Abstract
A panel of mouse T-cell lymphomas induced by SL3-3 murine leukemia virus (MLV) and three primer binding site mutants thereof (A. H. Lund, J. Schmidt, A. Luz, A. B. Sørensen, M. Duch, and F. S. Pedersen, J. Virol. 73:6117–6122, 1999) were analyzed for the occurrence of recombination between the exogenous input virus and endogenous MLV-like sequences within the 5′ leader region. Evidence of recombination within the region studied was found in 14 of 52 tumors analyzed. Sequence analysis of a ∼330-bp fragment of 44 chimeric proviruses, encompassing the U5, the primer binding site, and the upstream part of the 5′ untranslated region, enabled us to map recombination sites, guided by distinct scattered nucleotide differences. In 30 of 44 analyzed sequences, recombination was mapped to a 33-nucleotide similarity window coinciding with the kissing-loop stem-loop motif implicated in dimerization of the diploid genome. Interestingly, the recombination pattern preference found in replication-competent viruses from T-cell tumors is very similar to the pattern previously reported for retroviral vectors in cell culture experiments. The data therefore sustain the hypothesis that the kissing loop, presumably via a role in RNA dimer formation, constitutes a hot spot for reverse transcriptase-mediated recombination in MLV.
Recombination by template switching between heterologous copackaged viral transcripts during reverse transcription constitutes one mechanism by which retroviruses may diversify and/or overcome the presence of otherwise incapacitating mutations. Retroviruses are known to mutate at a high frequency (11). Since neither of the two polymerases involved in retroviral replication, RNA polymerase II or the virally encoded reverse transcriptase (RT), harbors 3′→5′ proofreading capability, point mutations due to nucleotide misincorporation occur frequently (1, 6, 21). Furthermore, the mechanism of reverse transcription involves two strand transfer reactions which are intrinsically erroneous (18, 20, 25). While these features may allow rapid retroviral diversification, they also result in the accumulation of defective viral genomes. Though unable to replicate independently, such defective viral genomes may still be transcribed and packaged into retroviral particles. Since retroviral particles contain a diploid genome, defective viral genomes may be rescued through recombinational patch repair with a heterologous, copackaged genome, resulting in the formation of a new chimeric provirus (3, 5, 22, 24). In previous studies, we have analyzed recombination events within the 5′ untranslated region, using retroviral vectors containing nonfunctional primer binding site (PBS) sequences (13, 14). By mutating a crucial viral cis element, such as the PBS, normal vector transduction is impaired, thereby facilitating the selection of rare transduction events, some of which result from recombination events. In these studies, a hot spot for recombination was found within the leader region (13, 14), coinciding precisely with the kissing-loop stem-loop structure involved in retroviral RNA dimer formation (7, 23).
In the present study, we have extended the analysis of 5′ leader recombination to replication-competent viruses by using an SL3-3 murine leukemia virus (MLV) pathogenesis model. During analysis of a panel of T-cell tumors derived from mice injected with wild-type (wt) and PBS-modified SL3-3 viruses, we previously noted a number of recombination events between the exogenous, injected SL3-3 virus and MLV-like sequences endogenous to the mouse genome (12). In this experiment, a wt SL3-3 MLV was compared to three virus mutants in which the PBS had been mutated to match the 3′ end of either tRNA1Gln, tRNA3Lys, or tRNA1,2Arg, and the viruses were compared in terms of mean latency period prior to lymphoma induction, tumor cell origin, and stability of the introduced mutations (12). To investigate the stability of the introduced PBS mutations, segments from tumor proviruses were PCR amplified and directly sequenced. In 14 of 52 analyzed tumors, the resulting sequence readouts were repeatedly highly ambiguous, with multiple double peaks consistent with simultaneous sequencing of different proviral templates (12). Interestingly, evidence of recombination was most often detected in tumors resulting from infection by the SL3-3-Lys3 mutant, since chimeric proviruses were detected in 12 of 13 analyzed tumors. Furthermore, the mean lymphoma latency period of this mutant was significantly longer than that of wt SL3-3 MLV (12). We ascribe this prolonged latency period, in combination with the high frequency of detection of recombinant proviruses, to a diminished replication capacity of the SL3-3-Lys3 mutant. However, the exact reasons for this finding remain unclear. Interestingly, recombination within the analyzed leader region was not limited to the apparently stunted SL3-3-Lys3 mutant but could also be detected in one tumor induced by the wt SL3-3-Pro as well as in one mutant harboring an arginine PBS sequence (12).
To analyze in greater detail the structures of the chimeric tumor proviruses from 13 of the tumors, PCR amplicons generated by using a U3-specific primer (primer 1; 5′-GATTCCCAGATGACCGGGGATC-3′) and a gag-specific primer (primer 2; 5′-TAGGGTCAGACTCAGAGGGGTGGT-3′) were cloned into pGEM-T (Promega) and individual subclones from each tumor were analyzed by sequencing of approximately 400 bp of the U5-PBS-5′ untranslated leader region, using primer 1, primer 3 (5′-CGCAGGCGCAAAAAGTAGATGC-3′; specific for the leader region), primer 4 (5′-TCCGAATCGTGGTCTCGCTGATCCTTGG-3′; specific for the U5 region), and primer 5 (5′-TTGCATCCGAATCGTGGTCWCGCT-3′; specific for the U5 region). All PCRs in this study were performed in 100 μl of PCR buffer (Perkin-Elmer) containing 25 pmol of each primer, 0.2 mM each deoxynucleoside triphosphate, and 3 U of AmpliTaq Gold polymerase (Perkin-Elmer). Sequencing was performed on both strands with an automated sequencer (ABI 373; Perkin-Elmer), using a Thermo Sequenase II dye terminator cycle sequencing kit (Amersham Pharmacia Biotech). The amplicons from each tumor were found to contain two types of proviruses, the virus originally used to infect the mouse and a novel chimeric provirus resulting from recombination between the injected virus and endogenous MLV-like sequences and characterized by having a PBS sequence matching the 3′ end of a glutamine tRNA molecule. Aside from the PBS-Gln, the chimeric proviruses contained a pattern of nucleotide substitutions, deletions, and insertions relative to the injected virus and exhibited a high degree of similarity to previously characterized viruses endogenous to the mouse genome (3, 4, 13, 19). To retrieve sufficient subclones for subsequent analysis of the pattern of recombination, the bacterial colonies from individual subclonings were PCR screened with primer 2 and primer 6 (5′-GGGGGTCTTTCATTTGGAGGT-3′; specific for PBS-Gln). From the resulting sequences, unique recombinants from each tumor were aligned and compared to the sequence of the injected SL3-3-Lys3 as well as to sequences of previously characterized endogenous MLVs.
As can be seen in Fig. 1, the recombinant viruses contain a pattern of alterations relative to SL3-3-Lys3. Within the ∼330-bp sequence studied, the 22 markers are relatively evenly distributed. Interestingly, whereas all of the recombinants contain nucleotide alterations in the U5 region and the upstream part of the 5′ untranslated leader region, most of the recombinants contain SL3-3-specific sequences in the downstream part of the analyzed sequence window. Hence, the distinct differences between the endogenous recombination partner and the injected virus may be used as molecular markers to map the actual site of recombination. While some of the genetic markers, such as IV, VIII, X, XI, and XII, are ubiquitous, other positions are highly variable (for example, VI, VII, and IX). Interestingly, some of the genetic markers are mutually exclusive (for example, I and II/III), thus providing evidence that several endogenous MLV loci are involved. Furthermore, marker XIV is present in five different forms which, in combination with the mutually exclusive markers I and II/III, give rise to eight different recombinants based on analysis of these four marker positions only. However, the analyzed chimeric proviruses may result from independent serial recombination events. Hence, the finding of a large number of different endogenous sequences frozen in the tumors at the time of analysis could have resulted from initial recombinations involving only two different endogenous MLV loci followed by additional secondary recombinations and genetic drift within hypervariable-sequence regions. Given the variation in the analyzed sequences and the fact that recombinants can be found in SL3-3-Lys3-, SL3-3-Pro-, and SL3-3-Arg1,2-induced tumors, it seems likely that recombination took place during viral spread in the mice. However, prior to injection into mice, the viruses used in this study were generated by transfection of plasmid DNA into NIH 3T3 cells. The viruses were allowed to spread in the cell culture, after which the stability of the introduced PBS mutations was analyzed by reverse transcription-PCR and direct sequencing of the resulting amplicon (12). While no evidence of recombination was detected at this point at the level of sensitivity of this assay, we cannot exclude the possibility of recombination during viral growth in cell culture.
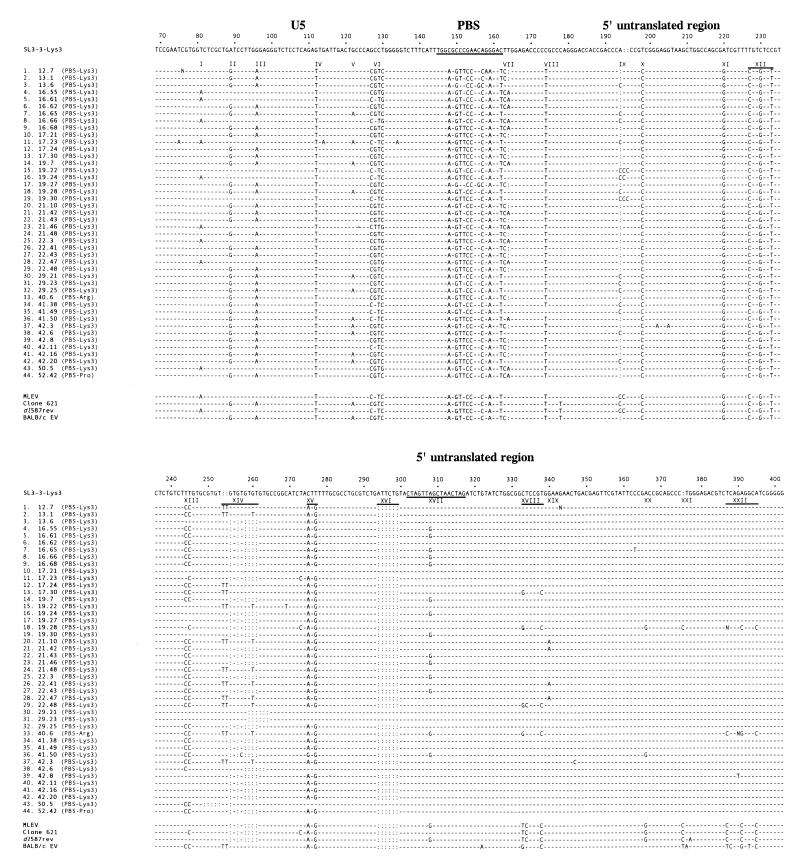
FIG. 1.
Nucleotide sequences of cloned fragments of tumor proviruses resulting from recombination within the leader region of either SL3-3-Lys3, SL3-3-Arg1,2, or wt SL3-3-pro and endogenous MLV-like sequences. The numbers refer to the distance from the transcription initiation site. For comparison, the sequence of the SL3-3-Lys3 U5-PBS-leader region is shown at the top. The sequences of the PBS (nucleotides 145 to 162) and the kissing stem-loop (nucleotides 302 to 317) are underlined. Similarly, at the bottom are shown the sequences of four different MLV-like sequences endogenous to the mouse genome (3, 4, 13, 19). Nucleotides homologous to SL3-3 MLV are indicated by hyphens, and deleted nucleotides are indicated by colons. Nucleotide differences between SL3-3 and the endogenous recombination partner (signified by Roman numerals) serve as molecular markers for identification of the site of recombination.
From the alignment of 44 sequenced proviruses containing patches of endogenous MLV sequences of variable length, the actual site of recombination can be deduced from the presence or absence of specific marker mutations in the sequences (Fig. 1). Of the 44 recombinations, 42 took place within seven of the similarity windows studied whereas 2 map to sequences downstream. Within the sequence window studied, the distribution of the recombinations is highly skewed, with 30 of 44 recombination events taking place between markers XVI and XVIII. Two recombination events took place after marker XVIII, four recombination events occurred after marker XIX, and individual cases of recombination were detected after markers X, XIV, and XX. In some proviral sequences, individual point mutations are detected that are not present in SL3-3 or in any known endogenous MLV sequence (for example, position 362 in chimeric provirus 16.65 or position 389 in provirus 42.8). These mutations may have arisen independently during viral replication or the subsequent PCR amplification and were not taken into consideration when the site of recombination was mapped. However, base substitutions also found in endogenous viruses (for example, position 365 in provirus 41.50) or occurring repeatedly in different tumors (for example, position 339 [marker XIX] in proviruses 21.10, 21.42, 22.41, and 22.47) were taken into account when mapping the site of recombination.
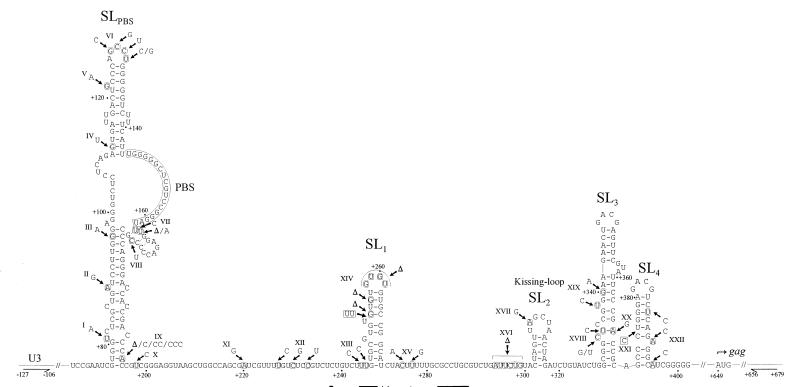
The positions of the marker mutations relative to a putative secondary structure of the U5-PBS-5′ leader region is shown in Fig. 2. Notably, most of the markers are in apparently nonpaired regions. An exception is a set of complementary mutations at positions 334 and 366, where a possible U-A base pair has been substituted for a C-G pair. Functional assignments have been established for stem-loops 2, (SL2), SL3, and SL4. SL2 contains the kissing-loop motif, a 16-nucleotide palindromic sequence thought to initiate the dimerization process (7). Marker XVII, an A-to-G substitution situated within the loop sequence of SL2, allows for the formation of an alternative dimer involving a G-U base pair. SL3 and SL4 constitute the core packaging signal of MLV (16). The structures of these SLs and the GACG loop sequences important for encapsidation (27) are unaffected by the marker mutations. Hence, none of the marker mutations in the 5′ leader region are likely to have significantly affected the replication capacity of the chimeric viruses, in accordance with previous findings (3, 9). Importantly, in 30 of the 44 analyzed viruses, the site of recombination was found to overlap with the kissing-loop motif of SL2, indicating a functional importance of close RNA-RNA interactions in this region in mediating recombination.
FIG. 2.
Simplified structural model of the U5-PBS-leader region of MLV (based on data from references 15 and 26). The nucleotides are numbered relative to the transcription initiation site. The positions of nucleotide markers are indicated by arrows and roman numerals. Nucleotide substitutions are shown in rounded boxes, and nucleotide insertions relative to SL3-3-PBS-Lys3 are shown in rectangles. Triangles represent deletions relative to SL3-3-PBS-Lys3. The positions of the U3- and gag-specific primers used to amplify chimeric proviruses from tumor DNA are indicated by horizontal arrows.
In previous reports, a site preference for recombination within the MLV leader region was detected by using crippled Akv-MLV-derived retroviral vectors harboring nonfunctional PBS sequences (13, 14). By this approach, normal vector replication is impaired, enabling easy detection of rare transduction events involving recombinational patch repair by endogenous viral RNA of murine packaging cells. Using several different vector constructs, a hot spot for recombination corresponding to the region between markers XVI and XVIII was identified. As shown in Fig. 3, the recombinational cluster found in tumor proviruses and the hot spot for recombination identified in cell culture by using retroviral vectors coincide exactly. Obviously, selective forces other than recombination frequencies per se, such as effects on RNA processing or translation initiation, may operate during virus replication in animals. However, the finding of similar recombination patterns in single-cycle studies using retroviral vectors and in replication-competent proviruses from T-cell tumors indicates that within the 5′ leader window studied here, the kissing-loop motif is particularly prone to recombination.
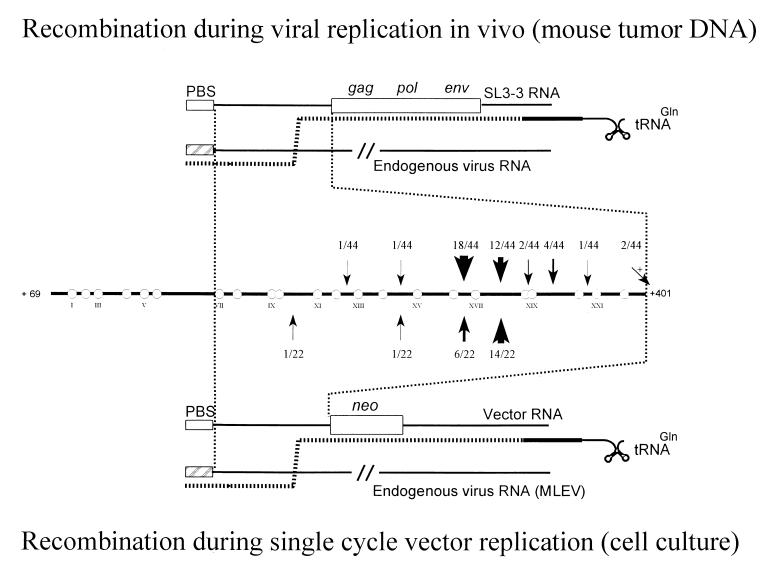
FIG. 3.
Model for RT-mediated recombination within the leader region. (Top) Model for recombination of replication-competent SL3-3 viruses in inbred NMRI mice. Recombinations result from copackaging of an SL3-3 genome with the genome of an endogenous virus and involve intermolecular first-strand transfer from the endogenous virus to the SL3-3 genome followed by RT-mediated template switching within the leader region. (Bottom) Recombinational rescue of PBS-modified retroviral vectors in cell culture (data are from reference 13). (Middle) Frequencies of recombinations within the leader region. Circles indicate molecular markers. Arrows (with frequencies) point to identified sites of recombination. neo indicates the neomycin resistance cassette.
To explain both the cell culture and animal results, we favor a model involving RT-mediated homologous recombination facilitated by direct RNA base pairing at the dimer linkage structure. In this model (Fig. 3), a copy of the endogenous recombination partner is copackaged in vivo with a copy of the injected SL3-3 MLV. Minus-strand strong-stop DNA generated on the endogenous viral RNA is transferred to the 3′ end of the SL3-3 genome (intermolecular strand transfer); this is followed by minus-strand synthesis of the SL3-3 coding region. An intermolecular strand transfer to the genome of the endogenous viral RNA sequence within the 5′ leader region then serves to incorporate a PBS sequence matching tRNAGln, thereby facilitating the second jump of reverse transcription. Whereas local sequence identity and the length of similarity windows have previously been proposed to be major determinants in recombinogenic strand transfer (10, 28), data from our studies, obtained using both replication-competent viruses and retroviral vectors, underline the importance of specific RNA structures in promoting recombination within the 5′ leader region. Similarly, recombination preferences mediated by pairing of RNA molecules have also been proposed for other RNA viruses (17).
Why is recombination preferentially seen in the kissing-loop motif? It can be speculated that RT pausing due to extended RNA base pairings in this region promotes template switching. Alternatively, recombination at the kissing-loop motif may be promoted by other factors dependent on base pairing, i.e., protein binding to the dimer linkage structure or forced copy choice template switching, perhaps even mediated by RT-mediated degradation of double-stranded RNA at the dimerization initiation site (2, 8). The presence of RNA structures mediating recombination may increase the speed of retroviral evolution by resulting in shuffling of existing mutations, thereby generate new combinations of variants, possibly with altered biological properties. The finding of kissing-loop-mediated recombinants in T-cell tumors induced by both wt and PBS-modified SL3-3 MLVs indicates that the kissing loop may be an important element in viral diversification.
Acknowledgments
The technical assistance of Jane Jensen is gratefully acknowledged.
This work was supported by contracts CT 95-100 (Biotechnology) and CT 95-0675 (Biomed 2) of the European Commission, the Karen Elise Jensen Foundation, the Danish Cancer Society, the Danish Biotechnology Program, and the Danish Natural Sciences and Medical Research Councils.
REFERENCES
- 1.Bebenek K, Kunkel T A. The fidelity of retroviral reverse transcriptases. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1993. pp. 85–102. [Google Scholar]
- 2.Blain S W, Goff S P. Nuclease activities of Moloney murine leukemia virus reverse transcriptase. Mutants with altered substrate specificities. J Biol Chem. 1993;268:23585–23592. [PubMed] [Google Scholar]
- 3.Colicelli J, Goff S P. Isolation of a recombinant murine leukemia virus utilizing a new primer tRNA. J Virol. 1987;57:37–45. doi: 10.1128/jvi.57.1.37-45.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colicelli J, Goff S P. Identification of endogenous retroviral sequences as potential donors for recombinational repair of mutant retroviruses: positions of cross-over point. Virology. 1987;160:518–522. doi: 10.1016/0042-6822(87)90030-4. [DOI] [PubMed] [Google Scholar]
- 5.DiFronzo N L, Holland C A. A direct demonstration of recombination between an injected virus and endogenous viral sequences, resulting in the generation of mink cell focus-inducing viruses in AKR mice. J Virol. 1993;67:3763–3770. doi: 10.1128/jvi.67.7.3763-3770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dougherty J P, Temin H M. Determination of the rate of base-pair substitution and insertion mutations in retrovirus replication. J Virol. 1988;62:2817–2822. doi: 10.1128/jvi.62.8.2817-2822.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Girard P-M, Bonnet-Mathoniére B, Muriaux D, Paoletti J. A short autocomplementary sequence in the 5′ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry. 1995;34:9785–9794. doi: 10.1021/bi00030a016. [DOI] [PubMed] [Google Scholar]
- 8.Götte M, Fackler S, Hermann T, Perola E, Cellai L, Gross H J, Le Grice S F, Heumann H. HIV-1 reverse transcriptase-associated RNase H cleaves RNA/RNA in arrested complexes: implications for the mechanism by which RNase H discriminates between RNA/RNA and RNA/DNA. EMBO J. 1995;14:833–841. doi: 10.1002/j.1460-2075.1995.tb07061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grez M, Akgün E, Hilberg F, Ostertag W. Embryonic stem cell virus, a recombinant murine retrovirus with expression in embryonic stem cells. Proc Natl Acad Sci USA. 1990;87:9202–9206. doi: 10.1073/pnas.87.23.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu W-S, Temin H M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 11.Katz R A, Skalka A M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- 12.Lund A H, Schmidt J, Luz A, Sørensen A B, Duch M, Pedersen F S. Replication and pathogenicity of primer binding site mutants of SL3-3 murine leukemia viruses. J Virol. 1999;73:6117–6122. doi: 10.1128/jvi.73.7.6117-6122.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen J G, Lund A H, Kristensen K D, Duch M, Sørensen M S, Jørgensen P, Pedersen F S. A preferred region for recombinational patch repair in the 5′ untranslated region of primer binding site-impaired murine leukemia virus vectors. J Virol. 1996;70:1439–1447. doi: 10.1128/jvi.70.3.1439-1447.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikkelsen J G, Lund A H, Duch M, Pedersen F S. Recombination in the 5′ leader of murine leukemia virus is accurate and influenced by sequence identity with a strong bias toward the kissing-loop dimerization region. J Virol. 1998;72:6967–6978. doi: 10.1128/jvi.72.9.6967-6978.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mougel M, Tounekti N, Darlix J-L, Paoletti J, Ehresmann B, Ehresmann C. Conformational analysis of the 5′ leader and the gag initiation site of Mo-MuLV RNA and allosteric transitions induced by dimerization. Nucleic Acids Res. 1993;21:4677–4684. doi: 10.1093/nar/21.20.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mougel M, Zhang Y, Barklis E. cis-active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J Virol. 1996;70:5043–5050. doi: 10.1128/jvi.70.8.5043-5050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy P D, Simon A E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 18.Olsen J C, Bova-Hill C, Grandgenett D P, Quinn T P, Manfredi J P, Swanstrom R. Rearrangements in unintegrated retroviral DNA are complex and are the result of multiple genetic determinants. J Virol. 1990;64:5475–5484. doi: 10.1128/jvi.64.11.5475-5484.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ou C-Y, Boone L R, Yang W K. A novel sequence segment and other nucleotide structural features in the long terminal repeat of a BALB/c mouse genomic murine leukemia virus-related DNA clone. Nucleic Acids Res. 1983;11:5603–5620. doi: 10.1093/nar/11.16.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pulsinelli G A, Temin H M. Characterization of large deletions occurring during a single round of retrovirus vector replication: novel deletion mechanism involving errors in strand transfer. J Virol. 1991;65:4786–4797. doi: 10.1128/jvi.65.9.4786-4797.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts J D, Preston B D, Johnston L A, Soni A, Loeb L A, Kunkel T A. Fidelity of two reverse transcriptases during DNA-dependent DNA synthesis in vitro. Mol Cell Biol. 1989;9:469–476. doi: 10.1128/mcb.9.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartzberg P, Colicelli J, Goff S P. Recombination between a defective retrovirus and homologous sequences in host DNA: reversion by patch repair. J Virol. 1985;53:719–726. doi: 10.1128/jvi.53.3.719-726.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stuhlmann H, Berg P. Homologous recombination of copackaged retrovirus RNAs during reverse transcription. J Virol. 1992;66:2378–2388. doi: 10.1128/jvi.66.4.2378-2388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tounekti N, Mougel M, Roy C, Marquet R, Darlix J-L, Paoletti J, Ehresmann B, Ehresmann C. Effect of dimerization on the conformation of the encapsidation Psi domain of Moloney murine leukemia virus RNA. J Mol Biol. 1992;223:205–220. doi: 10.1016/0022-2836(92)90726-z. [DOI] [PubMed] [Google Scholar]
- 27.Yang S, Temin H M. A double hairpin structure is necessary for the efficient encapsidation of spleen necrosis virus retroviral RNA. EMBO J. 1994;13:713–726. doi: 10.1002/j.1460-2075.1994.tb06311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J, Temin H M. Retrovirus recombination depends on the length of sequence identity and is not error prone. J Virol. 1994;68:2409–2414. doi: 10.1128/jvi.68.4.2409-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]