RNA has come a long way from a simple “messenger” or “translator” of canonical genic information during the production of proteins. A plethora of new types of noncoding RNAs have been discovered, including thousands of long noncoding RNAs (lncRNAs), many of which have no identified functions (1, 2). Throughout this “RNA revolution,” one property of RNA has been thought to be constant: RNAs are shortlived molecules that turn over, unlike DNA, which is much more stable. On page 53 of this issue, Zocher et al. (3) challenge that paradigm by showing that newly synthesized RNA labeled with 5-ethynyl uridine (EU) in early postnatal mice was still present in many brain cells 2 years later. The complex pattern of when and which cells are labeled suggests that EU that is incorporated into RNA in neural progenitor cells (NPCs) frequently remains in adult neurons. This suggests that a diversity of long and repeat-rich RNAs, collectively called long-lived RNAs (LL-RNAs), can be stable fixtures in postmitotic and quiescent neural cells.
There have been decades of studies that demonstrate that the half-lives of mRNA range from minutes to hours, with relatively “stable” ribosomal RNA persisting for days. So how could LL-RNAs not have been found before? A key difference is that Zocher et al. examined RNA in mouse brains filled with postmitotic neurons, whereas most studies have examined proliferative cells. The prior studies show that RNA turnover is dynamically regulated to meet cellular demands (4). Thus, because RNAs are not subject to unrestrained ribonucleases (RNases), if they are structurally protected in nuclei, perhaps they can persist indefinitely.
An important point is that the persistent EU label observed by Zocher et al. is distinctly nuclear. Why would this be, particularly because RNAs for protein production are in the cytoplasm? Answers to this question will require future research, but the authors provide one possibility focused on satellite RNAs, which are expressed from small tandem satellite repeats that form peri- and centromeric heterochromatin. Several studies have shown that brief transient satellite RNA expression in cycling cells plays a role in peri- and centromere structure in chromosomes (5, 6). Zocher et al. report that satellite RNA is enriched in LL-RNAs and further suggest that it continually serves to maintain repressive chromatin modifications, particularly histone H3 lysine 27 (H3K27) methylation, on centromeric heterochromatin.
Satellite RNA was assayed by quantitative polymerase chain reaction (qPCR) in adult mouse cells, but to characterize EU-labeled RNAs more broadly, Zocher et al. carried out RNA sequencing on NPCs that were induced to quiescence in vitro. NPCs were pulsed briefly with EU, and then RNA was extracted 8 days later (an extremely long time for most RNAs). RNA sequencing showed that EU-labeled LL-RNAs were mostly long transcripts, including thousands of protein-coding transcripts, lncRNAs, and intergenic transcripts. Interspersed repeats [short and long interspersed nuclear elements (SINEs and LINEs)] are also enriched in LL-RNA, although their prevalence may be higher, given their abundance in pre-mRNAs, lncRNAs, and intergenic RNAs. As shown by the sequencing data, it is notable that these diverse long transcripts dwarf the satellite RNA component of EU-labeled RNA. It is not clear why protein-coding RNAs would remain stable and in the nucleus, so it will be important for future studies to further investigate this. Given the nuclear signal, it is worth noting that prior work (7) and control experiments by Zocher et al. demonstrate that the EU is not incorporated into DNA.
Stable RNAs, potentially analogous to LL-RNAs, may have been found before. Evidence of long-lived structural RNAs in the nuclei of human cells was reported in 2014 (8). The highly repetitive “junk” RNA of the genome, called CoT-1 RNA, was detected in human cells by in situ hybridization. CoT-1 RNA labeled euchromatin, not heterochromatin, and had unusual properties. CoT-1 RNA tightly localized in cis to the chromosome territory (unlike mRNAs), and the bright RNA territory remained unperturbed for 16 to 32 hours after transcriptional inhibition. Although there are caveats to the use of transcriptional inhibitors, the results suggested that CoT-1 RNA may be part of a protein-RNA nuclear scaffold (also known as the matrix), the existence of which was reported in earlier studies (9, 10), although these results were debated.
Recently, a more-selective procedure was developed to isolate highly insoluble nuclear RNAs that remain after robust extraction and deoxyribonuclease (DNAse) digestion and that cofractionate with architectural RNAs [X-inactive specific transcript (XIST) and nuclear para-speckle assembly transcript 1 (NEAT1)] (11), which have established roles in forming nuclear structures (12). The 15% of insoluble scaffold RNAs (scaffRNAs) that remained with XIST and NEAT1 RNAs were CoT-1 RNAs. Sequencing showed that scaffRNAs were largely long and repeat-rich, primarily intron-rich pre-mRNAs, lncRNAs, and intergenic transcripts. There are strong similarities between these scaffRNAs and the EU-labeled LL-RNAs found by Zocher et al. Potentially relevant to LL-RNA functions, the removal of scaffRNAs, by numerous different means, rapidly resulted in condensation of euchromatin and delocalized specific nuclear matrix proteins (11), which other studies further support regulate chromatin packaging (13).
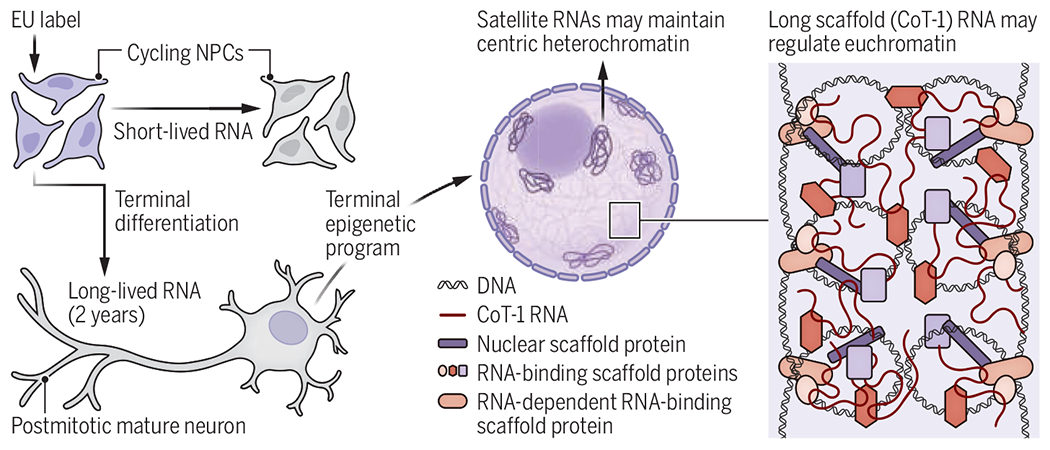
If LL-RNAs function in regulating chromatin, this could relate to the finding of Zocher et al. that adult neurons were labeled with EU only if it was injected in early development. Unlike adult neurons, NPCs express the pyrimidine salvage pathway that is needed to metabolize EU (14). NPCs that take up EU just before terminal differentiation may retain LL-RNAs in postmitotic nuclei, and these RNAs may act as a structural fixture of nuclear genome architecture (see the figure). In this sense, LL-RNAs may be more akin to lamin proteins (which form the nuclear lamina) and persist long-term in neurons (15). If LL-RNAs are related to scaffRNAs, they may maintain euchromatin that is prevalent in many neurons. The findings of Zocher et al., together with prior reports, raise the possibility that as neural stem cells terminally differentiate to long-lived neurons, they establish their epigenomic program in part through junk RNAs that act as structural components of interphase chromosomes.
Model of long-lived RNAs in neurons Labeling.
Labeling RNAs in neural progenitor cells (NPCs) with 5-ethynyl uridine (EU) reveals that some nuclear RNAs are retained for 2 years in postmitotic neurons. These heterogeneous long-lived RNAs contain satellite RNAs that maintain centric heterochromatin and long transcripts that are similar to CoT-1 scaffold RNAs that regulate euchromatin.
ACKNOWLEDGMENTS
The authors are supported by the National institutes of Health (grants R35GM122597, R01HD091357, and R01HD094788).
REFERENCES AND NOTES
- 1.Willingham AT, Gingeras TR, Cell 125, 1215 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Mattick JS et al. , Nat. Rev. Mol. Cell Biol 24, 430 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zocher S et al. , Science 384, 53 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisen TJ et al. , RNA 28, 808 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ninomiya K et al. , EMBO J. 42, e114331 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novo CL et al. , Nat. Commun 13, 3525 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jao CY, Salic A, Proc. Natl. Acad. Sci. U.S.A 105, 15779 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall LL et al. , Cell 156, 907 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fey EG et al. , J. Cell Sci. Suppl 5, 99 (1986). [DOI] [PubMed] [Google Scholar]
- 10.Nickerson JA et al. , Proc. Natl. Acad. Sci. U.S.A 86, 177 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creamer KM et al. , Mol. Cell 81, 3509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palihati M, Saitoh N, Curr. Opin. Genet. Dev 86, 102176 (2024). [DOI] [PubMed] [Google Scholar]
- 13.Marenda M et al. , Curr. Opin. Genet. Dev 72, 38 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Walter M, Herr P, Cells 11, 739 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchwalter A, Curr. Opin. Cell Biol 84, 102220 (2023). [DOI] [PubMed] [Google Scholar]


