, 6H) ppm; 19F NMR (376 MHz, MeOD) δ −82.36 (t, J = 10.1 Hz, 6F), −115.32 (t, J = 13.3 Hz), −122.69 (bs, 4F), −122.89 (bs, 8F), −123.74 (bs, 4F), −124.33 (bs, 4F), −127.22 to −127.36 (m, 4F) ppm. HRMS-ESI calculated: C23H9F34NO2, m/z = 977.0090; experimental = 976.0016 [M – H]−.
4.2.1.3. PD-10
Yield = 160 mg (90%); 1H NMR (400 MHz, MeOD) δ 2.49–2.28 (m, 2H), 2.30–2.07 (m, 6H) ppm; 19F NMR (376 MHz, MeOD) δ −82.37 (tt, J = 9.8 Hz, 6F), −115.58 to −115.67 (m, 4F), −125.34 to −125.43 (m, 4F), −127.22 to −127.34 (m, 4F) ppm; 13C NMR (100 MHz, CDCl3) δ 168.28, 117.44, 27.21, 26.97, 26.75 ppm. HRMS-ESI calculated: C15H9F18NO2, m/z = 577.0346; experimental = 576.0271 [M – H]−; 1153.0605 [2M – H]−.
4.2.2. General Synthetic Procedure for PD-7, PD-9, and PD-11
Bis((perfluorohexyl)ethyl) cyanoacetic acid derivative PD-6/PD-8/PD-10 (0.257 mmol, 1.0 equiv) was taken in a 250 mL round-bottomed flask containing concentrated HCl (10 mL). The reaction mixture was connected to a base trap and heated while being stirred at 100 °C for 2 h. The reaction mixture was then cooled to 0 °C by transferring to an ice bath followed by dropwise addition of concentrated H2SO4 using a pressure equalizing funnel while connected to the base trap. Once the concentrated H2SO4 was completely transferred to the reaction mixture, the solution was carefully heated while stirring at 160 °C. After overnight stirring, the reaction mixture was allowed to cool to room temperature. Water (50 mL) was added into the reaction mixture and extracted using diethyl ether (4 × 20 mL). The residual water in the ether layer was dried with Na2SO4. Concentration of the ether layer produced the product as a white solid (Supplementary Scheme S1). See corresponding 1H, 19F, and 13C NMR and mass spectra in Supplementary Figures S39–S42, S46–S48, and S53–S56.
4.2.2.1. PD-7
Yield = 150 mg (78%); 1H NMR (400 MHz, MeOD) δ 2.65–2.50 (sep, J = 4.8 Hz, 1H), 2.27 (tt, J = 17.8, 8.2 Hz, 4H), 2.00–1.82 (m, 4H) ppm; 19F NMR (376 MHz, MeOD) δ −82.45 (t, J = 10.3 Hz, 6F), −115.59 to −115.75 (m, 4F), −122.84 to −123.06 (m, 4F), −123.82 to −124.04 (m, 4F), −124.46 to −124.61 (m, 4F), −127.30 to −127.44 (m, 4F) ppm; 13C NMR (100 MHz, CDCl3) δ 175.79, 43.10, 28.18 (t, JCF = 21.7 Hz), 22.27 ppm. HRMS-ESI calculated: C18H10F26O2, m/z = 752.0266; experimental = 751.0195 [M – H]−; 797.0249 [M + HCOO]−.
4.2.2.2. PD-9
Yield = 170 mg (70%); 1H NMR (400 MHz, MeOD) δ 2.49–2.41 (sep, J = 4.4 Hz, 1H), 2.15 (tt, J = 17.7, 8.2 Hz, 4H), 1.92–1.69 (m, 4H) ppm; 19F NMR (376 MHz, MeOD) δ −82.37 (t, J = 10.2 Hz, 6F), −115.59 (d, J = 15.1 Hz, 4F), −122.73 (bs, 4F), −122.92 (bs, 8F), −123.76 (bs, 4F), −124.42 to −124.58 (m, 4F), −127.24 to −127.36 (m, 4F) ppm. HRMS-ESI calculated: C22H10F34O2, m/z = 952.0138; experimental = 951.0063 [M – H]−; 997.0115 [M + HCOO]−.
4.2.2.3. PD-11
Yield = 122 mg (82%); 1H NMR (400 MHz, MeOD) δ 2.60–2.53 (sep, J = 4.8 Hz, 1H), 2.33–2.19 (m, 4H), 2.01–1.81 (m, 4H) ppm; 19F NMR (376 MHz, MeOD) δ −82.69 (t, J = 9.8 Hz, 6F), −115.82 to −116.02 (m, 4F), −125.49 to −125.61 (m, 4F), −127.23 to −127.35 (m, 4F) ppm; 13C NMR (100 MHz, CDCl3) δ 175.80, 43.13, 28.09 (t, JCF = 22.0 Hz), 22.28 ppm. HRMS-ESI calculated: C14H10F18O2, m/z = 552.0393; experimental = 551.0320 [M – H]−; 1103.0704 [2M – H]−.
4.2.3. General Synthetic Procedure for PD-12, PD-13, and PD-14
Conjugate 3a/3b/3c (1.0 equiv, 0.237 mmol) and KOH (6.2 equiv, 1.47 mmol) were taken in a round-bottomed flask having 10 mL of ethanol/H2O (1:1) mixture (Supplementary Scheme S2). The resultant reaction mixture was stirred with heating at 80 °C for 3 h. During this time, the complete conversion of the 3a/3b/3c to the product was seen as evidenced by TLC. The reactant mixture was neutralized using 1 M HCl followed by extraction using diethyl ether. This ether layer was further extracted with brine solution. The ether layer was dried over Na2SO4 and concentrated to give monosubstituted malonic acid derivative PD-12/PD-13/PD-14 as a white solid in quantitative yield. See the corresponding 1H, 19F, and 13C NMR and mass spectra in Supplementary Figures S26–S34 and S57–S67.
4.2.3.1. PD-12
Yield = 100 mg (94%); 1H NMR (400 MHz, MeOD) δ 3.49 (t, J = 7.2 Hz, 1H), 2.38–2.25 (m, 2H), 2.17–2.11 (m, 2H) ppm; 19F NMR (376 MHz, MeOD) δ −82.42 (m, 3F), −115.72 to −115.81 (m, 2F), −122.88 to −123.04 (m, 2F), −123.86 to −124.02 (m, 2F), −124.53 to −124.67 (m, 2F), −127.30 to −127.42 (m, 2F) ppm; 13C NMR (100 MHz, CDCl3) δ 170.74, 50.14, 28.03 (t, JCF = 26.4 Hz), 19.45 ppm. HRMS-ESI calculated: C11H7F13O4, m/z = 450.0137; experimental = 449.0063 [M – H]−; 899.0197 [2M – H]−.
4.2.3.2. PD-13
Yield = 120 mg (92%); 1H NMR (400 MHz, MeOD) δ 3.47 (t, J = 7.2 Hz, 1H), 2.37–2.23 (m, 2H), 2.15–2.09 (m, 2H) ppm; 19F NMR (376 MHz, MeOD) δ −82.37 (t, J = 9.78 Hz, 3F), −115.70 to −115.79 (m, 2F), −122.74 (bs, 2F), −122.92 (bs, 4F), −123.74 (bs, 2F), −124.51 to −124.60 (m, 2F), −127.23 to −127.35 (m, 2F); 13C NMR (100 MHz, CDCl3) δ 170.72, 50.13, 28.03, 19.41 ppm. HRMS-ESI calculated: C13H7F17O4, m/z = 550.0073; experimental = 549.0004 [M – H]−.
4.2.3.3. PD-14
Yield = 75 mg (91%); 1H NMR (400 MHz, MeOD) δ 3.46 (t, J = 7.2 Hz, 1H), 2.36–2.22 (m, 2H), 2.15–2.09 (m, 2H) ppm; 19F NMR (376 MHz, MeOD) δ −82.68 (tt, J = 9.7, 3.4 Hz, 3F), −115.96 to −116.07 (m, 2F), −125.52 to −125.65 (m, 2F), −127.23 to −127.35 (m, 2F); 13C NMR (100 MHz, CDCl3) δ 170.81, 50.14, 27.94 (t, JCF = 21.9 Hz), 19.39 ppm. HRMS-ESI calculated: C9H7F9O4, m/z = 350.0201; experimental = 349.0124 [M – H]−; 699.0328 [2M – H]−.
4.3. Protein Dispersion and Elution
Protein dispersion into perfluorooctane (PFOc) was performed according to an established protocol previously reported.8,15 Briefly, PFOc containing 1 mM PD-7 was added to lyophilized protein stocks prepared in 1.5 mL centrifuge tubes to achieve final concentrations of 5–10 μM protein. Dispersion was performed via 30 s vortexing followed by a 1 min sonication and a final vortexing step. Samples were sealed via parafilm and rotisserie mixed overnight before centrifugation for 5 min at 1950g to remove insoluble aggregates. Dispersion percentage was quantified by transferring 100 μL of the supernatant to a 96-well plate and evaporating the solvent. Residual protein solids were resuspended in 100 μL of 1× PBS with equal parts Coomassie blue reagent for the Bradford assay. The protein prepared in PBS and blank PFOc (without dispersant) were included as positive and negative controls, respectively. Samples were shaken at room temperature for 10 min, and absorbance at 595 nm was read using a Biotek Cytation 3. Dispersion efficiency was calculated as follows:
| 1 |
Protein extraction from PFOc dispersions was performed by adding an equal amount of 1× PBS and vortexing the mixture for ≤10 s. The top, aqueous phase could then be removed from the bottom, PFOc phase via pipetting.
4.4. Cellular Toxicity
HepG2 cells were seeded in 96-well plates at 2000 cells/well and incubated overnight to adhere. One hundred microliters of treatment solutions containing 10 nM–1 mM of the PD dispersant in cell culture media was added to the wells. Experiments testing the toxicity of PFOc were performed by the addition of 2–128 μL of the fluorous solvent to the cell culture medium. Blank serum-free medium and 20% DMSO containing serum-free medium were included as negative and positive controls, respectively. After a 48 h treatment period, the supernatant was replaced by 100 μL of serum-free media containing 0.5 mg/mL MTT dye, and samples were incubated for 2 h. The supernatant was then aspirated, and 100 μL of DMSO was added to each well to dissolve the formazan product before reading absorbance at 540 nm using a BioTek Cytation 3 microplate reader. Cell viability was calculated as follows:
| 2 |
4.5. Spectroscopy
Circular dichroism (CD) spectroscopy was used to evaluate temperature-dependent changes in the protein structure. Proteins were diluted in PBS or dispersed in PFOc by using 1 mM PD-7. For aqueous samples, dissolved proteins were heated between 40 and 90 °C, and their corresponding CD spectra (Jasco J-1500; Easton, MD) at each temperature were measured. For PFOc samples, each of the PFOc dispersed proteins was subjected to heat treatment at 40, 55, 70, and 85 °C for 15 min. Heat-treated samples were then mixed with PBS to elute proteins before CD measurements. Replicates (n = 3) were performed for each condition, with representative spectra reported.
Nuclear magnetic resonance (NMR) spectroscopic titrations were accomplished with a PFOc solution of 1000:1 dispersant/BSA molar ratio ([PD-7 or PD-2] = 60 mM). Samples were added to a Wilmad thin wall precision tube. Norell coaxial inserts were inserted into NMR tubes and contained D2O (reference solvent to lock NMR signal). 1H NMR spectra were recorded on a Bruker NEO-400 spectrometer at 298 K with zg30 pulse sequence (32 scans, sweep width = 20.4850 ppm; origin point = 6.175 ppm), and zgig sequence was used for 19F NMR (512 scans, sweep width = 241.4836 ppm; origin point = −100.0 ppm). Data were analyzed with the Mnova software. Elution of BSA (30 μM) dispersed with PD-2 or PD-7 (30 mM) in PFOc (400 μL) was performed by adding an equal amount of PBS. The PBS layer was isolated and lyophilized. This lyophilized solid was dissolved in PBS (400 μL) to record 19F NMR spectra. 2-(Trifluoromethyl)acrylic acid was used as internal standard for 19F signal.
Fourier transform infrared (FTIR) spectrometry was performed on 1000:1 dispersant/protein molar ratio ([PD-7 or PD-2] = 1 mM) in PFOc (200 μL) and mixed on a rotisserie rotator overnight. Samples were eluted into an equal volume of PBS by vortexing briefly. The PBS layer was dried overnight before performing analysis using a Bruker VERTEX 70 FTIR spectrophotometer (Billerica, MA) employing an LN-MCT detector (4000–800 cm–1, backward output). FTIR spectra of the protein and PD-7 or PD-2 alone as controls were also recorded. Replicates (n = 3) were performed for each condition with representative spectra reported.
4.6. Protein Activity Assay
One and 10 μM concentrations of β-Gal and trypsin proteins, respectively, were diluted in PBS or PFOc containing 1 mM PD-7. Next, samples were incubated at 25 °C (room temperature) or 90 °C for 30 min. Samples were cooled before elution in PBS, as described above. An equal volume of 4 mg/mL ONPG in PBS for β-Gal or 1 mg/mL BAEE in PBS for trypsin was added to the proper sample and incubated following manufacturer’s instructions. Absorbance at 420 or 400 nm was read to measure conversion of the ONPG and BAEE substrates, respectively, using a BioTek Cytation 3 microplate reader. Relative activity was calculated by normalizing test sample data to the enzymatic activity of proteins at room temperature in their respective solvent environment.
4.7. Molecular Dynamics Simulations
All-atom molecular dynamics simulations were performed by using GROMACS 2020.2. Protein structure files were obtained from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) using the following codes: BSA: 4F5S, β-Gal: 6TBF, hemoglobin: 1A3N, GFP: 4OGS, and trypsin: 4DOQ. After heteroatoms and crystal water were removed from the protein structures, an all-atom OPLS force field with an SPC/E water model was chosen for generating a topology file for proteins. The OPLS topology of the ligand was generated using the LigParGen server from the Jorgensen group.18 A rhombic dodecahedron unit cell was defined with a 1 nm distance between the protein molecule and the box edge. This box was filled with water molecules using the solvate module, and appropriate ions were added to make it charge neutral. The molecular structures inside the assembled unit cell were relaxed by using the steepest descent energy minimization algorithm. The first phase of equilibration was performed using an NVT ensemble for 100 ps, which was followed by 100 ps of NPT equilibration. After the equilibration step, the final production MD run was performed for 10 ns (BSA, GFP, and trypsin) and 5 ns (β-Gal and hemoglobin) with a time step of 2 fs using the leapfrog integrator. An NPT ensemble was used to perform these MD runs using a velocity rescale thermostat and Parrinello–Rahman barostat, where the temperature and pressure were controlled at 300 K and 1 bar, respectively. After the simulation, interaction energy was calculated, and VMD was used for analysis and visualization of the H-bonds.
4.8. Transmission Electron Microscopy
BSA solutions in water (1 μM) or dispersed into PFOc with and without PD-7 (10 μM, following the dispersion protocol above) were deposited as a 5 μL aliquot onto a copper grid and dried overnight, and TEM imaging was performed at 200 kV using a FEI Tecnai LaB6 electron microscope (Hillsboro, OR).
4.9. Contamination Assays
For pathogen contamination studies, E. coli (101-1), P. aeruginosa (PAO1), K. pneumoniae (NCTC 9633), and methicillin-resistant S. aureus (MRSA; USA300) were cultured in MHB broth. C. albicans (3147) was cultured in YPD broth at 37 °C. All broth cultures were grown at 37 °C in a shaking incubator (200 rpm; plates were cultured in a static incubator as advised by the Clinical and Laboratory Standards Institute (CLSI). Bacterial lawns were prepared by streaking liquid cultures onto MHB (E. coli, P. aeruginosa, K. pneumoniae, and MR S. aureus) or YPD (C. albicans) agar and incubated until a lawn had developed. To contaminate PBS or PFOc liquid samples, a 21G needle was streaked in a single pass across the microbial lawn and dipped into liquid samples containing BSA (10 μM). Samples were then incubated at 37 °C for 1 day before streaking onto an agar plate. Plates were incubated at 37 °C overnight before qualitative visualization of colony formation.
Viral contamination studies were initiated by production of lentiviral particles. A total of 2 × 106 HEK293 cells/well were seeded in a six-well plate and allowed to adhere overnight. Cells were transfected with 5 μg of the GFP vector and 5 μg of the packaging plasmid in the presence of Hepes buffer and 15 mM CaCl2. The cells were incubated for 24 h, with medium change after the first 9 h, before collecting the lentiviral particle containing culture media. A PEG-based lentiviral concentrator (80% w/vol PEG600, 2.4 M NaCl in PBS) was used to extract the viral particles from culture supernatant via filtration through a 0.2 μm filter. The concentrated viral stock was stored overnight at 4 °C and then centrifuged at 1600g for 1 h before collection of the viral pellet. To contaminate test solutions, 500 μL of the viral stock was added to PBS or 1 mM PD-7 in PFOc and incubated for 12 h. PFOc samples were then extracted by gentle vortexing with an equal volume of added PBS. Collected solutions were then tested for their bioactivity via addition to HEK293 cultures and incubated for 24 h. Infected cells were then trypsinized and subjected to flow cytometry to measure successful GFP transduction. HEK293 treated with blank, sterile media was used as a negative control.
Enzymatic degradation was evaluated by preparing solutions of β-Gal (1 μM) and proteinase K (10 μM) in PBS or by dispersion in PFOc (1 mM PD-7). β-Gal was incubated at 37 °C for 24 h and extracted into an equal volume of PBS by a short burst of intense vortexing. Afterward, the ONPG assay of this β-Gal solution was performed to evaluate its enzymatic activity.
The environmental degradation assay was performed by preparing 1000:1 dispersant/β-Gal molar ratio ([PD-7] = 1 mM) solution in PBS or dispersed in PFOc. Contaminants (10% bleach = 1% v/v or 0.1 M HCl = 4% v/v) were added into this solution and incubated for 30 s at 37 °C. This solution was extracted into an equal volume of PBS by short and strong vortexing. Afterward, the ONPG assay of this β-Gal solution was performed. Relative activity was further calculated by using negative and positive controls, i.e., blank buffer or β-Gal stock solution (1 μM), respectively.
4.10. Animal Experiments
Animal studies were preapproved under IACUC protocol no. 202101978. Two treatment sets involving 100 μL of 1 μM β-Gal dissolved in sterile PBS or dispersed in PFOc (1 mM PD-7) followed by extraction into sterile PBS were administered via tail vein injection into two cohorts of C57BL/6J mice (n = 5/group, females). Mice were sacrificed at specified time points to collect whole blood via a cardiac puncture. Serum containing β-Gal protein was extracted from whole blood by centrifuging at 2000 rcf for 4 min. A 1:10 dilution (final volume = 200 μL) of this serum was mixed with the fluorescent substrate 4-methylumbelliferyl-α-d-galactopyranoside (μ-GAL, final concentration = 1 μM). Samples were quickly pipet mixed and incubated in the dark at room temperature for 5 min. Fluorescent intensity was measured with λex = 360 nm and λem = 440 nm using a BioTek Cytation 3 microplate reader (Winooski, VT). Data were normalized to the background fluorescent intensity of the blank serum.
Acute toxicity of PD-7 was assessed by intravenous injection (100 μL of 1 μM β-Gal, dissolved in sterile PBS or dispersed in PFOc (1 mM PD-7) followed by extraction into sterile PBS) to mice. Mice were sacrificed after 24 h to collect blood via cardiac puncture. EDTA was added to at least 100 μL of whole blood for a complete blood count. Serum, for chemistry analysis, was isolated from the leftover whole blood by centrifuging at 1000 rcf for 10 min. Serological analyses were performed by the Pennsylvania State University Animal Resources Program. Histologic organ analysis was conducted on lung, kidney, liver, and spleen tissue, excised 24 h after administration, from three treatment groups (control (saline), β-GalSaline, or β-GalExt. PFOc) of 24 mice followed by fixation with 10% buffered formalin. Each organ was microtomed into 5 μM sections. Four representatives from these sections per organ were mounted onto glass slides and stained with hematoxylin and eosin (H&E). Four fields were randomly selected in every section for examination under a microscope at 100× magnification. Each imaging group consisted of n = 4 mice.
4.11. Statistical Analysis
Unless stated otherwise, all data were collected with n ≥ 3 and represented as mean ± standard deviation. Significance between groups was determined by Student’s t test, where p < 0.05 was considered significant.
Acknowledgments
Penn State Microscopy and Cytometry Facility, University Park, PA, is acknowledged for access to transmission electron microscopy. NMR spectroscopic analysis was performed at the Penn State NMR spectroscopy core. The Huck Institutes X-Ray Crystallography Facility provided access to a CD spectrophotometer (NIH grant S10-OD025145). Fourier transform Infrared spectroscopy was done at the Penn State Materials Research Institute. Molecular graphics and analyses were performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311. We acknowledge USDA-NIFA Hatch project # PEN04771 to G.S.K. as well as NSF DMR-1845053, NIH 1R35-GM142902, and DARPA D21AP10182 to S.H.M for supporting this work.
Data Availability Statement
The data supporting this work are available for research purposes upon request to the corresponding authors.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.4c03724.
Synthetic diagrams for protein dispersants PD-6 to PD-14; synthetic methods for intermediates 2a–2c and 3a–3c; relationship of PD-7 dispersion efficiency to protein MW; cytotoxicity of dispersants and PFOc against HepG2 cells; bioactivity of proteins in heated PBS containing PD-7; representative TEM micrograph of BSA in water; 1H and 19F NMR spectra of PD-7 and PD-7/BSA complexes; 1H and 19F NMR spectra of PD-2 and PD-2/BSA complexes; FTIR spectra of PD-7 and PD-7/β-Gal/IgG complexes and PD-2 and PD-2/BSA complexes; 19F NMR spectra of PBS extracted PD-7/BSA and PD-2/BSA; protein residue-specific hydrogen bonding interactions; dispersant–protein interaction free energy; dispersant–dispersant interaction free energy; viral contamination of saline and fluorous formulations; and 1H, 19F, 13C NMR, and HRMS spectra of intermediates and PD-6 to PD-14 (PDF)
Author Contributions
H.S. and S.H.M. conceived this work and wrote the manuscript. H.S. and A.L. performed and analyzed most experiments. U. performed molecular dynamics simulations. S.P. assisted with collection of thermal data. A.D. performed in vivo studies. D.M.R. performed pathohistologic analyses. Correspondence and requests for materials should be addressed to S.H.M. (shm126@psu.edu).
The authors declare no competing financial interest.
Supplementary Material
References
- Wang W. Instability, stabilization, and formulation of liquid protein pharmaceuticals. Int. J. Pharm. 1999, 185 (2), 129–188. 10.1016/S0378-5173(99)00152-0. [DOI] [PubMed] [Google Scholar]
- Yu Y. B.; Briggs K. T.; Taraban M. B.; Brinson R. G.; Marino J. P. Grand Challenges in Pharmaceutical Research Series: Ridding the Cold Chain for Biologics. Pharm. Res. 2021, 38 (1), 3–7. 10.1007/s11095-021-03008-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok A.; Brison M.; LeTallec Y. Improving cold chain systems: Challenges and solutions. Vaccine 2017, 35 (17), 2217–2223. 10.1016/j.vaccine.2016.08.045. [DOI] [PubMed] [Google Scholar]
- Kumru O. S.; Joshi S. B.; Smith D. E.; Middaugh C. R.; Prusik T.; Volkin D. B. Vaccine instability in the cold chain: mechanisms, analysis and formulation strategies. Biologicals 2014, 42 (5), 237–259. 10.1016/j.biologicals.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Contreras-Montoya R.; Arredondo-Amador M.; Escolano-Casado G.; Mañas-Torres M. C.; González M.; Conejero-Muriel M.; Bhatia V.; Díaz-Mochón J. J.; Martínez-Augustin O.; de Medina F. S.; Lopez-Lopez M. T.; Conejero-Lara F.; Gavira J. A.; de Cienfuegos L. A. Insulin Crystals Grown in Short-Peptide Supramolecular Hydrogels Show Enhanced Thermal Stability and Slower Release Profile. ACS Appl. Mater. Interfaces 2021, 13 (10), 11672–11682. 10.1021/acsami.1c00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence P. B.; Price J. L. How PEGylation influences protein conformational stability. Curr. Opin. Chem. Biol. 2016, 34, 88–94. 10.1016/j.cbpa.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prashar D.; Cui D.; Bandyopadhyay D.; Luk Y. Y. Modification of proteins with cyclodextrins prevents aggregation and surface adsorption and increases thermal stability. Langmuir 2011, 27 (21), 13091–13096. 10.1021/la203271u. [DOI] [PubMed] [Google Scholar]
- Rahban M.; Zolghadri S.; Salehi N.; Ahmad F.; Haertlé T.; Rezaei-Ghaleh N.; Sawyer L.; Saboury A. A. Thermal stability enhancement: Fundamental concepts of protein engineering strategies to manipulate the flexible structure. Int. J. Biol. Macromol. 2022, 214, 642–654. 10.1016/j.ijbiomac.2022.06.154. [DOI] [PubMed] [Google Scholar]
- Tamasi M. J.; Patel R. A.; Borca C. H.; Kosuri S.; Mugnier H.; Upadhya R.; Murthy N. S.; Webb M. A.; Gormley A. J. Machine Learning on a Robotic Platform for the Design of Polymer–Protein Hybrids. Adv. Mater. 2022, 34 (30), 2201809. 10.1002/adma.202201809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K.; Kawai S.; Fujitani M.; Koikeda S.; Kato R.; Ema T. Enhancement of protein thermostability by three consecutive mutations using loop-walking method and machine learning. Sci. Rep. 2021, 11 (1), 11883. 10.1038/s41598-021-91339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawanprasert A.; Sloand J. N.; González Vargas M.; Singh H.; Eldor T.; Miller M. A.; Pimcharoen S.; Wang J.; Leighow S. M.; Pritchard J. R.; Dokholyan N. V.; Medina S. H. Deciphering the Mechanistic Basis for Perfluoroalkyl-Protein Interactions. ChemBioChem 2023, 24 (13), e202300159 10.1002/cbic.202300159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloand J. N.; Nguyen T. T.; Zinck S. A.; Cook E. C.; Zimudzi T. J.; Showalter S. A.; Glick A. B.; Simon J. C.; Medina S. H. Ultrasound-Guided Cytosolic Protein Delivery via Transient Fluorous Masks. ACS Nano 2020, 14 (4), 4061–4073. 10.1021/acsnano.9b08745. [DOI] [PubMed] [Google Scholar]
- Lawanprasert A.; Pimcharoen S.; Singh H.; Vargas M. G.; Dewan A.; Kirimanjeswara G. S.; Medina S.. Heat Stable and Intrinsically Sterile Liquid Protein Formulations. bioRxiv 2024, 581015. [DOI] [PMC free article] [PubMed]
- Miller M. A.; Sletten E. M. A General Approach to Biocompatible Branched Fluorous Tags for Increased Solubility in Perfluorocarbon Solvents. Org. Lett. 2018, 20 (21), 6850–6854. 10.1021/acs.orglett.8b02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H.; Zhou Z.; Wang L.; Liu G.; Sun Y.; Wang Y.; Wang T.; Liang Y. Screening of Potential PFOS Alternatives To Decrease Liver Bioaccumulation: Experimental and Computational Approaches. Environ. Sci. Technol. 2019, 53 (5), 2811–2819. 10.1021/acs.est.8b05564. [DOI] [PubMed] [Google Scholar]
- Bassler J.; Ducatman A.; Elliott M.; Wen S.; Wahlang B.; Barnett J.; Cave M. C. Environmental perfluoroalkyl acid exposures are associated with liver disease characterized by apoptosis and altered serum adipocytokines. Environ. Pollut. 2019, 247, 1055–1063. 10.1016/j.envpol.2019.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman R.; Lorton O.; Guillemin P. C.; Desgranges S.; Contino-Pépin C.; Salomir R. Perfluorocarbon emulsion contrast agents: A mini review. Front. Chem. 2022, 9, 810029 10.3389/fchem.2021.810029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodda L. S.; Cabeza de Vaca I.; Tirado-Rives J.; Jorgensen W. L. LigParGen web server: an automatic OPLS-AA parameter generator for organic ligands. Nucleic Acids Res. 2017, 45 (W1), W331–W336. 10.1093/nar/gkx312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this work are available for research purposes upon request to the corresponding authors.